Abstract
Toona ciliata var. pubescens is classified as Toona subgenus of Meliaceae family, which belongs to a large deciduous tree species. It is also a kind of precious timber tree species and has a certain medicinal value. Here, the first complete chloroplast genome (cpDNA) sequence of T. ciliata var. pubescens was determined using the Illumina sequencing platform. The cpDNA genome is 159,481 bp in length, containing a large single-copy region (LSC) of 87,176 bp and a small single-copy region (SSC) of 18,381 bp, which were separated by a pair of inverted repeats (IRs) regions of 26,962 bp. The genome contains 138 genes, including 90 protein-coding genes, eight ribosomal RNA genes, and 40 transfer RNA genes. The phylogenetic analysis based on 19 cpDNA genomes showed a close relationship with Toona ciliate.
Keywords: Meliaceae, Toona ciliata var. pubescens, chloroplast genome, phylogenetic analysis
Toona ciliata Roem. var. pubescens (Franch.) Hand.-Mazz (Toona ciliata var. pubescens, Hand.-Mazz, 1864) is a variety of T. ciliata, belonging to Toona subgenus of Meliaceae family. As large deciduous trees, it grows rapidly with straight trunks, hard texture, and produces red wood with a desirable grain (Li et al. 2018). Referred to ‘Chinese mahogany’, T. ciliata var. pubescens has high economic value in wood industry and certain medicinal value (Kundal et al. 2020). However, its numbers have gradually declined due to environmental changes, low natural regeneration, and over-exploitation (Liu et al. 2014). According to China plant red data book, this species became an endangered tree and listed as a level II national key protected wild plant endemic to China (Fu and Jin 1992). However, unavailable genome sequence of T. ciliata var. pubescens caused difficult research on genetic information. At present, the value of chloroplast genome (cpDNA) genome in species identification and phylogenetic studies has been widely concerned. Although the complete cp genome sequence of T. ciliata has been finished (Xin et al. 2018), it is still important to evaluate the cp genome sequence of T. ciliata var. pubescens as an endangered tree. Here, we assembled and characterized the complete cpDNA sequence of T. ciliata var. pubescens as a resource for future studies on this species.
Individual T. ciliata var. pubescens were collected at Research Institute of Subtropical Forestry, Chinese Academy of Forestry, Hangzhou, China (29°44’ -30°11’ N,119°25’-120°19.5’E), and the specimen had been preserved in Shanghai Chenshan Herbarium (http://cfh.ac.cn/subsite/default.aspx?siteid=CSH, Binjie Ge, gebinjie123@163.com) under the voucher number CSH0105415. A modified CTAB method was used for the extraction of genomic DNA from its tender leaves (Doyle 1987). The quality of the DNA was estimated by NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA) and 1% agarose gel electrophoresis. DNA with the concentration higher than 100 ng/µL and total mass higher than 100 mg was used for further Illumina sequencing. The Illumina libraries were constructed using TruSeq DNA sample Preparation kit (Vanzyme, Nanjing, China), with an average insert size of 300 bp and then sequenced using the Illumina HiSeq-2500 platform (Illumina, San Diego, CA) at Genesky Biotechnologies (Shanghai, China). Approximately, 3.7 G raw data were gained, and finally we retrieved 1,326,708 clean organelle reads and 194,758,681 bp clean bases after quality control using FastQC and R. The trimmed reads were assembled by metaSPAdes 3.13.0 and annotated using CPGAVAS2 with the cpDNA of T. ciliata as a reference (GenBank accession number: NC_039592). The annotated cpDNA sequence of T. ciliata var. pubescens was uploaded to GenBank (GenBank accession number: MZ926838).
In general, the complete cp genome sequence of T. ciliata var. pubescens is 159,481 bp in length with GC content of 37.92%, and the nucleotide composition exhibited an asymmetric distribution (30.7% A, 19.3% C, 18.6% G, and 31.4% T). The cp genome has a typical quadripartite circular structure including a large single-copy (LSC) region of 87,176 bp, a small single-copy (SSC) region of 18,381 bp, and two inverted repeat (IR) regions of 26,962 bp. A total of 138 genes are annotated, including 90 protein-coding genes (PCGs), 40 tRNA genes, and eight rRNA genes. Most of these genes are single copy genes, but 17 genes (seven PCGs, six tRNA genes, and four rRNA genes) are duplicated. In the genome, 10 PCGs (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, rps12, and rps16) have only one intron, and clpP and ycf3 gene contained two introns.
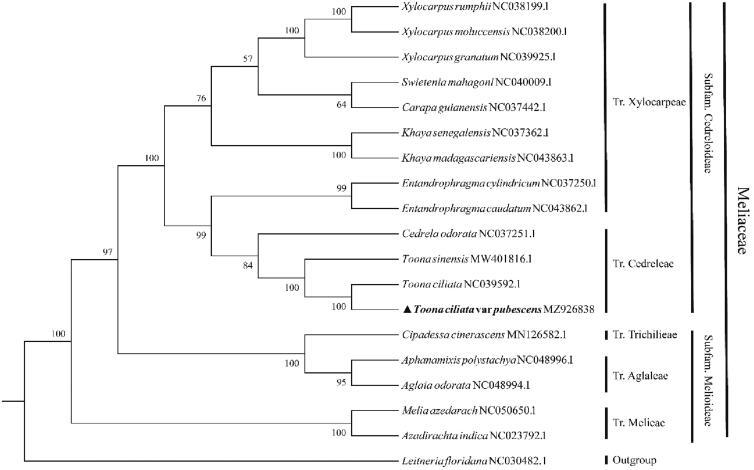
We performed a phylogenetic analysis using PCGs from 18 Meliaceae species, and an outgroup species Leitneria floridana. The GenBank accession numbers of the sequences used in this paper are given in Figure 1. Phylogenetic analysis of homologous genes was performed by Orthofinder 2.2.7 (Emms and Kelly 2018). The phylogenetic tree was constructed using the maximum-likelihood (ML) method of ggtree 1.14.6 (R package), and performed with 1000 bootstrap replicates (Yu et al. 2017). Consistent with the available evidence, T. ciliata var. pubescens was closely related to T. ciliata with 100% bootstrap support (Figure 1), and they both clustered with T. sinensis in the Cedreloideae Harms. The complete cp genome information reported in this study will be a valuable resource for future studies on conservation, phylogeny, and breeding of the T. ciliata var. pubescens.
Figure 1.
The phylogenetic tree of Toona ciliata var. pubescens and other related species.
Acknowledgements
Ethical approval: Experiment was performed in accordance with the Ethics Committee recommendations of Institute of Subtropical Forestry, Chinese Academy of Forestry.
Funding Statement
This study was supported by the National Nonprofit Institute Research Grant of Chinese Academy of Forestry [CAFYBB2020SZ004] and the Zhejiang Science and Technology Major Program on Agricultural New Variety Breeding [2021C02070-1 and 2021C02070-4].
Author contributions
JL and XH designed and conceived the experiments. XD collected the materials. ZL and XD used software to process data. ZL analyzed the corresponding results, drafted the manuscript. YF, WL, and JD helped in sample preparation, DNA extraction and contributed to the revision of manuscript. JL supervised this whole process and revised this paper. All authors read and approved the final manuscript.
Disclosure statement
The authors declare no conflict of interest.
Data availability statement
The assembled chloroplast genome sequence of T. ciliata var. pubescens has been submitted to GenBank under the accession number: MZ926838 (https://www.ncbi.nlm.nih.gov/nuccore/MZ926838.1). In NCBI Short Read Archive (SRA), the associated BioProject, SRA, and Bio-Sample numbers are PRJNA774927, SRR16686901, and SAMN22594705, respectively.
References
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Emms D, Kelly S.. 2018. OrthoFinder2: fast and accurate phylogenomic orthology analysis from gene sequences. BioRxiv: 466201. [Google Scholar]
- Fu LK, Jin J.. 1992. China plant red data book: rare and endangered plants. Vol. 1. New York: Science Press. [Google Scholar]
- Kundal M, Thakur S, Dhillon GPS.. 2020. Evaluation of growth performance of half sib progenies of Toona ciliata M. Roem under field conditions. Genetika. 52(2):651–660. [Google Scholar]
- Liu J, Jiang JM, Chen YT.. 2014. Genetic diversity of central and peripheral populations of Toona ciliata var. pubescens, an endangered tree species endemic to China. Genet Mol Res. 13(2):4579–4590. [DOI] [PubMed] [Google Scholar]
- Li P, Shang Y, Zhou W, Hu X, Mao W, Li J, Li J, Chen X.. 2018. Development of an efficient regeneration system for the precious and fast-growing timber tree Toona ciliata. Plant Biotechnol. 35(1):51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin G, Liu J, Liu J.. 2018. Complete chloroplast genome of an endangered tree species, Toona ciliata (Sapindales: Meliaceae). Mitochondrial DNA B Resour. 3(2):663–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Smith DK, Zhu H, Guan Y, Lam TTY.. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 8(1):28–36. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The assembled chloroplast genome sequence of T. ciliata var. pubescens has been submitted to GenBank under the accession number: MZ926838 (https://www.ncbi.nlm.nih.gov/nuccore/MZ926838.1). In NCBI Short Read Archive (SRA), the associated BioProject, SRA, and Bio-Sample numbers are PRJNA774927, SRR16686901, and SAMN22594705, respectively.