Atkins et al. create a PSC differentiation model for human yolk sac hematopoiesis and discover multipotent progenitors with erythro-myeloid and T lymphoid potential.
Abstract
Atkins et al. (2022. J. Exp. Med. https://doi.org/10.1084/jem.20211924) create a PSC differentiation model for human yolk sac hematopoiesis and discover multipotent progenitors with erythro-myeloid and T lymphoid potential. The multipotent progenitors emerge via hemogenic endothelium and share origin with primitive erythroid wave in KDR+CD235a/b+ mesoderm.
For decades, scientists have attempted to solve the puzzle of developmental hematopoiesis and understand its complex architecture that involves multiple anatomical sites and temporally distinct waves of blood cell production. Paradoxically, the first priority of developmental hematopoiesis is not to create the undifferentiated, self-renewing hematopoietic stem cells (HSCs), but to rush order differentiation primed progenitors that rapidly generate primitive red cells, platelets, and macrophages for the embryo’s needs. The early simplistic model of two developmental hematopoietic waves—“primitive” in the extra-embryonic yolk sac, followed by “definitive” in the embryo proper—has been repeatedly revised after the discoveries of multiple developmentally restricted progenitor types that precede HSC generation (Ghosn et al., 2019; Palis, 2016). Importantly, these intermediate populations not only possess erythroid and myeloid potential, but also include progenitors with lymphoid (NK, B, or T cell) differentiation capacity, previously thought to be a hallmark of the HSC lineage (Bian et al., 2020; Ghosn et al., 2019; Zeng et al., 2019b). As hematopoietic cells begin to circulate in the embryo and extraembryonic tissues after the onset of heartbeat, it has been challenging to conclusively define the precursors and products of the different developmental hematopoietic waves. Studying these developmental processes is especially challenging in human, as the early hemogenic tissues are not readily accessible for research and in vivo lineage tracing studies are not possible. Given the different macroscopic anatomy of human and mouse extraembryonic tissues, the vastly different durations of pregnancy (both in absolute and relative terms), and the poor conservation of HSC/progenitor surface markers between mouse and human, it is essential to define the origin and kinetics of these populations in each species. In addition to the long-standing quest to understand human HSC development, attention has recently been drawn to the HSC independent progenitor waves since it was discovered that yolk sac progenitors also establish long-lasting progeny such as the brain microglia, liver Kupffer cells, and other tissue resident macrophages (Ginhoux et al., 2010; Gomez Perdiguero et al., 2015). As these populations have been associated with the pathogenesis of neurodegenerative and other diseases and they cannot be replenished from bone marrow HSCs, understanding their development has become a high priority (Salter and Stevens, 2017).

Insights from Hanna K.A. Mikkola.
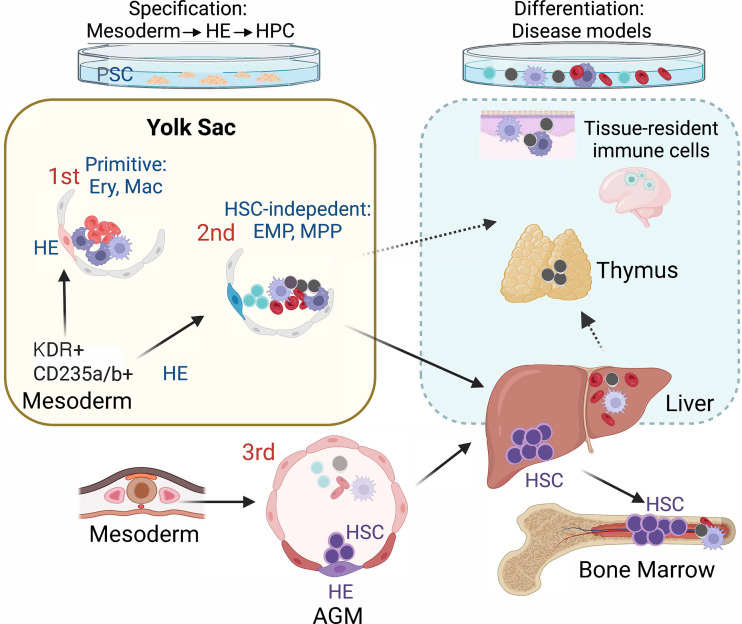
Since the derivation of human embryonic stem cells in 1998, pluripotent stem cell (PSC) in vitro differentiation has been viewed as a highly promising tool to study human developmental hematopoiesis, and it is essentially the only available model to access the earliest stages of this process. However, since PSC cultures lack the natural organization of intra- and extraembryonic tissues, these distinct and dynamically changing microenvironments have to be created in the culture dish by mimicking the developmental cues using growth factors, small molecules, and stroma cells. Despite over two decades of efforts, most differentiation protocols are not sophisticated enough to reproducibly generate the desired hematopoietic cell types, and the assays to determine their success lack sensitivity and specificity. The elegant study by Atkins et al. (2022) presents a human PSC in vitro differentiation model for yolk sac hematopoiesis that makes it possible to answer several long-standing questions about the cellular origin, developmental hierarchy, and differentiation potential of human yolk sac–type progenitors. By carefully guiding PSC differentiation with Activin A, BMP4, and FGF2 signaling, they generate KDR+CD235a/b+ mesoderm that, during a defined time window, gives rise to yolk sac–type primitive erythroid cells and macrophages via CD34+KDR+CD144+CD31+CD45− hemogenic endothelial (HE) intermediate. They further show that the next hematopoietic progenitor wave emerging from KDR+CD235a/b+ mesoderm, previously thought to consist exclusively of erythro-myeloid progenitors (EMPs), is in fact broadly multipotent, as many of the CD34+CD45+CD90+CD7− progenitors also have robust T lymphoid potential. The T cells generated from these precursors have unique features of fetal T lymphoid cells (Vδ2+ subset of γδ T cells; Tieppo et al., 2020) that are distinct from those derived from cord blood hematopoietic stem and progenitor cells. It remains to be shown whether human innate-like B1 B cells also develop from the same precursors. The challenges with establishing robust B cell differentiation protocols for human PSCs still make it difficult to evaluate B lymphoid potential reliably. The proposed developmental origin of B1 B cells from HSC independent progenitors in the yolk sac and embryo proper raises the possibility that they could also be linked to the multipotent progenitor (MPP) presented in this study (Ghosn et al., 2019; Yoshimoto et al., 2012).
The Keller group also validated these findings in mice, by sorting and culturing individual cells from CD41+kit+CD16/32+ immunophenotypic EMP population and conducting inducible lineage tracing using a Csf1r-Cre mouse line. These data showed that the embryonic day (E)9.25–9.5 yolk sac possesses similar multipotent progenitors with erythroid, myeloid, and/or T lymphoid potential as found in human embryonic stem cell cultures. Although the Csf1r-Cre–labeled cells had seeded E16.5 liver and thymus, they did not include fetal liver HSCs, confirming that these multiple differentiated cell types derive from HSC independent progenitors. Their study confirmed that the yolk sac–like progenitor hierarchy could also be generated from other PSC lines, such as human induced PSCs (iPSCs). Nevertheless, the authors emphasized the importance of optimizing the growth factor concentrations individually for the different PSC lines, which is an important factor to be taken into consideration when patient-specific iPSC lines are used for disease modeling or developing cell-based therapies. Notably, the PSC-derived KDR+CD235a/b+ mesoderm that gives rise to both YS primitive hematopoiesis and the EMP/MPP progenitor wave was created using the same protocol as the mesoderm that produces ventricular cardiomyocytes, the first cardiac lineage that develops in the embryo, witnessing another important link between the development of the cardiovascular and blood systems. Analysis of single cell RNA sequencing data from Carnegie stage 7 human embryos suggests the presence of KDR+CD235a/b+ mesoderm subset in gastrulating human embryos (Tyser et al., 2021), providing critical in vivo validation for the findings of mesoderm subtypes associated with the onset of hematopoiesis during human development.
Recreating human yolk sac hematopoiesis in a dish. Directing human PSC differentiation with Activin A, BMP4, and FGF2 generates KDR+CD235a/b+ mesoderm that gives rise to two consecutive waves of HE that represent the onset of yolk sac–type hematopoiesis. The first, primitive wave is characterized by the emergence of primitive erythroid cells (Ery) and macrophages (Mac). In the second wave, CD34+KDR+CD144+CD31+CD45− HE generates EMPs and MPPs that also differentiate to fetal T cells. In addition to seeding the liver and thymus to support blood and immune cell production during fetal life, these yolk sac progenitors generate long-lived, tissue-resident immune cells that populate various adult tissues and organs, such as the microglia in the brain. Differentiation of these yolk sac–type progenitors in culture provides unique opportunities for studying the relevance of these cells in disease processes.
This study tracked both the primitive and EMP/MPP waves to HE precursors, supporting the hypothesis that all human developmental hematopoietic waves originate from endothelial intermediates. Key questions for the future are: What makes these HEs different? What regulatory mechanisms in the HEs determine which type of hematopoietic cells to create? Recent studies have uncovered the importance of arterial signaling environment in the emergence of HSC from HE (Zeng, 2019a). As the HE in early human yolk sac and embryo are essentially inaccessible for robust functional studies, highly refined in vitro models that mimic yolk sac or aorta-gonad-mesonephros region hematopoiesis, such as those presented by Atkins et al. (2022), provide important tools to study these diverse HEs and their products both functionally and mechanistically (Ivanovs, 2017; Uenishi, 2018; Zeng, 2019a; Motazedian, 2020). These studies open up new opportunities to distinguish how priming to embryonic blood lineages (primitive wave), transient multipotency (EMP/MPP wave), or sustained multipotency and long-term self-renewal ability (HSC wave) are established in HEs at the molecular level. Complementing these studies with cutting-edge single-cell transcriptomics and multiomics to compare rigorous in vitro models to tissues that develop in vivo should provide valuable insights into these fundamental questions.
This work may also help decipher the connection of various developmental hematopoietic populations with malignant hematological disorders that originate before birth. Mixed lineage leukemia translocations lead to more aggressive, therapy-resistant leukemias if they occur in utero. Although these translocations are prime examples of strong oncogenes where one hit can confer self-renewal to progenitors and convert them to leukemia stem cells, it is unknown if translocation in HSC independent progenitors, or only in those that are downstream of HSC, can initiate this aggressive disease. Down’s syndrome, characterized by trisomy of chromosome 21, commonly results in imbalance of developmental hematopoietic populations during fetal life and a transient myeloproliferative disorder that also increases predisposition to leukemia. The fetal-like Vδ2+ γδ T cells were pinpointed as the cell of origin in aggressive cutaneous lymphomas. PSC-derived disease models provide new opportunities to uncover the molecular underpinnings of these disease processes in a tractable human model system. The work by Atkins et al. (2022) provides a missing piece in the puzzle that is needed to connect yolk sac developmental biology to understanding and treating human disease.
References
- Atkins, M.H., et al. 2022. J. Exp. Med. 10.1084/jem.20211924 [DOI] [Google Scholar]
- Bian, Z., et al. 2020. Nature. 10.1038/s41586-020-2316-7 [DOI] [Google Scholar]
- Ghosn, E., et al. 2019. Development. 10.1242/dev.170571 [DOI] [Google Scholar]
- Ginhoux, F., et al. 2010. Science. 10.1126/science.1194637 [DOI] [Google Scholar]
- Gomez Perdiguero, E., et al. 2015. Nature. 10.1038/nature13989 [DOI] [Google Scholar]
- Ivanovs, A., et al. 2017. Development. 10.1242/dev.134866 [DOI] [Google Scholar]
- Motazedian, A., et al. 2020. Nat. Cell Biol. 10.1038/s41556-019-0445-8 [DOI] [PubMed] [Google Scholar]
- Palis, J. 2016. FEBS Lett. 10.1002/1873-3468.12459 [DOI] [PubMed] [Google Scholar]
- Salter, M.W., and Stevens B.. 2017. Nat. Med. 10.1038/nm.4397 [DOI] [PubMed] [Google Scholar]
- Tieppo, P., et al. 2020. J. Exp. Med. 10.1084/jem.20190580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyser, R.C.V., et al. 2021. Nature. 10.1038/s41586-021-04158-y [DOI] [Google Scholar]
- Uenishi, G.I., et al. 2018. Nat. Commun. 10.1038/s41467-018-04134-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto, M., et al. 2012. Blood. 10.1182/blood-2011-12-397489 [DOI] [Google Scholar]
- Zeng, Y., et al. 2019a. Cell Res. 10.1038/s41422-019-0228-6 [DOI] [Google Scholar]
- Zeng, Y., et al. 2019b. Immunity. 10.1016/j.immuni.2019.09.008 [DOI] [Google Scholar]