Abstract
Niclosamide, a widely-used anthelmintic drug, inhibits SARS-CoV-2 virus entry through TMEM16F inhibition and replication through autophagy induction, but the relatively high cytotoxicity and poor oral bioavailability limited its application. We synthesized 22 niclosamide analogues of which compound 5 was found to exhibit the best anti-SARS-CoV-2 efficacy (IC50 = 0.057 M) and compounds 6, 10, and 11 (IC50 = 0.39, 0.38, and 0.49 M, respectively) showed comparable efficacy to niclosamide. On the other hand, compounds 5, 6, 11 contained higher stability in human plasma and liver S9 enzymes assay than niclosamide, which could improve bioavailability and half-life when administered orally. Fluorescence microscopy revealed that compound 5 exhibited better activity in the reduction of phosphatidylserine externalization compared to niclosamide, which was related to TMEM16F inhibition. The AI-predicted protein structure of human TMEM16F protein was applied for molecular docking, revealing that 4′-NO2 of 5 formed hydrogen bonding with Arg809, which was blocked by 2′-Cl in the case of niclosamide.
Keywords: SARS-CoV-2, Niclosamide, Stability test, TMEM16F, Entry inhibition
Abbreviations: DCM, dichloromethane; MERS, East respiratory syndrome coronavirus; ACE-2, angiotensin-converting enzyme 2; CPE, cytopathic effect; PAMPA, parallel artificial membrane permeability assay
Graphical abstract

1. Introduction
The global COVID-19 pandemic caused by SARS-CoV-2 has been an ongoing global economic and health burden since 2019. Different from the previous outbreaks related to other two coronavirus, SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV), SARS-CoV-2 exhibits higher transmission rate and lower fatality rate, resulting in a prolonged pandemic situation, extraordinary social and economic burdens and a big threat to public health [1].
With extensive efforts from pharmaceutical industry around the world, effective SARS-CoV-2 vaccines were produced and made a great success in controlling the global pandemic [2,3]. Currently available treatments for COVID-19 include neutralizing antibodies, corticosteroids, and viral RNA-dependent RNA polymerase (rdrp) inhibitor remdesivir (Fig. 1 A) [4]. However, remdesivir is only modestly effective and available in hospitalized patients with mild to moderate disease [5], and the antibodies are less effective against novel SARS-CoV-2 variants bearing a spike protein mutation such as B.1.1.7, B.1.351, and B.1.1.529. Therefore, novel small molecule therapeutics exhibiting good oral bioavailability are urgently needed [6]. Recently, clinical trial results of two orally administered anti-SARS-CoV-2 agents were reported. Molnupiravir (Fig. 1A), an inhibitor of viral RNA-dependent RNA polymerase (RdRp), was found to reduce the risk of hospitalization or death in non-hospitalized adult patients with mild-to-moderate COVID-19 by 30% in phase 3 study [7]. On the other hand, the combination of viral protease inhibitors, nirmatrelvir and ritonavir (Fig. 1A), showed 89% reduction of hospitalization or death [8]. These results indicated that the development of oral available anti-SARS-CoV-2 drugs with various mechanism is necessary for controlling future pandemics.
Fig. 1.
(A) Chemical structures of currently available small molecule anti-SARS-CoV-2 agents. (B) Design rationale for the synthesis of niclosamide analogues.
Niclosamide (Fig. 1B) is an oral anthelmintic drug which has been approved by FDA for the treatment of tapeworm infection in humans for several decades. Recently, multifactorial roles of niclosamide in human disease have been discussed, and its broad antiviral activity was reported [[9], [10], [11]]. The anti-SARS-CoV-2 activity of niclosamide was identified in several drug-repurposing studies, and evaluated in clinical trials [12,13]. Cellular experiments revealed that the inhibition mechanism is related to TMEM16F-mediated viral syncytia and SKP2 inhibition [14,15]. These results suggest that niclosamide is a promising lead for structure-activity relationship study to identify novel structures against SARS-CoV-2 infection. Although structure derivatization of niclosamide have been applied for the inhibition of SARS-CoV-2, the ratio between cytotoxicity (CC50) and antiviral efficacy (IC50) were unable to exceed 5, indicating that the compound selectivity is beyond satisfying [16,17].
In this study, we focused on the structure-activity relationships of niclosamide core, which was divided into benzoic acid and aniline moiety. The strategy for modification included aromatic substitution with electron donating and withdrawing group, chain elongation, and glycosylation (Fig. 1B). The antiviral efficacy of compound was primarily evaluated with cytopathic effect and antibody quantification in Vero E6 cells. The membrane penetration activity and cytotoxicity of compounds were obtained by PAMPA, clogP calculation and the MTT assay to identify the compound with ideal biological activity. The in vitro human plasma and human liver S9 assay were applied to identify compounds with better stability. The TMEM16F inhibition was confirmed by the phosphatidylserine externalization assay using fluorescence microscopy, and the computer simulation revealed the essential residue for the TMEM16F protein binding.
2. Results and discussion
2.1. Synthesis of niclosamide analogues
Niclosamide analogues were synthesized through a two-step amide coupling reaction using acyl chloride (Scheme 1 ). Compounds 1–15 were synthesized from 5-chloro-2-hydroxybenzoyl chloride and a variety of anilines substituted with electron withdrawing group (NO2, Cl, CF3) and electron donating group (F, tBu, OH, piperidine) on the meta or para positions. Compounds 16 and 17 containing an additional carbon between the amine and phenyl ring were also synthesized to explore the impact of structural flexibility of niclosamide analogues.
Scheme 1.
General synthesis route of niclosamide analogs. (a) i. SOCl2; ii. aniline derivatives or benzyl amine derivatives, DCM, 10–75% for 2 steps.
Similarly, compounds 18–22 were synthesized to evaluate the effect of substitution on the benzoic acid moiety. Benzoic acids with hydroxyl and methyl substitution were treated with thionyl chloride to give corresponding acyl chloride, followed by the addition of 2-chloro-4-nitroaniline in dichloromethane to afford niclosamide analogues 18–22.
To increase water solubility of the niclosamide analogues and hence alter their cellular distribution, analogue 14 was glycosylated with glucose (Scheme 2 ). Boron trifluoride diethyl etherate was added to a solution of tetra-O-acetyl-protected glucopyranosyl imidate 23 and 4-nitrophenol in DCM at 0 °C with stirring. After 3 h, compound 24 was obtained. Hydrogenation of the nitro group on the para position of 24 was accomplished using 10% Pd/C under a hydrogen atmosphere in MeOH/DCM and gave amine compound 25, which was reacted with acyl chloride 29, obtained from treatment of 5-chlorosalicylic acid 28 with SOCl2, to give 26. Compound 26 was dissolved in MeOH/DCM and the acetyl groups were removed by sodium methoxide to afford glycosylated analogue 27.
Scheme 2.
Synthesis of glycosylated niclosamide analogs. (a) 4-nitrophenol, BF3·OEt2, DCM, 0 °C to rt, 74%; (b) H2, Pd/C, MeOH/DCM, 93%; (c) i. 5-chlorosalicylic acid 28, SOCl2, 40 °C; ii. DCM, rt, 38% for 2 steps; (d) NaOMe, MeOH/DCM, rt, 61%.
2.2. Structure-activity relationships of niclosamide analogues against SARS-CoV-2
The antiviral efficacies of these niclosamide analogues were first evaluated by microscopic visualization of their cytopathic effect (CPE), as previously described [18]. Vero E6 cells, which are widely used in coronavirus assays due to their high expression levels of angiotensin-converting enzyme 2 (ACE2) receptor [19], were infected with SARS-CoV-2 virus strain obtained from Taiwan Centers for Disease Control (hCoV19/Taiwan/4/2020). Of the 23 niclosamide analogues tested, nine (2, 3, 4, 5, 6, 10, 11, 12, 18) exhibited levels of antiviral efficacy that were superior to their cytotoxicity (Fig. 2 ), and were selected for determination of the concentration of each to inhibit half virus replication (IC50).
Fig. 2.
Primary screening of antiviral efficacy by microscopic examination of virus-induced cytopathic effect and cytotoxicity in Vero E6 cells. The stock solutions of niclosamide analogues were 10 mM.
Since niclosamide has been reported to inhibit the entry of viruses into cells, the pretreat experiment was applied for the determination of antiviral IC50 in Vero E6 cells [14]. Niclosamide and 9 analogues were separately added to the culture medium and incubated with cells. After 2 h, 100 TCID50 virus was added to the medium, incubated for 24 h, and the virus titer was quantified with antibodies targeting viral nucleocapsid proteins (Fig. 3 ). Under these conditions, the IC50 of niclosamide was found to be 0.4 M, which is comparable with the literature (0.34 M) [14].
Fig. 3.
Dose-response relationships of niclosamide and 9 derivatives. Vero E6 cells were pretreated with compounds for 2 h, followed by addition of SARS-CoV-2 for 24 h. The inhibition efficacy evaluated with viral titer determined by viral nucleocapsid protein antibody (red curve) and cell viability (blue curve) were measured and presented as mean ± SD of at least three independent experiments.
The antiviral activity of compound 2 was greatly reduced compared to niclosamide (1) (IC50 = 8.59 M vs 0.4 M), indicating the importance of aromatic substitution on the aniline moiety. Compound 3, lacking a 2′-Cl compared to niclosamide (1) exhibited a slightly reduced antiviral activity (IC50 = 1.11 M vs 0.4 M) as did compound 4 (IC50 = 1.29 M vs 0.4 M), bearing a 3′-NO2 instead of a 4′-NO2. The compounds containing 3′-Cl (6, 0.39 M), 4′-CF3 (10, 0.38 M) and 3′-CF3 (11, 0.49 M) exhibited comparable antiviral IC50 to niclosamide. However, the IC50 of 3′-tBu (12, 1.51 M) was about three-fold higher than niclosamide, indicating that the lone pairs on aniline moiety were important for their antiviral activity. The IC50 of 3′-Cl analogue 6 was 0.39 M, whereas 4′-F (8) and 3′-F (9) were both inactive in the CPE assay, suggesting that larger valence orbital of Cl might be important for inhibition. Analogues bearing 3′-OH (13), 4′-OH (14), 4′-piperidine (15), and 4′-O-glucose (27) were all inactive in the CPE screening as well, suggests that mono-substituted electron withdrawing groups on the aniline moiety are more favorable than electron donating groups.
The IC50 of the di-substituted analogues with the highest anti-SARS-CoV-2 activity was compound 5 (0.057 M), bearing a 3′-Cl instead of the 2′-Cl presented in niclosamide. Surprisingly, the antiviral efficacy of 7 bearing a 4′-F in place of the 4′-NO2 was diminished. Compounds 16 and 17 did not show viral inhibition in the CPE screening, suggesting that elongated aniline moieties and/or methyl substitution at the 3′ and 4′ position might disfavor an antiviral effect. To investigate the benzoic acid moiety, the 5-Cl of niclosamide was removed, and the antiviral effect was decreased (18, 1.68 M). The antiviral activity disappeared when all the substituents were removed from benzoic acid moiety. On the other hand, the antiviral activity was also diminished when 5-Cl was substituted with 5-Me (20), 4-Me (21), and 5-F (22), respectively.
2.3. Drug-like properties and cytotoxicity evaluation of niclosamide analogues
Niclosamide is well tolerated when administered orally to patients infected with tapeworm, but its high cytotoxicity, low solubility, and low oral bioavailability (F = 10%) limited its usage [9]. Accordingly, we investigated the clogP, PAMPA permeability, and cytotoxicity against Vero E6 cells of the analogues synthesized (Table 1 , Table 2 , Fig. S1).
Table 1.
Biological activity of niclosamide analogues.
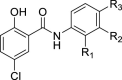
| Compounds | Anti-SARS-CoV-2 IC50 ( M) | Vero E6 48 h Cytotoxicity CC50 ( M)a | CC50/IC50 | ||||
|---|---|---|---|---|---|---|---|
 | |||||||
| R1 | R2 | R3 | |||||
| 1 | Cl | H | NO2 | 0.40 | 1.03 ± 0.08 | 2.6 | |
| 2 | H | H | H | 8.59 | 19.2 ± 0.77 | 2.2 | |
| 3 | H | H | NO2 | 1.11 | 2.28 ± 0.14 | 2.1 | |
| 4 | H | NO2 | H | 1.29 | 3.53 ± 0.34 | 2.7 | |
| 5 | H | Cl | NO2 | 0.057 | 1.51 ± 0.02 | 26.5 | |
| 6 | H | Cl | H | 0.39 | 2.46 ± 0.15 | 6.3 | |
| 7 | H | Cl | F | – | 2.44 ± 0.30 | – | |
| 8 | H | H | F | – | 6.78 ± 0.57 | – | |
| 9 | H | F | H | – | 3.63 ± 0.38 | – | |
| 10 | H | H | CF3 | 0.38 | 1.18 ± 0.04 | 3.1 | |
| 11 | H | CF3 | H | 0.49 | 5.95 ± 0.76 | 12.1 | |
| 12 | H | tBu | H | 1.51 | 2.36 ± 0.10 | 1.6 | |
| 13 | H | OH | H | – | >100 | – | |
| 14 | H | H | OH | – | >100 | – | |
| 15 | H | H | Piper | – | 65.9 ± 7.93 | – | |
|
27 |
H |
H |
OGlc |
– |
>100 |
– |
|
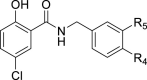 | |||||||
| R4 | R5 | ||||||
| 16 | Me | H | – | 47.7 ± 1.07 | – | ||
|
17 |
H |
Me |
– |
82.4 ± 2.00 |
– |
||
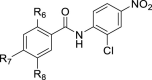 | |||||||
| R6 | R7 | R8 | |||||
| 18 | OH | H | H | 1.68 | 2.03 ± 0.09 | 1.2 | |
| 19 | H | H | H | – | >100 | – | |
| 20 | OH | H | Me | – | 5.68 ± 0.35 | – | |
| 21 | OH | Me | H | – | 21.6 ± 0.53 | – | |
| 22 | OH | H | F | – | 8.15 ± 1.04 | – | |
| Nelfinavir [6] | – | – | – | 3.3 | – | – | |
| Cepharanthine [6] | – | – | – | 2.8 | – | – | |
N.I.: No inhibition; a48 h MTT assay.
Table 2.
PAMPA GI permeability of niclosamide analogues.
| Compounds | PAMPA (10−6 cm/s) | Compounds | PAMPA (10−6 cm/s) |
|---|---|---|---|
| 1 | 8.5 | 13 | 15.9 |
| 2 | 16.0 | 14 | 33.1 |
| 3 | 12.4 | 15 | 43.3 |
| 4 | 19.5 | 16 | 42.6 |
| 5 | 12.3 | 17 | 28.4 |
| 6 | 8.6 | 18 | 15.3 |
| 7 | 11.5 | 19 | 42.2 |
| 8 | 18.2 | 20 | 20.9 |
| 9 | 3.7 | 21 | 6.1 |
| 10 | 6.9 | 22 | 30.7 |
| 11 | 4.2 | 27 | 1.9 |
| 12 | 18.7 |
clogP values were calculated in silico using Molecular Operating Environment (MOE) software and, with the exception of compound 12, were all found to be within the range of 0–5, which is preferred for oral absorption (Fig. S1). Permeability in the gastrointestinal (GI) tract was evaluated using the parallel artificial membrane permeability assay (PAMPA). Verapamil (114.8 × 10−6 cm/s) and ranitidine (1.6 × 10−6 cm/s) were used as positive and negative controls, respectively. Niclosamide exhibited a PAMPA permeability of 8.5 × 10−6 cm/s (Table 2). The permeability of three compounds 15, 16, and 19 was 5-fold higher, but none of them exhibited antiviral efficacy. The PAMPA permeabilities of all the compounds exhibiting anti-SARS-CoV-2 activity were similar to niclosamide suggesting that a prodrug strategy, such as acetylation, might be necessary for the improvement of oral bioavailability.
The cytotoxicity of each niclosamide analogue was firstly measured in conjunction with its antiviral efficacy over a 24 h period, and most compounds exhibited an IC50 higher than 100 M (Fig. 3). To elucidate the difference in the cytotoxicity of all the analogues, the inhibition assay of Vero E6 proliferation was prolonged to 48 h (Table 1). As a result, niclosamide exhibited the highest cytotoxicity against Vero E6 (IC50 = 1.03 M), and the selectivity between antiviral efficacy and cytotoxicity was 2.6. Two compounds, 5 and 11, exhibited selectivity greater than ten folds. Compound 5, bearing 3′-Cl and 4′-NO2 substituents on the aniline moiety, exhibited the best antiviral activity (IC50 = 0.057 M) and slightly less cytotoxic (IC50 = 1.51 M) than niclosamide and improved the selectivity ratio to 26.5. Compound 11 was less cytotoxic than niclosamide as 6.23 M, improving the selectivity to 12.7, however, low PAMPA permeability resulting from trifluoromethyl group might diminish the GI absorption.
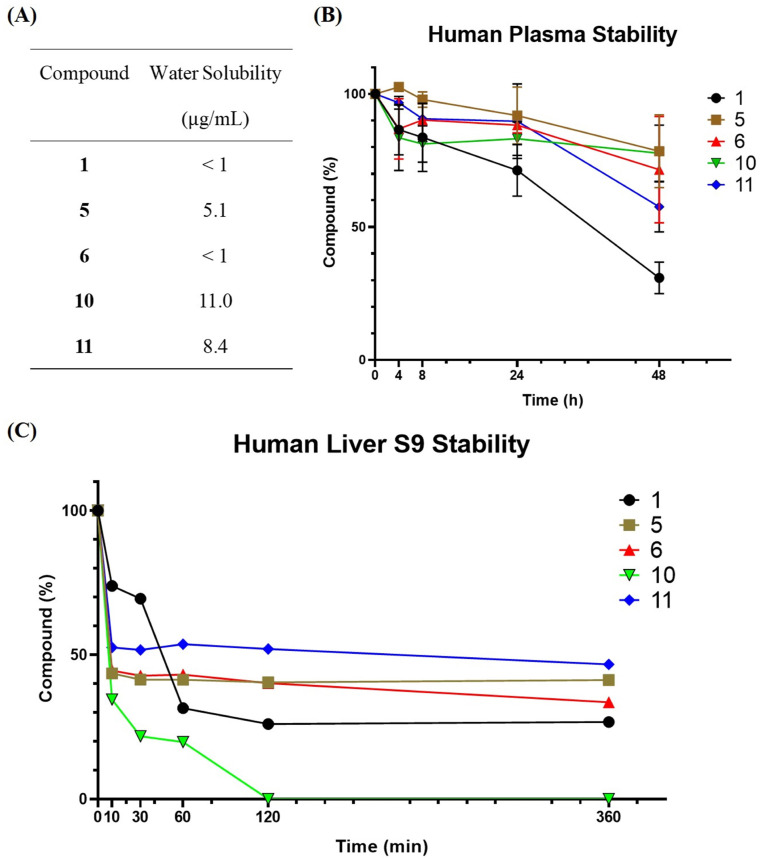
The low oral bioavailability of niclosamide in human body prevented it from being applied to broader use [20]. Therefore, the stability of compound was evaluated in human plasma and liver S9 enzymes to search for more stable analogues and the water solubility was also evaluated (Fig. 4 A). In human plasma, compounds 5, 6, 10, and 11 contained 78%, 71%, 77%, and 58% after 48 h under 37 °C while niclosamide (1) was only 40% remain (Fig. 4B). On the other hand, liver metabolism played an important role in orally administered drug, and compounds 5, 6, and 11 showed slightly better stability than niclosamide (1) (41%, 33%, 47% versus 27%) in liver S9 stability assay (Fig. 4C). These results showed that 5, 6, 11 were better anti-SARS-CoV-2 agents than niclosamide for oral delivery.
Fig. 4.
Water solubility and in vitro stability profiles of niclosamide analogues. (A) Water solubility profile of niclosamide and 4 analogues. (B) Stability analysis of niclosamide and 4 analogues (100 M) in human plasma at 37 °C. (C) Stability analysis of niclosamide and 4 analogues (50 M) with human liver S9 enzyme at 37 °C.
2.4. Fluorescence and in silico experiments for mechanism elucidation
Niclosamide, as a TMEM16F protein inhibitor, was able to prevent virus entry by reducing scramblase activity of TMEM16F, which can be quantified by staining the externalized phosphatidylserine with a fluorescence probe [14]. Subjection of either niclosamide or 5 for 1 h significantly reduced the signal of phosphatidylserine on cell surface triggered by ionomycin. On the other hand, nelfinavir, a SARS-CoV-2 inhibitor targeting viral protease [6], exhibited no inhibition toward phosphatidylserine externalization (Fig. 5 ). The results indicated that compound 5 was an inhibitor of TMEM16F scramblase activity in a similar way of niclosamide.
Fig. 5.
Phosphatidylserine externalization experiment. (A) The phosphatidylserine externalization triggered by ionomycin was visualized by annexin XII after treatment with 100 nM niclosamide and 5 or 500 nM nelfinavir in Vero E6 cells. Scale bar, 500 m. (B) Quantification result of annexin XII positive cells (n = 3).
To elucidate the TMEM16F inhibition mechanism of 5, the computer simulation was performed. Since the crystal structure of human TMEM16F (ANO6) was not available, predicted structure was obtained from AlphaFold protein structure database [21], and the potential ligand binding pockets of TMEM16F were identified using the PrankWeb (Fig. 6 A) [22]. The TMEM16K protein, an analogue protein of TMEM16F, was chose for the validation of AlphaFold-predicted protein structure. The predicted (Uniprot: Q9NW15) and crystal (PDB: 5OC9) structure of human TMEM16K protein were superimposed and showed very high similarity in quaternary protein structure, indicating the prediction was reliable (Fig. S4A). To validate the pocket identification process, crystal structure of TMEM16F from mus musculus (PDB: 6P46), which shared 90% similarity in protein sequence to human's, was subjected to PrankWeb and the top-ranking pocket was at the same place to the predicted human TMEM16F protein (Fig. S5). Moreover, the top-ranking pocket, existed at extracellular domain, was close to the previously reported binding pocket of TMEM16A, which is also a TMEM16F analogue [23]. Altogether, the binding pocket of predicted human TMEM16F protein was used for docking of niclosamide and compound 5.
Fig. 6.
MOE software analysis of niclosamide and 5 to predicted binding site of TMEM16F protein. (A) Alphafold and P2Rank prediction of human TMEM16F protein structure and the binding site at extracellular domain (UniProt: Q4KMQ2). (B) Interaction 2D map of the niclosamide with surrounding residues in the pocket of TMEM16F, (C) Interaction 2D map of the 5 with surrounding residues in the pocket of TMEM16F.
To evaluate the drug-protein interaction, niclosamide and 5 were docked into the predicted binding site by MOE software. The result showed that aniline moiety of niclosamide provided important interaction to the polar residue, such as Gly574, Lys575, and Asp810 inside the binding pocket, however, 2′-Cl was seemed to block the interaction on the other side, including Asp596, Cys600, and Arg809 (Fig. 6B). Nevertheless, the alteration of 2′-Cl to 3′-Cl provided the exposure of chloride atom to the Lys575 and Gly574, enhancing the affinity of 5 to the pocket by the interaction of 4′-NO2 and carbonyl group to Arg809 (Fig. 6C). The computer simulation provided a preliminary understanding of the TMEM16F inhibition, which was a potential target for virus entry inhibition.
3. Conclusion
In this study, 22 niclosamide analogues were synthesized and evaluated for the anti-SARS-CoV-2 activity. Compound 5 was found to exhibit the best anti-SARS-CoV-2 activity (IC50 = 0.057 M), which was 7-fold higher than niclosamide, and the selectivity of cytotoxicity versus antiviral potency was improved from 2.6 (niclosamide) to 26.5. The GI absorption efficacies of these analogues were evaluated by PAMPA assay, however, the effective compounds (2–6, 10–12, 18) contained low permeability in the range of 4–20 (x 10−6 cm/s). Human plasma and liver S9 enzyme assay revealed that compounds 5, 6, and 11 were more stable than niclosamide. The inhibition activity toward TMEM16F was confirmed by phosphatidylserine externalization experiment through fluorescence microscopy, and 5 exhibited the same inhibition pattern to niclosamide. The extracellular binding pocket of AI-predicted human TMEM16F protein was revealed by computer simulation and docking results showed that Arg809 was an important residue for drug-protein interaction. Accordingly, compound 5, 6, 11 are proposed as a better anti-SARS-CoV-2 agents than niclosamide, which are worth for advanced studies in the future.
4. Experimental
4.1. Chemistry
All reagents and solvents were reagent grade and used without further purification, unless otherwise stated. Molecular sieves were activated at 200 °C and cooled down to rt prior to use. Reactions were monitored by analytical TLC on 0.25 mm Merck Millipore silica gel 60 F254 using p-anisaldehyde, ninhydrin, and ceric ammonium molybdate as visualizing agents. Flash column chromatography was performed employing 230–400 mesh silica gel. NMR spectra were acquired by using Bruker-AV-400 (400 MHz). Chemical shifts (δ) are given in ppm relative to 1H: 7.26 ppm, 13C: 77.16 ppm for CDCl3; 1H: 3.31 ppm, 13C: 49.0 ppm for CD3OD; 1H: 2.50 ppm, 13C: 39.52 ppm for DMSO‑d 6. Splitting patterns are reported as s (singlet), br s (broad singlet) d (doublet), br d (broad doublet), t (triplet), q (quartet) and m (multiplet). Coupling constants (J) are given in Hertz (Hz). Purities of biologically tested compounds were carried out on a HITACHI D-2000 Elite HPLC system with pump L-2130. Exact mass measurements were performed on VG platform electrospray ESI/MS or BioTOF II. Melting points were measured on a FARGO melting point apparatus MP-1D (Mandarin Scientific Co., Ltd.).
4.1.1. General procedure of amide bond formation
The benzoic acid derivatives (0.75 mmol) was added to thionyl chloride (2.0 mL, 0.10 mol) and heated to 60 °C with stirring. After 2 h, the mixture was cooled to rt and concentrated under reduced pressure to give the solid acyl chloride which was advanced to the next step without further purification. The crude acyl chloride residue and the aniline derivative (1.5 mmol) were suspended in anhydrous DCM (5.0 mL) under nitrogen atmosphere, and the mixture was stirred overnight at rt. The mixture was diluted with DCM, washed with 1 M HCl solution, brine, dried over MgSO4 and concentrated under reduced pressure. The crude residue was purified by recrystallization or column chromatography to afford benzamide derivative.
4.1.2. 5-Chloro-N-(2-chloro-4-nitrophenyl)-2-hydroxybenzamide (1)
Compound 1 (61.2 mg, 31% 2-step yield), as a yellow solid, was synthesized according to the general procedure using 5-chlorosalicylic acid (103 mg, 0.597 mmol) and 2-chloro-4-nitroaniline (206 mg, 1.19 mmol), and recrystallized from EtOAc/hexanes = 1/4; m.p. 240 °C; 1H NMR (400 MHz, DMSO‑d 6) δ 11.58 (s, 1H, OH), 8.82 (d, J = 9.2 Hz, 1H), 8.43 (d, J = 2.5 Hz, 1H), 8.29 (dd, J = 9.2, 2.5 Hz, 1H), 7.95 (d, J = 2.7 Hz, 1H), 7.52 (dd, J = 8.8, 2.7 Hz, 1H), 7.07 (d, J = 8.8 Hz, 1H) ppm; 13C NMR (100 MHz, DMSO‑d 6) δ 162.8 (NC O), 155.8, 142.5, 141.4, 134.0, 130.0, 124.9, 124.0, 123.4, 122.4, 120.8, 119.4, 119.4 ppm; HRMS (ESI-TOF) calcd. for C13H7Cl2N2O4–H [M − H]-: 324.9788; found: 324.9792; HPLC purity 98.9% (tR: 10.6 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.3. 5-Chlorosalicylanilide (2)
Compound 2 (160 mg, 65% 2-step yield), as a pale yellow solid, was synthesized according to the general procedure using 5-chlorosalicylic acid (146 mg, 0.843 mmol) and aniline (0.15 mL, 1.7 mmol), and recrystallized from MeOH/DCM/hexanes = 1/1/4; m.p. 223 °C; 1H NMR (400 MHz, DMSO‑d 6) δ 11.89 (s, 1H), 10.48 (s, 1H), 7.95 (d, J = 2.7 Hz, 1H), 7.69 (d, J = 7.9 Hz, 2H), 7.46 (dd, J = 8.8, 2.7 Hz, 1H), 7.38 (t, J = 7.9 Hz, 2H), 7.15 (t, J = 7.5 Hz, 1H), 7.01 (d, J = 8.8 Hz, 1H) ppm; 13C NMR (100 MHz, DMSO‑d 6) δ 165.0 (NC O), 157.2, 138.1, 133.0, 128.8, 128.4, 124.3, 122.5, 120.8, 119.7, 119.2 ppm; HRMS (ESI-TOF) calcd. for C13H11ClNO2+H [M+H]+: 248.0473, found 248.0470; HPLC purity 97.1% (tR: 9.6 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.4. 5-Chloro-N-(4-nitrophenyl)-2-hydroxybenzamide (3)
Compound 3 (44 mg, 23% 2-step yield), a pale yellow solid, was synthesized according to the general procedure by using 5-chlorosalicylic acid (113 mg, 0.655 mmol) and 4-nitroaniline (181 mg, 1.31 mmol), and recrystallized from EtOAc/hexanes = 1/3; m.p. 275 °C; 1H NMR (400 MHz, DMSO‑d 6) δ 11.0 (s, 1H, NH), 8.27 (d, J = 9.2 Hz, 2H, H-3′, H-5′), 7.98 (d, J = 9.2 Hz, 2H, H-2′, H-6′), 7.81 (d, J = 2.7 Hz, 1H, H-6), 7.46 (dd, J = 8.8, 2.7 Hz, 1H, H-4), 7.02 (d, J = 8.8 Hz, 1H, H-3) ppm; 13C NMR (100 MHz, DMSO‑d 6) δ 165.1 (NC O), 156.2, 144.7, 142.7, 133.0, 128.8, 125.0, 122.6, 121.0, 120.0, 119.1 ppm; HRMS (ESI-TOF) calcd. for C13H8ClN2O4–H [M − H]-: 291.0178, found: 291.0177; HPLC purity 95.2% (tR: 9.6 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.5. 5-Chloro-N-(3-nitrophenyl)-2-hydroxybenzamide (4)
Compound 4 (65 mg, 38% 2-step yield), a pale yellow solid, was synthesized according to the general procedure using 5-chlorosalicylic acid (101 mg, 0.585 mmol) and 3-nitroaniline (162 mg, 1.17 mmol), and recrystallized from MeOH/DCM/hexanes = 1/5/50; m.p. 260 °C; 1H NMR (400 MHz, DMSO‑d 6) δ 10.89 (s, 1H), 8.77 (s, 1H), 8.07 (d, J = 7.9 Hz, 1H), 8.00 (d, J = 8.2 Hz, 1H), 7.88 (d, J = 2.5 Hz, 1H), 7.67 (t, J = 8.2 Hz, 1H), 7.47 (dd, J = 8.7, 2.5 Hz, 1H), 7.02 (d, J = 8.8 Hz, 1H) ppm; 13C NMR (100 MHz, DMSO‑d 6) δ 165.3 (NC O), 156.6, 148.0, 139.5, 133.1, 130.2, 128.6, 126.5, 122.5, 120.3, 119.1, 118.6, 114.6 ppm; HRMS (ESI-TOF) calcd. for C13H10ClN2O4+H [M+H]+: 293.0324, found: 293.0321; HPLC purity 97.7% (tR: 9.4 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.6. 5-Chloro-N-(3-chloro-4-nitrophenyl)-2-hydroxybenzamide (5)
Compound 5 (53 mg, 25% 2-step yield), a yellow solid, was synthesized according to the general procedure using 5-chlorosalicylic acid (113 mg, 0.655 mmol) and 3-chloro-4-nitroaniline (226 mg, 1.31 mmol), and recrystallized from EtOAc/DCM/hexanes = 1/2/10; m.p. 258 °C; 1H NMR (400 MHz, DMSO‑d 6) δ 10.97 (s, 1H, NH), 8.22–8.14 (m, 2H, H-2′, H-5′), 7.85 (dd, J = 9.1, 2.1 Hz, 1H, H-6′), 7.79 (d, J = 2.5 Hz, 1H, H-6), 7.47 (dd, J = 8.8, 2.5 Hz, 1H, H-4), 7.03 (d, J = 8.8 Hz, 1H, H-3) ppm; 13C NMR (100 MHz, DMSO‑d 6) δ 165.2 (NC O), 156.1, 143.5, 141.9, 133.1, 128.8, 127.6, 126.7, 122.6, 121.6, 120.9, 119.0, 118.9 ppm; HRMS (ESI-TOF) calcd. for C13H7Cl2N2O4–H [M − H]-: 324.9788, found: 324.9789; HPLC purity 96.5% (tR: 10.9 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.7. 5-Chloro-N-(3-chlorophenyl)-2-hydroxybenzamide (6)
Compound 6 (86 mg, 41% 2-step yield), a white solid, was synthesized according to the general procedure by using 5-chlorosalicylic acid (128 mg, 0.740 mmol) and 3-chloroaniline (189 mg, 1.48 mmol), and recrystallized from THF/hexanes = 1/5; m.p. 230 °C; 1H NMR (400 MHz, DMSO‑d 6) δ 10.63 (s, 1H, NH), 7.92 (s, 1H, H-2′), 7.87 (d, J = 2.6 Hz, 1H, H-6), 7.59 (br d, J = 7.4 Hz, 1H), 7.47 (dd, J = 8.8, 2.5 Hz, 1H, H-4), 7.40 (t, J = 8.1 Hz, 1H, H-5′), 7.20 (br d, J = 8.0 Hz, 1H), 7.01 (d, J = 8.8 Hz, 1H, H-3) ppm; 13C NMR (100 MHz, DMSO‑d 6) δ 165.0 (NC O), 156.7, 139.7, 133.1, 133.0, 130.5, 128.6, 123.9, 122.5, 120.2, 120.0, 119.1, 119.0 ppm; HRMS (ESI-TOF) calcd. for C13H10Cl2NO2+H [M+H]+: 282.0083, found: 282.0081; HPLC purity 99.1% (tR: 11.3 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.8. 5-Chloro-N-(3-chloro-4-fluorophenyl)-2-hydroxybenzamide (7)
Compound 7 (285 mg, 75% 2-step yield), a white solid, was synthesized according to the general procedure using 5-chlorosalicylic acid (219 mg, 1.27 mmol) and 3-chloro-4-fluoroaniline (369 mg, 2.53 mmol), and recrystallized from MeOH/DCM/hexanes = 1/1/4; m.p. 236 °C; 1H NMR (400 MHz, DMSO‑d 6) δ 11.62 (s, 1H, OH), 10.58 (s, 1H, NH), 8.03 (dd, J = 6.8, 2.4 Hz, 1H), 7.87 (d, J = 2.4 Hz, 1H, H-6), 7.68–7.61 (m, 1H), 7.50–7.39 (m, 2H), 7.01 (d, J = 8.8 Hz, 1H, H-3) ppm; 13C NMR (100 MHz, DMSO‑d 6) δ 165.1 (NC O), 153.5 (d, J = 238.9 Hz), 135.9 (d, J = 4.9 Hz), 132.9, 128.5, 121.9, 120.9, 120.8, 119.8, 119.3, 119.1, 117.1, 116.9 ppm; HRMS (ESI-TOF) calcd. for C13H7Cl2FNO2–H [M − H]-: 297.9843, found: 297.9835; HPLC purity 97.5% (tR: 11.5 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.9. 5-Chloro-N-(4-fluorophenyl)-2-hydroxybenzamide (8)
Compound 8 (81.2 mg, 40% 2-step yield), a white solid, was synthesized according to the general procedure using 5-chlorosalicylic acid (131 mg, 0.756) and 4-fluoroaniline (143 L, 1.51 mmol), and recrystallized from DCM/hexanes = 1/5; m.p. 252 °C; 1H NMR (400 MHz, DMSO‑d 6) δ 11.84 (s, 1H, OH), 10.48 (s, 1H, NH), 7.93 (d, J = 2.6 Hz, 1H, H-6), 7.78–7.68 (m, 2H, H-2′, H-6′), 7.46 (dd, J = 8.8, 2.6 Hz, 1H, H-4), 7.27–7.17 (m, 2H, H-3′, H-5′), 7.00 (d, J = 8.8 Hz, 1H, H-3) ppm; 13C NMR (100 MHz, DMSO‑d 6) δ 165.1 (NC O), 158.7 (d, J = 239.5 Hz), 157.0, 134.4 (d, J = 2.3 Hz), 133.1, 128.3, 122.8 (d, J = 7.9 Hz), 122.6, 119.6, 119.1, 115.4 (d, J = 22.2 Hz) ppm; HRMS (ESI-TOF) calcd. for C13H8ClFNO2–H [M − H]-: 264.0233, found: 264.0232; HPLC purity 98.2% (tR: 9.3 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.10. 5-Chloro-N-(3-fluorophenyl)-2-hydroxybenzamide (9)
Compound 9 (69.6 mg, 40% 2-step yield), a white solid, was synthesized according to the general procedure using 5-chlorosalicylic acid (113 mg, 0.655 mmol) and 3-fluoroaniline (126 L, 1.31 mmol), and recrystallized from DCM/hexanes = 2/1; m.p. 248 °C; 1H NMR (400 MHz, DMSO‑d 6) δ 11.67 (s, 1H, OH), 10.61 (s, 1H, NH), 7.87 (d, J = 2.7 Hz, 1H, H-6), 7.71 (dt, J = 11.6, 2.0 Hz, 1H), 7.49–7.44 (m, 2H), 7.44–7.36 (m, 1H), 7.01 (d, J = 8.8 Hz, 1H, H-3), 7.00–6.94 (m, 1H) ppm; 13C NMR (100 MHz, DMSO‑d 6) δ 164.9 (NC O), 163.6 (d, J = 239.9 Hz), 156.6, 140.0 (d, J = 11.1 Hz), 133.0, 130.5 (d, J = 9.5 Hz), 128.6, 122.6, 120.3, 119.1, 116.3 (d, J = 2.4 Hz), 110.7 (d, J = 21.0 Hz), 107.4 (d, J = 26.1 Hz) ppm; HRMS (ESI-TOF) calcd. for C13H8ClFNO2–H [M − H]-: 264.0233, found: 264.0233; HPLC purity 95.4% (tR: 10.2 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.11. 5-Chloro-N-(4-trifluoromethylphenyl)-2-hydroxybenzamide (10)
Compound 10 (157.5 mg, 66% 2-step yield), a white solid, was synthesized according to the general procedure using 5-chlorosalicylic acid (130 mg, 0.750 mmol) and 4-trifluoromethylaniline (189 L, 1.50 mmol), and recrystallized from DCM/hexanes = 1/4; m.p. 230 °C; 1H NMR (400 MHz, DMSO‑d 6) δ 10.85 (s, 1H, NH), 7.94 (br d, J = 8.5 Hz, 2H, H-3′, H-5′), 7.86 (d, 1H, J = 2.6 Hz, H-6), 7.74 (br d, J = 8.5 Hz, 2H, H-2′, H-6′), 7.46 (dd, 1H, J = 8.8, 2.6 Hz, H-4), 7.01 (d, 1H, J = 8.8 Hz, H-3) ppm; 13C NMR (100 MHz, DMSO‑d 6) δ 165.1 (NC O), 156.7, 142.0, 133.0, 128.7, 126.1, 126.1, 124.4 (q, J = 269.4 Hz), 124.0 (q, J = 32.0 Hz), 122.5, 120.5, 120.4, 119.2 ppm; HRMS (ESI-TOF) calcd. for C14H8ClF3NO2 [M − H]-: 314.0201, found: 314.0202; HPLC purity 95.4% (tR: 11.8 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.12. 5-Chloro-N-(3-trifluoromethylphenyl)-2-hydroxybenzamide (11)
Compound 11 (113.6 mg, 48% 2-step yield), a white solid, was synthesized according to the general procedure using 5-chlorosalicylic acid (130 mg, 0.750 mmol) and 3-trifluoromethylaniline (189 L, 1.50 mmol), and recrystallized from DCM/hexanes = 1/4; m.p. 207 °C; 1H NMR (400 MHz, DMSO‑d 6) δ 10.86 (s, 1H, NH), 8.21 (s, 1H, H-2′), 7.92 (br d, J = 8.2 Hz, 1H), 7.88 (d, J = 2.7 Hz, 1H, H-6), 7.61 (t, J = 8.0 Hz, 1H, H-5′), 7.52–7.42 (m, 2H), 7.01 (d, J = 8.8 Hz, 1H, H-3); 13C NMR (100 MHz, DMSO‑d 6) δ 165.3 (NC O), 157.0, 139.1, 133.1, 129.5 (q, J = 31.7 Hz), 128.5, 124.2, 124.1 (q, J = 270.6 Hz), 122.3, 120.4 (q, J = 3.6 Hz), 120.1, 119.2, 120.1, 116.7 (q, J = 3.6 Hz) ppm; HRMS (ESI-TOF) calcd. for C14H8ClF3NO2–H [M − H]-: 314.0201, found: 314.0202; HPLC purity 96.2% (tR: 11.5 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.13. 5-Chloro-N-(3-tert-butylphenyl)-2-hydroxybenzamide (12)
Compound 12 (44.3 mg, 20% 2-step yield), a pale yellow solid, was synthesized according to the general procedure by using 5-chlorosalicylic acid (126 mg, 0.730 mmol) and 3-tert-butylaniline (218 mg, 1.46 mmol), and recrystallized from DCM/hexanes = 1/4; m.p. 175 °C; 1H NMR (400 MHz, DMSO‑d 6) δ 11.94 (s, 1H, OH), 10.46 (s, 1H, NH), 7.97 (d, J = 2.6 Hz, 1H, H-6), 7.69 (s, 1H, H-2′), 7.57 (br d, J = 7.9 Hz, 1H), 7.46 (dd, 1H, J = 8.8, 2.6 Hz, H-4), 7.30 (t, J = 7.9 Hz, 1H, H-5′), 7.18 (br d, J = 7.9 Hz, 1H), 7.00 (d, J = 8.8 Hz, 1H, H-3), 1.29 (s, 9H, C(CH 3)3); 13C NMR (100 MHz, DMSO‑d 6) δ 165.2 (NC O), 157.4, 151.4, 137.8, 133.1, 128.5, 128.3, 122.5, 121.4, 119.5, 119.2, 118.2, 117.9, 34.5(C(CH3)3), 31.2 (C(CH3)3) ppm; HRMS (ESI-TOF) calcd. for C17H19ClNO2+H [M+H]+: 304.1099, found: 304.1095; HPLC purity 97.7% (tR: 13.6 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.14. 5-Chloro-N-(3-hydroxybenzyl)-2-hydroxybenzamide (13)
Compound 13 (37.0 mg, 19% 2-step yield), a white solid, was synthesized according to the general procedure by using 5-chlorosalicylic acid (130 mg, 0.750 mmol) and 3-hydroxybenzylamine (166 mg, 1.50 mmol), and was purified by column chromatography (silica gel; EtOAc/hexanes = 1/2); m.p. 251 °C; 1H NMR (400 MHz, CD3OD) δ 7.98 (d, J = 2.6 Hz, 1H), 7.39 (dd, J = 8.8, 2.6, 2H), 7.28 (t, J = 2.2, 1H), 7.16 (t, J = 8.0, 1H), 7.07–7.01 (m, 1H), 6.95 (d, J = 8.8 Hz, 1H), 6.63–6.57 (m, 1H); 13C NMR (100 MHz, CD3OD): δ 167.1 (NC O), 159.0, 159.0, 140.1, 134.5, 130.6, 129.6, 125.2, 120.1, 119.8, 113.4, 112.9, 109.5 ppm; HRMS (ESI-TOF) calcd. for C13H10ClNO3+H [M+H]+: 264.0422, found: 264.0419; HPLC purity 95.4% (tR: 6.8 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.15. 5-Chloro-N-(4-hydroxybenzyl)-2-hydroxybenzamide (14)
Compound 14 (65.7 mg, 17% 2-step yield), a white solid, was synthesized according to the general procedure using 5-chlorosalicylic acid (260 mg, 1.50 mmol) and 4-hydroxybenzylamine (330 mg, 3.00 mmol), and was purified by column chromatography (silica gel; EtOAc/hexanes = 1/2); m.p. 214 °C; 1H NMR (400 MHz, CD3OD) δ 7.96 (d, J = 2.6 Hz, 1H), 7.45–7.40 (m, 2H), 7.38 (dd, J = 8.8, 2.6, 1H), 6.94 (d, J = 8.8, 1H), 6.82–6.75 (m, 2H); 13C NMR (100 MHz, CD3OD): δ 167.3 (NC O), 159.2, 156.0, 134.4, 130.6, 129.2, 125.1, 124.5, 120.0, 119.3, 116.3 ppm; HRMS (ESI-TOF) calcd. for C13H10ClNO3–H [M − H]-: 262.0276, found: 262.0276; HPLC purity 95.1% (tR: 6.4 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.16. 5-Chloro-N-(4-(piperidin-1-yl)phenyl)-2-hydroxybenzamide (15)
Compound 15 (25.2 mg, 10% 2-step yield), a white solid, was synthesized according to the general procedure using 5-chlorosalicylic acid (130 mg, 0.750 mmol) and 4-piperidinoaniline (250 mg, 1.50 mmol), and was purified by column chromatography (silica gel; EtOAc/hexanes = 1/8); m.p. 219 °C; 1H NMR (400 MHz, DMSO‑d 6) δ 9.42 (br s, 1H, NH), 7.97 (d, J = 2.6 Hz, 1H, H-6), 7.43 (dd, J = 8.8, 2.6 Hz, 1H, H-4), 7.21 (d, J = 7.8 Hz, 2H), 7.14 (d, J = 7.8 Hz, 2H) 6.93 (d, J = 8.8 Hz, 1H, H-3), 4.45 (d, J = 5.4 Hz, 2H, OCNHCH 2), 2.27 (s, 3H, CH 3); 13C NMR (100 MHz, DMSO‑d 6) δ 167.4 (NC O), 158.6, 138.7, 137.6, 133.4, 128.4, 128.0, 127.7, 127.4, 124.5, 122.4, 119.4, 116.9, 42.5 (OCNHCH2), 21.1 (CH3) ppm; HRMS (ESI-TOF) calcd. for C18H19ClN2O2–H [M − H]-: 329.1062, found: 329.1061; HPLC purity 98.0% (tR: 10.7 min, Alltima C8, 150 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.17. 5-Chloro-N-(4-methylbenzyl)-2-hydroxybenzamide (16)
Compound 16 (134 mg, 67% 2-step yield), a white solid, was synthesized according to the general procedure using 5-chlorosalicylic acid (131 mg, 0.759 mmol) and 4-methylbenzylamine (193 L, 1.52 mmol), and was recrystallized from DCM/hexanes = 1/5; m.p. 169 °C; 1H NMR (400 MHz, DMSO‑d 6) δ 9.39 (t, J = 5.7 Hz, 1H, NH), 7.98 (d, J = 2.6 Hz, 1H, H-6), 7.45 (dd, J = 8.8, 2.6 Hz, 1H, H-4), 7.22 (t, J = 7.5 Hz, 1H), 7.16-7.04 (m, 3H) 6.95 (d, J = 8.8 Hz, 1H, H-3), 4.45 (d, J = 5.4 Hz, 2H, OCNHCH 2), 2.27 (s, 3H, CH 3); 13C NMR (100 MHz, DMSO‑d 6) δ 167.4 (NC O), 158.8, 136.1, 135.7, 133.3, 129.0, 127.4, 122.1, 119.4, 116.9, 42.3 (OCNHCH2), 20.7 (CH3) ppm; HRMS (ESI-TOF) calcd. for C15H13ClNO2–H [M − H]-: 274.0640, found: 274.0638; HPLC purity 98.1% (tR: 10.5 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.18. 5-Chloro-N-(3-methylbenzyl)-2-hydroxybenzamide (17)
Compound 17 (95.3 mg, 48% 2-step yield), a white solid, was synthesized according to the general procedure using 5-chlorosalicylic acid (131 mg, 0.759 mmol) and 3-methylbenzylamine (193 L, 1.52 mmol), and was recrystallized from DCM/hexanes = 1/5; m.p. 150 °C; 1H NMR (400 MHz, DMSO‑d 6) δ 9.42 (br s, 1H, NH), 7.97 (d, J = 2.6 Hz, 1H, H-6), 7.43 (dd, J = 8.8, 2.6 Hz, 1H, H-4), 7.21 (d, J = 7.8 Hz, 2H), 7.14 (d, J = 7.8 Hz, 2H) 6.93 (d, J = 8.8 Hz, 1H, H-3), 4.45 (d, J = 5.4 Hz, 2H, OCNHCH 2), 2.27 (s, 3H, CH 3); 13C NMR (100 MHz, DMSO‑d 6) δ 167.4 (NC O), 158.6, 138.7, 137.6, 133.4, 128.4, 128.0, 127.7, 127.4, 124.5, 122.4, 119.4, 116.9, 42.5 (OCNHCH2), 21.1 (CH3) ppm; HRMS (ESI-TOF) calcd. for C15H13ClNO2–H [M − H]-: 274.0640, found: 274.0635; HPLC purity 98.7% (tR: 10.5 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.19. N-(2-chloro-4-nitrophenyl)-2-hydroxybenzamide (18)
Compound 18 (63 mg, 31% 2-step yield), a yellow solid, was synthesized according to the general procedure using salicylic acid (96 mg, 0.69 mmol) and 2-chloro-4-nitroaniline (239 mg, 1.39 mmol), and was purified by column chromatography (silica gel; EtOAc/DCM/hexanes = 1/1/6); m.p. 242 °C; 1H NMR (400 MHz, DMSO‑d 6) δ 11.56 (s, 1H), 8.86 (d, J = 9.2 Hz, 1H), 8.43 (d, J = 2.4 Hz, 1H), 8.29 (dd, J = 9.2, 2.4 Hz, 1H), 8.04 (dd, J = 7.8, 1.5 Hz, 1H), 7.53–7.44 (m, 1H), 7.12–6.95 (m, 1H) ppm; 13C NMR (100 MHz, DMSO‑d 6) δ 164.3 (NC O), 156.7, 142.3, 141.7, 134.6, 131.2, 124.9, 124.0, 122.2, 120.6, 120.0, 117.9, 117.2 ppm; HRMS (ESI-TOF) calcd. for C13H10ClN2O4+H [M+H]+: 293.0324, found: 293.0320; HPLC purity 98.4% (tR: 13.8 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.20. N-(2-chloro-4-nitrophenyl)benzamide (19)
Compound 19 (48.5 mg, 20% yield), a yellow solid, was synthesized according to the general procedure using benzoic acid (107 mg, 0.876 mmol) and 2-chloro-4-nitroaniline (302 mg, 1.75 mmol), and was purified by column chromatography (silica gel; EtOAc/hexanes = 1/10); m.p. 197 °C; 1H NMR (400 MHz, DMSO‑d 6) δ 10.34 (s, 1H), 8.42 (d, J = 2.6 Hz, 1H), 8.27 (dd, J = 8.9, 2.6 Hz, 1H), 8.07 (d, J = 8.8 Hz, 1H), 8.04-7.98 (m, 2H), 7.69-7.62 (m, 1H), 7.61-7.54 (m, 2H); 13C NMR (100 MHz, DMSO‑d 6) δ 165.6 (NC O), 144.5, 141.6, 133.5, 132.4, 128.7, 128.2, 128.0, 126.8, 125.0, 123.0 ppm; HRMS (ESI-TOF) calcd. for C13H10ClN2O3+H [M+H]+: 277.0374, found: 277.0372; HPLC purity 96.9% (tR: 9.4 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.21. 5-Methyl-N-(2-chloro-4-nitrophenyl)-2-hydroxybenzamide (20)
Compound 20 (23 mg, 10% yield), a pale yellow solid, was synthesized according to the general procedure using 5-methylsalicylic acid (116 mg, 0.765 mmol) and 2-chloro-4-nitroaniline (264 mg, 1.53 mmol), and was recrystallized from ether; m.p. 233 °C; 1H NMR (400 MHz, DMSO‑d 6) δ 11.58 (s, 1H, NH), 8.85 (d, J = 9.3 Hz, 1H, H-6′), 8.42 (d, J = 2.6 Hz, 1H, H-3′), 8.28 (dd, J = 9.3, 2.6 Hz, 1H, H-5′), 7.82 (d, J = 2.0 Hz, 1H, H-6), 7.29 (dd, J = 8.3, 2.0 Hz, 1H, H-4), 6.96 (d, J = 8.3 Hz, 1H, H-3), 2.27 (s, 3H, CH 3) ppm; 13C NMR (100 MHz, DMSO‑d 6) δ 164.2 (NC O), 154.5, 142.3, 141.8, 135.3, 131.0, 128.7, 124.9, 124.0, 122.2, 120.6, 117.5, 117.2, 20.1 (CH3) ppm; HRMS (ESI-TOF) calcd. for C14H10ClN2O4–H [M − H]-: 305.0335, found: 305.0320; HPLC purity 98.6% (tR: 9.7 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.22. 4-Methyl-N-(2-chloro-4-nitrophenyl)-2-hydroxybenzamide (21)
Compound 21 (35 mg, 15% yield), a white solid, was synthesized according to the general procedure using 4-methylsalicylic acid (120 mg, 0.787 mmol) and 2-chloro-4-nitroaniline (272 mg, 1.58 mmol), and was recrystallized from THF/hexanes = 1/5; m.p. 275 °C; 1H NMR (400 MHz, DMSO‑d 6) δ 11.84 (s, 1H, NH), 8.87 (d, J = 9.3 Hz, 1H, H-6′), 8.43 (d, J = 2.6 Hz, 1H, H-3′), 8.28 (dd, J = 9.3, 2.6 Hz, 1H, H-5′), 7.92 (d, J = 8.0 Hz, 1H), 6.90–6.74 (m, 2H), 2.30 (s, 3H, CH 3) ppm; 13C NMR (100 MHz, DMSO‑d 6) δ 169.9 (NC O), 164.3, 145.0, 142.2, 131.1, 125.6, 124.8, 123.9, 122.2, 120.5, 118.8, 117.4, 115.5, 21.1 (CH3) ppm; HRMS (ESI-TOF) calcd. for C14H10ClN2O4–H [M − H]-: 305.0335, found: 305.0336; HPLC purity 95.9% (tR: 9.6 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 45/55–90/10, 1 mL/min, 20 min).
4.1.23. 5-Fluoro-N-(2-chloro-4-nitrophenyl)-2-hydroxybenzamide (22)
Compound 22 (22 mg, 10% yield), a pale yellow solid, was synthesized according to the general procedure using 5-fluorolsalicylic acid (120 mg, 0.769 mmol) and 2-chloro-4-nitroaniline (265 mg, 1.54 mmol), and was recrystallized from DCM; m.p. 250 °C; 1H NMR (400 MHz, DMSO‑d 6) δ 11.58 (s, 1H, NH), 8.83 (d, J = 9.2 Hz, 1H, H-6′), 8.42 (d, J = 2.4 Hz, 1H, H-3′), 8.28 (dd, J = 9.2, 2.4 Hz, 1H, H-5′), 7.71 (dd, J = 9.6, 3.2 Hz, 1H, H-6), 7.36 (td, J = 8.3, 3.2 Hz, 1H, H-4), 7.08 (dd, J = 8.9, 4.5 Hz, 1H, H-3) ppm; 13C NMR (100 MHz, DMSO‑d 6) δ 162.9 (NC O), 155.4 (d, J = 237.1 Hz), 153.1, 142.5, 141.4, 124.9, 124.0, 122.3, 121.5 (d, J = 23.3 Hz), 120.6, 118.8 (d, J = 7.6 Hz), 118.8 (d, J = 6.8 Hz), 116.1 (d, J = 24.3 Hz) ppm; HRMS (ESI-TOF) calcd. for C13H7Cl2N2O4–H [M − H]-: 309.0084, found: 309.0087; HPLC purity 97.9% (tR: 14.2 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 10/90–90/10, 1 mL/min, 20 min).
4.1.24. 4-Nitrophenyl-2,3,4,6-tetra-O-acetyl-β-d-glucopyranoside (24)
Glucosyl imidate 23 was synthesized as previously reported [24]. BF3·Et2O (2.3 L, 0.020 mmol) was added to a suspension of compound 23 (110 mg, 0.230 mmol), 4-nitrophenol (26 mg, 0,19 mmol) and activated 4 Å molecular sieves in DCM (2 mL) at 0 °C under a nitrogen atmosphere with stirring. After 2 h, the reaction was quenched by TEA, filtered, concentrated under reduced pressure and purified by column chromatography (silica gel; EtOAc/DCM/hexanes = 1/1/3) to give compound 24 (77 mg, 74%) as white solid; 1H NMR (400 MHz, CDCl3) δ 8.21 (d, J = 9.2 Hz, 2H, Ar–H), 7.07 (d, J = 9.2 Hz, 2H, Ar–H), 5.36–5.26 (m, 2H), 5.25–5.14 (m, 2H), 4.28 (dd, J = 12.4, 5.6 Hz, 1H), 4.18 (dd, J = 12.4, 2.4 Hz, 1H), 3.98–3.89 (m, 1H), 2.07 (s, 3H), 2.06 (s, 3H), 2.06 (s, 3H), 2.05 (s, 3H) ppm.
4.1.25. 4-Aminophenyl-2,3,4,6-tetra-O-acetyl-β-d-glucopyranoside (25)
A suspension of compound 24 (77 mg, 0.16 mmol) and 10% Pd/C (8 mg) in DCM (1.0 mL) and MeOH (2.0 mL) was stirred under an H2 atmosphere. After 3.5 h, the mixture was filtered, concentrated under reduced pressure, and purified by column chromatography (silica gel; EtOAc/hexanes = 7/3) to afford compound 25 (67 mg, 93%) as white foam; 1H NMR (200 MHz, CDCl3) δ 6.84 (d, J = 8.7 Hz, 2H, Ar–H), 6.62 (d, J = 8.7 Hz, 2H, Ar–H), 5.29–5.09 (m, 3H), 4.90 (dd, J = 5.5, 2.1 Hz, 1H), 4.34–4.09 (m, 2H), 3.86–3.70 (m, 1H), 2.08 (s, 3H), 2.07 (s, 3H), 2.04 (s, 3H), 2.03 (s, 3H) ppm.
4.1.26. 5-Chloro-N-(4-phenyl-2,3,4,6-tetra-O-acetyl-β-d-glucopyranoside)-2-hydroxybenzamide (26)
Compound 26 (39 mg, 38%), a white solid, was synthesized according to the general procedure using 5-chlorosalicylic acid (29.5 mg, 0.171 mmol) and compound 25 (149 mg, 0.340 mmol), and recrystallized from DCM/hexanes = 1/5; 1H NMR (400 MHz, CDCl3) δ 11.9 (s, 1H, OH), 8.02 (s, 1H, NH), 7.58–7.45 (m, 3H, Ar–H), 7.39 (dd, J = 11.3, 2.4 Hz, 1H, Ar–H), 7.07–6.96 (m, 3H, Ar–H), 5.34–5.23 (m, 2H), 5.17 (t, J = 9.7 Hz, 1H), 5.05 (d, J = 7.5 Hz, 1H, H-1″), 4.29 (dd, J = 12.3, 5.3 Hz, 1H), 4.17 (dd, J = 12.3, 2.3 Hz, 1H), 3.90–3.80 (m, 1H), 2.09 (s, 3H), 2.08 (s, 3H), 2.05 (s, 3H), 2.04 (s, 3H) ppm; 13C NMR (100 MHz, DMSO‑d 6) δ 170.8, 170.4, 169.6, 169.5, 160.5, 154.6, 134.7, 131.8, 125.2, 123.8, 123.2, 120.6, 117.9, 99.5 (C-1″), 72.8, 72.2, 71.2, 68.3, 62.0, 20.9, 20.8, 20.8, 20.8 ppm.
4.1.27. 5-Chloro-N-(4-phenyl-β-d-glucopyranoside)-2-hydroxybenzamide (27)
NaOMe (5 mg, 0.09 mmol) was added to a stirred suspension of compound 26 (15 mg, 0.025 mmol) in MeOH (1 mL) and DCM (1 mL). After 30 min, the reaction was neutralized by the addition of Amberlite H+, filtered, concentrated under reduced pressure, and recrystallized from MeOH/EtOAc/hexanes = 1/2/20 to give compound 27 (6.6 mg, 61%) as a white solid; m.p. 190 °C; 1H NMR (400 MHz, CD3OD) δ 7.98 (d, J = 2.6 Hz, 1H, H-6), 7.58 (d, J = 9.0 Hz, 2H), 7.39 (dd, J = 8.8, 2.6 Hz, 1H, H-4), 7.13 (d, J = 9.0 Hz, 2H), 6.95 (d, J = 8.8 Hz, 1H, H-3), 3.91 (dd, J = 12.0, 1.8 Hz, 1H), 3.71 (dd, J = 12.0, 5.4 Hz, 1H), 3.54-3.33 (m, 5H) ppm; 13C NMR (100 MHz, DMSO‑d 6) δ 167.4 (NC O), 159.4, 156.3, 134.5, 133.5, 129.3, 125.1, 123.9, 120.2, 119.5, 118.1, 102.6 (C-1″), 78.2, 78.0, 74.9, 71.3, 62.5 ppm; HRMS (ESI-TOF) calcd. for C19H20ClNO8+Na [M+Na]+: 448.0770, found: 448.0779; HPLC purity 99.0% (tR: 10.1 min, Hypersil BDS C18, 250 4.6 mm, 5 m, ACN/H2O = 1/99–90/10, 1 mL/min, 20 min).
4.2. Primary screening of anti-SARS-CoV-2 efficacy with cytopathic evaluation
All compounds were diluted in DMEM (2% FBS) to final concentrations of 10, 3.3, and 1 M (or lower for potent compounds). Vero E6 cells (1 × 104 per well) were cultured in a 96-well plate in DMEM supplemented with 10% FBS. The culture medium was removed after a 1-day incubation, when the cells reached 80–90% confluence. A solution of 100 L of DMEM, with 2% FBS containing the compound to be tested, was placed in each of three wells. Cells were incubated in a CO2 incubator at 37 °C with a SARS-CoV-2 strain from Taiwan CDC (hCoV-19/Taiwan/4/2020, isolated from the throat swab of a confirmed 39 y/o male patient from Taiwan) at a dose of 100 TCID50 per well; the cytopathic morphology of the cells was examined by using an inverted microscope at 72 h.
4.3. MTT cytotoxicity assay
Vero E6 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) with 10% FBS (v/v) and 1% penicillin (100 U/mL)/streptomycin (100 mg/mL). Cells were maintained in a humidified incubator at 37 °C in 5% CO2/95% air. The cytotoxicity of compounds to Vero E6 cells was assayed using mitochondrial MTT reduction assay to obtain absorbance density values according to the manufacturer's protocol. Briefly, after incubation with the indicated compounds at various concentrations for 48 h, 100 L of DMEM containing 10 L MTT reagent were added to each well of the 96-well plate and placed in a CO2 incubator for 1 h. After dissolving with DMSO, the absorbance was measured with enzyme-linked immunosorbent assay reader at 570 nm. Data are expressed as percentage of control cells (as 100%) cultured in the absence of compounds.
4.4. PAMPA permeability assay
The PAMPA evolution instrument from pION Inc. was used in this study. In PAMPA, a set of sandwich plates consists of a 96-well microtiter donor plate at the bottom and a 96-well filter acceptor plate on the top from Millipore (IPVH, 125 m thick filter, 0.45 m pore). The stock solutions of the drug samples were prepared at 10 mM concentrations in DMSO and stored at 0 °C before use. Before being added to a 96-well filter plate, the stock solution was diluted first with buffer to achieve a final concentration of 50 M.
Each well of donor plate was filled with a 200 L diluted drug solution (50 M), and the acceptor plate was then wetted with a 5 l 2% w/v phospholipid compound solution in n-dodecane and subsequently placed on the donor wells. Immediately, the acceptor wells were filled with 200 L of buffer solution and the PAMPA plate sandwich was sealed and incubated at 25 °C for 3 h. The PAMPA plate sandwich was separated and the solution was transferred to 96-well UV plates. The amounts of drug in both the donor and acceptor compartments were measured by comparing the experimental spectra with the UV spectrum (220–400 nm) obtained from reference standards, each result was calculated with at least 3 experiments.
4.5. Quantification of SARS-CoV-2 viral load via antibody staining
Vero E6 cells were seeded into 96-well plates (1 × 104 per well) and incubated for 24 h. Two hours before infection, the medium was replaced with 50 L of DMEM (2% FBS) containing the compound of interest at concentrations 2-fold greater than those indicated, including a DMSO control. Plates were then transferred into the BSL-3 facility and 100 TCID50 of SARS-CoV-2 was added in 50 L of DMEM (2% FBS), bringing the final compound concentration to those indicated. Plates were then incubated for 24 h at 37 °C. After infection, the supernatants were removed and cells were fixed with 4% formaldehyde for 24 h prior to removal from the BSL-3 facility. The cells were then immune-stained for the viral N protein (1:3000, antisera were kindly provided by Dr. Ma, Genomic Research Center, Acadmia Sinica, Taiwan) with a DAPI counterstain. Infected cells and total cells (DAPI) were quantified using the Celigo (Nexcelcom) imaging cytometer. Infectivity was measured by the accumulation of viral NP protein in the nucleus of the Vero E6 cells (fluorescence accumulation). Percent infection was quantified and the DMSO control was then set to 100% infection for analysis. The IC50 for each experiment was determined using the Prism software (GraphPad 8.0, San Diego, CA, USA).
4.6. Water solubility assay
To access water solubility of compound 1, 5, 6, 10, 11, niclosamide analogues were added to water continuously until the solid remain in the solution after sonication for 5 min. After centrifugation at 3000 rpm for 20 min, supernatants were collected and filtered through 0.22 m PTFE membranes into 12 × 32 mm vials. An aliquot of 10 L of the sample solution was injected into the HPLC each time and eluted with ACN/H2O = 45/55–90/10. Areas of corresponding peaks (λ = 210 nm) were plotted against concentrations.
4.7. Human plasma stability assay
To access human plasma stability, niclosamide analogues were dissolved in DMSO (10 mM), and the solution for analysis was prepared by diluting 3 L of the analogue stock with 97 L of human plasma to a final concentration of 300 M. After incubation for indicated time, proteins were denatured by the addition of 200 L ACN. Followed by vortexing and centrifugation at 13,000 rpm for 20 min, supernatants were collected and filtered through 0.22 m PTFE membranes into 12 × 32 mm vials. An aliquot of 10 L of the sample solution was injected into the HPLC each time and eluted with ACN/H2O = 45/55–90/10. Areas of corresponding peaks (λ = 210 nm) were plotted against concentrations.
4.8. Human liver S9 stability assay
The suspension for the S9 assay was prepared as follows: 2.2 mg/mL of human liver S9 fraction (20 donor pool, mixed gender, Corning Gentest), 1.1 mM EDTA, 3.67 mM MgCl2, and an NADPH regenerating system (1.43 mM NADP, 3.67 mM glucose-6-phosphate, and 0.44 U/mL glucose-6-phosphate dehydrogenase) at a total volume of 90 L. After the suspension was preincubated at 37 °C for 10 min, the niclosamide analogues (1, 5, 6, 10, 11, 0.5 mM in 10 L PBS) were added to the suspension and incubated for 0, 10, 30, 60, 120, and 360 min, after which experiments were halted by dilution with 200 L of ACN. Followed by vortexing and centrifugation at 13,000 rpm for 20 min, supernatants were collected and filtered through 0.22 m PTFE membranes into 12 × 32 mm vials. An aliquot of 10 L of the sample solution was injected into the HPLC each time and eluted with ACN/H2O = 45/55–90/10. Areas of corresponding peaks (λ = 210 nm) were plotted against concentrations.
4.9. Phosphatidylserine externalization assay
The assay was performed as previously reported method with slight modification [14]. Vero E6 cells were seeded in 96-well microplates with a clear bottom (TPP Culture Test Plate 6–96 wells, TPP) at a density of 8000 cells per well for 2 h, and the cells were treated with compounds for 1 h. After one wash with FBS-free medium, cells were incubated with or without 10 M ionomycin for 30 s, and the medium was removed. Cells were incubated with 100 L 1:100 annexin XII (ab129817, Abcam), and imaged by Olympus inverted microscope (CKX41) with reflected fluorescence system (U-RFLT50). Nine images per well were acquired at different wavelengths: (1) excitation 460–490 nm, emission 500–550 nm (annexin XII ‘green’); (2) excitation 530–560, emission 570–650 nm (PI ‘red’); (3) brightfield.
4.10. Computer simulation
The predicted structure of human TMEM16F and TMEM16K proteins were obtained from Alphafold protein structure database (http://alphafold.ebi.ac.uk) searching by UniProt number Q4KMQ2 and Q9NW15 [21]. The crystal structures of mus musculus mTMEM16F and human TMEM16K proteins were obtained from PDB database by PDB ID 6P46 and 5OC9. Structural issues presented in the protein were corrected using QuickPrep function in Molecular Operating Environment (MOE) software, which performed protein protonation and calculated the minimum energy conformation of the protein. Default parameters of MOE was applied for QuickPrep function. Binding pocket of human TMEM16F and mus musculus TMEM16F (PDB: 6P46) were calculated by PrankWeb (https://prankweb.cz/) binding site prediction tool with default setting [22]. The drug molecule of niclosamide was downloaded from The Cambridge Crystallographic Data Centre (https://www.ccdc.cam.ac.uk/) in CIF format. MOE software was used to dock the compounds with human TMEM16F protein. Docking was performed on the predicted binding pocket of TMEM16F, and 30 poses for London dG and 10 poses of GBVI/WSA dG were used for final docking. All the results were ordered by the binding energy using S Score function, and the interactions of compound in the pocket were visualized by ligand interaction function in MOE.
4.11. Statistical analysis
All data were obtained at least in triplicate, and results are reported as mean ± mean of standard deviation (S.E.M.). Comparisons among groups were analyzed via student t tests, one-way ANOVA, and two-way ANOVA analysis using GraphPad Prism 8. The statistical significance was determined: n.s., nonsignificant difference; ∗∗∗P < 0.001; ∗P < 0.05.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This research was financially supported by the Ministry of Science and Technology, Taiwan (MOST 111-2622-8-002-016-TB1; 110-2628-B-002-031; 107-2320-B-002-019-MY3; 104-2320-B-002-008-MY3). Mass spectrometry data were acquired at the Academia Sinica Common Mass Spectrometry Facilities for Proteomics. Yu-Pu is scholarship supported by Program of Research Performance Enhancement via Students Entering PhD Programs Straight from an Undergraduate/Master's Program from National Taiwan University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejmech.2022.114295.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Helmy Y.A., Fawzy M., Elaswad A., Sobieh A., Kenney S.P., Shehata A.A. The COVID-19 pandemic: a comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control. J. Clin. Med. 2020;9:1225. doi: 10.3390/jcm9041225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Covid-19 Treatment Guidelines Panel Coronavirus disease 2019 (COVID-19) treatment guidelines. https://www.covid19treatmentguidelines.nih.gov/ National Institutes of Health, Available at: Accessed [20210911] [PubMed]
- 5.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jan J.T., Cheng T.R., Juang Y.P., Ma H.H., Wu Y.T., Yang W.B., Cheng C.W., Chen X., Chou T.H., Shie J.J., et al. Identification of existing pharmaceuticals and herbal medicines as inhibitors of SARS-CoV-2 infection. Proc. Natl. Acad. Sci. U.S.A. 2021;118 doi: 10.1073/pnas.2021579118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jayk Bernal A., Gomes da Silva M.M., Musungaie D.B., Kovalchuk E., Gonzalez A., Delos Reyes V., Martin-Quiros A., Caraco Y., Williams-Diaz A., Brown M.L., et al. Molnupiravir for oral treatment of covid-19 in nonhospitalized patients. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahase E. Covid-19: Pfizer's paxlovid is 89% effective in patients at risk of serious illness, company reports, Bmj-Brit. Med. J. 2021;375:n2713. doi: 10.1136/bmj.n2713. [DOI] [PubMed] [Google Scholar]
- 9.Chen W., Mook R.A., Jr., Premont R.T., Wang J. Niclosamide: beyond an antihelminthic drug. Cell. Signal. 2018;41:89–96. doi: 10.1016/j.cellsig.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu J., Shi P.Y., Li H., Zhou J. Broad spectrum antiviral agent niclosamide and its therapeutic potential. ACS Infect. Dis. 2020;6:909–915. doi: 10.1021/acsinfecdis.0c00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu J.M., Berastegui-Cabrera J., Ye N., Carretero-Ledesma M., Pachon-Diaz J., Chen H.Y., Pachon-Ibanez M.E., Sanchez-Cespedes J., Zhou J. Discovery of novel substituted N-(4-Amino-2-chlorophenyl)-5-chloro-2-hydroxybenzamide analogues as potent human adenovirus inhibitors. J. Med. Chem. 2020;63:12830–12852. doi: 10.1021/acs.jmedchem.0c01226. [DOI] [PubMed] [Google Scholar]
- 12.Backer V., Sjobring U., Sonne J., Weiss A., Hostrup M., Johansen H.K., Becker V., Sonne D.P., Balchen T., Jellingso M., et al. A randomized, double-blind, placebo-controlled phase 1 trial of inhaled and intranasal niclosamide: a broad spectrum antiviral candidate for treatment of COVID-19. Lancet Reg. Health Eur. 2021;4:100084. doi: 10.1016/j.lanepe.2021.100084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeon S., Ko M., Lee J., Choi I., Byun S.Y., Park S., Shum D., Kim S. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00819-20. e00819-00820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braga L., Ali H., Secco I., Chiavacci E., Neves G., Goldhill D., Penn R., Jimenez-Guardeno J.M., Ortega-Prieto A.M., Bussani R., et al. Drugs that inhibit TMEM16 proteins block SARS-CoV-2 spike-induced syncytia. Nature. 2021;594:88–93. doi: 10.1038/s41586-021-03491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gassen N.C., Papies J., Bajaj T., Emanuel J., Dethloff F., Chua R.L., Trimpert J., Heinemann N., Niemeyer C., Weege F., et al. SARS-CoV-2-mediated dysregulation of metabolism and autophagy uncovers host-targeting antivirals. Nat. Commun. 2021;12:3818. doi: 10.1038/s41467-021-24007-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blake S., Shaabani N., Eubanks L.M., Maruyama J., Manning J.T., Beutler N., Paessler S., Ji H., Teijaro J.R., Janda K.D. Salicylanilides reduce SARS-CoV-2 replication and suppress induction of inflammatory cytokines in a rodent model. ACS Infect. Dis. 2021;7:2229–2237. doi: 10.1021/acsinfecdis.1c00253. [DOI] [PubMed] [Google Scholar]
- 17.Shamim K., Xu M., Hu X., Lee E.M., Lu X., Huang R., Shah P., Xu X., Chen C.Z., Shen M., et al. Application of niclosamide and analogs as small molecule inhibitors of Zika virus and SARS-CoV-2 infection. Bioorg. Med. Chem. Lett. 2021;40:127906. doi: 10.1016/j.bmcl.2021.127906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu C.Y., Jan J.T., Ma S.H., Kuo C.J., Juan H.F., Cheng Y.S., Hsu H.H., Huang H.C., Wu D., Brik A., et al. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10012–10017. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan X., Li H., Ding X., Zhang Q.Y. Contributions of hepatic and intestinal metabolism to the disposition of niclosamide, a repurposed drug with poor bioavailability. Drug Metab. Dispos. 2019;47:756–763. doi: 10.1124/dmd.119.086678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jendele L., Krivak R., Skoda P., Novotny M., Hoksza D. PrankWeb: a web server for ligand binding site prediction and visualization. Nucleic Acids Res. 2019;47:W345–W349. doi: 10.1093/nar/gkz424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo S., Chen Y., Shi S., Wang X., Zhang H., Zhan Y., An H. Arctigenin, a novel TMEM16A inhibitor for lung adenocarcinoma therapy. Pharmacol. Res. 2020;155:104721. doi: 10.1016/j.phrs.2020.104721. [DOI] [PubMed] [Google Scholar]
- 24.Babouri R., Clarion L., Rolland M., Van der Lee A., Kabouche Z., Volle J.N., Virieux D., Pirat J.L. Synthesis of oxaphosphinane-based pseudodisaccharides. Eur. J. Org Chem. 2017;2017:5357–5369. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.