Summary
Background
Childhood neurodevelopmental disorders, including autism spectrum disorder (ASD), attention-deficit hyperactivity disorder (ADHD), and Tourette syndrome (TS), comprise a major cause of health-related disabilities in children. However, biomarkers towards pathogenesis or novel drug targets are still limited. Our study aims to provide a comprehensive investigation of the causal effects of the plasma proteome on ASD, ADHD, and TS using the two-sample Mendelian Randomization (MR) approach.
Methods
Genetic associations with 2994 plasma proteins were selected as exposures and genome-wide association data of ASD, ADHD, TS were utilized as outcomes. MR analyses were carried out using the inverse-variance weighted method, and the MR-Egger and weighted median methods were used for sensitivity analysis.
Findings
Using single-nucleotide polymorphisms as instruments, the study suggested increased levels of MAPKAPK3 (OR: 1.09; 95% CI: 1.05–1.13; P = 1.43 × 10−6) and MRPL33 (OR: 1.07; 95% CI: 1.04–1.11; P = 5.37 × 10−6) were causally associated with a higher risk of ASD, and increased MANBA level was associated with a lower risk of ADHD (OR: 0.91; 95% CI: 0.88–0.95; P = 8.97 × 10−6). The causal associations were robust in sensitivity analysis, leave-one-out analysis and Multivariable MR, and no pleiotropy was observed. No significant risk protein was identified for TS.
Interpretation
The study findings support the idea that the MAPK/ERK signaling pathway and mitochondrial dysfunction are involved in the pathogenesis of ASD, while a deficiency in beta-mannosidase might play a role in the development of ADHD.
Keywords: Autism spectrum disorder, Attention-deficit hyperactivity disorder, MAPKAPK3, MRPL33, MANBA
Research in context.
Evidence before this study
Childhood neurodevelopmental disorders, including autism spectrum disorder, attention-deficit hyperactivity disorder, and Tourette syndrome, comprise a major cause of health-related disabilities, and are known to have a strong genetic component. Proteins are important intermediate phenotypes that can provide valuable insights into the complex processes influencing human biology and disease pathophysiology. A recent genome-wide association study has built a large genomic atlas of the human plasma proteome, which offers great opportunity to identify disease related risk proteins using genetic epidemiology strategies.
Added value of this study
In this two-sample Mendelian randomization study using genetic instruments from large-scale genome-wide association studies, increased levels of MAPKAPK3 and MRPL33 were associated with a higher risk of autism spectrum disorder, and increased levels of MANBA were associated with a lower risk for attention-deficit hyperactivity disorder.
Implications of all the available evidence
These findings support the idea that the MAPK/ERK signaling pathway and mitochondrial dysfunction have pathogenic roles in the autism spectrum disorder and that beta-mannosidase deficiency has a role in attention-deficit hyperactivity disorder.
Alt-text: Unlabelled box
Introduction
Childhood is a time of fun and games; however, it could be a frustrating period with suffering, for some families. Childhood neurodevelopmental disorders, including autism spectrum disorder (ASD), attention-deficit hyperactivity disorder (ADHD), and Tourette syndrome (TS), account for 15% to 30% of the disability-adjusted life-years (DALYs), and they are a major cause of health-related disability in this age group.1,2 Patients with such disorders frequently have learning difficulties, as well as attainment, work, and interpersonal problems; the symptoms persist in adulthood for a substantial proportion of the population.3, 4, 5, 6, 7 Recent technological advances, particularly genome-wide association studies (GWASs), have successfully identified a number of novel biomarkers for these disorders.8, 9, 10, 11, 12 However, further evidence regarding their pathogenesis and the development of drug targets is required.
Proteins are important intermediate phenotypes that can provide valuable insights into the complex processes influencing human biology and disease pathophysiology. With the recent advent of proteomic profiling technologies, more than 5,000 circulating plasma proteins in the human body can now be detected. This provides a window through which an individual's physiology and health status can be assessed.13,14 Moreover, a growing body of studies have focused on integrating information from genomics and proteomics to identify disease-related biomarkers, uncover potential drug targets, reveal biologically functional pathways, and elucidate disease pathogenesis.15, 16, 17, 18 The GWASs on the human plasma proteome have reported thousands of associations between single-nucleotide polymorphisms (SNPs) and circulating protein levels. These offer a great opportunity to investigate the causal effects of a large number of potential biomarkers on human diseases, using a Mendelian randomization (MR) approach.19
MR analysis is a novel genetic epidemiological study design that can elucidate the causal association between a modifiable exposure and disease outcome by using SNPs as instrument variables.20 Conceptually, MR has similarities with randomized controlled trials (RCTs), following the foundational principle that genetic variants are naturally randomly assigned to offspring before birth; therefore, the individuals are consequently randomized into different levels of exposure.21 Genotypes are assigned before birth and are not susceptible to environmental confounders; therefore, MR has the ability to expose unbiased causality of modifiable exposures on disease outcomes of interest.22 The MR study design is the most common, and it utilizes genetic associations with exposure and outcome generated from non-overlapping samples.23 In this study, we applied a MR to investigate the causal effects of the human plasma proteome on three childhood neurodevelopmental disorders, ASD, ADHD, and TS.
Methods
Study design and data sources
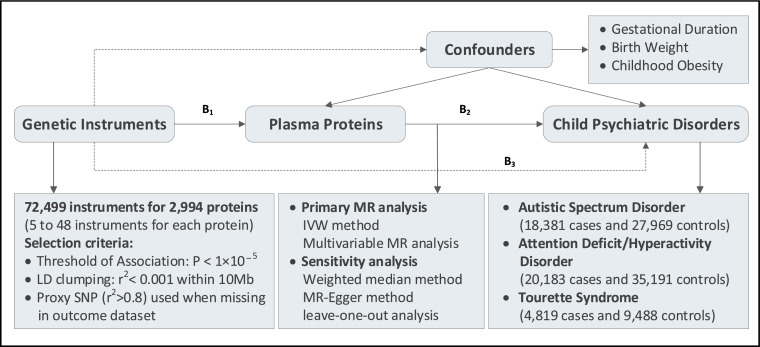
The overall design used for this work is illustrated in Figure 1. We employed a MR analysis to investigate the causal associations between genetically predicted plasma protein levels and three childhood psychiatric disorders. The basic principle of MR is that genetic instruments, which could predict the level of a modifiable exposure, should be causally associated with the exposure-related outcome.20 Three assumptions are required for a valid genetic instrument: (i) it is causally related with the exposure; (ii) it is independent of confounders; (iii) it is only associated with the outcome through the exposure. In this present study, genetic instruments for the plasma proteins were obtained from a large-scale GWAS on 3301 healthy adults of European ancestry.19 Plasma proteome were quantified using an aptamer-based multiplex protein assay (SOMAscan) and the robustness of protein measurements were further verified using several subsequent experiments. Quality control processes were performed at both the sample and the protein levels, leaving 3283 SOMA aptamers (SOMAmers) mapping to 2994 plasma proteins for inclusion in final GWAS analysis. Outcomes datasets included GWAS summary statistics of ASD (18,381 cases and 27,969 controls),9 ADHD (20,183 cases and 35,191 controls),11 and TS (4819 and 9488 controls)12 from the Psychiatric Genomics Consortium (PGC). We also extracted GWAS datasets of gestational duration,24 birth weight,25 and childhood obesity26 from the Early Growth Genetics (EGG) Consortium as potential confounders (Table S1).
Figure 1.
Study overview and MR model
All summary-level GWAS data were derived from participants of predominantly European ancestry. B2 is the causal association of interest (2,994 plasma proteins on three childhood neurodevelopmental disorders: ASD, ADHD, and TS), estimated using B2 =B1/B3. B1 and B3 are the direct associations of the genetic variants on the exposure (plasma proteins) and outcomes (ASD, ADHD, TS). IVW, inverse variance weighted.
Selection of genetic instruments for plasma proteins
Genetic instruments were extracted from the large genetic atlas of the human plasma proteome according to the unified standards. All relevant SNPs in each dataset met the significance threshold of P ≤ 1 × 10−5. The relatively relaxed threshold for the genetic instruments was used in the MR investigations when there were only a few significant and genome-wide SNPs (P < 5 × 10−8) available.27,28 We performed linkage disequilibrium (LD) clumping to identify independent SNPs (r2 < 0.001 within 10 Mb), using the 1000 Genomes Project Phase 3 (EUR) as the reference panel. We harmonized exposure and outcome datasets, obtained SNP effects and corresponding standard errors, and removed palindromic SNPs with intermediate allele frequencies. We used proxy SNPs (LD at r2 > 0.8) when no SNP was available for predicting a specific protein for the outcome. Two parameters, the proportion of variance (R2) explained by the SNPs and the F statistic, were used to evaluate the instrument strength for each protein.29 Typically, an F statistic >10 is considered sufficiently informative for MR analyses.30 In this present study, we extracted a range of 9–42 SNPs explaining an average R2 of 17.8% (range 5.7–82.2%), and the minimum F statistic was 21.83, suggesting all instrumental variables were sufficiently informative (F statistic >10) for MR analyses (Table S2).
Datasets of outcomes
The PGC unified much of the psychiatric genetics and aimed to enable rapid progress in elucidating the genetic basis of psychiatric disorders.31 They have performed statistically rigorous and comprehensive GWAS meta-analyses for most psychiatric disorders, including ASD, ADHD, and TS. GWASs of ASD and ADHD were both performed by combining samples from Integrative Psychiatric Research (iPSYCH) and the PGC.9,11 The iPSYCH samples were collected from a population-based cohort of all children born in Denmark between 1981 and 2005, whereas the PGC samples were extracted from several European cohorts. GWAS of TS was performed using samples from four European cohorts by the PGC.12 All GWAS analysis including quality control, imputation, and primary association analysis were conducted according to the PGC Ricopili GWAS pipeline.32
Statistical analysis
The inverse variance-weighted (IVW) method was used as the primary MR analysis. This method provided a high-powered estimate and relied on the assumption that all SNPs were valid genetic instruments.30 The weighted median method and MR-Egger method were adopted as sensitivity analyses to evaluate the robustness of causality and detect pleiotropy. The weighted median method could provide a consistent estimate if less than 50% of the SNPs were invalid instruments.33 The MR-Egger method was useful when up to 100% of the SNPs came from invalid instruments.34 We tested for pleiotropy by performing MR-Egger intercept test,35 Cochran's Q test,36 and leave-one-out analyses.37 What's more, to further avoid the bias introduced by pleiotropic outliers, we used the MR pleiotropy residual sum and outlier (MR-PRESSO) method to detect potential outliers and re-performed the MR analyses (using IVW, MR-Egger, and the weighted median methods) after removing these outliers.38 Multivariable MR analyses were performed to statistically adjust for potential confounders. Genetic instruments from three datasets were included, respectively, in the multivariable MR model: 1 genome-wide significant SNP with gestational duration,24 146 genome-wide significant SNPs with birth weight,25 and 18 genome-wide significant SNPs with childhood obesity.26 All analyses were carried out using the TwoSampleMR package in R Software 3.6.1. A multiple-testing threshold of P < 1.52 × 10−5 (0.05/3283) was adopted to declare a statistical significance using the Bonferroni method.
Ethics
All datasets were publicly available, and ethical approval was acquired for all original studies.
Role of funding source
The funding sources had no role in study design, data collection, analysis, or interpretation, or any aspect pertinent to the study.
Results
Causal estimates of genetically predicted proteins on ASD
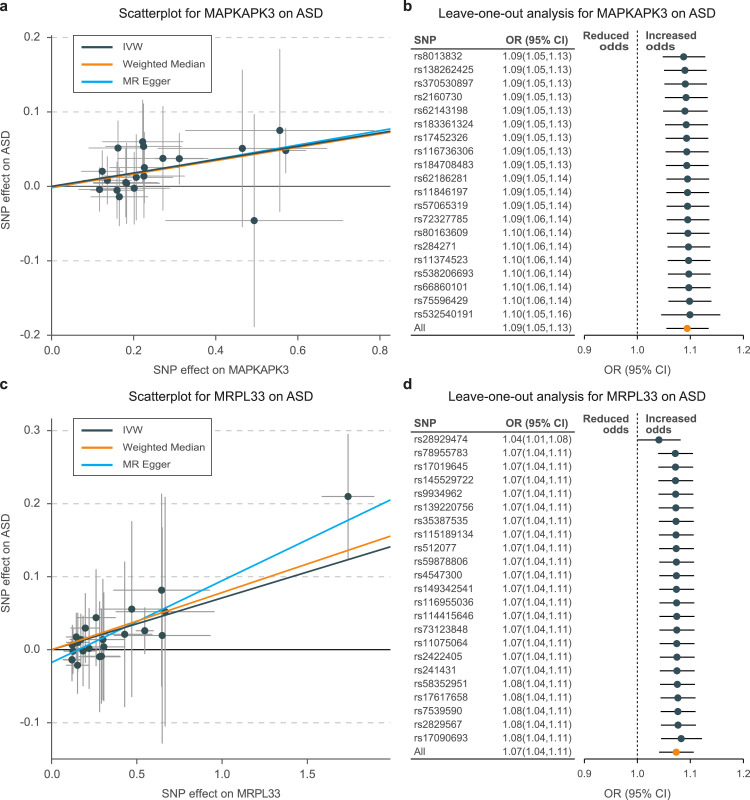
The MR estimates identified two causal risk proteins for ASD (Fig. S1) and one risk protein for ADHD (Fig. S2). Using 20 SNPs as genetic instruments, we observed that each 1-standard deviation (SD) increase in the level of MAPKAPK3 would result in an approximately 10% higher risk of ASD (IVW odds ratio [OR], 1.09 for per 1-SD increase in protein quantification; 95% CI: 1.05–1.13; P = 1.43 × 10−6; Tables 1, S3; Figure 2a). The causal estimates were broadly consistent, when using the additional methods for sensitivity analysis (MR-Egger OR: 1.10; 95% CI: 1.03–1.18; P = 7.61 × 10−3; weighted median OR: 1.09; 95% CI: 1.04–1.15; P = 3.03 × 10−4). Horizontal pleiotropies were not observed in the MR-Egger intercept test (P = 0.843), Cochran's Q test (IVW derived Q statistic = 15.2; P = 0.338), or leave-one-out analyses (Figure 2b). Similar results were observed for the causality of MRPL33 on ASD (IVW OR: 1.07; 95% CI: 1.04–1.11; P = 5.37 × 10−6; MR-Egger OR: 1.12; 95% CI: 1.07–1.17; P = 5.63 × 10−7; weighted median OR: 1.08; 95% CI: 1.03–1.13; P = 8.33 × 10−4; Tables 1, S4; Figure 2c). Bias from horizontal pleiotropies could be largely ruled out using the Cochran's Q test (IVW derived Q statistic = 16.3; P = 0.800) and leave-one-out analyses (Figure 2d).
Table 1.
Mendelian randomization results of causal risk proteins on childhood neurodevelopmental disorders.a,b
| Method | No. of SNPs | MR analysis |
Heterogeneity test |
MR-Egger intercept P | |||
|---|---|---|---|---|---|---|---|
| OR (95%) | P | Cochran's Q | I2 | P | |||
| MAPKAPK3 on ASD | |||||||
| IVW | 20 | 1.09(1.05,1.13) | 1.43e-06 | 15.2 | 0% | 0.710 | - |
| MR-Egger | 20 | 1.10(1.03,1.18) | 7.61e-03 | 15.2 | 0% | 0.711 | 0.843 |
| Weighted median | 20 | 1.09(1.04,1.15) | 3.03e-04 | - | - | - | - |
| MRPL33 on. ASD | |||||||
| IVW | 23 | 1.07(1.04,1.11) | 5.37e-06 | 16.3 | 0% | 0.800 | |
| MR-Egger | 23 | 1.12(1.07,1.17) | 5.63e-07 | 12.8 | 0% | 0.939 | 0.011 |
| Weighted median | 23 | 1.08(1.03,1.13) | 8.33e-04 | - | - | - | |
| MANBA on ADHD | |||||||
| IVW | 14 | 0.91(0.88,0.95) | 8.97e-06 | 15.1 | 13.6% | 0.304 | - |
| MR-Egger | 14 | 0.86(0.81,0.93) | 3.68e-05 | 11.2 | 0% | 0.596 | 0.072 |
| Weighted median | 14 | 0.89(0.85,0.94) | 3.65e-06 | - | - | - | - |
Abbreviations: MR, Mendelian Randomization; ASD, autism spectrum disorder; ADHD, attention-deficit hyperactivity disorder.
Results from 2-sample MR analysis; main analysis method: outliers identified and removed by MR PRESSO tool; estimated associations reported as OR of outcome per unit increase in log odds of the quantification of specific protein.
Genetic instruments selected from GWAS of plasma proteome, selection threshold P less than 1 × 10−5, pruned at linkage disequilibrium R2 less than .001 (10 megabytes pair window); No. of SNPs refers to the number of genetic instruments included in final MR analysis.
Figure 2.
Scatterplot and leave-one-out analysis of genetic risk of MAPKAPK3 and MRPL33 on ASD
Scatter plots of MR-derived associations between genetically predicted levels of MAPKAPK3 (a) and MRPL33 (c) with ASD, calculated using the inverse variance–weighted (IVW), weighted median, and MR-Egger methods. The slopes represent the causal association for each method. Leave-one-out analysis indicates fluctuant MR associations of MAPKAPK3 vs. ASD (b) and MRPL33 vs. ASD (d).
Causal estimates of genetically predicted proteins on ADHD and TS
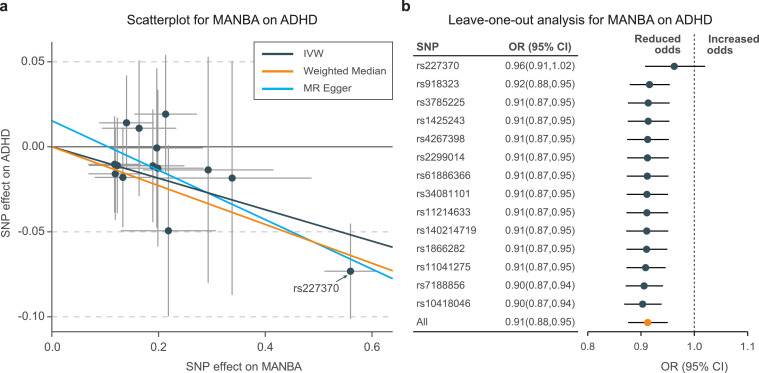
The genetically predicted levels of beta-mannosidase (MANBA) were associated with a decreased risk of ADHD (IVW OR: 0.91; 95% CI: 0.88–0.95; P = 8.97 × 10−6; MR-Egger OR: 0.86; 95% CI: 0.81–0.93; P = 3.68 × 10−5; Weighted median OR: 0.89; 95% CI: 0.85–0.94; P = 3.65 × 10−6; Tables 1, S5; Figure 3a). However, the causal estimate was largely affected by a single SNP, rs227370 (Figure 3b). The MR-Egger intercept test (P = 0.072) and Cochran's Q test (Q statistic = 15.1; P = 0.304) indicated no evidence of pleiotropy; therefore, it is necessary to verify whether rs227370 is a pleiotropic variant. On further analysis, rs227370 was found to be located in the intron region of MANBA. Therefore, the causal association between MANBA and ADHD was robust, and the rs227370 could have an essential role in determining this relationship. No significant risk protein was identified for TS (Fig. S3).
Figure 3.
Scatterplot and leave-one-out analysis of genetic risk of MANBA on ADHD
a. Scatter plots of MR-derived associations between genetically predicted levels of MANBA with ADHD, calculated using the inverse variance weighted (IVW), weighted median, and MR-Egger methods. The slopes represent the causal association for each method. b. Leave-one-out analysis shows the fluctuant MR associations of MANBA with ADHD.
Multivariable MR estimates
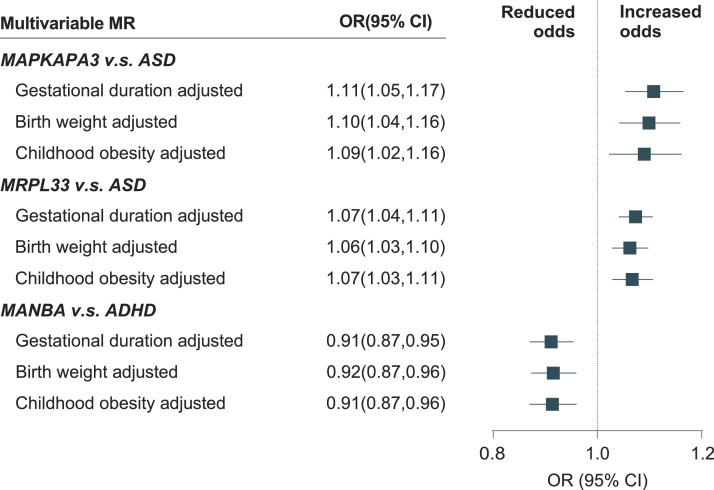
In the multivariable MR models, the causal effects of MAPKAPK3 on ASD were broadly consistent, when adjusted for gestational duration (OR: 1.11; 95% CI: 1.05–1.17; P = 1.80 × 10−3; Figure 4), birth weight (OR: 1.10; 95% CI: 1.04–1.16; P = 6.75 × 10−4), and childhood obesity (OR: 1.09; 95% CI: 1.02–1.16; P = 0.013). The results of the associations between MRPL33 and ASD were also robust, when adjusted for gestational duration (OR: 1.07; 95% CI: 1.04–1.11; P = 2.78 × 10−4), birth weight (OR: 1.06; 95% CI: 1.03–1.10; P = 3.99 × 10−4), and childhood obesity (OR: 1.07; 95% CI: 1.03–1.11; P = 1.41 × 10−3). Similar results were observed for the association between MANBA and ADHD, when adjusted for gestational duration (OR: 0.91; 95% CI: 0.87–0.95; P = 2.36 × 10−3), birth weight (OR: 0.92; 95% CI: 0.87–0.96; P = 3.50 × 10−4), and childhood obesity (OR: 0.91; 95% CI: 0.87–0.96; P = 1.22 × 10−3).
Figure 4.
Multivariate MR associations of causal risk proteins with childhood neurodevelopmental disorders adjusted for potential confounders
Estimates reported as odds ratios (OR) of childhood neurodevelopmental disorders per 1-SD increase in quantification of specific proteins, accounting for gestational duration, birth weight, and childhood obesity.
Discussion
We investigated the causal associations between the genetically predicted plasma proteome and three childhood neurodevelopmental disorders. We found genetic evidence that the increased levels of MAPKAPK3 and MRPL33 in human blood were associated with a higher risk of ASD, while increased levels of MANBA were associated with a lower risk of ADHD. No significant risk protein was identified for TS.
The MAPKAPK3 belongs to the Ser/Thr protein kinase family, which functions as a mitogen-activated protein (MAP) kinase. MAP kinases are also known as extracellular signal-regulated kinases (ERKs). The MAPK/ERK signaling pathway participates in important biological processes in the central nervous system, both temporally and spatially.39 It is involved in the pathogenesis of multiple neurological disorders, including ASD,40 Parkinson's disease,41 Alzheimer's disease,42 amyotrophic lateral sclerosis,43 and Huntington's disease.44 The role of the MAPK/ERK pathway in such diseases may be related to the glial cell function and inflammatory responses.39 During embryonic development and the early postnatal period, ERK participates in the maturation process of dendritic trees and synaptogenesis.39 Conditional inactivation of ERK signa.ls promote neurogenesis and repress gliogenesis.45,46 CD93 knockout mice showed an increase in astrocytes and a decrease in neurogenesis in the cerebral cortex, and they presented autism-like behavior.47 In addition, an ASD mouse model displayed autistic-like behaviors (impaired social interaction and communication as well as increased repetitive behaviors) and exhibited abnormal growth of its dendritic tree; this was associated with dysregulated MAPK/ERK signaling.48
MRPL33 is another protein that is causally associated with ASD. MRPL33 is a mitochondrial ribosomal protein involved in protein synthesis within the mitochondrion. The mitochondrial bioenergetics defects are a causal factor of ASD.49, 50, 51, 52 ASD could occur when mitochondrial function falls below the brain's minimum needs, because the human brain is a complex system and the organ with the highest mitochondrial energy demand.53 Specific biological mechanisms are involved in neurogenesis, neuronal migration, synaptic function, and signal transduction.54 There is a potential interaction between the mitochondrial function and the MAPK/ERK signaling pathway in neurologic disorders.55 In this study, genetic predictions for the level of MRPL33 were causally associated with ASD, which supported the hypothesis that mitochondrial dysfunction could be a cause of ASD. Furthermore, we identified a valuable target that could improve our understanding of the mechanisms underlying the mitochondrial dysfunction in the pathogenesis of ASD.
This study identified MANBA, as a causal risk protein for ADHD. Deficiency in MANBA is associated with hypotonia in the newborn, followed by global development delay, behavioral problems, and intellectual disability.56,57 Beta-mannosidosis is a disorder characterized by the accumulation of disaccharides resulting from insufficient lysosomal MANBA activity. Sedel et al. summarized the characteristics of 14 patients with MANBA; 78% presented with mental retardation, and 64% presented behavioral problems, including hyperactivity and/or aggressiveness.58 Furthermore, some cases with MANBA developed to ADHD.58,59 This study showed that lower levels of MANBA were associated ADHD, which was consistent with previous findings. Although evidence from large-scale epidemiological studies is still lacking, this study provides novel insights in improving our understanding of the pathogenesis of ADHD.
This study had several limitations. First, although genetic instruments for plasma proteins were extracted from the largest current GWAS source of plasma proteomes, there were still only a few genome-wide significant SNPs available for MR analysis. To address this, we adopted a relatively relaxed threshold for selecting genetic instruments, by referring to similar cases. We evaluated the strength for these genetic instruments and they were suitable for MR analysis. Second, all proteins in the human plasma were quantified in the GWAS proteome. Blood was a logical choice for the biomarker applications considering its convenience; however, we do not know whether these proteins had similar roles in specific brain regions, because of the existence of the blood-brain-barrier. Third, we performed an MR study and identified two causal risk proteins for ASD and one risk protein for ADHD. MR is an efficient tool for inferring causality; however, our findings need further experimental verification, and the mechanisms need further exploration. Nevertheless, our study might identify novel biomarkers for ASD and ADHD, and the results provide valuable information for better understanding the pathogenesis of childhood neurodevelopmental disorders.
In conclusion, our study supported the idea that increased levels of MAPKAPK3 and MRPL33 were causally associated with a higher risk of ASD, and increased level of MANBA was associated with a lower risk of ADHD. Our study highlighted the role of the MAPK/ERK signaling pathway and mitochondrial dysfunction in the pathogenesis of ASD, as well as the deficiency of MANBA in the development of ADHD. Our study might provid novel insight for understanding the pathogenesis and uncovering drug targets for childhood neurodevelopmental disorders.
Contributors
J.Y. and X.M. conceived the hypothesis and study design. J.Y. and X.H. analysed the data and drafted the manuscript. L.Q. and B.Z. participated in data extraction. Y.F. interpreted analysis results. F.G. was responsible for data management. B.Y., F.Z. and X.M. reviewed and revised the manuscript. All authors approved the final version of the manuscript.
Data sharing statement
Data can be obtained upon reasonable request to the corresponding author.
Declaration of Competing Interest
The authors have nothing to disclose.
Acknowledgments
The authors would like to thank the High-Performance Computing Cluster of the First Affiliated Hospital of Xi'an Jiaotong University for data computing support. The study was funded by the Natural Science Basic Research Program of Shaanxi (2021JQ-390).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103948.
Appendix. Supplementary materials
References
- 1.Kieling C., Baker-Henningham H., Belfer M., et al. Child and adolescent mental health worldwide: evidence for action. Lancet. 2011;378(9801):1515–1525. doi: 10.1016/S0140-6736(11)60827-1. [DOI] [PubMed] [Google Scholar]
- 2.Whiteford H.A., Degenhardt L., Rehm J., et al. Global burden of disease attributable to mental and substance use disorders: findings from the Global Burden of Disease Study 2010. Lancet. 2013;382(9904):1575–1586. doi: 10.1016/S0140-6736(13)61611-6. [DOI] [PubMed] [Google Scholar]
- 3.Gustavson K., Knudsen A.K., Nesvag R., Knudsen G.P., Vollset S.E., Reichborn-Kjennerud T. Prevalence and stability of mental disorders among young adults: findings from a longitudinal study. BMC Psychiatry. 2018;18(1):65. doi: 10.1186/s12888-018-1647-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel V., Flisher A.J., Hetrick S., McGorry P. Mental health of young people: a global public-health challenge. Lancet. 2007;369(9569):1302–1313. doi: 10.1016/S0140-6736(07)60368-7. [DOI] [PubMed] [Google Scholar]
- 5.Ogundele M.O. Behavioural and emotional disorders in childhood: a brief overview for paediatricians. World J Clin Pediatr. 2018;7(1):9–26. doi: 10.5409/wjcp.v7.i1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vander Stoep A., Beresford S.A., Weiss N.S., McKnight B., Cauce A.M., Cohen P. Community-based study of the transition to adulthood for adolescents with psychiatric disorder. Am J Epidemiol. 2000;152(4):352–362. doi: 10.1093/aje/152.4.352. [DOI] [PubMed] [Google Scholar]
- 7.Mojtabai R., Stuart E.A., Hwang I., Eaton W.W., Sampson N., Kessler R.C. Long-term effects of mental disorders on marital outcomes in the National Comorbidity Survey ten-year follow-up. Soc Psychiatry Psychiatr Epidemiol. 2017;52(10):1217–1226. doi: 10.1007/s00127-017-1373-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Autism Spectrum Disorders Working Group of The Psychiatric Genomics C Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol Autism. 2017;8:21. doi: 10.1186/s13229-017-0137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grove J., Ripke S., Als T.D., et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51(3):431–444. doi: 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neale B.M., Medland S.E., Ripke S., et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry. 2010;49(9):884–897. doi: 10.1016/j.jaac.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demontis D., Walters R.K., Martin J., et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51(1):63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu D., Sul J.H., Tsetsos F., et al. Interrogating the genetic determinants of Tourette's syndrome and other tic disorders through genome-wide association studies. Am J Psychiatry. 2019;176(3):217–227. doi: 10.1176/appi.ajp.2018.18070857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geyer P.E., Kulak N.A., Pichler G., Holdt L.M., Teupser D., Mann M. Plasma proteome profiling to assess human health and disease. Cell Syst. 2016;2(3):185–195. doi: 10.1016/j.cels.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Williams S.A., Kivimaki M., Langenberg C., et al. Plasma protein patterns as comprehensive indicators of health. Nat Med. 2019;25(12):1851–1857. doi: 10.1038/s41591-019-0665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suhre K., Arnold M., Bhagwat A.M., et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun. 2017;8:14357. doi: 10.1038/ncomms14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sjaarda J., Gerstein H.C., Kutalik Z., et al. Influence of genetic ancestry on human serum proteome. Am J Hum Genet. 2020;106(3):303–314. doi: 10.1016/j.ajhg.2020.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akiyama M. Multi-omics study for interpretation of genome-wide association study. J Hum Genet. 2021;66(1):3–10. doi: 10.1038/s10038-020-00842-5. [DOI] [PubMed] [Google Scholar]
- 18.Suhre K., McCarthy M.I., Schwenk J.M. Genetics meets proteomics: perspectives for large population-based studies. Nat Rev Genet. 2021;22(1):19–37. doi: 10.1038/s41576-020-0268-2. [DOI] [PubMed] [Google Scholar]
- 19.Sun B.B., Maranville J.C., Peters J.E., et al. Genomic atlas of the human plasma proteome. Nature. 2018;558(7708):73–79. doi: 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans D.M., Davey Smith G. Mendelian randomization: new applications in the coming age of hypothesis-free causality. Annu Rev Genom Hum Genet. 2015;16:327–350. doi: 10.1146/annurev-genom-090314-050016. [DOI] [PubMed] [Google Scholar]
- 21.Zheng J., Baird D., Borges M.C., et al. Recent developments in Mendelian randomization studies. Curr Epidemiol Rep. 2017;4(4):330–345. doi: 10.1007/s40471-017-0128-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burgess S., Small D.S., Thompson S.G. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2017;26(5):2333–2355. doi: 10.1177/0962280215597579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartwig F.P., Davies N.M., Hemani G., Davey Smith G. Two-sample Mendelian randomization: avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int J Epidemiol. 2016;45(6):1717–1726. doi: 10.1093/ije/dyx028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X., Helenius D., Skotte L., et al. Variants in the fetal genome near pro-inflammatory cytokine genes on 2q13 associate with gestational duration. Nat Commun. 2019;10(1):3927. doi: 10.1038/s41467-019-11881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warrington N.M., Beaumont R.N., Horikoshi M., et al. Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat Genet. 2019;51(5):804–814. doi: 10.1038/s41588-019-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradfield J.P., Vogelezang S., Felix J.F., et al. A trans-ancestral meta-analysis of genome-wide association studies reveals loci associated with childhood obesity. Hum Mol Genet. 2019;28(19):3327–3338. doi: 10.1093/hmg/ddz161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanna S., van Zuydam N.R., Mahajan A., et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet. 2019;51(4):600–605. doi: 10.1038/s41588-019-0350-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi K.W., Chen C.Y., Stein M.B., et al. Assessment of bidirectional relationships between physical activity and depression among adults: a 2-sample Mendelian randomization study. JAMA Psychiatry. 2019;76(4):399–408. doi: 10.1001/jamapsychiatry.2018.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brion M.J., Shakhbazov K., Visscher P.M. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol. 2013;42(5):1497–1501. doi: 10.1093/ije/dyt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgess S., Butterworth A., Thompson S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan P.F. The psychiatric GWAS consortium: big science comes to psychiatry. Neuron. 2010;68(2):182–186. doi: 10.1016/j.neuron.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ripke S. Ricopili pipeline and standards of GWAS analyses. Eur Neuropsychopharmacol. 2019;29:S713–S7S4. [Google Scholar]
- 33.Bowden J., Smith G., Haycock P.C., Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowden J., Smith G., Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bowden J., Del Greco M.F., Minelli C., Davey Smith G., Sheehan N., Thompson J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat Med. 2017;36(11):1783–1802. doi: 10.1002/sim.7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowden J., Del Greco M F., Minelli C., et al. Improving the accuracy of two-sample summary-data Mendelian randomization: moving beyond the NOME assumption. Int J Epidemiol. 2019;48(3):728–742. doi: 10.1093/ije/dyy258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hemani G., Bowden J., Davey Smith G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet. 2018;27(R2):R195–R208. doi: 10.1093/hmg/ddy163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verbanck M., Chen C.Y., Neale B., Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albert-Gasco H., Ros-Bernal F., Castillo-Gomez E., Olucha-Bordonau F.E. MAP/ERK signaling in developing cognitive and emotional function and its effect on pathological and neurodegenerative processes. Int J Mol Sci. 2020;21(12) doi: 10.3390/ijms21124471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wen Y., Alshikho M.J., Herbert M.R. Pathway network analyses for autism reveal multisystem involvement, major overlaps with other diseases and convergence upon MAPK and calcium signaling. PLoS One. 2016;11(4) doi: 10.1371/journal.pone.0153329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinhardt P., Schmid B., Burbulla L.F., et al. Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell. 2013;12(3):354–367. doi: 10.1016/j.stem.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 42.Zhu X., Castellani R.J., Takeda A., et al. Differential activation of neuronal ERK, JNK/SAPK and p38 in Alzheimer disease: the 'two hit' hypothesis. Mech Ageing Dev. 2001;123(1):39–46. doi: 10.1016/s0047-6374(01)00342-6. [DOI] [PubMed] [Google Scholar]
- 43.Perlson E., Jeong G.B., Ross J.L., et al. A switch in retrograde signaling from survival to stress in rapid-onset neurodegeneration. J Neurosci. 2009;29(31):9903–9917. doi: 10.1523/JNEUROSCI.0813-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bodai L., Marsh J.L. A novel target for Huntington's disease: ERK at the crossroads of signaling. The ERK signaling pathway is implicated in Huntington's disease and its upregulation ameliorates pathology. Bioessays. 2012;34(2):142–148. doi: 10.1002/bies.201100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samuels I.S., Karlo J.C., Faruzzi A.N., et al. Deletion of ERK2 mitogen-activated protein kinase identifies its key roles in cortical neurogenesis and cognitive function. J Neurosci. 2008;28(27):6983–6995. doi: 10.1523/JNEUROSCI.0679-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller F.D., Gauthier A.S. Timing is everything: making neurons versus glia in the developing cortex. Neuron. 2007;54(3):357–369. doi: 10.1016/j.neuron.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 47.Liang Q., Su L., Zhang D., Jiao J. CD93 negatively regulates astrogenesis in response to MMRN2 through the transcriptional repressor ZFP503 in the developing brain. Proc Natl Acad Sci U S A. 2020;117(17):9413–9422. doi: 10.1073/pnas.1922713117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyza K.Z., Defensor E.B., Jensen A.L., et al. The BTBR T+ tf/J mouse model for autism spectrum disorders-in search of biomarkers. Behav Brain Res. 2013;251:25–34. doi: 10.1016/j.bbr.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rossignol D.A., Frye R.E. Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Mol Psychiatry. 2012;17(3):290–314. doi: 10.1038/mp.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang G., Gutierrez Rios P., Kuo S.H., et al. Mitochondrial abnormalities in temporal lobe of autistic brain. Neurobiol Dis. 2013;54:349–361. doi: 10.1016/j.nbd.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guevara-Campos J., Gonzalez-Guevara L., Cauli O. Autism and intellectual disability associated with mitochondrial disease and hyperlactacidemia. Int J Mol Sci. 2015;16(2):3870–3884. doi: 10.3390/ijms16023870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goh S., Dong Z., Zhang Y., DiMauro S., Peterson B.S. Mitochondrial dysfunction as a neurobiological subtype of autism spectrum disorder: evidence from brain imaging. JAMA Psychiatry. 2014;71(6):665–671. doi: 10.1001/jamapsychiatry.2014.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pei L., Wallace D.C. Mitochondrial etiology of neuropsychiatric disorders. Biol Psychiatry. 2018;83(9):722–730. doi: 10.1016/j.biopsych.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castora F.J. Mitochondrial function and abnormalities implicated in the pathogenesis of ASD. Prog Neuropsychopharmacol Biol Psychiatry. 2019;92:83–108. doi: 10.1016/j.pnpbp.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 55.Zhu J.H., Kulich S.M., Oury T.D., Chu C.T. Cytoplasmic aggregates of phosphorylated extracellular signal-regulated protein kinases in Lewy body diseases. Am J Pathol. 2002;161(6):2087–2098. doi: 10.1016/S0002-9440(10)64487-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lund T.C., Miller W.P., Eisengart J.B., et al. Biochemical and clinical response after umbilical cord blood transplant in a boy with early childhood-onset beta-mannosidosis. Mol Genet Gen Med. 2019;7(7):e00712. doi: 10.1002/mgg3.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bedilu R., Nummy K.A., Cooper A., et al. Variable clinical presentation of lysosomal beta-mannosidosis in patients with null mutations. Mol Genet Metab. 2002;77(4):282–290. doi: 10.1016/s1096-7192(02)00172-5. [DOI] [PubMed] [Google Scholar]
- 58.Sedel F., Friderici K., Nummy K., et al. Atypical Gilles De La Tourette syndrome with beta-mannosidase deficiency. Arch Neurol. 2006;63(1):129–131. doi: 10.1001/archneur.63.1.129. [DOI] [PubMed] [Google Scholar]
- 59.Blomqvist M., Smeland M.F., Lindgren J., et al. Beta-mannosidosis caused by a novel homozygous intragenic inverted duplication in MANBA. Cold Spring Harb Mol Case Stud. 2019;5(3) doi: 10.1101/mcs.a003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.