Abstract
Objective
We aimed to explore the prognostic patterns of ferroptosis-related genes in papillary renal cell carcinoma (PRCC) and investigate the relationship between ferroptosis-related genes and PRCC tumor immune microenvironment.
Methods
We obtained the mRNA expression and corresponding clinical data of PRCC from the public tumor cancer genome atlas database (TCGA). The PRCC patients were randomly divided into two cohort, training cohort and verification cohort, respectively. Univariate Cox regression, LASSO Cox regression, multivariate Cox regression analysis were utilized to construct ferroptosis signature for PRCC patients. And then, risk prognostic model was established and verified. The correlation of ferroptosis-related signature with survival and immune microenvironment was systematically analyzed.
Results
A 4-genes ferroptosis signature (CDKN1A, MIOX, PSAT1, and RRM2) was constructed. Multivariate Cox regression assay indicates that the risk score of ferroptosis signature was an independent prognostic indicator (HR=1.391, p<0.001). The survival curve shows that the high-risk group has a poorer prognosis than the low-risk group (p<0.001). The risk prognostic model was established based on prognostic factors of clinical-stage, hemoglobin, and risk score. The time-dependent receiver operating characteristic curve (ROC) analysis proves the predictive capacity of the ferroptosis signature, the 3 years area under the curve (AUC) is 0.890, and the 5 years AUC is 0.733. Further analysis suggested that cell cycle, pentose phosphate pathway, P53 signaling pathway were significantly enriched in the high-risk group. The significantly different fractions of dendritic cells resting, macrophage cells, and T cells follicular helper were observed in risk groups.
Conclusion
This study implicates a ferroptosis signature which has a good predict capacity of the prognosis in PRCC patients. Ferroptosis-related genes may have a key role in the process of anti-tumor and serve as therapeutic targets for PRCC.
Keywords: papillary renal cell carcinoma, ferroptosis, gene signature, prognosis model, tumor immune microenvironment
Introduction
Renal cell carcinoma (RCC) is the most common cancer of the kidney. RCC has three subtypes based on histological and genetic features. They are clear cell renal cell carcinoma (CCRCC, 70–80%), Papillary renal cell carcinoma (PRCC, 10–20%), and chromophobe renal cell carcinoma (CHRCC).1,2 PRCC has been separated into two types on account of its histological and immunohistochemical characteristics, type 1 has basophilic cytoplasm, and type 2 has eosinophilic cytoplasm.3 Recently, more and more clinical studies were focused on CCRCC patients and discovered some efficacy therapeutic targets, such as vascular endothelial growth factor (VEGFR) and mammalian target of rapamycin (mTOR). However, these targets were less efficacy in PRCC patients, which might result from the different genetic mutations and molecular pathways in PRCC tumorigenesis was compared to CCRCC.1 Therefore, more specific biomarkers and efficacy treatment strategies need to be developed for PRCC.
Ferroptosis is a non-apoptotic form of regulated cell death. The characteristic of ferroptosis is an accumulation of reactive oxygen species and lipid peroxidation products that lead to cell death, and this process is oxidative and iron-dependent.4,5 Ferroptosis is involved in multiple biological processes, including development, immunity, senescence, and a variety of diseases, therefore sustaining the function of normal cells and tissues. Accumulation of iron can sensitize cancer cells to oxidative damage and ferroptosis.6 Cancer cells need to regulate the expression of iron metabolism-related genes to keep the balance of iron.7 Reprogramming of metabolic pathways to obtain energy and materials and sustain the high level of cancer cell proliferation is the main characteristic of cancer cells, including glycolysis, tricarboxylic acid cycle (TCA), β-oxidation. This process will break the redox homeostasis. To balance the redox homeostasis tumor cells must heavily rely on the intracellular antioxidant machinery.8,9 Intriguingly, ferroptosis can be activated by oxidative turbulence in the intracellular microenvironment. More recent studies indicated that ferroptosis-related genes have an important role in various tumor cells, such as in breast cancer cells,10 lung adenocarcinomas cells,11 renal carcinoma cells.12 The prognostic value of ferroptosis-related prognostic signature also has been studied in several cancers, such as colorectal cancer,13 breast cancer,14 hepatocellular carcinoma,15 head and neck squamous cell carcinoma.16 In clear cell renal cell carcinoma, the expression of ferroptosis-related signature was significantly correlated with immune infiltration and response to immunotherapy.17 However, the relationship between ferroptosis-related signature and treatment or prognosis of PRCC patients was remained unclear.
In this study, we download mRNA expression data and the corresponding clinical data of PRCC patients from the tumor cancer genome atlas (TCGA) database. Combining 259 ferroptosis-related genes, we established a powerful multigene ferroptosis-related signature for predicting the prognosis of PRCC patients. Furthermore, we performed gene set enrichment analysis (GSEA) and go analysis, and investigated the correlation of ferroptosis signature and tumor immune microenvironment. The present study revealed novel insights into the prognostic value of ferroptosis signature and explore the relationship with immune infiltration in PRCC patients.
Materials and Methods
Gene Expression Datasets and Ferroptosis Gene Set
In this study, we collected PRCC expression data and clinical data from TCGA (https://portal.gdc.cancer.gov/) public databases, which included 289 PRCC tissue samples and 32 normal tissue samples (total 321 samples). A total of 259 ferroptosis-related genes were downloaded from the FerrDb website (http://www.zhounan.org/ferrdb/legacy/index.html),18 which include driver 108 genes, suppressor 69 genes, and marker 111 genes.
Identifying Ferroptosis-Related Prognostic Differentially Expressed Genes
The mRNA-sequencing data was analyzed by GDCRNATools. 2904 genes were selected as differentially expressed genes (DEG) via the “limma” R package (log2-fold change>1 and P-value <0.001). Survival analysis showed 745 genes were prognosis-related (P-value<0.05). We used Jvenn (http://www.bioinformatics.com.cn), an interactive Venn diagram viewer,19 to examine overlapping genes in prognosis-related genes and ferroptosis-related genes.
The protein-protein interaction network of ferroptosis-related prognostic DEG was analyzed by STRING (https://string-db.org) database.
The Characteristics of Signature Genes Expression and Survival Analysis
We used gene expression profiling interactive analysis 2 (GEPIA2, http://gepia2.cancer-pku.cn/#analysis)20 to analyze the expression level of signature genes in different tumor clinical stages. Immunohistochemistry (IHC) analysis of signature proteins was utilized in the human protein atlas (https://www.proteinatlas.org).
Determination of Ferroptosis Prognostic Signature
The “survival” R package was used for performing univariate Cox regression analysis to screen out ferroptosis genes related to prognostic in patients with PRCC. Genes with a P-value <0.05 were incorporated into the subsequent LASSO Cox regression using the “glmnet” R package. Based on a multivariate Cox regression for these genes, we built a ferroptosis prognostic signature. The prognostic risk score = (exp gene1 * coef gene1) + (exp gene2 * coef gene2) + … + (exp gene4 * coef gene4). PRCC patients were divided into the high-risk group or low-risk group by the median of the risk score. The overall survival (OS) between different groups was compared by Kaplan-Meier analysis with the Log rank test.
Construction and Evaluation of Prognostic Model
Clinical factors and the risk score of ferroptosis signature were included in the univariate Cox regression analysis. And statistically significant (p-value< 0.05) were selected as prognostic factors to perform multivariate Cox regression analysis. And then the establishment of a prognostic risk model utilizes the above prognostic factors, provide the risk score, and plot the nomogram. The “survivalROC” R package was used to perform the time-dependent ROC and to evaluate the accuracy and specificity of the prognostic risk model.21 New high or low-risk groups were divided based on the new risk score. Kaplan-Meier analysis OS of risk groups.
Gene Set and Functional Enrichment Analysis
We used the “tinyarray and tidyverse” R package to obtain 19,712 genes mRNA expression data and divided them into two groups based on the risk score. Gene set enrichment analysis was performed by GSEA software (http://www.broadinstitute.org/gsea). The gene sets were filtered using the Kyoto encyclopedia of genes and genomes (KEGG) gene set sizes of 15 and 500 genes.
745 prognostic DEG (P-value<0.05) was performed Gene Ontology (Go) analysis using “GDCRNATools” R package. Top 10 enrichment was shown with a false discovery rate (FDR)<0.05.
Estimation of Immune Cells Type Fractions
The proportion of different tumor immune cells fractions was calculated by CIBERSORT.22 The normalized gene expression data was uploaded to the CIBERSORT web portal (http://cibersort.stanford.edu/), and the algorithm was based on the LM22 gene signature and 1000 permutations. The samples were filtered based on a P-value <0.05. The results produced by CIBERSORT were analyzed subsequently.
Statistical Analysis
The statistical analyses were conducted using R software (version 4.0.3). The prognostic analysis was conducted using the Kaplan-Meier method with the significance of differences identified using Log rank tests. Correlations between two variables were examined via Spearman correlation analysis. The risk score discrepancy between the subgroup of clinical characteristics was using the Wilcoxon test.
Results
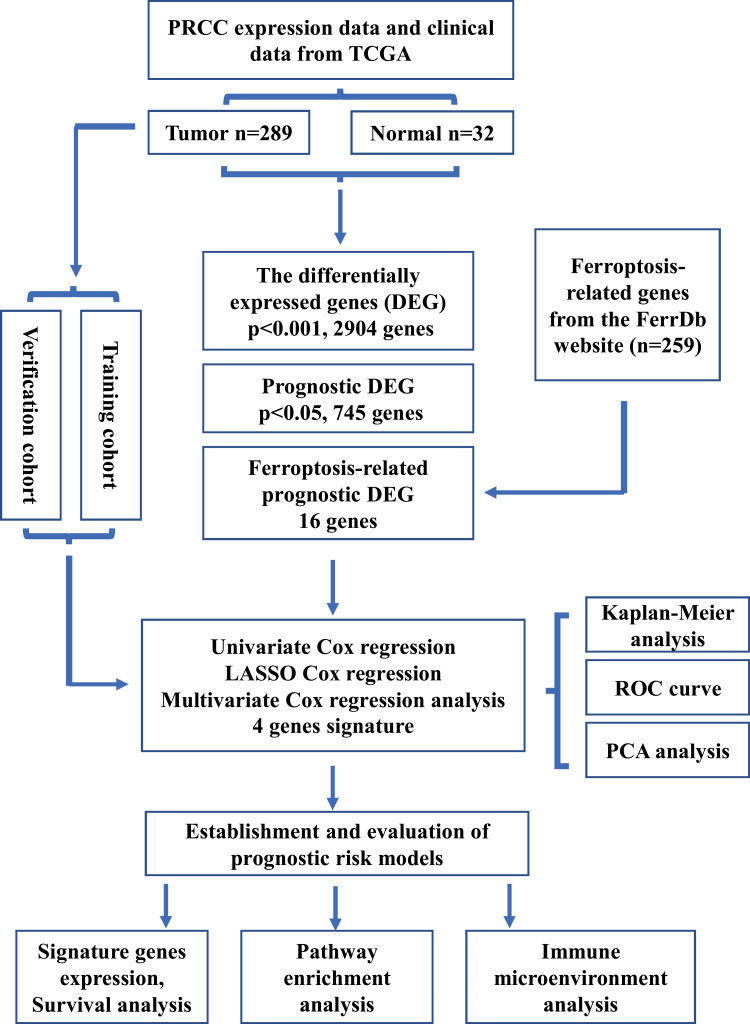
As shown in Figure 1, a total of 289 PRCC patients from the TCGA database were incorporated into this study.
Figure 1.
Overview of study design.
Abbreviations: TCGA, tumor cancer genome atlas database, DEG, differentially expressed genes, FerrDb, ferroptosis regulators and markers and ferroptosis-disease associations database, ROC, receiver operating characteristic, PCA, principal components analysis.
Identification of Prognostic Differentially Expressed Ferroptosis-Related Genes in PRCC
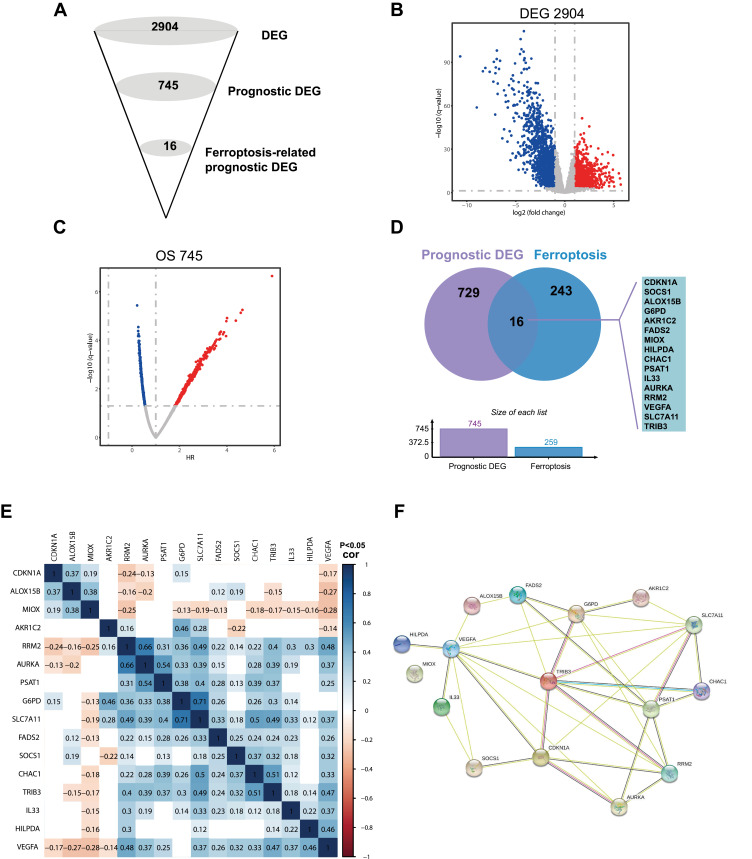
We took advantage of publicly available TCGA data and filtered ferroptosis-related prognostic DEG in a multi-step approach (Figure 2A). First, PRCC expression data and clinical data were downloaded from TCGA public databases, which included 289 PRCC tissue samples and 32 normal tissue samples (total 321 samples). Comparing tumor tissues with normal tissues 2094 genes were DEG in PRCC with log2-fold change>1 and P-value <0.001 (Figure 2B). Second, survival analysis of 2904 DEG showed 745 genes were prognostically related (P value<0.05, Figure 2C). Finally, we used Jvenn, an interactive Venn diagram viewer,19 to examine overlapping genes in prognosis-related DEG and 259 ferroptosis-related genes. The results suggested that 16 ferroptosis-related genes were overlapping with prognosis-related DEG, including cyclin dependent kinase inhibitor 1A (CDKN1A), suppressor of cytokine signaling 1 (SOCS1), arachidonate 15-lipoxygenase type B (ALOX15B), glucose-6-phosphate dehydrogenase (G6PD), aldo-keto reductase family 1 member C2 (AKR1C2), fatty acid desaturase 2 (FADS2), myo-inositol oxygenase (MIOX), hypoxia inducible lipid droplet associated (HILPDA), chaC glutathione specific gamma-glutamylcyclotransferase 1 (CHAC1), phosphoserine aminotransferase 1 (PSAT1), interleukin 33 (IL33), aurora kinase A (AURKA), ribonucleotide reductase regulatory subunit M2 (RRM2), vascular endothelial growth factor A (VEGFA), solute carrier family 7 member 11 (SLC7A11), and tribbles pseudokinase 3 (TRIB3). These genes were named ferroptosis-related prognostic DEG (FRPG, Figure 2D). The correlation between each member of FRPG was shown in Figure 2E, and the protein-protein interaction (PPI) network of FRPG analyzed by STRING was shown in Figure 2F.
Figure 2.
Identification of the candidate ferroptosis-related genes in PRCC. (A) Schematic description of the filtering process for identification of ferroptosis-related candidates. (B) Identified differentially expressed genes (DEG) between tumor and normal tissues, screening threshold (p<0.001, fold change>1). (C) Identified DEG that were related to prognosis, screening threshold (p<0.05). (D) Venn diagram to identify ferroptosis-related prognostic DEG. (E) the correlation of the candidate genes of ferroptosis-related prognostic DEG. (F) STRING database analysis the interactions among the candidate genes.
Establishment of a Prognostic Signature
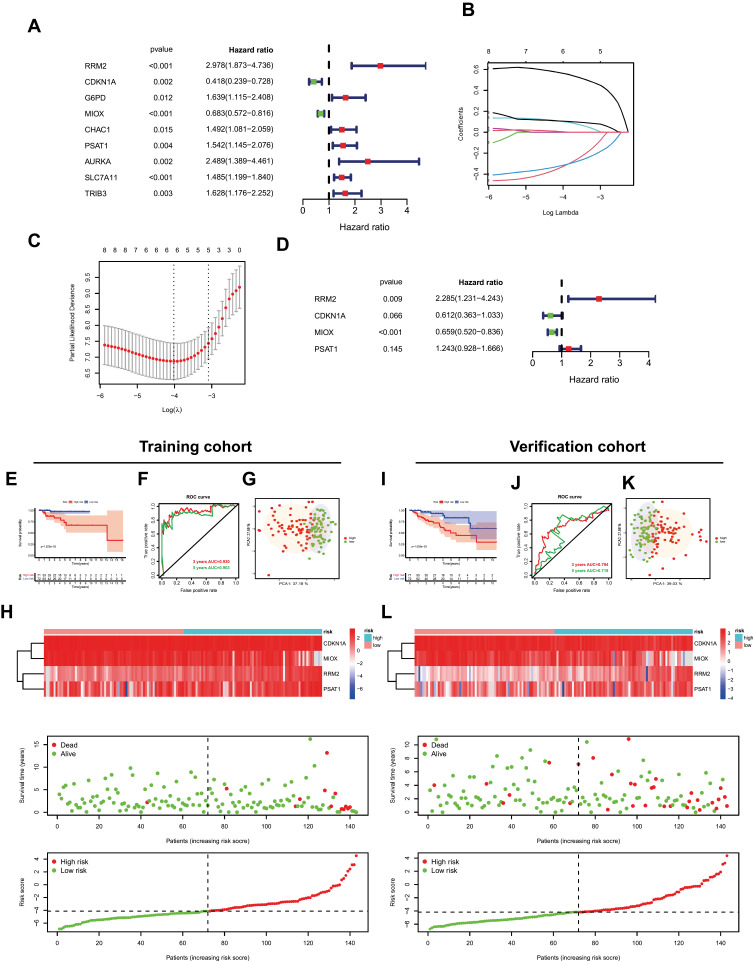
Through univariate Cox regression analysis of FRPG in patients with PRCC, 9 FRPG was shown prognostic related, including RRM2, CDKN1A, G6PD, MIOX, CHAC1, PSAT1, AURKA, SLC7A11, and TRIB3 (Figure 3A). These 9 FRPG were incorporated into the subsequent LASSO Cox regression analysis, and combined multivariate Cox regression analysis, we construct a ferroptosis prognostic signature, which contains 4 genes (CDKN1A, MIOX, PSAT1, and RRM2, Figure 3B–D). The expression level of signature genes in PRCC and immunohistochemistry results of kidney adenocarcinoma were shown in Figure S1. The OS and disease-free survival (DFS) of signature genes were shown in Figure S2. The prognostic Risk score was calculated. Risk score = (0.826584061278619*exp RRM2) + (−0.490534605327226*exp CDKN1A) + (−0.416576419121587*exp MIOX) + (0.217734038998151*exp PSAT1). Then, the PRCC patients were divided into the high-risk group or low-risk group by the median of the Risk score. The OS between different groups was compared by Kaplan-Meier analysis with the Log rank test. The results suggested that the high-risk group showed a poor prognosis compared with the low-risk group in two cohorts (both P<0.001, Figure 3E and I). The 3-years and 5-years ROC curves were analyzed to evaluate the predictive accuracy of the ferroptosis-related signature (Figure 3F and J). PCA test indicated that the patients in the two groups were stratified into two directions (Figure 3G and K). The heatmap of ferroptosis signature after risk score grouping and the distribution of risk status and risk score was shown in Figure 3H and L.
Figure 3.
Establishment of ferroptosis signature. (A) The univariate Cox regression analysis between gene expression and overall survival (OS). (B and C) LASSO Cox regression analysis of ferroptosis-related prognostic DEG. (D) Multivariate Cox regression analysis of ferroptosis signature. (E and I) The survival status in two cohorts. (F and J) Receiver operating characteristic (ROC) curve analysis in two cohorts. (G and K) Principal components analysis (PCA) analysis in two cohorts. (H and L) The heatmap of ferroptosis signature after risk score grouping, the distribution of OS status, and the distribution of risk score.
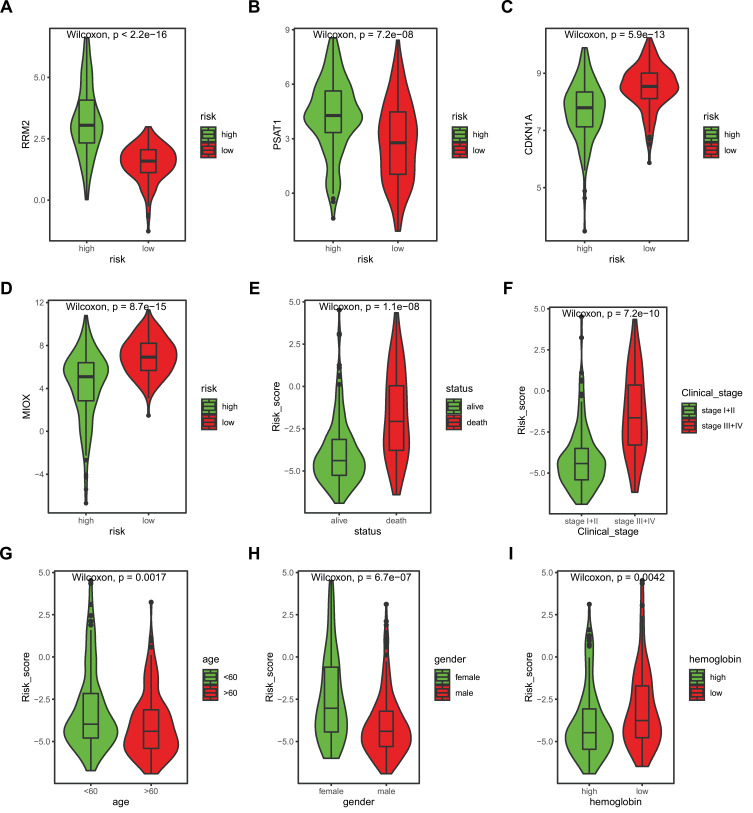
We first compared the expression of the 4 signature genes in risk groups (Figure 4A–D). Then, we analyzed the differences in the risk score in patients with different clinical characteristics, including clinical status (alive vs death), age (<60 vs >60), gender (female vs male), hemoglobin (low vs normal), and clinical_stage (stage I+II vs stage III+IV, Figure 4E–I). The risk score in death patients was significantly higher than in alive patients (p=1.1e-08), and the age<60 was significantly higher than the age>60 (p=0.0017). The risk score in clinical-stage III+IV was significantly higher in stage I+II patients (p=7.2e-10). The risk score in females was remarkably higher than male patients (p=6.7e-07), and hemoglobin low was significantly higher in hemoglobin normal groups (p=0.0042, Figure 4E–I).
Figure 4.
Signature gene expression and risk score discrepancy between the subgroup of clinical characteristics. (A–D) The expression of signature genes in the high-risk group and low-risk group, RRM2 (A), PSAT1 (B), CDKN1A (C), MIOX (D). E-I. Risk score discrepancy between the subgroup of clinical characteristics: status (E), age (F), gender (G), hemoglobin (H), and clinical_stage (I).
Abbreviations: RRM2, ribonucleotide reductase regulatory subunit M2, PSAT1, phosphoserine aminotransferase 1, CDKN1A, cyclin dependent kinase inhibitor 1A, MIOX, myo-inositol oxygenase.
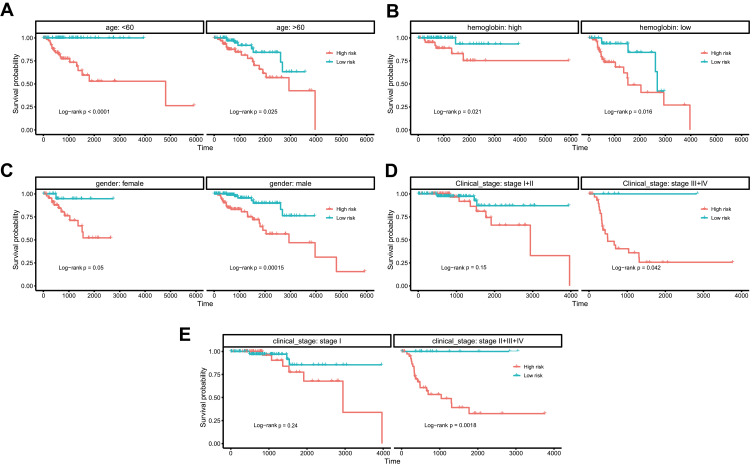
Further analysis was performed to explore whether the risk score correlated with the prognosis of patients in different subgroups of clinical factors. In an age (<60 and >60), gender (female and male), and hemoglobin (low and normal), patients in the high-risk group showed a poor prognosis (Figures 5A–C, log-rank P<0.05). In clinical_stage subgroups, patients in the high-risk group showed a poor prognosis compared with the stage II+III+IV or stage III+IV group (log-rank P=0.042 and log-rank P=0.0018, respectively Figure 5D and E).
Figure 5.
Kaplan-Meier survival plots of the risk score of ferroptosis signature with different clinical characteristics. Kaplan-Meier survival plots of the risk score of ferroptosis signature with different clinical characteristics: age (A), hemoglobin (B), gender (C), clinical_stage (D and E).
Establishment and Evaluation of Prognostic Risk Models
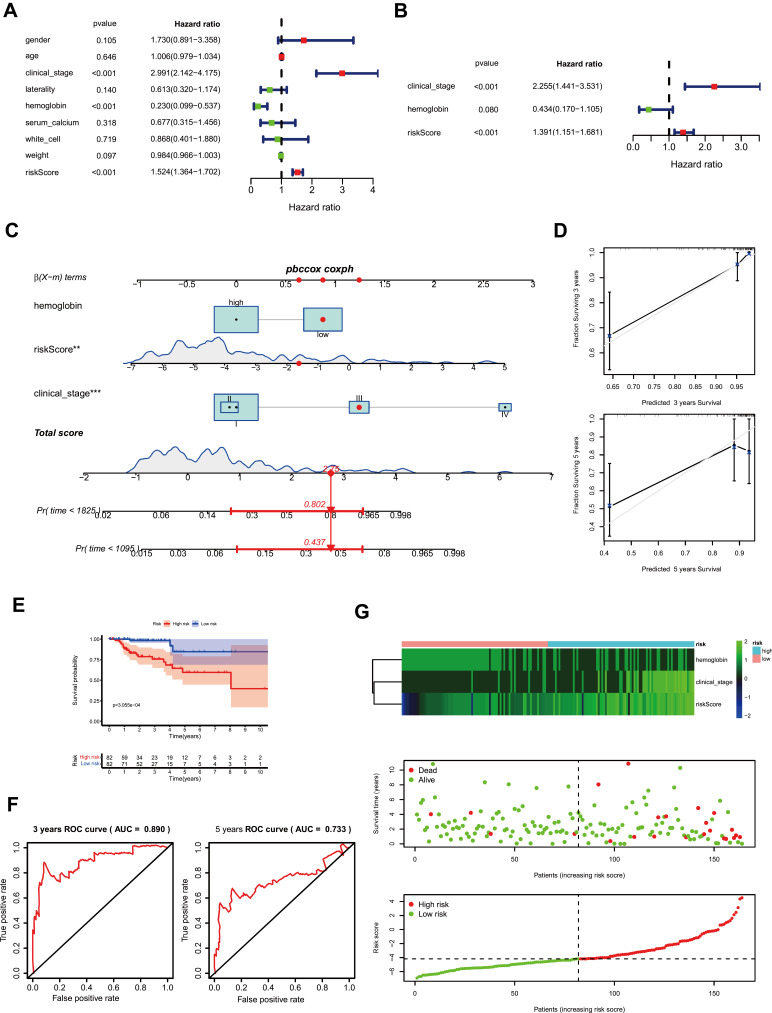
Clinical factors and the Risk score of ferroptosis signature were included in the univariate Cox regression analysis. Clinical_stage (HR=2.991, P<0.001), hemoglobin (HR=0.23, P<0.001), and Risk score (HR=1.524, P<0.001) were significantly correlated with prognostic of PRCC patients (Figure 6A). These 3 factors were selected as prognostic factors to perform multivariate Cox regression analysis. The results suggested that clinical_stage (HR=2.255, 95% CI=1.441–3.531, P<0.001), hemoglobin (HR=0.434, 95% CI=0.17–1.105, P=0.08), and Risk score (HR=1.391, 95% CI=1.151–1.681, P<0.001) were confirmed to be an independent predictive factor for predicting OS after calibration of other clinical characteristics, respectively (Figure 6B).
Figure 6.
Establishment and evaluation of prognostic risk models. (A) The univariate Cox regression analysis of clinical factors and the risk score of ferroptosis signature. (B) Multivariate Cox regression analysis of clinical factors and the risk score of ferroptosis signature. (C) The nomogram for 3-, or 5-year overall survival (OS). (D) The calibration plots for nomogram. (E) Survival analysis based on risk score according to multivariate regression analysis. (F) The receiver operating characteristic (ROC) curve is based on the risk score. (G) The heatmap of ferroptosis signature and clinical factors after risk score grouping, the survival status, and the distribution of risk score.
The risk prognostic model was established based on prognostic factors of clinical_stage, hemoglobin, and Risk score, and the 3- and 5-years survival were given (Figure 6C). The calibration curves of 3-years and 5-years survival and time-dependent ROC of risk indicated the model has a good predictive ability (Figure 6D and F). The prognostic risk score of the prognosis model was calculated. The PRCC patients were divided into the high-risk group or low-risk group by the median of the Risk score. The OS results indicated that the high-risk group showed a poor prognosis compared with the low-risk group (P<0.001, Figure 6E). The heatmap of clinical factors and Risk score of ferroptosis signature after risk grouping and the distribution of OS status and risk score was shown in Figure 6G. These results implicated that ferroptosis prognostic signature may play a key role and as a valuable prognostic factor in PRCC.
Discovery of Important Pathways by GSEA and GO
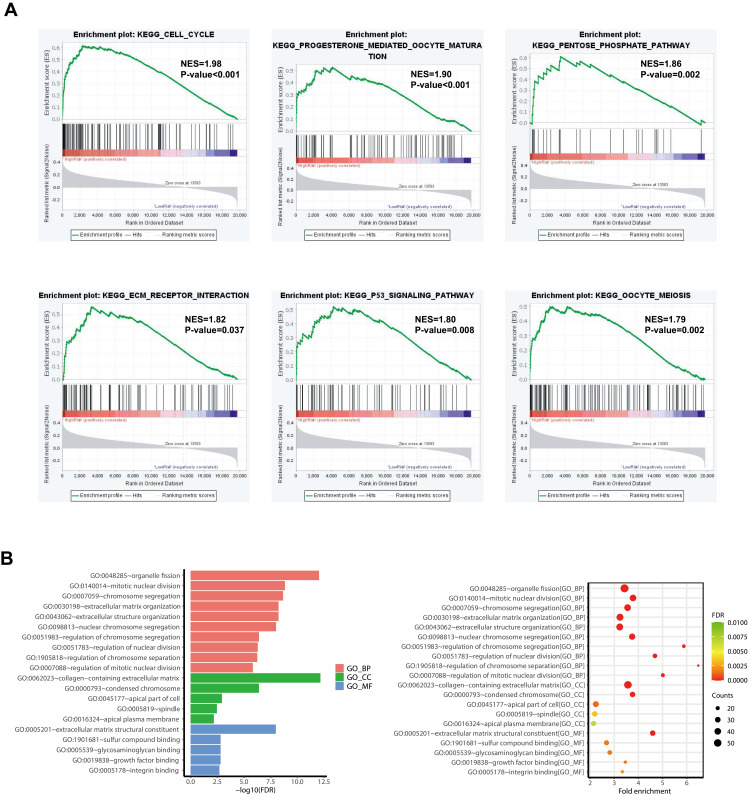
To further analyze the functions associated with the ferroptosis prognostic signature, we used the “tinyarray and tidyverse” R package to obtain 19,712 genes mRNA expression data in PRCC and divided them into two groups based on the risk score. Gene set enrichment analysis was performed by GSEA software. The gene sets were filtered using the KEGG gene set size of 15 and 500 genes. The top 6 significantly enriched pathways in the high-risk group are cell cycle, pentose phosphate pathway, P53 signaling pathway, ECM receptor interaction, progesterone mediated oocyte maturation, and oocyte meiosis (Figure 7A).
Figure 7.
Representative results of pathway analysis. (A) Functional annotation of the high-risks group of differentially expressed genes (DEG) by gene set enrichment analysis (GSEA). (B) Go analysis of 745 prognostic DEG.
Next, 745 prognostic DEG (P <0.05) was performed Gene Ontology (Go) analysis. The top 10 significant enrichment was shown with FDR<0.05. The results show that the top associated pathway is mainly involved in organelle fission, mitotic nuclear division, and chromosome segregation (Figure 7B).
The Immune Cell Infiltration Landscape in PRCC
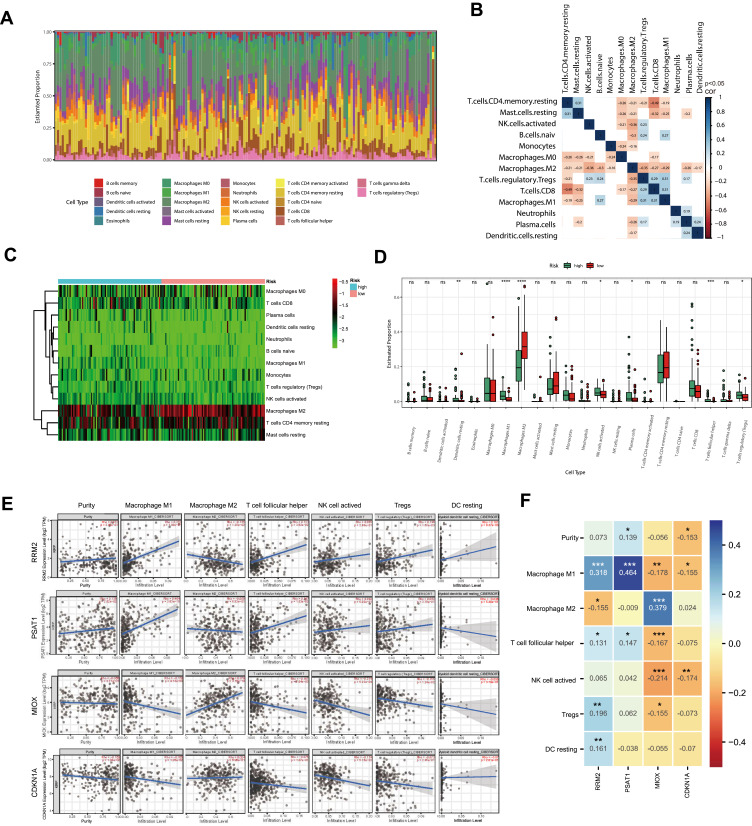
To further explore the relationship between ferroptosis prognostic signature and tumor immune microenvironment, we screened the RNA-seq datasets of 164 PRCC patients using the CIBERSORT algorithm,22 investigating the proportion of different tumor immune cell fractions. The proportion of different tumor immune cell fractions and the correlation of immune fraction was shown in Figure 8A and B. Wilcoxon rank test indicated that immune infiltration of dendritic cells resting, macrophage cells, NK cells activated, plasma cells, T cells follicular helper, and T cells regulatory were significantly different between the high-risk and the low-risk group (Figure 8C and D). Finally, we analyze the correlation between the expression of signature genes and immune infiltration cells, respectively (Figure 8E and F).
Figure 8.
Immune fraction in low-risk compared to high-risk patients. (A) Bar plot of the immune fraction denoted by different colors. (B) Correlation of immune fraction. (C) The heatmap of immune fraction based on risk score. (D) Box plots depicting the CIBERSORT scores of 22 immune cells of the high-risk patients compared with low-risk patients. (E and F) The correlation with the expression level of signature genes and the immune infiltration cells. Adjusted P-values were showed as: ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P<0.0001.
Discussion
PRCC is a malignant renal parenchymal tumor, but, specific clinical drugs and molecular immunotherapy strategies for PRCC patients are still limited. Therefore, it is an emergency to investigate credible molecular biomarkers for the prognosis of PRCC patients.
Ferroptosis is an iron-dependent regulatory pathway and is involved in multiple biological processes and diseases.5 Recently study indicated several ferroptosis-related signatures have prognostic value in different cancers. However, the relationship between ferroptosis-related signature and the prognosis of PRCC patients is still unknown. In this study, based on the mRNA expression data and clinical data of TCGA PRCC datasets, we are the first time to investigate and construct 4-genes ferroptosis-related signature (CDKN1A, MIOX, PSAT1, and RRM2) acting as independent prognostic factors in PRCC patients. The established prognostic risk models show a good capability in predicting PRCC patient survival. Recent study indicated that 11-genes ferroptosis-related signature accurately predicts clinical prognosis in patients with CCRCC.23 Intriguingly, these 4-genes ferroptosis-related signatures are specific for PRCC patients compared with the discovered ferroptosis-related signature in CCRCC,23,24 which is benefit to develop specific clinical drugs and diagnostic strategies for PRCC patients only.
According to the ferroptosis database, the 4-genes ferroptosis signature could be divided into three types, MIOX belongs to drivers, PSAT1 and RRM2, are belong to markers, CDKN1A belongs to suppressors. RRM2 is a catalytic subunit of ribonucleotide reductase and functions as a regulator for DNA replication and repair. RRM2 is overexpressed in several cancers and associated with poor prognoses, such as in breast cancer and sarcoma.25,26 RRM2 has been reported as an independent prognostic factor in lung adenocarcinoma.27,28 CDKN1A is a cell cycle regulator involved in genomic stability. Decreased expression of CDKN1A was correlated with poor prognosis in chromophobe renal cell carcinoma.29 A recent study suggested that CDKN1A as a member of ferroptosis-related signature could predict graft loss following renal allograft.30 PSAT1 is an enzyme catalyzing serine biosynthesis which regulates cyclin D1 degradation progress proliferation of non-small cell lung cancer.31 Myo-inositol oxygenase (MIOX) is a tubular enzyme, overexpression of MIOX results in impaired cellular energy homeostasis and exacerbates renal injury.32 Combined with these studies suggested the signature genes may involve in the metabolism of energy or nucleotide and cell cycles, therefore, regulating cells fate, as indicated by GSEA analysis. Notably, although these signature genes have been reported to have a large function or acted as prognostic factors in several cancers, almost no report on PRCC tumors.
GSEA analysis suggested P53 signaling pathway was enriched in the high-risk group. P53 involve in multiple regulatory pathways, such as cell cycle, apoptosis, metabolism, and plays a critical role in tumor suppression.33 A recent study suggested that p53 regulates ferroptosis by directly regulating the ferroptosis target gene. P53 sensitizes cancer cells to ferroptosis via suppressing the expression of SLC7A11, which is a cystine/glutamate transporter.34 Inactivation of ALOX12 blocked p53-mediated ferroptosis stimulated by reactive oxygen species.35 However, p53 also could suppress ferroptosis in some cases. P53 blocks the activity of dipeptidyl-peptidase-4 (DPP4) and limits elastin-induced ferroptosis in colorectal cancer. Plasma-membrane-associated DPP4 facilitates lipid peroxidation results in ferroptosis.36 These researches suggested that p53 regulated ferroptosis in a complicated manner which is cell type-specific.
Furthermore, we analyze the tumor immune microenvironment in different risk groups. A recent study indicated that macrophages affect cancer initiation and malignancy. Macrophage was divided into two subsets, M1 (classically activated) and alternatively activated M2.37 Macrophage M1 function as anti-tumorigenic properties, and macrophage M2 promote tissue repair and tumor growth.38 Intriguingly, we observed that macrophages M1 accumulated in high-risk group tumors while macrophages M2 accumulated in low-risk group tumors. Furthermore, our correlation results suggest the RRM2 and PSAT1 genes positively correlated with macrophage M1 and negatively correlated with macrophage M2, and the MIOX and CDKNIA genes positively correlated with macrophage M2 and negatively correlated with macrophage M1. This may be one of the reasons why the different abundance of macrophage was observed in different risk groups. The regulation of immune in tumors is complicated, in this case, the mechanism of the specific accumulation of macrophages in different risk groups is still unclear, and further research is needed to confirm this.
Conclusion
In summary, we constructed a 4-genes ferroptosis signature, which acts as an independent prognostic factor for PRCC. Furthermore, we established the prognostic risk models confirmed that the ferroptosis signature has a good capability in predicting PRCC patient survival. Our study provides new insight into the development of ferroptosis signature as a biomarker and therapeutic target in PRCC.
Acknowledgments
We thank TCGA and GEO database for providing available data. We express our sincere gratitude to reviewers for their constructive comments.
Funding Statement
This work was supported by the National Natural Science Foundation of China (Youth fund projects, 82103383); supported by the Basic and Applied Basic Research Foundation of Guangdong Province (Youth fund projects, 2019A1515110111); and supported by the Sanming Project of Medicine in Shenzhen (No. SZSM201612021).
Data Sharing Statement
We collected PRCC expression data and clinical data from TCGA (https://portal.gdc.cancer.gov/) public databases. A total 259 ferroptosis-related genes were download from FerrDb website (http://www.zhounan.org/ferrdb/legacy/index.html).18
Ethics Statement
This project was approved by the Research Ethics Committee of Peking University Shenzhen Hospital.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests in this work.
References
- 1.Courthod G, Tucci M, Di Maio M, Scagliotti GV. Papillary renal cell carcinoma: a review of the current therapeutic landscape. Crit Rev Oncol Hematol. 2015;96(1):100–112. doi: 10.1016/j.critrevonc.2015.05.008 [DOI] [PubMed] [Google Scholar]
- 2.Linehan WM, Ricketts CJ. The cancer genome atlas of renal cell carcinoma: findings and clinical implications. Nat Rev Urol. 2019;16(9):539–552. doi: 10.1038/s41585-019-0211-5 [DOI] [PubMed] [Google Scholar]
- 3.Delahunt B, Eble JN. Papillary renal cell carcinoma: a clinicopathologic and immunohistochemical study of 105 tumors. Mod Pathol. 1997;10(6):537–544. [PubMed] [Google Scholar]
- 4.Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang C, Zhang X, Yang M, Dong X. Recent progress in ferroptosis inducers for cancer therapy. Adv Mater. 2019;31(51):e1904197. doi: 10.1002/adma.201904197 [DOI] [PubMed] [Google Scholar]
- 6.Yang WS, Stockwell BR. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem Biol. 2008;15(3):234–245. doi: 10.1016/j.chembiol.2008.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou L, Zhao B, Zhang L, et al. Alterations in cellular iron metabolism provide more therapeutic opportunities for cancer. Int J Mol Sci. 2018;19(5):1545. doi: 10.3390/ijms19051545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li C, Zhang G, Zhao L, Ma Z, Chen H. Metabolic reprogramming in cancer cells: glycolysis, glutaminolysis, and Bcl-2 proteins as novel therapeutic targets for cancer. World J Surg Oncol. 2016;14(1):15. doi: 10.1186/s12957-016-0769-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Wei Z, Pan K, Li J, Chen Q. The function and mechanism of ferroptosis in cancer. Apoptosis. 2020;25(11–12):786–798. doi: 10.1007/s10495-020-01638-w [DOI] [PubMed] [Google Scholar]
- 10.Ma S, Henson ES, Chen Y, Gibson SB. Ferroptosis is induced following siramesine and lapatinib treatment of breast cancer cells. Cell Death Dis. 2016;7(7):e2307. doi: 10.1038/cddis.2016.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alvarez SW, Sviderskiy VO, Terzi EM, et al. NFS1 undergoes positive selection in lung tumours and protects cells from ferroptosis. Nature. 2017;551(7682):639–643. doi: 10.1038/nature24637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woo SM, Seo SU, Min KJ, et al. Corosolic acid induces non-apoptotic cell death through generation of lipid reactive oxygen species production in human renal carcinoma Caki cells. Int J Mol Sci. 2018;19(5):1309. doi: 10.3390/ijms19051309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shao Y, Jia H, Huang L, et al. An original ferroptosis-related gene signature effectively predicts the prognosis and clinical status for colorectal cancer patients. Front Oncol. 2021;11:711776. doi: 10.3389/fonc.2021.711776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang K, Ping L, Du T, et al. A ferroptosis-related lncRNAs signature predicts prognosis and immune microenvironment for breast cancer. Front Mol Biosci. 2021;8:678877. doi: 10.3389/fmolb.2021.678877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang B, Zhu J, Li J, et al. The ferroptosis and iron-metabolism signature robustly predicts clinical diagnosis, prognosis and immune microenvironment for hepatocellular carcinoma. Cell Commun Signal. 2020;18(1):174. doi: 10.1186/s12964-020-00663-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He F, Chen Z, Deng W, et al. Development and validation of a novel ferroptosis-related gene signature for predicting prognosis and immune microenvironment in head and neck squamous cell carcinoma. Int Immunopharmacol. 2021;98:107789. doi: 10.1016/j.intimp.2021.107789 [DOI] [PubMed] [Google Scholar]
- 17.Bai D, Feng H, Yang J, et al. Genomic analysis uncovers prognostic and immunogenic characteristics of ferroptosis for clear cell renal cell carcinoma. Mol Ther Nucleic Acids. 2021;25:186–197. doi: 10.1016/j.omtn.2021.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou N, Bao J. FerrDb: a manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database (Oxford). 2020;2020. doi: 10.1093/database/baaa021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bardou P, Mariette J, Escudie F, Djemiel C, Klopp C. jvenn: an interactive Venn diagram viewer. BMC Bioinform. 2014;15(1):293. doi: 10.1186/1471-2105-15-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi: 10.1093/nar/gkz430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song W, Shao Y, He X, et al. IGFLR1 as a novel prognostic biomarker in clear cell renal cell cancer correlating with immune infiltrates. Front Mol Biosci. 2020;7:565173. doi: 10.3389/fmolb.2020.565173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang K, Yuan C, Liu X. Ferroptosis-related gene signature accurately predicts survival outcomes in patients with clear-cell renal cell carcinoma. Front Oncol. 2021;11:649347. doi: 10.3389/fonc.2021.649347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong Y, Lin M, Ou D, Huang Z, Shen P. A novel ferroptosis-related 12-gene signature predicts clinical prognosis and reveals immune relevancy in clear cell renal cell carcinoma. BMC Cancer. 2021;21(1):831. doi: 10.1186/s12885-021-08559-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohmura S, Marchetto A, Orth MF, et al. Translational evidence for RRM2 as a prognostic biomarker and therapeutic target in Ewing sarcoma. Mol Cancer. 2021;20(1):97. doi: 10.1186/s12943-021-01393-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen WX, Yang LG, Xu LY, et al. Bioinformatics analysis revealing prognostic significance of RRM2 gene in breast cancer. Biosci Rep. 2019;39:BSR20182062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin CY, Du L, Nuerlan AH, Wang XL, Yang YW, Guo R. High expression of RRM2 as an independent predictive factor of poor prognosis in patients with lung adenocarcinoma. Aging. 2020;13(3):3518–3535. doi: 10.18632/aging.202292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma C, Luo H, Cao J, Gao C, Fa X, Wang G. Independent prognostic implications of RRM2 in lung adenocarcinoma. J Cancer. 2020;11:7009–7022. doi: 10.7150/jca.47895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohashi R, Angori S, Batavia AA, et al. Loss of CDKN1A mRNA and protein expression are independent predictors of poor outcome in chromophobe renal cell carcinoma patients. Cancers. 2020;12(2):465. doi: 10.3390/cancers12020465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan Z, Liu T, Huang H, Lin J, Zeng Z. A ferroptosis-related gene signature for graft loss prediction following renal allograft. Bioengineered. 2021;12(1):4217–4232. doi: 10.1080/21655979.2021.1953310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y, Wu J, Cai J, et al. PSAT1 regulates cyclin D1 degradation and sustains proliferation of non-small cell lung cancer cells. Int J Cancer. 2015;136(4):E39–50. doi: 10.1002/ijc.29150 [DOI] [PubMed] [Google Scholar]
- 32.Sharma I, Deng F, Kanwar YS. Modulation of renal injury by variable expression of Myo-Inositol Oxygenase (MIOX) via perturbation in metabolic sensors. Biomedicines. 2020;8(7):217. doi: 10.3390/biomedicines8070217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li T, Kon N, Jiang L, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149(6):1269–1283. doi: 10.1016/j.cell.2012.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang L, Kon N, Li T, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520(7545):57–62. doi: 10.1038/nature14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu B, Kon N, Chen D, et al. ALOX12 is required for p53-mediated tumour suppression through a distinct ferroptosis pathway. Nat Cell Biol. 2019;21(5):579–591. doi: 10.1038/s41556-019-0305-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie Y, Zhu S, Song X, et al. The tumor suppressor p53 limits ferroptosis by blocking DPP4 activity. Cell Rep. 2017;20(7):1692–1704. doi: 10.1016/j.celrep.2017.07.055 [DOI] [PubMed] [Google Scholar]
- 37.Rhee I. Diverse macrophages polarization in tumor microenvironment. Arch Pharm Res. 2016;39(11):1588–1596. doi: 10.1007/s12272-016-0820-y [DOI] [PubMed] [Google Scholar]
- 38.Ruffell B, Affara NI, Coussens LM. Differential macrophage programming in the tumor microenvironment. Trends Immunol. 2012;33(3):119–126. doi: 10.1016/j.it.2011.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]