Abstract
The Microbial Culturomics Project aiming to discover several bacterial species made it possible to isolate the strain Marseille-P4308T from a stool sample of a healthy indigenous Congolese volunteer. Strain Marseille-P4308T is a Gram-positive coccus shaped bacterium that optimally grows at 37 °C. The 16S rRNA gene sequence of the strain has a 96.2% sequence similarity to Peptostreptococcus anaerobius strain NCTC 11460T (GenBank accession number: NR_042847.1). In addition, the average nucleotide identity of strain Marseille-P4308T with its closest related species was 71.1%, which was far below the recommended threshold (>95–96%). The genome of the strain Marseille-P4308T has a length of 2.14 Mbp with G + C content of 30.4 mol%. Based on phenotypic, biochemical, genomic and phylogenetic analysis, strain Marseille-P4308T (= CSUR P4308 = CECT 9960) clearly appears to be a new species for which the name Peptostreptococcus faecalis sp. nov., is proposed.
Keywords: Peptostreptococcus faecalis, Gut microbiote, Indigenous congolese, Culturomics, Taxonogenomics
Peptostreptococcus faecalis; Gut microbiote; Indigenous congolese; Culturomics; Taxonogenomics
1. Introduction
Different genera of anaerobic cocci bacteria are involved in a broad range of infections, occurring in all parts of the human body [1]. In various environments, members of the Peptostreptococcaceae family belonging to the order Clostridiales, phylum Firmicutes can be found in the human body, manure, soil and sediment [2]. The closest phylogenetic neighbours to this family belong to the genera Alkaliphilus, Natronincola, and Tindallia [2]. The species of the genus Peptostreptococcus are commensal species that colonise almost every mucosal human tissue, forming a part of the intestinal, urinary, vaginal, oral tract microbiotas and also the skin. They are pathogenic under certain circumstances and can cause bacteraemia and abscesses in different organs [3].
The genus Peptostreptococcus is a group of Gram-positive, mostly anaerobic cocci species that are very diverse phenotypically and phylogenetically [4]. Cells measure between 0.3 and 2.0 μm and are arranged in chains, pairs, tetrads or masses. Some are aero-tolerant but do not form spores. Their ability to use carbohydrates varies considerably. The main source of energy would be the products of protein metabolism [1]. However, the pathogenic character of some members of the genus Peptostreptococcus is complex to determine, because some members are part of the normal microflora, while species such as Peptostreptococcus anaerobius and Peptostreptococcus stomatis have been frequently identified from clinical samples of diseased individuals [5, 6, 7]. Indeed, Peptostreptococcus russellii was isolated in environment as a storage pit [8].
Numerous previously uncultured members of the human microbiome have been isolated by the culturomics method [9]. Isolated species are then described by multiphasic approach including a MALDI-TOF MS identification, 16S rRNA gene sequencing and the phenotypic and biochemical analysis of the strain [10].We report here a taxonogenomic description of Peptostreptococcus faecalis sp. nov., strain Marseille-P4308T isolated from human gut microbiota.
2. Material and methods
2.1. Ethics and sample collection
The stool sample was collected from a healthy indigenous Congolese volunteer living in Bene Gain (1°59′24.7″S 15°52′18.1″E), in the Republic of the Congo, in August 2015 (supplementary Figure S1) [11]. Approval for this study was obtained from the Ministry for Health of the Republic of Congo (000208/MSP/CAB.15 du Ministère de la Santé et de la Population, 20 August 2015). The study was also approved by the ethics committee of the Institut Fédératif de Recherche IFR48, Faculty of Medicine, Marseille, France, under reference number 09–022. Prior to sampling, an informed consent form was obtained from each individual. In the presence of representatives from a local health centre and village elders, full information was given orally in the local language (Lingala) to ensure that the project was fully understood, as the participants were illiterate.
2.2. Strain isolation and growth conditions
One gram of the stool sample was pre-incubated in a blood culture bottle containing rumen and sheep blood, as described above for culturing human samples [12]. The culture medium was serially diluted and inoculated on Columbia agar with 5% sheep blood (BioMérieux, Marcy-L’Etoile, France). All pure colonies obtained by culture were identified by MALDI-TOF MS [13]. If, despite the good quality of the bacterial spectrum, the bacteria could not be identified, sequencing of the 16S rRNA gene of the bacterium was performed for identification.
2.3. 16S rRNA sequencing and phylogenetic analysis
The DNA of the bacterial strain was extracted using the EZ1 (Qiagen, Venlo, The Netherlands) DNA tissue kit on the EZ1 (Qiagen) automate. The universal primers fD1 and rP2 (Eurogentec, Angers, France) were used to amplify the16S rRNA gene sequence. Sequencing was performed using the BigDye® Terminator v1.1 Cycle Sequencing Kit and ABI Prism 3130xl Genetic Analyzer capillary sequencer (Thermo Fisher, Saint-Aubin, France), as previously described [14]. Using Codon Code Aligner software (http://www.codoncode.com), the 16S rRNA nucleotide sequences were assembled and corrected. Consensus sequence from 16S rRNA gene sequencing was compared by BLASTn within the NCBI 16S rRNA database (https://blast.ncbi.nlm.nih.gov/). In order to create a robust phylogenetic tree, the 16S rRNA sequences of species with a validly published name were downloaded from the LPSN website (https://lpsn.dsmz.de/). Using MEGA X software [15], the sequences were aligned and a phylogenetic tree was constructed with 1,000 bootstrap replicates.
2.4. Genome annotation and comparison
Genomes of closely related species were downloaded from the GenBank database and annotated with Prokka software v1.14.6 [16]. Coding sequences were predicted using Prodigal software 2.6 [17], then the predicted bacterial protein sequences were searched against the GenBank (https://www.ncbi.nlm.nih.gov/genbank/) database using BLASTp. ARAGORN software 1.2 [18] was used to find tRNA and mRNA genes, whereas rRNA genes were predicted with Barrnap software 0.4.
The genome of strain Marseille-P4308T was compared to the genomes of the following closely related species, including Peptostreptococcus anaerobius NCTC11460T (GenBank accession: UGTB00000000), Peptostreptococcus russellii RT-10BT (JYGE00000000), Peptostreptococcus stomatis DSM 17678T (ADGQ00000000), Peptostreptococcus canis DSM 27025T (JABGBW000000000), Asaccharospora irregularis DSM 2635T (FQWX00000000), Clostridioides difficile ATCC 9689T (AUOX00000000), Clostridioides mangenotii LM2 (JIAA00000000) and Clostridium hiranonis DSM 13275T (CP036523). The DNA-DNA hybridisation (dDDH) was calculated to assess similarity between studied genome sequences using the online server of Genome-to-Genome Distance Calculator (GGDC 2.1) (http://ggdc.dsmz.de/), taking into account the method and formula two, as suggested [19, 20]. Furthermore, the genomic average nucleotide identity (ANI) was determined using OrthoANI software v0.93.1 [21]. Clusters of Orthologous genes (COGs) were detected by performing BLASTp of genomes against the COG database [22].
2.5. Conditions of growth
The strain was grown under different conditions. In terms of temperature, the growth of the strain was evaluated at room temperature, 28 °C, 37 °C, 45 °C and 55 °C and incubated under aerobic, anaerobic and microaerophilic conditions on Colombia agar enriched with 5% sheep blood (BioMérieux), using the GENbag anaer and GENbag microaer systems (ThermoFisher Scientific, Basingstoke), respectively. To determine the tolerated salt concentration and the optimum pH for growth, the strain was cultured in different media with varied pH (6, 6.5, 7 and 8.5) and at different NaCl concentrations (5, 10, 50, 75 and 100 g/L NaCl) at 37 °C under anaerobic conditions.
2.6. Phenotypic characteristics
Motility test and Gram staining of the strain were verified using a DM1000 photomicroscope (Leica Microsystems, Nanterre, France) under a 100× objective. In addition, a bacterial suspension was fixed with a 2.5% glutaraldehyde solution at 0.1 mol/L with the aim of observing the morphology of cells using a Hitachi TM4000 electron microscope (Hitachi Group, Krefeld, Germany). Spore-forming was investigated by exposing a bacterial suspension for 10 min under thermal shock at 80 °C. Multiple biochemical criteria from strain Marseille-P4308T were revealed using API tests (50CH, ZYM and 20A; bioMérieux). Oxidase and catalase reactions were determined using a BD BBL™ DrySlide (Becton Dickinson, Le Pont-de-Claix, France).
2.7. Antibiotic susceptibility
The E test method was used to obtain an antimicrobial sensitivity profile and minimum inhibitory concentration (μg/mL) of the strain Marseille-P4308T [23]. Antimicrobial discs placed on a blood agar Petri dish were amikacin, amoxicillin, benzylpenicillin, ceftazidime, ceftriaxone, ciprofloxacin, clindamycin, daptomycin, erythromycin, imipenem, linezolid, metronidazole, minocycline, rifampicin, teicoplanin, tigecycline, trimethoprim/sulfamethoxazole, tobramycin and vancomycin.
3. Results
3.1. Identification and classification
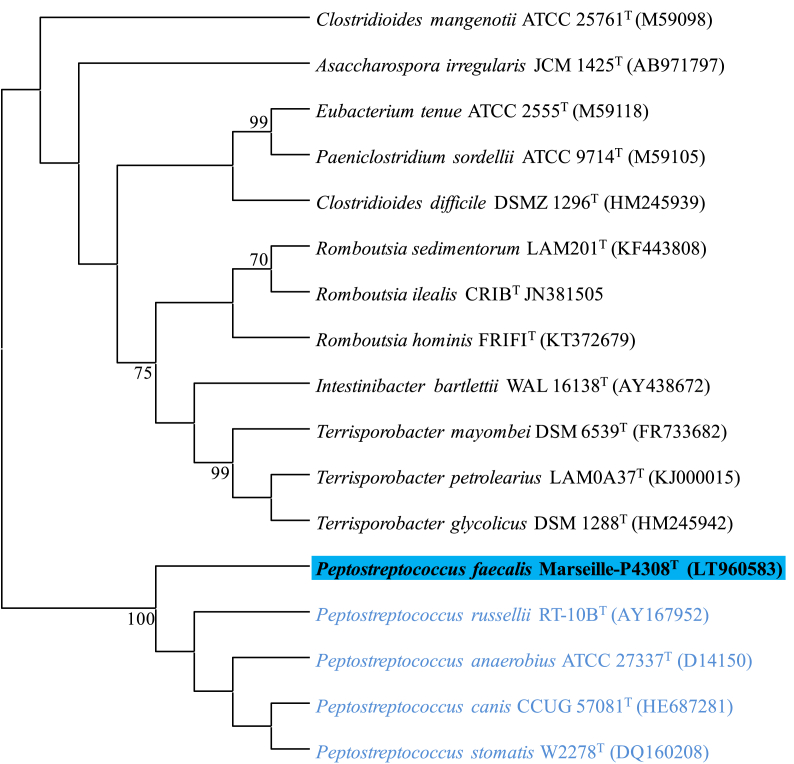
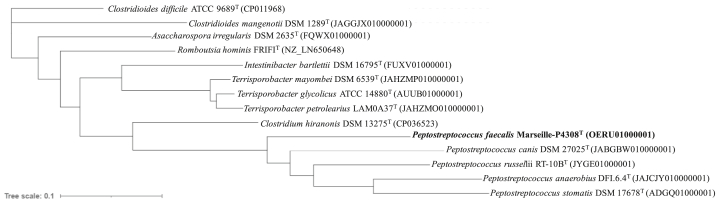
Analysis of the 16S rRNA gene sequence on NCBI Blast showed that the Marseille-P4308T strain has the highest similarity score of 96.2% to Peptostreptococcus anaerobius strain NCTC 11460T (GenBank accession number: NR_042847.1). This value obtained is below the threshold value recommended for delimiting a new species of prokaryote [24]. Therefore, the strain Marseille-P4308T is presumed to be a potential new bacterial species belonging to the genus Peptostreptococcus within the Peptostreptococcaceae family and phylum Firmicutes. The phylogenetic tree constructed on the basis of the sequences of the 16S rRNA gene shows the strain Marseille-P4308T anchored in the Peptostreptococcus sp group with closely related species (Figure 1). The position is confirmed with the phylogenomic tree basing on the genomic sequences of the closely related species (Figure 2).
Figure 1.
Phylogenetic tree with the 16S rRNA gene sequences indicating the position of Peptostreptococcus faecalis sp. nov. Marseille-P4308T among other closely related species. Sequences were aligned using MUSCLE, and phylogenetic inferences were obtained using the Neighbor-Joining method [29] within the MEGA X software [15]. Accession numbers are indicated in parenthesis.
Figure 2.
Phylogenomic tree basing on genomic sequences of Peptostreptococcus faecalis sp. nov., Marseille-P4308T and other its related species. Sequences were concatenated with union EMBOSS (https://www.bioinformatics.nl/cgi-bin/emboss/union) and aligned by Mugsy software [30]. The tree was built using iTOL an online bioinformatic tool [31].
3.2. Phenotypic characteristics
Strain Marseille-P4308T grows under anaerobic and micro-aerobic conditions between 25 °C and 42 °C, with an optimal growth at 37 °C. It is a Gram-positive coccus-shaped bacterium, non-motile, non-spore-forming, oxidase-negative and catalase-negative. Using a Hitachi electron microscope, the strain's morphology is highlighted; cells had a mean length of 0.84 μm and a mean diameter of 0.7 μm (supplementary Figure S2). Furthermore, this strain was able to grow on media at a pH between 6 and 8.5 and tolerated NaCl concentration up to 50 g/L. Colonies were smooth, convex and regular in appearance, with a mean diameter of 0.5 mm on Columbia Agar with 5% Sheep Blood (bioMérieux).
Using API 50CH, the following carbohydrates are fermented: glycerol, erythritol, D-arabinose, D-ribose, L-xylose, D-adonitol, methyl ß-D-xylopyranoside, D-galactose, D-fructose, D-mannose, L-sorbose, L-rhamnose, dulcitol, inositol, D-sorbitol, methyl α-D-mannopyranoside, methyl α-D-glucopyranoside, N-acetylglucosamine, amygdalin, arbutin, esculin ferric citrate, D-cellobiose, D-melibiose, D-saccharose, D-trehalose, inulin, D-melezitose, D-raffinose, starch, glycogen, xylitol, gentiobiose, D-lyxose, D-tagatose, L-fucose, D-arabitol, L-arabitol, potassium gluconate and potassium 5-ketogluconate. Negative reactions were observed for D-glucose, D-lactose, D-maltose, D-mannitol, D-turanose, D-xylose, salicin and sucrose. Indeed, the use of API ZYM strips revealed positive reactions for esterase, leucine arylamidase, cystine arylamidase, α-glucosidase and α-galactosidase, while the following tests, such as alkaline phosphatase, esterase lipase, lipase (C14), valine arylamidase, trypsin, α-chymotrypsin, acid phosphatase, β-galactosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase and α-fucosidase were negative. Finally, the use of API 20A strips shows that strain Marseille-P4308T was positive for gelatin and esculin ferric citrate, while it was negative for tryptophan, urea, D-glucose, D-mannitol, D-lactose, D-sucrose, D-maltose, salicin, and D-xylose. The comparison of the main phenotypic and chemical differences between strain Marseille-P4308T and its phylogenetically related species is given in Table 1.
Table 1.
Differential characteristics of Marseille-P4308T, Peptostreptococcus stomatis W2278T [5], Peptostreptococcus canis CCUG 57081T [27], and Peptostreptococcus anaerobius ATCC 27337T [28].
| Characteristics | Marseille-P4308T | W2278T | CCUG 57081T | ATCC 27337T |
|---|---|---|---|---|
| Oxygen requirement | Strictly anaerobic | Strictly anaerobic | Facultative anaerobic | Strictly anaerobic |
| Acid production from: | ||||
| D-Cellobiose | + | − | − | + |
| α-Galactosidase | + | + | − | + |
| D-Glucose | − | + | − | + |
| α-Glucosidase | + | + | NP | + |
| D-Lactose | − | − | − | + |
| D-Mannose | + | − | + | NP |
| D-Raffinose | + | − | − | NP |
| Sucrose | − | − | − | + |
| G + C content (mol%) | 30.4 | 36 | 30.8 | 34–36 |
| Habitat | Human | Human | Dog | Cat |
NP, not performed.
Antimicrobial susceptibility testing of strain Marseille-P4308T showed the following minimal inhibitory concentrations (in parenthesis) for the following antibiotics: ceftazidime (0.75 μg/mL), trimethoprim (0.25 μg/mL), daptomycin (0.016 μg/mL), tobramycin (8 μg/mL), benzylpenicillin (5 μg/mL), ciprofloxacin (1 μg/mL), ceftriaxone (0.5 μg/mL), linezolid (0.094 μg/mL), ciprofloxacin (0.75 μg/mL), clindamycin (0.064 μg/mL) and amikacin (16 μg/mL).
3.3. Genome properties and comparison
Strain Marseille-P4308T has a genome size of 2,145,294 bp, assembled into 29 contigs with 30.4 mol% G + C content. Its genome possesses 2,095 predicted genes, of which 2,031 are protein-coding genes and 64 code for RNAs (9 rRNA genes, 53 tRNA genes and 2 tmRNA gene). Of the 2,031 protein-coding genes, 1,442 were assigned to COG functional categories. The genomic structure of the strain is represented by a circular map showing the coding regions of the genome (supplementary Figure S3). The genome of strain Marseille-P4308T was compared with eight other genomes belonging to closely-related species in the Peptostreptococcaceae family (Table 2). Among the Peptostreptococcus species, strain Marseille-P4308T has the second largest genome (2.14 Mbp), shorter only than the genome of Peptostreptococcus anaerobius ATCC 27337T (2.25 Mbp). The distribution of genes among the different COG categories is almost the same proportion in all the genomes of the compared Peptostreptococcus species (supplementary Figure S4).
Table 2.
Comparison of the size, the content of G + C mol% and the number of proteins of the genome of Peptostreptococcus faecalis sp. nov., strain Marseille-P4308T with the other genomes of related species.
| Species | Strain Number | Genbank accession | Size (bp) | G + C mol% | Nb of proteins | |
|---|---|---|---|---|---|---|
| Peptostreptococcus faecalis | Marseille-P4308T | OERU00000000 | 2,145,294 | 30.4 | 2,031 | |
| Peptostreptococcus anaerobius | ATCC 27337T | UGTB00000000 | 2,256,756 | 35.7 | 2,070 | |
| Peptostreptococcus canis | CCUG 57081T | JABGBW000000000 | 2,064,240 | 30.2 | 1,834 | |
| Peptostreptococcus russellii | RT-10BT | JYGE00000000 | 2,082,949 | 30.9 | 1,756 | |
| Peptostreptococcus stomatis | W2278T | ADGQ00000000 | 1,988,044 | 36.6 | 1,799 | |
The DDH values obtained after genomic analysis vary for strain Marseille-P4308T from 18.9% with Asaccharospora irregularis DSM 2635T to 23.7% with Clostridioides mangenotii DSM 1289T. Among species within Peptostreptococcus genus, DDH values ranged from 19.5%, between P. faecalis Marseille-P4308T and P. canis CCUG 57081T, to 27.9% between P. stomatis W2278T and P. anaerobius ATCC 27337T (Table 3). These values are therefore lower than the recommended threshold value of 70% for predicting a new prokaryote species [19, 24]. However, on this basis, strain Marseille-P4308T is considered to be a new species in the genus Peptostreptococcus. In addition, among the Peptostreptococcus species studied, the lowest OrthoANI value was 71.16% (between P. faecalis Marseille-P4308T and P. anaerobius ATCC 27337T or P. stomatis W2278T) while 73.25% (between P. canis CCUG 57081T and P. russellii RT-10BT) was the highest OrthoANI value obtained in this analysis (Table 3). The mean nucleotide identity (ANI) analysis based on the genomes of species close to the Marseille-P4308T strain revealed genomic sequence similarities of less than 80%. Indeed, the cut-off value of 95–96% was recommended to delineate the species barrier in prokaryotes [25, 26]. Therefore, our strain Marseille-P4308T sharing with its all other related species is considered as a new bacterial species.
Table 3.
Average nucleotide identity (ANI) and dDDH values (%) obtained by pairwise comparison of the nine studied genomes. Compared genomes: 1, Peptostreptococcus faecalis Marseille-P4308T; 2, Peptostreptococcus anaerobius ATCC 27337T; 3, Peptostreptococcus canis CCUG 57081T; 4, Peptostreptococcus russellii RT-10BT; 5, Peptostreptococcus stomatis W2278T; 6, Asaccharospora irregularis DSM 2635T; 7, Clostridioides difficile DSM 1296T; 8, Clostridioides mangenotii DSM 1289T; 9, Clostridium hiranonis DSM 13275T. OrthoANI values are shown on right bottom (in bolt) and dDDH values calculated using GGDC formula 2 software (DDH estimates based on HSP identities/length) shown on upper left. The empty boxes between them are 100%.
| Strains | Marseille-P4308T | ATCC 27337T | CCUG 57081T | RT-10BT | W2278T | DSM 2635T | DSM 1296T | DSM 1289T | DSM 13275T |
|---|---|---|---|---|---|---|---|---|---|
| Marseille-P4308T | 22.6% | 19.5% | 21.2% | 20.7% | 18.9% | 19.3% | 23.7% | 23.1% | |
| ATCC 27337T | 71.16% | 24.0% | 22.5% | 27.9% | 20.4% | 21.3% | 28.3% | 26.0% | |
| CCUG 57081T | 71.17% | 71.68% | 21.3% | 25.1% | 18.9% | 18.8% | 25.8% | 25.7% | |
| RT-10BT | 73.25% | 72.26% | 73.83% | 23.3% | 25.7% | 19.4% | 27.1% | 22.6% | |
| W2278T | 71.16% | 73.83% | 71.63% | 72.29% | 19.4% | 21.2% | 28.4% | 27.8% | |
| DSM 2635T | 69.14% | 68.03% | 68.85% | 70.06% | 67.83% | 21.1% | 20.3% | 20.7% | |
| DSM 1296T | 69.16% | 68.00% | 69.31% | 69.40% | 67.60% | 75.83% | 19.8% | 21.6% | |
| DSM 1289T | 68.82% | 68.28% | 68.58% | 68.80% | 67.50% | 72.90% | 73.53% | 26.1% | |
| DSM 13275T | 69.71% | 69.54% | 69.13% | 69.66% | 68.38% | 72.03% | 71.95% | 71.65% |
4. Conclusion
Based on the results of phylogenetic, phenotypic and biochemical analyses, it appeared that Peptostreptococcus faecalis sp. nov., strain Marseille-P4308T, is formally considered as a new species within the genus Peptostreptococcus. The type strain of this new species is Marseille-P4803T.
4.1. Description of Peptostreptococcus faecalis sp. nov.
Peptostreptococcus faecalis (fae.ca'lis. N.L. fem. adj. faecalis of faeces). Bacterial colonies of strain Marseille-P4308T are convex, smooth and regular with a mean diameter of 0.7 mm on blood agar. Cells are non-motile and non-spore-forming. They are Gram-positive cocci bacteria and are oxidase and catalase negative. Glycerol, erythritol, D-arabinose, potassium gluconate, D-ribose, L-xylose, D-adonitol, methyl ß-D-xylopyranoside, D-galactose, D-fructose, D-mannose, L-sorbose, L-rhamnose, dulcitol, inositol, D-sorbitol, methyl α-D-mannopyranoside, methyl α-D-glucopyranoside, N-acetylglucosamine, amygdalin, arbutin, esculin ferric citrate, D-cellobiose, D-melibiose, D-saccharose, D-trehalose, inulin, D-melezitose, D-raffinose, starch, glycogen, xylitol, gentiobiose, D-lyxose, D-tagatose, L-fucose, D-arabitol, L-arabitol, potassium 5-ketogluconate, esterase, leucine arylamidase, cystine arylamidase, α-galactosidase, α-glucosidase and gelatin are fermented. The genome size of strain Marseille-P4308T is about 2.14 Mbp with a 30.4 mol% of G + C content. The 16S rRNA and genome sequences are deposited in the GenBank database under Accession numbers LT960583 and OERU00000000, respectively.
The type strain Marseille-P4308T (=CSUR P4308 = CECT 9960) was isolated from stool sample from a healthy indigenous Congolese volunteer.
Declarations
Author contribution statement
Pierre-Edouard Fournier: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Florence Fenollar: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Didier Raoult: Conceived and designed the experiments.
Oleg Mediannikov, Jean Akiana and Geor Mongo Ndombe: Performed the experiments.
Rita Zgheib: Analyzed and interpreted the data.
Fatima Mekhalif: Analyzed and interpreted the data; Wrote the paper.
Cheikh Ibrahima Lo: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Melhem Bilen and Stéphane Alibar: Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by the Institut Hospitalo-Universitaire (IHU) Méditerranée Infection, the National Research Agency under the “Investissements d'avenir” programme (reference ANR-10-IAHU-03), the Region Provence-Alpes-Côte d’Azur and European ERDF PRIMI funding.
Data availability statement
Data associated with this study has been deposited at CSUR collection under the accession number CSUR P4308, CECT collection under the accession number CECT 9960 and GenBank database under the accession numbers LT960583 and OERU00000000.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors thank Professor Henri-Joseph Parra, Doctor Bernard Davoust, and Doctor Cheikh Sokhna for their help in setting up the fieldwork, Ludivine Brechard for sequencing the genome and Aurelia Caputo for submitting the genomic sequence to GenBank. The authors would like to thank the Hitachi Company for supplying the TM4000 Plus tabletop microscope.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Murdoch D.A. Gram-positive anaerobic cocci. Clin. Microbiol. Rev. 1998;11:81–120. doi: 10.1128/cmr.11.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slobodkin A. In: The Prokaryotes: Firmicutes and Tenericutes. Rosenberg E., DeLong E.F., Lory S., Stackebrandt E., Thompson F., editors. Springer; Berlin, Heidelberg: 2014. The family Peptostreptococcaceae; pp. 291–302. [Google Scholar]
- 3.Berman J.J. Taxonomic Guide to Infectious Diseases. Elsevier; 2012. The magnitude and diversity of infectious diseases; pp. 3–5. [Google Scholar]
- 4.Murdoch D.A., Young K.A., Magee J.T. 1997. Description of Three New Species of the Genus Peptostreptococcus from Human Clinical Specimens: Peptostreptococcus Harei Sp. nov., Peptostreptococcus ivoni Sp. nov., and Peptostreptococcus octavius Sp. Nov. p. 7. [Google Scholar]
- 5.Downes J., Wade W.G. Peptostreptococcus stomatis sp. nov., isolated from the human oral cavity. Int. J. Syst. Evol. Microbiol. 2006;56:751–754. doi: 10.1099/ijs.0.64041-0. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y.-L., Tsai S.-H., Hsu K.-C., Chen C.-S., Hsu C.-W. Primary sternal osteomyelitis due to Peptostreptococcus anaerobius. Infection. 2012;40:195–197. doi: 10.1007/s15010-011-0171-z. [DOI] [PubMed] [Google Scholar]
- 7.Bolivar I., Whiteson K., Stadelmann B., Baratti-Mayer D., Gizard Y., Mombelli A., Pittet D., Schrenzel J. Geneva Study Group on Noma (GESNOMA), Bacterial diversity in oral samples of children in Niger with acute noma, acute necrotizing gingivitis, and healthy controls. PLoS Neglected Trop. Dis. 2012;6:e1556. doi: 10.1371/journal.pntd.0001556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitehead T.R., Cotta M.A., Falsen E., Moore E., Lawson P.A. Peptostreptococcus russellii sp. nov., isolated from a swine-manure storage pit. Int. J. Syst. Evol. Microbiol. 2011;61:1875–1879. doi: 10.1099/ijs.0.023762-0. [DOI] [PubMed] [Google Scholar]
- 9.Lagier J.-C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P., Caputo A., Cadoret F., Traore S.I., Seck E.H., Dubourg G., Durand G., Mourembou G., Guilhot E., Togo A., Bellali S., Bachar D., Cassir N., Bittar F., Delerce J., Mailhe M., Ricaboni D., Bilen M., Dangui Nieko N.P.M., Dia Badiane N.M., Valles C., Mouelhi D., Diop K., Million M., Musso D., Abrahão J., Azhar E.I., Bibi F., Yasir M., Diallo A., Sokhna C., Djossou F., Vitton V., Robert C., Rolain J.M., La Scola B., Fournier P.-E., Levasseur A., Raoult D. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Pagnier I., Croce O., Robert C., Raoult D., La Scola B. Non-contiguous finished genome sequence and description of Anaerococcus provenciensis sp. nov. Stand. Genomic. Sci. 2014;9:1198–1210. doi: 10.4056/sigs.5501035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angelakis E., Bachar D., Yasir M., Musso D., Djossou F., Gaborit B., Brah S., Diallo A., Ndombe G.M., Mediannikov O., Robert C., Azhar E.I., Bibi F., Nsana N.S., Parra H.-J., Akiana J., Sokhna C., Davoust B., Dutour A., Raoult D. Treponema species enrich the gut microbiota of traditional rural populations but are absent from urban individuals. New Microbes New Infect. 2018;27:14–21. doi: 10.1016/j.nmni.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagier J.-C., Hugon P., Khelaifia S., Fournier P.-E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin. Microbiol. Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seng P., Drancourt M., Gouriet F., La Scola B., Fournier P.-E., Rolain J.M., Raoult D. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 14.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J. Clin. Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 17.Hyatt D., Chen G.-L., LoCascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laslett D. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auch A.F., von Jan M., Klenk H.-P., Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genom. Sci. 2010;2:117–134. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meier-Kolthoff J.P., Auch A.F., Klenk H.-P., Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinf. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee I., Ouk Kim Y., Park S.-C., Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 22.Tatusov R.L., Galperin M.Y., Natale D.A., Koonin E.V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sader H.S., Pignatari A.C. E test: a novel technique for antimicrobial susceptibility testing. Sao Paulo Med. J. 1994;112:635–638. doi: 10.1590/s1516-31801994000400003. [DOI] [PubMed] [Google Scholar]
- 24.Kim M., Oh H.-S., Park S.-C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 25.Richter M., Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. Unit. States Am. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chun J., Oren A., Ventosa A., Christensen H., Arahal D.R., da Costa M.S., Rooney A.P., Yi H., Xu X.-W., De Meyer S., Trujillo M.E. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018;68:461–466. doi: 10.1099/ijsem.0.002516. [DOI] [PubMed] [Google Scholar]
- 27.Lawson P.A., Johnson C.N., Bengtsson L., Charalampakis G., Dahlén G., Moore E., Falsen E. Peptostreptococcus canis sp. nov., isolated from subgingival plaque from canine oral cavity. Anaerobe. 2012;18:597–601. doi: 10.1016/j.anaerobe.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 28.Gerritsen J., Fuentes S., Grievink W., van Niftrik L., Tindall B.J., Timmerman H.M., Rijkers G.T., Smidt H. Characterization of Romboutsia ilealis gen. nov., sp. nov., isolated from the gastro-intestinal tract of a rat, and proposal for the reclassification of five closely related members of the genus Clostridium into the genera Romboutsia gen. nov., Intestinibacter gen. nov., Terrisporobacter gen. nov. and Asaccharospora gen. nov. Int. J. Syst. Evol. Microbiol. 2014;64:1600–1616. doi: 10.1099/ijs.0.059543-0. [DOI] [PubMed] [Google Scholar]
- 29.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 30.Angiuoli S.V., Salzberg S.L. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics. 2011;27:334–342. doi: 10.1093/bioinformatics/btq665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Letunic I., Bork P. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293. doi: 10.1093/nar/gkab301. –W296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at CSUR collection under the accession number CSUR P4308, CECT collection under the accession number CECT 9960 and GenBank database under the accession numbers LT960583 and OERU00000000.