Summary
Mutations in the gene encoding DNA methyltransferase 3A (DNMT3A) are the most common cause of clonal hematopoiesis and are among the most common initiating events of acute myeloid leukemia (AML). Studies in germline and somatic Dnmt3a knockout mice have identified focal, canonical hypomethylation phenotypes in hematopoietic cells; however, the kinetics of methylation loss following acquired DNMT3A inactivation in hematopoietic cells is essentially unknown. Therefore, we evaluated a somatic, inducible model of hematopoietic Dnmt3a loss, and show that inactivation of Dnmt3a in murine hematopoietic cells results in a relatively slow loss of methylation at canonical sites throughout the genome; in contrast, remethylation of Dnmt3a deficient genomes in hematopoietic cells occurs much more quickly. This data suggests that slow methylation loss may contribute, at least in part, to the long latent period that characterizes clonal expansion and leukemia development in individuals with acquired DNMT3A mutations in hematopoietic stem cells.
Subject areas: Biological sciences, Molecular biology, Epigenetics
Graphical abstract
Highlights
-
•
Somatic inactivation of Dnmt3a in hematopoietic cells causes slow DNA methylation loss
-
•
Methylation loss occurs in an ordered fashion, at canonical sites in the genome
-
•
Many genomic regions that lose or gain methylation rapidly are overlapping
-
•
Methylation remodeling is an integrated process involving methylases and demethylases
Biological sciences; Molecular biology; Epigenetics
Introduction
The deposition and maintenance of methyl groups on mammalian DNA is mediated by DNA methyltransferases. Dysregulation and mutations of methyltransferase genes are well recognized contributors to the pathogenesis of many tumor types. For example, somatic mutations in DNMT3A are the most common cause of age-related clonal hematopoiesis (Xie et al., 2014; Jaiswal et al., 2014; Genovese et al., 2014). Furthermore, DNMT3A is mutated in approximately 20% of de novo acute myeloid leukemia (AML) and >30% of normal karyotype AML (Renneville et al., 2012; Marcucci et al., 2012) making it one of the most common AML initiating mutations. Over 60% of AML-associated DNMT3A mutations are heterozygous missense mutations at the codon for amino acid R882 (Ley et al., 2010; Gaidzik et al., 2013; Gale et al., 2015). Mechanistic studies have shown that the R882H mutation encodes a dominant negative protein that creates a methyltransferase “sink” that sequesters the WT protein into relatively inactive complexes. This inhibits formation of active homodimers, reducing its enzymatic activity by approximately 80% (Russler-Germain et al., 2014). Reduction of enzymatic activity leads to a focal, canonical hypomethylation phenotype both in non-leukemic blood cells (Smith et al., 2021) and in AML cells expressing the R882H mutation (Spencer et al., 2017). However, many aspects of this process are not yet understood, including the rate of methylation loss in HSPCs (and their progeny) over time. In this report, we show that somatic inactivation of Dnmt3a in the hematopoietic cells of adult mice results in a very slow loss of CpG methylation at predictable, canonical sites throughout the genome. These findings have implications for understanding the long latency and subtle phenotypes associated with DNMT3A mutations that cause clonal hematopoiesis and AML.
Results
Kinetics of DNA methylation loss in mouse bone marrow cells after somatic Dnmt3a inactivation
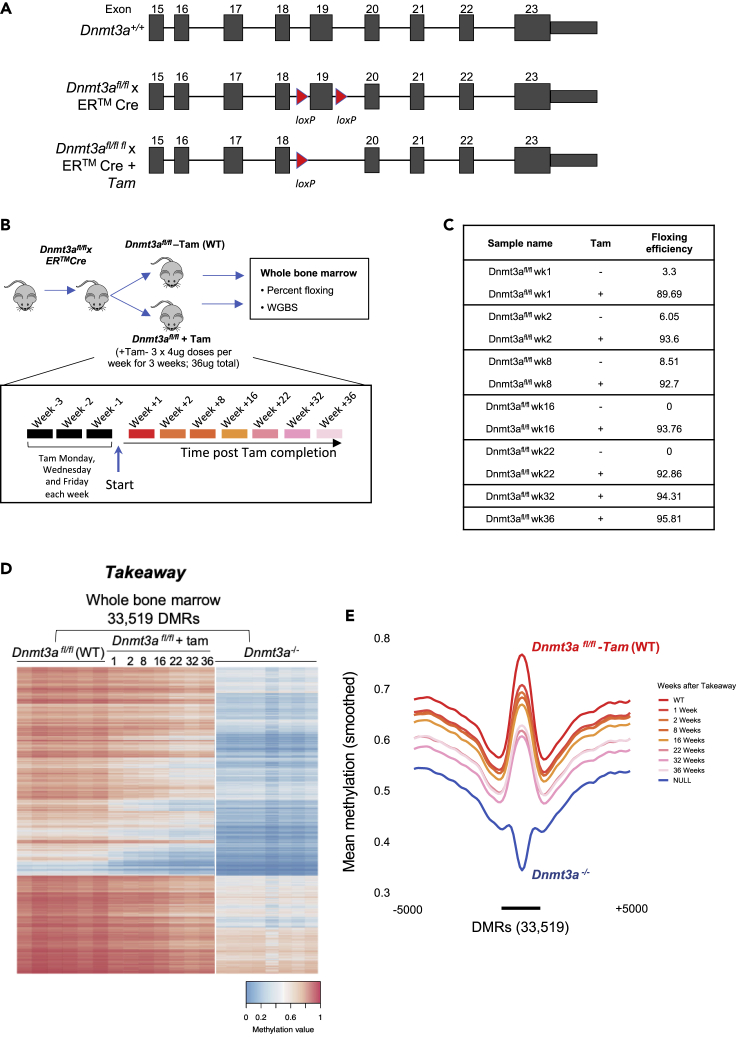
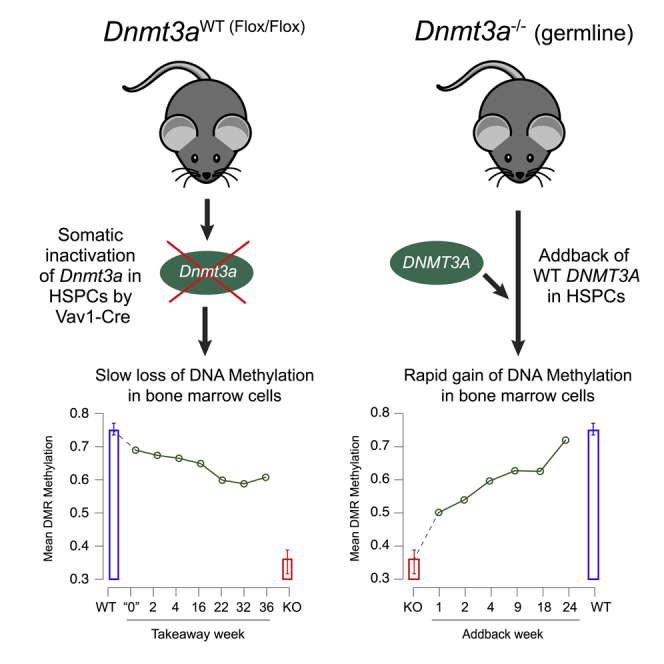
To better understand the kinetics of methylation loss following inactivation of Dnmt3a (Dnmt3a ‘takeaway’), we utilized an inducible Cre-ERTM system to delete both endogenous Dnmt3a alleles in transplanted hematopoietic cells (annotated Dnmt3afl/fl + Tam; wild-type controls annotated Dnmt3afl/fl − Tam (WT); Figure 1A). Cre-mediated deletion of exon 19 (flanked by loxP sites) leads to out-of-frame splicing of the mRNAs encoding all active isoforms (Kaneda et al., 2004). To limit the deletion to hematopoietic cells, we used a secondary transplant model, which allowed us to ameliorate any deleterious effects of deleting Dnmt3a in non-hematopoietic tissues (Figure 1B). Importantly, floxing of Dnmt3a was not performed until we confirmed engraftment at 4-weeks posttransplant, to minimize the stress of transplantation as a potential confounding variable. Floxing efficiency in whole bone marrow (WBM) following tamoxifen dosing (9 doses administered over 3 weeks) was established with RT-PCR at the time of sacrifice and was compared to no-tamoxifen controls for each timepoint, when available. Dnmt3a floxing efficiency was >90% in nearly all tested mice, and was rapidly achieved, because it had occurred in approximately 90% of measured alleles at “week 1”, which was four weeks after the first dose of tamoxifen was given (Figure 1C). Importantly, Dnmt3a deletion did not confer a disadvantage to the transplanted cells, because floxing efficiency was maintained above 90% until the end of the experiment, 36 weeks after the final dose of tamoxifen was given.
Figure 1.
Methylation loss following Dnmt3a deletion is time dependent and slow
(A) Schematic representation of the knockout strategy. Exons are represented as filled boxes and loxP sites as red triangles. Tam, tamoxifen.
(B) Schematic representation of the Dnmt3a takeaway experiment. Tam, tamoxifen; WGBS, whole genome bisulfite sequencing.
(C) Table of floxing efficiencies determined by quantitative RT-PCR.
(D) Heatmap of mean methylation values for 33,519 DMRs identified by comparing Dnmt3afl/fl − Tam (WT) vs. Dnmt3a−/− (without dox; null), with mean methylation values for Tam-treated takeaway samples plotted passively. Time of Tam (takeaway) treatment is shown in weeks.
(E) Methylation density of 33,519 DMRs defined in d, in Dnmt3afl/fl + Tam. DMR; differentially methylated region, tam; tamoxifen, WGBS; whole genome bisulfite sequencing. Time is shown in weeks. Data are presented as means over all DMRs and all samples from each treatment group: Dnmt3a−/− (N = 8), Dnmt3afl/fl + Tam (N = 6), N = 1 for each takeaway timepoint.
We defined differentially methylated regions (DMRs) from whole-genome bisulfite sequencing (WGBS) by comparing Dnmt3a−/− germline knockout bone marrow cells (from 6 independent mice), vs. non-floxed Dnmt3afl/fl marrow (from 8 independent mice), which corresponded to Dnmt3a null and Dnmt3a wild-type levels, respectively. DMRs were required to have >10 CpGs, a mean methylation difference between the two groups of >0.2, a false discovery rate (FDR) of <0.05 and within-group standard deviations in methylation levels <0.1, parameters established in previous work (Ketkar et al., 2020). Contiguous DMRs within 50 base pairs (bp) of each other were merged. Using these criteria, we identified 33,519 DMRs in the Dnmt3a−/− samples; 33,503 (99.96%) of these were hypomethylated (Figure 1D). The genomic locations and annotations for all of these DMRs (and how they were affected by the takeaway and addback experiments at each time point) are shown in Table S1. Passively plotting the methylation values of the Dnmt3afl/fl + Tam (takeaway) samples at these 33,519 DMRs revealed a progressive, time-dependent loss of methylation for some but not all regions, and the degree of methylation loss did not reach Dnmt3a−/− (knockout) levels even at the latest time points (Figures 1D and 1E). The methylation loss occurred in a canonical and ordered way, where hypomethylated DMRs in week 1 were also present in subsequent weeks; additional methylation loss progressed over time at the same sites, suggesting that DMR location or sequence context was deterministic for these patterns.
Characteristics of genomic regions that are sensitive or refractory to DNA methylation loss caused by Dnmt3a inactivation
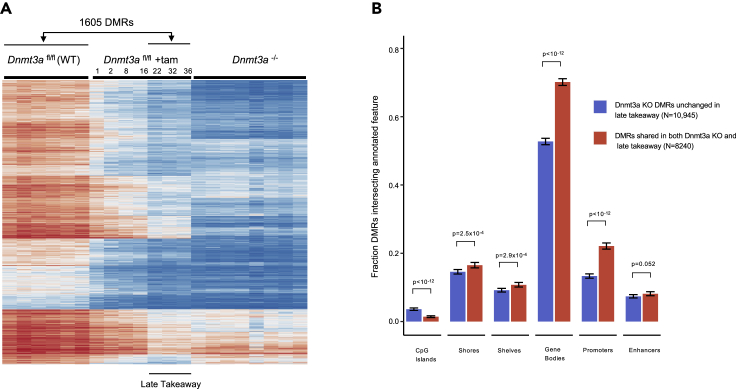
Next, we defined DMRs occurring in the late takeaway samples by comparing the same set of WT samples to the pooled takeaway samples from weeks 22, 32, and 36, which had similar methylation patterns. Using the same criteria described above for calling DMRs, we identified a total of 1,605 DMRs with this analysis. Nearly all (1603/1605, 99.88%) were hypomethylated, as shown in the heat map of Figure 2A, and Table S2. Of these late takeaway DMRs, 1328 (83%) overlapped with the Dnmt3a null DMRs defined above. The mean methylation beta value of the 277 nonoverlapping DMRs was 0.38 ± 0.17 SD, significantly less than that of the mean beta value for WT bone marrow cells at the same regions (0.73 ± 0.14 SD, p = 8.2 × 10−155, 2-sided paired t-test). Dnmt3a KO samples were also hypomethylated in the same 277 regions (mean beta value 0.46 ± 0.19 SD, p = 3.1 × 10−97 compared to WT values), only slightly less so. Of these 277 DMRs, 273 were hypomethylated in both the late takeaway and Dnmt3a KO samples.
Figure 2.
Comparison of WT vs Dnmt3a KO DMRs to Late Takeaway DMRs
(A) Heatmap of mean methylation values for 1605 DMRs identified by comparing WT bone marrow samples vs. the last 3 takeaway timepoints, i.e., Dnmt3afl/fl + Tamoxifen at weeks 22, 32, 36 (“Late Takeaway”). Mean methylation values for the additional takeaway timepoints and Dnmt3a−/− (without dox; null) are plotted passively. Time after the completion of Tamoxifen treatment is shown in weeks.
(B) Enrichment of annotated features in subsets of Dnmt3a KO DMRs that also changed in the late takeaway samples, vs. those that did not change. DMRs were classified based on the change in mean methylation values between WT bone marrow samples, and the last 3 takeaway timepoints (weeks 22, 32, and 36). DMRs with an absolute mean beta value change of <0.1 were classified as non-changing ('refractory'); DMRs with absolute mean beta value change of >0.2 were classified as changing. 95% Wilson score confidence intervals and Fisher’s exact p-values are shown.
We also investigated the characteristics of the DMRs that were “refractory” to methylation loss in the late takeaway samples to determine whether they had unique features or genomic locations that altered their methylation fates. Late takeaway DMRs were classified based on the change in mean beta methylation values between the WT samples and the last 3 takeaway timepoints (from weeks 22, 32, 36). Late takeaway DMRs with an absolute mean beta value change of <0.1 were classified as 'unchanged' (i.e., refractory to methylation loss, n = 10,945); DMRs with absolute mean change >0.2 were concordant with the Dnmt3a KO samples (n = 8,240). The genomic annotations of these regions are shown in Figure 2B; the refractory regions were enriched for CpG islands (which have the lowest levels of methylation of all the annotated regions). Conversely, the regions that did become hypomethylated in the late takeaway samples were significantly enriched for shores, shelves, gene bodies, and promoters.
Kinetics of remethylation in Dnmt3a deficient bone marrow cells with DNMT3A addback
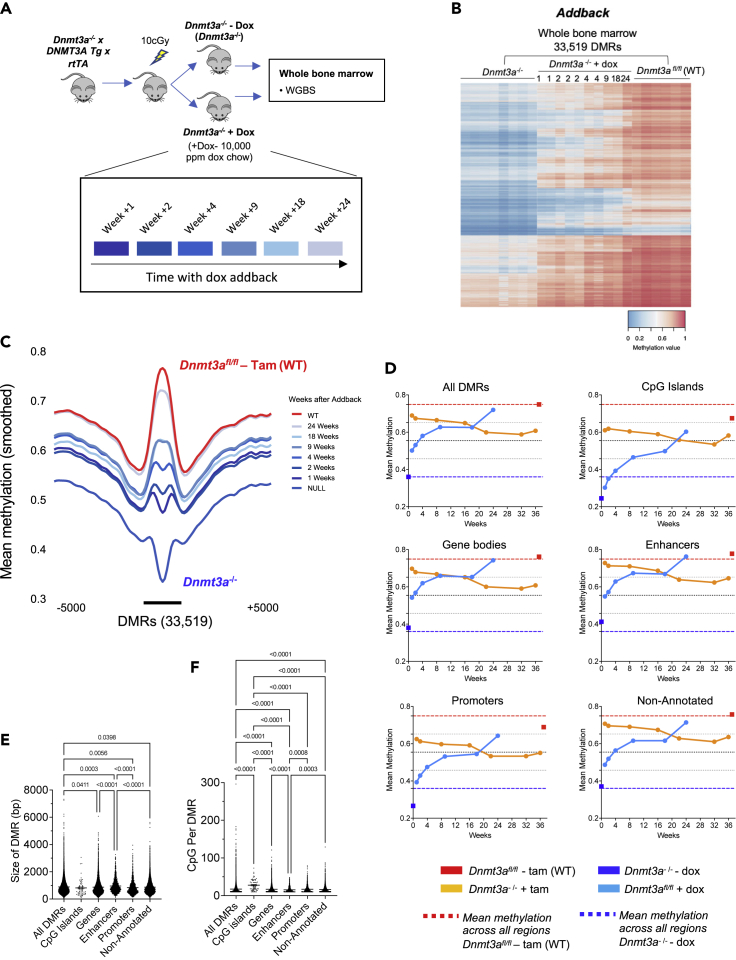
Next, we utilized previously published data from our lab that measured the kinetics and patterns of remethylation in Dnmt3a null mice that contained an ‘addback’ Dox-inducible DNMT3A transgene (Figure 3A) (Ketkar et al., 2020). By comparing the fate of DMRs in the takeaway and addback experiments, we were able to assess whether the changes were concordant. Remethylation of most of the 33,519 DMRs occurred rapidly following expression of DNMT3A transgene in Dnmt3a−/− mice treated with Dox, so that methylation levels approached that of WT mice by 24 weeks. This suggests that methylation loss after Dnmt3a inactivation occurs much more slowly than remethylation of Dnmt3a-dependent sites after restoration of DNMT3A activity (Figures 3B and 3C). This trend was true across all functionally annotated regions of the genome, including CpG islands, gene bodies, promoters, and enhancers (Figure 3D). However, there was variation in the level of methylation in different annotated regions of the genome in wild-type bone marrow cells. For example, enhancer regions and gene bodies had higher levels of baseline methylation (red square) compared to promoters and CpG islands (i.e., there is a higher level of methylation in these regions, even in the absence of Dnmt3a; blue square). This suggests that some regions of the genome have additional, Dnmt3a-independent mechanisms for methylating and/or maintaining CpG methylation. Interestingly, the baseline methylation levels did not appear to dictate rates of methylation loss with takeaway (or methylation gain with addback) because these rates were consistent across all genomic regions assessed (Figure 3D). Further, promoter DMRs were significantly smaller in size, compared to DMRs in genes and enhancers (Figure 3E), and CpG islands had significantly higher CpG density per DMR compared to other annotated regions (Figure 3F), factors which may influence their levels of basal methylation.
Figure 3.
Methylation gain following DNMT3A addback is time dependent and rapid
(A) Schematic representation of the DNMT3A addback experiment. Dox, doxycycline; WGBS, whole genome bisulfite sequencing.
(B) Heatmap of mean methylation values for 33,519 DMRs identified by comparing Dnmt3afl/fl – Tam (WT) vs. Dnmt3a−/− (without dox; null), with mean methylation values for dox-treated addback samples, plotted passively. Time of dox (addback) treatment is shown in weeks.
(C) Methylation density of 33,519 DMRs defined in (B), in Dnmt3a−/− + Dox samples obtained at different timepoints. Data are presented as means over all DMRs and all samples from each treatment group: Dnmt3a−/− (N = 8), Dnmt3afl/fl + Tam (N = 6), and addback weeks 1 (N = 2), 2 (N = 3), 4 (N = 2), 9 (N = 1), 18 (N = 1), 24 (N = 1).
(D) Mean methylation values for all 33,519 DMRs and DMRs exclusive to specific annotated genomic regions are shown. The red dotted line indicates mean methylation value of all DMRs in Dnmt3afl/fl – Tam (WT), blue dotted line indicates mean methylation value of all DMRs in Dnmt3a−/−, red square indicates mean methylation value of DMRs in specified annotated region for Dnmt3afl/fl – Tam (WT), blue square data point indicates mean methylation value of DMRs in specified annotated region for Dnmt3a−/−. For quartile calculations, Dnmt3afl/fl - Tam (red dotted line) and Dnmt3a−/− (blue dotted line) were defined as 100% and 0% methylation, respectively, and quartiles are represented by black dotted lines. Mean methylation values for all Dnmt3afl/fl + Tam (orange line) and Dnmt3a−/− + Dox (pale blue line) over time in weeks. Data are presented as means over all DMRs in each annotated category and all samples from each treatment group: Dnmt3a−/− (N = 8), Dnmt3afl/fl + Tam (N = 6), and addback weeks 1 (N = 2), 2 (N = 3), 4 (N = 2), 9 (N = 1), 18 (N = 1), 24 (N = 1).
(E) Size in bp (+/− Std Dev) of all 33,519 DMRs and DMRs exclusive to specific annotated genomic regions.
(F) CpGs per DMR for all 33,519 DMRs and DMRs exclusive to specific annotated genomic regions. Hypothesis testing in (E) and (F) was performed via one-way ANOVA with Tukey’s multiple comparison test between each annotated genomic region (p values are indicated for each comparison; nonsignificant values are not presented). Bp; base-pairs, DMR; differentially methylated region, Dox; doxycycline, tam; tamoxifen, WGBS; whole genome bisulfite sequencing. Time is shown in weeks.
Characteristics of dynamic DMRs
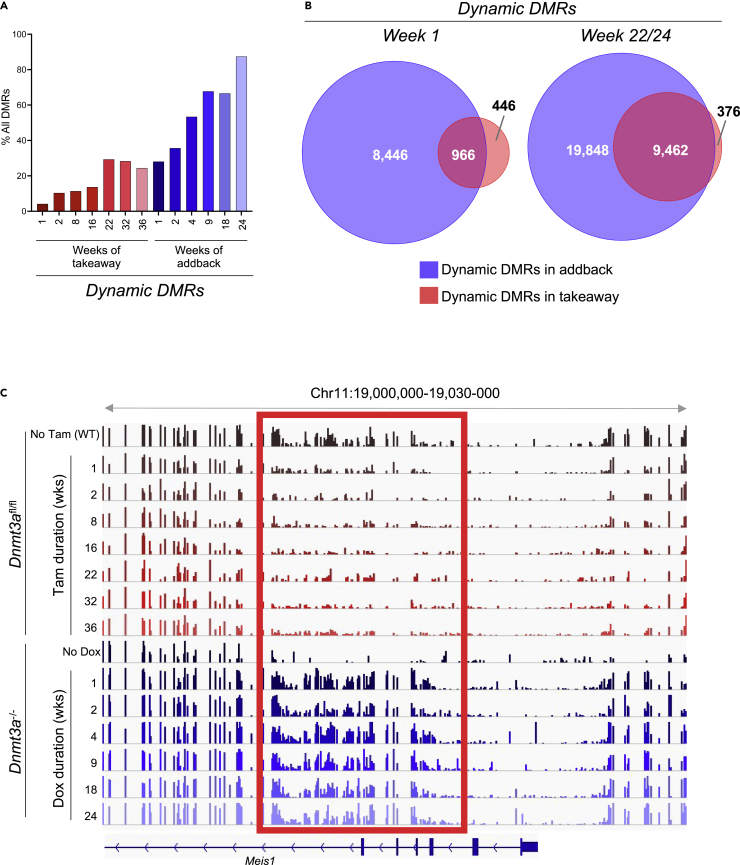
We next wanted to understand the characteristics of ‘dynamic DMRs’ — those DMRs that lose and gain methylation as a function of time — to determine whether these represent the same regions with takeaway and addback. We defined a dynamic DMR as one that moves >0.2 (i.e., a 20% change in methylation density) away from baseline at the assessed timepoint. For dynamic takeaway DMRs, the baseline was Dnmt3afl/fl – Tam (WT) methylation levels; for dynamic addback DMRs, the baseline was Dnmt3a−/− methylation levels. As time progressed, additional dynamic DMRs were observed (Figure 4A). However, even at week 36 after takeaway, only ∼1/3 of DMRs were dynamic. In contrast, nearly 100% of DMRs were dynamic by week 24 following addback. At the 1-week timepoint, 966/1,412 (68.4%) dynamic takeaway DMRs overlapped with the 9,412 (10.3%) dynamic addback DMRs. This overlap was magnified by week 22/24 (takeaway/addback), where 9,462/9,838 (96.2%) dynamic takeaway DMRs overlapped with 29,310 (32.3%) dynamic addback DMRs (Figure 4B). We classified the dynamic DMRs identified at week-1 as fast, because of the rapidity with which they gain and lose methylation. An example of a fast dynamic DMR in the Meis1 gene is shown in Figure 4C. Together, these data show that methylation gain and loss is canonical and ordered, implying that there are undefined factors that determine the rate of methylation loss at specific CpGs.
Figure 4.
Characteristics of dynamic DMRs
(A) Percent of DMRs that are dynamic at each time point in the whole bone marrow from Dnmt3afl/fl + Tam (takeaway) and Dnmt3a−/− + Dox (addback) mice. Dynamic DMRs are defined as those that have a change in beta value of >0.2 from baseline where baseline for takeaway and addback are defined as values in Dnmt3afl/fl – Tam (WT) and Dnmt3a−/- for individual DMRs, respectively.
(B) Venn diagram showing overlap between dynamic DMRs identified in Dnmt3afl/fl + Tam (takeaway; red) and Dnmt3a−/− + Dox (addback; blue) mice at week 1, and at weeks 22/24. The dynamic DMRs found in week 1 are classified as rapid dynamic DMRs.
(C) Example of a rapid dynamic DMR within the Meis1 gene. The gene structure is shown below, genomic location (assembly mm10) shown above, individual CpGs are represented by colored bars, y -axis is mean methylation of CpGs, time is given in weeks, and the DMR is highlighted in the gray box. DMR; differentially methylated region, Dox; doxycycline, tam; tamoxifen. Time is shown in weeks.
Contributions of DNA methylases and demethylases to methylation remodeling in hematopoietic cells
We next attempted to define the genes and pathways that are the most strikingly affected by fast dynamic DMRs. Defining the specific gene targets of DMR dysregulation is complex, because the proximity of a DMR to a gene does not reliably predict an effect on its expression (Spencer et al., 2017; Ketkar et al., 2020). Therefore, we interrogated dynamic vs. non-dynamic DMRs (both from week 1, and from week 22), and linked these DMRs to genes in two different ways: first, we required a DMR to be mapped to a gene promoter, gene body, or anywhere within 10 kb of a gene. The second analysis was more restrictive, requiring the DMR to map to a gene's promoter. Using these definitions, we performed gene ontology analysis to determine whether specific pathways related to GO biological processes were significantly enriched (DMR locations, genomic annotation, and enrichment analyses are found in Table S3, and graphical representations of the enrichment analyses are in Figures S1 and S2). Using a false-discovery-rate cutoff of 0.001, several pathways were enriched in the dynamic DMRs with both analyses, and at both timepoints; the GO terms for these pathways were extraordinarily diverse, and were difficult to connect to processes thought to be related to altered self-renewal and/or differentiation in Dnmt3a deficient hematopoietic cells. Clearly, additional studies will be required to better understand the physiologic consequences associated with fast vs. slow methylation loss at specific loci.
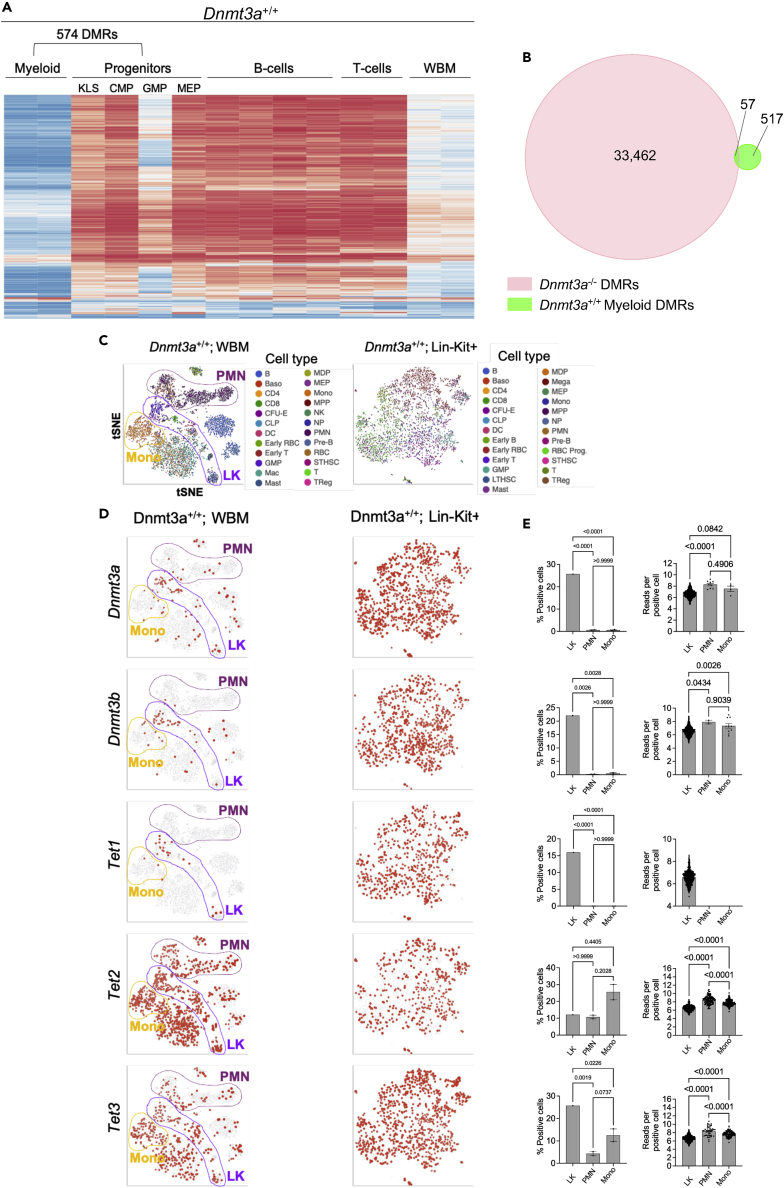
We explored the hypothesis that the slow loss of methylation with takeaway may also be influenced by the slow rates of hematopoietic stem/progenitor cell (HSPC) division, with passive methylation loss as cells repeatedly divide. Because Dnmt3a is primarily expressed in HSPCs (Ketkar et al., 2020; Tadokoro et al., 2007), methylation patterns may be established by Dnmt3a in HSPCs, maintained during subsequent cell divisions by Dnmt1, and remodeled by the Tet demethylases as cells commit to terminal differentiation. We tested this hypothesis by comparing the methylomes of unfractionated whole bone marrow cells, purified progenitor populations (KLS, CMP, GMP, and MEP; data grouped for analysis), and mature lineage populations (myeloid, B-cells, or T-cells). By comparing WGBS data from mature myeloid cells (CD11b+/Gr1+) to the progenitor populations of WT mice, we identified 574 DMRs (Figure 5A), of which 552 (96.2%) were hypomethylated in the mature myeloid cells; this supports the hypothesis that the Tet demethylases may actively remodel the methylome during lineage commitment. Passively plotting B-cell and T-cell WGBS data against the myeloid-defined DMRs showed that these regions are specific to the mature myeloid cells, retaining a progenitor-like signature in lymphoid cells. In addition, because myeloid cells are a major population in unfractionated whole bone marrow, the mature myeloid signature was also apparent in those samples. More committed GMPs were more hypomethylated than CMPs and MEPs, suggesting that remodeling is initiated before terminal differentiation, and may therefore play a role in commitment. Likewise, we identified specific B-cell (Figure S3A) and T-cell (Figure S3B) DMRs, which were methylated in myeloid cells.
Figure 5.
Differential methylation and expression signatures in hematopoietic progenitors vs. mature myeloid cells
(A) Heatmap of mean methylation values for DMRs identified by comparing flow-purified progenitors (KLS, MEP, CMP, and GMP; grouped for analysis) and mature myeloid cells (CD11b+/Gr1+) isolated by FACS sorting. Methylation values for those DMRs in B-cells, T-cells, and whole bone marrow are passively plotted.
(B) Venn diagram showing overlap of the 33,519 DMRs defined in Dnmt3a−/− bone marrow vs. the 574 DMRs from Dnmt3a+/+ myeloid cells. Overlap between DMRs is defined as a minimum of 1 bp.
(C) tSNE projections of scRNA-seq data from whole bone marrow (left) and lineage depleted, Kit positive (Lin-Kit+) progenitor cells (right) purified by FACS. Mature neutrophils (PMN), monocytes (mono), and lineage negative, KIT positive cells (LK) are highlighted on the whole bone marrow projection.
(D) Cells are colored according to levels of expression of Dnmt3a, Dnmt3b, Tet1, Tet2, and Tet3 (top to bottom) in Dnmt3a+/+ whole bone marrow (left panels) or lineage negative c-Kit positive cells (right panels).
(E) Quantitation of percent total cells positive for expression (left panels) and number of normalized reads per positive cell (right panels) for Dnmt3a, Dnmt3b, Tet1, Tet2, and Tet3 genes in the indicated cell type. LK; lineage negative, c-KIT positive, WBM; whole bone marrow. One-way ANOVA testing was used to define statistical differences between groups. For Tet1, no reads were detected in the PMNs and Monos, so statistical comparisons could not be performed.
Only 57/574 (10.3%) mature myeloid DMRs were detected in the 33,519 Dnmt3a dependent DMRs defined in Figure 1D (Figure 5B). In contrast, 416/2,031 (20.5%) B-cell DMRs and 619/2,271 (27.3%) T-cell DMRs overlapped with the 33,519 Dnmt3a−/− DMRs (Figures S3A–S3D). Passive plotting of the progenitor and mature lineage populations from Dnmt3a+/+ mice against the 33,519 Dnmt3a−/− DMRs confirmed that these Dnmt3a dependent regions are almost mutually exclusive from the majority of myeloid-specific remodeling sites (Figure S3E). As expected, the 6,435 DMRs defined in the earliest progenitors (KLS and CMP) were persistent in the GMP and MEP compartments, and also in whole bone marrow samples (Figure S4); this suggests that the methylation events mediated by Dnmt3a in HSPCs must be maintained throughout hematopoietic differentiation by Dnmt1 (Ketkar et al., 2020). In aggregate, these data suggest that lineage-specific methylation remodeling during hematopoietic differentiation must primarily depend on the actions of the Tet proteins, and not the actions of Dnmt3a or 3b in more mature cells (Farlik et al., 2015, 2016).
This hypothesis is also supported by single cell RNA sequencing data from unfractionated whole bone marrow and lineage-depleted, Kit positive (Lin-Kit+; LK) cells from WT mice (Ketkar et al., 2020). Dnmt3a and Dnmt3b are expressed in almost a third of all LK cells, but in less than 1% of mature myeloid and monocytic cells (PMN and Mono; Figures 5C–5E). Although Tet1 expression follows the same pattern, Tet2 and 3 gene expressions persist in the mature populations, suggesting they may play a more active role in methylome remodeling during differentiation (Figures 5D and 5E). Combined, these data suggest that the influence of Dnmt3a on the methylome is largely restricted to HSPCs; because these cells divide slowly and comprise only a tiny fraction of the bone marrow, this may help to explain, at least in part, the slow rate of methylation loss detected in the whole bone marrow samples.
Discussion
In this report, we describe the kinetics of DNA methylation loss following the inactivation of Dnmt3a in hematopoietic cells in vivo. A canonical, ordered pattern of DNA methylation loss was observed in bone marrow cells following Dnmt3a floxing, as early as one-week after a 3 weeks treatment course of Tamoxifen, and with progressive loss over the following 35 weeks. Even at the final time point, mean levels of methylation did not reach half that of DMRs defined in the bone marrow cells of Dnmt3a knockout mice. This contrasts to the speed of methylation gain following re-expression of physiologic levels of DNMT3A in constitutive null bone marrow cells, where a significant fraction of DMRs are remethylated within a week of initiating addback (Ketkar et al., 2020). Further, 94% of knockout DMRs are remethylated (to within 20% of Dnmt3a-WT values) by week 24 of addback. The contrasting kinetics of Dnmt3a dependent methylation loss and gain were highlighted by assessing dynamic DMRs (i.e., those DMRs that had a change in beta values of ≥ 0.2 from their baselines at a specific timepoint). Rapidly changing, dynamic DMRs with takeaway or addback had significant overlap, suggesting that Dnmt3a is active at very specific genomic sites. Taken together, these data suggest that the loss of DNA methylation following somatic Dnmt3a inactivation in hematopoietic cells is canonical and slow, and may take years to develop fully.
Several studies have evaluated aspects of DNA methylation changes after Dnmt3a inactivation in vivo (Challen et al., 2011, 2014; Jeong et al., 2018). For example, a study of serial transplantation of hematopoietic stem cells from young Dnmt3a flox/flox mice with Mx1-Cre (treated with polyI:polyC), showed that significant methylation loss (i.e., 5,000 DMRs) could be detected after three serial passages in vivo (over a period of about 4 months, faster than the timeframe of this study), which progressed with additional rounds of serial transplantation over a period of nearly two years (Jeong et al., 2018). The rate of methylation loss in that study may have been accelerated by serial transplantation, which places HSCs under proliferative stress, as they undergo repeated rounds of enforced repopulation of the hematopoietic system. In addition, Dnmt3a deficient HSCs have a competitive advantage in this model system, which may accelerate the apparent rate of methylation loss. Based on that study and our observations, we suggest that a high rate of HSC division may accelerate ‘passive’ methylation loss in the absence of Dnmt3a, as a consequence of extra HSC cell divisions. In contrast, the model used in the current study more closely resembles the events expected to transpire after somatic DNMT3A mutations occur in the HSPCs of humans, because both may have steady state hematopoiesis at the time that the mutation occurs (Ketkar et al., 2020; Smith et al., 2021). Indeed, patients with clonal hematopoiesis, and those with germline mutations in DNMT3A (DNMT3A Overgrowth Syndrome) can have relatively unperturbed blood cell counts and hematopoiesis for many years (or even decades) before transformation (Smith et al., 2021; Jaiswal et al., 2014).
Although the factors that contribute to the rate of DNA methylation loss after somatic inactivation of Dnmt3a in steady state hematopoiesis are unclear, they are almost certainly influenced by the relative activities of DNA methyltransferases and demethylases during different stages of hematopoietic development. Maintenance of the methylome involves a network of DNA methyltransferases (DNMT1, DNMT3A, and DNMT3B) that add methylation marks to specific CpG residues in the genome, and a set of enzymes that act to demethylate CpGs (TET1, TET2, TET3). The discovery of the TET enzymes has implied that DNA demethylation is an active, regulated process that shapes DNA methylation patterns in the genome. The slow rate of methylation loss presented here suggests that Tet-mediated demethylation may predominantly occur in genomic regions that are not methylated by Dnmt3a in hematopoietic cells. This may be related to the role of Tet-mediated demethylation in the commitment and differentiation of myeloid cells (Ostrander et al., 2020). Izzo et al. (2020) used conditional knockout models to identify the opposing roles of Tet2 and Dnmt3a in myelomonocytic and erythroid lineage differentiation, respectively, which were related to CpG enrichment in the DNA binding motifs of cell fate-determining transcription factors (Izzo et al., 2020). Further, we have shown that mice that are constitutively null for Dnmt3a display an expansion of myeloid cells over time, at the expense of the lymphoid lineage (Ketkar et al., 2020). This myeloid expansion was directly linked to Dnmt3a deficiency and focal hypomethylation, because DNMT3A addback restored methylation and reduced myeloid skewing and restored lymphoid populations (Ketkar et al., 2020). Further, we suggest that it is probably the activity of demethylases, not methyltransferases that define the unique methylomes of the mature myeloid compartment. The DMRs defined in Dnmt3a null mice vs. myeloid-specific DMRs are nearly mutually exclusive; this supports the idea that remodeling that drives differentiation is not occurring in the same sites where Dnmt3a is active. In contrast, lymphoid lineages have more differentiation-dependent DMRs that overlap with Dnmt3a DMRs. Tadokoro et al. likewise showed that Dnmt3a expression is high in lymphoid cells from the spleen and thymus (sites of B-cell and T-cell maturation), compared to mature myeloid cells from bone marrow (Tadokoro et al., 2007). Further, conditional knockout of Dnmt3a in Lineage-/Kit-/Sca1-/CD34- cells from Dnmt3aflox/flox mice, did not impair peripheral blood chimerism, but led to reductions in B- and T-cell chimerism compared to non-floxed controls. Again, this suggests that expression of Dnmt3a and maintenance of Dnmt3a-dependent methylation is required for the differentiation of lymphoid cells (Tadokoro et al., 2007). Finally, the conditional knockout of both Dnmt3a and Dnmt3b was shown to impair B-cell differentiation, which is mediated at least in part by an increase in ß-catenin signaling (Challen et al., 2014). The competition between the methyltransferase and demethylase enzymes, and their expression levels at different stages of development, probably play a major role in methylome remodeling during hematopoietic development.
Additional studies suggest that Dnmt3b can act in the stead of Dnmt3a after it is deleted, perhaps slowing methylation loss, but not preventing it entirely. In support of this idea, a recent study performed in human embryonic stem cells showed that differentially methylated regions (defined by DNMT3A/DNMT3B double knockouts) retained 93% of WT methylation values when only one of the DNMT3 genes was deleted. Further, when TET knockout preceded DNMT3A /DNMT3B double knockout, methylation levels did not progressively fall, even after 20 passages (Charlton et al., 2020). It remains to be determined whether the same rules apply to hematopoietic stem cells, lineage committed progenitors, and terminally differentiated cells. Although a ‘methyltransferase-dead’ splicing variant of Dnmt3b (Dnmt3b3) is the most abundant in mouse (and human) hematopoietic cells, some Dnmt3b1 is expressed and is active, as manifest by the more severe methylation phenotype of Dnmt3a × Dnmt3b double knockout mice (Challen et al., 2014). Further, recent evidence suggests that Dnmt3b3 can directly interact with Dnmt3a to increase its activity (Xu et al., 2020; Zeng et al., 2020). Combined, these data suggest that the methyltransferase activities of DNMT3A and B are somewhat redundant, and that one can maintain methylation in the absence of the other.
Somatic mutations in DNMT3A are common in acute myeloid leukemia and associated with a focal, canonical hypomethylation phenotype (Spencer et al., 2017). Recent data from patients with germline mutations in DNMT3A (DNMT3A Overgrowth Syndrome) have shown that the hypomethylation phenotype develops in the hematopoietic cells in very young patients with relatively normal blood counts (Smith et al., 2021). However, studies from patients with clonal hematopoiesis show that there is an increased risk of malignancy in these patient cohorts (Genovese et al., 2014; Jaiswal et al., 2014; Xie et al., 2014), and emerging data suggests the same is true for DNMT3A Overgrowth Syndrome patients (Smith et al., 2021). Although previous studies have suggested that the hypomethylation phenotype may be in part responsible for the increased fitness of these cells for transformation, the connection between the mutational event and overt disease has been difficult to understand because of the long latency of the premalignant state. The data from this study suggests that the slow methylation loss after Dnmt3a inactivation may be another contributing factor to long latency in clonal hematopoiesis patients, and that interventions designed to reactivate or restore DNMT3A activity during this latent period may be therapeutically attractive.
Limitations of the study
All of the experiments of this study were performed in mice; hence, the kinetics of DNA methylation loss in human hematopoietic cells that acquire loss-of-function mutations in DNMT3A can only be estimated from these data. Because we measured DNA methylation loss in single animals at different time points after Dnmt3a inactivation, it was not possible to define statistical differences among DMRs at individual time points in the experiment; only trends could be discerned. The model system used (homozygous inactivation of Dnmt3a) was chosen to accentuate methylation findings; in contrast, human AML patients almost always have heterozygous DNMT3A inactivating mutations, or dominant negative mutations (at amino acid R882) that reduce DNMT3A activity by ∼80%. Finally, we currently have a limited understanding of the factors that influence DNA methylation remodeling during hematopoietic development, and how these factors may contribute to patterns of methylation loss after somatic inactivation of DNMT3A in humans.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD11b (MI/70) | Becton Dickinson | 565080; RRID: AB_2722548 |
| Gr1 (TB6-8C5) | Biolegend | 108456; RRID: AB_2616737 |
| Ter-119 (Ter119) | Becton Dickinson | 740875; RRID: AB_2740526 |
| CD71 (C2) | Becton Dickinson | 563013; RRID: AB_2737950 |
| B220 (RA3-6B2) | Becton Dickinson | 612950; RRID: AB_2870227 |
| CD3e (145-2C11) | Becton Dickinson | 564618; RRID: AB_2738868 |
| Sca-1 (D7) | Biolegend | 108138; RRID: AB_2564042 |
| c-KIT (2B8) | Becton Dickinson | 562609; RRID: AB_11154585 |
| CD34 (RAM34) | Becton Dickinson | 553733; RRID: AB_395017 |
| FLT3 (A2F10) | Biolegend | 135306; RRID: AB_1877217 |
| CD150 (TC15-12F12.2) | Biolegend | 115941; RRID: AB_2629660 |
| CD48 (HM48-1) | Biolegend | 103420; RRID: AB_1089037 |
| CD45.1 (A20) | Becton Dickinson | 563983; RRID: AB_2738523 |
| CD45.2 (104) | Becton Dickinson | 740131; RRID: AB_2739888 |
| Chemicals, peptides, and recombinant proteins | ||
| Ammonium Chloride/KCl (ACK) red cell lysis buffer | Sigma | A9434, P9541 |
| SYBR green | Thermo Fisher | K0221 |
| RPMI media | Gibco | 11875034 |
| 15% FBS | Atlanta Biologicals | S11550H |
| Tamoxifen | Sigma | T5648 |
| Critical commercial assays | ||
| Chromium Single Cell 5′ Kits | 10x Genomics | #1000286, #1000263, #1000215 |
| Accel NGS Methyl-Seq DNA library kit | Swift Biosciences | #30096 |
| Deposited data | ||
| Aligned Sequence Data | This paper | NCBI: PRJNA797693 |
| Experimental models: Organisms/strains | ||
| C57Bl/6-CD45.1 (NCI B6-Ly5.1/Cr) | Charles River Laboratories. | 564 |
| C57Bl/6-CD45.2 Dnmt3afl/fl -ERTM-cre mice (B6.Cg-Tg(CAG-cre/Esr1∗)5Amc/J | Jackson Labs | 004682 |
| Oligonucleotides | ||
| Actin F1 primer: 5’-CGGGCTGTATTCCCCTCCATCG-3’ | this paper | N/A |
| Actin R1 primer: 5’-GCCATGTTCAATGGGGTACTTCAGGG-3’ | this paper | N/A |
| Dnmt3a F1 primer: 5′-CGGTCATTCCAGATGATTCCTC-3′ | this paper | N/A |
| Dnmt3a R1 primer: 5′-TGCTGTGGATGT-AGGAAAGCTG-3′ | this paper | N/A |
| Software and algorithms | ||
| WGBS analysis pipeline | https://github.com/genome/analysis-workflows/blob/master/definitions/pipelines/ | bisulfite.cwl |
| RNA-seq analysis pipeline | https://github.com/genome/analysis-workflows/blob/master/definitions/pipelines/ | rnaseq.cwl |
| Metilene | Juhling et al., 2016 | v0.2.6; quay.io/biocontainers/metilene:0.2.6--h470a237_1 |
| Partek Flow | Partek, Inc., | build 10.0.21.0411 |
| CellRanger | 10X Genomics | v3.1 |
| Winlist | Verity Software House | version 8 |
| Other | ||
| 10,000 ppm doxycycline chow | Gateway Lab Supply | 1815461-203 |
| EDTA capillary tubes | Sarstetdt | 20.1278.100 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Timothy J. Ley (timley@wustl.edu)
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animal procedures were approved by the Washington University Institutional Animal Care and Use Committee (IACUC) and conducted in accordance with all institutional guidelines.
Dnmt3afl/fl -ERTM-cre mice
Bone marrow from 6 week old male C57Bl/6-CD45.2 Dnmt3afl/fl -ERTM-cre mice (B6.Cg-Tg(CAG-cre/Esr1∗)5Amc/J Jackson Labs 004682) was transplanted into lethally irradiated male C57Bl/6-CD45.1 recipients, and allowed to engraft for 4 weeks. Engraftment was assessed 4 weeks post-transplant by flow cytometric determination of the ratio of CD45.1 (recipient; clone A20; Becton Dickinson) to CD45.2 (donor; clone 104, Becton Dickinson) in the peripheral blood of recipient mice. Peripheral blood was obtained from the retroorbital space with EDTA capillary tubes (Fisher Scientific) after adequate anesthesia with 2-Chloro-2-(difluoromethoxy)-1,1,1-trifluoroethane. Engraftment frequencies of >80% were detected in virtually all mice. Inactivation of Dnmt3a in mice transplanted with C57Bl/6-CD45.2 Dnmt3afl/fl -ERTM-cre bone marrow was induced with 9 doses of Tamoxifen (4 mg per dose per mouse in corn oil; Sigma) given by gavage on Monday, Wednesday, and Friday of each week for three consecutive weeks.
Dnmt3a-/- x rtTA x WT DNMT3A mice
Bone marrow from Dnmt3a-/- mice containing a tetracycline—inducible, WT human DNMT3A1 cDNA (Dnmt3a-/- x rtTA x WT DNMT3A as previously described (Ketkar et al., 2020)) was transplanted into lethally irradiated C57Bl/6-CD45.1 recipients and allowed to engraft for 4 weeks. Engraftment was assessed as outlined above. Fully engrafted recipients were then fed 10,000 ppm doxycycline chow (Gateway Lab Supply; St. Louis, MO) to induce DNMT3A transgene expression.
Method details
Collection and processing of whole bone marrow
Femur, tibia, and pelvis-derived bone marrow cells were collected from mice in RPMI media (Gibco) with 15% FBS (Atlanta Biologicals). Bone marrow and peripheral blood were treated with Ammonium Chloride/KCl (ACK) red cell lysis buffer and resuspended in the RPMI/FBS mixture as a working solution. Lethal irradiation was performed by exposing mice to two doses of 550 rads four hours apart. CD45.1 recipients were purchased from Charles River Laboratories.
Floxing efficiency assay
PCR was performed using a 40-cycle program of 20 s at 94°C, 20 s at 65°C, and 30 s at 72°C per cycle. For Dnmt3a amplification, F1 primer (5′-CGGTCATTCCAGATGATTCCTC-3′) and R1 primer (5′-TGCTGTGGATGT-AGGAAAGCTG-3′) were used. For Actin amplification F1 primer (5’-CGGGCTGTATTCCCCTCCATCG-3’) and R1 primer (5’-GCCATGTTCAATGGGGTACTTCAGGG-3’) were used. cDNAs were normalized with Actin copy numbers calculated based on quantitative PCR using SYBR green. Floxing efficiency in whole bone marrow was calculated using a standard curve where 100% floxing was defined as floxing in germline Dnmt3afl/fl x (CMV-Cre) mice and 0% floxing was defined as floxing in Dnmt3afl/fl x Cre negative samples.
Whole-genome bisulfite sequencing
WGBS was performed as described in Smith et al. (Smith et al., 2021), using the AccelNGS Methyl-Seq DNA library kit (Swift Biosciences, #30096). Sequence was generated on Illumina HiSeq or NovaSeq instruments and reads were mapped with biscuit (version 0.3.8), and DMRs were called using metilene (Juhling et al., 2016) as previously described (Cole et al., 2017; Spencer et al., 2017; Ketkar et al., 2020). The workflow for WGBS data is described in detail at https://github.com/genome/analysis-workflows/blob/master/definitions/pipelines/bisulfite.cwl (commit id: 174f3b2).
FACS sorting of cell populations
Bone marrow cells were isolated from femurs and tibias and treated with ACK red cell lysis buffer. Cells were stained with combinations of the following antibodies against cell-surface markers to identify indicated cell types (all antibodies are from Becton Dickinson unless indicated; clone indicated in parentheses): CD11b (MI/70), Gr1 (TB6-8C5; Biolegend), Ter-119 (Ter119), CD71 (C2), B220 (RA3-6B2), CD3e (145-2C11), Sca-1 (D7; Biolegend), c-KIT (2B8), CD34 (RAM34), FLT3 (A2F10; Biolegend), CD150 (TC15-12F12.2; Biolegend), CD48 (HM48-1; Biolegend), CD45.1 (A20), CD45.2 (104). Bone marrow and LinKit donor cells were isolated from transplanted mice using CD45.1-/CD45.2+ gating prior to selection for immunophenotypically defined populations. The following phenotypes were used to define stem and progenitor populations for sorting and Dnmt3a flow cytometry: Lineage (Lin): CD11b+, Gr1+, Ter119+, CD71+, B220+, CD3e+; LSK: Lin-, Sca-1+, c-KIT+; GMP: Lin-, Sca-1+, c-KIT+, CD34+, FLT3+; CMP: Lin-, Sca-1+, c-KIT+, CD34+, FLT3-; MEP: Lin-, Sca-1+, c-KIT+, CD34-, FLT3-. Sorting was performed on a modified Sony Synergy SY3200 (Sony Biotechnology, San Jose, CA) updated to 24 parameters. Winlist version 8 software was used for data acquisition and analysis (Verity Software House, Topsham, ME).
Single cell RNA sequencing
Single-cell RNA libraries were created using the 10x Genomics Chromium Single Cell 5′ Kit and aligned with CellRanger (version 3.1). Using a nearest neighbor algorithm implemented in R (version 3.5.1), cells were annotated according to the Haemopedia expression atlas of hematopoietic cell types (de Graaf et al., 2016) using code available here at https://github.com/genome/docker-scrna_lineage_inference. The characteristics of each sample are summarized in (Ketkar et al., 2020). Using Partek Flow software build 10.0.21.0411, we eliminated the analysis of cells that contained fewer than 250 expressed genes, less than 500 total reads, or more than 10% mitochondrial transcripts. For each cell, expression of each gene was normalized to the sequencing depth of the cell, scaled to a constant depth (10,000), and log-transformed. Dimensionality reduction and visualization were performed with the t-SNE algorithm. Full details and parameters used are described in (Ketkar et al., 2020).
Quantification and statistical analysis
Quantification was performed as described in the relevant method details sections above. Statistics were performed as described above using metilene (v0.2.6), bsseq (v1.18.0) Partek flow software (build 10.0.21.0411), R (v4.1.0) and GraphPad Prism (v9.3.1). Standard statistical tests used are described in the results and figure legends.
Acknowledgments
The authors thank Ms. Mieke Hoock for excellent animal husbandry. This work was supported by an ASH Fellow to Faculty Award (A.M.S.), NIH grants CA237727 (D.Y.C.), CA211782 (C.A.M.), CA101937 and CA197561 (T.J.L.), and the Barnes-Jewish Hospital Foundation (T.J.L.).
Author contributions
A.M.S., A.M.V., and T.J.L. designed research. A.M.S., A.M.V., D.Y.C., and E.R.L. performed research. A.M.S., H.J.A., S.K., and C.A.M., analyzed the data. A.M.S. and T.J.L. wrote the paper.
Declaration of interests
A.M.S. is an employee of Incyte Corporation, A.M.V. is an employee of Vitalant Corporation, and E.R.L. is the owner of Leight Medical Communications, LLC. All studies in the manuscript were performed before the authors joined these companies. The other authors declare no competing interests.
Published: April 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104004.
Supplemental information
Data and code availability
-
•
Sequencing data for all mouse datasets were deposited to the NCBI: NCBI BioProject PRJNA797693 (https://www.ncbi.nlm.nih.gov/bioproject/797693), and are available without restrictions. Data from previously published work from our group that are included in this study were deposited to the NCBI, https://www.ncbi.nlm.nih.gov/bioproject/483874 (BioProject ID PRJNA483874).
-
•
All pipeline code used for genomic data processing is available at https://github.com/genome/analysis-workflows (commit id: 174f3b2). The alignment and expression quantification workflow is described in pipelines/rnaseq.cwl, and bisulfite alignment and methylation inference is described in pipelines/bisulfite.cwl. Haemopedia expression atlas cell lineage assignment code is available at https://github.com/genome/docker-scrna_lineage_inference.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Challen G.A., Sun D., Jeong M., Luo M., Jelinek J., Berg J.S., Bock C., Vasanthakumar A., Gu H., Xi Y., et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 2011;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen G.A., Sun D., Mayle A., Jeong M., Luo M., Rodriguez B., Mallaney C., Celik H., Yang L., Xia Z., et al. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell. 2014;15:350–364. doi: 10.1016/j.stem.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton J., Jung E.J., Mattei A.L., Bailly N., Liao J., Martin E.J., Giesselmann P., Brandl B., Stamenova E.K., Muller F.J., et al. TETs compete with DNMT3 activity in pluripotent cells at thousands of methylated somatic enhancers. Nat. Genet. 2020;52:819–827. doi: 10.1038/s41588-020-0639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C.B., Russler-Germain D.A., Ketkar S., Verdoni A.M., Smith A.M., Bangert C.V., Helton N.M., Guo M., Klco J.M., O'laughlin S., et al. Haploinsufficiency for DNA methyltransferase 3A predisposes hematopoietic cells to myeloid malignancies. J. Clin. Invest. 2017;127:3657–3674. doi: 10.1172/JCI93041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf C.A., Choi J., Baldwin T.M., Bolden J.E., Fairfax K.A., Robinson A.J., Biben C., Morgan C., Ramsay K., Ng A.P., et al. Haemopedia: an expression atlas of murine hematopoietic cells. Stem Cell Rep. 2016;7:571–582. doi: 10.1016/j.stemcr.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farlik M., Sheffield N.C., Nuzzo A., Datlinger P., Schonegger A., Klughammer J., Bock C. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 2015;10:1386–1397. doi: 10.1016/j.celrep.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farlik M., Halbritter F., Muller F., Choudry F.A., Ebert P., Klughammer J., Farrow S., Santoro A., Ciaurro V., Mathur A., et al. DNA methylation dynamics of human hematopoietic stem cell differentiation. Cell Stem Cell. 2016;19:808–822. doi: 10.1016/j.stem.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidzik V.I., Schlenk R.F., Paschka P., Stolzle A., Spath D., Kuendgen A., Von Lilienfeld-Toal M., Brugger W., Derigs H.G., Kremers S., et al. Clinical impact of DNMT3A mutations in younger adult patients with acute myeloid leukemia: results of the AML Study Group (AMLSG) Blood. 2013;121:4769–4777. doi: 10.1182/blood-2012-10-461624. [DOI] [PubMed] [Google Scholar]
- Gale R.E., Lamb K., Allen C., El-Sharkawi D., Stowe C., Jenkinson S., Tinsley S., Dickson G., Burnett A.K., Hills R.K., Linch D.C. Simpson's paradox and the impact of different DNMT3A mutations on outcome in younger adults with acute myeloid leukemia. J. Clin. Oncol. 2015;33:2072–2083. doi: 10.1200/JCO.2014.59.2022. [DOI] [PubMed] [Google Scholar]
- Genovese G., Kahler A.K., Handsaker R.E., Lindberg J., Rose S.A., Bakhoum S.F., Chambert K., Mick E., Neale B.M., Fromer M., et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo F., Lee S.C., Poran A., Chaligne R., Gaiti F., Gross B., Murali R.R., Deochand S.D., Ang C., Jones P.W., et al. DNA methylation disruption reshapes the hematopoietic differentiation landscape. Nat. Genet. 2020;52:378–387. doi: 10.1038/s41588-020-0595-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S., Fontanillas P., Flannick J., Manning A., Grauman P.V., Mar B.G., Lindsley R.C., Mermel C.H., Burtt N., Chavez A., et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong M., Park H.J., Celik H., Ostrander E.L., Reyes J.M., Guzman A., Rodriguez B., Lei Y., Lee Y., Ding L., et al. Loss of Dnmt3a immortalizes hematopoietic stem cells in vivo. Cell Rep. 2018;23:1–10. doi: 10.1016/j.celrep.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhling F., Kretzmer H., Bernhart S.H., Otto C., Stadler P.F., Hoffmann S. metilene: fast and sensitive calling of differentially methylated regions from bisulfite sequencing data. Genome Res. 2016;26:256–262. doi: 10.1101/gr.196394.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M., Okano M., Hata K., Sado T., Tsujimoto N., Li E., Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- Ketkar S., Verdoni A.M., Smith A.M., Bangert C.V., Leight E.R., Chen D.Y., Brune M.K., Helton N.M., Hoock M., George D.R., et al. Remethylation of Dnmt3a (−/−) hematopoietic cells is associated with partial correction of gene dysregulation and reduced myeloid skewing. Proc. Natl. Acad. Sci. U S A. 2020;117:3123–3134. doi: 10.1073/pnas.1918611117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley T.J., Ding L., Walter M.J., Mclellan M.D., Lamprecht T., Larson D.E., Kandoth C., Payton J.E., Baty J., Welch J., et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G., Metzeler K.H., Schwind S., Becker H., Maharry K., Mrozek K., Radmacher M.D., Kohlschmidt J., Nicolet D., Whitman S.P., et al. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. J. Clin. Oncol. 2012;30:742–750. doi: 10.1200/JCO.2011.39.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander E.L., Kramer A.C., Mallaney C., Celik H., Koh W.K., Fairchild J., Haussler E., Zhang C.R.C., Challen G.A. Divergent effects of Dnmt3a and Tet2 mutations on hematopoietic progenitor cell fitness. Stem Cell Rep. 2020;14:551–560. doi: 10.1016/j.stemcr.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renneville A., Boissel N., Nibourel O., Berthon C., Helevaut N., Gardin C., Cayuela J.M., Hayette S., Reman O., Contentin N., et al. Prognostic significance of DNA methyltransferase 3A mutations in cytogenetically normal acute myeloid leukemia: a study by the Acute Leukemia French Association. Leukemia. 2012;26:1247–1254. doi: 10.1038/leu.2011.382. [DOI] [PubMed] [Google Scholar]
- Russler-Germain D.A., Spencer D.H., Young M.A., Lamprecht T.L., Miller C.A., Fulton R., Meyer M.R., Erdmann-Gilmore P., Townsend R.R., Wilson R.K., Ley T.J. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014;25:442–454. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.M., Lavalle T.A., Shinawi M., Ramakrishnan S.M., Abel H.J., Hill C.A., Kirkland N.M., Rettig M.P., Helton N.M., Heath S.E., et al. Functional and epigenetic phenotypes of humans and mice with DNMT3A overgrowth syndrome. Nat. Commun. 2021;12:4549. doi: 10.1038/s41467-021-24800-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer D.H., Russler-Germain D.A., Ketkar S., Helton N.M., Lamprecht T.L., Fulton R.S., Fronick C.C., O'laughlin M., Heath S.E., Shinawi M., et al. CpG island hypermethylation mediated by DNMT3A is a consequence of AML progression. Cell. 2017;168:801–816.e13. doi: 10.1016/j.cell.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro Y., Ema H., Okano M., Li E., Nakauchi H. De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. J. Exp. Med. 2007;204:715–722. doi: 10.1084/jem.20060750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M., Lu C., Wang J., Mclellan M.D., Johnson K.J., Wendl M.C., Mcmichael J.F., Schmidt H.K., Yellapantula V., Miller C.A., et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T.H., Liu M., Zhou X.E., Liang G., Zhao G., Xu H.E., Melcher K., Jones P.A. Structure of nucleosome-bound DNA methyltransferases DNMT3A and DNMT3B. Nature. 2020;586:151–155. doi: 10.1038/s41586-020-2747-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Ren R., Kaur G., Hardikar S., Ying Z., Babcock L., Gupta E., Zhang X., Chen T., Cheng X. The inactive Dnmt3b3 isoform preferentially enhances Dnmt3b-mediated DNA methylation. Genes Dev. 2020;34:1546–1558. doi: 10.1101/gad.341925.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Sequencing data for all mouse datasets were deposited to the NCBI: NCBI BioProject PRJNA797693 (https://www.ncbi.nlm.nih.gov/bioproject/797693), and are available without restrictions. Data from previously published work from our group that are included in this study were deposited to the NCBI, https://www.ncbi.nlm.nih.gov/bioproject/483874 (BioProject ID PRJNA483874).
-
•
All pipeline code used for genomic data processing is available at https://github.com/genome/analysis-workflows (commit id: 174f3b2). The alignment and expression quantification workflow is described in pipelines/rnaseq.cwl, and bisulfite alignment and methylation inference is described in pipelines/bisulfite.cwl. Haemopedia expression atlas cell lineage assignment code is available at https://github.com/genome/docker-scrna_lineage_inference.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.