Abstract
With the improvement of people's living standards and rice trade worldwide, the demand for high-quality rice is increasing. Therefore, breeding high quality rice is critical to meet the market demand. However, progress in improving rice grain quality lags far behind that of rice yield. This might be because of the complexity of rice grain quality research, and the lack of consensus definition and evaluation standards for high quality rice. In general, the main components of rice grain quality are milling quality (MQ), appearance quality (AQ), eating and cooking quality (ECQ), and nutritional quality (NQ). Importantly, all these quality traits are determined directly or indirectly by the structure and composition of the rice seeds. Structurally, rice seeds mainly comprise the spikelet hull, seed coat, aleurone layer, embryo, and endosperm. Among them, the size of spikelet hull is the key determinant of rice grain size, which usually affects rice AQ, MQ, and ECQ. The endosperm, mainly composed of starch and protein, is the major edible part of the rice seed. Therefore, the content, constitution, and physicochemical properties of starch and protein are crucial for multiple rice grain quality traits. Moreover, the other substances, such as lipids, minerals, vitamins, and phytochemicals, included in different parts of the rice seed, also contribute significantly to rice grain quality, especially the NQ. Rice seed growth and development are precisely controlled by many genes; therefore, cloning and dissecting these quality-related genes will enhance our knowledge of rice grain quality and will assist with the breeding of high quality rice. This review focuses on summarizing the recent progress on cloning key genes and their functions in regulating rice seed structure and composition, and their corresponding contributions to rice grain quality. This information will facilitate and advance future high quality rice breeding programs.
Keywords: Rice grain quality, Seed structure, Seed size, Grain component, Starch, Protein, Gene cloning, Molecular function, Regulatory network
Introduction
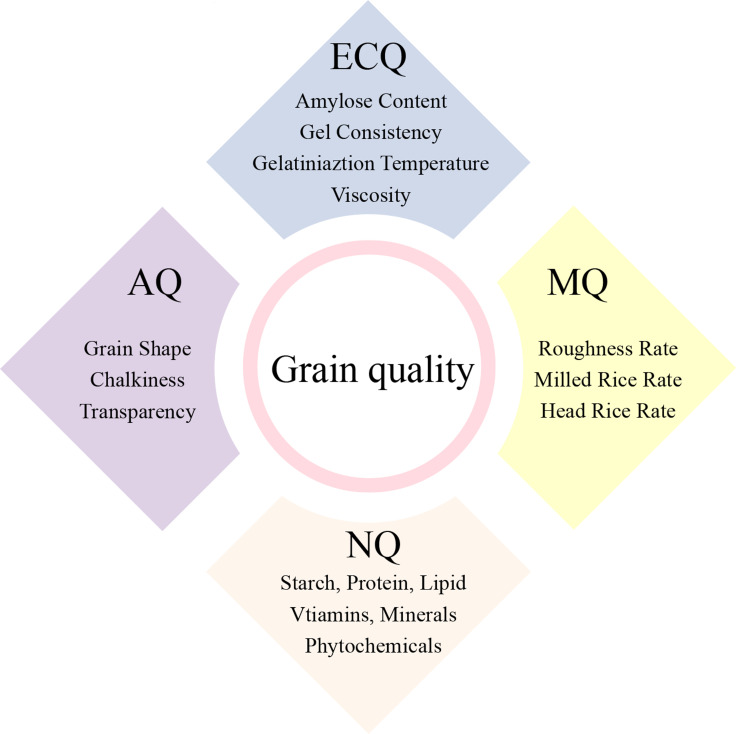
Rice is the major food crop for more than half of the world's population. Breeding elite rice with high yield and quality are the major goals of crop geneticists and rice breeders. In recent decades, benefiting from the discovery and application of "green revolution gene" and "heterosis", rice yield has improved greatly (Evenson and Gollin 2003; Pingali 2012; Chen et al. 2019; Liu et al. 2020; Wu et al. 2020). However, the progress of rice quality-related research and breeding practices lag far behind that of rice yield. This might reflect the emphasis placed on rice yield in the past, the complexity of rice quality studies, and the lack of consensus definitions and evaluation standards of rice quality. As people's living standards and the worldwide rice trade improve, the demand for high-quality rice is increasing. Therefore, breeding high quality rice is critical to meet market demand. A series of physical and chemical indexes are used for the comprehensive evaluation of rice quality during the rice processing and cooking. In general, rice quality is divided into four main sections: milling quality (MQ), appearance quality (AQ), nutritional quality (NQ), and eating and cooking quality (ECQ) (Fig. 1) (Bao 2014). Rice MQ refers to the integrity of rice during processing, including the roughness rate, milled rice rate, and head rice rate. AQ usually includes grain shape, chalkiness, transparency, and other indicators. Rice NQ is influenced by the quantity and quality of starch, protein, vitamins, minerals, and other phytochemicals that are beneficial to human health. ECQ mainly reflects the characteristics and palatability of cooked rice. Amylose content (AC), gel consistency (GC), gelatinization temperature (GT), and the results gained using a rapid viscosity analyzer (RVA) are usually used as indirect indicators to estimate rice ECQ (Zhang et al. 2016).
Fig. 1.
The scope of rice quality includes the following four parts, appearance quality (AQ), milling quality (MQ), nutritional quality (NQ), eating and cooking quality (ECQ)
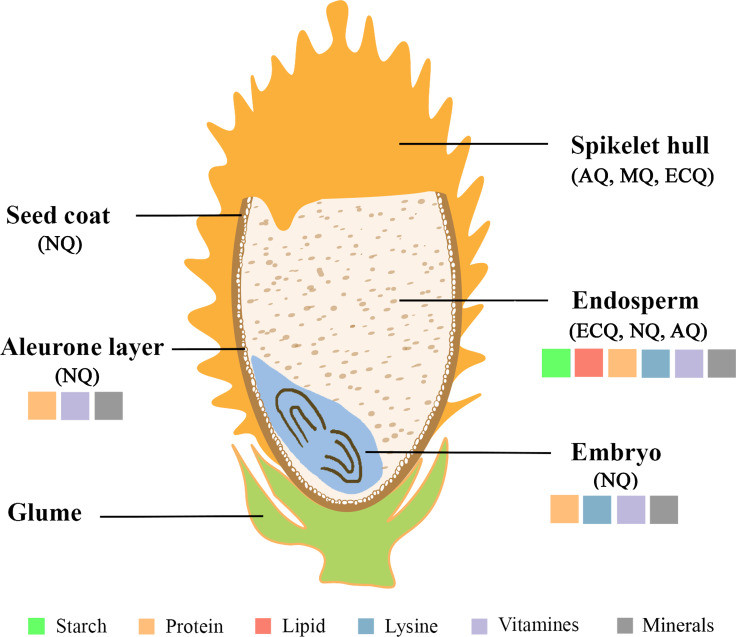
In general, rice grain quality should be closely correlated with rice seed characteristics. In brief, the rice seed structure mainly includes the spikelet hull, seed coat, aleurone layer, embryo, and endosperm (Fig. 2). The size, composition, and quality of these seed structures affect rice quality markedly. For example, in most cases, the size of the spikelet hull determines the grain size. Grain size is an important rice agronomic trait, which is not only a key element of rice yield, but also is a direct index for rice quality. Rice grain size includes grain length, width, thickness, and the ratio of length to width, which usually affects rice AQ, MQ, and ECQ. Recently, a series of grain size-related genes have been cloned successfully and functionally dissected (Li et al. 2019). The endosperm is the major edible part of rice seed. Therefore, the composition and proportion of endosperm components are the most important determinants of rice quality. In general, the rice endosperm includes starch, storage protein, lipids, minerals, and other trace elements. Among them, starch and protein are two major components of rice endosperm, accounting for approximately 80% and 10%, respectively, of the weight of milled rice (Wang et al. 2020a). As the largest constituent of rice endosperm, starch consists of two glucose polymers, amylose and amylopectin. The amylose content (AC) and amylopectin structure correlate tightly with rice ECQ (Leng et al. 2014). In addition, the arrangement of starch granules will affect rice AQ. For example, a looser arrangement of starch grains will form cavities inside or between the starch granules, thus decreasing the transparency of the rice grain and resulting in the so-called opaque or chalky endosperm (Zhang et al. 2017). As for proteins, the second largest component of rice endosperm, their content and amino acid composition will affect almost all aspects of rice quality (Duan and Sun 2005). Moreover, other substances in the endosperm, including lipids, free amino acids, minerals, vitamins, and other phytochemicals, albeit present in lower in quantities, are crucial for rice NQ. The other structures of rice seeds, including the embryo, seed coat, and aleurone layer, are rich in protein, fat, vitamins, and minerals. For example, functional metabolites, such as gamma aminobutyric acid (GABA), are enriched in embryo and aleurone layer compared with that in the endosperm (Pereira et al. 2021).
Fig. 2.
Overview of the main structure and components based on the longitudinal section of mature rice seeds. AQ, appearance quality; MQ, milling quality; NQ, nutritional quality; ECQ, eating and cooking quality. Not drawn in proportion
Each structure of the rice seed correlates with various rice quality traits, and rice grain development is directly controlled by many genes; hence, cloning and dissecting these quality-genes will aid the breeding of high quality rice. Therefore, this review focused on summarizing the recently cloned key genes involved in regulating rice seed structure and composition, and their roles in modulation of rice qualities (Table 1), which will facilitate and advance future high quality rice breeding programs.
Table 1.
List of genes involved in controlling rice seed structures and grain qualities
| Gene ID | Gene name | Protein category | Effect the quality | References |
|---|---|---|---|---|
| Spikelet hull & grain size & grain quality | ||||
| G protein pathway | ||||
| Os03g0407400 | GS3 | Gγ subunit | Grain length (AQ) |
Fan et al. (2006) Mao et al. (2010) Sun et al. (2018a) |
| Os09g0441900 | DEP1/qPE9 | Gγ subunit | Grain length (AQ) |
Zhou et al. (2009) Huang et al. (2009) |
| Os03g0635100 | RGG1 | Gγ subunit | Grain length (AQ) | Tao et al. (2020) |
| Os02g0137800 | RGG2 | Gγ subunit | Grain length (AQ) | Miao et al. (2019) |
| Os08g0456600 | GGC2 | Gγ subunit | Grain length (AQ) | Sun et al. (2018a) |
| Os05g0333200 | D1/RGA1 | Gα subunit | Grain length (AQ) |
Fujisawa et al. (1999) Ashikari et al. (1999) Sun et al. (2018a) |
| Os03g0669200 | RGB1 | Gβ subunit | Grain length (AQ) |
Utsunomiya et al. (2011) Zhang et al. (2021b) |
| The ubiquitin–proteasome pathway | ||||
| Os02g0244100 | GW2 | E3 ubiquitin ligase | Grain width (AQ) | Song et al. (2007) |
| Os02g0244300 | LG1/OsUBP15 | Ubiquitin specific protease | Grain width (AQ) | Shi et al. (2019) |
| Os03g0232600 | TUD1 | U-box E3 ubiquitin ligase | Grain length (AQ) | Hu et al. (2013) |
| Os02g0512400 | OsGRX8/WG1 | CC-type glutaredoxin | Grain length, grain width (AQ) | Hao et al. (2021) |
| Os06g0265400 | OsbZIP47 | bZIP transcription factor | Grain width (AQ) | Hao et al. (2021) |
| Os08g0537800 | WTG1/OsOTUB1 | Deubiquitinating enzyme | Grain width, grain thickness (AQ) | Huang et al. (2017) |
| Os06g0650300 | GW6a/OslHAT1 | GNAT-like Protein | Grain length (AQ) |
Song et al. (2015) Gao et al. (2021) |
| Os05g0551000 | CLG1/OsHRZ2 | RING E3 ubiquitin ligase | Grain length (AQ) | Yang et al. (2021b) |
| Mitogen-activated protein kinase (MAPK) signaling | ||||
| Os02g0787300 | OsMKK4/SMG1 | Mitogen-activated protein kinase kinase | Grain length, grain width (AQ) |
Duan et al. (2014) Guo et al. (2018) |
| Os06g0154500 | OsMAPK6/DSG1 | Mitogen activated protein kinase | Grain length, grain width (AQ) | Liu et al. (2015b) |
| Os04g0559800 | SMG2/OsMKKK10 | Mitogen activated protein kinase kinase kinase | Grain length, grain width (AQ) | Xu et al. (2018) |
| Os01g0699500 | OsMKKK70 | Mitogen activated protein kinase kinase kinase | Grain length, grain width (AQ) | Liu et al. (2021) |
| Os01g0699600 | OsMKKK62 | Mitogen activated protein kinase kinase kinase | Grain length, grain width (AQ) | Liu et al. (2021) |
| Os05g0115800 | OsMKP1/GSN1 | MAPK phosphatase | Grain length, grain width (AQ) |
Guo et al. (2018) Xu et al. (2018) |
| Phytohormone perception and homeostasis | ||||
| Brassinosteroids | ||||
| Os03g0602300 | BRD1/OsDWARF | Brassinosteroid biosynthetic enzyme | Grain length (AQ) |
Hong et al. (2002) Mori et al. (2002) |
| Os10g0397400 | BRD2 | Brassinosteroid biosynthetic enzyme | Grain length (AQ) | Hong et al. (2005) |
| Os04g0469800 | D11/ CYP724B1 | Brassinosteroid biosynthetic enzyme | Grain length (AQ) |
Tanabe et al. (2005) Zhu et al. (2015) Wu et al. (2016b) Zhou et al. (2017c) |
| Os05g0187500 | GW5/GSE5/qGW5 | Calmodulin binding protein |
Grain width, chalkiness (AQ) Milled rice rate (MQ) |
Weng et al. (2008) Shomura et al. (2008) Liu et al. (2017) Duan et al. (2017) |
| Os05g0207500 | OsGSK2 | GSK3/SHAGGY-like kinase | Grain length (AQ) | Tong et al. (2012) |
| Os02g0236200 | OsGSK3 | GSK3/SHAGGY-like kinase | Grain length (AQ) | Gao et al. (2019) |
| Os07g0580500 | OsBZR1 | Transcription factor in BR pathway | Grain length, grain width, grain thickness (AQ) | Zhu et al. (2015) |
| Os02g0517531 | LARGE1/ OML4 | MEI2-like protein | Grain length, grain width (AQ) | Lyu et al. (2020) |
| Os06g0127800 | DLT/D62/GS6 | GRAS family protein | Grain width (AQ) |
Tong et al. (2009) Sun et al. (2013) |
| Os05g0158500 | GS5 | Putative serine carboxypeptidase | Grain width (AQ) |
Li et al. (2011) Xu et al. (2015) |
| Os05g0343400 | OsWRKY53 | WRKY transcription factor | Grain length, grain width (AQ) | Tian et al. (2017) |
| Os08g0174700 | OsBAK1 | BRI1-associated receptor kinase | Grain length, grain width (AQ) | Li et al. (2009) |
| Os03g0646900 | GL3.1/GL3-1/qGL3/OsPPKL1 | Ser/Thr phosphatase | Grain width (AQ) |
Hu et al. (2012) Qi et al. (2012) Zhang et al. (2012a) Gao et al. (2019) |
| Os04g0674500 | OsmiR396d | MicroRNA | Grain length, grain width (AQ) | Miao et al. (2020) |
| Os02g0701300 | GS2/GL2/OsGRF4/PT2/ LGS1 | Growth-regulating factor | Grain length, grain width (AQ) |
Che et al. (2015) Duan et al. (2015) Hu et al. (2015) Sun et al. (2016) Li et al. (2016) Sun et al. (2016) |
| Os05g0458600 | OsLAC | Laccase-like protein | Grain length, grain width (AQ) |
Zhang et al. (2013) Zhong et al. (2020) |
| Os02g0169400 | OsAGO17 | Argonaute protein | Grain length, grain width (AQ) | Zhong et al. (2020) |
| Os09g0448500 | GS9 | Transcriptional activator | Grain width, chalkiness (AQ) | Zhao et al. (2018) |
| Os01g0718300 | OsBRI1/D61 | BR receptor kinase | Grain length, grain width (AQ) | Yamamuro et al. (2000) |
| Os01g0226700 | OFP1 | OVATE family protein | Grain length, grain width (AQ) | Xiao et al. (2017) |
| Os01g0732300 | OFP3 | OVATE family protein | Grain length, grain width (AQ) | Xiao et al. (2020) |
| Os01g0864000 | OFP8 | OVATE family protein | Grain length, grain width (AQ) | Yang et al. (2016a, b) |
| Os05g0324600 | OFP19 | OVATE family protein | Grain length (AQ) | Yang et al. (2018a) |
| Os05g0477200 | OFP22 | OVATE family protein | Grain length, grain width (AQ) | Chen et al. (2021) |
| Os10g0515400 | GW10 | Cytochrome P450 subfamily protein | Grain length, grain width (AQ) | Zhan et al. (2021) |
| Os07g0175100 | POW1 | Homeodomain-like protein | Grain length, grain width (AQ) | Zhang et al. (2021c) |
| Auxin | ||||
| Os06g0623700 | TGW6 | Protein with IAA-glucose hydrolase activity | Grain length, grain width (AQ) |
Ishimaru et al. (2013) Akabane et al. (2021) Kabir and Nonhebel (2021) |
| Os03g0175800 | BG1 | Positive regulator of auxin response and transport | Grain length, grain width (AQ) | Liu et al. (2015a) |
| Os03g0841800 | qTGW3/GL3.3/qGL6 | GSK3/SHAGGY-like kinase | Grain length, grain width, grain thickness (AQ) |
Hu et al. (2018) Xia et al. (2018) Ying et al. (2018) |
| Os02g0164900 | OsARF6 | Auxin response factor | Grain length (AQ) | Qiao et al. (2021) |
| Os05g0447200 | OsAUX3/qGL5 | Auxin influx carrier | Grain length (AQ) | Qiao et al. (2021) |
| GA | ||||
| Os06g0266800 | OsGSR1/GW6/OsGASR7 | GA-stimulated protein | Grain length, grain width (AQ) | Shi et al. (2020) |
| Cytokinin | ||||
| Os01g0680200 | OsPUP4/ BG3 | Purine permease | Grain length, grain width, grain thickness (AQ) |
Xiao et al. (2019) Yin et al. (2020) |
| Os04g0615700 | OsAGO2 | AGO family protein | Grain length (AQ) | Yin et al. (2020) |
| Transcriptional regulatory factors | ||||
| SQUAMOSA promoter binding protein-like (SPL) family | ||||
| Os07g0505200 | OsSPL13/GLW7 | Squamosa promoter-binding-like protein | Grain length, grain width (AQ) |
Si et al. (2016) Segami et al. (2017) |
| Os11g0247300 | SRS5/TID1 | Alpha-tubulin protein | Grain length, grain width (AQ) |
Sunohara et al. (2009) Segami et al. (2012) Segami et al. (2017) |
| Os08g0531600 | qGW8/OsSPL16 | Squamosa promoter binding-like protein |
AC, GC (ECQ)) Protein content (NQ) |
Wang et al. (2012) Wang et al. (2015a) |
| Os07g0603300 | GL7/GW7 | TRM-containing protein |
Grain length, grain width, chalkiness (AQ) AC, GC (ECQ)) Protein content (NQ) |
Wang et al. (2015c) |
| APETALA2-type (AP2) transcription factors | ||||
| Os05g0389000 | SMOS1/SHB | AP2-type transcription factor | Grain length, grain width (AQ) | Aya et al. (2014) |
| Os07g0235800 | SSH1 | AP2-like transcription factor | Grain length, grain width (AQ) | Jiang et al. (2019) |
| Basic helix-loop-helix (bHLH) family | ||||
| Os04g0350700 | An-1 | Basic helix-loop-helix protein | Grain length (AQ) | Luo et al. (2013) |
| Os03g0171300 | PGL1 | Atypical non-DNA-binding bHLH protein | Grain length (AQ) | Heang and Sassa (2012) |
| Other transcription factors | ||||
| Os06g0666100 | GL6/SG6 | PLATZ transcription factor | Grain length (AQ) |
Wang et al. (2019) Zhou and Xue (2020) |
| Os03g0215400 | OsMADS1/qLGY3 | MADS-domain transcription factor | Grain length (AQ) |
Yu et al. (2018) |
| Os03g0333200 | FLR1 | Receptor-like kinase | Grain width, chalkiness (AQ) | Wang et al. (2021) |
| Os01g0769700 | FLR2 | Receptor-like kinase | Grain length (AQ) | Wang et al. (2021) |
| Other functional proteins | ||||
| Os08g0485500 | GAD1 | Secretory signal peptide | Grain length (AQ) | Jin et al. (2016) |
| ORGLA04G0254300 | GL4 | Myb like protein similar to SH4/SHA1 | Grain length (AQ) | Wu et al. (2017) |
| Os07g0214300 | RAG2 | 16-kDa α-amylase/trypsin inhibitor |
Grain length, grain width (AQ) Protein content, lipid content (NQ) |
Zhou et al. (2017b) |
| Endosperm components and grain quality | ||||
| Starch | ||||
| Os06g0133000 | Wx | Granule-bound starch synthase |
AC, GC, GT (ECQ) Transparency, chalkiness (AQ) Digestion, RS (NQ) |
Wang et al. (2010) Zhang et al. (2019a) Huang et al. (2020) Zhang et al. (2021a) Zhou et al. (2021a) |
| Os02g0744700 | OsSSIIb/SSII-2 | Soluble starch synthase |
AC, GC, GT (ECQ) Transparency (AQ) Protein content (NQ) |
Xu et al. (2020) |
| Os06g0229800 | OsSSIIa/SSII-3/ALK | Soluble starch synthase | GT (ECQ) |
Umemoto et al. (2002) Gao et al. (2003) Chen et al. (2020) Zhang et al. (2020) |
| Os08g0191433 | OsSSIIIa/FLO5 | Soluble starch synthase |
Chalkiness (AQ) AC, GC (ECQ) Digestion, RS (NQ) |
Fujita et al. (2007) Zhou et al. (2016) |
| Os02g0528200 | OsSBEIIb/SBE3 | Starch branching enzyme |
Chalkiness (AQ) AC, GC (ECQ) Digestion, RS (NQ) |
Zhu et al. (2012) Baysal et al. (2020) Miura et al. (2021) |
| Os07g0182000 | OsbZIP58/ RISBZ1 | bZIP transcription factor |
Chalkiness (AQ) AC (ECQ) |
Yamamoto et al. (2006) Kawakatsu et al. (2009) Wang et al. (2013) |
| Os02g0725900 | OsNF-YB1 | Component of the NF-Y/HAP transcription factor complex |
Grain length, grain width, chalkiness (AQ) AC (ECQ) Protein content, lipid content (NQ) |
Bello et al. (2019) Xu et al. (2021) |
| Os10g0191900 | OsNF-YC12/OsNF-YC11 | Nuclear transcription factor Y subunit C |
Grain length, grain width, chalkiness (AQ) AC (ECQ) Protein content, lipid content (NQ) |
Bello et al. (2019) Xiong et al. (2019) |
| Os12g0189500 | OsYUC11 | Flavin-containing monooxygenase | Grain length, grain width, chalkiness (AQ) Protein content, lipid content (NQ) | Xu et al. (2021) |
| Os02g0682200 | OsMADS6/AFG1 | MADS-box protein |
Grain length, grain width (AQ) AC, GC (ECQ) Protein content (NQ) |
Zhang et al. (2010) |
| Os02g0170300 | OsMADS29 | MADS-box protein |
Shrunken seeds (AQ) AC (ECQ) |
Yin and Xue (2012) Nayar et al. (2013) |
| Os08g0531700 | OsMADS7/OsMADS45 | MADS-box protein | AC (ECQ) | Zhang et al. (2018a) |
| Os01g0104500 | OsNAC20/ONAC020 | NAC transcription factor |
Grain width, grain thickness (AQ) Starch content (ECQ) Protein content (NQ) |
Wang et al. (2020a) |
| Os01g0393100 | OsNAC26/ONAC026 | NAC transcription factor |
Grain width, grain thickness (AQ) Starch content (ECQ) Protein content (NQ) |
Wang et al. (2020a) |
| Os03g0686900 | FLO6 | CBM48 domain-containing protein |
Transparency, chalkiness (AQ) Starch content (ECQ) Protein content, lipid content (NQ) |
Peng et al. (2014) |
| Os07g0688100 | FLO18 | PPR protein |
Transparency, chalkiness (AQ) AC, starch content (ECQ) Protein content, lipid content(NQ) |
Yu et al. (2021) |
| Os03g0168400 | FLO10 | PPR protein | Transparency (AQ) Protein content, lipid content (NQ) | Wu et al. (2019) |
| Os08g0290000 | FGR1 | Nuclear-localized PPR protein | Grain thickness, transparency (AQ) AC, starch content (ECQ) | Hao et al. (2019) |
| Os03g0728200 | FLO14 | Nuclear-localized PPR protein | Chalkiness (AQ) | Xue et al. (2019) |
| Os07g0181000 | OsPK2/PKpα1 | Plastidic pyruvate kinase | Starch content (ECQ) Protein content, lipid content (NQ) | Cai et al. (2018) |
| Protein | ||||
| Os10g0400200 | OsGluA2/qGPC-10 | Glutelin type-A2 precursor |
GC, starch content (ECQ) Protein content (NQ) |
Yang et al. (2019) |
| Os05g0499100 | Glb1 | 26 kDa α-gloubulin | Seed storage protein (NQ) | Wu et al. (1998) |
| Os01g0762500 | GluA1 | Glutelin type-A | Seed storage protein (NQ) | Qu et al. (2008) |
| Os02g0249800 | GluB1a | Glutelin type-B | Seed storage protein (NQ) | Wu et al. (1998) |
| Os02g0249900 | GluB1b | Glutelin type-B | Seed storage protein (NQ) | Wu et al. (1998) |
| Os02g0268300 | GluB4 | Glutelin type-B | Seed storage protein (NQ) | Qu and Takaiwa (2004) |
| Os02g0268100 | GluB5 | Glutelin type-B | Seed storage protein (NQ) | Qu et al. (2008) |
| Os02g0722400 | OsAAP10 | Amino acid permease |
AC, RVA (ECQ) Protein content (NQ) |
Wang et al. (2020b) |
| Os12g0631100 | OsRab5a/gpa1 | Small GTPase | Chalkiness (AQ) Protein content (NQ) | Ren et al. (2020) |
| Os06g0643000 | OsGPA5 | Rab5a effector |
Chalkiness (AQ) Protein content (NQ) |
Ren et al. (2020) |
| Os03g0835800 | GPA3 | Regulator of post-Golgi vesicular Traffic |
AC (ECQ) Protein content (NQ) |
Ren et al. (2014) |
| Os08g0127100 | OsHT/OsLHT1 | Amino acid transporter |
AC, GC (ECQ) Protein content (NQ) |
Guo et al. (2020a) Guo et al. (2020b) |
| Os02g0252400 | RPBF/OsDof3 | Prolamin box binding factor | Starch content (ECQ) Protein content, lipid content (NQ) | Kawakatsu et al. (2009) |
| Os07g0668600 | OsGZF1 | CCCH‐type zinc‐finger transcription factor | Glutelin content (NQ) | Chen et al. (2014) |
| Lipid/fat | ||||
| Os02g0716500 | OsFAD2-1 | Fatty acid desaturase | Lipid content (NQ) | Shi et al. (2012) |
| Os12g0104400 | OsFAD3 | Fatty acid desaturase | Lipid content (NQ) | Liu et al. (2012) |
| Os03g0369100 | OsLTP36 | Lipid transfer protein | Lipid content (NQ) | Wang et al. (2015b) |
| Os04g0436100 | OsACOT | Acyl-CoA thioesterase |
Grain length, grain width (AQ) Lipid content (NQ) |
Zhao et al. (2019) |
| Os08g0110700 | FSE1 | Phospholipase-like protein | Lipid content (NQ) | Long et al. (2018) |
| Os01g0172400 | OsPLDα1 | Phospholipase Dα |
Phytic acid content (NQ) AC,GT, setback viscosity, viscosity profiles (ECQ) |
Khan et al. (2019) Khan et al. (2020) |
| Lysine (Amino acids) | ||||
| Os01g0927900 | AK2 | Aspartate kinase | Lysine content (NQ) | Yang et al. (2020) |
| Os04g0574800 | DHPS | Dihydrodipicolinate synthase | Lysine content (NQ) |
Yang et al. (2020) Yang et al. (2021a) |
| Os02g0783700 | OsLKR/SDH | Lysine ketoglutarate reductase | Lysine content (NQ) | Yang et al. (2020) |
| Fe, Zn (Minerals) | ||||
| Os08g0207500 | OsZIP4 | Zinc-regulated transporter | Zinc content (NQ) | Ishimaru et al. (2007b) |
| Os05g0472700 | OsZIP5 | Zinc-regulated transporter | Zinc content (NQ) | Lee et al. (2010a) |
| Os07g0232800 | OsZIP8 | Zinc-regulated transporter | Zinc content (NQ) | Lee et al. (2010b) |
| Os03g0667500 | OsIRT1 | Iron-regulated transporter | Iron content (NQ) | Lee and An (2009) |
| Os03g0667300 | OsIRT2 | Iron-regulated transporter | Iron content (NQ) | Nakanishi et al. (2006) |
| Os11g0106700 | OsFER1 | Rice ferritin protein | Iron content (NQ) | Stein et al. (2009) |
| Os12g0106000 | OsFER2 | Rice ferritin protein | Iron content (NQ) | Stein et al. (2009) |
| Os03g0307300 | OsNAS1 | Nicotianamine synthase | Iron content (NQ) | Inoue et al. (2003) |
| Os03g0307200 | OsNAS2 | Nicotianamine synthase | Iron content (NQ) | Inoue et al. (2003) |
| Os07g0689600 | OsNAS3 | Nicotianamine synthase | Iron content (NQ) | Inoue et al. (2003) |
| Os04g0542200 | OsYSL9 | Probable metal-nicotianamine transporter | Iron content (NQ) | Senoura et al. (2017) |
| Os04g0463400 | OsVIT1 | Vacuolar membrane transporter | Iron content (NQ) | Zhang et al. (2012b) |
| Os09g0396900 | OsVIT2 | Vacuolar membrane transporter | Iron content (NQ) | Zhang et al. (2012b) |
| Phytochemicals | ||||
| Os08g0424500 | OsBadh2 | Betaine aldehyde dehydrogenasea | 2-AP content (ECQ) |
Chen et al. (2006) Chen et al. (2008) Kovach et al. (2009) Bradbury et al. (2010) Hui et al. (2021) |
| Embryo and grain quality | ||||
| Os07g0603700 | OsGE | Cytochrome P450 | Embryo size (NQ) | Nagasawa et al. (2013) |
| Os04g0447800 | OsGAD2 | Glutamate decarboxylase | GABA content (NQ) |
Akama and Takaiwa (2007) Akama et al. (2009) |
| Os02g0112900 | OsGABA-T | γ-aminobutyrate transaminase | GABA content (NQ) | Shimajiri et al. (2013) |
| Os03g0236200 | OsGAD3 | Glutamate decarboxylase | GABA content (NQ) | Akama et al. (2020) |
| Aleurone layer and grain quality | ||||
| Os05g0509700 | TA1/OsmtSSB1 | Mitochondrion-targeted single-stranded DNA binding protein | Thick aleurone (NQ) | Li et al. (2021) |
| Os01g0218032 | TA2-1/OsROS1 | DNA demethylase | Thick aleurone (NQ) | Liu et al. (2018a) |
| Seed coat and grain quality | ||||
| Os07g0211500 | Rc | bHLH protein | Anthocyanin content (NQ) | Sweeney et al. (2006) |
| Os01g0633500 | Rd/OsDFR | Dihydroflavonol reductase | Anthocyanin content (NQ) | Furukawa et al. (2007) |
| Os04g0557500 | Kala4/OsB2 | bHLH transcription factor | Anthocyanin content (NQ) | Oikawa et al. (2015) |
| Os06g0205100 | OsC1 | MYB transcriptional activator | Anthocyanin content (NQ) | Sun et al. (2018b) |
| Os02g0682500 | OsTTG1 | WD40 repeat protein | Anthocyanin content (NQ) | Yang et al. (2021c) |
Genes Regulating Spikelet Hull Development and Their Roles in Grain Qualities
In general, rice MQ correlates negatively with the grain length, grain length–width ratio, and grain length–thickness ratio, and positively with the grain width, grain thickness, and grain width–thickness ratio. Hence, reducing the length and increasing the width and thickness of rice grains are beneficial to improving rice MQ. Rice AQ is usually related to grain size, chalkiness, and transparency. The chalky grain percentage correlates positively with the 1000-grain weight, grain width, grain thickness, and grain width–thickness ratio. There is a significant positive correlation between the grain width and chalkiness. The grain filling rate of a wide grain is too fast, which leads to a looser arrangement of granules and the subsequent formation of chalkiness. Chalkiness is a negative element of rice quality, the higher the chalkiness, the poorer AQ, MQ, and ECQ of rice (Cheng et al. 2005; Yamakawa et al. 2007). For example, chalky rice breaks more easily during processing, resulting in less head rice and a decreased MQ. In contrast, smaller or slender seeds have less chalkiness because of the short distance for grain filling. Therefore, decreasing the rice grain width could promote rice AQ. Importantly, grain weight loss resulting from reduced grain width could be compensated by increasing the grain length. In conclusion, breeding elite rice with a slender grain shape is a practical strategy to improve rice quality without sacrificing its yield.
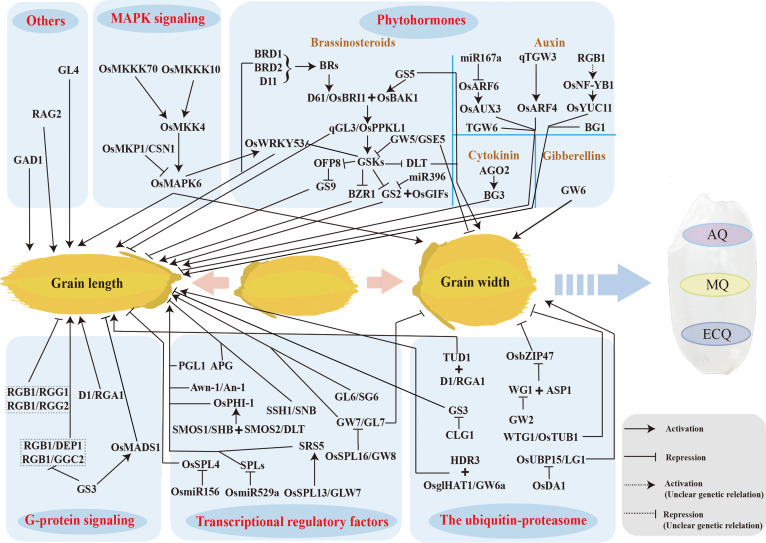
Therefore, the study of grain size has become a hot research topic among rice geneticists and breeders. At present, more than 400 quantitative trait loci (QTLs) linked to grain size have been mapped on all 12 chromosomes of rice, and over 80 grain size-related genes have been cloned (Huang et al. 2013; Zuo and Li 2014). Most importantly, several grain shape genes have been used in rice breeding practice, such as GS3 and DEP1/qPE9 (Huang et al. 2009; Zhou et al. 2009). This topic is well documented by several review papers (Li and Li 2016; Azizi et al. 2019; Li et al. 2019). Therefore, we have only summarized the functions and regulatory mechanisms of the key cloned genes related to grain size, especially the newly identified genes (Fig. 3).
Fig. 3.
The cloned genes and the major regulatory networks controlling rice size. Rice seed size is regulated by multiple signaling pathways, including G protein pathways, the ubiquitin–proteasome pathway, MAPK signaling pathways, phytohormones, and transcription regulatory factors. A dotted line indicates that the genetic relationship needs to be further verified. MAPK, Mitogen-Activated Protein Kinase; AQ, appearance quality; MQ, milling quality; ECQ, eating and cooking quality
G Protein Pathways
The heterotrimeric G-protein complex consists of Gα, Gβ, and Gγ subunits. GS3, encoding a non-canonical Gγ subunit, is a main QTL controlling grain length and is a negative regulator of grain size (Fan et al. 2006). The gs3 allele corresponds to long rice grains. The GS3 protein contains four functional domains, and the organ size regulation (OSR) domain is both necessary and sufficient to limit grain size (Mao et al. 2010). DEP1/qPE9, encoding another noncanonical Gγ subunit, is a major QTL for rice panicle architecture and grain size (Huang et al. 2009; Zhou et al. 2009). The dep1/qpe9-1 allele, encoding a truncated protein, leads to erect panicles and smaller grains. Consistently, overexpression of DEP1 increases the grain size, while knockout of the gene makes the grains smaller (Sun et al. 2018a). DEP1/qPE9-1 regulates starch accumulation positively, mainly through promoting the expression of starch biosynthesis-related genes, thus prolonging the duration of the grain filling process, which finally affects the grain size (Zhang et al. 2012b).
In addition to GS3 and DEP1, the G protein γ subunits also include RGG1, RGG2 and GGC2. Grain size is regulated positively by GGC2 and negatively by RGG1 and RGG2 (Kato et al. 2004; Sun et al. 2018a; Miao et al. 2019; Xu et al. 2019). Recently, RGG1 was reported to be involved in regulating the cytokinin content, thus forming a G protein-cytokinin module to control rice grain size (Tao et al. 2020). In addition, genetic analysis indicated that these Gγ proteins require Gα (RGA1) and Gβ (RGB1) subunits to control grain length (Sun et al. 2018a). A recent report indicated that RGB1 not only controls the grain size, but also controls the grain filling process by regulating the expression of OsNF-YB1, encoding a critical downstream effector of RGB1. In addition, OsNF-YB1 directly interacts with the OsYUC11 promoter to stimulate its expression, thus altering auxin homeostasis, starch biosynthesis and grain size (Zhang et al. 2021b).
The Ubiquitin–Proteasome Pathway
The ubiquitin proteasome pathway (UPP) is an important system in eukaryotes that regulates protein stability and activity. GW2, a main QTL controlling grain width and weight, encodes a nuclear ring E3 ubiquitin ligase (Song et al. 2007). A loss of function allele of GW2 promotes the proliferation of spikelet shell cells and produces wide grains. Importantly, this mutation notably enlarges the grain size and increases rice yield, but has little effect on rice AQ and ECQ. Large grain1-D (lg1-D) encodes a ubiquitin-specific protease 15 (OsUBP15). Loss-of-function of OsUBP15 or suppressing its expression generates narrower rice seeds, while OsUBP15 overexpression increases rice grain width significantly. Moreover, OsUBP15 and GW2 genetically interact with each other to co-regulate grain width (Shi et al. 2019).
WG1 encodes a CC glutaredoxin that promotes grain growth by enhancing cell proliferation, while OsbZIP47 negatively regulates grain width and weight by suppressing cell proliferation. WG1 interacts with and inhibits the transcriptional activity of transcription factor OsbZIP47 by recruiting ASP1, a transcriptional co-repressor. Moreover, WG1 is ubiquitinated by E3 ubiquitin ligase GW2 and is subsequently degraded. Genetic analysis demonstrated that the three proteins form a GW2-WG1-OsbZIP47 molecular regulatory module to coordinate grain size and weight (Hao et al. 2021).
TUD1 (Taihu Dwarf1) encodes a functional U-box E3 ubiquitin ligase, which interacts directly with D1/RGA1 to regulate plant height and grain size. In addition, TUD1 and D1 work together to regulate brassinosteroid (BR) signaling and produce short grains by reducing cell division (Hu et al. 2013). OsTUB1/WIDE AND THICK GRAIN (WTG1), encoding a deubiquitinase, controls grain size by affecting cell proliferation. Knockout of OsTUB1/WTG1 resulted in wider grains (Huang et al. 2017). In addition, two recent studies revealed novel regulators involved in UPP-mediated grain size regulation. One is HOMOLOG OF DA1 ON RICE CHROMOSOME 3 (HDR3), a ubiquitin interacting motif (UIM) type active ubiquitin receptor, can interact with and stabilize GW6a to slow down its degradation, thus promoting cell division and increasing the grain filling rate, which ultimately regulates grain size positively (Gao et al. 2021). GW6a is a histone acetyltransferase, whose overexpression increases grain weight and yield by increasing the cell number and accelerating grain filling (Gao et al. 2021). The other protein is Chang Li Geng1-1 (CLG1-1), an E3 ubiquitin ligase, which can ubiquitinate and mediate the degradation of GS3, thus changing G protein signaling and regulating the grain length (Yang et al. 2021b).
Mitogen-Activated Protein Kinase (MAPK) Signaling
The MAPK cascade signaling pathway also plays an important role in regulating rice grain size. Typical MAPK pathways are usually composed of MAPKs, MAPK kinases (MKKs) and MKK kinases (MKKKs) (Zhang et al. 2018b). The OsMKKK10-OsMKK4-OsMAPK6 molecular cascade positively regulates rice grain size and weight (Xu et al. 2018). Disruption the expression of either member leads to smaller rice grains, while overexpression of these genes produces larger rice grains (Duan et al. 2014; Liu et al. 2015b). Furthermore, OsMAPK6 phosphorylates the transcription factor OsWRKY53 and enhances its activity (Tian et al. 2017), while GSK2 directly phosphorylates WRKY53 and lowers its stability (Tian et al. 2021).
In addition, OsMKKK70 also functions through the established OsMKK4–OsMAPK6–OsWRKY53 module (Liu et al. 2021). Overexpression of OsMKKK70 leads to longer grain length and increased rice leaf angle. Moreover, overexpression of the genes encoding constitutively active OsMKK4, OsMAPK6, and OsWRKY53 in the context of osmkk62/70 double mutation can partially restore its phenotype of grain size and leaf angle, implying that these elements operate in the same regulatory pathway.
GSN1 encodes the mitogen activated protein kinase phosphatase, OsMKP1. Suppression of GSN1 expression induces the proliferation of rice glume cells, resulting in larger but fewer rice grains. GSN1 directly interacts with and inactivates OsMAPK6 through dephosphorylation, thus playing an opposite role to OsMKK4 (Guo et al. 2018; Xu et al. 2018). Therefore, the key to controlling rice grain size is to accurately regulate OsMAPK6 activity through reversible phosphorylation.
Phytohormone Perception and Homeostasis
Plant hormones, as central regulators of plant growth and development, not only orchestrating intrinsic developmental programs, but also conveying environmental inputs. Recently, a series of publications reported that some phytohormones, including BR, auxin, gibberellic acid (GA), and cytokinin, also play essential roles in regulating seed size via multiple molecular mechanisms.
BR is Involved in the Regulation of Grain Size
BRs, a group of plant-specific polyhydroxylated steroidal hormones, control a wide range of growth and developmental events, including grain size (Li et al. 2018b). Some BR mutants with defects in both BR biosynthesis or signaling, such as brd1 (Hong et al. 2002; Mori et al. 2002), dwaf2 (Hong et al. 2005), dwaf11 (Tanabe et al. 2005; Zhu et al. 2015; Wu et al. 2016b; Zhou et al. 2017c), OsBRI1/D61 (Yamamuro et al. 2000), OsBAK1 (Li et al. 2009), GS6/DLT/D62 (Tong et al. 2009; Sun et al. 2013), and OsBZR1 (Zhu et al. 2015), usually exhibit shorter plants and smaller grains. One exception is GSK2, which, as a GSK3/SHAGGY like kinase homologous to Arabidopsis BIN2 in rice, is a central negative regulator of the BR pathway. Therefore, suppressing GSK2 expression increased both grain size and leaf angles (Tong et al. 2012). Therefore, BR is a growth-promoting hormone with positive roles in regulating rice grain size.
GW5 is a main QTL for grain width with three haplotypes (Zhou et al. 2017a). GW5 participates in the BR pathway and regulates grain width and weight by inhibiting the function of GSK2 kinase, thus releasing the active forms of OsBZR1 and DLT transcription factors (Liu et al. 2017). Meanwhile, GW5 also affects rice quality, including the chalkiness rate, brown rice rate, and milled rice rate. GS5, encoding a serine carboxypeptidase (Yu et al. 2000), is a QTL controlling rice grain width. Increased expression of GS5 inhibits the endocytosis of OsBAK1-7 and subsequently increases BR signaling and promotes grain size (Xu et al. 2015).
GL3.1/GL3-1/qGL3/OsPPKl1 encodes a protein phosphatase OsPPKl1, which regulates rice grain length negatively by dephosphorylating OsGSK3 and inhibiting BR signaling (Hu et al. 2012; Qi et al. 2012; Zhang et al. 2012a; Gao et al. 2019). OsmiR396d regulates grain size by repressing the growth regulator GS2/GL2/OsGRF4, and GSK2 plays a similar inhibitory function, thereby affecting cell proliferation and grain shape (Che et al. 2015; Duan et al. 2015; Hu et al. 2015; Li et al. 2016; Sun et al. 2016). A recent study also showed that OsAGO17 forms an RNA-induced silencing complex (RISC) with OsmiR397b, which then affects rice development by inhibiting the expression of OsLAC, encoding a negative regulator of both grain size and grain number per panicle (Zhang et al. 2013; Zhong et al. 2020). Moreover, GSK2 interacts with and phosphorylates OML4, a negative regulator of grain size, thus modulating OML4 protein stability. Therefore, GSK2 and OML4 act in the same genetic pathway to regulate rice grain size (Lyu et al. 2020).
The mutation of GS9, encoding a novel transcriptional activator, leads to slender rice grains and reduced chalkiness, without affecting other major agronomic traits (Zhao et al. 2018). GS9 directly interacts with OVATE family proteins, thus forming a transcriptional complex to regulate glume cell division. Moreover, OFP8 and OFP14 inhibit the transcriptional activation activity of GS9, while OFP8 is directly suppressed by OsGSK2 in the BR pathway (Yang et al. 2016a). In addition to OFP8 and OFP14, several other OFP proteins also participate in grain size regulation via the BR pathway, including OFP1 (Xiao et al. 2017), OFP3 (Xiao et al. 2020), OFP19 (Yang et al. 2018a), and OFP22 (Chen et al. 2021). Among them, OFP1 is a positive regulator of BR signaling and seed size, while the others are all negative regulators.
Furthermore, two novel BR-related grain size genes were reported recently. One is GW10, encoding a P450 subfamily 89A2 homology protein, which plays a positive role in controlling grain size through the BR pathway (Zhan et al. 2021). The other is POW1 (Put On Weight 1), encoding an unknown protein. The pow1 mutant enhanced the endogenous BR content, leading to an enlarged leaf angle and grain size. Interestingly, downregulating the expression of BR biosynthesis or signaling genes could only restore the leaf angle phenotype of pow1 mutant, not the grain size. Further analysis shows that POW1 regulates grain size by inhibiting the transactivation activity of TAF2, its interacting protein. Hence, two regulatory modules, POW1-TAF2 and POW1-BR, are established that specifically regulate the grain size and leaf angle, respectively (Zhang et al. 2021c).
Auxin
Auxin is an important classical phytohormone with essential roles in many aspects of plant growth and development events, including grain size. qTGW6, as a main QTL controlling rice grain weight, encodes indole-3-acetic acid (IAA)-glucose hydrolase to generate free IAA. A loss-of-function TGW6 allele promoted grain length and weight (Ishimaru et al. 2013). Recent research indicated that TGW6 is exclusively expressed in pre-emergent inflorescences, suggesting that TGW6 might play important roles in regulating pollen development (Akabane et al. 2021; Kabir and Nonhebel 2021).
BG1, an auxin primary response gene, encodes a protein that participates in the regulation of auxin transport. It affects grain size by promoting cell division and elongation (Liu et al. 2015a). qTGW3/GL3.3 is a major QTL for grain weight, encoding a SHAGGY-like kinase 41 (OsSK41) (Hu et al. 2018; Xia et al. 2018; Ying et al. 2018). OsSK41 directly interacts with and phosphorylates OsARF4, a transcription repressor in the auxin pathway. Moreover, the OsSK41-OsARF4 module regulates rice grain size negatively by modulating the expression of a number of auxin-response genes. Recently, another transcription factor, OsARF6, was reported to bind directly to the OsAUX3 promoter to increase its expression (Qiao et al. 2021). OsARF6 and OsAUX3 regulate rice grain length and weight negatively by modulating the auxin content and distribution in glume cells, consequently affecting rice grain longitudinal elongation. miR167a, a positive regulator of grain length and weight, directly silences OsARF6 mRNA. Hence, a novel miR167a-OsARF6-OsAUX3 regulatory module is established successfully.
GA
The tetracyclic diterpenoid phytohormone GA has multiple roles in plant growth and development. However, the participation of GA in regulating rice grain size is rarely reported. Recently, Shi et al. (2020) indicated that a QTL for grain size, designated as GW6 (GRAIN WIDTH 6), was successfully cloned. GW6, encoding a GA-induced GAST family protein, plays positive roles in regulating grain size and weight. Moreover, knockout of GW6 reduced the GA content in young panicles. Importantly, a natural variation in the CAAT-box of the GW6 promoter determines its transcript abundance, as well as the grain width and weight, thus providing valuable natural genetic resources for rice breeding programs.
Cytokinin
A dominant mutant big grain 3 (bg3-D) was isolated, which featured larger rice grains. BG3, encoding a purine permease, OsPUP4, regulates grain size positively by controlling both the long-distance transport and local allocation of cytokinin (Xiao et al. 2019). Recently, Yin et al. (2020) reported that ARGONAUTE (AGO) proteins are essential to assemble RNA-induced silencing complexes to silence target genes. Overexpression of AGO2 boosts both the salt-stress resistance and grain length of rice by modulating the histone methylation level of BG3, hence promoting its expression. Salt treatment results in a similar cytokinin distribution pattern to AGO2 overexpression rice, implying that the cytokinin distribution pattern is critical to regulate stress tolerance and rice grain size.
Transcription Factors
Transcription factors (TFs) play vital roles in regulating plant growth and development by responding to upstream signals and modulating downstream transcriptional networks. Certain TFs are also involved in controlling seed size.
SQUAMOSA Promoter Binding Protein-Like (SPL) Transcription Factor Family
SPL transcription factors are involved in controlling the tiller, panicle configuration, and grain size of rice. A genome-wide association analysis (GWAS) indicated that a major QTL GLW7, encoding the TF OsSPL13, is a key element leading to larger grains and more panicles (Si et al. 2016). GLW7 binds directly to the promoter of SRS5, a positive regulator of grain length, and activates its expression (Segami et al. 2017). Suppressing the expression of OsSPL16/GW8 produces long grains, and thus decreased chalkiness and improved transparency of rice seeds (Wang et al. 2012). GW8 directly binds to the GW7/GL7 promoter and inhibits its expression (Wang et al. 2015a). GW7, encoding a TONNEAU1-recruiting motif protein, plays a positive role in producing slender grains via differential regulation of cell division in both longitudinal and transverse directions. Importantly, a semi-dominant GW7 allele enhances rice grain quality without any yield penalty (Wang et al. 2015c). Therefore, the identified OsSPL16/GW8-GW7 regulatory module should be useful in future elite rice breeding programs to improve both rice quality and yield. SPLs are known to be regulated by microRNAs, such as miR156 (Xie et al. 2006). Recently, two regulatory modules, miR529a-SPLs and OsmiR156-SPL4, are revealed and their essential roles in controlling rice grain size are studied (Hu et al. 2021; Yan et al. 2021).
APETALA2-Type (AP2) Transcription Factors
SMOS1/SHB encodes an APETALA2 (AP2) transcription factor with an incomplete AP2 domain. The grains and other organs of smos1 mutants are smaller because of the smaller cells and an abnormal microtubule orientation. SMOS1 directly modulates the transcription of a cell expansion regulator, phosphorylation inducible protein 1 (OsPHI-1) (Aya et al. 2014). SMOS1 interacts with SMOS2/DLT to form a key protein complex to orchestrate BR and auxin signaling, thereby coordinating rice growth and development, including grain size (Hirano et al. 2017). Moreover, Suppression of Shattering 1 (SSH1) is a novel allele of SUPERNUMERARY BRACT (SNB), encoding an AP2 transcription factor. A point mutation in the ninth intron of SNB alters its mRNA splicing and decreases SNB expression, consequently reducing rice shattering and increasing grain size (Jiang et al. 2019).
Basic Helix-Loop-Helix (bHLH) Family
Awn-1(An-1) encodes a bHLH protein that regulates awn development, grain size, and grain number in rice. Increased An-1 expression causes long awns and grains, but decreases the grain number per panicle (Luo et al. 2013). PGL1 is an atypical bHLH protein without DNA binding activity. Overexpression of PGL1 increases the grain length and weight while, APG exerts an opposite effect. PGL1 interacts directly with APG to regulate grain size antagonistically (Heang and Sassa 2012).
Other Transcription Factors
SHORT GRAIN 6 (SG6)/GL6 encodes a plant specific PLTAZ transcription factor which regulates grain length positively by promoting cell proliferation in young panicles and grains (Wang et al. 2019; Zhou and Xue 2020). GL6 interacts with RPC53 and OsTFC1 to participate in the RNA polymerase III transcription machinery and regulates the expression of genes involved in rice grain development and the cell cycle, thus regulates grain length positively (Wang et al. 2019). FLR family proteins, such as FLR1, FLR2, and FLR8, play negative roles, while FLR15 plays a positive role, in regulating grain size. Although the grains of the flr8 mutant are larger, their quality remains the same. Moreover, FLR1 can modulate the number of glume cells and the expression of starch metabolism genes, thus affecting seed size and grain filling (Wang et al. 2021). qLGY3/OsLG3b, a QTL for grain length, encodes the MADS-domain transcription factor, OsMADS1. qLGY3/ OsLG3b allele leads to the alternative splicing of OsMADS1, which is artificially selected and corresponds to a long grain phenotype (Liu et al. 2018b; Yu et al. 2018). GS3 and DEP1 directly interact with MADS1 to promote its transcriptional activity and hence inhibit grain growth (Liu et al. 2018b).
Other Functional Proteins
GAD1 encodes a small secretary signal peptide and its mutation leads to reduced grain numbers, shorter grains, and awnless rice. Mechanistically, GAD1 regulates the length of the grain and awn by modulating cell numbers (Jin et al. 2016). GL4, a QTL for grain length originating from African rice, encodes a Myb-like protein similar to SH4/SHA1. GL4 controls grain length by regulating the elongation of longitudinal cells of both the outer and inner glumes. A single nucleotide polymorphism (SNP) mutation in GL4 leads to a premature stop codon and consequently a truncated protein, which results in small seeds and the loss of seed shattering during African rice domestication (Wu et al. 2017). In addition, overexpression of RAG2, encoding a 16-kDa α-amylase/trypsin inhibitor, significantly increases the grain size and 1000-grain weight, as well as the protein and total lipid contents (Zhou et al. 2017b).
Genes Regulating Endosperm Components and Their Roles in Grain Quality
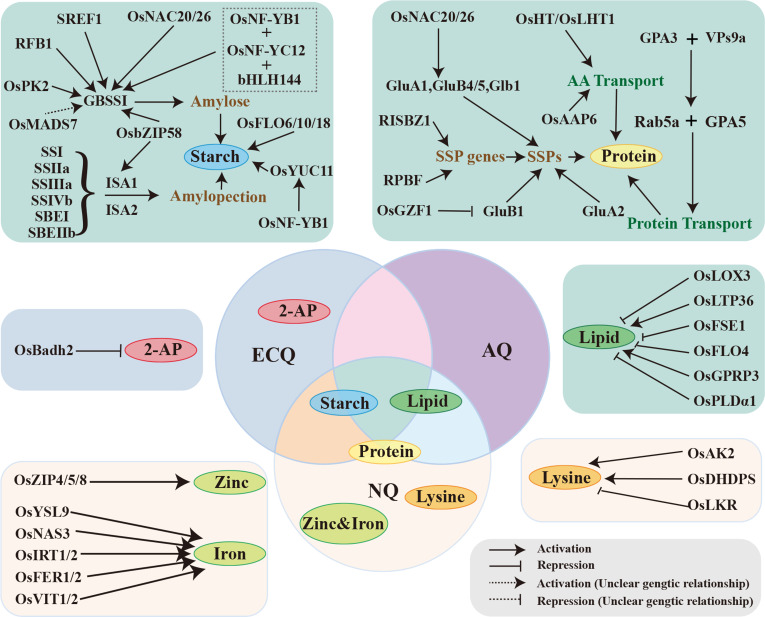
The components of the rice endosperm include starch, proteins, amino acids, lipids, vitamins, minerals, and other metabolites. Among them, starch and protein account for about 80% and 10% of the dry weight of rice endosperm, respectively (Wang et al. 2020a). Therefore, the constitution and quality of starch and protein contribute majorly to rice grain quality (Fig. 4). In general, starch is the primary determinant of rice ECQ and AQ (Li et al. 2018a). For example, rice AC influences a series of quality related parameters, including hardness, viscosity, and transparency. Proteins are considered as the most important elements of rice NQ. Their content and quality determine rice NQ. Protein quality is mainly evaluated by assessing essential amino acids, such as lysine. In contrast to starch and protein, the content of lipid is low in rice, accounting for only about 0.3–0.6% of rice weight (Morrison 1988). The lipid content, similar to the protein content, correlates negatively with rice ECQ. Nevertheless, lipids have a great influence on the storage, processing, and consumption of rice. In addition to these three components, some micronutrients and phytochemicals are also important in rice quality, such as vitamins and minerals (Buttery et al. 1983; Kovach et al. 2009).
Fig. 4.
The genes involved in the synthesis and regulation of the main components of the endosperm, and their contributions to corresponding rice quality traits. The dotted line indicates that an indirect relationship that is supported by genetic evidence, which needs to be further verified. AQ, appearance quality; NQ, nutritional quality; ECQ, eating and cooking quality
Genes Controlling Starch Biosynthesis, Their Transcriptional Regulation, and Their Effects on Rice Quality
Starch Biosynthesis Enzymes
Starch is mainly stored in the form of starch granules in endosperm cells. According to their different glycosidic bond connections, starch is usually classified into two groups, amylose and amylopectin. The amylose content (AC) is the most important effector of rice ECQ (Duan and Sun 2005). According to their different ACs, rice varieties can be divided into waxy (< 2%), very low (3–9%), low (10–19%), intermediate (20–25%), and high (> 25%) amylose types (Zhang et al. 2017). In general, the higher the AC, the harder the texture of the cooked rice. Meanwhile, the AC is also related closely to rice transparency (Li et al. 2018a). Glutinous rice, lacking almost all amylose, has a completely opaque, waxy endosperm.
To date, almost all the genes encoding key enzymes involved in starch biosynthesis have been cloned and studied, such as ADP-glucose pyrophosphorylases (AGPases), granule-bound starch synthases (GBSSs), soluble starch synthases (SSSs), starch branching enzymes (SBEs), debranching enzymes (DBEs), and starch phosphorylase (PHO) (Zemach et al. 2010; Seung and Smith 2019). Detailed information related to starch biosynthesis is well summarized by some excellent reviews (Huang et al. 2020, 2021a); therefore, we will only focus on several key genes that determine rice quality.
AGPase uses glucose-1-phosphate (Glu-1-P) as a substrate to generate ADPglucose, which is then used to synthesize amylose via GBSSI, and to generate amylopectin via the cooperation of a series of other enzymes, including SSSs, SBEs, and DBEs (Jeon et al. 2010; Pfister and Zeeman 2016). Among them, GBSSI, encoded by the Waxy (Wx) gene, is the sole enzyme directly controlling amylose synthesis, and is thus the primary determinant of rice AC, GC, and pasting property (Wang et al. 2010). Therefore, Wx has been studied extensively and used widely to improve rice ECQ. Hence, a number of useful natural Wx alleles have been cloned and applied in rice breeding practice. Up to now, about ten natural Wx alleles have been reported, including the newly cloned Wxlv and Wxmp/Wxla (Zhang et al. 2019a, 2021a; Zhou et al. 2021a). The wx allele, a null allele that does not encode a functional GBSSI, exists in glutinous rice, AC < 2% (Wanchana et al. 2003). Wxa and Wxb are two major Wx alleles that are distributed widely in most indica rice and japonica rice varieties, respectively, corresponding to high and low ACs.
The discovery of the Wx ancestor gene, Wxlv, and the differentiation of its functional sites might explain the evolutionary trend of the AC, from high to low during rice domestication (Zhang et al. 2019a). The AC decreased slightly because of the variation in amino acid sequence caused by a SNP mutation in exon 6 of Wxin. Rice with the Wxop/Wxhp (AC ~ 12.8%), Wxmp (AC ~ 10.5%), and Wxmq (AC ~ 10%) alleles are the so called soft rice (Hiroyuki et al. 2002; Mikami et al. 2008; Liu et al. 2009; Zhang et al. 2021a; Zhou et al. 2021a, b), which are famous for their good taste and high ECQ. The rare allele Wxmw/Wxla, derived from the homologous recombination of Wxin and Wxb, has a low AC, high transparency, good taste, and excellent ECQ (Zhang et al. 2021a; Zhou et al. 2021a). In addition to the identification of natural Wx alleles, the CRISPR/Cas9 gene editing strategy was also applied for accurate editing and to generate novel and excellent Wx alleles (Huang et al. 2020; Zeng et al. 2020).
The synthesis of amylopectin is complex and is coordinately regulated by several groups of enzymes. Moreover, each group contains several different types of enzymes. For example, SSS comprises SSSI, SSSII, SSSIII, and SSSIV. Except for SSSI, the other type of SSS all contain more than one isoform. Each enzyme isoform plays a distinct role in amylopectin biosynthesis. In general, SSSI prolongs the amylopectin by synthesizing short chains, while SSSII synthesizes medium-length amylopectin. SSSIIa/ALK is the major gene regulating rice GT (Gao et al. 2003). The different expression levels and allele types of ALK are the main cause for the differential amylopectin structure between indica and japonica subspecies (Umemoto et al. 2002). Two SNP-induced amino acid mutations affect the function of SSSIIa, resulting in a decrease in the branch chain length and GT (Bao et al. 2006). Recently, a detailed analysis of various ALK alleles, including a new identified ALKd allele, was performed, which clarified their roles in regulating rice GT, AC, and general taste values, demonstrating ALK is a crucial molecular target to improve rice ECQ (Chen et al. 2020; Zhang et al. 2020). Another SSSII isoform, SSSII-2, was reported to have the potential to improve rice quality. Suppressing SSSII-2 expression produced a novel soft rice with a low AC, improved taste, and transparent endosperm (Li et al. 2018a). Furthermore, simultaneous modulation of SSSII-2, SSSIIa, and Wx coordinated the biosynthesis of amylose and amylopectin, hence successfully improving rice ECQ (Huang et al. 2021b).
SSSIII synthesizes long amylopectin chains and SSSIIIa is an important target to study amylopectin biosynthesis and breed healthy rice. Resistant starch (RS) can reduce the incidence of type 2 diabetes and reduce the probability of obesity; therefore, high RS rice is considered to be a healthy food. SSIIIa affects the structure of amylopectin, the amylose content, and the physicochemical properties of starch grains in indica rice together with the Wxa allele, resulting in a higher AC and an increased lipid content, subsequently increasing the amount of amylose–lipid complex and RS starch (Zhou et al.2016). That study suggested that modulating the SSSIIIa and Wx genes could benefit future breeding of high RS rice (Zhou et al. 2016).
Transcription Factors
The biosynthesis of starch is crucial to both seed development and propagation, and a number of transcription factors are involved in regulating the expression of starch synthesis-related genes (SSRGs). OsbZIP58/RISBZ1 binds specifically to the ACGT motif in the Wx gene promoter (Wang et al. 2013), thus enhancing its expression. In addition, OsbZIP58 also binds to the promoters of AGPL3, SSSIIa, SBE1, and ISA2 to regulate their expression levels. bZIP58 is a core regulator of starch synthesis and its null mutant is chalky and its total starch and AC are decreased (Wang et al. 2013). In addition, OsbZIP58 interacts with RPBF, a Dof family transcription factor, to participate in the synthesis of storage substances, including starch, protein, and lipid, during rice endosperm development (Kawakatsu et al. 2009).
As an NF-Y transcription factor, OsNF-YB1 regulates endosperm sucrose transport and grain filling. Suppression of OsNF-YB1 expression leads to development defects of rice seeds with increased grain chalkiness and a decreased AC, resulting in a decline in rice quality (Sun et al. 2014). Knockout of OsNF-YB1 resulted in an increased protein content and decreased grain size and contents of amylose, total starch, crude fiber, and lipid, subsequently altering rice quality (Bello et al. 2019; Xu et al. 2021). In addition, OsNF-YB1 binds directly to the promoter of OsYUC11 and activates its expression. As a key element in auxin biosynthesis, OsYUC11 affects grain filling and the accumulation of endosperm storage products in rice (Xu et al. 2021). Another NF-Y member, NF-YC12, coordinates various pathways to regulate endosperm development and the accumulation of seed storage substances in rice. The phenotype of the osnf-yb12 mutant is similar to that of the osnf-yb1 mutant, with changed grain weight, starch, and protein accumulations (Bello et al. 2019; Xiong et al. 2019). Furthermore, NF-YB1 combines with NF-YC12 and bHLH144 to form an NF-YB1-YC12-bHLH144 heterotrimeric complex that coordinates grain development and rice quality. Hence, the mutation of any gene in the complex would change starch synthesis in the rice endosperm (Bello et al. 2019).
Several members of MADS box family transcription factors participate in the regulation of starch biosynthesis. OsMADS6 is highly expressed in the endosperm and regulates the expression of SSRGs. Its mutation leads to decreased starch plumpness and abnormal endosperm development (Zhang et al. 2010). Suppression of OsMADS29 expression caused abnormal seed development, such as shrunken seeds, a low grain-filling rate, and insufficient starch accumulation. Evidences indicates that OsMADS29 modulates the expression of genes related to programmed cell death (PCD), thus affecting the early development of rice seeds (Yin and Xue 2012). Another study revealed that OsMADS29 affects embryo and endosperm development, including starch biosynthesis, by modulating cytokinin signaling and biosynthesis (Nayar et al. 2013). As a high temperature induced gene, OsMADS7 encodes a protein involved in stabilization of the AC in response to high temperature, mainly by enhancing the expression of GBSSI, the key enzyme controlling amylose biosynthesis. Therefore, OsMADS7 is a valuable molecular target for breeding elite rice with ideal thermal tolerance and ECQ (Zhang et al. 2018a).
NAC transcription factors are plant-specific and participate in various processes of plant development, including biosynthesis of storage substance of the rice endosperm. Mutation of OsNAC20 or OsNAC26 alone does not have any effect on rice grains. However, the contents of starch and storage proteins in osnac20/26 double mutants are decreased. Further evidence demonstrated that OsNAC20 and OsNAC26 can promote the expression of multiple genes involved in starch and storage protein biosynthesis directly, such as those encoding SSSI, Pul, glutelin A1 (GluA1), glutelin B4/5 (GluB4/5), α globulin, and 16 kDa prolamin, thus regulating the synthesis of both starch and storage proteins (Wang et al. 2020a). ONAC127 and ONAC129 are not directly involved in starch synthesis in the rice endosperm. They regulate grain filling and starch accumulation by forming heterodimers, and participating in cytoplasmic transport and heat stress translation (Ren et al. 2021).
Other Proteins
In addition to the above mentioned starch synthesis-related enzymes and transcription factors, other proteins are involved in regulating starch biosynthesis. In general, the mutation of these genes, such as FLO6 (Peng et al. 2014), FLO10 (Wu et al. 2019), FLO14 (Xue et al. 2019), FLO18 (Yu et al. 2021), FGR1 (Hao et al. 2019) and OsPK2 (Cai et al. 2018), lead to defects in starch biosynthesis and the formation of abnormal starch granules, resulting in opaque, chalky, or powdery grains. Interestingly, only FLO6 shows a direct correlation with starch biosynthesis-related enzymes. FLO6 binds directly to starch through a CBM48 domain at the C terminus and to ISA1 through a domain at the N terminus, suggesting its role as a bridge between ISA1 and starch during starch synthesis (Peng et al. 2014).
Storage Proteins, Transporters of Amino Acids and Proteins, and Other Regulators of Proteins
Proteins are the secondary major components of rice endosperm, which could be divided into three categories, storage proteins, structural proteins, and protective proteins. The protein content and its amino acid constitution affect rice NQ directly. Moreover, the protein content is also involved in the regulation of rice ECQ. In general, a negative correlation exists between the protein content and ECQ in rice (Hori et al. 2016).
Storage Proteins
The rice storage proteins (SSPs) can be divided into four categories, albumin, globulin, prolamine, and glutelin. Among them, glutelin, as the most abundant SSP, has the highest nutritional value because of its high digestibility and lysine content (He et al. 2021). There are 15 glutelin encoding genes in the rice genome, which are classified into four subfamilies, GluA, GluB, GluC, and GluD (Kawakatsu and Takaiwa 2010). Glu genes encode 57 kDa pro-glutelin, consisting of a signal peptide, a 37 kDa acidic subunit, and a 20 kDa basic subunit. A SNP in the GluA2 promoter leads to a difference in the total protein content between indica and japonica rice. Hence, all haplotypes can be divided into two expression types, OsGluA2LET and OsGluA2HET. OsGluA2LET mainly exists in japonica rice, with low expression of OsGlu. Meanwhile, OsGluA2HET is highly expressed in indica rice. Therefore, the expression of OsGlu genes correlate closely with the grain total protein content and NQ (Yang et al. 2019).
Transporters of Amino Acids and Proteins
Efficient amino acid transfer, depending on amino acid transporters (AATs), is essential for protein biosynthesis in rice grains. Lysine-Histidine-type Transporter 1 (OsLHT1) can transport a broad spectrum of amino acids effectively. OsLHT1 mutation leads to declined root uptake and consequent transfer of amino acids to rice shoots (Guo et al. 2020b). Moreover, Oslht1 mutant rice has a reduced panicle length, seed setting rate, grain number per panicle, and total grain weight. More N and free amino acids are retained in the flag leaf of the Oslht1 mutant than in the wild-type at maturation, implying its essential roles in transferring amino acids from leaves to seeds, thus ensuring grain development and rice nutrition quality (Guo et al. 2020a). Furthermore, a number of amino acid permeases (AAPs), a main type of AAT, are responsible for amino acid loading in rice seeds. Mutation of OsAAP10 decreases the content of both protein and amylose in rice seeds. In addition, the RVA profile of osaap10 mutant seeds exhibits a higher peak viscosity, disintegration value, and lower recovery value, thus improving the ECQ (Wang et al. 2020b). GPA1/OsRab5a, a small GTPase, regulates the transport of glutelin to protein body II (PBII) and affects the protein content. GPA3 interacts directly with Rab5a and guanine exchange factor VPS9a, forming a regulatory complex with them. These three proteins regulate dense vesicle (DV)-mediated post-Golgi transport synergistically in rice. The gpa3 mutant showed a powdery endosperm, abnormal accumulation of glutelin precursors, irregularly arranged starch grains, decreased amylose content, and increased levels protein and lipids (Ren et al. 2014). GPA5, an effector of Rab5a, is also required for post-Golgi trafficking of storage proteins (Ren et al. 2020). GPA5 mutation also leads to a powdery white endosperm, resulting from the abnormal accumulation of glutelin precursors and a reduction in α globulin, consequently forming loosely arranged and round compound starch granules. GPA5 interacts with class C core vacuole/endosome tethering (CORVET) complex and VAMP727-containing soluble N-ethylmaleimide sensitive factor attachment protein receptor (SNARE) complex to promote the fusion of DVs and protein storage vacuoles to complete glutelin transportation.
Other Regulators
In addition to the above mentioned storage or functional proteins, other regulators also participate in regulating the protein content in rice endosperm. For example, RISBZ1/OsbZIP58 and RPBF regulate the expression of seed storage protein genes and the consequent protein content (Kawakatsu et al. 2009). OsGZF1, a CCCH type zinc finger protein, binds specifically to the core promoter region of GluB-1, thereby suppressing its expression and the accumulation of glutelins (Chen et al. 2014). Moreover, transcription factors NAC20 and NAC26 also bind directly to the promoters of SSP genes and regulate their expression in rice (Wang et al. 2020a). In addition to transcriptional regulation, control of glutelin mRNA localization is another important mechanism that modulates glutelin accumulation. The zip codes of storage protein mRNAs require assistance from RNA‐binding proteins (RBPs) for their correct localization. RBP‐A, RBP‐P, RBP‐L, and Tudor‐SN are reported to bind to the mRNAs of both glutelin and prolamin to aid their localization (Wang et al. 2008; Doroshenk et al. 2014; Chou et al. 2017, 2019; Tian et al. 2018; Tian et al. 2019a). A recent study showed that a quaternary protein complex, including RBP‐P, RBP‐L, Rab5a, and a membrane fusion protein NSF, cooperates to coordinate the transport of glutelin mRNAs in the rice endosperm (Tian et al. 2020).
Lipids
The main fatty acids in rice are palmitic acid (C16:0), oleic acid (C18:1), and linoleic acid (C18:2). Among them, linoleic acid is relatively good for human health. Lipids not only affect the rice NQ, but also influence the AQ and ECQ. Phospholipids and glycolipids can interact with starch in rice, thus reducing the water absorption and expansibility of starch, and can increase its GT.
The genes involved in carbon flow, lipid biosynthesis, transport, and oxidation affect the quantity and quality of lipids in rice. Pyruvate phosphate dikinase (encoded by OsPPDKB) regulates both carbon metabolism and carbon flow for starch and fat biosynthesis during rice filling. Its mutation produces a white powdery endosperm and a significantly increased fat content (Zhao et al. 2017). Another gene Floury Shrunken Endosperm1 (FSE1), encoding a phospholipase-like protein, regulates galactolipid biosynthesis in the rice endosperm. FSE1 mutation not only reduced the total galactolipid content significantly, but also caused abnormal amyloplast development in the developing endosperm, thus providing a novel connection between lipid metabolism and starch synthesis in rice (Long et al. 2018).
Fatty acid desaturase (FAD) genes, including OsFAD2 and OsFAD3, participate directly in different steps of fatty acid synthesis (Liu et al. 2012; Shi et al. 2012; Ding et al. 2015). OsACOT, a major target of miR1432-mediated cleavage, encodes acyl-CoA thioesterase, which is involved in the biosynthesis of medium-chain fatty acids. Suppression of miR1432 expression or overexpression of miR1432-resistant form of OsACOT (OXmACOT) promoted rice grain filling rate and grain weight significantly. Moreover, the contents of palmitic acid and stearic acid (18:0) in OXmACOT transgenic rice decreased, while those of the oleic acid and linoleic acid increased (Zhao et al. 2019).
Suppressing the expression of OsLTP36, encoding a lipid transporter, resulted in decreased contents of fatty acids and proteins, smaller and loose starch grains, and some other growth defects, including the seed setting rate, 1000-grain weight, chalkiness, and seed germination rate (Wang et al. 2015b). Moreover, lipoxygenase (LOX) catalyzes lipid oxidation, which leads to aging and a decrease of nutrient level in rice (Cho and Lim 2016). LOX-2 and LOX-3 regulate the degradation of fatty acids negatively, and suppression of their expression or loss-of-function mutation effectively prolonged the storage time and maintained a high nutritional value of rice (Long et al. 2013). Meanwhile, reducing the expression of LOX-3 could effectively reduce the degradation of β-carotene in golden rice (Huang et al. 2014; Zhou et al. 2014). A recent study showed that mutation of OsPLDα1, encoding a phospholipase, changed lipid metabolites and reduced the phytic acid content strikingly (Khan et al. 2019). Further analysis indicated that the mutant brown rice shows some changes in ECQ properties, including a decreased AC, setback viscosity, and gelatinization temperature (GT), as well as an increased disintegration rate, corresponding to improved ECQ and NQ (Khan et al. 2020).
Lysine
The types of amino acids and the proportion of essential amino acids also determine the nutritional quality of rice. Lysine (Lys) is considered as the first limiting essential amino acid in humans; however, its content in milled rice is quite low. Several strategies have been used to enhance the lysine content in rice, including overexpression of lysine-rich proteins and modulation of lysine metabolism pathways (Yang et al. 2021a). For example, overexpression of lysine-rich histone proteins, RLRH1 and RLRH2, increased the lysine content of rice by 35% (Wong et al. 2015). With regards to modulation of lysine metabolism pathway, one method is to enhance the expression of lysine biosynthesis genes, another is to block the catabolism of lysine, thus promoting the lysine content of rice. AK and DHPS, two rate-limiting enzymes in the lysine biosynthesis pathway, are under strict feedback inhibition by lysine. By overexpressing modified Lys-insensitive AK or DHPS, the free lysine content increase by 6.6- to 21.7-fold. When simultaneously expressing these two enzymes, the level of free lysine increased by 58.5-fold (Yang et al. 2020, 2021a). Suppressing the expression of LKR/SDH gene, encoding lysine ketoglutaric acid reductase/saccharopine dehydropine dehydrogenase (LKR/SDH), attenuated lysine catabolism and remarkably promoted the free lysine content in rice (Wu et al. 2016a; Yang et al. 2016b; Yang et al. 2018b; Zheng and Wang 2014). The rice varieties with high free lysine content had no significant differences in yield and other main agronomic traits except for plant height and grain color (Yang et al. 2016b). The changed endosperm color of high-lysine rice is mainly caused by activation of the jasmonic acid pathway by the high abundance of free lysine and subsequent enhanced serotonin biosynthesis (Yang et al. 2018b).
Carotenoids
Regardless of the fact that more than 700 kinds of carotene have been found in nature, only α-carotene, β-carotene, lutein, lycopene, zeaxanthin, and astaxanthin have been shown to be beneficial for health (Federico and Schmidt 2016). Carotenoids are important phytonutrients with antioxidant properties, and are used widely in foods and feedstuffs as supplements. In addition, carotenoids can also be used as antioxidants to prevent seed aging and promote seed vigor, leading to successful germination (Federico and Schmidt 2016). The synthesis of carotenoids in seeds is closely related to the ABA biosynthesis pathway, the dominant pathway for seed dormancy.
Rice carotenoids biosynthesis is blocked in the first enzymatic step. Biofortification is an effective way to produce and accumulate carotenoids in rice grains. Therefore, the major objective of golden rice (GR) development is to improve its carotenoids content. Driven by the Gt1 promoter, the daffodils-originating PSY gene and Erwinia uredovora-originating CRTI gene were transformed into rice, thus generating rice grains with β-carotene accumulation (Ye et al. 2000). Golden rice 2 (GR2) was developed by transferring the maize phytene synthase gene Zmpac1 and the carotene desaturase gene CrtI from soil bacteria Pantoea ananatis into japonica Kaybonnet rice. The content of β-carotene in the rice grain was 23 times higher than that in the first generation of GR (Paine et al. 2005). Recently, astaxanthin biosynthesis was bioengineered in the rice endosperm by introducing sZmPSY1, sPaCrtI, sCrBKT, and sHpBHY, four genes encoding the enzymes phytoene synthase, phytoene desaturase, β-carotene ketolase, and β-carotene hydroxylase, respectively, thus generating multiple healthy rice germplasms, including β-carotene-enriched Golden Rice, Canthaxanthin Rice, and Astaxanthin Rice (Zhu et al. 2018). Another study indicated that co-expression of tHMG1, ZmPSY1, and PaCRTI could boost the carotenoids flux through the MVA pathway, thus increasing the accumulation of carotenoids in the rice endosperm markedly (Tian et al. 2019b). The promotion of carotenoids in rice relies on insertions of target genes at random sites via conventional transgenic methods; therefore, the marker gene still exists in the genome of transgenic rice. Alternatively, an expression cassette including two carotenoid biosynthesis genes were introduced using targeted insertion at a safe site in the rice genome via CRISPR/Cas9, thus generating marker-free carotenoid-enriched rice (Dong et al. 2020).
Minerals, Taking Fe and Zn as Examples
At present, more than 90 Fe-related QTLs have been identified in the rice genome, among which 17 are stable and 25 harbor Fe-related genes nearby or within the QTL (Swamy et al. 2021). A common mechanism of transporters and chelators mediates iron and zinc absorption and transport; therefore, most instances of increased iron content in rice are accompanied by a parallel increase in zinc (Kawakami and Bhullar 2018).
There are two sources of iron in seeds, absorption by the roots from soil solution, followed by direct transfer to seeds and reactivation from different tissues and organs during seed development (Ishimaru et al. 2006, 2007a). Two Ferritin (FER) genes, OsFER1 and OsFER2, have been identified in rice (Stein et al. 2009). OsFER2 is more sensitive to external iron supply than OsFER1, implying its major role in rice FE chelation. Seed-specific overexpression of OsFER2 promotes the accumulation of iron and zinc in milled rice seeds by 2.1 and 1.37-fold, respectively (Paul et al. 2012).
Nicotinamide (NA) is a ubiquitous metal-chelated non-protein amino acid in terrestrial plants with important role in metal transport. Increasing the expression of the NA synthase gene (NAS) is a useful biofortification method to promote the Fe and Zn contents of rice. There are three NAS genes in rice, OsNAS1, OsNAS2, and OsNAS3 (Nozoye et al. 2019). Knockout of OsNAS3 reduced the iron content in rice flag leaves and seeds, while OsNAS3 overexpression had the opposite phenotype (Lee et al. 2009). Yellow Stripe 1-Like 9 (encoded by OsYSL9) transports the iron-sodium/DMA complex from the endosperm to the embryo during seed development. An OsYSL9 null mutant showed a decreased iron content in embryos, but an increased iron content in the endosperm (Senoura et al. 2017).
OsVIT1 and OsVIT2 are another two transporters that regulate the iron content in rice seeds. Knockout of the two genes promotes the amount of Fe/Zn in rice seeds, but decreases contents of these metals in rice leaves, suggesting that OsVIT1 and OsVIT2 play important roles in controlling Fe/Zn translocation between source and sink organs (Zhang et al. 2012b).
ZIP transporters include zinc regulated transporters (ZRT) and iron regulated transporters (IRT). The rice ZIP gene family contains 16 members, including 14 ZRT genes and two IRT genes (Sasaki et al. 2015). Overexpression of OsIRT1 reduces plant height, tiller, and yield of rice, but increases the content of Fe and Zn in rice grains (Lee and An 2009). In addition, overexpression of OsZIPs, such as OsZIP4, OsZIP5, and OsZIP8, promotes the Zn content in roots, but reduces the content in shoots and grains (Ishimaru et al. 2007b; Lee et al. 2010a, b).
In general, three major strategies can be used to enhance the iron content in rice, including overexpression of NAS genes, endosperm-specific expression of FERs, and promoting source-to-endosperm Fe remobilization (Kawakami and Bhullar 2021).
2-Acetyl-1-Pyrroline (2-AP)
2-AP is the main aroma substance in scented rice (Kovach et al. 2009; Bradbury et al. 2010). The Badh2 gene, encoding betaine aldehyde dehydrogenase, inhibits the biosynthesis of 2-AP by exhausting γ-aminobutyraldehyde (AB-ald), a presumed 2AP precursor. The significant increase of 2-AP levels in fragrant rice varieties greatly improves the aroma of milled rice. The null badh2 alleles, with a protein frameshift mutation, enhance 2-AP biosynthesis and hence the aroma of rice (Chen et al. 2008).
Genes Regulating Other Seed Structures and Their Roles in Grain Qualities
The Embryo
The rice embryo, containing most of the genetic information of rice, has the highest concentration of nutrients, including proteins, fatty acids, vitamins, and minerals. Giant embryo rice is a rice mutant whose embryo is about two to three times larger than the normal embryo. The phenotype of giant embryo rice is determined by the GIANT EMBRYO (GE) gene, encoding cytochrome P450 protein CYP78A13. GE mutation leads to large embryo and small endosperm, while overexpression of GE had the opposite phenotype (Nagasawa et al. 2013).
In plants, GABA is mainly catalyzed by glutamate decarboxylase (GAD), producing decarboxylate glutamate (Akama et al. 2009; Shelp et al. 2012). However, the higher GABA content in the grains of developing giant embryo rice mainly comes from the upregulation of the polyamine (PA) derivative pathway and the downregulation of GABA catabolism activity (Zhao et al. 2017). The C-terminal extension of OsGAD2 acts as a powerful self-inhibitory domain, and truncation of this domain caused the enzyme to function constitutively and actively. Rice endosperm specific expression of OsGAD2ΔC enhanced the GABA content by about tenfold (Shelp et al. 2012). In addition, the combination of seed-specific overexpression of truncated GAD with suppression of GABA-T, enhanced the GABA content strikingly (75–350 mg/100 g) in milled rice (Shimajiri et al. 2013). Recently, the coding region of CaMBD from the OsGAD3 gene was knocked out using CRISPR/Cas9 technology, thus improving rice GABA content by 7 times, accompanied by increased grain weight and protein content (Akama et al. 2020).
The Aleurone Layer
Brown rice is unpolished rice after shelling, which is composed of the endosperm, embryo, and rice bran. Compared with milled rice, brown rice contains more macronutrients, vitamins, minerals, and other functional substances, such as GABA, is thus more beneficial to human health. The proportion of nutrients among various rice components are different, thus optimization of rice structure is expected to change the nutritional quality of rice (Zheng and Wang 2014). Most nutrients of the seeds are stored in the endosperm. The triploid endosperm of rice develops from the fertilized polar nucleus. The endosperm at the filling stage consists of the aleurone layer, subaleurone layer, and starch endosperm, from outside to inside, respectively (Wu et al. 2016a). The cells of the aleurone layer of the mature endosperm are living, whereas the cells in the starch endosperm are dead. As a transitional cell type, the subaleurone layer cells accumulate both starch and protein in the early stage of development and differentiate into the starch endosperm in the late stage of endosperm development. The aleurone layer contains a large number of nutrient elements, such as proteins, vitamins, and minerals (Becraft and Yi 2011).
TA1, encoding a single-stranded DNA binding protein, OsmtSSB1 (located in mitochondria), is highly expressed in the aleurone layer, subaleurone layer and the caryopsis embryo, but not in the starch endosperm. The thickness of the aleurone layer of the mutant ta1 was about twice as thick as that of the wild-type, and the nutritional quality was greatly improved. In the ta1 mutant, the content of all nutrient elements increased (Li et al. 2021). In the mutant ta2-1, its grain transparency, 1000-grain weight, seed setting rate, total starch and AC were decreased; however, its nutritional composition of shelled grains increased, including the total protein, lipid, iron, zinc, calcium, dietary fiber, antioxidants, phenols, and vitamins (Liu et al. 2018a).
Although brown rice is rich in nutrients, it is accepted by few consumers in the market because of its poor taste. How to balance taste and nutrition is an urgent problem that should be solved in the future. Currently, using germinated brown rice seems to be a reasonable alternative, which maintains the nutritional quality of rice but improves its edible quality (Cho and Lim 2016).
The Seed Coat
Brown rice or dehulled rice refers to rice with the hull removed, and further polishing generates milled white rice, which is generally sold in the market and consumed by people. However, brown rice contains most of the nutritional substances, including dietary fiber, vitamins, and phenolics, most of which are lost during the polishing process. Therefore, brown rice, as the whole grain, contains more nutritional components. Moreover, brown rice with a red, purple, or black pericarp is more beneficial to human health than traditional white pericarp rice because of the accumulation of more antioxidant compounds. Red rice pigmentation is controlled by the joint action of two genes Rc and Rd (Sweeney et al. 2006; Furukawa et al. 2007). Rc, encoding a basic helix ring helix (bHLH) transcription factor, is responsible for the accumulation of the pigment, and Rd/OsDFR, encoding a dihydroflavonol 4-reductase, enhances the accumulation of procyanidin in the brown grain pericarp (Sweeney et al. 2006; Furukawa et al. 2007). Rc and Rd together cause the peel to be red and most cultivated rice varieties that produce white grains have a 14 bp frameshift deletion of the seventh exon of Rc (Furukawa et al. 2007). Recently, the recessive Rc allele with 14 bp deletion was functionally restored to an in-frame mutation via the CRISPR/Cas9 method, which successfully transformed three excellent white grain varieties into red grains, thus remarkably promoting the content of procyanidin and anthocyanin (Zhu et al. 2019).
The black rice pigment is also regulated by the joint action of two genes, Pb and Pp (Hu et al. 1996; Wang and Shu 2007). The Pb locus is composed of two genes, encoding a MYC transcription factor and a bHLH16 transcription factor, which are involved in the synthesis of anthocyanin and procyanidins, respectively. The expression of Pb seems to be the cause of pigment accumulation in the pericarp of brown grains, whereas the expression of Pp increases the content of pigment, resulting in purple grains. The copy number of the Pp gene correlates with the intensity of purple pigmentation. In the absence of Pp, Pb plants produce brown grains, while Pp plants without Pb have white grains (Tanaka et al. 2008; Rahman et al. 2013; Mbanjo et al. 2020).
The gene corresponding to black rice is Kala4/OsB2, encoding a bHLH transcription factor. The structural rearrangement of its promoter region leads to its ectopic expression, resulting in a black pericarp. OsB2 regulates a number of genes encoding anthocyanin synthesis related enzymes, including F3H, DFR, and ANS (Oikawa et al. 2015).
Recently, a C-S-A gene model of rice husk pigmentation was proposed. C1 and A1 jointly determine the color change, while S1 diversifies the pigment tissue. C1 encodes an R2R3-MYB transcription factor and acts as a color producing gene. S1 encodes a bHLH protein that functions in a tissue-specific manner. C1 interacts with S1 and activates the expression of A1, which encodes dihydroflavonol reductase, thus generating purple rice pigment. The involvement of functional A1 leads to high accumulation of cyano 3-o-glucoside, while A1 mutation results in a brown pigment. Instead of C1, rice pigment color is produced by the synergistic regulation of S1 and other MYB transcription factors (Sun et al. 2018b).
More recently, a study showed that OsTTG1, the WD40 gene in rice, is an important regulator of anthocyanin biosynthesis. The OsTTG1 protein is located in the nucleus and directly interacts with Kala4, OsC1, OsDFR, and Rc, which are determinants of pigments or anthocyanin biosynthesis. Knockout of OsTTG1 reduced the anthocyanin content of the mutant grain to only 0.15% of that of wild-type plants, suggesting that OsTTG1 is a vital regulator of rice anthocyanin biosynthesis (Yang et al. 2021c).
Conclusion and Perspectives
Rice grain quality, a typical quantitative trait, is influenced by complex genetic regulation and environmental factors. Rice quality generally includes milling quality, appearance quality, nutritional quality, and eating and cooking quality. The structure and composition of the rice seed are correlate closely with different aspects of rice quality. For example, grain size not only affects rice AQ, but also influences rice MQ and ECQ. Moreover, the rice endosperm, as the edible part of rice, is composed of starch, protein, lipids, and other micronutrients. The quantity and quality of these component are major contributors to rice ECQ, AQ, and NQ. Therefore, modifying and optimizing the structure and composition of rice seed are crucial to promoting its quality. Considering that rice growth and development are controlled by numerous genes and complicated regulation networks, the cloning of key genes involved in regulating specific grain trait and the dissection their molecular mechanisms will provide valuable gene resources and essential information for breeding high quality rice. In the present review, we summarized the cloned genes and their molecular functions that determine each part of rice seeds and their corresponding roles in regulating rice quality. In particular, the genes involved in controlling seed size and endosperm components are highlighted, which are also the two major determinants of rice quality.
Although impressive progress has been made in rice quality research, few cloned genes are suitable for use in high quality rice breeding programs. In addition, a huge gap still exists in our understanding of the regulatory network of rice grain quality. Therefore, a number of urgent problems remain to be solved. First, there is a contradiction between grain yield and rice quality; therefore, how to improve rice quality without sacrificing rice yield is an important issue. Second, it is a great challenge to explore the upstream and downstream components of known rice quality regulators, as well as the crosstalk between various regulation pathways. Third, the evaluation of the effects of most cloned rice quality genes or their elite alleles are limited and fragmented, being based on the analysis in a only a few rice varieties, especially some so-called model varieties. The performance of quality traits are genetic and environment-dependent, which often causes the silencing of some reported high quality genes in breeding practice. Fourth, some rice quality determinants often play contradictory roles in the context of different quality traits. For example, the protein content correlates positively with rice NQ. However, the higher the protein content, the more protein bodies accumulate among starch grains, resulting in increased chalkiness and the deterioration of ECQ. For lipids, the oxidation of unsaturated fatty acids helps to improve the flavor of rice. However, it is not conducive to long-term storage because the hydrolysis and oxidation of lipids will worsen the appearance and taste of rice. Finally, we lack universal and unified rice quality evaluation standards. Rice quality is relative and context-specific; thus the evaluation of rice quality depends on their application mode and consumer background, including region, age, or even religion.
Although rice quality research remains challenging, certain strategies should be employed in future studies. Most importantly, more genes encoding proteins with specific roles in the control of rice quality should be cloned, sequenced, and functionally analyzed, thus enlarging the high quality gene pool. In addition, powerful and accurate technologies can be used in basic research and in breeding practices of high quality rice, such as genome-wide selection, enhanced selection of major genes for grain quality, and precise gene knock-in or knock-out. For example, based on re-sequencing data of 200 japonica rice varieties in central China, Xiao et al. (2021) revealed a number of superior alleles for rice quality and rice blast resistance by using genome-wide association mapping and selection approach. Next, the alleles related to blast-resistance and excellent rice ECQ, such as Wxmp allele, corresponding to low amylose content, were successfully introduced into two high-yield rice cultivars. Hence, two elite rice lines, XY99 and JXY1, with both ECQ and blast resistance improved were efficiently developed (Xiao et al. 2021). In addition, the corresponding breeding character targets, as well as universal and objective rice quality evaluation standards according to regional or population preferences of consumers, should be established as soon as possible, which will help to accelerate high quality rice research.
Acknowledgements
Not applicable.
Abbreviations
- AAP10
Amino acid permease 10
- ABA
Abscisic acid
- AC
Amylose content
- ACOT
Acyl-CoA thioesterase
- AGPases
ADP-glucose pyrophosphorylases
- AK2
Aspartate kinase 2
- ALK
Alkali digestion
- AQ
Appearance quality
- AP2
APETALA2
- Badh2
Betaine aldehyde dehydrogenase 2
- BAK1
BRI1-ASSOCIATED RECEPTOR KINASE
- BG3
Big grain 3
- BR
Brassinosteroid
- BRD1
BR-deficient dwarf1
- BRI1
BRASSINOSTEROID-INSENSITIVE 1
- BZR1
BRASSINAZOLE-RESISTANT 1
- CaMBD
Calmodulin-binding domain
- CLG1-1
Chang Li Geng1-1
- CORVET
The class C core vacuole/endosome tethering
- D1
Dwarf 1
- DBE
Debranching enzyme
- DEP1
Dense and erect panicle 1
- DHPS
Dihydrodipicolinate synthase
- DLT
Dwarf and low-tillering
- ECQ
Eating and cooking quality
- FAD
Fatty acid desaturase
- FER1
Ferritin gene 1
- FGR1
Floury and growth retardation 1
- FLO6
FLOURY ENDOSPERM 6
- FLR
FERONIA-like receptor
- FSE1
Floury Shrunken Endosperm1
- GA
Gibberellin
- GABA
γ-Aminobutyric acid
- GAD
Glutamate decarboxylase
- GAD1
Grain length and awn development 1
- GBSS
Granule-bound starch synthase
- GC
Gel consistency
- GE
GIANT EMBRYO
- GGC
G protein gamma subunit
- GL3.1
Grain length 3.1
- GluA1
Glutelin type-A1
- GPA1
GLUTELIN PRECURSOR ACCUMULATION1
- GR
Golden rice
- GRF4
Growth-regulating factor 4
- GS3
QTL for grain size on chromosome 3
- GS5
Grain size on chromosome 5
- GS9
Grain shape gene on chromosome 9
- GSK2
GSK3/SHAGGY-like kinase 2
- GSN1
Grain size and number1
- GT
Gelatinization temperature
- GW2
Grain weight and width on chromosome 2
- GW5
QTL for grain width and weight on chromosome 5
- GW6
Grain width 6
- GW6a
Grain weight on chromosome 6-a
- GZF1
GluB-1-binding Zinc Finger 1
- HDR3
Homolog of DA1 on rice chromosome 3
- IRT
Iron regulated transporters
- ISA
Isoamylase
- LAC
Laccase
- LG1
Large grain 1
- LHT1
Lysine-histidine-type transporter 1
- LKR
Lysine ketoglutaric acid reductase
- LOX-2
Lipoxygenase 2
- Lys
Lysine
- MADS6
MADS-box transcription factor 6
- MAPK
Mitogen-activated protein kinase
- MKKs
MAPK kinases
- MKKKs
MKK kinases
- MQ
Milling quality
- NAC20
NAC domain-containing protein 20
- NAS
Nicotinamide synthase
- NF-YB1
Nuclear transcription factor Y subunit B1
- NQ
Nutrition quality
- OFP
OVATE family protein
- PA
Polyamine
- PGL1
Positive regulator of grain length 1
- PHI-1
Phosphorylation inducible protein 1
- PHO
Starch phosphorylase
- pk2
Plastidic pyruvate kinase 2
- PLDα1
Phospholipase Dα 1
- POW1
Put on weight 1
- PPDKB
Pyruvate phosphate dikinase
- PPKL1
Protein phosphatase with Kelch-like repeat domain 1
- QTL
Quantitative trait locus
- Rab5a
Small GTPase RAT BRAIN 5a
- RBP
RNA‐binding protein
- RGA
G protein alpha subunit
- RGB
G protein beta subunit
- RGG
Heterotrimeric G protein γ subunit
- RISC
RNA-induced silencing complex
- RLRH1
Rice lysine rich histone proteins 1
- RVA
Rapid visco analyzer
- SBE
Starch branching enzyme
- SDH
Saccharo-pine dehydropine dehydrogenase
- SMOS1
Small organ size 1
- SNARE
Soluble N-ethylmaleimide sensitive factor attachment protein receptor
- SNB
SUPERNUMERARY BRACT
- SPL
SQUAMOSA promoter binding protein-like
- SRS5
Small and round seed 5
- SSH1
Suppression of shattering 1
- SSRGs
Starch synthesis related genes
- SSS
Soluble starch synthase
- TA1
Thick aleurone 1
- TF
Transcription factor
- TGW6
Thousand-grain weight 6
- TTG1
TRANSPARENT TESTA GLABRA1
- TUD1
Taihu Dwarf 1
- UBP15
Ubiquitin-specific protease 15
- UPP
Ubiquitin proteasome pathway
- VIT1
Vacuolar iron transporter 1
- WTG1
Wide and thick grain 1
- Wx
Waxy
- YSL9
Yellow stripe1-like 9
- YUC11
YUCCA (YUC) flavin-containing monooxygenase 11
- ZRT
Zinc regulated transporters
Authors' Contributions
Q.F.L. and Q.Q.L. proposed the concept; P.L., Y.H.C., J.L., C.Q.Z., Q.Q.L. and Q.F.L. contributed to the writing of the manuscript; Q.F.L. and Q.Q.L. revised and finalized the manuscript. All the authors listed have participated in the work sufficiently. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32071984), the Hainan Yazhou Bay Seed Lab (B21HJ8105), and the Programs from Jiangsu Province Government (BK20200045, JBGS[2021]001, SWYY-154 and PAPD).
Availability of Data and Materials
Not applicable.
Declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
The manuscript has been approved by all authors.
Competing interests
The authors declare no competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Pei Li and Yu-Hao Chen have contributed equally to this work
Contributor Information
Qiao-Quan Liu, Email: qqliu@yzu.edu.cn.
Qian-Feng Li, Email: qfli@yzu.edu.cn.
References
- Akabane T, Suzuki N, Tsuchiya W, et al. Expression, purification and crystallization of TGW6, which limits grain weight in rice. Protein Expr Purif. 2021;188:105975. doi: 10.1016/j.pep.2021.105975. [DOI] [PubMed] [Google Scholar]
- Akama K, Takaiwa F. C-terminal extension of rice glutamate decarboxylase (OsGAD2) functions as an autoinhibitory domain and overexpression of a truncated mutant results in the accumulation of extremely high levels of GABA in plant cells. J Exp Bot. 2007;58:2699–2707. doi: 10.1093/jxb/erm120. [DOI] [PubMed] [Google Scholar]
- Akama K, Kanetou J, Shimosaki S, et al. Seed-specific expression of truncated OsGAD2 produces GABA-enriched rice grains that influence a decrease in blood pressure in spontaneously hypertensive rats. Transgenic Res. 2009;18:865–876. doi: 10.1007/s11248-009-9272-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akama K, Akter N, Endo H, et al. An in vivo targeted deletion of the calmodulin-binding domain from rice glutamate decarboxylase 3 (OsGAD3) increases γ-aminobutyric acid content in grains. Rice. 2020;13:20. doi: 10.1186/s12284-020-00380-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikari M, Wu J, Yano M, et al. Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the alpha-subunit of GTP-binding protein. Proc Natl Acad Sci USA. 1999;96:10284–10289. doi: 10.1073/pnas.96.18.10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aya K, Hobo T, Sato-Izawa K, et al. A novel AP2-type transcription factor, SMALL ORGAN SIZE1, controls organ size downstream of an auxin signaling pathway. Plant Cell Physiol. 2014;55:897–912. doi: 10.1093/pcp/pcu023. [DOI] [PubMed] [Google Scholar]
- Azizi P, Osman M, Hanafi M, et al. Molecular insights into the regulation of rice kernel elongation. Crit Rev Biotechnol. 2019;39:904–923. doi: 10.1080/07388551.2019.1632257. [DOI] [PubMed] [Google Scholar]
- Bao J. Genes and QTLs for rice grain quality improvement. Rice Germplasm Genet Improv. 2014;9:239–278. [Google Scholar]
- Bao JS, Corke H, Sun M. Nucleotide diversity in starch synthase IIa and validation of single nucleotide polymorphisms in relation to starch gelatinization temperature and other physicochemical properties in rice (Oryza sativa L.) Theor Appl Genet. 2006;113:1171–1183. doi: 10.1007/s00122-006-0355-6. [DOI] [PubMed] [Google Scholar]
- Baysal C, He W, Drapal M, et al. Inactivation of rice starch branching enzyme IIb triggers broad and unexpected changes in metabolism by transcriptional reprogramming. Proc Natl Acad Sci U S A. 2020;117:26503–26512. doi: 10.1073/pnas.2014860117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft PW, Yi G. Regulation of aleurone development in cereal grains. J Exp Bot. 2011;62:1669–1675. doi: 10.1093/jxb/erq372. [DOI] [PubMed] [Google Scholar]
- Bello BK, Hou Y, Zhao J, et al. NF-YB1-YC12-bHLH144 complex directly activates Wx to regulate grain quality in rice (Oryza sativa L.) Plant Biotechnol J. 2019;17:1222–1235. doi: 10.1111/pbi.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury L, Fitzgerald TL, Henry RJ, et al. The gene for fragrance in rice. Plant Biotechnol J. 2010;3:363–370. doi: 10.1111/j.1467-7652.2005.00131.x. [DOI] [PubMed] [Google Scholar]
- Buttery R, Ling L, Juliano B, et al. Cooked rice aroma and 2-acetyl-1-pyrroline. J Agric Food Chem. 1983;31:823–826. [Google Scholar]
- Cai Y, Li S, Jiao G, et al. OsPK2 encodes a plastidic pyruvate kinase involved in rice endosperm starch synthesis, compound granule formation and grain filling. Plant Biotechnol J. 2018;16:1878–1891. doi: 10.1111/pbi.12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che R, Tong H, Shi B, et al. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat Plants. 2015;2:15195. doi: 10.1038/nplants.2015.195. [DOI] [PubMed] [Google Scholar]
- Chen S, Wu J, Yang Y, et al. The fgr gene responsible for rice fragrance was restricted within 69 kb. Plant Sci. 2006;171:505–514. doi: 10.1016/j.plantsci.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Chen S, Yang Y, Shi W, et al. Badh2, encoding betaine aldehyde dehydrogenase, inhibits the biosynthesis of 2-acetyl-1-pyrroline, a major component in rice fragrance. Plant Cell. 2008;20:1850–1861. doi: 10.1105/tpc.108.058917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sun A, Wang M, et al. Functions of the CCCH type zinc finger protein OsGZF1 in regulation of the seed storage protein GluB-1 from rice. Plant Mol Biol. 2014;84:621–634. doi: 10.1007/s11103-013-0158-5. [DOI] [PubMed] [Google Scholar]
- Chen E, Huang X, Tian Z, et al. The genomics of oryza species provides insights into rice domestication and heterosis. Annu Rev Plant Biol. 2019;70:639–665. doi: 10.1146/annurev-arplant-050718-100320. [DOI] [PubMed] [Google Scholar]
- Chen Z, Lu Y, Feng L, et al. Genetic dissection and functional differentiation of ALKa and ALKb, two natural alleles of the ALK/SSIIa gene, responding to low gelatinization temperature in rice. Rice. 2020;13:39. doi: 10.1186/s12284-020-00393-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Yu H, Jiang W, et al. Overexpression of ovate family protein 22 confers multiple morphological changes and represses gibberellin and brassinosteroid signalings in transgenic rice. Plant Sci. 2021;304:110734. doi: 10.1016/j.plantsci.2020.110734. [DOI] [PubMed] [Google Scholar]
- Cheng F, Zhong LJ, Wang F, et al. Differences in cooking and eating properties between chalky and translucent parts in rice grains. Food Chem. 2005;90:39–46. [Google Scholar]
- Cho DH, Lim ST. Germinated brown rice and its bio-functional compounds. Food Chem. 2016;196:259–271. doi: 10.1016/j.foodchem.2015.09.025. [DOI] [PubMed] [Google Scholar]
- Chou HL, Tian L, Kumamaru T, et al. Multifunctional RNA binding protein OsTudor-SN in storage protein mRNA transport and localization. Plant Physiol. 2017;175:1608–1623. doi: 10.1104/pp.17.01388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HL, Tian L, Fukuda M, et al. The role of RNA-binding protein OsTudor-SN in post-transcriptional regulation of seed storage proteins and endosperm development. Plant Cell Physiol. 2019;60:2193–2205. doi: 10.1093/pcp/pcz113. [DOI] [PubMed] [Google Scholar]
- Ding W, Lin L, Zhang B, et al. OsKASI, a β-ketoacyl-[acyl carrier protein] synthase I, is involved in root development in rice (Oryza sativa L.) Planta. 2015;242:203–213. doi: 10.1007/s00425-015-2296-2. [DOI] [PubMed] [Google Scholar]
- Dong OX, Yu S, Jain R, et al. Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR-Cas9. Nat Commun. 2020;11:1178. doi: 10.1038/s41467-020-14981-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroshenk KA, Tian L, Crofts AJ, et al. Characterization of RNA binding protein RBP-P reveals a possible role in rice glutelin gene expression and RNA localization. Plant Mol Biol. 2014;85:381–394. doi: 10.1007/s11103-014-0191-z. [DOI] [PubMed] [Google Scholar]
- Duan M, Sun S. Profiling the expression of genes controlling rice grain quality. Plant Mol Biol. 2005;59:165–178. doi: 10.1007/s11103-004-7507-3. [DOI] [PubMed] [Google Scholar]
- Duan P, Rao Y, Zeng D, et al. SMALL GRAIN 1, which encodes a mitogen-activated protein kinase kinase 4, influences grain size in rice. Plant J. 2014;77:547–557. doi: 10.1111/tpj.12405. [DOI] [PubMed] [Google Scholar]
- Duan P, Ni S, Wang J, et al. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat Plants. 2015;2:15203. doi: 10.1038/nplants.2015.203. [DOI] [PubMed] [Google Scholar]
- Duan P, Xu J, Zeng D, et al. Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol Plant. 2017;10:685–694. doi: 10.1016/j.molp.2017.03.009. [DOI] [PubMed] [Google Scholar]
- Evenson RE, Gollin D. Assessing the impact of the green revolution, 1960 to 2000. Science. 2003;300:758–762. doi: 10.1126/science.1078710. [DOI] [PubMed] [Google Scholar]
- Fan C, Xing Y, Mao H, et al. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet. 2006;112:1164–1171. doi: 10.1007/s00122-006-0218-1. [DOI] [PubMed] [Google Scholar]
- Federico ML, Schmidt MA. Modern breeding and biotechnological approaches to enhance carotenoid accumulation in seeds. Subcell Biochem. 2016;79:345–358. doi: 10.1007/978-3-319-39126-7_13. [DOI] [PubMed] [Google Scholar]
- Fujisawa Y, Kato T, Ohki S, et al. Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc Natl Acad Sci USA. 1999;96:7575–7580. doi: 10.1073/pnas.96.13.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Kondo T, et al. Characterization of SSIIIa-deficient mutants of rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol. 2007;144:2009–2023. doi: 10.1104/pp.107.102533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Maekawa M, Oki T, et al. The Rc and Rd genes are involved in proanthocyanidin synthesis in rice pericarp. Plant J. 2007;49:91–102. doi: 10.1111/j.1365-313X.2006.02958.x. [DOI] [PubMed] [Google Scholar]
- Gao Z, Zeng D, Cui X, et al. Map-based cloning of the ALK gene, which controls the gelatinization temperature of rice. Sci China C Life Sci. 2003;46:661–668. doi: 10.1360/03yc0099. [DOI] [PubMed] [Google Scholar]
- Gao X, Zhang J, Zhang X, et al. Rice qGL3/OsPPKL1 functions with the GSK3/SHAGGY-like kinase OsGSK3 to modulate brassinosteroid signaling. Plant Cell. 2019;31:1077–1093. doi: 10.1105/tpc.18.00836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Zhang N, Wang W, et al. The ubiquitin-interacting motif-type ubiquitin receptor HDR3 interacts with and stabilizes the histone acetyltransferase GW6a to control the grain size in rice. Plant Cell. 2021;33:3331–3347. doi: 10.1093/plcell/koab194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Chen K, Dong N, et al. GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell. 2018;30:871–888. doi: 10.1105/tpc.17.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo N, Gu M, Hu J, et al. Rice OsLHT1 Functions in leaf-to-panicle nitrogen allocation for grain yield and quality. Front Plant Sci. 2020;11:1150. doi: 10.3389/fpls.2020.01150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo N, Hu J, Yan M, et al. Oryza sativa Lysine-Histidine-type Transporter 1 functions in root uptake and root-to-shoot allocation of amino acids in rice. Plant J. 2020;103:395–411. doi: 10.1111/tpj.14742. [DOI] [PubMed] [Google Scholar]
- Hao Y, Wang Y, Wu M, et al. The nuclear-localized PPR protein OsNPPR1 is important for mitochondrial function and endosperm development in rice. J Exp Bot. 2019;70:4705–4720. doi: 10.1093/jxb/erz226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, Wang D, Wu Y, et al. The GW2-WG1-OsbZIP47 pathway controls grain size and weight in rice. Mol Plant. 2021;14:1266–1280. doi: 10.1016/j.molp.2021.04.011. [DOI] [PubMed] [Google Scholar]
- He W, Wang L, Lin Q, et al. Rice seed storage proteins: Biosynthetic pathways and the effects of environmental factors. J Integr Plant Biol. 2021;63:1999–2019. doi: 10.1111/jipb.13176. [DOI] [PubMed] [Google Scholar]
- Heang D, Sassa H. Antagonistic actions of HLH/bHLH proteins are involved in grain length and weight in rice. PLoS ONE. 2012;7:e31325. doi: 10.1371/journal.pone.0031325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Yoshida H, Aya K, et al. SMALL ORGAN SIZE 1 and SMALL ORGAN SIZE 2/DWARF AND LOW-TILLERING form a complex to integrate Auxin and Brassinosteroid signaling in rice. Mol Plant. 2017;10:590–604. doi: 10.1016/j.molp.2016.12.013. [DOI] [PubMed] [Google Scholar]
- Hiroyuki S, Yasuhiro S, Makoto S, et al. Molecular characterization of Wx-mq, a novel Mutant gene for low-amylose content in endosperm of rice (Oryza sativa L.) Breed Sci. 2002;2:131–135. [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Shimizu-Sato S, et al. Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 2002;32:495–508. doi: 10.1046/j.1365-313x.2002.01438.x. [DOI] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Fujioka S, et al. The Rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell. 2005;17:2243–2254. doi: 10.1105/tpc.105.030973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Suzuki K, Iijima K, et al. Variation in cooking and eating quality traits in Japanese rice germplasm accessions. Breed Sci. 2016;66:309–318. doi: 10.1270/jsbbs.66.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Anderson B, Wessler SR. Isolation and characterization of rice R genes: evidence for distinct evolutionary paths in rice and maize. Genetics. 1996;142:1021–1031. doi: 10.1093/genetics/142.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, He H, Zhang S, et al. A Kelch motif-containing serine/threonine protein phosphatase determines the large grain QTL trait in rice. J Integr Plant Biol. 2012;54:979–990. doi: 10.1111/jipb.12008. [DOI] [PubMed] [Google Scholar]
- Hu X, Qian Q, Xu T, et al. The U-box E3 ubiquitin ligase TUD1 functions with a heterotrimeric G α subunit to regulate Brassinosteroid-mediated growth in rice. PLoS Genet. 2013;9:e1003391. doi: 10.1371/journal.pgen.1003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Wang Y, Fang Y, et al. A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant. 2015;8:1455–1465. doi: 10.1016/j.molp.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Hu Z, Lu S, Wang M, et al. A novel QTL qTGW3 encodes the GSK3/SHAGGY-Like Kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol Plant. 2018;11:736–749. doi: 10.1016/j.molp.2018.03.005. [DOI] [PubMed] [Google Scholar]
- Hu J, Huang L, Chen G, et al. The elite alleles of OsSPL4 regulate grain size and increase grain yield in rice. Rice. 2021;14:90. doi: 10.1186/s12284-021-00531-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Qian Q, Liu Z, et al. Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet. 2009;41:494–497. doi: 10.1038/ng.352. [DOI] [PubMed] [Google Scholar]
- Huang R, Jiang L, Zheng J, et al. Genetic bases of rice grain shape: so many genes, so little known. Trends Plant Sci. 2013;18:218–226. doi: 10.1016/j.tplants.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Huang J, Cai M, Long Q, et al. OsLOX2, a rice type I lipoxygenase, confers opposite effects on seed germination and longevity. Transgenic Res. 2014;23:643–655. doi: 10.1007/s11248-014-9803-2. [DOI] [PubMed] [Google Scholar]
- Huang K, Wang D, Duan P, et al. WIDE AND THICK GRAIN 1, which encodes an otubain-like protease with deubiquitination activity, influences grain size and shape in rice. Plant J. 2017;91:849–860. doi: 10.1111/tpj.13613. [DOI] [PubMed] [Google Scholar]
- Huang L, Li Q, Zhang C, et al. Creating novel Wx alleles with fine-tuned amylose levels and improved grain quality in rice by promoter editing using CRISPR/Cas9 system. Plant Biotechnol J. 2020;18:2164–2166. doi: 10.1111/pbi.13391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Tan H, Zhang C, et al. Starch biosynthesis in cereal endosperms: an updated review over the last decade. Plant Commun. 2021;2:100237. doi: 10.1016/j.xplc.2021.100237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Gu Z, Chen Z, et al. Improving rice eating and cooking quality by coordinated expression of the major starch synthesis-related genes, SSII and Wx, in endosperm. Plant Mol Biol. 2021;106:419–432. doi: 10.1007/s11103-021-01162-8. [DOI] [PubMed] [Google Scholar]
- Hui S, Li H, Mawia AM, et al. Production of aromatic three-line hybrid rice using novel alleles of BADH2. Plant Biotechnol J. 2021;20:59–74. doi: 10.1111/pbi.13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Higuchi K, Takahashi M, et al. Three rice nicotianamine synthase genes, OsNAS1, OsNAS2, and OsNAS3 are expressed in cells involved in long-distance transport of iron and differentially regulated by iron. Plant J. 2003;36:366–381. doi: 10.1046/j.1365-313x.2003.01878.x. [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Suzuki M, Tsukamoto T, et al. Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+ Plant J. 2006;45:335–346. doi: 10.1111/j.1365-313X.2005.02624.x. [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Kim S, Tsukamoto T, et al. Mutational reconstructed ferric chelate reductase confers enhanced tolerance in rice to iron deficiency in calcareous soil. Proc Natl Acad Sci USA. 2007;104:7373–7378. doi: 10.1073/pnas.0610555104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y, Masuda H, Suzuki M, et al. Overexpression of the OsZIP4 zinc transporter confers disarrangement of zinc distribution in rice plants. J Exp Bot. 2007;58:2909–2915. doi: 10.1093/jxb/erm147. [DOI] [PubMed] [Google Scholar]
- Ishimaru K, Hirotsu N, Madoka Y, et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet. 2013;45:707–711. doi: 10.1038/ng.2612. [DOI] [PubMed] [Google Scholar]
- Jeon JS, Ryoo N, Hahn TR, et al. Starch biosynthesis in cereal endosperm. Plant Physiol Biochem. 2010;48:383–392. doi: 10.1016/j.plaphy.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Jiang L, Ma X, Zhao S, et al. The APETALA2-Like Transcription Factor SUPERNUMERARY BRACT Controls Rice Seed Shattering and Seed Size. Plant Cell. 2019;31:17–36. doi: 10.1105/tpc.18.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Hua L, Zhu Z, et al. GAD1 Encodes a secreted peptide that regulates grain number, grain length, and awn development in rice domestication. Plant Cell. 2016;28:2453–2463. doi: 10.1105/tpc.16.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir M, Nonhebel H. Reinvestigation of THOUSAND-GRAIN WEIGHT 6 grain weight genes in wheat and rice indicates a role in pollen development rather than regulation of auxin content in grains. Theor Appl Genet. 2021;134:2051–2062. doi: 10.1007/s00122-021-03804-3. [DOI] [PubMed] [Google Scholar]
- Kato C, Mizutani T, Tamaki H, et al. Characterization of heterotrimeric G protein complexes in rice plasma membrane. Plant J. 2004;38:320–331. doi: 10.1111/j.1365-313X.2004.02046.x. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Bhullar NK. Molecular processes in iron and zinc homeostasis and their modulation for biofortification in rice. J Integr Plant Biol. 2018;60:1181–1198. doi: 10.1111/jipb.12751. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Bhullar NK. Delineating the future of iron biofortification studies in rice: challenges and future perspectives. J Exp Bot. 2021;72:2099–2113. doi: 10.1093/jxb/eraa446. [DOI] [PubMed] [Google Scholar]
- Kawakatsu T, Takaiwa F. Cereal seed storage protein synthesis: fundamental processes for recombinant protein production in cereal grains. Plant Biotechnol J. 2010;8:939–953. doi: 10.1111/j.1467-7652.2010.00559.x. [DOI] [PubMed] [Google Scholar]
- Kawakatsu T, Yamamoto MP, Touno SM, et al. Compensation and interaction between RISBZ1 and RPBF during grain filling in rice. Plant J. 2009;59:908–920. doi: 10.1111/j.1365-313X.2009.03925.x. [DOI] [PubMed] [Google Scholar]
- Khan MSS, Basnet R, Islam SA, et al. Mutational analysis of OsPLDalpha1 reveals its involvement in phytic acid biosynthesis in rice grains. J Agric Food Chem. 2019;67:11436–11443. doi: 10.1021/acs.jafc.9b05052. [DOI] [PubMed] [Google Scholar]
- Khan MSS, Basnet R, Ahmed S, et al. Mutations of OsPLDa1 increase lysophospholipid content and enhance cooking and eating quality in rice. Plants. 2020;9:390–400. doi: 10.3390/plants9030390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach M, Calingacion M, Fitzgerald M, et al. The origin and evolution of fragrance in rice (Oryza sativa L.) Proc Natl Acad Sci USA. 2009;106:14444–14449. doi: 10.1073/pnas.0904077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Qu Q, Takaiwa F. Evaluation of tissue specificity and expression strength of rice seed component gene promoters in transgenic rice. Plant Biotechnol J. 2004;2:113–125. doi: 10.1111/j.1467-7652.2004.00055.x. [DOI] [PubMed] [Google Scholar]
- le Qu Q, Xing YP, Liu WX, et al. Expression pattern and activity of six glutelin gene promoters in transgenic rice. J Exp Bot. 2008;59:2417–2424. doi: 10.1093/jxb/ern110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, An G. Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant Cell Environ. 2009;32:408–416. doi: 10.1111/j.1365-3040.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- Lee S, Chiecko JC, Kim SA, et al. Disruption of OsYSL15 leads to iron inefficiency in rice plants. Plant Physiol. 2009;150:786–800. doi: 10.1104/pp.109.135418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Jeong HJ, Kim SA, et al. OsZIP5 is a plasma membrane zinc transporter in rice. Plant Mol Biol. 2010;73:507–517. doi: 10.1007/s11103-010-9637-0. [DOI] [PubMed] [Google Scholar]
- Lee S, Kim SA, Lee J, et al. Zinc deficiency-inducible OsZIP8 encodes a plasma membrane-localized zinc transporter in rice. Mol Cells. 2010;29:551–558. doi: 10.1007/s10059-010-0069-0. [DOI] [PubMed] [Google Scholar]
- Leng Y, Xue D, Yang Y, et al. Mapping of QTLs for eating and cooking quality-related traits in rice (Oryza sativa L.) Euphytica. 2014;197:99–108. [Google Scholar]
- Li N, Li Y. Signaling pathways of seed size control in plants. Curr Opin Plant Biol. 2016;33:23–32. doi: 10.1016/j.pbi.2016.05.008. [DOI] [PubMed] [Google Scholar]
- Li Y, Fan C, Xing Y, et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet. 2011;43:1266–1269. doi: 10.1038/ng.977. [DOI] [PubMed] [Google Scholar]
- Li D, Wang L, Wang M, et al. Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol J. 2009;7:791–806. doi: 10.1111/j.1467-7652.2009.00444.x. [DOI] [PubMed] [Google Scholar]
- Li S, Gao F, Xie K, et al. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol J. 2016;14:2134–2146. doi: 10.1111/pbi.12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Huang L, Chu R, et al. Down-regulation of SSSII-2 gene expression results in novel low-amylose rice with soft, transparent grains. J Agric Food Chem. 2018;66:9750–9760. doi: 10.1021/acs.jafc.8b02913. [DOI] [PubMed] [Google Scholar]
- Li Q, Lu J, Yu J, et al. The brassinosteroid-regulated transcription factors BZR1/BES1 function as a coordinator in multisignal-regulated plant growth. Biochim Biophys Acta Gene Regul Mech. 2018;1861:561–571. doi: 10.1016/j.bbagrm.2018.04.003. [DOI] [PubMed] [Google Scholar]
- Li N, Xu R, Li Y. Molecular networks of seed size control in plants. Annu Rev Plant Biol. 2019;70:435–463. doi: 10.1146/annurev-arplant-050718-095851. [DOI] [PubMed] [Google Scholar]
- Li DQ, Wu XB, Wang HF, et al. Defective mitochondrial function by mutation in THICK ALEURONE 1 encoding a mitochondrion-targeted single-stranded DNA-binding protein leads to increased aleurone cell layers and improved nutrition in rice. Mol Plant. 2021;14:1343–1361. doi: 10.1016/j.molp.2021.05.016. [DOI] [PubMed] [Google Scholar]
- Liu L, Ma X, Liu S, et al. Identification and characterization of a novel Waxy allele from a Yunnan rice landrace. Plant Mol Biol. 2009;71:609–626. doi: 10.1007/s11103-009-9544-4. [DOI] [PubMed] [Google Scholar]
- Liu HL, Yin ZJ, Xiao L, et al. Identification and evaluation of ω-3 fatty acid desaturase genes for hyperfortifying α-linolenic acid in transgenic rice seed. J Exp Bot. 2012;63:3279–3287. doi: 10.1093/jxb/ers051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Tong H, Xiao Y, et al. Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc Natl Acad Sci USA. 2015;112:11102–11107. doi: 10.1073/pnas.1512748112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Hua L, Dong S, et al. OsMAPK6, a mitogen-activated protein kinase, influences rice grain size and biomass production. Plant J. 2015;84:672–681. doi: 10.1111/tpj.13025. [DOI] [PubMed] [Google Scholar]
- Liu J, Chen J, Zheng X, et al. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat Plants. 2017;3:17043. doi: 10.1038/nplants.2017.43. [DOI] [PubMed] [Google Scholar]
- Liu J, Wu X, Yao X, et al. Mutations in the DNA demethylase OsROS1 result in a thickened aleurone and improved nutritional value in rice grains. Proc Natl Acad Sci USA. 2018;115:11327–11332. doi: 10.1073/pnas.1806304115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Han R, Wu K, et al. G-protein βγ subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat Commun. 2018;9:852. doi: 10.1038/s41467-018-03047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Li M, Zhang Q, et al. Exploring the molecular basis of heterosis for plant breeding. J Integr Plant Biol. 2020;62:287–298. doi: 10.1111/jipb.12804. [DOI] [PubMed] [Google Scholar]
- Liu Z, Mei E, Tian X, et al. OsMKKK70 regulates grain size and leaf angle in rice through the OsMKK4-OsMAPK6-OsWRKY53 signaling pathway. J Integr Plant Biol. 2021;63:2043–2057. doi: 10.1111/jipb.13174. [DOI] [PubMed] [Google Scholar]
- Long Q, Zhang W, Wang P, et al. Molecular genetic characterization of rice seed lipoxygenase 3 and assessment of its effects on seed longevity. J Plant Biol. 2013;56:232–242. [Google Scholar]
- Long W, Wang Y, Zhu S, et al. FLOURY SHRUNKEN ENDOSPERM1 connects phospholipid metabolism and amyloplast development in rice. Plant Physiol. 2018;177:698–712. doi: 10.1104/pp.17.01826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Liu H, Zhou T, et al. An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell. 2013;25:3360–3376. doi: 10.1105/tpc.113.113589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu J, Wang D, Duan P, et al. Control of grain size and weight by the GSK2-LARGE1/OML4 pathway in rice. Plant Cell. 2020;32:1905–1918. doi: 10.1105/tpc.19.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H, Sun S, Yao J, et al. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc Natl Acad Sci U S A. 2010;107:19579–19584. doi: 10.1073/pnas.1014419107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbanjo EGN, Kretzschmar T, Jones H, et al. The genetic basis and nutritional benefits of pigmented rice grain. Front Genet. 2020;11:229. doi: 10.3389/fgene.2020.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J, Yang Z, Zhang D, et al. Mutation of RGG2, which encodes a type B heterotrimeric G protein γ subunit, increases grain size and yield production in rice. Plant Biotechnol J. 2019;17:650–664. doi: 10.1111/pbi.13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao C, Wang D, He R, et al. Mutations in MIR396e and MIR396f increase grain size and modulate shoot architecture in rice. Plant Biotechnol J. 2020;18:491–501. doi: 10.1111/pbi.13214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami I, Uwatoko N, Ikeda Y, et al. Allelic diversification at the wx locus in landraces of Asian rice. Theor Appl Genet. 2008;116:979–989. doi: 10.1007/s00122-008-0729-z. [DOI] [PubMed] [Google Scholar]
- Miura S, Koyama N, Crofts N, et al. Generation and starch characterization of non-transgenic BEI and BEIIb double mutant rice (Oryza sativa) with ultra-high level of resistant starch. Rice. 2021;14:3. doi: 10.1186/s12284-020-00441-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Nomura T, Ooka H, et al. Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol. 2002;130:1152–1161. doi: 10.1104/pp.007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison W. Lipids in cereal starches: a review. J Cereal Sci. 1988;8:1–15. [Google Scholar]
- Nagasawa N, Hibara K, Heppard EP, et al. GIANT EMBRYO encodes CYP78A13, required for proper size balance between embryo and endosperm in rice. Plant J. 2013;75:592–605. doi: 10.1111/tpj.12223. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Ogawa I, Ishimaru Y, et al. Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Sci Plant Nutr. 2006;52:464–469. [Google Scholar]
- Nayar S, Sharma R, Tyagi AK, et al. Functional delineation of rice MADS29 reveals its role in embryo and endosperm development by affecting hormone homeostasis. J Exp Bot. 2013;64:4239–4253. doi: 10.1093/jxb/ert231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozoye T, von Wirén N, Sato Y, et al. Characterization of the nicotianamine exporter ENA1 in rice. Front Plant Sci. 2019;10:502. doi: 10.3389/fpls.2019.00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T, Maeda H, Oguchi T, et al. The birth of a black rice gene and its local spread by introgression. Plant Cell. 2015;27:2401–2414. doi: 10.1105/tpc.15.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine JA, Shipton CA, Chaggar S, et al. Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat Biotechnol. 2005;23:482–487. doi: 10.1038/nbt1082. [DOI] [PubMed] [Google Scholar]
- Paul S, Ali N, Gayen D, et al. Molecular breeding of Osfer 2 gene to increase iron nutrition in rice grain. GM Crops Food. 2012;3:310–316. doi: 10.4161/gmcr.22104. [DOI] [PubMed] [Google Scholar]
- Peng C, Wang Y, Liu F, et al. FLOURY ENDOSPERM6 encodes a CBM48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm. Plant J. 2014;77:917–930. doi: 10.1111/tpj.12444. [DOI] [PubMed] [Google Scholar]
- Pereira C, Lourenço VM, Menezes R, et al. Rice compounds with impact on diabetes control. Foods. 2021;10:1992. doi: 10.3390/foods10091992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister B, Zeeman SC. Formation of starch in plant cells. Cell Mol Life Sci. 2016;73:2781–2807. doi: 10.1007/s00018-016-2250-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingali PL. Green revolution: impacts, limits, and the path ahead. Proc Natl Acad Sci USA. 2012;109:12302–12308. doi: 10.1073/pnas.0912953109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi P, Lin Y, Song X, et al. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 2012;22:1666–1680. doi: 10.1038/cr.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J, Jiang H, Lin Y, et al. A novel miR167a-OsARF6-OsAUX3 module regulates grain length and weight in rice. Mol Plant. 2021;14:1683–1698. doi: 10.1016/j.molp.2021.06.023. [DOI] [PubMed] [Google Scholar]
- Rahman MM, Lee KE, Lee ES. The genetic constitutions of complementary genes Pp and Pb determine the purple color variation in pericarps with cyanidin-3-O-glucoside depositions in black rice. J Plant Biol. 2013;56:24–31. [Google Scholar]
- Ren Y, Wang Y, Liu F, et al. GLUTELIN PRECURSOR ACCUMULATION3 encodes a regulator of post-Golgi vesicular traffic essential for vacuolar protein sorting in rice endosperm. Plant Cell. 2014;26:410–425. doi: 10.1105/tpc.113.121376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Wang Y, Pan T, et al. GPA5 encodes a Rab5a effector required for post-golgi trafficking of rice storage proteins. Plant Cell. 2020;32:758–777. doi: 10.1105/tpc.19.00863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Huang Z, Jiang H, et al. A heat stress responsive NAC transcription factor heterodimer plays key roles in rice grain filling. J Exp Bot. 2021;72:2947–2964. doi: 10.1093/jxb/erab027. [DOI] [PubMed] [Google Scholar]
- Sasaki A, Yamaji N, Mitani-Ueno N, et al. A node-localized transporter OsZIP3 is responsible for the preferential distribution of Zn to developing tissues in rice. Plant J. 2015;84:374–384. doi: 10.1111/tpj.13005. [DOI] [PubMed] [Google Scholar]
- Segami S, Kono I, Ando T, et al. Small and round seed 5 gene encodes alpha-tubulin regulating seed cell elongation in rice. Rice. 2012;5:4. doi: 10.1186/1939-8433-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segami S, Takehara K, Yamamoto T, et al. Overexpression of SRS5 improves grain size of brassinosteroid-related dwarf mutants in rice (Oryza sativa L.) Breed Sci. 2017;67:393–397. doi: 10.1270/jsbbs.16198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoura T, Sakashita E, Kobayashi T, et al. The iron-chelate transporter OsYSL9 plays a role in iron distribution in developing rice grains. Plant Mol Biol. 2017;95:375–387. doi: 10.1007/s11103-017-0656-y. [DOI] [PubMed] [Google Scholar]
- Seung D, Smith AM. Starch granule initiation and morphogenesis-progress in Arabidopsis and cereals. J Exp Bot. 2019;70:771–784. doi: 10.1093/jxb/ery412. [DOI] [PubMed] [Google Scholar]
- Shelp BJ, Bozzo GG, Trobacher CP, et al. Hypothesis/review: contribution of putrescine to 4-aminobutyrate (GABA) production in response to abiotic stress. Plant Sci. 2012;193–194:130–135. doi: 10.1016/j.plantsci.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Shi J, Cao Y, Fan X, et al. A rice microsomal delta-12 fatty acid desaturase can enhance resistance to cold stress in yeast and Oryza sativa. Mol Breed. 2012;29:743–757. [Google Scholar]
- Shi C, Ren Y, Liu L, et al. Ubiquitin Specific Protease 15 has an important role in regulating grain width and size in rice. Plant Physiol. 2019;180:381–391. doi: 10.1104/pp.19.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, Dong N, Guo T, et al. A quantitative trait locus GW6 controls rice grain size and yield through the gibberellin pathway. Plant J. 2020;103:1174–1188. doi: 10.1111/tpj.14793. [DOI] [PubMed] [Google Scholar]
- Shimajiri Y, Oonishi T, Ozaki K, et al. Genetic manipulation of the γ-aminobutyric acid (GABA) shunt in rice: overexpression of truncated glutamate decarboxylase (GAD2) and knockdown of γ-aminobutyric acid transaminase (GABA-T) lead to sustained and high levels of GABA accumulation in rice kernels. Plant Biotechnol J. 2013;11:594–604. doi: 10.1111/pbi.12050. [DOI] [PubMed] [Google Scholar]
- Shomura A, Izawa T, Ebana K, et al. Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet. 2008;40:1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- Si L, Chen J, Huang X, et al. OsSPL13 controls grain size in cultivated rice. Nat Genet. 2016;48:447–456. doi: 10.1038/ng.3518. [DOI] [PubMed] [Google Scholar]
- Song X, Huang W, Shi M, et al. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet. 2007;39:623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- Song XJ, Kuroha T, Ayano M, et al. Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc Natl Acad Sci USA. 2015;112:76–81. doi: 10.1073/pnas.1421127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein RJ, Ricachenevsky FK, Fett JP. Differential regulation of the two rice ferritin genes (OsFER1 and OsFER2) Plant Sci. 2009;177:563–569. [Google Scholar]
- Sun L, Li X, Fu Y, et al. GS6, a member of the GRAS gene family, negatively regulates grain size in rice. J Integr Plant Biol. 2013;55:938–934. doi: 10.1111/jipb.12062. [DOI] [PubMed] [Google Scholar]
- Sun X, Ling S, Lu Z, et al. OsNF-YB1, a rice endosperm-specific gene, is essential for cell proliferation in endosperm development. Gene. 2014;551:214–221. doi: 10.1016/j.gene.2014.08.059. [DOI] [PubMed] [Google Scholar]
- Sun P, Zhang W, Wang Y, et al. OsGRF4 controls grain shape, panicle length and seed shattering in rice. J Integr Plant Biol. 2016;58:836–847. doi: 10.1111/jipb.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Wang L, Mao H, et al. A G-protein pathway determines grain size in rice. Nat Commun. 2018;9:851. doi: 10.1038/s41467-018-03141-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Zhang Z, Chen C, et al. The C-S-A gene system regulates hull pigmentation and reveals evolution of anthocyanin biosynthesis pathway in rice. J Exp Bot. 2018;69:1485–1498. doi: 10.1093/jxb/ery001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunohara H, Kawai T, Shimizu-Sato S, et al. A dominant mutation of TWISTED DWARF 1 encoding an alpha-tubulin protein causes severe dwarfism and right helical growth in rice. Genes Genet Syst. 2009;84:209–218. doi: 10.1266/ggs.84.209. [DOI] [PubMed] [Google Scholar]
- Swamy BPM, Marathi B, Ribeiro-Barros AIF, et al. Iron biofortification in rice: an update on quantitative trait loci and candidate genes. Front Plant Sci. 2021;12:647341. doi: 10.3389/fpls.2021.647341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney MT, Thomson MJ, Pfeil BE, et al. Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell. 2006;18:283–294. doi: 10.1105/tpc.105.038430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe S, Ashikari M, Fujioka S, et al. A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell. 2005;17:776–790. doi: 10.1105/tpc.104.024950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 2008;54:733–749. doi: 10.1111/j.1365-313X.2008.03447.x. [DOI] [PubMed] [Google Scholar]
- Tao Y, Miao J, Wang J, et al. RGG1, involved in the cytokinin regulatory pathway, controls grain size in rice. Rice. 2020;13:76. doi: 10.1186/s12284-020-00436-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Li X, Zhou W, et al. Transcription factor OsWRKY53 positively regulates Brassinosteroid signaling and plant architecture. Plant Physiol. 2017;175:1337–1349. doi: 10.1104/pp.17.00946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Chou HL, Zhang L, et al. RNA-Binding Protein RBP-P is required for glutelin and prolamine mRNA localization in rice endosperm cells. Plant Cell. 2018;30:2529–2552. doi: 10.1105/tpc.18.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Chou HL, Zhang L, et al. Targeted endoplasmic reticulum localization of storage protein mRNAs requires the RNA-Binding Protein RBP-L. Plant Physiol. 2019;179:1111–1131. doi: 10.1104/pp.18.01434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian YS, Wang B, Peng RH, et al. Enhancing carotenoid biosynthesis in rice endosperm by metabolic engineering. Plant Biotechnol J. 2019;17:849–851. doi: 10.1111/pbi.13059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Doroshenk KA, Zhang L, et al. Zipcode RNA-Binding Proteins and membrane trafficking proteins cooperate to transport glutelin mRNAs in rice endosperm. Plant Cell. 2020;32:2566–2581. doi: 10.1105/tpc.20.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, He M, Mei E, et al. WRKY53 integrates classic brassinosteroid signaling and the mitogen-activated protein kinase pathway to regulate rice architecture and seed size. Plant Cell. 2021;33:2753–2775. doi: 10.1093/plcell/koab137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H, Jin Y, Liu W, et al. DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009;58:803–816. doi: 10.1111/j.1365-313X.2009.03825.x. [DOI] [PubMed] [Google Scholar]
- Tong H, Liu L, Jin Y, et al. DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell. 2012;24:2562–2577. doi: 10.1105/tpc.112.097394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemoto T, Yano M, Satoh H, et al. Mapping of a gene responsible for the difference in amylopectin structure between japonica-type and indica-type rice varieties. Theor Appl Genet. 2002;104:1–8. doi: 10.1007/s001220200000. [DOI] [PubMed] [Google Scholar]
- Utsunomiya Y, Samejima C, Takayanagi Y, et al. Suppression of the rice heterotrimeric G protein β-subunit gene, RGB1, causes dwarfism and browning of internodes and lamina joint regions. Plant J. 2011;67:907–916. doi: 10.1111/j.1365-313X.2011.04643.x. [DOI] [PubMed] [Google Scholar]
- Wanchana S, Toojinda T, Tragoonrung S, et al. Duplicated coding sequence in the waxy allele of tropical glutinous rice (Oryza sativa L.) Plant Sci. 2003;165:1193–1199. [Google Scholar]
- Wang C, Shu Q. Fine mapping and candidate gene analysis of purple pericarp gene Pb in rice (Oryza sativa L.) Chin Sci Bull. 2007;52:3097–3104. [Google Scholar]
- Wang C, Washida H, Crofts AJ, et al. The cytoplasmic-localized, cytoskeletal-associated RNA binding protein OsTudor-SN: evidence for an essential role in storage protein RNA transport and localization. Plant J. 2008;55:443–454. doi: 10.1111/j.1365-313X.2008.03516.x. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Zheng FQ, Shen GZ, et al. The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J. 2010;7:613–622. doi: 10.1046/j.1365-313x.1995.7040613.x. [DOI] [PubMed] [Google Scholar]
- Wang S, Wu K, Yuan Q, et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet. 2012;44:950–954. doi: 10.1038/ng.2327. [DOI] [PubMed] [Google Scholar]
- Wang JC, Xu H, Zhu Y, et al. OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J Exp Bot. 2013;64:3453–3466. doi: 10.1093/jxb/ert187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Li S, Liu Q, et al. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat Genet. 2015;47:949–954. doi: 10.1038/ng.3352. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhou W, Lu Z, et al. A lipid transfer protein, OsLTPL36, is essential for seed development and seed quality in rice. Plant Sci. 2015;239:200–208. doi: 10.1016/j.plantsci.2015.07.016. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xiong G, Hu J, et al. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat Genet. 2015;47:944–948. doi: 10.1038/ng.3346. [DOI] [PubMed] [Google Scholar]
- Wang A, Hou Q, Si L, et al. The PLATZ transcription factor GL6 affects grain length and number in rice. Plant Physiol. 2019;180:2077–2090. doi: 10.1104/pp.18.01574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chen Z, Zhang Q, et al. The NAC transcription factors OsNAC20 and OsNAC26 regulate starch and storage protein synthesis. Plant Physiol. 2020;184:1775–1791. doi: 10.1104/pp.20.00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Yang Y, Guo M, et al. Targeted mutagenesis of amino acid transporter genes for rice quality improvement using the CRISPR/Cas9 system. Crop J. 2020;3:457–464. [Google Scholar]
- Wang L, Wang D, Yang Z, et al. Roles of FERONIA-like receptor genes in regulating grain size and quality in rice. Sci China Life Sci. 2021;64:294–310. doi: 10.1007/s11427-020-1780-x. [DOI] [PubMed] [Google Scholar]
- Weng J, Gu S, Wan X, et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008;18:1199–1209. doi: 10.1038/cr.2008.307. [DOI] [PubMed] [Google Scholar]
- Wong HW, Liu Q, Sun SS. Biofortification of rice with lysine using endogenous histones. Plant Mol Biol. 2015;87:235–248. doi: 10.1007/s11103-014-0272-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CY, Adach T, Hatano T, et al. Promoters of rice seed storage protein genes direct endosperm-specific gene expression in transgenic rice. Plant Cell Physiol. 1998;39:885–889. [Google Scholar]
- Wu X, Liu J, Li D, et al. Rice caryopsis development II: Dynamic changes in the endosperm. J Integr Plant Biol. 2016;58:786–798. doi: 10.1111/jipb.12488. [DOI] [PubMed] [Google Scholar]
- Wu Y, Fu Y, Zhao S, et al. CLUSTERED PRIMARY BRANCH 1, a new allele of DWARF11, controls panicle architecture and seed size in rice. Plant Biotechnol J. 2016;14:377–386. doi: 10.1111/pbi.12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Liu X, Wang M, et al. A single-nucleotide polymorphism causes smaller grain size and loss of seed shattering during African rice domestication. Nat Plants. 2017;3:17064. doi: 10.1038/nplants.2017.64. [DOI] [PubMed] [Google Scholar]
- Wu M, Ren Y, Cai M, et al. Rice FLOURY ENDOSPERM10 encodes a pentatricopeptide repeat protein that is essential for the trans-splicing of mitochondrial nad1 intron 1 and endosperm development. New Phytol. 2019;223:736–750. doi: 10.1111/nph.15814. [DOI] [PubMed] [Google Scholar]
- Wu K, Wang S, Song W, et al. Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science. 2020;367:eaaz2046. doi: 10.1126/science.aaz2046. [DOI] [PubMed] [Google Scholar]
- Xia D, Zhou H, Liu R, et al. GL3.3, a novel QTL encoding a GSK3/SHAGGY-like Kinase, epistatically interacts with GS3 to produce extra-long grains in rice. Mol Plant. 2018;11:754–756. doi: 10.1016/j.molp.2018.03.006. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Liu D, Zhang G, et al. Brassinosteroids Regulate OFP1, a DLT interacting protein, to modulate plant architecture and grain morphology in rice. Front Plant Sci. 2017;8:1698. doi: 10.3389/fpls.2017.01698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Liu D, Zhang G, et al. Big Grain3, encoding a purine permease, regulates grain size via modulating cytokinin transport in rice. J Integr Plant Biol. 2019;61:581–597. doi: 10.1111/jipb.12727. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Zhang G, Liu D, et al. GSK2 stabilizes OFP3 to suppress brassinosteroid responses in rice. Plant J. 2020;102:1187–1201. doi: 10.1111/tpj.14692. [DOI] [PubMed] [Google Scholar]
- Xiao N, Pan C, Li Y, et al. Genomic insight into balancing high yield, good quality, and blast resistance of japonica rice. Genome Biol. 2021;22:283. doi: 10.1186/s13059-021-02488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Wu C, Xiong L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006;142:280–293. doi: 10.1104/pp.106.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Ren Y, Li W, et al. NF-YC12 is a key multi-functional regulator of accumulation of seed storage substances in rice. J Exp Bot. 2019;70:3765–3780. doi: 10.1093/jxb/erz168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Liu Y, Li Y, et al. Differential expression of GS5 regulates grain size in rice. J Exp Bot. 2015;66:2611–2623. doi: 10.1093/jxb/erv058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Duan P, Yu H, et al. Control of grain size and weight by the OsMKKK10-OsMKK4-OsMAPK6 signaling pathway in rice. Mol Plant. 2018;11:860–873. doi: 10.1016/j.molp.2018.04.004. [DOI] [PubMed] [Google Scholar]
- Xu R, Li N, Li Y. Control of grain size by G protein signaling in rice. J Integr Plant Biol. 2019;61:533–540. doi: 10.1111/jipb.12769. [DOI] [PubMed] [Google Scholar]
- Xu Z, Yu M, Yin Y, et al. Generation of selectable marker-free soft transgenic rice with transparent kernels by downregulation of SSSII-2. Crop J. 2020;8:53–61. [Google Scholar]
- Xu X, E Z, Zhang D, et al. OsYUC11-mediated auxin biosynthesis is essential for endosperm development of rice. Plant Physiol. 2021;185:934–950. doi: 10.1093/plphys/kiaa057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M, Liu L, Yu Y, et al. Lose-of-function of a rice nucleolus-localized pentatricopeptide repeat protein is responsible for the floury endosperm14 mutant phenotypes. Rice. 2019;12:100. doi: 10.1186/s12284-019-0359-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa H, Hirose T, Kuroda M, et al. Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray. Plant Physiol. 2007;144:258–277. doi: 10.1104/pp.107.098665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto MP, Onodera Y, Touno SM, et al. Synergism between RPBF Dof and RISBZ1 bZIP activators in the regulation of rice seed expression genes. Plant Physiol. 2006;141:1694–1707. doi: 10.1104/pp.106.082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, et al. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell. 2000;12:1591–1606. doi: 10.1105/tpc.12.9.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Wei M, Li Y, et al. MiR529a controls plant height, tiller number, panicle architecture and grain size by regulating SPL target genes in rice (Oryza sativa L.) Plant Sci. 2021;302:110728. doi: 10.1016/j.plantsci.2020.110728. [DOI] [PubMed] [Google Scholar]
- Yang C, Shen W, He Y, et al. OVATE family protein 8 positively mediates brassinosteroid signaling through interacting with the GSK3-like kinase in rice. PLoS Genet. 2016;12:e1006118. doi: 10.1371/journal.pgen.1006118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang QQ, Zhang CQ, Chan ML, et al. Biofortification of rice with the essential amino acid lysine: molecular characterization, nutritional evaluation, and field performance. J Exp Bot. 2016;67:4285–4296. doi: 10.1093/jxb/erw209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Ma Y, He Y, et al. OsOFP19 modulates plant architecture by integrating the cell division pattern and brassinosteroid signaling. Plant J. 2018;93:489–501. doi: 10.1111/tpj.13793. [DOI] [PubMed] [Google Scholar]
- Yang QQ, Zhao DS, Zhang CQ, et al. A connection between lysine and serotonin metabolism in rice endosperm. Plant Physiol. 2018;176:1965–1980. doi: 10.1104/pp.17.01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Guo M, Sun S, et al. Natural variation of OsGluA2 is involved in grain protein content regulation in rice. Nat Commun. 2019;10:1949. doi: 10.1038/s41467-019-09919-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Zhao D, Liu Q. Connections between amino acid metabolisms in plants: lysine as an example. Front Plant Sci. 2020;11:928. doi: 10.3389/fpls.2020.00928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang QQ, Yu WH, Wu HY, et al. Lysine biofortification in rice by modulating feedback inhibition of aspartate kinase and dihydrodipicolinate synthase. Plant Biotechnol J. 2021;19:490–501. doi: 10.1111/pbi.13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Wu K, Wang B, et al. The RING E3 ligase CLG1 targets GS3 for degradation via the endosome pathway to determine grain size in rice. Mol Plant. 2021;14:1699–1713. doi: 10.1016/j.molp.2021.06.027. [DOI] [PubMed] [Google Scholar]
- Yang X, Wang J, Xia X, et al. OsTTG1, a WD40 repeat gene, regulates anthocyanin biosynthesis in rice. Plant J. 2021;107:198–214. doi: 10.1111/tpj.15285. [DOI] [PubMed] [Google Scholar]
- Ye X, Al-Babili S, Klöti A, et al. Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science. 2000;287:303–305. doi: 10.1126/science.287.5451.303. [DOI] [PubMed] [Google Scholar]
- Yin LL, Xue HW. The MADS29 transcription factor regulates the degradation of the nucellus and the nucellar projection during rice seed development. Plant Cell. 2012;24:1049–1065. doi: 10.1105/tpc.111.094854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Xiao Y, Niu M, et al. ARGONAUTE2 enhances grain length and salt tolerance by activating BIG GRAIN3 to modulate cytokinin distribution in rice. Plant Cell. 2020;32:2292–2306. doi: 10.1105/tpc.19.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying J, Ma M, Bai C, et al. TGW3, a major QTL that negatively modulates grain length and weight in rice. Mol Plant. 2018;11:750–753. doi: 10.1016/j.molp.2018.03.007. [DOI] [PubMed] [Google Scholar]
- Yu S, Xu C, Tan Y, et al. Analyzing quantitative trait loci for yield using a vegetatively replicated F 2 population from a cross between the parents of an elite rice hybrid. TAG Theor Appl Genet. 2000;101:248–254. [Google Scholar]
- Yu J, Miao J, Zhang Z, et al. Alternative splicing of OsLG3b controls grain length and yield in japonica rice. Plant Biotechnol J. 2018;16:1667–1678. doi: 10.1111/pbi.12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Wu M, Ren Y, et al. Rice FLOURY ENDOSPERM 18 encodes a pentatricopeptide repeat protein required for 5' processing of mitochondrial nad5 messenger RNA and endosperm development. J Integr Plant Biol. 2021;63:834–847. doi: 10.1111/jipb.13049. [DOI] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Silva P, et al. Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci U S A. 2010;107:18729–18734. doi: 10.1073/pnas.1009695107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng D, Liu T, Ma X, et al. Quantitative regulation of Waxy expression by CRISPR/Cas9-based promoter and 5'UTR-intron editing improves grain quality in rice. Plant Biotechnol J. 2020;18:2385–2387. doi: 10.1111/pbi.13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan P, Wei X, Xiao Z, et al. GW10, a member of P450 subfamily regulates grain size and grain number in rice. Theor Appl Genet. 2021;134:3941–3950. doi: 10.1007/s00122-021-03939-3. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nallamilli BR, Mujahid H, et al. OsMADS6 plays an essential role in endosperm nutrient accumulation and is subject to epigenetic regulation in rice (Oryza sativa) Plant J. 2010;64:604–617. doi: 10.1111/j.1365-313X.2010.04354.x. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang J, Huang J, et al. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc Natl Acad Sci USA. 2012;109:21534–21539. doi: 10.1073/pnas.1219776110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu YH, Yi HY, et al. Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J. 2012;72:400–410. doi: 10.1111/j.1365-313X.2012.05088.x. [DOI] [PubMed] [Google Scholar]
- Zhang YC, Yu Y, Wang CY, et al. Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat Biotechnol. 2013;31:848–852. doi: 10.1038/nbt.2646. [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhou L, Zhu Z, et al. Characterization of grain quality and starch fine structure of two japonica rice (Oryza Sativa) cultivars with good sensory properties at different temperatures during the filling stage. J Agric Food Chem. 2016;64:4048–4057. doi: 10.1021/acs.jafc.6b00083. [DOI] [PubMed] [Google Scholar]
- Zhang C, Chen S, Ren X, et al. Molecular structure and physicochemical properties of starches from rice with different amylose contents resulting from modification of OsGBSSI activity. J Agric Food Chem. 2017;65:2222–2232. doi: 10.1021/acs.jafc.6b05448. [DOI] [PubMed] [Google Scholar]
- Zhang H, Xu H, Feng M, et al. Suppression of OsMADS7 in rice endosperm stabilizes amylose content under high temperature stress. Plant Biotechnol J. 2018;16:18–26. doi: 10.1111/pbi.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Su J, Zhang Y, et al. Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr Opin Plant Biol. 2018;45:1–10. doi: 10.1016/j.pbi.2018.04.012. [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhu J, Chen S, et al. Wxlv, the ancestral allele of rice Waxy Gene. Mol Plant. 2019;12:1157–1166. doi: 10.1016/j.molp.2019.05.011. [DOI] [PubMed] [Google Scholar]
- Zhang D, Zhang M, Zhou Y, et al. The rice G protein γ subunit DEP1/qPE9-1 positively regulates grain-filling process by increasing auxin and cytokinin content in rice grains. Rice. 2019;12:91. doi: 10.1186/s12284-019-0344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Yang Y, Chen Z, et al. Characteristics of grain physicochemical properties and the starch structure in rice carrying a mutated ALK/SSIIa gene. J Agric Food Chem. 2020;68:13950–13959. doi: 10.1021/acs.jafc.0c01471. [DOI] [PubMed] [Google Scholar]
- Zhang C, Yang Y, Chen S, et al. A rare Waxy allele coordinately improves rice eating and cooking quality and grain transparency. J Integr Plant Biol. 2021;63:889–901. doi: 10.1111/jipb.13010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zhang M, Liang J. RGB1 regulates grain fevelopment and starch accumulation through its effect on OsYUC11-mediated auxin biosynthesis in rice endosperm cells. Front Plant Sci. 2021;12:585174. doi: 10.3389/fpls.2021.585174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang R, Xing Y, et al. Separable regulation of POW1 in grain size and leaf angle development in rice. Plant Biotechnol J. 2021;19:2517–2531. doi: 10.1111/pbi.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao GC, Xie MX, Wang YC, et al. Molecular mechanisms underlying γ-aminobutyric acid (GABA) accumulation in giant embryo rice seeds. J Agric Food Chem. 2017;65:4883–4889. doi: 10.1021/acs.jafc.7b00013. [DOI] [PubMed] [Google Scholar]
- Zhao D, Li Q, Zhang C, et al. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat Commun. 2018;9:1240. doi: 10.1038/s41467-018-03616-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YF, Peng T, Sun HZ, et al. miR1432-OsACOT (Acyl-CoA thioesterase) module determines grain yield via enhancing grain filling rate in rice. Plant Biotechnol J. 2019;17:712–723. doi: 10.1111/pbi.13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wang Z. Protein accumulation in aleurone cells, sub-aleurone cells and the center starch endosperm of cereals. Plant Cell Rep. 2014;33:1607–1615. doi: 10.1007/s00299-014-1651-2. [DOI] [PubMed] [Google Scholar]
- Zhong J, He W, Peng Z, et al. A putative AGO protein, OsAGO17, positively regulates grain size and grain weight through OsmiR397b in rice. Plant Biotechnol J. 2020;18:916–928. doi: 10.1111/pbi.13256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Xue H. The rice PLATZ protein SHORT GRAIN6 determines grain size by regulating spikelet hull cell division. J Integr Plant Biol. 2020;62:847–864. doi: 10.1111/jipb.12851. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhu J, Li Z, et al. Deletion in a quantitative trait gene qPE9-1 associated with panicle erectness improves plant architecture during rice domestication. Genetics. 2009;183:315–324. doi: 10.1534/genetics.109.102681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Ren N, Qi J, et al. The 9-lipoxygenase Osr9-LOX1 interacts with the 13-lipoxygenase-mediated pathway to regulate resistance to chewing and piercing-sucking herbivores in rice. Physiol Plant. 2014;152:59–69. doi: 10.1111/ppl.12148. [DOI] [PubMed] [Google Scholar]
- Zhou H, Wang L, Liu G, et al. Critical roles of soluble starch synthase SSIIIa and granule-bound starch synthase Waxy in synthesizing resistant starch in rice. Proc Natl Acad Sci USA. 2016;113:12844–12849. doi: 10.1073/pnas.1615104113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Li P, Xie W, et al. Genome-wide association analyses reveal the genetic basis of stigma exsertion in rice. Mol Plant. 2017;10:634–644. doi: 10.1016/j.molp.2017.01.001. [DOI] [PubMed] [Google Scholar]
- Zhou W, Wang X, Zhou D, et al. Overexpression of the 16-kDa α-amylase/trypsin inhibitor RAG2 improves grain yield and quality of rice. Plant Biotechnol J. 2017;15:568–580. doi: 10.1111/pbi.12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Tao Y, Zhu J, et al. GNS4, a novel allele of DWARF11, regulates grain number and grain size in a high-yield rice variety. Rice. 2017;10:34. doi: 10.1186/s12284-017-0171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Xia D, Zhao D, et al. The origin of Wxla provides new insights into the improvement of grain quality in rice. J Integr Plant Biol. 2021;63:878–888. doi: 10.1111/jipb.13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Li X, Liu Q, et al. DNA demethylases remodel DNA methylation in rice gametes and zygote and are required for reproduction. Mol Plant. 2021;14:1569–1583. doi: 10.1016/j.molp.2021.06.006. [DOI] [PubMed] [Google Scholar]
- Zhu L, Gu M, Meng X, et al. High-amylose rice improves indices of animal health in normal and diabetic rats. Plant Biotechnol J. 2012;10:353–362. doi: 10.1111/j.1467-7652.2011.00667.x. [DOI] [PubMed] [Google Scholar]
- Zhu X, Liang W, Cui X, et al. Brassinosteroids promote development of rice pollen grains and seeds by triggering expression of Carbon Starved Anther, a MYB domain protein. Plant J. 2015;82:570–581. doi: 10.1111/tpj.12820. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Zeng D, Yu S, et al. From golden rice to aSTARice: Bioengineering astaxanthin biosynthesis in rice endosperm. Mol Plant. 2018;11:1440–1448. doi: 10.1016/j.molp.2018.09.007. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Lin Y, Chen S, et al. CRISPR/Cas9-mediated functional recovery of the recessive rc allele to develop red rice. Plant Biotechnol J. 2019;17:2096–2105. doi: 10.1111/pbi.13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Li J. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu Rev Genet. 2014;48:99–118. doi: 10.1146/annurev-genet-120213-092138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.