Abstract
The mammalian circadian clock regulates a wide variety of physiological and behavioral processes. In turn, its disruption is associated with sleep deficiency, metabolic syndrome, neurological and psychiatric disorders, and cancer. At the turn of the century, the circadian clock was determined to be regulated by a transcriptional negative feedback mechanism composed of a dozen core clock genes. More recently, large-scale genomic studies have expanded the clock into a complex network composed of thousands of gene outputs and inputs. A major task of circadian research is to utilize systems biological approaches to uncover the governing principles underlying cellular oscillatory behavior and advance understanding of biological functions at the genomic level with spatiotemporal resolution. This review focuses on the genes and pathways that provide inputs to the circadian clock. Several emerging examples include AMP-activated protein kinase AMPK, nutrient/energy sensor mTOR, NAD+-dependent deacetylase SIRT1, hypoxia-inducible factor HIF1α, oxidative stress-inducible factor NRF2, and the proinflammatory factor NF-κB. Among others that continue to be revealed, these input pathways reflect the extensive interplay between the clock and cell physiology through the regulation of core clock genes and proteins. While the scope of this crosstalk is well-recognized, precise molecular links are scarce, and the underlying regulatory mechanisms are not well understood. Future research must leverage genetic and genomic tools and technologies, network analysis, and computational modeling to characterize additional modifiers and input pathways. This systems-based framework promises to advance understanding of the circadian timekeeping system and may enable the enhancement of circadian functions through related input pathways.
Keywords: circadian clock, systems biology, cell homeostasis, mTOR, NF-κB
Circadian Systems Biology
Physiology and behavior are the product of complex systems rather than simple outputs of single genes. Thus, their integrative understanding should be achieved using a systems approach at the genome-wide level with both spatial and temporal resolution. The Human Genome Project highlights this, as it utilized an interdisciplinary approach to generate a large volume of high throughput data and helped pave the way for systems biology. While a concise and universal definition of systems biology remains elusive, it can be thought of as an iterative process: identifying the components of a system, determining how the components fit together, and developing models to describe the emergent properties of the system; for example, the circadian oscillatory behavior in cells and organisms [1–3].
Understanding how gene networks produce emergent properties in physiology and behavior is a fundamental question in biology. The mammalian circadian timekeeping system represents an ideal model for this research [4,5]. The cellular level holds a particularly important position in circadian timekeeping, as the underlying genetic program is ubiquitous amongst nearly all cell types, and circadian oscillation is cell-autonomous [6–8], although different tissues exhibit physiologically distinctive functions [9–11]. Cellular clock models have been used extensively for mechanistic studies and systematic identification of novel components as the first step toward a systems understanding of circadian timekeeping. Circadian systems biology research has led the way in genome-wide analyses of the clock outputs in multiple cell and tissue types, identified additional clock modifiers, and uncovered the extensive interplay between the circadian clock and cell physiology. This systems biology approach is being used to uncover circadian functions at the higher biological levels, particularly the varying organ and tissue types. These efforts are ongoing, but future research requires more focused network analysis and computational modeling to uncover the missing connections linking genes to complex oscillatory behavior.
Current Understanding of the Clock Mechanism
The circadian system consists of a hierarchy of oscillators at the cellular, tissue, and organismal levels. The suprachiasmatic nucleus (SCN) of the anterior hypothalamus acts as the central clock to coordinate the extra-SCN and peripheral oscillators into a coherent timekeeping system [8,12]. The SCN clock is reset by environmental cues, with photic input being the most dominant by transmitting input about the light-dark cycle via the retina to align with the 24-h rotation of the Earth.
The E-box-centered Core Negative Feedback Loop
Although each tissue is distinctive at the physiological level, they share a similar molecular mechanism at the cellular level: an autoregulatory transcriptional/translational negative feedback loop [11] (Fig. 1). In mammals, the positive arm consists of BMAL1 and CLOCK, two bHLH-PAS transcriptional activators that heterodimerize and bind to E-box cis-elements to upregulate the transcription of Period (Per1, Per2, Per3) and Cryptochrome (Cry1, Cry2). Thesegenes are transcribed and subsequently translated, after which they dimerize in the cytoplasm. After accumulation in the cytoplasm, the PER/CRY complex is translocated into the nucleus and represses the transcriptional activity of BMAL1-CLOCK, thereby inhibiting their own transcription. During the early repression phase, PER represses E-box transcription by binding to CRY1 and displacing the BMAL1-CLOCK-CRY complex from the E-box in a CRY-dependent manner. This is followed by the late repression phase, in which CRY1 represses BMAL1-CLOCK, extending CRY1 repression into the next cycle [13,14].
Fig. 1. The mammalian circadian clock.
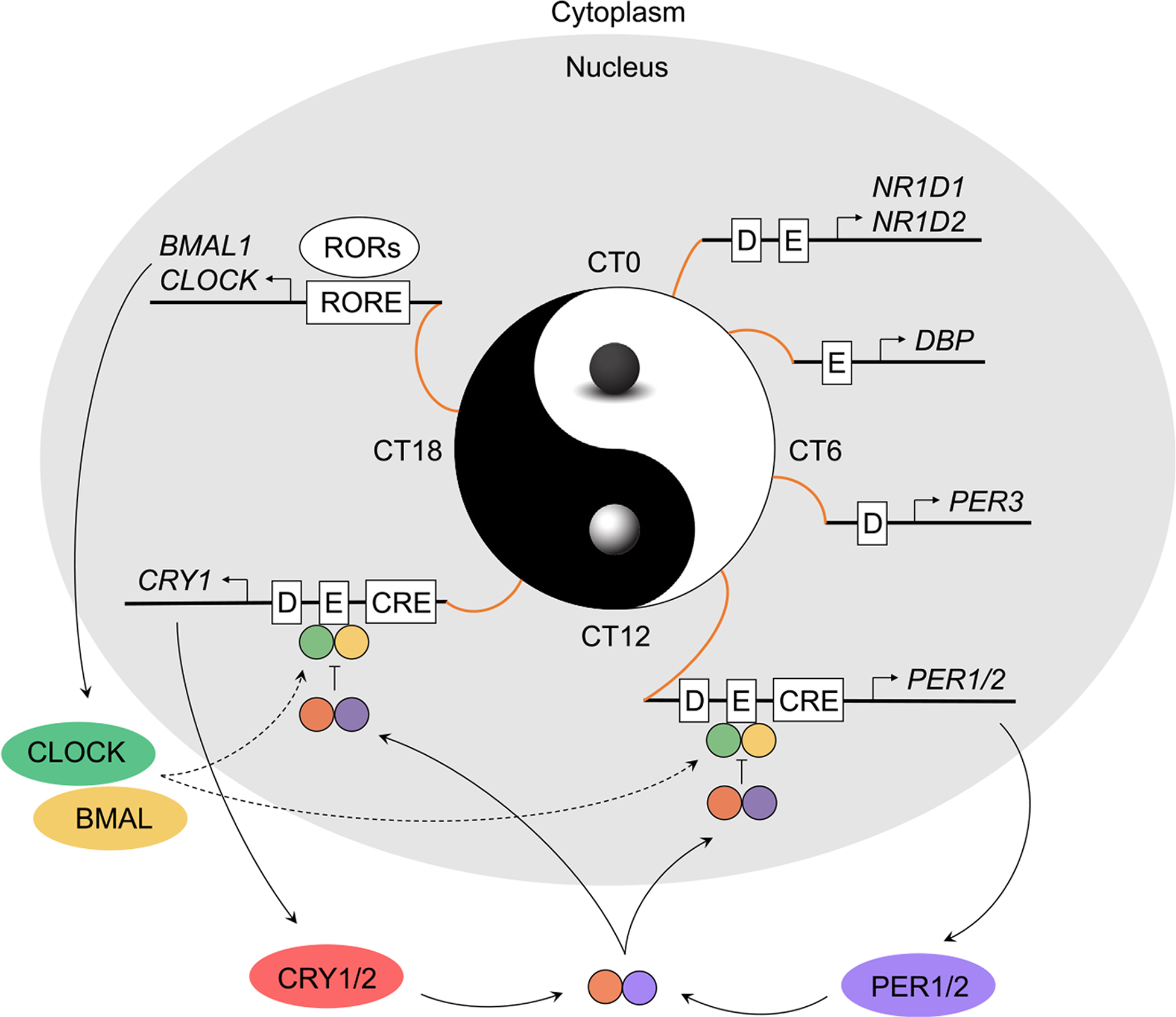
The circadian clock is a highly conserved mechanism that synchronizes numerous oscillating proteins into the endogenous timekeeping system. At the core of the circadian clock is a transcriptional negative feedback loop, in which clock activators BMAL1 and CLOCK dimerize to act upon the respective E-Box elements of Per and Cry. In turn, PER and CRY dimerize within the cytoplasm and return to the nucleus to repress BMAL1-CLOCK and, subsequently, their own transcription. Additionally, BMAL1-CLOCK acts upon the respective E-Boxes of NR1D1/NR1D2 and DBP to promote their expression, while NR1D1/NR1D2 and DBP act to repress the transcription of Bmal1 and activate the transcription of RORs, respectively.
The D-box and RORE-centered Interlocking Loops
Two additional feedback loops are integrated with the E-box-centered core clock mechanism, mediated by the retinoic acid-related orphan receptor-binding element (RORE) and the D-box [11,15] (Fig. 1). Within the RORE loop, ROR activators (RORα, RORβ, and RORγ) and repressors (REV-ERBα, or NR1D1; REV-ERBβ, or NR1D2) act to regulate rhythmic transcription of BMAL1, CLOCK, and NFIL3 (or E4BP4). The D-box transcription is regulated by the NFIL3 repressor and DBP, TEF, and HLF activators [16]. Circadian transcription mediated by the D-box, E-box, and RORE cis-elements forms a clockwork more complex than the E-box loop alone. Within the clockwork, circadian DBP and NR1D1 expression are regulated primarily by the E-box, PER3 by the D-box, and BMAL1 and CLOCK by RORE with each exhibiting a distinct phase. PER1 and PER2, through regulation of the E-box by a still-to-be-defined mechanism, display a delayed phase in comparison to DBP and NR1D1. Interestingly, CRY1 (as well as RORs) is regulated by a combinatorial mechanism involving all three cis-elements, rendering a significant phase delay in comparison to DBP and NR1D1 [17]. These loops, individually and in combination, underlie the rhythmic expression of a large number of genes at several distinct circadian phases [10,13,15]. The core and secondary interlocking loops function to impart robustness against genetic perturbation, as well as offer additional nodes to regulate additional circadian outputs [18].
Over the past two decades, several major genome-wide studies have identified thousands of clock output genes and hundreds of clock input genes. Thus, the relatively simple clockwork centered on the three cis-elements has further expanded to a more complex circadian gene network. This network complexity appears to be the norm in biological systems and disease states, not the exception [19–22].
Post-translational Regulation of PER and CRY
Post-translational modifications and clock protein turnover rates play critical roles in the regulation of circadian oscillation [11,23,24]. The PER and CRY repressors, in particular, have a dominant role in circadian transcription. In principle, PER/CRY repression involves protein accumulation, post-translational modifications, translocation into the nucleus, repression of BMAL1-CLOCK, and eventual degradation. The PER protein stability and turnover, regulated by its respective phosphorylation and ubiquitination states, in turn regulates the speed of the clock. PER1 and PER2 have been shown to be phosphorylated by CK1D and CK1E kinases, dephosphorylated by phosphatase PP1, and subsequently ubiquitinated by the bTrCP (aka FBXW1) E3 ubiquitin ligase complex for proteasome degradation [25,26]. In CK1D/E deficient cells and mouse models, elevated PER slows transcriptional progression and produces a longer period. Conversely, a CK1E gain-of-function (tau) mutation led to the acceleration of PER degradation and shortened period length [27,28]. This is showcased in familial advanced sleep phase syndrome, which is caused by a missense mutation in PER2 that interferes with its phosphorylation by CK1E, and thereby shortens the period length [29].
Similarly, CRY is regulated by the coordinated actions of FBXL3 and FBXL21. They act as CRY recognition F-box proteins in the SKP1/CUL1/F-box (SCF) E3 ligase complex for CRY ubiquitination and proteasomal degradation. Loss-of-function mutations in FBXL3 compromise its ubiquitination activity, allowing CRY stabilization, and subsequently causing lengthened circadian periods [30,31]. FBXL21 is predominantly localized in the cytosol and functions in concert with FBXL3 to regulate the normal period length. FBXL3 functions as the primary CRY E3 ligase to slow down CRY buildup. Interestingly, FBXL21 binds to CRY with a stronger affinity but is less effective in ubiquitination. In the nucleus, FBXL21 counteracts FBXL3 activity, which results in less efficient CRY ubiquitination, thereby stabilizing CRY in the nucleus. With a missense mutation in FBXL21, CRY proteins are degraded faster, and a shortened period is produced [32,33].
While it is generally accepted that these factors regulate the nuclear translocation and accumulation of the PER/CRY repressor complex, the precise steps by which they do so remain elusive. A recent study implicated a perinuclear protein complex (including CK1) assembled by a long noncoding RNA titled NRON [34]. Perturbation of the NRON scaffolding complex affects PER/CRY nuclear translocation, dampens amplitude, and alters period length. An active area of research is the elucidation of the relative nuclear/cytoplasmic levels of PER and CRY repressors, nucleocytoplasmic ratio of BMAL1/CLOCK and PER/CRY, and relative potential of transcriptional activation versus repression.
The Outputs: Tissue-specific Circadian Output Networks
The circadian clock extends well beyond the core and secondary loops mediated by the three circadian cis-elements. Circadian regulated genes have been profiled in a number of organ/tissue and cell types, including adrenal gland, aorta, brainstem, brown fat, cerebellum, colon, heart, hypothalamus, kidney, liver, lung, pancreas, pituitary, SCN, skeletal muscle, spleen, white adipose, pancreatic beta cells, fibroblasts, hepatocytes, and osteosarcoma cells [10,35–41]. In the most comprehensive genome-wide study to date, protein-coding genes in mice were screened for circadian rhythms using RNA-sequencing and DNA arrays across twelve tissue types. Nearly half of all genes screened were found to exhibit rhythmic transcription in at least one organ [10]. Nearly all organs studied were found to have two primary periods of high transcription: dawn and dusk. These studies demonstrate the pervasiveness of circadian regulation in cellular and organismal physiology. Further, most core clock genes were found to be rhythmically expressed in-phase across all the tissue types, with the majority of these genes highly specific to both cell and tissue type. For instance, the liver contains rhythmically expressed genes that are often involved in the rate-limiting steps of various metabolic pathways. While these studies advanced our understanding of circadian regulation of biological systems and contribute significantly to systems biology of the circadian clock, major questions remain. In particular, future studies should elucidate how tissue-specific circadian gene expression and phases are achieved.
Noncoding and regulatory RNA has also been shown to oscillate in a circadian manner. In a study of the liver transcriptome, lincRNAs, and miRNAs (several of which have known functions in the liver) were found to have circadian expression patterns [10,42]. Nascent RNA transcripts have been demonstrated to loosely overlap with mRNA transcripts in amplitude and timing, which emphasizes consideration of post-transcriptional regulation in studying circadian timing [43].
Studies have emerged that profiled the cistrome and epigenome to uncover the genome-wide target sites of core clock components [13,44–46]. In the most comprehensive study to date, Koike and colleagues examined transcriptional regulation at the genome-wide level with a temporal focus on transcription factors, RNA polymerase II (RNAPII) recruitment, nascent RNA expression, and chromatin remodeling. The transcriptional circadian cycle of the clock can be understood in three distinct phases. During the poised phase, CRY1 represses BMAL1-CLOCK, followed by a decline in occupancy of CRY1 and an increase in binding of the coactivator p300. During the transcriptional activation phase, BMAL1 and CLOCK heterodimerize and bind to E-box cis-elements to upregulate the transcription of PER and CRY. Lastly, during the repression phase, the PER/CRY complex is translocated into the nucleus and begins to repress the transcriptional activity of BMAL1-CLOCK. This final phase encompasses a larger period of repression than that of the first, as PER and CRY must accumulate to levels that are sufficient to repress BMAL1-CLOCK. Chromatin remodeling also is regulated by the clock. Histone modifications, such as H3K4me3 and H3K36me3, are rhythmic and vary in amplitude among genes. RNAPII was also found to demonstrate rhythmic recruitment. RNAPII recruitment appeared 3 h prior to H3K36me3 and mRNA accumulation and 1 h prior to H3K4me3. Therefore, the recruitment and initiation of RNAPII, in conjunction with histone modification, appears to be regulated by the circadian clock. These studies provided insight into the genome-wide circadian transcriptional mechanisms on the chromatin level.
A similar circadian gene expression paradigm can be seen across several recent studies utilizing human tissues [47–51]. Powered by the cyclic ordering by periodic structure (CYCLOPS) algorithm, designed to reconstruct human sample temporal order in the absence of time-of-day information, these studies identified thousands of genes exhibiting rhythmic expression in the body. More specifically, nearly half of protein-coding genes were shown to be cycling in at least one of the thirteen tissues from several hundred human donors. Collectively, these studies reveal the prevalence of clock-regulated genes in both mice and humans. Further, the majority of all manufactured drugs target genes with circadian expression, contributing to the relatively novel practice of circadian medicine that calls for prospective chronotherapeutics in translational research and clinical medicine [52,53].
The Inputs: Circadian Networking with Cell Physiology
Intercellular coupling in the SCN confers robustness against genetic perturbation. However, peripheral clocks function autonomously and lack strong intercellular coupling, making them more susceptible to genetic manipulation [8,12]. Thus, cell-based assays allow for the discovery of cell-autonomous circadian defects, and strategical cell-based models are more tractable for phenotypic screening and rapid discovery of clock function. Several groups, including our own, have developed peripheral cell-based clock model systems for clock studies [12,54–56]. In a collaborative effort to gain a systems-level understanding of the molecular clock network, we carried out a high-throughput genome-wide siRNA screen in a U2OS (human osteosarcoma) cell line and were able to identify nearly a thousand clock modifiers [55]. The identified genes were additionally screened for “network effects” and were found to have a dose-dependent impact on BMAL1, CLOCK, PER1, PER2, CRY1, CRY2, DBP, and NR1D1 expression levels. Pathway and network analyses have revealed a deep integration between the circadian clock and many cellular functions [55]. Many of the clock modifiers, through direct and indirect pathways, associate with known core clock components. In particular, while inhibition of multiple components of the insulin signaling pathway resulted in circadian oscillation perturbation, many components of the pathway are regulated at the transcriptional level by the circadian clock [10,55]. Thus, insulin signaling and the circadian clock are functionally integrated. Network analysis in several additional studies revealed dynamic circadian protein-protein interaction networks, which enabled the prediction of temporal organization of cellular functions and identification of additional clock factors [57,58]. Other small-scale screens have identified post-translational modifiers, such as kinases, phosphatases, F-box proteins, and E3 ubiquitin ligases that target NR1D1 and affect circadian rhythms by often targeting a protein for proteasomal degradation [59,60]. These studies revealed that the circadian clock gene network is far more extensive than previously recognized (Fig. 2).
Fig. 2. Input cellular pathways to the circadian clock.

The core clock is further interconnected with additional cellular pathways that play a role in regulating circadian oscillations. Depicted here are several recently characterized input pathways. The tonic signaling pathway mediated by cAMP-CREB contributes to both photic resetting and cell-autonomous circadian oscillations. AMPK and mTOR act as homeostatic energy sensors to affect the clock function through nutrient sensing and CRY1 protein regulation. While SIRT1 alters clock function via post-translational modifications of PER2 and PGC1α, PARP1 functions by modifying CLOCK. In response to oxygen level fluctuations, HIF1α and NFR2 modify the transcription of several clock genes. NF-κB has been shown to regulate many clock genes, including DBP and NR1D1. Most of these input pathways are under the control of the circadian clock, and conversely, as depicted above, also provide input to and regulate the clock function, highlighting the extensive interplay between the circadian clock and cell physiological processes.
AMPK and mTOR
AMP-activated protein kinase (AMPK) is a rhythmically expressed kinase that functions as a tissue-specific energy sensor to maintain homeostasis [61,62]. AMPK regulates the cellular circadian clock based upon energy needs and nutrient availability, with its dysregulation linked to chronic diseases including obesity, inflammation, diabetes, and cancer [63]. The activity level of AMPK has also proven to be rhythmic [64,65]. When the energy state of the cell is low, and AMP/ATP is high, AMPK is activated to inhibit catabolic consumption of ATP and induce its anabolic production. Research over the last decade has identified diverse molecular mechanisms that regulate AMPK activity, which, in turn, regulate various metabolic and physiological processes in the central nervous system and peripheral tissues [63]. As a clock regulator, AMPK was shown to phosphorylate CK1E (i.e., increased activity), resulting in enhanced phosphorylation and degradation of PER2 [66,67]. In fibroblasts, CRY1 was shown to be phosphorylated by AMPK at the highly conserved Ser71, leading to its ubiquitination by FBXL3 and subsequent proteasomal degradation [64,67,68]. However, a recent study showed that CRY1 Ser71Ala mutation altered voluntary activity but failed to affect circadian rhythms in vivo [69]. Therefore, while phosphorylation of CRY1 by AMPK is not required for the regulation of circadian rhythms under normal physiological conditions, it is unknown as to whether it is critical under conditions of AMPK activation.
Recent findings also suggest that AMPK could regulate the clock function via the AMPK-mTOR-autophagy pathway to effect CRY1. Unlike AMPK, which is activated by low energy states, the mechanistic target of rapamycin (mTOR) is an intracellular nutrient sensor for high cellular energy states. mTOR associates with several components to form two mTOR complexes, mTORC1 and mTORC2. mTORC1 is regulated by growth factors, insulin, and certain amino acids. Activation of mTORC1 can result in increased protein synthesis, lipid synthesis, and inhibits autophagy to promote cell growth [70]. Key players of the PI3K-AKT-mTOR pathway (e.g., Pik3ca, Pik3r1, mTOR, Rictor, 4EBP1) are rhythmically expressed [10,55], and mTOR activity (signified by phosphorylated S6 and 4EBP1) displays a circadian pattern in the SCN, liver, skeletal muscles, and adipose tissues [71,72]. PER2 acts as a scaffold to recruit TSC1 and suppress mTORC1 activity, thereby contributing to the circadian regulation of mTOR [73].
Conversely, recent studies identified a critical role for mTOR in regulation by photic entrainment, synchronization of the SCN clock [74–76], and regulation of cell- and tissue-autonomous circadian clock function [71,77,78]. We identified mTOR as a clock modifier in our functional genomic screen, where RNAi knockdown led to a long period and low amplitude oscillations in human U2OS cells [55]. Using genetic and pharmacological approaches, we extended this finding and showed that mTOR perturbation altered circadian rhythms in fibroblasts, hepatocytes, adipocytes, liver, SCN, and animal behavior [71]. For example, mTOR inhibitors caused a long period and low amplitude in mouse hepatocytes and SCN explants. Further, mTOR heterozygous mice showed more scattered activity patterns, longer duration in the active phase, and less precise activity onset and offset, indicative of a compromised clock [71,75]. In summary, while mTOR activation results in shorter periods and a faster clock, its inhibition causes both lengthened periods and a dampened amplitude. These phenotypes are consistent across cell and tissue types, including the SCN, and are supported by several recent reports that identify the role of mTOR in maintaining circadian function [77–81]. Further, genetic alteration of mTOR also impacted circadian rhythms in Drosophila [82,83].
It has been shown that mTOR modifies the circadian clock through CRY1 regulation, in which mTOR signaling upregulates CRY1 through increased translation and decreased autophagic degradation (Fig. 2). In Tsc2 knockout cells (i.e., constitutively activated mTOR), BMAL1, CLOCK, and CRY1 expression levels are elevated [71,77]. However, only CRY1 induction by serum is affected by rapamycin, suggesting mTORC1 dependence, and is independent of the Bmal1 or Per genes [71]. Further, mTOR-autophagy underlies CRY1 accumulation, as CRY1 was elevated in autophagy-deficient Atg7 deficient mouse liver samples [84]. CRY1 is known to be ubiquitinated by FBXL3 and degraded by the proteasome [32,33]. However, FBXL3 levels were higher in Atg7 deficient cells [84], which does not explain the elevated CRY1 levels. CRY1 was shown to be the only clock protein that is targeted by autophagy and elevated in Atg7 knockout mouse liver. More specifically, CRY1 was shown to be degraded by autophagy via LC3 ligation; CRY1 contains four LC3 interaction region (LIR) motifs, and LIR mutations abolished its autophagic degradation [84].
PARP-1 and SIRT1
Nicotinamide adenine dinucleotide (NAD+) production is rhythmic and regulated by the clock [85,86]. Conversely, NAD+ and NADP both act as clock regulators. In an in vitro assay, their respective oxidized forms (NAD+ and NADP+) inhibit the binding of BMAL1-CLOCK to the E-box; however, their reduced forms (NADH and NADPH) enhance E-box binding [87]. NADP+/NADPH can also affect the ability of the CLOCK paralog NPAS2 and BMAL1 complex and bind to DNA [88].
Poly(ADP-ribose) Polymerase 1 (PARP-1) and Sirtuin 1 (SIRT1), known NAD+ sensors, have been shown to regulate the clock [89,90]. Previously, PARP-1 has been associated with transcriptional regulation in an NAD+-dependent manner [91]. PARP-1 is a multifunctional enzyme that, upon activation, catalyzes the transfer of ADP from NAD to synthesize Poly(ADP-ribose). As this polymer acts to facilitate DNA repair, intracellular levels of NAD can become substantially depleted [92]. ADP-ribosylation mediated by PARP-1 has been shown to have rhythmic activity, peaking at ZT4 [89]. This study further confirmed that PARP-1 acts on the BMAL1-CLOCK heterodimer. Poly(ADP-ribosyl) ation of CLOCK limits the DNA-binding ability of BMAL1-CLOCK to the E-Box and results in delayed interaction with PER and CRY. Conversely, PARP-1 deficiency upregulates BMAL1-CLOCK binding to the E-box and leads to a phase-shift in negative regulatory interactions [93]. As such, PARP-1 can modify the clock in response to nutrient status, providing a mechanism for metabolic coordination with circadian function.
There is a mutual regulation between the circadian clock and the NAD+-SIRT1 pathway. SIRT1 is an NAD+-dependent deacetylase that has been shown to bind to both BMAL1-CLOCK and PER2 [89,90,94,95]. SIRT1 recruitment has a direct effect on BMAL1-CLOCK’s regulation of nicotinamide phosphoribosyltransferase (NAMPT), the rate-limiting enzyme of the NAD+ salvage pathway [85,86]. NAMPT was shown to be activated by BMAL1-CLOCK and display strong circadian expression. As NAMPT upregulates NAD+, SIRT1 is upregulated and binds to the BMAL1-CLOCK complex, promoting the expression of NAMPT. SIRT1 was shown to deacetylate core clock proteins BMAL1 and PER2, as well as the transcriptional coactivator PGC-1α.The perturbation of SIRT1 activity altered circadian amplitudes of PER2 and DBP mRNA expression levels [89,94,96]. Likewise, Sirt1 knockout mice exhibited an increase in Dbp and Per2 expression. SIRT1 also regulates the SCN and may play a role in aging. Levels of BMAL1, PER2, and SIRT1 in the SCN decreased with age in wild-type mice [97]. SCN knockout of SIRT1 in young mice mimicked the phenotypic effects of aged mice, while overexpression of SIRT1 enhanced BMAL1 and PER2 expression. Knockdown, knockout, and overexpression of PGC-1α demonstrated similar expression patterns in response to nullified and overexpressed SIRT1 [96].
CREB
While the E-box is the predominant site for circadian clock gene regulation, rhythmic gene expression in the SCN is also under the control of the cAMP response element-binding protein (CREB) [98,99]. CREB is a basic leucine-zipper transcription factor that can be phosphorylated in response to increases in intracellular Ca2+ and cAMP levels. The cAMP-CREB pathway plays roles in entrainment and synchronization of the SCN clock, as well as in cell- and tissue-autonomous oscillations. Neurons in the SCN must synchronize not only to the light/dark cycle but also within the SCN ensemble. The SCN inputs can be categorized into photic signals and nonphotic signals. The light-induced pathway originates at the rods and cones of the retina [100]. Intrinsically photosensitive retinal ganglion cells (ipRGCs) containing melanopsin receive the input through bipolar cells and amacrine cells. The ipRGCs can be categorized into five different classes (M1–M5), with M1 ipRGCs having the highest melanopsin expression [101]. The ipRGCs extend to the ventral neurons in the SCN through the retinohypothalamic tract (RHT). The postsynaptic signaling network partially converges on the Ca2+/cAMP-CREB pathway to acutely induce PER1 and PER2 transcription, which resets the phase of postsynaptic neurons [100]. Photic induction of PER1 and PER2 and phase resetting is rapid and circadian time-dependent, with early and late subjective night causing phase delay and advancement of locomotor rhythms, respectively. Upon light exposure, PER1 and PER2 transcription is induced through the binding of CREB to CRE upstream of their promoter, which causes the phase shifting of circadian rhythms. Compromised CREB signaling results in a reduced PER1 and PER2 amplitude. Through a similar mechanism of CREB regulation of PER1 and PER2, the recurring synaptic transmission also underlies stabilization and amplification of neuronal rhythms to generate coherent and persistent circadian oscillations in the SCN.
Previous studies show that the SCN displays robust circadian oscillations of intracellular [Ca2+] and Ca2+/cAMP response element (CRE)-mediated transcription [98,102], suggesting the Ca2+/cAMP-CREB signaling pathway is regulated by the circadian clock. The circadian rhythm of CREB activity may be enabled through the repression of CRY1 activity in the nucleus and the cytoplasm. In the nucleus, CREB is known to activate gene transcription through its association with CREB-binding protein (CBP), a histone acetylase and transcriptional coactivator. CRY1 exhibits strong repression on CREB-CBP transcription of target genes, including PER1 and PER2 [103]. Intriguingly, CRY1 was shown to inhibit cAMP production in response to G protein-coupled receptor activation through the interaction and interference with Gsα, thereby reducing the activity of adenyl cyclase [104]. The rhythmic cAMP-CREB signaling and control of PER1 and PER2 may form negative feedback in the SCN to sustain autonomous oscillations in the SCN, in which the CREB output provides regulatory input to subsequent clock cycles. Inhibition of AC by MDL, a potent inhibitor, was able to reduce [cAMP] levels significantly [98]. This perturbation caused a dose-dependent suppression of the amplitude of circadian transcription and translation in the SCN. This additional regulation likely contributes to the robust function of the SCN.
Recently, CREB has been shown to play a role in the resetting of the peripheral clock in response to nutrient signaling. During fasting, glucagon upregulates CREB signaling to induce PER2 gene transcription. PER2 was shown to be recruited to the mTORC1 complex to inhibit its activity, resulting in reduced overall protein synthesis in the liver [73]. CREB-mediated PER2 expression links circadian clock function to fasting states and metabolism. The interconnectedness with other signaling pathways may allow for the fine-tuning of the clock to changing nutrient and metabolic inputs.
HIF1α and NRF2
Circadian rhythms allow for temporal segregation of oxidative events and cellular proliferation to minimize damage to the cell and thereby enable the diurnal coordination of cell cycle and metabolism [105]. Under stress conditions, cellular and tissue level responses are mediated by two specific transcription factors: hypoxia-inducible factor 1α (HIF1α) and oxidative stress-inducible nuclear factor erythroid-derived 2-related factor 2 (NRF2). These factors work to maintain homeostatic levels of oxygen in the cell and minimize cellular damage. Recent studies reveal bidirectional interactions between the circadian and HIF-mediated pathways that influence metabolic adaptation to hypoxia. HIF1α contains a canonical E-box within its promoter region and can be bound by BMAL1 and CLOCK in the mouse liver [106,107]. Genetic disruption of BMAL1 in skeletal myotubes and fibroblasts led to increased levels of HIF1α under hypoxic conditions, whereas abrogation of CRY stabilized HIF1α [107].These studies suggest that HIF1α transcription is under the control of the circadian clock. Conversely, HIF1α has been shown to bind to the E-box on CRY1, PER1, and PER2 genes to upregulate their transcription [107,108]. HIF1α deficient cells displayed lower levels of PER1, PER2, CRY1, CRY2, and NR1D1 expression compared to controls cells. As such, hypoxia lengthens the period and attenuates the amplitude of oscillations in a dose-dependent manner [106,107]. Oxygen can function as a resetting cue for circadian clocks, with tissue oxygenation in living animals showcasing a daily rhythm. Physiological range oxygen cycles were able to alter core clock gene expression and synchronize cellular clocks in a HIF1α-dependent manner, suggesting that oxygen resets the clock via HIF1α activation [108].
Recent studies uncovered a reciprocal relationship between NRF2 and the circadian clock. NRF2 has been shown to be expressed rhythmically, which is expected to play an important role in anticipating redox stress and maintaining redox homeostasis [109,110]. BMAL1-CLOCK was shown to bind to the E-box of NRF2 to upregulate its transcription [109]. In line with this notion, it was shown that Bmal1 knockout mouse tissues accumulated higher levels of reactive oxygen species than wild type mice [111]. NADPH was also shown to regulate circadian oscillations [112]. Genetic and pharmacological inhibition of the pentose phosphate pathway, which interferes with NADPH production, and therefore, redox homeostasis, led to lengthened periods. Mechanistically, this regulation involves NRF2 activity and increased coactivator CBP/p300 interaction with BMAL1-CLOCK [110,112]. NRF2 was shown to bind to the E-box of PER2, CRY2, and NR1D1 to increase their transcription [109,110,113]. Oxidative stress caused an NRF2-dependent reduction of circadian amplitude [110]. Thus, maintenance of redox homeostasis is shown to alter circadian clock function in a HIF1α- and NRF2-dependent manner, while the circadian clock also alters the expression of both transcription factors.
NF-κB
The Nuclear factor kappa-light chain enhancer of activated B cells (NF-κB) family of transcription factors plays a critical role in inflammation, immunity, and cell proliferation and survival [114–117]. Proinflammatory stimuli, such as tumor necrosis factor alpha (TNFα), interleukin 1 (IL-1), and bacterial lipopolysaccharides (LPS) activate the canonical NF-κB pathway. LPS, IL-1, and TNFα were shown to alter clock gene expression in the SCN, lung, liver, and fibroblasts, as well as alter locomotor activity [118–124]. LPS-induced lung and liver inflammation resulted in a reprogramming of core clock genes and outputs in the liver and lung, which led to a shortened period [120,121]. In a mouse model of chronic obstructive pulmonary disease (COPD), cigarette smoke exposure combined with viral infection disrupts clock function and leads to enhanced inflammatory responses in the lung and reduced locomotor activity [125]. Further, endotoxin was shown to suppress clock gene expression in human peripheral blood leukocytes [126]. The NF-κB RELB subunit was shown to interact with BMAL1-CLOCK and repress DBP expression [127]. Conversely, CLOCK was shown to interact with the NF-κB subunit pRELA/p65 and promote NF-κB-related transcription [128]. These results, taken together, support an interplay between the circadian clock and the inflammatory NF-κB pathway. This interplay is expected to have important implications in patho-physiology and disease states, such as asthma, a prominent disease type with a strong circadian variation of symptoms. Despite these findings, the mechanism behind NF-κB’s repression of clock gene expression remains unclear.
Computational Modeling
Network analysis and computational modeling are essential to gaining a systems-level understanding of the circadian network. Several clock models have been developed over recent years that incorporate core clock components and their respective interactions, followed by the quantification of these interactions in the feedback loops that yield cellular oscillatory behavior [129–134]. These models have helped to understand the relatively less complex clockwork of multiple positive and negative feedback loops. For example, mathematical modeling was used to predict a highly specific gain-of-function tau mutation of CK1E that increased kinase activity for phosphorylation and degradation of PER1 and PER2 [135]. More recently, the same group combined phosphoswitch mathematical modeling with experimental validation to clarify that CK1D/E are the PER2 priming kinases and suggest that multisite phosphorylation is the key step to determine periodicity [25].
The growing list of input pathways showcases the highly interactive cellular homeostasis network. These cellular functions are regulated by the clock and provide inputs to regulate the clock function in the context of a physiologically relevant cellular homeostatic state. Understanding the complexity requires network analyses and computational models that incorporate these input pathways into the clockwork model. For example, a recent mathematical model of the circadian clock in the liver links the feeding cycle to clock function [136]. This model incorporated SIRT1 and AMPK in response to NAD+/NADH and AMP/ATP ratios, respectively, mimicking different cellular energy states and feeding cycles. The model reproduced the dampened circadian rhythms under a high-fat diet mediated by AMPK and predicted that this effect might be pharmacologically rescued by timed REV-ERB agonist administration.
As discussed earlier, SIRT1 plays a role in regulating the circadian clock. However, while SIRT1 deficiency was shown to increase amplitude via BMAL1 acetylation in one study [94], another group showed decreased amplitude through an increase in acetylated PER2 [89]. To resolve this issue, a more recent study combined mathematical modeling with experimental validation and clarified mechanistic insights into the effects of SIRT1 on the circadian cycle [137]. In this study, they developed a circadian enzymatic model that incorporates the core clockwork and the SIRT1 input pathway. Predictions from this model were experimentally validated using previously developed approaches, such as cellular genetic methods, network analysis, and quantitative kinetic bioluminescence measurements [18,54,55]. The data suggest that PER2, not BMAL1, is the primary target of SIRT1. Further, SIRT1 was also shown to regulate PGC1α, leading to elevated ROR, and as a consequence, enhanced rhythmic BMAL1 expression. The combined computational and experimental approaches suggest that SIRT1 positively regulates clock function through its actions on PER2 and PGC1α, and SIRT1 deficiency causes low amplitude oscillations. Thus, the SIRT1 dual effect on PER2 and PGC1α contributes to circadian amplitude regulation.
Future modeling is expected to offer mechanistic insights into how the core clock works, as well as its integration within local cell physiology. For example, with the basic clockwork model as the foundation, future work should incorporate other well-understood pathways, such as the AMPK and mTOR signaling nexus and the redox sensing pathway. It is noted that the vast majority of circadian clock research has focused on the regulation of period length and phase. However, virtually all clock genes regulate amplitude in addition to period and phase. Despite its importance, much less research has been done on the mechanisms that underlie amplitude. The precise regulatory mechanism governing period and amplitude, and the relationship between the two, are not well understood. Although all the computational models produce 24 h periodicity, they have limited predictive capacity, especially under varying genetic and environmental conditions. At this time, it is not intuitive as to how the clock is integrated with multiple distinct signaling inputs. The insights from the modeling can make predictions for mechanistic testing, model iteration, and possibly inform how asynchronous and incongruous genetic and environmental conditions alter circadian properties.
To more accurately model the clockwork, future modeling must integrate the intracellular network features of the mammalian clock [18]. The network features act in concert as a genetic buffering system to maintain robust functions of the clock against genetic and environmental perturbation. Alteration of clock function is ultimately reflected at the level of transcription. Thus, mechanistic insights can be obtained from studying the effects of genetic and environmental perturbations on the expression patterns of the core clock genes. For endogenous clock gene expression, we must examine the entire clock network rather than a few outputs. A previous study established a gene dosage network analysis (GDNA) with which to determine how gene perturbation affects the clockwork. This is best exemplified by the effects of PER1 knockdown on PER2 and PER3, CRY1 knockdown on CRY2, and BMAL1 knockdown on NR1D1 and NR1D2, and as a result, on CLOCK and CRY1 [18,54]. The clock is a dynamic system, so a dynamic GDNA assay that incorporates a temporal dimension must be developed. Results from these novel GDNA studies will then be used as training datasets to integrate into computational models of the circadian clock, enabling the probe into the mechanistic basis of novel clock modulators.
It is worth noting that studies over the past two decades also demonstrated cell- and tissue-type specific clock modifiers and inputs. The cell-type-specific function of clock genes may result from their differential tissue expression, alternative splice variants, post-translational modifications, and cell-type-specific compensatory mechanisms (see Ref. [56] and references cited therein). Thus, cell-type-specific clock modifier function starts to become clear when local physiology is considered as an input to the clock. Thus, systems-level understanding will offer deeper insights into the mechanistic basis of tissue-specific clocks, akin to the realization that cyclin-dependent kinase networks in the cell cycle control program are tissue-specific [138].
The circadian function represents a fascinating field of study in which a complex behavior has a genetic basis and the underlying molecular mechanism is cell-autonomous. For example, CRY2 deletion caused long period length in peripheral cell and tissue models, the SCN, and circadian locomotor behavior [12]. A gain-of-function mutation in CRY1 caused stronger E-box repression activity and lengthened the period of circadian rhythms in cells and mice, which underlies the familial delayed sleep phase syndrome in humans [139]. However, system robustness increases from the autonomous cell to the organismal level. Highlighting this is the SCN ensemble, a network of neurons coupled by intercellular signaling mechanisms that act to protect against genetic and environmental perturbations. For example, the long period length phenotype in CRY2 knockout cells is much stronger than in SCN explants and more so in animal behavior. CRY1 knockout cells are only transiently rhythmic, but Cry1 knockout SCN explants are robustly rhythmic [12]. Similar observations were made in the case of Clock and Bmal1 deficiency [132,140,141]. There appear to be distinct levels of robustness at different biological levels, with each exhibiting varying degrees of circadian phenotypes. Future work must consider the inclusion of the biological levels in modeling. Given the complexity of the system, this represents a unique and unprecedented opportunity to advance our understanding of the circadian timekeeping system and its position within the larger physiological schematic.
Perspectives
While much is known of the circadian network, a full understanding remains elusive. To gain insights into this complex biological network, systems biology approaches promise the best results. In this context, we need to continue this systems-based framework to study how the clock gene network is assembled and how network interactions culminate in emergent clock properties. Intriguingly, it is evident that the circadian and other metabolic networks exhibit remarkable parallels with those of metabolic diseases, aging, and cancer, which underscores the interconnectedness of all cellular functions.
Future research should focus on characterizing more cellular functions that input to the clock network in a cell and tissue-type dependent manner. Identification of these clock modifiers has raised important questions about their spatiotemporal location within the network and their role in regulating the clock mechanism and circadian behavior. While past screens have utilized RNAi, which has inherent drawbacks, future genome-wide screens may leverage the power of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 genome editing to screen for clock modifiers and provide a complete answer to these questions. It is now possible to readily manipulate a larger number of genes simultaneously to validate their functions, in a manner comparable to the yeast system decades ago. CRISPR/Cas9 genome editing uses homology-directed repair (HDR) with an exogenous template for genetic knock-ins and non-homologous end joining (NHEJ) for frame-shift mutations and premature stop codons through insertions and deletions (indels) for genetic knock-outs [142]. CRISPR has been utilized in lentivirus to knockout single genes, such as FBXL3 in U2OS BMAL1-luciferase reporter cells, leading to increased protein levels and decreased mRNA transcripts of CRY1 and CRY2 and matched previous low amplitude, long period phenotypes previously demonstrated in mice [143]. CRISPR has also been used in lentivirus for double knockouts, such as both CRY1 and CRY2 in U2OS BMAL1-luciferase reporter cells [144]. CRY1 knockouts had low amplitude, short period phenotypes while CRY2 knockouts had long period phenotypes. Knockout of both CRY1 and CRY2 resulted in arrhythmicity, which, along with the other two phenotypes, is consistent with previous studies in cells and mice [12,55,145,146]. By utilizing a streamlined cloning and all-in-one adenoviral strategy, a recent study used CRISPR to knock out both BMAL1 and PER2 in a U2OS BMAL1-luciferase reporter cell line [147]. The generated BMAL1 knockouts were arrhythmic and were able to be rescued, while the PER2 knockouts demonstrated low amplitude. These studies showcase the potential that CRISPR/Cas9 has in delineating the circadian clock and its extensive input pathways; however, examples are still relatively scarce. Here we present several recently characterized input genes and pathways to highlight this concept and potential challenges. CRISPR has been utilized for more than simply knockouts, for example, generating site mutations and fusion proteins across species at the cellular to organismal levels. When the catalytic activity of Cas9 is rendered inactive (dCas9), CRISPR can be used as an RNA-guided site-selective DNA recognition tool [148]. For example, dCas9 proteins fused with a repressor can be coupled with small guide RNA (sgRNA) to silence gene expression (CRISPRi), similar to RNA interference (RNAi), but with minimizing the off-target effects of RNAi [149]. Likewise, dCas9 proteins fused with a transcriptional activator (CRISPRa) can activate gene expression. These are just a few of the tools that can be developed utilizing dCas9 fusion proteins, and the possibilities are endless. These approaches make it possible to study multiple players within the same pathway or across pathways to expand upon our understanding of the clock network.
Further, systems biology research requires more focused efforts on network analysis and computational modeling to help uncover design principles of circadian function at multiple levels of the circadian organization, with a central focus on representative cellular clock models. Several prominent cellular models (i.e., 3T3 fibroblasts, U2OS osteosarcomas, and MMH-D3 hepatocytes) are cell-autonomous, without systemic input otherwise present in vivo. Conversely, given the prevalence of cell- and tissue-specific circadian functions, findings from one cell type should be extrapolated to another with both experimental validation and caution.
Small molecules can also be used as a tool in future studies to gain a further understanding of the clock network. Given the strong correlation between disease states, such as aging and circadian decline, recent studies have started to explore pharmacological agents to enhance dampened clocks [150,151]. Mice with high-amplitude rhythms show improved longevity [97,152]. Time-restricted feeding (i.e., limiting food access only to active phase) can improve age-related or diet-induced circadian disruption and have beneficial effects [153–155]. Many small molecules target the clock directly through input pathways and oscillatory proteins or indirectly through feedback from output pathways [150]. Phenotypic kinetic reporter assays, in which luciferase expression is under the control of a circadian expression promoter protein, can be used to observe changes in amplitude, period, and phase to identify small molecule clock modifiers. Several small molecules (e.g., rapamycin, resveratrol, nobiletin, and metformin) have shown great potential for increasing lifespan and healthspan [150,151,156–161]. For example, resveratrol (which is found in grapes and red wine) has been reported to activate SIRT1 [160,162]. Resveratrol was found to decrease transcription in both E-box-luciferase and Per1-luciferase reporter cell lines [163], and likely affects the clock via its effect on mTOR and autophagy [71,159,160,162]. Rapamycin’s antiaging affect involves mTORC1’s role in regulating CRY1 and clocks [71,156,164]. Nobiletin was shown to increase rhythm amplitude in aged mice by targeting RORs [158,165]. Another example of a small molecule modulator of the clock is the type 2 diabetes mellitus drug, metformin. As an AMPK activator, metformin creates a phase advance in most clock genes in the liver and both phase advance and phase delay in clock genes in the muscle [166]. These are just a few examples of the many small molecules that have been found to modulate the clock. Future studies will further explore identifying small molecules that effect the clock and discover new therapeutic uses for circadian clock-related disorders.
The handful of clock modifiers discussed here showcase that the circadian clock is intimately linked to cellular homeostasis, and deregulation can manifest in perturbed clock function. Thus, normal circadian performance can serve as a biomarker for cellular homeostasis. Intriguingly, it is now possible to alter clock function by tweaking the modifiers outside the core clock loop, which offers new opportunities for enhancing circadian function through the input pathways. Further, as many drugs target rhythmic genes and clock modifiers, they may affect circadian timekeeping and alter cellular homeostasis, resulting in altered sleep/wake cycle and physiological processes. Circadian medicine offers a novel and important approach to patient care based on temporal precision and personalization.
Acknowledgements
A.C.L. acknowledges funding from the National Science Foundation (NSF) Division of Integrative Organismal Systems (IOS) Award Number 1656647 and the National Institutes of Health (NIH) National Institute of Neurological Disorders and Stroke (NINDS) R01 NS054794.
References
- [1].Nurse P, Hayles J, The cell in an era of systems biology, Cell 144 (2011) 850–854, 10.1016/j.cell.2011.02.045. [DOI] [PubMed] [Google Scholar]
- [2].Nurse P, Life, logic and information, Nature 454 (2008) 424–426, 10.1038/454424a. [DOI] [PubMed] [Google Scholar]
- [3].Sauer U, Heinemann M, Zamboni N, Genetics. Getting closer to the whole picture, Science 316 (2007) 550–551, 10.1126/science.1142502. [DOI] [PubMed] [Google Scholar]
- [4].Ukai H, Ueda HR, Systems biology of mammalian circadian clocks, Annu. Rev. Physiol 72 (2010) 579–603, 10.1146/annurev-physiol-073109-130051. [DOI] [PubMed] [Google Scholar]
- [5].Hogenesch JB, Ueda HR, Understanding systems-level properties: timely stories from the study of clocks, Nat. Rev. Genet 12 (2011) 407–416, 10.1038/nrg2972. [DOI] [PubMed] [Google Scholar]
- [6].Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U, Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells, Cell 119 (2004) 693–705, 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- [7].Welsh DK, Yoo S-H, Liu AC, Takahashi JS, Kay SA, Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression, Curr. Biol 14 (2004) 2289–2295, 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mohawk J.a., Green CB, Takahashi JS, Central and peripheral circadian clocks in mammals, Annu. Rev. Neurosci 35 (2012) 445–462, 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Young MW, Kay SA, Time zones: a comparative genetics of circadian clocks, Nat. Rev. Genet 2 (2001) 702–715, 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- [10].Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB, A circadian gene expression atlas in mammals: implications for biology and medicine, Proc. Natl. Acad. Sci. U. S. A 111 (2014) 16219–16224, 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Takahashi JS, Transcriptional architecture of the mammalian circadian clock, Nat. Rev. Genet 18 (2017) 164–179, 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest A.a., Buhr ED, Singer O, Meeker K, Verma IM, Doyle FJ 3rd, Takahashi JS, Kay S.a., Doyle FJ 3rd, Takahashi JS, Kay S.a., Doyle FJ, Takahashi JS, Kay S.a., Doyle FJ 3rd, Takahashi JS, Kay S.a., Intercellular coupling confers robustness against mutations in the SCN circadian clock network, Cell 129 (2007) 605–616, 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Koike N, Yoo S-H, Huang H-C, Kumar V, Lee C, Kim T-K, Takahashi JS, Transcriptional architecture and chromatin landscape of the core circadian clock in mammals, Science 338 (2012) 349–354, 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chiou Y-Y, Yang Y, Rashid N, Ye R, Selby CP, Sancar A, Mammalian Period represses and de-represses transcription by displacing CLOCK–BMAL1 from promoters in a Cryptochrome-dependent manner, Proc. Natl. Acad. Sci 113 (2016) E6072–E6079, 10.1073/pnas.1612917113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S, System-level identification of transcriptional circuits underlying mammalian circadian clocks, Nat. Genet 37 (2005) 187–192, 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- [16].Gachon F, Olela FF, Schaad O, Descombes P, Schibler U, The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification, Cell Metabol 4 (2006) 25–36, 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- [17].Ukai-Tadenuma M, Yamada RG, Xu H, Ripperger JA, Liu AC, Ueda HR, Delay in feedback repression by cryptochrome 1 Is required for circadian clock function, Cell 144 (2011) 268–281, 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
- [18].Baggs JE, Price TS, DiTacchio L, Panda S, FitzGerald GA, Hogenesch JB, Network features of the mammalian circadian clock, PLoS Biol 7 (2009) e52, 10.1371/journal.pbio.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G, The hallmarks of aging, Cell 153 (2013) 1194–1217, 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hanahan D, Weinberg RA, Hallmarks of cancer: the next generation, Cell 144 (2011) 646–674, 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- [21].Zhao Y, Forst CV, Sayegh CE, Wang I-M, Yang X, Zhang B, Molecular and genetic inflammation networks in major human diseases, Mol. Biosyst 12 (2016) 2318–2341, 10.1039/c6mb00240d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zelezniak A, Pers TH, Soares S, Patti ME, Patil KR, Metabolic network topology reveals transcriptional regulatory signatures of type 2 diabetes, PLoS Comput. Biol 6 (2010), e1000729, 10.1371/journal.-pcbi.1000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hirano A, Fu Y-H, Ptáček LJ, The intricate dance of post-translational modifications in the rhythm of life, Nat. Struct. Mol. Biol 23 (2016) 1053–1060, 10.1038/nsmb.3326. [DOI] [PubMed] [Google Scholar]
- [24].Gallego M, Virshup DM, Post-translational modifications regulate the ticking of the circadian clock, Nat. Rev. Mol. Cell Biol 8 (2007) 139–148, 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- [25].Narasimamurthy R, Hunt SR, Lu Y, Fustin J-M, Okamura H, Partch CL, Forger DB, Kim JK, Virshup DM, CK1δ/ϵ protein kinase primes the PER2 circadian phosphoswitch, Proc. Natl. Acad. Sci 115 (2018) 5986–5991, 10.1073/pnas.1721076115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lee H, Chen R, Lee Y, Yoo S, Lee C, Essential roles of CKIδ and CKIϵ in the mammalian circadian clock, Proc. Natl. Acad. Sci 106 (2009) 21359–21364, 10.1073/pnas.0906651106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS, Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau, Science 288 (2000) 483–492, 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Meng QJ, Logunova L, Maywood ES, Gallego M, Lebiecki J, Brown TM, Sládek M, Semikhodskii AS, Glossop NRJ, Piggins HD, Chesham JE, Bechtold D.a., Yoo SH, Takahashi JS, Virshup DM, Boot-Handford RP, Hastings MH, Loudon ASI, Setting clock speed in mammals: the CK1ϵ tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins, Neuron 58 (2008) 78–88, 10.1016/j.neuron.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptácek LJ, Fu YH, An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome, Science 291 (2001) 1040–1043, 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- [30].Godinho SIH, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, Pagano M, Kendall R, Quwailid MM, Romero MR, O’neill J, Chesham JE, Brooker D, Lalanne Z, Hastings MH, Nolan PM, The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period, Science 316 (2007) 897–900, 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- [31].Siepka SMSM, Yoo S-HHS-H, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JSJS, Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression, Cell 129 (2007) 1011–1023, 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yoo S-H, Mohawk JA, Siepka SM, Shan Y, Huh SK, Hong H-K, Kornblum I, Kumar V, Koike N, Xu M, Nussbaum J, Liu X, Chen Z, Chen ZJ, Green CB, Takahashi JS, Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm, Cell 152 (2013) 1091–1105, 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hirano A, Yumimoto K, Tsunematsu R, Matsumoto M, Oyama M, Kozuka-Hata H, Nakagawa T, Lanjakornsiripan D, Nakayama KI, Fukada Y, FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes, Cell 152 (2013) 1106–1118, 10.1016/j.cell.2013.01.054. [DOI] [PubMed] [Google Scholar]
- [34].Lee Y, Shen Y, Francey LJ, Ramanathan C, Sehgal A, Liu AC, Hogenesch JB, The NRON complex controls circadian clock function through regulated PER and CRY nuclear translocation, Sci. Rep 9 (2019) 11883, 10.1038/s41598-019-48341-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB, Harmonics of circadian gene transcription in mammals, PLoS Genet 5 (2009), e1000442, 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Perelis M, Marcheva B, Moynihan Ramsey K, Schipma MJ, Hutchison AL, Taguchi A, Peek CB, Hong H, Huang W, Omura C, Allred AL, Bradfield CA, Dinner AR, Barish GD, Bass J, Pancreatic cell enhancers regulate rhythmic transcription of genes controlling insulin secretion, Science 350 (2015), 10.1126/science.aac4250 aac4250–aac4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Atwood A, DeConde R, Wang SS, Mockler TC, Sabir JSM, Ideker T, Kay SA, Cell-autonomous circadian clock of hepatocytes drives rhythms in transcription and polyamine synthesis, Proc. Natl. Acad. Sci. U. S. A 108 (2011) 18560–18565, 10.1073/pnas.1115753108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Keller M, Mazuch J, Abraham U, Eom GD, Herzog ED, Volk H-D, Kramer A, Maier B, A circadian clock in macrophages controls inflammatory immune responses, Proc. Natl. Acad. Sci 106 (2009) 21407–21412, 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, Iino M, Shigeyoshi Y, Hashimoto S, A transcription factor response element for gene expression during circadian night, Nature 418 (2002) 534–539, 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- [40].Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB, Coordinated transcription of key pathways in the mouse by the circadian clock, Cell 109 (2002) 307–320. http://www.ncbi.nlm.nih.gov/pubmed/12015981. [DOI] [PubMed] [Google Scholar]
- [41].Storch K-F, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ, Extensive and divergent circadian gene expression in liver and heart, Nature 417 (2002) 78–83, 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- [42].Vollmers C, Schmitz RJ, Nathanson J, Yeo G, Ecker JR, Panda S, Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome, Cell Metabol 16 (2012) 833–845, 10.1016/j.cmet.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Menet JS, Rodriguez J, Abruzzi KC, Rosbash M, Nascent-Seq reveals novel features of mouse circadian transcriptional regulation, Elife 1 (2012), e00011, 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F, Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver, PLoS Biol 9 (2011), 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yoshitane H, Ozaki H, Terajima H, Du N-H, Suzuki Y, Fujimori T, Kosaka N, Shimba S, Sugano S, Takagi T, Iwasaki W, Fukada Y, CLOCK-controlled polyphonic regulation of circadian rhythms through canonical and noncanonical E-boxes, Mol. Cell Biol 34 (2014) 1776–1787, 10.1128/MCB.01465-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Le Martelot G, Canella D, Symul L, Migliavacca E, Gilardi F, Liechti R, Martin O, Harshman K, Delorenzi M, Desvergne B, Herr W, Deplancke B, Schibler U, Rougemont J, Guex N, Hernandez N, Naef F, CycliX consortium, genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles, PLoS Biol 10 (2012), e1001442, 10.1371/journal.pbio.1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wu G, Ruben MD, Schmidt RE, Francey LJ, Smith DF, Anafi RC, Hughey JJ, Tasseff R, Sherrill JD, Oblong JE, Mills KJ, Hogenesch JB, Population-level rhythms in human skin with implications for circadian medicine, Proc. Natl. Acad. Sci 115 (2018) 12313–12318, 10.1073/pnas.1809442115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ruben MD, Wu G, Smith DF, Schmidt RE, Francey LJ, Lee YY, Anafi RC, Hogenesch JB, A database of tissue-specific rhythmically expressed human genes has potential applications in circadian medicine, Sci. Transl. Med 10 (2018), eaat8806, 10.1126/scitranslmed.aat8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Anafi RC, Francey LJ, Hogenesch JB, Kim J, CYCLOPS reveals human transcriptional rhythms in health and disease, Proc. Natl. Acad. Sci 114 (2017) 5312–5317, 10.1073/pnas.1619320114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Christou S, Wehrens SMT, Isherwood C, Möller-Levet CS, Wu H, Revell VL, Bucca G, Skene DJ, Laing EE, Archer SN, Johnston JD, Circadian regulation in human white adipose tissue revealed by transcriptome and metabolic network analysis, Sci. Rep 9 (2019) 2641, 10.1038/s41598-019-39668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Braun R, Kath WL, Iwanaszko M, Kula-Eversole E, Abbott SM, Reid KJ, Zee PC, Allada R, Universal method for robust detection of circadian state from gene expression, Proc. Natl. Acad. Sci 115 (2018) E9247–E9256, 10.1073/pnas.1800314115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Cederroth CR, Albrecht U, Bass J, Brown SA, Dyhrfjeld-Johnsen J, Gachon F, Green CB, Hastings MH, Helfrich-Förster C, Hogenesch JB, Lévi F, Loudon A, Lundkvist GB, Meijer JH, Rosbash M, Takahashi JS, Young M, Canlon B, Medicine in the fourth dimension, Cell Metabol 30 (2019) 238–250, 10.1016/j.cmet.2019.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ruben MD, Smith DF, FitzGerald GA, Hogenesch JB, Dosing time matters, Science 365 (2019) 547–549, 10.1126/science.aax7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA, Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms, PLoS Genet 4 (2008), e1000023, 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, Liu X, Atwood A, Huss JW, Janes J, Su AI, Hogenesch JB, Kay SA, A genome-wide RNAi screen for modifiers of the circadian clock in human cells, Cell 139 (2009) 199–210, 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Ramanathan C, Xu H, Khan SK, Shen Y, Gitis PJ, Welsh DK, Hogenesch JB, Liu AC, Cell type-specific functions of period genes revealed by novel adipocyte and hepatocyte circadian clock models, PLoS Genet 10 (2014), e1004244, 10.1371/journal.pgen.1004244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Wallach T, Schellenberg K, Maier B, Kalathur RKR, Porras P, Wanker EE, Futschik ME, Kramer A, Dynamic circadian protein-protein interaction networks predict temporal organization of cellular functions, PLoS Genet 9 (2013), e1003398, 10.1371/journal.pgen.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Anafi RC, Lee Y, Sato TK, Venkataraman A, Ramanathan C, Kavakli IH, Hughes ME, Baggs JE, Growe J, Liu AC, Kim J, Hogenesch JB, Machine learning helps identify CHRONO as a circadian clock component, PLoS Biol 12 (2014), e1001840, 10.1371/journal.pbio.1001840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Maier B, Wendt S, Vanselow JT, Wallach T, Reischl S, Oehmke S, Schlosser A, Kramer A, A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock, Genes Dev 23 (2009) 708–718, 10.1101/gad.512209.understood. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].DeBruyne JP, Baggs JE, Sato TK, Hogenesch JB, Ubiquitin ligase Siah2 regulates RevErbα degradation and the mammalian circadian clock, Proc. Natl. Acad. Sci 112 (2015) 12420–12425, 10.1073/pnas.1501204112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hardie DG, Ross FA, Hawley SA, AMPK: a nutrient and energy sensor that maintains energy homeostasis, Nat. Rev. Mol. Cell Biol 13 (2012) 251–262, 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Inoki K, Kim J, Guan K-L, AMPK and mTOR in cellular energy homeostasis and drug targets, Annu. Rev. Pharmacol. Toxicol 52 (2012) 381–400, 10.1146/annurev-pharmtox-010611-134537. [DOI] [PubMed] [Google Scholar]
- [63].Jeon S-M, Regulation and function of AMPK in physiology and diseases, Exp. Mol. Med 48 (2016), 10.1038/emm.2016.81 e245–e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].a Lamia K, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, Thompson CB, Evans RM, AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation, Science 326 (2009) 437–440, 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Um J-H, Pendergast JS, Springer DA, Foretz M, Viollet B, Brown A, Kim MK, Yamazaki S, Chung JH, AMPK regulates circadian rhythms in a tissue- and isoform-specific manner, PLoS One 6 (2011), e18450, 10.1371/journal.pone.0018450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Um JH, Yang S, Yamazaki S, Kang H, Viollet B, Foretz M, Chung JH, Activation of 5’-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPer2, J. Biol. Chem 282 (2007), 10.1074/jbc.C700070200, 20794–8. [DOI] [PubMed] [Google Scholar]
- [67].Sahar S, Sassone-Corsi P, Regulation of metabolism: the circadian clock dictates the time, Trends Endocrinol Metabol 23 (2012) 1–8, 10.1016/j.tem.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Jordan SD, Lamia KA, AMPK at the crossroads of circadian clocks and metabolism, Mol. Cell. Endocrinol 366 (2013) 163–169, 10.1016/j.mce.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Vaughan M, Jordan SD, Duglan D, Chan AB, Afetian M, Lamia KA, Phosphorylation of CRY1 serine 71 alters voluntary activity but not circadian rhythms in vivo, J. Biol. Rhythm 34 (2019) 401–409, 10.1177/0748730419858525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Saxton RA, Sabatini DM, mTOR signaling in growth, metabolism, and disease, Cell 169 (2017) 361–371, 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- [71].Ramanathan C, Kathale ND, Liu D, Lee C, Freeman A, Hogenesch JB, Cao R, Liu AC, mTOR signaling regulates central and peripheral circadian clock function, PLoS Genet 14 (2018), e1007369, 10.1371/journal.pgen.1007369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Jouffe C, Cretenet G, Symul L, Martin E, Atger F, Naef F, Gachon F, The circadian clock coordinates ribosome biogenesis, PLoS Biol 11 (2013), e1001455, 10.1371/journal.pbio.1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Wu R, Dang F, Li P, Wang P, Xu Q, Liu Z, Li Y, Wu Y, Chen Y, Liu Y, The circadian protein Period2 suppresses mTORC1 activity via recruiting Tsc1 to mTORC1 complex, Cell Metabol 29 (2019) 653–667, 10.1016/j.cmet.2018.11.006, e6. [DOI] [PubMed] [Google Scholar]
- [74].Cao R, Obrietan K, mTOR signaling and entrainment of the mammalian circadian clock, Mol. Cell. Pharmacol 2 (2010) 125–130, 10.4255/mcpharmacol.10.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Cao R, Robinson B, Xu H, Gkogkas C, Khoutorsky A, Alain T, Yanagiya A, Nevarko T, Liu AC, Amir S, Sonenberg N, Translational control of entrainment and synchrony of the suprachiasmatic circadian clock by mTOR/4E-BP1 signaling, Neuron 79 (2013) 712–724, 10.1016/j.neuron.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Liu D, Stowie A, de Zavalia N, Leise T, Pathak SS, Drewes LR, Davidson AJ, Amir S, Sonenberg N, Cao R, mTOR signaling in VIP neurons regulates circadian clock synchrony and olfaction, Proc. Natl. Acad. Sci. U. S. A 115 (2018) E3296–E3304, 10.1073/pnas.1721578115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Lipton JO, Boyle LM, Yuan ED, Hochstrasser KJ, Chifamba FF, Nathan A, Tsai PT, Davis F, Sahin M, Aberrant proteostasis of BMAL1 underlies circadian abnormalities in a paradigmatic mTOR-opathy, Cell Rep 20 (2017) 868–880, 10.1016/j.celrep.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Feeney KA, Hansen LL, Putker M, Olivares-Yañez C, Day J, Eades LJ, Larrondo LF, Hoyle NP, O’Neill JS, van Ooijen G, Daily magnesium fluxes regulate cellular timekeeping and energy balance, Nature 532 (2016) 375–379, 10.1038/nature17407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Cornu M, Oppliger W, Albert V, Robitaille AM, Trapani F, Quagliata L, Fuhrer T, Sauer U, Terracciano L, Hall MN, Hepatic mTORC1 controls locomotor activity, body temperature, and lipid metabolism through FGF21, Proc. Natl. Acad. Sci. U. S. A 111 (2014) 11592–11599, 10.1073/pnas.1412047111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Lipton JO, Yuan ED, Boyle LM, Ebrahimi-Fakhari D, Kwiatkowski E, Nathan A, Güttler T, Davis F, Asara JM, Sahin M, The circadian protein BMAL1 regulates translation in response to S6K1-mediated phosphorylation, Cell 161 (2015) 1138–1151, 10.1016/j.cell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Walton ZE, Patel CH, Brooks RC, Yu Y, Ibrahim-Hashim A, Riddle M, Porcu A, Jiang T, Ecker BL, Tameire F, Koumenis C, Weeraratna AT, Welsh DK, Gillies R, Alwine JC, Zhang L, Powell JD, Dang CV, Acid suspends the circadian clock in hypoxia through inhibition of mTOR, Cell 174 (2018) 72–87, 10.1016/j.cell.2018.05.009, e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kijak E, Pyza E, TOR signaling pathway and autophagy are involved in the regulation of circadian rhythms in behavior and plasticity of L2 interneurons in the brain of Drosophila melanogaster, PLoS One 12 (2017), e0171848, 10.1371/journal.pone.0171848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Zheng X, Sehgal A, AKT and TOR signaling set the pace of the circadian pacemaker, Curr. Biol 20 (2010) 1203–1208, 10.1016/j.cub.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Toledo M, Batista-Gonzalez A, Merheb E, Aoun ML, Tarabra E, Feng D, Sarparanta J, Merlo P, Botrè F, Schwartz GJ, Pessin JE, Singh R, Autophagy regulates the liver clock and glucose metabolism by degrading CRY1, Cell Metabol 28 (2018) 268–281, 10.1016/j.cmet.2018.05.023, e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P, Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1, Science 324 (2009) 654–657, 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong H-KH-K, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S.-I.S. -i., Bass J, Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis, Science 324 (2009) 651–654, 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Rutter J, Reick M, Wu LC, McKnight SL, Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors, Science 293 (2001) 510–514, 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- [88].Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA, McKnight SL, NPAS2: a gas-responsive transcription factor, Science 298 (2002) 2385–2387, 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- [89].Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U, SIRT1 regulates circadian clock gene expression through PER2 deacetylation, Cell 134 (2008) 317–328, 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- [90].Nemoto S, Fergusson MM, Finkel T, SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha}, J. Biol. Chem 280 (2005) 16456–16460, 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- [91].Hassa PO, Haenni SS, Elser M, Hottiger MO, Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol. Mol. Biol. Rev 70 (2006) 789–829, 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Berger NA, Poly(ADP-ribose) in the cellular response to DNA damage, Radiat. Res 101 (1985) 4–15. http://www.ncbi.nlm.nih.gov/pubmed/3155867. (Accessed 2 November 2019). [PubMed] [Google Scholar]
- [93].Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U, Poly(ADP-Ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding, Cell 142 (2010) 943–953, 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- [94].Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P, The NAD+-Dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control, Cell 134 (2008) 329–340, 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Asher G, Schibler U, Crosstalk between components of circadian and metabolic cycles in mammals, Cell Metabol 13 (2011) 125–137, 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- [96].Liu C, Li S, Liu T, Borjigin J, Lin JD, Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism, Nature 447 (2007) 477–481, 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- [97].Chang H-C, Guarente L, SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging, Cell 153 (2013) 1448–1460, 10.1016/j.cell.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].O’Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH, cAMP-dependent signaling as a core component of the mammalian circadian pacemaker, Science 320 (2008) 949–953, 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Impey S, Fong AL, Wang Y, Cardinaux JR, Fass DM, Obrietan K, Wayman GA, Storm DR, Soderling TR, Goodman RH, Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV, Neuron 34 (2002) 235–244, 10.1016/s0896-6273(02)00654-2. [DOI] [PubMed] [Google Scholar]
- [100].Liu AC, Lewis WG, Kay SA, Mammalian circadian signaling networks and therapeutic targets, Nat. Chem. Biol 3 (2007) 630–639, 10.1038/nchem-bio.2007.37. [DOI] [PubMed] [Google Scholar]
- [101].Lazzerini Ospri L, Prusky G, Hattar S, Mood, the circadian system, and melanopsin retinal ganglion cells, Annu. Rev. Neurosci 40 (2017) 539–556, 10.1146/annurev-neuro-072116-031324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Brancaccio M, Maywood ES, Chesham JE, Loudon ASI, Hastings MH, A Gq-Ca2+ axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus, Neuron 78 (2013) 714–728, 10.1016/j.neuron.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Xu H, Gustafson CL, Sammons PJ, Khan SK, Parsley NC, Ramanathan C, Lee H-W, Liu AC, Partch CL, Cryptochrome 1 regulates the circadian clock through dynamic interactions with the BMAL1 C terminus, Nat. Struct. Mol. Biol 22 (2015) 476–484, 10.1038/nsmb.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y, Brenner DA, Montminy M, Kay SA, Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis, Nat. Med 16 (2010) 1152–1156, 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Milev NB, Reddy AB, Circadian redox oscillations and metabolism, Trends Endocrinol Metabol 26 (2015) 430–437, 10.1016/j.tem.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Wu Y, Tang D, Liu N, Xiong W, Huang H, Li Y, Ma Z, Zhao H, Chen P, Qi X, Zhang EE, Reciprocal regulation between the circadian clock and hypoxia signaling at the genome level in mammals, Cell Metabol 25 (2017) 73–85, 10.1016/j.cmet.2016.09.009. [DOI] [PubMed] [Google Scholar]
- [107].Peek CB, Levine DC, Cedernaes J, Taguchi A, Kobayashi Y, Tsai SJ, Bonar NA, McNulty MR, Ramsey KM, Bass J, Circadian clock interaction with HIF1α mediates oxygenic metabolism and anaerobic glycolysis in skeletal muscle, Cell Metabol 25 (2017) 86–92, 10.1016/j.cmet.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Adamovich Y, Ladeuix B, Golik M, Koeners MP, Asher G, Rhythmic oxygen levels reset circadian clocks through HIF1α, Cell Metabol 25 (2017) 93–101, 10.1016/j.cmet.2016.09.014. [DOI] [PubMed] [Google Scholar]
- [109].Pekovic-Vaughan V, Gibbs J, Yoshitane H, Yang N, Pathiranage D, Guo B, Sagami A, Taguchi K, Bechtold D, Loudon A, Yamamoto M, Chan J, van der Horst GTJ, Fukada Y, Meng Q-J, The circadian clock regulates rhythmic activation of the NRF2/glutathione-mediated antioxidant defense pathway to modulate pulmonary fibrosis, Genes Dev 28 (2014) 548–560, 10.1101/gad.237081.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Wible RS, Ramanathan C, Sutter CH, Olesen KM, Kensler TW, Liu AC, Sutter TR, NRF2 regulates core and stabilizing circadian clock loops, coupling redox and timekeeping inMus musculus, Elife 7 (2018), 10.7554/eLife.31656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].V Kondratov R, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP, Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock, Genes Dev 20 (2006) 1868–1873, 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Rey G, Valekunja UK, Feeney KA, Wulund L, Milev NB, Stangherlin A, Ansel-Bollepalli L, Velagapudi V, O’Neill JS, Reddy AB, The pentose phosphate pathway regulates the circadian clock, Cell Metabol 24 (2016) 462–473, 10.1016/j.cmet.2016.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Yang G, Wright CJ, Hinson MD, Fernando AP, Sengupta S, Biswas C, La P, Dennery PA, Oxidative stress and inflammation modulate rev-erbα signaling in the neonatal lung and affect circadian rhythmicity, Antioxidants Redox Signal 21 (2014) 17–32, 10.1089/ars.2013.5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Oeckinghaus A, Hayden MS, Ghosh S, Crosstalk in NF-κB signaling pathways, Nat. Immunol 12 (2011) 695–708, 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- [115].Oeckinghaus A, Ghosh S, The NF- B family of transcription factors and its regulation, Cold Spring Harbor Perspect. Biol 1 (2009), 10.1101/cshperspect.a000034 a000034–a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Wu Z-H, Shi Y, When ubiquitin meets NF-κB: a trove for anti-cancer drug development, Curr. Pharmaceut. Des 19 (2013) 3263–3275. http://www.ncbi.nlm.nih.gov/pubmed/23151140. (Accessed 18 July 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Zhang Q, Lenardo MJ, Baltimore D, 30 Years of NF-κB: a blossoming of relevance to human pathobiology, Cell 168 (2017) 37–57, 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Cavadini G, Petrzilka S, Kohler P, Jud C, Tobler I, Birchler T, Fontana A, TNF-alpha suppresses the expression of clock genes by interfering with E-box-mediated transcription, Proc. Natl. Acad. Sci. U. S. A 104 (2007) 12843–12848, 10.1073/pnas.0701466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Okada K, Yano M, Doki Y, Azama T, Iwanaga H, Miki H, Nakayama M, Miyata H, Takiguchi S, Fujiwara Y, Yasuda T, Ishida N, Monden M, Injection of LPS causes transient suppression of biological clock genes in rats, J. Surg. Res 145 (2008) 5–12, 10.1016/j.jss.2007.01.010. [DOI] [PubMed] [Google Scholar]
- [120].Haspel JA, Chettimada S, Shaik RS, Chu J-H, Raby BA, Cernadas M, Carey V, Process V, Hunninghake GM, Ifedigbo E, Lederer JA, Englert J, Pelton A, Coronata A, Fredenburgh LE, Choi AMK, Circadian rhythm reprogramming during lung inflammation, Nat. Commun 5 (2014) 4753, 10.1038/ncomms5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Hong H-K, Maury E, Ramsey KM, Perelis M, Marcheva B, Omura C, Kobayashi Y, Guttridge DC, Barish GD, Bass J, Requirement for NF-κB in maintenance of molecular and behavioral circadian rhythms in mice, Genes Dev 32 (2018) 1367–1379, 10.1101/gad.319228.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Leone MJ, Marpegan L, Bekinschtein TA, Costas MA, Golombek DA, Suprachiasmatic astrocytes as an interface for immune-circadian signalling, J. Neurosci. Res 84 (2006) 1521–1527, 10.1002/jnr.21042. [DOI] [PubMed] [Google Scholar]
- [123].Marpegán L, Bekinschtein TA, Costas MA, Golombek DA, Circadian responses to endotoxin treatment in mice, J. Neuroimmunol 160 (2005) 102–109, 10.1016/j.jneuroim.2004.11.003. [DOI] [PubMed] [Google Scholar]
- [124].Paladino N, Mul Fedele ML, Duhart JM, Marpegan L, Golombek DA, Modulation of mammalian circadian rhythms by tumor necrosis factor-α, Chronobiol. Int 31 (2014) 668–679, 10.3109/07420528.2014.886588. [DOI] [PubMed] [Google Scholar]
- [125].Sundar IK, Ahmad T, Yao H, Hwang J, Gerloff J, Lawrence BP, Sellix MT, Rahman I, Influenza A virus-dependent remodeling of pulmonary clock function in a mouse model of COPD, Sci. Rep 4 (2015) 9927, 10.1038/srep09927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Haimovich B, Calvano J, Haimovich AD, Calvano SE, Coyle SM, Lowry SF, In vivo endotoxin synchronizes and suppresses clock gene expression in human peripheral blood leukocytes, Crit. Care Med 38 (2010), 10.1097/CCM.0b013e3181cd131c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Bellet MM, Zocchi L, Sassone-Corsi P, The RelB subunit of NFκB acts as a negative regulator of circadian gene expression, Cell Cycle 11 (2012) 3304–3311, 10.4161/cc.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Spengler ML, Kuropatwinski KK, Comas M, Gasparian a. V., Fedtsova N, Gleiberman a.S., Gitlin II, Artemicheva NM, Deluca K.a., Gudkov a. V., Antoch MP, Core circadian protein CLOCK is a positive regulator of NF-kB-mediated transcription, Proc. Natl. Acad. Sci 109 (2012) E2457–E2465, 10.1073/pnas.1206274109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Mirsky HP, Liu AC, Welsh DK, Kay SA, Doyle FJ, A model of the cell-autonomous mammalian circadian clock, Proc. Natl. Acad. Sci. U. S. A 106 (2009) 11107–11112, 10.1073/pnas.0904837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Forger DB, Peskin CS, Stochastic simulation of the mammalian circadian clock, Proc. Natl. Acad. Sci. U. S. A 102 (2005) 321–324, 10.1073/pnas.0408465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Yamada Y, Forger D, Multiscale complexity in the mammalian circadian clock, Curr. Opin. Genet. Dev 20 (2010) 626–633, 10.1016/j.gde.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Ko CH, Yamada YR, Welsh DK, Buhr ED, Liu AC, Zhang EE, Ralph MR, Kay SA, Forger DB, Takahashi JS, Ko CH, Yamada YR, Welsh DK, Buhr ED, Liu AC, Zhang EE, Ralph MR, Kay SA, Forger DB, Takahashi JS, Emergence of noise-induced oscillations in the central circadian pacemaker, PLoS Biol 8 (2010), e1000513, 10.1371/journal.-pbio.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Leloup J-C, Goldbeter A, Modelling the dual role of Per phosphorylation and its effect on the period and phase of the mammalian circadian clock, IET Syst. Biol 5 (2011) 44–49, 10.1049/iet-syb.2009.0068. [DOI] [PubMed] [Google Scholar]
- [134].Kim JK, Forger DB, A mechanism for robust circadian timekeeping via stoichiometric balance, Mol. Syst. Biol 8 (2012) 630, 10.1038/msb.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Gallego M, Eide EJ, Woolf MF, Virshup DM, Forger DB, An opposite role for tau in circadian rhythms revealed by mathematical modeling, Proc. Natl. Acad. Sci. U. S. A 103 (2006) 10618–10623, 10.1073/pnas.0604511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Woller A, Duez H, Staels B, Lefranc M, A mathematical model of the liver circadian clock linking feeding and fasting cycles to clock function, Cell Rep 17 (2016) 1087–1097, 10.1016/j.celrep.2016.09.060. [DOI] [PubMed] [Google Scholar]
- [137].Foteinou PT, Venkataraman A, Francey LJ, Anafi RC, Hogenesch JB, Doyle FJ, Computational and experimental insights into the circadian effects of SIRT1, Proc. Natl. Acad. Sci 115 (2018) 11643–11648, 10.1073/pnas.1803410115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Pagano M, Jackson PK, Wagging the dogma, Cell 118 (2004) 535–538, 10.1016/j.cell.2004.08.013. [DOI] [PubMed] [Google Scholar]
- [139].Patke A, Murphy PJ, Onat OE, Krieger AC, Özçelik T, Campbell SS, Young MW, Mutation of the human circadian clock gene CRY1 in familial delayed sleep phase disorder, Cell 169 (2017) 203–215, 10.1016/j.cell.2017.03.027, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Debruyne JP, Noton E, Lambert CM, Maywood ES, Weaver DR, Reppert SM, A clock shock: mouse CLOCK is not required for circadian oscillator function, Neuron 50 (2006) 465–477, 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- [141].DeBruyne JP, Weaver DR, Reppert SM, Peripheral circadian oscillators require CLOCK, Curr. Biol 17 (2007). R538–9, http://www.ncbi.nlm.nih.gov/entrez/query.fcgid=Retrieve&db=PubMed&dopt=Citation&list_uids=17637349. [DOI] [PubMed] [Google Scholar]
- [142].Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F, Multiplex genome engineering using CRISPR/Cas systems, Science 339 (2013) 819–823, 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Korge S, Grudziecki A, Kramer A, Highly efficient genome editing via CRISPR/Cas9 to create clock gene knockout cells, J. Biol. Rhythm 30 (2015) 389–395, 10.1177/0748730415597519. [DOI] [PubMed] [Google Scholar]
- [144].Börding T, Abdo AN, Maier B, Gabriel C, Kramer A, Generation of human CRY1 and CRY2 knockout cells using duplex CRISPR/Cas9 technology, Front. Physiol 10 (2019) 577, 10.3389/fphys.2019.00577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker a P., van Leenen D, Buijs R, Bootsma D, Hoeijmakers JH, Yasui A, Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms, Nature 398 (1999) 627–630, 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- [146].Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, Takahashi JS, Sancar A, Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2, Proc. Natl. Acad. Sci. U. S. A 96 (1999) 12114–12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Jin Y-H, Joo H, Lee K, Kim H, Didier R, Yang Y, Shin H, Lee C, Streamlined procedure for gene knockouts using all-in-one adenoviral CRISPR-Cas9, Sci. Rep 9 (2019) 277, 10.1038/s41598-018-36736-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA, Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression, Cell 152 (2013) 1173–1183, 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS, CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes, Cell 154 (2013) 442–451, 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Chen Z, Yoo S-H, Takahashi JS, Development and therapeutic potential of small-molecule modulators of circadian systems, Annu. Rev. Pharmacol. Toxicol 58 (2018) 231–252, 10.1146/annurev-pharmtox-010617-052645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Gloston GF, Yoo S-H, Chen Z. (Jake), Clock-enhancing small molecules and potential applications in chronic diseases and aging, Front. Neurol 8 (2017), 10.3389/fneur.2017.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Gutman R, Genzer Y, Chapnik N, Miskin R, Froy O, Long-lived mice exhibit 24 h locomotor circadian rhythms at young and old age, Exp. Gerontol 46 (2011) 606–609, 10.1016/j.exger.2011.02.015. [DOI] [PubMed] [Google Scholar]
- [153].Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J, High-fat diet disrupts behavioral and molecular circadian rhythms in mice, Cell Metabol 6 (2007) 414–421, 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- [154].Manoogian ENC, Panda S, Circadian rhythms, time-restricted feeding, and healthy aging, Ageing Res. Rev 39 (2017) 59–67, 10.1016/j.arr.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JAJ, Ellisman MH, Panda S, Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet, Cell Metabol 15 (2012) 848–860, 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Bitto A, Ito TK, Pineda VV, Letexier NJ, Huang HZ, Sutlief E, Tung H, Vizzini N, Chen B, Smith K, Meza D, Yajima M, Beyer RP, Kerr KF, Davis DJ, Gillespie CH, Snyder JM, Treuting PM, Kaeberlein M, Transient rapamycin treatment can increase lifespan and healthspan in middle-aged mice, Elife 5 (2016) 1–17, 10.7554/eLife.16351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Martin-Montalvo A, Mercken EM, Mitchell SJ, Palacios HH, Mote PL, Scheibye-Knudsen M, Gomes AP, Ward TM, Minor RK, Blouin M-JJ, Schwab M, Pollak M, Zhang Y, Yu Y, Becker KG, Bohr VA, Ingram DK, Sinclair DA, Wolf NS, Spindler SR, Bernier M, De Cabo R, Metformin improves healthspan and lifespan in mice, Nat. Commun 4 (2013) 2192, 10.1038/ncomms3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Nohara K, Mallampalli V, Nemkov T, Wirianto M, Yang J, Ye Y, Sun Y, Han L, Esser KA, Mileykovskaya E, D’Alessandro A, Green CB, Takahashi JS, Dowhan W, Yoo S-H, Chen Z, Nobiletin fortifies mitochondrial respiration in skeletal muscle to promote healthy aging against metabolic challenge, Nat. Commun 10 (2019) 3923, 10.1038/s41467-019-11926-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA, Resveratrol improves health and survival of mice on a high-calorie diet, Nature 444 (2006) 337–342, 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Armour SM, Baur JA, Hsieh SN, Land-Bracha A, Thomas SM, Sinclair DA, Inhibition of mammalian S6 kinase by resveratrol suppresses autophagy, Aging (Albany. NY) 1 (2009) 515–528, 10.18632/aging.100056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Bass J, Targeting time in metabolic therapeutics, Cell Metabol 23 (2016) 575–577, 10.1016/j.cmet.2016.03.011. [DOI] [PubMed] [Google Scholar]
- [162].Park D, Jeong H, Lee MN, Koh A, Kwon O, Yang YR, Noh J, Suh P-G, Park H, Ryu SH, Resveratrol induces autophagy by directly inhibiting mTOR through ATP competition, Sci. Rep 6 (2016) 21772, 10.1038/srep21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Park I, Lee Y, Kim H-D, Kim K, Effect of resveratrol, a SIRT1 activator, on the interactions of the CLOCK/BMAL1 complex, Endocrinol. Metab. (Seoul, Korea) 29 (2014) 379–387, 10.3803/EnM.2014.29.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Khapre RV, Kondratova AA, Patel S, Dubrovsky Y, Wrobel M, Antoch MP, V Kondratov R, BMAL1-dependent regulation of the mTOR signaling pathway delays aging, Aging (Albany. NY) 6 (2014) 48–57, 10.18632/aging.100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].He B, Nohara K, Park N, Park Y-S, Guillory B, Zhao Z, Garcia JM, Koike N, Lee CC, Takahashi JS, Yoo S-H, Chen Z, The small molecule nobiletin targets the molecular oscillator to enhance circadian rhythms and protect against metabolic syndrome, Cell Metabol 23 (2016) 610–621, 10.1016/j.cmet.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Barnea M, Haviv L, Gutman R, Chapnik N, Madar Z, Froy O, Metformin affects the circadian clock and metabolic rhythms in a tissue-specific manner, Biochim. Biophys. Acta - Mol. Basis Dis 1822 (2012) 1796–1806, 10.1016/j.bbadis.2012.08.005. [DOI] [PubMed] [Google Scholar]


