Abstract
Background: At present, radical total mesorectal excision after neoadjuvant chemoradiotherapy is crucial for locally advanced rectal cancer. Therefore, the use of histopathological images analysis technology to predict the efficacy of neoadjuvant chemoradiotherapy for rectal cancer is of great significance for the subsequent treatment of patients. Methods: In this study, we propose a new pathological images analysis method based on multi-instance learning to predict the efficacy of neoadjuvant chemoradiotherapy for rectal cancer. Specifically, we proposed a gated attention normalization mechanism based on the multilayer perceptron, which accelerates the convergence of stochastic gradient descent optimization and can speed up the training process. We also proposed a bilinear attention multi-scale feature fusion mechanism, which organically fuses the global features of the larger receptive fields and the detailed features of the smaller receptive fields and alleviates the problem of pathological images context information loss caused by block sampling. At the same time, we also designed a weighted loss function to alleviate the problem of imbalance between cancerous instances and normal instances. Results: We evaluated our method on a locally advanced rectal cancer dataset containing 150 whole slide images. In addition, to verify our method’s generalization performance, we also tested on two publicly available datasets, Camelyon16 and MSKCC. The results show that the AUC values of our method on the Camelyon16 and MSKCC datasets reach 0.9337 and 0.9091, respectively. Conclusion: Our method has outstanding performance and advantages in predicting the efficacy of neoadjuvant chemoradiotherapy for rectal cancer. Clinical and Translational Impact Statement—This study aims to predict the efficacy of neoadjuvant chemoradiotherapy for rectal cancer to assist clinicians quickly diagnose and formulate personalized treatment plans for patients.
Keywords: Pervasive computing, neoadjuvant chemoradiotherapy, internet of things, pathological images, rectal cancer
I. Introduction
Colorectal cancer is the third leading cause of cancer globally, and rectal cancer accounts for about 30%-35% of colorectal cancer cases [1], [2]. More than 100,000 people worldwide are diagnosed with rectal cancer each year, 70% of which are locally advanced rectal cancer (LARC). Neoadjuvant chemoradiation (NCRT) and radical surgery are the best treatments recommended for locally advanced rectal cancer [3] because preoperative radiotherapy and chemotherapy can reduce recurrence and improve survival [4]. Many studies have shown that the response to neoadjuvant therapy affects the prognosis, especially the pathological complete response has a good effect on the prognosis [5]–[7]. Approximately 15% to 27% of patients will show a pathological complete response (PCR) to neoadjuvant chemoradiotherapy [8], [9]. However, a considerable number of patients will not respond to neoadjuvant therapy. Based on the resection specimens, these non-responders can be defined as patients with no changes in tumor regression after neoadjuvant therapy. These non-responders may benefit little from neoadjuvant therapy but still have related toxicity. More importantly, tumor progression may occur in some patients during treatment. Therefore, it is very important to accurately predict non-response before implementing neoadjuvant therapy to develop a personalized treatment plan, including avoiding overtreatment and timely selection of alternative treatments. In addition, it can also help patients avoid the risks and uncertain consequences of surgery. However, in addition to the pathological evaluation after neoadjuvant therapy, there is currently no reliable method to accurately divide patients into non-good response (non-GR) groups and good response (GR) groups. Due to the heterogeneity of tumors [10], accurately predicting a non-response to neoadjuvant therapy remains challenging. Therefore, it is of great significance to predict the efficacy of neoadjuvant chemoradiotherapy for rectal cancer using histopathological images. In this study, we established a deep learning model based on the pathological images of patients with locally advanced rectal cancer to accurately predict the patient’s response to neoadjuvant chemoradiotherapy so as to assist clinicians in formulating personalized treatment plans for patients.
Histopathological images analysis is an important step in cancer or disease diagnosis. In recent years, the diagnosis of cancer has been improved by using a deep learning based histopathological images analysis framework [11]. These advances have promoted the progress of diagnostic and computational methods for gastrointestinal diseases [12], [13]. However, many issues remain to be addressed in the field, including the variability of histopathological features across diseases, limited data, and high resolution of whole slide images (WSIs), making it difficult for the model to use WSIs for training directly. To match traditional input sizes for traditional feed-forward CNN models, typical WSIs would need to be down-sampled by a factor of
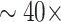 , resulting in the loss of cellular and structural details, which are critical for prediction. To overcome this bottleneck, state-of-the-art methods for WSIs analysis adopt a two-stage approach [14]–[21]. First, patches are extracted by using block sampling on high-resolution WSIs, and the patches are used to train the CNN model, and the patches are encoded as prediction scores or low-dimensional feature vectors. Second, learn an aggregation model to integrate the slice-level information obtained for the entire slide prediction. Although the two-stage training method has achieved good results in the field of pathological images analysis, it still has defects; that is, the parameters in the feature extraction stage cannot be updated, resulting in the inability to obtain the specific features of pathological tissues. Therefore, recently, some researchers proposed an end-to-end framework to solve this problem [22], [23], but the end-to-end training method has the problem of very slow training.
, resulting in the loss of cellular and structural details, which are critical for prediction. To overcome this bottleneck, state-of-the-art methods for WSIs analysis adopt a two-stage approach [14]–[21]. First, patches are extracted by using block sampling on high-resolution WSIs, and the patches are used to train the CNN model, and the patches are encoded as prediction scores or low-dimensional feature vectors. Second, learn an aggregation model to integrate the slice-level information obtained for the entire slide prediction. Although the two-stage training method has achieved good results in the field of pathological images analysis, it still has defects; that is, the parameters in the feature extraction stage cannot be updated, resulting in the inability to obtain the specific features of pathological tissues. Therefore, recently, some researchers proposed an end-to-end framework to solve this problem [22], [23], but the end-to-end training method has the problem of very slow training.
In addition to the above problems, in the study of predicting the efficacy of neoadjuvant chemoradiotherapy for rectal cancer, there are the following limitations and challenges: First of all, since pathological images are all gigapixel-level data, the data training process is slow. Secondly, in pathological images analysis, the pathological images are usually first divided into slices of the same size, and then each slice is sent to the model for separate processing, and finally, the predictions of all patches are aggregated to obtain patient-level predictions, but this method often leads to the loss of global information of pathological images. Additionally, in the pathological image of the tumor, the normal tissue is much larger than the cancer tissue. After the slice sampling, the number of normal tissue slices is much larger than the number of cancer tissue slices, which will cause the problem of imbalance between positive and negative instances. Finally, the traditional pathological image analysis methods often require pathologists to manually divide the decision boundary between tumor and normal tissue, which is time-consuming and laborious, and greatly increases the burden on oncologists. Therefore, how to use pathological images without pixel-level annotation to predict the efficacy of neoadjuvant chemoradiotherapy for rectal cancer is also the limitation and challenges faced by this study.
To address these challenges, we propose a new based on weakly-supervised learning classification framework to classify rectal cancer histological images inspired by the previous work [24], [25], which uses multi-scale features. The experiments revealed that the proposed method could be directly applied to WSIs classification without delineating the decision boundary of cancerous tissues, greatly reducing the labeling burden of pathologists and outperforming the traditional multi-instance learning (MIL) methods. The main contributions of this paper are as below:
-
1)
We designed a gated attention weight normalization mechanism based on the multilayer perceptron by reparameterizing the weight vectors in a gated attention network that decouples the length of those weight vectors from their direction, thereby speeding up the training process and the convergence speed of stochastic gradient descent.
-
2)
We propose a bilinear attention multi-scale feature fusion mechanism to alleviate the problem of global information loss. This mechanism makes full use of the given vision-language information by learning the bilinear attention distributions of pathological images and can better integrate the global features provided by the larger receptive field patches and the detailed features provided by the smaller receptive field patches.
-
3)
We designed a weighted loss function to alleviate the problem of instance imbalance by optimizing the instance-level and bag-level loss functions simultaneously.
-
4)
Our method directly uses pathological images without pixel-level annotations for training and has excellent performance on both the private dataset and the publicly available Camelyon16 and MSKCC datasets.
We applied the proposed method to predict the efficacy of neoadjuvant chemoradiotherapy for rectal cancer and proved the outstanding performance of this method. We also verified the proposed algorithm on the Camelyon16 and MSKCC public datasets. The experimental results show that the proposed method performs better than the conventional method on the weakly labeled dataset without any processing. The rest of this paper is organized as follows. Section II briefly reviews related work of the early diagnosis of rectal cancer and histopathology images analysis. In Section III, we describe our proposed method in detail. The experiments and comparison results are given in Section IV, followed by discussions and conclusions in Section V.
II. Related Works
Our research is mainly to directly use the pathological tissue images that do not delineate the decision boundary of cancerous tissue to predict the pathological response of neoadjuvant chemoradiotherapy for rectal cancer. In this section, we briefly reviewed the latest advances in the prediction of pathological response of neoadjuvant chemoradiotherapy for rectal cancer and the related work of deep learning models for pathological images analysis.
A. Prediction of Pathological Response to Neoadjuvant Chemoradiotherapy in Rectal Cancer
Currently, the research on the pathological response of neoadjuvant chemoradiotherapy of rectal cancer mainly uses the radiomic characteristics of rectal cancer and the histological characteristics of WSIs to predict. Radiomics is a combination of quantitative images analysis and machine learning methods, and it is considered a form of AI. Histological features are quantitative images features that can provide tumor intensity, shape, size, volume, and texture features. Different imaging modes (such as MRI, CT, PET, etc.) can be used as the basis for feature extraction in imaging omics. All the features extracted from the images are “radiomics”, and those feature sets with the predictive value selected after feature selection are usually called “radiomic signature”. At present, the basic function of radiomics is to quantitatively analyze tumor regions of interest through a large number of radiomics characteristics, which can provide valuable diagnostic, prognostic or predictive information. Its purpose is to explore and use these information resources to develop diagnostic, predictive, or prognostic imaging omics models to support personalized clinical decision-making and improve individualized treatment options [26]–[28]. Histopathology is the gold standard of clinical tumor diagnosis, directly related to the development of treatment and the evaluation of prognosis. Recently, many studies have shown that it is feasible and effective to use the features of digital pathological images to predict the treatment response of neoadjuvant chemoradiotherapy [29], [30]. Therefore, this study is of great research significance.
B. Deep Learning Models for WSIs Analysis
At present, deep learning methods have been widely used to predict the prognosis of various cancers. For example, some researchers use deep convolutional neural networks to assess the human tumor microenvironment and directly predict the prognosis from histopathological images [31]. At the same time, there is also evidence that deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer [32]. In addition, some scholars use deep learning based tissue analysis to predict the prognosis of colorectal cancer [33]. In recent years, pathological image analysis methods based on deep learning mainly rely on multi-instance learning and weakly supervised learning. Since only coarse-grained labels are available in pathological images analysis problems, after down-sampling WSIs into patches, it is not easy to model the patches with real labels. We can characterize this problem as an inexact supervision problem, so many studies use multi-instance learning to solve this problem. In the two-class multi-instance learning, the bag is marked as a positive bag only if it contains at least one positive instance; otherwise, it is a negative bag. In pathological images analysis research, the researcher regards the down-sampled patches as instances and WSIs as bags. MIL has been successfully applied to the classification of histopathological images [16], [25], [34]. Recently, authors in [14] proposed a MIL-RNN aggregation operator comprising patch-level training, top-k instance selection, and RNN-based aggregation for patient-level prediction. In addition, some researchers use multi-scale convolutional layers on pre-trained CNNs to capture scale-invariant patterns and use top-k pools to aggregate feature maps for patient-level prediction. Also, some researchers [24] proposed a weight normalization method by reparameterizing the weight vectors in a neural network that decouples the length of those weight vectors from their direction, thereby accelerating the convergence speed of stochastic gradient descent. Authors in [25] proposed a clustering-constrained attention multiple instance learning method (CLAM) that uses attention-based learning to automatically identify sub-regions of high diagnostic value to classify the whole slide images accurately. Furthermore, authors in [35] proposed bilinear attention networks (BAN) that find bilinear attention distributions to utilize given vision-language information seamlessly. At the same time, some researchers have also explored the application of ensemble learning and transfer learning in medical research and have achieved outstanding results [36]–[39]. At present, although these algorithms have achieved good results in different tasks, they rely too much on the way pathologists manually divide the decision-making boundary of pathological tissues [40], [41]. The motivation of our proposed method is to directly learn pathological images without any pre-processing by introducing attention modules for use in complex WSIs analysis tasks.
III. Methods
A. Attention-Based Weight Normalization MIL
Compared with inputting a series of individually labeled instances, under the MIL framework, the input is a series of labeled “bags”, and each “bag” includes many instances, and these instances have permutation invariance. In multiple instance learning, the images are described as a “bag”:
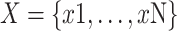 , each
, each
 is a feature vector extracted from the corresponding i-th region in the images (instance), and N is the number of regions (instance) where the images are segmented. Take two-classification as an example. A bag contains multiple instances. If all instances are marked negative, the bag is negative; otherwise, the bag is positive. The current state-of-the-art attention-based MIL method [42] uses a permutation invariant aggregation operator called “Attention-based MIL pooling”. Encode each instance
is a feature vector extracted from the corresponding i-th region in the images (instance), and N is the number of regions (instance) where the images are segmented. Take two-classification as an example. A bag contains multiple instances. If all instances are marked negative, the bag is negative; otherwise, the bag is positive. The current state-of-the-art attention-based MIL method [42] uses a permutation invariant aggregation operator called “Attention-based MIL pooling”. Encode each instance
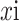 as a low-dimensional embedding through CNN, then use the attention mechanism to assign attention scores to each low-dimensional embedding (instance), and use their weighted average to generate bag-level prediction z. Let
as a low-dimensional embedding through CNN, then use the attention mechanism to assign attention scores to each low-dimensional embedding (instance), and use their weighted average to generate bag-level prediction z. Let
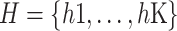 be a bag containing K low-dimensional embeddings, and then the bag-level prediction is defined as follows:
be a bag containing K low-dimensional embeddings, and then the bag-level prediction is defined as follows:
 |
where
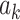 is defined as follows:
is defined as follows:
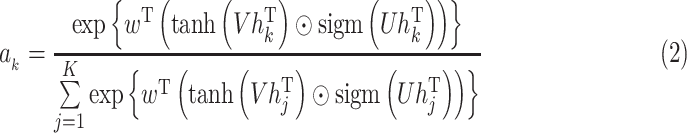 |
where
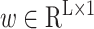 ,
,
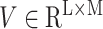 and
and
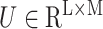 are trainable parameters. Attention-based MIL pooling is trainable and allows the network to identify discriminative instances. Since our problem only contains slide-level labels, using the attention mechanism in the MIL pooling will help achieve better results. Although the mil pool based on attention can help us find examples closely related to diagnosis, this method has a clear disadvantage. That is, it does not consider the optimization of attention weight.
are trainable parameters. Attention-based MIL pooling is trainable and allows the network to identify discriminative instances. Since our problem only contains slide-level labels, using the attention mechanism in the MIL pooling will help achieve better results. Although the mil pool based on attention can help us find examples closely related to diagnosis, this method has a clear disadvantage. That is, it does not consider the optimization of attention weight.
Gated attention weight normalization mechanism. We propose to normalize the attention weight by weight normalization [24] and use the multi-layer perceptron to calculate the attention score that accelerates the convergence of the stochastic gradient descent optimization and alleviates the problem of “vanishing gradient”. Specifically, we decompose
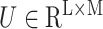 and
and
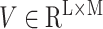 into a parameter vector
into a parameter vector
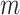 and a parameter scalar
and a parameter scalar
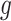 :
:
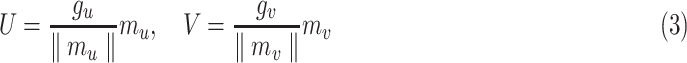 |
In summary, we can get:
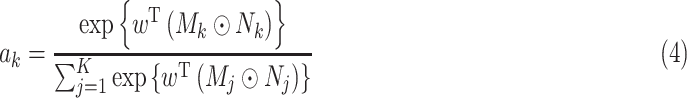 |
Among these:
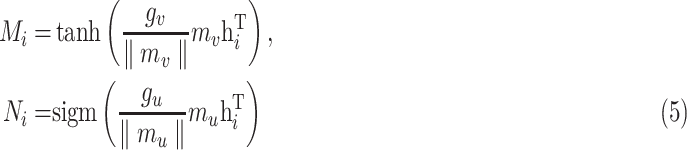 |
During training,
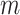 and
and
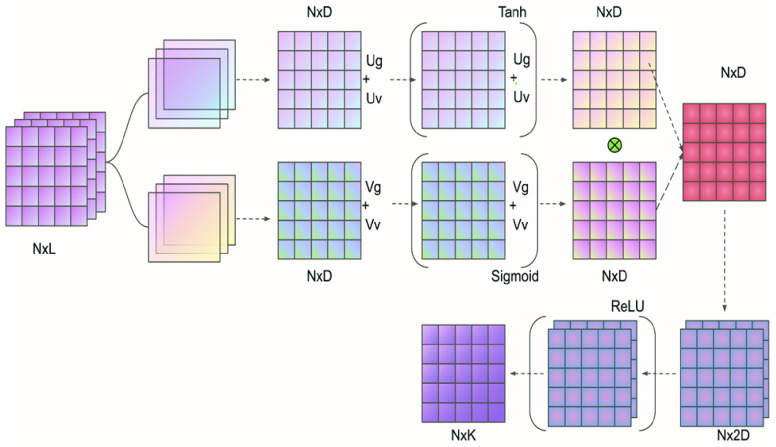
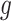 were updated, respectively. The experiments indicated that the proposed Gated attention weight normalization mechanism is superior to the traditional attention-based MIL method. The normalization mechanism of gated attention weight is shown in Fig. 1.
were updated, respectively. The experiments indicated that the proposed Gated attention weight normalization mechanism is superior to the traditional attention-based MIL method. The normalization mechanism of gated attention weight is shown in Fig. 1.
FIGURE 1.
Overview of MLP-based attention gating weight normalization. Among them, Ug, Uv, and Vg, Vv represent network parameters. N, L, D, and K respectively represent the number of patches included in each slide, the input feature dimension, the hidden layer dimension, and the number of classes.
B. Bilinear Attention Multi-Scale Feature Fusion
To alleviate the problem of contextual information loss caused by single-scale sampling, we propose a bilinear attention multi-scale feature fusion mechanism. Firstly, we extract
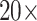 and
and
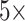 magnification patches from pathological images by sliding window strategy. Then, the attention matrix of WSIs under the different field of view sampling is obtained through feature extraction and attention mechanism. Finally, we get the final WSIs attention matrix by bilinear transformation. The bilinear transformation follows the following formula:
magnification patches from pathological images by sliding window strategy. Then, the attention matrix of WSIs under the different field of view sampling is obtained through feature extraction and attention mechanism. Finally, we get the final WSIs attention matrix by bilinear transformation. The bilinear transformation follows the following formula:
 |
Among these,
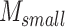 is the attention matrix corresponding to the small field of view patches,
is the attention matrix corresponding to the small field of view patches,
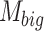 is the attention matrix corresponding to the big field of view patches,
is the attention matrix corresponding to the big field of view patches,
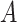 is the weight,
is the weight,
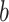 is the bias. Attention-based bilinear multi-scale feature fusion mechanism can effectively improve the classification performance of pathological images.
is the bias. Attention-based bilinear multi-scale feature fusion mechanism can effectively improve the classification performance of pathological images.
C. Network Framework
We follow the feature extraction and classification process shown in Fig. 2. In this study, we use the Otsu binarization algorithm to separate tissue regions, and use the residual neural network (ResNet18) [43] as feature extractors, then use the method proposed in this study to aggregate the patch-level predictions into slide-level predictions. Since the tumor area is much smaller than the normal tissue area, the number of positive and negative instances is seriously unbalanced, which leads to the wrong division of instances in the model. Therefore, in the instance classification stage, we introduce dual focal loss [44] to alleviate this problem. The loss function of the classification network is defined as follows:
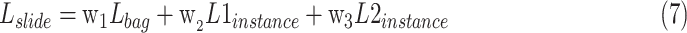 |
where
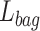 means cross entropy loss,
means cross entropy loss,
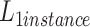 means cross entropy loss and
means cross entropy loss and
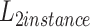 means dual focal loss. We use the weighted loss function as the objective function of network optimization to obtain better performance.
means dual focal loss. We use the weighted loss function as the objective function of network optimization to obtain better performance.
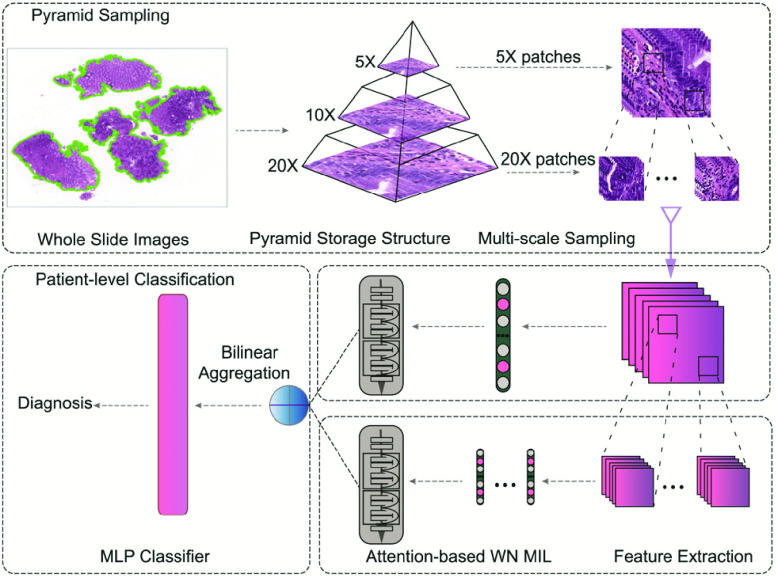
FIGURE 2.
Overview of our model. Patches extracted from each magnification of the WSIs are used for feature extractor separately. The trained feature extractors are used to compute embeddings of patches. The bilinear attention multi-scale feature fusion mechanism is used to connect WSIs embeddings of different scales to obtain the input of the final classifier.
IV. Experiment and Results
A. Data Description
We demonstrated our method and compared it with the standard method several times. Our locally advanced rectal cancer (LARC) dataset consists of 150 high-resolution WSIs from Yunnan cancer hospital. At present, the most commonly used clinical pathology evaluation criteria include TNM staging, histological subtype classification, tumor regression grade, pathological morphology, etc. In this study, the tumor regression grade was used as the qualitative standard for the treatment response of rectal cancer after neoadjuvant chemoradiotherapy. The data were first stained by H&E and then labeled by pathologists according to the regression grading system (TRG) of the American Joint Commission on Cancer (AJCC). There are four TRG groups: TRG 0, no residual tumor cells; TRG 1, single or small tumor remnant; TRG 2, part of the cancer tissue remains; TRG 3, a large number of cancer cells remain. Then, according to the AJCC TRG system, the treatment response is divided into two groups: good response (GR, TRG 0-1) and non-good response (non-GR, TRG 2-3). That is, in the model training stage, we use the pathological image of rectal cancer as the original data, extract the histopathological features such as the intensity, shape, and texture features of the tumor, and use the treatment response after neoadjuvant chemoradiotherapy as a label to establish a deep learning model. Then, the model can be used to predict the patient’s response to neoadjuvant chemoradiotherapy accurately. We divided the training set, validation dataset, and test dataset with the ratio of 60%-20%-20%. The specific data are shown in Table 1.
TABLE 1. The LARC Dataset Distribution.
| Total | GR | non-GR | |
|---|---|---|---|
| Training | 90 | 44 | 46 |
| Validation | 30 | 15 | 15 |
| Testing | 30 | 15 | 15 |
We also reported the performance of our model on the publicly available Camelyon16 [11] and MSKCC [14] breast cancer metastasis detection datasets. The Camelyon16 dataset contains 399 WSIs, including 270 for training and 129 for testing. The MSKCC dataset contains 130 WSIs, including 100 for training and 30 for testing. We extracted two groups of pathological patches with an area of more than 10% from each WSIs according to
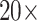 and
and
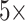 magnification for standby. In the Camelyon16 and MSKCC datasets, although the tumor area has complete pixel-level annotations on each slide, to verify the effectiveness of our weakly supervised model, we ignore pixel-level annotations during training and only consider slide-level labels. The specific data are shown in Table 2 and Table 3.
magnification for standby. In the Camelyon16 and MSKCC datasets, although the tumor area has complete pixel-level annotations on each slide, to verify the effectiveness of our weakly supervised model, we ignore pixel-level annotations during training and only consider slide-level labels. The specific data are shown in Table 2 and Table 3.
TABLE 2. The Camelyon16 Dataset Distribution.
| Total | Normal | Tumor | |
|---|---|---|---|
| Training | 270 | 159 | 111 |
| Testing | 129 | 80 | 49 |
TABLE 3. The MSKCC Dataset Distribution.
| Total | Normal | Tumor | |
|---|---|---|---|
| Training | 100 | 76 | 24 |
| Testing | 30 | 18 | 12 |
B. Results
In this section, we briefly introduce the evaluation results of our proposed algorithm. For a comprehensive evaluation, we employed two standard metrics for evaluating classification quality: area under the receiver operating characteristic curve (AUC) and accuracy. We carefully conducted ablation experiments and observed positive results. The ablation experiment aims to explore the role of the gated attention weight normalization mechanism based on the multilayer perceptron, the multi-scale feature fusion method based on bilinear attention, and the weighted loss function in improving the performance of pathological images classification. We first conducted experiments on the prediction dataset of neoadjuvant chemoradiotherapy for rectal cancer collected from Yunnan Cancer Hospital. Beyond that, to verify the generalization ability of the model, we compare the proposed algorithm with CLAM [25], MIL-CE [22], MIL-RNN [14], DSMIL [45] four state-of-the-art algorithms are evaluated on Camelyon16 and MSKCC datasets. We reimplemented all the previous methods based on the literature and open source code. The backbone of the CNN used in our proposed model is ResNet18 [43].
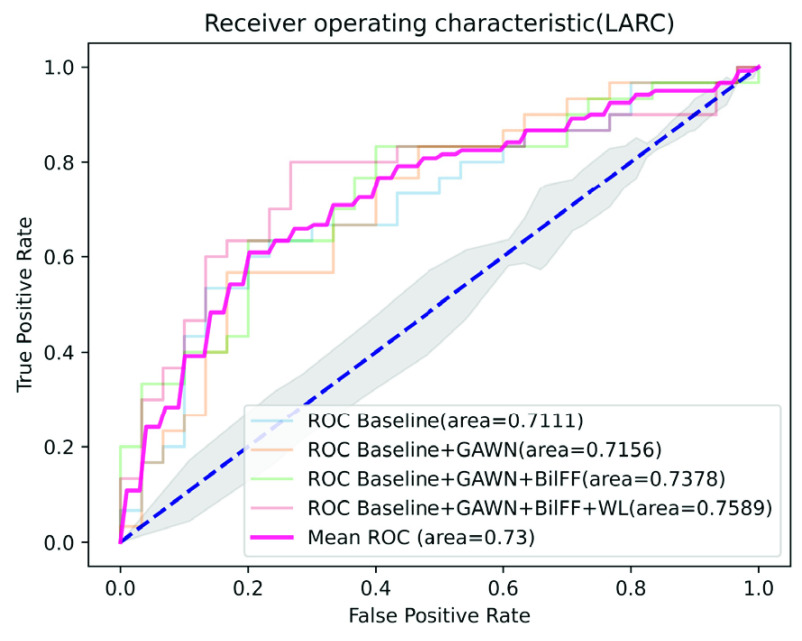
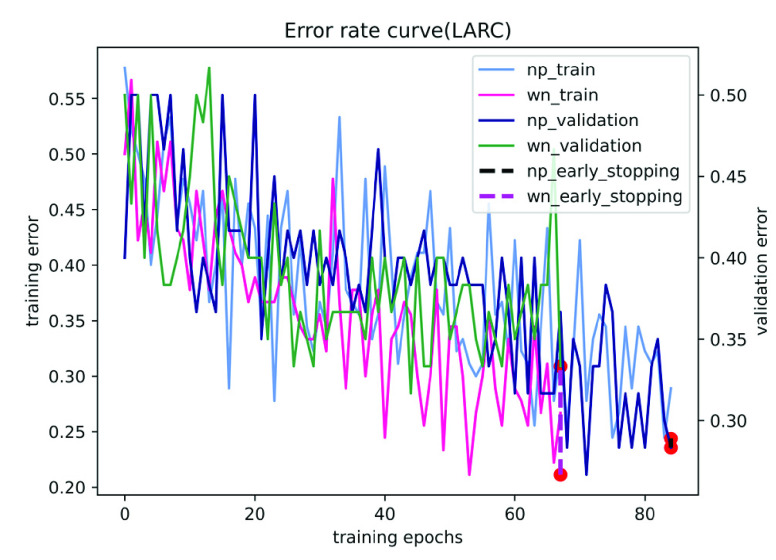
On the LARC dataset, we only use 90 pathological images as training data to obtain a 0.7589 AUC score on the test data set, achieving performance comparable to that of pathologists. Fig. 3 and Table 4 shows the performance of our method on the LARC dataset. It can be seen from Fig. 4 that our model is easier to learn the best parameters than the traditional model, and the number of iterations is significantly reduced, which proves the effectiveness of our gated attention weight normalization mechanism.
FIGURE 3.
Ablation experiment on LARC dataset, among them, GAWN represents the gated attention weight normalization mechanism, BiFF represents the bilinear attention multi-scale feature fusion mechanism, and WL represents the weighted loss function.
TABLE 4. Results on LARC Dataset.
FIGURE 4.
The training and validation error for LARC using different network parameterizations. Where np and wn represent normal parameters and weight normalization parameters, respectively. The red dot is the early stop.
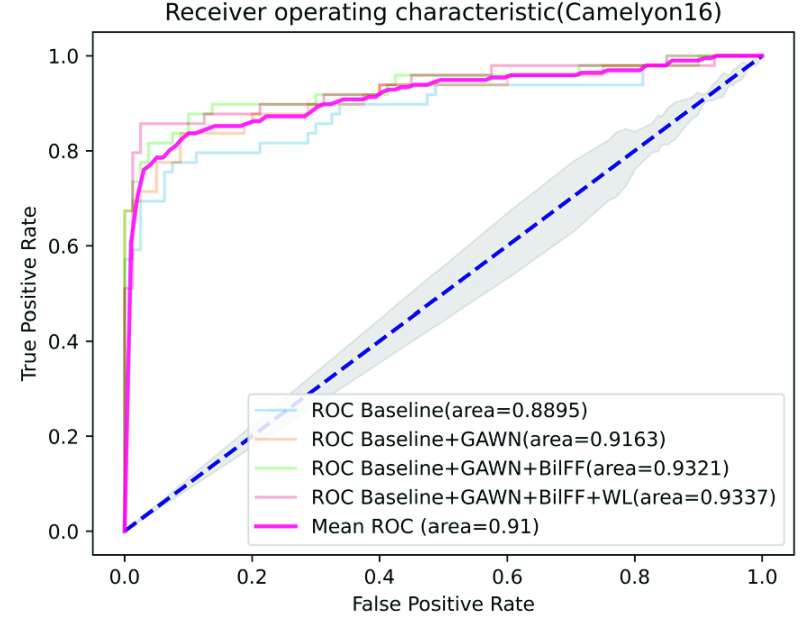
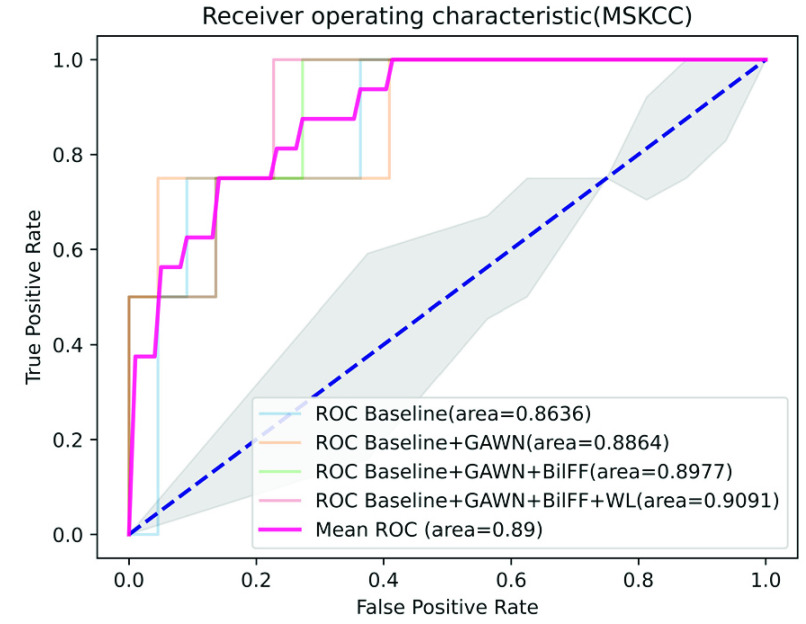
On the Camelyon16 and MSKCC datasets, by training only the slide-level labels, the AUC scores of our method on the test dataset reached 0.9337 and 0.9091, respectively. Among the other three models, DSMIL and MIL-RNN have the best performance, with AUC scores of 0.9225 and 0.8846, respectively.
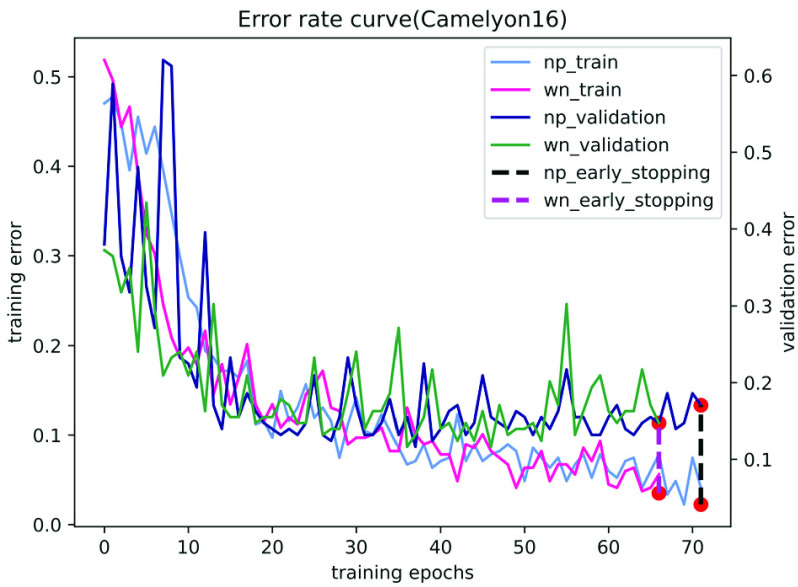
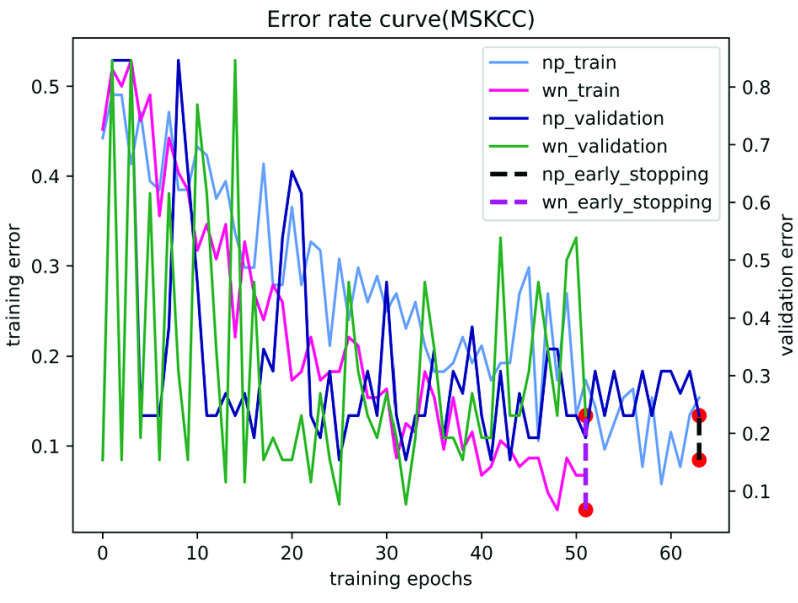
In contrast, our performance is quite competitive. Fig. 5 and Fig. 6 show the results of the ablation study of our algorithm on the two datasets of Camelyon16 and MSKCC. We observe that our method can accurately identify patches with tumor areas and give them higher attention weights (see Table 5 and Table 6). Compared with traditional attention-based multi-instance learning methods, our method accelerates the convergence of stochastic gradient descent optimization (see Fig. 7 and Fig. 8). Experiments show that our method is more advanced than other algorithms and has excellent performance on both private and publicly available datasets.
FIGURE 5.
Ablation experiment on Camelyon16 dataset, among them, GAWN represents the gated attention weight normalization mechanism, BiFF represents the bilinear attention multi-scale feature fusion mechanism, and WL represents the weighted loss function.
FIGURE 6.
Ablation experiment on MSKCC dataset, among them, GAWN represents the gated attention weight normalization mechanism, BiFF represents the bilinear attention multi-scale feature fusion mechanism, and WL represents the weighted loss function.
TABLE 5. Results on Camelyon16 Dataset.
TABLE 6. Results on MSKCC Dataset.
FIGURE 7.
The training and validation error for Camelyon16 using different network parameterizations. Where np and wn represent normal parameters and weight normalization parameters, respectively. The red dot is the early stop.
FIGURE 8.
The training and validation error for MSKCC using different network parameterizations. Where np and wn represent normal parameters and weight normalization parameters, respectively. The red dot is the early stop.
V. Discussions and Conclusion
In this study, to simulate the actual diagnosis process of pathologists and provide personalized treatment plans for patients with rectal cancer, we propose a new classification framework for weakly-supervised pathological images. The framework only uses patient-level labels to predict the response to neoadjuvant chemoradiotherapy for rectal cancer, without the need for pathologists to manually outline the decision boundary between cancerous tissues and normal tissues, which greatly reduces the burden of labeling for pathologists.
In addition, it can be seen from the result figures of the ablation experiment that each method we propose can effectively improve the performance of the model. Among them, the gated attention weight normalization mechanism speeds up the training process and effectively alleviates the problem of gradient diffusion. The bilinear attention multi-scale feature fusion mechanism can better integrate pathological image features, which can effectively alleviate the problem of global information loss caused by block sampling of WSIs. Finally, because the cancerous area in the pathological images is much larger than the normal area, it is easy to cause the imbalance of positive and negative instances after block sampling, and the weighted loss function just alleviates this problem. Experimental results show that our algorithm is superior to other weakly supervised learning algorithms on the private dataset and publicly available Camelyon16 and MSKCC datasets. Therefore, the method we propose is an effective method to predict the efficacy of neoadjuvant chemoradiotherapy for rectal cancer using histopathological images, and also has an excellent performance in breast cancer metastasis detection. In the future, we will further explore the feasibility of our method in other cancer prognosis studies and apply this technology to the clinic to assist clinicians in rapid diagnosis.
Funding Statement
This work was supported in part by the Chinese Natural Science Foundation under Grant 61876166 and Grant 61663046, in part by the Yunnan Provincial Major Science and Technology Special Plan Project under Grant 202002AD080001, in part by the Yunnan Basic Research Program for Distinguished Young Youths Project under Grant 202101AV070003, in part by the Yunnan Provincial Major Science and Technology Special Plan Projects: Digitization Research and Application Demonstration of Yunnan Characteristic Industry under Grant 202002AD080001, and in part by the Open Foundation of Key Laboratory in Software Engineering of Yunnan Province under Grant 2020SE304.
Contributor Information
Jing Guo, Email: guojing@mail.ynu.edu.cn.
Yaowei Wang, Email: 12019202407@mail.ynu.edu.cn.
Yun Yang, Email: yangyun@ynu.edu.cn.
Zhenhui Li, Email: lizhenhui621@qq.com.
References
- [1].Siegel R. L., “Colorectal cancer statistics, 2020,” CA: Cancer J. Clinicians, vol. 70, no. 3, pp. 145–164, 2020. [DOI] [PubMed] [Google Scholar]
- [2].Glynne-Jones R., “Rectal cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up,” Ann. Oncol., vol. 28, pp. iv22–iv40, Jul. 2017. [DOI] [PubMed] [Google Scholar]
- [3].Scott N. A., Susnerwala S., Gollins S., Myint A. S., and Levine E., “Preoperative neo-adjuvant therapy for curable rectal cancer–reaching a consensus 2008,” Colorectal Disease, vol. 11, no. 3, pp. 245–248, 2009. [DOI] [PubMed] [Google Scholar]
- [4].Sauer R.et al. , “Adjuvant versus neoadjuvant radiochemotherapy for locally advanced rectal cancer a progress report of a phase-III randomized trial (protocol CAO/ARO/AIO-94): A progress report of a phase-III randomized trial (protocol CAO/ARO/AIO-94),” Strahlentherapie und Onkologie, vol. 177, no. 4, pp. 173–181, Mar. 2001. [DOI] [PubMed] [Google Scholar]
- [5].Park I. J., You Y. N., Agarwal A., and Skibber J. M., “Neoadjuvant treatment response as an early response indicator for patients with rectal cancer,” J. Clin. Oncol., vol. 30, no. 15, p. 1770, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Patel U. B., Taylor F., Blomqvist L., and George C., “Magnetic resonance imaging–detected tumor response for locally advanced rectal cancer predicts survival outcomes: MERCURY experience,” J. Clin. Oncol., vol. 29, no. 28, pp. 3753–3760, 2011. [DOI] [PubMed] [Google Scholar]
- [7].Fokas E.et al. , “Tumor regression grading after preoperative chemoradiotherapy as a prognostic factor and individual-level surrogate for disease-free survival in rectal cancer,” JNCI: J. Nat. Cancer Inst., vol. 109, no. 12, Dec. 2017, djx095. [DOI] [PubMed] [Google Scholar]
- [8].Sanghera P., Wong D. W. Y., McConkey C. C., Geh J. I., and Hartley A., “Chemoradiotherapy for rectal cancer: An updated analysis of factors affecting pathological response,” Clin. Oncol., vol. 20, no. 2, pp. 176–183, Mar. 2008. [DOI] [PubMed] [Google Scholar]
- [9].Maas M.et al. , “Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: A pooled analysis of individual patient data,” Lancet Oncol., vol. 11, no. 9, pp. 835–844, Sep. 2010. [DOI] [PubMed] [Google Scholar]
- [10].Bedard P. L., Hansen A. R., Ratain M. J., and Siu L. L., “Tumour heterogeneity in the clinic,” Nature, vol. 501, no. 7467, pp. 355–364, Sep. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bejnordi B. E.et al. , “Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer,” JAMA, vol. 318, no. 22, pp. 2199–2210, Dec. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].van der Sommen F.et al. , “Machine learning in GI endoscopy: Practical guidance in how to interpret a novel field,” Gut, vol. 69, no. 11, pp. 2035–2045, Nov. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Syed S. and Stidham R. W., “Potential for standardization and automation for pathology and endoscopy in inflammatory bowel disease,” Inflammatory Bowel Diseases, vol. 26, no. 10, pp. 1490–1497, Sep. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Campanella G.et al. , “Clinical-grade computational pathology using weakly supervised deep learning on whole slide images,” Nature Med., vol. 25, no. 8, pp. 1301–1309, Aug. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang X.et al. , “Weakly supervised deep learning for whole slide lung cancer image analysis,” IEEE Trans. Cybern., vol. 50, no. 9, pp. 3950–3962, Sep. 2020. [DOI] [PubMed] [Google Scholar]
- [16].Hou L., Samaras D., Kurc T. M., Gao Y., Davis J. E., and Saltz J. H., “Patch-based convolutional neural network for whole slide tissue image classification,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR), Jun. 2016, pp. 2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li S.et al. , “Multi-instance multi-scale CNN for medical image classification,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervent. Cham, Switzerland: Springer, 2019, pp. 531–539. [Google Scholar]
- [18].Yao J., Zhu X., and Huang J., “Deep multi-instance learning for survival prediction from whole slide images,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervent. Cham, Switzerland: Springer, 2019, pp. 496–504. [Google Scholar]
- [19].Chen H.et al. , “Rectified cross-entropy and upper transition loss for weakly supervised whole slide image classifier,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervent. Cham, Switzerland: Springer, 2019, pp. 351–359. [Google Scholar]
- [20].Zhu X., Yao J., Zhu F., and Huang J., “WSISA: Making survival prediction from whole slide histopathological images,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR), Jul. 2017, pp. 7234–7242. [Google Scholar]
- [21].Xu Y.et al. , “Parallel multiple instance learning for extremely large histopathology image analysis,” BMC Bioinf., vol. 18, no. 1, pp. 1–15, Dec. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chikontwe P., Kim M., Nam S. J., Go H., and Park S. H., “Multiple instance learning with center embeddings for histopathology classification,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervent. Cham, Switzerland: Springer, 2020, pp. 519–528. [Google Scholar]
- [23].Xie C.et al. , “Beyond classification: Whole slide tissue histopathology analysis by end-to-end part learning,” in Medical Imaging With Deep Learning. PMLR, 2020, pp. 843–856. [Google Scholar]
- [24].Salimans T. and Kingma D. P., “Weight normalization: A simple reparameterization to accelerate training of deep neural networks,” in Proc. Adv. Neural Inf. Process. Syst., vol. 29, 2016, pp. 901–909. [Google Scholar]
- [25].Lu M. Y., Williamson D. F. K., Chen T. Y., Chen R. J., Barbieri M., and Mahmood F., “Data-efficient and weakly supervised computational pathology on whole-slide images,” Nature Biomed. Eng., vol. 5, no. 6, pp. 555–570, Jun. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhou X.et al. , “Radiomics-based pretherapeutic prediction of non-response to neoadjuvant therapy in locally advanced rectal cancer,” Ann. Surgical Oncol., vol. 26, no. 6, pp. 1676–1684, Jun. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Liu Z.et al. , “Radiomics analysis for evaluation of pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer,” Clin. Cancer Res., vol. 23, no. 23, pp. 7253–7262, Dec. 2017. [DOI] [PubMed] [Google Scholar]
- [28].Shao L.et al. , “Multiparametric MRI and whole slide image-based pretreatment prediction of pathological response to neoadjuvant chemoradiotherapy in rectal cancer: A multicenter radiopathomic study,” Ann. Surgical Oncol., vol. 27, no. 11, pp. 4296–4306, Oct. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Skrede O.-J.et al. , “Deep learning for prediction of colorectal cancer outcome: A discovery and validation study,” Lancet, vol. 395, no. 10221, pp. 350–360, 2020. [DOI] [PubMed] [Google Scholar]
- [30].Zhang F.et al. , “Predicting treatment response to neoadjuvant chemoradiotherapy in local advanced rectal cancer by biopsy digital pathology image features,” Clin. Translational Med., vol. 10, no. 2, p. e110, Jun. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kather J. N.et al. , “Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study,” PLOS Med., vol. 16, no. 1, Jan. 2019, Art. no. e1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kather J. N.et al. , “Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer,” Nature Med., vol. 25, no. 7, pp. 1054–1056, Jul. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bychkov D.et al. , “Deep learning based tissue analysis predicts outcome in colorectal cancer,” Sci. Rep., vol. 8, no. 1, pp. 1–11, Dec. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sudharshan P. J., Petitjean C., Spanhol F., Oliveira L. E., Heutte L., and Honeine P., “Multiple instance learning for histopathological breast cancer image classification,” Expert Syst. Appl., vol. 117, pp. 103–111, Mar. 2018. [Google Scholar]
- [35].Kim J.-H., Jun J., and Zhang B.-T., “Bilinear attention networks,” 2018, arXiv:1805.07932.
- [36].Yang Y., Hu Y., Zhang X., and Wang S., “Two-stage selective ensemble of CNN via deep tree training for medical image classification,” IEEE Trans. Cybern., early access, Mar. 11, 2021, doi: 10.1109/TCYB.2021.3061147. [DOI] [PubMed]
- [37].Yang Y. and Jiang J., “Adaptive Bi-weighting toward automatic initialization and model selection for HMM-based hybrid meta-clustering ensembles,” IEEE Trans. Cybern., vol. 49, no. 5, pp. 1657–1668, May 2019. [DOI] [PubMed] [Google Scholar]
- [38].Yang Y. and Jiang J., “Bi-weighted ensemble via HMM-based approaches for temporal data clustering,” Pattern Recognit., vol. 76, pp. 391–403, Apr. 2018. [Google Scholar]
- [39].Yang Y., Li X., Wang P., Xia Y., and Ye Q., “Multi-source transfer learning via ensemble approach for initial diagnosis of Alzheimer’s disease,” IEEE J. Transl. Eng. Health Med., vol. 8, 2020, Art. no. 1400310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhao Y.et al. , “Predicting lymph node metastasis using histopathological images based on multiple instance learning with deep graph convolution,” in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit. (CVPR), Jun. 2020, pp. 4837–4846. [Google Scholar]
- [41].Wang S.et al. , “RMDL: Recalibrated multi-instance deep learning for whole slide gastric image classification,” Med. Image Anal., vol. 58, Dec. 2019, Art. no. 101549. [DOI] [PubMed] [Google Scholar]
- [42].Mormont R., Geurts P., and Maree R., “Comparison of deep transfer learning strategies for digital pathology,” in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit. Workshops (CVPRW), Jun. 2018, pp. 2262–2271. [Google Scholar]
- [43].He K., Zhang X., Ren S., and Sun J., “Deep residual learning for image recognition,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR), Jun. 2016, pp. 770–778. [Google Scholar]
- [44].Hossain M. S., Paplinski A. P., and Betts J. M, “Adaptive class weight based dual focal loss for improved semantic segmentation,” 2019, arXiv:1909.11932.
- [45].Li B., Li Y., and Eliceiri K. W., “Dual-stream multiple instance learning network for whole slide image classification with self-supervised contrastive learning,” in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit. (CVPR), Jun. 2021, pp. 14318–14328. [DOI] [PMC free article] [PubMed] [Google Scholar]