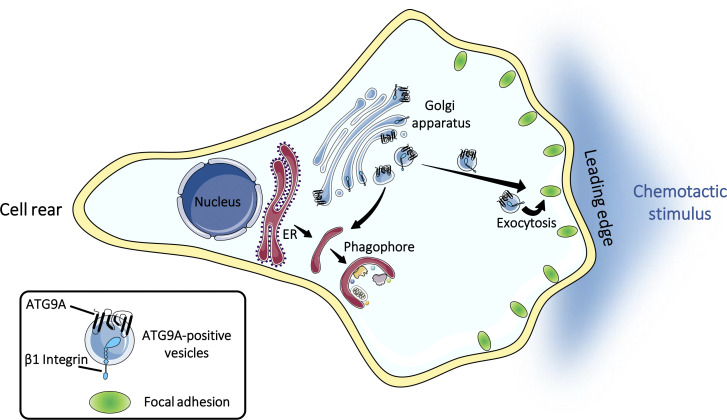
Campisi et al. uncover a new function of the core autophagy protein ATG9A in cell migration by demonstrating that ATG9A-positive vesicles are mobilized during chemotactic stimulation to participate in the extension and stabilization of leading-edge protrusions.
Abstract
Chemotactic migration is a fundamental cellular behavior relying on the coordinated flux of lipids and cargo proteins toward the leading edge. We found here that the core autophagy protein ATG9A plays a critical role in the chemotactic migration of several human cell lines, including highly invasive glioma cells. Depletion of ATG9A protein altered the formation of large and persistent filamentous actin (F-actin)–rich lamellipodia that normally drive directional migration. Using live-cell TIRF microscopy, we demonstrated that ATG9A-positive vesicles are targeted toward the migration front of polarized cells, where their exocytosis correlates with protrusive activity. Finally, we found that ATG9A was critical for efficient delivery of β1 integrin to the leading edge and normal adhesion dynamics. Collectively, our data uncover a new function for ATG9A protein and indicate that ATG9A-positive vesicles are mobilized during chemotactic stimulation to facilitate expansion of the lamellipodium and its anchorage to the extracellular matrix.
Graphical Abstract
Introduction
Chemotactic cell migration is critical for a variety of cellular processes, including cell movement during normal development, immune responses, and pathological processes such as tumor cell invasion and metastasis (Keller, 2005; Bravo-Cordero et al., 2012). It is a highly coordinated and dynamic process that involves polarization of the cell, followed by the extension of an actin-dependent membrane protrusion, the lamellipodium, toward the chemotactic stimulus. Formation of adhesions to the extracellular matrix, via the integrin family of transmembrane receptors, then stabilizes the lamellipodium and acts as a molecular clutch, allowing the cell body to pull itself forward (Ridley et al., 2003).
Although many studies have indicated that biased vesicle trafficking toward the leading edge is crucial for the different steps of cell migration (Fletcher and Rappoport, 2010), the origin and diversity of the vesicles, as well as their modes of transport to the cell front, are still unclear. Mounting evidence suggests a prominent role of the secretory pathway in cell migration. Disruption of Golgi integrity and failure of the Golgi to reorient toward the leading edge of migrating cells have been shown to block directional cell movement (Bisel et al., 2008; Millarte and Farhan, 2012; Pallesi-Pocachard et al., 2016). Accordingly, using time-lapse total internal reflection fluorescence (TIRF) microscopy, post-Golgi vesicles were found to exhibit polarized exocytosis toward the cell front of migrating fibroblasts and astrocytes (Schmoranzer et al., 2003; Letinic et al., 2009). Data obtained from several cell types also indicate that recycling endosomes constitute an important source of internal membrane and cargo proteins that are incorporated at the leading edge during cell migration (Kean et al., 2009; Sneeggen et al., 2019; Proux-Gillardeaux et al., 2005; Riggs et al., 2012; Tayeb et al., 2005; Veale et al., 2011; Veale et al., 2010).
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved lysosomal pathway involved in the degradation of long-lived proteins and cytoplasmic organelles. Several highly conserved autophagy-related (ATG) proteins that function at key steps in the autophagy process have been identified. Among them, ATG9A is the only transmembrane protein. In mammalian cells, ATG9A cycles primarily between the TGN, the plasma membrane, and endosomal compartments (Young et al., 2006; Ravikumar et al., 2010; Puri et al., 2013; Søreng et al., 2018; Longatti and Tooze, 2012). Extensive studies have demonstrated the critical role of ATG9A protein in autophagosome formation, proposedly by functioning in vesicular delivery to the phagophore initiation site and by translocating lipids from the outer to the inner leaflet of the phagophore membrane to enable its expansion (Orsi et al., 2012; Maeda et al., 2020; Matoba et al., 2020). In addition to this well-recognized function of ATG9A in phagophore expansion, recent studies point to the fact that it may have a wider role than anticipated and could act as a general regulator of vesicular trafficking. Hence, Jia et al. (2017) found that ATG9A was required for the post-Golgi transport and maturation of lysosomal hydrolases. In cortical neurons, the loss of ATG9A, but not that of other autophagy genes (ATG7 or ATG16L1), markedly reduced neurite extension (Yamaguchi et al., 2018), a process that strongly relies on intracellular membrane trafficking (Sann et al., 2009).
In this framework, we found here that ATG9A-containing vesicles are essential components of directed cell migration. ATG9A-depleted cells were unable to form large and stable F-actin–rich lamellipodia. By the design of a pH-sensitive fluorescently tagged ATG9A, together with live-cell TIRF imaging, we established that exocytosis of ATG9A-containing vesicles is highly polarized toward the cell front, induced by chemotactic stimuli, and synchronized with protrusive activity. Finally, we further demonstrated that ATG9A is critical for the delivery of β1 integrin to the leading edge and normal adhesion dynamics.
Results
Chemotactic migration depends on ATG9A protein
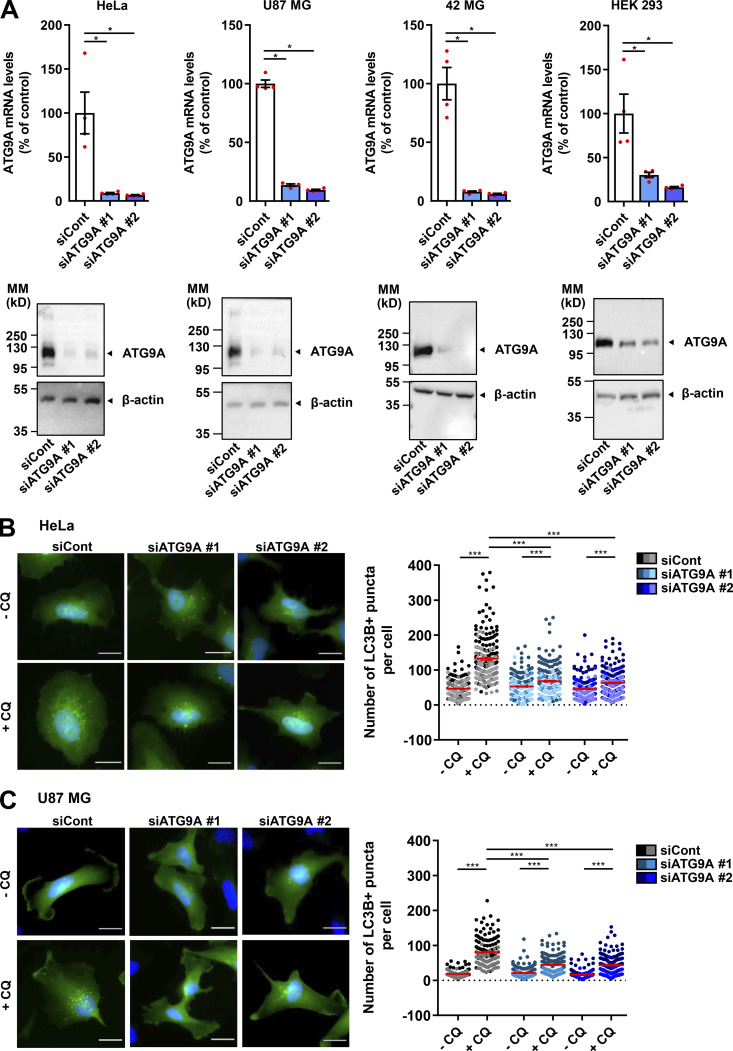
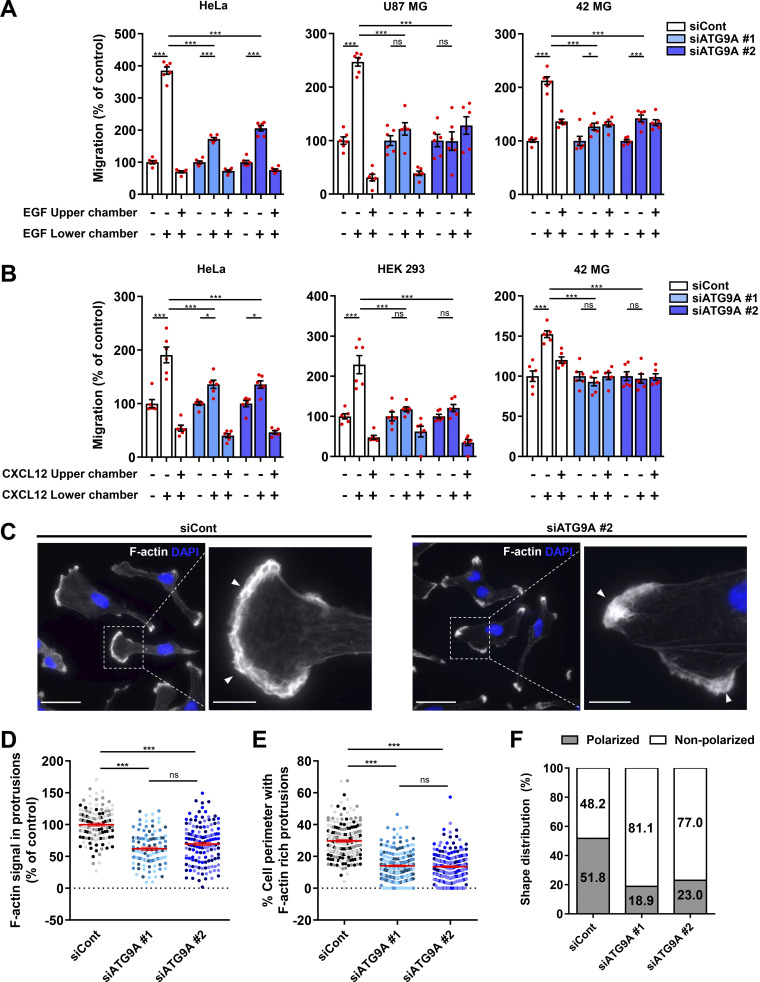
We tested the role of ATG9A during chemotactic migration by targeting the ATG9A transcript by siRNA in several human cell lines, i.e., HeLa, HEK 293, and the highly invasive U87 MG and 42 MG glioma cells. These cell lines were chosen because of their well-described responsiveness to various chemotactic stimuli (Dillenburg-Pilla et al., 2015; Lecointre et al., 2015; Coly et al., 2016) and the already-characterized localization and trafficking of ATG9A during autophagy induction in both HeLa and HEK 293 cells (Takahashi et al., 2016; Zhou et al., 2017; Kakuta et al., 2017; Davies et al., 2018).To make sure that siRNA effects would not be due to off-target effects, we performed experiments with two independent siRNAs interacting with different regions of the ATG9A transcript. Effectiveness of each siRNA was verified by quantitative RT-PCR (RT-qPCR) and Western blot (Fig. S1 A). As a control, we also tested the effect of each siRNA on autophagic activity. In agreement with the well-documented role of ATG9A in phagophore expansion, the knockdown of ATG9A strongly reduced autophagosome biogenesis, as evaluated by the accumulation of EGFP–microtubule associated protein 1 light chain 3β (LC3B) puncta under chloroquine (CQ) treatment (Fig. S1, B and C). We next assessed the role of ATG9A in chemotactic migration using a Transwell chemotaxis assay, in which cells loaded in the upper chamber of the Transwell migrated toward the bottom chamber containing the chemoattractant. We first examined the effects of EGF, a potent factor that has been shown to be critical for normal and cancer cell chemotaxis (Adelmann-Grill et al., 1990; Segall et al., 1996; Kim et al., 2008; Mendoza et al., 2011; Biswenger et al., 2018). A chemotactic gradient elicited by EGF stimulated the migration of HeLa, U87 MG, and 42 MG cells (Fig. 1 A). EGF produced no effect when added in both chambers of the Transwell, stressing the fact that it stimulates chemotaxis rather than chemokinesis (random motility). Transfection of cells with each siRNA targeting the ATG9A transcript abrogated or markedly reduced chemotactic cell migration induced by EGF (Fig. 1 A). The experiments were repeated using C-X-C motif chemokine ligand 12 (CXCL12), the ligand of the prototypical chemokine receptor CXCR4 (Chatterjee et al., 2014). Similar to data obtained with EGF, ATG9A depletion abolished or markedly reduced CXCL12-induced chemotactic migration of HeLa, HEK 293, and 42 MG cells (Fig. 1 B). Together, these data suggest that, regardless of the cell type and the nature of the stimulus, ATG9A protein is a critical component of directed cell migration.
Figure S1.
Validation of ATG9A knockdown. (A) The indicated cell lines were transfected with nontargeting siRNA (siCont) or one of the two siRNAs targeting ATG9A (siATG9A #1, siATG9A #2). Upper panel: RT-qPCR analysis of ATG9A mRNA levels. Data show means ± SEM (n = 4). Lower panel: Western blot analysis of ATG9A protein levels. The blot was cut, and the upper and the lower parts were probed with antibodies against ATG9A and β-actin, respectively. MM, molecular mass. (B and C) Effect of siRNA knockdown of ATG9A on autophagosome biogenesis in HeLa cells (B) or U87 MG cells (C). Cells were transfected with nontargeting siRNA (siCont) or one of the two siRNAs targeting ATG9A (siATG9A #1, siATG9A #2), together with a construct encoding the EGFP-LC3B protein (marker of autophagosomes). Transfected cells were placed in serum-free medium for 6 h, in the presence or absence of CQ (5 × 10−5 M), as indicated. Cells were fixed, and the number of autophagosomes (EGFP-LC3B fluorescent dots) per cell was quantified. Data represent means ± SEM (n = 140–221 cells per group; cells from independent experiments were color-coded. Scale bar, 20 µm. Statistical significance was evaluated using Mann–Whitney U test (A) or one-way ANOVA followed by Sidak post hoc test (B and C). *, P < 0.05; ***, P < 0.001.
Figure 1.
Depletion of ATG9A impairs chemotactic migration and the formation of F-actin–rich protrusions. (A) Effect of siRNA knockdown of ATG9A on chemotactic migration induced by EGF. HeLa cells (left), U87 MG cells (middle), and 42 MG cells (right) were transfected with nontargeting siRNA (siCont) or one of the two siRNAs targeting ATG9A (siATG9A #1, siATG9A #2). Transfected cells were loaded in the upper chamber of Transwells, with or without EGF (50 ng/ml) in the upper or lower chamber, as indicated. After 24 h, cells that migrated onto the lower surface of the membrane were fixed, stained, and counted. Data represent means and SEM (n = 6 Transwells). (B) Effect of siRNA knockdown of ATG9A on chemotactic migration induced by CXCL12. Experiment was performed as described in C, using CXCL12 (10−8 M), on HeLa cells (left), HEK 293 cells (middle), and 42 MG cells (right). (C) Representative epifluorescence images showing F-actin (rhodamine phalloidin labeling; gray levels) and nuclei (DAPI labeling; blue) of U87 MG cells transfected with the indicated siRNAs. Control cells generally develop large F-actin–rich lamellipodia (arrowheads), whereas ATG9A-depleted cells display smaller and irregular protrusions (arrowheads). Scale bars, 20 µm; magnified views, 5 µm. (D) Quantification of F-actin intensities, after background subtraction, in protrusions of U87 MG cells transfected with the indicated siRNAs. For each cell, values correspond to cumulated signal of all protrusions. Data represent means and SEM (n = 106–135 cells per group; cells from independent experiments were color-coded). (E) Percentage of the cell perimeter containing F-actin–rich protrusions, in U87 MG cells transfected with the indicated siRNAs. Data represent means and SEM (n = 209–219 cells per group; cells from independent experiments were color-coded). (F) Effect of siRNA knockdown of ATG9A on cell polarization of U87 MG cells (n = 212–226 cells per group; from three independent experiments). Statistical significance was evaluated using a one-way ANOVA followed by Tukey post hoc test. *, P < 0.05; ***, P < 0.001.
Depletion of ATG9A protein alters the formation of F-actin–rich lamellipodia driving efficient cell migration
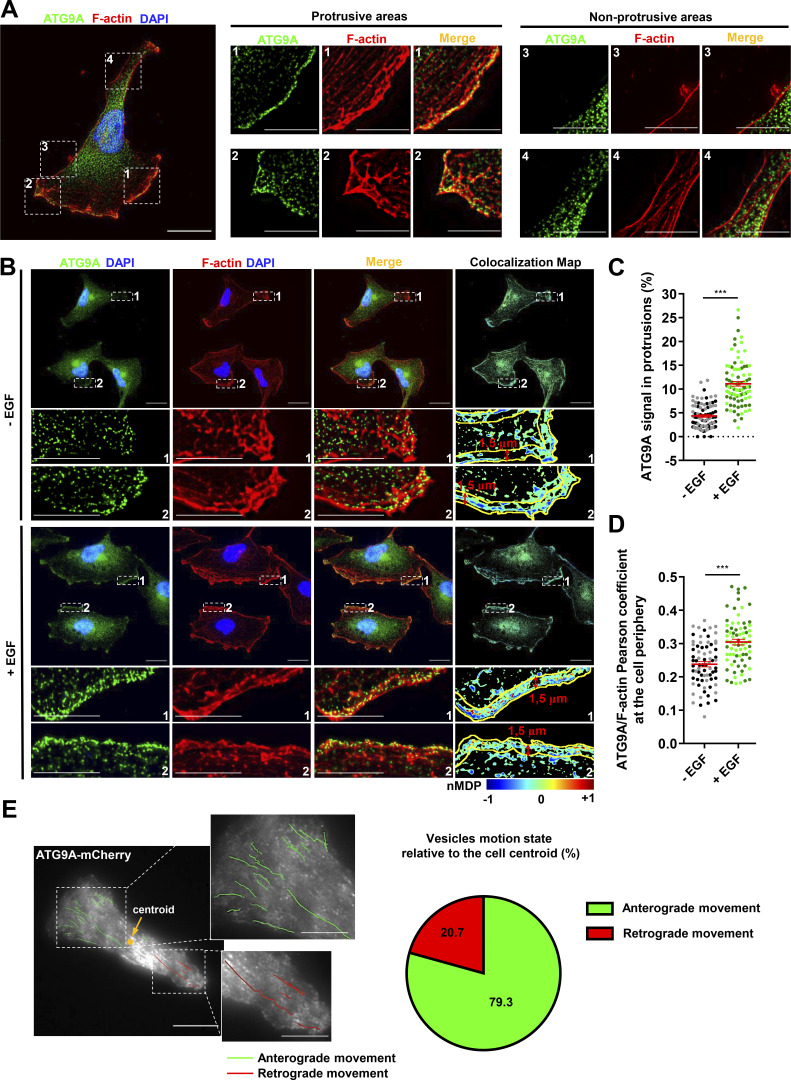
Because lamellipodia/lamella are considered the main drivers of chemotactic migration, we carefully examined the impact of ATG9A depletion on the morphology of cell protrusions. Experiments were done in U87 MG cells, since they consistently develop well-defined protrusions, even in the absence of chemotactic stimulation. Rhodamine phalloidin labeling of U87 MG cells transfected with irrelevant siRNA indeed revealed prominent lamellipodia, with intense F-actin staining (Fig. 1 C). In ATG9A-depleted cells, most of the protrusions exhibited an abnormal morphology, as they were narrow and often resembled small ruffles (Fig. 1 C), and broad F-actin–rich lamellipodia were rarely observed. Quantitative analysis of the rhodamine phalloidin fluorescence signal was then used to measure relative amounts of peripheral F-actin. Filament levels in protrusions of ATG9A-depleted cells were reduced >38% in comparison to control cells (Fig. 1 D). Moreover, we estimated that F-actin–rich protrusions occupied ∼14% of the cell perimeter in ATG9A-depleted cells, compared with ∼30% in control cells (Fig. 1 E). In agreement with their inability to form large lamellipodia, we noticed that most ATG9A-depleted cells (≃80%) displayed a nonpolarized phenotype, with no clear front–rear axis (Fig. 1 F).
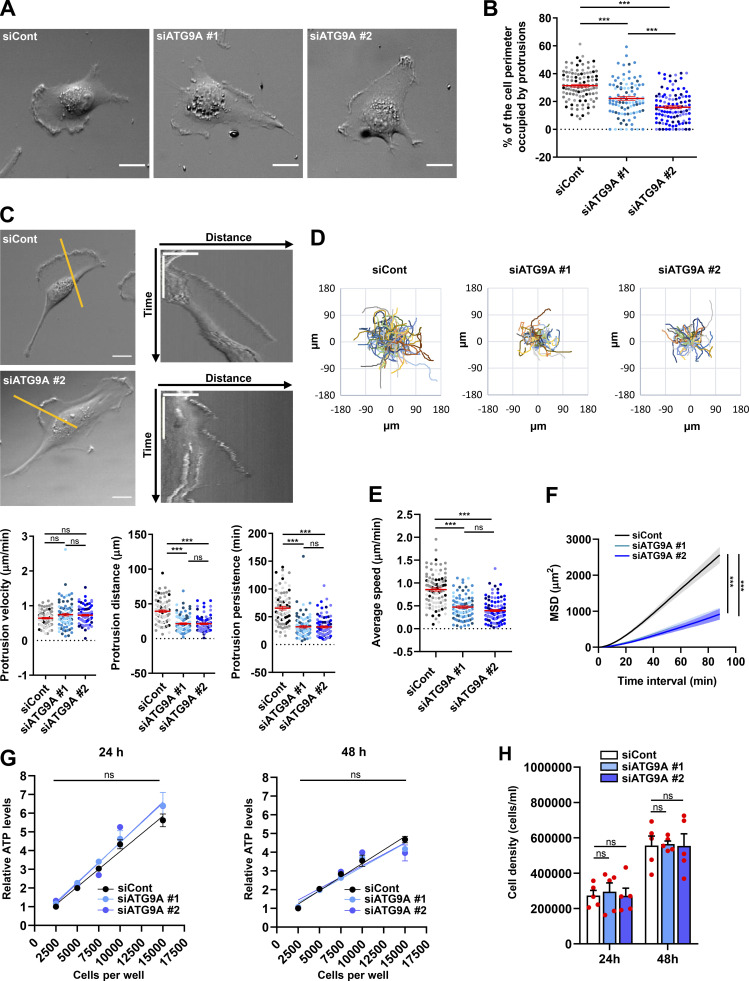
The altered morphology of protrusions on fixed ATG9A-depleted cells led us to evaluate their dynamics, using phase-contrast time-lapse microscopy. Live-cell videos of U87 MG cells transfected with irrelevant siRNA indicated that cells developed large protrusions associated with typical ruffles (Innocenti, 2018), accounting for 31% of the cell perimeter (Fig. 2, A and B). In agreement with experiments done on fixed cells, siATG9A-transfected cells displayed smaller protrusions, developing on ∼18% of the cell perimeter (Fig. 2, A and B). Analyses of the live-cell videos (Video 1) using kymographs indicated that, although the protrusion velocities were similar in the two groups (Fig. 2 C), siATG9A-transfected cells displayed protrusions with reduced distance and persistence (Fig. 2 C). We next monitored the intrinsic movement of cells transfected with siRNAs against ATG9A or irrelevant siRNA over a 3-h period. The single-cell trajectories are shown in Fig. 2 D. Quantitative analysis revealed that, whereas control cells migrated with a speed of 0.85 µm/min, the speed of ATG9A-depleted cells was reduced to ∼0.40 µm/min (Fig. 2 E). Accordingly, depletion of ATG9A also markedly reduced the mean square displacement (Fig. 2 F), which is a measure of the surface area explored by the cell over time. Measurement of ATP levels and cell growth indicated that these effects of ATG9A depletion were not due to a global modification of the metabolic or proliferation states (Fig. 2, G and H). Collectively, our data indicate that ATG9A protein is necessary for the formation of large and polarized F-actin–rich protrusions and efficient cell migration.
Figure 2.
Depletion of ATG9A protein impairs the dynamics of cell protrusions and intrinsic cell speed. (A) Representative images from phase-contrast time-lapse sequences acquired from U87 MG cells transfected with nontargeting siRNA (siCont) or one of the two siRNAs targeting ATG9A (siATG9A #1, siATG9A #2). (B) Percentage of the cell perimeter occupied by ruffling protrusions (n = 94–106 cells per group; cells from independent experiments were color-coded). (C) Phase-contrast time-lapse sequences acquired from U87 MG cells transfected with nontargeting siRNA (siCont) or one of the two siRNAs targeting ATG9A (siATG9A #1 or siATG9A #2). Upper panels: Single images from representative sequences of U87 MG cells transfected with nontargeting siRNA (siCont) and siATG9A #2 (upper left; scale bar, 20 µm), and kymographs (upper right) generated from 1-pixel-wide lines drawn on the cell edges. The scale bars in the kymographs are 20 µm (horizontal) and 40 min (vertical). Lower panels: Quantification, from kymographs, of protrusion velocity (left), protrusion distance (middle), and protrusion persistence (right; n = 84–87 protrusions per group; cells from independent experiments were color-coded). (D) Cell trajectories over a 3-h period (one frame every 40 s) of randomly migrating U87 MG cells transfected with nontargeting siRNA (siCont, n = 101 cells) or siRNA targeting ATG9A (siATG9A #1, n = 88 cells; siATG9A #2, n = 97 cells). (E and F) Analysis of average cell speed (E) and mean square displacement (MSD; F), calculated from the cell trajectories presented in D. (G) Relative ATP levels from lysates of serial amounts of U87 MG cells transfected with nontargeting siRNA (siCont) or one of the two siRNAs targeting ATG9A (siATG9A #1, siATG9A #2). Analyses (n = 3 wells for each cell number) were performed at 24 h (left panel) or 48 h (right panel) after transfection. Results were normalized to that of cells transfected with nontargeting siRNA (2,500 cells). (H) Measurement of cell density at 24 and 48 h after transfection, from cells transfected with nontargeting siRNA (siCont) or one of the two siRNAs targeting ATG9A (siATG9A #1, siATG9A #2). Data represent means and SEM. Statistical significance was evaluated using one-way ANOVA followed by Tukey post hoc test (B, C, and E), two-way ANOVA followed by Dunnett post hoc test (F), two-way ANOVA followed by Bonferroni post hoc test (G), or one-way ANOVA followed by Sidak post hoc test (H). ***, P < 0.001.
Video 1.
Depletion of ATG9A protein impairs the formation of cell protrusions. Representative live-cell phase-contrast images showing U87 MG cells transfected with nontargeting siRNA (siCont, left) or siRNA targeting ATG9A (siATG9A #1, right). Control cells develop large and stable protrusions, whereas ATG9A-depleted cells display smaller and unstable protrusions. Video plays at 25 frames/s. Scale bar, 20 µm.
ATG9A-positive vesicles concentrate in F-actin–rich protrusions and display anterograde trafficking toward the leading edge
To evaluate whether ATG9A-positive vesicles could potentially deliver bulk membrane or essential proteins for the formation/anchorage of cell protrusions, we first examined the subcellular distribution of endogenous ATG9A. The ATG9A signal, whose specificity was verified using siRNAs (data not shown), displayed a punctuated pattern, and vesicles mainly localized in the perinuclear area (Fig. 3 A). Importantly, ∼5% of the total ATG9A signal was also concentrated in F-actin–rich protrusions, at the cell membrane, and in small vesicles localized beneath the plasma membrane (Fig. 3 A). Chemotactic stimulation of cells with EGF increased the fraction of ATG9A signal localized in protrusions (+156%), as well as the colocalization score (Pearson correlation coefficient) of peripheral ATG9A with F-actin (Fig. 3, B–D). To examine the dynamics of ATG9A-positive vesicles, we next used live-cell TIRF microscopy on cells expressing the ATG9A-mCherry fusion protein (De Pace et al., 2018). Live-cell videos of U87 MG cells revealed that many small (<500-nm) motile or nonmotile ATG9A-positive puncta were located in the evanescent field of the TIRF microscope (110-nm penetration depth), in the perinuclear region, or at the tips of cell protrusions. Tracking experiments revealed that most of the motile vesicles (≃80%) located in protrusions displayed an anterograde movement toward the cell edge (Fig. 3 E). This prompted us to evaluate whether ATG9A-positive vesicles could indeed reach the plasma membrane and accomplish polarized exocytosis at the migration front.
Figure 3.
ATG9A-positive vesicles concentrate in F-actin–rich protrusions and display anterograde trafficking toward the leading edge. (A) Endogenous ATG9A localizes in F-actin–rich cell protrusions. Left: U87 MG cells colabeled for endogenous ATG9A (green), F-actin (rhodamine phalloidin, red) and nuclei (DAPI labeling, blue). Scale bar, 20 µm. Middle and right: Magnified views of cell presented on the left panel, showing protrusive (middle) and nonprotrusive (right) areas. Scale bar for magnified views, 10 µm. (B) Chemotactic stimulation with EGF increases peripheral localization of ATG9A. U87 MG cells were starved (30 min) in serum-free medium and incubated (30 min) with or without EGF (50 ng/ml), as indicated. Cells were fixed and labeled for endogenous ATG9A (green), F-actin (rhodamine phalloidin, red), and DAPI (nuclei, blue). For each condition, a merged image and an ATG9A/F-actin colocalization map are shown. The association of ATG9A with F-actin was representatively shown with Colocalization Colormap tool, where normalized mean deviation product (nMDP) shows the correlation between intensities of corresponding pixels. Scale bar, 20 µm; magnified views, 10 µm. (C) Quantification from images shown in B of the fraction of the ATG9A signal located in F-actin–rich protrusions. For each cell, values correspond to the cumulated signal of all protrusions (−EGF, n = 88 cells; +EGF, n = 89 cells; from two independent experiments). (D) Quantification from images shown in B of the ATG9A/F-actin colocalization score (Pearson coefficient) at the cell periphery, in an ROI 1.5-µm width from the cell membrane. Data represent means ± SEM (−EGF, n = 73 cells; +EGF, n = 74 cells; from two independent experiments). (E) ATG9A-mCherry–positive vesicles display anterograde trafficking toward the leading edge. Left: Map of all observed (5-min period) ATG9A-mCherry vesicle trajectories from a representative polarized U87 MG cell. Trajectories were color-coded and defined as anterograde (green) or retrograde (red), as a function of vesicle displacement relative to the cell centroid. Scale bar, 20 µm; magnified views, 10 µm. Right: Pie chart showing the percentage of ATG9A-mCherry vesicles displaying anterograde or retrograde movement in cell protrusions (total number of 87 trajectories, recorded from four U87 MG polarized cells). Statistical significance was evaluated using Mann–Whitney U test. ***, P < 0.001.
Design and validation of ATG9A-pHluorin as a new probe to visualize ATG9A exocytosis
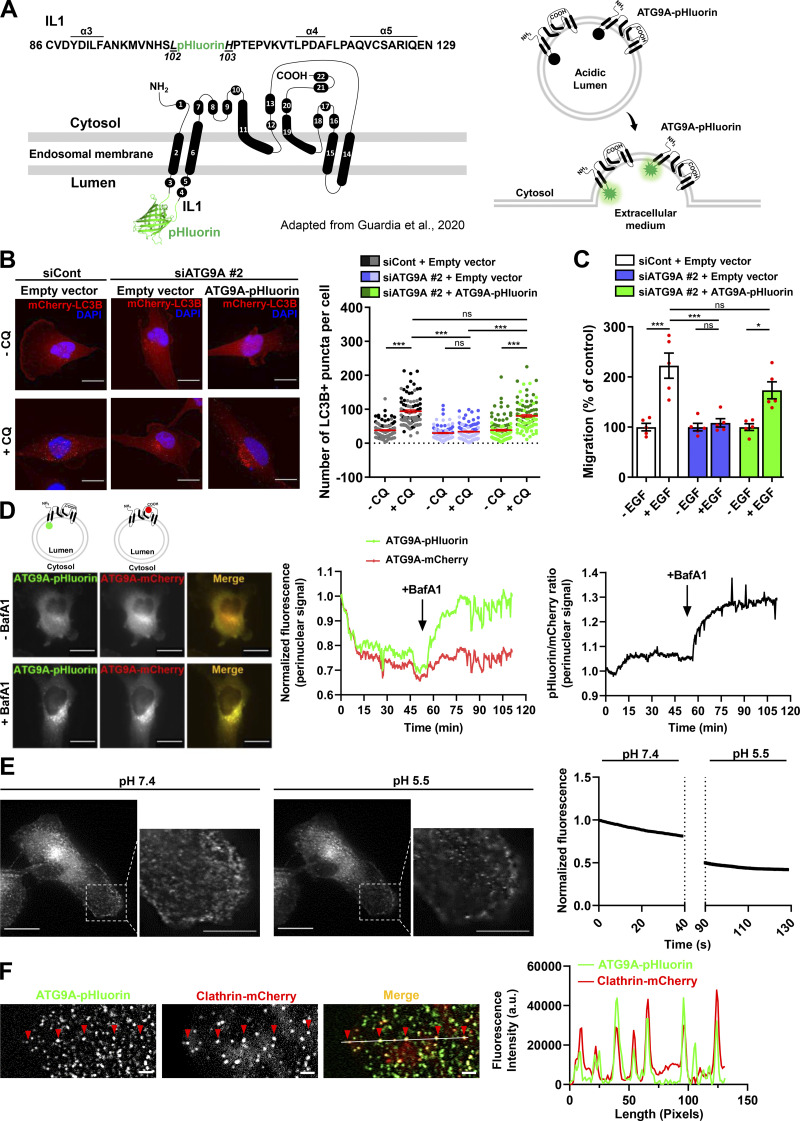
To record exocytosis of ATG9A-positive vesicles, we designed an ATG9A construct containing the highly pH-sensitive GFP variant superecliptic pHluorin (Fig. S2 A, left panel). The fluorescence of pHluorin is quenched at the acidic endosomal pH, but upon fusion with the plasma membrane, low luminal pH is immediately neutralized, resulting in a sudden increase in fluorescence intensity (Fig. S2 A, right panel; Miesenböck et al., 1998; Sankaranarayanan et al., 2000). We designed the ATG9A-pHluorin based on the predicted topology of mammalian ATG9A, displaying six transmembrane domains, with both N- and C-termini located in the cytosol (Young et al., 2006). Based on these preliminary data, we tentatively inserted the pHluorin sequence into intraluminal loop 1 (IL1) of ATG9A, since alignment of mammalian ATG9A and yeast ATG9 sequences indicated significantly lower conservation in IL1 than IL2 or IL3 predicted domains. During the course of our study, Guardia et al. (2020) solved the structure of human ATG9A to a 2.9-Å resolution using cryo-EM. Unlike the predicted structure, they showed that ATG9A displayed in fact four transmembrane helices (α2, α6, α14, and α15; Fig. S2 A), with an IL1 identical to that predicted but devoid of the predicted IL2 and IL3. Reinforcing the rationale of our strategy, the solved structure of ATG9A further indicated that the selected intraluminal insertion site for pHluorin (between amino acids Leu102 and His103) has no defined secondary structures (Fig. S2 A).
Figure S2.
Structure and functional validation of ATG9A-pHluorin construct. (A) Left: Topology of human ATG9A protein, adapted from Guardia et al. (2020), displaying four transmembrane domains (α2, α6, α14, and α15) and two domains (α11 and α19) that are partially embedded in the membrane. The ATG9A-pHluorin fusion protein was produced by inserting the pHluorin sequence (depicted by a cartoon of GFP) into the first luminal domain, between amino acids Leu102 and His103 of the human sequence. Right: The pHluorin fluorescence is expected to be quenched at the acidic endosomal pH. Upon fusion of ATG9A-containing endosomes with the plasma membrane, pHluorin fluorescence will sharply increase at the contact of extracellular physiological pH. (B) Left: Validation of functional activity of ATG9A-pHluorin on autophagosome biogenesis. U87 MG cells were transfected with nontargeting siRNA (siCont) or siRNA targeting ATG9A (siATG9A #2; this siARN specifically targets endogenous ATG9A, without affecting the expression of recombinant ATG9A-pHluorin, due to the codon-optimization procedure of the recombinant sequence introducing several siRNA/target mRNA mismatches), together with an empty vector or the ATG9A-pHluorin construct and a construct encoding the mCherry-LC3B protein (marker of autophagosomes). Transfected cells were placed in serum-free DMEM for 6 h, in the presence or absence of CQ (5 × 10−5 M), as indicated. Cells were fixed, and the number of autophagosomes (mCherry-LC3B fluorescent dots) per cell was quantified. Data represent means and SEM (n = 85–111 cells per group; cells from independent experiments were color-coded). Scale bar, 20 µm. Note that the marked reduction of autophagosome biogenesis induced by the knockdown of endogenous ATG9A was totally rescued by overexpression of recombinant ATG9A-pHluorin, indicating that insertion of the pHluorin sequence within the IL1 loop of ATG9A does not preclude its pro-autophagic function. (C) Validation of functional activity of ATG9A-pHluorin on cell migration. U87 MG cells were transfected with nontargeting siRNA (siCont) or siRNA targeting ATG9A (siATG9A #2), together with an empty vector or the vector encoding ATG9A-pHluorin. Transfected cells were loaded in the upper chamber of Transwells, with or without EGF (50 ng/ml) in the lower chamber, as indicated. After 24 h, cells that migrated onto the lower surface of the membrane were fixed, stained, and counted. Data represent means and SEM (n = 5 Transwells). Note that inhibition of chemotactic migration induced by the knockdown of endogenous ATG9A was rescued by overexpression of recombinant ATG9A-pHluorin. (D) Validation of ATG9A-pHluorin topology. Representative U87 MG cell coexpressing ATG9A-pHluorin and ATG9A-mCherry was imaged by epifluorescence (one frame every 30 s) before and after treatment with BafA1 (100 nM), a selective inhibitor of the vacuolar-type ATPase, an essential proton pump for maintaining vesicular acidic pH. Left: Images extracted from the time-lapse sequence before (−BafA1) and at the end (+BafA1; 1-h incubation) of BafA1 treatment. Scale bar, 20 µm. Middle: mCherry and pHluorin fluorescence signal intensities measured in the perinuclear area and plotted against time. Right: Analysis of the pHluorin/mCherry fluorescence intensity ratio, plotted against time. The increased ratio value following BafA1 treatment indicates dequenching of the pHluorin signal in endosomes, suggesting that the ATG9A-pHluorin fusion protein has the expected topology, with the pHluorin facing the vesicular lumen. (E) Left: Representative U87 MG cell expressing ATG9A-pHluorin recorded using TIRF microscopy (one frame every 390 ms), before (pH 7.4) and after (pH 5.5) acidification of the extracellular medium. Note that most ATG9A-pHluorin puncta rapidly dimmed after extracellular acidification, indicating that ATG9A-pHluorin proteins concentrating in these puncta are located at the plasma membrane, with the pHluorin facing the extracellular medium. Right: pHluorin fluorescence signal intensity measured before and after extracellular acidification and plotted against time. Scale bar, 20 µm; magnified views, 10 µm. (F) ATG9A static puncta colocalize with the endocytic marker clathrin. Left: Representative TIRF images extracted from a time-lapse sequence showing a U87 MG cell coexpressing ATG9A-pHluorin (green) and clathrin-mCherry (red). Note that ATG9A-pHluorin puncta frequently localize with clathrin (red arrowheads), suggesting that they likely represent ATG9A-pHluorin proteins trapped in forming endocytic structures. Right: Line profile plot indicates the fluorescence intensity distribution of green and red channels, through the white line shown in the merged image. Scale bar, 2 µm. Statistical significance was evaluated using one-way ANOVA followed by Tukey post hoc test (B and C). *, P < 0.05; ***, P < 0.001.
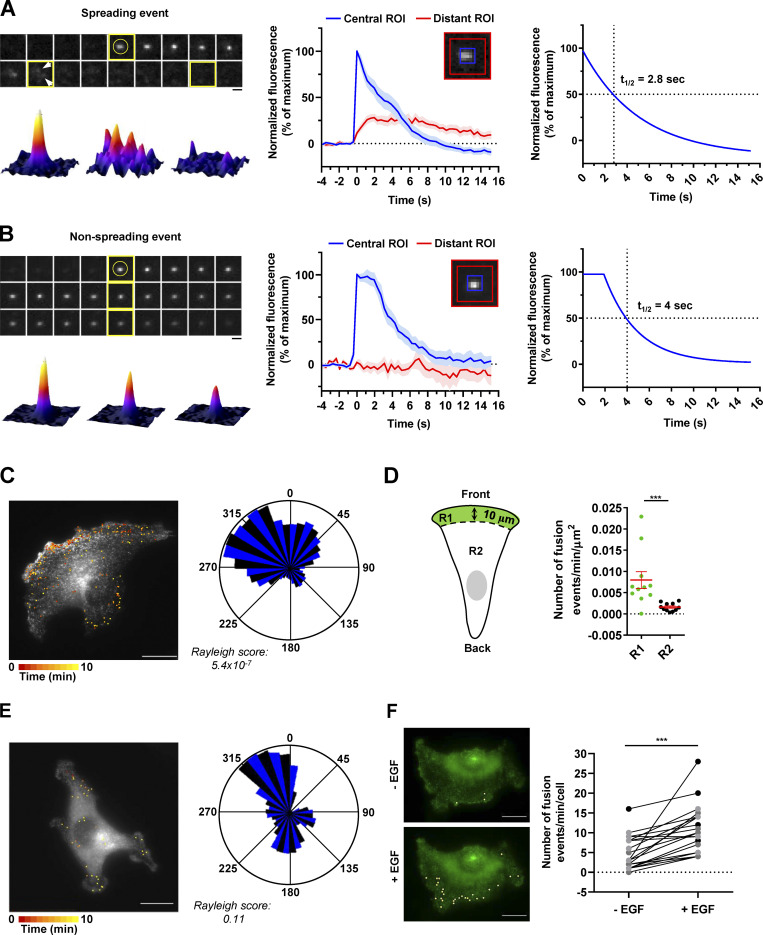
After having validated our construct by demonstrating that the ATG9A-pHluorin fusion protein retains autophagic activity (Fig. S2 B) and promigratory properties (Fig. S2 C) and has the expected topology (Fig. S2 D), we next recorded ATG9A-pHluorin exocytotic events by using live-cell TIRF microscopy (one frame every 390 ms). Besides exocytotic events, we first noticed that, under TIRF configuration, most of the ATG9A-pHluorin signal appears scattered in small and relatively static puncta decorating the bottom surface of the cell (Fig. S2 E). These puncta likely represent ATG9A-pHluorin proteins trapped in endocytic structures, since (a) they rapidly dimmed after extracellular acidification (Fig. S2 E), indicating that the protein is at the plasma membrane, with the pHluorin facing the extracellular medium, and (b) they frequently localized with the endocytic marker clathrin (Fig. S2 F). We next focused on exocytotic events, which were monitored as bright spots suddenly appearing (within a frame). Interestingly, visual examination of the events suggested heterogeneity. The majority (≃70%) of the events showed a clear signature of full fusion exocytosis, i.e., (a) a rapid increase (<0.390 s) of the pHluorin signal in a central region of interest (ROI), likely due to fusion pore opening, and (b) diffusion of the ATG9A-pHluorin cargo into the plasma membrane, revealed by the delayed increase of fluorescence in a distant ROI (Fig. 4 A and Video 2). The average profile of these fusion events (n = 68), which were time-aligned to fusion, allowed the estimation of a half-life of ∼2.8 s (Fig. 4 A). The remaining events were characterized by the absence of significant diffusion of the fluorescence signal in areas of the plasma membrane surrounding the insertion site, and generally displayed a slower intensity decay (mean decay half-life 4 s; Fig. 4 B and Video 3). Although in-depth analysis would be required to make definite conclusions, the latter events likely represent fusion of ATG9A-loaded vesicles with the plasma membrane according to a kiss-and-run mechanism (Alabi and Tsien, 2013). Such a mechanism has been described for the delivery of lipids and/or cargo proteins from ATG9A-positive vesicles to the expanding phagophores, without retention of ATG9A protein in mature autophagosomes (Orsi et al., 2012).
Figure 4.
Exocytosis of ATG9A-positive vesicles is polarized toward the cell front and induced by chemotactic stimulation. (A) U87 MG cells expressing ATG9A-pHluorin were recorded (one frame every 390 ms) using TIRF microscopy. Upper left: Time-lapse sequence of a representative spreading event. Yellow circle indicates the rapid appearance of the pHluorin signal, likely due to de-acidification upon fusion pore opening. Arrows indicate diffusion of the fluorescence signal in areas of the plasma membrane surrounding the insertion sites. Scale bar, 1 µm. Bottom left: 3D fusion profiles of the indicated images (framed in yellow) from the time-lapse sequence. Middle: Normalized fluorescence intensities, from the analysis of 68 spreading events. For each event, measurements were performed in parallel into a central ROI (5 × 5 pixels; 0.64 × 0.64 µm) and a distant ROI (delimited by 13 × 13–pixel and 17 × 17–pixel squares). Data represent means and SEM. Right: Nonlinear regression analysis (single exponential decay) from the profile obtained in the central ROI, for the determination of spreading event half-life (2.8 s). (B) U87 MG cells expressing ATG9A-pHluorin were recorded as in A. Upper left: Time-lapse sequence of a representative nonspreading event. Scale bar, 1 µm. Bottom left: 3D fusion profiles of the indicated images (framed in yellow) from the time-lapse sequence. Middle: Normalized fluorescence intensities, from the analysis of 13 nonspreading events. For each event, measurements were performed in a central ROI and a distant ROI, as in A. Data represent means and SEM. Right: Nonlinear regression analysis (plateau followed by single exponential decay) from the profile obtained in the central ROI, for the determination of nonspreading event half-life (4 s). (C) Left: Map of all observed (n = 248) ATG9A-pHluorin fusion events from a representative, polarized U87 MG cell, during a 10-min time-lapse sequence. Scale bar, 20 µm. Right: Rose plot from the cell shown on the left, depicting the angular distribution of the fusion events relative to the cell centroid. (D) Quantification, from U87 MG cells displaying clear polarization with unique and large lamellipodia (n = 11 cells), of the number of fusion events occurring at a distance inferior (R1 region) or superior (R2 region) to 10 µm from the leading edge. For each cell, data were normalized to the areas of the respective regions. Data represent means and SEM. (E) Left: Map of all observed (n = 40) ATG9A-pHluorin fusion events from a representative, nonpolarized U87 MG cell, during a 10-min time-lapse sequence. Scale bar, 20 µm. Right: Rose plot from the cell shown on the left, depicting the angular distribution of the fusion events relative to the cell centroid. (F) Left: Representative U87 MG cell with projections of all observed ATG9A-pHluorin fusion events, during 1-min time-lapse sequences, before (−EGF) and 2-min after (+EGF) treatment with EGF (50 ng/ml). Scale bar, 20 µm. Right: Quantification of the effect of EGF, as depicted on the left, on the number ATG9A-pHluorin fusion events (n = 23 cells from three independent experiments; cells from independent experiments were color-coded). Statistical significance was evaluated using Mann–Whitney U test (D), paired t test (F), and Rayleigh test for fusion events distribution (C and E). ***, P < 0.001.
Video 2.
Spreading ATG9A-pHluorin exocytotic events. Time-lapse TIRF images of U87 MG cells expressing ATG9A-pHluorin. Arrows indicate the rapid appearance of the pHluorin signal upon fusion pore opening, followed by signal diffusion into the plasma membrane. Video plays at 15 frames/s. Scale bar, 1 µm.
Video 3.
Nonspreading ATG9A-pHluorin exocytotic events. Time-lapse TIRF images of U87 MG cells expressing ATG9A-pHluorin. Arrows indicate the rapid appearance of the pHluorin signal upon fusion pore opening, followed by signal decay without diffusion into the plasma membrane. Video plays at 15 frames/s. Scale bar, 1 µm.
Exocytosis of ATG9A-positive vesicles is highly polarized toward the leading edge
We next evaluated the spatial distribution of ATG9A-pHluorin fusion events in cells displaying a clear rear–front axis, with a unique and well-developed protrusion. We found that ATG9A-pHluorin exocytotic events were highly polarized, with clear clustering near the cell front (Fig. 4 C). Quantitative analysis indicated that the frequency of events, after normalization by the areas of the ROIs, was approximately fivefold higher at the cell front (<10 µm from the leading edge) than at the other parts of the cell (Fig. 4 D). In sharp contrast, TIRF analysis of nonpolarized cells, devoid of large lamellipodia, showed that fusion events were relatively uniformly distributed, with no obvious hotspot (Fig. 4 E).
We next tested the impact of chemotactic stimulation with EGF. For each cell, fusion events were recorded over 1-min periods, just before and 2 min after EGF treatment. Analyses of the time-lapse movies indicated that EGF exposure significantly increases the fusion activity ATG9A-positive vesicles (Fig. 4 F). Altogether, our data suggest that ATG9A-positive vesicles are targeted toward the leading edge for exocytosis, to deliver lipids and/or cargo proteins for cell expansion.
Peripheral ATG9A-pHluorin signal correlates with protrusive activity
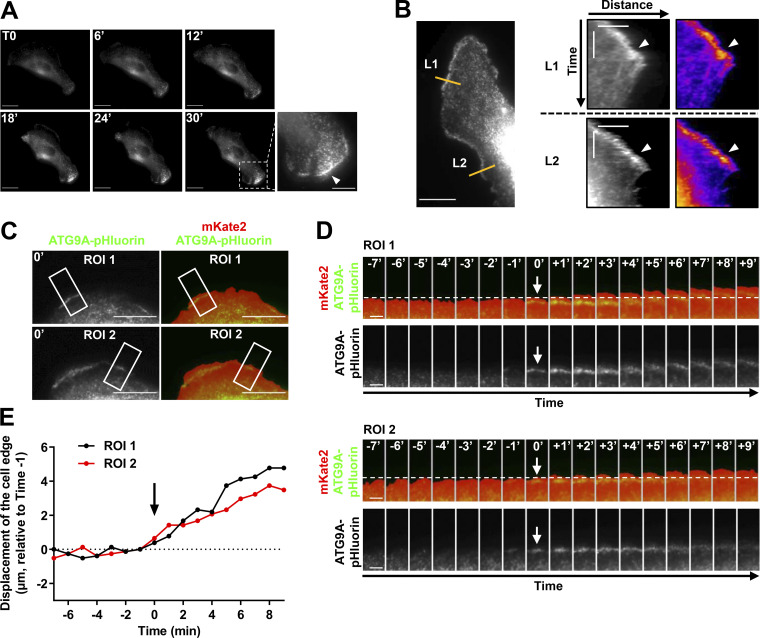
Cells expressing ATG9A-pHluorin were recorded at a low frame rate (one frame every minute) using TIRF, during the spontaneous formation of protrusions. The formation of broad protrusions was systematically marked by the appearance of a bright ATG9A-pHluorin signal close to the leading edge (Fig. S3 A and Video 4). The vesicular pattern of the signal likely represents ATG9A-pHluorin proteins, which rapidly relocalize in endocytic structures after focal bursts of exocytosis. Notably, analyses of the videos using kymographs indicated that the ATG9A-pHluorin signal is sustained at the edges of expanding protrusions but sharply decreased during protrusion collapse (Fig. S3 B). To precisely define the cell edges and establish a temporal correlation between edge advance and appearance of ATG9A signal, we used mKate2, a red fluorescent protein diffusely expressed in the cytosol. A representative time-lapse sequence (Fig. S3, C–E) shows that appearance of ATG9A-pHluorin signal near the cell edge was synchronized with protrusive activity.
Figure S3.
ATG9A-pHluorin signal at the cell periphery correlates with protrusive activity. (A) Time-lapse TIRF images of a U87 MG cell expressing ATG9A-pHluorin. The development of a large cell protrusion correlates with the appearance of a marked pHluorin signal at the leading edge. A magnified image of the leading edge (dashed rectangle) is presented in the lower right and shows numerous ATG9A-pHluorin positive vesicles (arrowhead). Scale bar, 20 µm; magnified view, 10 µm. (B) Left: TIRF image extracted from a time-lapse sequence of a U87 MG cell expressing ATG9A-pHluorin. Scale bar, 20 µm. Right: Kymographs were made from 10-pixel-wide lines (L1 and L2) indicated in the left image. The scale bars in the kymograph are 5 µm (horizontal) and 10 min (vertical). A clear ATG9A-pHluorin signal was observed near the cell edge during the formation of protrusions (arrowheads). The signal sharply decreased during protrusion collapses. (C) TIRF image of a U87 MG cell expressing mKate2 (red) and ATG9A-pHluorin (green). The mKate2 protein, diffusely expressed in the cytosol, was used to precisely delineate the cell edges. Scale bar, 20 µm. (D) Time-lapse montage of ROI 1 and ROI 2 shown in C. For each time-lapse, time 0 represents the time of appearance of ATG9A-pHluorin–positive vesicles (arrows) near to the cell edge. The position of the cell edge before the appearance of the ATG9A-pHluorin signal is marked by a dashed line. Scale bar, 5 µm. (E) Displacements of the cell edges shown in ROI 1 and 2 were plotted against time. The position of the cell edge 1 min before the appearance of the ATG9A-pHluorin signal was set at 0, and positive values in the y axis represent cell protrusion.
Video 4.
ATG9A-pHluorin signal appearance at the leading edge correlates with protrusive activity. Time-lapse TIRF images of a U87 MG cell expressing ATG9A-pHluorin. Video plays at 15 frames/s. Scale bar, 20 µm.
ATG9A regulates delivery of TGN46 and β1 integrin to the leading edge through its N-terminal adaptor protein (AP) sorting signal
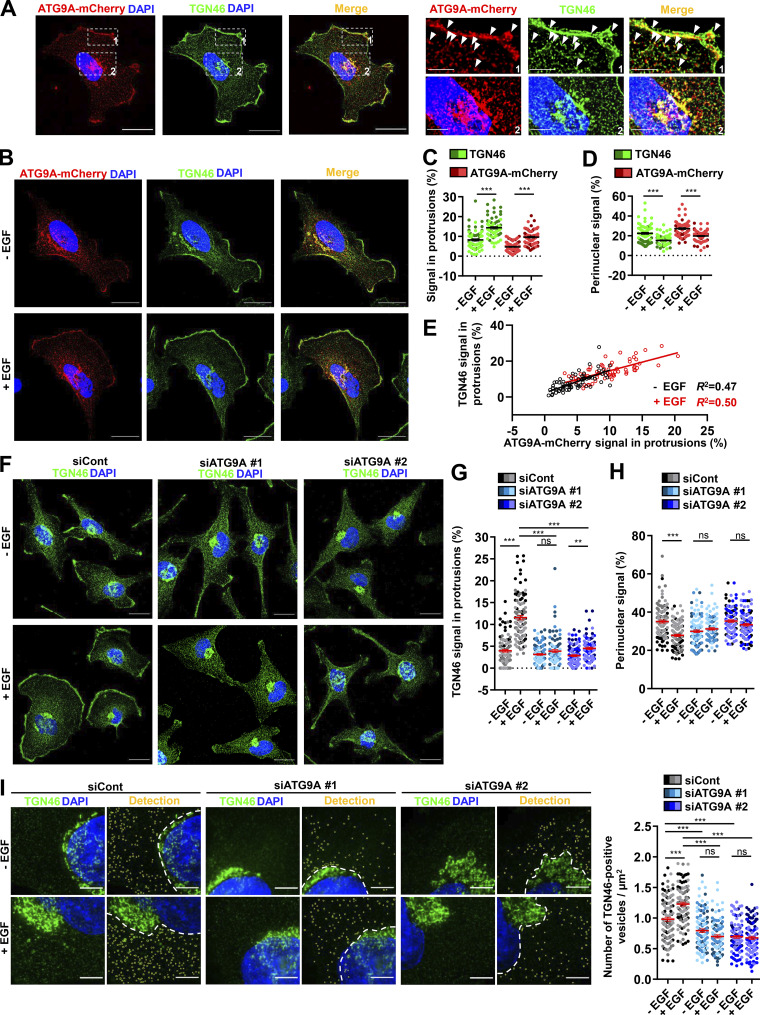
Delivery of cargoes from the TGN to the leading edge is critical for directional migration (Millarte and Farhan, 2012; Riggs et al., 2012; Shafaq-Zadah et al., 2016; Mana et al., 2016; Hao et al., 2020), and several studies, using live-cell TIRF microscopy, indeed demonstrated polarized exocytosis of post-Golgi carriers at the cell front (Schmoranzer et al., 2003; Letinic et al., 2009). This led us to evaluate whether ATG9A proteins transported toward cell protrusions essentially emanate from the TGN. We performed colocalization experiments with the Golgi marker TGN46, an integral membrane protein mainly residing in the TGN, that has been shown to cycle between the TGN and the plasma membrane (Banting and Ponnambalam, 1997). In agreement with previous studies (Orsi et al., 2012), we found that ATG9A-mCherry and TGN46 strongly colocalize in typical TGN cisternae, as well as in more peripheral post-Golgi carriers (Fig. S4 A). Importantly, TGN46 and ATG9A-mCherry also displayed a marked colocalization in cell protrusions, near or at the cell membrane (Fig. S4 A). A 30-min chemotactic stimulation of cells with EGF increased the fractions of ATG9A and TGN46 signals localized in protrusions (TGN46: +74.3%; ATG9A-mCherry: +104.4%), with a concomitant decrease of their signals in the perinuclear area (Fig. S4, B–E).
Figure S4.
ATG9A protein regulates delivery of TGN46-positive post-Golgi carriers to cell protrusions. (A) Left: Representative U87 MG cell expressing ATG9A-mCherry (red) and colabeled for TGN46 (green) and nuclei (DAPI labeling, blue). Scale bar, 20 µm. Right: Magnified views of the cell presented on the left. Note that ATG9A colocalizes with TGN46 in typical TGN cisternae (upper panels) and at the cell membrane or vesicles close to the cell membrane (lower panels). Scale bar for magnified views, 5 µm. (B–E) Chemotactic stimulation induces redistribution of TGN46 and ATG9A-mCherry toward the cell membrane. (B) U87 MG cells expressing ATG9A-mCherry were starved (30 min) in serum-free medium and incubated (30 min) with or without EGF (50 ng/ml), as indicated. Cells were fixed and labeled for TGN46 (green) and nuclei (DAPI labeling, blue). Scale bar, 20 µm. (C) Quantification from images shown in B of the TGN46 and ATG9A-mCherry signals located in protrusions. For each cell, values correspond to the cumulated signal of all protrusions. (D) Quantification from images shown in B of the TGN46 and ATG9A-mCherry signals located in the perinuclear area. Data represent means ± SEM (n = 68 cells per group; cells from independent experiments were color-coded). (E) Cell-to-cell correlation between TGN46 and ATG9A-mCherry signals located in cell protrusions, in both untreated (black dots) and EGF-treated (red dots) cells. (F–H) EGF-induced redistribution of TGN46 to the cell protrusions depends on ATG9A. (F) U87 MG cells were transfected with nontargeting siRNA (siCont) or one of the two siRNA targeting ATG9A (siATG9A #1, siATG9A #2). Transfected cells were starved (30 min) in serum-free medium and incubated (30 min) with or without EGF (50 ng/ml), as indicated. Cells were fixed and labeled for TGN46 (green) and nuclei (DAPI labeling, blue). Scale bar, 20 µm. (G) Quantification from images shown in F of the TGN46 signal located in protrusions. (H) Quantification from images shown in F of the TGN46 signal located in the perinuclear area. Data represent means ± SEM (n = 104–113 cells per group; cells from independent experiments were color-coded). (I) Detection (left) and quantification (right), from cells treated as in F, of the number of cytosolic TGN46-positive vesicles. Data represent means ± SEM (n = 99–112 cells per group; cells from independent experiments were color-coded). Statistical significance was evaluated using Mann–Whitney U test (C and D), one-way ANOVA followed by Tukey post hoc test (G and I), or one-way ANOVA followed by Sidak post hoc test (H), **, P < 0.01; ***, P < 0.001.
ATG9A protein, through its interaction with the AP complex AP-1, plays an active role in the TGN-to-lysosome transport of hydrolases (Jia et al., 2017), putting forward the idea that ATG9A could be a constitutive component of cargo sorting/trafficking from the TGN. We therefore investigated the impact of ATG9A on TGN46 localization and found that depletion of ATG9A protein levels using two independent siRNAs markedly reduces EGF-induced redistribution of TGN46 from the perinuclear area toward the cell periphery (Fig. S4, F–H). To evaluate whether this effect of ATG9A may depend on its ability to regulate the biogenesis of post-Golgi carriers, we also monitored the number of TGN46-positive vesicles dispersed in the cytosol. We found that, in control cells, EGF treatment increased the number of TGN46-positive post-Golgi carriers. Depletion of ATG9A reduced the number of vesicles in unstimulated cells and totally blocked the effect of EGF (Fig. S4 I).
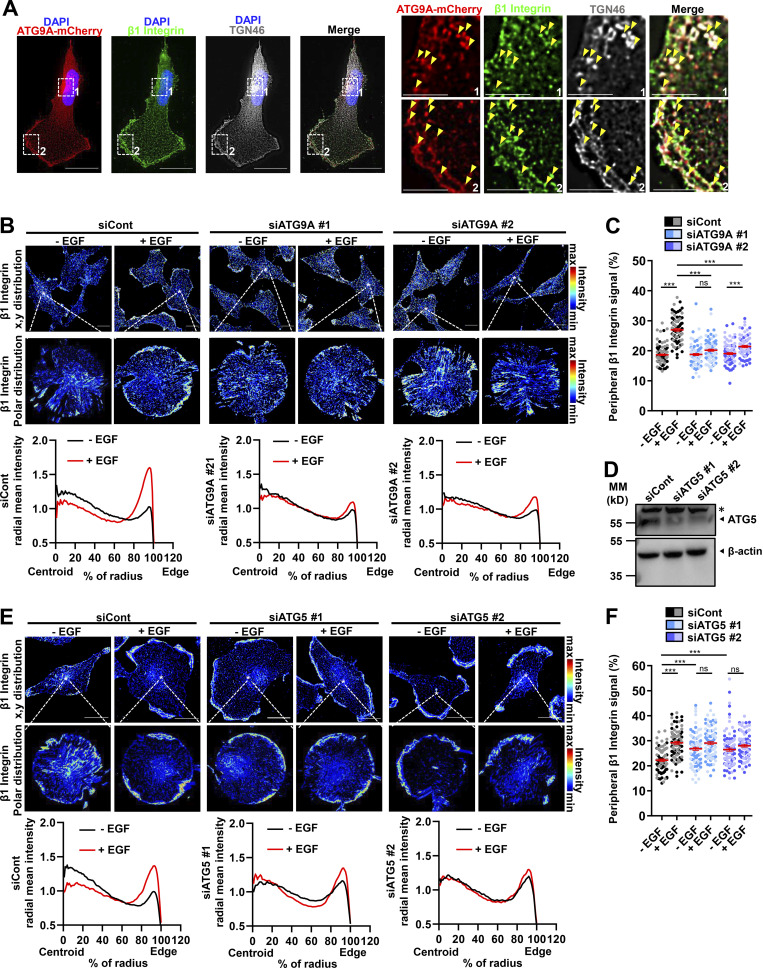
Through its luminal domain, the rat homologue of TGN46 interacts with β1 integrin, thereby regulating the sorting of this major adhesion molecule from the TGN to the plasma membrane (Wang and Howell, 2000). Notably, several reports demonstrated that retrograde transport of β1 integrin to the TGN, followed by its polarized reshuffling from the TGN to the leading edge, is essential for the establishment of nascent adhesions and directional cell migration (Riggs et al., 2012; Shafaq-Zadah et al., 2016; Mana et al., 2016). This prompted us to assess whether ATG9A also regulates EGF-induced delivery of β1 integrin to the cell protrusions. Fluorescently immunostained endogenous β1 integrin partially colocalized with TGN46 and ATG9A in the perinuclear area and in puncta dispersed throughout the cytosol or near the cell membrane, likely representing bona fide post-Golgi carriers (Fig. 5 A). In cells transfected with control siRNAs, 30-min incubation with EGF induced a marked redistribution of β1 integrin toward the periphery, with a concomitant decrease of its relative amounts in the cytosol (Fig. 5, B and C). Similar to data obtained with TGN46, depletion of ATG9A using each siRNA strongly inhibited EGF-induced redistribution of β1 integrin toward cell protrusions (Fig. 5, B and C). To verify that this effect of ATG9A was not due to a modulation of the autophagic status, we also used siRNAs targeting the core autophagy protein ATG5 (Fig. 5 D). In contrast to ATG9A depletion, which blocked EGF-induced transport of β1 integrin toward the periphery, ATG5 depletion induced a marked accumulation of β1 integrin near or at the cell membrane, even in the absence of EGF (Fig. 5, E and F). These data are consistent with a previous report demonstrating that ATG5 regulates cell migration through the disassembly of adhesion complexes (Sharifi et al., 2016) and stress the fact that ATG proteins may regulate different steps of the migration process, independently of the overall autophagic status.
Figure 5.
ATG9A protein regulates delivery of β1 integrin to cell protrusions. (A) U87 MG cells expressing ATG9A-mCherry were labeled for β1 integrin (green), TGN46 (white), and nuclei (DAPI labeling, blue). Scale bar, 20 µm. Note, from the magnified views on the right panels, that ATG9A-mCherry, β1 integrin, and TGN46 signals partially colocalize in tubular or vesicular structures (arrowheads) near the cell membrane or in the perinuclear area. Scale bar for magnified views, 6 µm. (B) U87 MG cells were transfected with nontargeting siRNA (siCont) or one of the two siRNAs targeting ATG9A (siATG9A #1, siATG9A #2). Transfected cells were starved (30 min) in serum-free medium, incubated (30 min) with or without EGF (50 ng/ml), fixed, and labeled for β1 integrin. Upper images: β1 integrin immunoreactive signal (false-color scale) in representative fields of each experimental group. Scale bar, 20 µm. Middle images: Polar distribution of the β1 integrin immunoreactive signal (false-color scale; Clock Scan plug-in) from the cell marked by an asterisk in the upper image. Lower panels: Radial intensity profile of the β1 integrin signal, from the cell centroid to the cell edge (Clock Scan plug-in). For each experimental group, profiles were normalized and averaged from 76–98 cells per group (two independent experiments). (C) Measurement of peripheral β1 integrin signal (signal located in the 80–100% interval of the cell radius, normalized to the total signal) from images shown in B. Cells from independent experiments were color-coded. (D) Validation of ATG5 knockdown. Western blot analysis of ATG5 protein levels from U87 MG cells transfected with nontargeting siRNA (siCont) or one of the two siRNAs targeting ATG5 (siATG5 #1, siATG5 #2). MM, molecular mass. Asterisk, nonspecific band. (E) U87 MG cells transfected with nontargeting siRNA (siCont) or one of the two siRNAs targeting ATG5 (siATG5 #1, siATG5 #2) were incubated with or without EGF, and distribution of the β1 integrin immunoreactive signal was analyzed and presented as in B. For each experimental group, profiles were normalized and averaged from 85–103 cells per group (two independent experiments). Scale bar, 20 µm. (F) Measurement of peripheral β1 integrin signal (signal located in the 80–100% interval of the cell radius, normalized to the total signal) from images shown in E. Cells from independent experiments were color-coded. Data represent means and SEM. Statistical significance was evaluated using one-way ANOVA followed by Tukey post hoc test (C and F). ***, P < 0.001.
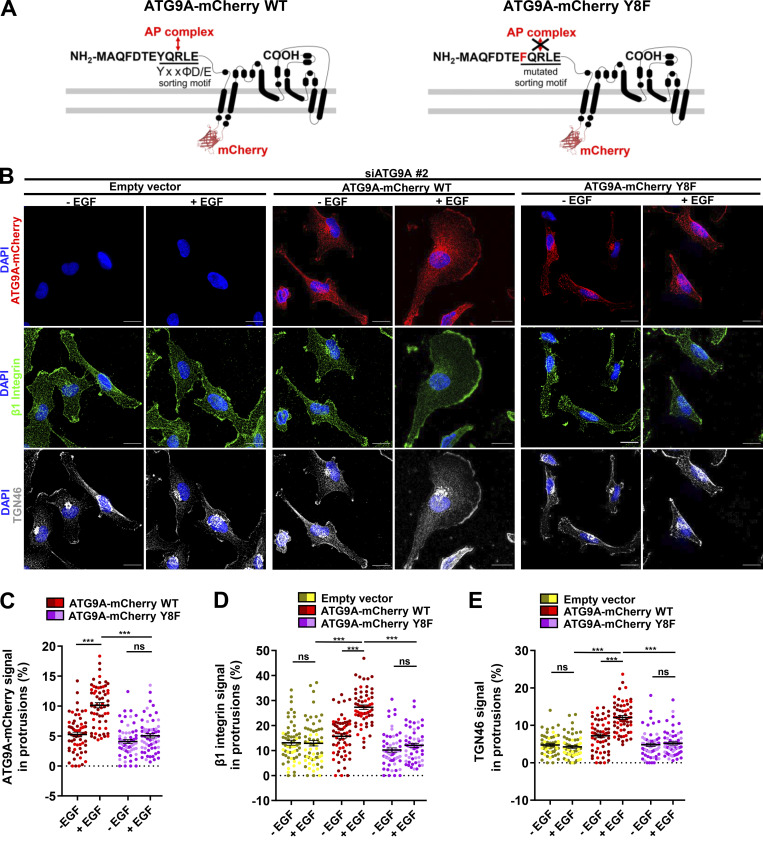
ATG9A trafficking from the TGN, recycling endosomes or the plasma membrane, is tightly controlled by clathrin adaptor complexes. Hence, ATG9A contains a canonical AP sorting signal (8YXXØD/E12) within its N-terminal domain (Fig. 6 A), which confers binding to AP-1 (endosome- and TGN-localized), AP-2 (plasma membrane–localized), and AP-4 (TGN-localized) complexes (Imai et al., 2016; Zhou et al., 2017; Jia et al., 2017; Mattera et al., 2017; Ivankovic et al., 2020). Within this motif, tyrosine 8 residue is critical, since its mutation abolishes AP binding, resulting in altered ATG9A trafficking (Zhou et al., 2017; Mattera et al., 2017). We demonstrated that, in cells depleted of endogenous ATG9A, overexpression of wild-type recombinant ATG9A-mCherry efficiently rescued EGF-induced delivery of β1 integrin and TGN46 to the leading edge (Fig. 6, B–E). As expected, mutant ATG9A-mCherry, in which tyrosine 8 was replaced by a phenylalanine, did not show increased localization in cell protrusions after EGF stimulation, nor did it rescue β1 integrin and TGN46 trafficking to the cell periphery (Fig. 6, B–E). Together, our data indicate that ATG9A, through its interaction with AP, is critical for EGF-induced delivery of both β1 integrin and TGN46 toward cell protrusions.
Figure 6.
ATG9A regulates delivery of TGN46 and β1 integrin to the leading edge through its N-terminal AP sorting signal. (A) Left: Scheme depicting the topology of the ATG9A-mCherry fusion protein and the canonical AP sorting signal (8YQRLE12) located in the N-terminal cytosolic domain. Right: Scheme depicting the mutant ATG9A-mCherry protein, for which replacement of the critical tyrosine residue at position 8 by a phenylalanine abolishes AP binding (Zhou et al., 2017; Mattera et al., 2017). (B–E) Rescue experiments with wild-type and mutant ATG9A-mCherry proteins. (B) After depletion of endogenous ATG9A using interfering RNA (siATG9A #2), U87 MG cells were transfected with an empty vector, a vector encoding wild-type ATG9A-mCherry, or a vector encoding mutant ATG9A-mCherry Y8F. Transfected cells were starved (30 min) in serum-free medium and incubated (30 min) with or without EGF (50 ng/ml), as indicated. Cells were fixed and labeled for β1 integrin (green), TGN46 (white), and nuclei (DAPI, blue). Scale bar, 20 µm. (C) Quantification from images shown in B of the ATG9A-mCherry (wild-type or mutant) signal located in protrusions. (D) Quantification from images shown in B of the β1 integrin signal located in protrusions. (E) Quantification from images shown in B of the TGN46 signal located in protrusions. Data represent means and SEM (n = 60–63 cells per group; cells from independent experiments were color-coded). Statistical significance was evaluated using one-way ANOVA followed by Tukey post hoc test. ***, P < 0.001.
ATG9A protein controls the dynamics of adhesion complexes
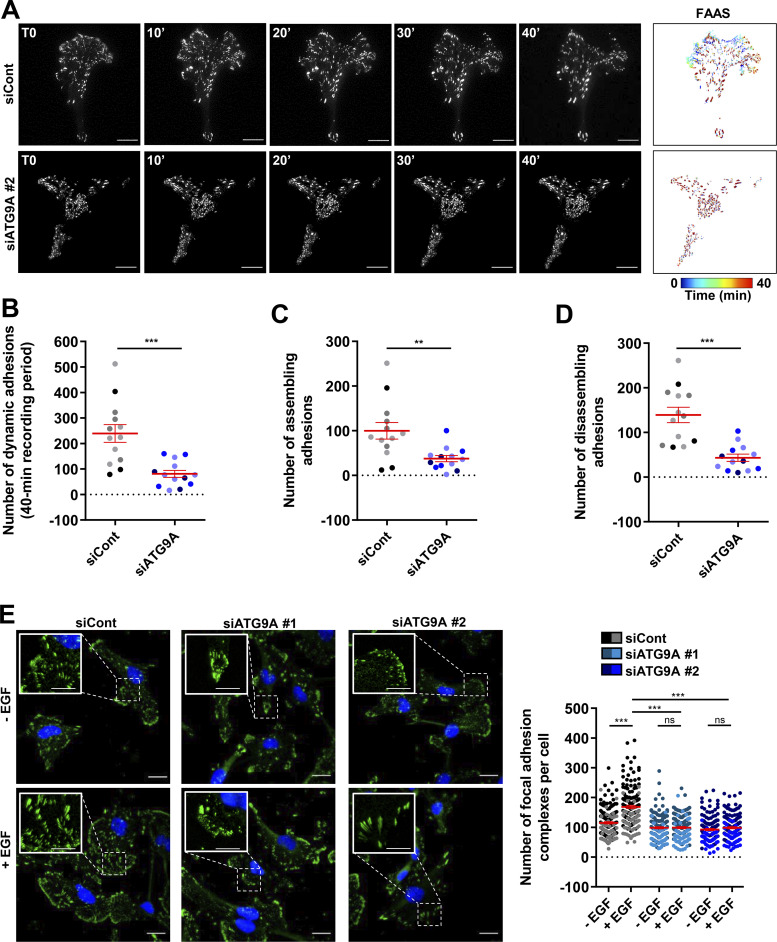
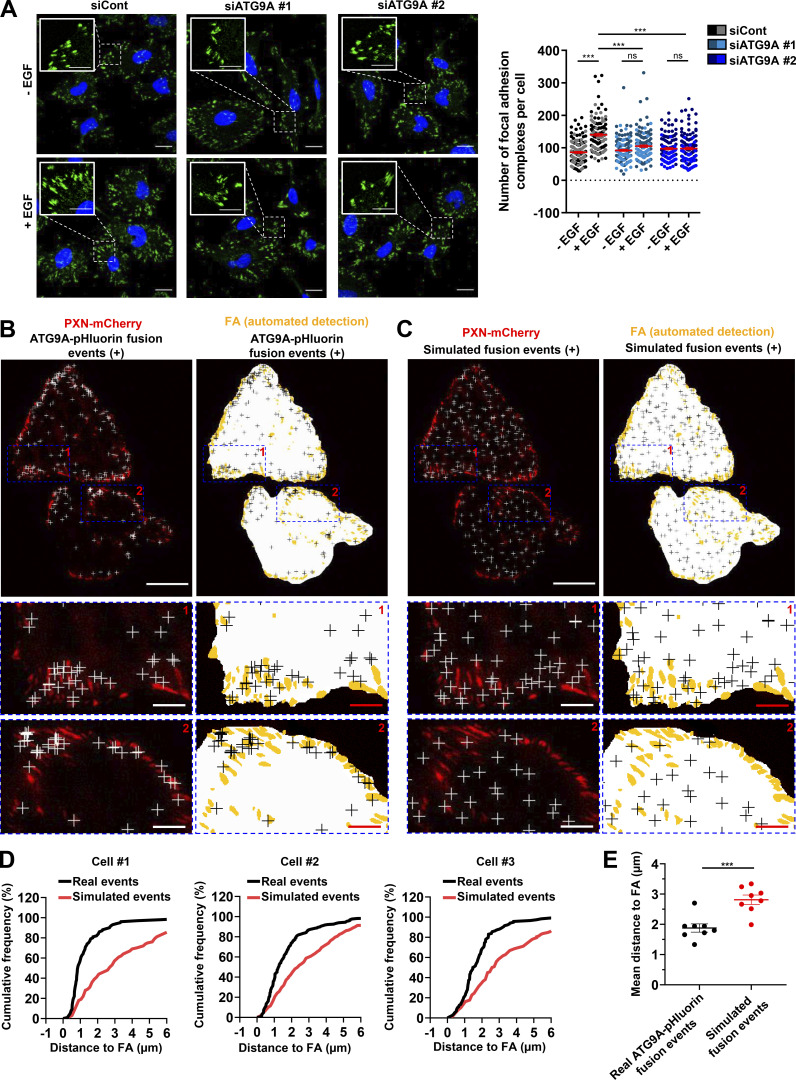
Clustering of β1 integrin at the plasma membrane triggers the recruitment of several cytoplasmic proteins, including paxillin (PXN), that participates to the formation and maturation of adhesion complexes (Deakin and Turner, 2008). We tested the impact of ATG9A protein on the formation and turnover of adhesion complexes in cells expressing PXN-EGFP (Stehbens et al., 2014; Stehbens and Wittmann, 2014). Live-cell TIRF imaging of cells transfected with irrelevant siRNA revealed a dynamic adhesion turnover. Automated analysis using the Focal Adhesion Analysis Server (FAAS) allowed the detection of a mean of ∼100 assembling adhesions and a mean of 139 disassembling adhesions during the 40-min recording period (Fig. 7, A–D and Video 5). In ATG9A-depleted cells, adhesions appeared much more static (Fig. 7, A–D and Video 5), and automated analysis of the live-cell videos revealed a marked reduction in the number of both assembling and disassembling adhesions (Fig. 7, A–D). We next tested whether ATG9A is also involved in the formation of adhesions induced by EGF treatment. As expected, 1-h incubation with EGF induced an increase in the number of PXN-positive adhesion complexes in U87 MG cells (+46%; Fig. 7 E) and HeLa cells (+66%; Fig. S5 A). Knockdown of ATG9A using each siRNA targeting the ATG9A transcript totally abolished the effects of EGF in both cell lines (Fig. 7 E and Fig. S5 A).
Figure 7.
ATG9A protein regulates adhesion dynamics. (A–D) ATG9A depletion reduces intrinsic adhesion dynamics. (A) Representative live-cell TIRF images of U87 MG cells expressing PXN-EGFP together with the indicated siRNAs. Image sequences show adhesion changes over a 40-min period. Righthand panels: Color scale output generated from FAAS, representing early (blue) to late (red) adhesions. Scale bar, 20 µm. (B–D) FAAS output of the number of dynamic adhesions (B), assembling adhesions (C), and disassembling adhesions (D). Data represent means and SEM (siCont, n = 13 cells; siATG9A #2, n = 13 cells; from five independent experiments). (E) ATG9A depletion inhibits EGF-induced formation of adhesion complexes. U87 MG cells were transfected with the indicated siRNAs. After starvation (1 h) in serum-free DMEM, cells were treated (1 h) with or without EGF (50 ng/ml), fixed, and labeled with an anti-PXN antibody (green) and DAPI (nuclei, blue). The number of adhesion complexes was quantified for each cell. Data represent means and SEM (n = 144–158 cells per group; cells from independent experiments were color-coded). Scale bar, 20 µm; inset magnifications, 10 µm. Statistical significance was evaluated using Mann–Whitney U test (B–D) or one-way ANOVA followed by Sidak post hoc test (E). **, P < 0.01; ***, P < 0.001.
Video 5.
ATG9A protein regulates adhesion dynamics. Representative live-cell TIRF images of U87 MG cells expressing PXN-EGFP together with nontargeting siRNA (siCont, left) or siRNA targeting ATG9A (siATG9A #2, right). Image sequences show adhesions changes over a 40-min period. Video plays at 15 frames/s. Scale bar, 20 µm.
Figure S5.
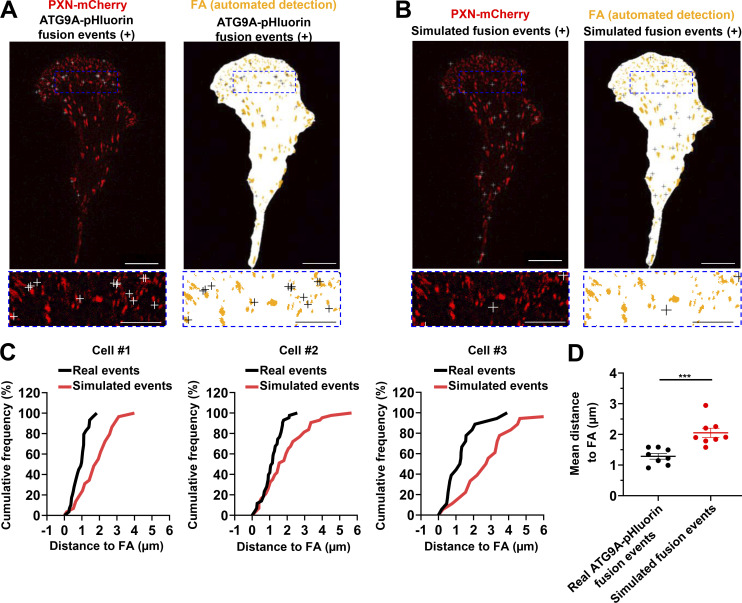
AT9A regulates the formation of adhesion complexes in HeLa cells. (A) Depletion of ATG9A protein inhibits EGF-induced formation of adhesion complexes in HeLa cells. HeLa cells were transfected with nontargeting siRNA (siCont) or one of the two siRNAs targeting ATG9A (siATG9A #1, siATG9A #2). After starvation (1 h) in serum-free medium, cells were treated (1 h) with or without EGF (50 ng/ml), fixed, and labeled with an anti-PXN antibody (green) and nuclei (DAPI labeling, blue). The number of adhesion complexes was quantified for each cell. Data represent means ± SEM (n = 125–131 cells per group; cells from independent experiments were color-coded). Scale bar, 20 µm; inset magnification, 10 µm. Statistical significance was evaluated using one-way ANOVA followed by Sidak post hoc test. ***, P < 0.001. (B–E) ATG9A-positive vesicles target adhesion sites in HeLa cells. HeLa cells expressing PXN-mCherry and ATG9A-pHluorin were recorded using live-cell TIRF microscopy. (B) Left: Distribution of all observed ATG9A-pHluorin fusion events during the recording period (crosses), overlaid on the PXN-mCherry signal (red). Right: Derived synthetic image depicting the ventral cell surface area (white), the PXN-positive adhesion complexes (orange, automated detection using ImageJ), and the ATG9A-pHluorin fusion events (crosses). Note the promiscuity between the adhesion complexes and the fusion events in the magnified view. Scale bar, 20 µm; magnified views, 5 µm. (C) Left: Map of randomly simulated fusion events (crosses), overlaid on the PXN-mCherry signal (red). Right: Derived synthetic image depicting the ventral cell surface area (white), the PXN-positive adhesion complexes (orange) and the simulated fusion events (crosses). Scale bar, 20 µm; magnified views, 5 µm. (D) Cumulative frequency charts, from the cells shown in B (cell #1 and cell #2) and one other representative cell (cell #3), demonstrating the difference in distance to focal adhesions (FA) between real ATG9A-pHluorin fusion events (black line) and simulated events (red line). (E) Quantification of the mean distance to the centroid of closest focal adhesion for real ATG9A-pHluorin events or simulated events (n = 8 cells, for a total number of 1,297 events). Data represent means ± SEM. Statistical significance was evaluated using one-way ANOVA followed by Sidak post hoc test (A) and Mann–Whitney U test (E). ***, P < 0.001.
To further support a role of ATG9 protein in adhesion dynamics, we next assessed whether the exocytosis of ATG9A-positive vesicles occurs in close proximity to focal adhesions. Live-cell TIRF imaging of cells expressing both ATG9A-pHluorin and PXN-mCherry allowed us to generate spatial maps of exocytotic events, overlaid on a focal adhesion map (Fig. 8 A and Fig. S5 B). To statistically test our data, a parallel analysis was performed by producing simulated events randomly scattered on the ventral surface of the cell (Fig. 8 B and Fig. S5 C). An algorithm was then used to compute the distance of each fusion event, real or simulated, to the closest focal adhesion. Experiments done in U87 MG (Fig. 8, C and D) or HeLa cells (Fig. S5, D and E) revealed an obvious clustering, with a mean distance between real ATG9A-pHluorin events and focal adhesions of 1.3 µm (365 events from eight U87 MG cells) and 1.9 µm (1,297 events from eight HeLa cells), respectively. Randomly simulated events occurred farther away from focal adhesions (Fig. 8, C and D; and Fig. S5, D and E).
Figure 8.
ATG9A-positive vesicles target adhesion sites. U87 MG cells expressing PXN-mCherry and ATG9A-pHluorin were recorded using live-cell TIRF microscopy. (A) Left: Distribution of all observed ATG9A-pHluorin fusion events during the recording period (crosses), overlaid on the PXN-mCherry signal (red). Right: Derived synthetic image depicting the ventral cell surface area (white), the PXN-positive adhesion complexes (orange, automated detection using ImageJ), and the ATG9A-pHluorin fusion events (crosses). Scale bar, 20 µm; magnified views, 10 µm. (B) Left: Map of randomly simulated fusion events (crosses), overlaid on the PXN-mCherry signal (red). Right: Derived synthetic image depicting the ventral cell surface area (white), the PXN-positive adhesion complexes (orange), and the simulated fusion events (crosses). Scale bar, 20 µm; magnified views, 10 µm. (C) Cumulative frequency charts, from the cell shown in A (cell #1) and two other representative cells (cells #2 and #3), demonstrating the difference in distance to focal adhesions (FA) between real ATG9A-pHluorin fusion events (black line) and simulated events (red line). (D) Quantification of the mean distance to the centroid of closest focal adhesion for real ATG9A-pHluorin events or simulated events (n = 8 cells, for a total number of 365 events). Statistical significance was evaluated using Mann–Whitney U test. ***, P < 0.001.
Discussion
Accumulating reports indicate that several components of the autophagy machinery mediate one or multiple cellular functions, which may not directly depend on autophagosome biogenesis (Subramani and Malhotra, 2013). Among studies specifically related to cell shape dynamics, it was demonstrated that Atg1 and Ref(2)P, the Drosophila homologs of mammalian ULK 1/2 and SQSTM1, respectively, were essential for the extension of protrusions in blood cells (Kadandale et al., 2010). Depletion of the autophagy proteins ULK1 and beclin-1 severely reduced the ability of mouse macrophages to spread and elongate (Kadandale et al., 2010). A recent report pointed to a non-autophagic function of Drosophila ATG9, indicating that loss of ATG9 caused aberrant cortical actin network organization in nurse cells surrounding the oocyte and female sterility (Kiss et al., 2020). Another study demonstrated that ATG9A was required for the post-Golgi transport and maturation of lysosomal hydrolases (Jia et al., 2017). The present work adds to these data and stresses the fact that, in addition to its well-documented role during phagophore expansion, ATG9A should be viewed as an essential component of vesicular trafficking driving cell migration.
We found that ATG9A signal localized in cell protrusions, at the plasma membrane, or in small vesicles located beneath the plasma membrane, displayed a marked colocalization with TGN46, suggesting that peripheral ATG9A mainly originates from the TGN. The Golgi apparatus plays a well-appreciated role during cell migration and invasion (Millarte and Farhan, 2012). In polarized cells, the TGN faces the leading edge and, as such, represents an optimally positioned sorting platform to ensure efficient delivery of promigratory proteins. Hence, Golgi-associated microtubules, as a special subgroup of microtubules, have been found to serve as fast tracks to support anterograde trafficking of post-Golgi cargoes to the leading edge (Hao et al., 2020). It should be noted that only a few studies have monitored polarized exocytosis of post-Golgi vesicles at the single-vesicle level, using live-cell TIRF microscopy. This includes data from Schmoranzer et al. (2003) showing that exocytosis of post-Golgi vesicles, loaded with low-density lipoprotein receptor–GFP, was polarized toward the leading edge of migrating fibroblasts. In polarized astrocytes, post-Golgi vesicles carrying the YFP-tagged vesicular stomatitis virus glycoprotein exhibited biased exocytosis toward the cell front, whereas exocytotic events appeared uniformly distributed in nonpolarized cells (Letinic et al., 2009). In line with these observations, spatial maps from our live-cell TIRF experiments demonstrated that most ATG9A-pHluorin events were clustered near (within <10 µm) the leading edge of polarized cells, while fusion events did not display obvious hotspots in nonpolarized cells. Critically, most of the recorded events were characterized by the dispersal of ATG9A-pHluorin into the plasma membrane, in a manner that shows a clear signature of full vesicle fusion. This raises the possibility that, in addition to the delivery of specific cargoes, ATG9A-positive vesicles may primarily act as a reservoir that dynamically delivers bulk lipids for lamellipodial expansion.
Integrin receptors are αβ heterodimers, in which 8 β subunits can assort with 18 α subunits to form 24 distinct heterodimers having varied affinities for extracellular matrix proteins (Hynes, 2002; Fu et al., 2012). By far the most commonly found subunit in integrin heterodimers is β1 integrin, which has been shown to pair with a variety of α subunits to form 12 different heterodimers. Several studies have focused on the recycling of specific heterodimers, leading to the demonstration of an antagonistic relationship between the recycling of αvβ3 and α5β1 during cell migration. Hence, the αvβ3 integrin heterodimer was found to be implicated in persistent cell migration, through a “short-loop” endosomal recycling pathway involving the small GTPase Rab4 (Danen et al., 2005). Endosomal recycling of the α5β1 heterodimer, through a Rab11-dependent, “long-loop” pathway, promotes random cell migration (White et al., 2007). Shafaq-Zadah et al. (2016) established a new paradigm by demonstrating that retrograde trafficking of β1 integrin, followed by its polarized re-secretion from the TGN, also supports directional cell migration. Interestingly, we found that depletion of ATG9A altered both chemotactic migration (Transwell assays) and intrinsic cell speed, evaluated from cell trajectories, in the absence of stimulus. Although the present study focused on the colocalization of ATG9A with the TGN marker TGN46, previous reports indicated that ATG9A also partially colocalizes with the Rab11-positive perinuclear compartment (Orsi et al., 2012; Puri et al., 2013). It is then likely that ATG9A participates in the trafficking of integrins from both the TGN and the perinuclear endosomal compartment. ATG9A protein may, as such, constitute a core component of random and directional migrations.
Could ATG9A protein be an intrinsic regulator of vesicle biogenesis from the endosomal or the Golgi compartments? A specific role of ATG9A in the budding of clathrin-coated pits from the TGN has been recently reported. It was demonstrated that ATG9A, through its interaction with the clathrin adaptor complex AP-1, was required for the transport of cathepsins from the TGN to lysosomes (Jia et al., 2017). Mechanistically, the authors found that the association of ATG9A with AP-1 facilitated subsequent interaction of AP-1 with the cathepsin receptor CIMPR, resulting in the polymerization of AP-1 and vesicle budding. We found from our rescue experiments that the N-terminal AP-binding site of ATG9A was critical for the transport of β1 integrin and TGN46 to the cell protrusions. These data are in agreement with the reported role of AP-1 in delivering β1 integrin to the cell membrane (Mana et al., 2016) and support the idea that association of ATG9A with AP-1 is a priming event, allowing the sorting/trafficking of a variety of cargoes. In addition to AP-1, it was found that ATG9A interacts with phosphatidylinositol 4-kinase III β (PI4KIIIβ), a phosphatidylinositol 4-phosphate (PI4P)–producing enzyme that is required for the budding of ATG9A-containing vesicles from the Golgi during autophagy induction (Judith et al., 2019). Interestingly, Golgi-localized PI4KIIIβ and PI4P, well-known regulators of membrane dynamics, have also been recognized as essential components of cell adhesion and migration (Tokuda et al., 2014; Bilodeau et al., 2020). Hence, functional studies established that PI4P produced at TGN microdomains triggers the recruitment of GOLPH3 protein which, by inducing local curvature of the TGN membrane, promotes the biogenesis of post-Golgi carriers and their transport to the leading edge (Xing et al., 2016; Rahajeng et al., 2019). In this framework, in-depth identification of the molecular mechanisms driven by ATG9A and GOLPH3 for the biogenesis of particular post-Golgi carriers, during chemotactic stimulation or autophagy induction, is a critical subject for further studies.
In summary, our work sheds light on a previously unappreciated role for ATG9A during chemotactic migration. This constitutes an additional mechanism linking Atg proteins and cell migration (Coly et al., 2016; Kenific et al., 2016a; Kenific et al., 2016b; Coly et al., 2017) and may help to fully characterize the protumoral functions of ATG9A already reported in triple-negative breast cancer (Claude-Taupin et al., 2018) and glioblastoma (Abdul Rahim et al., 2017). According to our model, we suspect that ATG9A, in addition to β1 integrin, may drive the delivery of an ensemble of cargoes to the leading edge of migrating cells. Future work is required to identify these critical cargoes.
Materials and methods
Reagents and chemicals
Antibodies were as follows: rabbit monoclonal anti-PXN (Y31; Ab32115; Abcam), mouse monoclonal &anti–c-myc (sc-40; Santa Cruz), sheep polyclonal TGN46 (AHP500GT; Bio-Rad), rabbit monoclonal anti-ATG9A (Ab108338; Abcam), mouse monoclonal β1 integrin (Ab24693; Abcam), and rabbit polyclonal anti-ATG5 (2630; Cell Signaling). Secondary antibodies used were Alexa Fluor 488–conjugated antibody against rabbit IgG (A21206; Invitrogen), Alexa Fluor 594–conjugated antibody against mouse IgG (A21203; Invitrogen), and Alexa Fluor 647–conjugated antibody against sheep IgG (Ab150179; Abcam). Other reagents in this study were recombinant human EGF (PHG0314; Gibco), human CXCL12 (350-NS; R&D Systems), CQ (C6628; Sigma-Aldrich), bafilomycin A1 (BafA1; B1793; Sigma-Aldrich), DAPI (D8417; Sigma-Aldrich), rhodamine phalloidin (R415; Thermo Fisher Scientific), hematoxylin (HHS32; Sigma-Aldrich), fibronectin (FN1; F0895; Sigma-Aldrich), Mowiol (475904; Merck Millipore), normal donkey serum (D9663; Sigma-Aldrich), formaldehyde (F8775; Sigma-Aldrich), Dulbecco’s PBS without salts (DPBS; D8537; Sigma-Aldrich), Triton X-100 (T3751/08; Thermo Fisher Scientific), BSA fraction V (10735108001; Roche), and methanol (M4056/17; Thermo Fisher Scientific).
Plasmid constructs and siRNAs
The expression vectors encoding the fusion protein EGFP-LC3B (#24920, deposited by Toren Finkel) and mCherry-Clathrin LC-15 (#55019, deposited by Michael Davidson) are available from Addgene. The pmKate2-N vector was purchased from Evrogen (#FP182). All the following expression vectors were purchased from GenScript. The expression vector encoding ATG9A-pHluorin was obtained by inserting codon-optimized human ATG9A (Q7Z3C6; Uniprot) and superecliptic pHluorin sequences into a pcDNA3.1(+) plasmid. The pHluorin sequence was fused in-frame into the first luminal domain of human ATG9A protein, between the amino acids Leu102 and His103. The expression vector encoding wild-type ATG9A-mCherry used in Figs. 5, 6, and S4 was obtained by inserting codon-optimized human ATG9A sequence (Q7Z3C6; Uniprot) and mCherry sequence (MK160997.1; GenBank) into a pcDNA3.1(+) plasmid. The mCherry sequence was fused in-frame into the first luminal domain of human ATG9A protein, between the amino acids Leu102 and His103. The ATG9A-mCherry Y8F mutant was obtained by replacing the TAC codon specifying tyrosine at position 8 by a TTT codon specifying a phenylalanine. An alternative expression vector encoding ATG9A-mCherry was used in Fig. 3 E and Fig. S2 D; it was generated by inserting a codon-optimized human ATG9A sequence, fused in-frame with the mCherry sequence (MK160997.1; GenBank) at its C-terminus, into a pcDNA3.1(+) plasmid, using HindIII and EcoRI cloning sites. The expression vector encoding ATG9A-myc was obtained by inserting the sequence of human ATG9A (GenScript clone ID OHu04028C; NM_001077198.2; GenBank) into a pcDNA3.1(+)-C-myc plasmid, using XhoI and ApaI cloning sites; the myc epitope is located at the C-terminus of the ATG9A protein. The expression vector encoding mCherry-LC3B was generated by inserting the human LC3B sequence (GenScript clone ID OHu18146C; NM_022818.5; GenBank), fused in-frame with the mCherry sequence at its N-terminus, into a pcDNA3.1(+) plasmid. The expression vector encoding PXN-EGFP was obtained by inserting the sequence of human PXN (GenScript clone ID OHu13990C; NM_002859.3; GenBank) into a pcDNA3.1(+)-N-EGFP plasmid, using KpnI and BamHI cloning sites; the EGFP sequence is located at the N-terminus of the PXN protein. The expression vector encoding PXN-mCherry was obtained by inserting the sequence of human PXN (GenScript clone ID OHu13990C; NM_002859.3; GenBank) into a pcDNA3.1(+) plasmid, using KpnI and BamHI cloning sites; the mCherry sequence is located at the N-terminus of the PXN protein. For ATG9A or ATG5 knockdown experiments, control (D-001810-03, 5′-UCAGAAAACAUGUAAACCA-3′), ATG9A #1 (J-014294-09, 5′-UUCUGCGUCUGCACGAUCC-3′), ATG9A #2 (J-014294-10, 5′-UUUCGGAAGAAGUCUAUAA-3′), ATG5 #1 (J-004374-07, 5′-GGCAUUAUCCAAUUGGUUU-3′), and ATG5 #2 (J-004374-08, 5′-GCAGAACCAUACUAUUUGC-3′) siRNAs were purchased from Dharmacon Horizon Discovery. Cells were subject to two rounds of transfection to ensure adequate knockdown of the proteins.
Cell culture and transfections
The glioblastoma multiform cell line U87 MG (WHO grade IV) was purchased from American Type Culture Collection (ATCC; HTB-14), and the glioblastoma multiform cell line 42 MG (WHO grade IV) from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ; ACC 431) was generously provided by Pr. J. Honnorat (Center for Research in Neuroscience in Lyon, Lyon, France). HeLa cell line (ATCC, CCL-2) was generously provided by Dr. P. Gaildrat (University of Rouen-Normandy, Rouen, France). HEK 293 cell line (CRL1573; ATCC) was generously provided by Drs. L. Prézeau, C. Barrère, and I. Bidaud (Institut de Génomique Fonctionnelle, Montpellier, France). These cells were routinely maintained in our laboratory according to the instructions from ATCC and cultured in DMEM (41965-039; Gibco), supplemented with 10% FBS (S181H-500; Dutscher), 1% antibiotic-antimycotic solution (A5955; Sigma-Aldrich), and 1% sodium pyruvate (S8636; Sigma-Aldrich). For all experiments, cells were placed in either serum-free DMEM supplemented with 1% of antibiotic-antimycotic solution and 1% of sodium pyruvate or serum- and phenol red–free Leibovitz’s L-15 medium (21083027; Gibco), supplemented with 1% D-glucose (G8644; Sigma-Aldrich), 1% antibiotic-antimycotic solution, and 1% sodium pyruvate. Transient transfections were performed using the Amaxa Cell Line Nucleofector Kit V (VCA-1003; Lonza) according to the manufacturer’s protocol, using the A-023 program for U87 MG and 42 MG cells, the I-013 program for HeLa cells, and the Q-001 program for HEK 293 cells.
RT-qPCR
Total RNA was extracted using TRI Reagent (T9424; Sigma-Aldrich) and purified on NucleoSpin RNA II columns (740955-250; Macherey-Nagel) according to the manufacturer’s instructions. 0.3 µg of total RNA was used for cDNA synthesis using the ImProm-II Reverse Transcription System (A3800; Promega). Real-time PCR was performed in a FAST SYBR green master mix (4385612; Applied Biosystems) using the QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific). The fold change in expression of ATG9A mRNA relative to GAPDH mRNA was calculated based on the threshold cycle (Ct) as 2−ΔCt, where ΔCt = CtATG9A − CtGAPDH. ATG9A primers: sense, 5′-CTGCCCTTCCGTATTGCAC-3′; antisense, 5′-CTCACGTTTGTGGATGCAGAT-3′; GAPDH primers: sense, 5′-CTGGGCTACACTGAGCACC-3′; antisense, 5′-AAGTGGTCGTTGAGGGCAATG-3′.
SDS-PAGE and Western blot
Cell lysis was performed in ice-cold lysis buffer (25 mM Tris HCl, pH 7.6 [BP153-1; Thermo Fisher Scientific], 150 mM NaCl [S3160/60; Thermo Fisher Scientific], 1% Nonidet P40 [74385; Sigma-Aldrich], 1% sodium deoxycholate [30970; Sigma-Aldrich], 0.1% SDS [BP2436; Thermo Fisher Scientific], 0.6 mM DTT [ALX-280-001-G050], 1× protease inhibitor cocktail [S8340; Sigma-Aldrich], and 1× phosphate inhibitor cocktails 2 [P5726; Sigma-Aldrich] and 3 [P0044; Sigma-Aldrich]) on ice for 10 min and was completed by passing cells through a syringe tip. After centrifugation (16,000g, 20 min, 4°C), cytoplasmic proteins were collected. Equal amounts of proteins (30 µg) were mixed with a volume of Laemmli buffer containing 2-mercaptoethanol (21985-023; Gibco) and loaded onto a precast 4–20% gradient SDS polyacrylamide gel (NB17-420; NuSep). A prestained protein ladder (26619; Thermo Fisher Scientific) was loaded onto the gel along with protein samples. After separation, proteins were transferred onto a polyvinylidene difluoride membrane within a transfer buffer using the Trans-blot Turbo RTA Transfer Kit (1704272; Bio-Rad) and the Trans-blot turbo system (1704150; Bio-Rad) according to the manufacturer’s protocol. Membranes were blocked with 5% nonfat milk (70166; Sigma-Aldrich; 1 h, room temperature) and incubated (overnight, 4°C) with the primary antibody (rabbit monoclonal anti-ATG9A [Ab108338; Abcam], rabbit polyclonal anti-ATG5 [2630; Cell Signaling], or mouse monoclonal anti–β-actin [sc-47778; Santa Cruz Biotechnology] as equal loading control). Membranes were then incubated (2 h, room temperature) with HRP-conjugated secondary antibodies raised against rabbit (31460; Thermo Fisher Scientific) or mouse (31430; Thermo Fisher Scientific) IgG. Immunoreactive bands were visualized with the enhanced chemiluminescence Western blotting substrate (RPN2236; GE Healthcare).
Autophagosome biogenesis
Autophagic activity was evaluated in cells expressing EGFP-LC3B or mCherry-LC3B fusion proteins. LC3B protein is a well-established effector of autophagy and a bona fide marker for autophagosomes (Klionsky et al., 2021). Punctate EGFP-LC3B or mCherry-LC3B staining provides a measure of ongoing autophagy, because it marks the successful processing of the cytosolic form LC3B-I to LC3B-II, a phospholipid-conjugated form that is targeted to phagophore membranes. Cells were transfected and placed in complete DMEM. 24 h after transfection, cells were rinsed twice with warm DPBS and placed for 6 h in serum-free DMEM in the absence or presence of CQ (10−5 M). After incubation, cells were rinsed with warm DPBS, fixed (4% formaldehyde), mounted on glass slides with Mowiol, and randomly photographed using the Leica DMI6000 B inverted microscope. The number of autophagosomes was calculated using the ComDet plug-in of ImageJ software.
ATP measurement and proliferation assay
ATP measurement
ATP levels were measured in cell lysates using the CellTiter-Glo 3D assay (G9682; Promega) according to the manufacturer’s instructions. Briefly, cells transfected with nontargeting siRNA (siCont) or one of the two siRNAs targeting ATG9A (siATG9A #1 or siATG9A #2) were incubated in complete DMEM (10% FBS) for 24 h, and then placed in serum-free DMEM 24–48 h after transfection. At each time point (24 or 48 h after transfection), cells were suspended and seeded at increasing densities onto a 96-well plate (2.5, 5.0, 7.5, 10, or 15 × 103 cells/well). After 5 h, the CellTiter-Glo 3D solution (100 μl) was added to each well. After orbital agitation (30 s) and incubation (25 min, room temperature), luminescence was recorded from each well using the TECAN Infinite PRO plate reader.
Proliferation assay
Cells transfected with nontargeting siRNA (siCont) or one of the two siRNAs targeting ATG9A (siATG9A #1 or siATG9A #2) were seeded onto a 24-well plate (50 × 103 cells/well). Cells were incubated in complete DMEM (10% FBS) for 24 h, then placed in serum-free DMEM 24–48 h after transfection. At each time point (24 or 48 h after transfection), cell counting was performed by removing the medium and incubating cells (20 min, 37°C) in a Cell-Tracker Green (10 µM) solution (C2925; Thermo Fisher Scientific). Cells were then suspended in a Trypsin-EDTA solution, and aliquots of each suspension were loaded onto a Chemometec A8 slide, for counting at 488 nm, using the Chemometec NC-3000 Nucleocounter.
Migration assay
HEK 293 cells (80,000/insert), U87 MG, 42 MG, or HeLa cells (20,000/insert) were seeded on FN1-coated Transwell inserts (8-μm pores, 24 wells; 3422; Corning). The gradient of ligands was generated by addition of EGF or CXCL12 in the lower chamber. After 24 h, cells on the upper surface of the insert were removed using a cotton swab. Cells that migrated onto the lower surface were fixed with four successive baths of methanol (50%, 70%, and 90% diluted in DPBS and 100% methanol) and then stained with hematoxylin. Insert membranes were cut out and mounted on glass slides with Mowiol and randomly photographed with a Nikon inverted microscope (Nikon France; 10 random fields/membrane). Cells in the photographic fields were counted by using the Cell counter plug-in of ImageJ software.
Immunocytochemistry and image analysis
Cells were fixed with formaldehyde (4%, 10 min), permeabilized with Triton X-100 (0.05%, 5 min), and blocked for 1 h with a mixture of normal donkey serum (2%) and fetal bovine serum (10%) diluted in DPBS. Cells were incubated overnight with the adequate primary antibody at 4°C followed by incubation for 2 h at room temperature with the adequate secondary antibody. Cells were counterstained with DAPI (1 µg/ml, 10 min) to label nuclei, and imaged using the Leica DMI6000 B inverted microscope, the Leica SP8 MP confocal multiphoton microscope or the Leica THUNDER Imager 3D Tissue. In PXN-labeled cells (Fig. 7 E and Fig. S5 A), the number of focal adhesion complexes was evaluated after image background subtraction using the Find Maxima function of the ImageJ software. Radial intensity profile of the β1 integrin signal (Fig. 5) was produced using the Clock-scan plug-in (Dobretsov and Romanovsky, 2006). The number of TGN46-positive vesicles was obtained using the ComDet plug-in of the ImageJ software.
Live-cell imaging
Transfected cells were seeded on FN1-coated 35-mm glass dishes (P35G-1.5-14-C, MatTek Corporation) for TIRF and epifluorescence experiments, or on FN1-coated 24 multi-well glass plate (P24G-1.5-13-F; MatTek Corporation) for phase-contrast experiments. 24 h after transfection, live-cell imaging was performed in serum-free Leibovitz’s L-15 medium supplemented with 1% D-glucose, 1% antibiotic-antimycotic, and 1% sodium pyruvate (TIRF and epifluorescence) or in serum-free DMEM supplemented with 1% antibiotic-antimycotic and 1% sodium pyruvate (phase contrast).
TIRF
TIRF images were acquired using a system that was built around a Leica DMI6000 B inverted microscope equipped with a ×100 oil objective with a numerical aperture of 1.46 (HCX PL APO; Leica), a Hamamatsu Orca-Flash 4.0 camera, and 488-nm and 561-nm lasers. The TIRF angle was set for the 488-nm and 561-nm lasers to achieve an evanescent field with a characteristic penetration depth of 110 nm. In the experiments involving dual-channel recording, 488- and 561-nm lasers were used consecutively for each image of the time series. Time intervals were recorded as follows. ATG9A-mCherry vesicle tracking experiments (Fig. 3 E): time series of images at 500-ms intervals; ATG9A-pHluorin exocytosis experiments (Figs. 4, 8, and S5): time series of images at 390-ms intervals; ATG9A-pHluorin and mKate2 dual-channel experiments (Fig. S3, C–E): time series of images at 1-min intervals and a time delay of 2.2 s between the two channels; ATG9A-pHluorin experiments for kymograph analyses (Fig. S3 B): time series of images at 30-s intervals; ATG9A-pHluorin and clathrin-mCherry dual-channel experiments (Fig. S2 F): time series of images at 3.4-s intervals (having a time delay of 1.6 s between the two channels); and PXN-EGFP experiments (Fig. 7, A–D): time series of images at 30-s intervals. All TIRF live-cell movies provided with this manuscript are shown at a frame rate of 15 frames/s.
Tracking of moving ATG9A-mCherry vesicles was performed using the Manual Tracking tool of ImageJ software. In PXN-EGFP transfected cells, the number of assembling and disassembling adhesions during a 40-min imaging session was determined using FAAS (Berginski et al., 2011). The results include only focal adhesions with a minimum size of 2 pixels (0.25 µm), and with assembly or disassembly phase lengths of ≥10 images (5 min). Simulated fusion events for uniform distributions were performed with R software (v.4.0.5). The function runif() was used to generate uniformly spaced points within the cell whose ROI coordinates defined the minimum and maximum xy interval for the position of the random points. For each cell, the number of simulated fusion events produced by the algorithm was identical to the number of real ATG9A-pHluorin events.
Epifluorescence
Images were acquired using a Leica DMI6000 B inverted microscope equipped with a ×63 oil objective with a numerical aperture of 1.32 (HCX PL APO CS; Leica), a Hamamatsu Orca-Flash 4.0 camera, and 488- and 561-nm lasers. For ATG9A-pHluorin and ATG9A-mCherry dual channel experiments (Fig. S2 D), time series of images at 30-s intervals were recorded (having a time delay of 1.7 s between the two channels).
Phase contrast
Cells were placed in a temperature- and CO2-controlled (37°C, 5% CO2) environment of a Zeiss Cell Discoverer 7 microscope system. Live-cell phase-contrast images of the individual field regions inside each well were automatically acquired using ZEN Blue v2.6 software with a time series of images at 40-s intervals. Kymographs of the cell protrusions were generated using the Multi Kymograph tool of Fiji software from 1-pixel-wide lines manually traced perpendicularly to the ruffling cell edges, and the protrusion distances (Δx), persistences (Δy), and velocities (Δx/Δy) were evaluated. Cell trajectories (Fig. 2 D) were obtained using a semiautomatic tracking macro of ImageJ software, generously provided by Federico Saltarin (Theodor Kocher Institute, Bern, Switzerland), and trajectory plots were rendered using the DiPer Excel macro (Gorelik and Gautreau, 2014). Average speeds (Fig. 2 E, average of the instantaneous speed) and mean square displacements (Fig. 2 F) were calculated using the DiPer Excel macro (Gorelik and Gautreau, 2014). Representative figure images were selected and additional image postprocessing steps (contrast adjustment, field selection, and scale bar addition) were performed using ImageJ software. Phase-contrast live-cell Video 1 is shown at a frame rate of 25 frames/s.
Statistical analysis
Data are presented as the mean ± SEM for each group of samples. Statistical analyses were performed using the Prism 8 software (GraphPad Software). Mann–Whitney U test was used for nonparametric analyses. Paired t test, one-way ANOVA, and two-way ANOVA were used for parametric analyses (data distribution was assumed to be normal, but this was not formally tested). For all tests, differences were considered statistically significant if P values were <0.05 (as indicated by *, P < 0.05; **, P < 0.01; and ***, P < 0.001). Details of the individual tests are included in the figure legends.
Online supplemental material
Fig. S1 shows validation of ATG9A knockdown. Fig. S2 shows structure and functional validation of ATG9A-pHluorin construct. Fig. S3 shows that ATG9A-pHluorin signal at the cell periphery correlates with protrusive activity. Fig. S4 shows that ATG9A protein regulates delivery of TGN46-positive post-Golgi carriers to cell protrusions. Fig. S5 shows that AT9A regulates the formation of adhesion complexes in HeLa cells. Video 1 shows that depletion of ATG9A protein impairs the formation of cell protrusions. Video 2 shows spreading ATG9A-pHluorin exocytotic events. Video 3 shows nonspreading ATG9A-pHluorin exocytotic events. Video 4 shows that ATG9A-pHluorin signal appearance at the leading edge correlates with protrusive activity. Video 5 shows that ATG9A protein regulates adhesion dynamics.
Supplementary Material
Acknowledgments
We thank Inès Drissa (Institut national de la santé et de la recherche médicale 1239, Mont Saint Aignan, France) for her generous technical support with ImageJ software and Federico Saltarin (Theodor Kocher Institute, Bern, Switzerland) for kindly providing us with the ImageJ macro for semiautomatic cell tracking. We are grateful to the Plate-Forme de Recherche en Imagerie Cellulaire de Normandie team (https://primacen.crihan.fr, the Cell Imaging Platform of Normandy, Institute for Research and Innovation in Biomedecine, Faculty of Sciences, University of Rouen-Normandy, Mont-Saint Aignan) for technical support on imaging experiments.
This work was supported by Institut National de la Santé et de la Recherche Médicale, Groupement des Entreprises Françaises dans la lutte contre le Cancer, TC2N network, la Ligue contre le Cancer Normandie, the French Agence Nationale de la Recherche, and the University of Rouen Normandy. D. Campisi is recipient of a fellowship from Normandy.
The authors declare no competing financial interests.
Author contributions: D. Campisi conducted experiments, designed the work, analyzed and interpreted data, and wrote the manuscript. L. Desrues conducted experiments and revised the manuscript. A. Mutel conducted experiments. K.-P. Dembélé conducted experiments. R. Parment analyzed data and edited the manuscript. P. Gandolfo contributed to discussion and reviewed and edited the manuscript. H. Castel contributed to discussion and reviewed and edited the manuscript. F. Morin designed and validated the work, analyzed and interpreted data, and wrote the manuscript.
References
- Abdul Rahim, S.A., Dirkse A., Oudin A., Schuster A., Bohler J., Barthelemy V., Muller A., Vallar L., Janji B., Golebiewska A., and Niclou S.P.. 2017. Regulation of hypoxia-induced autophagy in glioblastoma involves ATG9A. Br. J. Cancer. 117:813–825. 10.1038/bjc.2017.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelmann-Grill, B.C., Wach F., Cully Z., Hein R., and Krieg T.. 1990. Chemotactic migration of normal dermal fibroblasts towards epidermal growth factor and its modulation by platelet-derived growth factor and transforming growth factor-beta. Eur. J. Cell Biol. 51:322–326. [PubMed] [Google Scholar]
- Alabi, A.A., and Tsien R.W.. 2013. Perspectives on kiss-and-run: Role in exocytosis, endocytosis, and neurotransmission. Annu. Rev. Physiol. 75:393–422. 10.1146/annurev-physiol-020911-153305 [DOI] [PubMed] [Google Scholar]
- Banting, G., and Ponnambalam S.. 1997. TGN38 and its orthologues: Roles in post-TGN vesicle formation and maintenance of TGN morphology. Biochim. Biophys. Acta Mol. Cell Res. 1355:209–217. 10.1016/S0167-4889(96)00146-2 [DOI] [PubMed] [Google Scholar]
- Berginski, M.E., Vitriol E.A., Hahn K.M., and Gomez S.M.. 2011. High-resolution quantification of focal adhesion spatiotemporal dynamics in living cells. PLoS One. 6:e22025. 10.1371/journal.pone.0022025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau, P., Jacobsen D., Law-Vinh D., and Lee J.M.. 2020. Phosphatidylinositol 4-kinase III beta regulates cell shape, migration, and focal adhesion number. Mol. Biol. Cell. 31:1904–1916. 10.1091/mbc.E19-11-0600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisel, B., Wang Y., Wei J.-H., Xiang Y., Tang D., Miron-Mendoza M., Yoshimura S., Nakamura N., and Seemann J.. 2008. ERK regulates Golgi and centrosome orientation towards the leading edge through GRASP65. J. Cell Biol. 182:837–843. 10.1083/jcb.200805045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswenger, V., Baumann N., Jürschick J., Häckl M., Battle C., Schwarz J., Horn E., and Zantl R.. 2018. Characterization of EGF-guided MDA-MB-231 cell chemotaxis in vitro using a physiological and highly sensitive assay system. PLoS One. 13:e0203040. 10.1371/journal.pone.0203040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Cordero, J.J., Hodgson L., and Condeelis J.. 2012. Directed cell invasion and migration during metastasis. Curr. Opin. Cell Biol. 24:277–283. 10.1016/j.ceb.2011.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, S., Behnam Azad B., and Nimmagadda S.. 2014. The intricate role of CXCR4 in cancer. Adv. Cancer Res. 124:31–82. 10.1016/B978-0-12-411638-2.00002-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claude-Taupin, A., Fonderflick L., Gauthier T., Mansi L., Pallandre J.-R., Borg C., Perez V., Monnien F., Algros M.-P., Vigneron M., et al. 2018. ATG9A is overexpressed in triple negative breast cancer and its in vitro extinction leads to the inhibition of pro-cancer phenotypes. Cells. 7:248. 10.3390/cells7120248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coly, P.-M., Gandolfo P., Castel H., and Morin F.. 2017. The autophagy machinery: A new player in chemotactic cell migration. Front. Neurosci. 11:78. 10.3389/fnins.2017.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coly, P.-M., Perzo N., Le Joncour V., Lecointre C., Schouft M.-T., Desrues L., Tonon M.-C., Wurtz O., Gandolfo P., Castel H., and Morin F.. 2016. Chemotactic G protein-coupled receptors control cell migration by repressing autophagosome biogenesis. Autophagy. 12:2344–2362. 10.1080/15548627.2016.1235125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danen, E.H.J., van Rheenen J., Franken W., Huveneers S., Sonneveld P., Jalink K., and Sonnenberg A.. 2005. Integrins control motile strategy through a Rho–cofilin pathway. J. Cell Biol. 169:515–526. 10.1083/jcb.200412081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, A.K., Itzhak D.N., Edgar J.R., Archuleta T.L., Hirst J., Jackson L.P., Robinson M.S., and Borner G.H.H.. 2018. AP-4 vesicles contribute to spatial control of autophagy via RUSC-dependent peripheral delivery of ATG9A. Nat. Commun. 9:3958. 10.1038/s41467-018-06172-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pace, R., Skirzewski M., Damme M., Mattera R., Mercurio J., Foster A.M., Cuitino L., Jarnik M., Hoffmann V., Morris H.D., et al. 2018. Altered distribution of ATG9A and accumulation of axonal aggregates in neurons from a mouse model of AP-4 deficiency syndrome. PLoS Genet. 14:e1007363. 10.1371/journal.pgen.1007363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin, N.O., and Turner C.E.. 2008. Paxillin comes of age. J. Cell Sci. 121:2435–2444. 10.1242/jcs.018044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillenburg-Pilla, P., Patel V., Mikelis C.M., Zárate-Bladés C.R., Doçi C.L., Amornphimoltham P., Wang Z., Martin D., Leelahavanichkul K., Dorsam R.T., et al. 2015. SDF-1/CXCL12 induces directional cell migration and spontaneous metastasis via a CXCR4/Gαi/mTORC1 axis. FASEB J. 29:1056–1068. 10.1096/fj.14-260083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobretsov, M., and Romanovsky D.. 2006. “Clock-scan” protocol for image analysis. Am. J. Physiol.-Cell Physiol. 291:C869–C879. 10.1152/ajpcell.00182.2006 [DOI] [PubMed] [Google Scholar]
- Fletcher, S.J., and Rappoport J.Z.. 2010. Moving forward: Polarised trafficking in cell migration. Trends Cell Biol. 20:71–78. 10.1016/j.tcb.2009.11.006 [DOI] [PubMed] [Google Scholar]
- Fu, G., Wang W., and Luo B.-H.. 2012. Overview: Structural biology of integrins. Methods Mol. Biol. 757:81–99. 10.1007/978-1-61779-166-6_7 [DOI] [PubMed] [Google Scholar]
- Gorelik, R., and Gautreau A.. 2014. Quantitative and unbiased analysis of directional persistence in cell migration. Nat. Protoc. 9:1931–1943. 10.1038/nprot.2014.131 [DOI] [PubMed] [Google Scholar]
- Guardia, C.M., Tan X.-F., Lian T., Rana M.S., Zhou W., Christenson E.T., Lowry A.J., Faraldo-Gómez J.D., Bonifacino J.S., Jiang J., and Banerjee A.. 2020. Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery. Cell Rep. 31:107837. 10.1016/j.celrep.2020.107837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, H., Niu J., Xue B., Su Q.P., Liu M., Yang J., Qin J., Zhao S., Wu C., and Sun Y.. 2020. Golgi-associated microtubules are fast cargo tracks and required for persistent cell migration. EMBO Rep. 21:e48385. 10.15252/embr.201948385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes, R.O. 2002. Integrins: Bidirectional, allosteric signaling machines. Cell. 110:673–687. 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Imai, K., Hao F., Fujita N., Tsuji Y., Oe Y., Araki Y., Hamasaki M., Noda T., and Yoshimori T.. 2016. Atg9A trafficking through the recycling endosomes is required for autophagosome formation. J. Cell Sci. 129:3781–3791. 10.1242/jcs.196196 [DOI] [PubMed] [Google Scholar]
- Innocenti, M. 2018. New insights into the formation and the function of lamellipodia and ruffles in mesenchymal cell migration. Cell Adhes. Migr. 12:401–416. 10.1080/19336918.2018.1448352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivankovic, D., Drew J., Lesept F., White I.J., López Doménech G., Tooze S.A., and Kittler J.T.. 2020. Axonal autophagosome maturation defect through failure of ATG9A sorting underpins pathology in AP-4 deficiency syndrome. Autophagy. 16:391–407. 10.1080/15548627.2019.1615302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, S., Wang Y., You Z., Liu B., Gao J., and Liu W.. 2017. Mammalian Atg9 contributes to the post-Golgi transport of lysosomal hydrolases by interacting with adaptor protein-1. FEBS Lett. 591:4027–4038. 10.1002/1873-3468.12916 [DOI] [PubMed] [Google Scholar]
- Judith, D., Jefferies H.B.J., Boeing S., Frith D., Snijders A.P., and Tooze S.A.. 2019. ATG9A shapes the forming autophagosome through Arfaptin 2 and phosphatidylinositol 4-kinase IIIβ. J. Cell Biol. 218:1634–1652. 10.1083/jcb.201901115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadandale, P., Stender J.D., Glass C.K., and Kiger A.A.. 2010. Conserved role for autophagy in Rho1-mediated cortical remodeling and blood cell recruitment. Proc. Natl. Acad. Sci. USA. 6:1194–1195. 10.1073/pnas.0914168107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuta, S., Yamaguchi J., Suzuki C., Sasaki M., Kazuno S., and Uchiyama Y.. 2017. Small GTPase Rab1B is associated with ATG9A vesicles and regulates autophagosome formation. FASEB J. 31:3757–3773. 10.1096/fj.201601052R [DOI] [PubMed] [Google Scholar]
- Kean, M.J., Williams K.C., Skalski M., Myers D., Burtnik A., Foster D., and Coppolino M.G.. 2009. VAMP3, syntaxin-13 and SNAP23 are involved in secretion of matrix metalloproteinases, degradation of the extracellular matrix and cell invasion. J. Cell Sci. 122:4089–4098. 10.1242/jcs.052761 [DOI] [PubMed] [Google Scholar]
- Keller, R. 2005. Cell migration during gastrulation. Curr. Opin. Cell Biol. 17:533–541. 10.1016/j.ceb.2005.08.006 [DOI] [PubMed] [Google Scholar]
- Kenific, C.M., Stehbens S.J., Goldsmith J., Leidal A.M., Faure N., Ye J., Wittmann T., and Debnath J.. 2016a. NBR1 enables autophagy-dependent focal adhesion turnover. J. Cell Biol. 212:577–590. 10.1083/jcb.201503075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenific, C.M., Wittmann T., and Debnath J.. 2016b. Autophagy in adhesion and migration. J. Cell Sci. 129:3685–3693. 10.1242/jcs.188490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.-D., Guo T.W., Wu A.P., Wells A., Gertler F.B., and Lauffenburger D.A.. 2008. Epidermal growth factor-induced enhancement of glioblastoma cell migration in 3D arises from an intrinsic increase in speed but an extrinsic matrix- and proteolysis-dependent increase in persistence. Mol. Biol. Cell. 19:4249–4259. 10.1091/mbc.e08-05-0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss, V., Jipa A., Varga K., Takáts S., Maruzs T., Lőrincz P., Simon-Vecsei Z., Szikora S., Földi I., Bajusz C., et al. 2020. Drosophila Atg9 regulates the actin cytoskeleton via interactions with profilin and Ena. Cell Death Differ. 27:1677–1692. 10.1038/s41418-019-0452-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D.J., Abdel-Aziz A.K., Abdelfatah S., Abdellatif M., Abdoli A., Abel S., Abeliovich H., Abildgaard M.H., Abudu Y.P., Acevedo-Arozena A., et al. 2021. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy. 17:1–382. 10.1080/15548627.2020.1797280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecointre, C., Desrues L., Joubert J.E., Perzo N., Guichet P.-O., Le Joncour V., Brulé C., Chabbert M., Leduc R., Prézeau L., et al. 2015. Signaling switch of the urotensin II vasosactive peptide GPCR: Prototypic chemotaxic mechanism in glioma. Oncogene. 34:5080–5094. 10.1038/onc.2014.433 [DOI] [PubMed] [Google Scholar]
- Letinic, K., Sebastian R., Toomre D., and Rakic P.. 2009. Exocyst is involved in polarized cell migration and cerebral cortical development. Proc. Natl. Acad. Sci. USA. 106:11342–11347. 10.1073/pnas.0904244106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longatti, A., and Tooze S.A.. 2012. Recycling endosomes contribute to autophagosome formation. Autophagy. 8:1682–1683. 10.4161/auto.21486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, S., Yamamoto H., Kinch L.N., Garza C.M., Takahashi S., Otomo C., Grishin N.V., Forli S., Mizushima N., and Otomo T.. 2020. Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat. Struct. Mol. Biol. 27:1194–1201. 10.1038/s41594-020-00520-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mana, G., Clapero F., Panieri E., Panero V., Böttcher R.T., Tseng H.-Y., Saltarin F., Astanina E., Wolanska K.I., Morgan M.R., et al. 2016. PPFIA1 drives active α5β1 integrin recycling and controls fibronectin fibrillogenesis and vascular morphogenesis. Nat. Commun. 7:13546. 10.1038/ncomms13546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba, K., Kotani T., Tsutsumi A., Tsuji T., Mori T., Noshiro D., Sugita Y., Nomura N., Iwata S., Ohsumi Y., et al. 2020. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat. Struct. Mol. Biol. 27:1185–1193. 10.1038/s41594-020-00518-w [DOI] [PubMed] [Google Scholar]
- Mattera, R., Park S.Y., De Pace R., Guardia C.M., and Bonifacino J.S.. 2017. AP-4 mediates export of ATG9A from the trans-Golgi network to promote autophagosome formation. Proc. Natl. Acad. Sci. USA. 114:E10697–E10706. 10.1073/pnas.1717327114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza, M.C., Er E.E., Zhang W., Ballif B.A., Elliott H.L., Danuser G., and Blenis J.. 2011. ERK-MAPK drives lamellipodia protrusion by activating the WAVE2 regulatory complex. Mol. Cell. 41:661–671. 10.1016/j.molcel.2011.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesenböck, G., De Angelis D.A., and Rothman J.E.. 1998. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 394:192–195. 10.1038/28190 [DOI] [PubMed] [Google Scholar]
- Millarte, V., and Farhan H.. 2012. The golgi in cell migration: Regulation by signal transduction and its implications for cancer cell metastasis. Sci. World J. 2012:498278–498311. 10.1100/2012/498278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi, A., Razi M., Dooley H.C., Robinson D., Weston A.E., Collinson L.M., and Tooze S.A.. 2012. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell. 23:1860–1873. 10.1091/mbc.e11-09-0746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallesi-Pocachard, E., Bazellieres E., Viallat-Lieutaud A., Delgrossi M.-H., Barthelemy-Requin M., Le Bivic A., and Massey-Harroche D.. 2016. Hook2, a microtubule-binding protein, interacts with Par6α and controls centrosome orientation during polarized cell migration. Sci. Rep. 6:33259. 10.1038/srep33259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proux-Gillardeaux, V., Gavard J., Irinopoulou T., Mege R.-M., and Galli T.. 2005. Tetanus neurotoxin-mediated cleavage of cellubrevin impairs epithelial cell migration and integrin-dependent cell adhesion. Proc. Natl. Acad. Sci. USA. 102:6362–6367. 10.1073/pnas.0409613102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri, C., Renna M., Bento C.F., Moreau K., and Rubinsztein D.C.. 2013. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 154:1285–1299. 10.1016/j.cell.2013.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahajeng, J., Kuna R.S., Makowski S.L., Tran T.T.T., Buschman M.D., Li S., Cheng N., Ng M.M., and Field S.J.. 2019. Efficient Golgi forward trafficking requires GOLPH3-driven, PI4P-dependent membrane curvature. Dev. Cell. 50:573–585.e5. 10.1016/j.devcel.2019.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar, B., Moreau K., Jahreiss L., Puri C., and Rubinsztein D.C.. 2010. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 12:747–757. 10.1038/ncb2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley, A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., and Horwitz A.R.. 2003. Cell migration: Integrating signals from front to back. Science. 302:1704–1709. 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- Riggs, K.A., Hasan N., Humphrey D., Raleigh C., Nevitt C., Corbin D., and Hu C.. 2012. Regulation of integrin endocytic recycling and chemotactic cell migration by syntaxin 6 and VAMP3 interaction. J. Cell Sci. 125:3827–3839. 10.1242/jcs.102566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanan, S., De Angelis D., Rothman J.E., and Ryan T.A.. 2000. The use of pHluorins for optical measurements of presynaptic activity. Biophys. J. 79:2199–2208. 10.1016/S0006-3495(00)76468-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sann, S., Wang Z., Brown H., and Jin Y.. 2009. Roles of endosomal trafficking in neurite outgrowth and guidance. Trends Cell Biol. 19:317–324. 10.1016/j.tcb.2009.05.001 [DOI] [PubMed] [Google Scholar]
- Schmoranzer, J., Kreitzer G., and Simon S.M.. 2003. Migrating fibroblasts perform polarized, microtubule-dependent exocytosis towards the leading edge. J. Cell Sci. 116:4513–4519. 10.1242/jcs.00748 [DOI] [PubMed] [Google Scholar]
- Segall, J.E., Tyerech S., Boselli L., Masseling S., Helft J., Chan A., Jones J., and Condeelis J.. 1996. EGF stimulates lamellipod extension in metastatic mammary adenocarcinoma cells by an actin-dependent mechanism. Clin. Exp. Metastasis. 14:61–72. 10.1007/BF00157687 [DOI] [PubMed] [Google Scholar]
- Shafaq-Zadah, M., Gomes-Santos C.S., Bardin S., Maiuri P., Maurin M., Iranzo J., Gautreau A., Lamaze C., Caswell P., Goud B., and Johannes L.. 2016. Persistent cell migration and adhesion rely on retrograde transport of β1 integrin. Nat. Cell Biol. 18:54–64. 10.1038/ncb3287 [DOI] [PubMed] [Google Scholar]
- Sharifi, M.N., Mowers E.E., Drake L.E., Collier C., Chen H., Zamora M., Mui S., and Macleod K.F.. 2016. Autophagy promotes focal adhesion disassembly and cell motility of metastatic tumor cells through the direct interaction of paxillin with LC3. Cell Rep. 15:1660–1672. 10.1016/j.celrep.2016.04.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneeggen, M., Pedersen N.M., Campsteijn C., Haugsten E.M., Stenmark H., and Schink K.O.. 2019. WDFY2 restrains matrix metalloproteinase secretion and cell invasion by controlling VAMP3-dependent recycling. Nat. Commun. 10:2850. 10.1038/s41467-019-10794-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søreng, K., Munson M.J., Lamb C.A., Bjørndal G.T., Pankiv S., Carlsson S.R., Tooze S.A., and Simonsen A.. 2018. SNX18 regulates ATG9A trafficking from recycling endosomes by recruiting Dynamin-2. EMBO Rep. 19. 10.15252/embr.201744837e44837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehbens, S.J., Paszek M., Pemble H., Ettinger A., Gierke S., and Wittmann T.. 2014. CLASPs link focal-adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nat. Cell Biol. 16:561–573. 10.1038/ncb2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehbens, S.J., and Wittmann T.. 2014. Analysis of focal adhesion turnover: A quantitative live-cell imaging example. Methods Cell Biol. 123:335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani, S., and Malhotra V.. 2013. Non-autophagic roles of autophagy-related proteins. EMBO Rep. 14:143–151. 10.1038/embor.2012.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y., Tsotakos N., Liu Y., Young M.M., Serfass J., Tang Z., Abraham T., and Wang H.-G.. 2016. The Bif-1-Dynamin 2 membrane fission machinery regulates Atg9-containing vesicle generation at the Rab11-positive reservoirs. Oncotarget. 7:20855–20868. 10.18632/oncotarget.8028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayeb, M., Skalski M., Cha M., Kean M., Scaife M., and Coppolino M.. 2005. Inhibition of SNARE-mediated membrane traffic impairs cell migration. Exp. Cell Res. 305:63–73. 10.1016/j.yexcr.2004.12.004 [DOI] [PubMed] [Google Scholar]
- Tokuda, E., Itoh T., Hasegawa J., Ijuin T., Takeuchi Y., Irino Y., Fukumoto M., and Takenawa T.. 2014. Phosphatidylinositol 4-phosphate in the golgi apparatus regulates cell–cell adhesion and invasive cell migration in human breast cancer. Cancer Res. 74:3054–3066. 10.1158/0008-5472.CAN-13-2441 [DOI] [PubMed] [Google Scholar]
- Veale, K.J., Offenhäuser C., and Murray R.Z.. 2011. The role of the recycling endosome in regulating lamellipodia formation and macrophage migration. Commun. Integr. Biol. 4:44–47. 10.4161/cib.4.1.13569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veale, K.J., Offenhäuser C., Whittaker S.P., Estrella R.P., and Murray R.Z.. 2010. Recycling endosome membrane incorporation into the leading edge regulates lamellipodia formation and macrophage migration. Traffic. 11:1370–1379. 10.1111/j.1600-0854.2010.01094.x [DOI] [PubMed] [Google Scholar]
- Wang, J., and Howell K.E.. 2000. The luminal domain of TGN38 interacts with integrin β1 and is involved in its trafficking. Traffic. 1:713–723. 10.1034/j.1600-0854.2000.010904.x [DOI] [PubMed] [Google Scholar]
- White, D.P., Caswell P.T., and Norman J.C.. 2007. αvβ3 and α5β1 integrin recycling pathways dictate downstream Rho kinase signaling to regulate persistent cell migration. J. Cell Biol. 177:515–525. 10.1083/jcb.200609004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, M., Peterman M.C., Davis R.L., Oegema K., Shiau A.K., and Field S.J.. 2016. GOLPH3 drives cell migration by promoting Golgi reorientation and directional trafficking to the leading edge. Mol. Biol. Cell. 27:3828–3840. 10.1091/mbc.E16-01-0005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, J., Suzuki C., Nanao T., Kakuta S., Ozawa K., Tanida I., Saitoh T., Sunabori T., Komatsu M., Tanaka K., et al. 2018. Atg9a deficiency causes axon-specific lesions including neuronal circuit dysgenesis. Autophagy. 14:764–777. 10.1080/15548627.2017.1314897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, A.R.J., Chan E.Y.W., Hu X.W., Köchl R., Crawshaw S.G., High S., Hailey D.W., Lippincott-Schwartz J., and Tooze S.A.. 2006. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119:3888–3900. 10.1242/jcs.03172 [DOI] [PubMed] [Google Scholar]
- Zhou, C., Ma K., Gao R., Mu C., Chen L., Liu Q., Luo Q., Feng D., Zhu Y., and Chen Q.. 2017. Regulation of mATG9 trafficking by Src- and ULK1-mediated phosphorylation in basal and starvation-induced autophagy. Cell Res. 27:184–201. 10.1038/cr.2016.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.