Abstract
Objective
Ferroptosis is a specific subtype of programmed cell death, which plays an essential role in the immune-associated disease, atherosclerosis (AS). The purpose of this study was to identify potential ferroptosis-related gene biomarkers and its association with immune infiltration characteristics in atherosclerosis with bioinformatics methods.
Methods
Differentially expressed genes (DEGs) between AS and control groups were screened from GSE40231, analyzed for functional enrichment and then intersected with ferroptosis-related genes. Then, a random forest model was constructed based on these differentially expressed ferroptosis-related genes (DE-FRGs) and validated with dataset GSE132651. The performance of the models was evaluated with the area under receiver operating characteristic curves (AUC). Finally, we analyzed the correlation between DE-FRGs above and the characteristics of immune infiltration via CIBERSORT method.
Results
Six DE-FRGs (IL6, ANGPTL7, CDKN1A, AKR1C3, NOX4 and VLDLR) were detected based on dataset of GSE40231. Furthermore, a random forest model was constructed based on them with a compelling diagnostic performance of AUC = 0.8974 in the validation dataset GSE132651. In addition, the proportion of follicular helper T (Tfh) cells was significantly higher in AS group (P < 0.001). And we found significant correlation relationship between Tfh and expression level of ANGPTL7 (R = 0.35, P < 0.01), CDKN1A (R = 0.4, P < 0.0001), AKR1C3 (R = 0.64, P < 0.0001), NOX4 (R = 0.32, P < 0.01) and VLDLR (R = −0.43, P < 0.0001).
Conclusion
This study identified 6 DE-FRGs and validated a predicted model for the early prediction of AS, which also proved the close relationship between ferroptosis and immunity in the pathogenesis of AS.
Keywords: ferroptosis, atherosclerosis, differentially expressed genes, random forest model, follicular helper T cell
Introduction
Coronary artery disease (CAD) resulting from AS is a leading cause of mortality worldwide. In 2017, it was estimated that CAD affects around 126 million individuals all over the world, accounting for 1.72% of the global population.1 AS is characterized by endothelial dysfunction or death in the early stage, among which oxidative stress is an important initiating factor.2 In the end, large amounts of lipids and several types of immune cells, including macrophages, T and B lymphocytes accumulated and narrowed down the arterial lumen by forming the atherosclerotic plaques in the arterial wall.3,4 Experimental studies have demonstrated that large amounts of macrophages are activated by different cytokines and mediators and become foam cells in the lumen lesion.5 T cells, despite its minority proportion, also have a very significant function in the immune regulation of AS.6 In addition, all subsets of T cells (including CD4+, CD8+, NK and follicular helper T cells) have been identified in human atherosclerotic plaque.6,7 Inflammatory cells released specific cytokines and enzymes, which finally promote plaque erosion and rupture, leading to partial or total occlusion of the affected artery.8
Studies over the past several decades have defined and characterized several cell death types such as apoptosis, pyroptosis, necroptosis, and ferroptosis, which have all been reported to accelerate the process of atherosclerotic lesion formation.9–13 The roles of ferroptosis in AS are attracting increasingly attention at present. Ferroptosis is characterized by overwhelming, iron-dependent accumulation of lipid hydroperoxides. Glutathione peroxidase (GPX4) is an essential enzyme that defends cells against oxidative damage. Specifically, it could reverse lipid peroxidation and ferroptosis by consuming glutathione. GPX4 inactivation and the depletion of intracellular glutathione blocked the metabolic pathway of lipid oxides, which induced iron homeostatic limits and occurrence of severe oxidative damage, thereby resulting in the progression of ferroptosis.14 Nowadays, more and more literatures support an important role of ferroptosis in the pathogenesis of AS. For instance, Yang et al reported that overexpression of prenyl diphosphate synthase subunit 2 (PDSS2) suppressed the ferroptosis of human coronary artery endothelial cells by promoting the activation of nuclear factor erythroid 2-related factor-2 (Nrf2) pathways.15 Moreover, trapping ferroptosis by effective and specific small molecular ferroptosis inhibitors such as deferoxamine (DFO) could alleviate AS through attenuating lipid peroxidation and endothelial dysfunction.13,16
The underlying possible molecular mechanisms of ferroptosis and its association with immunity in the field of AS are still in the veil. To identify key ferroptosis-related genes, we used integrated bioinformatic analysis methods to screen differentially expressed genes (DEGs) between AS and control artery tissue of CAD patients. Meanwhile, functional enrichment analysis was exhibited to uncover biological functions and enriched pathways for these genes. Besides, six DE-FRGs were obtained by intersecting these DEGs with the ferroptosis gene dataset from FerrDb database. Additionally, we established a random forest predictive model based on these DE-FRGs and externally verified it using another data set (GSE132651). Pearson correlation coefficient was also calculated to exhibit the association between immune cells and DE-FRGs. In this study, systematic bioinformatics analyses are applied to screen hub ferroptosis-related genes and its relationship with immunity in AS. Our results will help to shed light on new thoughts in the mechanism of CAD.
Materials and Methods
Microarray Data Download and Preprocessing
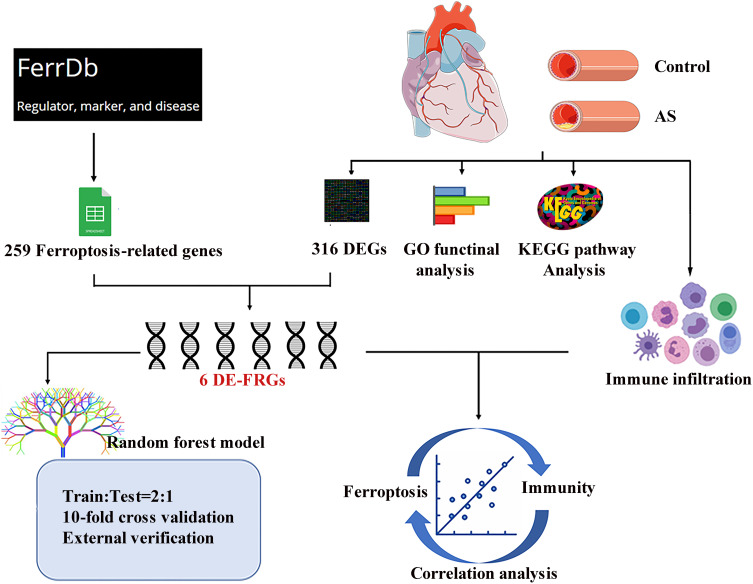
We downloaded the dataset of GSE40231 from GEO database, which was original from the Stockholm Atherosclerosis Gene Expression (STAGE) study. The primary aim of the study was to illustrate functionally associated genes in CAD. In this gene expression study, samples of atherosclerotic and unaffected arterial wall (distal part of mammary artery which was used as a graft) were isolated from 40 CAD patients undergoing coronary artery bypass grafting (CABG) surgery.17 After the microarray raw data were downloaded successfully, they were normalized by using a robust multichip average (RMA) background correction algorithm provided by Affy and limma package. Details of the data analysis workflow for the study design were described (Figure 1).
Figure 1.
Flow chart for the whole study.
Abbreviations: AS, atherosclerosis; DEGs, differentially expressed genes; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DE-FRGs, differentially expressed ferroptosis-related genes.
Functional Enrichment Analysis
GO term enrichment analysis, which contains subsections of biological processes, cellular component and molecular functions,18 was conducted through EnrichR web tool (https://maayanlab.cloud/Enrichr/).19 Critical signaling pathways enriched in DEGs were identified from the database of KEGG (Kyoto Encyclopedia of Genes and Genomes)20 on the same platform. P < 0.05 was considered statistically significant for both GO and KEGG analysis. Top 10 listed pathways with the smallest P value are displayed.
Recognition of Differentially Expressed Genes
The expression profiles of AS and unaffected arterial wall were compared to screen the DEGs. The cut-off criteria is as following: |log2(fold-change)|>1 and adjusted P-value <0.05. Volcano plot and heat map were drawn to display the expression of DEGs between AS and control groups with the “limma” package.21
Identification of DE-FRGs
We obtained a gene dataset which included 269 genes from the Ferroptosis Database (http://zhounan.org/ferrdb).22 Intersection of the ferroptosis genes and DEGs from GSE40231 was exhibited using the VennDiagram package in R. Principal component analysis (PCA) was applied for dimensionality reduction to distinguish the AS and control groups using the ggfortify package in R. Finally, the ggpaired package in R was utilized to exhibit the expression of these DE-FRGs in different groups using a paired sample t-test as the statistical analysis method. The P-value <0.05 was considered statistically significant.
Construction of Random Forest Model
The random forest is a machine learning technique of training and predicting samples with high accuracy via constructing a multitude of decision trees.23 The prediction model has been widely used for identifying potential predictors with the randomForest package24 in R. To achieve less biased or less optimistic performance of the training set, K-fold cross-validation with cv = 10 was applied. In the ongoing study, six DE-FRGs and disease state of control and AS were viewed as the classification feature and variables, respectively. The samples in GSE40231 (n = 80) were randomly assigned into the nonoverlapping training set and test set with the ratio of 2:1. Suitable values for key parameters were specified, such as mtry and ntree. The model built by the training set was verified by the test set further. The error rate was calculated to evaluate the combined classification through Out-of-bag (OOB) algorithm.25 The mean decrease accuracy (MDA) and the mean decrease Gini (MDG) were positively correlated with the importance of variables.26 Finally, GSE132651 was used to externally verify the model and demonstrate the prediction ability of this diagnostic model. ROC curves were visualized through pROC package.
Immune Infiltration Characteristic and Its Correlation with DE-FRGs
Immune infiltration characteristics of 22 types of immune cells were assessed by the CIBERSORT algorithm,27 including macrophages, T cells, natural killer (NK) cells, mast cells, B cells, dendritic cells (DC), monocytes, plasma cells, neutrophils, basophils and eosinophils. After obtaining the expression proportion matrix of immune cell subtypes with the guide of the CIBERSORT website, we use “ggplot2” package to draw boxplot to depict the distribution of immune cells. The Pearson correlation coefficient between each kind of immune cells and DE-FRGs was calculated, respectively, and the results with correlation coefficient and P value were exhibited using “ggpubr” package in R software.
Result
Identification and Functional Analysis of DEGs
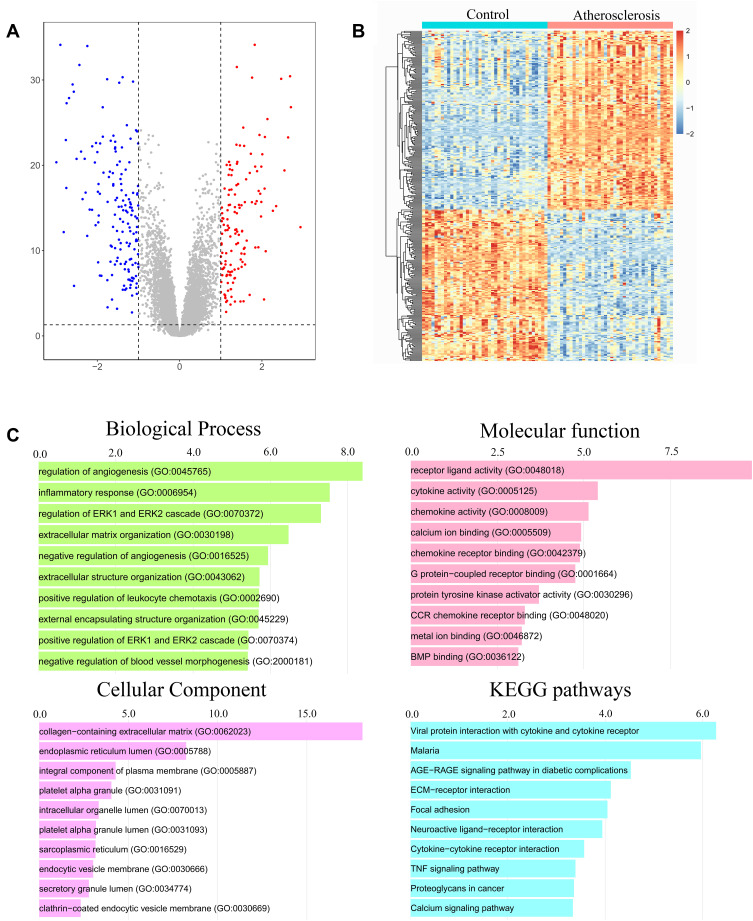
The 80 samples which contained the AS and control groups were normalized via RMA algorithm. Three hundred and sixteen DEGs were identified after setting the cutoff values (|log2(fold-change)|>1 and adjusted P-value <0.05), including 145 upregulated genes and 171 downregulated genes. They were displayed in a volcano plot (Figure 2A). Results have also revealed the top five upregulated genes (CARTPT, SCG2, CYTL1, SLC10A4, HAPLN1) and top five downregulated genes (HOXC6, EMX2, HOXA5, RBPMS2, HOXA9) in the AS group. In addition, these DEGs were exhibited in a heat map (Figure 2B).
Figure 2.
Identification of differentially expressed genes (DEGs) and functional enrichment analysis. (A) The DEGs identified were displayed in a volcano plot. Up-regulated genes are marked in red, and down-regulated genes are labeled in blue. (B) Heat map of the DEGs. (C) Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis among these DEGs.
Functional enrichment analysis was carried out with Enrichr web tool. We analyzed GO terms and KEGG pathway enriched in 316 DEGs. A collection of GO terms and pathways according to -log10 (P value) was displayed through bar plot (Figure 2C). For biological pathway analysis, the most enrichment terms were regulation of angiogenesis and inflammatory response. Molecular function subsection data indicated that receptor ligand, cytokine and chemokine activity involved in the DEGs significantly. Cellular component study displayed significant enrichment of these genes in collagen-containing extracellular matrix. KEGG pathway analysis mapped these genes into viral protein interaction with cytokine and cytokine receptor.
Collection and Analysis of DE-FRGs
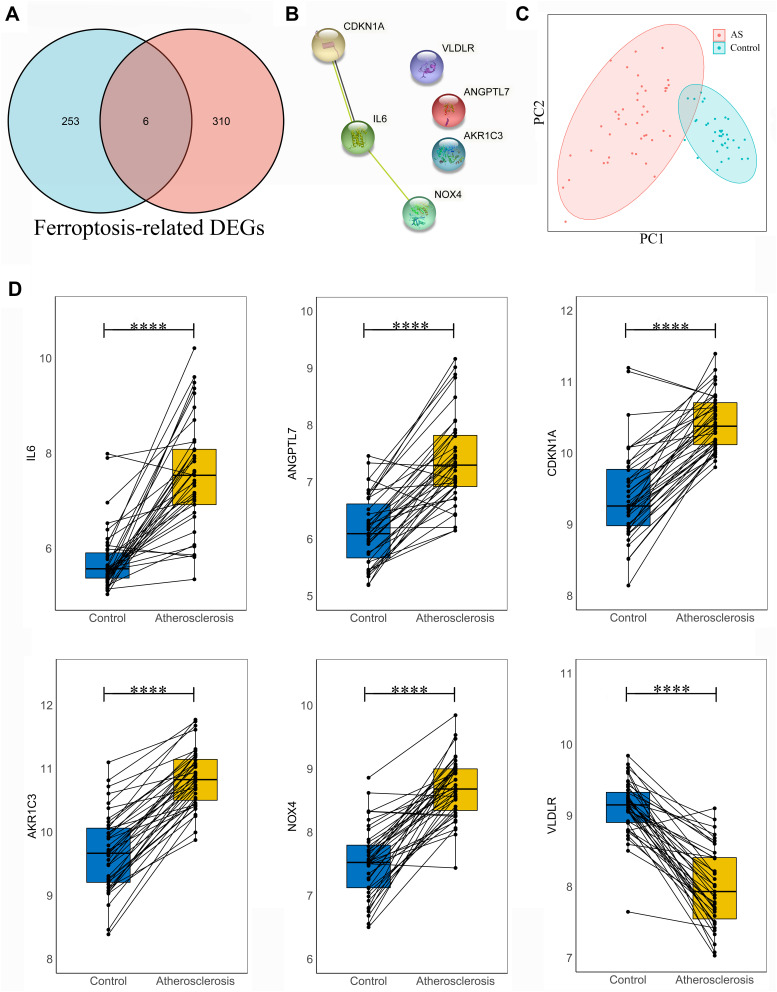
We collected a dataset of 259 genes after deduplication from the Ferroptosis Database (FerrDb), which included 108 drivers, 69 suppressors and 111 markers of ferroptosis. The common genes between ferroptosis genes from FerrDb and DEGs of GSE40231 were visualized through a Venn diagram. Six DE-FRGs were identified, including Interleukin 6 (IL6), Angiopoietin Like 7 (ANGPTL7), Cyclin Dependent Kinase Inhibitor 1A (CDKN1A), aldo-keto Reductase Family 1 Member C3 (AKR1C3), NADPH Oxidase 4 (NOX4) and Very Low Density Lipoprotein Receptor (VLDLR) (Figure 3A). The relevant information of these screened ferroptosis-related genes is elaborated in Table 1. A protein-protein-interaction (PPI) network is shown in Figure 3B. Results from PCA revealed that genes mentioned above could successfully distinguish AS patients from those healthy people (Figure 3C). All these DE-FRGs were exhibited to be greatly changed in the AS group in GSE40231. P-value <0.0001 was considered statistically significant (Figure 3D).
Figure 3.
Analysis of the differentially expressed ferroptosis-related genes (DE-FRGs). (A) Overlapped DE-FRGs in Venn diagram; (B) protein-protein-interaction (PPI) network of 6 DE-FRGs. (C) principal component analysis of these DE-FRGs; (D) gene expression level of 6 DE-FRGs was compared in AS and control groups with paired t-test, respectively. Significance markers: ****P < 0.0001.
Abbreviations: PC1, principal component 1; PC2, principal component 2; CDKN1A, cyclin dependent kinase inhibitor 1A; IL-6, interleukin 6; NOX4, NADPH oxidase 4; VLDLR, very low density lipoprotein receptor; ANGPTL7, angiopoietin like 7; AKR1C3, aldo-keto reductase family 1 member C3.
Table 1.
DE-FRGs of Coronary Artery Atherosclerosis
| Gene | Full Name | Role in Ferroptosis | logFC | P-value | adj.P.Val |
|---|---|---|---|---|---|
| IL6 | Interleukin 6 | Marker | 2.092407 | 3.22E-12 | 1.19E-10 |
| ANGPTL7 | Angiopoietin Like 7 | Marker | 1.257568 | 2.92E-12 | 1.09E-10 |
| CDKN1A | Cyclin Dependent Kinase Inhibitor 1A | Suppressor | 1.018134 | 3.16E-13 | 1.42E-11 |
| AKR1C3 | Aldo-keto Reductase Family 1 Member C3 | Suppressor | 1.169341 | 1.80E-15 | 1.19E-13 |
| NOX4 | NADPH Oxidase 4 | Driver | 1.162017 | 2.94E-16 | 2.15E-14 |
| VLDLR | Very Low Density Lipoprotein Receptor | Marker | −1.15079 | 2.22E-17 | 1.97E-15 |
Abbreviations: logFC, log fold change; adj.P.Val, adjusted P value.
Construction of Random Forest Model
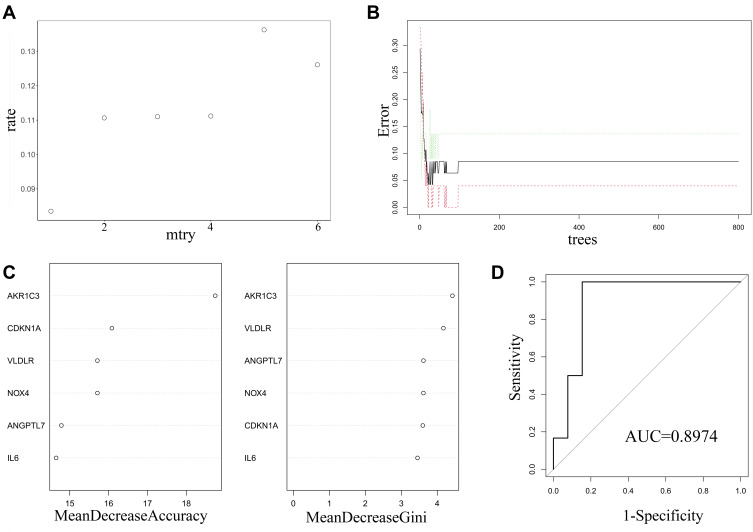
The random forest regression classification model was constructed based on the 6 DE-FRGs. When mtry = 1, the false-positive rate was the lowest (about 8.35%) (Figure 4A). The error rate became stable when the number of decision trees reached about 200 (Figure 4B). The importance of variables was weighed according to two parameters, mean decrease accuracy and mean decrease Gini. It was found that AKR1C3 was the most important variables for AS prediction (Figure 4C). After we established the random forest model with train data, the testing data sets were verified to prove its high prediction cost with AUC of 0.996 (Figure 4D). Additionally, GSE132651 was selected to externally verify the predictive model with a compelling performance of AUC = 0.8974.
Figure 4.
Construction and evaluation of random forest model based on the 6 DE-FRGs. (A) The scatter plot exhibited the association between the false-positive rate and mtry index (from 1 to 6). (B) Trend of the related errors according to the number of decision trees. (C) The relationship between variables and two parameters, mean decrease accuracy (the left panel) and mean decrease Gini (the right panel), respectively. (D) Evaluation of the prediction efficiency of the prediction model using ROC analysis in GSE132651.
Abbreviations: MeanDecreaseAccuracy, mean decrease accuracy; MeanDecreaseGini, mean decrease Gini; ROC, receiver operating characteristic; AUC, area under the ROC curve.
Relationships Between DE-FRGs and Immune Infiltration
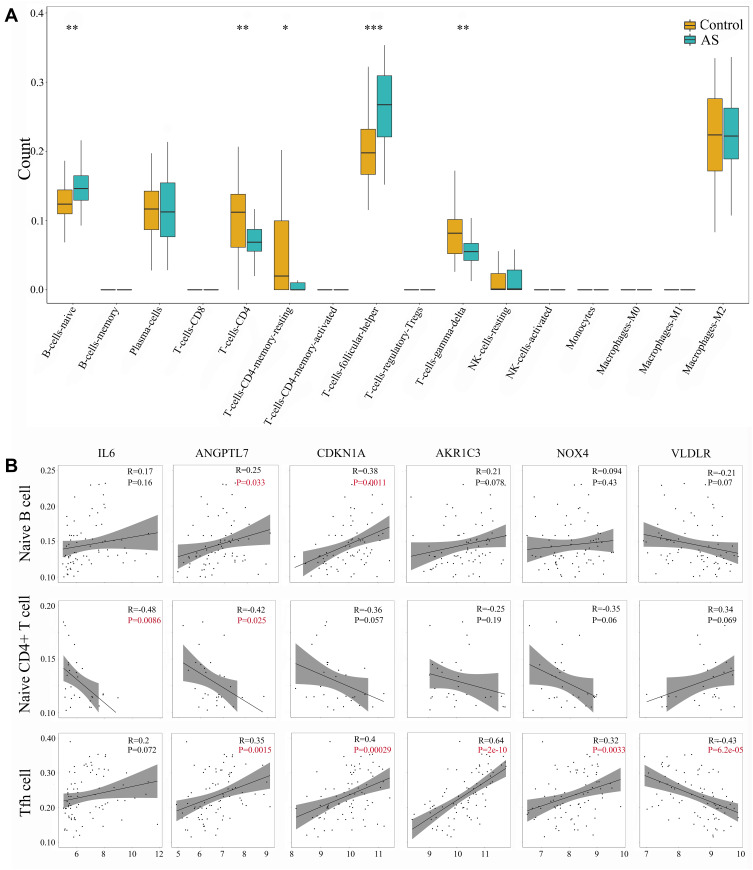
CIBERSORT algorithm was used to evaluate the immune infiltration characteristics of AS. The relative proportion of immune cell subtypes was displayed in the boxplot. As shown in Figure 5A, follicular helper T cells (Tfh) showed higher proportion significantly in AS group, compared with control group (P < 0.001). Meanwhile, there was significant difference in naïve B cells, naïve CD4+ T cells, gamma-delta T cells (P < 0.01) and resting CD4+ memory T cells (P < 0.05) between AS and control. Further analysis about Pearson correlation coefficient demonstrated close relationship between DE-FRGs and immune infiltration (Figure 5B). We found significant association between Tfh and expression level of ANGPTL7 (R = 0.35, P < 0.01), CDKN1A (R = 0.4, P < 0.0001), AKR1C3 (R = 0.64, P < 0.0001), NOX4 (R = 0.32, P < 0.01) and VLDLR (R = −0.43, P < 0.0001). Besides, naïve B cells were shown to be related to ANGPTL7 (R = 0.25, P = 0.033) and CDKN1A (R = 0.38, P = 0.0011). And naïve CD4+ T cells was related to IL6 (R = −0.48, P = 0.0086) and ANGPTL7 (R = −0.42, P = 0.0025). The correlation relationship between other immune cell subtypes and 6 DE-FRGs did not show significant association (which was not shown). Overall, these 6 DE-FRGs were in varying degrees related to immune cells, especially Tfh.
Figure 5.
Identification of immune infiltration and its correlation relationship with DE-FRGs. (A) Proportion of immune cell subtypes exhibited in boxplot diagram with Wilcoxon test. Significance markers: *P < 0.05; **P < 0.01; ***P < 0.001. (B) Pearson correlation coefficient analysis between immune cells and DE-FRGs. P values of P < 0.05 were labeled with red color.
Abbreviations: AS, atherosclerosis; IL-6, interleukin 6; ANGPTL7, angiopoietin like 7; CDKN1A, cyclin dependent kinase inhibitor 1A; AKR1C3, aldo-keto reductase family 1 member C3; NOX4, NADPH oxidase 4; VLDLR, very low density lipoprotein receptor; Tfh cell, follicular helper T cell.
Discussion
The term ferroptosis was newly proposed in 2012, which was defined as a specific cell death type induced by the small molecule erastin.14 It has been reported to be closely involved in the pathophysiological process of diseases characterized by sterile inflammation, including cancers, degenerative brain diseases and cigarette smoke-induced chronic obstructive pulmonary disease.28 However, definite evidence for the underlying molecular mechanisms of ferroptosis in the field of atherosclerosis is still limited. In the current study, we used integrated bioinformatics methods to identify key ferroptosis-related genes in the pathogenesis of atherosclerosis. The six ferroptosis-related genes (IL6, ANGPTL7, CDKN1A, AKR1C3, NOX4 and VLDLR) were speculated to play a vital role in CAD.
A random forest model with 10-fold cross-validation was constructed base on these hub genes. Cross-validation is a resampling procedure used to evaluate the machine learning model when sample sizes are limited. It usually results in a less biased or less optimistic generalization performance.29 The random forest model constructed in this study was verified further in GSE132651, which was published by Hebbel et al.30 In GSE132651, blood outgrowth endothelial cells (BOEC) from subjects with abnormal and normal coronary endothelial function were used for gene expression analysis. Our study addresses the feasibility of applying the model above to predict endothelial function with a satisfying diagnostic performance of AUC = 0.8974. Since coronary endothelial dysfunction is the earliest clinically detectable form of atherosclerosis,31 our study may also have practical clinical implications in diagnosis of atherosclerosis at an early stage.
Previous literatures reviewed the relationship of these screened ferroptosis-related hub genes with atherosclerosis. Interleukin-6 (IL-6) is a widely used cytokine that causes inflammation and oxidative stress. A study in 1994 mentioned that IL-6 was expressed locally in coronary atherosclerotic plaques, with expression level of 10- to 40-fold in the atherosclerotic arteries compared with the non-atherosclerotic artery.32 Not only IL-6 receptor antibody but also IL-6 inhibitor has been proved to be useful to alleviate atherosclerotic lesion development.33 Li et al reported that inhibition of ANGPTL7 significantly reversed tumor necrosis factor-α induced oxidative stress and endothelial cell adhesion,34 which identified Angptl7 as a potential target in atherosclerosis. CDKN1A, also named p21, encodes a potent cyclin-dependent kinase inhibitor. However, the role of p21 in atherosclerosis is controversial. The study carried out by Merched and Chan revealed the proatherogenic effect of p21, since loss of p21 protected against atherosclerosis in apoE-/- mice.35 Some studies considered p21 to be as an antiatherogenic target that can be used to treat or prevent atherosclerosis and postangioplasty restenosis.36,37 AKR1C3 is one of aldo/keto reductase superfamily. While AKR1C3 is expressed mainly in human steroid-hormone-targeting tissues including prostate and lung, it is also constitutively expressed in artery endothelial cells.38 Moreover, AKR1C3 was involved in the process of reactive oxygen species (ROS) accumulation in endothelial cells.38 NOX4, which belongs to the NADPH oxidase family, is constitutively active in smooth muscle cells (SMCs) and endothelial cells.39 Tong et al reported that Nox4 plays a pro-atherosclerosis role by stimulating SMC proliferation, migration and inflammation.39 VLDLR belongs to LDL receptor family, and its association with atherosclerosis is complicated. On the one hand, VLDLR is involved in peripheral triglyceride uptake, exerting a proatherogenic effect. On the other hand, VLDLR in SMC likely has a protective role against atherosclerosis. Besides, both pro- and anti-atherogenic roles of VLDLR in macrophages have been reported.40
Accumulated evidences point to a functional role for Tfh cells in AS. Since the inducible T cell costimulatory ligand (ICOSL) signal provided by DCs or B cells is critical for the differentiation and maintenance of Tfh cells, αICOSL antibody can be used to downregulate Tfh.41 Gaddis et al reported that αICOSL antibody-treated group showed a 10-fold reduction in the numbers of Tfh cells derived from Treg in the aorta root of AopE−/− mice, accompanied by a 30% reduction in lesion area of atherosclerosis.42 Besides, atherosclerosis development was alleviated in Tfh cell-deficient mice (Bcl6fl/flcreCd4),42 which proved that Tfh cells are indeed directly pro-atherogenic, instead of through other immune cell population impacted by ICOS/ICOSL interactions indirectly. Moreover, it was shown that oxidized low-density lipoprotein (ox-LDL) contributed to plaque formation via facilitating the generation of Tfh cells, which were original from nTreg cells, via reduction of IL2Rα and enhancement of IL6R expression.42 However, reports about the relationship between ferroptosis and Tfh cells are limited, let alone in the field of AS. A newest related study was just published, which illustrated selenium-GPX4-ferroptosis axis, instead of apoptosis or pyroptosis, played a key role in regulating Tfh homeostasis in immunized mice and young adults after influenza vaccination.43 In our study, we found significant correlation relationship between Tfh and expression level of ANGPTL7, CDKN1A, AKR1C3, NOX4 and VLDLR. The direct evidences about the association were not mentioned in papers published previously. Further experiment research is needed to classify whether these ferroptosis-related genes and proteins participate in the pathogenesis of AS by regulating local immune cells in artery wall, especially Tfh.
In this study, we have constructed and validated a ferroptosis-related genes associated model in AS. Our results revealed the powerful prediction ability of ferroptosis gene biomarkers and reflected their relationship with the immune infiltration. Nevertheless, the shortcomings of this study should also be pointed out. Firstly, our results were dependent on existing data by bioinformatic analysis, suggesting that scant analytical methods may limit the predictive capability of the constructed model and new proof from future studies may renew current results. Secondly, our analyses are based on public datasets, and further researches about the detailed molecular mechanism are necessary.
Conclusion
In the current study, we screened six DE-FRGs (IL6, ANGPTL7, CDKN1A, AKR1C3, NOX4 and VLDLR) in artery wall between the AS and control groups. Furthermore, a random forest model of AS was developed with these hub genes and verified in dataset GSE132651 (AUC = 0.8974). Immune cell infiltration and its correlation relationship with DE-FRGs were recognized. The DE-FRGs associated model provided a simple tool for the early prediction of AS, which improved our understanding of the association of ferroptosis and immunity in the pathogenesis of AS further.
Funding Statement
There is no funding for this work.
Data Sharing Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.
Ethics Approval Statement
The research has been reviewed by the Ethics Committee at the Wuhan Union Hospital of Huazhong University of Science and Technology in 2021 (No. 0959).
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Khan MA, Hashim MJ, Mustafa H, et al. Global epidemiology of ischemic heart disease: results from the global burden of disease study. Cureus. 2020;12(7):e9349. doi: 10.7759/cureus.9349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kattoor AJ, Pothineni NVK, Palagiri D, Mehta JL. Oxidative stress in atherosclerosis. Curr Atheroscler Rep. 2017;19(11):42. doi: 10.1007/s11883-017-0678-6 [DOI] [PubMed] [Google Scholar]
- 3.Emini Veseli B, Perrotta P, De Meyer GRA, et al. Animal models of atherosclerosis. Eur J Pharmacol. 2017;816:3–13. doi: 10.1016/j.ejphar.2017.05.010 [DOI] [PubMed] [Google Scholar]
- 4.Abdolmaleki F, Gheibi Hayat SM, Bianconi V, Johnston TP, Sahebkar A. Atherosclerosis and immunity: a perspective. Trends Cardiovasc Med. 2019;29(6):363–371. doi: 10.1016/j.tcm.2018.09.017 [DOI] [PubMed] [Google Scholar]
- 5.Shashkin P, Dragulev B, Ley K. Macrophage differentiation to foam cells. Curr Pharm Des. 2005;11(23):3061–3072. doi: 10.2174/1381612054865064 [DOI] [PubMed] [Google Scholar]
- 6.Lahoute C, Herbin O, Mallat Z, Tedgui A. Adaptive immunity in atherosclerosis: mechanisms and future therapeutic targets. Nat Rev Cardiol. 2011;8(6):348–358. doi: 10.1038/nrcardio.2011.62 [DOI] [PubMed] [Google Scholar]
- 7.Andersson J, Libby P, Hansson GK. Adaptive immunity and atherosclerosis. Clin Immunol. 2010;134(1):33–46. doi: 10.1016/j.clim.2009.07.002 [DOI] [PubMed] [Google Scholar]
- 8.Libby P. Molecular bases of the acute coronary syndromes. Circulation. 1995;91(11):2844–2850. doi: 10.1161/01.cir.91.11.2844 [DOI] [PubMed] [Google Scholar]
- 9.Green DR. The coming decade of cell death research: five riddles. Cell. 2019;177(5):1094–1107. doi: 10.1016/j.cell.2019.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paone S, Baxter AA, Hulett MD, Poon IKH. Endothelial cell apoptosis and the role of endothelial cell-derived extracellular vesicles in the progression of atherosclerosis. Cell Mol Life Sci. 2019;76(6):1093–1106. doi: 10.1007/s00018-018-2983-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu YJ, Zheng L, Hu YW, Wang Q. Pyroptosis and its relationship to atherosclerosis. Clin Chim Acta. 2018;476:28–37. doi: 10.1016/j.cca.2017.11.005 [DOI] [PubMed] [Google Scholar]
- 12.Karunakaran D, Geoffrion M, Wei L, et al. Targeting macrophage necroptosis for therapeutic and diagnostic interventions in atherosclerosis. Sci Adv. 2016;2(7):e1600224. doi: 10.1126/sciadv.1600224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bai T, Li M, Liu Y, Qiao Z, Wang Z. Inhibition of ferroptosis alleviates atherosclerosis through attenuating lipid peroxidation and endothelial dysfunction in mouse aortic endothelial cell. Free Radic Biol Med. 2020;160:92–102. doi: 10.1016/j.freeradbiomed.2020.07.026 [DOI] [PubMed] [Google Scholar]
- 14.Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang K, Song H, Yin D. PDSS2 inhibits the ferroptosis of vascular endothelial cells in atherosclerosis by activating Nrf2. J Cardiovasc Pharmacol. 2021;77(6):767–776. doi: 10.1097/FJC.0000000000001030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Y, Yan Y, Niu F, et al. Ferroptosis: a cell death connecting oxidative stress, inflammation and cardiovascular diseases. Cell Death Discov. 2021;7(1):193. doi: 10.1038/s41420-021-00579-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hägg S, Skogsberg J, Lundström J, et al. Multi-organ expression profiling uncovers a gene module in coronary artery disease involving transendothelial migration of leukocytes and LIM domain binding 2: the Stockholm Atherosclerosis Gene Expression (STAGE) study. PLoS Genet. 2009;5(12):e1000754. doi: 10.1371/journal.pgen.1000754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doms A, Schroeder M. GoPubMed: exploring PubMed with the gene ontology. Nucleic Acids Res. 2005;33(WebServer issue):W783–W786. doi: 10.1093/nar/gki470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou N, Bao J. FerrDb: a manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database (Oxford). 2020;2020:baaa021. doi: 10.1093/database/baaa021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Speiser JL, Durkalski VL, Lee WM. Random forest classification of etiologies for an orphan disease. Stat Med. 2015;34(5):887–899. doi: 10.1002/sim.6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rigatti SJ. Random forest. J Insur Med. 2017;47(1):31–39. doi: 10.17849/insm-47-01-31-39.1 [DOI] [PubMed] [Google Scholar]
- 25.Degenhardt F, Seifert S, Szymczak S. Evaluation of variable selection methods for random forests and omics data sets. Brief Bioinform. 2019;20(2):492–503. doi: 10.1093/bib/bbx124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Yang F, Luo Z. An experimental study of the intrinsic stability of random forest variable importance measures. BMC Bioinform. 2016;17:60. doi: 10.1186/s12859-016-0900-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao H, Zhao Y, Li H, Lei L. Ferroptosis as an emerging target in inflammatory diseases. Prog Biophys Mol Biol. 2020;155:20–28. doi: 10.1016/j.pbiomolbio.2020.04.001 [DOI] [PubMed] [Google Scholar]
- 29.Pérez-Guaita D, Quintás G, Kuligowski J. Discriminant analysis and feature selection in mass spectrometry imaging using constrained repeated random sampling - Cross validation (CORRS-CV). Anal Chim Acta. 2020;1097:30–36. doi: 10.1016/j.aca.2019.10.039 [DOI] [PubMed] [Google Scholar]
- 30.Hebbel RP, Wei P, Milbauer L, et al. Abnormal endothelial gene expression associated with early coronary atherosclerosis. J Am Heart Assoc. 2020;9(14):e016134. doi: 10.1161/JAHA.120.016134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gutiérrez E, Flammer AJ, Lerman LO, Elízaga J, Lerman A, Fernández-Avilés F. Endothelial dysfunction over the course of coronary artery disease. Eur Heart J. 2013;34(41):3175–3181. doi: 10.1093/eurheartj/eht351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seino Y, Ikeda U, Ikeda M, et al. Interleukin 6 gene transcripts are expressed in human atherosclerotic lesions. Cytokine. 1994;6(1):87–91. doi: 10.1016/1043-4666(94)90013-2 [DOI] [PubMed] [Google Scholar]
- 33.Su JH, Luo MY, Liang N, et al. Interleukin-6: a novel target for cardio-cerebrovascular diseases. Front Pharmacol. 2021;12:745061. doi: 10.3389/fphar.2021.745061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Liang T, Wang Y, et al. Angiopoietin-like protein 7 mediates TNF-α-induced adhesion and oxidative stress in human umbilical vein epithelial cell. Gen Physiol Biophys. 2020;39(3):285–292. doi: 10.4149/gpb_2019062 [DOI] [PubMed] [Google Scholar]
- 35.Merched AJ, Chan L. Absence of p21Waf1/Cip1/Sdi1 modulates macrophage differentiation and inflammatory response and protects against atherosclerosis. Circulation. 2004;110(25):3830–3841. doi: 10.1161/01.CIR.0000148681.01282.89 [DOI] [PubMed] [Google Scholar]
- 36.Yang ZY, Simari RD, Perkins ND, et al. Role of the p21 cyclin-dependent kinase inhibitor in limiting intimal cell proliferation in response to arterial injury. Proc Natl Acad Sci U S A. 1996;93(15):7905–7910. doi: 10.1073/pnas.93.15.7905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Condorelli G, Aycock JK, Frati G, Napoli C. Mutated p21/WAF/CIP transgene overexpression reduces smooth muscle cell proliferation, macrophage deposition, oxidation-sensitive mechanisms, and restenosis in hypercholesterolemic apolipoprotein E knockout mice. FASEB J. 2001;15(12):2162–2170. doi: 10.1096/fj.01-0032com [DOI] [PubMed] [Google Scholar]
- 38.Matsunaga T, Arakaki M, Kamiya T, Endo S, El-Kabbani O, Hara A. Involvement of an aldo-keto reductase (AKR1C3) in redox cycling of 9, 10-phenanthrenequinone leading to apoptosis in human endothelial cells. Chem Biol Interact. 2009;181(1):52–60. doi: 10.1016/j.cbi.2009.05.005 [DOI] [PubMed] [Google Scholar]
- 39.Tong X, Khandelwal AR, Wu X, et al. Pro-atherogenic role of smooth muscle Nox4-based NADPH oxidase. J Mol Cell Cardiol. 2016;92:30–40. doi: 10.1016/j.yjmcc.2016.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mineo C. Lipoprotein receptor signalling in atherosclerosis. Cardiovasc Res. 2020;116(7):1254–1274. doi: 10.1093/cvr/cvz338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Dwyer R, Kovaleva M, Zhang J, et al. Anti-ICOSL new antigen receptor domains inhibit T cell proliferation and reduce the development of inflammation in the collagen-induced mouse model of rheumatoid arthritis. J Immunol Res. 2018;2018:4089459. doi: 10.1155/2018/4089459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaddis DE, Padgett LE, Wu R, et al. Apolipoprotein AI prevents regulatory to follicular helper T cell switching during atherosclerosis. Nat Commun. 2018;9(1):1095. doi: 10.1038/s41467-018-03493-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao Y, Chen Z, Zhang H, et al. Selenium-GPX4 axis protects follicular helper T cells from ferroptosis. Nat Immunol. 2021;22(9):1127–1139. doi: 10.1038/s41590-021-00996-0 [DOI] [PubMed] [Google Scholar]