Abstract
Seven complete genes and one incomplete gene for the biosynthesis of the glycopeptide antibiotic balhimycin were isolated from the producer, Amycolatopsis mediterranei DSM5908, by a reverse-cloning approach and characterized. Using oligonucleotides derived from glycosyltransferase sequences, a 900-bp glycosyltransferase gene fragment was amplified and used to identify a DNA fragment of 9,882 bp. Of the identified open reading frames, three (oxyA to -C) showed significant sequence similarities to cytochrome P450 monooxygenases and one (bhaA) showed similarities to halogenase, and the genes bgtfA to -C showed similarities to glycosyltransferases. Glycopeptide biosynthetic mutants were created by gene inactivation experiments eliminating oxygenase and glycosyltransferase functions. Inactivation of the oxygenase gene(s) resulted in a balhimycin mutant (SP1-1) which was not able to synthesize an antibiotically active compound. Structural analysis by high-performance liquid chromatography–mass spectrometry, fragmentation studies, and amino acid analysis demonstrated that these oxygenases are involved in the coupling of the aromatic side chains of the unusual heptapeptide. Mutant strain HD1, created by inactivation of the glycosyltransferase gene bgtfB, produced at least four different compounds which were not glycosylated but still antibiotically active.
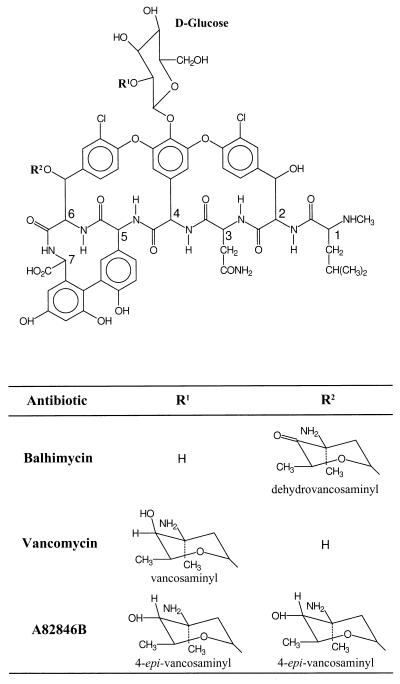
Amycolatopsis mediterranei DSM5908 is a member of the family Pseudonocardiaceae and is the producer of the vancomycin-like glycopeptide antibiotic balhimycin (17). The chemical structure of the heptapeptide backbone of balhimycin (Fig. 1) is identical with that of vancomycin and A82846B (also known as chloroeremomycin), which are synthesized by two different Amycolatopsis orientalis strains (28). The antibiotic structures differ only in the glycosylation pattern. Both balhimycin and vancomycin contain two sugars, whereas A82846B contains three sugars. Balhimycin contains a glucose residue at the aromatic ring of aa (amino acid) 4 and the rare aminosugar dehydrovancosamine (4) at aa 6. Vancomycin carries a glucose-vancosamine disaccharide at the aromatic ring of aa 4. In contrast, A82846B is modified by three sugar moieties: a glucose-epivancosamine disaccharide at aa 4 and an epivancosamine residue at aa 6. Minor compounds (balhimycin V and dechloro-balhimycin V) with three sugars were also detected in the balhimycin producer (35).
FIG. 1.
Chemical structures of the glycopeptide antibiotics balhimycin (A. mediterranei DSM5908), vancomycin (A. orientalis C329.4), and A82846B (A. orientalis A82846).
Vancomycin is a valuable and important antibiotic, because it is the drug of choice and sometimes the drug of last resort for the treatment of severe infections caused by antibiotic-resistant gram-positive bacteria, especially by methicillin-resistant staphylococci (42). The in vivo and in vitro antibacterial activity of balhimycin is comparable to that of vancomycin, but its activity against anaerobes, particularly Clostridium, is superior to that of vancomycin (4). The occurrence of numerous vancomycin-resistant bacteria in the past 10 years (1) has demonstrated the need for new glycopeptide antibiotics. One way to produce new and more potent glycopeptide antibiotics is by chemical modification (19). Another way is the production of new hybrid glycopeptide antibiotics. Solenberg et al. (28) described the identification of two glycosyltransferase genes from the vancomycin producer A. orientalis C329.4 and three glycosyltransferase genes from the A82846B producer, A. orientalis A82846. They showed that some of these genes can be used to produce hybrid glycopeptide antibiotics in vitro as well as by the genetically modified Streptomyces toyocaensis, a producer of a teicoplanin glycopeptide that lacks sugar residues.
In order to manipulate the biosynthesis of glycopeptide antibiotics and to create new antibiotics by rational design, it is important to identify the biosynthetic gene cluster for glycopeptide biosynthesis. Recently, the DNA sequence of genes which are likely to be involved in the biosynthesis of the vancomycin group antibiotic A82846B (chloroeremomycin) in A. orientalis was obtained (34). However, the function of the putative enzymes in glycopeptide antibiotic biosynthesis has not yet been shown.
To prove whether the identified putative biosynthetic genes are involved in the biosynthesis of an antibiotic, it is important to inactivate the respective genes in vivo by gene disruption. Gene cloning systems were described for the vancomycin producer A. orientalis ATCC 19795 (14) and the producer of the teicoplanin-like antibiotic A47934, S. toyocaensis (15). However, to date, there are no reports of generating glycopeptide antibiotic mutants by gene disruption experiments in these strains. For the balhimycin producer, A. mediterranei, however, a gene cloning system was developed (23), and its successful application was shown in several gene disruption and gene replacement experiments (23).
Here we report the isolation of balhimycin biosynthetic genes from A. mediterranei by a reverse-cloning approach. Using the established gene cloning system, balhimycin mutants were created by gene disruption experiments. We furthermore describe the chemical structure analysis of balhimycin precursors, which were synthesized by the mutants.
MATERIALS AND METHODS
Strains and cultures.
The strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference |
|---|---|---|
| E. coli | ||
| XL1-blue | recA1 hsdR17[F′ lacIqZΔM15] | 3 |
| JM110 | lacY dam dcm [F′ lacIqZΔM15] | 41 |
| A. mediterranei | ||
| DSM5908 | Balhimycin-producing wild type | 17 |
| HD1 | Balhimycin mutant constructed by integration of pHD1 in the bgtfB gene | This study |
| SP1 | Balhimycin mutant constructed by integration of pSP1.5Pst between oxyA and oxyB | This study |
| Plasmid | ||
| pUC18 | bla | 36 |
| pSP1 | Gene disruption vector; ermE | 23 |
| pOJ446 | Shuttle cosmid vector; am | 2 |
| pSP1.5Pst | pSP1 derivative containing the 1,459-bp PstI-fragment of cosmid 16.1 encoding parts of the balhimycin oxygenase genes oxyA and oxyB in the PstI site | This study |
| pHD12 | pUC18 derivative containing a 797-bp XmaI fragment of a PCR product including a part of the bgtfB gene | This study |
| pHD1 | pSP1 derivative containing the 797-bp XmaI fragment of pHD12 in the XmaI site of the MCS | This study |
| Cosmid 16.1 | pOJ446 derivative containing a 36-kb chromosomal insert of A. mediterranei including a part of the balhimycin biosynthetic gene cluster | This study |
Multiple cloning site.
Media and culture conditions.
Escherichia coli and A. mediterranei strains were grown as described previously (23).
Direct transformation of A. mediterranei DSM5908.
For transformation of A. mediterranei, a modified direct transformation method was used as described previously (23).
Preparation, manipulation, and sequencing of DNA.
Isolation and manipulation of DNA was performed according to the methods of Sambrook et al. (25) and Hopwood et al. (9). Isolation of plasmid DNA from E. coli for sequencing was performed with ion-exchange columns from Qiagen (Hilden, Germany). PCR fragments were isolated from an agarose gel with the Qiaquick gel extraction kit (Qiagen). Restriction endonucleases were used according to the specifications of the suppliers (New England Biochemical, Beverly, Mass., and Promega, Madison, Wis.).
For preparation of a gene library, genomic DNA of the balhimycin producer, A. mediterranei DSM5908, was partially digested with Sau3AI and fragments in a 30- to 40-kb size range were cloned into the BamHI/HpaI-digested vector pOJ446 (2) as described by Gaisser et al. (6). The gene library consisted of 30,000 primary clones, with an average size of 35 kb.
The nucleotide sequences of both strands of a 9,882-bp region of cosmid 16.1, encoding different oxygenase and glycosyltransferase genes involved in balhimycin biosynthesis, was determined by the chain termination method (26). Different sequencing strategies, like construction of deletion clones (using the Double-stranded Nested Deletion Kit [Pharmacia, Uppsala, Sweden]), construction of specific subclones, and primer walking (using custom-synthesized oligonucleotides as primers), were applied. For sequencing reactions, the ALFexpress AutoRead sequencing kit (Pharmacia) or the Thermo Sequenase fluorescent labeled primer cycle sequencing kit with 7-deaza-dGTP (Amersham Pharmacia Biotech AB, Uppsala, Sweden) were used according to the specifications of the suppliers. Sequencing data were generated by using the ALFexpress DNA sequencer (Pharmacia).
Alignment of sequence contigs and examination for open reading frames (ORFs) were performed by applying the codon usage program of Staden and McLachlan (29). The programs BLAST (7) and FASTA (22) were used for homology searches. For the alignment of protein sequences and phylogenetic analysis, the programs ClustalW (33) and Treeview (21) were applied.
PCR protocol for the amplification of the glycosyltransferase fragment.
PCR was performed on a RoboCycler Gradient 40 thermocycler from Stratagene (La Jolla, Calif.) with the Expand High Fidelity PCR System (Boehringer, Mannheim, Germany). The PCR mixture (100 μl) contained 50 pmol of each primer (Gly1 and Gly7), 0.1 μg of genomic DNA of A. mediterranei DSM5908, deoxyribonucleoside 5′ triphosphates at a final concentration of 200 μM each (DNA Polymerization Mix; Pharmacia), 10× reaction buffer, 1.5 mM MgCl2, and 3.5 U of Taq DNA polymerase. Dimethyl sulfoxide at a final concentration of 3% was added to the reaction mixtures to enhance the specificity of hybridization. Several variations of the thermocycling parameters for amplification of the glycosyltransferase gene fragment led to the following PCR procedure: initial denaturation (95°C; 5 min) before addition of the polymerase (72°C; 10 min); 35 cycles of denaturation (94°C; 70 s), annealing (59 to 63°C; 70 s), and polymerization (72°C; 90 s); and an additional polymerization step (72°C; 10 min) at the end of the program. The sequences of the primers were as follows: Gly1, 5′-TCCCCCCGGGIWSSCGCGGIGACGTSGA-3′; Gly7, 5′-TCCCCCCGGGTGGTGGATSRCSGCSGCSACSCGICCGAA-3′ (I, Inosin; S, C/G; W, A/T).
Southern hybridization.
Southern hybridizations with the digoxigenin (DIG) DNA labeling and detection kit from Boehringer were performed as described previously (23). As a size standard, the DIG-labeled DNA Molecular Weight Marker VII (Boehringer) with the following fragment lengths (in base pairs) was used: 81, 359, 492, 710, 718, 992, 1,164, 1,482, 1,515, 1,882, 1,953, 2,799, 3,639, 4,899, 6,106, 7,427, and 8,576. DNA colony hybridization experiments were performed as described by Sambrook et al. (25).
Construction of plasmids pHD1 and pSP1.5Pst.
Plasmids were constructed for gene disruption of the glycosyltransferase gene bgtfB (pHD1) and inactivation of oxygenase genes (pSP1.5Pst).
A 797-bp XmaI fragment, a subfragment of the 900-bp PCR product encoding a part of the balhimycin glycosyltransferase gene bgtfB, was cloned into the XmaI site of pUC18, resulting in the plasmid pHD12. This bgtfB fragment was then cloned as an EcoRI/SphI-fragment into the single EcoRI/SphI site of the vector pSP1 (23), resulting in the gene disruption plasmid pHD1. To construct the gene disruption plasmid pSP1.5Pst, a 1,459-bp PstI fragment of cosmid 16.1, encoding 355 aa of the C terminus of the balhimycin oxygenase gene oxyA (391 aa) and 116 aa of the N terminus of the balhimycin oxygenase gene oxyB (398 aa), was cloned into the single PstI site of vector pSP1 (23).
Determination of balhimycin biosynthesis.
Balhimycin production was determined by bioassays with Bacillus subtilis ATCC6633 as the test organism after growth on R5 medium (9) or by high-performance liquid chromatography (HPLC) as described previously (23).
HPLC purification.
For daughter ion experiments and for amino acid analysis, peptides were purified from fermentor broth by preparative HPLC (Nucleosil; C18; 250 by 20 mm; 5 μm; Grom, Herrenberg, Germany) with a solvent delivery system 600 (Waters, Eschborn, Germany) with a flow rate of 10 ml/min. As a detector, a Lambda Max model 481 (Waters) with a detection wavelength of 214 nm was used. Peptides were separated with a linear gradient: 10% B to 50% B in 60 min (solvent A, 0.1% trifluoroacetic acid in water; solvent B, 0.1% trifluoroacetic acid in acetonitrile).
Electrospray-MS.
Mass spectra were recorded on an API III TAGA triple-quadrupole mass spectrometer (Perkin-Elmer, Thornhill, Ontario, Canada) equipped with an electrospray (ES) ion source. The orifice voltage was set to 80 V in positive mode. Argon was used as the collision gas for daughter ion experiments.
Samples were diluted in acetonitrile-water (1:1) to give a concentration of 0.1 mg/ml. For single measurements, the sample was introduced into the ion source via a Harvard Apparatus (South Natick, Mass.) syringe infusion pump 22. For HPLC-MS analysis, a double syringe pump (model 140A; Applied Biosystems, Weiterstadt, Germany) was used at a flow rate of 200 μl/min split down to 80 μl/min after the column. Separations were performed on a Nucleosil C18 column (2 by 100 mm; 5 μm) (Grom). Crude fermentor broth was injected with a linear gradient: 10% B to 100% B in 30 min (solvent A, 0.1% trifluoroacetic acid in water; solvent B, 0.1% trifluoroacetic acid in acetonitrile).
Amino acid analysis.
Peptides were hydrolyzed in 6 N HCl under a nitrogen atmosphere for 12 h at 110°C. The amino acid samples were derivatized with isopropanol-trifluoroacetic anhydride. Separation was performed with gas chromatography (GC)-MS (MAT 112S; Finnigan, Bremen, Germany) on a Chirasil-Val column (20 m by 0.3 mm; film thickness [df] = 0.13 μm).
Nucleotide sequence accession number.
The nucleotide sequences of the balhimycin biosynthetic genes described in this paper are available from the EMBL data library under accession no. Y16952.
RESULTS
Identification of a glycosyltransferase gene fragment of A. mediterranei possibly involved in balhimycin biosynthesis.
The glycopeptide antibiotic balhimycin consists of a heptapeptide backbone to which two sugar molecules (glucose and dehydrovancosamine) are attached (Fig. 1). To identify the balhimycin glycosyltransferase genes which are involved in the glycosylation reaction, a reverse-genetic approach was used. Comparison of sequences of the five glycosyltransferases GtfA to -C and GtfD and -E, which are probably involved in the biosynthesis of A82846B and vancomycin in A. orientalis A82846 and C329.4, respectively, revealed several regions with significant similarity (28). To design PCR primers for the amplification of balhimycin-specific glycosyltransferase gene fragments, two conserved regions were chosen. Based on these sequences, two oligonucleotide primers (Gly1 and Gly7) which were adapted to the Streptomyces codon usage (40) were synthesized. Each primer contained an additional XmaI recognition site at the 5′ end to enable cloning of the amplified fragment into the XmaI site of pUC18.
Using these primers in an optimized PCR protocol (see Materials and Methods), a DNA fragment of about 900 bp was amplified from genomic DNA of the balhimycin producer. The PCR fragment was cloned into the XmaI site of pUC18, resulting in plasmid pHD1. Sequencing of the fragment revealed an insert size of 797 bp. The nucleotide sequence did not include the sequence of primer Gly7, because the PCR fragment has an internal XmaI recognition site approximately 100 bp upstream from the Gly7 primer sequence.
Comparison of the deduced amino acid sequence of the cloned fragment (′bgtfB′) with databases revealed remarkable similarities to the glycosyltransferases GtfA to -C and GtfD and -E. The highest similarities found were to GtfB (85.7% identity over 258 aa) from the A82846B producer and GtfE (80.6% identity over 258 aa) from the vancomycin producer. We therefore concluded that bgtfB may be involved in balhimycin biosynthesis.
Isolation and analysis of cosmid 16.1, a clone carrying several putative balhimycin biosynthetic genes.
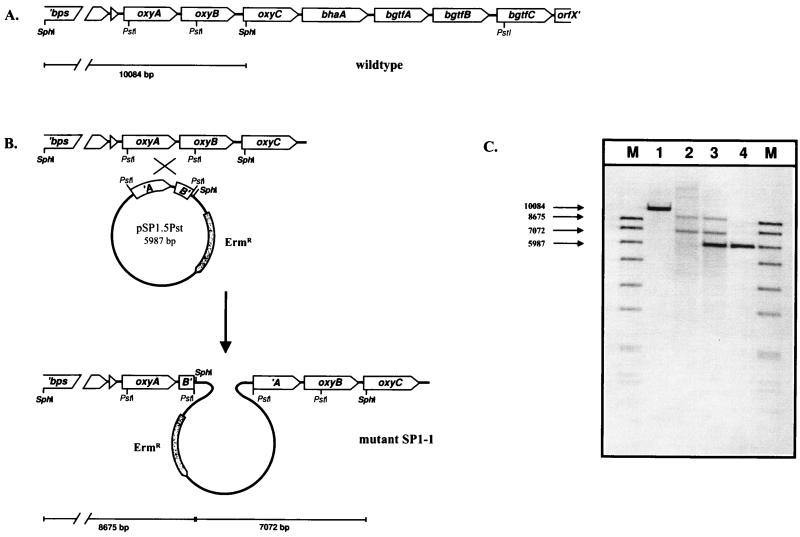
To isolate the balhimycin biosynthetic gene cluster, a cosmid library was screened with the ′bgtfB′ probe. In colony hybridization experiments, 12 of 1,408 cosmid clones with homology to the glycosyltransferase gene fragment ′bgtfB′ were identified. A 9,882-bp region of cosmid 16.1 with homology to the ′bgtfB′ probe was sequenced. The nucleotide sequence had an overall G+C content of 70%, a characteristic of actinomycetes (40). An ORF analysis revealed seven complete ORFs and one incomplete ORF (Fig. 2A). All ORFs were transcribed in the same orientation. Putative start codons were assigned by choosing the ATG, GTG, and TTG furthest upstream in the probable ORFs, which were characterized by a coding probability of >50%.
FIG. 2.
Integration of the plasmid pSP1.5Pst into the balhimycin gene cluster and Southern hybridization. (A) The gene organization of a part of the balhimycin biosynthetic cluster in the A. mediterranei wild type is shown. Downstream of peptide synthetase genes (′bps), the oxygenase genes (oxyA to -C), a halogenase gene (bhaA), the glycosyltransferase genes (bgtfA to -C), and orfX′ are located. (B) Construction of the gene inactivation mutant SP1-1 obtained after integration of plasmid pSP1.5Pst into the chromosome by a single-crossover event. The relevant PstI and SphI restriction sites are indicated. The size of the expected hybridizing SphI fragments with the 1,459-bp ′oxyA-oxyB′ (A′-B′) PstI fragment as a probe is drawn below the clusters. (C) Genomic DNA of the A. mediterranei wild type (lane 1) and of the oxygenase mutants SP1-1 (lane 2) and SP1-3 (lane 3) and DNA of the plasmid pSP1.5Pst (lane 4) were digested with SphI and probed with the 1,459-bp ′oxyA-oxyB′ (A′-B′) PstI fragment in a Southern hybridization. Lane M, DIG-labeled DNA Molecular Weight Marker VII (Boehringer). The sizes (in base pairs) of the characteristic fragments are shown.
ORFs 1 to 8 were designated oxyA, oxyB, oxyC, bhaA, bgtfA, bgtfB, bgtfC, and orfX′ (Fig. 2A). The characteristics of the putative genes (start and stop codons), the putative regulatory elements, and the sizes and molecular masses of the deduced gene products are shown in Table 2. The most frequently used start codon was ATG (five times). Two genes probably start with a GTG start codon, and the unusual TTG start codon is found in the oxyB gene. Possible ribosome binding site (RBS) sequences with the typical distance to the putative start codons were identified upstream of seven genes (Table 2). No typical Streptomyces-like consensus promoter (31) was present in the intergenic regions of the whole sequence. Sequence comparisons revealed that the sequence of the PCR fragment (′bgtfB′) is identical with the sequence of an internal part of bgtfB.
TABLE 2.
Characteristics of the eight identified genes, localized on the 9,882-bp fragment
| Gene | Putative RBS | Distance (nt) from the start codon | Start codon (position) | Stop codon (position) | Size/mass of the gene product (aa/kDa) | Distance to the following gene (nt) |
|---|---|---|---|---|---|---|
| oxyA | AGGA | 8 | GTG (224) | TGA (1399) | 391/43.5 | 49 |
| oxyB | AAGAGG | 11 | TTG (1449) | TGA (2645) | 398/44.0 | 148 |
| oxyC | AAGGAG | 12 | ATG (2795) | TAG (4015) | 406/44.8 | 59 |
| bhaA | AGAGG | 11 | ATG (4076) | TGA (5551) | 491/53.9 | 82 |
| bgtfA | AGAGG | 7 | ATG (5634) | TGA (6824) | 396/41.1 | 63 |
| bgtfB | AGGA | 11 | GTG (6889) | TAA (8118) | 409/43.3 | 146 |
| bgtfC | −a | −a | ATG (8266) | TGA (9495) | 409/42.6 | 32 |
| orfX′ | AAGAGG | 7 | ATG (9529) | −b | 13.1 (partial) | −c |
No RBS detected.
No stop codon, because only the N-terminal part of OrfX is located on this fragment.
No gene follows orfX′ on this fragment.
Features of the deduced gene products OxyA to -C, BhaA, BgtfA to -C, and OrfX′.
Computer-assisted homology searches with the sequences of the deduced gene products of oxyA to -C, bhaA, bgtfA to -C, and orfX′ revealed significant similarities to the following enzymes, whose functions have been determined.
The putative gene products of oxyA to -C significantly resemble cytochrome P450 monooxygenases. Data bank searches with OxyA to -C gene products revealed similarities (ca. 50%) to a great number of characterized P450-related monooxygenases which are either involved in the biosynthesis of antibiotics, as, for example, a P450-like mycinamycin biosynthetic gene product (11), RapN from rapamycin biosynthesis (16), and EryF from erythromycin biosynthesis (38), or in catabolite degradation, like sulphonylurea herbicide-inducible P450 monooxygenases from Streptomyces griseolus (20).
The three putative enzymes OxyA to -C all contain a characteristic sequence motif around a cysteine residue at the C terminus (consensus sequence, FGHGXHXCLG), which is typical for cytochrome P450 monooxygenases and is known to be involved in providing a pocket for cysteine heme coordination (20).
In addition, OxyA showed a very high percentage of identity and similarity to ORF7 (85 and 92%, respectively, over 391 aa), OxyB showed high identity and similarity to ORF8 (87 and 92%, respectively, over 396 aa), and OxyC showed high identity and similarity to ORF9 (91 and 95%, respectively, over 404 aa), which are described as being similar to P450-related oxidases and are supposed to be involved in the biosynthesis of the glycopeptide antibiotic A82846B from A. orientalis A82846 (34).
The gene product of bhaA is similar to the halogenase PrnC from Pseudomonas fluorescens (27% identity and 42% similarity over 355 aa), which is responsible for the chlorination of an aromatic precursor of pyrrolnitrin biosynthesis (12). No similarity with enzymes of the cytochrome P450 monooxygenase type was found. The BhaA enzyme contains a motif located at the N terminus of the protein (the “ADP-binding βαβ fold”; most highly conserved residues, GXGXXG/AXXXG/A), which is known to be involved in the binding of ADP and to be common to a number of FAD- and NAD(P)H-dependent enzymes (39). There is a weak similarity to a second motif (residues 298 to 350), which seems to be important for binding FAD. At amino acid position 304, a highly conserved Asp was identified, which possibly forms hydrogen bonds with the 3-hydroxy group of the ribityl chain of the flavine moiety of FAD, as in other FAD-dependent enzymes (5).
BhaA showed high identity and similarity to ORF10 (95 and 97%, respectively, over 491 aa) of A. orientalis A82846 and to hydroxylase A (93 and 97%, respectively, over 491 aa) of A. orientalis C329.4. As a result of sequence comparisons, ORF10 was described as being a nonheme halogenase and was supposed to be involved in the biosynthesis of the glycopeptide antibiotic A82846B (34). The hydroxylase A gene is part of the presumptive vancomycin biosynthetic gene cluster and was also only assigned by sequence comparison (28).
The deduced products of the three genes bgtfA to -C showed a high sequence homology with glycosyltransferases, which are probably involved in the biosynthesis of the glycopeptide antibiotics A82846B and vancomycin (28).
BgtfA revealed the highest similarity to GtfA (78% identity and 85% similarity over 396 aa), BgtfB showed the highest similarity to GtfB (85% identity and 88% similarity over 407 aa), and BgtfC showed the highest similarity to GtfC (80% identity and 87% similarity over 409 aa), which are proposed to be involved in the glycosylation of A82846B (34). There was also a striking similarity between BgtfB and the glycosyltransferase GtfE (81% identity and 86% similarity over 407 aa), which may be responsible for glucose transfer to the vancomycin aglycone (28). It is remarkable that the proteins BgtfA to -C also have similarities to other glycosyltransferases of eucaryotic and procaryotic origin, but even other glycosyltransferases which are involved in antibiotic biosynthesis, like glycosyltransferases from erythromycin biosynthesis (i.e., EryBV [32]) have only weak similarities (ca. 30% identity over 110 aa).
The orfX′ gene encodes 118 aa of the N-terminal part of a protein. OrfX′ showed the highest similarity to two gene products of the presumptive biosynthetic gene clusters of the glycopeptides A82846B (ORF14) and vancomycin of the A. orientalis (hydroxylase B) producer strains described above. Moreover, there is a high similarity to the daunorubicin-doxorubicin biosynthetic enzyme DnrX from Streptomyces peucetius (63% identity and 71% similarity over 113 aa), which was published recently (13). The exact role of DnrX in daunorubicin-doxorubicin biosynthesis remains unclear.
Evidence for the participation of oxygenase and glycosyltransferase genes in balhimycin biosynthesis.
To prove that the identified gene cluster of A. mediterranei is involved in balhimycin biosynthesis, gene inactivation experiments were performed.
To inactivate the putative oxygenase gene(s), the plasmid pSP1.5Pst (see Materials and Methods) was used to transform the balhimycin producer by a modified direct transformation method (23). Two of several erythromycin-resistant transformants (SP1-1 and SP1-3) were analyzed in detail by Southern hybridization experiments (Fig. 2). SphI-digested genomic DNA from the wild type, SP1-1, and SP1-3 were probed with the 1,459-bp PstI fragment. Whereas in the wild type a 10-kb SphI fragment was detected, both transformants showed signals at 8.7 and 7.1 kb with the same probe, which is in accordance with an integration of plasmid pSP1.5Pst into the chromosome via homologous recombination with the 1,459-bp PstI fragment (Fig. 2). However, the hybridization pattern of transformant SP1-3 showed an additional strong signal at 6 kb, most likely representing the linearized vector pSP1.5Pst, indicating multiple or tandem integrations.
To test, whether the integration of the plasmid pSP1.5Pst affected balhimycin biosynthesis, the balhimycin phenotype was analyzed in bioassays with B. subtilis as an indicator. Neither of the transformants (SP1-1 and SP1-3) induced an inhibition zone, indicating that the transformants were not able to synthesize an antibiotically active compound. The results of the bioassays clearly showed that the identified gene region is involved in balhimycin biosynthesis.
In the second gene activation experiment the role of the putative glycosyltransferase gene bgtfB in balhimycin biosynthesis was examined. The plasmid pHD1 was used to transform A. mediterranei by direct transformation. Several erythromycin-resistant colonies were detected. Southern hybridizations of transformants revealed that the integrations of pHD1 into the chromosome occurred in the bgtfB locus via homologous recombination (data not shown). In bioassays, these transformants produced clear inhibition zones, which were not distinguishable from those of the wild type. Hence, the glycosyltransferase mutant was still able to synthesize antibiotically active molecules.
Biochemical characterization of the synthesized balhimycin precursors in the oxygenase mutant SP1-1 and the glycosyltransferase mutant HD1.
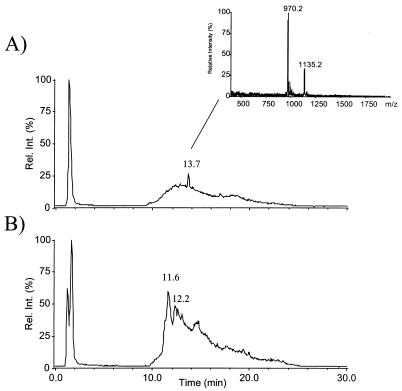
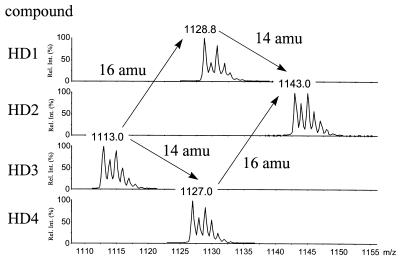
Using the HPLC-electrospray ionization (ESI)-MS coupling technique, several compounds of the culture broth of the oxygenase mutant SP1-1 and the glycosyltransferase mutant HD1 were identified. Two coeluting compounds with molecular masses of 969 (SP969) and 1,134 (SP1134) Da, respectively, were detected in the culture broth of the oxygenase mutant SP1-1 (Fig. 3), and four compounds with molecular masses of 1,112 (HD3), 1,126 (HD4), 1,128 (HD1), and 1,142 (HD2) Da were detected in the culture broth of the glycosyltransferase mutant HD1 (Fig. 4).
FIG. 3.
Reconstructed total-ion chromatogram of a liquid chromatography-MS run of crude culture broth of SP1-1 mutant (13.7 min) (A) and wild type (balhimycin, 11.6 min; desvancosamine-balhimycin, 12.2 min) (B). The corresponding mass spectrum of panel A shows two peptides, SP969 and SP1134, coeluting at 13.7 min. Neither peptide mass was detected in the wild type. Rel. int., relative intensity.
FIG. 4.
Isotopically resolved mass spectra of HD mutants. The mass distribution of the isotopic patterns indicates the presence of two covalently bound chlorine atoms in the molecules. The characteristic mass differences of 16 Da (HD1-HD3 and HD2-HD4) and 14 Da (HD1-HD2 and HD3-HD4) between the derivatives suggest hydroxylation and methylation as major structural differences. Rel. int., relative intensity.
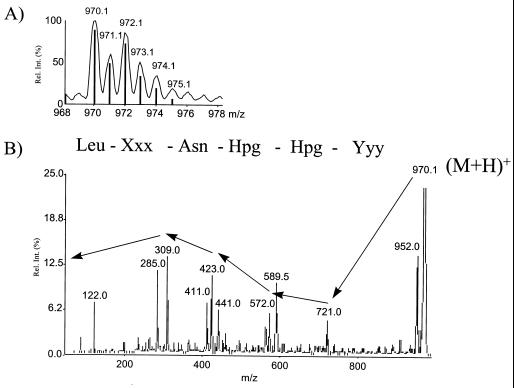
High-resolution MS revealed that all of these compounds contained two chlorine atoms due to their characteristic isotopic patterns. This was proved by the comparison of the calculated isotopic patterns with the mass spectra (Fig. 5A).
FIG. 5.
MS analysis of peptide SP969. (A) Isotopically resolved mass spectrum. The observed isotopic pattern is consistent with the theoretical, calculated isotopic pattern (bars) for a molecule containing two chlorine atoms. (B) The daughter-ion spectrum reveals the partial sequence Leu-Xxx-Asn-Hpg-Hpg-Yyy. Rel. int., relative intensity.
Moreover, MS-MS experiments, amino acid analysis, and GC-MS experiments for the SP-type as well as for the HD-type compounds were performed. In the case of bridged or partially bridged amino acids, no sequence information could be obtained in fragmentation studies for the bridged region of the molecule.
SP compounds.
For SP969 and SP1134, fragments suitable for sequence analysis were obtained. As a daughter ion scan (ESI-MS/MS) demonstrated (Fig. 5B), continuous fragmentation of SP969 could be observed, indicating a linear peptide structure for SP969.
Moreover, qualitative amino acid analysis of SP969 was performed, and in addition to the amino acids d-Leu and l-Asp (hydrolyzed l-Asn), d-hydroxyphenylglycine (d-Hpg) was detected. This confirms the assumption that SP969 has a linear peptide structure. However, chlorine-containing tyrosine-like amino acids (positions 2 and 6), as demonstrated by high-resolution MS, were not found.
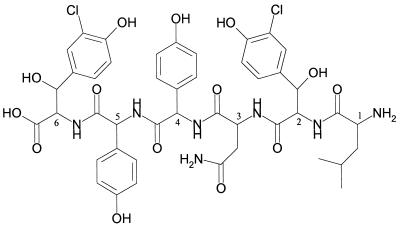
Sugar analysis with GC-MS after hydrolysis revealed no glycosylation of the peptide. The results of fragmentation studies and amino acid analysis led to the partial sequence Leu-Xxx-Asn-Hpg-Hpg-Yyy (Fig. 5B).
Fragmentation of the second compound, SP1134, led to the same fragments as shown for SP969 (Fig. 5B), indicating that SP1134 is identical in most of its structural moieties to SP969, with the exception of the fragment representing the seventh amino acid, which could not be determined by amino acid analysis. Moreover, no glycosylation of SP1134 was observed.
HD-compounds.
For the HD-type compounds, which were synthesized by the glycosyltransferase mutant HD1, the same analytical methods were applied as for SP-type compounds. Under MS/MS conditions, glycosylated balhimycin showed characteristic fragments which correlate to the loss of sugar residues. For the HD-type compounds, none of these characteristic fragments were observed. This lack of glycosylation was also confirmed by GC-MS experiments.
Furthermore, fragmentation with ESI-MS/MS showed only two characteristic signals: one with a molecular mass/charge ratio of 86 m/z (HD1 and HD3) and one with 100 m/z (HD2 and HD4). As demonstrated by amino acid analysis, these fragments correspond to the amino acids d-Leu and N-Me–d-Leu, respectively, which represent the N-terminal amino acids. N-Me–d-Leu or d-Leu is the only amino acid not included in the bridged part of the molecule and therefore is accessible to fragmentation. This clearly implies a bridged molecule structure for all HD type compounds, as is known for balhimycin (Fig. 1).
As expected, the amino acid l-Asp (hydrolyzed Asn), which is connected via peptide bonds with the rest of the molecule, was detected by amino acid analysis after total hydrolyzation of the bridged HD-type compounds.
DISCUSSION
In the far-reaching area of antibiotic biosynthesis, one of the topics that remains to be explored is the analysis on the molecular-genetic level of the biosynthesis of the important vancomycin-type antibiotics. Although there were some reports on genes and clusters possibly involved in the biosynthesis of vancomycin and A82846B (28, 34), the function of the genes was not shown by gene inactivation experiments. In this report, the identification and the targeted inactivation of genes essential for the synthesis of the glycopeptide antibiotic balhimycin from A. mediterranei is described for the first time.
Balhimycin biosynthetic genes were identified by analyzing a DNA fragment of 9,882 bp. Sequencing revealed seven complete ORFs and one incomplete ORF; one of those, the oxyB gene, most likely starts with a TTG start codon. This is an unusual feature, since there are only a few other examples of TTG starts, which have been proposed to be involved in antibiotic biosynthesis: the Streptomyces griseus strE gene (24), the Saccharopolyspora erythraea P450 hydroxylase gene eryK (30), the Streptomyces antibioticus phsA gene (10), and the most similar gene, ORF8 of the A82846B producer A. orientalis A82846 (34). For the Streptomyces glaucescens oxygenase gene tcmG (27), a TTG start codon was confirmed by N-terminal protein sequencing of the purified enzyme.
The significant similarities of the seven identified complete genes to known sequences provided the first clues to their possible functions. Interestingly, the organization of the genes of the identified region is identical to the gene organization described for the biosynthesis of the glycopeptide antibiotic A82846B and genes involved in vancomycin biosynthesis. Random sequencing experiments revealed that all of the genes on cosmid 16.1 (data not shown) are organized very similarly to the A82846B gene cluster (34), indicating that both clusters are involved in the biosynthesis of a similar glycopeptide antibiotic and may originate from a common ancestor. The gene disruption mutant SP1-1 was not able to synthesize an antibiotically active balhimycin derivative, thus proving that the identified gene cluster is involved in balhimycin biosynthesis. The structural analysis demonstrated clearly that the derivatives SP969 and SP1134 are linear, because they are accessible to fragmentation, sequencing studies, and amino acid analysis. Although fragmentation studies propose a linear structure, aa 7 of SP1134, as well as aa 2 and 6 of SP969 and SP1134, which carry a chlorine atom, were not found by amino acid analysis. This contradiction can be resolved if we assume for positions 2 and 6 β-hydroxy-amino acids, as found in the final structure of balhimycin, which are dehydrated under hydrolytic conditions. Such β-hydroxy-amino acids are not accessible for detection under total hydrolytic conditions due to dehydration (11a).
It remains to be shown why the SP1-1 mutant produced two peptidic derivatives, SP969, which is truncated before the seventh amino acid, and, simultaneously, a second, untruncated peptide, SP1134. A possible explanation could be that peptide SP1134 is unstable and the SP969 derivative is a degradation product.
MS/MS experiments and amino acid and sugar analyses also showed that these two peptides are not glycosylated and that Leu is not methylated. The missing glycosylation may be due to nonrecognition of a linear derivative as a substrate by the glycosyltransferases. According to our analytical results, we propose the chemical structure for the derivative SP969 (Fig. 6).
FIG. 6.
Proposed structure for the linear peptide SP969 deduced from the experimental data of MS and amino acid analysis.
The common feature of both balhimycin precursors, SP969 (Fig. 6) and SP1134, is their linear structure without oxidatively coupled but hydroxylated tyrosine residues 2 and 6. The organization of the balhimycin biosynthetic genes after integration of plasmid pSP1.5Pst into the chromosome (Fig. 2) indicates that the oxyA gene should not be influenced by the integration event, but the transcription of oxyB and probably oxyC may be affected. A more far-reaching effect of the integration can be excluded, as the following gene, bhaA, carries its own promoter (22a). Since the mutant SP1-1 is not able to synthesize a cyclic balhimycin molecule, one could therefore assume that oxygenase B and/or C is involved in the coupling of the aromatic side chains. The loss of activity can well be explained by the linear, unbridged structure, because the tyrosine coupling is important to hold the peptide backbone in a rigid, cup-shaped form, which is able to interact with the d-Ala–d-Ala terminus of the cell wall precursor (42).
The BhaA gene product of the balhimycin gene cluster could be involved in halogenating balhimycin. BhaA showed significant similarities to PrnC, a halogenase of P. fluorescens, which acts in the chlorinating of pyrrolnitrin (12). PrnC is a new example of the NADH-dependent halogenases, which are involved in the specific biosynthesis of halometabolites (8). Instrumental analysis showed that the balhimycin precursors synthesized by mutant SP1-1 were still halogenated. This confirms the fact that the polar effect of the integration of plasmid pSP1.5Pst in mutant SP1-1 does not affect the activity of the bhaA gene.
The BgtfA-C enzymes all have significant similarities to glycosyltransferases. It is surprising that the balhimycin biosynthetic gene cluster comprises three glycosyltransferase genes, like the A82846B gene cluster (34), although balhimycin contains only two sugars, one at aa 4 and the other at aa 6 (Fig. 1). A possible explanation for this phenomenon is the fact that the balhimycin producer creates at least 10 different balhimycin analogs (35). The minor compounds balhimycin V and dechloro-balhimycin V carry three sugar residues, with a glucose-dehydrovancosamine disaccharide at aa 4, representing a glycosylation pattern which requires three glycosyltransferases.
Solenberg et al. (28) reported that only the glycosyltransferases GtfB from the A82846B producer and GtfE from the vancomycin producer are able to add glucose specifically to the vancomycin heptapeptide. Thus, it is most likely that the glycosyltransferase BgtfB from the balhimycin producer, which showed the highest similarity to GtfB and GtfE, has the same function. The existence of the bgtfB mutant HD1, which produced only balhimycin derivatives (HD-type compounds) which were not glycosylated, confirmed that BgtfB is indeed involved in the glycosylation of balhimycin.
Because GtfE is responsible for the addition of glucose to vancomycin, it is most likely that the other glycosyltransferase, GtfD, of the vancomycin producer adds the second sugar, vancosamine, to the glucose residue. Since BgtfC from the balhimycin producer showed the highest identity to GtfD, it can be assumed that this enzyme catalyzes the addition of the dehydrovancosamine residue to the glucose residue in the two minor compounds of balhimycin.
The third glycosyltransferase, BgtfA, revealed the highest similarity to GtfA from the A82846B producer. Solenberg et al. (28) speculated that GtfA most likely adds 4-epivancosamine to aa 6 of A82846B. If this is true, BgtfA should have a similar function, namely, the addition of dehydrovancosamine to aa 6 of balhimycin.
However, the bgtfB mutant, in which the bgtfA gene was not inactivated, did not contain a sugar residue. One explanation could be that the glycosyltransferase BgtfA is only able to recognize the substrate glycosylated at aa 4. A similar observation of a glycosylation that follows a special order was described for erythromycin biosynthesis. Here, the sugar mycarose must be added to the 3-OH of erythronolide B before desosamine is added at the 5-OH position (37). A second explanation could be an antisense-like influence on the expression of the bgtfA gene in the bgtfB mutant, caused by the promoter of the ermE resistance gene of the integrated pHD1, which is transcribed in the opposite direction to bgtfA.
The results of tandem MS experiments and amino acid analysis clearly demonstrated that all HD compounds have bridged structures. This implies that the biphenyl and the biphenyl ether linkages are all formed before glycosylation occurs. Small differences among the four compounds are the N-methylation of Leu (Δm = 14 atomic mass units [amu]; HD1-HD2 and HD3-HD4 [Fig. 4]) as well as a possible hydroxylation (Δm = 16 amu; HD1-HD3 and HD2-HD4 [Fig. 4]). However, this hydroxylation, as one possible cause, could not be located.
Since it is known that vancomycin derivatives that lack sugars are still active (18), it is not surprising that the unglycosylated derivatives still show antibiotic activity.
The inactivation of glycopeptide antibiotic genes created a variety of new substances, which have novel properties or represent the basis for chemical modifications to synthesize new semisynthetic derivatives. Moreover, all of the genes described in this paper are modification genes and excellent tools for the creation of hybrid antibiotics by rational combinatorial biosynthesis.
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (Sonderforschungsbereich 323) and the BMBF (ZSP 0316500C).
We thank G. Nicholson for chiral amino acid analysis, H. P. Fiedler for fermentation and HPLC analysis, K.-H. van Pée, C. R. Hutchinson, E. Cundliffe, and A. Bechthold for communicating results prior to publication and for helpful discussions, E. Nußbaum for help in sequencing, the students M. Elbs and U. Baisch for help in isolating the peptides, and M. Labes for critical reading of the manuscript.
REFERENCES
- 1.Arthur M, Depardieu F, Reynolds P, Courvalin P. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol Microbiol. 1996;21:33–44. doi: 10.1046/j.1365-2958.1996.00617.x. [DOI] [PubMed] [Google Scholar]
- 2.Bierman M, Logan R, O’Brien K, Seno E T, Rao R N, Schoner B E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 3.Bullock W O, Fernandez J M, Short J M. XL1-Blue, a high efficiency plasmid transforming recA Escherichia coli strain with beta galactosidase selection. Focus. 1987;5:376–378. [Google Scholar]
- 4.Chatterjee S, Vijayakumar E K S, Nadkarni S R, Patel M V, Blumbach J, Ganguli B N. Balhimycin, a new glycopeptide antibiotic with an unusual hydrated 3-amino-4-oxoaldopyranose sugar moiety. J Org Chem. 1994;59:3480–3484. [Google Scholar]
- 5.Eggink G, Engel H, Vriend G, Terpstra P, Witholt B. Rubredoxin reductase of Pseudomonas oleovorans: structural relationship to other flavoprotein oxidoreductases based on one NAD and two FAD fingerprints. J Mol Biol. 1990;212:135–142. doi: 10.1016/0022-2836(90)90310-I. [DOI] [PubMed] [Google Scholar]
- 6.Gaisser S, Trefzer A, Stockert S, Kirschning A, Bechthold A. Cloning of an avilamycin biosynthetic gene cluster from Streptomyces viridochromogenes Tü57. J Bacteriol. 1997;179:6271–6278. doi: 10.1128/jb.179.20.6271-6278.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gish W, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 8.Hohaus K, Altmann A, Burd W, Fischer I, Hammer P E, Hill D S, Ligon J M, van Pée K-H. NADH-dependent halogenases are more likely to be involved in halometabolite biosynthesis than haloperoxidases. Angew Chem Int Ed Engl. 1997;36:2012–2013. [Google Scholar]
- 9.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser D J, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 10.Hsieh C-J, Jones G H. Nucleotide sequence, transcriptional analysis, and glucose regulation of the phenoxazinone synthase gene (phsA) from Streptomyces antibioticus. J Bacteriol. 1995;177:5740–5747. doi: 10.1128/jb.177.20.5740-5747.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inouye M, Takada Y, Muto N, Horinouchi S, Beppu T. Characterization and expression of a P-450-like mycinamycin biosynthesis gene using a novel Micromonospora-Escherichia coli shuttle cosmid vector. Mol Gen Genet. 1994;245:456–464. doi: 10.1007/BF00302258. [DOI] [PubMed] [Google Scholar]
- 11a.Jung, G. Unpublished results.
- 12.Kirner S, Hammer P E, Hill D S, Altmann A, Fischer I, Weislo L J, Lanahan M, van Pée K-H, Ligon J M. Functions encoded by pyrrolnitrin biosynthetic genes from Pseudomonas fluorescens. J Bacteriol. 1998;180:1939–1943. doi: 10.1128/jb.180.7.1939-1943.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lomovskaya N, Doi-Katayama Y, Filippini S, Nastro C, Fonstein L, Gallo M, Colombo A L, Hutchinson C R. The Streptomyces peucetius dpsY and dnrX genes govern early and late steps of daunorubicin and doxorubicin biosynthesis. J Bacteriol. 1998;180:2379–2386. doi: 10.1128/jb.180.9.2379-2386.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsushima P, McHenney M, Baltz R H. Efficient transformation of Amycolatopsis orientalis (Nocardia orientalis) protoplasts by Streptomyces plasmids. J Bacteriol. 1987;169:2298–2300. doi: 10.1128/jb.169.5.2298-2300.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsushima P, Baltz R H. A gene cloning system for Streptomyces toyocaensis. Microbiology. 1996;142:261–267. doi: 10.1099/13500872-142-2-261. [DOI] [PubMed] [Google Scholar]
- 16.Molnar I, Aparicio J F, Haydock S F, Khaw L E, Schwecke T, Koenig A, Staunton J, Leadlay P F. Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of genes flanking the polyketide synthase. Gene. 1996;169:1–7. doi: 10.1016/0378-1119(95)00799-7. [DOI] [PubMed] [Google Scholar]
- 17.Nadkarni S R, Patel M V, Chaterjee S, Vijayakumar E K S, Desikan K R, Blumbach J, Ganguli B N. Balhimycin, a new glycopeptide antibiotic produced by Amycolatopsis sp. Y-86,21022. J Antibiot. 1994;47:334–341. doi: 10.7164/antibiotics.47.334. [DOI] [PubMed] [Google Scholar]
- 18.Nagarajan R. Structure-activity relationships of vancomycin-type glycopeptide antibiotics. J Antibiot. 1993;46:1181–1195. doi: 10.7164/antibiotics.46.1181. [DOI] [PubMed] [Google Scholar]
- 19.Nicas T I, Mullen D L, Flokowitsch J E, Snyder N S, Zweifel M J, Wilkie S C, Rodriguez M J, Thompson R C, Cooper R D G. Semisynthetic glycopeptide antibiotics derived from LY264826 active against vancomycin-resistant enterococci. Antimicrob Agents Chemother. 1996;40:2194–2199. doi: 10.1128/aac.40.9.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Keefe D P, Harder P A. Occurrence and biological function of cytochrome P450 monooxygenases in the actinomycetes. Mol Microbiol. 1991;5:2099–2105. doi: 10.1111/j.1365-2958.1991.tb02139.x. [DOI] [PubMed] [Google Scholar]
- 21.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 22.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Pelzer, S. Unpublished results.
- 23.Pelzer S, Reichert W, Huppert M, Heckmann D, Wohlleben W. Cloning and analysis of a peptide synthetase gene of the balhimycin producer Amycolatopsis mediterranei DSM5908 and development of a gene disruption/replacement system. J Biotechnol. 1997;56:115–128. doi: 10.1016/s0168-1656(97)00082-5. [DOI] [PubMed] [Google Scholar]
- 24.Pissowotzki K, Mansouri K, Piepersberg W. Genetics of streptomycin production in Streptomyces griseus: molecular structure of genes strE, L, M, B3, N. Mol Gen Genet. 1991;231:113–123. doi: 10.1007/BF00293829. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-termination inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen B, Hutchinson C R. Triple hydroxylation of tetracenomycin A2 to tetracenomycin C in Streptomyces glaucescens. J Biol Chem. 1994;269:30726–30733. [PubMed] [Google Scholar]
- 28.Solenberg P J, Matsushima P, Stack D R, Wilkie S C, Thompson R C, Baltz R H. Production of hybrid glycopeptide antibiotics in vitro and in Streptomyces toyocaensis. Chem Biol. 1997;4:195–202. doi: 10.1016/s1074-5521(97)90288-x. [DOI] [PubMed] [Google Scholar]
- 29.Staden R, McLachlan A D. Codon preference and its use in identifying protein coding regions in large DNA sequences. Nucleic Acids Res. 1982;10:141–156. doi: 10.1093/nar/10.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stassi D, Donadio S, Staver M J, Katz L. Identification of a Saccharopolyspora erythraea gene required for the final hydroxylation step in erythromycin biosynthesis. J Bacteriol. 1993;175:182–189. doi: 10.1128/jb.175.1.182-189.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strohl W R. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992;20:961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Summers R G, Donadio S, Staver M J, Wendt-Pienkowski E, Hutchinson C R, Katz L. Sequencing and mutagenesis of genes from the erythromycin biosynthetic gene cluster of Saccharopolyspora erythraea that are involved in L-mycarose and D-desosamine production. Microbiology. 1997;143:3251–3262. doi: 10.1099/00221287-143-10-3251. [DOI] [PubMed] [Google Scholar]
- 33.Thompson J D, Higgins D G, Gibson T J. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Wageningen A M A, Kirkpatrick P N, Williams D H, Harris B R, Kershaw J K, Lennard N J, Jones M, Jones S J M, Solenberg P J. Sequencing and analysis of genes involved in the biosynthesis of a vancomycin group antibiotic. Chem Biol. 1998;5:155–162. doi: 10.1016/s1074-5521(98)90060-6. [DOI] [PubMed] [Google Scholar]
- 35.Vértesy L, Fehlhaber H W, Kogler H, Limbert M. New 4-oxovancosamine-containing glycopeptide antibiotics from Amycolatopsis sp. Y-86,21022. J Antibiot. 1996;49:115–118. doi: 10.7164/antibiotics.49.115. [DOI] [PubMed] [Google Scholar]
- 36.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–269. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 37.Weber J M, Wierman C K, Hutchinson C R. Genetic analysis of erythromycin production in Streptomyces erythraeus. J Bacteriol. 1985;164:425–433. doi: 10.1128/jb.164.1.425-433.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weber J M, Leung J O, Swanson S J, Idler K B, McAlpine J B. An erythromycin derivative produced by targeted gene disruption in Saccharopolyspora erythraea. Science. 1991;252:114–117. doi: 10.1126/science.2011746. [DOI] [PubMed] [Google Scholar]
- 39.Wierenga R K, Terpstra P, Hol W G J. Prediction of the occurrence of the ADP-binding βαβ-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- 40.Wright F, Bibb M J. Codon usage in the G+C-rich Streptomyces genome. Gene. 1992;113:55–65. doi: 10.1016/0378-1119(92)90669-g. [DOI] [PubMed] [Google Scholar]
- 41.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 42.Yao R C, Crandall L W. Glycopeptides: classification, occurrence, and discovery. In: Nagarajan R, editor. Glycopeptide antibiotics. New York, N.Y: Marcel Dekker Inc.; 1994. pp. 1–28. [Google Scholar]