Abstract
Common genetic variants in more than forty loci modulate risk for Alzheimer’s disease (AD). AD risk alleles are enriched within enhancers active in myeloid cells, suggesting that microglia, the brain-resident macrophages, may play a key role in the etiology of AD. A major genetic risk factor for AD is Apolipoprotein E (APOE) genotype, with the ε4/ε4 (E4) genotype increasing risk for AD by approximately 15 fold compared to the most common ε3/ ε3 (E3) genotype. However, the impact of APOE genotype on microglial function has not been thoroughly investigated. To address this, we cultured primary microglia from mice in which both alleles of the mouse Apoe gene have been humanized to encode either human APOE ε3 or APOE ε4. Relative to E3 microglia, E4 microglia exhibit altered morphology, increased endolysosomal mass, increased cytokine/chemokine production, and increased lipid and lipid droplet accumulation at baseline. These changes were accompanied by decreased translation and increased phosphorylation of eIF2ɑ and eIF2ɑ-kinases that participate in the integrated stress response, suggesting that E4 genotype leads to elevated levels of cellular stress in microglia relative to E3 genotype. Using live-cell imaging and flow cytometry, we also show that E4 microglia exhibited increased phagocytic uptake of myelin and other substrates compared to E3 microglia. While transcriptomic profiling of myelin-challenged microglia revealed a largely overlapping response profile across genotypes, differential enrichment of genes in interferon signaling, extracellular matrix and translation-related pathways was identified in E4 versus E3 microglia both at baseline and following myelin challenge. Together, our results suggest E4 genotype confers several important functional alterations to microglia even prior to myelin challenge, providing insight into the molecular and cellular mechanisms by which APOE4 may increase risk for AD.
Keywords: Alzheimer’s disease, APOE genotype, Microglia, Phagocytosis, Interferon signaling, eIF2 signaling, Translation inhibition, Lipid accumulation, Lipid droplets, Myelin
1. Introduction
Alzheimer’s disease (AD) is a devastating neurodegenerative disorder and the most common cause of dementia in the elderly, affecting approximately 5.8 million Americans. Due to an aging population and the absence of an effective treatment or prevention strategy, this number is projected to increase to 13.8 million by 2050, thus representing an urgent public health crisis (Alzheimer’s Association, 2021; Hebert et al., 2013). AD is characterized by severe neurodegeneration, abnormal deposition of extracellular amyloid-β (Aβ) plaques, and widespread formation of intracellular neurofibrillary tangles. In addition to these classical neuropathological hallmarks, glial activation (gliosis) and lipid accumulation (lipidosis) have also been described (Foley, 2010).
Most AD cases are classified as late-onset AD (LOAD) and have a complex genetic architecture (Neuner et al., 2020; Zhang et al., 2020). Large-scale genome-wide association studies (GWAS) have so far identified 40 genomic regions associated with LOAD (Andrews et al., 2020). The strongest association with AD risk and age at onset is consistently observed on chromosome 19 in the genomic region harboring the apolipoprotein E (APOE) gene. The association between APOE genotype and AD risk was first identified using linkage studies in families with multiple instances of LOAD (Corder et al., 1993) and it has since been replicated in multiple large-scale analyses (Reiman et al., 2020). APOE is a secreted glycoprotein that binds cholesterol and other lipids to form lipoproteins, facilitating lipid transport in plasma and brain interstitial fluid (Ulrich et al., 2013). In humans, there are three major APOE isoforms designated as APOE2, APOE3 and APOE4 that are encoded by three common alleles (ε2, ε3, and ε4, respectively). These isoforms differ at only two amino acid residues; APOE3 has a cysteine at position 112 and arginine at position 158, while APOE4 has two arginines and APOE2 has two cysteines at these positions (Mahley et al., 2009). APOE ε4 increases risk in a semi-dominant manner, with one copy increasing risk ~3 fold and two copies increasing risk ~15 fold relative to the ε3/ε3 genotype (Genin et al., 2011), which is the most common in all populations studied (Farrer et al., 1997; Liu et al., 2013). In contrast, individuals with two copies of the ε2 allele exhibit an approximately seven-fold decreased risk for AD (Farrer et al., 1997; Liu et al., 2013; Reiman et al., 2020).
Given the strong link between APOE genotype and AD risk, a better understanding of how the different APOE isoforms promote or protect against the disease is critical. Multiple studies have shown that APOE4 can exacerbate Aβ aggregation (Bales et al., 2009; Liao et al., 2018), reduce Aβ clearance in mice (Castellano et al., 2011), and that APOE genotype is associated with the onset and extent of Aβ deposition in humans (Holtzman et al., 2012). However, it is likely that the role of APOE in disease pathogenesis extends beyond its effect on amyloid accumulation (Guerreiro et al., 2012). Post-GWAS studies have strongly implicated myeloid cells such as microglia in the etiology of LOAD (Huang et al., 2017; Nott et al., 2019). Coding variants in genes that are specifically expressed in myeloid cells including microglia (e.g., TREM2, PLCG2, and ABI3) have been associated with AD (Guerreiro et al., 2013; Jonsson et al., 2013; Sims et al., 2017). Further, AD risk alleles have been shown to be enriched in active enhancers specific to monocytes, macrophages and microglia, suggesting that gene expression regulation in these myeloid cell types may be critical to modifying disease susceptibility (Huang et al., 2017; Nott et al., 2019; Novikova et al., 2021). While APOE is primarily produced by astrocytes in the brain, it is one of the most upregulated genes during the response of microglia to damage associated with aging, demyelination or neurodegeneration (Keren-Shaul et al., 2017; Krasemann et al., 2017). Given the causal role of both APOE and microglia in AD, investigating the impact of differing APOE genotypes on microglial function is likely to advance our understanding of AD pathogenesis.
Recent single cell transcriptomic studies have identified a subset of microglia in the aging and diseased brain, referred to as damage- or disease- associated microglia (DAM), which exhibit downregulation of several genes enriched in surveillant microglia (such as P2ry12 and Tgfbr1) and upregulation of genes associated with phagocytosis and lipid metabolism (e.g., Axl and Lpl) (Keren-Shaul et al., 2017; Podlésny-Drabiniok et al., 2020). Transition to the DAM state occurs in a stepwise fashion, with loss of either Apoe or Trem2 preventing microglia from converting into early- or late-stage DAM state, respectively (Damisah et al., 2020; Krasemann et al., 2017). In addition, loss of either Trem2 or Apoe leads to abnormal accumulation of cholesterol esters or formation of cholesterol crystals in microglia from mice subjected to treatments that induce demyelination, suggesting that both TREM2 and APOE are necessary for microglia to appropriately handle excess cholesterol resulting from phagocytic uptake of myelin debris (Cantoni et al., 2015; Cantuti-Castelvetri et al., 2018; Nugent et al., 2020). Similar phenotypes are observed upon loss of another AD risk gene, PLCG2 (Andreone et al., 2020), demonstrating that APOE and other AD risk genes play a critical role in the response of microglia to aging- and disease-associated conditions. However, most of these studies were conducted using Apoe knockout mice, and how human APOE genotypes differentially impact microglial function in these contexts remains to be elucidated.
As human microglia are difficult to obtain due to a general lack of access to living brain tissue, mouse microglia have emerged as a useful model to experimentally investigate the complex function of these cells. However, as mouse Apoe exhibits only 70% homology with human APOE, and mice do not express APOEε2/3/4 isoforms, mice in which the endogenous Apoe sequence has been humanized by targeted insertion of exons 2–4 of either human APOE2, APOE3, or APOE4 have been generated (Sullivan et al., 1997). These targeted replacement (TR) mice have provided important insights into the functional effects of APOE isoforms in vivo. Given the utility of this TR mouse model to investigate the effect of human APOE genotype on cellular functions, here we investigated the impact of APOE genotype on cultured primary microglia at baseline and following acute challenge with an excess of cellular debris in the form of myelin fragments. Using live-cell imaging, flow cytometry, immunocytochemistry, immunoblotting, and RNA-sequencing, we demonstrate that compared to APOE ε3/ε3, APOE ε4/ε4 microglia display amoeboid morphology, increased levels of select cytokines/chemokines, decreased translation, increased phosphorylation of eIF2ɑ and eIF2ɑ-kinases that participate in the integrated stress response, increased phagocytic uptake of myelin fragments and other substrates, increased lipid and lipid droplet accumulation, and increased endolysosomal mass. Our results provide new insight into the effect of APOE4 on microglial function and nominate candidate pathways for therapeutic targeting.
2. Materials and Methods
2.1. Animals
Human APOE targeted replacement mice, E3 and E4 (C57BL/6, Taconic, 1548, 1549) or Apoe knockout mice (C57BL/6, Jackson Laboratory, 002052) and wild type mice (C57BL/6, Jackson Laboratory, 000664) were bred to obtain postnatal day 1–3 (P1–3) pups. All animals were housed under a standard light dark cycle and received food and water ad libitum. All animal procedures and experiments were conducted in compliance with guidelines by James J. Peters Veteran Affairs Medical Center (protocol #CAI-10–044a) and Icahn School of Medicine at Mount Sinai (protocol #2014–0172, 2017–0296) Institutional Animal Care and Use (IACUC) committees. In addition, all experiments involving animals were carried out in compliance with ARRIVE guidelines and in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No. 8023, revised 1978).
2.2. Preparation of microglial cultures
Mixed glial cultures were prepared by pooling forebrains from 8–10 pups aged P1–3 from a single litter. Each litter was considered an experimental replicate; experiments were repeated in 4–5 independent litters as denoted in the corresponding figure legends. Pups were not genotyped for sex chromosomes prior to pooling. Specifically, the whole brain was removed and placed into cold Hanks Balanced Salt Solution (HBSS, Invitrogen #14025092). Meninges were removed and forebrain tissue was cut into small pieces and digested in 0.25% trypsin (GIBCO #25200114) at 37 °C for 15 mins. DNase I (0.2mg/ml, Sigma, D4527– 20KU) was added and incubated at 37 °C for 10 mins, followed by centrifugation at 1300 rpm for 5 min. Homogenate was washed and resuspended in DMEM (Invitrogen, 10565) and filtered through a pre- soaked 40μm nylon mesh. Mixed glial cultures were prepared by plating dissociated cells (60,000 cells/cm2) in T75 flasks (Corning #354537) and growing them at 37°C, 5% CO2 in glial media [DMEM containing 10% fetal bovine serum (FBS, Gibco #16140), and 100μg/ml penicillin/streptomycin (Pen/Strep, Gibco #15140). Cultures were maintained by replacing the growth media once per week. After 4 weeks, cell cultures were enriched for microglia by washing with DMEM for one minute, followed by incubation with 1:3 dilution trypsin in warm DMEM at 37°C, 5% CO2 for 35 mins with occasional shaking. The detached astrocyte-rich layer was carefully removed and discarded. The remaining microglial cells were washed with DMEM and incubated with trypsin for 10min. Cells were collected and pooled into glial medium and seeded into tissue culture plates and allowed to adhere for 24 hours prior to experimentation. Routine testing of cell lines using MycoAlert PLUS mycoplasma detection kit (Lonza #LT07–118) showed that all cells were negative for mycoplasma contamination.
2.3. Immunofluorescence microscopy and image analysis
Microglia were plated on round coverslips coated with polyD-lysine in a 24-well plate. Cells were allowed to adhere overnight and select wells were treated with myelin according to the phagocytosis protocol described below. Cells were fixed in 4% paraformaldehyde in PBS at 4°C for 10 min. Cells were permeabilized (1.0% Triton in PBS) at room temperature for 15 min and blocked in 5% bovine serum albumin (BSA) at room temperature for 1 hours. Cells were incubated in primary antibodies as follows: 1:500 anti-IBA1 (Abcam #5076), 1:1000 anti-CD68 (Bio-Rad #1957), 1:1000 anti-Perilipin3 (Abcam #47639), 1:2000 anti- RAB5 (Cell Signaling #3547), and 1:1000 anti-RAB7 (Abcam #198337). Secondary antibodies donkey anti-goat Alexa 488 (Invitrogen #A11055), goat anti-rat Alexa 647 (Abcam #150159) or donkey anti- rabbit Alexa 488 (Life Technologies #21206) were used at 1:500. For BODIPY staining, cells were incubated with 2 μM BODIPY (Invitrogen #D3861) for 15 mins at 37°C. DAPI staining (1 μg/ml for 5 min, Life Technologies #D1306) was used to visualize nuclei. Images were acquired using an Olympus IX51 Fluorescence Microscope or a Zeiss LSM780 confocal microscope. Purity of microglial cultures was assessed by quantifying the number of DAPI-labeled cells also expressing the microglial marker IBA1. Percentage of microglia in each culture was determined by dividing the number of IBA1-positive cells to the total number of cells. For quantification of specific markers, fluorescence intensity was calculated using the processing software FIJI Is Just ImageJ (Schindelin et al., 2012). Briefly, a region of interest was drawn around each cell in a given field of view and integrated density (the product of Area and Mean Value) over the selected regions of interest was measured. Average integrated density per image was calculated by normalizing the integrated density to the number of cells. To quantify the degree of colocalization between BODIPY and either RAB5 or RAB7, the Pearson’s correlation coefficient between the two fluorescent signals from a single field of view was calculated using ZEN software according to the Zeiss protocol Acquiring and analyzing data from colocalization experiments in AIM or ZEN software (https://www.zeiss.com/content/dam/Microscopy/Downloads/Pdf/FAQs/zen-aim_colocalization.pdf). To compare correlation coefficients, Fisher’s r-to-z transformation was used (Eid et al., 2017). Pearson’s correlation coefficients were transformed into z scores using Fisher’s r-to-z transformation for statistical analyses. A two sided z-test was then used to compare correlations across groups. In all cases, quantification was performed on at least 60 cells per genotype from 4 independent experiments. All immunofluorescence microscopy image acquisition and quantification were performed by an experimenter blind to genotype/treatment.
2.4. Morphometric image analysis using Ilastik
In order to quantitatively assess morphological differences across E3 and E4 microglia, we utilized the publicly available machine-learning image analysis software Ilastik (Berg et al., 2019). A set of ~10 representative phase contrast images was uploaded as training data and the Pixel Classification + Object Classification workflow was followed according to the developer’s protocol (www.ilastik.org). Specifically, training features specifying which aspects of phase images corresponded to cells vs background were optimized across training images. Hysteresis thresholding (threshold = 0.65) was used across images such that cells were clearly segmented from the background using pixel classification. Subsequent object classification was used to identify microglia and extract relevant feature information for each identified cell. Specifically, Ilastik uses the Vision with Genetic Algorithms (VIGRA) library (https://ukoethe.github.io/vigra/) to compute object features. Features reported here include object area, length (defined as the longest path between two terminals of a given object’s skeleton), and branch length (computed by deriving the average length of a given object’s skeleton’s branches following default pruning algorithms). Skeletons, or one pixel- wide representations of segmented objects, were generated using the default ilastik algorithm which relies on the successive pruning via the skeletonizeImage() algorithm as defined in the VIGRA image analysis library (https://ukoethe.github.io/vigra/).
2.5. Preparation of pHrodo red-labeled myelin fragments
Myelin fragments were isolated from two month-old C57BL/6 wild-type mouse brain. All solutions were used ice-cold and all centrifugations were performed at 4C unless otherwise noted. Hemi brains (n=4) were homogenized and sonicated for 10 min in 5 ml of 0.32M sucrose (Sigma-Aldrich #S1888) and additional 10ml of 0.32M sucrose and 1:100 protease inhibitor (Cell Signaling Technology #5872S) was added. A sucrose step-gradient was prepared in an ultracentrifuge tube (Beckman Coulter #344058) by overlaying 15ml of 0.85M sucrose and 15ml of 0.32M sucrose containing tissue homogenate. The tube was then centrifuged in a SW32Ti rotor at 24,4000 rpm for 40 min using a Beckman L70 ultracentrifuge, with low acceleration and deceleration. The gradient interface containing the fragmented myelin was collected using a Pasteur pipette and washed with MilliQ water at 24,4000 rpm for 15 min. The pellet was given two osmotic shocks by resuspending in water and centrifuging at 9800 rpm for 15 min and then resuspended in 0.32M sucrose and combined with 0.85M sucrose in a 1:1 ratio, followed by centrifugation at 24,4000 rpm for 40 min. The interface was again collected and resuspended in 0.1M sodium bicarbonate (Sigma-Aldrich #S8761). The volume was adjusted to obtain a final protein concentration of 1μg/ml. Endotoxin levels were measured using a commercial kit following manufacturer instructions (GenScript #L00350C) and myelin fragments were conjugated with 1:100 pHrodo red dye (Invitrogen #P36600) according to the manufacturer’s protocol and stored at −80 °C.
2.6. Preparation of pHrodo red-labeled early apoptotic Jurkat cells
Jurkat human T cell leukemia cells were kindly provided by Dr. Benjamin Chen (MSSM). Jurkat cells were cultured in RPMI (Gibco #11965) supplemented with 10% FBS (Gibco #16140), and 1:100 Pen/ Strep (Gibco #15140). Cells were treated with 1μM staurosporine for 3 hours to obtain early apoptotic Jurkat (EAJ) cells. Cells were collected and washed with PBS and stained with pHrodo red 1:100 in PBS for 1 hour in the dark at room temperature. Cells were washed once with PBS, resuspended at 1×106 cells/ml in PBS and added to plated primary microglia for phagocytosis assay as described below.
2.7. Phagocytosis assay
Microglial cells were seeded at a density of 60,000 cells/well in a 96 well plate and allowed to adhere overnight. The following day, plates were placed inside an IncuCyte live imaging system housed inside a tissue culture incubator at 37 °C/5% CO2 and a baseline image was captured prior to treatment with phagocytic substrate. Cells were treated with one the following substrates for three hours: 10μg/ml of pHrodo-red labeled myelin, 10μg/ml pHrodo-red labeled zymosan (Thermo Fisher #P35364), 1:100 pHrodo-red labeled early apoptotic Jurkat (EAJ) cells, and 1:100 red fluorescent carboxylate-modified polystyrene latex beads (Millipore Sigma #L3030, 0.9 μm). To inhibit phagocytosis, select control wells were pre-treated with 2 μM cytochalasin D (CD, Sigma-Aldrich #C8273) for 30 min and during the 3 hours incubation with the substrate. As additional negative controls, some wells were left untreated (i.e., no myelin was added). After 3 hours, cells were washed with PBS and fresh glial medium was added. Live imaging scans continued every hour for the duration of the assay. No myelin wells were monitored for the duration of the experiment while CD wells were only monitored for 48 hours, after which a majority of cells had detached. Specifically, red fluorescent (acquisition time 800 ms) and phase contrast images were acquired using a 20x objective. Three images per well from two technical replicate wells were taken and then analyzed using the IncuCyte ZOOM software. For phase contrast images, cell segmentation was performed such that a mask applied to the image clearly differentiated cells from background. An area filter was applied to exclude objects below 10μm. The obtained cell mask was used to export raw “percent phase confluence” values for each image. Thresholding was performed to reliably identify regions of red fluorescence within a given image (Kapellos et al., 2016) and a second mask corresponding to red fluorescence was obtained. Raw values of total red integrated fluorescence intensity (red calibrated units, RCU, red calibrated units x μm2/image) were obtained. These raw values were normalized to % phase confluence. In order to estimate the degradation rate of internalized myelin, we performed non-linear regression of normalized red fluorescence intensity over time using GraphPad Prism software ((GraphPad Software, n.d); Stewart et al., 2011) and the one-phase exponential decay model as specified by the equation: Y = (Y0 - Plateau) e(-KX) + Plateau, where X = Time, Y = normalized red fluorescent intensity, Y0 = Y value at 6 hours, K = rate constant, and Plateau = Y value at infinite times. Half-life was computed as ln(2)/K.
For measurement of phagocytosis by flow cytometry 24 hours post- challenge, cells were trypsinized (Gibco #25200), pelleted, stained with DAPI for 5 mins to identify live versus dead cells, and geometric mean fluorescence intensity (gMFI) was measured using an Attune NxT flow cytometer (ThermoFisher Scientific). To distinguish microglia from EAJ during flow cytometry, samples were pre-stained with an APC-conjugated antibody against macrophage-specific cell surface marker CD11b (eBioscience #17–0112-81) at 1:400 dilution for 30 minutes on ice. Data were quantified using FCS Express 7 (De Novo Software) and GraphPad Prism 9 (GraphPad Software) and the average phagocytic index of each replicate was calculated as the percentage of red fluorescent-positive and DAPI-negative cells multiplied by the geometric mean of red fluorescence intensity divided by 106. APC-positive, pHrodo red-positive and DAPI-negative cells were gated and used to calculate the phagocytic index of EAJ internalization.
2.8. Lysosomal mass and pH assays
To assess lysosomal mass, microglia were stained with 100nM LysoTracker Red DND-99 (Invitrogen #L7528) for 30 mins. To assess lysosomal pH, microglia were washed with PBS and treated with 2μM LysoSensor Green DND-189 (Invitrogen #L7535) for 10 mins. Microglia were then washed with PBS three times, trypsinized, pelleted, resuspended in 100ul PBS, and stained with 0.1ug/ml DAPI for 5 min. Cells were again pelleted, washed three times in PBS, and resuspended in 100ul PBS at 60,000 cells/μL and analyzed using an Attune NxT flow cytometer. Cells were gated on DAPI to measure fluorescence intensity of LysoTracker and LysoSensor only in live cells. Calibration curve was prepared by incubating cells in LysoSensor pHrodo Green AM intracellular pH indicator (ThermoFisher Scientific #P35373) according to the manufacturer’s protocol.
2.9. Cholesterol loading assay
To assess cellular cholesterol levels, microglial cells were incubated with 20μg/ml [N- [(7-nitro-2–1,3-benzoxadiazol-4-yl) methyl] amino] (NBD)-cholesterol (Abcam #236212) in serum free medium for 24 hours at 37 °C. Cells were trypsinized, pelleted, resuspended in 100ul PBS and stained with 0.1ug/ml DAPI for 5 min. Cells were pelleted, washed three times in PBS and resuspended in 100ul PBS at 60,000 cells/μL and analyzed using an Attune NxT flow cytometer. Cells were gated on DAPI to measure fluorescence intensity of NBD.
2.10. RNA sequencing
RNA sequencing was performed on microglial cell lysates collected as described above for phagocytosis assay. Twenty-four hours after myelin treatment, 120,000 cells per genotype/treatment were harvested, pelleted, and stored at −80 °C. RNA was extracted using the RNeasy (Qiagen) kit following manufacturer’s guidelines. Libraries were prepared from polyA-enriched mRNA using the universal low-input kit (SMARTer #634940), validated using Agilent TapeStation (Agilent Technologies, Palo Alto, CA, USA), quantified by using Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA) as well as by quantitative PCR (KAPA Biosystem, Wilmington, MA, USA), and normalized to ensure equal amounts of each sample were used for sequencing. These normalized libraries were sequenced as paired-end 150bp reads (15–20 million reads/sample) on the Illumina HiSeq 4000. Reads were screened and trimmed for adaptor contamination and paired-end sequencing reads were pseudoaligned to the Gencode mm10 transcriptome (version M20) and expression was quantified and normalized as transcripts per million (TPM) using Salmon (version 0.13.1) (Patro et al., 2017). Next, differentially expressed protein-coding genes (DEGs) across comparisons of interest were identified by using the limma (version 3.40.6) package (Ritchie et al., 2015) in the R programming environment (version 3.6.3). An FDR adj. p-value < 0.05 was used to identify genes significantly differentially expressed throughout the manuscript unless noted otherwise.
2.11. Overrepresentation enrichment analysis
Genes differentially expressed at an adjusted p value < 0.05 (Fig. 4c) were used for overrepresentation enrichment analysis. Specifically, lists of significantly upregulated or downregulated genes were uploaded to WebGestalt [WEB-based GEneSeT AnaLysis Toolkit (www.webgestalt.org] (Liao et al., 2019). Overrepresentation of genes belonging to Reactome pathways was identified using the protein-coding mouse genome as the reference set within WebGestalt. Pathways with FDR adj. p-value < 0.05 were identified. Redundancy in pathways was reduced using WebGestalt’s “Affinity Propagation” option for graphing in Fig. 4c.
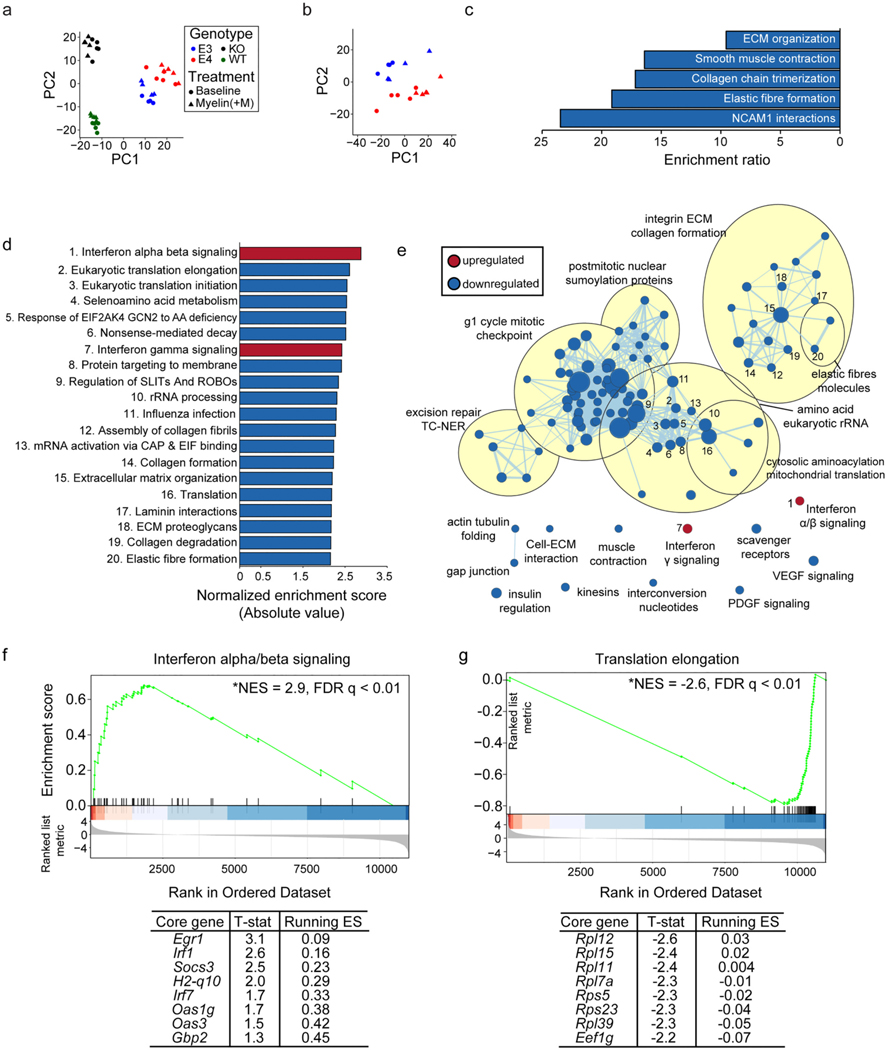
Fig. 4.
E4 microglia exhibit differential enrichment of genes in interferon signaling, extracellular matrix, and translation-related pathways at baseline. Principal component analysis (PCA) using the top 500 most variable genes from RNA-sequencing was performed using a) all samples or b) E3 and E4 samples only in order to examine clustering among samples. c) Reactome pathways significantly enriched (FDR adj. p-value < 0.05) for genes significantly downregulated (FDR adj. p-value < 0.05) in E4 vs E3 microglia, as identified by overrepresentation enrichment analysis performed using WebGestalt (Liao et al., 2019). d) The top 20 significantly enriched (FDR adj. p-value < 0.01) Reactome pathways enriched for genes differentially expressed in E4 vs E3 microglia, as identified by gene set enrichment analysis (GSEA) (Subramanian et al., 2005). e) Cytoscape Enrichment Map (Merico et al., 2010) and AutoAnnotate packages (Kucera et al., 2016) were used to identify relationships among all significantly enriched Reactome pathways (FDR adj. p-value < 0.01). Pathways enriched for upregulated genes are shown in red while pathways enriched for downregulated genes are shown in blue. Node size directly corresponds to the size of the plotted gene set. Edge width represents the number of genes that overlap between a pair of gene sets. Nodes corresponding to the top 20 pathways plotted in (d) are numbered appropriately. f) GSEA enrichment plot for “interferon alpha/beta signaling”, the most significantly enriched pathway at the top end of the ranked list (i.e., with a positive fold-change, see Methods). The mouse orthologues of the top 8 core genes identified by GSEA are listed below, along with the t-statistic of their differential expression and running enrichment score (ES) at their position. G) GSEA plot and list of core genes for “translation elongation”, the most significantly enriched pathway among genes at the bottom of the ranked list (i.e., with a negative fold change, see Methods). Detailed results are available in Table S3.
2.12. Gene set enrichment analysis (GSEA)
Ranked lists were generated from differential gene expression analyses by ordering genes according to the test statistic generated by limma (Ritchie et al., 2015). This metric was chosen as it takes into account both the fold change across conditions as well as the standard error of that fold change. Ranked lists were analyzed using the “GSEA Preranked” module using default GSEA settings (Subramanian et al., 2005) including 1000 permutations. Ensembl IDs of mouse genes were converted to human orthologues using the “Mouse_ENSEMBL_Gene_ID_Human_Orthologs_MSigDB.v7.1” chip file. Our preranked lists were tested for enrichment against gene sets from the Molecular Signatures Database (MSigDB, Broad Institute) v7.1 and 1000 permutations were performed. Enrichment scores representing the degree of overrepresentation of a single gene set at the top or bottom of a ranked list were calculated for each individual gene set. Enrichment scores were normalized by gene set size to generate ‘normalized enrichment scores’ according to standard protocols (Subramanian et al., 2005) and these normalized scores were used to determine significance of enrichment. Core genes driving observed enrichments were identified according to standard GSEA protocols, with core genes annotated as those contributing most significantly to the leading-edge subset within a given gene set. As genes within the top and bottom sections of ranked gene lists are given priority weighting, this “core” set of genes can also be described as the subset of genes that contributes most to the enrichment result. Several core genes were identified and plotted throughout our analyses. At the pathway level, those with FDR adj. p-value < 0.25 were considered to be statistically significantly enriched among upregulated or downregulated genes (Subramanian et al., 2005), although we utilized more stringent cutoffs throughout the paper in order to facilitate graphical representation of enriched pathways. For example, to visualize enriched pathways across genotypes, we utilized Cytoscape’s Enrichment Map (Merico et al., 2010) to plot the relationships between pathways enriched at FDR adj. p-value < 0.01. AutoAnnotate (Kucera et al., 2016) was further used on pathways enriched at FDR adj. p-value < 0.01 to identify clusters in an unbiased manner and visually annotate them based on word frequency across pathway annotation. Specific cutoffs used are noted in figure legends where appropriate. For plots in Fig. 7, measures of enrichment that accounted for both strength and direction of enrichment were calculated by log10 transforming the FDR adj. p-value (plus 0.001 to account for pathways where FDR adj. p-value = 0.0) and multiplying this value by the absolute value of the enrichment score (Neuner et al., 2019; Raj et al., 2017).
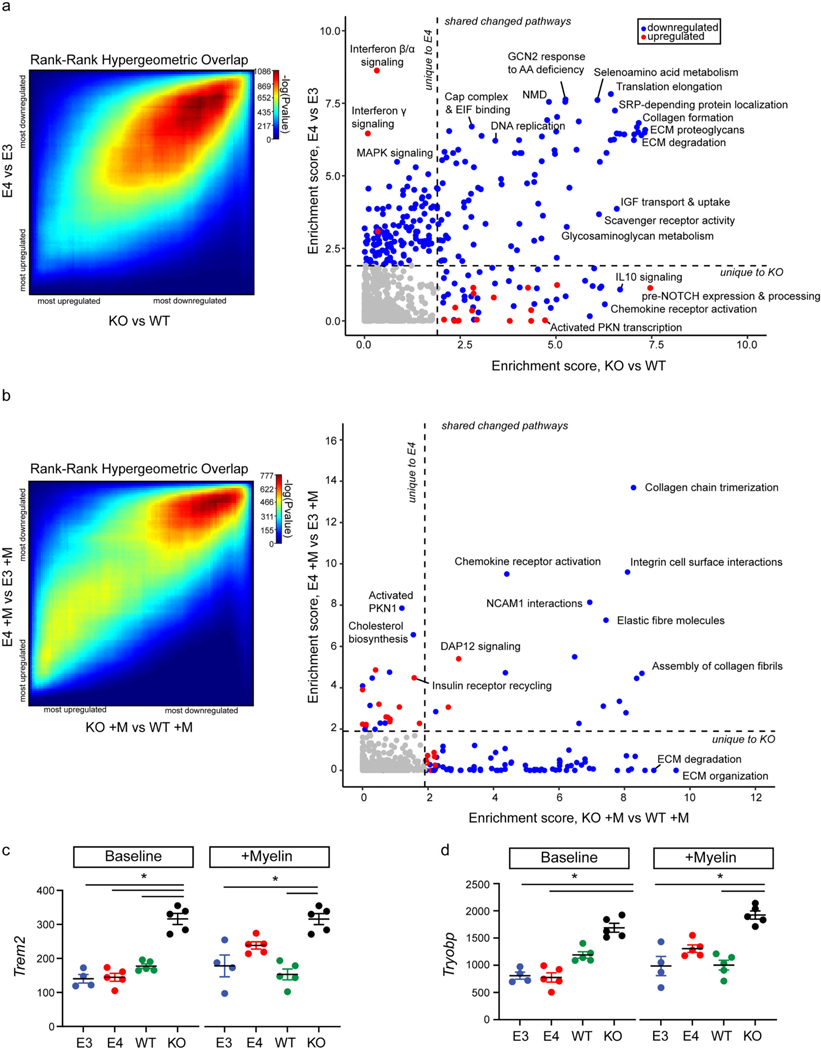
Fig. 7.
Transcriptional signatures of E4 and KO microglia relative to E3 and WT microglia exhibit both shared and unique features. a) Left, rank-rank hypergeometric overlap testing was used to identify how similar or divergent the transcriptional signatures of E4 vs E3 microglia and KO vs WT microglia are at baseline. Right, graph of enrichment strength of Reactome pathways across (y axis) KO vs WT microglia or (x axis) E4 vs E3 microglia. Specifically, FDR adj. p-values were transformed to obtain a measure of enrichment strength (see Methods) and scores were plotted against each other to identify unique and/ or common differentially enriched Reactome pathways. As such, each axis can be thought of as a measure of statistical significance; the upper left quadrant highlights pathways that are uniquely significantly altered in E4 microglia relative to E3 microglia, and the bottom right quadrant highlights pathways that are uniquely significantly altered in KO microglia relative to WT microglia. Dots are colored based on directionality of enrichment score calculated by GSEA: red, genes belonging to this pathway were significantly positively enriched; blue, genes belonging to this category were significantly negatively enriched. Dotted lines represent enrichment score cutoff values for FDR = 0.05 and absolute value of GSEA normalized enrichment score = 1.5. b) The above analysis was repeated following myelin challenge. c, d) mRNA abundance of Trem2 and Tyrobp across groups (within each graph, left = baseline and right = post-myelin). Each dot corresponds to an independent experiment. FDR adj. p-values (q) from limma differential expression analysis: baseline Trem2; KO vs E3: t = 3.7, logFC = 1.2, q = 0.0006; KO vs E4: t = 3.8, logFC = 1.1, q = 0.002; KO vs WT: t = 2.7, logFC = 0.8, q = 0.04. Post-myelin Trem2, KO vs E3: t = 3.3, logFC = 1.1, q = 0.01; KO vs WT: t = 4.0, logFC = 1.2, q = 0.002. Baseline Tyrobp; KO vs E3: t = 3.0, logFC = 1.1, q = 0.004; KO vs E4: t = 3.5, logFC = 1.2, q = 0.004. Post-myelin Tyrobp; KO vs E3: t = 2.9, logFC= 1.03, q = 0.005; KO vs WT: t = 2.9 logFC = 0.96, q = 0.006. c-d) Each dot corresponds to an independent experiment (n = 4/group). Black horizontal bars indicate the mean and error bars represent SE.
2.13. Cytokine/chemokines proteome profiler and ELISA assays
To measure cytokines/chemokines and APOE in conditioned media, microglia were plated at 60,000 cells/well in a 96 well plate and cultured for 24 hours. Conditioned media was collected, cleared of cell debris with 10 min centrifugation at 3,000 rpm. Cytokines/chemokines were measure using Proteome Profiler Mouse Cytokine Array Kit, Panel A (R&D Systems #ARY006) according to the manufacturers’ instructions. APOE levels were measured using a mouse APOE ELISA kit (Mabtech #3752–1HP-2) according to the manufacturers’ protocol. To measure myelin basic protein (MBP) levels in myelin treated cells and conditioned media, cells were treated with myelin as described for the phagocytosis assay. Conditioned media was collected as stated above and pellets were resuspended in 100ul PBS and one freeze-thaw cycle was used to lyse cells. MBP levels in cell lysate and conditioned media were measured using mouse MBP ELISA kit (Mybiosource #MBS161123) according to the manufacturer’s protocol.
2.14. Western blotting
Microglia were lysed in RIPA buffer (Sigma-Aldrich #R0278) supplemented with protease/phosphatase Inhibitor Cocktail (Cell Signaling Technology #5872) followed by 30 min incubation on ice. Lysates were cleared with 30 min centrifugation at 15,000g. Protein concentration was measured using the BCA kit (Thermo Fisher #PI-23225) and 40ug protein was used for western blotting. Samples were resolved by electrophoresis with Bolt 4–12% Bis-Tris Plus Gels (Invitrogen #NP0326BOX) in Bolt MES SDS running buffer (Invitrogen #B0002) and transferred using iBlot 2 nitrocellulose transfer stacks (Invitrogen #IB24002). Membranes were blocked for 1 hour and probed with the following antibodies dissolved in either 5% non-fat dry milk in PBS/ 0.1% Tween-20 buffer or 5% Bovine serum overnight at +40C: 1:1000 eIF2α (Cell Signaling Technology #2103), 1:500 Phospho-eIF2α (Cell Signaling #3398), 1:1000 PKR (Abcam #184257), 1:1000 Phospho-PKR (Invitrogen #44668G), 1:1000 PERK (Cell Signaling Technology #3192), 1:1000 Phospho-PERK (Cell Signaling Technology #3179), 1:1000 GCN2 (Invitrogen #PA5–17523), 1:1000 Phospho-GCN2 (Invitrogen #PA5–105886), 1:1000 Anti-apolipoprotein E (Sigma-Aldrich #AB947), 1:5000 β-actin (Sigma-Aldrich #A5441). Membranes were washed, then incubated with secondary antibody 1:2000 for 1 hour at RT and visualized with WesternBright ECL HRP Substrate Kit (Advansta, #K-12045) and UVP ChemiDoc-ItTS2 Imager (UVP). Images were quantified using ImageJ (NIH).
2.15. Quantitative RT-PCR
To perform quantitative reverse transcriptase PCR, microglia were seeded at 60,000 cells/well in a 96 well plate and cultured for 24 hours. Cell pellets were collected and mRNA was extracted using RNeasy mini kit (QIAgen #74106) including the DNase I treatment step with RNase-Free DNase set (QIAgen #79254) according to the manufacturer’s protocol. Nanodrop 8000 (Thermo Scientific) was used to quantify mRNA, and 1000ng of RNA was used for reverse transcription reaction using the High-Capacity RNA-to-cDNA kit (Thermo Fisher #4387406). cDNA (10ng) was used in the qPCR reaction with Power SYBR Green Master Mix (Applied Biosystems #4368706) and amplified by using QuantStudio 7 Flex Real- Time PCR System (Thermo Fisher Scientific). Primers were designed using the Primer-BLAST program (NCBI) (Table S1). Two technical replicates for each gene were used, and Ct values were averaged. Gapdh Ct values were used for normalization. Gene expression levels were quantified using the 2-ddCt method relative to the average dCT value from all four E3 samples. A total of 25 genes were evaluated using RT-qPCR, results were appropriately corrected for multiple comparisons using the Benjamini-Hochberg method. The fold change of a subset of genes are plotted in Fig. 5a. A list of primers used is available in Table S1.
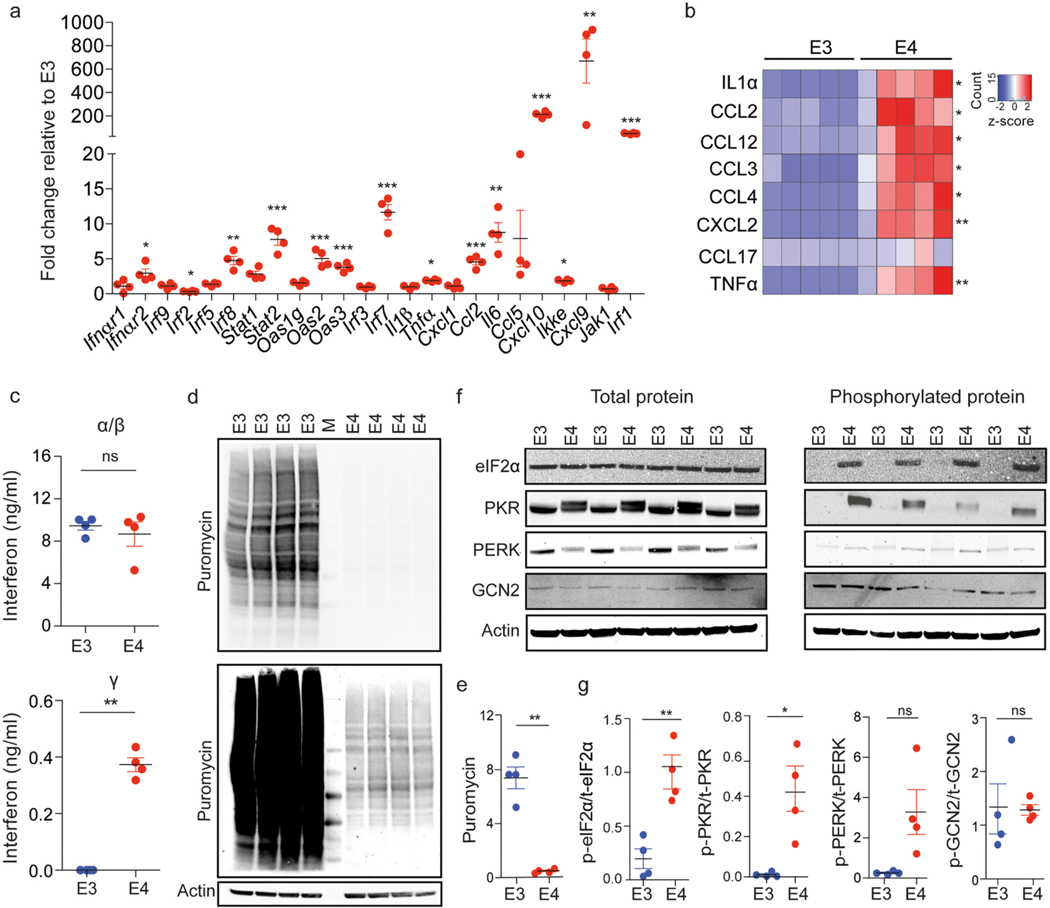
Fig. 5.
E4 microglia exhibit increased interferon signaling, increased eIF2 signaling, and decreased protein translation at baseline. a) Expression of several genes from the interferon α/β and interferon ɣ signaling pathways (Fig. 4d) were measured from cell lysates of E3 and E4 microglia using qRT-PCR. E4 microglia exhibited an increase in the expression of several of these genes compared to E3 microglia; unpaired Welch’s t-test followed by Benjamini-Hochberg (BH) correction for multiple comparisons, Ifnαr1 avg. fold change (FC) ± SEM = 1.1 ± 0.4, adj. p = 1.0; Ifnαr2 FC = 3.0 ± 0.6, adj. p = 0.02; Irf9 FC = 1.1 ± 0.2, adj.p = 1.0; Irf2 FC = 0.31 ± 0.04, adj. p = 0.005; Irf5 FC = 1.4 ± 0.1, adj. p = 0.08, Irf8 FC = 4.8 ± 0.6, adj. p = 0.002; Stat1 FC = 2.8 ± 0.40, adj. p = 0.06; Stat2 avg. FC = 7.8 ± 0.85, adj. p = 0.0003; Oas1g FC = 1.6 ± 0.2, adj.p = 0.06; Oas2 avg. FC = 5.1 ± 0.60, adj. p = 0.0004; Oas3 avg. FC = 3.8 ± 0.29, adj p = 0.0004; Irf3 FC = 1.0 ± 0.08, adj. p = 1.0; Irf7 avg. FC = 11.7 ± 1.1, adj. p = 5.7 × 10−05; Il1β FC = 1.0 ± 0.1, adj. p = 1.0; Tnfα avg. FC = 1.90 ± 0.07, adj. p = 0.02; Cxcl1 FC = 1.1 ± 0.2, adj. p = 1.0; Ccl2 avg. FC = 4.5 ± 0.41, adj. p = 0.0003; Il6 avg. FC = 8.8 ± 1.4, adj. p = 0.003, Ccl5 FC = 7.9 ± 4.0, adj. p = 0.06; Cxcl10 avg. FC = 213.5 ± 12.3, adj. p = 3.1 × 10−07; Ikke avg. FC = 1.9 ± 0.1, adj. p = 0.01; Cxcl9: 669.5 ± 187.9, adj. p = 0.004; Jak1 FC = 0.7 ± 0.1, adj. p = 0.3; Irf1 avg. FC = 50.3 ± 3.1, adj. p = 6.05 × 10−05. b) Levels of select secreted cytokines/chemokines and other inflammatory mediators were measured in the conditioned media from E3 and E4 microglial cultures using a Cytokine Proteome Profiler array kit. E4 microglia showed significantly increased levels of specific cytokines/chemokines compared to E3: unpaired Student’s t-test followed by BH correction for multiple comparisons, IL1α t(8) = 4.7, adj. p = 0.01, d = 3.0 CI95% [1.0, 4.8]; CCL2 t(8) = 4.2, adj. p = 0.02, d = 2.7 CI95% [0.8, 4.4]; CCL12 t(8) = 4.1, adj. p = 0.02, d = 2.6 CI95% [0.8, 4.3]; CCL3 t(8) = 5.8, adj. p = 0.007, d = 3.6 CI95% [1.5, 5.7]; CCL4 t(8) = 5.3, adj. p = 0.007, d = 3.4 CI95% [1.3, 5.4]; CXCL2 t(8) = 5.3, adj. p = 0.007, d = 3.4, CI95% [1.3, 5.4]; CCL17 t(8) = 1.5, adj. p = 0.8, d = 1.0 CI95% [−0.4, 2.3]; TNFα t(8) = 5.3, adj. p = 0.007, d = 3.3 CI95% [1.2, 5.3], n = 5 independent experiments. c) Levels of secreted interferons in the supernatant of E3 and E4 microglia cultures were measured using ELISA. E4 microglia showed no significant change in interferon-α/β; t(3.8) = −0.6, p = 0.6, d = −0.5 CI95% [−1.8, 1.0], but exhibited a significant increase in interferon-ɣ t(2) = 10.8, p = 0.008, d = 8.8 CI95% [2.8, 14.9] relative to E3 microglia. d) Protein synthesis was measured in E3 and E4 microglia using the SUnSET assay, which detects levels of puromycin incorporated into newly synthesized proteins. A representative western blot is shown. M indicates the molecular weight ladder and both short exposure (top) and longer exposure (bottom) are displayed. e) Original non-over exposed western blot (top) was quantified and shows decreased puromycin incorporation in E4 microglia at 1 hour compared to E3, t(3.0) = −8.6, p = 0.003, d = −6.1 CI95% [−10.9, −1.4]. f) Representative Western blots showing the protein levels of both total and phosphorylated eIF2α, PRK, PERK, and GCN2 protein. g) Quantification of western blots shows E4 microglia exhibit a significant increase in the ratio of phosphorylated to total eIF2, t(4.8) = 4.6, p = 0.007, d = 3.2 CI95% [0.8, 5.6] and PKR; t(3.0) = 8.6, p = 0.04, d = 2.5 CI95% [0.1, 4.9] compared to E3 microglia. The ratio of p-PERK to total PERK was increased but did not reach statistical significance, t(3.0) = 2.7, p = 0.07, d = 1.9 CI95% [−0.2, 3.9] while the percentage of GCN2 that was phosphorylated remained unchanged, t(3.3) = 0.1, p = 0.92, d = 0.1 CI95% [−1.3, 1.5]. a, c, e, g) Each dot corresponds to an independent experiment (n = 4/group). Black horizontal bars indicate the mean and error bars represent SE. c, e,g) Unpaired Welch’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001.
2.16. Interferon ELISA assay
To perform ELISA assays from conditioned media, microglia were plated at 60,000 cells/well in a 96 well plate and cultured for 24 hours. Conditioned media was collected after 24 hours, cleared of cell debris with 10 min centrifugation at 3,000 rpm. Interferon levels were measured using a mouse interferon-α/β (Thermo Fisher # 23225) and interferon-ɣ (Thermo Fisher # EM39RB) ELISA kits according to the manufacturers’ protocols.
2.17. Surface sensing of translation (SUnSET) assay
To measure protein synthesis in primary microglia, puromycin (2 μg/ ml) (Sigma-Aldrich # P9620) was applied for 1 hour (Schmidt et al., 2009). Cultured microglia were collected, lysed and used for Western blot. Puromycin incorporation into proteins was detected using mouse anti-puromycin antibody (1:1000, Millipore, MABE343).
2.18. Statistics
Data were analyzed and visualized using GraphPad Prism 9 (GraphPad Software, n.d) and R programming environment (version 3.6.3). For statistical inference, three to five independent experiments were performed for each genotype, using microglia isolated and pooled from all pups in a single litter for each experiment. Two technical replicates were averaged within each experiment to obtain a single value. Statistical tests used for given comparisons are detailed in the corresponding figure legend.
3. Results
3.1. E4 microglia exhibit altered morphology, increased lipid accumulation, and increased endolysosomal mass relative to E3 microglia at baseline
To investigate the impact of APOE ε4/ε4 (E4) genotype on microglial phenotypes relative to ε3/ε3 (E3) genotype, we cultured primary microglia from forebrains of postnatal day 1–3 (P1–3) homozygous APOE4 and APOE3 TR mice. A highly pure microglia population was obtained after 28 days in culture (Lian et al., 2016), with no statistically significant differences in percentage of cells expressing the microglial marker IBA1 between genotypes (Figure S1a). Similar numbers of viable cells were obtained across genotypes (Figure S1b). As previously reported (Vitek et al., 2009), E4 microglia exhibited a smaller overall size, with E3 microglia having a significantly larger area, length, and average branch length (Figure S1c). In addition, E4 microglia exhibited significantly higher levels of intracellular APOE protein and reduced levels of secreted APOE detectable in the supernatant as compared to E3 microglia (Figure S2).
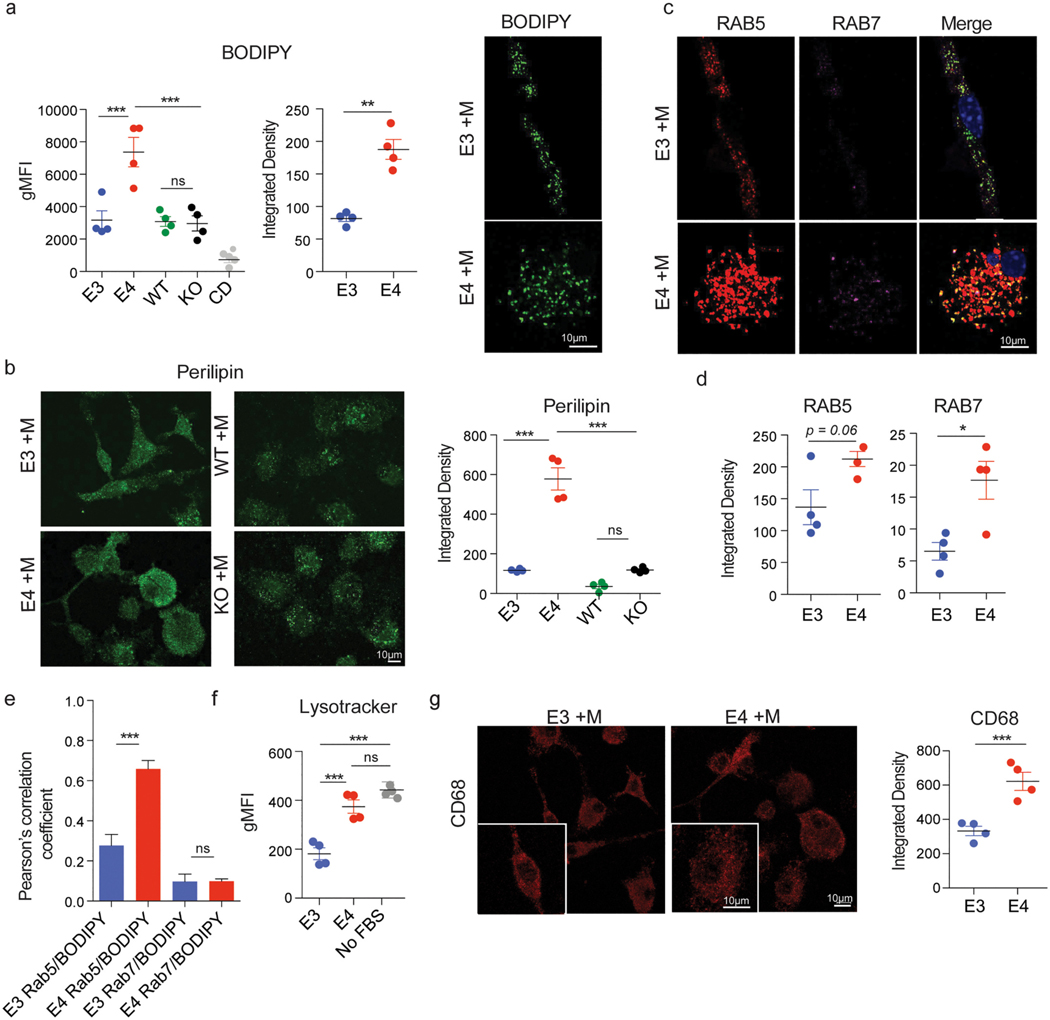
APOE is known to play a central role in lipid/cholesterol metabolism. To investigate whether APOE genotype alters the levels of neutral lipids (e.g., cholesterol esters and triglycerides) in primary microglia, we first stained them using the lipophilic dye BODIPY [see Methods, (Qiu and Simon, 2016)]. Quantification by either fluorescent microscopy staining or flow cytometry indicated E4 microglia have significantly higher levels of neutral lipids when compared to E3 microglia (Fig. 1a, b). Additionally, E4 microglia incorporated significantly higher levels of a fluorescently labeled cholesterol analog (Fig. 1c) in serum-free media conditions, suggesting these baseline alterations were not due to a genotype-dependent differential uptake of serum-derived lipids.
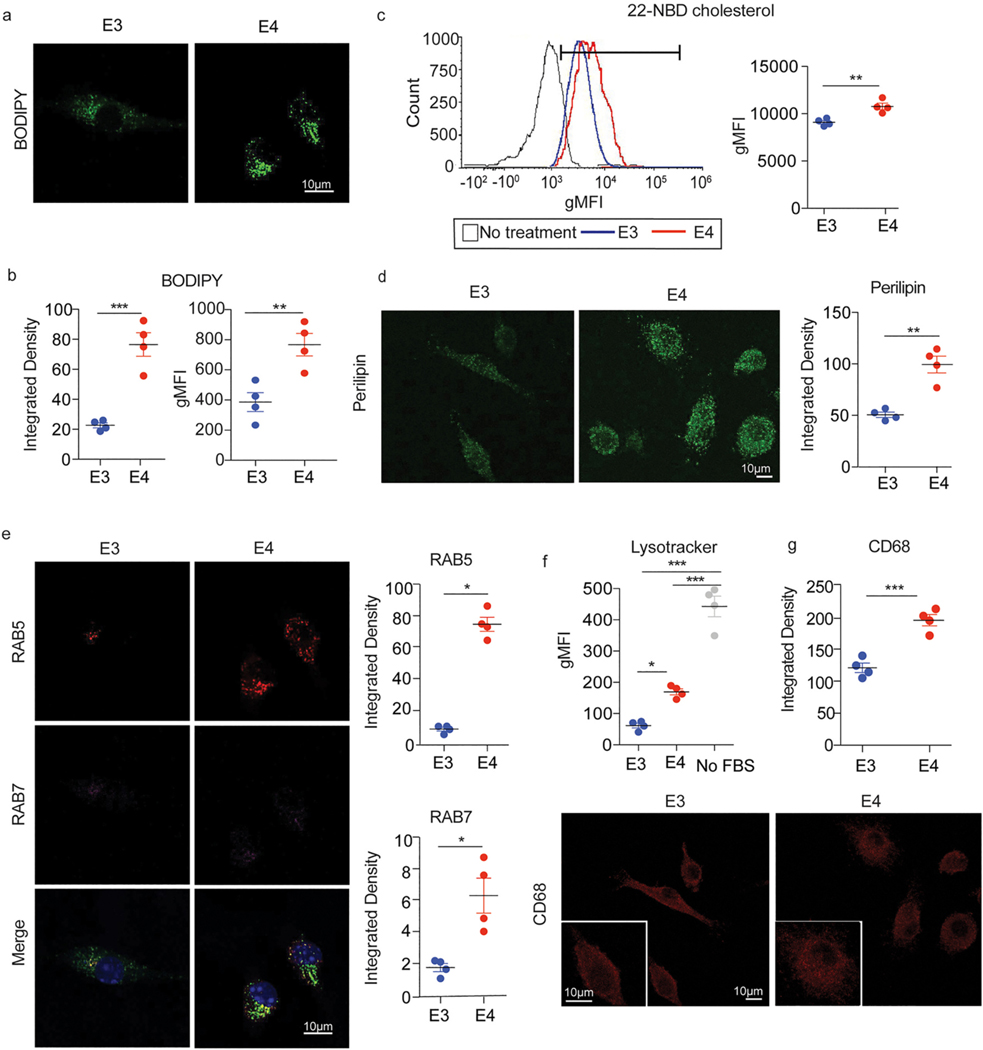
Fig. 1.
E4 microglia exhibit increased lipid accumulation and endolysosomal mass relative to E3 microglia at baseline. a) BODIPY staining was used to visualize the levels of neutral lipids and b) the average integrated density of BODIPY staining per cell was quantified in ImageJ (left, t(4.1) = 8.4, p = 0.0009, d = 6.0, CI95% [1.8, 10.1]) as well as by measuring geometric mean fluorescence intensity (gMFI) by flow cytometry (right, t(5.8) = 3.9, p = 0.008, d = 2.8, CI95% [0.6, 4.8]). c) Cellular cholesterol was measured by treating cells with a fluorescently labeled cholesterol analog (22-NBD cholesterol) overnight in a serum free condition (see Methods). Cholesterol loading was quantified by measuring gMFI via flow cytometry, t(4.7) = 4.3, p = 0.009, d = 3.1, CI95% [0.7, 5.4]. d) Average integrated density of perilipin immunostaining per cell was used to quantify the levels of lipid droplets within microglia, t(3.6) = 5.7, p = 0.006, d = 4.0 CI95% [0.9, 7.1]. e) Levels of early endosome or late endosome markers RAB5 and RAB7, respectively, were measured by quantifying the average integrated density of immunostaining per cell. RAB5: t (5.4) = 2.8, p = 0.04, d = 10.1 CI95% [2.9, 17.5], RAB7: t(3.3) = 4.0, p = 0.02, d = 2.8 CI95% [0.3, 5.3]. Merged image is an overlay of RAB5 (red), RAB7 (magenta), DAPI (blue), and BODIPY from (a) in green. f) The mass of lysosomes within microglia was quantified by measuring the gMFI of LysoTracker dye by flow cytometry, one-way ANOVA main effect of APOE genotype: F(2, 9) = 93.6, p = 9.5 × 10−07 followed by Tukey’s post-hoc corrected t-test E4 vs E3: p.adj = 0.01, d = 6.1 CI95% [2.5, 9.8]. Serum-starved (no FBS) cells were used as a positive control (see Methods). g) CD68 immunostaining was used as an alternative measure of lysosomal mass, t(5.8) = 6.4, p = 0.0007, d = 4.6, CI95% [1.6, 7.4]). b-e, g) Unpaired Welch’s t-test. b-g) Each dot corresponds to an independent experiment (n = 4/group). Black horizontal bars indicate the mean and error bars represent SE. *p < 0.05, **p < 0.01, ***p < 0.001.
We next investigated whether increased levels of neutral lipids corresponded to increased levels of lipid droplets, organelles where neutral lipids are normally stored (Olzmann and Carvalho, 2019). Indeed, E4 microglia exhibited significantly higher levels of lipid droplets relative to E3 microglia, as indicated by increased integrated density of fluorescent immunostaining of the lipid droplet surface protein perilipin (Fig. 1d), suggesting at least some of the neutral lipids were being sequestered within these organelles (Marschallinger et al., 2020). In addition, the levels of both early endosome (RAB5) and late endosome (RAB7) markers were increased in E4 microglia relative to E3 microglia (Fig. 1e), which may be reflective of an increased number and/or size of endosomes. These increased levels of endosomal markers were accompanied by a significant increase in lysosomal mass as quantified by LysoTracker flow cytometry [Fig. 1f, (Luzio et al., 2007)]. Serum-starved cells were used as a positive control for this experiment, as removal of FBS from the media is known to induce autophagic processing and increase lysosomal mass (Bampton et al., 2005). In addition, integrated density of fluorescent immunostaining of the lysosome-associated protein CD68 was increased in E4 microglia relative to E3 microglia, further demonstrating an increase in lysosomal mass (Fig. 1g). We then further used LysoSensor dye to measure the pH of lysosomes, which were significantly more acidic in E4 microglia when compared to E3 microglia (Figure S3). These data demonstrate E4 microglia have higher load of neutral lipids and lipid droplets relative to E3 microglia, together with increased mass and acidification of endolysosomes, at baseline.
3.2. E4 microglia exhibit increased phagocytic uptake of diverse substrates
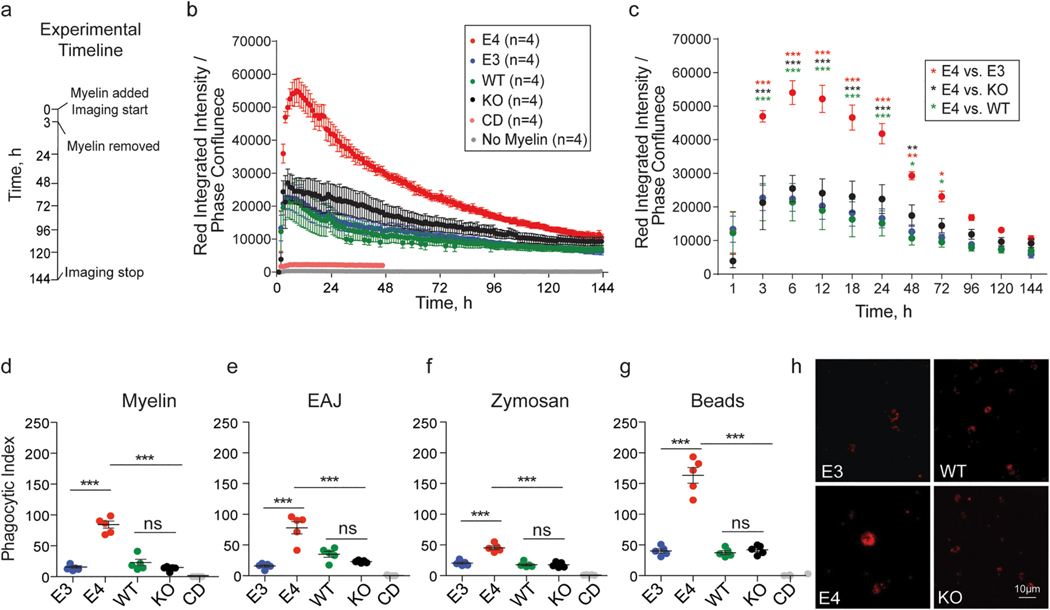
To examine how these baseline changes altered the functionality of E4 microglia relative to E3 microglia, we next assessed the ability of these cells to phagocytose and clear lipid/cholesterol-rich cellular debris. Specifically, we treated both E3 and E4 microglia as well as microglia with mouse Apoe (wild-type, WT) or lacking Apoe (knock-out, KO) with myelin fragments (see Methods) labeled with pHrodo red (Sullivan et al., 1997). pHrodo is a pH-sensitive dye which increases in fluorescence intensity as it enters acidic environments such as endosomes and lysosomes (Czaplinska et al., 2019´ ), allowing for the internalization of labeled substrates to be visualized over time. pHrodo- labeled myelin was applied to microglia for 3 hours and the red fluorescent signal was captured using the IncuCyte live imaging system every hour for 6 days (Fig. 2a), a time frame that allowed us to monitor acute uptake as well as longer-term degradation of internalized myelin. E4 microglia exhibited significantly higher integrated intensity of red fluorescence relative to E3 microglia as well as microglia from mice homozygous for the WT or KO allele of mouse Apoe (Fig. 2b). Specifically, a significant effect of time and genotype was observed on the integrated intensity of red fluorescence. To determine the time point(s) at which this difference occurs, we performed post-hoc testing on E4 vs E3 at select time points (Fig. 2c). We observed increased red fluorescence in E4 microglia as early as 3 hours post myelin application, immediately following myelin washout, suggestive of increased phagocytic uptake of myelin debris by E4 microglia. Notably, red fluorescence was also increased in E4 microglia relative to KO microglia, suggesting the E4 allele does not phenocopy loss-of-APOE function in this assay (Fig. 2c). Increased red fluorescence was confirmed using flow cytometry at 24 hours (Fig. 2d). Levels of myelin basic protein (MBP) were also increased at 24 hours in cell lysates from E4 microglia (Figure S4a), further confirming our findings. We also observe a significantly longer half-life of red fluorescence decay in E4 microglia relative to E3, WT and KO microglia, which were not significantly different from one another (Figure S4b, see Methods). This suggests that in addition to exhibiting increased phagocytic uptake, E4 microglia may exhibit impaired ability to degrade internalized myelin.
Fig. 2.
E4 microglia exhibit increased phagocytic uptake of diverse substrates. a) Experimental timeline: E3, E4, mouse Apoe (wild-type, WT), and homozygous null Apoe (KO) microglia were treated with substrate (or left untreated) for three hours. Imaging continued for up to six days. b) Red fluorescence was measured using the IncuCyte ZOOM live imaging system every hour. Integrated intensity of the red fluorescent signal [measured as red calibrated units (RCU) x μm2/image] was normalized to phase confluence to obtain a quantitative readout of red fluorescent substrate internalization. Cytochalasin D (CD) treatment in WT cells was used as a negative control. Mixed-effects analysis (restricted maximum likelihood as implemented in GraphPad Prism 9; fixed factors: time and genotype, random factor: experiment) identified a main effect of time F(1.7, 18.4) = 62.0, p = 3.1 × 10−07, a main effect of APOE genotype F(3,11) = 14.7, p = 0.0004, and an interaction between the two F(432, 1538) = 7.3, p = 1.7 × 10−187. c) Post-hoc t-tests at select time points identified significant differences (FDR adj. p-value = q < 0.05) between E4 microglia and microglia of other genotypes as early as 3 hours (E4 vs E3: q = 2.2 × 10−06, d = 3.7 CI95% [0.7, 6.6]; E4 vs KO: q = 1.1 × 10−06, d = 2.2 CI95% [0.3, 4.0], E4 vs WT, q = 2.6 × 10−06, d = 3.9 CI95% [1.1, 6.6]) and up to 72 hours (E4 vs E3: q = 0.001, d = 4.7 CI95% [1.7, 7.6], E4 vs KO, q = 0.1, d = 2.3 CI95% [0.4, 4.1], E4 vs WT, q = 0.03, d = 4.6 CI95% [1.4, 7.7]) post-treatment. d-g) Phagocytic index at 24 hours post-treatment was measured using flow cytometry (see Methods). pHrodo red-conjugated myelin: F(4, 20) = 80.1, p= 5.1 × 10−12, E4 vs E3: adj. p = 1.8 × 10−10, d = 7.2 CI95% [3.5, 10.9], E4 vs KO: adj. p = 1.4 × 10−10, d = 7.4 CI95% [3.7, 11.2], E4 vs WT: adj. p = 1.4 × 10−09, d = 5.0 CI95% [2.3, 7.7]; early apoptotic Jurkat cells (EAJ): F(4, 20) = 33.4, p = 1.3 × 10−08, E4 vs E3: adj. p = 1.8 × 10−10, d = 3.9 CI95% [1.6, 6.1], E4 vs KO: adj. p = 1.4 × 10−10, d = 3.5 CI95% [1.4, 5.5], E4 vs WT: adj. p = 1.4 × 10−09, d = 2.5 CI95% [0.7, 4.1]; zymosan bioparticles: F(4, 20) = 64.2, p = 4.0 × 10−11, E4 vs E3: adj. p = 2.0 × 10−07, d = 4.5 CI95% [2.0, 7.0], E4 vs KO: adj. p = 3.7 × 10−08, d = 5.0 CI95% [2.3, 7.7], E4 vs WT: adj. p = 4.4 × 10−08, d = 5.0 CI95% [2.2, 7.6]; red fluorescent carboxylate-modified polystyrene latex beads: F(4, 20) = 99.2, p = 6.8 × 10−13, E4 vs E3: adj. p = 8.6 × 10−11, d = 5.9 CI95% [2.8, 9.0], E4 vs KO: adj. p = 1.1 × 10−10, d = 5.8 CI95% [2.7, 8.9], E4 vs WT: adj. p = 5.8 × 10−11, d = 6.0 CI95% [2.9, 9.2]. h) Representative images of red fluorescent beads internalized 24 hours post-treatment. Scale bar 10μm. b-c) Each dot corresponds to the average of 3–5 independent experiments, error bars represent ± SE. d-g) One-way ANOVA followed by Tukey’s post-hoc corrected t-test. Each dot corresponds to an independent experiment (n = 5/group). Black horizontal bars indicate the mean and error bars represent SE. *p < 0.05, **p < 0.01, ***p < 0.001.
To examine whether increased levels of phagocytosed substrate in E4 microglia is specific to myelin, we treated microglia with pHrodo red- conjugated early apoptotic Jurkat (EAJ) cells, pHrodo red-conjugated zymosan particles, or 0.9 μm red fluorescent polystyrene carboxylated latex beads. As we observed with myelin, E4 microglia exhibited significantly higher levels of red fluorescence at 24 hours post-treatment relative to E3, WT and KO microglia (Fig. 2e–g) across all substrates, suggesting a mechanism that is not target- or receptor-specific. Furthermore, since latex beads cannot be degraded, these results further suggest that the observed increase in red fluorescent signal is not solely due to differences in degradation across genotypes and is, at least in part, due to an increase in phagocytic uptake in E4 microglia.
3.3. E4 microglia exhibit increased lipid accumulation and endolysosomal mass following myelin challenge
We next sought to test whether E4 microglia exhibit differences in accumulation of phagocytosed materials, particularly myelin, compared to E3 microglia. We again used BODIPY fluorescent staining to measure neutral lipid levels in E4 and E3 microglia following a 3-hour myelin challenge identical to that described above for the phagocytosis assay. At 24 hours post-myelin application, E4 microglia showed significantly higher levels of neutral lipids relative to E3 microglia as indicated by increased BODIPY fluorescence, measured by both flow cytometry and fluorescence microscopy (Fig. 3a). Notably, the intensity of BODIPY staining was significantly increased from pre-myelin treatment levels across both genotypes (Fig. 1a, BODIPY gMFI E3 +M vs E3: t(3.1) = 4.7, p = 0.02, d = 3.4 CI95% [0.5, 6.2], E4 +M vs E4 t(3.0) = 7.3, p = 0.005, d = 5.1 CI95% [1.1, 9.3]), further confirming an accumulation of neutral lipids intracellularly as a result of myelin challenge.
Fig. 3.
E4 microglia exhibit increased neutral lipid accumulation and endolysosomal mass 24h after myelin challenge. a) Left, geometric mean fluorescence intensity (gMFI) of BODIPY was quantified by flow cytometry, F(4, 15) = 19.3, p = 8.8×10−06, E4 vs E3: adj. p = 0.0006, d = 2.8, CI95% [0.7, 4.8]. Middle, BODIPY staining was used to visualize neutral lipids and the average integrated density per cell was quantified in ImageJ, t(3.6) = 6.6, p = 0.004, d = 4.7 CI95% [1.1, 8.2]. Right, representative image of BODIPY staining. b) Average integrated density of perilipin immunostaining per cell was used to quantify levels of lipid droplets, F(3, 12) = 73.4, p = 5.5 × 10−08, E4 vs E3: adj. p = 5.0 × 10−07, d = 5.8 CI95% [2.3, 9.2]. c, d) Average integrated density per cell of RAB5 and RAB7 immunostaining was used to quantify the mass of early and late endosomes, respectively. RAB5: t(4.1) = 2.5, p = 0.06, d = 1.8 CI95% [0.1, 3.6], RAB7: t(4.2) = 3.4, p = 0.02, d = 2.4 CI95% [0.3, 4.4]. Merged image is an overlay of RAB5 (red), RAB7 (magenta), DAPI (blue), and BODIPY from (a) in green. e) Colocalization analysis (see Methods) was used to assess the overlap between BODIPY staining and immunostaining of RAB5 and RAB7. Pearson’s correlation coefficients were converted to Z scores using the Fisher’s r-to-z transformation and compared across groups using two-sample z-test, RAB5/BODIPY: E4 vs E3, Z = −5.3, p = 1.2 × 10−07, d = 3.8 CI95% [1.2, 6.2], RAB7/ BODIPY: E4 vs E3 Z = −0.08, p = 0.97, d = 0.02, CI95% [−1.4, 1.4,]. f) The mass of lysosomes within microglia was quantified by measuring the gMFI of LysoTracker dye by flow cytometry, F(2, 9) = 22.8, p = 0.0003, E4 vs E3: p = 0.002, d = 3.7 CI95% [1.2, 6.2]. g) CD68 immunostaining was used as an alternative measure of lysosome mass. Average integrated density of CD68 staining per cell was quantified in ImageJ, t(5.8) = 4.7, p = 0.004, d = 3.3, CI95% [1.0, 5.6]. a-g) Each dot corresponds to an independent experiment (n = 4/group). Black horizontal bars indicate the mean and error bars represent SE.a (left), b, e-f) One-way ANOVA followed by Tukey’s post-hoc corrected t-test. a (middle), d, g) Unpaired Welch’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001.
To further investigate the localization of these accumulated lipids, we again measured levels of the lipid droplet protein perilipin using immunostaining. Similar to what was observed at baseline, E4 microglia exhibited significantly higher levels of perilipin following myelin challenge, reflecting increased levels of lipid droplets in E4 microglia as compared to E3, WT, or KO microglia (Fig. 3b). E4 microglia again displayed higher levels of both RAB5 and RAB7 relative to E3 microglia (Fig. 3c, d). Levels of both proteins were significantly increased from pre-treatment levels in E3 and E4 microglia (RAB5 E3 +M vs E3: t(3) =4.6, p = 0.02, d = 3.3 CI95% [0.4, 6.1]; E4 +M vs E4: t(3.9) = 10.3, p =0.0006, d = 7.3 CI95% [2.3, 12.4]; RAB7 E3 +M vs E3: t(3.2) = 3.2, p =0.04, d = 2.3, CI95% [0.1, 4.4]; E4 +M vs E4: t(3.9) = 3.4, p = 0.03, d = 2.4 CI95% [0.2, 4.5]), suggesting microglia of both genotypes were able to increase the mass of early and late endosomes following myelin challenge. A significantly higher degree of colocalization of BODIPY with RAB5 was observed in E4 microglia relative to E3 microglia (Fig. 3e), suggesting E4 microglia accumulated neutral lipids in early endosomes or other RAB5-positive organelles to a greater extent than E3 microglia after challenge. Low levels of colocalization between BODIPY and RAB7 were observed and no statistically significant difference across genotypes could be inferred (Fig. 3e), suggesting a portion of neutral lipids accumulated in early endosomes or other RAB5-positive organelles rather than RAB7-positive organelles.
We next measured the mass and acidity of microglial lysosomes following myelin challenge. E4 microglia again exhibited increased lysosomal mass relative to E3 microglia as measured by both Lyso-Tracker gMFI via flow cytometry (Fig. 3f) and CD68 immunostaining integrated density via fluorescence microscopy (Fig. 3g). Lysosomal pH was then measured using LysoSensor dye (Ma et al., 2017). Following treatment with myelin, E3 microglia significantly acidified their lysosomes (Figure S3). Interestingly, a similar decrease in lysosomal pH was observed in E4 microglia relative to E3 microglia at baseline. As lysosome function is optimal at lower pH, these effects may reflect activation of the lysosomal degradation machinery to cope with the increased burden of internalized debris following myelin challenge in E3 microglia, or in E4 vs E3 microglia even before myelin challenge. E4 microglia did not exhibit further acidification following myelin challenge but rather a slight (albeit not significant) increase of their lysosomal pH (Figure S3). As lipid overload is known to impair lysosomal function by multiple mechanisms, including by increasing lysosomal pH (Jaishy et al., 2015; Jaishy and Abel, 2016), this increase may reflect an impairment of E4 microglia to digest internalized debris as suggested by increased half-life of internalized myelin (Figure S4).
3.4. E4 microglia exhibit differential enrichment of genes in interferon signaling, extracellular matrix, and translation-related pathways at baseline
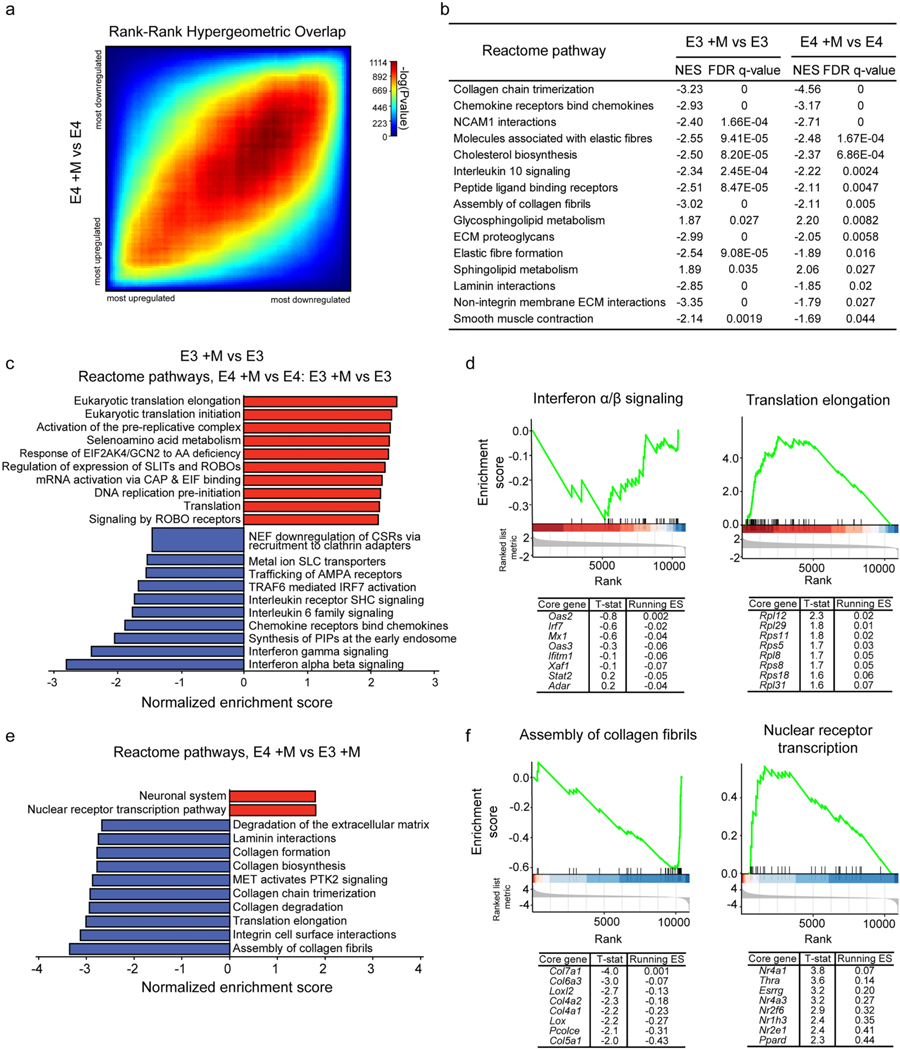
In order to investigate the molecular mechanisms by which E4 genotype may lead to the cellular alterations described above, we performed RNA sequencing of our primary microglia cultures. Specifically, microglia were either left untreated or were challenged with myelin for 3 hours as in our phagocytosis experiment and were collected 21 hours later for RNA-sequencing. Principal component analysis (PCA) of the top 500 most variable genes showed first that E3 and E4 microglia cluster distantly from those with mouse Apoe (WT) or no Apoe (KO), demonstrating a significant impact of mouse and human APOE genotypes on microglial gene expression profiles (Fig. 4a). Even when considering E3 and E4 microglia alone, samples still clearly clustered by genotype (Fig. 4b), suggesting APOE genotype alters the transcriptional profile of microglia. To test this hypothesis, we performed differential gene expression analysis across genotypes at baseline (Ritchie et al., 2015). A relatively small number of genes were significantly differentially expressed in E4 vs E3 microglia at an FDR adj. p-value < 0.05 (Table S2). Overrepresentation enrichment analysis (Liao et al., 2019) using the 125 genes significantly downregulated in E4 microglia relative to E3 microglia identified a significant overrepresentation (FDR adj. p- value < 0.05) of genes belonging to Reactome pathways “ECM organization”, “smooth muscle contraction”, “collagen chain trimerization”, “elastic fibre formation”, and “NCAM1 interactions” (Fig. 4c, (Croft et al., 2014). No significant functional enrichment among the 21 significantly upregulated genes was observed.
Next, we performed gene set enrichment analysis (GSEA) in order to gain a more complete understanding of the biological pathways altered by APOE genotype at the transcriptional level (Subramanian et al., 2005). A large number of Reactome pathways emerged as significantly impacted (FDR adj. p-value < 0.25) by APOE genotype (Table S3, Fig. 4d), so we used the Cytoscape Enrichment Map (Merico et al., 2010) and AutoAnnotate (Kucera et al., 2016) applications to unbiasedly cluster pathways based on member genes (Fig. 4e). Pathways enriched for genes at the top of the GSEA ranked list (i.e., genes with a positive fold change) included “interferon alpha/beta signaling” and “interferon gamma signaling” (Fig. 4d), suggesting E4 microglia exhibit an increase in interferon signaling relative to E3 microglia. Interestingly, genes significantly differentially expressed in E4 vs E3 microglia at a nominal p-value < 0.05 significantly overlapped with those identified as representative of an interferon-enriched subcluster of microglia in vivo [hypergeometric test, p = 0.02, data from Supplementary Data 5 from (McQuade et al., 2020)]. Examination of the GSEA output in more detail identified genes identified by GSEA as core drivers of “interferon alpha/ beta signaling” pathway [Fig. 4f, see Methods and (Subramanian et al., 2005)], including Oas1g and Oas3 which are antiviral proteins as well as interferon-responsive genes (Lee et al., 2019). Oas1g and Oas3 are known to alter expression of chemokines (Lee et al., 2019). In addition, OAS1 was recently identified as a putative AD risk gene based on its localization to an amyloid-responsive network in mouse microglia and association with AD risk variants identified by GWAS (Salih et al., 2019).
Similar to our previous overrepresentation enrichment analysis (Fig. 4c), GSEA identified pathways related to the extracellular matrix, including “collagen formation”, “collagen degradation” as well as “ECM organization” as enriched for genes at the bottom of the ranked list (i.e., those with a negative fold change). Additionally, pathways related to translation, including “translation elongation” and “translation initiation” were identified as those most significantly enriched (FDR adj. p-value < 0.01) among genes downregulated in E4 vs E3 microglia (Fig. 4d). This enrichment was driven by a large number of genes encoding ribosomal proteins (Fig. 4g) and components of the translation machinery (i.e., elongation factor 1-gamma, Eef1g). Translation is an energetically expensive process, which is often inhibited in times of cellular stress and/or inflammation (Advani and Ivanov, 2019; Mazumder et al., 2010), further suggesting E4 microglia are stressed relative to E3 microglia even under basal conditions, as reflected by their amoeboid morphology (Figure S1) (Martina et al., 2016) and increased lysosomal mass (Fig. 1f and Figure S3b, c) (Jurga et al., 2020).
3.5. E4 microglia exhibit increased interferon signaling, increased eIF2 signaling, and decreased translation at baseline
We next wanted to validate our top findings from RNA-sequencing. As the top upregulated pathways in E4 vs E3 microglia at baseline included both interferon-α/β and γ signaling, we first selected key genes from each pathway to measure via RT-qPCR. As expected, several genes involved in interferon-γ signaling including Oas2 and Oas3 showed significant increases in mRNA levels in E4 microglia compared to E3 microglia (Fig. 5a). In addition, general interferon responsive genes (e.g. Irf1, Irf8, Irf7) which are downstream of both types of interferon signaling were also upregulated in E4 microglia compared to E3 microglia (Fig. 5a). Alterations in interferon signaling can induce a wide range of effects, including altered secretion of various pro- or anti- inflammatory cytokines and chemokines (Bhat et al., 2018; Bolívar et al., 2018). To investigate the functional impact of elevated interferon signaling in E4 vs E3 microglia, we collected the supernatant from E3 and E4 microglia and used a proteome profiler array kit to measure the levels of 40 cytokines, chemokines and other inflammatory mediators in E4 vs E3 microglia. Compared to E3 microglia, E4 microglia showed significantly increased secretion of specific pro-inflammatory cytokines such as TNFα (Fig. 5b) as well as CC family chemokines which are known to be induced by interferon signaling (Lehmann et al., 2016; Raport and Gray, 2010; Rauch et al., 2013). Specifically, E4 microglia showed increased secretion of chemokine ligands including CCL3, CCL4, and CXCL2 (Fig. 5b). Finally, we measured levels of interferon proteins themselves in order to determine which specific interferon may be driving these observed changes. We measured interferon-⍺/β and interferon-ɣ levels using ELISA in the supernatant collected from both E3 and E4 microglia following 24 hours in culture. Interestingly, E4 microglia showed no difference in interferon-α/β levels relative to E3 microglia (Fig. 5c, top). However, interferon-ɣ levels were significantly elevated (Fig. 5c, bottom) in E4 microglia compared to E3 microglia, suggesting that interferon-ɣ is the cytokine most likely responsible for the increased inflammatory state observed in E4 microglia compared to E3 microglia at baseline.
The most significantly negatively enriched pathways in E4 vs E3 microglia at baseline were related to translation, including “translation elongation” and “translation initiation”. To test the hypothesis that these transcriptional changes did indeed lead to changes in protein synthesis, we used the SUnSET assay which measures incorporation of puromycin into newly synthesized polypeptides (Schmidt et al., 2009). Specifically, E3 and E4 microglia were treated with puromycin for 1 hour, after which cells were collected, proteins isolated, and levels of puromycin incorporation measured by western blot. While puromycin incorporation was dramatically decreased in E4 microglia compared to E3 microglia (Fig. 5d, top and 5e), prolonged exposure of the blot clearly showed some puromycin incorporation in E4 microglia (Fig. 5d, bottom). Together, these results demonstrate E4 microglia synthesize proteins at a lower rate compared to E3 microglia under baseline culture conditions.
To further investigate the cause of this decrease in protein synthesis, we next measured the activation state of the alpha subunit of eukaryotic initiation factor-2 (eIF2α), as phosphorylation of this regulatory subunit precludes the recruitment and binding of other eIF subunits, thereby inhibiting general initiation of translation (Adomavicius et al., 2019). The ratio of phosphorylated eIF2α (p-eIF2α) to total eIF2α was significantly higher in E4 microglia compared to E3 microglia, suggestive of activation of the integrated stress response via eIF2α (Taniuchi et al., 2016) (Fig. 5f, g). We next investigated the activation state of three kinases known to phosphorylate eIF2α: protein kinase R (PKR), PKR-like endoplasmic reticulum kinase (PERK), and general control non- depressible 2 (GCN2) (Gal-Ben-Ari et al., 2018). As each kinase is activated by phosphorylation (Adomavicius et al., 2019; Chesnokova et al., 2017), we measured levels of both the total and phosphorylated forms of each protein. Among the three kinases, the ratio of phosphorylated PKR (p-PKR) to total PKR was significantly elevated in E4 microglia compared to E3 microglia (Fig. 5f, g). The ratio of phosphorylated PERK (p-PERK) to total PERK was also elevated in E4 microglia compared to E3 microglia, however the mean difference was not statistically significant (Fig. 5f, g). On the contrary, the ratio of phosphorylated GCN2 (p- GCN2) to total GCN2 was not elevated in E4 microglia compared to E3 microglia (Fig. 5f, g). Together, these results validate our RNA sequencing findings and suggest E4 microglia exhibit higher levels of cellular stress relative to E3 microglia at baseline as indicated by reduced translation coupled with activation of eIF2α kinases.
3.6. E4 and E3 microglia exhibit both shared and unique transcriptional responses to myelin challenge
We next sought to characterize the transcriptional response of microglia from each genotype to myelin treatment. Differential gene expression analysis comparing the within-genotype response to myelin was performed (Ritchie et al., 2015). Rank-rank hypergeometric overlap (RRHO) testing (Plaisier et al., 2010) (see Methods) revealed a large degree of similarity in the response of E4 and E3 microglia to myelin (Fig. 6a, Spearman rho = 0.6, p < 2.2 × 10−16). To evaluate the pathways commonly altered after exposure to myelin across genotypes, we compared GSEA results from each of the response profiles. Several pathways were significantly enriched across both E4 and E3 microglia responses to myelin challenge (FDR adj. p-value < 0.05) (Fig. 6b). Notably, microglia of both genotypes downregulated the expression of “cholesterol biosynthesis” genes, suggesting they are appropriately responding to overload of myelin-derived cholesterol by reducing internal biosynthesis. Several ECM-related pathways were also enriched for downregulated genes, including “collagen chain trimerization”, “assembly of collagen fibrils”, and “ECM proteoglycans” (Fig. 6b).
Fig. 6.
E4 and E3 microglia exhibit both shared and unique transcriptional responses to myelin challenge. a) Rank-rank hypergeometric overlap plot comparing the response of each genotype to myelin treatment (E4 +M vs E4 and E3 +M vs E3). b) GSEA-identified pathways significantly enriched (FDR adj. p-value < 0.05 and normalized enrichment score, NES, < −1.5 or > 1.5) in both comparisons. c) GSEA was performed on a ranked list generated from a two-factor differential gene expression analysis (E4 +M vs E3 +M relative to E4 vs E3) aimed at identifying genes for which expression changes in response to myelin challenge showed a significant interaction with APOE genotype. The top 10 pathways enriched for genes either at the top or bottom of the ranked list are plotted here. D) GSEA enrichment plots for “interferon alpha/beta signaling” and “translation elongation”, the most significantly enriched pathways at the bottom and top of the ranked list, respectively. The mouse orthologues of the top 8 core genes identified by GSEA are listed below, along with the t-statistic of their differential expression and running enrichment score (ES) at their position. e) GSEA was performed on the ranked list from one-factor differential gene expression analysis following myelin challenge (E4 +M vs E3 +M) and significantly enriched pathways (FDR adj. p-value < 0.25) were identified and plotted. f) GSEA enrichment plots and top core genes for the “assembly of collagen fibrils” and “nuclear receptor transcription” pathways as in (d). Detailed results available in Table S4 and Table S5.
While E3 and E4 microglia responded similarly to myelin challenge, we wanted to better understand which, if any, pathways were differentially regulated in E4 vs E3 microglia in response to myelin. To identify genes differentially regulated by genotype in response to myelin, we performed a two-factor differential gene expression analysis (Ritchie et al., 2015). Specifically, we compared the genes differentially expressed post-myelin treatment (E4 +M vs E3 +M) to those differentially expressed pre-myelin treatment (E4 vs E3) to identify statistical interactions between genotype and myelin treatment on gene expression (Table S2). Since no genes showed an FDR adj. p-value < 0.05, we again performed GSEA analysis using the ranked two-factor DEG list to gain insight into pathways that may be differentially regulated in E4 vs E3 microglia in response to myelin. Negatively enriched pathways included interferon response-related pathways (“interferon alpha/beta signaling”, and “interferon gamma signaling”) as well as “PIP synthesis in the early endosome” (Fig. 6c and Table S4). Given the nature of the two-factor analysis, there are a few scenarios in which genes are considered downregulated: 1) genes downregulated in E4 microglia in response to myelin but upregulated in E3 microglia, 2) those upregulated to lesser extent in E4 microglia relative to E3 microglia, and 3) those more downregulated in E4 microglia than E3 microglia. Investigation of GSEA-identified core drivers of these enrichments identified several genes (e.g., Irf7, Oas2, Fig. 6d) which were slightly upregulated in E4 microglia at baseline and increased mainly in E3 microglia following myelin challenge (Table S1). These results, particularly the inflammatory-related pathways, are reminiscent of changes in E4 microglia relative to E3 microglia at baseline (Fig. 4d).
Positively enriched pathways included “translation elongation”, “translation initiation”, “response to amino acid deficiency”, and several DNA replication-related pathways (Fig. 6c and Table S4). Similar to those genes annotated as downregulated, there are three scenarios in which genes may have been annotated as upregulated in this two factor analysis: 1) genes upregulated in E4 microglia in response to myelin but downregulated in E3 microglia, 2) those downregulated to a lesser extent in E4 microglia relative to E3 microglia, or 3) those upregulated to a greater extent in E4 microglia relative to E3 microglia. Again, the differentially enriched pathways were highly reminiscent of those altered in E4 microglia relative to E3 microglia at baseline. Investigation of GSEA-identified core drivers of translation-related pathway enrichments (Fig. 6d) identified several ribosomal protein-encoding genes downregulated to a greater extent in E3 microglia than E4 microglia in response to myelin (Table S2). Together, these transcriptional changes suggest that challenging E3 microglia with myelin can produce an E4- like transcriptional state, and that E4 microglia exist in this “challenged” state even at baseline. This is supported by RRHO testing between genes altered in E4 relative to E3 microglia at baseline (E4 vs E3) and those altered in E3 microglia in response to myelin (E3 +M vs E3), which revealed a significant overlap in the genes altered in each condition (Spearman rho = 0.4, p < 2.2 × 10−16, Figure S5).
A final differential expression analysis comparing E4 and E3 post-myelin treatment (E4 +M vs E3 +M) suggest ECM and collagen-related pathways are downregulated to a greater extent in E4 microglia than E3 microglia following myelin challenge (Fig. 6e and Table S5). In contrast, genes belonging to the nuclear receptor transcription pathway were upregulated (Table S5), which is significant as nuclear receptors such as liver X receptors (LXRs) have previously been implicated in AD pathogenesis (Sandoval-Hernandez et al., 2015´; Fitz et al., 2020). Specific drivers of these pathways identified by GSEA include Nr1h3, Nr4a1 and Nr4a3 (Fig. 6f). We next investigated the expression of several phagocytic and lipoprotein receptors to identify which, if any, may be responsible for the increased uptake behavior in E4 microglia (Figure S6). Specifically, we looked at a variety of receptors known to be involved in phagocytosis, including non-opsonic receptors dectin-2 (also known as Clec4n) and Cd33 (Li and Underhill, 2020; Uribe-Querol and Rosales, 2020), opsonic receptors including Fc receptors (e.g., Fcgr1, Fcgr2) and complement proteins (Itgam, Vsig4) (Uribe-Querol and Rosales, 2020), as well as other receptors known to mediate uptake of a variety of substrates such as low-density lipoprotein (LRP) receptors (e.g. Lrp1, Lrp10, Lrp11) (Lillis et al., 2008), Trem2 (Gratuze et al., 2018), Mertk (Healy et al., 2017), and Axl (Fujimori et al., 2015), among others. We also included toll-like receptors, which can cooperate with phagocytic receptors in the uptake of either pathogens or host-derived cellular debris (Iwasaki and Medzhitov, 2015; Kawai and Akira, 2011). Only adhesion G protein-coupled receptor B1 (Adgrb) (Mazaheri et al., 2014; Park et al., 2007) showed an FDR adj. p- value < 0.05 for differential expression following myelin challenge (Figure S6), although Lrp11 was modestly upregulated in E4 microglia relative to E3 microglia following myelin challenge. However, a number of receptors were modestly increased in E4 microglia treated with myelin (E4 +M) relative to untreated E4 microglia (E4, Figure S6), highlighting several candidate receptors that may mediate the increased internalization of myelin and other substrates observed in E4 microglia.
3.7. Transcriptional signatures of E4 and KO microglia relative to E3 and WT microglia exhibit both shared and unique features
A key outstanding question is whether the APOE4 isoform leads to increased AD risk by gain of a novel toxic function, loss of a normal protective function, or other mechanisms (Kim et al., 2009). To begin exploring this question in terms of transcriptional profiles, we took advantage of our WT and KO microglia and performed differential gene expression analysis comparing KO to WT microglia both at baseline and following myelin challenge. We then compared the transcriptional changes induced by Apoe KO (KO vs WT) and E4 genotype (E4 vs E3) using RRHO (Fig. 7a, left). Significant overlap was identified, particularly among downregulated genes (Spearman rho =0.5, p < 2.2 × 10−16). To better understand the functions affected by either Apoe KO or E4 genotype relative to WT or E3, respectively, we next compared GSEA enrichment results across contrasts (Fig. 7a, right). Several pathways already highlighted to be enriched among genes downregulated in E4 vs E3 microglia were also enriched among genes downregulated in KO vs WT microglia, including “translation elongation”, “response to amino acid deficiency”, and “collagen formation”. However, both E4 and KO microglia also exhibited several uniquely enriched pathways, demonstrating that the E4 genotype does not completely phenocopy a total loss of APOE function. Notably, inflammatory pathways including “interferon alpha/beta signaling” and “interferon gamma signaling” were uniquely upregulated in E4 microglia, suggesting interferon activation observed in E4 microglia is a unique property conferred by the E4 isoform and not the result of loss of an APOE function.
To further understand the similarities and differences across APOE genotypes in response to myelin treatment, this analysis was repeated using ranked gene lists generated from DEG analysis of myelin-treated cells (Fig. 7b). There was again a significant, though slightly weaker, correlation between KO and E4 transcriptional profiles (Spearman rho = 0.4, p < 2.2 × 10−16). Similar enrichments to those observed under baseline culture conditions were observed, including a shared downregulation of ECM-related functions such as “integrin cell surface interactions”, “assembly of collagen fibrils”, and “collagen chain trimerization”. Interestingly, following myelin challenge both E4 and KO microglia significantly upregulated genes in the “DAP12 signaling” pathway (Fig. 7c, d). This is significant as DAP12 (also known as Tyrobp) forms a signaling complex with TREM2, a microglial receptor linked to AD risk (Gratuze et al., 2018; Konishi and Kiyama, 2018). E4 microglia also exhibited a unique downregulation of genes in the “cholesterol biosynthesis” pathway, and of genes in the “protein kinase N1 (PKN1) transcriptional pathway”, a pathway linked to the regulation of the actin cytoskeleton and cell migration (Manser et al., 2008; Mashud et al., 2017). Together, these data help elucidate the extent to which the E4 isoform may be influencing cellular functions beyond those typically associated with the loss of APOE function.
4. Discussion
Here, we take advantage of a humanized mouse model (Knouff et al., 1999; Sullivan et al., 1997) to better understand the molecular and cellular mechanisms by which APOE genotype alters the function of primary mouse microglia and may ultimately leads to increased risk for AD (Kloske and Wilcock, 2020). Using live-cell imaging, flow cytometry, immunocytochemistry, immunoblotting, and RNA-sequencing we show that relative to E3 microglia, E4 microglia exhibit altered morphology, increased lipid and lipid droplet accumulation, increased endolysosomal mass, increased cytokine/chemokine production, and increased phosphorylation of EIF2ɑ and EIF2ɑ-kinases coupled with decreased protein synthesis. We also show that E4 microglia exhibit increased phagocytic uptake of a variety of substrates, including myelin debris, apoptotic cells, zymosan, and latex beads. Together, our results suggest E4 genotype confers several important functional alterations to microglia, the implications of which we discuss in more detail below.
4.1. Functional alterations in E4 microglia reflect increased immune reactivity and cellular stress at baseline
Under basal conditions, we observed APOE genotype-dependent changes in cell morphology, with E4 microglia showing a smaller overall cell size and shorter average branch length compared to E3 microglia (see Methods, Figure S1). These morphological features have been previously reported in primary microglia from TR mice (Vitek et al., 2009) as well as immortalized murine microglial-like N9 cells transfected with E3 and E4 expression constructs (Muth et al., 2019) and human induced pluripotent stem cell (iPSC)-derived microglia with either E3 or E4 genotype (Lin et al., 2018), demonstrating this effect of E4 genotype on microglia morphology is not unique to our study. Our RNA-sequencing analyses identified genes related to elastic fibre formation, collagen formation, collagen degradation, and ECM structure and organization downregulated in E4 microglia relative to E3 microglia (Fig. 4c, d), providing candidate pathways that may contribute to the altered morphology we observed (Milner and Campbell, 2003). The smaller, more rounded morphology observed in E4 microglia is thought to be indicative of a more reactive state of microglia responding to damage or pathogens in the aged, disease, injured, or infected brain (Davis et al., 1994; Hanisch and Kettenmann, 2007; Prinz et al., 2011; Ransohoff and Perry, 2009). Our RNA-sequencing analyses and subsequent validation work support this hypothesis, with E4 microglia upregulating interferon signaling as well as specific cytokines/chemokines known to be activated by interferon signaling relative to E3 microglia. This immune-related phenotype has been observed in human iPSC-derived microglia harboring two copies of the ε4 allele (Konttinen et al., 2019; Lin et al., 2018), suggesting this is a general and not a species-specific effect.
Pathway analysis of RNA sequencing data from E4 microglia relative to E3 microglia revealed downregulation of genes involved in eukaryotic translation initiation and elongation, both critical steps in the regulation of protein synthesis (Adomavicius et al., 2019). Similar changes, including alterations in other translation-related pathways such as tRNA charging have been shown in brain lysates from E4 TR mice as compared to E3 (Zhao et al., 2020). At the protein level in our study, E4 microglia show an increased ratio of phosphorylated to total eIF2α. Phosphorylated eIF2α is a well-established marker of the integrated stress response (Pakos-Zebrucka et al., 2016) and is known to lead to repression of translation (Adomavicius et al., 2019). Phosphorylation of eIF2α and its upstream kinases are well-documented in AD, with increased phosphorylation of eIF2α itself observed in postmortem brain tissue (Oliveira et al., 2021) and increased phosphorylation of eIF2-kinases such as PKR observed in cerebrospinal fluid of AD patients (Mouton-Liger et al., 2015). We additionally validated that activation of these kinases did indeed lead to a dramatic decrease in protein synthesis in E4 microglia relative to E3 microglia (Fig. 5d, e). As translation regulation has been shown to play a critical role in microglial function such as phagocytosis of myelin debris in vitro and synaptic pruning in vivo (Xu et al., 2020), this activation of the integrated stress response may be responsible for a number of functional differences we observed in E4 microglia relative to E3 microglia.
While our results strongly suggest the E4 microglia exist in a state of cellular stress relative to E3 microglia at baseline, the instigator(s) of the stress response in these cells remains unclear. We showed that several kinases directly upstream of eIF2 are more highly activated in E4 microglia than E3 microglia, including PERK and PKR. These kinases are known to be activated by treatment with free fatty acids, neutral or oxidized lipids (Nakamura et al., 2010; Haberzettl and Hill, 2013; Volmer et al., 2013). PERK can also be activated by ER stress (Onat et al., 2019), which is a characteristic of lipid-laden macrophages in human and mouse (Erbay et al., 2009). This suggests the increased lipid accumulation observed in E4 microglia at baseline and reported in TREM2 or PLCG2 microglia in response to cuprizone-induced demyelination (Andreone et al., 2020; Nugent et al., 2020) may provide a mechanistic explanation for the observed activation of these kinases. In addition, ER stress and downstream activation of PERK can also be triggered by mis-or unfolded proteins (Gal-Ben-Ari et al., 2018). It has been hypothesized that the E4 isoform is more prone to unfolding, thus possibly leading to increased ER stress and activation of PERK, leading to increased phosphorylation of eIF2α and global repression of translation (Segev et al., 2013; Zhong and Weisgraber, 2009). Alternatively, interferon-γ-induced inflammation has been shown to induce phosphorylation of both PKR and PERK (Carret-Rebillat et al., 2015; Sadler and Williams, 2008), suggesting the increased interferon signaling we observed in E4 microglia relative to E3 microglia could also be contributing to increased stress in these cells.
4.2. Increased accumulation of phagocytosed myelin fragments reflects an impaired balance of uptake and degradation in E4 microglia
Phagocytosis of a diverse array of substrates including myelin, zymosan, latex beads, and apoptotic cells was markedly and rapidly increased in E4 microglia relative to microglia with all other genotypes we tested (E3, KO, and WT). Notably, APOE4 was recently shown to increase phagocytosis of apoptotic cells by murine microglia-like N9 cells (Muth et al., 2019), suggesting this effect of APOE4 is reproducible across experimental systems. Here, we expand upon these findings and show that these results generalize to a wide variety of substrates in primary mouse microglia. While phagocytic clearance of cellular debris by macrophages is critical to maintain tissue homeostasis, microglia have been shown to cause neuronal death by phagocytosis of stressed but still viable neurons in vitro (Neher et al., 2012). Therefore, increased phagoptosis by E4 microglia may be a mechanism by which APOE4 increases AD risk.
In addition to increased phagocytic uptake, we observed myelin internalized by E4 microglia to have a longer half-life than myelin internalized by microglia from other genotypes, suggesting impaired digestion of phagocytosed material (Figure S4). The specific mechanism by which APOE4 may alter the ability of microglia to degrade phagocytosed cellular debris remains unclear, but chronic lipid overload has been linked to lysosome dysfunction via a variety of mechanisms (Jaishy and Abel, 2016), including generation of reactive oxygen species, alteration of lysosomal membrane composition (Koga et al., 2010) which impairs fusion to other organelles, and lysosomal membrane permeabilization leading to increased pH (Emanuel et al., 2014; Feldstein et al., 2006). Given the increased levels of lipids present in E4 microglia prior to myelin challenge (Fig. 1), these cells may be dealing with low levels of lipid overload even at baseline. When challenged with additional lipid-rich debris such as myelin, lysosomes from E4 microglia may already be chronically activated and unable to efficiently deal with the additional lipid load. While this hypothesis remains to be tested, it is supported by our observation that lysosomes of E4 microglia have lower pH compared to those of E3 microglia at baseline and fail to further acidify (and perhaps even slightly increase their pH) following myelin challenge (Figure S3).
4.3. Functional alterations in E4 microglia may provide insight into mechanisms leading to increased disease risk in E4 carriers
As APOE ε4 is the strongest genetic risk factor for AD, some of the functional alterations observed in microglia harboring this allele may provide insight into mechanisms by which disease risk is increased in APOE ε4 carriers. There is evidence that APOE4 increases microgliosis in a dose-dependent manner, particularly in the frontal and temporal cortices (Egensperger et al., 1998; Tzioras et al., 2019), and increases the production of pro-inflammatory cytokines and chemokines in human microglia (Fernandez et al., 2019; Friedberg et al., 2020). Here we highlight several specific cytokines/chemokines altered by E4 genotype. For example, several of the inflammatory mediators that accumulated to a greater extent in conditioned media from E4 microglia relative to E3 microglia belong to the CC class of chemokines, including CCL3 and CCL4 (Fig. 5b). This class of chemokines have emerged as putative drug targets (White et al., 2013), suggesting they may be harnessed to resolve the pro-inflammatory effects of E4 genotype in future studies. Alternatively, these chemokines are known to play important roles in chemoattraction and recruitment of immune cells (White et al., 2013). In vivo, most chemokines are thought to bind to glycosaminoglycans, including proteoglycans such as heparin sulfate and chondroitin sulfate expressed on the surface of cells (White et al., 2013). Given the robust downregulation of ECM proteins observed in E4 microglia (Fig. 4c, d), the robust upregulation of select cytokines/chemokines may be secondary to ECM alterations in E4 microglia, a hypothesis that will be tested in future studies.
In addition to changes in specific cytokines and chemokines, E4 microglia showed transcriptional alterations positively enriched for interferon signaling. Here, we identified several core genes driving this enrichment, including Oas (oligoadenylate synthase) family members Oas1g, Oas2, and Oas3 (Figs. 4 and 6). Oas1 was recently nominated as a potential AD risk gene in the GWAS locus identified on chromosome 12 based on 1) its co-expression with known risk genes in mouse microglia, and 2) regulation of its expression in human monocytes and human macrophages by variants located within the chromosome 12 GWAS loci (Salih et al., 2019). Notably, the association of OAS1 expression and AD risk variants was only observed in cells stimulated with either interferon γ or salmonella (Salih et al., 2019), suggesting OAS1 effect on disease risk may be conditional to microglia within a pro-inflammatory milieu. Additionally, Oas genes are known to regulate the expression of cytokines and chemokines (Lee et al., 2019), suggesting they may contribute to the increased levels of cytokines and chemokines we measured in E4 vs E3 microglia conditioned media.
The effect of APOE genotype on immune reactivity is not limited to brain microglia but has also been observed in peripheral macrophages (Vitek et al., 2009). For example, altered interferon signaling has been shown to perturb lipid metabolism, specifically the levels of particular genes involved in lipid droplet formation (Piln1) (Shao et al., 2013), cholesterol ester synthesis (Soat), and cholesterol efflux (Abca1, Abcg1) in mouse peripheral macrophages (Panousis and Zuckerman, 2000) as well as human THP-1 macrophages (Reiss et al., 2004). Given that AD is a neurodegenerative disorder, it has been hypothesized that microglia, as the brain-resident macrophages, are the myeloid cells critical for modifying disease susceptibility (Bachiller et al., 2018). However, genetic studies have identified an enrichment of AD risk alleles in regulatory elements active in peripheral monocytes and macrophages in addition to microglia (Novikova et al., 2019), highlighting the possibility that these peripheral cells also play a key role in modulating disease risk.
4.4. Microglial phenotypes associated with AD risk variants suggest alterations in lipid metabolism and protein translation may be central to AD pathogenesis
Here we showed E4 microglia exhibited increased accumulation of neutral lipids as well as lipid droplets under both baseline culture conditions and following treatment with myelin fragments. Similar results have been observed in iPSC-derived microglia carrying two copies of the APOE ε4 allele, which display increased cholesterol biosynthesis and decreased cholesterol efflux (Tcw et al., 2019; Lin et al., 2018; Podlésny- Drabiniok et al., 2020; Sienski et al., 2021) as well as numbers of lipid droplets (Sienski et al., 2021), suggesting our findings are not unique to primary mouse microglia. Moreover, other AD risk genes confer lipid handling deficits in microglia similar to those observed in E4 microglia (Andreone et al., 2020; Nugent et al., 2020) suggesting that defective lipid metabolism in microglia may be a mechanism central to AD pathogenesis. Specifically, loss of either TREM2 or PLCG2 in microglia has been shown to cause accumulation of cholesterol esters following myelin challenge, both in vitro and in vivo following cuprizone-induced demyelination (Andreone et al., 2020; Nugent et al., 2020).
In neurodegenerative diseases including AD, microglia have been shown to adopt a disease-or damage-associated profile (Krasemann et al., 2017; Keren-Shaul et al., 2017). This profile is a transcriptional program thought to enable microglia to enhance their phagocytic and lipid handling properties in order to facilitate effective metabolism of cholesterol and other cellular debris (Loving and Bruce, 2020). Apoe and Trem2 have been shown to play critical roles in the transition of microglia from a homeostasis-associated state to this damage-associated state, with Apoe being robustly upregulated early in the transition period and Trem2 being necessary for the final stages of the transition (Keren-Shaul et al., 2017; Krasemann et al., 2017; Pimenova et al., 2017). Lipidated APOE has recently been identified as a ligand for TREM2 (Atagi et al., 2015), and the AD risk-increasing R47H TREM2 variant significantly decreases binding affinity (Cheng-Hathaway et al., 2018), suggesting a complex interplay between these two risk genes. A better understanding of how APOE interacts with TREM2 and other AD risk genes is important for elucidating AD pathogenesis. A number of studies aimed at addressing these outstanding questions have been carried out in mouse models harboring a global loss of Apoe (Van Giau et al., 2015). As we showed here, the E4 allele does not fully phenocopy a loss of APOE function and the isoform-specific differences in the interaction between Apoe and other AD risk genes should be investigated in more detail in future studies.
Both increased lipid accumulation and inflammatory signaling can disrupt cellular homeostasis and directly activate kinases involved in the integrated stress response as described above. Activation of these kinases leads to a global repression of translation, which has been observed in various models of AD and other neurodegenerative diseases. For example, differential expression of genes in translation-related pathways, particularly the EIF2 signaling pathway, has been reported across various models of AD. These include E4 microglia (Tcw et al., 2019), mouse microglia with reduced expression of the AD risk factor PU.1 (Pimenova et al., 2017), as well as mouse models with global loss of Trem2 (Fitz et al., 2020; Ulland and Colonna, 2018). In addition, increased phosphorylation of eIF2α and its upstream kinases has been reported in neurons from sporadic AD patients (Chang et al., 2002) as well as mouse models harboring mutations that cause early-onset autosomal dominant AD and exhibiting amyloid pathology (Devi and Ohno, 2010; Mouton-Liger et al., 2015; Segev et al., 2013). Finally, integrative analyses of AD genetics and myeloid genomics datasets have identified EIF2B2 as a candidate AD risk gene, implicating translation regulation in microglia (and possibly other myeloid cells) in the etiology of AD (Novikova et al., 2021).
Together, our results suggest that increased levels of cellular stress leading to global repression of translation and activation of the integrated stress response may be a putative mechanism by which the E4 genotype (and possibly, other high-risk genotypes) may increase susceptibility to AD. A small molecule known as ISRIB (integrated stress response inhibitor) has been shown to reduce the effects of eIF2α phosphorylation on eIF2b-mediated translation and thus prevent the activation of the integrated stress response (Zyryanova et al., 2021). Delivery of ISRIB to animal models of neurological diseases has proven beneficial under a variety of conditions. For example, ISRIB has been shown to rescue memory deficits in AD mouse model (Oliveira et al., 2021) as well as exert beneficial effects (e.g. improve motor deficits, ameliorate white matter pathology) in both chronic traumatic brain injury (Chou et al., 2017) and chronic demyelination in vanishing white matter disease (Abbink et al., 2019; Wong et al., 2018). It remains unknown whether neurons or other cell types such as microglia mediate the beneficial effects of ISRIB. Our study provides support for the hypothesis that targeting eIF2α or eIF2α kinases (e.g. PKR, PERK) in microglia may be especially beneficial, as we showed increased phosphorylation of eIF2α and several eIF2α kinases, and globally reduced protein synthesis in E4 microglia compared to E3 microglia. These results may contribute to our understanding of how E4 and other genotypes lead to high-risk of AD and may motivate future development of ISR-targeting therapeutics for AD.
4.5. Limitations of the current study
Despite being an in-depth characterization of the functional and transcriptional effects of APOE genotypes in microglia, several important limitations of the current study should be considered when interpreting its results. One major limitation of our study is that it does not account for the differential impact of APOE genotype across males and females (Altmann et al., 2014; Dumitrescu et al., 2019; Hsu et al., 2019). In order to obtain sufficient numbers of microglia for downstream experiments, our microglial cultures were obtained by pooling brain samples from multiple pups born in the same litter. As a result, our cultures are likely mixtures of both male and female microglia. This may have contributed to some of the variability in our studies, particularly within the RNA sequencing results, as APOE genotype has been shown to differentially alter the transcriptional profile in males and females (Hsu et al., 2019). In addition, APOE genotype has been shown to have differential effects on AD risk based on sex (Altmann et al., 2014; Neu et al., 2017), with exacerbated risk for disease observed in female vs male carriers of APOE ε4. Finally, several important differences have been described across male and female microglia (Villa et al., 2018). The importance of sex in relation to both APOE genotype and microglial function make the consideration of sex in future studies aimed at elucidating the role of APOE in microglia critical to fully understanding how APOE isoforms act in microglia to modulate disease risk across individuals.
Second, this study was conducted in an in vitro culture system using mouse cells genetically modified to express human APOE. There are many benefits to working in an in vitro culture setting, including the ability to utilize well-defined experimental conditions. However, as microglia are exquisitely primed to respond to their environment (Hanamsagar and Bilbo, 2017), studies conducted outside the brain should be interpreted with caution as isolation procedures (Ayata et al., 2018) and culture conditions alter their transcriptional profile and functional state (Gosselin et al., 2017). Our findings should be replicated in vivo using TR mice combined with a challenge condition such as cuprizone-induced demyelination or amyloid deposition (Elder et al., 2010; Gudi et al., 2014). However, even these experiments would need to be interpreted with caution, as several important differences between the mouse and human immune system have been documented (Hasselmann et al., 2019; Smith and Dragunow, 2014). Given the robust effect of APOE genotype on immune-related pathways (Figs. 4–5), findings from these TR mouse models should ultimately be replicated using human cells. Finally, in these TR mice, human APOE is functioning in the context of mouse biology and under the control of mouse regulatory elements. APOE has been documented to interact with several microglial receptors, including TREM2 (Shi and Holtzman, 2018; Wolfe et al., 2018), which in these mice have not been humanized. Despite an overall high degree of homology in most proteins, several important differences exist that are likely to impact these protein-protein interactions in important ways (Smith and Dragunow, 2014).
5. Conclusions
Given the crucial roles of both APOE and microglia in AD, a better understanding of human APOE isoform-dependent effects on microglial function will likely provide insight into AD pathogenesis. Our work highlights several important functions including lipid metabolism, cytokine production, translation, and phagocytosis which are altered in E4 microglia relative to E3 microglia. We highlight several candidate genes and proteins that may mediate these effects, including eIF2α-kinases PERK and PKR. Notably, downregulation of PKR has been shown to rescue cognitive deficits and prevent amyloid accumulation in AD mouse models (Hwang et al., 2017; Mouton-Liger et al., 2015; Zhu et al., 2011), pointing to potential leads for future therapeutic investigation. In addition, it will be important in future studies to elucidate which of the multiple changes we observed occur first – for example, whether heightened cytokine production and inflammatory signaling leads to an exacerbated stress response and reduced translation or vice versa. As the integrated stress response can be targeted by a variety of small molecules (Romero-Ramírez et al., 2017), a better understanding of how this pathway contributes to altered microglial physiology in AD downstream of APOE4 is likely to yield important leads for therapeutic intervention. When combined with previous findings from models with other AD risk- increasing genotypes (Andreone et al., 2020; Nugent et al., 2020), our results also point to altered microglial lipid metabolism as a possible major contributor to disease pathogenesis. Future studies should be aimed at expanding on these results, both in vivo and using human cells, with the ultimate goal of deriving therapeutics based on APOE genotype and its downstream cellular effects.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Anna Podlesny-Drabiniok, Dr. Francesca Garretti, Dr. Julia TCW for critical reading during the manuscript preparation as well as Dr. John Crary and Dr. Nikos Robakis for contributing to data discussion. All data generated or analyzed during this study will be made available upon request. The RNA-Seq data generated in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series access number GSE171280. This study was funded by the JPB Foundation (AMG), BrightFocus Foundation (EM) and (JMC), NIA/NIH AG061382 (JMC) and NIA/NIH K99/R00 AG051711 (JMC). These funding organizations had no direct involvement in the study design, collection, analysis, or interpretation of results.
Abbreviations:
- AD
Alzheimer’s disease
- LOAD
Late-onset AD
- GWAS
Genome wide association studies
- APOE
Apolipoprotein E
- E3
APOE ε3
- E4
APOE ε4
- Aβ
Amyloid beta
- DAM
Damage- or disease- associated microglia
- TR
Targeted replacement
- EAJ
Early apoptotic Jurkat
- MBP
Myelin basic protein
- WT
Wild-type
- KO
Knock-out
- NBD
N-[(7-nitro-2–1,3-benzoxadiazol-4-yl)methyl]amino
- GSEA
Gene set enrichment analysis
- ECM
Extracellular matrix
- OAS1
2’−5’-oligoadenylate synthetase 1
- TNFα
Tumor necrosis factor
- CXCL2
C-X-C motif chemokine ligand 2
- ER
Endoplasmic reticulum
- eIF2α
Eukaryotic initiation factor-2
- p-eIF2α
phosphorylated eukaryotic initiation factor-2
- PKR
Protein kinase R
- p-PKR
phosphorylated protein kinase R
- PERK
PKR-like endoplasmic reticulum kinase
- p- PERK
phosphorylated PKR-like endoplasmic reticulum kinase
- GCN2
General control non-depressible 2
- GCN2
phosphorylated general control non-depressible 2
- RRHO
Rank-rank hypergeometric overlap
- LXRs
Liver X receptors
- TLRs
Toll-like receptors
- TREM2
Triggering receptor expressed on myeloid cells 2
- PLCG2
Phospholipase C gamma 2
- DAP12
DNAX- activating protein of 12 kDa
- PKN1
Protein kinase N1
- TYROBP
TYRO protein tyrosine kinase-binding protein
Footnotes
Declaration of Competing Interest
AMG has consulted for Eisai, Biogen, Pfizer, AbbVie, Cognition Therapeutics and GSK; she also serves on the Scientific Advisory Board of Genentech. JMC is a co-inventor listed on issued patents licensed by Alkahest, Inc. SIM, SMN, BMH, RK, MH, JDZ, DC, JMC, EM report no conflicts of interest.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nbd.2022.105615.
References
- Abbink TEM, Wisse LE, Jaku E, Thiecke MJ, Voltolini-Gonzalez D, Fritsen H, Bobeldijk S, Ter Braak TJ, Polder E, Postma NL, Bugiani M, Struijs EA, Verheijen M, Straat N, van der Sluis S, Thomas AAM, Molenaar D, van der Knaap MS, 2019. Vanishing white matter: deregulated integrated stress response as therapy target. Ann. Clin. Transl. Neurol. 6, 1407–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adomavicius T, Guaita M, Zhou Y, Jennings MD, Latif Z, Roseman AM, Pavitt GD, 2019. The structural basis of translational control by eIF2 phosphorylation. Nat. Commun. 10, 2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Advani VM, Ivanov P, 2019. Translational Control under Stress: Reshaping the Translatome. Bioessays 41, e1900009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann A, Tian L, Henderson VW, Greicius MD, Alzheimer’s Disease Neuroimaging Initiative Investigators, 2014. Sex modifies the APOE-related risk of developing Alzheimer disease. Ann. Neurol. 75, 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzheimer’s Association, 2021. Alzheimer’s disease facts and figures. Alzheimer’s Dement 17, 327–406. 10.1002/alz.12328. [DOI] [PubMed] [Google Scholar]
- Andreone BJ, Przybyla L, Llapashtica C, Rana A, Davis SS, van Lengerich B, Lin K, Shi J, Mei Y, Astarita G, Di Paolo G, Sandmann T, Monroe KM, Lewcock JW, 2020. Alzheimer’s-associated PLCγ2 is a signaling node required for both TREM2 function and the inflammatory response in human microglia. Nat. Neurosci. 23, 927–938. [DOI] [PubMed] [Google Scholar]
- Andrews SJ, Fulton-Howard B, Goate A, 2020. Interpretation of risk loci from genome-wide association studies of Alzheimer’s disease. Lancet Neurol. 19, 326–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atagi Y, Liu C-C, Painter MM, Chen X-F, Verbeeck C, Zheng H, Li X, Rademakers R, Kang SS, Xu H, Younkin S, Das P, Fryer JD, Bu G, 2015. Apolipoprotein E Is a Ligand for Triggering Receptor Expressed on Myeloid Cells 2 (TREM2). J. Biol. Chem. 290, 26043–26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayata P, Badimon A, Strasburger HJ, Duff MK, Montgomery SE, Loh Y-HE, Ebert A, Pimenova AA, Ramirez BR, Chan AT, Sullivan JM, Purushothaman I, Scarpa JR, Goate AM, Busslinger M, Shen L, Losic B, Schaefer A, 2018. Epigenetic regulation of brain region-specific microglia clearance activity. Nat. Neurosci. 21, 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachiller S, Jimenez-Ferrer I, Paulus A, Yang Y, Swanberg M, Deierborg T, Boza- Serrano A, 2018. Microglia in neurological diseases: a road map to brain-disease dependent-inflammatory response. Front. Cell. Neurosci. 12, 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bales KR, Liu F, Wu S, Lin S, Koger D, DeLong C, Hansen JC, Sullivan PM, Paul SM, 2009. Human APOE isoform-dependent effects on brain beta-amyloid levels in PDAPP transgenic mice. J. Neurosci. 29, 6771–6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bampton ETW, Goemans CG, Niranjan D, Mizushima N, Tolkovsky AM, 2005. The dynamics of autophagy visualised in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy 1, 23–36. [DOI] [PubMed] [Google Scholar]
- Berg S, Kutra D, Kroeger T, Straehle CN, Kausler BX, Haubold C, Schiegg M, Ales J, Beier T, Rudy M, Eren K, Cervantes JI, Xu B, Beuttenmueller F, Wolny A, Zhang C, Koethe U, Hamprecht FA, Kreshuk A, 2019. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods. 10.1038/s41592-019-0582-9. [DOI] [PubMed] [Google Scholar]
- Bhat MY, Solanki HS, Advani J, Khan AA, Keshava Prasad TS, Gowda H, Thiyagarajan S, Chatterjee A, 2018. Comprehensive network map of interferon gamma signaling. J. Cell Commun. Signal. 12, 745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolívar S, Anfossi R, Humeres C, Vivar R, Boza P, Munoz C, Pardo-Jimenez V, Olivares-Silva F, Díaz-Araya G, 2018. IFN-β plays both pro- and anti-inflammatory roles in the rat cardiac fibroblast through differential STAT protein activation. Front. Pharmacol. 9, 1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantoni C, Bollman B, Licastro D, Xie M, Mikesell R, Schmidt R, Yuede CM, Galimberti D, Olivecrona G, Klein RS, Cross AH, Otero K, Piccio L, 2015. TREM2 regulates microglial cell activation in response to demyelination in vivo. Acta Neuropathol. 129, 429–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L, Fitzner D, Bosch-Queralt M, Weil M-T, Su M, Sen P, Ruhwedel T, Mitkovski M, Trendelenburg G, Lütjohann D, Mobius W, Simons M, 2018. Defective cholesterol clearance limits remyelination in the aged central nervous system. Science 359, 684–688. [DOI] [PubMed] [Google Scholar]
- Carret-Rebillat A-S, Pace C, Gourmaud S, Ravasi L, Montagne-Stora S, Longueville S, Tible M, Sudol E, Chang RC-C, Paquet C, Mouton-Liger F, Hugon J, 2015. Neuroinflammation and Aβ accumulation linked to systemic inflammation are decreased by genetic PKR down-regulation. Sci. Rep. 5, 8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, Patterson BW, Fagan AM, Morris JC, Mawuenyega KG, Cruchaga C, Goate AM, Bales KR, Paul SM, Bateman RJ, Holtzman DM, 2011. Human apoE isoforms differentially regulate brain amyloid-β peptide clearance. Sci. Transl. Med. 3, 89ra57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang RCC, Wong AKY, Ng H-K, Hugon J, 2002. Phosphorylation of eukaryotic initiation factor-2α (eIF2α) is associated with neuronal degeneration in Alzheimer’s disease. Neuroreport 13, 2429. [DOI] [PubMed] [Google Scholar]
- Cheng-Hathaway PJ, Reed-Geaghan EG, Jay TR, Casali BT, Bemiller SM, Puntambekar SS, von Saucken VE, Williams RY, Karlo JC, Moutinho M, Xu G, Ransohoff RM, Lamb BT, Landreth GE, 2018. The Trem2 R47H variant confers loss-of-function-like phenotypes in Alzheimer’s disease. Mol. Neurodegener. 13, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokova E, Bal N, Kolosov P, 2017. Kinases of eIF2a Switch Translation of mRNA Subset during Neuronal Plasticity. Int. J. Mol. Sci. 18 10.3390/ijms18102213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou A, Krukowski K, Jopson T, Zhu PJ, Costa-Mattioli M, Walter P, Rosi S, 2017. Inhibition of the integrated stress response reverses cognitive deficits after traumatic brain injury. Proc. Natl. Acad. Sci. U. S. A. 114, E6420–E6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA, 1993. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261, 921–923. [DOI] [PubMed] [Google Scholar]
- Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, Jassal B, Jupe S, Matthews L, May B, Palatnik S, Rothfels K, Shamovsky V, Song H, Williams M, Birney E, Hermjakob H, Stein L, D’Eustachio P, 2014. The Reactome pathway knowledgebase. Nucleic Acids Res. 42, D472–D477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinska B, Malarz K, Mrozek-Wilczkiewicz A, Musiol R, 2019. Acid selective pro-dye for cellular compartments. Sci. Rep. 9, 15304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damisah EC, Rai A, Grutzendler J, 2020. TREM2: Modulator of Lipid Metabolism in Microglia. Neuron. 105, 759–761. [DOI] [PubMed] [Google Scholar]
- Davis EJ, Foster TD, Thomas WE, 1994. Cellular forms and functions of brain microglia. Brain Res. Bull. 34, 73–78. [DOI] [PubMed] [Google Scholar]
- Devi L, Ohno M, 2010. Phospho-eIF2α level is important for determining abilities of BACE1 reduction to rescue cholinergic neurodegeneration and memory defects in 5XFAD mice. PLoS One 5, e12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitrescu L, Mayeda ER, Sharman K, Moore AM, Hohman TJ, 2019. Sex Differences in the Genetic Architecture of Alzheimer’s Disease. Curr. Genet. Med. Rep. 7, 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE, 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egensperger R, Kosel S, von Eitzen U, Graeber MB, 1998. Microglial activation in¨ Alzheimer disease: Association with APOE genotype. Brain Pathol. 8, 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid M, Gollwitzer M, Schmitt M, 2017. Statistik und forschungsmethoden. [Google Scholar]
- Elder GA, Gama Sosa MA, De Gasperi R, 2010. Transgenic mouse models of Alzheimer’s disease. Mt Sinai J. Med. 77, 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel R, Sergin I, Bhattacharya S, Turner J, Epelman S, Settembre C, Diwan A, Ballabio A, Razani B, 2014. Induction of lysosomal biogenesis in atherosclerotic macrophages can rescue lipid-induced lysosomal dysfunction and downstream sequelae. Arterioscler. Thromb. Vasc. Biol. 34, 1942–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbay E, Babaev VR, Mayers JR, Makowski L, Charles KN, Snitow ME, Fazio S, Wiest MM, Watkins SM, Linton MF, Hotamisligil GS, 2009. Reducing endoplasmic reticulum stress through a macrophage lipid chaperone alleviates atherosclerosis. Nat. Med. 15, 1383–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM, 1997. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA 278, 1349–1356. [PubMed] [Google Scholar]
- Feldstein AE, Werneburg NW, Li Z, Bronk SF, Gores GJ, 2006. Bax inhibition protects against free fatty acid-induced lysosomal permeabilization. Am. J. Physiol. Gastrointest. Liver Physiol. 290, G1339–G1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez CG, Hamby ME, McReynolds ML, Ray WJ, 2019. The Role of APOE4 in Disrupting the Homeostatic Functions of Astrocytes and Microglia in Aging and Alzheimer’s Disease. Front. Aging Neurosci. 11, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitz NF, Wolfe CM, Playso BE, Biedrzycki RJ, Lu Y, Nam KN, Lefterov I, Koldamova R, 2020. Trem2 deficiency differentially affects phenotype and transcriptome of human APOE3 and APOE4 mice. Mol. Neurodegener. 15, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley P, 2010. Lipids in Alzheimer’s disease: A century-old story. Biochim. Biophys. Acta 1801, 750–753. [DOI] [PubMed] [Google Scholar]
- Friedberg JS, Aytan N, Cherry JD, Xia W, Standring OJ, Alvarez VE, Nicks R, Svirsky S, Meng G, Jun G, Ryu H, Au R, Stein TD, 2020. Associations between brain inflammatory profiles and human neuropathology are altered based on apolipoprotein E ε4 genotype. Sci. Rep. 10, 2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori T, Grabiec AM, Kaur M, Bell TJ, Fujino N, Cook PC, Svedberg FR, MacDonald AS, Maciewicz RA, Singh D, Hussell T, 2015. The Axl receptor tyrosine kinase is a discriminator of macrophage function in the inflamed lung. Mucosal Immunol. 8, 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Ben-Ari S, Barrera I, Ehrlich M, Rosenblum K, 2018. PKR: A Kinase to Remember. Front. Mol. Neurosci. 11, 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin E, Hannequin D, Wallon D, Sleegers K, Hiltunen M, Combarros O, Bullido MJ, Engelborghs S, De Deyn P, Berr C, Pasquier F, Dubois B, Tognoni G, Fiévet N, Brouwers N, Bettens K, Arosio B, Coto E, Del Zompo M, Mateo I, Epelbaum J, Frank-Garcia A, Helisalmi S, Porcellini E, Pilotto A, Forti P, Ferri R, Scarpini E, Siciliano G, Solfrizzi V, Sorbi S, Spalletta G, Valdivieso F, Vepsälainen S, Alvarez V, Bosco P, Mancuso M, Panza F,Nacmias B, Bossù P, Hanon O, Piccardi P, Annoni G, Seripa D, Galimberti D, Licastro F, Soininen H, Dartigues J-F, Kamboh MI, Van Broeckhoven C, Lambert JC, Amouyel P, Campion D, 2011. APOE and Alzheimer disease: a major gene with semi-dominant inheritance. Mol. Psychiatry 16, 903–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, Jaeger BN, O’Connor C, Fitzpatrick C, Pasillas MP, Pena M, Adair A, Gonda DD, Levy ML, Ransohoff RM, Gage FH, Glass CK, 2017. An environment-dependent transcriptional network specifies human microglia identity. Science 356. 10.1126/science.aal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GraphPad Software, LLC, n.d. GraphPad Prism 9 Curve Fitting Guide - Equation: One phase decay [WWW Document]. URL https://www.graphpad.com/guides/prism/latest/curve-fitting/reg_exponential_decay_1phase.htm (accessed 2.4.21).
- Gratuze M, Leyns CEG, Holtzman DM, 2018. New insights into the role of TREM2 in Alzheimer’s disease. Mol. Neurodegener. 13, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudi V, Gingele S, Skripuletz T, Stangel M, 2014. Glial response during cuprizone- induced de- and remyelination in the CNS: lessons learned. Front. Cell. Neurosci. 8, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro RJ, Gustafson DR, Hardy J, 2012. The genetic architecture of Alzheimer’s disease: beyond APP. PSENs APOE. Neurobiol. Aging 33, 437–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro RJ, Lohmann E, Bras JM, Gibbs JR, Rohrer JD, Gurunlian N,´ Dursun B, Bilgic B, Hanagasi H, Gurvit H, Emre M, Singleton A, Hardy J, 2013. Using exome sequencing to reveal mutations in TREM2 presenting as a frontotemporal dementia-like syndrome without bone involvement. JAMA Neurol. 70, 78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberzettl P, Hill BG, 2013. Oxidized lipids activate autophagy in a JNK-dependent manner by stimulating the endoplasmic reticulum stress response. Redox Biol. 1, 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamsagar R, Bilbo SD, 2017. Environment matters: microglia function and dysfunction in a changing world. Curr. Opin. Neurobiol. 47, 146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch U-K, Kettenmann H, 2007. Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat. Neurosci. 10, 1387–1394. [DOI] [PubMed] [Google Scholar]
- Hasselmann J, Coburn MA, England W, Figueroa Velez DX, Kiani Shabestari S, Tu CH, McQuade A, Kolahdouzan M, Echeverria K, Claes C, Nakayama T, Azevedo R, Coufal NG, Han CZ, Cummings BJ, Davtyan H, Glass CK, Healy LM, Gandhi SP, Spitale RC, Blurton-Jones M, 2019. Development of a Chimeric Model to Study and Manipulate Human Microglia In Vivo. Neuron 103, 1016–1033.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy LM, Jang JH, Won S-Y, Lin YH, Touil H, Aljarallah S, Bar-Or A, Antel JP, 2017. MerTK-mediated regulation of myelin phagocytosis by macrophages generated from patients with MS. Neurol. Neuroimmunol. Neuroinflamm. 4, e402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert LE, Weuve J, Scherr PA, Evans DA, 2013. Alzheimer disease in the United States (2010–2050) estimated using the 2010 census. Neurology 80, 1778–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DM, Herz J, Bu G, 2012. Apolipoprotein E and apolipoprotein E receptors: normal biology and roles in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2, a006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M, Dedhia M, Crusio WE, Delprato A, 2019. Sex differences in gene expression patterns associated with the APOE4 allele. F1000Res. 8, 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K-L, Marcora E, Pimenova AA, Di Narzo AF, Kapoor M, Jin SC, Harari O, Bertelsen S, Fairfax BP, Czajkowski J, Chouraki V, Grenier- Boley B, Bellenguez C, Deming Y, McKenzie A, Raj T, Renton AE, Budde J, Smith A, Fitzpatrick A, Bis JC, DeStefano A, Adams HHH, Ikram MA, van der Lee S, Del-Aguila JL, Fernandez MV, Ibanez L, International Genomics of˜ Alzheimer’s Project, Alzheimer’s Disease Neuroimaging Initiative, Sims R, Escott- Price V, Mayeux R, Haines JL, Farrer LA, Pericak-Vance MA, Lambert JC, van Duijn C, Launer L, Seshadri S, Williams J, Amouyel P, Schellenberg GD, Zhang B, Borecki I, Kauwe JSK, Cruchaga C, Hao K, Goate AM, 2017. A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat. Neurosci. 20, 1052–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang K-D, Bak MS, Kim SJ, Rhee S, Lee Y-S, 2017. Restoring synaptic plasticity and memory in mouse models of Alzheimer’s disease by PKR inhibition. Mol. Brain 10, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R, 2015. Control of adaptive immunity by the innate immune system. Nat. Immunol. 16, 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaishy B, Abel ED, 2016. Lipids, lysosomes, and autophagy. J. Lipid Res. 57, 1619–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaishy B, Zhang Q, Chung HS, Riehle C, Soto J, Jenkins S, Abel P, Cowart LA, Van Eyk JE, Abel ED, 2015. Lipid-induced NOX2 activation inhibits autophagic flux by impairing lysosomal enzyme activity. J. Lipid Res. 56, 546–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, Rujescu D, Hampel H, Giegling I, Andreassen OA, Engedal K, Ulstein I, Djurovic S, Ibrahim- Verbaas C, Hofman A, Ikram MA, van Duijn CM, Thorsteinsdottir U, Kong A, Stefansson K, 2013. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 368, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurga AM, Paleczna M, Kuter KZ, 2020. Overview of general and discriminating markers of differential microglia phenotypes. Front. Cell. Neurosci. 14, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapellos TS, Taylor L, Lee H, Cowley SA, James WS, Iqbal AJ, Greaves DR, 2016. A novel real time imaging platform to quantify macrophage phagocytosis. Biochem. Pharmacol. 116, 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S, 2011. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34, 637–650. [DOI] [PubMed] [Google Scholar]
- Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, Itzkovitz S, Colonna M, Schwartz M, Amit I, 2017. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169, 1276–1290.e17. [DOI] [PubMed] [Google Scholar]
- Kim J, Basak JM, Holtzman DM, 2009. The role of apolipoprotein E in Alzheimer’s disease. Neuron 63, 287–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloske CM, Wilcock DM, 2020. The Important Interface Between Apolipoprotein E and Neuroinflammation in Alzheimer’s Disease. Front. Immunol. 11, 754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knouff C, Hinsdale ME, Mezdour H, Altenburg MK, Watanabe M, Quarfordt SH, Sullivan PM, Maeda N, 1999. Apo E structure determines VLDL clearance and atherosclerosis risk in mice. J. Clin. Invest. 103, 1579–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga H, Kaushik S, Cuervo AM, 2010. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 24, 3052–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi H, Kiyama H, 2018. Microglial TREM2/DAP12 Signaling: A Double-Edged Sword in Neural Diseases. Front. Cell. Neurosci. 12, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konttinen H, Cabral-da-Silva MEC, Ohtonen S, Wojciechowski S, Shakirzyanova A, Caligola S, Giugno R, Ishchenko Y, Hernandez D,´Fazaludeen MF, Eamen S, Budia MG, Fagerlund I, Scoyni F, Korhonen P, Huber N, Haapasalo A, Hewitt AW, Vickers J, Smith GC, Oksanen M, Graff C, Kanninen KM, Lehtonen S, Propson N, Schwartz MP, Pebay A,´ Koistinaho J, Ooi L, Malm T, 2019. PSEN1ΔE9, APPswe, and APOE4 Confer Disparate Phenotypes in Human iPSC-Derived Microglia. Stem Cell Rep. 13, 669–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, Beckers L, O’Loughlin E, Xu Y, Fanek Z, Greco DJ, Smith ST, Tweet G, Humulock Z, Zrzavy T, Conde-Sanroman P, Gacias M, Weng Z, Chen H, Tjon E, Mazaheri F, Hartmann K, Madi A, Ulrich JD, Glatzel M, Worthmann A, Heeren J, Budnik B, Lemere C, Ikezu T, Heppner FL, Litvak V, Holtzman DM, Lassmann H, Weiner HL, Ochando J, Haass C, Butovsky O, 2017. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 47, 566–581.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucera M, Isserlin R, Arkhangorodsky A, Bader GD, 2016. AutoAnnotate: A Cytoscape app for summarizing networks with semantic annotations. F1000Res. 5, 1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W-B, Choi WY, Lee D-H, Shim H, Kim-Ha J, Kim Y-J, 2019. OAS1 and OAS3 negatively regulate the expression of chemokines and interferon-responsive genes in human macrophages. BMB Rep. 52, 133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann MH, Torres-Domínguez LE, Price PJR, Brandmüller C, Kirschning CJ, Sutter G, 2016. CCL2 expression is mediated by type I IFN receptor and recruits NK and T cells to the lung during MVA infection. J. Leukoc. Biol. 99, 1057–1064. [DOI] [PubMed] [Google Scholar]
- Li K, Underhill DM, 2020. C-Type Lectin Receptors in Phagocytosis. Curr. Top. Microbiol. Immunol. 429, 1–18. [DOI] [PubMed] [Google Scholar]
- Lian H, Roy E, Zheng H, 2016. Protocol for Primary Microglial Culture Preparation. Bio Protoc. 6 10.21769/BioProtoc.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao F, Li A, Xiong M, Bien-Ly N, Jiang H, Zhang Y, Finn MB, Hoyle R, Keyser J, Lefton KB, Robinson GO, Serrano JR, Silverman AP, Guo JL, Getz J, Henne K, Leyns CE, Gallardo G, Ulrich JD, Sullivan PM, Lerner EP, Hudry E, Sweeney ZK, Dennis MS, Hyman BT, Watts RJ, Holtzman DM, 2018. Targeting of nonlipidated, aggregated apoE with antibodies inhibits amyloid accumulation. J. Clin. Invest. 128, 2144–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B, 2019. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47, W199–W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillis AP, Greenlee MC, Mikhailenko I, Pizzo SV, Tenner AJ, Strickland DK, Bohlson SS, 2008. Murine low-density lipoprotein receptor-related protein 1 (LRP) is required for phagocytosis of targets bearing LRP ligands but is not required for C1q-triggered enhancement of phagocytosis. J. Immunol. 181, 364–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-T, Seo J, Gao F, Feldman HM, Wen H-L, Penney J, Cam HP, Gjoneska E, Raja WK, Cheng J, Rueda R, Kritskiy O, Abdurrob F, Peng Z, Milo B, Yu CJ, Elmsaouri S, Dey D, Ko T, Yankner BA, Tsai L-H, 2018. APOE4 Causes Widespread Molecular and Cellular Alterations Associated with Alzheimer’s Disease Phenotypes in Human iPSC-Derived Brain Cell Types. Neuron 98, 1141–1154.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C-C, Liu C-C, Kanekiyo T, Xu H, Bu G, 2013. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat. Rev. Neurol. 9, 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loving BA, Bruce KD, 2020. Lipid and Lipoprotein Metabolism in Microglia. Front. Physiol. 11, 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzio JP, Pryor PR, Bright NA, 2007. Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 8, 622–632. [DOI] [PubMed] [Google Scholar]
- Ma L, Ouyang Q, Werthmann GC, Thompson HM, Morrow EM, 2017. Live-cell Microscopy and Fluorescence-based Measurement of Luminal pH in Intracellular Organelles. Front. Cell Dev. Biol. 5, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW, Weisgraber KH, Huang Y, 2009. Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer’s disease to AIDS. J. Lipid Res. 50 (Suppl), S183–S188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser C, Stevenson A, Banner S, Davies J, Tudor EL, Ono Y, Leigh PN, McLoughlin DM, Shaw CE, Miller CCJ, 2008. Deregulation of PKN1 activity disrupts neurofilament organisation and axonal transport. FEBS Lett. 582, 2303–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschallinger J, Iram T, Zardeneta M, Lee SE, Lehallier B, Haney MS, Pluvinage JV, Mathur V, Hahn O, Morgens DW, Kim J, Tevini J, Felder TK, Wolinski H, Bertozzi CR, Bassik MC, Aigner L, Wyss-Coray T, 2020. Lipid- droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 23, 194–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Diab HI, Brady OA, Puertollano R, 2016. TFEB and TFE 3 are novel components of the integrated stress response. EMBO J. 35, 479–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashud R, Nomachi A, Hayakawa A, Kubouchi K, Danno S, Hirata T, Matsuo K, Nakayama T, Satoh R, Sugiura R, Abe M, Sakimura K, Wakana S, Ohsaki H, Kamoshida S, Mukai H, 2017. Impaired lymphocyte trafficking in mice deficient in the kinase activity of PKN1. Sci. Rep. 7, 7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazaheri F, Breus O, Durdu S, Haas P, Wittbrodt J, Gilmour D, Peri F, 2014. Distinct roles for BAI1 and TIM-4 in the engulfment of dying neurons by microglia. Nat. Commun. 5, 4046. [DOI] [PubMed] [Google Scholar]
- Mazumder B, Li X, Barik S, 2010. Translation control: a multifaceted regulator of inflammatory response. J. Immunol. 184, 3311–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuade A, Kang YJ, Hasselmann J, Jairaman A, Sotelo A, Coburn M, Shabestari SK, Chadarevian JP, Fote G, Tu CH, Danhash E, Silva J, Martinez E, Cotman C, Prieto GA, Thompson LM, Steffan JS, Smith I, Davtyan H, Cahalan M, Cho H, Blurton-Jones M, 2020. Gene expression and functional deficits underlie TREM2-knockout microglia responses in human models of Alzheimer’s disease. Nat. Commun. 11, 5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD, 2010. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5, e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner R, Campbell IL, 2003. The extracellular matrix and cytokines regulate microglial integrin expression and activation. J. Immunol. 170, 3850–3858. [DOI] [PubMed] [Google Scholar]
- Mouton-Liger F, Rebillat A-S, Gourmaud S, Paquet C, Leguen A, Dumurgier J, Bernadelli P, Taupin V, Pradier L, Rooney T, Hugon J, 2015. PKR downregulation prevents neurodegeneration and β-amyloid production in a thiamine-deficient model. Cell Death Dis. 6, e1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth C, Hartmann A, Sepulveda-Falla D, Glatzel M, Krasemann S, 2019. Phagocytosis of Apoptotic Cells Is Specifically Upregulated in ApoE4 Expressing Microglia in vitro. Front. Cell. Neurosci. 13, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Furuhashi M, Li P, Cao H, Tuncman G, Sonenberg N, Gorgun CZ, Hotamisligil GS, 2010. Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell 140, 338–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher JJ, Neniskyte U, Brown GC, 2012. Primary phagocytosis of neurons by inflamed microglia: potential roles in neurodegeneration. Front. Pharmacol. 3, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu SC, Pa J, Kukull W, Beekly D, Kuzma A, Gangadharan P, Wang L-S, Romero K, Arneric SP, Redolfi A, Orlandi D, Frisoni GB, Au R, Devine S, Auerbach S, Espinosa A, Boada M, Ruiz A, Johnson SC, Koscik R, Wang JJ, Hsu W-C, Chen Y-L, Toga AW, 2017. Apolipoprotein E Genotype and Sex Risk Factors for Alzheimer Disease: A Meta-analysis. JAMA Neurol. 74, 1178–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuner SM, Heuer SE, Huentelman MJ, O’Connell KMS, Kaczorowski CC, 2019. Harnessing Genetic Complexity to Enhance Translatability of Alzheimer’s Disease Mouse Models: A Path toward Precision Medicine. Neuron 101 (399–411), e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuner SM, Tcw J, Goate AM, 2020. Genetic architecture of Alzheimer’s disease. Neurobiol. Dis. 143, 104976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Holtman IR, Coufal NG, Schlachetzki JCM, Yu M, Hu R, Han CZ, Pena M, Xiao J, Wu Y, Keulen Z, Pasillas MP, O’Connor C, Nickl CK, Schafer ST, Shen Z, Rissman RA, Brewer JB, Gosselin D, Gonda DD, Levy ML, Rosenfeld MG, McVicker G, Gage FH, Ren B, Glass CK, 2019. Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Science 366, 1134–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova G, Kapoor M, Abud EM, Efthymiou AG, Cheng H, Fullard JF, Bendl J, Roussos P, Poon WW, Hao K, Marcora E, Goate AM, 2019. Integration of Alzheimer’s disease genetics and myeloid cell genomics identifies novel causal variants, regulatory elements, genes and pathways. Cold Spring Harbor Lab. 10.1101/694281. [DOI] [Google Scholar]
- Novikova G, Kapoor M, Tcw J, Abud EM, Efthymiou AG, Chen SX, Cheng H, Fullard JF, Bendl J, Liu Y, Roussos P, Bjorkegren JL, Liu Y, Poon WW,¨ Hao K, Marcora E, Goate AM, 2021. Integration of Alzheimer’s disease genetics and myeloid genomics identifies disease risk regulatory elements and genes. Nat. Commun. 12, 1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent AA, Lin K, van Lengerich B, Lianoglou S, Przybyla L, Davis SS, Llapashtica C, Wang J, Kim DJ, Xia D, Lucas A, Baskaran S, Haddick PCG, Lenser M, Earr TK, Shi J, Dugas JC, Andreone BJ, Logan T, Solanoy HO, Chen H, Srivastava A, Poda SB, Sanchez PE, Watts RJ, Sandmann T, Astarita G, Lewcock JW, Monroe KM, Di Paolo G, 2020. TREM2 Regulates Microglial Cholesterol Metabolism upon Chronic Phagocytic Challenge. Neuron 105, 837–854.e9. [DOI] [PubMed] [Google Scholar]
- Oliveira MM, Lourenco MV, Longo F, Kasica NP, Yang W, Ureta G, Ferreira DDP, Mendonça PHJ, Bernales S, Ma T, De Felice FG, Klann E, Ferreira ST, 2021. Correction of eIF2-dependent defects in brain protein synthesis, synaptic plasticity, and memory in mouse models of Alzheimer’s disease. Sci. Signal. 14 10.1126/scisignal.abc5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann JA, Carvalho P, 2019. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20, 137–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onat UI, Yildirim AD, Tufanli O, Çimen I, Kocatürk B, Veli Z, Hamid SM,Shimada K, Chen S, Sin J, Shah PK, Gottlieb RA, Arditi M, Erbay E, 2019. Intercepting the lipid-induced integrated stress response reduces atherosclerosis. J. Am. Coll. Cardiol. 73, 1149–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM, 2016. The integrated stress response. EMBO Rep. 17, 1374–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panousis CG, Zuckerman SH, 2000. Regulation of cholesterol distribution in macrophage-derived foam cells by interferon-gamma. J. Lipid Res. 41, 75–83. [PubMed] [Google Scholar]
- Park D, Tosello-Trampont A-C, Elliott MR, Lu M, Haney LB, Ma Z, Klibanov AL, Mandell JW, Ravichandran KS, 2007. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 450, 430–434. [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C, 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimenova AA, Marcora E, Goate AM, 2017. A Tale of Two Genes: Microglial Apoe and Trem2. Immunity. 47, 398–400. [DOI] [PubMed] [Google Scholar]
- Plaisier SB, Taschereau R, Wong JA, Graeber TG, 2010. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 38, e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlésny-Drabiniok A, Marcora E, Goate AM, 2020. Microglial Phagocytosis: A Disease-Associated Process Emerging from Alzheimer’s Disease Genetics. Trends Neurosci. 43, 965–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz M, Priller J, Sisodia SS, Ransohoff RM, 2011. Heterogeneity of CNS myeloid cells and their roles in neurodegeneration. Nat. Neurosci. 14, 1227–1235. [DOI] [PubMed] [Google Scholar]
- Qiu B, Simon MC, 2016. BODIPY 493/503 Staining of Neutral Lipid Droplets for Microscopy and Quantification by Flow Cytometry. Bio Protoc. 6 10.21769/BioProtoc.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj T, Chibnik LB, McCabe C, Wong A, Replogle JM, Yu L, Gao S, Unverzagt FW, Stranger B, Murrell J, Barnes L, Hendrie HC, Foroud T, Krichevsky A, Bennett DA, Hall KS, Evans DA, De Jager PL, 2017. Genetic architecture of age-related cognitive decline in African Americans. Neurol. Genet. 3, e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff RM, Perry VH, 2009. Microglial physiology: unique stimuli, specialized responses. Annu. Rev. Immunol. 27, 119–145. [DOI] [PubMed] [Google Scholar]
- Raport CJ, Gray PW, 2010. Chapter 24 - Chemokines and Chemokine Receptors: Structure and Function. In: Bradshaw RA, Dennis EA (Eds.), Handbook of Cell Signaling, Second edition. Academic Press, San Diego, pp. 157–161. [Google Scholar]
- Rauch I, Müller M, Decker T, 2013. The regulation of inflammation by interferons and their STATs. JAKSTAT 2, e23820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Arboleda-Velasquez JF, Quiroz YT, Huentelman MJ, Beach TG, Caselli RJ, Chen Y, Su Y, Myers AJ, Hardy J, Paul Vonsattel J, Younkin SG, Bennett DA, De Jager PL, Larson EB, Crane PK, Keene CD, Kamboh MI, Kofler JK, Duque L, Gilbert JR, Gwirtsman HE, Buxbaum JD, Dickson DW, Frosch MP, Ghetti BF, Lunetta KL, Wang L-S, Hyman BT, Kukull WA, Foroud T, Haines JL, Mayeux RP, Pericak-Vance MA, Schneider JA, Trojanowski JQ, Farrer LA, Schellenberg GD, Beecham GW, Montine TJ, Jun GR, Alzheimer’s Disease Genetics Consortium, 2020. Exceptionally low likelihood of Alzheimer’s dementia in APOE2 homozygotes from a 5,000-person neuropathological study. Nat. Commun. 11, 667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss AB, Patel CA, Rahman MM, Chan ESL, Hasneen K, Montesinos MC, Trachman JD, Cronstein BN, 2004. Interferon-gamma impedes reverse cholesterol transport and promotes foam cell transformation in THP-1 human monocytes/macrophages. Med. Sci. Monit. 10, BR420–5. [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK, 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Ramírez L, Nieto-Sampedro M, Barreda-Manso MA, 2017. Integrated Stress Response as a Therapeutic Target for CNS Injuries. Biomed. Res. Int. 2017, 6953156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler AJ, Williams BRG, 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8, 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salih DA, Bayram S, Guelfi S, Reynolds RH, Shoai M, Ryten M, Brenton JW, Zhang D, Matarin M, Botia JA, Shah R, Brookes KJ, Guetta-Baranes T, Morgan K, Bellou E, Cummings DM, Escott-Price V, Hardy J, 2019. Genetic variability in response to amyloid beta deposition influences Alzheimer’s disease risk. Brain Commun. 1, fcz022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Hernandez AG, Buitrago L, Moreno H, Cardona-Gómez GP, Arboleda G,´ 2015. Role of Liver X Receptor in AD Pathophysiology. PLoS One 10, e0145467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, Pierre P, 2009. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277. [DOI] [PubMed] [Google Scholar]
- Segev Y, Michaelson DM, Rosenblum K, 2013. ApoE ε4 is associated with eIF2α phosphorylation and impaired learning in young mice. Neurobiol. Aging 34,863–872. [DOI] [PubMed] [Google Scholar]
- Shao S, Yang Y, Yuan G, Zhang M, Yu X, 2013. Signaling molecules involved in lipid-induced pancreatic beta-cell dysfunction. DNA Cell Biol. 32, 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Holtzman DM, 2018. Interplay between innate immunity and Alzheimer disease: APOE and TREM2 in the spotlight. Nat. Rev. Immunol. 18, 759–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Narayan P, Bonner JM, Kory N, Boland S, Arczewska AA, Ralvenius WT, Akay L, Lockshin E, He L, Milo B, Graziosi A, Baru V, Lewis CA, Kellis M, Sabatini DM, Tsai L-H, Lindquist S, 2021. APOE4 disrupts intracellular lipid homeostasis in human iPSC-derived glia. Sci. Transl. Med. 13 10.1126/scitranslmed.aaz4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims R, van der Lee SJ, Naj AC, Bellenguez C, Badarinarayan N, Jakobsdottir J, Kunkle BW, Boland A, Raybould R, Bis JC, Martin ER, Grenier-Boley B, Heilmann-Heimbach S, Chouraki V, Kuzma AB, Sleegers K, Vronskaya M, Ruiz A, Graham RR, Olaso R, Hoffmann P, Grove ML, Vardarajan BN, Hiltunen M, Nothen MM, White CC, Hamilton-Nelson KL, Epelbaum J,Maier W, Choi S-H, Beecham GW, Dulary C, Herms S, Smith AV, Funk CC, Derbois C, Forstner AJ, Ahmad S, Li H, Bacq D, Harold D, Satizabal CL, Valladares O, Squassina A, Thomas R, Brody JA, Qu L, Sanchez-Juan P,´ Morgan T, Wolters FJ, Zhao Y, Garcia FS, Denning N, Fornage M, Malamon J, Naranjo MCD, Majounie E, Mosley TH, Dombroski B, Wallon D, Lupton MK, Dupuis J, Whitehead P, Fratiglioni L, Medway C, Jian X, Mukherjee S, Keller L, Brown K, Lin H, Cantwell LB, Panza F, McGuinness B, Moreno-Grau S, Burgess JD, Solfrizzi V, Proitsi P, Adams HH, Allen M, Seripa D, Pastor P, Cupples LA, Price ND, Hannequin D, Frank- García A, Levy D, Chakrabarty P, Caffarra P, Giegling I, Beiser AS, Giedraitis V, Hampel H, Garcia ME, Wang X, Lannfelt L, Mecocci P, Eiriksdottir G, Crane PK, Pasquier F, Boccardi V, Henandez I, Barber RC,´ Scherer M, Tarraga L, Adams PM, Leber M, Chen Y, Albert MS, Riedel- Heller S, Emilsson V, Beekly D, Braae A, Schmidt R, Blacker D, Masullo C, Schmidt H, Doody RS, Spalletta G, Longstreth WT Jr., Fairchild TJ, Bossù P, Lopez OL, Frosch MP, Sacchinelli E, Ghetti B, Yang Q, Huebinger RM, Jessen F, Li S, Kamboh MI, Morris J, Sotolongo-Grau O, Katz MJ, Corcoran C, Dunstan M, Braddel A, Thomas C, Meggy A, Marshall R, Gerrish A, Chapman J, Aguilar M, Taylor S, Hill M, Fairen MD, Hodges A,´ Vellas B, Soininen H, Kloszewska I, Daniilidou M, Uphill J, Patel Y, Hughes JT, Lord J, Turton J, Hartmann AM, Cecchetti R, Fenoglio C, Serpente M, Arcaro M, Caltagirone C, Orfei MD, Ciaramella A, Pichler S, Mayhaus M, Gu W, Lleo A, Fortea J, Blesa R, Barber IS, Brookes K, Cupidi C, Maletta RG, Carrell D, Sorbi S, Moebus S, Urbano M, Pilotto A, Kornhuber J, Bosco P, Todd S, Craig D, Johnston J, Gill M, Lawlor B, Lynch A, Fox NC, Hardy J, Consortium ARUK, Albin RL, Apostolova LG, Arnold SE, Asthana S, Atwood CS, Baldwin CT, Barnes LL, Barral S, Beach TG, Becker JT, Bigio EH, Bird TD, Boeve BF, Bowen JD, Boxer A, Burke JR, Burns JM, Buxbaum JD, Cairns NJ, Cao C, Carlson CS, Carlsson CM, Carney RM, Carrasquillo MM, Carroll SL, Diaz CC, Chui HC, Clark DG, Cribbs DH, Crocco EA, DeCarli C, Dick M, Duara R, Evans DA, Faber KM, Fallon KB, Fardo DW, Farlow MR, Ferris S, Foroud TM, Galasko DR, Gearing M, Geschwind DH, Gilbert JR, Graff-Radford NR, Green RC, Growdon JH, Hamilton RL, Harrell LE, Honig LS, Huentelman MJ, Hulette CM, Hyman BT, Jarvik GP, Abner E, Jin L-W, Jun G, Karydas A, Kaye JA, Kim R, Kowall NW, Kramer JH, LaFerla FM, Lah JJ, Leverenz JB, Levey AI, Li G, Lieberman AP, Lunetta KL, Lyketsos CG, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Morris JC, Murrell JR, Myers AJ, O’Bryant S, Olichney JM, Pankratz VS, Parisi JE, Paulson HL, Perry W, Peskind E, Pierce A, Poon WW, Potter H, Quinn JF, Raj A, Raskind M, Reisberg B, Reitz C, Ringman JM, Roberson ED, Rogaeva E, Rosen HJ, Rosenberg RN, Sager MA, Saykin AJ, Schneider JA, Schneider LS, Seeley WW, Smith AG, Sonnen JA, Spina S, Stern RA, Swerdlow RH, Tanzi RE, Thornton-Wells TA, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Van Eldik LJ, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Wilhelmsen KC, Williamson J, Wingo TS, Woltjer RL, Wright CB, Yu C-E, Yu L, Garzia F, Golamaully F, Septier G, Engelborghs S, Vandenberghe R, De Deyn PP, Fernadez CM, Benito YA, Thonberg H, Forsell C, Lilius L, Kinhult- Stahlbom A, Kilander L, Brundin R, Concari L, Helisalmi S, Koivisto AM, Haapasalo A, Dermecourt V, Fievet N, Hanon O, Dufouil C, Brice A, Ritchie K, Dubois B, Himali JJ, Keene CD, Tschanz J, Fitzpatrick AL, Kukull WA, Norton M, Aspelund T, Larson EB, Munger R, Rotter JI, Lipton RB, Bullido MJ, Hofman A, Montine TJ, Coto E, Boerwinkle E, Petersen RC, Alvarez V, Rivadeneira F, Reiman EM, Gallo M, O’Donnell CJ, Reisch JS, Bruni AC, Royall DR, Dichgans M, Sano M, Galimberti D, St George-Hyslop P, Scarpini E, Tsuang DW, Mancuso M, Bonuccelli U, Winslow AR, Daniele A, Wu C-K, GERAD/PERADES, CHARGE, ADGC, EADI, Peters O, Nacmias B, Riemenschneider M, Heun R, Brayne C, Rubinsztein DC, Bras J, Guerreiro R, Al-Chalabi A, Shaw CE, Collinge J, Mann D, Tsolaki M, Clarimon J, Sussams R, Lovestone S, O’Donovan MC, Owen MJ, Behrens TW, Mead S, Goate AM, Uitterlinden AG, Holmes C, Cruchaga C, Ingelsson M, Bennett DA, Powell J, Golde TE, Graff C, De Jager PL, Morgan K, Ertekin-Taner N, Combarros O, Psaty BM, Passmore P, Younkin SG, Berr C, Gudnason V, Rujescu D, Dickson DW, Dartigues J-F, DeStefano AL, Ortega-Cubero S, Hakonarson H, Campion D, Boada M, Kauwe JK, Farrer LA, Van Broeckhoven C, Ikram MA, Jones L, Haines JL, Tzourio C, Launer LJ, Escott-Price V, Mayeux R, Deleuze J-F, Amin N, Holmans PA, Pericak-Vance MA, Amouyel P, van Duijn CM, Ramirez A, Wang L-S, Lambert J-C, Seshadri S, Williams J, Schellenberg GD, 2017. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat. Genet. 49, 1373–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Dragunow M, 2014. The human side of microglia. Trends Neurosci. 37, 125–135. [DOI] [PubMed] [Google Scholar]
- Stewart DJ, Behrens C, Roth J, Wistuba II, 2011. Exponential decay nonlinear regression analysis of patient survival curves: preliminary assessment in non-small cell lung cancer. Lung Cancer 71, 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PM, Mezdour H, Aratani Y, Knouff C, Najib J, Reddick RL, Quarfordt SH, Maeda N, 1997. Targeted Replacement of the Mouse Apolipoprotein E Gene with the Common Human APOE3 Allele Enhances Diet-induced Hypercholesterolemia and Atherosclerosis*. J. Biol. Chem. 272, 17972–17980. [DOI] [PubMed] [Google Scholar]
- Taniuchi S, Miyake M, Tsugawa K, Oyadomari M, Oyadomari S, 2016. Integrated stress response of vertebrates is regulated by four eIF2α kinases. Sci. Rep. 6, 32886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcw J, Liang SA, Qian L, Pipalia NH, Chao MJ, 2019. Cholesterol and matrisome pathways dysregulated in human APOE ε4 glia. BioRxiv. preprint 713362. [Google Scholar]
- Tzioras M, Davies C, Newman A, Jackson R, Spires-Jones T, 2019. Invited Review: APOE at the interface of inflammation, neurodegeneration and pathological protein spread in Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 45, 327–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulland TK, Colonna M, 2018. TREM2 - a key player in microglial biology and Alzheimer disease. Nat. Rev. Neurol. 14, 667–675. [DOI] [PubMed] [Google Scholar]
- Ulrich JD, Burchett JM, Restivo JL, Schuler DR, Verghese PB, Mahan TE, Landreth GE, Castellano JM, Jiang H, Cirrito JR, Holtzman DM, 2013. In vivo measurement of apolipoprotein E from the brain interstitial fluid using microdialysis. Mol. Neurodegener. 8, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe-Querol E, Rosales C, 2020. Phagocytosis: Our Current Understanding of a Universal Biological Process. Front. Immunol. 11, 1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Giau V, Bagyinszky E, An SSA, Kim SY, 2015. Role of apolipoprotein E in neurodegenerative diseases. Neuropsychiatr. Dis. Treat. 11, 1723–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa A, Gelosa P, Castiglioni L, Cimino M, Rizzi N, Pepe G, Lolli F, Marcello E, Sironi L, Vegeto E, Maggi A, 2018. Sex-Specific Features of Microglia from Adult Mice. Cell Rep. 23, 3501–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitek MP, Brown CM, Colton CA, 2009. APOE genotype-specific differences in the innate immune response. Neurobiol. Aging 30, 1350–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volmer R, van der Ploeg K, Ron D, 2013. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc. Natl. Acad. Sci. U. S. A. 110, 4628–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White GE, Iqbal AJ, Greaves DR, 2013. CC chemokine receptors and chronic inflammation–therapeutic opportunities and pharmacological challenges. Pharmacol. Rev. 65, 47–89. [DOI] [PubMed] [Google Scholar]
- Wolfe CM, Fitz NF, Nam KN, Lefterov I, Koldamova R, 2018. The Role of APOE and TREM2 in Alzheimer’s Disease-Current Understanding and Perspectives. Int. J. Mol. Sci. 20 10.3390/ijms20010081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YL, LeBon L, Edalji R, Lim HB, Sun C, Sidrauski C, 2018. The small molecule ISRIB rescues the stability and activity of Vanishing White Matter Disease eIF2B mutant complexes. Elife 7. 10.7554/eLife.32733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z-X, Kim GH, Tan J-W, Riso AE, Sun Y, Xu EY, Liao G-Y, Xu H, Lee SH, Do N-Y, Lee CH, Clipperton-Allen AE, Kwon S, Page DT, Lee KJ, Xu B, 2020. Elevated protein synthesis in microglia causes autism-like synaptic and behavioral aberrations. Nat. Commun. 11, 1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Sidorenko J, Couvy-Duchesne B, Marioni RE, Wright MJ, Goate AM, Marcora E, Huang K-L, Porter T, Laws SM, Australian Imaging Biomarkers and Lifestyle (AIBL) Study, Sachdev PS, Mather KA, Armstrong NJ, Thalamuthu A, Brodaty H, Yengo L, Yang J, Wray NR, McRae AF, Visscher PM, 2020. Risk prediction of late-onset Alzheimer’s disease implies an oligogenic architecture. Nat. Commun. 11, 4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, Ren Y, Yamazaki Y, Qiao W, Li F, Felton LM, Mahmoudiandehkordi S, Kueider-Paisley A, Sonoustoun B, Arnold M, Shue F, Zheng J, Attrebi ON, Martens YA, Li Z, Bastea L, Meneses AD, Chen K, Thompson JW, St John- Williams L, Tachibana M, Aikawa T, Oue H, Job L, Yamazaki A, Liu C-C, Storz P, Asmann YW, Ertekin-Taner N, Kanekiyo T, Kaddurah-Daouk R, Bu G, 2020. Alzheimer’s risk factors age, APOE genotype, and sex drive distinct molecular pathways. Neuron 106 (727–742.e6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong N, Weisgraber KH, 2009. Understanding the basis for the association of apoE4 with Alzheimer’s disease: opening the door for therapeutic approaches. Curr. Alzheimer Res. 6, 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu PJ, Huang W, Kalikulov D, Yoo JW, Placzek AN, Stoica L, Zhou H, Bell JC, Friedlander MJ, Krnjevíc K, Noebels JL, Costa-Mattioli M, 2011. Suppression of PKR promotes network excitability and enhanced cognition by interferon-γ-mediated disinhibition. Cell 147, 1384–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyryanova AF, Kashiwagi K, Rato C, Harding HP, Crespillo-Casado A, Perera LA, Sakamoto A, Nishimoto M, Yonemochi M, Shirouzu M, Ito T, Ron D, 2021. ISRIB Blunts the Integrated Stress Response by Allosterically Antagonising the Inhibitory Effect of Phosphorylated eIF2 on eIF2B. Mol. Cell 81, 88–103.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.