Abstract
Membrane proteins have long been a challenge for biochemical and functional studies. Absent a bilayer environment, individual proteins and critical macromolecular complexes can be insoluble and display altered or absent activities. Nanodisc technology provides important advantages for the isolation, purification, structural resolution and functional characterization of membrane proteins. In addition, the ability to precisely control the composition of the Nanodisc provides a nanoscale membrane surface for investigating molecular recognition events.
Introduction
Membrane proteins are central to life processes. They are the key conduit for communications between cells, conduct the vital transformations that produce energy, provide channels for the transport of molecules between the inside and outside of cells and compartments and are the workhorses for a plethora of enzyme catalyzed metabolic transformations. Due to their role in the regulation of vital cellular function, membrane proteins (MPs) are the target for a majority of currently marketed therapeutics. Despite these crucial roles, MPs have been notoriously difficult to work with as, absent their place in a phospholipid environment, they often display altered or loss of activity and function. Understanding the molecular mechanisms of membrane protein action is impossible without detailed knowledge of their structure and how they interact with other proteins, nucleic acids and lipids. Thus, the structural biology of MPs occupies a central place in current biophysics, biochemistry and cell biology investigations.
Ideally, a MP structural study must provide not only information on the conformations and protein-protein interactions in supramolecular complexes, but also as to their topology in the lipid bilayer and the details of protein-lipid interface, which often are highly specific with respect to lipid composition. For many systems and projects a planar bilayer model system ~ 10 nm in diameter would be ideal, providing space for one or more MPs and allowing access to both sides for the assay of signaling events. By providing such a native membrane environment, Nanodiscs have proven to be an invaluable tool for revealing the structure and function of isolated membrane proteins as well as their complexes with other proteins and lipids.
Nanodiscs are discoidal lipid bilayers, 8–16 nm in diameter, which are stabilized and rendered soluble in aqueous solutions by two encircling amphipathic helical proteins belts, termed membrane scaffold proteins 1,2. The size of Nanodiscs is determined by the length of the membrane scaffold protein and the stoichiometry of lipids used in the self-assembly process. The resultant discoidal bilayers can be made homogeneous and monodisperse and obtained with high yield 2,3.
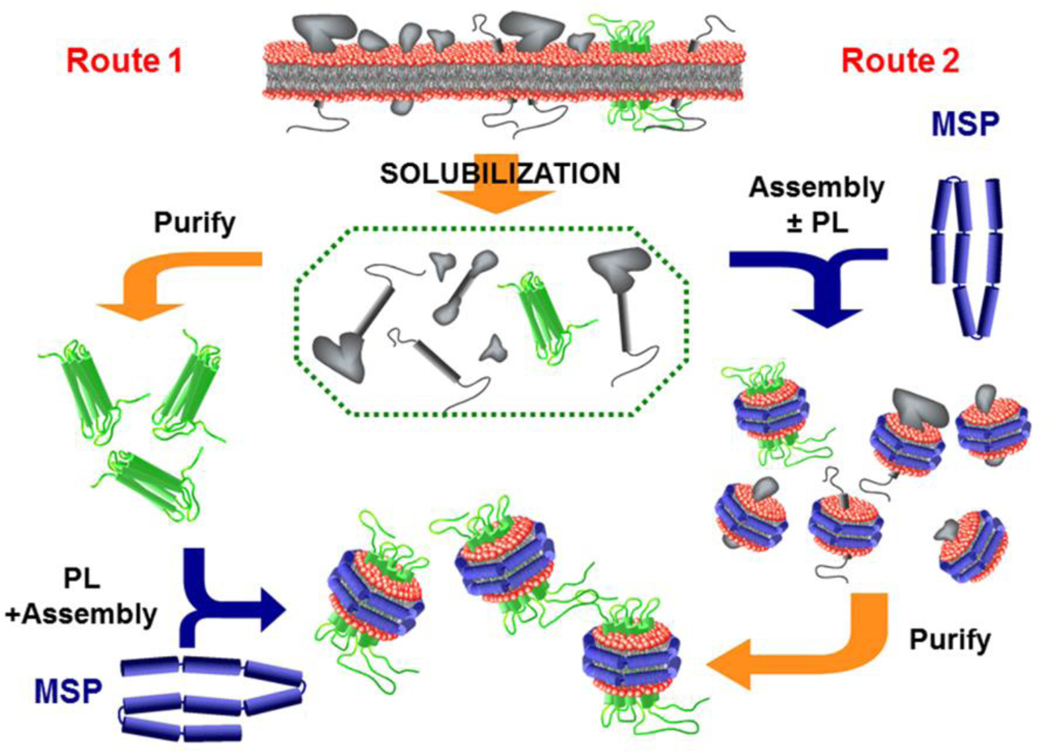
A large body of published work has used Nanodiscs as a vehicle to incorporate a recalcitrant membrane protein target into the bilayer in order to preserve its structure and activity. MPs of all types and topologies have been successfully self-assembled into a Nanodisc starting from a detergent solubilized mixture of all components; target, lipid mixture of choice and membrane scaffold proteins. The molar stoichiometric ratio of lipid to scaffold protein is important for optimized yield 2,3, while the size and ratio of scaffold protein to the membrane protein may be selected in order to favor incorporation of predominantly monomeric (large excess of scaffold protein and lipid) or oligomeric target into the Nanodisc 3,4. This simple approach provides a detailed control over the composition and homogeneity of the resulting assembly and can be optimized to maximize the yield of MP incorporation. If the MP under investigation is unstable to purification in detergent, or one desires to generate a library of all MP in a specific tissue, incorporation into Nanodiscs can be achieved via direct solubilization of cell membranes using detergents5 or membrane-active polymers 6,7. This pathway allows one to bypass the purification step and accelerates the entire process of Nanodisc self-assembly, which is sometimes critically important for preservation of the native and functional form of the target MP, as illustrated in Figure 1. For successful incorporation of purified membrane proteins (Route 1), the most important factor is efficient solubilization and prevention of aggregation of the target protein, which can be assisted by using of excess lipid and scaffold protein while maintaining the correct overall stoichiometry 3. Parameters that control the branch point that leads to successful incorporation of a target MP include choice of detergent, speed of detergent removal and the identity of lipid and target 3.
Figure 1.
Assembling membrane proteins into Nanodiscs. The standard method for self-assembling a membrane protein into a Nanodisc is shown in Route 1 on the left. A purified MP is detergent solubilized together with the membrane scaffold protein and lipid at the correct stoichiometry, followed by detergent removal though incubation with hydrophobic beads. Often, however, the MP is not stable in detergent for the extended times needed for purification. Alternatively, the starting membrane or tissue can be directly solubilized with excess lipid and scaffold protein with rapid detergent removal resulting in the target, together with other MP in the tissue, being placed into the Nanodisc. Subsequent purification, often with an affinity tag, then takes place where the target is stabilized in the Nanodisc environment. This latter route is also used to generate a soluble membrane protein library that faithfully represents that in the starting tissue.
The number of laboratories using Nanodiscs is growing rapidly, as evidenced by the published literature. Nanodiscs are robust, can be frozen or lyophilized, with our without an incorporated MP 8, and provide a precise control over the lipid composition. Scaffold proteins are available with a wide range of specific tags for isolation, in vivo targeting, imaging or reversible / irreversible surface immobilization. They are finding increasing use as “cassettes” that allow a MP to be assayed without denaturation in a variety of analytical methods including surface plasmon resonance 9–11, electrochemistry and optical waveguides. A partial compendium of Nanodisc uses in the study of MP is presented Table 1, with a focus on structural investigations. Successful approaches include X-ray crystallography, electron microscopy (EM), various spectroscopic methods, single molecule studies as well as solution x-ray (SAXS) and neutron (SANS) scattering. The processes illustrated often take advantage of the ability of Nanodiscs to provide a homogeneous sample with a desired target oligomerization state or supramolecular complex topology. Many utilize the ability to control lipid, fatty acid or cholesterol composition, investigate protein-membrane interaction specificity or use affinity tags on Nanodiscs for surface immobilization for sensing and detection.
Table 1.
Methods and tools used with membrane proteins incorporated into Nanodiscs.
| Features and applications | Experimental methods | Membrane proteins | References |
|---|---|---|---|
| Solubilization and stabilization | X-ray crystallography | GPCR | 23 |
| NMR | Various membrane proteins | 8,14-18 | |
| Resonance Raman | Cytochrome P450, rhodopsins | 56 | |
| Optical absorption | 53-57 | ||
| EPR | Transporters, rhodopsin | 36, 37 | |
| Cell-free expression | Various | 12, 42 | |
| Surface immobilization using affinity tags on scaffold protein or lipids | Single-molecule studies SPR, LSPR, sensing and detection | P450, GPCR | 9-11, 47-50 |
| Factor VII | |||
| Isolation of monomers, oligomers, and functional complexes of multiple proteins | Structural and functional studies | Receptors, rhodopsins, cytochrome P450 with CPR, ATPase |
4, 53-55, 59,60 57,58,61, 67 |
| Structurally homogeneous preparations | SAXS, SANS | Cytochrome P450 and reductase Receptors, channels |
4,42,43,45 20,21,26-31 |
| EM | |||
| Fractionation by size, charge, density. | Chromatography, Electrophoresis, Ultracentrifugation | 3,4,15, 59,60 | |
| Protein-membrane binding | SPR | K-ras protein, TF:VIIa, PgP transporter |
9-11 |
| Membrane effect on protein properties | Redox potential, Kinetics | Cytochrome P450 reductase Factor VIII Respiratory Complex | 39 29 40 |
Nanodiscs offer significant advantages over more classical approaches. For example, MP solubilization in proteoliposomes often results in turbid and viscous samples, which may be especially troublesome for cell-free expression systems12 and many biophysical methods. Nanodisc preparations remain fluid and intact, even at the high concentrations needed for NMR experiments 13–18. Nanodiscs do not suffer from the well documented perturbation of MP native structure under non-native conditions, i.e. when present in detergent micelles19, and offer the advantage maintaining higher structural stability and much longer life-time in comparison to bicelles. The high kinetic stability of Nanodiscs when assembled on trans-membrane fragments of large MPs and their complexes has proved to be very useful for the prevention of aggregation during sample preparation for EM studies 20,21, as evidenced by the example in Figure 2. Recently various amphipathic polymers, e.g. SMALPs6 or amphipols 7, have had success in solubilizing MPs from tissue and forming soluble particles on the nanoscale. Although useful for MP extraction, in general, these entities do not provide the same control over size and composition as the Nanodisc approach.
Figure 2.
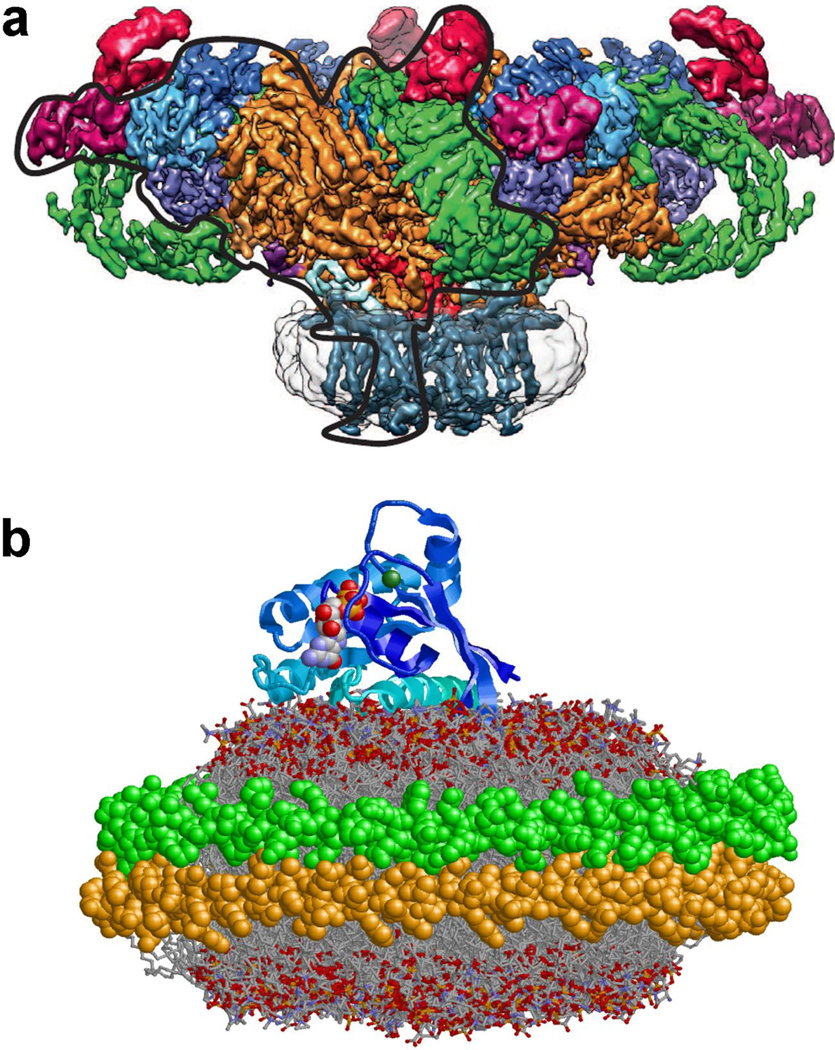
Examples of membrane protein structure in Nanodiscs. (A) The structure of the ryanodine receptor in Nanodisc is resolved to ~6.1 Å using cryo-electron microscopy. The Nanodisc is shown as a light gray envelope with 24 transmembrane helices forming a square structure similar to that of the incorporated voltage-gated sodium channel Nav (reproduced with permission from21). (B) The structure of KRas4b bound to the surface of a Nanodisc as determined by NMR spectroscopy 38 (2MSC.pdb file from the Protein Data Bank).
Structural Biology with Nanodiscs
Several examples from the recent literature can be used as examples of the expanding use of Nanodisc technology in structural biology. Nanodiscs are often used for the isolation and purification of MP for structural studies, including X-ray crystallography22,23 and EM 20,21. Often a challenge is to collect enough material for structure determination if the MP is unstable in detergent our in the absence of lipids. Self-assembling the MP target into Nanodiscs, where it is stable and remains in its native conformation, offers a way to accumulate and store material23. Clearly, any type of compositional diversity leads to additional complications in structural studies, often prohibiting successful analysis at high resolution. Nanodiscs with incorporated MP can be generated with controlled composition and very high degree of size homogeneity2–4. Nevertheless, we have yet to see a complete x-ray structure of a membrane protein in a Nanodisc bilyer. A potential challenge is the translational and azimuthal mobility of incorporated target which may lead to difficulties in crystallization. Such motion could be potentially be reduced by association with a bi-functional antibody that recognizes both the target and membrane scaffold protein 24 or by reducing the size of the Nanodisc by shortening the scaffold protein belt and using less phospholipid. One might also realize a spectacular success if a membrane protein with large globular domains on either side of the bilayer were studied where crystal contacts could be made between the target MP, rendering the Nanodisc blurred, but the structure of the membrane protein in a bilayer revealed.
Electron microscopy
Recent advances in electron microscopy (EM) have brought near atomic resolution to the tool-kit of the structural biologist. Because of the high homogeneity of Nanodiscs, they are being widely used for the EM of isolated MP, functional MP complexes20,25–28 as well as MP binding on the surface of the bilayer29,30. Three- dimensional reconstructions, based on combined analysis of multiple 2D images, have been reported 21,27,31. The incorporation of a MP or membrane domain(s) into Nanodiscs prevents aggregation of the MP and significantly improves the sample quality20. Atomic level detail is rapidly becoming a reality as evidenced by the recent structure of the ryanodine receptor in Nanodiscs 21 (resolution 6.1 Å) where the positions of individual trans-membrane helices are resolved and the 3.8 A structure of Mg2+ channel by Subramaniam and colleagues 31.
NMR and EPR Methods
One of the most useful advantages of Nanodiscs for structural studies of MPs is that they enable work at high concentrations without aggregation or loss of integrity – often a problem in using the powerful tools of NMR for determining the structure of MPs. The first NMR spectra of the membrane scaffold protein in Nanodiscs was obtained by Rienstra and colleagues using magic angle spinning solid state NMR32 and was quickly followed by acquisition of high resolution 2D-NMR signals for a large MP assembled in the Nanodisc bilayer 8 33. Many laboratories have recently reported impressive solution NMR of MPs in Nanodiscs14,16,34,35. The pioneering work of Wagner and colleagues provided the first complete atomic-resolution structures of a membrane protein (OmpX) in the Nanodisc bilayer 14, revealing significant differences between the structure of a MP in solution from when it is in its native phospholipid environment. Nanodiscs have also been used for individual distance measurements. For instance, electron paramagnetic resonance (EPR) has been extensively used to monitor conformational changes and dynamics of ligand binding in maltose transporter MalFGK2 36 where EPR probes were placed at selected sites on the target, with the functionally important conformational changes observed in Nanodiscs but not detergent solubilized protein. In another study, direct distance measurements using DEER spectroscopy was used to monitor the conformational changes of a ND incorporated G-protein coupled receptor upon binding of its signaling partner37.
The Nanodisc as a Controlled Membrane Surface
The membrane surface is the site for the recruitment of many signaling proteins, and is recently been viewed as an important player in dictating the structure of signaling molecules and their faithful formation of specific multi-protein complexes that are required for activity. Noteworthy, therefore, is the recent use of Nanodiscs by investigators at the Frederick National Laboratory for Cancer Research to document the affinity of the oncogenic protein K-Ras4B for Nanodisc surfaces 9 and the use of solution NMR by Ikura and colleagues to define the conformational changes of K-Ras4B bound to Nanodiscs 38. Here the NMR of a MP in Nanodiscs provided important information about orientation of the protein with respect to the lipid bilayer, specific protein-lipid membrane interactions involved in molecular recognition and the dynamics of small molecule binding sites at the protein-lipid interface. Other systems that use Nanodiscs to reveal specific lipid compositions that are necessary to recruit proteins to the membrane surface include extensive work on the blood coagulation cascade 10 and the activation of integrins 28. Nanodiscs have been extensively used to reveal the role of specific phospholipid type in MP function. Published work includes documenting the upshift of redox potential of redox transfer proteins due to the presence of negatively charged lipids39 and the critical role of cardiolipin in the assembly, activity and production of reactive oxygen species by the mitochondrial respiratory Complex II 40. The ability to precisely control the mixture of lipids in the Nanodisc has enabled the determination of lipid specificity in the binding of blood coagulation factors 10, integrin activators28 and Ras oncogenes38.
X-Ray and Neutron Scattering
Both the structure of, and phase transition in, lipid bilayer Nanodiscs have been extensively characterized using SAXS, fluorescence and calorimetry 2,41. SAXS of several MP in Nanodiscs was used to confirm faithful incorporation in the lipid bilayer and to characterize the topology of the target protein. Examples include bR trimers 4, curdlan synthase 42 and cytochrome P45043. The latter reference demonstrated for the first time the potential of ab initio reconstruction of a low-resolution MP structure solely based on SAXS data. Such analysis can be significantly improved and refined if SAXS is analyzed in combination with SANS data collected using various contrasting methods, such as partially deuterated solvent and lipids 44, made possible by contrast matching to render the scaffold protein and lipid invisible. Neutron reflectometry has been used to directly measure large conformational changes of a MP in Nanodiscs, in one case the redox dependent movement needed for efficient electron transfer by NADPH cytochrome P450 reductase45. Differences in the conformation of monomers and dimers of vertebrate visual rhodopsins reconstituted in Nanodiscs was directly monitored during photoactivation using high-angle X-ray scattering 46. Comparison with the X-ray scattering calculated for the known structures of photocycle intermediates suggests a dimeric form of rhodopsin in the membrane, and that this dimerization modulates the observed functionally important structural changes 46.
Single Molecule Measurements
Single-molecule studies are increasingly being used to avoid masking details of protein function when only ensemble averages are measured. Nanodiscs are ideally suited for this type of investigation as they can be affixed to a surface without impairing the subtle conformational changes happening in the MP. Either scaffold proteins or lipids have been labeled with affinity tags to successfully immobilize Nanodiscs on a surface while allowing access to both sides of the bilayer for the association of substrates or signaling partners. This label-free approach avoids suggestions that deritivization of the target alters the function or conformation. Single-molecule Nanodisc studies have been reported for cytochrome P450 reductase47, cytochrome P450 CYP3A448 and the β2-adrenergic receptor 49. Using the natural affinity of lipids for atomically flat silicon oxide or mica surfaces allows the Nanodisc to sit flat, enabling atomic force microscopy (AFM) to directly measure depth of insertion of proteins in the bilayer50 and linear dichroism dichroism optical spectroscopy to quantitate the orientation of a target protein relative to the Nanodisc surface51.
Spectroscopic Assay Techniques
Compared to other membrane mimetics, Nanodiscs provide significant advantages for optical spectroscopy due to the low viscosity and lack of turbidity. This allows rapid mixing methods, for example stopped- or continuous flow, to be used for monitoring structural and functional changes on the millisecond time range, while preserving the target MP in a lipid bilayer environment 52. Laser flash techniques were used in the study of the monomer and trimer of bacteriorhodopsin (bR) 4,53 and various rhodopsins 54,55. Optical spectroscopy and rapid mixing was also used to probe functional intermediates in human cytochromes P450, including stopped flow 52 and resonance Raman spectroscopy 56. In all cases, biophysical studies were enabled by the high stability and structural homogeneity of monomeric proteins and their functional complexes with redox partners in Nanodiscs57,58. Other ligand binding events, such as the kinetics of oxygen binding and autoxidation, also show clean monoexponential behavior when measured for monomers in Nanodiscs, as opposed to solution aggregates. As with rhodopsin monomer and dimer preparations 59,60, Nanodiscs helped avoid ambiguities in the interpretation of experimental data and to assign the observed functional properties of the main drug-metabolizing human P450 enzyme CYP3A4 to the intrinsic behavior of its monomeric form. Allosteric regulation in this enzyme is determined by the properties of the peripheral high-affinity binding site at the protein-lipid interface. 61 Importantly, this binding pocket is formed when CYP3A4 is incorporated into the membrane of Nanodiscs, and cannot be probed in detergent solubilized preparations. New enzyme intermediates can be trapped using Nanodiscs. For instance, Resonance Raman spectroscopy of human CYP17A1 made possible by cryogenic methods revealed a previously unobserved functional intermediate in the last step of androstenedione biosynthesis, answering a long-standing question as to the mechanism of this important drug target 56.
Control of Oligomerization State, Macromolecular Machines and the Epitaxial Presentation of Membrane Proteins
Antibodies are extensively used in the study of membrane proteins, often as an aid in crystallization or in the development of therapeutic agents. Raising antibodies to MPs is inherently difficult as an aggregated state can both occlude the antigenic site and lower the overall immune response. In vitro phage and yeast display technologies provide significant advantages, yet still need to have a MP in its native conformation, ideally in the absence of detergents. Pioneering work in the Kossiakoff laboratory have used Nanodiscs to develop protocols for competitive and subtractive selection in phage display approaches62. Many membrane proteins operate in higher order complexes, ranging from homo-/hetero dimers to large supra-molecular complexes containing many subunits. Large molecular machines, such as cytochrome oxidases63, photosynthetic reaction center 64 and peptide translocases25 can be functionally self-assembled into Nanodiscs. In the case of signaling entities, individual monomers or oligomers of specific composition can be prepared, allowing measurement of activity as a function of oligomerization state to be determined. For example, the first demonstrations that a monomeric GPCR had full signaling activity utilized Nanodiscs containing either a rhodopsin monomer or dimer 59,60. More complex signaling pathways, for example those involved in chemotaxis signaling, made use of Nanodiscs to separate the role of dimers and trimers-of-dimers in specific activities 65. Other examples of controlled oligomerization through the use of Nanodiscs include studies of many rhodopsins55,66 and the plasma membrane H+-ATPase from Arabidopsis67. Because Nanodisc assemblies display high stability, these fractions can be isolated and purified to homogeneity using size-exclusion chromatography or density gradient ultracentrifugation 3.
Conclusion and perspectives
The use of Nanodiscs is rapidly spreading, made easy by the ready access of the genes for the membrane scaffold proteins through AddGene® and the purified proteins from Sigma (www.sigmaaldrich.com) or BioNanoConLLC (www.bionanocon.com). In addition to the already established applications of Nanodisc technology, some recent developments suggest new ideas, such as generation of therapeutic antibodies against antigens incorporated into Nanodiscs 68 and various in vivo uses of Nanodiscs including drug delivery 69, imaging 70 and vaccine development. A most exciting application is the ability to generate soluble membrane protein libraries that faithfully represent a starting membrane protein composition, with individual Nanodiscs carrying a specific target5. By allowing high throughput screening of membrane proteins, one can potentially discover new approaches to therapeutic intervention.
Acknowledgement:
Nanodisc research in our laboratory is supported by the National Institutes of Health.
References
- 1.Bayburt TH, Grinkova YV & Sligar SG Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2, 853–856 (2002). [Google Scholar]
- 2.Denisov IG, Grinkova YV, Lazarides AA & Sligar SG Directed Self-Assembly of Monodisperse Phospholipid Bilayer Nanodiscs with Controlled Size. J. Am. Chem. Soc. 126, 3477–3487 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Bayburt TH & Sligar SG Membrane protein assembly into nanodisks. FEBS Lett. 584, 1721–1727 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayburt TH, Grinkova YV & Sligar SG Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch. Biochem. Biophys. 450, 215–222 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Wilcox KC et al. Nanoscale Synaptic Membrane Mimetic Allows Unbiased High Throughput Screen That Targets Binding Sites for Alzheimer’s-Associated Abeta Oligomers. PLoS One 10, e0125263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doerr JM et al. The styrene-maleic acid copolymer: a versatile tool in membrane research. Eur. Biophys. J. 45, 3–21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Planchard N. et al. The Use of Amphipols for Solution NMR Studies of Membrane Proteins: Advantages and Constraints as Compared to Other Solubilizing Media. J. Membr. Biol. 247, 827–842 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Kijac AZ, Li Y, Sligar SG & Rienstra CM Magic-Angle Spinning Solid-State NMR Spectroscopy of Nanodisc-Embedded Human CYP3A4. Biochemistry 46, 13696–13703 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillette WK et al. Farnesylated and methylated KRAS4b: high yield production of protein suitable for biophysical studies of prenylated protein-lipid interactions. Sci Rep 5, 15916 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw AW, Pureza VS, Sligar SG & Morrissey JH The Local Phospholipid Environment Modulates the Activation of Blood Clotting. J. Biol. Chem. 282, 6556–6563 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Ritchie TK, Kwon H. & Atkins WM Conformational Analysis of Human ATP-binding Cassette Transporter ABCB1 in Lipid Nanodiscs and Inhibition by the Antibodies MRK16 and UIC2. J. Biol. Chem. 286, 39489–39496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Proverbio D, Henrich E, Orbán E, Dötsch V, Bernhard F. Membrane Protein Quality Control in Cell-Free Expression Systems: Tools, Strategies and Case Studies. in Membrane Proteins Production for Structural Analysis (ed. Mus-Veteau I) 45–70 (Springer, New York, 2014). [Google Scholar]
- 13.Catoire LJ, Warnet XL, Warschawski DE Micelles, Bicelles, Amphipols, Nanodiscs, Liposomes, or Intact Cells: The Hitchhiker’s Guide to the Study of Membrane Proteins by NMR. in Membrane Proteins Production for Structural Analysis (ed. Mus-Veteau I) 315–345 (Springer, New York, 2014). [Google Scholar]
- 14.Hagn F, Etzkorn M, Raschle T. & Wagner G. Optimized Phospholipid Bilayer Nanodiscs Facilitate High-Resolution Structure Determination of Membrane Proteins. J. Am. Chem. Soc. 135, 1919–1925 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malhotra K. & Alder NN Advances in the use of nanoscale bilayers to study membrane protein structure and function. Biotechnol. Genet. Eng. Rev. 30, 79–93 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Morgado L, Zeth K, Burmann BM, Maier T. & Hiller S. Characterization of the insertase BamA in three different membrane mimetics by solution NMR spectroscopy. J Biomol NMR 61, 333–45 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Shenkarev ZO et al. Lipid-protein nanodiscs offer new perspectives for structural and functional studies of water-soluble membrane-active peptides. Acta Naturae 6, 84–94 (2014). [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Mu Z, Li Y, Bi Y. & Wang Y. Smaller Nanodiscs are Suitable for Studying Protein Lipid Interactions by Solution NMR. Protein J 34, 205–11 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Zhou HX & Cross TA Influences of membrane mimetic environments on membrane protein structures. Annu Rev Biophys 42, 361–92 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akkaladevi N. et al. Following Natures Lead: On the Construction of Membrane-Inserted Toxins in Lipid Bilayer Nanodiscs. J Membr Biol 248, 595–607 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Efremov RG, Leitner A, Aebersold R. & Raunser S. Architecture and conformational switch mechanism of the ryanodine receptor. Nature 517, 39–43 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Chung KY, Day PW, Velez-Ruiz G, Sunahara RK & Kobilka BK Identification of GPCR-interacting cytosolic proteins using HDL particles and mass spectrometry-based proteomic approach. PLoS One 8, e54942 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasmussen SG et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature 469, 175–80 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dominik PK et al. Conformational Chaperones for Structural Studies of Membrane Proteins Using Antibody Phage Display with Nanodiscs. Structure 24, 300–309 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frauenfeld J. et al. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 18, 614–621 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gogol EP et al. Three dimensional structure of the anthrax toxin translocon-lethal factor complex by cryo-electron microscopy. Protein Sci. 22, 586–594 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katayama H. et al. Three-dimensional structure of the anthrax toxin pore inserted into lipid nanodiscs and lipid vesicles. Proc. Natl. Acad. Sci. U. S. A. 107, 3453–3457, S3453/1-S3453/3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye F. et al. Recreation of the terminal events in physiological integrin activation. J. Cell Biol. 188, 157–173 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grushin K, Miller J, Dalm D. & Stoilova-McPhie S. Factor VIII organisation on nanodiscs with different lipid composition. Thromb Haemost 113, 741–9 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Zhang P. et al. An Isoform-Specific Myristylation Switch Targets Type II PKA Holoenzymes to Membranes. Structure 23, 1563–72 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthies D. et al. Cryo-EM Structures of the Magnesium Channel CorA Reveal Symmetry Break upon Gating. Cell 164, 747–756 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Kijac AZ, Sligar SG & Rienstra CM Structural analysis of nanoscale self-assembled discoidal lipid bilayers by solid-state NMR spectroscopy. Biophys. J. 91, 3819–3828 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding Y, Fujimoto LM, Yao Y. & Marassi FM Solid-state NMR of the Yersinia pestis outer membrane protein Ail in lipid bilayer nanodiscs sedimented by ultracentrifugation. J Biomol NMR 61, 275–86 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kucharska I, Edrington TC, Liang B. & Tamm LK Optimizing nanodiscs and bicelles for solution NMR studies of two beta-barrel membrane proteins. J Biomol NMR 61, 261–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mineev KS, Goncharuk SA, Kuzmichev PK, Vilar M. & Arseniev AS NMR Dynamics of Transmembrane and Intracellular Domains of p75NTR in Lipid-Protein Nanodiscs. Biophys J 109, 772–82 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alvarez FJ et al. Full engagement of liganded maltose-binding protein stabilizes a semi-open ATP-binding cassette dimer in the maltose transporter. Mol. Microbiol. 98, 878–894 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang Y. et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 523, 561–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mazhab-Jafari MT et al. Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proc Natl Acad Sci U S A 112, 6625–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Das A. & Sligar SG Modulation of the Cytochrome P450 Reductase Redox Potential by the Phospholipid Bilayer. Biochemistry 48, 12104–12112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwall CT, Greenwood VL & Alder NN The stability and activity of respiratory Complex II is cardiolipin-dependent. Biochim. Biophys. Acta, Bioenerg 1817, 1588–1596 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Denisov IG, McLean MA, Shaw AW, Grinkova YV & Sligar SG Thermotropic Phase Transition in Soluble Nanoscale Lipid Bilayers. J. Phys. Chem. B 109, 15580–15588 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Periasamy A. et al. Cell-free protein synthesis of membrane (1,3)-β-D-glucan (curdlan) synthase: Co-translational insertion in liposomes and reconstitution in nanodiscs. Biochim. Biophys. Acta, Biomembr. 1828, 743–757 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Skar-Gislinge N. et al. Small-angle scattering determination of the shape and localization of human cytochrome P450 embedded in a phospholipid nanodisc environment. Acta Crystallogr D Biol Crystallogr 71, 2412–21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maric S. et al. Stealth carriers for low-resolution structure determination of membrane proteins in solution. Acta Crystallogr D Biol Crystallogr 70, 317–28 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Wadsaeter M. et al. Monitoring Shifts in the Conformation Equilibrium of the Membrane Protein Cytochrome P450 Reductase (POR) in Nanodiscs. J. Biol. Chem. 287, 34596–34603 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imamoto Y, Kojima K, Oka T, Maeda R. & Shichida Y. Helical rearrangement of photoactivated rhodopsin in monomeric and dimeric forms probed by high-angle X-ray scattering. Photochem Photobiol Sci 14, 1965–1973 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Laursen T. et al. Single molecule activity measurements of cytochrome P450 oxidoreductase reveal the existence of two discrete functional states. ACS Chem Biol 9, 630–4 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Nath A, Koo PK, Rhoades E. & Atkins WM Allosteric Effects on Substrate Dissociation from Cytochrome P450 3A4 in Nanodiscs Observed by Ensemble and Single-Molecule Fluorescence Spectroscopy. J. Am. Chem. Soc. 130, 15746–15747 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lamichhane R. et al. Single-molecule view of basal activity and activation mechanisms of the G protein-coupled receptor β2AR. Proc. Natl. Acad. Sci. U. S. A. 112, 14254–14259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bayburt TH & Sligar SG Single-molecule height measurements on microsomal cytochrome P450 in nanometer-scale phospholipid bilayer disks. Proc. Natl. Acad. Sci. U. S. A. 99, 6725–6730 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baylon JL, Lenov IL, Sligar SG & Tajkhorshid E. Characterizing the Membrane-Bound State of Cytochrome P450 3A4: Structure, Depth of Insertion, and Orientation. J. Am. Chem. Soc. 135, 8542–8551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Denisov IG, Grinkova YV, McLean MA & Sligar SG The One-electron Autoxidation of Human Cytochrome P450 3A4. J. Biol. Chem. 282, 26865–26873 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Johnson PJM et al. The photocycle and ultrafast vibrational dynamics of bacteriorhodopsin in lipid nanodiscs. Phys. Chem. Chem. Phys. 16, 21310–20 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Tsukamoto H, Szundi I, Lewis JW, Farrens DL & Kliger DS Rhodopsin in Nanodiscs Has Native Membrane-like Photointermediates. Biochemistry 50, 5086–5091 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ranaghan MJ, Schwall CT, Alder NN & Birge RR Green proteorhodopsin reconstituted into nanoscale phospholipid bilayers (nanodiscs) as photoactive monomers. J Am Chem Soc 133, 18318–18327 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mak PJ, Gregory MC, Denisov IG, Sligar SG & Kincaid JR Unveiling the crucial intermediates in androgen production. Proc Natl Acad Sci U S A 112, 15856–15861 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Denisov IG, Baas BJ, Grinkova YV & Sligar SG Cooperativity in cytochrome P450 3A4 - Linkages in substrate binding, spin state, uncoupling, and product formation. J. Biol. Chem. 282, 7066–7076 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Khatri Y, Gregory MC, Grinkova YV, Denisov IG & Sligar SG Active site proton delivery and the lyase activity of human CYP17A1. Biochem. Biophys. Res. Commun. 443, 179–184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bayburt TH, Leitz AJ, Xie G, Oprian DD & Sligar SG Transducin Activation by Nanoscale Lipid Bilayers Containing One and Two Rhodopsins. J. Biol. Chem. 282, 14875–14881 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Bayburt TH et al. Monomeric Rhodopsin is Sufficient for Normal Rhodopsin Kinase (GRK1) Phosphorylation and Arrestin-1 Binding. J. Biol. Chem. 286, 1420–1428 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Denisov IG, Grinkova YV, Baylon JL, Tajkhorshid E. & Sligar SG Mechanism of drug-drug interactions mediated by human cytochrome P450 CYP3A4 monomer. Biochemistry 54, 2227–39 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dominik PK & Kossiakoff AA Phage display selections for affinity reagents to membrane proteins in nanodiscs. Methods Enzymol 557, 219–45 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Näsvik Öjemyr L, von Ballmoos C, Gennis RB, Sligar SG & Brzezinski P. Reconstitution of respiratory oxidases in membrane nanodiscs for investigation of proton-coupled electron transfer. FEBS Lett. 586, 640–645 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ham M-H et al. Photoelectrochemical complexes for solar energy conversion that chemically and autonomously regenerate. Nat. Chem. 2, 929–936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li M. & Hazelbauer GL Selective allosteric coupling in core chemotaxis signaling complexes. Proc. Natl. Acad. Sci. U. S. A. 111, 15940–15945 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moers K. et al. Modified lipid and protein dynamics in nanodiscs. Biochim. Biophys. Acta, Biomembr. 1828, 1222–1229 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Justesen BH et al. Active plasma membrane P-type H+-ATPase reconstituted into nanodisks is a monomer. J. Biol. Chem. 288, 26419–26429 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reichart TM et al. Trimerization of the HIV Transmembrane Domain in Lipid Bilayers Modulates Broadly Neutralizing Antibody Binding. Angew. Chem., Int. Ed, Ahead of Print (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Numata M. et al. Nanodiscs as a therapeutic delivery agent: inhibition of respiratory syncytial virus infection in the lung. Int. J. Nanomed. 8, 1417–1427 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carney CE et al. Nanodiscs as a Modular Platform for Multimodal MR-Optical Imaging. Bioconjug Chem 26, 899–905 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]