Abstract
Background
Chiari malformation type 1 (C1M) is a neurological disease characterized by herniation of the cerebellar tonsils below the foramen magnum. Cranial bone constriction is suspected to be its main cause. To date, genes related to bone development (e.g. DKK1 or COL1A2) have been associated with C1M, while some bone diseases (e.g. Paget) have been found to cosegregate with C1M. Nevertheless, the association between bone mineral density (BMD) and C1M has not been investigated, yet. Here, we systematically investigate the association between C1M and BMD, and between bone related genes and C1M.
Methods
We have recruited a small cohort of C1M patients (12 unrelated patients) in whom we have performed targeted sequencing of an in-house bone-related gene panel and BMD determination through non-invasive DXA.
Results
In the search for association between the bone related genes and C1M we have found variants in more than one C1M patient in WNT16, CRTAP, MYO7A and NOTCH2. These genes have been either associated with craniofacial development in different ways, or previously associated with C1M (MYO7A). Regarding the potential link between BMD and C1M, we have found three osteoporotic patients and one patient who had high BMD, very close to the HBM phenotype values, although most patients had normal BMD.
Conclusions
Variants in bone related genes have been repeatedly found in some C1M cases. The relationship of bone genes with C1M deserves further study, to get a clearer estimate of their contribution to its etiology. No direct correlation between BMD and C1M was observed.
Abbreviations: C1M, Chiari Malformation type 1; HBM, High Bone Mass; BMD, Bone Mineral Density
Keywords: Chiari malformation type 1, Bone mineral density, NOTCH2, WNT16, CRTAP, MYO7A
Highlights
-
•
We used an in-house bone gene panel to investigate a small cohort of C1M patients.
-
•
Variants in WNT16, CRTAP, MYO7A and NOTCH2 were found in more than one C1M patient.
-
•
No clear relationship was found between C1M and BMD in this small C1M cohort.
1. Introduction
Chiari malformation type 1 (C1M; OMIM 118420) is characterized by downward herniation of the cerebellar tonsils through the foramen magnum by at least 5 mm without involving the brain stem (Barkovich et al., 1986). This herniation results in compression of the neural tissue at the craniovertebral junction leading to neurological dysfunction, which may or may not be accompanied by syringomyelia or hydrocephalus (Milhorat et al., 1999; Urbizu et al., 2013; Speer et al., 2003). C1M presents in many cases as asymptomatic, and in many others with pain or headache within the occipital or upper cervical region, which can also be accompanied by ocular or otoneurological disturbances, lower cranial nerve signs, cerebellar ataxia or spasticity (McVige and Leonardo, 2014; Piper et al., 2019; Speer et al., 2003). The diagnosis is made through magnetic resonance imaging (MRI) or ultrasonography for the prenatal diagnosis (Frič and Eide, 2020; Holly and Batzdorf, 2019). Although it has been described that some cases of C1M may be the consequence of trauma, many cases can be congenital (Speer et al., 2003). It has been proposed that classical C1M is generated via an insufficient development of the paraxial mesoderm involving occipital bone sclerotomes, resulting in a smaller and shallow posterior fossa (PF), too small to accommodate the normal size of the cerebellum (Nishikawa et al., 1997; Marin-Padilla and Marin-Padilla, 1981). C1M is likely to have a genetic basis, based on familial aggregation, monozygotic twin studies and its overlap with known genetic conditions (Markunas et al., 2014; Mavinkurve et al., 2005; Szewka et al., 2006; Speer et al., 2003; Abbott et al., 2018; Speer et al., 2000). The main recent strategy to find the genetic basis of the disease and the inheritance pattern has involved the use of next generation sequencing (NGS) techniques such as whole exome sequencing (WES). These works have found several genes, including chromodomain genes (Sadler et al., 2021) and chromatin remodeling genes (Provenzano et al., 2021).
The fact that the cranial bone constriction is suspected to be the most common biologic mechanism leading to C1M suggests that variants in genes related with bone metabolism may be contributing to the malformation through an incorrect development of the cranial bone. This hypothesis is reinforced by the findings of several recent works which have found association of C1M with rare variants in genes involved in collagen metabolism (Urbizu et al., 2021), in Wnt pathway genes (Merello et al., 2017), or in cranial bone sutures (Provenzano et al., 2021).
In this work, we have further investigated the relationship between C1M and bone metabolism, searching for rare variants in bone related genes through an in-house bone gene panel and, for the first time, exploring the association between bone mineral density and C1M, in a small series of C1M patients.
2. Material and methods
2.1. Study cohort, brain MRI and bone mineral density measurements
C1M cases, previously diagnosed at the Neurosurgery Service of the Hospital del Mar, were invited to participate in the study. Twelve patients agreed to participate, and in three cases, additional family members were also recruited (n = 6), four of whom were also affected with C1M. Diagnosis of C1M was based on brain-MRI (Achieva 3.0 T, Philips, Amsterdam, Netherlands) demonstration of downward herniation >5 mm of the cerebellar tonsils on a mid-sagittal T1-weighted image in the presence of signs or symptoms indicating neural compression at the cranio-vertebral junction, syringo-hydromyelia, cerebellar dysfunction or intracranial hypertension. All C1M cases and family members underwent brain MRI. The cohort characterization includes femoral neck and lumbar spine bone mineral density measurements obtained through dual-energy X-ray absorptiometry scans (DXA; QDR 4500 SL; Hologic, Waltham, MA, USA) and collection of blood samples (Table 1). All DXA measurements were performed prior to any treatment that could increase bone mass. For individuals CH7a and CH10 no DXA scan was available, for CH4 no T-score was available and for CH7 no Z-score was available (Table 1).
Table 1.
Description of the Spanish C1M case series.
| ID |
Sex |
Age |
Family relationships |
Disease status |
BMD (T-score) |
BMD (Z-Score) |
|||
|---|---|---|---|---|---|---|---|---|---|
| LS | FN | LS | FN | TOTAL | |||||
| CH1 | F | 59 | A | a | −2.6 | a | −1.3 | ||
| CH2 | F | 65 | A | −3.2 | −1.5 | −0.9 | −0.9 | −1.8 | |
| CH2a | F | 35 | daughter of CH2 | A | −0.5 | −1.1 | −0.4 | −0.8 | −1.2 |
| CH2b | F | 68 | sister of CH2 | A | −2.1 | −1.6 | 0 | −0.4 | −0.4 |
| CH2c | F | 71 | sister of CH2 | N | −1.4 | −1.6 | +0.8 | +0.3 | +1.1 |
| CH2d | F | 72 | sister of CH2 | A | −4.1 | −3.8 | −1.8 | −0.1 | −1.9 |
| CH3 | F | 46 | A | −0.3 | −1.1 | +0.1 | −0.1 | 0 | |
| CH4 | M | 24 | A | NA | NA | +0.2 | 0 | +0.2 | |
| CH5 | F | 45 | A | +1.4 | +1.5 | +1.8 | +2 | +3.8 | |
| CH6 | M | 46 | A | −0.6 | −0.6 | −0.4 | +0.5 | +0.1 | |
| CH7 | F | 61 | A | −2.5 | −3.2 | NA | NA | NA | |
| CH7a | F | 63 | sister of CH7 | A | NA | NA | NA | NA | NA |
| CH8 | M | 47 | A | −2 | −1.9 | −1.7 | −0.6 | −2.3 | |
| CH9 | F | 45 | A | +0.9 | +0.1 | +1.3 | +0.6 | +1.9 | |
| CH10 | M | 74 | A | NA | NA | NA | NA | NA | |
| CH10a | F | NA | sister of CH10 | N | −1.1 | −0.5 | +0.8 | +1.4 | +2.2 |
| CH11 | F | 36 | A | +0.8 | −0.1 | +0.8 | +0.2 | +1 | |
| CH12 | F | 83 | A | −2.4 | −1.1 | +0.3 | +1.3 | +1.6 | |
F: Female; M: Male; A: Affected N: Non-affected; FN: Femoral Neck; LS: Lumbar Spine; NA: Not available.
Dysmorphic lumbar.
2.2. Genetic analyses
The genomic DNA of 12 unrelated C1M and their 6 relatives was isolated from peripheral blood leukocytes using Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, USA), according to the manufacturer's instructions. The 12 unrelated C1M cases underwent targeted sequencing of the exonic and flanking intronic regions of 127 genes related with BMD in GWAS studies or associated with monogenic bone diseases included in an in-house bone related gene panel (IRN:1000011543, KAPA HyperChoice MAX 0.5 Mb T4, 12 rxn; Supplementary Table 1). Sequencing was performed at CNAG (Barcelona, Spain). Shortly, DNA was enzymatically fragmented and libraries were constructed, pooled and hybridized to the probesets. Captured fragments were sequenced in an Illumina NovaSeq 6000 sequencer. The reads were then aligned to the hg38 reference genome with BWA-mem, duplicate-marked, recalibrated and sorted before calling variants with GATK's haplotype caller (V4) following GATK standard parameters. After quality-filtering following GATK recommended hard filters (https://gatk.broadinstitute.org/hc/en-us/articles/360035890471-Hard-filtering-germline-short-variants), variants were annotated with Varaft (Desvignes et al., 2018), and prioritized under the hypothesis of an autosomal dominant segregation.
Filtering included the exclusion of: variants located outside of the coding region (intergenic, 5′ and 3′ UTR, upstream, downstream, ncRNA or unknown variants); intronic variants not predicted to affect the splice site; synonymous variants; variants with a minor allele frequency > 0.02; missense variants with less than two damaging or potentially damaging prediction scores out or four tools (CADD (>20), http://cadd.gs.washington.edu; PROVEAN, http://provean.jcvi.org/, Polyphen-2, http://genetics.bwh.harvard.edu/pph2/; SIFT, https://sift.bii.a-star.edu.sg/); or, for indel variants, those with a neutral SIFT indel score (https://sift.bii.a-star.edu.sg/). For the remaining variants, cosegregation in available family members (for CH2, CH7 and CH10) was examined by Sanger sequencing at the CCiTUB genomics service (Genòmica, Parc Cientific, Barcelona, Spain) using BigDye ™ Terminator v3.1 Cycle Sequencing Kit, followed by detection on automated capillary sequencer models 3730 Genetic Analyzer and 3730xl Genetic Analyzer.
3. Results
3.1. Relationship between C1M and BMD
To study the relationship between C1M and BMD we have determined the BMD of a small group of C1M patients (Table 1). The average LS and FN BMD Z-score for the cases with available data was 0.16 and −0.058, respectively. Seven unrelated C1M cases had normal BMD according to the WHO diagnostic criteria based on the T-scores and Z-scores. Instead, following the same criteria, three unrelated C1M cases (CH1, CH2, CH7) and one of their relatives (Ch2d) are classified as osteoporotic. On the other hand, a patient is considered to have high bone mass (HBM) phenotype when the sum of the LS and FM BMD Z-score values is higher than 4 (Little et al., 2002). According to this definition, only one patient (CH5) had values close to this threshold (sum Z-scores = 3.8), while the mean sum Z-score value for all 9 cases was +0.5 (Table 1).
3.2. Relationship between bone-related genes and C1M
To elucidate the relationship between variants in bone related genes and C1M, we sequenced the coding regions and intron boundaries of 127 genes through an in-house bone panel in 12 unrelated patients with C1M. After filtering (see Material and Methods) we identified 32 variants in 24 different genes in the 12 unrelated C1M patients (Supplementary Table 2). Seven of these 24 genes contain variants in more than one C1M patient (Table 2) and only one variant was present in two unrelated patients (p.P582H in DAAM2; Table 2).
Table 2.
Genes with variants in more than one Chiari patient.
| Gene | BMD GWAS |
OMIM Disease |
Variant | C1M patient | Coseg. | CADD | PP | PV | SIFT | MAF gnomAD |
rs number |
|---|---|---|---|---|---|---|---|---|---|---|---|
| WNT16a | Y | N | p.R222C | CH10 | Y | 27.8 | D | D | D | 0.00001995 | rs773948299 |
| p.S260P | CH4 | NA | 23.8 | D | N | T | 0.002061 | rs116444834 | |||
| CRTAP | N | OI | p.C243G | CH12 | NA | 25.1 | D | D | D | NA | NA |
| p.L347F | CH2 | IP | 24.3 | D | D | D | 0.001987 | rs115198029 | |||
| DMP1b | Y | ARHR1 | p.D478A | CH2 | N | 22.4 | D | D | D | 0.0003853 | rs148156611 |
| p.L282I | CH10 | Y | 8.14 | P | N | D | 0.005413 | rs141979823 | |||
| DAAM2c | Y | NPHS24 | p.R1049X | CH8 | NA | 44 | NA | NA | NA | 0.00002014 | rs759817118 |
| p.P582H | CH7 | N | 22.2 | P | N | D | 0.001564 | rs150676991 | |||
| CH11 | NA | ||||||||||
| FKBP10 | Y | BRKS1, OI | p.R556C | CH5 | NA | 29 | D | D | D | NA | NA |
| p.I436T | CH7 | N | 28.8 | D | D | D | 0.002465 | rs61749879 | |||
| MYO7Ad | N | DFN; USH1 | p.M2008fs | CH6 | NA | NA | NA | NA | D+ | NA | NA |
| p.R654C | CH11 | NA | 29.1 | D | D | D | 0.0001296 | rs201928014 | |||
| NOTCH2 | N | ALGS2; HJCYS | p.V1064M | CH3 | NA | 23.1 | B | N | D | 0.000003978 | rs373969789 |
| p.R91L | CH4 | NA | 22.1 | P | N | T | 0.008614 | rs143195893 |
In bold genes not discarded for cosegregation. BMD GWAS: Genes associated with BMD in GWAS; Y: Yes; N: No; OMIM DISEASE: Gene associated with human diseases according to OMIM; N: No; OI: Osteogenesis imperfecta; ARHR1: Hypophosphatemic rickets, autosomal recessive, 1; NPHS24: Nephrotic syndrome, type 24; BRKS1: Bruck syndrome 1; DFN: Deafness; USH1: Usher syndrome, type 1B; ALGS2: Alagille syndrome 2; HJCYS: Hajdu-Cheney syndrome; Coseg: Cosegregation in the available families (CH2, CH7, CH10): Y: Yes: Variant present in other C1M cases in the family and/or absent in healthy family members; IP: incomplete penetrance: variant found in a healthy family member; N: NO; variant absent in other C1M cases in the family; CADD: http://cadd.gs.washington.edu; PP: Polyphen-2 http://genetics.bwh.harvard.edu/pph2/; D: Probably damaging; P: Possibly damaging; B: Benign; NA: Not available; PV: PROVEAN http://provean.jcvi.org/; D: Deleterious; N: Neutral; NA: No available; SIFT: https://sift.bii.a-star.edu.sg/; D: Deleterious; T: Tolerated; NA: Not available; +: Tested with SIFT indels https://sift.bii.a-star.edu.sg/www/SIFT_indels2.html; MAF: minor allele frequency from gnomAD V2.1.1.
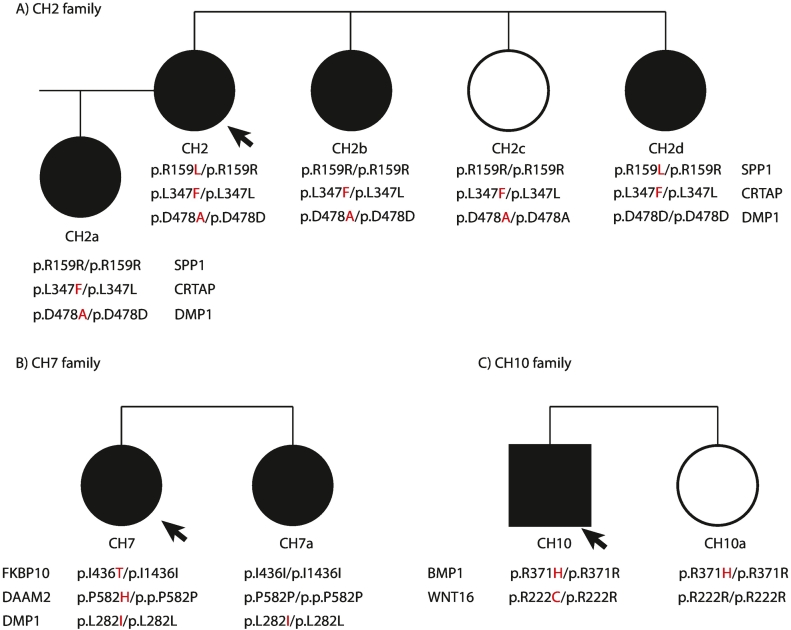
Next, in three cases (CH2, CH7, CH10) with available DNA from family members, we tested the variants' cosegregation with the C1M phenotype (n = 6; Fig. 1 and “Coseg” column in Table 2 and Supplementary Table 2).
Fig. 1.
Pedigree of the three C1M families. In black the C1M members, the arrow indicates the proband, the alterantive allele of the variant is highlighted in red color.
Two variants were consistent with cosegregation with the C1M phenotype in the families, p.R222C in WNT16 and p.L282I in DMP1, two were compatible with cosegregation only if assuming incomplete penetrance (i.e. they were present in both affected and healthy individuals; p.L347F in CRTAP and p.R371H in BMP1), and four did not cosegregate with the phenotype (i.e. they were absent in other affected family members; p.D478A in DMP1, p.I436T in FKBP10, p.P582H in DAAM2 and p.R159L in SPP1; Table 2 and Supplementary Table 2). Unfortunately, the variants present in the remaining C1M cases could not be tested for cosegration for lack of available relatives' samples.
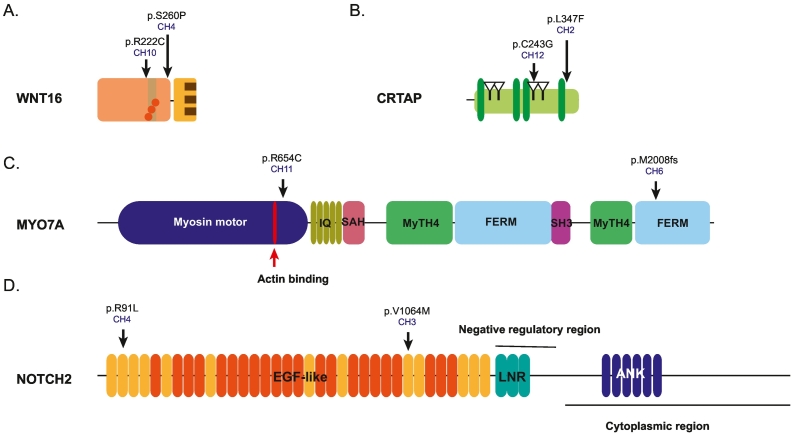
After excluding variants that do not cosegregate with C1M, 26 variants in 23 different genes were present in at least one of the 12 unrelated C1M patients (Supplementary Table 2) and four genes presented variants in more than one C1M patient. These are WNT16, CRTAP, MYO7A and NOTCH2 (highlighted in bold in Table 2 and Fig. 2).
Fig. 2.
Linear representation of the WNT16 (A), CRTAP (B), MYO7A (C) and NOTCH2 (D) with its domains and regions. For WNT16 in orange N-terminal domain, in yellow the CRD domain, in light brown thumb domain, in dark brown finger and in orange circles the O-palmitoleoyl serine modification (reviewed in Martínez-Gil et al., 2022). For CRTAP in green the tetratico repeat motif, triangles show the four CXXXC domains from (Ishikawa and Bächinger, 2013). For MYO7A in dark blue the Myosin motor and in red the actin binding site, in dark yellow the isoleucine-glutamine (IQ) motif, in pink the Single alpha-helix (SAH) domain, in green the Myosin Tail Homology 4 (MyTH4) domains, in light blue the FERM domains and in purple the SRC Homology 3 (SH3) domain from uniprot. For NOTCH2 in yellow the EGF-like domain, in orange the EGF-like with calcium binding, in green the cysteine-rich Lin-12/Notch (LNR) Repeats and in dark blue the Ankyrin (ANK) domains from uniprot.The black arrow signals the position of the variants found and ID patient where is found.
4. Discussion
Here, we present a first study on the possible relationship among C1M, variants in bone related genes and BMD, through the sequencing of a bone in-house panel and BMD determination in 12 unrelated patients with C1M.
We did not find a clear positive association between BMD and C1M. Even so, we have found 3 unrelated C1M patients with osteoporosis while one patient (CH5) presented a sum Z-score of 3.8, very close to the accepted threshold for HBM. Knowing that C1M can occur as an isolated malformation or as a part of a broader genetic entity (Speer et al., 2003), and many Mendelian diseases have been described to coexist with C1M, we may speculate that these extreme values of BMD (Osteoporotic or HBM) can be somehow related to C1M phenotype. The CH5 patient is of special interest because given the reported prevalence of the HBM phenotype of 0.2–1% (Sarrión et al., 2014; Gregson et al., 2012, Gregson et al., 2013), under the hypothesis of no association it would have been highly unlikely to find a HBM case in this small cohort. Nevertheless, this could also be a spurious finding, as it is only one individual. In order to elucidate the relationship between the C1M and bone mass, it would be crucial to measure the LS and FN BMD of the C1M patients from previous publications (Boyles et al., 2006; Markunas et al., 2013; Urbizu et al., 2013; Markunas et al., 2014; Rosenblum et al., 2019; Gonçalves et al., 2019; Merello et al., 2017; Urbizu et al., 2021; Provenzano et al., 2021; Sadler et al., 2021), thus increasing the size of the cohort of patients with C1M and BMD.
Regarding the relationship between variants in bone-related genes and C1M, we have found 4 such genes each with variants in two unrelated C1M patients. These are WNT16, CRTAP, MYO7A and NOTCH2 (Table 2 and Fig. 2). In WNT16, two missense variants, p.S260P and p.R222C, have been found in patients CH4 and CH10, respectively, the latter absent in CH10's healthy sister (CH10a), thus co-segregating with the phenotype in this small family (Fig. 1). WNT16 is a ligand of the Wnt pathway, both canonical and non-canonical, in a cell-type specific manner (reviewed in Martínez-Gil et al., 2022). WNT16 locus is one of the most consistent bone-related GWAS signals, and has been associated with different skeletal phenotypes, including BMD, bone strength, geometric parameters, cortical bone thickness and fracture risk (reviewed in Martínez-Gil et al., 2022). No variants in WNT16 have been previously associated with C1M, but this pathology has been associated with other members of the Wnt pathway, such as DKK1, a Wnt pathway inhibitor, in which three missense variants, whose partial loss-of-function has been recently demonstrated by our group (Martínez-Gil et al., 2020), were identified in C1M cases (Provenzano et al., 2021; Merello et al., 2017). In addition, Whyte et al. (2004) described one HBM woman with gain-of-function variants in LRP5, a canonical Wnt pathway coreceptor, who also presented with a headache and C1M. Interestingly, we also find a LRP5 variant (p.R1036Q) in CH3 and a DAAM2 variant (p.R1049*) in CH8 (Supplementary Table 2). DAAM2 is an important positive regulator of Wnt signaling and is required for various processes during development, such as dorsal patterning or determination of left/right symmetry (Lee and Deneen, 2012; Lee et al., 2015; Welsh et al., 2013). Taken together, and due to the important and widely described role played by the Wnt pathway in the correct development of the head and face (Mishina and Snider, 2014; Mani et al., 2010), it would be possible to speculate that rare variants in WNT16, LRP5 and DAAM2, alone or in combination with other variants, could be responsible for C1M in these individual cases.
Another gene present in two C1M cases is CRTAP (variants p.C243G and p.L347F, in patients CH12 and CH2, respectively). Nevertheless, the p.L347F variant is present in all available CH2 family members (Fig. 1), including the healthy sister (CH2c), which would require the assumption of incomplete penetrance. CRTAP, a member of the multifunctional complex P3H1·CRTAP·CypB, is involved in posttranslational modifications of fibrillar collagens. CRTAP loss-of-function variants are associated with osteogenesis imperfecta type VII in humans, with a phenotype that includes craniofacial bone defects (Morello et al., 2006; Baldridge et al., 2010; Xu et al., 2020; Barbirato et al., 2015; Valli et al., 2012). Interestingly, the p.C243G variant is modifying the first cysteine of the third CXXXC domain of the CRTAP protein, an important domain that could have disulfide isomerase activity (Fig. 2; Ishikawa and Bächinger, 2013). The involvement of this gene in osteogenesis imperfecta could partially explain the low BMD present in all the studied individuals with CRTAP variants. In family CH2, where all family members carry the p.L347F variant, two of them show a clear osteoporosis and the remaining three show osteopenia (Table 1) while patient the CH12, carrying variant p.C243G, shows an osteopenia phenotype (Table 1). Considering all this together, it is tempting to speculate that the variants in CRTAP may be contributing to a decrease in BMD and possibly to C1M, too.
In MYO7A, we have found a frameshift variant (p.M2008fs) in CH6 and a missense variant (p.R654C) in CH11 (Table 2 and Fig. 2). MYO7A encodes an unconventional myosin which is an actin-based motor molecule with ATPase activity, important in intracellular movement (reviewed in Williams and Lopes, 2011). Variants in the MYO7A gene have been associated with different forms of inherited deaf-blindness or deafness (Weil et al., 1995, Gibson et al., 1995; Hildebrand et al., 2010; Table 2). The variants found affect the myosin motor and the second FERM domain, which has been described as a regulator of MYO7A activation (Fig. 2; Yang et al., 2009). Interestingly, MYO7A has been already associated with C1M under the autosomal recessive inheritance pattern in the work of Sadler et al. (2021), who found two missense variants in MYO7A (p.R1229Q and p.R605W) in a C1M patient. Actually, in the search for the genetic bases of C1M, 207 different genes have been found to date associated in some way with the phenotype under different segregation hypotheses (Boyles et al., 2006; Markunas et al., 2013; Urbizu et al., 2013; Markunas et al., 2014; Rosenblum et al., 2019; Gonçalves et al., 2019; Merello et al., 2017; Urbizu et al., 2021; Provenzano et al., 2021; Sadler et al., 2021). Of these 207 genes, 5 were included in the bone panel that we used in the present study. These are MYO7A, BMP1, COL1A2, DKK1 and LRP4 (Merello et al., 2017; Urbizu et al., 2021; Sadler et al., 2021). Interestingly, in addition to MYO7A, we also have found a variant in BMP1 in one C1M patient (p.R371H in CH10, Supplementary Table 2). The BMP1 variant is also present in the healthy sister CH10a, indicating that the cosegregation is only possible if incomplete penetrance is assumed. Interestingly, as in the case of CRTAP, variants in BMP1 are also associated with osteogenesis imperfecta. Unfortunately, no BMD information for CH10 is available. Our findings in MYO7A and BMP1 replicate the results by others and reinforce the importance of these two genes in the development of C1M.
In NOTCH2, two missense variants p.V1064M and p.R91L have been found, in patients CH3 and CH4, respectively. These two variants are located in the extracellular EGF-like domains (EGF-like 2 and EGF-like 28; Fig. 2). Interestingly, two variants in the extracellular EGF-like domains (p.T294M in EGF-like 7 and p.S978R in EGF-like 25) in NOTCH3, another receptor of the Notch family, have been previously found in a C1M patient (Sadler et al., 2021). These EGF-like domains are responsible for the interaction with cognate ligands (Cordle et al., 2008; Zanotti and Canalis, 2016). Thus, the variants found may produce a lower affinity with the ligand that may lead to less activation of the signaling pathway. The Notch signaling pathway plays an important role in skeletal development, chondrogenesis, osteoblastogenesis and osteoclastogenesis. Specifically, Notch suppresses cell differentiation in cells of immature osteoblastic lineage through inhibition of the Wnt pathway and by interactions with Runx2, while in osteocytes, Notch decreases SOST expression, thus increasing the Wnt pathway activity. In addition, Notch2 induces osteoclast differentiation through its interaction with NFκB. Two diseases associated with variants in NOTCH2 have been described, the Alagille syndrome, associated with loss-of-function variants, and the Hajdu-Cheney syndrome, associated with gain-of-function variants. Alagille syndrome is characterized by cardiovascular defects, cholestatic liver disease, kidney anomalies and abnormalities of the craniofacial skeleton and vertebrae (Alagille et al., 1987; Zanotti and Canalis, 2016). Problems in craniofacial development can cause craniosynostosis and characteristic facial features, and developmental problems in vertebrae cause a characteristic structure of the vertebrae called “butterfly” that is observed in radiographs (Emerick et al., 1999; Zanotti and Canalis, 2016). Hadju-Cheney syndrome is characterized by acroosteolysis of the hands and feet and developmental defects of bones, teeth and joints producing craniofacial and skull changes, osteoporosis and short stature (Hajdu and Kauntze, 1948; Zanotti and Canalis, 2016). These patients' also present platybasia and basilar invagination that can produce severe neurological complications (reviewed in Zanotti and Canalis, 2016). Interestingly, the Hadju-Cheney syndrome has been previously associated with C1M (Sawin and Menezes, 1997; Speer et al., 2003).
5. Conclusions
In conclusion, while we did not find a clear association of BMD with the C1M, we have found variants in more than one C1M patient in four interesting genes -WNT16, CRTAP, MYO7A and NOTCH2- involved in craniofacial development or previously associated with C1M. Altogether, we consider the relationship of C1M with bone biology a very interesting issue, and we provide a few pieces of evidence supporting it. We encourage the C1M research community to pursue this effort.
Funding
Funds for the study include grants PID2019-107188RB-C21 (Spanish MICINN) and CIBERER (U720). NMG and JDP were recipients of FI and FI-SDUR predoctoral fellowships from Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR), respectively.
Availability of data and material
All the data are available upon request.
Ethics approval and consent to participate
All procedures performed were in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Both the Bioethics Committee of Universitat de Barcelona and the Clinical Research Ethics Committee of Parc de Salud MAR have emitted a favourable bioethical statement regarding the present research. Written informed consents were obtained from the participants in both instances.
CRediT authorship contribution statement
NMG, DG, SB were involved in the study conception and design. LM and DMLG recruited the Chiari 1 malformation cohort and made bone mineral density measurements. NMG, MC, JDP and RR were involved in the design of the bone gene panel and the sequencing of the patients and relatives. The first draft of the manuscript was written by NMG, DG, SB, RR and all authors commented on previous versions of the manuscript and all authors read and approved the final version.
Declaration of competing interest
Núria Martínez-Gil, Leonardo Mellibovsky, Demian Manzano-Lopez Gonzalez, Juan David Patiño, Monica Cozar, Raquel Rabionet; Daniel Grinberg and Susanna Balcells, authors of the above-mentioned manuscript, jointly declare they hold no conflicts of interest regarding the research and results presented in it.
Acknowledgements
We thank Nurgül Atalay for technical assistance and the patients with Chiari malformation type 1 and their family for their enthusiastic participation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bonr.2022.101181.
Appendix A. Supplementary data
Supplementary tables
References
- Abbott D., et al. Population-based description of familial clustering of Chiari malformation Type I. J. Neurosurg. 2018;128:460–465. doi: 10.3171/2016.9.JNS161274. [DOI] [PubMed] [Google Scholar]
- Alagille D., et al. Syndromic paucity of interlobular bile ducts (Alagille syndrome or arteriohepatic dysplasia): review of 80 cases. J. Pediatr. 1987;110:195–200. doi: 10.1016/s0022-3476(87)80153-1. [DOI] [PubMed] [Google Scholar]
- Baldridge D., et al. Generalized connective tissue disease in Crtap-/- mouse. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbirato C., et al. Mutational characterization of the P3H1/CRTAP/CypB complex in recessive osteogenesis imperfecta. Genet. Mol. Res. 2015;14:15848–15858. doi: 10.4238/2015. [DOI] [PubMed] [Google Scholar]
- Barkovich A.J., et al. Significance of cerebellar tonsillar position on MR. AJNR Am. J. Neuroradiol. 1986;7:795–799. [PMC free article] [PubMed] [Google Scholar]
- Boyles A.L., et al. Phenotypic definition of Chiari type I malformation coupled with high-density SNP genome screen shows significant evidence for linkage to regions on chromosomes 9 and 15. Am. J. Med. Genet. A. 2006;140:2776–2785. doi: 10.1002/ajmg.a.31546. [DOI] [PubMed] [Google Scholar]
- Cordle J., et al. Localization of the delta-like-1-binding site in human Notch-1 and its modulation by calcium affinity. J. Biol. Chem. 2008;283:11785–11793. doi: 10.1074/jbc.M708424200. [DOI] [PubMed] [Google Scholar]
- Desvignes J.P., et al. VarAFT: a variant annotation and filtration system for human next generation sequencing data. Nucleic Acids Res. 2018;46:W545–W553. doi: 10.1093/nar/gky471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerick K.M., et al. Features of Alagille syndrome in 92 patients: frequency and relation to prognosis. Hepatology. 1999;29:822–829. doi: 10.1002/hep.510290331. [DOI] [PubMed] [Google Scholar]
- Frič R., Eide P.K. Chiari type 1-a malformation or a syndrome? A critical review. Acta Neurochir. (Wien) 2020;162:1513–1525. doi: 10.1007/s00701-019-04100-2. [DOI] [PubMed] [Google Scholar]
- Gibson F., et al. A type VII myosin encoded by the mouse deafness gene shaker-1. Nature. 1995;374:62–64. doi: 10.1038/374062a0. [DOI] [PubMed] [Google Scholar]
- Gonçalves D., et al. Chiari malformation type I in a patient with a novel NKX2-1 mutation. J. Pediatr. Neurosci. 2019;14:169–172. doi: 10.4103/jpn.JPN_108_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregson C.L., et al. Sink or swim': an evaluation of the clinical characteristics of individuals with high bone mass. Osteoporos. Int. 2012;23:643–654. doi: 10.1007/s00198-011-1603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregson C.L., et al. The high bone mass phenotype is characterised by a combined cortical and trabecular bone phenotype: findings from a pQCT case-control study. Bone. 2013;52:380–388. doi: 10.1016/j.bone.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu N., Kauntze R. Cranio-skeletal dysplasia. Br. J. Radiol. 1948;21:42–48. doi: 10.1259/0007-1285-21-241-42. [DOI] [PubMed] [Google Scholar]
- Hildebrand M.S., et al. Variable hearing impairment in a DFNB2 family with a novel MYO7A missense mutation. Clin. Genet. 2010;77:563–571. doi: 10.1111/j.1399-0004.2009.01344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holly L.T., Batzdorf U. Chiari malformation and syringomyelia. J. Neurosurg. Spine. 2019;31:619–628. doi: 10.3171/2019.7.SPINE181139. [DOI] [PubMed] [Google Scholar]
- Ishikawa Y., Bächinger H.P. An additional function of the rough endoplasmic reticulum protein complex prolyl 3-hydroxylase 1·cartilage-associated protein·cyclophilin B: the CXXXC motif reveals disulfide isomerase activity in vitro. J. Biol. Chem. 2013;288:31437–31446. doi: 10.1074/jbc.M113.498063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Deneen B. Daam2 is required for dorsal patterning via modulation of canonical Wnt signaling in the developing spinal cord. Dev. Cell. 2012;22:183–196. doi: 10.1016/j.devcel.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., et al. Daam2-PIP5K is a regulatory pathway for Wnt signaling and therapeutic target for remyelination in the CNS. Neuron. 2015;85:1227–1243. doi: 10.1016/j.neuron.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little R.D., et al. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am. J. Hum. Genet. 2002;70:11–19. doi: 10.1086/338450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani P., et al. Visualizing canonical Wnt signaling during mouse craniofacial development. Dev. Dyn. 2010;239:354–363. doi: 10.1002/dvdy.22072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Padilla M., Marin-Padilla T.M. Morphogenesis of experimentally induced Arnold-Chiari malformation. J. Neurol. Sci. 1981;50:29–55. doi: 10.1016/0022-510x(81)90040-x. [DOI] [PubMed] [Google Scholar]
- Markunas C.A., et al. Stratified whole genome linkage analysis of Chiari type I malformation implicates known Klippel-Feil syndrome genes as putative disease candidates. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markunas C.A., et al. Genetic evaluation and application of posterior cranial fossa traits as endophenotypes for Chiari type I malformation. Ann. Hum. Genet. 2014;78:1–12. doi: 10.1111/ahg.12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Gil N., et al. Functional assessment of coding and regulatory variants from the DKK1 locus. JBMR Plus. 2020;4 doi: 10.1002/jbm4.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Gil N., et al. Wnt pathway extracellular components and their essential roles in bone homeostasis. Genes (Basel) 2022;13:138. doi: 10.3390/genes13010138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavinkurve G.G., et al. Familial Chiari type I malformation with syringomyelia in two siblings: case report and review of the literature. Childs Nerv. Syst. 2005;21:955–959. doi: 10.1007/s00381-005-1146-0. [DOI] [PubMed] [Google Scholar]
- McVige J.W., Leonardo J. Imaging of Chiari type I malformation and syringohydromyelia. Neurol. Clin. 2014;32:95–126. doi: 10.1016/j.ncl.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Merello E., et al. Exome sequencing of two Italian pedigrees with non-isolated Chiari malformation type I reveals candidate genes for cranio-facial development. Eur. J. Hum. Genet. 2017;25:952–959. doi: 10.1038/ejhg.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milhorat T.H., Chou M.W., Trinidad E.M., Kula R.W., Mandell M., Wolpert C., Speer M.C. Chiari I malformation redefined: clinical and radiographic findings for 364 symptomatic patients. Neurosurgery. 1999;44(5):1005–1017. doi: 10.1097/00006123-199905000-00042. [DOI] [PubMed] [Google Scholar]
- Mishina Y., Snider T.N. Neural crest cell signaling pathways critical to cranial bone development and pathology. Exp. Cell Res. 2014;325:138–147. doi: 10.1016/j.yexcr.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morello R., et al. CRTAP is required for prolyl 3- hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell. 2006;127:291–304. doi: 10.1016/j.cell.2006.08.039. [DOI] [PubMed] [Google Scholar]
- Nishikawa M., et al. Pathogenesis of Chiari malformation: a morphometric study of the posterior cranial fossa. J. Neurosurg. 1997;86:40–47. doi: 10.3171/jns.1997.86.1.0040. [DOI] [PubMed] [Google Scholar]
- Piper R.J., et al. Chiari malformations: principles of diagnosis and management. BMJ. 2019;8(365) doi: 10.1136/bmj.l1159. [DOI] [PubMed] [Google Scholar]
- Provenzano A., et al. Chiari 1 malformation and exome sequencing in 51 trios: the emerging role of rare missense variants in chromatin-remodeling genes. Hum. Genet. 2021;140:625–647. doi: 10.1007/s00439-020-02231-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum J.S., et al. Chiari malformation type 1 in EPAS1-associated syndrome. Int. J. Mol. Sci. 2019;20:2819. doi: 10.3390/ijms20112819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler B., et al. Rare and de novo coding variants in chromodomain genes in Chiari I malformation. Am. J. Hum. Genet. 2021;108:100–114. doi: 10.1016/j.ajhg.2020.12.001. Erratum. In: Am J Hum Genet. 2021;108, 368. Erratum in: Am J Hum Genet. 2021;108, 530-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrión P., et al. Genetic analysis of high bone mass cases from the BARCOS cohort of Spanish postmenopausal women. PLoS One. 2014;9 doi: 10.1371/journal.pone.0094607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin P.D., Menezes A.H. Basilar invagination in osteogenesis imperfecta and related osteochondrodysplasias: medical and surgical management. J. Neurosurg. 1997;86:950–960. doi: 10.3171/jns.1997.86.6.0950. [DOI] [PubMed] [Google Scholar]
- Speer M.C., et al. A genetic hypothesis for Chiari I malformation with or without syringomyelia. Neurosurg. Focus. 2000;8:E12. doi: 10.3171/foc.2000.8.3.12. [DOI] [PubMed] [Google Scholar]
- Speer M.C., et al. Review article: Chiari type I malformation with or without syringomyelia: prevalence and genetics. J. Genet. Couns. 2003;12:297–311. doi: 10.1023/A:1023948921381. [DOI] [PubMed] [Google Scholar]
- Szewka A.J., et al. Chiari in the family: inheritance of the Chiari I malformation. Pediatr. Neurol. 2006;34:481–485. doi: 10.1016/j.pediatrneurol.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Urbizu A., et al. Chiari malformation type I: a case-control association study of 58 developmental genes. PLoS One. 2013;8 doi: 10.1371/journal.pone.0057241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbizu A., et al. Rare functional genetic variants in COL7A1, COL6A5, COL1A2 and COL5A2 frequently occur in Chiari malformation type 1. PLoS One. 2021;16 doi: 10.1371/journal.pone.0251289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli M., et al. Deficiency of CRTAP in non-lethal recessive osteogenesis imperfecta reduces collagen deposition into matrix. Clin. Genet. 2012;82:453–459. doi: 10.1111/j.1399-0004.2011.01794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil D., et al. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995;374:60–61. doi: 10.1038/374060a0. [DOI] [PubMed] [Google Scholar]
- Welsh I.C., et al. Integration of left-right Pitx2 transcription and Wnt signaling drives asymmetric gut morphogenesis via Daam2. Dev. Cell. 2013;26:629–644. doi: 10.1016/j.devcel.2013.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte M.P., Reinus W.H., Mumm S. High-bone-mass disease and LRP5. N. Engl. J. Med. 2004;350:2096–2099. doi: 10.1056/NEJM200405133502017. author reply 2096–9. [DOI] [PubMed] [Google Scholar]
- Williams D.S., Lopes V.S. The many different cellular functions of MYO7A in the retina. Biochem. Soc. Trans. 2011;39:1207–1210. doi: 10.1042/BST0391207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., et al. Dental and craniofacial defects in the Crtap-/- mouse model of osteogenesis imperfecta type VII. Dev. Dyn. 2020;249:884–897. doi: 10.1002/dvdy.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., et al. A FERM domain autoregulates Drosophila myosin 7a activity. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4189–4194. doi: 10.1073/pnas.0808682106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti S., Canalis E. Notch signaling and the skeleton. Endocr. Rev. 2016;37:223–253. doi: 10.1210/er.2016-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables
Data Availability Statement
All the data are available upon request.