Abstract
DNA methylation plays a pivotal role in the progression of renal fibrosis. Methyl-CpG–binding domain protein 2 (MBD2), a protein reader of methylation, is involved in the development of acute kidney injury (AKI) caused by vancomycin. However, the role and mechanism of action of MBD2 in renal remain unclear. In this study, MBD2 mediated extracellular matrix (ECM) production induced by TGF-β1 in Boston University mouse proximal tubule (BUMPT) cells,and upregulated the expression EGR1 to promote ECM production in murine embryonic NIH 3T3 fibroblasts. ChIP analysis demonstrated that MBD2 physically interacted with the promoter region of the CpG islands of EGR1 genes and then activated their expression by inducing hypomethylation of the promoter region. In vivo, PT-MBD2-KO attenuated unilateral ureteral obstruction (UUO)-induced renal tubulointerstitial fibrosis via downregulation of EGR1, which was demonstrated by the downregulation of fibronectin (FN), collagen I and IV, α-SMA, and EGR1. Injection of MBD2-siRNA attenuated the UUO- and I/R-induced renal fibrosis. Those molecular changes were verified by biopsies from patients with obstructive nephropathy (OB). These data collectively demonstrated that inhibition of MBD2 reduces renal fibrosis via downregulating EGR1, which could be a target for treatment of fibrotic kidney disease.
Keywords: MT: Bioinformatics, MBD2, EGR1, renal fibrosis, UUO, I/R
Graphical abstract
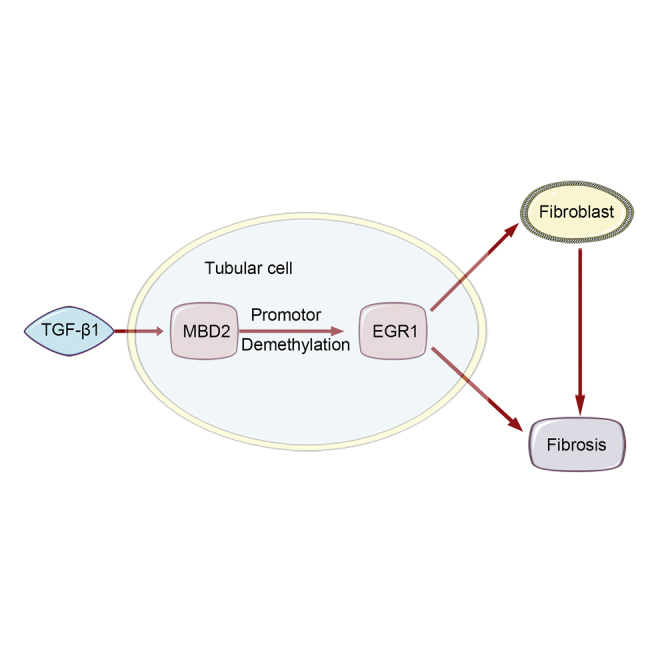
We verified that MBD2 mediated TGF-β1-induced ECM production by BUMPT cells and induced EGR1 to promote ECM production by murine embryonic NIH 3T3 fibroblasts. In vivo, PT-MBD2-KO attenuated UUO- and I/R-induced renal tubulointerstitial fibrosis via downregulation of EGR1. Injection of MBD2-siRNA attenuated the UUO- and I/R-induced renal fibrosis.
Introduction
Epigenetic modifications play essential roles in various biological processes and diseases,1 and DNA methylation is an essential epigenetic mechanism. DNA methyltransferases (DNMT1, DNMT3a, and DNMT3b) are involved in generating and maintaining the epigenome. Specifically, DNMT3a and DNMT3b are responsible for establishing DNA methylation, while DNMT1 maintains methylation.2,3 A recent study has demonstrated that DNMT1 inhibition ameliorated renal fibrosis.4 Thus, the regulation of DNA methylation levels has become a research focal point in the study of renal fibrosis.5
Methyl-CpG–binding domain protein 2 (MBD2) might be involved in modulating gene transcriptional activity and has a critical role in various diseases.6, 7, 8, 9, 10, 11, 12 Our recent study demonstrated that global MBD2 knock out (KO) attenuated AKI caused by sepsis, rhabdomyolysis, and vancomycin.13, 14, 15 In addition, recent studies reported that tubular epithelial cells (TECs) were involved in the progression of renal fibrosis progression.16, 17, 18, 19, 20 However, the role of tubular MBD2 in renal fibrosis remains unclear. Early growth response protein 1 (EGR1), a member of the immediate–early response gene family, has been shown to mediate renal fibrosis induced by multiple growth factors.21, 22, 23, 24, 25, 26 Interestingly, another study reported that MeCP2 regulated EGR1 expression through binding to its CpG islands.27 Thus, the role and mechanism of regulation for MBD2 in the expression of EGR1 remain unclear. We hypothesized that tubular MBD2 might promote renal fibrosis by upregulating the expression of EGR1.
In this study, we verified that PT-MBD2-KO or MBD2-small interfering RNA (siRNA) attenuated transforming growth factor β1 (TGF-β1)-, unilateral ureteral obstruction (UUO)-, and ischemia/reperfusion (I/R)-induced renal fibrosis. We also discovered that the molecular mechanism underlying that MBD2-induced renal fibrosis in UUO and I/R involved upregulation of the expression of EGR1. These markers also were increased in fibrotic human kidneys, indicating that this mechanism might be involved in human fibrotic kidney disease.
Results
TGF-β1 and UUO induced the expression of MBD2 in BUMPT cells and mouse kidneys
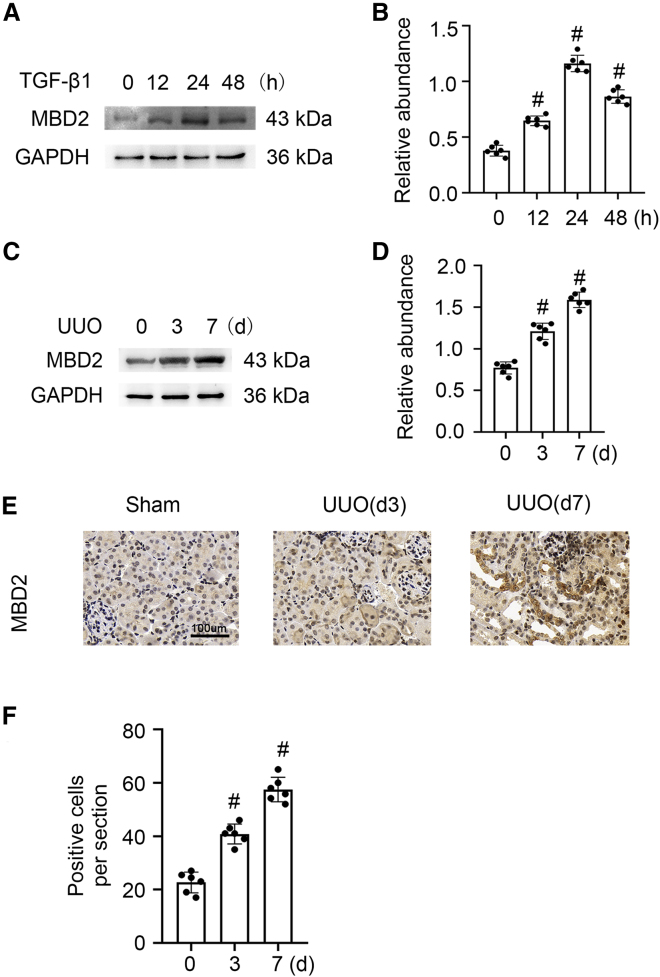
We explored whether MBD2 was induced after treatment with TGF-β1 in Boston University mouse proximal tubule (BUMPT) cells and UUO in C57BL/6J mice. The western blot results demonstrated that MBD2 was increased somewhat in BUMPT cells at 12 h, reached peak levels at 24 h, and declined by 48 h (Figures 1A and 1B). Compared with the sham group, MBD2 also was markedly upregulated in the mouse UUO model at days 3 and 7 (Figures 1C and 1D), which was confirmed by the immunohistochemical staining results (Figures 1E and 1F). We concluded that MBD2 expression was induced by TGF-β1 and UUO and was associated with renal fibrosis progression.
Figure 1.
MBD-2 was induced by TGF-β1 or UUO in BUMPT cells or mice kidneys
BUMPT cells were treated with 5 ng/mL TGF-β1 for 0–48 h. Male C57BL/6J mice were subjected to UUO for examination on days 0–7. Whole-cell lysates were collected at the time points and analyzed by western blotting with antibodies to MBD2. Left kidneys were removed for immunoblot and immunohistochemistry analysis of MBD2 at the indicated time points. (A and C) Immunoblot analysis of MBD2 at indicated time points. (B and D) Analysis of the gray-scale image between them. (E and F) Immunohistochemistry of MBD2. These data are representative of at least four separate experiments shown as means ± SD (n = 6). #p < 0.05 versus hour or day 0 or sham group. Original magnification, ×400. Scale bar: 100μm.
MBD2 mediated the TGF-β1-induced ECM production by BUMPT cells
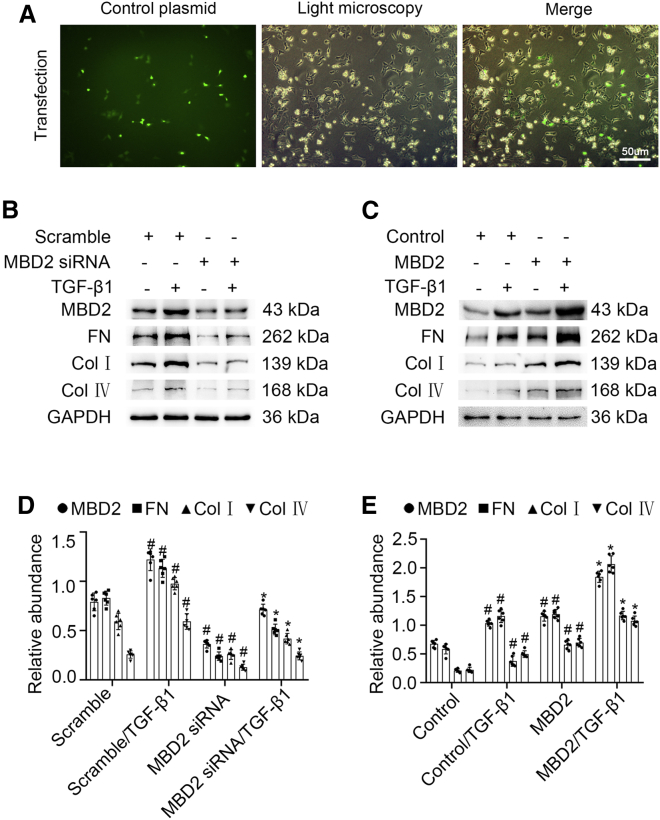
Next, we determined if MBD2 mediated the renal fibrosis induced by TGF-β1. BUMPT cells were transfected with MBD2 siRNA with or without TGF-β1. The expression of MBD2 induced by TGF-β1 was silenced by MBD2 siRNA. The loss of MBD2 expression was accompanied by reduced fibronectin (FN) and collagen (Col) I and IV expression (Figure 2B). These protein changes were verified using gray-level analysis (Figure 2D). To confirm the role of MBD2 in renal fibrosis induced by TGF-β1, MBD2 plasmid was constructed (Figure S5). BUMPT cells were transfected with the MBD2 plasmid in the absence or presence of TGF-β1; immunofluorescence showed the success of transfection (Figure 2A). These results indicated that overexpression of MBD2 enhanced the TGF-β1 induction of FN and Col I and IV expression (Figures 2C and 2E). These data suggested that MBD2 mediated the renal fibrosis caused by exposure to TGF-β1.
Figure 2.
MBD2 was involved in ECM accumulation induced by TGF-β1 in BUMPT cells
BUMPT cells were transfected with MBD2 siRNA or MBD2 plasmid and then treated with 5 ng/mL TGF-β1 for 24 h to collect total cell lysates for immunoblot analysis of MBD2, FN, and Col I and IV. (A) transfection of control plasmid. (B and C) Representative immunoblots. (D and E) Densitometry analysis of proteins signals, and normalized to internal control of GAPDH. These data are representative of at least four separate experiments shown as means ± SD (n = 6). #p < 0.05 versus scramble or control group. ∗p < 0.05 versus TGF-β1 group. Original magnification, ×200. Scale bar: 50μm.
MBD2 mediated the TGF-β1-induced renal fibrosis via upregulation of EGR1 in supernatant of BUMPT cells cultured with or without murine embryonic NIH 3T3 fibroblasts
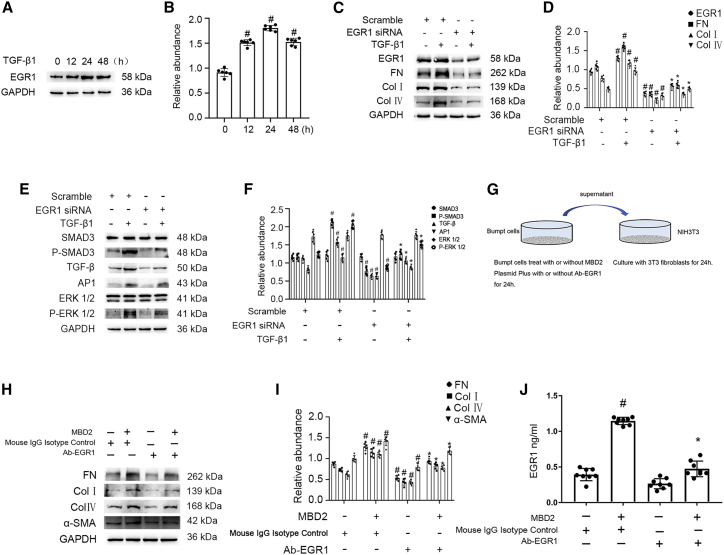
Recent studies demonstrated that EGR1 was involved in the production of renal fibrosis.24,26 This study verified that EGR1 expression was observed within 12 h of TGF-β1 treatment, peaked at 24 h, and then decreased significantly by 48 h (Figures 3A and 3B). Subsequently, the knockdown of EGR1 using EGR1 siRNA markedly suppressed the expression of EGR1, FN, and Col I and IV that was induced by TGF-β1 (Figures 3C and 3D). Furthermore, we also found that EGR1 siRNA noticeably inhibited the TGF-β1-induced activation of Smad3, Ap-1, and ERK1/2 and the increasing of TGF-β (Figures 3E and 3F). In addition, the control or MBD2 plasmid was transfected into BUMPT cells and then treated with mouse immunoglobulin G (IgG) isotype control or Ab-EGR1 for 24 h; the supernatant was collected to culture murine embryonic NIH 3T3 fibroblasts for another 24 h (Figure 3G). The immunoblot results indicated that the supernatant from BUMPT cells transfected with MBD2 induced the increasing of FN, Col I and IV, and α-SMA, which was markedly suppressed by neutralizing the expression of EGR1 (Figures 3H and 3I). The ELISA results also showed that MBD2 promoted the secretion of EGR1 in supernatant of BUMPT cells, which was suppressed by the Ab-EGR1 (Figure 3J). These data suggested that the MBD2 mediated the TGF-β1-induced fibrosis through regulation of EGR1 expression.
Figure 3.
EGR1 mediated the promoting fibrosis role of MBD2 during BUMPT cells treated with TGF-β1 or co-culture of murine embryonic NIH 3T3 fibroblasts, and BUMPT cells
BUMPT cells were treated with 5 ng/mL TGF-β1 for 0–48 h or transfected with EGR1 siRNA and then treated with 5 ng/mL TGF-β1 for 24 h. (A) Immunoblot analysis of EGR1 at indicated time points. (B) Analysis of the gray-scale image between them. (C) Immunoblot analysis of EGR1, FN, and Col I and IV. (D) Analysis of the gray-scale image between them. (E) Immunoblot analysis of SMAD3, p-SMAD3, AP-1, TGF-β, ERK1/2, and p-ERK1/2. (F) Analysis of the gray-scale image between them. (G) The murine embryonic NIH 3T3 fibroblasts were treated with the supernatant from BUMPT cells transfected with plasmid of MBD2 plus with or without EGR1 neutralizing antibody for 24 h, which indicated by the co-culture model diagram of BUMPT cells and murine embryonic NIH 3T3 fibroblasts. (H) Immunoblot analysis of FN, Col I and IV, and α-SMA. (I) Analysis of the gray-scale image between them. (J) Concentration of EGR1 by ELISA. Data are expressed as means ± SD (n = 6). #p < 0.05 versus scramble or control group. ∗p < 0.05 versus TGF-β1 or MBD2 group.
MBD2 activated the expression of EGR1 by inhibiting methylation of its promoter
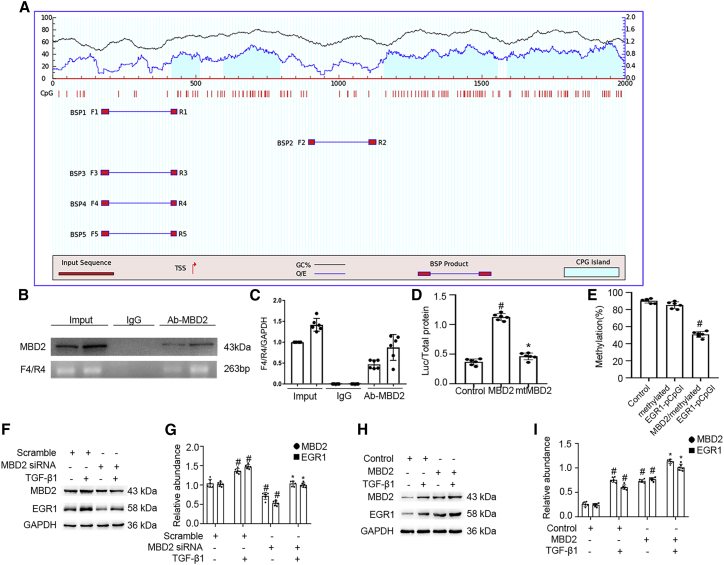
To investigate how MBD2 regulates the EGR1 expression, the MethPrimer Promoter 2.0 (http://www.urogene.org/cgi-bin/methprimer2/MethPrimer.cgi) was used to predict the CpG islands for EGR1, and primers were designed. The results indicated that the promoter sequence of EGR1 contained four islands (Figure 4A). Furthermore, a chromatin immunoprecipitation (ChIP) assay was applied to determine whether MBD2 interacted with the promoter region of EGR1 in BUMPT cells. MBD2 physically interacted with the F/R4-4 binding sites of the EGR1 promoter; the results of qPCR further confirmed this (Figures 4B and 4C). Furthermore, the CG DNA methylation target sequences in the EGR1 promoter region were cloned into the CpG-free pCpGI luciferase reporter plasmid. The transcriptional activity was markedly increased by the MBD2 plasmid but not the mutant plasmid with the MBD2 methylated DNA binding domain deletion (Figure 4D). We observed that MBD2 affected the methylation levels of the promoter of the ectopic EGR1. The level of methylated pCpGI of EGR1 is reduced very slightly by endogenous MBD2-bound DNA compared with the control group and was markedly inhibited by ectopic MBD2 expression (Figure 4E). Finally, EGR1 induced by TGF-β1 was reduced by MBD2 siRNA. In contrast, this effect was enhanced by the MBD2 plasmid (Figures 4F–4I). These data demonstrated that MBD2 activated the expression of EGR1 through hypomethylation of the promoter region.
Figure 4.
MBD2 directly binds to CpG islands of promoter of EGR1 and positively activates transcription of them by hypomethylation of promoter
(A) The patterns of CpG islands of EGR1 promoter and five pairs of primer were predicted by the software of MethPrimer 2.0. (B) ChIP assays represents the binding sites of MBD2 interaction with CpG islands of promoter of EGR1. (C) qRT-PCR confirmed the expression of CpG islands of promoter of EGR1. (D) Relative luciferase activity of MBD2 or MBD2 mutation plasmids co-transfected with methylated EGR1 pCpGI plasmid in BUMPT cells. (E) CpG-DNA methylation of the EGR1 promoter region. (F and H) Representative immunoblots of MBD2 and EGR1. (G and I) Densitometry analysis of proteins levels, and normalized to internal control of GAPDH. Data are expressed as means ± SD (n = 6). #p < 0.05 versus scramble or control group. ∗p < 0.05 versus TGF-β1 group.
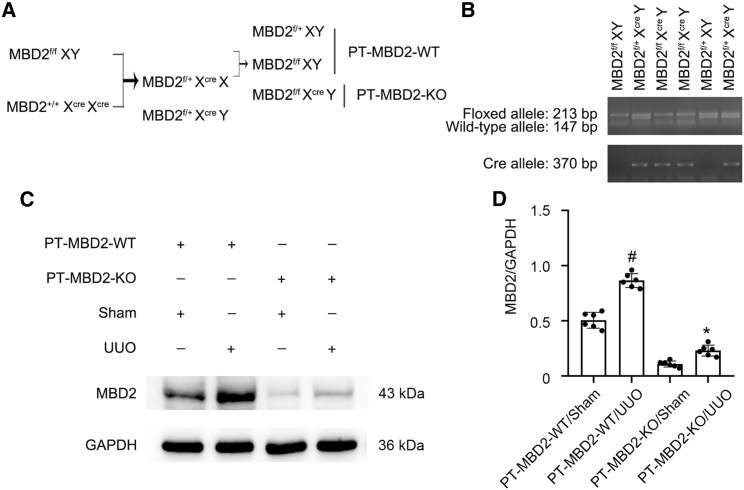
PT-MBD2-KO mice were established
To clarify the role of tubular MBD2, we established a mouse model of MBD2-KO specifically in proximal kidney tubules. Briefly, MBD2flox/floxXY male mice were crossed with MBD2+/+XcreXcre female mice. After two generations, littermates of the proximal tubule MBD2 wild-type (WT) (PT-MBD2-WT) and PT-MBD2-KO (MBD2flox/floxXcreY) mice were produced (Figure 5A). RT-qPCR results demonstrated that the genotype of the PT-MBD2-KO mice was characterized by a 213 bp DNA fragment floxed allele and a 370 bp DNA fragment of the Cre gene (Figure 5B, lanes 3 and 4). The lack of a Cre gene indicated WT (PT-MBD2-WT) mice (Figure 5B, lanes 1 and 5). Western blot analysis revealed that the MBD2 protein levels in the kidney cortices of PT-MBD2-KO mice were considerably lower than that in PT-MBD2-WT mice with or without UUO injury (Figures 5C and 5D). Thus, proximal tubular MBD2 deletion mice were constructed.
Figure 5.
Generation and features of the PT-MBD2-KO mouse model
(A) Breeding procedure for the creation of PT-MBD2-KO mice. (B) PCR-based genotyping of wild-type and floxed alleles of MBD2 and PEPCK-Cre allele. (C) Representative immunoblots for the expression of MBD2 and GAPDH. (D) Densitometry analysis of proteins signals, and normalized to internal control of GAPDH. Data are expressed as means ± SD (n = 6). #p < 0.05 versus sham group. ∗p < 0.05 versus PT-MBD2-WT with UUO group.
UUO- and I/R-induced renal fibrosis that was attenuated in PT-MBD2-KO mice
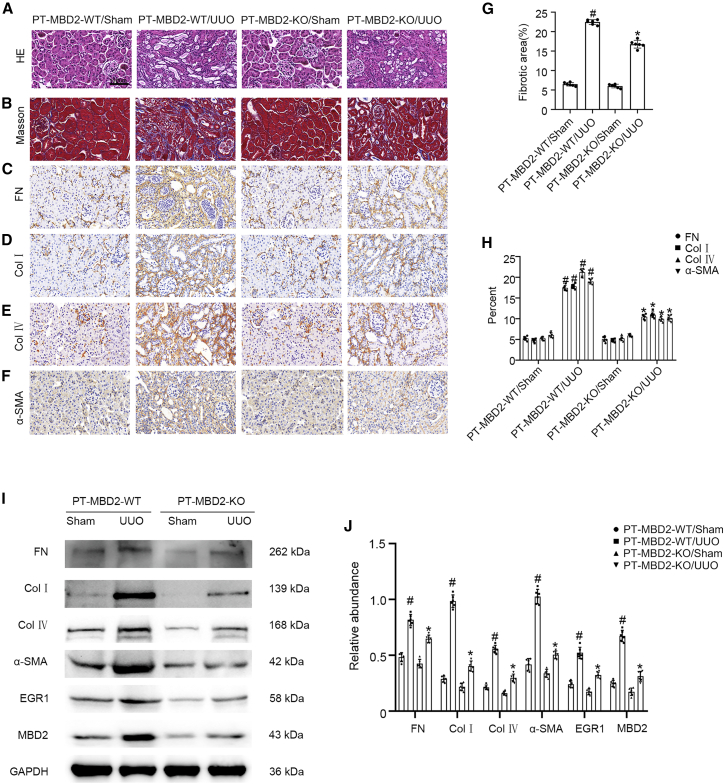
Littermates of PT-MBD2-WT and PT-MBD2-KO mice were subjected to UUO or I/R for 7 or 21 days. PT-MBD2-KO mice exhibited markedly attenuated UUO- or I/R-induced tubular damage (Figures 6A and S1C). In addition, the examination of serum creatinine (SCR) and blood urea nitrogen (BUN) showed that PT-MBD2-KO mice also notably ameliorated the I/R-induced renal function deterioration (Figures S1A and S1B). Masson trichome staining demonstrated that UUO- or I/R-induced accumulation of extracellular matrix (ECM) was significantly ameliorated in PT-MBD2-KO mice (Figures 6B and S1D); this effect was further confirmed by the fibrotic area analysis (Figures 6G and S1I). The immunohistochemical staining results revealed that PT-MBD2-KO mice exhibited markedly reduced UUO- or I/R-induced expression of FN, Col I and IV, and α-SMA (Figures 6C–6F and S1E–S1H), which was further demonstrated by immunohistochemistry staining analysis (Figures 6H and S1J). The western blot results confirmed that the PT-MBD2-KO mice exhibited considerable suppression of the UUO- or I/R-induced expression of FN, Col I and IV, and α-SMA due to downregulation of EGR1 (Figures 6I, 6J, S1K, and S1L). These data demonstrated that MBD2 in the proximal kidney tubules mediated the progression of renal fibrosis during UUO and I/R injury, which supported the in vitro observations previously described.
Figure 6.
PT-MBD2-KO mice attenuated UUO-induced renal fibrosis
The left ureter of PT-MBD2-WT and PT-MBD2-KO littermate mice was ligated to establish UUO model for 7 days. (A) Representative H&E staining. (B) Masson trichrome staining shows interstitial collagen deposition (blue). (C–F) Immunohistochemistry of FN, Col I and IV, and α-SMA. (G) Quantify tubulointerstitial fibrosis in the kidney cortex. (H) Quantification of immunohistochemistry staining. (I) Representative immunoblots for the expression of FN, Col I and IV, α-SMA, EGR1, and GAPDH. (J) Densitometry analysis of proteins signals, and normalized to internal control of GAPDH. Data are expressed as means ± SD (n = 6). #p < 0.05 versus sham group. ∗p < 0.05 versus PT-MBD2-WT UUO group. Original magnification, ×400. Scale bar: 100μm.
MBD2 siRNA ameliorated UUO- and IR-induced renal fibrosis through suppression of EGR1
To explore the potential therapeutic value of MBD2 in renal fibrosis disease. We injected MBD2 siRNA at 15 mg/kg through the tail vein before C57BL/6J mice were subjected to UUO and I/R. The data showed that MBD2 siRNA attenuated UUO- and IR-induced tubular damage and accumulation of ECM (Figures S2A, S2B, S2G, S3C, S3D, and S3I). In addition, I/R-induced renal function deterioration was attenuated by the MBD2 siRNA (Figures S3A and S3B). In addition, UUO- and IR-induced expression of FN, Col I and IV, and α-SMA also was attenuated by injection of MBD2 siRNA (Figures S2C–S2F, S2H, S3E–S3H and S3J). The immunoblot analysis further verified MBD2 siRNA markedly reduced expression of FN, Col I and IV, andα-SMA as well as MBD2 and EGR1 (Figures S2I, S2J, S3K and S3L). In summary, MBD2-siRNA attenuated UUO- and I/R-induced renal fibrosis via suppression of EGR1.
Increased expression of MBD2 regulated genes in human kidneys
We verified the in vitro and in vivo findings by measuring the renal expression of MBD2, ECM-related proteins expression, and EGR1 in obstructive nephropathy (OB). Tubulointerstitial injury and fibrosis were markedly elevated in OB kidneys compared with patients diagnosed with minimal change disease (MCD) (Figures S4A, S4B, and S4I). Furthermore, nuclear staining for MBD2 and EGR1, and the expression of FN, Col I and IV, and α-SMA, were significantly higher in kidneys from patients with OB compared with the MCD group (Figures S4C–S4H). Data are represented in histograms of semi-quantitative immunostaining scores (Figures S4J and S4K). These observations were consistent with the western blots obtained for these proteins (Figures S4L and S4M). These results demonstrated that MBD2 was involved in the regulation of fibrosis in human OB.
Discussion
Although MBD2 has been considered the most promising target for DNA demethylation therapy in tumor disease,28,29 its role in renal fibrosis remained unclear. In the present study, for the first time, we demonstrated that MBD2 siRNA or PT-MBD2-KO attenuated renal fibrosis that was induced by TGF-β1, UUO, and I/R treatment. MBD2 directly resulted in the increased expression of EGR1 to induce renal fibrosis. Similar changes in these molecules were detected in human kidney samples taken from patients with OB. Together, these results provided substantial evidence to support the theory that renal tubular MBD2 is a therapeutic target for renal fibrosis.
Recent advances in understanding the functional role of DNA methylation in renal fibrosis have been reported.4 Furthermore, one study reported that MBD2 in macrophages mediated the progression of lung fibrosis.30 However, the role of the MBD family in renal fibrosis is unknown. In the current study, we demonstrated that MBD2 was induced by TGF-β1 and UUO (Figure 1), and MBD2 positively regulated the TGF-β1- induced expression of FN and Col I and IV in renal tubular cells (Figure 2). Consistently, proximal tubular deletion of MBD2 mice also reduced the UUO- or I/R-induced expression of renal fibrosis (Figures 6 and S1), which was confirmed by the injection of MBD2 siRNA (Figures S2 and S3). The results supplied the strong evidence to support that tubular MBD2 also mediated the expression of renal fibrosis.
To clarify the regulation mechanism of MBD2 on renal fibrosis, we focused on EGR1, which is a zinc finger nuclear phosphoprotein and transcription factor.31, 32, 33, 34, 35 Several studies have demonstrated that EGR1 mediated the progress of renal fibrosis.24, 25, 26 In the current study, we confirmed and extended the finding that EGR1 mediated TGF-β1-induced renal fibrosis (Figures 3A–3D). In addition, we also found that inhibition of EGR1 noticeably attenuated TGF-β1-induced activation of Smad2/3 and ERK1/2 and expression of AP1 (Figures 3E and 3F), which was consistent with the previous studies.36, 37, 38 Furthermore, we found that EGR1 was a key mediator that tubular cells promote NIH 3T3 fibroblasts to promote ECM accumulation (Figures 3G–3J). Interestingly, the ChIP and methylated DNA precipitation (MeDIP) assays demonstrated that one of the five primers (F/R4-4) was amplified (Figures 4A–4C), which suggested that MBD2 physically interacted with that region of the EGR1 promoter. Furthermore, MBD2 activated the expression of EGR1 through induction of hypomethylation of the promoter region (Figures 4D and 4E). EGR1 was positively regulated by MBD2 (Figures 4F–4I). Taken together, these data demonstrated that MBD2 promoted the occurrence of renal fibrosis in part by upregulating the expression of EGR1.
In summary, to our knowledge, this is the first report that tubular MBD2 could act as an inducer of renal fibrosis. Furthermore, we described a novel mechanism by which MBD2 induced the expression of EGR1. These changes resulted in increased renal fibrosis by inducing the hypomethylation of the promotor. Thus, this study suggested that tubular MBD2 might be an attractive therapeutic target for renal fibrosis.
Materials and methods
Reagents and antibodies
Antibodies were obtained from multiple sources: anti-glyceraldehyde 3-phosphate dehydrogenase (anti-GAPDH) and anti-AP1 from Santa Cruz Biotechnology (Santa Cruz, CA, USA); anti-MBD2, anti-collagen I, anti-collagen IV, anti-TGF-β, anti-ERK1/2, anti-p-ERK1/2, and anti-fibronectin from Abcam (Cambridge, MA, USA); and anti-EGR1, anti-SMAD3, and anti-p-SMAD3 from Proteintech Group (Rosemont, IL, USA),NCM Universal Antibody Diluent (New Cell and Molecular Biotech,Suzhou, China). The recombinant human TGF-β1 was obtained from Proteintech Group. The plasmids containing the methylation promoter of EGR1 CpG-free pCpGI luciferase reporter, MBD2 and mtMBD2 (the deletion of the methylated DNA binding domain), were constructed by the Ruqi Biology (Guangdong, Guangzhou, China), according to previously published reports.28,39
Animals
The proximal tubule-specific MBD2-deletion mice were generated by crossing MBD2flox/flox mice (obtained from Shanghai Model Organisms, Shanghai, China) with PEPCK-Cre mice (provided by Dr. Volker Haase (University of Pennsylvania, Philadelphia, PA), as previously described.20 The UUO model was produced by ligating the left ureter, as previously described.40, 41, 42 The ischemic acute kidney injury (AKI) (I/R) model was induced by the duration of bilateral renal artery was continuously clipped for 28 min and followed by reperfusion for 21 days.20 The C57BL/6J mice with ischemia-reperfusion (I/R) operation were injected with MBD2-siRNA or control at 15 mg/kg via the tail vein once a week. Animal experiments were performed in accord with the guidelines of the Institutional Committee for the Care and Use of Laboratory Animals of the Second Xiangya Hospital, People's Republic of China, and approved by the Institutional Animal Use Committee of Second Xiangya Hospital(number 20200310. The mice were housed under a 12-h light/dark cycle and given free access to clean food and drinking water.
Human samples
After the experimental protocol was approved by the institutional review board (IRB) (number 20200320) of the Second Xiangya Hospital, the People's Republic of China, IRB sections were obtained from kidney biopsies from patients with OB (n = 8) and MCD (n = 8). The sections were examined using H&E, Masson stains, and immunohistochemistry staining as well as Western blots.
Cell culture and treatments
BUMPT cells were cultured with 10% fetal bovine serum (FBS), 0.5% penicillin DMEM (Thermo Fisher Scientific, Waltham, MA, USA), and streptomycin in a 5% CO2 incubator at 37°C.43,44 At 24 h after transfection with MBD2 siRNA, a negative control, or MBD2 plasmid, BUMPT cells were subjected to nutrient deprivation in a serum-free medium overnight. Then, the cells were or were not treated with 0.1% BSA (control) or TGF-β1 (5 ng/mL) for an additional 24 h.
Cell transfection experiments
BUMPT cells (1×105 cells) were seeded into 6-well plates in complete DMEM and cultured overnight at 37°C. Medium was removed and Lipofectamine 2000 (Thermo Fisher Scientific) was used for cell transfection according to manufacturer's instructions. Si-MBD2, si-Egr1, and si-NC were synthesized by Shanghai GenePharma and then transfected with at final concentrations of 50 nM using 0.25% Lipofectamine 2000. After 4–6 h of transfection, the culture medium was replaced with fresh medium, and then the cells were collected by continuous culture for 48 h. The sequences of siRNA were described as follows: si-MBD2: 5′-GAAGAUGAUGCCAGUAAUUUU-3′; si- NC: 5′-UUCUCCGAACGUGUCAC GUTT-3′; si-Egr1: 5′-CCAAGGCCGAGAUGCAAUUTT-3′; and si- NC: 5′-ACGUGACACGUUCGGAGAATT-3′.
Vivo injection siRNA
MBD2 siRNA used for vivo injection is a chemically modified siRNA, consisted of a 21-nucleotide length with the following modifications: Chol-si-MBD2 F,5′-AGAcGAuGccAGuAAUUUUTT-3′and Chol-si-MBD2 R,5′-AAAAuuACuGGCauCAucuuc-3′. The lower-case letters represent 2′-O-methyl (OMe)-modified nucleotides. Chol represents cholesterol linked through a hydroxyprolinol linkage; it is injected once on the first and fourth days after the operation. The mouse tail was wiped with alcohol and iodophor. Afterward, the tail of the mouse was placed in warm water, followed by injection of siRNA solution (200 μL) into the root of the tail. In addition, normal saline was injected with the equal volume in the control mice. After injection, mice were maintained in the cage.
ChIP analysis and transcriptional activation activity
ChIP assays were performed using anti-MBD2, according to the instruction described for the ChIP kit (Millipore, USA).13,42,45, 46, 47 The sequence of EGR1 promoter (−2000–0) was input into MethPrimer Promoter 2.0 (http://www.urogene.org/cgi-bin/methprimer2/MethPrimer.cgi) to predict CpG islands and design primers. Precipitated DNA was amplified by PCR using CpG island binding primers for promoters of EGR1: F1:5′-GTTATAGAGGGATTAAGTTTAAGAAAG-3′; R1: 5′-CAATCAATTACTCACCCCAAAA-3′; F2: 5′-TTATTTGGATTGGATAAAGGGG-3′; R2: 5′-TAAACTACCTTACCTCTACCACCTAT-3′; F3: 5′-AGTTATAGAGGGATTAAGTTTAAGAAAG-3′; R3: 5′-CAATCAATTACTCACCCCAAAA-3′; F4: 5′-TTATAGAGGGATTAAGTTTAAGAAAG-3′; R: 5′-AATCAATTACTCACCCCAAAAA-3′; F5: 5′-GTTATAGAGGGATTAAGTTTAAGAAAG-3′; and R5: 5′-AATCAATTACTCACCCCAAAAA-3′. The transcriptional activation activity of EGR1 was evaluated using the relative activity of luciferase by employing the Promega kit, as described previously.48
Methylated CpG-DNA immunoprecipitation
The methylated CpG-DNA immunoprecipitation assay was performed according to the manufacturer's protocol (Zymo Research, Irvine, CA, USA), as previously described.49 The enriched genome was eluted and purified, and the obtained genomic DNA was used for methylated CpG immunoprecipitation, Supernatants were transferred to fresh microcentrifuge tubes. The DNA was purified through a spin column (QIAGEN, Hilden, Germany). DNA was resuspended in 50 μL of H2O. We used the DNA as a template with TB Green (TaKaRa, Tokyo, Japan,RR820A) and a LightCycler 96(Roche, Basal, Switzerland), with the F4/R4 primer and then performed qPCR analysis.
Bisulfite sequencing
Gene-specific DNA methylation was assessed by a next-generation bisulfite sequencing PCR (Bisulfite sequencing PCR,BSP). In brief. 1 μg of genomic DNA was bisulfite converted using the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA, USA), and one-twentieth of the elution products were used as templates for PCR amplification with 35 cycles using HiFi HotStart Uracil and ReadyMix PCR Kit (Kapa Biosystems, Wilmington, MA, USA). For each sample, BSP products of multiple genes were pooled equally, 5′-phosphorylated, 3′-dA-tailed, and ligated to barcoded adapter using T4 DNA ligase (New England BioLabs,Frankfurt am Main, Germany). Barcoded libraries from all samples were sequenced on Illumina platform. BSP primers were designed using the online MethPrimer software, EGR1 promoter primer sequence: Forward, 5′-TAGGTAGTGTTTTAAGAATTAGTAGTTAAATG-3′, and Reverse, 5′-TTCTACAACCRTCATCCTAAACTCAC-3′.
Histology, immunohistochemistry, and immunoblot analyses
Harvested kidney tissues were fixed in 4% buffered paraformaldehyde, embedded in paraffin blocks, and sectioned at 4 μm thickness. Sections were stained with H&E and Masson trichrome stains.42,50 Immunohistochemical analyses were carried out using anti-MBD2, Col I and IV, FN, and EGR1, according to previously published protocols.42 The details for quantification have been described in our recently published study.40 For western blot analysis, tissue lysates from kidneys or BUMPT cells were subjected to SDS-PAGE gel electrophoresis, membrane transfer, and immunodetection using anti-MBD2, Col I and IV, FN, and EGR1 antibodies, according to published standard procedures.
Statistical analysis
Data were expressed as means ± SD. A two-tailed Student’s t test was used for two-group comparison. One-way ANOVA was used for multiple comparisons. Kruskal-Wallis test was used for the non-parametric tests for multiple-group comparison. Statistical significance was set at p < 0.05.
Acknowledgments
The study was supported in part by grants from the National Natural Science Foundation of China (81870475, 81570646, 81770951, and 81770692), Changsha Science and Technology Bureau Project (kq2001039), key project of Hunan Provincial Science and Technology Innovation (2020SK1014). Department of Science and Technology of Hunan Province Project of International Cooperation and Exchanges (2020WK2009), Natural Science Foundation of Hunan Province of China (2018JJ2566 and 2021JJ40818), Fundamental Research Funds for the Central Universities of Central South University (2020zzts282 and 2021zzts0362), Hunan Provincial Innovation Foundation for Postgraduate (CX20200290 and CX20210364), and China Hunan Provincial Science and Technology Department (2021SK4004).
Author contributions
D.Z. conceived and designed the experiments; K.A., X.L., P.Z., and J.P. carried out the experiments; Z.H., H.Z., L.Y., Y.K., J.C., and H.L. analyzed the data; Y.W., X.C., Y.L., and X.X. contributed reagents/materials/analysis tools; and D.Z. wrote the main manuscript text but all authors reviewed the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.02.015.
Supplemental information
References
- 1.Hamidi T., Singh A.K., Chen T. Genetic alterations of DNA methylation machinery in human diseases. Epigenomics. 2015;7:247–265. doi: 10.2217/epi.14.80. [DOI] [PubMed] [Google Scholar]
- 2.Jaenisch R., Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 3.Okano M., Bell D.W., Haber D.A., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 4.Bechtel W., McGoohan S., Zeisberg E.M., Muller G.A., Kalbacher H., Salant D.J., Muller C.A., Kalluri R., Zeisberg M. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat. Med. 2010;16:544–550. doi: 10.1038/nm.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larkin B.P., Glastras S.J., Chen H., Pollock C.A., Saad S. DNA methylation and the potential role of demethylating agents in prevention of progressive chronic kidney disease. FASEB J. 2018;32:5215–5226. doi: 10.1096/fj.201800205R. [DOI] [PubMed] [Google Scholar]
- 6.Cheng J., Song J., He X., Zhang M., Hu S., Zhang S., Yu Q., Yang P., Xiong F., Wang D.W., et al. Loss of Mbd2 protects mice against high-fat diet-induced obesity and insulin resistance by regulating the homeostasis of energy storage and expenditure. Diabetes. 2016;65:3384–3395. doi: 10.2337/db16-0151. [DOI] [PubMed] [Google Scholar]
- 7.Zhou M., Zhou K., Cheng L., Chen X., Wang J., Wang X.M., Zhang Y., Yu Q., Zhang S., Wang D., et al. MBD2 ablation impairs lymphopoiesis and impedes progression and maintenance of T-ALL. Cancer Res. 2018;78:1632–1642. doi: 10.1158/0008-5472.CAN-17-1434. [DOI] [PubMed] [Google Scholar]
- 8.May S., Owen H., Phesse T.J., Greenow K.R., Jones G.R., Blackwood A., Cook P.C., Towers C., Gallimore A.M., Williams G.T., et al. Mbd2 enables tumourigenesis within the intestine while preventing tumour-promoting inflammation. J. Pathol. 2018;245:270–282. doi: 10.1002/path.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng L., Tang Y., Chen X., Zhao L., Liu S., Ma Y., Wang N., Zhou K., Zhou J., Zhou M. Deletion of MBD2 inhibits proliferation of chronic myeloid leukaemia blast phase cells. Cancer Biol. Ther. 2018;19:676–686. doi: 10.1080/15384047.2018.1450113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng Z., Li M., Chen J., Li Q., Ning Q., Zhao J., Xu Y., Xie J., Yu J. Reduced MBD2 expression enhances airway inflammation in bronchial epithelium in COPD. Int. J. Chron. Obstruct Pulmon Dis. 2018;13:703–715. doi: 10.2147/COPD.S148595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia A., Wang Y., Sun W., Xiao B., Wei Y., Qiu L., Mu L., Xu L., Li J., Zhang X., et al. MBD2 regulates Th17 cell differentiation and experimental severe asthma by affecting IRF4 expression. Mediators Inflamm. 2017;2017:6249685. doi: 10.1155/2017/6249685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stirzaker C., Song J.Z., Ng W., Du Q., Armstrong N.J., Locke W.J., Statham A.L., French H., Pidsley R., Valdes-Mora F., et al. Methyl-CpG-binding protein MBD2 plays a key role in maintenance and spread of DNA methylation at CpG islands and shores in cancer. Oncogene. 2017;36:1328–1338. doi: 10.1038/onc.2016.297. [DOI] [PubMed] [Google Scholar]
- 13.Wang J., Li H., Qiu S., Dong Z., Xiang X., Zhang D. MBD2 upregulates miR-301a-5p to induce kidney cell apoptosis during vancomycin-induced AKI. Cell Death Dis. 2017;8:e3120. doi: 10.1038/cddis.2017.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie Y., Liu B., Pan J., Liu J., Li X., Li H., Qiu S., Xiang X., Zheng P., Chen J., et al. MBD2 mediates septic AKI through activation of PKCeta/p38MAPK and the ERK1/2 Axis. Mol. Ther. Nucleic Acids. 2021;23:76–88. doi: 10.1016/j.omtn.2020.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun T., Liu Q., Wang Y., Deng Y., Zhang D. MBD2 mediates renal cell apoptosis via activation of Tox4 during rhabdomyolysis-induced acute kidney injury. J. Cell Mol. Med. 2021;25:4562–4571. doi: 10.1111/jcmm.16207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang H.M., Ahn S.H., Choi P., Ko Y.A., Han S.H., Chinga F., Park A.S., Tao J., Sharma K., Pullman J., et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat. Med. 2015;21:37–46. doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L., Liu L., Bai M., Liu M., Wei L., Yang Z., Qian Q., Ning X., Sun S. Hypoxia-induced HE4 in tubular epithelial cells promotes extracellular matrix accumulation and renal fibrosis via NF-kappaB. FASEB J. 2020;34:2554–2567. doi: 10.1096/fj.201901950R. [DOI] [PubMed] [Google Scholar]
- 18.Liu B.C., Tang T.T., Lv L.L. How tubular epithelial cell injury contributes to renal fibrosis. Adv. Exp. Med. Biol. 2019;1165:233–252. doi: 10.1007/978-981-13-8871-2_11. [DOI] [PubMed] [Google Scholar]
- 19.Li H., Peng X., Wang Y., Cao S., Xiong L., Fan J., Wang Y., Zhuang S., Yu X., Mao H. Atg5-mediated autophagy deficiency in proximal tubules promotes cell cycle G2/M arrest and renal fibrosis. Autophagy. 2016;12:1472–1486. doi: 10.1080/15548627.2016.1190071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X., Pan J., Li H., Li G., Liu X., Liu B., He Z., Peng Z., Zhang H., Li Y., et al. DsbA-L mediated renal tubulointerstitial fibrosis in UUO mice. Nat. Commun. 2020;11:4467. doi: 10.1038/s41467-020-18304-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sukhatme V.P. The Egr family of nuclear signal transducers. Am. J. Kidney Dis. 1991;17:615–618. doi: 10.1016/s0272-6386(12)80333-7. [DOI] [PubMed] [Google Scholar]
- 22.Sukhatme V.P. The Egr transcription factor family: from signal transduction to kidney differentiation. Kidney Int. 1992;41:550–553. doi: 10.1038/ki.1992.79. [DOI] [PubMed] [Google Scholar]
- 23.Mozes M.M., Szoleczky P., Rosivall L., Kokeny G. Sustained hyperosmolarity increses TGF-ss1 and Egr-1 expression in the rat renal medulla. BMC Nephrol. 2017;18:209. doi: 10.1186/s12882-017-0626-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho L.C., Sung J.M., Shen Y.T., Jheng H.F., Chen S.H., Tsai P.J., Tsai Y.S. Egr-1 deficiency protects from renal inflammation and fibrosis. J. Mol. Med. (Berl) 2016;94:933–942. doi: 10.1007/s00109-016-1403-6. [DOI] [PubMed] [Google Scholar]
- 25.Wang D., Guan M.P., Zheng Z.J., Li W.Q., Lyv F.P., Pang R.Y., Xue Y.M. Transcription factor Egr1 is involved in high glucose-induced proliferation and fibrosis in rat glomerular mesangial cells. Cell Physiol. Biochem. 2015;36:2093–2107. doi: 10.1159/000430177. [DOI] [PubMed] [Google Scholar]
- 26.Sun S., Ning X., Zhai Y., Du R., Lu Y., He L., Li R., Wu W., Sun W., Wang H. Egr-1 mediates chronic hypoxia-induced renal interstitial fibrosis via the PKC/ERK pathway. Am. J. Nephrol. 2014;39:436–448. doi: 10.1159/000362249. [DOI] [PubMed] [Google Scholar]
- 27.Kemme C.A., Marquez R., Luu R.H., Iwahara J. Potential role of DNA methylation as a facilitator of target search processes for transcription factors through interplay with methyl-CpG-binding proteins. Nucleic Acids Res. 2017;45:7751–7759. doi: 10.1093/nar/gkx387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sansom O.J., Maddison K., Clarke A.R. Mechanisms of disease: methyl-binding domain proteins as potential therapeutic targets in cancer. Nat. Clin. Pract. Oncol. 2007;4:305–315. doi: 10.1038/ncponc0812. [DOI] [PubMed] [Google Scholar]
- 29.Sansom O.J., Berger J., Bishop S.M., Hendrich B., Bird A., Clarke A.R. Deficiency of Mbd2 suppresses intestinal tumorigenesis. Nat. Genet. 2003;34:145–147. doi: 10.1038/ng1155. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y., Zhang L., Wu G.R., Zhou Q., Yue H., Rao L.Z., Yuan T., Mo B., Wang F.X., Chen L.M., et al. MBD2 serves as a viable target against pulmonary fibrosis by inhibiting macrophage M2 program. Sci. Adv. 2021;7:eabb6075. doi: 10.1126/sciadv.abb6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sukhatme V.P., Cao X.M., Chang L.C., Tsai-Morris C.H., Stamenkovich D., Ferreira P.C., Cohen D.R., Edwards S.A., Shows T.B., Curran T., et al. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell. 1988;53:37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- 32.Lee C.G., Cho S.J., Kang M.J., Chapoval S.P., Lee P.J., Noble P.W., Yehualaeshet T., Lu B., Flavell R.A., Milbrandt J., et al. Early growth response gene 1-mediated apoptosis is essential for transforming growth factor beta1-induced pulmonary fibrosis. J. Exp. Med. 2004;200:377–389. doi: 10.1084/jem.20040104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhattacharyya S., Wu M., Fang F., Tourtellotte W., Feghali-Bostwick C., Varga J. Early growth response transcription factors: key mediators of fibrosis and novel targets for anti-fibrotic therapy. Matrix Biol. 2011;30:235–242. doi: 10.1016/j.matbio.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhattacharyya S., Sargent J.L., Du P., Lin S., Tourtellotte W.G., Takehara K., Whitfield M.L., Varga J. Egr-1 induces a profibrotic injury/repair gene program associated with systemic sclerosis. PLoS One. 2011;6:e23082. doi: 10.1371/journal.pone.0023082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derdak Z., Villegas K.A., Wands J.R. Early growth response-1 transcription factor promotes hepatic fibrosis and steatosis in long-term ethanol-fed Long-Evans rats. Liver Int. 2012;32:761–770. doi: 10.1111/j.1478-3231.2012.02752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fortin J., Bernard D.J. SMAD3 and EGR1 physically and functionally interact in promoter-specific fashion. Cell Signal. 2010;22:936–943. doi: 10.1016/j.cellsig.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 37.Nakashima A., Ota A., Sabban E.L. Interactions between Egr1 and AP1 factors in regulation of tyrosine hydroxylase transcription. Brain Res. Mol. Brain Res. 2003;112:61–69. doi: 10.1016/s0169-328x(03)00047-0. [DOI] [PubMed] [Google Scholar]
- 38.Dahal B., Lin S.C., Carey B.D., Jacobs J.L., Dinman J.D., van Hoek M.L., Adams A.A., Kehn-Hall K. EGR1 upregulation following Venezuelan equine encephalitis virus infection is regulated by ERK and PERK pathways contributing to cell death. Virology. 2020;539:121–128. doi: 10.1016/j.virol.2019.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsunaga N., Ikeda E., Kakimoto K., Watanabe M., Shindo N., Tsuruta A., Ikeyama H., Hamamura K., Higashi K., Yamashita T., et al. Inhibition of G0/G1 switch 2 ameliorates renal inflammation in chronic kidney disease. EBioMedicine. 2016;13:262–273. doi: 10.1016/j.ebiom.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L., Xu X., Yang R., Chen J., Wang S., Yang J., Xiang X., He Z., Zhao Y., Dong Z., et al. Paclitaxel attenuates renal interstitial fibroblast activation and interstitial fibrosis by inhibiting STAT3 signaling. Drug Des. Devel Ther. 2015;9:2139–2148. doi: 10.2147/DDDT.S81390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang R., Xu X., Li H., Chen J., Xiang X., Dong Z., Zhang D. p53 induces miR199a-3p to suppress SOCS7 for STAT3 activation and renal fibrosis in UUO. Sci. Rep. 2017;7:43409. doi: 10.1038/srep43409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang D., Sun L., Xian W., Liu F., Ling G., Xiao L., Liu Y., Peng Y., Haruna Y., Kanwar Y.S. Low-dose paclitaxel ameliorates renal fibrosis in rat UUO model by inhibition of TGF-beta/Smad activity. Lab. Invest. 2010;90:436–447. doi: 10.1038/labinvest.2009.149. [DOI] [PubMed] [Google Scholar]
- 43.Ge Y., Wang J., Wu D., Zhou Y., Qiu S., Chen J., Zhu X., Xiang X., Li H., Zhang D. lncRNA NR_038323 suppresses renal fibrosis in diabetic nephropathy by targeting the miR-324-3p/DUSP1 Axis. Mol. Ther. Nucleic Acids. 2019;17:741–753. doi: 10.1016/j.omtn.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J., Pan J., Li H., Long J., Fang F., Chen J., Zhu X., Xiang X., Zhang D. lncRNA ZEB1-AS1 was suppressed by p53 for renal fibrosis in diabetic nephropathy. Mol. Ther. Nucleic Acids. 2018;12:741–750. doi: 10.1016/j.omtn.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang D., Pan J., Xiang X., Liu Y., Dong G., Livingston M.J., Chen J.K., Yin X.M., Dong Z. Protein kinase cdelta suppresses autophagy to induce kidney cell apoptosis in cisplatin nephrotoxicity. J. Am. Soc. Nephrol. 2017;28:1131–1144. doi: 10.1681/ASN.2016030337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang D., Li Y., Liu Y., Xiang X., Dong Z. Paclitaxel ameliorates lipopolysaccharide-induced kidney injury by binding myeloid differentiation protein-2 to block Toll-like receptor 4-mediated nuclear factor-kappaB activation and cytokine production. J. Pharmacol. Exp. Ther. 2013;345:69–75. doi: 10.1124/jpet.112.202481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun L., Zhang D., Liu F., Xiang X., Ling G., Xiao L., Liu Y., Zhu X., Zhan M., Yang Y., et al. Low-dose paclitaxel ameliorates fibrosis in the remnant kidney model by down-regulating miR-192. J. Pathol. 2011;225:364–377. doi: 10.1002/path.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klug M., Rehli M. Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics. 2006;1:127–130. doi: 10.4161/epi.1.3.3327. [DOI] [PubMed] [Google Scholar]
- 49.Alvarado S., Wyglinski J., Suderman M., Andrews S.A., Szyf M. Methylated DNA binding domain protein 2 (MBD2) coordinately silences gene expression through activation of the microRNA hsa-mir-496 promoter in breast cancer cell line. PLoS One. 2013;8:e74009. doi: 10.1371/journal.pone.0074009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang P., Yi L., Qu S., Dai J., Li X., Liu B., Li H., Ai K., Zheng P., Qiu S., et al. The biomarker TCONS_00016233 drives septic AKI by targeting the miR-22-3p/AIFM1 signaling Axis. Mol. Ther. Nucleic Acids. 2020;19:1027–1042. doi: 10.1016/j.omtn.2019.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.