Summary
Intestinal intraepithelial lymphocytes (IELs), the first line of defense against microbial and dietary antigens, are classified as natural or induced based on their origin and receptor expression. Induced CD4+CD8αα+TCRβ+ T cells (double positive, DPIELs) originated from CD4+CD8α−TCRβ+ T cells (single positive, SPIELs) increase with aging. However, the metabolic requirements and the metabolic-related genes in IEL development remain unclear. We determined that the intraepithelial compartment is hypoxic in the presence of microbes and DPIELs increased more than natural IELs in this location. Moreover, DPIELs consumed less oxygen and glucose and exhibited unique alterations in mitochondria. Using inhibitors and genetically modified mice, we revealed that DPIELs adapt to their surrounding oxygen-deprived environment in peripheral tissues by modulating specific genes, including hypoxia-inducible factor, mammalian target of rapamycin complexes (mTORC), phosphorylated ribosomal protein S6 (pS6), and other glycolytic factors. Our findings provide valuable insight into the metabolic properties of IELs.
Subject areas: Biological sciences, Immunology, Components of the immune system, Cell biology
Graphical abstract

Highlights
-
•
Microbes induce hypoxic conditions in the intraepithelial compartment
-
•
Induced IELs show a lower OCR, glucose uptake, and mitochondrial membrane potential
-
•
DPIELs show reduced expression of Hif1α/Hif2α during development
-
•
Downregulation of Rptor and Hif1α/Hif2α expression in CD4+ T cells induces DPIELs
Biological sciencesImmunologyComponents of the immune systemCell biology
Introduction
The intestinal mucosal barrier, which has a poor blood supply, is formed by a dense mucus layer, organized epithelial cells, and intraepithelial lymphocytes (IELs) (Cheroutre et al., 2011). IELs are located in the intraepithelial compartment and are the first line of defense against bacterial or food antigens. IELs are classified as natural IELs and induced IELs. Natural IELs, including TCRγδ+ cells and CD8αα+ T cells, develop in the thymus, and induced IELs, such as CD8αβ+TCRβ+ cells and CD4+CD8αα+TCRβ+ T cells (double positive cells, DPIELs), develop in the periphery (Olivares-Villagomez and Van Kaer, 2018; Van Kaer and Olivares-Villagomez, 2018). DPIELs originate from CD4+CD8α−TCRβ+ T cells (single positive IELs, SPIELs) and accumulate with aging (Konkel et al., 2011). DPIELs express both CD4 and CD8αα via the downregulation of T helper POZ/Kruppel like factor (Thpok also known as Zbtb7b) and upregulation of Runt-related transcription factor (Runx3) during their development (Bilate et al., 2016; Cervantes-Barragan et al., 2017; Sujino et al., 2016). IELs express activation markers such as CD44 and CD69 (Cheroutre et al., 2011), also they express enzymes and cytokines, such as granzyme B and interleukin (IL)-17A, depending on the cell type. They proliferate less compared with other T cells, and originate from long-lived tissue-resident effector memory cells (Groux et al., 1997; Konjar et al., 2018; Mucida et al., 2013; Reis et al., 2013; Sydora et al., 1993; Vandereyken et al., 2020; Yu et al., 2008).
Metabolic reprogramming is essential for T cell activation and function. The activation of naïve CD4+ T cells in lymphoid tissues leads to metabolic reprogramming from oxidative phosphorylation (OXPHOS) to anabolic metabolism. For instance, Foxp3-positive regulatory T cells (Tregs) preferentially use OXPHOS, and effector T cell subsets [T helper type (Th) 1), Th2, Th17, and CD8+ T cells] preferentially undergo glycolysis with lactate production (Jung et al., 2019; MacIver et al., 2013; Pearce and Pearce, 2013; Shi et al., 2011). The metabolic requirements of T cell subsets were previously studied using in vitro cultured cells or cells isolated from spleen and lymphoid tissues. In addition, cells alter their energy usage in response to environmental factors. Tregs mainly use OXPHOS in oxygen-rich conditions, whereas in glucose-deprived and oxygen-deprived environments, such as cancer or peripheral tissues, Tregs rely on lactate as an alternate energy source (Angelin et al., 2017; Newton et al., 2016). Hypoxia inducible factor 1 subunit alpha (Hif1α) is a key gene in the regulation of glycolysis and mitochondrial respiration in hypoxic tissue environments. Under hypoxic conditions, Tregs switch their metabolic status to glycolysis and upregulate Hif1α. This metabolic shift is essential for Tregs to migrate and survive in hypoxic tissues (Dodd et al., 2015; He et al., 2018). The oxygen and glucose consumption levels of natural and peripheral IELs and how metabolic-related genes affect the development of induced IELs remain unclear.
Here, we first show that the intraepithelial compartment is hypoxic in the presence of microbes. Induced IELs, DPIELs, and SPIELs showed a reduced oxygen consumption rate (OCR), glucose uptake, and extracellular acidification rate (ECAR). DPIELs exhibited an increased number of mitochondria compared with naive CD4+ T cells, and SPIELs and DPIELs showed enhanced expression of fission related-genes and a low mitochondrial membrane potential compared with naive CD4+ T cells. Although DPIELs showed reduced glucose uptake compared with naive CD4+ T cells, 2-deoxy-D-glucose (2DG), which blocks glucose uptake, inhibited the development of DPIELs. Under hypoxic conditions, cells expressed Hif1α/Hif2α to adapt to the tissue environment. Interestingly, DPIELs showed reduced expression levels of Hif1α/Hif2α and phosphorylated ribosomal protein S6 (pS6) during development. Genetic ablation of Rptor or Hif1α/Hif2α in CD4+ T cells increased the percentage of DPIELs. We uncovered that DPIELs develop as a unique population and adapt to low oxygen circumstances by decreasing oxygen and glucose consumption, but the expression levels of Hif1α/Hif2α and phosphorylation level of S6 are reduced during their development from SPIELs. Our findings indicate that peripheral tissue-resident cells develop by adapting to the environmental niche, such as hypoxic conditions, with alterations in metabolic-related gene expression.
Results
Intestinal intraepithelial compartment is hypoxic in SPF mice
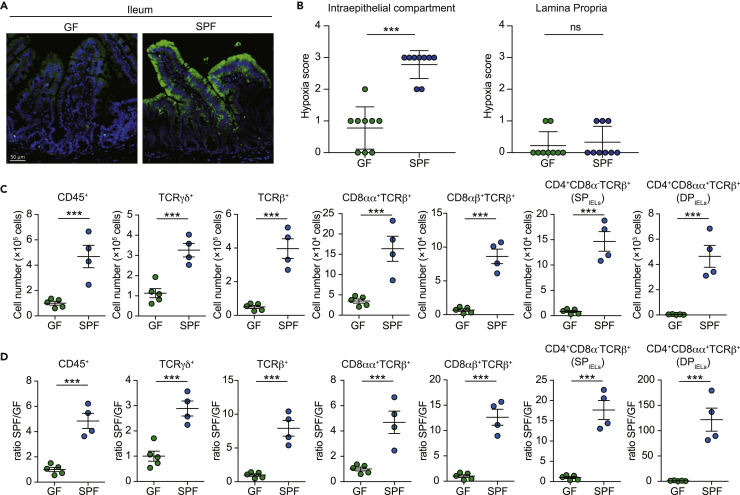
We first analyzed the intestinal oxygen supply in the presence of microbes. Although the oxygen concentration of the lamina propria compartment was equivalent in specific-pathogen-free (SPF) mice and germ-free (GF) mice, the oxygen concentration of the intraepithelial compartment was decreased in SPF mice compared with GF mice (Figures 1A and 1B). These data indicate that the intraepithelial compartment is deprived of oxygen in the presence of microbes. Intraepithelial cells develop and reside in a suitable location with low oxygenic conditions in SPF mice. Therefore, we questioned which cell types develop in the low oxygenic intraepithelial compartment in SPF mice. The numbers of cells in the intraepithelial compartment in GF mice and SPF mice were analyzed (Figure 1C). We divided the cell number in SPF mice and GF mice in each population. The numbers of natural IELs, including TCRγδ+ T cells and CD8αα+ TCRβ+ T cells, were increased by 3-fold to 5-fold in SPF mice compared with GF mice (Figure 1D). Furthermore, induced IELs, such as CD4+TCRβ+ T cells and CD8αβ+TCRβ+ T cells, were increased by 15-fold to 20-fold in SPF mice compared with GF mice (Figure 1D). CD4+TCRβ+ IELs are divided into two populations: terminally differentiated IELs that express CD4+CD8αα+TCRβ+ (DPIELs) and CD4+CD8αα−TCRβ+ cells (SPIELs). Significantly, DPIELs were increased by more than a thousand times in SPF mice compared with GF mice. These data indicate induced IELs, especially DPIELs, develop in low oxygenic conditions.
Figure 1.
Intestinal intraepithelial compartment is hypoxic in SPF mice
(A) Immunohistochemical visualization (green) of “physiological hypoxia” in intestinal mucosa of wild-type mice by Hypoxyprobe-1 staining. Mice were administered pimonidazole 30 min before sacrifice. PFA-fixed small intestine samples were sectioned and stained according to the manufacturer’s instructions and counterstained with DAPI.
(B) Epithelium and lamina propria regions were scored for intensity of hypoxia staining (0, no hypoxia; 1, mild focal hypoxia; 2, moderate multifocal hypoxia; 3, intense diffuse hypoxia). Each dot represents a hypoxia score of 9 individual images (n = 2 GF and n = 2 SPF).
(C and D) Percentage (C) and ratio between GF and SPF mice (D) of CD45+, TCRγδ+, TCRβ+, CD8αα+TCRβ+, CD8αβ+TCRβ+, CD4+CD8α−TCRβ+ (SPIELs), and CD4+CD8αα+TCRβ+ (DPIELs) cells in small intestine epithelium. Data are expressed as mean ± SD of individual mice (n = 5 GF and n = 4 SPF). Results represent one of two independent experiments. ns: not significant, ∗∗∗p < 0.001 (unpaired Student’s t test with Welch’s correction).
CD4+CD8αα+TCRβ+ IELs exhibit a reduced OCR and ECAR
We then determined the OCR of IELs in the intraepithelial compartment because CD4+TCRβ+ T cells, especially DPIELs, develop in low oxygen conditions. We measured the OCR in splenic naive CD4+ T cells, TCRγδ+ T cells, CD8αβ+ T cells, SPIELs, and DPIELs from small intestine epithelium. SPIELs and DPIELs showed a reduced OCR compared with naive CD4+ T cells and TCRγδ+ T cells. DPIELs displayed a slightly lower OCR compared with SPIELs, but the result was not significant (Figure 2A). CD8αβ+ T cells tended to show a higher OCR than SPIELs and DPIELs. We next measured glycolysis by determining the ECAR and glucose intake using 2-NBDG in each cell type. SPIELs and DPIELs showed a lower ECAR than naïve CD4+ T cells and TCRγδ+ T cells (Figure 2B). The ECAR in CD8αβ+ T cells was higher than in SPIELs and DPIELs, although not significantly. Glucose uptake in naïve CD4+ T cells was higher than in IEL cells. Among IELs, glucose uptake was increased in TCRγδ+ T cells compared with CD8αβ+ T cells, SPIELs, and DPIELs (Figure 2C). These data indicate that induced IELs exhibit reduced oxygen consumption, glycolysis, and glucose uptake.
Figure 2.
Induced CD4+CD8αα+TCRβ+ IELs exhibit a reduced OCR and ECAR
(A and B) Summaries of the oxygen consumption rate (OCR) (A) and extracellular acidification rate (ECAR) (B) of TCRγδ+, CD8αβ+TCRβ+, SPIELs (CD4+CD8α−TCRβ+), and DPIELs (CD4+CD8αα+TCRβ+) cells in small intestine epithelium and naive CD4+ (splenic naive CD4+ T) cells isolated from wild-type mice.
(C) Quantification of glucose uptake into TCRγδ+, CD8αβ+TCRβ+, SPIELs, and DPIELs cells in small intestine epithelium and naïve CD4+ T cells isolated from wild-type mice.
(D) Transmission electron microscopy (TEM) electron micrographs of sorted SPIELs, DPIELs cells in small intestine epithelium and naïve CD4+ T cells isolated from wild-type mice. Black arrows indicate each mitochondrion. Results represent two independent experiments. Scale bar, 0.5 μm.
(E and F) Quantitation of TEM micrographs of sorted SPIELs, DPIELs in small intestine epithelium and naïve CD4 T+ cells isolated from wild-type mice showing difference in the number (E) (n = 15 to 25) and size (F) (n = 40 to 118) of the mitochondria.
(G) The mRNA level of Drp1 and Fis1 were evaluated by RT-qPCR with sorted fresh SPIELs and DPIELs in small intestine epithelium and naïve CD4+ cells of wild-type mice (n = 4). The data were shown as fold change normalized to naïve CD4+ T cells. Results represent one of two independent experiments. ns: not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (one-way ANOVA with Tukey’s multiple comparisons).
Given that CD8+ IELs showed an increased number of mitochondria with altered lipid metabolism compared with naïve CD4+ T cells (Konjar et al., 2018), we next counted the number and size of mitochondria in naïve CD4+ T cells and induced IELs, SPIELs, and DPIELs. The total number of mitochondria was increased in SPIELs and DPIELs compared with naïve CD4+ T cells (Figures 2D and 2E). Interestingly, DPIELs displayed a higher number of mitochondria than SPIELs. Of note, the area of each mitochondrion in DPIELs was smaller than in naïve CD4+ T cells and SPIELs (Figures 2D and 2F). Mitochondrial fission controls the number and the size of mitochondria in memory T cells (Buck et al., 2016). To support our findings, we analyzed the mitochondrial fission-related genes dynamin-related protein 1 (Drp1) and mitochondria fission 1 gene (Fis1). The expression levels of Drp1 and Fis1 were increased in SPIELs and DPIELs compared with naïve CD4+ T cells (Figure 2G). The expression levels of Drp1 and Fis1 were comparable in DPIELs and SPIELs. These data indicate that SPIELs and DPIELs have a lower OCR with an increased number of small mitochondria, and mitochondrial fission genes are upregulated in SPIELs and DPIELs.
CD4+CD8αα+TCRβ+ IELs are a mitochondrial membrane potentiallo population
We then measured mitochondrial size and membrane potential in SPIELs and DPIELs using MitoTracker. Among CD4+ T cells in spleen and mesenteric lymph node (mLN) cells, CD4+CD62L+ cells were classified as mitochondria sizehi and membrane potentialhi (gated as Q1) populations, and CD4+CD44+ effector and memory cells were identified as mitochondria sizehi and membrane potentiallo (Q2) populations (Figure S1A). We analyzed the size and membrane potential of mitochondria in natural IELs and induced IELs from 12-week-old GF mice and SPF mice. Most TCRγδ+ T cells were mitochondria sizelo and membrane potentiallo (Q4) compared with naïve CD4+ T cells. Of note, the percentage of each fraction (Q1–4) in TCRγδ+ T cells was not significantly different between GF and SPF mice (Figures 3A and 3B). Next, we measured the mitochondrial size and membrane potential of CD4+ T cells in the intraepithelial (CD4IELs) and lamina propria (CD4LPLs) compartment. The percentage of Q1 and Q3 in CD4IELs of SPF mice was reduced compared with GF mice, whereas the percentage of Q1 and Q3 in CD4LPLs was comparable between SPF mice and GF mice (Figures 3C–3F). In CD4LPLs, the percentage of Q2 was slightly increased in GF mice than in SPF mice. These data indicate that CD4IELs but not CD4LPLs have a lower mitochondrial potential in SPF mice compared with GF mice. We then divided CD4IELs into two populations (SPIELs and DPIELs) because DPIELs were increased in SPF mice (Figures 1C and 1D). The percentage of Q1 and Q2 in DPIELs was decreased compared with that in SPIELs (Figures 3G and 3H). We also confirmed that DPIELs exhibited a reduced mitochondrial membrane potential compared with SPIELs by tetramethylrhodamine ethyl ester (TMRE) staining (Figures 3I and 3J). Taken together, induced IELs, especially DPIELs, are a population of cells with small mitochondria and low mitochondrial membrane potential.
Figure 3.
Induced CD4+CD8αα+TCRβ+ IELs are a mitochondrial membrane potentiallo population
(A and B) Flow cytometric analysis of mitochondrial membrane potential and size monitored by MitoTracker staining (A) and frequencies of Q1, Q2, Q3, and Q4 on TCRγδ+ cells (B) among CD45 + cells from small intestine epithelium of GF or SPF mice.
(C–F) Representative flow cytometric analysis and frequencies Q1, Q2, Q3, and Q4 of mitochondrial membrane potential and size monitored by MitoTracker staining of CD4IELs (C and D) and CD4LPLs (E and F) of wild-type mice reared under GF (n = 4) or SPF (n = 4) condition.
(G and H) Representative flow cytometric analysis (G) and frequencies Q1, Q2, Q3, and Q4 (H) of MitoTracker staining of SPIELs and DPIELs cells from 12W wild-type mice. n = 5.
(I and J) Representative flow cytometric analysis (I) and frequencies (J) of TMRE positive cells in SPIELs and DPIELs cells from small intestine epithelium of 12W wild-type mice. Data are composed of triplicate analysis from individual 2 mice. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (one-way ANOVA with Tukey’s multiple comparisons for B, D, F, and H; unpaired Student’s t test with Welch’s correction for J).
2DG inhibits the induction of CD4+CD8αα+TCRβ+ IELs
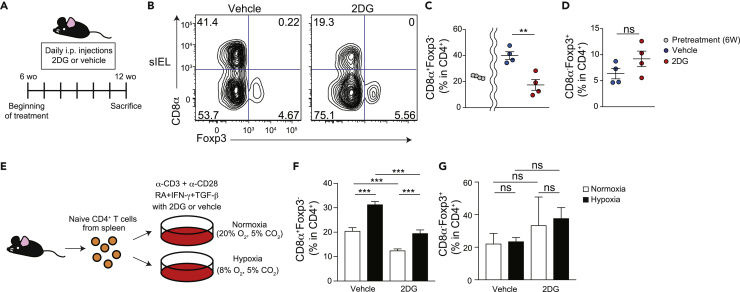
Because DPIELs consume less glucose than naïve CD4+ T cells (Figure 2C), we next investigated whether glucose uptake affects the development of DPIELs. We administered 6-week-old mice with or without 2DG via daily intraperitoneal injection for 5 weeks (Figure 4A) (Hamanaka and Chandel, 2012). 2DG administration did not reduce the percentage of TCRγδ+ T cells in CD45+ cells but reduced the percentage of TCRβ+ T cells in CD45+ cells which contained the DPIELs cells (Figure S2A). Among TCRβ+ T cells, the percentage of CD8α+CD8β− T cells was slightly increased in 2DG mice and the percentage of CD8αβ+ T cells was comparable between with or without 2DG administration. Of note, the percentage of total CD4+ T cells in TCRβ+ T cells was reduced in 2DG administration. The proportion of DPIELs among total CD4+ cells before 2DG treatment was approximately 20%. The percentage of DPIELs was ∼40% without 2DG treatment and decreased to ∼20% in 2DG treated mice (Figures 4B and 4C). Moreover, the percentage of DPIELs in total TCRβ+ cells and CD45+ cells was decreased with 2DG treatment (Figure S2B).
Figure 4.
2DG inhibits the induction of CD4+CD8αα+TCRβ+ IELs
(A) Schematic diagram showing the experimental design to investigate the effect of 2DG treatment on wild-type mice in vivo.
(B–D) Representative flow cytometric figures of surface CD8α and intracellular Foxp3 (B) and frequencies of CD8α+Foxp3- (C) and CD8α−Foxp3+ cells (D) among TCRβ+CD4+CD8β− T cells in small intestine epithelium from wild-type mice treated with or without 2DG via daily intraperitoneal injection for 5 weeks. Data are expressed as mean ± SD of individual mice (n = 4), representative of two independent experiments.
(E) Schematic diagram showing the experimental design to investigate the effect of 2DG treatment and hypoxic condition on development of naïve CD4+ T cells into CD4+CD8αα+ cells in vitro.
(F and G) Frequencies of CD8α+Foxp3- (F) and CD8α−Foxp3+ cells (G) among TCRβ+CD4+CD8β− cells after in vitro CD4+CD8αα+ cell induction in normoxic and hypoxic condition with or without 2DG treatment. Splenic naïve CD4+ T cells isolated from wild-type mice are cultured for 4 days with plate-bound α-CD3 and α-CD28 in the presence of TGF-β+RA + IFN-γ. ns: not significant, ∗∗p < 0.01, ∗∗∗p < 0.001 (unpaired Student’s t test with Welch’s correction).
These data indicate that 2DG inhibited the development of DPIELs. 2DG might affect Treg populations because some ex-Tregs differentiate into DPIELs. However, the percentages of Tregs were comparable between mice with and without 2DG (Figure 4D). These data indicate that 2DG inhibits the development of DPIELs without changing the Treg population. To exclude the possibility of other immune cells being involved, we then questioned the oxygen concentration and 2DG directly affecting the development of DPIELs. We cultured splenic naïve CD4+ T cells with or without 2DG in the presence of TGF-β, retinoic acid (RA), and IFN-γ (DPIEL condition) in vitro under normoxic (O2 20%) or hypoxic conditions (O2 8%) (Figures 4E–4G). As expected, the proportion of CD4+CD8αα+ T cells in total CD4+ T cells was increased under the hypoxic condition compared with normoxia. These data indicate the hypoxia induces CD4+CD8αα+ T cells in DPIEL condition in vitro. 2DG decreased the proportion of CD4+CD8αα+ T cells in CD4+ T cells when they cultured under both normoxic and hypoxic condition. Of note, 2DG tends to increase the proportion of Tregs in CD4+ T cells in DPIEL condition but not significantly under both normoxic and hypoxic conditions. These data indicate that the prevention of glucose uptake inhibits the development of DPIELs, and they consume less glucose than naïve CD4+ T cells.
Hif1α/Hif2α are reduced during the development of CD4+CD8αα+TCRβ+ IELs
The intestinal epithelium is an oxygen-poor environment with no blood vessels separating anaerobic intestinal bacteria from the intestinal epithelium. Hif1α and Hif2α are known to be upregulated under hypoxic conditions. We questioned whether the expression of HIF was upregulated in CD4IELs. We FACS sorted naïve CD4+ T cells, SPIELs, and DPIELs and analyzed the expression of HIF-related genes (Figure 5A). As previously reported, DPIELs expressed higher levels of Runx3 than SPIELs and naïve CD4+ T cells. Interestingly, the RNA expression levels of Hif1α and Hif2α in DPIELs were lower than in SPIELs. Von Hippel–Lindau (Vhl) is a negative regulator of HIF. The RNA expression level of Vhl was comparable between DPIELs and SPIELs. These data indicate that DPIELs exhibit reduced Hif1α/Hif2α expression during development.
Figure 5.
Hif/Vhl gene expression regulates the development of CD4+CD8αα+TCRβ+ IELs
(A) The mRNA level of Runx3, Hif1α, Hif2α and Vhl were evaluated by RT-qPCR with sorted fresh naïve CD4+ T cells from spleen and SPIELs, DPIELs cells from small intestine epithelium of wild-type mice (n = 4). The data were shown as fold change normalized to naïve CD4+ T cells.
(B and C) Representative flow cytometric analysis of surface CD8α and intracellular Foxp3 (B) and frequencies of CD8α+Foxp3- and CD8α−Foxp3+ cells (C) among TCRβ+CD4+CD8β− T cells in small intestine epithelium from Hif1αfl/flHif2αfl/fl and Cd4creHif1αfl/flHif2αfl/fl (HifΔCD4) mice. Data are expressed as mean ± SD of individual mice (n = 3 to 9), representative of two independent experiments.
(D and E) Representative flow cytometric analysis of surface CD8α and intracellular Foxp3 (D) and frequencies of CD8α+Foxp3- and CD8α−Foxp3+ cells (E) among TCRβ+CD4+CD8β− T cells in small intestine epithelium from Vhlfl/fl and Cd4creVhlfl/fl (VhlΔCD4) mice. Data are expressed as mean ± SD of individual mice (n = 3 to 4), representative of two independent experiments.
(F and G) Representative flow cytometric analysis of surface CD8α and intracellular Foxp3 (F) and frequencies of CD8α+Foxp3- and CD8α−Foxp3+ cells (G) among TCRβ+CD4+CD8β− T cells in small intestine epithelium from Hif1αfl/flHif2αfl/flVhlfl/fl and Cd4creHif1αfl/flHif2αfl/flVhlfl/fl (TKO) mice. Data are expressed as mean ± SD of individual mice (n = 4 to 7), representative of two independent experiments. ns: not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (one-way ANOVA with Tukey’s multiple comparisons for A; unpaired Student’s t test with Welch’s correction for C, E, and G).
Hif/Vhl gene expression regulates the development of CD4+CD8αα+TCRβ+ IELs
We next questioned whether the downregulation of Hif1α/Hif2α expression was essential for the development of DPIELs. We crossed Hif1αfl/flHif2αfl/fl mice with Cd4cre mice to generate mice lacking Hif1α and Hif2α in CD4+ T cells (HifΔCD4). The total number of IELs in HifΔCD4 mice was not increased compared with Hif1αfl/flHif2αfl/fl mice (Figure S3A). The percentage of DPIELs was increased by almost 2-fold in HifΔCD4 mice compared with Hif1αfl/flHif2αfl/fl mice, whereas the percentage of Tregs was decreased in HifΔCD4 mice compared with Hif1αfl/flHif2αfl/fl mice (Figures 5B and 5C). As HIF1α also regulates retinoic acid-related orphan receptor gamma t (RORγt) expression (Shi et al., 2011), we confirmed that the proportion of RORγt+Foxp3- IELs was decreased in HifΔCD4 mice compared with Hif1αfl/flHif2αfl/fl mice (Figures S3B and S3C). Among LPLs, the percentages of Tregs and RORγt+ Foxp3- T cells were comparable between HifΔCD4 mice and Hif1αfl/flHif2αfl/fl mice (Figures S3D and S3E). These data indicate that the downregulation of Hif1α/Hif2α expression in CD4 T cells induces DPIELs. Then, we crossed Vhlfl/fl mice with Cd4cre mice to generate Cd4cre:Vhlfl/fl (VhlΔCD4) mice and analyzed the cell populations at the age of 12 weeks. The percentage and total number of CD4+ T cells in the thymus were comparable in Vhlfl/fl mice and VhlΔCD4 mice (Figures S4A–S4C), as previously reported (Zhu et al., 2019). The total number of CD4+ T cells in IELs was also comparable in Vhlfl/fl mice and VhlΔCD4 mice (Figure S4D). As expected, the proportion of DPIELs but not Tregs was decreased in VhlΔCD4 mice compared with Vhlfl/fl mice (Figures 5D and 5E). We also found that the percentages of RORγt+Foxp3- IELs and LPLs were increased in VhlΔCD4 mice (Figures S4E–S4H). These data indicate that the downregulation of vhl expression in CD4+ T cells inhibits the development of DPIELs. Because Vhl regulates Hif1α/Hif2α and vice versa, we further investigated whether Hif1 or Vhl was important for the induction of DPIELs by crossing Vhlfl/fl/Hif1αfl/fl/Hif2αfl/fl mice with Cd4cre mice [referred to as triple KO (TKO) mice]. Interestingly, TKO mice showed an increased proportion of DPIELs compared with Vhlfl/fl/Hif1αfl/fl/Hif2αfl/fl mice, similar to HifΔCD4 mice (Figures 5F and 5G). Taken together, these data suggest that the downregulation of Hif1α/Hif2α expression in CD4+ T cells regulates the development of DPIELs.
CD4+CD8αα+TCRβ+ IELs show reduced phosphorylation level of S6 compared with CD4+CD8α−TCRβ+ IELs
As DPIELs exhibit reduced Hif1α/Hif2α expression during development, and the downregulation of Hif1α/Hif2α expression is important for the development of DPIELs, we hypothesized that the downregulation of Hif1α/Hif2α expression in CD4IELs is not only regulated by hypoxia but also by mTORC signaling, which controls Hif1α/Hif2α expression. mTORC exists as mTORC1 and mTORC2 complexes. Interestingly, although the expression of Hif1α/Hif2α was reduced in DPIELs compared with SPIELs, the expression of the mTORC1 gene Rptor in DPIELs was comparable to that in SPIELs (Figure 6A). Both DPIELs and SPIELs expressed higher levels of Rptor than splenic naive CD4+ T cells. We then analyzed the RNA expression of Rptor-related genes, including isoform 2 of the glycolytic enzyme hexokinase (Hk2) (enhances glucose metabolism) and arginase-2 (Arg2) (regulates L-arginine in the urea cycle). The expression of Hk2 in DPIELs was slightly but not significantly increased compared with SPIELs. The expression of Arg2 was comparable in DPIELs and SPIELs. The expression of Rictor was also comparable in DPIELs and SPIELs. In addition, mTORC regulates sterol regulatory binding protein (SREBP), which mediates fatty acid metabolism. The expression of Srebp1 and Srebp2 in DPIELs was higher than in naïve CD4+ T cells and SPIELs (Figure 6A). These data indicate that the expression level of mTORC in DPIELs was comparable to that in SPIELs. However, DPIELs displayed reduced Hif1α/Hif2α and increased Srebp1 and Srebp2 expression. Given that T cell receptor signaling induces the Akt/mTOR/pS6 pathway, we next analyzed pS6 in CD4IELs. DPIELs expressed lower phosphorylation levels of S6 than SPIELs (Figure 6B). Of note, CD4+CD8αα+ LPLs and CD4+CD8α− LPLs expressed similar levels of pS6. Our findings are consistent and show reduced mTORC/pS6 signaling in DPIELs.
Figure 6.
Downregulation of Rptor but not Rictor induces CD4+CD8αα+TCRβ+ IELs
(A) The mRNA level of Rptor, Hk2, Arg2, Rictor, Srebp1, and Srebp2 were evaluated by RT-qPCR with sorted fresh SPIELs and DPIELs in small intestine epithelium and naïve CD4+ cells of wild-type mice (n = 4). The data were shown as fold change normalized to naïve CD4+ T cells.
(B) Histogram demonstrating phosphorylation of S6 ribosomal protein in CD4+CD8α− and CD4+CD8αα+ within CD4+ cells from small intestine epithelium (CD4IELs) or lamina propria (CD4LPLs) compartment of wild-type mice. A representative of two independent experiments.
(C) Schematic diagram showing the experimental design to investigate the effect of Rapamycin treatment on wild-type mice in vivo.
(D and E) Representative flow cytometric figures of surface CD8α and intracellular Foxp3 (D) and frequencies of CD8α+Foxp3- and CD8α−Foxp3+ cells (E) among TCRβ+CD4+CD8β− T cells in small intestine epithelium from wild-type mice treated with or without Rapamycin via intraperitoneal injection for 5 weeks. Data are expressed as mean ± SD of individual mice (n = 3 to 9), representative of two independent experiments.
(F and G) Representative flow cytometric figures of surface CD8α and intracellular Foxp3 (F) and frequencies of CD8α+Foxp3- and CD8α−Foxp3+ cells (G) among TCRβ+CD4+CD8β− T cells in small intestine epithelium from Rptorfl/fl and Cd4creRptorfl/fl (iRaptorΔCD4) treated with tamoxifen via oral administration for 5 weeks. Data are expressed as mean ± SD of individual mice (n = 3 to 9), representative of two independent experiments.
(H and I) Representative flow cytometric figures of surface CD8α and intracellular Foxp3 (H) and frequencies of CD8α+Foxp3- and CD8α−Foxp3+ cells (I) among TCRβ+CD4+CD8β− T cells in small intestine epithelium from Rictorfl/fl and Cd4creRictorfl/fl (RictorΔCD4) mice. Data are expressed as mean ± SD of individual mice (n = 4), representative of two independent experiments.
(J and K) Representative flow cytometric figures of surface CD8α and intracellular Foxp3 (J) and frequencies of CD8α+Foxp3- (K) cells among TCRβ+CD4+CD8β− cells after in vitro CD4+CD8αα+ cell induction with or without Torin 1 treatment. Naïve CD4+ T cells isolated from RictorΔCD4 mice are cultured for 4 days with plate-bound α-CD3 and α-CD28 in the presence of TGF-β+RA + IFN-γ. ns: not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (one-way ANOVA with Tukey’s multiple comparisons for A and K; unpaired Student’s t test with Welch’s correction for E, G, and I).
Downregulation of Rptor but not rictor induces CD4+CD8αα+TCRβ+ IELs
We next questioned whether mTORC1 or 2 was important for the induction of DPIELs. We first administered rapamycin, which modulates mTORC1 signaling, to 6-week-old mice for 5 weeks (Figure 6C) (Neff et al., 2013). Interestingly, the percentage of DPIELs was increased in mice treated with rapamycin compared with control mice, whereas the percentages of Tregs in IEL were comparable in both groups (Figures 6D and 6E). The percentage and total number of Rorγt+Foxp3- cells in LPL were decreased in rapamycin-treated mice, whereas the percentage and total number of Rorγt−Foxp3+ cells were increased in rapamycin-treated mice (Figures S5A and S5B). The total number of Rorγt−Foxp3+ cells in rapamycin-treated mice was reduced, but the percentage was comparable. These data indicate that the inhibition of mTORC1 function is important for the development of DPIELs. To confirm mTORC1 involvement in the induction of DPIELs, we generated Cd4creERT2:Rptorfl/fl (referred to as iRaptorΔCD4) mice with CD4-specific Rptor downregulation after tamoxifen administration because the downregulation of Rptor in the thymus impairs peripheral CD4+ T cells (Hoshii et al., 2014). We administered tamoxifen to mice at the age of 6 weeks and analyzed them 5 weeks later. No spontaneous intestinal inflammation was observed in Rptorfl/fl or iRaptorΔCD4 mice (data not shown). DPIELs accounted for approximately 60% of CD4IELs, and the proportion of DPIELs was significantly increased in iRaptorΔCD4 mice (Figures 6F and 6G). We then generated Cd4cre:Rictorfl/fl mice (referred to as RictorΔCD4). The proportion of DPIELs was comparable between Rictorfl/fl and RictorΔCD4 mice (Figures 6H and 6I). Moreover, we cultured naive CD4+ T cells from Rictorfl/fl mice and RictorΔCD4 mice under the DPIEL condition with or without Torin 1. As observed in vivo, the percentage of DPIELs in CD4+ T cells in the absence of Torin 1 was comparable between Rictorfl/fl and RictorΔCD4 cells in vitro. Torin 1 increased the percentage of DPIELs development in both Rictorfl/fl and RictorΔCD4 cells (Figures 6J and 6K). Taken together, the inhibition of mTORC1 functionally but not mTORC2 is important for the development of DPIELs, whereas the expression levels of Rptor and Rictor are not downregulated during the development of DPIELs.
Discussion
CD4-induced IELs, especially CD4+CD8αα+ T cells, in the intestinal epithelium are not increased in GF mice, indicating that the involvement of bacteria in the intestinal epithelium plays a major role in the induction of CD4-induced IELs (Sujino et al., 2016). CD4+CD8αα+ T cells are induced by aryl hydrocarbon receptor ligands from food or intestinal bacteria and the surrounding environment, such as IL-15, food antigens, and MHC class ll molecules in the intestinal epithelium (Bilate et al., 2016; Mucida et al., 2013). Environmental cues shape induced IELs; however, how the cells adapt in oxygen rich or poor circumstances remains poorly understood. Similarly, how induced IELs adapt to their environmental conditions by altering gene expression related to intracellular metabolism is unknown (Almeida et al., 2016; Buck et al., 2015; Dumitru et al., 2018; Konjar et al., 2018; Phan et al., 2016; Raud et al., 2018; Shi et al., 2011). Induced CD4IELs exist as long-lived effector memory cells and express enzymes, such as granzyme B. Effector T cells are induced by elevated glycolysis, and they upregulate Hif1α/Hif2α expression. In contrast, memory T cells survive long-term in peripheral tissue by using OXPHOS. Here, we determined that induced CD4IELs, especially DPIELs, adapt to their unique metabolic conditions and exhibit a low OCR with reduced glucose uptake and anabolic glycolysis.
We first show that oxygen saturation is decreased in organs with poor blood flow, including the intestinal epithelium, not only because of cell oxygen consumption but also the presence of intestinal bacteria. Previously the number of TCRγδ+ cells was comparable in the GF and SPF conditions (Hoytema van Konijnenburg et al., 2017). As we counted the intestinal live cells in GF and SPF condition, we observed the increased number of both TCRγδ+ cells and TCRβ+ cells in SPF condition. Moreover, the total number of CD4+ T cells, especially DPIELs T cells, increased dramatically In the SPF condition.
In the small intestine, the presence of several commensal anaerobes that consume oxygen is consistent with the results. Long-lived memory CD8+ T cells exhibit an increased mitochondrial size and membrane potential, resulting in a higher level of oxygen consumption (Buck et al., 2016). CD4IELs consume less oxygen with increased mitochondrial fission and low mitochondrial membrane potential during development. Some IELs show an increased mitochondrial size and membrane potential after anti-CD3 antibody injection (Konjar et al., 2018). Anti-CD3 antibodies induce severe inflammation in the small intestine. As a result, cells use mitochondrial respiration because the oxygen supply might be increased in the epithelial compartment of the small intestine. Because DPIELs are diminished after anti-CD3 antibody injection, we cannot conclude that the size of mitochondria and membrane potential are increased in DPIELs after anti-CD3 antibody stimulation. We demonstrate hypoxic conditions induce more CD4+CD8αα+ T cells in DPIEL condition in vitro. However, the effect of oxygen saturation itself on the differentiation and maintenance of CD4IELs remains to be investigated. Our data suggest that induced CD4IELs are the population of cells that adapt to the hypoxic intraepithelial compartment.
DPIELs display enhanced gene expression of mTORC, which regulate HIF, mitochondrial biosynthesis, and lipid metabolism. However, DPIELs exhibit reduced HIF and pS6 expression during development, which are downstream factors of mTORC. Although DPIELs reduce glucose uptake compared with naïve CD4+ T cells, complete prevention of glucose intake with 2DG did not increase DPIELs, indicating that glucose intake is indispensable for the development of DPIELs.
DPIELs do not show an increase in NUR77, indicating reduced T cell receptor signaling, but they express TCRβ (Bilate et al., 2020). In addition, previous studies have shown that MHC-II signaling in the epithelium is required for the induction of DPIELs but dispensable for their expansion (Bilate et al., 2020; Mucida et al., 2013). Once naïve CD4+ T cells activated via T cell receptor signaling import glucose, T cell activation continues in the late phase via Akt/pS6 and HIF upregulation with increased glucose uptake (Frauwirth et al., 2002; Jacobs et al., 2008; Menk et al., 2018). Interestingly, DPIELs show reduced pS6 expression, HIF, and glucose uptake during their development from SPIELs. Thus, DPIELs might develop from cells that are activated temporally and quickly adapt to the tissue environments by reducing the expression of pS6 and HIF.
DPIELs display reduced pS6 signaling and Hif1α expression but not decreased mTORC expression during their development from SPIELs. The downregulation of mTORC1 or HIF in CD4+ T cells induces more DPIELs. These data indicate that mTORC gene expression in DPIELs may play a role other than regulating glucose uptake or HIF. One possibility is that memory T cells residing in the peripheral tissue consume fatty acids (Fahrer et al., 2001; Konkel et al., 2011; Pan et al., 2017). Therefore, mTORC in DPIELs might regulate lipid metabolism. In fact, the expression levels of the lipid biosynthesis genes Srebp1 and Srebp2 were increased in DPIELs compared with naïve CD4+ T cells and SPIELs. The mechanisms by which mTORC regulates the balance between HIF, glucose uptake, and lipid metabolism in SPIELs and DPIELs remain unknown.
Compared with Hif2α, Hif1α is dominantly expressed in CD4+ T cells. Hif1α expression might be a key factor that regulates DPIELs. The specific factor that reduces mTORC1 or HIF expression and promotes DPIELs development is still unknown. Further studies are needed because we cannot exclude the possibility that environmental autocrine or paracrine signals, such as cytokine production by the downregulation of mTORC1 or hypoxia in CD4+ T cells, might induce DPIELs.
Here, we demonstrate that CD4IELs alter their metabolic status and express unique genes to adapt to hypoxic conditions, and the downregulation of Rptor or Hif1α/Hif2α expression induces DPIELs (Figure S6). TCRγδ cells rapidly use glycolysis and migrate to protect the epithelium following bacterial invasion via MYD88 signaling. However, DPIELs, which increase in number with aging, are dual function cells with the potential to suppress intestinal inflammation, produce IL-10, and prevent bacterial invasion (Bilate et al., 2020; Mucida et al., 2013). The presence of diverse cell populations dependent on various metabolic pathways and metabolic genes in the IEL may be significant in the intestinal epithelium, which is at the forefront of immune tolerance and immune responses against antigens.
Limitations of the study
We measure the OCR and ECAR in splenic naïve CD4+ T cells and IELs. The OCR and ECAR in this study do not fully capture in each organ, because we analyze them after isolating each cell from peripheral tissue and the localization of each cell is different peripherally. We analyzed the frequencies and numbers of each immune cell in the intestine, but the right control of the metabolic study is not determined yet. DPIELs are differentiated by transcriptional factors, such as Runx3, Zbtb7b, Tbet and by the environmental factors, such as IL-15 and IFN-γ produced by CD4+ T cells or the other immune cells, but it is unknown how these transcriptional factor and environmental factors affect the metabolic status to develop DPIELs. We measured mitochondrial size, membrane potential with fission related genes, but the mitochondrial content and the precise mechanism of mitochondrial fission related genes on DPIELs are not elucidated yet.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD4 (clone: RM4-5; BV421) | Biolegend | Cat# 100544; RRID: AB_11219790 |
| CD8α (clone: 53-6.7; PE-Cy7) | BD Bioscience | Cat# 552877; RRID: AB_394506 |
| CD8β (clone: eBioH35-17.2; APC) | eBioscience | Cat# 17-0083-81; RRID: AB_657760 |
| CD44 (clone: IM7; APC) | Biolegend | Cat# 103012; RRID: AB_312963 |
| CD45 (clone: 30-F11; BV510) | Biolegend | Cat# 103138; RRID: AB_2563061 |
| CD62L (clone: MEL-14; FITC) | Biolegend | Cat# 104406; RRID: AB_313093 |
| TCRβ (clone: H57-597; APC-Cy7) | Biolegend | Cat# 109220; RRID: AB_893624 |
| TCRγδ (clone: GL3; PerCP-Cy5.5) | Biolegend | Cat# 118117; RRID: AB_10612572 |
| Foxp3 (clone: FJK-16s; PE) | eBioscience | Cat# 12-5773-82; RRID: AB_465936 |
| Rorγt (clone: Q31-378; BV421) | BD Bioscience | Cat# 562894; RRID: AB_2687545 |
| IL-17A (clone: eBio17B7; PE) | eBioscience | Cat# 12-7177-81; RRID: AB_763582 |
| IFNγ (clone: XMG1.2; FITC) | eBioscience | Cat# 11-7311-82; RRID: AB_465412 |
| CD16/32 (clone: 2.4G2) | BD Bioscience | Cat# 553142; RRID: AB_394657 |
| CD3ε (clone: 145-2C11) | Biolegend | Cat# 100359; RRID: AB_2616673 |
| CD28 (clone: 37.51) | Biolegend | Cat# 102116; RRID: AB_11147170 |
| Phospho-S6 ribosomal protein (Ser235/236) (clone: D57.2.2E; Alexa647) | Cell Signaling | Cat# 4851; RRID: AB_10695457 |
| Chemicals, peptides, and recombinant proteins | ||
| Tamoxifen | Sigma-Aldrich | Cat# T5648 |
| Corn oil | Wako | Cat# 032-17016 |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma-Aldrich | Cat# P8139 |
| Ionomycin | Sigma-Aldrich | Cat# I9657 |
| Rapamycin | Sigma-Aldrich | Cat# R0395 |
| Torin-1 | Selleckchem | Cat# S2827 |
| 2-DG | Sigma-Aldrich | Cat# D8375 |
| Fixable Viability Dye eFluor 780 | eBioscience | Cat# 65-0865-14 |
| Dithiothreitol (DTT) | Thermo Fisher Scientific | Cat# P2325 |
| EDTA | Nacalai Tesque | Cat# 06894-85 |
| Collagenase | Wako | Cat# 032-22364 |
| DNase I | Sigma-Aldrich | Cat# DN25 |
| Percoll | GE Healthcare | Cat# 1789101 |
| Ammonium chloride | Nacalai Tesque | Cat# 02424-55 |
| 4% Paraformaldehyde Phosphate Buffer Solution | Wako | Cat# 163-20145 |
| 8% Glutaraldehyde | Wako | Cat# 533-08681 |
| Agarose LM | Nacalai Tesque | Cat# 01161-54 |
| Bovine serum albumin (BSA) | Nacalai Tesque | Cat# 01863-48 |
| Triton-X100 | Polysciences | Cat# 04605 |
| DAPI | Dojindo | Cat# D523 |
| Mouse Naïve CD4+ T cell isolation kit | Miltenyi Biotech | Cat# 130-104-453 |
| TGF-β | R&D systems | Cat# 7666-MB-005 |
| Retinoic acid (RA) | Tokyo Chemical Industry | Cat# 0064-1G |
| IFN-γ | Peprotech | Cat# 315-05 |
| Fetal bovine serum (FBS) | Thermo Fisher Scientific | Cat# 10270-106 |
| HBSS | Nacalai Tesque | Cat# 17460-15 |
| RPMI1640 | Nacalai Tesque | Cat# 30264-85 |
| DPBS | Nacalai Tesque | Cat# 14249-24 |
| Penicillin/streptmycin | Nacalai Tesque | Cat# 09367-34 |
| Sodium pyruvate | Thermo Fisher Scientific | Cat# 11360070 |
| MEM/NEAA | Thermo Fisher Scientific | Cat# 11140050 |
| HEPES | Thermo Fisher Scientific | Cat# 15630080 |
| β-mercaptoethanol | Thermo Fisher Scientific | Cat# 21985023 |
| XF media | Agilent Technologies | Cat# 102353-100 |
| TRIzol | Invitrogen | Cat# 15596018 |
| Critical commercial assays | ||
| iScript cDNA Synthesis Kit | BioRad | Cat# 170-8891 |
| SYBR Green FAST qPCR Master Mix kit | Kapa Biosystems | Cat# KK4602 |
| HypoxyprobeTM-1 Plus Kit | Hypoxyprobe I | Cat# HP2-100KIT |
| XF Cell Mito Stress Test Kit | Agilent Technologies | Cat# 103015-100 |
| MitoTracker Green | Invitrogen | Cat# M7512 |
| MitoTracker CMXROS | Invitrogen | Cat# M7152 |
| TMRE-Mitochondrial Membrane Potential Assay Kit | Abcam | Cat# ab113852 |
| Foxp3/transcription factor staining buffer set | eBioscience | Cat# 00-5523-00 |
| 2-NBDG Glucose Uptake Assay Kit | Biovision | Cat# K682-50 |
| Anaeropack | Mitsubishi Gas Company | Cat# A-26 |
| GoldiStop Protein Transport Inhibitor | BD Bioscience | Cat# 554724; RRID: AB_2869012 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | JAX:000664 |
| Mouse: Cd4cre | The Jackson Laboratory | JAX:022071 |
| Mouse: Cd4creERT2 | The Jackson Laboratory | JAX:022356 |
| Mouse: Hif1αfl/fl | Ryan et al., 2000 | N.A. |
| Mouse: Hif2αfl/fl | Gruber et al., 2007 | N.A. |
| Mouse: Vhlfl/fl | Haase et al., 2001 | N.A. |
| Mouse: Rptorfl/fl | Hoshii et al., 2014 | N.A. |
| Mouse: Rictorfl/fl | Magee et al., 2012 | N.A. |
| Oligonucleotides | ||
| QPCR primers | Hokkaido System Science | Table S1 |
| Software and algorithms | ||
| GraphPad Prism 8 | GraphPad Software | https://www.graphpad.com/ |
| FlowJo Vx software | TreeStar | https://www.flowjo.com/ |
| ImageJ | ImageJ | https://imagej.net/Welcome |
| Imaris 8.4 | Bitplane | https://imaris.oxinst.com |
Resource availability
Lead contact
Further information and requests for the resources and reagents should be directed and will be fulfilled by the lead contact, Tomohisa Sujino (tsujino1224@keio.jp).
Materials availability
All the mouse lines used in this study are available upon request.
This study did not generate new unique reagents.
Experimental model and subject details
Mice
Mice used in this study were of a C57BL/6 background. Cd4cre, Cd4creERT2, Rptorfl/fl, and Rictorfl/fl, Hif1αfl/fl, Hif2αfl/fl, and Vhlfl/fl mice were previously described (Gruber et al., 2007; Haase et al., 2001; Hoshii et al., 2014; Magee et al., 2012; Ryan et al., 2000) and were maintained under specific pathogen-free (SPF) conditions in the Animal Care Facility of Keio University School of Medicine. Germ-free (GF) mice (C57BL/6 background strain) were purchased from Sankyo Lab Service Corporation and were kept in the GF Facility of Keio University School of Medicine. Both female and male mice were used for this study and were aged between 6 and 12 weeks at the time of experiments. All experiments were approved by the Institutional Review Board for Animal Experiments of Keio University and were performed according to the institutional guidelines and home office regulations.
Method details
Preparation of intraepithelial lymphocytes and lamina propria mononuclear cells
Mice were euthanized by cervical dislocation, and the small intestines were collected. After removal of residual fat tissue and Peyer’s patches, the small intestine tissue was opened longitudinally and washed with Ca2+, Mg2+-free Hank’s balanced salt solution (HBSS) (Nacalai Tesque) to remove fecal content. After washing, the small intestine was further cut into small pieces and incubated with HBSS containing 1 mM dithiothreitol (Invitrogen) and 5 mM EDTA (Nacalai Tesque) for 30 min at 37°C to remove the epithelial layer. After removal of the epithelial layer, the mucosal pieces were washed with HBSS and digested by incubation with HBSS containing 1.5% fetal bovine serum (Thermo Fisher Scientific), 1 mg/mL collagenase (Wako), and 0.1 mg/mL DNase (Sigma-Aldrich) for 30 min at 37°C. The digested solution was centrifuged at 1,700 rpm for 5 min. The pellet was resuspended in 40% Percoll (GE healthcare) and overlaid on 75% Percoll. Percoll gradient separation was performed by centrifugation at 2,000 rpm for 20 min at 20°C. Cells at the interphase were collected as LPL. For intraepithelial lymphocytes, the supernatant containing the epithelial layers was collected and centrifuged at 1,700 rpm for 5 min. The pellet was resuspended in 40% Percoll and overlaid on 75% Percoll. Percoll gradient separation was performed by centrifugation at 2,000 rpm for 20 min at 20°C. Cells at the interphase were collected as IEL.
Preparation of spleen cell suspensions
Spleens were harvested from the mice after death and homogenized manually in HBSS. The lysates were filtered through a cell strainer. The passed cells were hemolyzed with 0.84% (vol/wt) ammonium chloride (Nacalai Tesque), washed with HBSS, and collected for analysis.
Flow cytometry
Nonspecific staining was inhibited by incubation with anti-CD16/32 (BD Bioscience) for 20 min. The surface antigens of isolated single-cell suspensions were stained with the following antibodies: CD4 (RM4-5), CD8α (53-6.7), CD8β (eBioH35-17.2), CD45 (30-F11), TCRβ (H57-597), TCRγδ (GL3). For intracellular Foxp3 (FJK-16s) and RORγt (Q31-378) staining, cells were permeabilized using fixation/permeabilization solution (eBioscience) before intracellular staining with antibodies. For IL-17A (eBio17B7) and IFNγ (XMG1.2) staining, cells were incubated in Roswell Park Memorial Institute (RPMI)-1640 medium containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Nacalai Tesque) with 50 ng/mL PMA (Sigma-Aldrich), 500 ng/mL ionomycin (Sigma-Aldrich), and GoldiStop (BD Bioscience). Cells were stained with antibodies against the indicated cell surface markers, and dead cells were stained with Fixable Viability Dye eFluor 780 (eBioscience). Cells were permeabilized using fixation/permeabilization solution before IL-17A and IFNγ staining. Spleen cell suspensions were stained with the following antibodies: CD45 (30-F11), TCRβ (H57-597), CD4 (RM4-5), CD8α (53-6.7), CD44 (IM7), and CD62L (MEL-14). For flow cytometry analysis, the FACS Canto system (BD Biosciences) was used. Data were analyzed by FlowJo Vx software (Tree Star). Cell sorting was performed using the BD FACS Aria II sorter (BD Biosciences). For measurement of mitochondrial mass and membrane potential, purified cells were washed with prewarmed RPMI-1640 medium (Nacalai Tesque) supplemented with 10% FBS (staining buffer). Approximately, 1.5 × 106 cells were resuspended in 1 mL of staining buffer containing MitoTracker Green (20 nM; M7514; Invitrogen) and MitoTracker CMXROS (20 nM; M7152; Invitrogen) and incubated in a CO2 incubator at 37 °C for 30 min. Stained cells were washed with 1 mL of prewarmed staining buffer and used for FACS analysis or further staining with antibodies. For pS6 staining, CD4IELs, CD4LPLs and spleen cells were stimulated for 30 min with soluble α-CD3 mAb (1 μg/mL), then fixed in phosphate-buffered saline (PBS)/2% paraformaldehyde for 20 min, permeabilized with 90% methanol for 30 min on ice, and then stained for CD4, CD45, TCRβ, TCRγδ, CD8α, CD8β and pS6 in PBS. All the antibodies were used at 1:200 dilution. Alternatively, TMRE (Abcam) was used to monitor mitochondrial membrane potential. Cells were incubated with 5 μM TMRE for 30 min at 37°C and washed in PBS.
Metabolic assays
OCR and ECAR were measured with an XF-24 analyzer (Seahorse Bioscience) according to the manufacturer’s protocols. FACS-sorted 5 × 105 T cells were cultured in XF media (non-buffered Dulbecco’s modified Eagle’s medium containing 25 mM glucose, 2 mM L-glutamine, 1 mM sodium pyruvate, and 2% fetal bovine serum, Agilent Technologies). The assay was performed using XF Cell Mito Stress Test Kit (Agilent Technologies). Three baseline recordings were taken, and this was followed by the sequential injection of the ATP synthase inhibitor oligomycin (2.5 μM), the mitochondrial uncoupler carbonyl cyanide-4-(trifluoromethoxy)phenyl-hydrazone (FCCP; 0.5 μM), and the respiratory chain inhibitors antimycin A (1 μM) and rotenone (1 μM).
Quantitative real-time polymerase chain reaction
RNA was isolated using TRIzol (Invitrogen) according to the manufacturer’s instructions. Complementary DNA was synthesized from the extracted RNA using the iScript cDNA Synthesis Kit (Bio-Rad). The obtained complementary DNA was amplified by quantitative real-time polymerase chain reaction (PCR) using primer sets and the SYBR Green FAST qPCR Master Mix kit (Kapa Biosystems). The house-keeping gene Rpl32 was used for normalization of samples. Primer sequences used were listed in Table S1.
Transmission electron microscopy
Freshly sorted naïve CD4+ T cells, SPIELs and DPIELs were immediately fixed with an equal amount of 4% glutaraldehyde and 4% paraformaldehyde in 0.1 M phosphate buffer pH 7.4 at 4°C overnight. Fixed samples were dehydrated and transferred to a fresh 100% resin to polymerize at 60°C for 48 h. The polymerized samples were ultra-thin sectioned at 70 nm with an ultramicrotome (Ultracut UCT, Leica) and observed by a transmission electron microscope (JEM-1400Plus, JEOL Ltd.).
In vivo tamoxifen treatment
Mice were gavaged for two consecutive days at the beginning of the week with 5 mg of tamoxifen (Sigma-Aldrich) dissolved in corn oil (Wako) at 50 mg/ml. After 5 weeks from first administration, mice were used for experiments.
In vivo treatment of rapamycin and 2DG
Mice were injected intraperitoneally with vehicle, rapamycin (8 mg/kg, Sigma-Aldrich) or 2DG (500 mg/kg, Sigma-Aldrich) daily for 5 weeks. Rapamycin and 2DG were first reconstituted in DMSO and then diluted with PBS.
Hypoxyprobe-1 staining
Mice were administrated pimonidazole (Hypoxyprobe, Burlington, MA) by intraperitoneal injection 30 min prior to sacrifice. Dissected pieces of mouse small intestine were washed in PBS and then fixed in 4% PFA for overnight, 4°C. Tissue was embedded in 4% low melting temperature agarose (Nacalai Tesque) and cut into 200 μM sections using a Vibratome (Leica). Vibratome sections were washed in PBS and incubated with Blocking buffer (1% BSA, 1% mouse serum, 0.1% Triton-X100 in PBS) for 2 hours, room temperature. Sections were incubated with FITC conjugated α-pimonidazole monoclonal antibody (Hypoxyprobe, 1:100) for 1 hour, room temperature. PBS washed sections were counterstained with DAPI and mounted to analyze by confocal microscopy (Olympus FV3000)
In vitro CD4+CD8αα+ T cell differentiation
Naïve CD4+ T cells were isolated by negative selection with mouse Naïve CD4+ T cell isolation kit (Miltenyi). 1 × 105 cells were cultured for 4 days in 96-well plates precoated with 1 μg/ml of α-CD3ε (145-2C11; Biolegend) and 1 μg/ml of soluble α-CD28 (37.51; Biolegend) in RPMI 1640 media supplemented with 10% FBS, 1% penicillin/streptomycin, 1% pyruvate (11360070; Thermo Fisher Scientific), 1% MEM/NEAA (11140050; Thermo Fisher Scientific), 2.5% HEPES (15630080; Thermo Fisher Scientific), and 55 μM β-mercaptoethanol (21985023; Thermo Fisher Scientific) under CD4+CD8αα+ T cell conditions: RA (10 nM, 0064-1G, Tokyo Chemical Industry), TGFβ (2 ng/ml, 7666-MB-005, R&D systems), IFNγ (20 ng/ml, 315-05, Peprotech). For the treatment with inhibitors, Rapamycin (1 μM, Sigma-Aldrich), Torin-1 (1 μM, Selleckchem) and 2DG (1 mM, Sigma-Aldrich) were added into the cell culture. In some experiments, cells were placed inside a sealed air tight container which contains an anaeropack (Mitsubishi Gas Company, Tokyo, Japan). The anaeropack contains a gas-controlling reagent which absorbs oxygen and generates carbon dioxide resulting in a hypoxic atmosphere (8% O2).
Analysis of glucose uptake
Glucose uptake was determined with a fluorescently labeled deoxyglucose analog (2-NBDG) using a commercially available kit (Biovision, Milpitas, CA, USA). Purified cells were washed twice with RPMI 1640 media supplemented with 0.5% FBS. Cells were resuspended in 400 μl of glucose-free medium supplemented with 150 μg/ml 2-NBDG and incubated at 37°C for 30 min. The cells were washed with assay buffer, and glucose uptake was quantified by FACS analysis.
Quantification and statistical analysis
Statistical analysis was performed using GraphPad Prism software. Data were analyzed by applying one-way analysis of variance or unpaired Student’s t-test whenever necessary. A P value < 0.05 was considered significant.
Acknowledgments
This work was supported by Grants-in-Aid from the Japanese Society for the Promotion of Science (JSPS) (19K17503 to Y.H., 17K19668, 17H05082, 19K22624, 20H03665, 21K18272 to T.S., 19K08402 to N.H., 21H02905 to H.O., 21K07084 K.O.), the Japan Agency for Medical Research and Development (19ek0109214 to T.S., 21gm1510002h0001 to T.K.), Mochida Memorial Foundation (T.S.), Takeda Science Foundation (T.S.), GSK Science Foundation (T.S.), and Yakult Bioscience Research Foundation (T.S.). We thank Jeremy Allen, PhD, and Melissa Crawford, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Author contributions
Y.H., E.N., K.M., and T.S. performed experiments. T.S. designed and supervised the experiments and wrote the manuscript. Y.Y., S.T., S.U., K.O., Y.M., N.N., K.T., N.H., and H.O. assisted with experiments. T.I., A.H., and Y.K. provided mice and assisted with generating mice. T.S. and T.K. conceived the paper.
Declaration of interests
Kentaro Miyamoto is an employee of Miyarisan Pharm. The other authors declare no competing interests.
Published: April 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104021.
Contributor Information
Tomohisa Sujino, Email: tsujino1224@keio.jp.
Takanori Kanai, Email: takagast@keio.jp.
Supplemental information
Data and code availability
All the detailed data in this paper are available upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Almeida L., Lochner M., Berod L., Sparwasser T. Metabolic pathways in T cell activation and lineage differentiation. Semin. Immunol. 2016;28:514–524. doi: 10.1016/j.smim.2016.10.009. [DOI] [PubMed] [Google Scholar]
- Angelin A., Gil-de-Gomez L., Dahiya S., Jiao J., Guo L., Levine M.H., Wang Z., Quinn W.J., 3rd, Kopinski P.K., Wang L., et al. Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab. 2017;25:1282–1293.e1287. doi: 10.1016/j.cmet.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilate A.M., Bousbaine D., Mesin L., Agudelo M., Leube J., Kratzert A., Dougan S.K., Victora G.D., Ploegh H.L. Tissue-specific emergence of regulatory and intraepithelial T cells from a clonal T cell precursor. Sci. Immunol. 2016;1 doi: 10.1126/sciimmunol.aaf7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilate A.M., London M., Castro T.B.R., Mesin L., Bortolatto J., Kongthong S., Harnagel A., Victora G.D., Mucida D. T cell receptor is required for differentiation, but not maintenance, of intestinal CD4(+) intraepithelial lymphocytes. Immunity. 2020;53:1001–1014. doi: 10.1016/j.immuni.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M.D., O'Sullivan D., Klein Geltink R.I., Curtis J.D., Chang C.H., Sanin D.E., Qiu J., Kretz O., Braas D., van der Windt G.J., et al. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell. 2016;166:63–76. doi: 10.1016/j.cell.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M.D., O'Sullivan D., Pearce E.L. T cell metabolism drives immunity. J. Exp. Med. 2015;212:1345–1360. doi: 10.1084/jem.20151159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragan L., Chai J.N., Tianero M.D., Di Luccia B., Ahern P.P., Merriman J., Cortez V.S., Caparon M.G., Donia M.S., Gilfillan S., et al. Lactobacillus reuteri induces gut intraepithelial CD4(+)CD8alphaalpha(+) T cells. Science. 2017;357:806–810. doi: 10.1126/science.aah5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheroutre H., Lambolez F., Mucida D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat. Rev. Immunol. 2011;11:445–456. doi: 10.1038/nri3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd K.M., Yang J., Shen M.H., Sampson J.R., Tee A.R. mTORC1 drives HIF-1alpha and VEGF-A signalling via multiple mechanisms involving 4E-BP1, S6K1 and STAT3. Oncogene. 2015;34:2239–2250. doi: 10.1038/onc.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitru C., Kabat A.M., Maloy K.J. Metabolic adaptations of CD4(+) T cells in inflammatory disease. Front Immunol. 2018;9:540. doi: 10.3389/fimmu.2018.00540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrer A.M., Konigshofer Y., Kerr E.M., Ghandour G., Mack D.H., Davis M.M., Chien Y.H. Attributes of gammadelta intraepithelial lymphocytes as suggested by their transcriptional profile. Proc. Natl. Acad. Sci. U S A. 2001;98:10261–10266. doi: 10.1073/pnas.171320798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauwirth K.A., Riley J.L., Harris M.H., Parry R.V., Rathmell J.C., Plas D.R., Elstrom R.L., June C.H., Thompson C.B. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16:769–777. doi: 10.1016/s1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- Groux H., O'Garra A., Bigler M., Rouleau M., Antonenko S., de Vries J.E., Roncarolo M.G. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature. 1997;389:737–742. doi: 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- Gruber M., Hu C.J., Johnson R.S., Brown E.J., Keith B., Simon M.C. Acute postnatal ablation of Hif-2alpha results in anemia. Proc. Natl. Acad. Sci. U S A. 2007;104:2301–2306. doi: 10.1073/pnas.0608382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase V.H., Glickman J.N., Socolovsky M., Jaenisch R. Vascular tumors in livers with targeted inactivation of the von Hippel-Lindau tumor suppressor. Proc. Natl. Acad. Sci. U S A. 2001;98:1583–1588. doi: 10.1073/pnas.98.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka R.B., Chandel N.S. Targeting glucose metabolism for cancer therapy. J. Exp. Med. 2012;209:211–215. doi: 10.1084/jem.20120162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Gomes A.P., Wang X., Yoon S.O., Lee G., Nagiec M.J., Cho S., Chavez A., Islam T., Yu Y., et al. mTORC1 promotes metabolic reprogramming by the suppression of GSK3-dependent Foxk1 phosphorylation. Mol. Cell. 2018;70:949–960.e944. doi: 10.1016/j.molcel.2018.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshii T., Kasada A., Hatakeyama T., Ohtani M., Tadokoro Y., Naka K., Ikenoue T., Ikawa T., Kawamoto H., Fehling H.J., et al. Loss of mTOR complex 1 induces developmental blockage in early T-lymphopoiesis and eradicates T-cell acute lymphoblastic leukemia cells. Proc. Natl. Acad. Sci. U S A. 2014;111:3805–3810. doi: 10.1073/pnas.1320265111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoytema van Konijnenburg D.P., Reis B.S., Pedicord V.A., Farache J., Victora G.D., Mucida D. Intestinal epithelial and intraepithelial T cell crosstalk mediates a dynamic response to infection. Cell. 2017;171:783–794.e713. doi: 10.1016/j.cell.2017.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S.R., Herman C.E., Maciver N.J., Wofford J.A., Wieman H.L., Hammen J.J., Rathmell J.C. Glucose uptake is limiting in T cell activation and requires CD28-mediated Akt-dependent and independent pathways. J. Immunol. 2008;180:4476–4486. doi: 10.4049/jimmunol.180.7.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J., Zeng H., Horng T. Metabolism as a guiding force for immunity. Nat. Cell Biol. 2019;21:85–93. doi: 10.1038/s41556-018-0217-x. [DOI] [PubMed] [Google Scholar]
- Konjar S., Frising U.C., Ferreira C., Hinterleitner R., Mayassi T., Zhang Q., Blankenhaus B., Haberman N., Loo Y., Guedes J., et al. Mitochondria maintain controlled activation state of epithelial-resident T lymphocytes. Sci. Immunol. 2018;3 doi: 10.1126/sciimmunol.aan2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel J.E., Maruyama T., Carpenter A.C., Xiong Y., Zamarron B.F., Hall B.E., Kulkarni A.B., Zhang P., Bosselut R., Chen W. Control of the development of CD8alphaalpha+ intestinal intraepithelial lymphocytes by TGF-beta. Nat. Immunol. 2011;12:312–319. doi: 10.1038/ni.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIver N.J., Michalek R.D., Rathmell J.C. Metabolic regulation of T lymphocytes. Annu. Rev. Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee J.A., Ikenoue T., Nakada D., Lee J.Y., Guan K.L., Morrison S.J. Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell Stem Cell. 2012;11:415–428. doi: 10.1016/j.stem.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menk A.V., Scharping N.E., Moreci R.S., Zeng X., Guy C., Salvatore S., Bae H., Xie J., Young H.A., Wendell S.G., et al. Early TCR signaling induces rapid aerobic glycolysis enabling distinct acute T cell effector functions. Cell Rep. 2018;22:1509–1521. doi: 10.1016/j.celrep.2018.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucida D., Husain M.M., Muroi S., van Wijk F., Shinnakasu R., Naoe Y., Reis B.S., Huang Y., Lambolez F., Docherty M., et al. Transcriptional reprogramming of mature CD4(+) helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nat. Immunol. 2013;14:281–289. doi: 10.1038/ni.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff F., Flores-Dominguez D., Ryan D.P., Horsch M., Schroder S., Adler T., Afonso L.C., Aguilar-Pimentel J.A., Becker L., Garrett L., et al. Rapamycin extends murine lifespan but has limited effects on aging. J. Clin. Invest. 2013;123:3272–3291. doi: 10.1172/JCI67674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton R., Priyadharshini B., Turka L.A. Immunometabolism of regulatory T cells. Nat. Immunol. 2016;17:618–625. doi: 10.1038/ni.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares-Villagomez D., Van Kaer L. Intestinal intraepithelial lymphocytes: sentinels of the mucosal barrier. Trends Immunol. 2018;39:264–275. doi: 10.1016/j.it.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Tian T., Park C.O., Lofftus S.Y., Mei S., Liu X., Luo C., O'Malley J.T., Gehad A., Teague J.E., et al. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature. 2017;543:252–256. doi: 10.1038/nature21379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce E.L., Pearce E.J. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan A.T., Doedens A.L., Palazon A., Tyrakis P.A., Cheung K.P., Johnson R.S., Goldrath A.W. Constitutive glycolytic metabolism supports CD8(+) T cell effector memory differentiation during viral infection. Immunity. 2016;45:1024–1037. doi: 10.1016/j.immuni.2016.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raud B., McGuire P.J., Jones R.G., Sparwasser T., Berod L. Fatty acid metabolism in CD8(+) T cell memory: challenging current concepts. Immunol. Rev. 2018;283:213–231. doi: 10.1111/imr.12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis B.S., Rogoz A., Costa-Pinto F.A., Taniuchi I., Mucida D. Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4(+) T cell immunity. Nat. Immunol. 2013;14:271–280. doi: 10.1038/ni.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan H.E., Poloni M., McNulty W., Elson D., Gassmann M., Arbeit J.M., Johnson R.S. Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer Res. 2000;60:4010–4015. [PubMed] [Google Scholar]
- Shi L.Z., Wang R., Huang G., Vogel P., Neale G., Green D.R., Chi H. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujino T., London M., Hoytema van Konijnenburg D.P., Rendon T., Buch T., Silva H.M., Lafaille J.J., Reis B.S., Mucida D. Tissue adaptation of regulatory and intraepithelial CD4(+) T cells controls gut inflammation. Science. 2016;352:1581–1586. doi: 10.1126/science.aaf3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydora B.C., Mixter P.F., Holcombe H.R., Eghtesady P., Williams K., Amaral M.C., Nel A., Kronenberg M. Intestinal intraepithelial lymphocytes are activated and cytolytic but do not proliferate as well as other T cells in response to mitogenic signals. J. Immunol. 1993;150:2179–2191. [PubMed] [Google Scholar]
- Van Kaer L., Olivares-Villagomez D. Development, homeostasis, and functions of intestinal intraepithelial lymphocytes. J. Immunol. 2018;200:2235–2244. doi: 10.4049/jimmunol.1701704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandereyken M., James O.J., Swamy M. Mechanisms of activation of innate-like intraepithelial T lymphocytes. Mucosal Immunol. 2020;13:721–731. doi: 10.1038/s41385-020-0294-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Bruce D., Froicu M., Weaver V., Cantorna M.T. Failure of T cell homing, reduced CD4/CD8alphaalpha intraepithelial lymphocytes, and inflammation in the gut of vitamin D receptor KO mice. Proc. Natl. Acad. Sci. U S A. 2008;105:20834–20839. doi: 10.1073/pnas.0808700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Zhao Y., Zou L., Zhang D., Aki D., Liu Y.C. The E3 ligase VHL promotes follicular helper T cell differentiation via glycolytic-epigenetic control. J. Exp. Med. 2019;216:1664–1681. doi: 10.1084/jem.20190337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the detailed data in this paper are available upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.