Abstract
The deep sea (> 200 m) is considered as the largest and most remote biome, which characterized by low temperatures, low oxygen level, scarce food, constant darkness, and high hydrostatic pressure. The sea stars (class Asteroidea) are ecologically important and diverse echinoderms in all of the world’s oceans, occurring from the intertidal to the abyssal zone (to about 6000 m). To date, the phylogeny of the sea stars and the relationships of deep-sea and shallow water groups have not yet been fully resolved. Here, we recovered five mitochondrial genomes of deep-sea asteroids. The A+T content of the mtDNA in deep-sea asteroids were significantly higher than that of the shallow-water groups. The gene orders of the five new mitogenomes were identical to that of other asteroids. The phylogenetic analysis showed that the orders Valvatida, Paxillosida, Forcipulatida are paraphyletic. Velatida was the sister order of all the others and then the cladeValvatida-Spinulosida-Paxillosida-Notomyotida versus Forcipulatida-Brisingida. Deep-sea asteroids were nested in different lineages, instead of a well-supported clade. The tropical Western Pacific was suggested as the original area of asteroids, and the temperate water was initially colonized with asteroids by the migration events from the tropical and cold water. The time-calibrated phylogeny showed that Asteroidea originated during Devonian-Carboniferous boundary and the major lineages of Asteroidea originated during Permian–Triassic boundary. The divergence between the deep-sea and shallow-water asteroids coincided approximately with the Triassic-Jurassic extinction. Total 29 positively selected sites were detected in fifteen mitochondrial genes of five deep-sea lineages, implying a link between deep-sea adaption and mitochondrial molecular biology in asteroids.
Subject terms: Evolution, Genetics, Molecular biology
Introduction
The Asteroidea (sea star) is the second most diverse class of echinoderms after the Ophiuroidea (~ 2100 species), with approximately 1900 accepted extant species worldwide, grouped into 36 families 370 extant genera1,2. Asteroids are widely spread in all of the world’s oceans, but reveal highest levels of species richness in the Indo-Pacific and tropical Atlantic regions1,3–5. They present at all depths from the shallow-water to the deep sea (~ 6000 m)1,6. Out of the 36 families of living Asteroidea, 15 of those occur exclusively in deep-sea settings and 14 families have deep-sea members1. However, to date, the relationships of the starfish have not yet been fully resolved1,7–10. The monophyly of the order Valvatida still not supported7,8,10. Another most important issue in asteroid phylogeny is the phylogenetic position of the order Paxillosida. Two contrasting hypotheses have been proposed with regard to the phylogenetic position of Paxillosida. Some morphological and molecular-based evidences supported that the Paxillosida was a primitive asteroid group11–19, as they lacks an anus and suckers on the tube feet. However, an opposite opinion argued that the paxillosidans diverged later during asteroid evolution1,8,10,20–28. The controversy highlights the need for a well sampled and well-supported phylogenetic relationship for asteroids, to distinguish the early divergence asteroid orders and locate the problematic and morphologically unusual taxa.
The deep sea (> 200 m), which is considered as the largest and most remote biome of the world, occupyed about 66% of the bottom of the global ocean29. The deep sea was characterized by low temperatures, high hydrostatic pressure, low oxygen level, food supply scarcity and constant darkness30,31. If the marine invertebrates originated in shallow-water or the deep sea have been debated for a long time. The studies of different taxa, e.g. molluscan, crustacean, corals and holothurians, have revealed a variety of different relationships. An analysis of the deep-sea molluscan distribution and taxonomy revealed that the deep sea was initially colonized with molluscans by migration events from the shallow water32. The phylogenetic trees inferred with respectively the mitogenomes, the nuclear, and the combined datasets suggested that multiple transitions between shallow water and deep-sea habitats occurred during the evolution of the vetigastropods molluscan33. Using complete 18S and partial 28S rDNA, Raupach et al.34 found that the deep-sea asellote isopods probably shaped by invasions from adjacent shallow-water regions34. The shallow to deep pattern has also been proposed to explain the evolution of deep-sea decapod crustaceans based on mitogenomes35. Lindner et al.35 found the stylasterid corals originated and diversified extensively in the deep sea, and subsequently invaded shallow waters36. The mitogenomic phylogenetic relationship of the holothurians showed that the deep-sea species formed the basal clades in the phylogenetic trees37.
The mitochondrial genomes (mitogenomes) are characterized by maternal inheritance, small genome size, fast nucleotide substitution rate and high mutation rate38,39. Therefore, mitochondrial DNA (mtDNA) is widely used as an informative molecular marker for study of phylogenetic relationships40–42, phylogeography43 and genomic evolution44–46. Furthermore, the complete mitogenomes show greater accuracy than single mtDNA markers, and could obtain more accurate phylogenetic relationships47,48. In addition, all the 13 mitochondrial PCGs that involved in oxidative phosphorylation play key roles in oxygen usage and energy metabolism. Despite strong functional constraints, mtDNA may evolve under positively selection in response to pressures from extreme harsh environment49. Variation in mitochondrial mtDNA can directly influence metabolic performance, an increasing number of studies related to adaptive evolution of mtDNA have been reported35,50–52.
Prior to our study, only twenty-five mitogenomes from Asteroidea are available, which distributed within fourteen families (Acanthasteridae, Archasteridae, Asterinidae, Asteriidae, Astropectinidae, Echinasteridae, Freyellidae, Goniasteridae, Luidiidae, Ophidiasteridae, Oreasteridae, Pterasteridae, Porcellanasteridae, Solasteridae). Compared to their diversity and abundance, the mitogenome sequences are still limited in the class Asteroidea. Among all available mitogenomes of asteroids, data from deep-sea environments are so scarce that only two mitogenomes have been reported to date53,54. The studies on them would provide statistically meaningful data for various aspects, such as origin and evolution and even adaptive mechanisms in deep-sea extreme environments.
In this study, we newly sequenced and annotated five complete mitogenomes of the deep-sea sea stars (Cheiraster sp. Studer, 1883, Zoroaster ophiactis Fisher, 1916, Brisinga sp. Asbjørnsen, 1856, Paulasterias sp. Mah et al.55, Asthenactis papyraceus Fisher, 1916) from the families Benthopectinidae, Zoroasteridae, Brisingidae, Paulasteriidae, and Myxasteridae, respectively. Currently, no mitogenome has been reported in these families. This paper aims to improve our understanding of the higher-order phylogeny and evolution history of deep-sea asteroid by the following: (1) comparing the deep-sea asteroid mitogenomes (the bias of strand nucleotide composition and the codon usage) with that from shallower habitats; (2) evaluating the phylogenetic relationships and estimating the divergence time of asteroids; (3) reconstructing the ancestral geographic area and habitat of asteroids; (4) estimating the selective pressures operating on the deep-sea asteroids mitogenomes in order to understand the genetic basis of deep-sea adaptation in sea stars.
Results and discussion
Mitochondrial genome assemblies and organization
The sequencing output data for the five species of deep-sea sea stars were summarized in Table 1. There were about 4.07–8.38 Gb clean reads, 97.33–98.50% of the reads passed Q20. The mitogenomes of the five asteroids were all circularized using NOVOPlasty from the reads, with 16,042 (Paulasterias sp.) to 16,426 bp (Cheiraster sp.) in length. The size range primarily due to the size variation of intergenic regions. The annotations, length and strand position of all PCGs and RNA genes of the five mitogenomes were summarized in Supplementary Table S1. For all the five mitogenomes, 37 genes (13 protein-coding genes (PCGs), 2 ribosomal RNA genes (rRNAs) and 22 transfer RNA genes (tRNAs)) were detected as typical in other metazoans56. Within these genes, nad1, nad2, nad6, rrnL and eleven tRNA genes (tRNA-SerUCN, tRNA-Ile, tRNA-LeuUUR, tRNA-Gly, tRNA-Tyr, tRNA-Met, tRNA-Cys, tRNA-Trp, tRNA-LeuCUN, tRNA-Asn, tRNA-Pro) were encoded by the minority strand, and the remaining ones were encoded by the major strand. The gene order of the five new mitogenomes were identical to that of other asteroids.
Table 1.
New mitochondrial genomes analyzed in present study.
| Species | Genbank accession no | Mitogenome size (bp) | Clean data (Mb) | Clean data Q20 (%) | Location | Positioning | Coordinates | Depth (m) | Cruise details | Collect time |
|---|---|---|---|---|---|---|---|---|---|---|
| Cheiraster sp. | MZ702701 | 16,426 | 8,581.0 | 97.58 | Tropical Western Pacific Ocean | FX-Dive219 | 10.1114 N, 140.2422E | 899 | Remotely Operated Vehicle (ROV) | 06/07/19 |
| Paulasterias sp. | MZ702702 | 16,042 | 4,938.9 | 97.82 | Tropical Western Pacific Ocean | FX-Dive222 | 10.0781 N, 140.1519E | 2141 | Remotely Operated Vehicle (ROV) | 06/10/19 |
| Asthenactis papyraceus | MZ702703 | 16,076 | 4,718.7 | 97.33 | Tropical Western Pacific Ocean | FX-Dive63 | 11.2726 N, 139.3702E | 1569 | Remotely Operated Vehicle (ROV) | 03/21/16 |
| Zoroaster ophiactis | MZ702704 | 16,193 | 4,164.7 | 98.28 | Tropical Western Pacific Ocean | FX-Dive16 | 8.8608 N, 137.7344E | 1108 | Remotely Operated Vehicle (ROV) | 12/15/14 |
| Brisinga sp. | MZ702705 | 16,147 | 6,988.6 | 98.50 | Tropical Western Pacific Ocean | FX-Dive63 | 11.2830 N, 139.3770E | 1126 | Remotely Operated Vehicle (ROV) | 03/21/16 |
For all the five species, most PCGs start with the codon ATN. Deviations occurred in nad1 (Z. ophiactis and Brisinga sp.) and nad2 (Paulasterias sp. and Z. ophiactis), which started with GTG. Most PCGs were stopped by the complete stop codons (TAG and TAA), but in Cheiraster sp. and A. papyraceus the cox2 gene, and in Z. ophiactis, A. papyraceus and Paulasterias sp. the cytb gene were ended with a single T nucleotide. The cytb gene in Brisinga sp. and Cheiraster sp. were ended with TA. The gene orders of the five new asteroids mitogenomes were identical to these of other sea stars, but distinct from the echinoderm ground pattern52.
Base composition and codon usage of mtDNA in Asteroidea
The five new mitogenomes were AT rich (> 60%), with A. papyraceus having the highest (68.30%) and Paulasterias sp. having the lowest (62.94%) AT content. Among the reported asteroids mitogenomes, the highest A+T content 68.07%, 68.23% and 68.30% occurred in the deep-sea species F. benthophila (MG563681), Brisinga sp. and A. papyraceus, respectively. In order to explore if the A+T content between the deep-sea and shallow-water species was different, the A+T content at complete mtDNA, PCG, tRNA, and rRNA genes were compared. Statistical t-tests showed that the A+T content of the complete mtDNA (two-tailed t-test, p = 0.007), PCG (p = 0.012), tRNA (p = 0.001), and rRNA (p = 0.012) in deep-sea asteroids were significantly higher than those of the shallow-water species (Supplementary Table S2). This phenomenon highlights the divergence of base composition between deep-sea and shallow water asteroids.
Among PCGs, the Leucine (15.40–16.70%) and Serine (9.11–10.66%) were the most frequently used amino acids, whereas Cysteine (0.86–1.21%), Arginine (1.65–2.04%) and Aspartic Acid (1.50–1.88%) were rarely used ones (Supplementary Table S3). As all the 13 mitochondrial PCGs were transmembrane proteins57, and the membrane-based processes were sensitive to the lowered temperature and increased hydrostatic pressure of deep sea58. Considering that the amino acid composition and properties may influence the protein functions59–61, we compared these two parameters between the deep-sea and shallow water asteroids. However, we failed to find significant differences in the percentages of non-polar and polar-uncharged, as well as the charged amino acids between the deep-sea and shallow water groups, which may be owing to the bias of sample sizes.
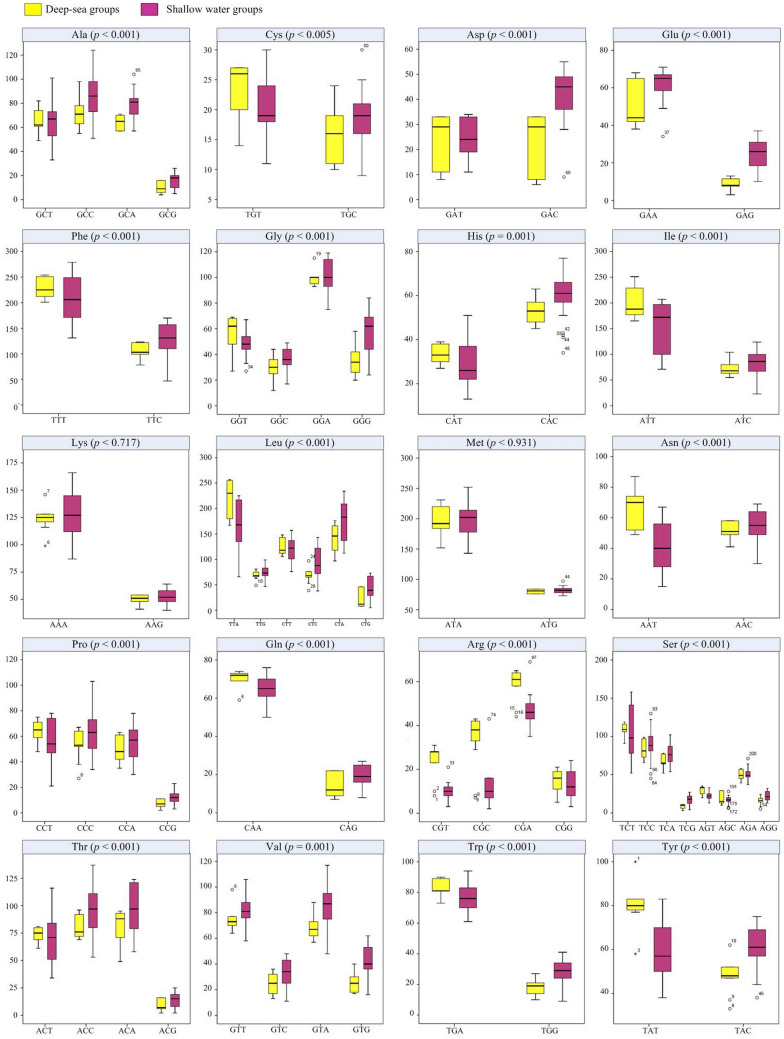
Figure 1 showed the strong A+T bias in codon usage across 30 asteroids mitogenomes. Among the 62 available codons, the six most used codons were TTT (Phe), TTA (Leu), CTA (Leu), ATT (Ile), ATA (Met) and AAA (Lys), almost all of them made up of solely A and T nucleotides. Even so, in the deep-sea species, the preference for A+T codons was stronger than the shallow water group. Statistical t-tests showed that for every amino acid, the proportions of the codons were significantly different between the deep-sea and shallow water asteroids (p-values in Fig. 1). The evolution of eukaryotic genome demonstrates a universal mutational bias toward A+T composition, leading to the hypothesis that genome-wide nucleotide composition generally evolves to a point at which the selective power in favor of G+C is approximately balanced by the power of random genetic drift, therefore variation in equilibrium genome-wide nucleotide composition is largely defined by variation in mutation biases62. Based on this hypothesis, the differences in nucleotide composition between the deep-sea and shallow-sea asteroids not only have functional implications, but also evolved as a result of a random genetic drift. In addition, the vast majority of the structures and components of the mitochondrion is encoded by nuclear genes63. This means that the functions of every product from mitogenome are association with products of the nuclear genome64,65. In other words, nuclear genome DNA may be a potential driver for mtDNA differences between the deep-sea and shallow-sea asteroids.
Figure 1.
Comparisons of codon usage (y-axis) between the deep-sea and shallow water asteroids. x-axis indicated the synonymous codons. The amino acid and its corresponding p-value were shown at the top of each box plot.
Phylogenetic analysis
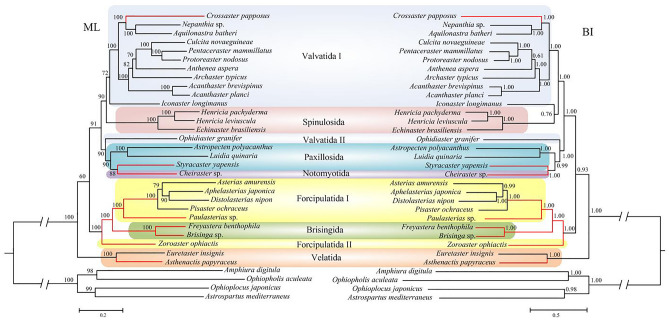
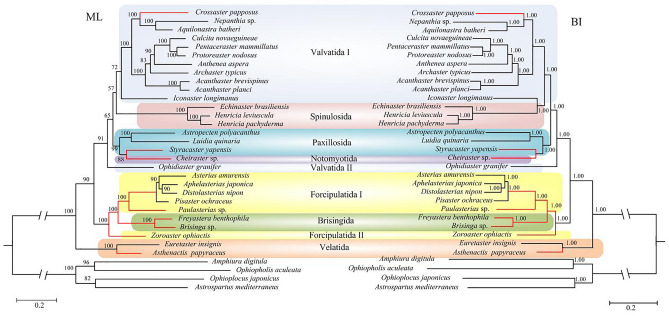
Although previous studies investigated the relationships within the class Asteroidea using molecular data1,7–10,66, the relationships has remained controversial up to now, with no consensus emerged. Our trees represented one of the well-sampled (for both taxa and molecular marker) and well-supported phylogenomic evidences for asteroid phylogeny. In this study, the phylogenetic analyses (ML and BI) were performed for the 30 species of sea stars based on the nucleotide (Fig. 2) and amino acid sequences (Fig. 3) of 13 mitochondrial PCGs. All the analysis produced phylogenies that differed only slightly. These topologies recovered that deep-sea asteroids were nested in different lineages, instead of a well-supported clade. Velatida was the sister Order of all the others and then the clade Valvatida-Spinulosida-Paxillosida-Notomyotida versus Forcipulatida-Brisingida. Our study suggested monophyly only for four asteroid orders, Spinulosida, Notomyotida, Brisingida and Velatida. However, the phylogenetic analysis revealed several discrepancies between our molecular results and current taxonomy at order levels. The unwell-supported clades included Valvatida, Paxillosida, Forcipulatida. These results were not perfectly consistent with the previous works based on transcriptomics datasets9,10, and limited molecular markers1,7–9. This discrepancy may due to the heterogeneity of data. These results were also different from the recent mitogenomic studies of Asteroidea in some orders66, which contained limited taxonomic sampling. Some primarily deep-sea taxa form orders Brisingida, Forcipulatida and Notomyotida have not been sampled in study of Zheng et al.66.
Figure 2.
Phylogenetic analysis of ML and BI based on the nucleotide sequencces of the 13 concatenated protein-coding genes. The bootstrap probability and the Bayesian posterior probability were shown at each node. The deep-sea branchs were marked by red.
Figure 3.
Phylogenetic analysis of ML and BI based on the amino acid sequencces of the 13 concatenated protein-coding genes. The bootstrap probability and the Bayesian posterior probability were shown at each node. The deep-sea branchs were marked by red.
The Valvatida, the most diversified Order within the Asteroidea, was never recovered as monophyletic in any of previous studies1,10. Our results concerning the taxonomic relationships within Valvatida are sensitive to the choice of dataset and analytical methods. Ophidiasteridae (n = 106) is the largest asteroid family throughout the tropical shallow-water Atlantic, and Indo-Pacific1,4. Our phylogenetic trees based on nucleotide sequence showed that the Ophidiasteridae (represented by Ophidiaster granifer) was placed in a branch (Valvatida II) as a sister group to Paxillosida + Notomyotida, then grouped with Valvatida I + Spinulosida (Fig. 2). However, the amino acid sequences based trees supported Ophidiasteridae (Valvatida II) as a sister clade to Valvatida I + Spinulosida + Paxillosida + Notomyotida (Fig. 3). The family Goniasteridae (n = 256) contained the largest number of species within the Asteroidea1. In the ML trees, Goniasteridae (represented by Iconaster longimanus) was the sister family to the remaining families of Valvatida (except Ophidiasteridae) (Fig. 2). While in the BI trees, Goniasteridae was more closely related to Spinulosida, instead of other species of Valvatida (Fig. 3). Thus, the Valvatida is paraphyletic, and the phylogenetic positions of Ophidiasteridae and Goniasteridae are still need to be resolved.
As summarized in the introduction, the phylogenetic status of the order Paxillosida, primitive or derived, is a controversial issue in asteroid phylogeny67. Our trees showed that the Paxillosida + Notomyotida lineages later diverged within the Valvatida + Spinulosida + Paxillosida + Notomyotida clade (Figs. 2, 3). This result was consistent with a “Paxillosida is derived” perspective1,8,10,20–28, which countered the “Paxillosida is primitive” scenario11–19. In present study, the deep-sea paxillosidan Styracaster yapensis and notomyotidan Cheiraster sp. formed a well-supported clade, making the Paxillosida paraphyletic. This relationship has never been observed in prior literature10, which may benefited from the well-sampled asteroid taxa in our study.
An earlier cladogenetic event among extant asteroids separated Forcipulatida + Brisingida from the Valvatida + Spinulosida + Paxillosida + Notomyotida clade (Figs. 2, 3). The close relationship between the Forcipulatida and Brisingida has also been suggested by Gale15, Knott and Wray7, Linchangco et al.10, Zheng et al.66. In this study, the deep-sea brisingidans Freyastera benthophila and Brisinga sp. nested within Forcipulatida, and closely related with the Forcipulatida species (Forcipulatida I in Figs. 2, 3). Thus, the monophyletic of Forcipulatida was not supported. Our results were similar to those of Knott and Wray7. The family Zoroasteridae is a small but widespread group of forcipulatidan asteroids distributed throughout the deep sea from bathyal to abyssal habitats (200–6000 m)6. In our trees, the Zoroasteridae (represented by Zoroaster ophiactis) was recovered as the basal forcipulatidan lineage (Forcipulatida II), as suggested by Knott and Wray7 and Mah and Foltz68. The Paulasteriidae is a new family described from deep-sea settings, representing the first new taxon within the Forcipulatacea since the 1800s55. Based on the molecular evidence presented in Figs. 2, 3, the deep-sea species Paulasterias sp. from the Paulasteriidae showed the closest relationship with the species from Forcipulatida, which is consistent with the phylogenetic status revealed by three-gene data set (12S rDNA, 16S rDNA, and histone H3 genes)55.
Velatida was represented here by two species from the family Pterasteridae and the deep-sea Myxasteridae that formed a clade, occupying a basal position separated from other asteroids (Figs. 2, 3). This is consistent with the research of Janies et al.8, placing the Velatida as the earliest branch in the evolution of Asteroidea. However, our study differs from the previous placement of Velatida as a sister group to Forcipulatida10, and sister to Forcipulatida+Brisingida66. So a more representative sampling is in need to better assess the relationships among the asteroid orders.
Geographical and habitat origin of the Asteroidea
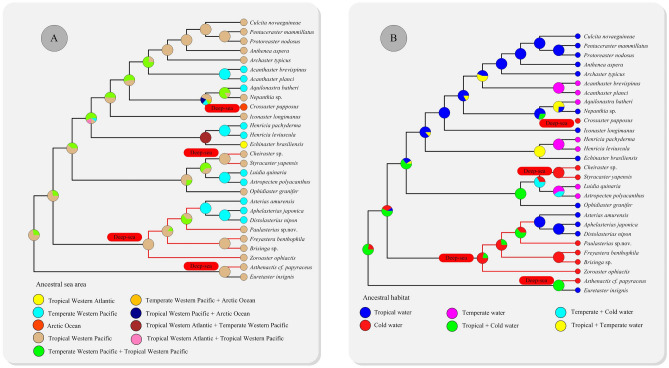
Asteroids widely distribute in all of the world’s oceans and present at all depths from the intertidal to the abyssal (to about 6000 m)1,3. Combining the phylogenetic analyses with the ancestral geographic area can provide new insights into early geographical origin of the Asteroidea. The ancestral geographic area reconstruction provided evidences that the Most Recent Common Ancestor (MRCA) of asteroids may occurred in tropical Western Pacific, and the Western-Pacific ancestor results in seven difierent lineages invading the temperate Western Pacific, tropical Western Atlantic and Arctic regions (Fig. 4A). It is intriguing to note that the clades of the tropical Western Pacific were distributed with tropical and cold asteroids from both shallow water and deep-sea. The Western Pacific is also the source of diversity for asteroids3–5. As the main component of the modern Indo-Australian Archipelago (IAA), the Western Pacific is a modern hotspot of marine biodiversity69. Based on the species-energy framework, thermal energy shape the marine shallow water biodiversity, while chemical energy availability (export productivity) and connectivity to shallower habitats drive deep-sea diversity6. The IAA hotspot lies between 30° N and 30° S latitudes, with strong thermal energy input from the sun. Thus, higher seasonal surface productivity exist in it’s shelf and slope regions6. The radiation hypothesis also suggested that deep-water richness is maintained by immigration from shallower regions69. The IAA hotspot locates in the region of convergence between Eurasia, Australia, and the Pacific-Philippine Sea plates, where exist a complex mosaic of shallow-seas, arc, and microcontinental fragments6. Thus, the IAA region was marked by higher export productivity and proximity to shallower communities, which were the unique geographical and biological advantages to maintain high biodiversity in the deep sea.
Figure 4.
Historical biogeography (A) and habitat (B) of Asteroidea. The scenario was inferred from three different models (S-DIVA, S-DEC and DEC) based on the ML tree constructed in our study. Pie charts near nodes indicated the probabilities of certain ancestral geographic areas and habitat. Red labels indicated the deep-sea species.
The asteroids were broadly categorized as occurring in “cold”, “temperate”, and “tropical” based on sea-surface temperatures1,70. Generally, deep-sea and high-latitude settings are treated as “cold” temperatures1,70. In our study, the “cold” temperature mainly refers to the deep-sea settings, as only one high-latitude clade included here. The ancestral habitat reconstruction suggested that the temperate water was initially colonized with asteroids by the migration events from the tropical and cold water (Fig. 4B). Controversial hypotheses have been proposed to describe the evolutionary origin for the marine invertebrates, e.g. shallow-water origin32,35,71–73, deep-sea origin36 or in both directions74,75. The new results presented herein was sufficient to illustrate that, not conclusive, the deep sea setting was an ancestral habitat of the asteroids. Our recent evolutionary study of Holothuroidea (sea cucumbers) revealed a similar phenomenon37. More samples are required to clarify their origin and evolution.
Divergence time estimation
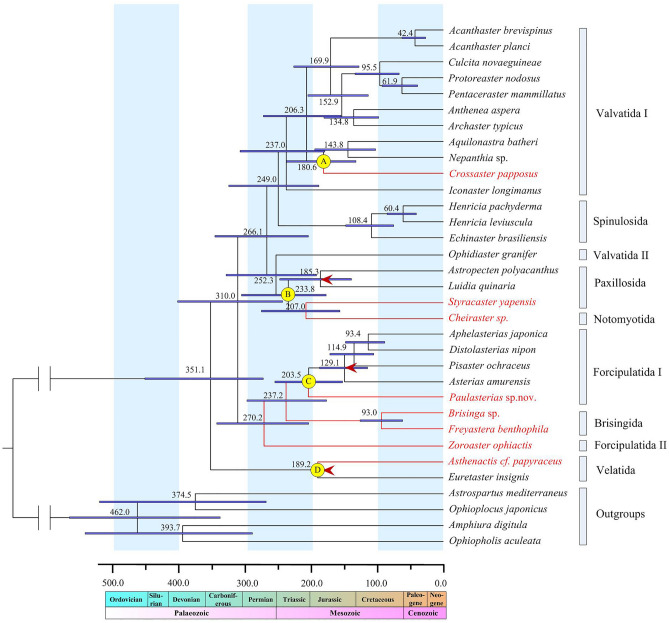
Species of the class Asteroidea have an old fossil record, which can be dated back to the post-Paleozoic15,76. The fossil record and current accounts suggested that most of the asteroids diversification occurred during the Permian–Triassic transition interval1,77. Based on our time-calibrated phylogeny, the MRCA of Asteroidea originated around the boundary of Devonian and Carboniferous (351.1 Ma with 95% highest posterior density interval [HPD] 275.9–454.9 Ma), which supported the first important faunal shift of Paleozoic asteroid evolution the Late Devonian78. The diversification times of major lineages of Asteroidea are estimated as follows: Forcipulatida + Paulasteriidae + Brisingida, 270.2 Ma (95% HPD 208.2–347.1 Ma); Paxillosida + Notomyotida + Valvatacea II, 252.3 Ma (95% HPD 191.5–327.9 Ma); Valvatacea I, 237.0 Ma (95% HPD 181.5–310.4 Ma); Velatida, 189.2 Ma (95% HPD 179.5–198.8 Ma); Spinulosida, 108.4 Ma (95% HPD 74.3–147.5 Ma) (Fig. 5). These major lineages of Asteroidea likely originated around the boundary of Permian–Triassic (240–270 Ma). This result was consistent with second important faunal shift of the Paleozoic asteroid evolution, during the late Paleozoic to early Mesozoic transition to modem asteroids78. Twitchett and Oji77 suggested that the Permian–Triassic interval was a bottleneck in the evolutionary history of the echinoderms (including asteroids). Paleozoic lineages of asterozoans and early asteroids underwent extinction during this interval and was followed by re-diversification of at least one surviving lineage.
Figure 5.
Time-calibrated mitogenomic phylogeny of Asteroidea estimated by the Bayesian relaxed-molecular clock method. The 95% HPD for each node were indicated in light purple bars. Calibrated nodes are indicated by red arrows. The divergence between the deep-sea and shallow-water species were marked by yellow circles (Nodel A, Nodel B, Nodel C and Nodel C).
The deep-sea and shallow-water Valvatida species split at 180.6 Ma (95% HPD 108.9–199.7 Ma) (Node A). The deep-sea species of Paxillosida and Notomyotida diverged with its closest shallow-water relative at 233.8 Ma (95% HPD 176.7–304.6 Ma) (Node B). The divergence between the deep-sea species of Paulasteriidae and shallow-water Forcipulatida was located at 203.5 Ma (95% HPD 157.5–260.1 Ma) (Node C). The deep-sea and shallow-water species of Velatida split at 189.2 Ma (95% HPD 179.5–198.8 Ma) (Node D). The results indicated that the divergence between the deep-sea and shallow-water asteroids coincided approximately with the Triassic-Jurassic transition, which was marked by mass extinction event79,80. The widespread extinction created new opportunities for marine faunas, and contributed to the rapid divergence of both deep-sea and shallow water taxa.
Positive selection analysis
We evaluated the potential positive selection pressures in deep-sea asteroids lineages, because colonisation may impact the function of mitochondrial oxidative phosphorylation genes. When the ω ratio for the 13 concatenated mitochondrial PCGs were compared between the deep-sea and shallow water sea stars, we failed to find a significant difference in their ω (Ks/Ks) ratios (chi-square: p > 0.05) (Table 2), implying that the ω ratio of deep-sea asteroids lineages (ω1 = 0.09158) have no significantly difference with the shallow water ones (ω0 = 0.09132). This may be biased by the sample sizes.
Table 2.
Selective pressure analyses of the mitochondrial OXPHOS genes of asteroids.
| Branch-specific models | |||||
|---|---|---|---|---|---|
| Models | lnL | Parameter estimates | Model compared | 2ΔlnL | p-values |
| Free-ratio model (M1) | − 204017.5186 | M1 versus M0 | − 1823.43544** | 0.0000 | |
| Two-ratio model (M2) | − 204929.2363 | ω0 = 0.09129 ω1 = 0.09158 | M2 versus M0 | 0.00565 | 0.9402 |
| One-ratio model (M0) | − 204929.2391 | ω = 0.09132 | |||
| Branch | Branch-sits models | ||||
|---|---|---|---|---|---|
| Species | Gene | Positively selected sites | |||
| Foreground-lineage I | C. papposus | atp8 | 45 K 0.966* | ||
| nad2 | 229 F 0.957* | ||||
| 279 G 0.960* | |||||
| nad6 | 110 W 0.997** | ||||
| 156 S 0.973* | |||||
| Foreground-lineage II | S. yapensis + Cheiraster sp. | nad2 | 45 C 0.960* | ||
| nad4 | 375 K 0.957* | ||||
| Foreground-lineage III | Paulasterias sp. | nad2 | 158 G 0.974* | ||
| 201 G 0.961* | |||||
| nad3 | 20 A 0.970* | ||||
| nad4 | 417 K 0.961* | ||||
| Foreground-lineage IV | F. benthophila + Brisinga sp. | cox3 | 145 A 0.979* | ||
| cytb | 324 T 0.953* | ||||
| nad1 | 88 C 0.962* | ||||
| 277 S 0.972* | |||||
| nad2 | 42 P 0.958* | ||||
| 196 Y 0.965* | |||||
| 280 G 0.960* | |||||
| nad4 | 11 T 0.976* | ||||
| 78 L 0.981* | |||||
| 101 L 0.984* | |||||
| 188 S 0.986* | |||||
| 254 S 0.983* | |||||
| 453 K 0.976* | |||||
| nad4l | 42 T 0.987* | ||||
| 81 S 0.989* | |||||
| Foreground-lineage V | Z. ophiactis | nad2 | 21 V 0.952* | ||
| 146 V 0.972* | |||||
| 213 S 0.989* | |||||
| nad4l | 41 N 0.992** | ||||
| Foreground-lineage VI | A. papyraceus | nad2 | 281 L 0.957* | ||
| nad4 | 45 H 0.985* | ||||
| 362 L 0.982* | |||||
| 405 A 0.974* | |||||
* 0.001 < p < 0.01 ** p < 0.001
It is the positive selection that is the basis of adaptation evolution81. Although the mitochondrial genes have been considered to be under purifying (negative) selection56, the signatures of positive selection may act on only a few sites in a gene sequence in some extreme environments53,82–84. In our results, BEB analysis of individual genes identified well-supported (PP ≥ 0.95) positively selected sites in two genes (nad2, nad4: total of 2 sites) along the S. yapensis + Cheiraster sp. lineage, three genes (nad2, nad3, nad4: total of 4 sites) along the Paulasterias sp. lineage, six genes (cox3, cytb, nad1, nad2, nad4, nad4l: total of 15 sites) along the F. benthophila + Brisinga sp. lineage, two genes (nad2, nad4l: total of 4 sites) along the Z. ophiactis lineage, and two genes (nad2, nad4: total of 4 sites) along the A. papyraceus lineage (Table 2). However, no positively selected sites were found along the C. papposus lineage. Although the positively selected genes vary for different deep-sea asteroids lineages, most of the positively selected loci were located in the NADH dehydrogenase (Complex I). Currently, the roles of NADH dehydrogenase in deep-sea adaptation still unclear, however, changes in these amino acid have functional implications and are a part of the adaptive response. Researchers have discovered that NADH dehydrogenase was the repeated target of positive selection in diverse deep-sea animals53,82,83. Under the extreme environment of deep sea, survival needs a modified and adapted energy metabolism53,82,83. As the first and largest enzyme complex, NADH dehydrogenase is supposed to act as proton-pumping devices in the mitochondrial OXPHOS process57,85. Therefore, mutations in the NADH dehydrogenase genes may effect metabolic efficiency57,86. So we predicted that the positively selected NADH dehydrogenase genes may play an important part in the adaptation of asteroids to deep-sea environment.
Materials and methods
Specimen collection and mitochondrial genome sequencing
Specimen collection information was shown in Table 1. All specimens were immediately preserved in 95% ethanol following collection. Total genomic DNA was extracted using the E.Z.N.A Tissue DNA kit (OMEGA, Wuhan, China) following manufacture’s protocols. The paired-end libraries were constructed using TruSeq Nano DNA Sample Prep Kit (Illumina, San Diego, CA, USA) with an insert size of 450 bp. The above libraries were sequenced by an Illumina (San Diego, CA, USA) HiSeq 4000 platform (2 × 150 bp paired-end reads).
Mitochondrial genome assembly
The paired-end raw sequences for the five samples were trimmed using Trimmomatic 0.3987 with the following parameters: ILLUMINACLIP:TruSeq3-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:75. The obtained clean reads were assembled de novo by NOVOPlasty software88 with default parameters. In the seed extension algorithm achieved via NOVOPlasty, the cox1 or 16S rDNA gene fragment of their closed related species (cox1 of C. dawsoni (JQ918233) for Cheiraster sp., 16S rDNA of Paulasterias sp. AAR (KT258987) for Paulasterias sp., cox1 of Asthenactis sp. CLM-2012 (JQ918238) for A. papyraceus, cox1 of Z. spinulosus (JQ918246) for Z. ophiactis, and cox1 of Brisinga sp. 2 RSIOAS023 (MN885902) for Brisinga sp.) was used as a seed sequence. The mtDNA will be circularized when the length is in the expected range and both ends overlap by at least 200 bp. Considering the influence of the nuclear mitochondrial DNA (NUMT) sequences89, we checked the obtained mitochondrial genome sequences completely, as significantly higher coverage may pinpoint to sequences derived from nuclear DNA or duplicated regions. The evenness and similarity for the coverage of each individual gene and control region (Supplementary Table S4) mean that the NUMT sequences may not confound the results from the Illumina sequencing. The control region was usually considered as “difficult genomic regions”. The fragments located in the large non-coding regions, were also verified by PCR amplification and Sanger sequencing90. The PCR Primer sequences and positions were shown in Supplementary Table S5.
Mitochondrial genome sequence annotation and analysis
MITOS web server91 was preliminarily used to annotate the PCGs, 2 rRNAs and tRNAs under default settings and the invertebrate mitochondrial genetic code. Boundaries of the PCGs and rRNAs genes were determined by comparing them with the homologous genes of other asteroids. The tRNA genes and their secondary structures were further confirmed by the ARWEN 1.2.3.c92. The gene orders of asteroids were compared with that of the echinoderm ground pattern. The CREx webserver (http://pacosy.informatik.uni-leipzig.de/crex/)93 was used to compare the mitochondrial gene order and deduce the gene rearrangement scenarios based on common intervals. The arrangement of all the genes (PCGs, rRNAs and tRNAs) were considered. The nucleotide composition and codon usage counts were computed by MEGA 594 based on the invertebrate mitochondrial genetic code (genetic code = 5). The differences and significant levels (p-value) for nucleotide composition and amino acid contents between the deep-sea species and the asteroids from shallow waters were performed by a two-tailed t-test and the chi-square test in IBM SPSS Statistics, release 19.0.0.1, respectively. The complete mtDNA sequences have been deposited in the GenBank database with the accession numbers MZ702701-MZ702705.
Phylogenetic inference
Thirty-three asteroids mitogenomes including the five new sequenced in this study were used for phylogenetic analysis, with four species of ophiuroids used as outgroups. The detailed informations of these available sequences were listed in Supplementary Table S6. In the following study, the species collected below 200 m are treated herein as “deep-sea group”, and these from 0 to 200 m are considered as “shallow water group”. The nucleotide and amino acid sequences from each of the 13 PCGs were aligned separately using MAFFT95 with default settings. The ambiguously aligned and variable regions were eliminated using the program Gblocks96 with default stringent parameters. Then, the aligned homologous nucleotide and amino acid sequences were concatenated into a single supermatrix with FASconCAT97. The best-fit substitution models for each nucleotide and amino acid partition were selected by PartitionFinder98.
Phylogenetic analysis was carried out using maximum likelihood (ML) and Bayesian inference (BI). ML analysis was performed in IQ-TREE web server99 with partition models. The node reliability was assessed using 1000 ultrafast bootstrap replicates100. BI analysis was employed using MrBayes 3.1 software101 with partition models. A total of 10,000,000 generations (Markov chain Monte Carlo, MCMC) were run, with a sampling frequency of 1,000 generations to allow adequate time for convergence. The run was stopped when the standard deviation of split frequencies was less than 0.01. The software Tracer v1.6102 was used to checked the parameters of the two independent runs (effective sampling size for all parameters > 200). The first 5000 trees were burnin. The 50% majority rule consensus tree was constructed using the remaining 5000 sampled trees.
Ancestral geographic area and habitat reconstructions
The ancestral geographic area and habitat of the asteroids were reconstructed by RASP v. 3.2103. The RASP provides a graphical user interface (GUI) to specify a phylogenetic tree and geographic distributional information, draws pie charts on the nodes of a phylogenetic tree to indicate levels of uncertainty, and generates high-quality exportable graphical results to reconstruct ancestral geographical distributions. Three models, i.e., Statistical Dispersal-Vicariance Analysis (S-DIVA), Statistical Dispersal-Extinction-Cladogenesis (S-DEC) and Dispersal-Extinction-Cladogenesis (DEC) were used. In the model S-DIVA, the occurrence of an ancestral range at a node was calculated by the frequency of all of the alternative reconstructions generated by the DIVA algorithm for each tree in the data set. The DEC and S-DEC models export the likelihood of all possible biogeographic scenarios estimated at a given node. Akaike weights were then calculated and interpreted as the relative probability of different ancestral ranges, which were displayed as pie charts on the nodes of a phylogenetic tree.
The mtDNA of Pisaster ochraceus was submitted directly in NCBI with no specimen collection information. So the location of the species was equivocal, which was not used in this analysis. Distribution data were compiled from the specimen collection records (Supplementary Table S6) and from the summary of Mah and Blake1. For the ancestral geographic area reconstruction, four large-scale biogeographic patterns were coded as follows: (A) Tropical Western Atlantic, (B) Temperate Western Pacific, (C) Tropical Western Pacific Ocean, and (D) Arctic Ocean. For the ancestral habitat reconstruction, the asteroids were categorized into (A) tropical, (B) temperate, and (C) cold.
Divergence time estimation
BEAST v2.3.3104 estimated the divergence times of major clades of Asteroidea using the following parameters: the uncorrelated relaxed lognormal clocks, random starting trees and a the Yule process. The divergence times were sampled once every 1,000 generations from 10 million MCMC generations. Tracer v1.6102 examined the effective sample sizes (ESS) of parameters (ESS > 200). The initial 50% of cycles were burn-in. TreeAnnotator (BEAST software) summarized the 95% highest posterior densities (HPD) and the maximum-clade-credibility tree topology with median ages. FigTree 1.4105 was used to view the the tree, divergence times and 95% HPD around the node ages. The divergence times were calibrated with three fossil taxa. The age of order Velatida was constrained with a lognormal distribution between 182.0 and 201.6 Ma based on Plesiastropecten hallovensis106,107. The age of family Astropectinidae was constrained with a lognormal distribution between 161.2 and 171.6 Ma108 based on Tethyaster jurassicus. The age of family Goniasteridae was constrained between 167.7 and 171.6 Ma, based on Comptoniaster sharpii109.
Positive selection analysis
The codon-based likelihood approach implemented in the CODEML program of PAMLx package110 was used to evaluate the changes in selective pressure between the deep-sea asteroids and other shallow water ones. The ML and BI tree topology inferred in this study was used. Non-synonymous to synonymous substitution rates (Ka/Ks, ω) was considered as the measure of selective pressure at the protein level.
The branch-specific model were used to test if the selective pressure between the deep-sea and shallow water starfish were different. For branch-specific model, the comparison between “free-ratio” (M1) and “one-ratio” models (M0), “two-ratio” (M2) and “one-ratio” models (M0) were conducted in the combined dataset of 13 protein-coding genes using the likelihood ratio test (LRT). The Chi-square test111 was applied for testing p-values.
The branch-site model was used to detect the sites under positive selection in each mitochondrial gene of the foreground-lineages (deep-sea asteroids lineages), which allowed ω to vary across branches and sites. For this model, the asteroids phylogeny was partitioned into foreground and background lineages in multiple ways. As the branch-site model does not allow for multiple foreground lineages, we considered the branches leading to the deep-sea asteroids as foreground-lineage separately. Firstly, the lineage of C. papposus was set as the foreground-lineage I. Secondly, we set the monophyletic branch leading to S. yapensis and Cheiraster sp. as the foreground-lineage II. Thirdly, we consider the branch Paulasterias sp. as the foreground-lineage III. Fourthly, the monophyletic branch leading to F. benthophila and Brisinga sp. was set as the foreground-lineage IV. Fifthly, the branch leading to Z. ophiactis was set as the foreground-lineage V. Lastly, the branch leading to A. papyraceus was set as the foreground-lineage VI. LRT were conducted to test if the alternative model (MA) fits the data significantly better than the corresponding null model (MA0). All the positively selected sites were determined by Bayes empirical Bayes (BEB) method112 with posterior probabilities of ≥ 0.95.
Supplementary Information
Acknowledgements
We sincerely thank the assistance of the crew of ROV FAXIAN and RV KEXUE for sample collection. This study was supported by research grants from the National Science Foundation for Distinguished Young Scholars (42025603), the Key Research Program of Frontier Sciences, Chinese Academy of Sciences (QYZDB-SSWDQC036), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB42000000), the Biological Resources Program, Chinese Academy of Sciences (KFJ-BRP-017-38), and the Science & Technology Basic Resources Investigation Program of China (2017FY100804).
Author contributions
Data curation: S. S. Funding acquisition: Z. S., N. X. Software: S. S. Writing original draft: S. S. Writing review and editing: S. S., Z. S., N. X.
Data availability
All relevant data are within the manuscript and its Supporting information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-08644-9.
References
- 1.Mah CL, Blake DB. Global diversity and phylogeny of the asteroidea (Echinodermata) PLoS ONE. 2012;7:e35644. doi: 10.1371/journal.pone.0035644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WoRMS. Asteroidea. Accessed at: http://www.marinespecies.org/aphia.php?p=taxdetails&id=123080. on 2021-08-02.
- 3.Blake DB. Adaptive zones of the class Asteroidea (Echinodermata) Bull. Mar. Sci. 1990;46:701–718. [Google Scholar]
- 4.Clark AM, Rowe FWE. Monograph of shallow-water indo-west pacific echinoderms. Br. Mus. Nat. Hist. 1971;690:1–230. [Google Scholar]
- 5.Clark AM, Downey ME. Starfishes of the ATLANTIC. Chapman & Hall; 1992. p. 794. [Google Scholar]
- 6.Mah CL. Phylogeny of the Zoroasteridae (Zorocallina; Forcipulatida): Evolutionary events in deep-Sea Asteroidea displaying Paleozoic features. Zool. J. Linn. Soc. 2007;150:177–210. doi: 10.1111/j.1096-3642.2007.00291.x. [DOI] [Google Scholar]
- 7.Knott KE, Wray GA. Controversy and consensus in asteroid systematics: new insights to ordinal and familial relationships. Am. Zool. 2000;40:382–392. doi: 10.1093/icb/40.3.382. [DOI] [Google Scholar]
- 8.Janies DA, Voight JR, Daly M. Echinoderm phylogeny including Xyloplax, a progenetic asteroid. Syst. Biol. 2011;60:420–438. doi: 10.1093/sysbio/syr044. [DOI] [PubMed] [Google Scholar]
- 9.Reich A, Dunn C, Akasaka K, Wessel G. Phylogenomic analyses of Echinodermata support the sister groups of Asterozoa and Echinozoa. PLoS ONE. 2015;10:e0119627. doi: 10.1371/journal.pone.0119627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linchangco GV, et al. The phylogeny of extant starfish (Asteroidea: Echinodermata) including Xyloplax, based on comparative transcriptomics. Mol. Phylogenet. Evol. 2017;115:161–170. doi: 10.1016/j.ympev.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 11.Mortensen T. Echinoderm larvae and their bearing on classification. Nature. 1923;111:322–323. doi: 10.1038/111322b0. [DOI] [Google Scholar]
- 12.Fell HB. A surviving somasteroid from the Eastern Pacific Ocean. Science. 1962;136:633–636. doi: 10.1126/science.136.3516.633. [DOI] [PubMed] [Google Scholar]
- 13.Fell HB. A living somasteroid. Platasterias latiradiata Gray Univ. Kansas Paleontol. Contrib. Art. 1962;6:1–16. [Google Scholar]
- 14.Fell HB. The phylogeny of sea-stars. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1963;246:381–435. doi: 10.1098/rstb.1963.0010. [DOI] [Google Scholar]
- 15.Gale AS. Phylogeny and classification of the Asteroidea (Echinodermata) Zool. J. Linn. Soc. 1987;89:107–132. doi: 10.1111/j.1096-3642.1987.tb00652.x. [DOI] [Google Scholar]
- 16.Gale, A. S. The Phylogeny of Post-Palaeozoic Asteroidea (Neoasteroidea, Echinodermata). Spec. Pap. Palaeontol. 5–112 (2011).
- 17.Gale AS. Phylogeny of the Asteroidea. In: Lawrence JM, editor. Starfish: Biology and Ecology of the Asteroidea. Johns Hopkins University Press; 2013. pp. 3–14. [Google Scholar]
- 18.Lafay B, Smith AB, Christen R. A combined morphological and molecular approach to the phylogeny of asteroids (Asteroidea: Echinodermata) Syst. Biol. 1995;44:190–208. doi: 10.2307/2413706. [DOI] [Google Scholar]
- 19.Wada H, Komatsu M, Satoh N. Mitochondrial rDNA phylogeny of the Asteroidea suggests the primitiveness of the Paxillosida. Mol. Phylogenet. Evol. 1996;6:97–106. doi: 10.1006/mpev.1996.0062. [DOI] [PubMed] [Google Scholar]
- 20.Macbride EW. Echinoderm larvæ and their bearing on classification. Nature. 1921;108:529–530. doi: 10.1038/108529c0. [DOI] [Google Scholar]
- 21.Macbride EW. Echinoderm larvae and their bearing on classification. Nature. 1923;111:47. doi: 10.1038/111047a0. [DOI] [Google Scholar]
- 22.Macbride EW. Echinoderm larvae and their bearing on classification (reply) Nature. 1923;111:323–324. doi: 10.1038/111323a0. [DOI] [Google Scholar]
- 23.Madsen FJ. The recent sea-star platasterias and the fossil Somasteroidea. Nature. 1966;209:1367. doi: 10.1038/2091367a0. [DOI] [Google Scholar]
- 24.Blake DB. Sea star Platastenras: Ossicle morphology and taxonomic position. Science. 1972;176:306–307. doi: 10.1126/science.176.4032.306. [DOI] [PubMed] [Google Scholar]
- 25.Blake DB. A classification and phylogeny of post-Palaeozoic sea stars (Asteroidea: Echinodermata) J. Nat. Hist. 1987;21:481–528. doi: 10.1080/00222938700771141. [DOI] [Google Scholar]
- 26.Blake D. Paxillosidans are not Primitive Asteroids: A Hypothesis Based on Functional Considerations. In: Burke R, Mladenov P, Lambert P, editors. Echinoderm Biology. Balkema; 1988. pp. 309–314. [Google Scholar]
- 27.Matsubara M, Komatsu M, Wada H. Close Relationship between Asterina and Solasteridae (Asteroidea) supported by both nuclear and mitochondrial gene molecular phylogenies. Zool. Sci. 2004;21:785–793. doi: 10.2108/zsj.21.785. [DOI] [PubMed] [Google Scholar]
- 28.Blake DB, Mah CL. The phylogeny of post-Palaeozoic Asteroidea (Neoasteroidea, Echinodermata)” by A.S. Gale and perspectives on the systematics of the Asteroidea. Zootaxa. 2014;3779:177–194. doi: 10.11646/zootaxa.3779.2.4. [DOI] [PubMed] [Google Scholar]
- 29.Woolley SN, et al. Deep sea diversity patterns are shaped by energy availability. Nature. 2016;533:393–396. doi: 10.1038/nature17937. [DOI] [PubMed] [Google Scholar]
- 30.Sanders HL, Hessler RR. Ecology of the deep-sea benthos. Science. 1969;163:1419–1424. doi: 10.1126/science.163.3874.1419. [DOI] [PubMed] [Google Scholar]
- 31.Naganoa Y, Nagahama T. Fungal diversity in deep-sea extreme environments. Fungal Ecol. 2012;5:463–471. doi: 10.1016/j.funeco.2012.01.004. [DOI] [Google Scholar]
- 32.Clarke AH. On the composition, zoogeography, origin and age of the deep-sea mollusk fauna. Deep-Sea Res. 1962;9:291–306. doi: 10.1016/0011-7471(62)90012-8. [DOI] [Google Scholar]
- 33.Lee H, Chen WJ, Puillandre N, Aznar-Cormano L, Tsai MH, Samadi S. Incorporation of deep-sea and small-sized species provides new insights into gastropods phylogeny. Mol. Phylogenet. Evol. 2019;135(136–147):2019. doi: 10.1016/j.ympev.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Raupach MJ, Mayer C, Malyutina M, Wägele JW. Multiple origins of deep-sea Asellota (Crustacea: Isopoda) from shallow waters revealed by molecular data. Proc. Royal Soc. B. 2009;276:799–808. doi: 10.1098/rspb.2008.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun S, Sha Z, Wang Y. Divergence history and hydrothermal vent adaptation of decapod crustaceans: A mitogenomic perspective. PLoS ONE. 2019;14:e0224373. doi: 10.1371/journal.pone.0224373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindner, A., Cairns, S. D. & Cunningham, C. W. From off shore to onshore: multiple origins of shallow-water corals from deep-sea ancestors. PLoS One, 3, e2429, (2008) [DOI] [PMC free article] [PubMed]
- 37.Sun S, Sha Z, Xiao N. The first two complete mitogenomes of the order Apodida from deep-sea chemoautotrophic environments: New insights into the gene rearrangement, origin and evolution of the deep-sea sea cucumbers. Comp. Biochem. Physiol. Part D. 2021;39:100839. doi: 10.1016/j.cbd.2021.100839. [DOI] [PubMed] [Google Scholar]
- 38.Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hao W, Richardson AO, Zheng Y, Palmer JD. Gorgeous mosaic of mitochondrial genes created by horizontal transfer and gene conversion. Proc. Natl. Acad. Sci. USA. 2010;107:21576–21581. doi: 10.1073/pnas.1016295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osigus HJ, Eitel M, Bernt M, Donath A, Schierwater B. Mitogenomics at the base of Metazoa. Mol. Phylogenet. Evol. 2013;69:339–351. doi: 10.1016/j.ympev.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 41.Bourguignon T, et al. The evolutionary history of termites as inferred from 66 mitochondrial genomes. Mol. Biol. Evol. 2015;32:406–421. doi: 10.1093/molbev/msu308. [DOI] [PubMed] [Google Scholar]
- 42.Tihelka E, Cai CY, Pisani D, Donoghue PCJ. Mitochondrial genomes illuminate the evolutionary history of the Western honey bee (Apis mellifera) Sci. Rep. 2020;10:14515. doi: 10.1038/s41598-020-71393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gvoždík V, Moravec J, Klütsch C, Kotlík P. Phylogeography of the Middle Eastern tree frogs (Hyla, Hylidae, Amphibia) as inferred from nuclear and mitochondrial DNA variation, with a description of a new species. Mol. Phylogen. Evol. 2010;55:1146–1166. doi: 10.1016/j.ympev.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 44.Gissi C, Iannelli F, Pesole G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity. 2010;101:301–320. doi: 10.1038/hdy.2008.62. [DOI] [PubMed] [Google Scholar]
- 45.Eo SH, DeWoody JA. Evolutionary rates of mitochondrial genomes correspond to diversification rates and to contemporary species richness in birds and reptiles. Proc. R Soc. Lond. B Biol. Sci. 2010;277:3587–3592. doi: 10.1098/rspb.2010.0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun YB, Shen YY, Irwin DM, Zhang YP. Evaluating the roles of energetic functional constraints on teleost mitochondrial encoded protein evolution. Mol. Biol. Evol. 2010;28:39–44. doi: 10.1093/molbev/msq256. [DOI] [PubMed] [Google Scholar]
- 47.Boore JL, Collins TM, Stanton D, Daehler LL, Brown WM. Deducing the pattern of arthropod phylogeny from mitochondrial DNA rearrangements. Nature. 1995;376:163–165. doi: 10.1038/376163a0. [DOI] [PubMed] [Google Scholar]
- 48.Boore JL, Brown WM. Big trees from little genomes: Mitochondrial gene order as a phylogenetic tool. Curr. Opin. Genet. Dev. 1998;8:668–674. doi: 10.1016/S0959-437X(98)80035-X。. [DOI] [PubMed] [Google Scholar]
- 49.Tomasco IH, Lessa EP. The evolution of mitochondrial genomes in subterranean caviomorph rodents: Adaptation against a background of purifying selection. Mol. Phylogenet. Evol. 2011;61:64–70. doi: 10.1016/j.ympev.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 50.Sun S, Li Q, Kong L, Yu H. Limited locomotive ability relaxed selective constraints on molluscs mitochondrial genomes. Sci. Rep. 2017;7:10628. doi: 10.1038/s41598-017-11117-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun YB, Shen YY, Irwin MD, Zhang YP. Evaluating the roles of energetic functional constraints on teleost mitochondrial-encoded protein evolution. Mol. Biol. Evol. 2011;28:39–44. doi: 10.1093/molbev/msq256. [DOI] [PubMed] [Google Scholar]
- 52.Shen YY, Shi P, Sun YB, Zhang YP. Relaxation of selective constraints on avian mitochondrial DNA following the degeneration of flight ability. Genome Res. 2009;19:1760–1765. doi: 10.1101/gr.093138.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mu W, Liu J, Zhang H. The first complete mitochondrial genome of the Mariana trench Freyastera benthophila (Asteroidea: Brisingida: Brisingidae) allows insights into the deep-sea adaptive evolution of Brisingida. Ecol. Evol. 2018;8:10673–10686. doi: 10.1002/ece3.4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mu W, Liu J, Zhang H. The complete mitochondrial genome of Styracaster yapensis (Paxillosida: Porcellanasteridae): Characterization and phylogenetic position. Mitochondrial DNA. 2018;4:81–82. doi: 10.1080/23802359.2018.1536468. [DOI] [Google Scholar]
- 55.Mah C, et al. Description of a new family, new genus, and two new species of deep-sea Forcipulatacea (Asteroidea), including the first known sea star from hydrothermal vent habitats. Zool. J. Linn. Soc. 2015;174:93–113. doi: 10.1111/zoj.12229. [DOI] [Google Scholar]
- 56.Castellana S, Vicario S, Saccone C. Evolutionary patterns of the mitochondrial genome in Metazoa: Exploring the role of mutation and selection in mitochondrial protein-coding genes. Genome Biol. Evol. 2011;3:1067–1079. doi: 10.1093/gbe/evr040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.da Fonseca RR, Johnson WE, O’Brien SJ, Ramos MJ, Antunes A. The adaptive evolution of the mammalian mitochondrial genome. BMC Genom. 2008;9:119. doi: 10.1186/1471-2164-9-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siebenaller JF, Garrett DJ. The effects of the deep-sea environment on transmembrane signaling. Comp. Biochem. Physiol. Part B. 2002;131:675–694. doi: 10.1016/S1096-4959(02)00027-1. [DOI] [PubMed] [Google Scholar]
- 59.Doerks T, Copley RR, Schultz J, Ponting CP, Bork P. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 2002;12:47–56. doi: 10.1101/gr.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Metpally RP, Reddy BV. Comparative proteome analysis of psychrophilic versus mesophilic bacterial species: Insights into the molecular basis of cold adaptation of proteins. BMC Genom. 2009;10:11. doi: 10.1186/1471-2164-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang LL, Tang SK, Huang Y, Zhi XY. Low temperature adaptation is not the opposite process of high temperature adaptation in terms of changes in amino acid composition. Genome Biol. Evol. 2015;7:3426–3433. doi: 10.1093/gbe/evv232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lynch M. Rate, molecular spectrum, and consequences of human mutation. P Natl. Acad. Sci. USA. 2010;107:961–968. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Calvo SE, Mootha VK. The mitochondrial proteome and human disease. Annu. Rev. Genom. Hum. Genet. 2010;11:25–44. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Woodson JD, Chory J. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 2008;9:383–395. doi: 10.1038/nrg2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lane N. Mitonuclear match: Optimizing fitness and fertility over generations drives ageing within generations. BioEssays. 2011;33:860–869. doi: 10.1002/bies.201100051. [DOI] [PubMed] [Google Scholar]
- 66.Zheng B, Jia J, Yin C, Yong K, Huang D. Mitogenomes reveal alternative initiation codons and lineage-specific gene order conservation in echinoderms. Mol. Biol. Evol. 2020;38:981–985. doi: 10.1093/molbev/msaa262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsubara M, Komatsu M, Araki T, Asakawa S, Wada H. The phylogenetic status of Paxillosida (Asteroidea) based on complete mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2005;36:598–605. doi: 10.1016/j.ympev.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 68.Mah C, Foltz D. Molecular phylogeny of the Forcipulatacea (Asteroidea: Echinodermata): Systematics and biogeography. Zool. J. Linn. Soc. 2011;162:646–660. doi: 10.1111/j.1096-3642.2010.00688.x. [DOI] [Google Scholar]
- 69.Renema W, et al. Hopping hotspots: Global shifts in marine biodiversity. Science. 2008;321:654–657. doi: 10.1126/science.1155674. [DOI] [PubMed] [Google Scholar]
- 70.Duxbury, A. C., Duxbury, A. B. & Sverdrup, K. A. An Introduction to the World’s Oceans 10th edition McGraw Hill Science/Education/Math Publishers. 528 p. (2010).
- 71.Raupach MJ, Mayer C, Malyutina M, Wagele JW. Multiple origins of deep-sea Asellota (Crustacea: Isopoda) from shallow waters revealed by molecular data. Proc. Biol. Sci. 2009;276:799–808. doi: 10.1098/rspb.2008.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsang LM, Chan TY, Cheung MK, Chu KH. Molecular evidence for the Southern Hemisphere origin and deep-sea diversification of spiny lobsters (Crustacea: Decapoda: Palinuridae) Mol. Phylogenet. Evol. 2009;51:304–311. doi: 10.1016/j.ympev.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 73.Yang C-H, Sha ZL, Chan TY, Liu RY. Molecular phylogeny of the deep-sea penaeid shrimp genus Parapenaeus (Crustacea: Decapoda: Dendrobranchiata) Zool. Scr. 2015;44:312–323. doi: 10.1111/zsc.12097. [DOI] [Google Scholar]
- 74.Lipps JH, Hickman CS. The Environment of the Deep Sea. Prentice Hall; 1982. Origin, Age and Evolution of Antarctic and Deep-Sea Faunas; pp. 324–356. [Google Scholar]
- 75.Hayward BW. Global deep-sea extinctions during the Pleistocene ice ages. Geology. 2001;29:599–602. doi: 10.1130/0091-7613(2001)029<0599:GDSEDT>2.0.CO;2. [DOI] [Google Scholar]
- 76.Blake DB. Classification and phylogeny of post-Paleozoic sea stars (Asteroidea: Echinodermata) J. Nat. Hist. 1987;21:481–528. doi: 10.1080/00222938700771141. [DOI] [Google Scholar]
- 77.Twitchett RJ, Oji T. Early Triassic recovery of echinoderms. C. R. Palevol. 2005;4:531–542. doi: 10.1016/j.crpv.2005.02.006. [DOI] [Google Scholar]
- 78.Blake DB, Hagdorn H. The Asteroidea (Echinodermata) of the muschelkalk (middle triassic of germany) Pal. Z. 2003;77:23–58. doi: 10.1007/BF03004558. [DOI] [Google Scholar]
- 79.Hesselbo SP, Robinson SA, Surlyk F, Piasecki S. Terrestrial and marine extinction at the Triassic-Jurassic boundary synchronized with major carbon-cycle perturbation: A link to initiation of massive volcanism? Geology. 2002;30:251–254. doi: 10.1130/0091-7613(2002)0302.0.CO;2. [DOI] [Google Scholar]
- 80.Percival LME, et al. Mercury evidence for pulsed volcanism during the end-Triassic mass extinction. Proc. Natl. Acad. Sci. USA. 2017;114:7929–7934. doi: 10.1073/pnas.1705378114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bigham AW, et al. Identifying positive selection candidate loci for high-altitude adaptation in Andean populations. Hum. Genomics. 2009;4:79–90. doi: 10.1186/1479-7364-4-2-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun S, Hui M, Wang Y, Sha Z. The complete mitochondrial genome of the alvinocaridid shrimp Shinkaicaris leurokolos (Decapoda, Caridea): Insight into the mitochondrial genetic basis of deep-sea hydrothermal vent adaptation in the shrimp. Comp. Biochem. Physiol. Part D. 2018;25:42–52. doi: 10.1016/j.cbd.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 83.Mu W, Liu J, Zhang H. Complete mitochondrial genome of Benthodytes marianensis (Holothuroidea: Elasipodida: Psychropotidae): Insight into deep sea adaptation in the sea cucumber. PLoS ONE. 2018;13:e0208051. doi: 10.1371/journal.pone.0208051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Y, et al. Phylogeny, evolution and mitochondrial gene order rearrangement in scale worms (Aphroditiformia, Annelida) Mol. Phylogenet. Evol. 2018;125:220–231. doi: 10.1016/j.ympev.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 85.Brandt U. Energy converting NADH: Quinone oxidoreductase (complex I) Annu. Rev. Biochem. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- 86.Zhang B, Zhang YH, Wang X, Zhang HX, Lin Q. The mitochondrial genome of a sea anemone Bolocera sp. exhibits novel genetic structures potentially involved in adaptation to the deep-sea environment. Ecol. Evol. 2017;7:4951–4962. doi: 10.1002/ece3.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017;45:e18. doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Skujina I, McMahon R, Hegarty M. Re-interpreting mitogenomes: Are nuclear/mitochondrial sequence duplications correctly characterised in published sequence databases? Insights Genet. Genom. 2017;1:1. [Google Scholar]
- 90.Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol. Biol. 1975;94:441–446. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- 91.Bernt M, et al. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013;69:313–319. doi: 10.1016/j.ympev.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 92.Laslett D, Canbäck B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 2007;24:172–175. doi: 10.1093/bioinformatics/btm573. [DOI] [PubMed] [Google Scholar]
- 93.Bernt M, et al. CREx: Inferring genomic rearrangements based on common intervals. Bioinformatics. 2007;23:2957–2958. doi: 10.1093/bioinformatics/btm468. [DOI] [PubMed] [Google Scholar]
- 94.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 97.Kück P, Meusemann K. FASconCAT: Convenient handling of data matrices. Mol. Phylogenet. Evol. 2010;56:1115–1118. doi: 10.1016/j.ympev.2010.04.024. [DOI] [PubMed] [Google Scholar]
- 98.Lanfear R, Calcott B, Ho SYW, Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 99.Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Minh BQ, Nguyen MAT, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference undermixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 102.Rambaut, A. & Drummond, A. Tracer v1.6. Available from. http://tree.bio.ed.ac.uk/software/tracer/ (2013).
- 103.Yu Y, Harris AJ, Blair C, He X. RASP (Reconstruct ancestral state in phylogenies): A tool for historical biogeography. Mol. Phylogenet. Evol. 2015;87:46–49. doi: 10.1016/j.ympev.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 104.Himeno H, et al. Unusual genetic codes and a novel gene structure for tRNASerAGY in starfish mitochondrial DNA. Gene. 1987;56:219–230. doi: 10.1016/0378-1119(87)90139-9. [DOI] [PubMed] [Google Scholar]
- 105.Bouckaert R, et al. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rambaut, A. FigTree v1.4.0: tree figure drawing tool. Available from http://tree.bio.ed.ac.uk/software/figtree (2012).
- 107.Peyer B. Beiträge zur Kenntnis von Rhät und Lias, Contributions to knowledge of the Rhaetian and Lias] Eclogae Geol. Helv. 1944;36:303–326. [Google Scholar]
- 108.Gale AS. Origin and phylogeny of velatid asteroids (Echinodermata, Neoasteroidea)-new evidence from the Jurassic. Swiss J. Palaeontol. 2018;137:279–318. doi: 10.1007/s13358-018-0155-z. [DOI] [Google Scholar]
- 109.Blake DB. Some new post-Paleozoic sea stars (Asteroidea: Echinodermata) and comments on taxon endurance. J. Paleontol. 1986;60:1103–1119. doi: 10.1017/S0022336000022630. [DOI] [Google Scholar]
- 110.Wright T. Monograph on the British fossil echinodermata of the oolitic formations: Volume II.—The Asteroidea and Ophiuroidea. Monographs Palaeontographical Soc. 1880;34(158):i–203. doi: 10.1080/02693445.1880.12027961. [DOI] [Google Scholar]
- 111.Storey JD, Tibshirani R. Statistical significance for genome wide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yang Z, Wong WS, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the manuscript and its Supporting information files.