Abstract
Coagulase-negative staphylococcal isolates (n = 188) were screened for susceptibility to oxacillin, ciprofloxacin, and trovafloxacin, a new fluoroquinolone. At an oxacillin concentration of ≥4 μg/ml, 43% were methicillin resistant; of these, 70% were ciprofloxacin resistant (MIC, ≥4 μg/ml). Of the methicillin-resistant, ciprofloxacin-resistant isolates, 46% were susceptible to ≤2 μg of trovafloxacin per ml and 32% were susceptible to ≤1 μg of trovafloxacin per ml. Sixteen isolates, including twelve that expressed fluoroquinolone resistance, were chosen for detailed analysis. Identification of species by rRNA sequencing revealed a preponderance of Staphylococcus haemolyticus and S. hominis among fluoroquinolone-resistant strains. Segments of genes (gyrA and grlA) encoding DNA gyrase and DNA topoisomerase IV were sequenced. Considerable interspecies variation was noted, mainly involving noncoding nucleotide changes. Intraspecies variation consisted of coding changes associated with fluoroquinolone resistance. As for S. aureus, ciprofloxacin resistance (MIC, ≥8 μg/ml) and increased trovafloxacin MICs (0.25 to 2 μg/ml) could be conferred by the combined presence of single mutations in each gyrA and grlA gene. Trovafloxacin MICs of ≥8 μg/ml also occurred, but these required an additional mutation in grlA.
Coagulase-negative staphylococci (CoNS) are frequently recovered from blood cultures and are a leading cause of infections associated with prosthetic implants (28, 33, 41). Such infections may pose serious therapeutic dilemmas because of the tendency of CoNS to develop resistance to multiple antibiotics (2).
The fluoroquinolones norfloxacin and ciprofloxacin came into widespread use in the 1980s, and resistance developed rapidly in Staphylococcus aureus isolates, especially methicillin-resistant S. aureus (MRSA) (1, 4). Ciprofloxacin resistance has been studied intensively in this species and is due in most isolates to single mutations in each of the dual targets of fluoroquinolones: DNA gyrase and DNA topoisomerase IV (11, 19, 34, 38, 40). Resistance mutations most often occur within a stretch of ca. 50 nucleotides, the so-called quinolone-resistance-determining regions (QRDRs), which are located in the genes for the A subunits of the respective enzymes, gyrA and grlA. The resistance mutations generally involve gyrA codons 84 or 88 and grlA codons 80 or 84 (6). Although the emergence of ciprofloxacin resistance in CoNS paralleled or even surpassed that of S. aureus (21, 25, 28), less is known about ciprofloxacin resistance in CoNS. An S. epidermidis isolate with high-level ciprofloxacin resistance (MIC, 16 μg/ml) was found to have a mutation at gyrA codon 84 (36), as did four S. haemolyticus isolates with high-level resistance (MIC, ≥6 μg/ml) to ciprofloxacin, norfloxacin, and ofloxacin (42).
The present study was aimed at characterizing resistance among CoNS isolates from four New Jersey hospitals to methicillin, ciprofloxacin, and trovafloxacin, a new fluoroquinolone with good activity against gram-positive bacteria (5). The study was conducted in parallel with a similar study of S. aureus in which we found a high incidence of ciprofloxacin resistance (MIC, ≥8 μg/ml) among strains of MRSA (12).
(Portions of this work were presented at the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy [8].)
MATERIALS AND METHODS
Strains.
Clinical isolates characterized as coagulase-negative staphylococci were collected during the latter part of 1996 from the diagnostic microbiology laboratories of four hospitals in the central New Jersey area. Isolates were subcultured onto Mueller-Hinton agar, stored at −70°C in equal parts tryptic soy broth and fetal calf serum containing 2% yeast extract, and resuscitated in batches for susceptibility assays or DNA preparation.
The following strains were purchased from the American Type Culture Collection (ATCC), Manassas, Va.: S. capitis 35661, S. epidermidis 35547, S. hominis 27844, S. haemolyticus 29970, S. saprophyticus 15305, S. simulans 27851 and ATCC strain 29885. This last strain was listed as S. hominis but our rRNA sequence was the same as that reported in GenBank for S. warneri ATCC 27836, and thus its designation has been changed by ATCC to S. warneri (39a).
Antibiotic susceptibility assays.
Initial assays for susceptibility to oxacillin, ciprofloxacin, and trovafloxacin were performed on 188 consecutive independent isolates by agar dilution on Mueller-Hinton plates (12); antibiotic concentrations ranged from 0.125 to 8 μg/ml in twofold increments. Selected isolates were subjected to additional liquid microdilution assays for determining susceptibilities to ciprofloxacin, trovafloxacin, sparfloxacin, and levofloxacin over the range of 0.015 to 256 μg/ml (12). Oxacillin powder was purchased from Sigma Chemical Co., St. Louis, Mo., and ciprofloxacin was obtained from Bayer Corp., West Haven, Conn. Levofloxacin was provided by the Robert Wood Johnson Research Institute, Raritan, N.J.; sparfloxacin was provided by Rhone-Poulenc-Rorer, Collegeville, Pa.; and trovafloxacin was provided by Pfizer, Inc., New York, N.Y.
In categorizing isolates as methicillin-resistant or ciprofloxacin-resistant, we used National Committee for Clinical Laboratory Standards (NCCLS) oxacillin and ciprofloxacin criteria in effect during analysis of our data and prior to 1999 (30) (namely, an MIC of ≥4 μg/ml). We also reevaluated the data using the new criterion for methicillin resistance, i.e., an MIC of ≥0.5 μg/ml (31), as noted in the Results section.
DNA preparation, amplification, and sequencing.
DNA was prepared by using Instagene Matrix (Bio-Rad Laboratories, Hercules, Calif.) or the QIAamp Tissue Kit (Qiagen, Inc., Santa Clarita, Calif.), and PCR amplifications were performed as previously described (12).
Early studies on topoisomerase genes were done with PCR primers with 5′ T3 or T7 promoter tags to permit subsequent sequencing with a LiCor 4000L system and standard fluorescent primers (Li-Cor, Lincoln, Neb.). Initial sets of primers were based on gyrA subsequences conserved between S. aureus and S. epidermidis (gyrt3 and gyrt7s, Table 1) or grlA subsequences relatively conserved between S. aureus and S. pneumoniae (topot3s and topot7s, Table 1). These primers were tested with an S. epidermidis isolate (cn96), an S. haemolyticus isolate (cn134), and an S. hominis isolate (cn182) (see Table 3 and below), and they worked well except for S. epidermidis grlA. However, the grlA primer pair previously used for S. aureus (12), topot3 and topot7 (Table 1), was found to work with S. epidermidis. Using sequence results from the above primers, we designed primers corresponding to subsequences relatively conserved among S. epidermidis, S. haemolyticus, and S. hominis, which would amplify 0.3 kb around the respective QRDRs (gcn1, gcn2, tcn1, and tcn2; Table 1). These primers proved to amplify appropriate segments from all CoNS examined, including strains of S. simulans, S. lugdunensis, S. saprophyticus, S. capitis, and S. warneri, as well as S. aureus. The primers were used also as sequencing primers in an ABI377 system (PE Applied Biosystems, Foster City, Calif.) using fluorescent terminators, thus precluding the need for T3 or T7 tags.
TABLE 1.
Primers
| Primer | Sequencea | Gene or 16S RNA (coordinates)b | Application |
|---|---|---|---|
| gyrt3 | ATGGCTGAATTACCTCAATC | gyrA (1–20) | Forward primer for S. aureus and CoNS gyrA |
| gyrt7s | CATCATAGTTATCGATGAAATC | gyrA (454–433) | Reverse primer for CoNS gyrA |
| topot3 | GTGAGTGAAATAATTCAAGATT | grlA (1–22) | Forward primer for S. aureus and S. epidermidis grlA |
| topot3s | CCAGATITTCGTGATGG | grlA (94–110) | Forward primer for CoNS grlA |
| topot7 | TAATAATTAACTGTTTACGTCC | grlA (772–751) | Reverse primer for S. aureus and S. epidermidis grlA |
| topot7s | TCAGCTAAATTATGIGG | grlA (557–541) | Reverse primer for CoNS grlA |
| gcn1 | ATGCGTGAATCATTTTTAGACTATGC | gyrA (53–88) | Forward primer for CoNS gyrA |
| gcn2 | GAGCCAAAGTTACCTTGACC | gyrA (336–317) | Reverse primer for CoNS gyrA |
| tcn1 | AATACGTATGATAAGAATTTCCG | grlA (159–182) | Forward primer for CoNS grlA |
| tcn2 | GTTGTGTCATCATAGTTTGG | grlA (449–430) | Reverse primer for CoNS grlA |
| st1f | GTTAGCGGCGGACGGGTGAG | 16S RNA (95–114) | Forward primer for 16S RNA |
| st2r | CAACATCTCACGACACGAGC | 16S RNA (1096–1077) | Reverse primer for 16S RNA |
| st3r | CGTATTACCGCGGCTGCTGG | 16S RNA (546–526) | Reverse primer for 16S RNA |
Sequences are in 5′-to-3′ direction. T3 or T7 promoter tags for “gyr” and “topo” sets are not shown. “I” in topot3s and topot7s = inosine.
TABLE 3.
Species, QRDR sequence, and MICs of selected CoNS strains
| Straina | Species | Amino acid/nucleotidesb
|
MICs (μg/ml)
|
||||||
|---|---|---|---|---|---|---|---|---|---|
|
gyrA codon
|
grlA codon
|
Antibioticc
|
|||||||
| 84 | 88 | 80 | 84 | Cipro | Trova | Spar | Levo | ||
| Ciprofloxacin susceptible, trovafloxacin susceptible | |||||||||
| cn7 | S. epidermidis | S/TCT | E/GAA | S/TCT | D/GAT | 0.125 | 0.03 | 0.06 | 0.125 |
| cn9 | S. epidermidis | S/TCT | E/GAA | S/TCT | D/GAT | 0.125 | 0.03 | 0.06 | 0.125 |
| cn96 | S. epidermidis | S/TCT | E/GAA | S/TCT | D/GAT | 0.125 | 0.03 | 0.06 | 0.125 |
| cn85 | S. lugdunensis | S/TCG | D/GAT | S/TCA | D/GAT | 0.25 | 0.03 | 0.125 | 0.25 |
| ATCC 29213 | S. haemolyticus | S/TCA | D/GAT | S/TCA | D/GAT | 0.125 | 0.015 | 0.06 | 0.125 |
| ATCC 27844 | S. hominis | S/TCT | D/GAT | S/TCT | D/GAT | 0.25 | 0.03 | 0.125 | 0.25 |
| ATCC 27851 | S. simulans | S/TCA | E/GAA | S/TCT | D/GAT | 0.125 | 0.03 | 0.125 | 0.125 |
| Ciprofloxacin resistant, trovafloxacin susceptible | |||||||||
| cn90 | S. haemolyticus | L/TTA | D/GAT | S/TCA | Y/TAT | 8 | 0.25 | 8 | 4 |
| cn138 | S. haemolyticus | L/TTA | D/GAT | S/TCA | Y/TAT | 16 | 0.25 | 8 | 4 |
| cn168 | S. haemolyticus | L/TTA | D/GAT | L/TTA | D/GAT | 48 | 2 | 8 | 8 |
| cn182 | S. hominis | A/GCT | D/GAT | F/TTT | D/GAT | 32 | 1 | 1 | 4 |
| cn128d | S. hominis | F/TTT | D/GAT | Y/TAT | D/GAT | 3 | 0.5 | 4 | 2 |
| cn17 | S. simulans | L/TTA | E/GAA | F/TTT | D/GAT | 12 | 2 | 8 | 8 |
| Ciprofloxacin resistant, trovafloxacin resistant | |||||||||
| cn78 | S. haemolyticus | L/TTA | D/GAT | L/TTA | G/GGT | 128 | 16 | 8 | 16 |
| cn81 | S. haemolyticus | L/TTA | D/GAT | L/TTA | G/GGT | 128 | 32 | 8 | 16 |
| cn134 | S. haemolyticus | L/TTA | D/GAT | L/TTA | G/GGT | 64 | 16 | 8 | 16 |
| cn143 | S. haemolyticus | L/TTA | D/GAT | L/TTA | G/GGT | 64 | 16 | 8 | 16 |
| cn164 | S. hominis | F/TTT | D/GAT | Y/TAT | N/AAT | 64 | 16 | 8 | 16 |
| cn95 | S. epidermidis | F/TTT | E/GAA | Y/TAT | Y/TAT | 64 | 8 | 8 | 8 |
Strains designated “cn” are clinical isolates collected as part of our survey.
Cipro, ciprofloxacin; Trova, trovafloxacin; Spar, sparfloxacin; Levo, levofloxacin. MICs are the average of duplicate determinations, which did not differ by more than one twofold dilution.
This strain had a ciprofloxacin MIC of >8 μg/ml and a trovafloxacin MIC of 2 μg/ml in the agar screen.
Primers for 16S rRNA amplification were designed by scanning alignments of sequences for 18 CoNS species plus S. aureus, as obtained from GenBank. Primers st1f and st2r were used to obtain a 1-kb amplicon. A 410-bp segment was sequenced by using st3r as primer and the labelled terminators as described above.
Sequences are available in GenBank under accession numbers AF127630-52, Af128292-314, and AF128276-291.
RESULTS
Survey of oxacillin, ciprofloxacin, and trovafloxacin susceptibilities.
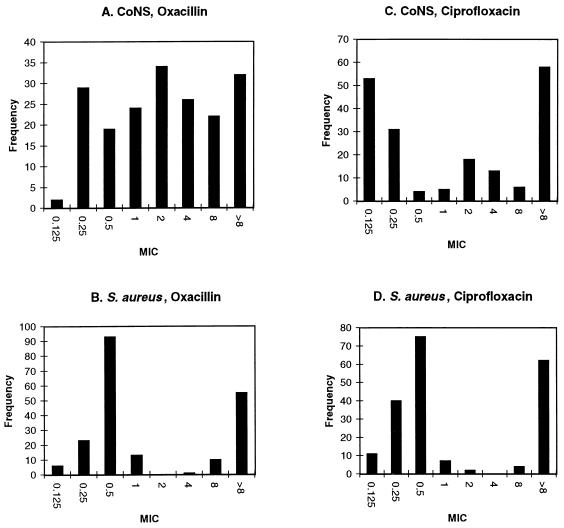
The distribution of oxacillin MICs among the CoNS isolates was quite broad (Fig. 1A), differing from the bimodal curve obtained for S. aureus isolates collected in parallel (Fig. 1B). Methicillin-resistant CoNS (MRCoNS), at the oxacillin resistance breakpoint used in the present study (≥4 μg/ml [12, 30]), amounted to 43% of the total. This figure rose to 83.5% for the new oxacillin breakpoint, ≥0.5 μg/ml (31).
FIG. 1.
Oxacillin and ciprofloxacin susceptibility distributions of coagulase-negative staphylococcal isolates compared to contemporary S. aureus isolates. The data are from the present study (A and C) and from a parallel study on S. aureus (12) (B and D). Here and in Fig. 2 the columns designated 0.125 represent MICs of ≤0.125 μg/ml.
The distribution of CoNS ciprofloxacin MICs was trimodal (Fig. 1C), again in contrast to S. aureus (Fig. 1D). The overall frequency of ciprofloxacin resistance (MIC, ≥4 μg/ml) among CoNS was 41%, while that among MRCoNS was 70%, higher, as expected, than that for methicillin-susceptible CoNS (MSCoNS), i.e., 19%.
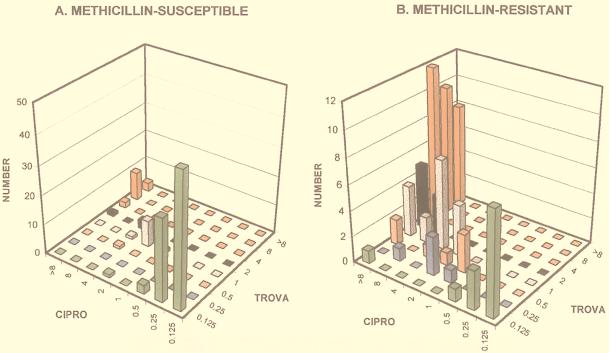
The distribution of CoNS trovafloxacin susceptibilities as a function of ciprofloxacin susceptibilities is plotted for MSCoNS in Fig. 2A and for MRCoNS in Fig. 2B. As shown in the figure, all ciprofloxacin-susceptible isolates (MIC, ≤1 μg/ml [31]) were also susceptible to trovafloxacin (MIC, ≤1 μg/ml). The ciprofloxacin-resistant MRCoNS (MIC, ≥4 μg/ml) showed a broad distribution of trovafloxacin MICs. The trovafloxacin breakpoints for staphylococci are somewhat controversial. The susceptibility breakpoint of ≤2 μg/ml recommended by the U.S. Food and Drug Administration (see references 13 and 20) results in 46% of the ciprofloxacin-resistant MRCoNS isolates being scored as trovafloxacin susceptible and 21% being scored as trovafloxacin intermediate (MIC, 4 μg/ml). The more conservative alternative breakpoint of ≤1 μg/μl (9, 20, 35) results in 32% of this subclass of isolates being scored as trovafloxacin susceptible and 14% being scored as trovafloxacin intermediate (MIC, 2 μg/ml). Curiously, almost all (83%) of the ciprofloxacin-resistant (MIC, ≥4 μg/ml) MSCoNS also had trovafloxacin MICs of ≥4 μg/ml (Fig. 2B). The CoNS that would be newly categorized as methicillin resistant by the 1999 NCCLS revision (oxacillin MIC of 0.5 to 2 μg/ml) (31) resembled the group that was methicillin susceptible by the criterion used above (oxacillin MIC of ≤2 μg/ml) in the distribution of fluoroquinolone MICs.
FIG. 2.
Relationships between trovafloxacin and ciprofloxacin susceptibilities for methicillin-susceptible and -resistant coagulase-negative staphylococcal isolates. Ciprofloxacin MICs (ascending right to left) are plotted against trovafloxacin MICs (ascending front-to-back) for methicillin-susceptible (A) and methicillin-resistant (B) CoNS. Trovafloxacin-resistant and -intermediate isolates (MIC, ≥4 μg/ml) are indicated by the red columns.
Detailed analysis of selected isolates: species determination.
Sixteen isolates were chosen for further detailed examination on the basis of their ciprofloxacin and trovafloxacin MICs from the agar dilution screening. These 16 isolates included 4 isolates that were susceptible to both agents (MIC, ≤0.125 μg/ml), 6 that were resistant to ciprofloxacin (MIC, ≥8 μg/ml) and susceptible to trovafloxacin (MIC, ≤2 μg/ml), and 6 that were resistant to both agents (MIC, ≥8 μg/ml). The species of these strains were identified, and the strains were then subjected to topoisomerase gene sequencing and more extensive MIC determinations. Analogous studies are under way on some of the many isolates that fell into intermediate- or marginal-susceptibility categories.
The API STAPH kit (BioMerieux Vitek, Hazelwood, Mo.) was used to perform a series of 19 biochemical reactions on 10 isolates: 4 isolates that were shown by 16S rRNA sequencing to be S. epidermidis, 1 isolate that was an S. epidermidis type strain (ATCC 35547), 2 isolates that proved to be S. hominis, and 3 isolates that proved to be S. haemolyticus. The S. epidermidis strains were correctly identified by the reactions. The S. hominis and S. haemolyticus isolates, however, yielded ambiguous results, as has been reported by others (15, 41). We therefore relied on 16S RNA sequences for speciation. A segment corresponding to nucleotides 115 to 526 of Bacillus subtilis 16S RNA (24) was found to discriminate among human CoNS species (see reference 22). Interspecies variation among the CoNS species listed in Table 3 averaged ca. 5%, while intraspecies variation was zero, except for some S. hominis strains which differed by a single residue (0.2%) over this stretch.
Of the four ciprofloxacin-susceptible, trovafloxacin-susceptible isolates examined, three were S. epidermidis and one was S. lugdunensis. Of the six ciprofloxacin-resistant, trovafloxacin-susceptible isolates, three were S. haemolyticus and two were S. hominis. Of the six ciprofloxacin-resistant, trovafloxacin-resistant isolates, four were S. haemolyticus and one was S. hominis. There was one ciprofloxacin-resistant, trovafloxacin-susceptible S. simulans and one ciprofloxacin-resistant, trovafloxacin-resistant S. epidermidis isolate.
Detailed analysis of selected isolates: QRDR sequences and MICs.
Using the stepwise approach described in Materials and Methods, we generated primers that amplified segments of ∼0.3 kb around the QRDRs of the gyrA and grlA genes of the five CoNS species described above, as well as three others: S. capitis, S. saprophyticus, and S. warneri. Differences among CoNS species were comparable to differences between CoNS and S. aureus. Nucleotide differences among the eight CoNS species examined were considerably higher than for 16S RNA, ranging from 13 to 22% for gyrA (average, 18%) and 13 to 28% for grlA (average, 19%). Amino acid differences ranged from 0 to 8% for gyrA (average, 2.8%) and 0 to 16% for grlA (average, 5.7%). The results for topoisomerase IV are shown in Table 2, which includes also corresponding S. aureus and B. subtilis sequences for comparison. It is noteworthy that, despite the substantial nucleotide divergence among the CoNS species, all wild-type sequences retained the HinfI restriction sites used to screen for mutations at gyrA codon 84 and grlA codon 80 (see, for example, references 36 and 40).
TABLE 2.
Sequence differences for grlA genesa
| Strain | Sequence differences (% amino acids or nucleotides) with strain:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S. epidermidis | S. warneri | S. capitis | S. haemolyticus | S. hominis | S. simulans | S. aureus | S. lugdunensis | S. saprophyticus | B. subtilis | |
| S. epidermidis | 0 | 1.22 | 3.75 | 1.22 | 2.45 | 3.71 | 6.29 | 10.34 | 23.98 | |
| S. warneri | 13.0 | 1.22 | 3.75 | 1.22 | 2.45 | 3.71 | 6.29 | 10.34 | 23.98 | |
| S. capitis | 14.5 | 12.1 | 2.48 | 2.45 | 3.71 | 4.99 | 7.62 | 10.34 | 23.98 | |
| S. haemolyticus | 14.8 | 17.8 | 16.8 | 2.48 | 3.75 | 7.71 | 7.71 | 11.90 | 24.32 | |
| S. hominis | 19.5 | 14.0 | 13.5 | 19.9 | 3.71 | 4.99 | 7.62 | 11.74 | 25.65 | |
| S. simulans | 18.9 | 18.9 | 18.9 | 20.4 | 19.5 | 6.29 | 6.29 | 11.74 | 23.98 | |
| S. aureus | 17.4 | 16.4 | 17.9 | 19.9 | 17.9 | 21.0 | 7.62 | 13.17 | 20.73 | |
| S. lugdunensis | 16.9 | 17.4 | 15.4 | 19.9 | 17.9 | 20.5 | 19.5 | 16.11 | 22.34 | |
| S. saprophyticus | 23.7 | 21.5 | 20.0 | 24.3 | 27.6 | 24.3 | 25.9 | 22.1 | 29.11 | |
| B. subtilis | 41.3 | 39.0 | 40.3 | 36.4 | 41.7 | 43.8 | 42.4 | 41.7 | 44.6 | |
Differences are given as percentages of amino acids (above the diagonal) or nucleotides (below) over the 248 bp between primers tcn1 and tcn2 (Table 1), as determined by using the GCG Program “Distances” with the default parameters. All species are wild type and ciprofloxacin susceptible and are CoNS, aside from S. aureus and B. subtilis. The S. aureus sequence is that of Ferrero et al. (11), and the B. subtilis sequence is that of Kunst et al. (24) (GenBank Z99104). The S. lugdunensis sequence is that of the fluoroquinolone-susceptible isolate cn85. The other sequences were determined on the ATCC strains listed in Materials and Methods and as described there.
QRDRs of fluoroquinolone-resistant isolates showed no changes from the corresponding wild type, except for mutations at gyrA codon 84 and grlA codons 80 and 84 as summarized in Table 3. The ciprofloxacin-resistant, trovafloxacin-susceptible isolates examined had mutations involving gyrA codon 84 and grlA codon 80 or 84. The ciprofloxacin-resistant, trovafloxacin-resistant isolates had a second mutation in grlA, at codon 84 or 80, depending on the position of the first mutation. Strikingly, four of the six trovafloxacin-resistant isolates were S. haemolyticus with Ser-to-Leu mutations at gyrA codon 84 and grlA codon 80, plus the unusual mutation Asp-to-Gly at grlA codon 84.
Table 3 also summarizes results of more extensive MIC testing. The four isolates examined that were scored as ciprofloxacin and trovafloxacin susceptible in the agar dilution screening had liquid dilution MICs of 0.125 to 0.25 μg/ml for ciprofloxacin, 0.03 μg/ml for trovafloxacin, and 0.06 to 0.125 μg/ml for two other fluoroquinolones, sparfloxacin and levofloxacin.
The six isolates examined that were scored as ciprofloxacin resistant, trovafloxacin susceptible in the agar screening had ciprofloxacin liquid dilution MICs of 8 to 48 μg/ml, aside from a seemingly aberrant result for an S. hominis isolate (MIC, 3 μg/ml). These isolates, all of which had single mutations in both the gyrA and grlA genes, had liquid dilution MICs of 0.25 to 2 μg/ml for trovafloxacin, which was significantly higher than for the four susceptible isolates lacking QRDR mutations but still in the range often considered susceptible. These isolates were scored for the most part as resistant to sparfloxacin and levofloxacin (MICs of mainly 4 to 8 μg/ml).
The six isolates examined that were scored as ciprofloxacin resistant, trovafloxacin resistant in the agar screening had the highest ciprofloxacin liquid dilution MICs (64 to 128 μg/ml) and were also highly resistant (MICs, 8 to 32 μg/ml) to the other three fluoroquinolones tested.
DISCUSSION
The high frequency of methicillin resistance among CoNS isolates is in accord with results of other studies (29, 32), as are the association between methicillin resistance and ciprofloxacin resistance (28, 32) and the absence of trovafloxacin resistance in ciprofloxacin-susceptible isolates (13, 29).
The overall frequency of trovafloxacin susceptibility among MRCoNS in the present study amounted to 63% at the trovafloxacin susceptibility breakpoint of ≤2 μg/ml, which was similar to that reported in a recent survey of isolates from 10 other North American centers (77.5%) (13). The frequency of trovafloxacin-susceptible CoNS isolates among ciprofloxacin-resistant MRCoNS, i.e., 46%, at this trovafloxacin susceptibility breakpoint is lower than the 86% we found for MRSA (7, 12); this is also in accord with other reports (13, 29). The utility of trovafloxacin for treating infections due to ciprofloxacin-resistant MRCoNS warrants further study.
Although most of the isolates screened in the present study were not identified to the species level, one of the contributing laboratories reported ca. 70% S. epidermidis and 10% each of S. haemolyticus and S. hominis, a finding resembling the distribution in an earlier collection from that laboratory (41). The results presented here thus clearly indicate the preferential occurrence of S. haemolyticus and S. hominis among the fluoroquinolone-resistant isolates. The relative enrichment for S. haemolyticus is in accord with earlier studies (3, 29, 42). The enrichment for S. hominis may reflect the emergence of a recently described subspecies, S. hominis subsp. novobiosepticus, isolates of which are likely to be ciprofloxacin resistant (23).
Our QRDR sequencing results indicate that the major mechanisms of fluoroquinolone resistance in clinical CoNS isolates resemble those of S. aureus. Ciprofloxacin resistance (MIC, ≥8 μg/ml) in S. aureus is generally associated with one mutation in gyrA, at codon 84 or 88, and one in grlA, at codon 80 or 84 (11, 12, 34, 37, 40). Such isolates are characteristically also resistant to levofloxacin and sparfloxacin (34, 39, 40) but are susceptible at a breakpoint of ≤2 μg/ml to the newer fluoroquinolones moxifloxacin (34) and DU-6859a (39, 40). They also remain susceptible to trovafloxacin at this breakpoint, albeit with higher MICs than wild-type S. aureus (12). S. aureus isolates have recently been described that are resistant to trovafloxacin (MIC, ≥8 μg/ml) (12) or DU-6859a (an MIC90 of ca. 6 μg/ml) (40) associated with a second mutation in grlA (at codon 80 or 84, depending on the position of the first mutation). The CoNS isolates sequenced in the present study (Table 3) follow this pattern, with the qualification that S. hominis ciprofloxacin-resistant, trovafloxacin-susceptible isolates were scored as susceptible or intermediate to sparfloxacin and levofloxacin; the clinical importance of this distinction remains to be determined.
The particular mutations seen at gyrA codon 84 and the homologous codon (codon 80) of grlA are similar or identical to those seen in S. aureus, with the mutation being Ser-to-Phe or Ser-to-Tyr on the one hand or Ser-to-Leu on the other, depending on whether the wild-type codons are TCT or TCA.
The Ser-to-Ala mutation at gyrA codon 84 in S. hominis isolate cn182 has not been reported before for staphylococci. It is possible that the presence of Ala, rather than the more hydrophobic Phe or Leu seen in other resistant mutants, is related to the relative susceptibility of this isolate to sparfloxacin. Ser-to-Ala changes at the homologous position have been associated with decreased fluoroquinolone susceptibility in Enterobacteriaceae (17, 18) and in some mycobacterial species (16).
The Asp(GAT)→Gly(GGT) change at grlA codon 84 in fluoroquinolone-resistant S. haemolyticus isolates resembles a mutation, Glu(GAA)→Gly(GGA), seen in local fluoroquinolone-resistant S. aureus isolates (12) but not otherwise reported in staphylococci. In other studies (37, 40), most fluoroquinolone-resistant S. aureus isolates with grlA codon 84 changes have Glu(GAA)→Lys(AAA) mutations here.
Although we have not explored additional resistance mechanisms such as permeability changes, the present results are compatible with the stepwise evolution of fluoroquinolone resistance in CoNS isolates via sequential mutations in the genes for topoisomerase and gyrase, as described for in vitro S. aureus mutants (10, 14, 39). According to a recent version of this scenario (14), an initial mutation causing low-level ciprofloxacin resistance (MIC, 1 to 2 μg/ml) occurs in topoisomerase IV, the primary target of fluoroquinolones in S. aureus. These strains generate high-level ciprofloxacin resistance (MIC, ≥6 μg/ml) under continuing fluoroquinolone selective pressure via a mutation in gyrase. Further selection yields variants with yet higher ciprofloxacin MICs and with MICs of new fluoroquinolones increased 2- to 32-fold, which are now associated with second mutations in topoisomerase IV. After this paper was submitted, Li et al. (26) reported sequences for S. epidermidis isolates that support such a scenario for the development of increasing resistance to ciprofloxacin, norfloxacin, and ofloxacin. In addition, they reported a grlA sequence from S. epidermidis ATCC 14990 which corresponds to a portion of our grlA amplicon and which is identical to our grlA sequence for S. epidermidis ATCC 35547.
In conclusion, we found a high level of ciprofloxacin resistance among CoNS isolates, especially MRCoNS and S. haemolyticus and S. hominis. We found that 46 or 32% of the ciprofloxacin-resistant MRCoNS were susceptible to trovafloxacin at breakpoints of ≤2 or ≤1 μg/ml, respectively. The mechanisms of fluoroquinolone resistance among CoNS resembled those of S. aureus isolates. The simultaneous presence of two mutations, one each in gyrA and grlA, was associated with ciprofloxacin resistance (MIC, ≥8 μg/ml). Resistance to trovafloxacin (MIC, ≥8 μg/ml) was less frequent, presumably because of the apparent requirement for an additional mutation in grlA.
ACKNOWLEDGMENTS
This work was supported in part by a grant from Pfizer, Inc., New York, N.Y.
We thank the Robert Wood Johnson Medical School DNA Laboratory for providing oligonucleotides and DNA sequencing; J. DeLucia for technical assistance; and D. Alcid, N. Gornish, K. Joho, K. Paz, and M. Weinstein for contributing isolates.
REFERENCES
- 1.Acar J F, Goldstein F W. Trends in bacterial resistance to fluoroquinolones. Clin Infect Dis. 1997;24(Suppl. 1):S67–S73. doi: 10.1093/clinids/24.supplement_1.s67. [DOI] [PubMed] [Google Scholar]
- 2.Archer G L, Climo M W. Antimicrobial susceptibility of coagulase-negative staphylococci. Antimicrob Agents Chemother. 1994;38:2231–2237. doi: 10.1128/aac.38.10.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannerman T L, Wadiak D L, Kloos W E. Susceptibility of Staphylococcus species and subspecies to fleroxacin. Antimicrob Agents Chemother. 1991;35:2135–2139. doi: 10.1128/aac.35.10.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumberg H M, Rimland D, Carroll D J, Terry P, Wachsmuth I K. Rapid development of ciprofloxacin resistance in methicillin-susceptible and -resistant Staphylococcus aureus. J Infect Dis. 1991;163:1279–1285. doi: 10.1093/infdis/163.6.1279. [DOI] [PubMed] [Google Scholar]
- 5.Brighty K E, Gootz T D. The chemistry and biological profile of trovafloxacin. J Antimicrob Chemother. 1997;39(Suppl. B):1–14. doi: 10.1093/jac/39.suppl_2.1. [DOI] [PubMed] [Google Scholar]
- 6.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubin, D. T., J. E. Fitzgibbon, and J. L. John. 1998. Unpublished observations.
- 8.Dubin D T, Fitzgibbon J E, John J F. Program and abstracts of the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1998. Topoisomerase mutations in fluoroquinolone (Fq)-resistant coagulase-negative staphylococci (CNS), abstr. C-44; p. 81. [Google Scholar]
- 9.Entenza J M, Marchetti O, Glauser M P, Moreillon P. Y-688, a new quinolone active against quinolone-resistant Staphylococcus aureus: lack of in vivo efficacy in experimental endocarditis. Antimicrob Agents Chemother. 1998;42:1889–1894. doi: 10.1128/aac.42.8.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrero L, Cameron B, Crouzet J. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39:1554–1558. doi: 10.1128/aac.39.7.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrero L, Cameron B, Manse B, Lagneaux D, Crouzet J, Famechon A, Blanche F. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol Microbiol. 1994;13:641–653. doi: 10.1111/j.1365-2958.1994.tb00458.x. [DOI] [PubMed] [Google Scholar]
- 12.Fitzgibbon J E, John J F, Delucia J L, Dubin D T. Topoisomerase mutations in trovafloxacin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1998;42:2122–2124. doi: 10.1128/aac.42.8.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs P C, Barry A L, Brown S D. In vitro activity of trovafloxacin against ciprofloxacin-susceptible and -resistant clinical bacterial isolates and assessment of the trovafloxacin disk test. Diagn Microbiol Infect Dis. 1999;33:33–38. doi: 10.1016/s0732-8893(98)00134-5. [DOI] [PubMed] [Google Scholar]
- 14.Fukuda H, Hori S, Hiramatsu K. Antibacterial activity of gatifloxacin (AM-1155, CG5501, BMS-206584), a newly developed fluoroquinolone, against sequentially acquired quinolone-resistant mutants and the norA transformant of Staphylococcus aureus. Antimicrob Agents Chemother. 1998;42:1917–1922. doi: 10.1128/aac.42.8.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gribaldo S, Cookson B, Saunders N, Marples R, Stanley J. Rapid identification by specific PCR of coagulase-negative staphylococcal species important in hospital infection. J Med Microbiol. 1997;46:45–53. doi: 10.1099/00222615-46-1-45. [DOI] [PubMed] [Google Scholar]
- 16.Guillemin I, Jarlier V, Cambau E. Correlation between quinolone susceptibility patterns and sequences in the A and B subunits of DNA gyrase in mycobacteria. Antimicrob Agents Chemother. 1998;42:2084–2088. doi: 10.1128/aac.42.8.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hallett P, Maxwell A. Novel quinolone resistance mutations of the Escherichia coli DNA gyrase A protein: enzymatic analysis of the mutant proteins. Antimicrob Agents Chemother. 1991;35:335–340. doi: 10.1128/aac.35.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heisig P, Kratz B, Halle E, Graser Y, Altwegg M, Rabsch W, Faber J P. Identification of DNA gyrase A mutations in ciprofloxacin-resistant isolates of Salmonella typhimurium from men and cattle in Germany. Microb Drug Resist. 1995;1:211–218. doi: 10.1089/mdr.1995.1.211. [DOI] [PubMed] [Google Scholar]
- 19.Hooper D C. Bacterial topoisomerases, anti-topoisomerases, and anti-topoisomerase resistance. Clin Infect Dis. 1998;27:S54–S63. doi: 10.1086/514923. [DOI] [PubMed] [Google Scholar]
- 20.Jones R N. Preliminary interpretive criteria for in vitro susceptibility testing of CP-99219 by dilution and disk diffusion methods. Diagn Microbiol Infect Dis. 1994;20:167–170. doi: 10.1016/0732-8893(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 21.Jones R N, Barry A L, Gardiner R V, Packer R R. The prevalence of staphylococcal resistance to penicillinase-resistant penicillins. A retrospective and prospective national surveillance trial of isolates from 40 medical centers. Diagn Microbiol Infect Dis. 1989;12:385–394. doi: 10.1016/0732-8893(89)90108-9. [DOI] [PubMed] [Google Scholar]
- 22.Kloos W. Taxonomy and systematics of staphylococci indigenous to humans. In: Crossley J B, Archer G L, editors. The staphylococci in human disease. New York, N.Y: Churchill Livingstone; 1997. pp. 113–137. [Google Scholar]
- 23.Kloos W E, George C G, Olgiate J S, Vanpelt L, Mckinnon M L, Zimmer B L, Muller E, Weinstein M P, Mirrett S. Staphylococcus hominis subsp. novobiosepticus, a novel trehalose- and N-acetyl-d-glucosamine-negative, novobiocin- and multiple-antibiotic-resistant subspecies isolated from human blood cultures. Int J Syst Bacteriol. 1998;48:799–812. doi: 10.1099/00207713-48-3-799. [DOI] [PubMed] [Google Scholar]
- 24.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 25.Laverdiere M, Weiss K, Rivest R, Delorme J. Trends in antibiotic resistance of staphylococci over an eight-year period—differences in the emergence of resistance between coagulase-positive and coagulase-negative staphylococci. Microb Drug Resist. 1998;4:119–122. doi: 10.1089/mdr.1998.4.119. [DOI] [PubMed] [Google Scholar]
- 26.Li Z, Deguchi T, Yasuda M, Kawamura T, Kanematsu E, Nishino Y, Ishihara S, Kawada Y. Alteration in the GyrA subunit of DNA gyrase and the ParC subunit of DNA topoisomerase IV in quinolone-resistant clinical isolates of Staphylococcus epidermidis. Antimicrob Agents Chemother. 1998;42:3293–3295. doi: 10.1128/aac.42.12.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Margerrison E E, Hopewell R, Fisher L M. Nucleotide sequence of the Staphylococcus aureus gyrB-gyrA locus encoding the DNA gyrase A and B proteins. J Bacteriol. 1992;174:1596–1603. doi: 10.1128/jb.174.5.1596-1603.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marshall S A, Wilke W W, Pfaller M A, Jones R N. Staphylococcus aureus and coagulase-negative staphylococci from blood stream infections: frequency of occurrence, antimicrobial susceptibility, and molecular (mecA) characterization of oxacillin resistance in the SCOPE program. Diagn Microbiol Infect Dis. 1998;30:205–214. doi: 10.1016/s0732-8893(97)00212-5. [DOI] [PubMed] [Google Scholar]
- 29.Montanari M P, Prenna M, Mingoia M, Ripa S, Varaldo P E. In vitro antibacterial activity of trovafloxacin and five other fluoroquinolones. Chemotherapy. 1998;44:85–93. doi: 10.1159/000007097. [DOI] [PubMed] [Google Scholar]
- 30.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing: 8th informational supplement. M100-S8. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1998. [Google Scholar]
- 31.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing: 9th informational supplement, p. 37–40. M100-S9. Villanova, Pa: National Committee for Clinical Laboratory Standards.; 1999. [Google Scholar]
- 32.Pegues D A, Colby C, Hibberd P L, Cohen L G, Ausubel F M, Calderwood S B, Hooper D C. The epidemiology of resistance to ofloxacin and oxacillin among clinical coagulase-negative staphylococcal isolates: analysis of risk factors and strain types. Clin Infect Dis. 1998;26:72–79. doi: 10.1086/516270. [DOI] [PubMed] [Google Scholar]
- 33.Rupp M E. Infections of intravascular catheters and vascular devices. In: Crossley J B, Archer G L, editors. The staphylococci in human disease. New York, N.Y: Churchill Livingstone; 1997. pp. 379–399. [Google Scholar]
- 34.Schmitz F J, Hofmann B, Hansen B, Scheuring S, Luckefahr M, Klootwijk, Verhoef J, Fluit A, Heinz H P, Kohrer K, Jones M E. Relationship between ciprofloxacin, ofloxacin, levofloxacin, sparfloxacin and moxifloxacin (BAY 12-8039) MICs and mutations in grlA, grlB, gyrA and gyrB in 116 unrelated clinical isolates of Staphylococcus aureus. J Antimicrob Chemother. 1998;41:481–484. doi: 10.1093/jac/41.4.481. [DOI] [PubMed] [Google Scholar]
- 35.Soussy C J, Chardon H, Drugeon H, Kitzis M D, Phippon A, Courvalin P. Program and abstracts of the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1998. In vitro antibacterial activity of trovafloxacin against hospital isolates and determination of breakpoints: a multicenter study, abstr. E-28; p. 175. [Google Scholar]
- 36.Sreedharan S, Peterson L R, Fisher L M. Ciprofloxacin resistance in coagulase-positive and -negative staphylococci: role of mutations at serine 84 in the DNA gyrase A protein of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 1991;35:2151–2154. doi: 10.1128/aac.35.10.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi H, Kikuchi T, Shoji S, Fujimura S, Lutfor A B, Tokue Y, Nukiwa T, Watanabe A. Characterization of gyrA, gyrB, grlA and grlB mutations in fluoroquinolone-resistant clinical isolates of Staphylococcus aureus. J Antimicrob Chemother. 1998;41:49–57. doi: 10.1093/jac/41.1.49. [DOI] [PubMed] [Google Scholar]
- 38.Takahata M, Yonezawa M, Kurose S, Futakuchi N, Matsubara N, Watanabe, Narita H. Mutations in the gyrA and grlA genes of quinolone-resistant clinical isolates of methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 1996;38:543–546. doi: 10.1093/jac/38.3.543. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka M, Onodera Y, Ushida Y, Sato K, Hayakawa I. Inhibitory activities of quinolones against gyrase and topoisomerase IV purified from Staphylococcus aureus. Antimicrob Agents Chemother. 1997;41:2362–2366. doi: 10.1128/aac.41.11.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.Tang, J. Personal communication.
- 40.Wang T, Tanaka M, Sato K. Detection of gyrA and grlA mutations in 344 Staphylococcus aureus strains. Antimicrob Agents Chemother. 1998;42:236–240. doi: 10.1128/aac.42.2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinstein M P, Mirrett S, Vanpelt L, Mckinnon M, Zimmer B L, Kloos W, Reller L B. Clinical importance of identifying coagulase-negative staphylococci isolated from blood cultures—evaluation of microscan rapid and dried overnight gram-positive panels versus a conventional reference method. J Clin Microbiol. 1998;36:2089–2092. doi: 10.1128/jcm.36.7.2089-2092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yonezawa M, Takahata M, Banzawa-Futakuchi N, Matsubara N, Watanabe Y, Narita H, Matsunaga T, Igarashi H, Kawahara M, Onodera S. DNA gyrase gyrA mutations in quinolone-resistant clinical isolates of Staphylococcus haemolyticus. Antimicrob Agents Chemother. 1996;40:1065–1066. doi: 10.1128/aac.40.4.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]