Abstract
Purpose of review:
Metabolomics enables rapid interrogation of widespread metabolic processes making it well-suited for studying diabetes. Here, we review the current status of metabolomic investigation in diabetes, highlighting its applications for improving risk prediction and mechanistic understanding.
Recent findings:
Findings of metabolite associations with type 2 diabetes risk have confirmed experimental observations (e.g., branched-chain amino acids) and also pinpointed novel pathways of diabetes risk (e.g., dimethylguanidino valeric acid). In type 1 diabetes, abnormal metabolite patterns are observed prior to the development of autoantibodies and hyperglycemia. Diabetes complications display specific metabolite signatures that are distinct from the metabolic derangements of diabetes and differ across vascular beds. Lastly, metabolites respond acutely to pharmacologic treatment, providing opportunities to understand inter-individual treatment responses.
Summary:
Metabolomic studies have elucidated biological mechanisms underlying diabetes development, complications, and therapeutic response. While not yet ready for clinical translation, metabolomics is a powerful and promising precision medicine tool.
Keywords: Metabolomics, type 1 diabetes, type 2 diabetes, risk prediction
Introduction
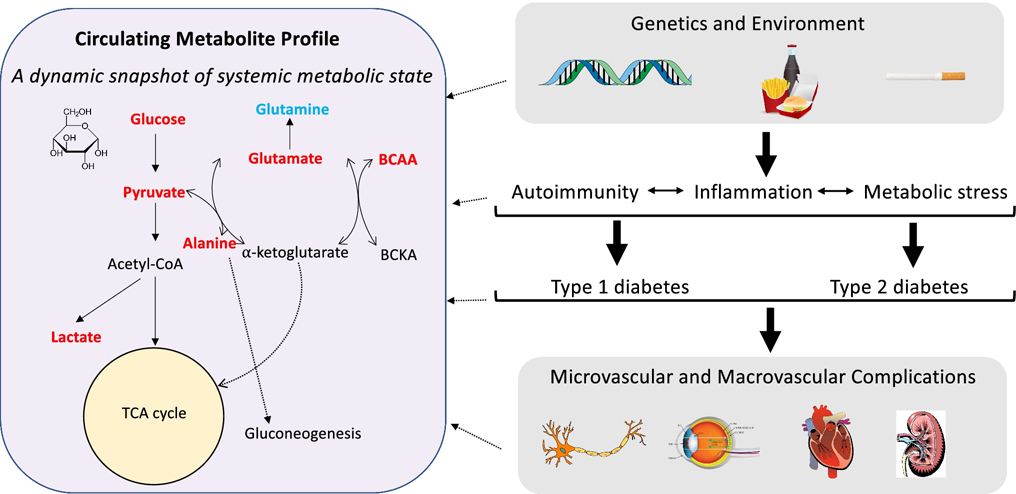
Despite substantial progress over recent decades in uncovering the pathophysiology and mechanisms underlying type 1 (T1D) and type 2 diabetes (T2D), and improved treatment options, their incidence and prevalence continue to rise(1). Diabetes is defined by hyperglycemia and almost invariably results from progressive loss of pancreatic β-cell mass and/or function(2). However, there are many different processes that culminate in the β-cell destruction and dysfunction that characterize diabetes and this pathophysiological heterogeneity may have implications for treatment and prognosis. Molecular signatures of metabolic dysfunction in diabetes are therefore highly relevant to identify disease subtypes with distinct risk profiles and treatment responses and to identify risk of diabetes at earlier, potentially more modifiable, stages. Quantification of small molecule “metabolites” in the blood (“metabolomics”) is a promising avenue for metabolic interrogation due to its proximal role in reflecting metabolic dysfunction at the organ tissue level(3) prior to the development of overt hyperglycemia(4). Accordingly, metabolomic investigation for risk prediction and mechanistic understanding has been an active area of diabetes research for the last decade, since the advent of high-throughput metabolomic techniques capable of measuring hundreds of small molecules in a single blood sample(5). In this review, we provide an overview of human metabolomic investigation into the development, progression of complications, and response to therapy of both T1D and T2D, seeking to highlight mechanistic insights that have emerged from these human studies (Figure 1).
Figure 1. Metabolomics as a snapshot of diabetes pathophysiology.
Diabetes is the culmination of metabolic insults that originate from a combination of genetic and environmental factors. These factors cause metabolic dysfunction, inflammation, and autoimmunity (in the case of type 1 diabetes), and eventually lead to overt hyperglycemia and diabetes, of which the potential microvascular and macrovascular complications are well-described. At each stage of this process, distinct molecular signatures can be identified in the blood through metabolomic profiling. Shown here is an overview of select metabolite pathways that have been linked with the risk of incident diabetes in human epidemiologic studies. Higher levels of metabolites in red are associated with higher diabetes risk and metabolites in blue are inversely related to diabetes risk. Each metabolite displayed here is also involved in numerous other biological pathways that are not shown in this figure for the sake of simplicity.
Metabolites and risk of incident diabetes
Type 2 diabetes
High-throughput metabolite profiling techniques have been applied over the last decade to better understand mechanisms and risk markers for T2D. Initial efforts demonstrated associations of branched-chain amino acids (BCAAs; e.g., valine, leucine, isoleucine) and aromatic amino acids (e.g., phenylalanine, tyrosine) with incident diabetes(6) and with its precursor states, insulin resistance (IR) and obesity(3). The observational finding of higher circulating BCAAs and aromatic amino acids being related to higher risk of developing T2D has been well-replicated(3, 7–13), and is in concert with mechanistic studies that have elucidated the pathobiological links between BCAAs and diabetes. Higher levels of circulating BCAAs are a consequence of decreased rates of oxidation in adipose tissue and reduced hepatic BCAA uptake from inactivation of the branched-chain ketoacid dehydrogenase (BCKDH) complex(14). While it remains uncertain whether BCAAs are truly causal in the development of diabetes, as opposed to a reflection of other metabolic processes, several lines of evidence point to potential causal contributions. Accumulation of BCAAs in the circulation may activate mammalian target of rapamycin (mTOR), which increases gluconeogenesis through downregulation of Akt signaling(15). It has also been demonstrated that higher circulating BCAA levels occur from over nutrition-induced activation of the transcription factor carbohydrate-response element binding protein (ChREBP)(16). ChREBP drives the expression of enzymes involved in de novo lipogenesis, an increasingly recognized contributor to diabetes pathobiology(17). Further support of a causal mechanism for BCAAs in diabetes is provided from human genetics studies demonstrating that genetic variants related to higher circulating BCAA levels (including modulators of the BCKDH complex) are also associated with a higher risk of incident diabetes(18).
In addition to BCAAs, numerous other metabolites have been linked to diabetes and IR (summarized in Table 1). Glutamine and glutamate, for example, have been independently associated with IR and T2D(19, 20). Glutamine is involved in regulating insulin secretion and pancreatic β-cell function, while glutamate promotes gluconeogenesis, glucagon release, and other processes relevant to energy metabolism(21, 22). Accordingly, glutamine is inversely associated with diabetes risk, while glutamate is directly related(4, 23). Molecular substrates of the tricarboxylic acid (TCA) cycle have also been shown to be indicative of IR and diabetes risk. Higher levels of lactate, pyruvate, and alanine are related to obesity(3) and T2D(24). These associations partly reflect mitochondrial dysfunction and reduced oxidative capacity exhibited by skeletal muscle in the setting of IR(25), but lactate may also contribute to T2D development directly via stimulation of hepatic gluconeogenesis and by interfering with glucose uptake(26).
Table 1.
Summary of metabolite associations with diabetes risk and with diabetes complications.
| Metabolite class/pathway | Example metabolite(s) | Associated phenotypes | Adverse direction | Potential mechanisms and biological implications |
|---|---|---|---|---|
| Branched-chain & aromatic amino acids | Branched-chain: Val, Leu, Ile | T2D, T1D, IR, retinopathy | ↑ (↓ for tyrosine and retinopathy) | From diet or microbial metabolism; ↑ levels in blood from ↓ adipose and liver metabolism; lead to ↑ gluconeogenesis (via mTOR), ↑ de novo lipogenesis (via ChREBP)(15, 16) |
| Aromatic: Phe, Tyr | ||||
| Glutamate/glutamine cycle | Glutamate | T2D, T1D, retinopathy | ↑ (↓ for retinopathy) | Glutamate promotes gluconeogenesis, muscle proteolysis, inflammation (via glutathione synthase)(20) Glutamine regulates insulin secretion, pancreatic β-cell function(4); contributes to cellular metabolism (anaplerosis) |
| Glutamine | ↓ (↑ for retinopathy) | |||
| Amino acid | Glycine | T2D, IR, hepatic steatosis | ↓ | Glycine involved in many homeostatic processes; protective; depletion may reflect oxidative stress and ↑ gluconeogenesis(9) |
| Glycolysis & cellular respiration | Lactate, pyruvate, alanine | T2D, T1D, kidney disease, CVD | ↑ | Pyruvate formed via glycolysis, leads to aerobic or anaerobic (lactate producing) respiration; alanine a substrate for TCA, Cahill cycles; ↑ levels due to mitochondrial dysfunction, ↓ oxidative capacity(3, 25) |
| ADMA metabolism | DMGV | Hepatic steatosis, fitness, T2D, CVD | ↑ | Formed by transamination of ADMA; dynamic to lifestyle changes(28, 29) |
| ADMA | Retinopathy, CVD | ADMA is an inhibitor of nitric oxide (NO) synthase, noted to be elevated in diabetes | ||
| NO metabolism | Arginine, citrulline | Retinopathy | ↑ | Arginine and citrulline are NO precursors; ↑ levels in microvascular complications reflects altered NO metabolism |
| Lysine metabolism | Lysine | T2D, CVD | ↑ (T2D)/ ↓ (CVD) | Amino acid, theorized to be elevated in T2D as response to upregulation of 2-AAA (for which it is precursor) |
| 2-alpha aminoadipic acid (2-AAA) | T2D | ↑ | Function is incompletely understood; produced by oxidative stress; promotes adipogenesis(33) | |
| Vitamin C metabolism | Dehydroascorbic acid (DHAA) | T1D | ↑ | Oxidized form of vitmin C; substrate for glutathione; may inhibit insulin secretion(42) |
| Hydroxy acids and derivatives | 3,4 dihydroxybutyric acid (DHBA) | Retinopathy | ↑ | Related to ketone bodies, may be higher in high fat diet; related to dyslipidemia(55) |
| Triglyceride metabolism | Triacylglycerols (TAGs) with ↓ carbon and double bond numbers | Hepatic steatosis, T2D, T1D, retinopathy, neuropathy, kidney disease | ↑ | Hypothesized to primarily reflect IR in the liver, may contribute to lipotoxicity |
| Additional lipid species | Phospthatidylcholines (PCs) and lysoPCs | T2D, kidney disease | ↑/↓ | Similar to TAGs, ↓carbon number and double bond content are linked with adverse outcomes; choline containing lipids, components of cell membrane and lipid metabolism(9) |
| Acylcarnitines | T2D, neuropathy, CVD | ↑ | Reflection of impaired mitochondrial function, reduced oxidative capacity |
This table is not an exhaustive list of all metabolites that have been associated with diabetes or its complications. It is intended to highlight metabolites discussed in this review and to demonstrate their links to relevant metabolic pathways.
Abbreviations: Val, valine; Leu, leucine; Ile, isoleucine; Phe, phenylalanine; Tyr, tyrosine; T2D, type 2 diabetes; T1D, type 1 diabetes; IR, insulin resistance; TCA, ticarboxylic acid; DMGV, dimethylguanidino valeric acid; ADMA, asymmetric dimethylarginine;
An important strength of metabolomic interrogation is the ability to measure both known molecular compounds and chemical structures with unknown curation, enabling novel discovery. In one such example, untargeted metabolomics was used to identify novel metabolite associations with liver fat. Through genetic analyses, the top metabolite association was identified as dimethylguanidino valeric acid (DMGV)(27). DMGV has subsequently been shown to be an important marker of cardiometabolic health that is associated with incident diabetes(27), poor cardiorespiratory fitness(28, 29), and future CVD(30). DMGV is formed via transamination of asymmetric dimethylarginine (ADMA) and it is theorized that higher circulating levels in cardiometabolic disease may be a reflection of greater transamination by alanine glyoxylate aminotransferase 2 in the context of reduced competition from β-aminoisobutyric acid (a protective compound linked to adipose tissue browning and leptin response)(27, 31). The metabolite 2-alpha aminoadipic acid (2-AAA) is another poorly understood metabolic intermediary that was identified via metabolomic quantification to be related to future T2D risk(32). Further investigation demonstrated that 2-AAA promotes adipogenesis, contributing to lipotoxicity, and may impair insulin signaling across different tissues(33).
Abnormalities in circulating lipid levels are well described to occur in the pathogenesis of diabetes, and elevated triglyceride levels are frequently observed in individuals with IR(34). Through profiling of individual triglyceride (triacylglycerol; “TAG”) species, investigators have demonstrated that TAGs with lower carbon number and double bond content (e.g., saturated or monounsaturated fatty acids) are more closely related to IR and diabetes risk(35). After a glucose challenge, TAGs with lower carbon number and double bond content tend to decrease, while other TAGs increase, likely reflecting the direct effects of insulin. Acylcarnitines may also be increased in IR due to impaired mitochondrial function and reduced beta-oxidation(36). Several additional lipid species have been associated with diabetes risk (e.g., phosphatidylcholines(9, 37, 38) and lysophosphatidylcholines(35)), but their mechanisms remain unclear.
Type 1 diabetes
T1D is speculated to typically result from specific factors (e.g., autoantibodies directed to the pancreatic β-cells), and is characterized by insulin deficiency. In the setting of insulin withdrawal in patients with T1D, the expected increase in proteolysis, lipolysis, ketogenesis, and other catabolic functions are observed and reflected by elevated circulating levels of BCAAs, glycerol, and beta-hydroxybutyrate(39). Intriguingly, however, it is increasingly recognized that widespread metabolic dysfunction may be apparent prior to the development of autoantibodies in T1D. Lamicchane and colleagues characterized circulating metabolite profiles of children who went on to develop T1D(40). They observed amino acid dysregulation to precede detectable islet autoantibodies, with lower levels of glutamic acid, aspartic acid, and tryptophan observed even in infancy in individuals who developed T1D. Metabolites of microbial origin also differed between individuals who developed T1D and controls, consistent with evolving evidence for a contribution of the gut microbiome to the pathogenesis of T1D(40). Abnormalities in circulating lipid profiles may also precede T1D diagnosis, with higher levels of several sphyngomyelins and lower levels of specific TAG and phosphatidylcholine species observed in individuals who eventually developed diabetes(41). In infants at increased risk of developing T1D, distinct metabolomic signatures comprised of dehydroascorbic acid (DHAA), amino acids, BCAAs and fatty acids were associated prospectively with autoantibody development(42). These findings suggest the possibility that autoimmunity may be a response to earlier metabolic dysfunction in T1D(43), presenting new opportunities for screening and prevention.
Metabolite profiling for predicting and understanding diabetes complications
It is well established that complications from diabetes—especially CVD, the leading cause of death in diabetic individuals—are not solely a consequence of elevated blood glucose levels(44–46). Additional metabolic insults occurring in the setting of diabetes are likely to contribute to complications and may vary across different tissues and organ systems. Metabolomic investigation is being increasingly utilized to uncover risk markers and mechanisms underlying diabetes complications, with potential applications for screening, prevention, and possible therapeutic modulation(44).
Retinopathy
While chronicity of disease, hyperglycemia, and blood pressure control are established risk factors for diabetic retinopathy (DR), there remains substantial unexplained variation in who develops DR despite similar levels of these clinical factors(47). Metabolites have shown promise both as biomarkers of increased risk for DR and as molecular snapshots that can be used to improve understanding of mechanisms underlying its pathogenesis. For example, circulating levels of ADMA and dimethylarginine (DMA) are higher in individuals with diabetes and DR versus those with diabetes but no DR(48). A unique advantage of metabolomic interrogation is that levels in the blood can be compared with levels in other complementary biofluids. In this case, higher ADMA levels have also been reported in the aqueous humor of individuals with severe diabetic retinopathy(49). As higher circulating levels of ADMA can limit NO bioavailability through inhibition of NO synthase, these observational findings build on experimental evidence suggesting a key role of deranged nitric oxide (NO) metabolism in the pathogenesis of DR(50). Higher blood levels of the NO precursors arginine and citrulline have also been described in individuals with DR, which may further reflect dysregulated NO metabolism(51). Moreover, arginine metabolism may be integral to the development of DR, as experimental model systems suggest arginase as a biological mediator of DR through its effects of reducing NO bioavailability and increasing oxidative stress(52).
Numerous other metabolites have been associated with DR severity in mostly cross-sectional studies, including amino acids (higher phenylalanine(53), lower histidine(53) and asparagine(54)) and several oxidated lipid species(54). While many of these studies have focused on patients with T2D, circulating metabolite levels have also been linked to DR in patients with T1D. In a study of 648 individuals with T1D, 3,4 dihydroxybutyric acid (DHBA) and several triglyceride species were associated with poorer DR grading(55). DHBA is a regulator of lipid metabolism and these findings, therefore, indicate that adverse lipid metabolism may be involved in DR development in T1D.
Recent efforts have sought to leverage multi-metabolite scores, often developed through machine learning approaches, to improve clinical prediction of DR(54, 56). While the initial data are promising, these models require validation against future (rather than cross-sectional) outcomes and broad validation across different population before they can be considered for clinical use.
Intriguingly, many of the metabolite associations that have been observed with DR risk so far are distinct from the metabolites that have been identified for future IR or diabetes itself. One notable example is the aromatic amino acid tyrosine. While higher tyrosine levels predict future diabetes development in apparently healthy individuals(6), lower tyrosine levels have been associated with increased risk of DR in individuals with T2D enrolled in the ADVANCE trial(53). Similarly, glutamine/glutamate metabolism has been related to DR(57), with lower glutamate and higher glutamine demonstrating associations with DR(58), which is opposite to the directionality observed for these metabolites and the risk of diabetes. While the biological processes underlying the different metabolomic signatures of diabetes and DR are not yet fully elucidated, metabolomic investigation of DR has clearly implicated separate metabolic mechanisms for diabetes and for DR, with important implications for screening/treatment(59).
Neuropathy
Similar to DR, investigations of metabolomic markers of diabetic neuropathy (DN) have largely implicated distinct metabolic signatures (and biological pathways) when compared to diabetes itself. Largely, these studies have focused on lipid metabolism given the extensive observational and experimental data establishing dysregulated lipid metabolism as being integral to the development of DN(60, 61). Circulating levels of several specific lipid species have been related to DN in diabetic individuals: higher 1-deoxydihydroceramides, potentially reflecting serine deficiency(62); higher ursodeoxycholate, which modulates lipid absorption(63); lower long-chain acylcarnitines(63), among others. These findings from lipidomic/metabolomic analyses support prior experimental models implicating dysregulation of peripheral nerve lipid metabolism as being central to neurotoxicity and DN pathogenesis(64, 65). Reflecting the accumulating evidence linking lipid metabolism to DN, an initial clinical trial of dietary supplementation with omega-3 polyunsaturated fatty acids (PUFA) to prevent lipotoxicity of sensory nerves in DN has shown improvement in DN symptoms and more definitive clinical trials are pending(66).
Kidney disease
Metabolic dysregulation is known to play an important role in the pathogenesis of diabetic kidney disease (DKD)(67). Accordingly, metabolites reflecting diverse biological processes have been related to DKD in mostly small cross-sectional studies(68). Lower levels of pyruvate and higher levels of glycolytic and TCA-cycle metabolites in individuals who develop DKD are in concert with findings from kidney biopsy samples implicating glycolytic flux as a potential mediator of DKD(69). In these studies, upregulation of enzymes and substrate for glycolysis was observed in individuals who did not develop DKD despite longstanding diabetes, suggesting that the ability to increase glycolysis in response to hyperglycemia was integral to glomerular cell integrity(69, 70). Altered tryptophan metabolism, reflected by higher levels of L-tryptophan and serotonin, has also been described in individuals with DKD, although the mechanisms are less clear(71). Dysfunctional lipid metabolism may contribute to progressive kidney disease in diabetes(72). In a study of 79 individuals with chronic kidney disease of whom ≈50% had diabetes, lower circulating levels of diacylglycerols (DAGs), cholesteryl esters, and phosphatidylcholines, and higher monoacylglycerols and phosphatidylethanolamine were associated with progression to end-stage renal disease(72).
Profiling of blood metabolites can be complemented by urinary metabolite profiling to provide additional information regarding the kidney’s metabolic responses in diabetes. This has been recently applied to identify urinary biomarkers of progression to DKD in T1D(73). Investigators evaluated 51 urinary metabolites from 24-hour urine samples in >2000 individuals with T1D in the Finnish Diabetic Nephropathy study and identified seven metabolites to be associated with progression of DKD: higher BCAAs (leucine, valine, isoleucine), pseudouridine, threonine, and 2-hydroxyisobutyrate, and lower citrate(73). The findings of higher urinary BCAA levels preceding DKD in T1D is intriguing and supports the hypothesis that development of IR contributes to DKD in T1D(73, 74).
Cardiovascular disease
It is well established that the high risk of CVD events in individuals with diabetes is not fully explained by hyperglycemia and that additional mechanisms such as inflammation, platelet hyperreactivity, endothelial dysfunction, autonomic nerve dysfunction, and membrane instability in various cell types play important roles(75–77). Additional biomarkers are, therefore, necessary to improve prediction of CVD events in individuals with diabetes and to improve preventative strategies.
In >700 individuals with T2D followed for over 5 years, a number of metabolites were associated with future CVD risk. These included associations of CVD with higher levels of DMGV (which has also been associated with hepatic steatosis and incident diabetes), N,N2-dimethylguanosine (a nucleoside derivative linked with cardiometabolic disease and CVD risk in those without diabetes(78)), homocitrulline, 1-methyladenosine, acylcarnitine C10:3, and urobilin and lower levels of lysine, hippurate, tryptophan, and threonine(79). Blood levels of BCAAs and aromatic amino acids have also been positively associated with the risk of CVD(13). Therefore, CVD-associated metabolites in diabetes have mostly implicated similar pathways as being involved with the pathogenesis of CVD and diabetes in individuals without diabetes (e.g., BCAAs, DMGV, N,N2-dimethylguanosine), while also indicating several metabolites with distinct associations that might reflect more complex biological regulation. Lysine, for example, is directly associated with diabetes risk, but is inversely associated with CVD risk in diabetes(80). Additional studies in larger numbers of individuals across both T1D and T2D are necessary to identify biomarkers and pathways most closely linked with CVD risk.
Dynamic changes in the metabolome in response to perturbation or therapy
A unique aspect of the circulating metabolome is that it can change rapidly in response to shifts in environmental factors (e.g., external stressors) or physiological changes. This feature can be leveraged to examine the metabolic response to a discrete stressor (e.g., glucose challenge or exercise) to evaluate disease susceptibility and biological pathways. It can also be used to provide insight into treatment responses, which may potentially enable the development of individualized and tailored, precision approaches to diabetes treatment.
Metabolic response to perturbation
Substantial changes are observed in the circulating metabolome in response to the increased metabolic demands imposed by acute exercise(29, 81). Favorable shifts in many metabolites previously linked to T2D risk (e.g., BCAAs, glutamate, DMGV, TAGs) are observed with an acute bout of exercise and these salutary changes may be blunted in the setting of obesity, implicating improved insulin sensitivity with exercise (and relative IR in obesity) as a potential unifying mechanism(29, 81). Indeed, chronic exercise training also exerts favorable responses in certain metabolites (e.g., DMGV and BAIBA(28, 31)) pointing to improvement in their representative metabolic pathways with habitual exercise. Future studies are needed to clarify optimal exercise training regimens for diabetes prevention and potential treatment.
The circulating metabolome also changes in response to a dietary stress, such as an oral glucose tolerance test (OGTT). The post-OGTT metabolic response is complex, with changes in metabolites with known roles in glucose and insulin metabolism as well as in metabolites with less-defined roles(82). In addition, large inter-individual differences can be observed in response to the same meal(83, 84). This observation was used in a recent study to identify novel genetic loci associated with the circulating lipid response to a mixed meal challenge(85). In turn, these genetic loci were also linked to body composition and clinical diabetes in large genetic consortia. Taken together, these findings suggest that inter-individual differences in the metabolic response to dietary perturbation can be assessed through metabolomic changes, and that these studies may improve understanding of diabetes susceptibility.
Weight loss and lifestyle interventions
Metabolites signatures demonstrate promise for uncovering the metabolic effects of different weight loss treatment strategies and, potentially, for targeting specific weight loss approaches towards patients that might derive the most benefit. For example, in obese individuals without diabetes, higher levels of BCAAs predict the degree of improvement in IR with weight loss(86). In the Diabetes Prevention Program, heterogeneity in response to lifestyle interventions versus metformin was observed across different metabolomic signatures, with higher levels of several phospholipids at baseline identifying individuals who appeared to benefit most from lifestyle modifications(87). Metabolite changes with different weight loss treatment strategies have also been investigated. The fall in levels of BCAAs and 2-hydroxybutyric acid following gastric bypass surgery correlate with improvements in IR, indicating potential mechanisms for metabolic benefits of surgical weight loss therapies(86, 88). Moreover, BCAA levels are noted to decrease more rapidly with surgical obesity therapies rather than weight loss from caloric restriction, mirroring the cardiometabolic health benefits reported for bariatric surgery(89).
Metabolic response to drug therapy for T2D
It is increasingly recognized that there are different subgroups of T2D that may benefit from different pharmacologic treatment approaches(90). Metabolites offer an attractive option for use in precision phenotyping as they (1) may enable the assigning of specific therapies to appropriate subgroups; (2) change rapidly in response to medication treatment; and, (3) can be assessed serially. While such approaches are not yet ready for clinical use, recent advances in employing metabolomic profiling to understand distinct metabolic effects of T2D medications or differences in inter-individual treatment responses make this an intriguing prospect.
In the Diabetes Prevention Program, heterogeneity in response to metformin treatment (in addition to variation in the effect of lifestyle modifications) was observed according to baseline metabolite levels(87). Notably, individuals with higher levels of AMP (a master cell energy regulator and potential mediator of metformin’s positive effects(91)) appeared to benefit more from metformin therapy, suggesting the potential of using metabolomics to assign specific drugs to specific patients(87). In a separate study, specific metabolites were observed to predict the therapeutic response to metformin or sulfonylureas(92). Higher levels of liver metabolites 2-hydroxybutanoic acid and 3-hydroxybutanoic acid were associated with a larger decrease in HbA1c with metformin treatment versus sulfonylurea treatment over the next 5 years(92). The acute metabolic effects of metformin have also been evaluated with a metabolomic approach to better understand its beneficial effects revealing changes in 36 metabolites after a single dose. Metabolites reflecting lipid metabolism, BCAA metabolism, energy homeostasis, and gut microbiota functions were among those with the greatest change in response to metformin administration(93).
Substantial recent interest has focused on using metabolomics to unravel the metabolic effects of sodium/glucose cotransporter-2 inhibitors (SGLT2i) due to their benefit across a broad array of cardiometabolic outcomes and uncertainty regarding their underlying mechanisms. In one study, untargeted metabolomics was performed on 25 individuals with T2D and CVD treated with empagliflozin for one month(94). After one month of treatment, 162 of 1269 assayed metabolites were altered including enrichment of TCA cycle intermediates that were not explained by increased rates of glycolysis(94). In a separate study, after 12 weeks of treatment with dapagliflozin, enhanced glycine degradation (better mitochondrial function), TCA cycle (energy metabolism), L-carnitine biosynthesis (energy metabolism), and super-pathway of citrulline metabolism (NO synthase and endothelial function) were identified in plasma and kidneys of individuals with T2D(95). Overall, these studies have demonstrated that the beneficial effects of SGLT2i may be partly explained by optimization of fuel substrate for end organs (such as the heart and kidneys). This effect of SGLT2i is theorized to act largely through SIRT and AMPK/mTOR, which stimulate a fasting/starvation metabolic response that results in preferential shift towards more optimal energy sources(96).
Metabolic response to drug therapy for T1D
The metabolic response to glycemic control in T1D is complex, with many metabolite levels corresponding to hyperglycemia itself, but metabolic profiles remain altered in T1D (when compared to controls without diabetes) even in the context of optimal blood glucose control(97). Levels of BCAAs and alanine appear to correspond with blood glucose levels in T1D, similar to T2D(98). With poor glycemic control, BCAAs, glucogenic amino acids such as glycine, serine, arginine, ketoacids, and others are noted to be elevated. While appropriate glycemic control can normalize certain pathways (e.g., nucleotide metabolism, lysine, histidine-glutamate-glutamine, cortisol, urea cycle), others remain elevated, suggesting widespread metabolic dysfunction in T1D beyond what can be explained by insulin deficiency alone(97).
Conclusions and future directions
Human metabolomic studies have identified numerous blood metabolites to be related to disease pathogenesis, risk of complications, and therapeutic response in both T1D and T2D (Table 1). In several instances (e.g., BCAAs), these findings point to pathways that may be biological mediators of diabetes and have served to either corroborate prior work in non-human experimental models or to discover novel pathways for further mechanistic interrogation. In this review, we have highlighted features of circulating metabolites that render them especially powerful tools for translational investigation in diabetes: (1) untargeted metabolomics can be used for discovery of novel compounds and biology; (2) metabolites are dynamic with controlled perturbations (e.g., exercise or dietary challenge) and may therefore provide a “readout” of the physiological response to stress; (3) metabolites can also be measured in complementary biofluids reflecting features of key end-organs (e.g., eyes, kidneys); and, (4) metabolites respond to lifestyle and pharmacologic interventions, and can therefore be used to guide precision therapeutics.
While metabolomic investigation in diabetes has already yielded important insights, the translation of these findings to clinical care will require several additional steps. Prior to clinical use, risk prediction models will need to be validated in larger samples, with representation across the age rage and encompassing racial and ethnic diversity(99). These prediction models will benefit from uniform approaches to metabolite quantification with assays developed for clinical use (rather than discovery). Studies of larger numbers of individuals are necessary also to explore the potential of using metabolomics to help define specific T2D subgroups that have implications for prognosis and treatment response(90). Metabolomics are likely to be an important part of the toolkit for precision diabetes care, but approaches that integrate multiple “omic” techniques (e.g., genomics, transcriptomics, epigenomics, microbiome, proteomics(100)) may have the greatest potential to advance the screening, prevention, and treatment of patients with diabetes.
Funding
Dr. Nayor is supported by NIH grants K23-HL138260 and R01-HL156975 and from a Career Investment Award from the Department of Medicine, Boston University School of Medicine.
Abbreviations:
- BCAA
branched-chain amino acids
- BCKA
branched-chain ketoacid
- TCA
tricarboxylic
Footnotes
Disclosures
The authors have no relevant disclosures to report.
Human and Animal Rights
All reported studies/experiments with human or animal subjects performed by the authors have been previously published and complied with all applicable ethical standards (including the Helsinki declaration and its amendments, institutional/national research committee standards, and international/national/institutional guidelines).
Publisher's Disclaimer: This AM is a PDF file of the manuscript accepted for publication after peer review, when applicable, but does not reflect post-acceptance improvements, or any corrections. Use of this AM is subject to the publisher's embargo period and AM terms of use. Under no circumstances may this AM be shared or distributed under a Creative Commons or other form of open access license, nor may it be reformatted or enhanced, whether by the Author or third parties. See here for Springer Nature's terms of use for AM versions of subscription articles: https://www.springernature.com/gp/open-research/policies/accepted-manuscript-terms
References
- 1.Ogurtsova K, da Rocha Fernandes JD, Huang Y, Linnenkamp U, Guariguata L, Cho NH, et al. IDF Diabetes Atlas: Global estimates for the prevalence of diabetes for 2015 and 2040. Diabetes Res Clin Pract. 2017;128:40–50. [DOI] [PubMed] [Google Scholar]
- 2.Skyler JS, Bakris GL, Bonifacio E, Darsow T, Eckel RH, Groop L, et al. Differentiation of Diabetes by Pathophysiology, Natural History, and Prognosis. Diabetes. 2017;66(2):241–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9(4):311–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng S, Rhee EP, Larson MG, Lewis GD, McCabe EL, Shen D, et al. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation. 2012;125(18):2222–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng S, Shah SH, Corwin EJ, Fiehn O, Fitzgerald RL, Gerszten RE, et al. Potential Impact and Study Considerations of Metabolomics in Cardiovascular Health and Disease: A Scientific Statement From the American Heart Association. Circ Cardiovasc Genet. 2017;10(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, et al. Metabolite profiles and the risk of developing diabetes. Nature medicine. 2011;17(4):448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walford GA, Ma Y, Clish C, Florez JC, Wang TJ, Gerszten RE, et al. Metabolite Profiles of Diabetes Incidence and Intervention Response in the Diabetes Prevention Program. Diabetes. 2016;65(5):1424–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamakado M, Nagao K, Imaizumi A, Tani M, Toda A, Tanaka T, et al. Plasma Free Amino Acid Profiles Predict Four-Year Risk of Developing Diabetes, Metabolic Syndrome, Dyslipidemia, and Hypertension in Japanese Population. Sci Rep. 2015;5:11918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Floegel A, Stefan N, Yu Z, Muhlenbruch K, Drogan D, Joost HG, et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 2013;62(2):639–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrannini E, Natali A, Camastra S, Nannipieri M, Mari A, Adam KP, et al. Early metabolic markers of the development of dysglycemia and type 2 diabetes and their physiological significance. Diabetes. 2013;62(5):1730–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wurtz P, Tiainen M, Makinen VP, Kangas AJ, Soininen P, Saltevo J, et al. Circulating metabolite predictors of glycemia in middle-aged men and women. Diabetes Care. 2012;35(8):1749–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah SH, Bain JR, Muehlbauer MJ, Stevens RD, Crosslin DR, Haynes C, et al. Association of a peripheral blood metabolic profile with coronary artery disease and risk of subsequent cardiovascular events. Circ Cardiovasc Genet. 2010;3(2):207–14. [DOI] [PubMed] [Google Scholar]
- 13.Magnusson M, Lewis GD, Ericson U, Orho-Melander M, Hedblad B, Engstrom G, et al. A diabetes-predictive amino acid score and future cardiovascular disease. Eur Heart J. 2013;34(26):1982–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White PJ, Newgard CB. Branched-chain amino acids in disease. Science. 2019;363(6427):582–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao H, Zhang F, Sun D, Wang X, Zhang X, Zhang J, et al. Branched-Chain Amino Acids Exacerbate Obesity-Related Hepatic Glucose and Lipid Metabolic Disorders via Attenuating Akt2 Signaling. Diabetes. 2020;69(6):1164–77. [DOI] [PubMed] [Google Scholar]
- 16. White PJ, McGarrah RW, Grimsrud PA, Tso SC, Yang WH, Haldeman JM, et al. The BCKDH Kinase and Phosphatase Integrate BCAA and Lipid Metabolism via Regulation of ATP-Citrate Lyase. Cell Metab. 2018;27(6):1281–93 e7. **This study demonstrated that branched-chain ketoacid dehydrogenase is an important regulator of the effects of BCAAs on glucose tolerance and elucidate some of the mechanisms responsible for the association of BCAAs and diabetes risk.
- 17.Qureshi W, Santaren ID, Hanley AJ, Watkins SM, Lorenzo C, Wagenknecht LE. Risk of diabetes associated with fatty acids in the de novo lipogenesis pathway is independent of insulin sensitivity and response: the Insulin Resistance Atherosclerosis Study (IRAS). BMJ Open Diabetes Res Care. 2019;7(1):e000691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lotta LA, Scott RA, Sharp SJ, Burgess S, Luan J, Tillin T, et al. Genetic Predisposition to an Impaired Metabolism of the Branched-Chain Amino Acids and Risk of Type 2 Diabetes: A Mendelian Randomisation Analysis. PLoS medicine. 2016;13(11):e1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felig P, Marliss E, Cahill GF Jr. Plasma amino acid levels and insulin secretion in obesity. N Engl J Med. 1969;281(15):811–6. [DOI] [PubMed] [Google Scholar]
- 20.Wopereis S, Rubingh CM, van Erk MJ, Verheij ER, van Vliet T, Cnubben NH, et al. Metabolic profiling of the response to an oral glucose tolerance test detects subtle metabolic changes. PLoS One. 2009;4(2):e4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Zheng Y, Guasch-Ferré M, Ruiz-Canela M, Toledo E, Clish C, et al. High plasma glutamate and low glutamine-to-glutamate ratio are associated with type 2 diabetes: Case-cohort study within the PREDIMED trial. Nutrition, Metabolism and Cardiovascular Diseases. 2019;29(10):1040–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fahien LA, Macdonald MJ. The complex mechanism of glutamate dehydrogenase in insulin secretion. Diabetes. 2011;60(10):2450–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guasch-Ferre M, Hruby A, Toledo E, Clish CB, Martinez-Gonzalez MA, Salas-Salvado J, et al. Metabolomics in Prediabetes and Diabetes: A Systematic Review and Meta-analysis. Diabetes Care. 2016;39(5):833–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crawford SO, Hoogeveen RC, Brancati FL, Astor BC, Ballantyne CM, Schmidt MI, et al. Association of blood lactate with type 2 diabetes: the Atherosclerosis Risk in Communities Carotid MRI Study. Int J Epidemiol. 2010;39(6):1647–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008;7(1):45–56. [DOI] [PubMed] [Google Scholar]
- 26.DiGirolamo M, Newby FD, Lovejoy J. Lactate production in adipose tissue: a regulated function with extra-adipose implications. Faseb j. 1992;6(7):2405–12. [DOI] [PubMed] [Google Scholar]
- 27.O’Sullivan JF, Morningstar JE, Yang Q, Zheng B, Gao Y, Jeanfavre S, et al. Dimethylguanidino valeric acid is a marker of liver fat and predicts diabetes. J Clin Invest. 2017;127(12):4394–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robbins JM, Herzig M, Morningstar J, Sarzynski MA, Cruz DE, Wang TJ, et al. Association of Dimethylguanidino Valeric Acid With Partial Resistance to Metabolic Health Benefits of Regular Exercise. JAMA Cardiol. 2019;4(7):636–43. **A study characterizing the response of the metabolite DMGV to exercise training.
- 29. Nayor M, Shah RV, Miller PE, Blodgett JB, Tanguay M, Pico AR, et al. Metabolic Architecture of Acute Exercise Response in Middle-Aged Adults in the Community. Circulation. 2020;142(20):1905–24. **This study described the response of the circulating metabolome to exercise, demonstrating favorable effects on broad metabolic pathways.
- 30.Ottosson F, Ericson U, Almgren P, Smith E, Brunkwall L, Hellstrand S, et al. Dimethylguanidino Valerate: A Lifestyle-Related Metabolite Associated With Future Coronary Artery Disease and Cardiovascular Mortality. Journal of the American Heart Association. 2019;8(19):e012846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts LD, Bostrom P, O’Sullivan JF, Schinzel RT, Lewis GD, Dejam A, et al. beta-Aminoisobutyric acid induces browning of white fat and hepatic beta-oxidation and is inversely correlated with cardiometabolic risk factors. Cell Metab. 2014;19(1):96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang TJ, Ngo D, Psychogios N, Dejam A, Larson MG, Vasan RS, et al. 2-Aminoadipic acid is a biomarker for diabetes risk. J Clin Invest. 2013;123(10):4309–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee HJ, Jang HB, Kim WH, Park KJ, Kim KY, Park SI, et al. 2-Aminoadipic acid (2-AAA) as a potential biomarker for insulin resistance in childhood obesity. Sci Rep. 2019;9(1):13610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aslam M, Aggarwal S, Sharma KK, Galav V, Madhu SV. Postprandial Hypertriglyceridemia Predicts Development of Insulin Resistance Glucose Intolerance and Type 2 Diabetes. PLoS One. 2016;11(1):e0145730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhee EP, Cheng S, Larson MG, Walford GA, Lewis GD, McCabe E, et al. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest. 2011;121(4):1402–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams SH, Hoppel CL, Lok KH, Zhao L, Wong SW, Minkler PE, et al. Plasma acylcarnitine profiles suggest incomplete long-chain fatty acid beta-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic African-American women. J Nutr. 2009;139(6):1073–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang-Sattler R, Yu Z, Herder C, Messias AC, Floegel A, He Y, et al. Novel biomarkers for pre-diabetes identified by metabolomics. Mol Syst Biol. 2012;8:615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cole LK, Vance JE, Vance DE. Phosphatidylcholine biosynthesis and lipoprotein metabolism. Biochim Biophys Acta. 2012;1821(5):754–61. [DOI] [PubMed] [Google Scholar]
- 39.Dutta T, Chai HS, Ward LE, Ghosh A, Persson XM, Ford GC, et al. Concordance of changes in metabolic pathways based on plasma metabolomics and skeletal muscle transcriptomics in type 1 diabetes. Diabetes. 2012;61(5):1004–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lamichhane S, Kemppainen E, Trost K, Siljander H, Hyoty H, Ilonen J, et al. Circulating metabolites in progression to islet autoimmunity and type 1 diabetes. Diabetologia. 2019;62(12):2287–97. **A comprehensive report of changes in metabolites preceding the development of type 1 diabetes, supporting the hypothesis that more widespread metabolic dysfunction is responsible for type 1 diabetes pathogenesis.
- 41.Lamichhane S, Ahonen L, Dyrlund TS, Kemppainen E, Siljander H, Hyoty H, et al. Dynamics of Plasma Lipidome in Progression to Islet Autoimmunity and Type 1 Diabetes - Type 1 Diabetes Prediction and Prevention Study (DIPP). Sci Rep. 2018;8(1):10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li Q, Parikh H, Butterworth MD, Lernmark A, Hagopian W, Rewers M, et al. Longitudinal Metabolome-Wide Signals Prior to the Appearance of a First Islet Autoantibody in Children Participating in the TEDDY Study. Diabetes. 2020;69(3):465–76. **Another study describing distinct metabolic signatures that can be identified prior to autoantibody formation in type 1 diabetes. Here, the investigators compared different types of autoantibodies.
- 43.Oresic M, Simell S, Sysi-Aho M, Näntö-Salonen K, Seppänen-Laakso T, Parikka V, et al. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J Exp Med. 2008;205(13):2975–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eid S, Sas KM, Abcouwer SF, Feldman EL, Gardner TW, Pennathur S, et al. New insights into the mechanisms of diabetic complications: role of lipids and lipid metabolism. Diabetologia. 2019;62(9):1539–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patel A, MacMahon S, Chalmers J, Neal B, Billot L, Woodward M, et al. Intensive blood glucose control and vascular outcomes in patients with type 2 diabetes. N Engl J Med. 2008;358(24):2560–72. [DOI] [PubMed] [Google Scholar]
- 46.Reaven PD, Emanuele NV, Wiitala WL, Bahn GD, Reda DJ, McCarren M, et al. Intensive Glucose Control in Patients with Type 2 Diabetes - 15-Year Follow-up. N Engl J Med. 2019;380(23):2215–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hou XW, Wang Y, Pan CW. Metabolomics in Diabetic Retinopathy: A Systematic Review. Invest Ophthalmol Vis Sci. 2021;62(10):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yun JH, Kim JM, Jeon HJ, Oh T, Choi HJ, Kim BJ. Metabolomics profiles associated with diabetic retinopathy in type 2 diabetes patients. PLoS One. 2020;15(10):e0241365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugai M, Ohta A, Ogata Y, Nakanishi M, Ueno S, Kawata T, et al. Asymmetric dimethylarginine (ADMA) in the aqueous humor of diabetic patients. Endocr J. 2007;54(2):303–9. [DOI] [PubMed] [Google Scholar]
- 50.Malecki MT, Undas A, Cyganek K, Mirkiewicz-Sieradzka B, Wolkow P, Osmenda G, et al. Plasma asymmetric dimethylarginine (ADMA) is associated with retinopathy in type 2 diabetes. Diabetes Care. 2007;30(11):2899–901. [DOI] [PubMed] [Google Scholar]
- 51.Sumarriva K, Uppal K, Ma C, Herren DJ, Wang Y, Chocron IM, et al. Arginine and Carnitine Metabolites Are Altered in Diabetic Retinopathy. Invest Ophthalmol Vis Sci. 2019;60(8):3119–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patel C, Rojas M, Narayanan SP, Zhang W, Xu Z, Lemtalsi T, et al. Arginase as a mediator of diabetic retinopathy. Front Immunol. 2013;4:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Welsh P, Rankin N, Li Q, Mark PB, Wurtz P, Ala-Korpela M, et al. Circulating amino acids and the risk of macrovascular, microvascular and mortality outcomes in individuals with type 2 diabetes: results from the ADVANCE trial. Diabetologia. 2018;61(7):1581–91. **A substudy of the ADVANCE trial demonstrating associations of baseline amino acid levels with long-term microvascular and macrovascular outcomes.
- 54.Rhee SY, Jung ES, Suh DH, Jeong SJ, Kim K, Chon S, et al. Plasma amino acids and oxylipins as potential multi-biomarkers for predicting diabetic macular edema. Sci Rep. 2021;11(1):9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Curovic VR, Suvitaival T, Mattila I, Ahonen L, Trost K, Theilade S, et al. Circulating Metabolites and Lipids Are Associated to Diabetic Retinopathy in Individuals With Type 1 Diabetes. Diabetes. 2020;69(10):2217–26. **A large, prospective study of individuals with type 1 diabetes, demonstrating novel metabolite associations with diabetic retinopathy.
- 56.Zuo J, Lan Y, Hu H, Hou X, Li J, Wang T, et al. Metabolomics-based multidimensional network biomarkers for diabetic retinopathy identification in patients with type 2 diabetes mellitus. BMJ Open Diabetes Res Care. 2021;9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu XR, Yang FY, Lu J, Zhang HR, Sun R, Zhou JB, et al. Plasma metabolomic profiling of proliferative diabetic retinopathy. Nutr Metab (Lond). 2019;16:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rhee SY, Jung ES, Park HM, Jeong SJ, Kim K, Chon S, et al. Plasma glutamine and glutamic acid are potential biomarkers for predicting diabetic retinopathy. Metabolomics. 2018;14(7):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen L, Cheng CY, Choi H, Ikram MK, Sabanayagam C, Tan GS, et al. Plasma Metabonomic Profiling of Diabetic Retinopathy. Diabetes. 2016;65(4):1099–108. [DOI] [PubMed] [Google Scholar]
- 60.Kwon B, Lee HK, Querfurth HW. Oleate prevents palmitate-induced mitochondrial dysfunction, insulin resistance and inflammatory signaling in neuronal cells. Biochim Biophys Acta. 2014;1843(7):1402–13. [DOI] [PubMed] [Google Scholar]
- 61.Wiggin TD, Sullivan KA, Pop-Busui R, Amato A, Sima AA, Feldman EL. Elevated triglycerides correlate with progression of diabetic neuropathy. Diabetes. 2009;58(7):1634–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fridman V, Zarini S, Sillau S, Harrison K, Bergman BC, Feldman EL, et al. Altered plasma serine and 1-deoxydihydroceramide profiles are associated with diabetic neuropathy in type 2 diabetes and obesity. J Diabetes Complications. 2021;35(4):107852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rumora AE, Guo K, Alakwaa FM, Andersen ST, Reynolds EL, Jorgensen ME, et al. Plasma lipid metabolites associate with diabetic polyneuropathy in a cohort with type 2 diabetes. Ann Clin Transl Neurol. 2021;8(6):1292–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O’Brien PD, Guo K, Eid SA, Rumora AE, Hinder LM, Hayes JM, et al. Integrated lipidomic and transcriptomic analyses identify altered nerve triglycerides in mouse models of prediabetes and type 2 diabetes. Dis Model Mech. 2020;13(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Freeman OJ, Unwin RD, Dowsey AW, Begley P, Ali S, Hollywood KA, et al. Metabolic Dysfunction Is Restricted to the Sciatic Nerve in Experimental Diabetic Neuropathy. Diabetes. 2016;65(1):228–38. [DOI] [PubMed] [Google Scholar]
- 66.Duran AM, Salto LM, Camara J, Basu A, Paquien I, Beeson WL, et al. Effects of omega-3 polyunsaturated fatty-acid supplementation on neuropathic pain symptoms and sphingosine levels in Mexican-Americans with type 2 diabetes. Diabetes Metab Syndr Obes. 2019;12:109–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reidy K, Kang HM, Hostetter T, Susztak K. Molecular mechanisms of diabetic kidney disease. J Clin Invest. 2014;124(6):2333–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roointan A, Gheisari Y, Hudkins KL, Gholaminejad A. Non-invasive metabolic biomarkers for early diagnosis of diabetic nephropathy: Meta-analysis of profiling metabolomics studies. Nutr Metab Cardiovasc Dis. 2021;31(8):2253–72. [DOI] [PubMed] [Google Scholar]
- 69.Qi W, Keenan HA, Li Q, Ishikado A, Kannt A, Sadowski T, et al. Pyruvate kinase M2 activation may protect against the progression of diabetic glomerular pathology and mitochondrial dysfunction. Nat Med. 2017;23(6):753–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gordin D, Shah H, Shinjo T, St-Louis R, Qi W, Park K, et al. Characterization of Glycolytic Enzymes and Pyruvate Kinase M2 in Type 1 and 2 Diabetic Nephropathy. Diabetes Care. 2019;42(7):1263–73. **A translational investigation in plasma and kidney biopsy samples demonstrating that upregulation of glycolytic enzymes is associated with preservation of kidney function in both type 1 and type 2 diabetes.
- 71.Zhang F, Guo R, Cui W, Wang L, Xiao J, Shang J, et al. Untargeted serum metabolomics and tryptophan metabolism profiling in type 2 diabetic patients with diabetic glomerulopathy. Ren Fail. 2021;43(1):980–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Afshinnia F, Rajendiran TM, Wernisch S, Soni T, Jadoon A, Karnovsky A, et al. Lipidomics and Biomarker Discovery in Kidney Disease. Semin Nephrol. 2018;38(2):127–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mutter S, Valo E, Aittomaki V, Nybo K, Raivonen L, Thorn LM, et al. Urinary metabolite profiling and risk of progression of diabetic nephropathy in 2670 individuals with type 1 diabetes. Diabetologia. 2022. Jan;65(1):140–149. **A prospective study of the association of urinary metabolites with the development of future diabetic nephropathy in >2000 individuals with type 1 diabetes.
- 74.Ekstrand AV, Groop PH, Grönhagen-Riska C. Insulin resistance precedes microalbuminuria in patients with insulin-dependent diabetes mellitus. Nephrol Dial Transplant. 1998;13(12):3079–83. [DOI] [PubMed] [Google Scholar]
- 75.Action to Control Cardiovascular Risk in Diabetes Study G, Gerstein HC, Miller ME, Byington RP, Goff DC Jr., Bigger JT, et al. Effects of intensive glucose lowering in type 2 diabetes. N Engl J Med. 2008;358(24):2545–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Group AC, Patel A, MacMahon S, Chalmers J, Neal B, Billot L, et al. Intensive blood glucose control and vascular outcomes in patients with type 2 diabetes. N Engl J Med. 2008;358(24):2560–72. [DOI] [PubMed] [Google Scholar]
- 77.Haas AV, McDonnell ME. Pathogenesis of Cardiovascular Disease in Diabetes. Endocrinol Metab Clin North Am. 2018;47(1):51–63. [DOI] [PubMed] [Google Scholar]
- 78.Ottosson F, Smith E, Gallo W, Fernandez C, Melander O. Purine Metabolites and Carnitine Biosynthesis Intermediates Are Biomarkers for Incident Type 2 Diabetes. J Clin Endocrinol Metab. 2019;104(10):4921–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ottosson F, Smith E, Fernandez C, Melander O. Plasma Metabolites Associate with All-Cause Mortality in Individuals with Type 2 Diabetes. Metabolites. 2020;10(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ottosson F, Smith E, Melander O, Fernandez C. Altered Asparagine and Glutamate Homeostasis Precede Coronary Artery Disease and Type 2 Diabetes. J Clin Endocrinol Metab. 2018;103(8):3060–9. [DOI] [PubMed] [Google Scholar]
- 81.Contrepois K, Wu S, Moneghetti KJ, Hornburg D, Ahadi S, Tsai MS, et al. Molecular Choreography of Acute Exercise. Cell. 2020;181(5):1112–30 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ho JE, Larson MG, Vasan RS, Ghorbani A, Cheng S, Rhee EP, et al. Metabolite profiles during oral glucose challenge. Diabetes. 2013;62(8):2689–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morris C, O’Grada C, Ryan M, Roche HM, Gibney MJ, Gibney ER, et al. Identification of differential responses to an oral glucose tolerance test in healthy adults. PLoS One. 2013;8(8):e72890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fiamoncini J, Rundle M, Gibbons H, Thomas EL, Geillinger-Kastle K, Bunzel D, et al. Plasma metabolome analysis identifies distinct human metabotypes in the postprandial state with different susceptibility to weight loss-mediated metabolic improvements. FASEB J. 2018;32(10):5447–58. [DOI] [PubMed] [Google Scholar]
- 85. Li-Gao R, Hughes DA, van Klinken JB, de Mutsert R, Rosendaal FR, Mook-Kanamori DO, et al. Genetic Studies of Metabolomics Change After a Liquid Meal Illuminate Novel Pathways for Glucose and Lipid Metabolism. Diabetes. 2021;70(12):2932–46. **Genetic analyses of the individual responses to a mixed meal tolerance test, identifying genetic variants associated with the post-meal response to lipid handling.
- 86.Shah SH, Crosslin DR, Haynes CS, Nelson S, Turer CB, Stevens RD, et al. Branched-chain amino acid levels are associated with improvement in insulin resistance with weight loss. Diabetologia. 2012;55(2):321–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chen ZZ, Liu J, Morningstar J, Heckman-Stoddard BM, Lee CG, Dagogo-Jack S, et al. Metabolite Profiles of Incident Diabetes and Heterogeneity of Treatment Effect in the Diabetes Prevention Program. Diabetes. 2019;68(12):2337–49. **A report from the Diabetes Prevention Program illustrating how heterogeneity in metabolic profiles may predict response to pharmacologic versus lifestyle treatments.
- 88.Shantavasinkul PC, Muehlbauer MJ, Bain JR, Ilkayeva OR, Craig DM, Newgard CB, et al. Improvement in insulin resistance after gastric bypass surgery is correlated with a decline in plasma 2-hydroxybutyric acid. Surg Obes Relat Dis. 2018;14(8):1126–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Laferrere B, Reilly D, Arias S, Swerdlow N, Gorroochurn P, Bawa B, et al. Differential metabolic impact of gastric bypass surgery versus dietary intervention in obese diabetic subjects despite identical weight loss. Science translational medicine. 2011;3(80):80re2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chung WK, Erion K, Florez JC, Hattersley AT, Hivert MF, Lee CG, et al. Precision medicine in diabetes: a Consensus Report from the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD). Diabetologia. 2020;63(9):1671–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108(8):1167–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.den Ouden H, Pellis L, Rutten G, Geerars-van Vonderen IK, Rubingh CM, van Ommen B, et al. Metabolomic biomarkers for personalised glucose lowering drugs treatment in type 2 diabetes. Metabolomics. 2016;12:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dahabiyeh LA, Mujammami M, Arafat T, Benabdelkamel H, Alfadda AA, Abdel Rahman AM. A Metabolic Pattern in Healthy Subjects Given a Single Dose of Metformin: A Metabolomics Approach. Front Pharmacol. 2021;12:705932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kappel BA, Lehrke M, Schutt K, Artati A, Adamski J, Lebherz C, et al. Effect of Empagliflozin on the Metabolic Signature of Patients With Type 2 Diabetes Mellitus and Cardiovascular Disease. Circulation. 2017;136(10):969–72. [DOI] [PubMed] [Google Scholar]
- 95.Mulder S, Hammarstedt A, Nagaraj SB, Nair V, Ju W, Hedberg J, et al. A metabolomics-based molecular pathway analysis of how the sodium-glucose co-transporter-2 inhibitor dapagliflozin may slow kidney function decline in patients with diabetes. Diabetes Obes Metab. 2020;22(7):1157–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Packer M. Autophagy-dependent and -independent modulation of oxidative and organellar stress in the diabetic heart by glucose-lowering drugs. Cardiovasc Diabetol. 2020;19(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dutta T, Kudva YC, Persson XM, Schenck LA, Ford GC, Singh RJ, et al. Impact of Long-Term Poor and Good Glycemic Control on Metabolomics Alterations in Type 1 Diabetic People. J Clin Endocrinol Metab. 2016;101(3):1023–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Knebel B, Strassburger K, Szendroedi J, Kotzka J, Scheer M, Nowotny B, et al. Specific Metabolic Profiles and Their Relationship to Insulin Resistance in Recent-Onset Type 1 and Type 2 Diabetes. J Clin Endocrinol Metab. 2016;101(5):2130–40. [DOI] [PubMed] [Google Scholar]
- 99.Nayor M, Brown KJ, Vasan RS. The Molecular Basis of Predicting Atherosclerotic Cardiovascular Disease Risk. Circulation Research. 2021;128(2):287–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nayor M, Shah SH, Murthy V, Shah RV. Molecular Aspects of Lifestyle and Environmental Effects in Patients With Diabetes: JACC Focus Seminar. J Am Coll Cardiol. 2021;78(5):481–95. [DOI] [PubMed] [Google Scholar]