Dear Editor,
Mitochondrial DNA (mtDNA) is encapsulated by the organelle membrane1 forming the barrier for the access of CRISPR-based gene-editing tools to the mtDNA. Furthermore, mitochondrial genome lacks similar repair systems for the protection of nuclear genome from DNA damage after induction of double-strand break by programmable nuclease such as ZFN, TALEN etc., which results in the elimination of target mtDNA2–5 instead of mutation installation on mtDNA in contrast to the outcome of indel formation on nuclear DNA6. Recent studies positioned DdCBE as a promising technology to install targeted mutations or introduce transmissible mutations of base conversion in mammalian mtDNA7–10 rather than eliminate them with previous ZF- or TALE-based nuclease2–5. Thus, DdCBE has the potential to model mitochondrial disease mutations, correct pathogenic variants, and expand our knowledge of mitochondrial biology. However, it is worth mentioning that these studies have found that DdCBE can cause low-frequent off-target events on mtDNA9,10. As indicated, the off-target profile of DdCBE remained to be comprehensively investigated by additional research for their systematic effect on mtDNA as well as nuclear genome.
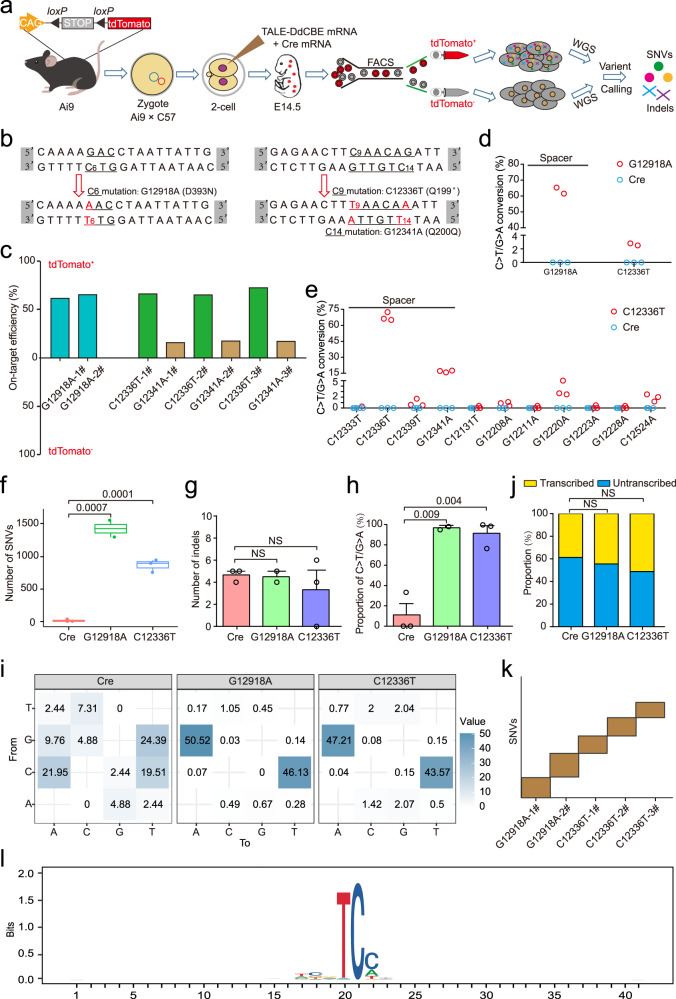
In the current study, we performed the GOTI (genome-wide off-target analysis by two-cell embryo injection) method previously developed by us11,12 to evaluate the off-target effect of DdCBE on both mtDNA and nuclear DNA modification. At first, we in vitro transcribed two pairs of DdCBE mRNA targeting the mtDNA ND5 gene (G12918 and C12336) and injected them with Cre mRNA into one blastomere of two-cell embryos derived from Ai9 background leaving another blastomere uninjected (Fig. 1a and Supplementary Fig. S1a). Thereby, Cre-activated tdTomato fluorescence will distinguish DdCBE-injected cells from non-fluorescent uninjected cells derived from the same two-cell Ai9 embryos. At 14.5 days after transferring injected 2-cell embryos into surrogate female mice, we collected E14.5 embryos to sort tdTomato+ and tdTomato– cells for genotyping base conversion outcomes on two targeted loci of mtDNA as well as whole-genome sequencing (WGS) analysis (Fig. 1a). Sanger sequencing and Targeted deep sequencing results showed efficient mtDNA editing by DdCBE with m.G12918A and m.C12336T conversion rate of up to 46% in both non-sorted and sorted tdTomato+ tissues, contrasting with the only wild-type alleles detected in tdTomato– tissues (Supplementary Fig. S1b, c and Table S1). For G12918 and C12336-targeting DdCBE, we also observed higher editing rate of up to 72% for sorted cells than unsorted ones on the basis of WGS (Fig. 1b, c). Furthermore, there are several unintended and sequence-independent C-to-T editing events identified by WGS analysis with lower than 5% frequency centered around m.G12918A or m.C12336T on mtDNA (Supplementary Fig. S2a). For all unintended editing events, some fall within spacer sequence between two recognition sequences for the TALE pair while several reside out of the spacer sequence, indicating low-frequent and sequence-independent off-target editing on mtDNA for DdCBE (Fig. 1d, e). We further performed sequence enrichment analysis on off-target sites and revealed a cognate 5′-TC-3′ motif preference of DdCBE for non-specific sequences editing (Supplementary Fig. S2b). These results collectively demonstrate that DdCBE pairs can produce efficient on-target base editing and low-frequent non-specific editing near the target loci of mtDNA.
Fig. 1. Off-target analysis of DdCBE for mitochondrial and nuclear genome editing with GOTI.
a GOTI workflow for analyzing off-target profile of DdCBE. b The DdCBE target for generating the m.G12918A point mutation (D393N), m.C12336T nonsense mutation (Q199stop), and m.G12341A silent mutation (Q200Q) in the ND5 protein. Translation triplets are underlined and target sequences with possible editing loci are shown in red. c On-target efficiency of ND5-DdCBE (m.G12918A and m.C12336T) for tdTomato+ and tdTomato– cells on the basis of WGS. Distribution pattern of off-target sites in m.G12918A (d) and m.C12336T (e) E14.5 fetuses (red dots) with Cre fetuses as control (blue dots). Spacer represents region between recognition sequences of the TALE pair. Each dotted box indicates a single off-target event. f Comparison of the total number of identified off-target SNVs in Cre and ND5-DdCBE (m.G12918A and m.C12336T) injected groups by WGS. g Number of indels identified in Cre and ND5-DdCBE (m.G12918A and m.C12336T) injected groups by WGS. h Proportion of C·G to T·A mutations among all identified SNVs for Cre and ND5-DdCBE (m.G12918A and m.C12336T) injected groups. i Distribution of mutation types. The number in each cell indicates the proportion of a certain type of mutation among all mutations. j The distribution of off-target SNVs in the transcribed and untranscribed regions. k SNVs identified from all DdCBE-edited samples did not overlap, suggesting that off-targets on the nuclear genome were mainly caused by the sequence-independent activity of DdCBE. l Sequence logos generated from off-target sequences with C·G to T·A conversions by ND5-DdCBE in nuclear genome. Bits reflect sequence conservation at a given position. Data are presented as the means ± SEM. P values were evaluated with the unpaired Student’s t-test (two-tailed).
To evaluate the potential influence of DdCBE activity on nuclear DNA in mammalian cells, we performed WGS for both edited cells marked by tdTomato fluorescence and unedited cells derived from the same embryo created as the previous GOTI protocol11,12. Unexpectedly, we identified about 1500 and 1000 single-nucleotide variants (SNVs) with significant confidence as potential off-target editing events for G12918 and C12336-targeting DdCBE in edited cells using unedited cells as SNV calling references (Fig. 1f). To exclude the possibility of SNVs caused by Cre injection, we also analyzed the SNV number for Cre-only samples without DdCBE injection and found less than 20 SNVs in Cre-only samples (Fig. 1f). In addition to SNVs analysis, indel (insertion and deletion) frequency was further checked in WGS datasets with less than 5 indels detected for both Cre-only and Cre-plus-DdCBE samples (Fig. 1g). Our results showed that DdCBE could significantly increase SNVs formation in edited cells while exhibit undetectable effect on indel frequency (Fig. 1f, g). Since SNVs identified by GOTI showed random distribution in nuclear genome11,12, we analyzed all SNVs identified in G12918 and C12336-targeting samples to find significantly strong enrichment of C-to-T/G-to-A conversion among five different base conversion outcomes, consistent with cytosine deaminase activity of DdCBE (Fig. 1h, i and Supplementary Fig. S3a). In contrast, SNVs identified in Cre-only samples exhibited no enrichment for any type of base conversion, corroborating off-target activity of DdCBE on nuclear DNA (Fig. 1h, i and Supplementary Fig. S3a). By examining SNVs distribution within transcribed and untranscribed regions, we found non-significant correlation of DdCBE-affected SNVs with genomic transcription features similar to the trend found in Cre-only samples (Fig. 1j). Moreover, off-target SNVs caused by DdCBE exhibited even and random distribution in the entire genome as SNVs identified in Cre-only samples (Supplementary Fig. S3b). In addition, there are no overlapping SNVs among all DdCBE-edited samples, suggesting sequence-independent off-target activity of DdCBE (Fig. 1k). Besides, we also checked the potential sequence-dependent off-target bias towards nuclear mitochondrial DNA segments (NUMTs) or other sequence similar to ND5 target loci. Our analysis revealed neither off-target editing nor efficiency bias towards similar sequences or NUMTs contrasted to the majority of off-target sequences (Supplementary Fig. S4). Lastly, we put together off-target sequences identified in all DdCBE-edited samples to find again the enrichment of 5′-TC-3′ motif for DdCBE-affected SNVs as the results on mitochondrial DNA (Fig. 1l). To verify SNVs identified in DdCBE-edited samples, we performed Sanger sequencing on random selected regions and found all of regions were edited in tdTomato+ cells while remained unedited in tdTomato– cells (Supplementary Fig. S5). Overall, our GOTI results demonstrated notable sequence-independent off-target activity of DdCBE on nuclear DNA of edited tissue derived from DdCBE-injected blastomere.
In summary, we showed for the first time that DdCBE cause thousands of off-target SNVs enriched for C-to-T/G-to-A conversion in the entire nuclear genome, which is two times the SNV number resulted from low-fidelity base editor BE3. Unlike the substrate preference of cytosine deaminase APOBEC1 in BE3 protein for ssDNA, DddAtox in DdCBE protein is a unique type of cytosine deaminase with dsDNA as substrate7. It might explain more off-target SNVs observed for DdCBE than BE3. Since DdCBE was designed to localize in mitochondria guided by mitochondrial targeting signal (MTS) in N-terminal region of DdCBE protein, our results imply that MTS seems to fail in blocking the entry of DdCBE into nuclear in mammalian cells. It would be interesting in the future to examine whether extra or different MTS could reduce off-target editing of DdCBE on nuclear DNA. In addition, expressing DdCBE in embryos may result in different results from when editing is done in differentiated cells. Therefore, it would be necessary when the technique available in the future to clarify different propensity of DdCBE off-target activity in somatic cells than embryonic cells. Taken together, our finding on off-target activity of DdCBE towards nuclear genome necessitate the strong need to optimize DdCBE for specific base editing on mtDNA especially before being used for treating mitochondrial diseases.
Supplementary information
Acknowledgements
We thank H. Wu and L. Quan from FACS facility of the Center for Excellence in Brain Science and Intelligence Technology, Chinese Academy of Sciences for their technical support. This work was supported by the Chinese National Science and Technology major project R&D Program of China (2018YFC2000101), Strategic Priority Research Program of Chinese Academy of Science (XDB32060000), the National Natural Science Foundation of China (31871502, 31901047, 31925016, 91957122, 82021001, 31922048), Basic Frontier Scientific Research Program of Chinese Academy of Sciences From 0 to 1 original innovation project (ZDBS-LYSM001), Shanghai Municipal Science and Technology Major Project (2018SHZDZX05), Shanghai City Committee of Science and Technology Project (18411953700, 18JC1410100, 19XD1424400 and 19YF1455100), Applied Basic Research Foundation of Guangdong Province (2020A1515110188 to Z.L.), International Partnership Program of Chinese Academy of Sciences (153D31KYSB20170059), Basic and Central Public-interest Scientific Institution Basal Research Fund (to E.Z).
Author contributions
Y.W., E.Z., C.X., H.Y., and M.Z. jointly conceived the project. Y.W., E.Z., C.X., H.Y., and M.Z. designed and conducted experiments. Y.W., C.X., Z.L., K.X., L.X., and D.L. assisted with plasmids construction, PCR, FACS, and other molecular experiments. Y.W., Z.L., and Z.Z. assisted with embryonic experiments. H.F. performed bioinformatics analysis. E.Z., H.Y., C.X., and M.Z. co-supervised the whole project. Y.W., E.Z., H.Y., and C.X. wrote the manuscript.
Data availability
The whole-genome sequencing data have been deposited to the NCBI Sequence Read Archive (SRA) database (accession ID, PRJNA786071).
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yinghui Wei, Zhifang Li, Kui Xu, Hu Feng, Long Xie
Contributor Information
Meiling Zhang, Email: zhml1119@sina.com.
Chunlong Xu, Email: xucl@bsbii.cn.
Hui Yang, Email: huiyang@ion.ac.cn.
Erwei Zuo, Email: zuoerwei@caas.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41421-022-00391-5.
References
- 1.Hopper RK, et al. Mitochondrial matrix phosphoproteome: effect of extra mitochondrial calcium. Biochemistry. 2006;45:2524–2536. doi: 10.1021/bi052475e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gammage PA, Rorbach J, Vincent AI, Rebar EJ, Minczuk M. Mitochondrially targeted ZFNs for selective degradation of pathogenic mitochondrial genomes bearing large-scale deletions or point mutations. EMBO Mol. Med. 2014;6:458–466. doi: 10.1002/emmm.201303672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bacman SR, Williams SL, Pinto M, Peralta S, Moraes CT. Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nat. Med. 2013;19:1111–1113. doi: 10.1038/nm.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reddy P, et al. Selective elimination of mitochondrial mutations in the germline by genome editing. Cell. 2015;161:459–469. doi: 10.1016/j.cell.2015.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gammage PA, et al. Genome editing in mitochondria corrects a pathogenic mtDNA mutation in vivo. Nat. Med. 2018;24:1691–1695. doi: 10.1038/s41591-018-0165-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adli M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018;9:1911. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mok BY, et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature. 2020;583:631–637. doi: 10.1038/s41586-020-2477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee H, et al. Mitochondrial DNA editing in mice with DddA-TALE fusion deaminases. Nat. Commun. 2021;12:1190. doi: 10.1038/s41467-021-21464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo J, et al. Precision modeling of mitochondrial diseases in zebrafish via DdCBE-mediated mtDNA base editing. Cell Discov. 2021;7:78. doi: 10.1038/s41421-021-00307-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi X, et al. Precision modeling of mitochondrial disease in rats via DdCBE-mediated mtDNA editing. Cell Discov. 2021;7:95. doi: 10.1038/s41421-021-00325-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuo E, et al. GOTI, a method to identify genome-wide off-target effects of genome editing in mouse embryos. Nat. Protoc. 2020;15:3009–3029. doi: 10.1038/s41596-020-0361-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuo E, et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019;364:289–292. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole-genome sequencing data have been deposited to the NCBI Sequence Read Archive (SRA) database (accession ID, PRJNA786071).