Summary:
The long-range interactions of cis-regulatory elements (cREs) play a central role in gene regulation. cREs can be characterized as accessible chromatin sequences. However, it remains technically challenging to comprehensively identify their spatial interactions. Here, we report a new method HiCAR (Hi-C on Accessible Regulatory DNA), which utilizes Tn5 transposase and chromatin proximity ligation, for the analysis of open chromatin anchored interactions with low-input cells. By applying HiCAR in human embryonic stem cells and lymphoblastoid cells, we demonstrate that HiCAR identifies high-resolution chromatin contacts with an efficiency comparable to that of in situ Hi-C over all distances range. Interestingly, we found that the “poised” gene promoters exhibit silencer-like function to repress the expression of distal genes via promoter-promoter interactions. Lastly, we applied HiCAR to 30,000 primary human muscle stem cells, and demonstrated that HiCAR is capable of analyzing chromatin accessibility and looping using low-input primary cells and clinical samples.
eTOC:
HiCAR utilizes Tn5 transposase and chromatin proximity ligation to capture open chromatin anchored interactions with low-input cells. It requires <10% sequencing depth of Hi-C to call high-resolution chromatin interactions. Interestingly, we found that the “poised” gene promoters exhibit silencer-like function to repress the expression of distal genes via promoter-promoter interactions.
Introduction:
Cis-regulatory elements (cREs) play a critical role in regulating spatial-temporal gene expression in development and disease. cREs are characterized by the presence of “open” or accessible chromatin that can be identified by ATAC-Seq (Buenrostro et al., 2013), DNase-Seq (Boyle et al., 2008), and FAIRE-Seq (Simon et al., 2012). A growing body of evidence suggests that cREs function in concert with dynamic changes in chromatin organization to precisely control the expression of distant target genes (Bonev et al., 2017; Freire-Pritchett et al., 2017; Greenwald et al., 2019; Jerkovic and Cavalli, 2021; Lu et al., 2020; Oudelaar et al., 2020; Rao et al., 2014; Rowley and Corces, 2018; Song et al., 2019; Vilarrasa-Blasi et al., 2021; Yu and Ren, 2017; Zheng and Xie, 2019). Therefore, a comprehensive view of spatial interactions of accessible cREs is key to unveiling gene regulation mechanisms.
Chromosome conformation capture (3C) techniques have greatly improved our understanding of high-order chromatin organization. Particularly, Hi-C has been widely used to map genome-wide chromatin architecture, but it requires several billions of reads to resolve cRE interactions at a resolution of 5- to 10-kilobase (Bonev et al., 2017; Lieberman-Aiden et al., 2009; Rao et al., 2014). To enrich cRE-associated chromatin interactions, Capture-C or Capture Hi-C utilize pre-designed probes to enrich cRE sequences (Dryden et al., 2014; Hughes et al., 2014; Javierre et al., 2016; Mifsud et al., 2015). However, it is impractical to synthesize a large pool of capture probes that comprehensively cover hundreds of thousands of cRE sequences genome-wide. Methods such as ChIA-PET, HiChIP, and PLAC-seq employ protein-centric strategies to pull down chromatin sequences that are associated with specific proteins or histone modifications (Davies et al., 2016; Fang et al., 2016; Fullwood et al., 2009; Mumbach et al., 2016), but they cannot identify all the interactions associated with distinct epigenome features. More importantly, in many model organisms, due to the lack of high-quality ChIP-grade antibodies, such protein-centric strategies are not even feasible. These limitations highlight the urgent need for a robust and sensitive method to study cRE-interactions in a cost-effective and comprehensive manner.
Here, we report a novel method that we call Hi-C on Accessible Regulatory DNA (HiCAR), that leverages principles of Tn5-mediated open chromatin transposition and high-throughput chromosome conformation capture (3C) to enable genome-wide profiling of chromatin accessibility and cRE-anchored chromatin interactions. HiCAR does not require antibodies or capture probes to pulldown cRE sequences. Compared to in situ Hi-C, HiCAR faithfully identifies high-resolution chromatin contacts with an efficiency comparable to that of in situ Hi-C over all genomic distance ranges, but only requires <10% of the sequencing depth to call high-confident cRE interaction at 5kb resolution. We also provide a user-friendly nextflow pipeline (https://nf-co.re/hicar) for HiCAR data processing (Ou et al., 2022). Applying HiCAR to hESCs and a human lymphoblastoid cell line (GM12878), we identify open chromatin anchored interactions at 5kb resolution. Interestingly, we find that “poised” gene promoters can act as silencer-like elements to repress expression of distal genes via long-range promoter-promoter looping. These findings add a new dimension to previous ideas about the regulatory function of gene promoters as distal cREs (Dao et al., 2017; Diao et al., 2017; Engreitz et al., 2016; Li et al., 2012; Ren et al., 2021).
Design:
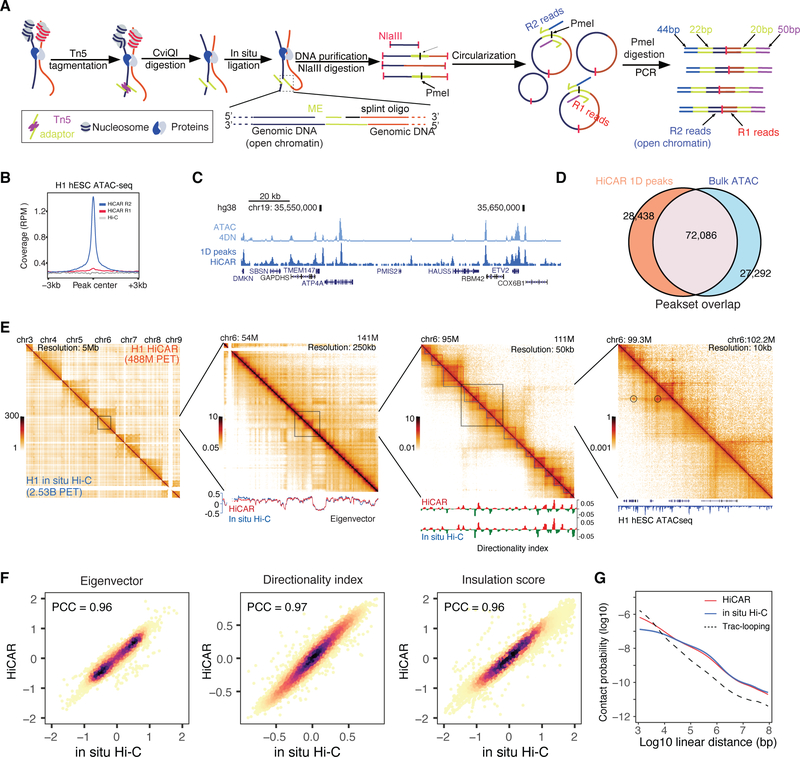
HiCAR was designed to capture the long-range chromatin interactions anchored on accessible regulatory DNA sequences. To perform HiCAR, 30,000 to 100,000 cells were crosslinked and treated with Tn5 transposase assembled with an engineered DNA adaptor (Fig 1A). The Tn5 adaptor contains a Mosaic End (ME) sequence for Tn5 recognition (Reznikoff, 2003) and a single-stranded flanking sequence that can be ligated to the genomic DNA digested by the 4-base cutter, CviQI, with a splint oligonucleotide (Table S1). After tagmentation, restriction enzyme digestion was performed using CviQI, followed by in situ proximity ligation to ligate the Tn5 adaptor to spatially proximal genomic DNA. Next, cross-links were reversed, and the purified DNA was digested with another 4-base cutter, NlaIII, and then circularized by intramolecular ligation. Using a 4C-seq like library preparation strategy (Noordermeer et al., 2011; Simonis et al., 2006; van de Werken et al., 2012; Zhao et al., 2006), PCR amplification was then performed to generate HiCAR libraries for Next-Generation-Sequencing (NGS). The forward and reverse PCR primers (Table S1) anneal specifically to the ME and splint oligo sequences, respectively. Therefore, the resulting PCR-amplified chimeric DNA fragments contain one end derived from genomic DNA and one derived from the Tn5-tagmented open chromatin (Fig 1A). Sequencing reads are referred to as R1 reads (CviQI digested genomic DNA, Fig 1A, “R1 reads” in red) and R2 reads (Tn5-tagemented open chromatin, Fig 1A, “R2 reads” in blue), respectively.
Figure 1. Overview of HiCAR experimental design.
(A) HiCAR workflow. The R2 reads of HiCAR libraries are derived from Tn5 insertion sites and can be used to call 1D open chromatin peaks. (B) The sequence depth normalized (reads per million, RPM) HiCAR R1 (red), R2 (blue) and in situ Hi-C (grey) reads was plotted as signal coverage surrounding +/− 3kb of each ATAC-seq peaks of H1 hESC. (C) A representative genome browser view showing the ATAC-seq data (top, light blue) and HiCAR R2 reads (bottom, dark blue) of H1 hESC. (D) Venn diagram showing overlap of MACS2 open chromatin peaks called from HiCAR R2 reads (orange) and ATAC-seq (blue) in H1 hESC. (E) H1 hESC chromatin contact matrices of HiCAR (above the diagonal) and in situ Hi-C (under the diagonal) at successive zoom-in views. Bottom tracks: the Eigenvector, Directionality index, and ATAC-seq track were plotted underneath the contact matrices as indicated. Color key: sequencing depth normalized reads counts (RPM). (F) Scatter plots comparing the Eigenvector, Directionality index, and Insulation score computed from HiCAR versus in situ Hi-C of H1 hESC. PCC: Pearson correlation coefficient. (G) Chromatin contact frequency (y-axis) was plotted as a function of linear genomic distance (x-axis) measured by HiCAR (red) and in situ Hi-C (blue) in H1 hESC and Trac-looping (dashed/black) in CD4 T cells. See also Figure S1, Figure S2, Table S1, and Table S3.
Results:
HiCAR faithfully captures chromatin accessibility and the key features of genome organization.
As a proof-of-principle, we tested HiCAR on H1 hESCs and human lymphoblastoid GM12878 cells. Each HiCAR library was made from 100,000 input cells and sequenced to ~300 million paired-end raw reads (Table S2). We took advantage of the publicly available genomic datasets for these two cell types generated by ENCODE (Davis et al., 2018; ENCODE Project Consortium, 2012), Epigenome Roadmap (Roadmap Epigenomics Consortium et al., 2015), and 4DN Consortium (Dekker et al., 2017), as well as previous studies (Juric et al., 2019; Lyu et al., 2018; Rao et al., 2014) to thoroughly benchmark our HiCAR data (Table S3 for public datasets used here). In H1 hESCs, the HiCAR R2 reads were indeed highly enriched at open chromatin regions defined by H1 hESC ATAC-seq data, whereas the R1 reads and the in situ Hi-C reads showed no enrichment (Fig 1B). Next, we confirmed that the HiCAR R2 reads are highly similar to the ATAC-seq reads by genome browser visualization (Fig 1C). We also called the 1D open chromatin peaks called by MACS2 (Zhang et al., 2008) using HiCAR R2 reads and ATAC-seq reads. As shown in Fig 1D, about 72% of HiCAR 1D peaks (total 100,524) overlapped with ATAC-seq peaks. The 27,292 ATAC-seq unique peaks were less confident peaks, as indicated by the larger P-value calculated by MACS2 (Fig S1A, left). Of the 28,438 HiCAR unique peaks, 61% are associated with epigenome features including CTCF/Cohesin binding sites (42.0%), DNase hypersensitivity (7.4%), enhancer/promoter marks H3K4me1/me3 (8.1%), and the active chromatin mark H3K27ac (3.7%) (Fig S1B, left). In GM12878 cells, we carried out the same benchmarking analysis by comparing R2 reads with ATAC-seq and ChIP-seq datasets. The results in GM12878 are consistent with those in H1 hESC regarding the performance of HiCAR (Fig S1A - S1E). We conclude that HiCAR accurately identifies 1D open chromatin peaks.
Next, we asked if HiCAR can identify the key features of genome architecture. For “gold standards” we used a deeply sequenced in situ Hi-C dataset from H1 hESC (2.5 billion PET) generated by the 4DN consortium (Krietenstein et al., 2020) and a deeply sequenced in situ Hi-C dataset from GM12878s (4.9 billion PET) (Rao et al., 2014). For HiCAR datasets, we generated 488 million PET for H1 hESC and 463 million PET for GM12878. Despite only using 9% to 19% of Hi-C sequencing depth, HiCAR generated a chromatin contact matrix similar to that of Hi-C at chromosome, compartment, topological associated domain (TAD), and 10kb-bin resolutions (Fig 1E, H1 hESC; Fig S1F, GM12878). Next, we employed HiCRep (Yang et al., 2017) to quantitatively assess HiCAR and Hi-C libraries. We found that HiCAR libraries are: (1) highly reproducible among biological replicates (Fig S1G, SCC = 0.94 to 0.95); and (2) more similar to the in situ Hi-C libraries generated from the same cell type (Fig S1G, SCC = 0.85 to 0.88 in H1 hESC; SCC = 0.77 to 0.79 in GM12878) but less similar to the HiCAR or Hi-C libraries made from different cell types (Fig S1G, SCC = 0.59 to 0.67). Furthermore, the A/B compartment PC1 eigenvector score (compartment score), insulation score, and directionality index calculated from HiCAR and in situ Hi-C data significantly correlate with each other (Fig 1F, PCC = 0.96, 0.97, 0.96 in H1 hESC; Fig S1H - S1J, PCC =0.96, 0.93, 0.89 in GM12878). Importantly, HiCAR identifies chromatin contacts over all distance ranges with an efficiency comparable to that seen in the in situ- Hi-C (Fig 1G, H1 hESCs; Fig S1K, GM12878). Taken together, we conclude that HiCAR faithfully captures the key features of genome architecture in the distinct cell types evaluated.
HiCAR outperforms existing methods in detecting open chromatin anchored long-range interactions.
Recently, a method called Trac-looping was developed to measure genome architecture and chromatin accessibility (Lai et al., 2018). Trac-looping relies on Tn5-transposition to recognize open chromatin sequences and utilizes a bivalent Mosaic End linker that favors the formation of a Tn5 tetramer complex to capture two pieces of spatially proximal open chromatin sequence. Compared to Trac-looping, HiCAR requires 1,000 fold less input cells (Fig 2A), yields more complex libraries (Fig 2B, 55.6% versus 13.4% uniquely mapped reads), and identifies about 18-fold more long-range (>20kb) paired-end tags (PETs) in cis (Fig 2B, orange bars). Compared to HiCAR and the “gold standard” in situ Hi-C data, Trac-looping identifies very few chromatin contacts over 10kb (Fig 1G). Therefore, HiCAR offers a significant technological advance over Trac-looping to measure chromatin accessibility and high-order genome structure.
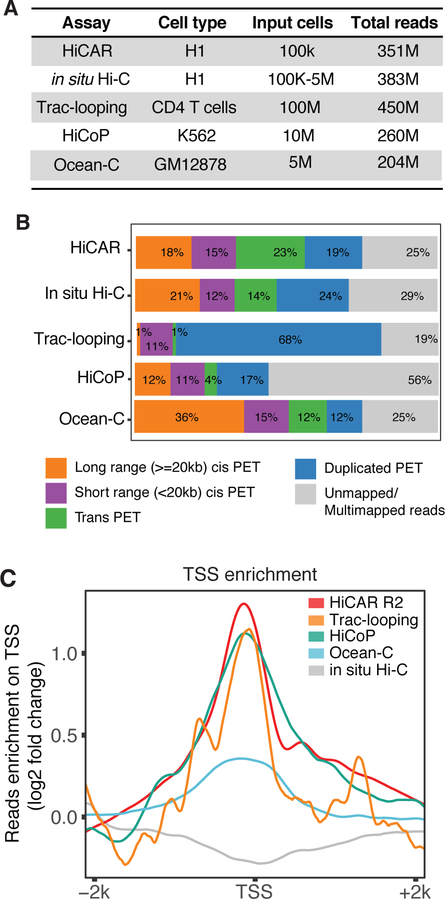
Figure 2. HiCAR outperforms existing methods to capture open chromatin anchored interactions.
(A) The number of input cells and the sequencing depth (total reads) of representative libraries of HiCAR, in situ Hi-C, Trac-looping, HiCoP, and Ocean-C. The representative H1 hESC in situ Hi-C data is obtained from 4DN data portal: 4DNFIYPLRRSZ. The Trac-looping, HiCoP, and Ocean-C data are obtained from previous studies (Lai et al., 2018; Zhang et al., 2020)(Li et al., 2018). (B) The percentage of PETs that are long range cis (>=20kb), short range cis (<20kb), trans (interchromosomal), PCR duplicates, and unmapped in the indicated dataset shown in (A). (C) The reads counts of HiCAR R2 (red), HiCoP (green), Ocean-C (blue), Trac-looping (orange), and in situ Hi-C (grey) are normalized against library sequence depth (counts per million), then aggregated within +/− 2kb window of human gene TSS. The fold change along the x-axis was calculated by comparing the reads counts of 100bp bin versus the mean reads counts of TSS +/− 2kb. See also Table S2 and Table S3.
In addition to Trac-looping, methods known as Ocean-C (Li et al., 2018) and HiCoP (Zhang et al., 2020) were also developed to enrich the chromatin interactions between DNA regions free of proteins. In Ocean-C and HiCoP, the protein-bound DNAs are removed by phenol/chloroform extraction or DNA purification columns, so the chromatin interactions associated with protein-free DNA sequences are enriched. Because HiCAR, Trac-looping, HiCoP, and Ocean-C were performed in different cell lines, we decided to assess the open chromatin enrichment efficiency of each method by examining transcription start site (TSS) signal enrichment, a metric widely used as a quality control of signal-to-noise ratios in ATAC-seq data (Corces et al., 2017). We found that HiCAR, HiCoP and Trac-looping reads show comparable TSS enrichment efficiency (Fig 2C), while Ocean-C reads show much weaker enriched signal on TSS (Fig 2C, light blue curve). Taken together, HiCAR outperforms existing methods regarding open chromatin enrichment efficiency, requirement of input cells, library complexity and the ratio of long-range cis-contacts.
HiCAR utilizes a 4C-seq-like library preparation strategy to capture “open-to-all” interactions.
The HiCAR library construction approach is similar to the 4C-seq protocol (Lu et al., 2020; van de Werken et al., 2012) (Fig 1A, Fig S2A). Therefore, HiCAR data can be analyzed from this perspective. Specifically, the accessible cREs can be treated as individual 4C “viewpoints” or “baits”, and all the PETs with R2 sequence overlapping the “bait” can be considered as 4C reads of the “bait” (Fig 2A, Fig S2B). Applying this approach, we examined the virtual 4C (v4C) contact profiles of HiCAR and Hi-C centered on the same set of 2kb “baits” in H1 hESCs. As shown in Fig S2C, the v4C contact profiles of HiCAR and Hi-C are very similar, despite the much lower sequencing depth used in HiCAR (488 million PET) versus in Hi-C (2.53 billion PET). These results further demonstrate that HiCAR can accurately define the chromatin contact profile centered on specific gene promoters.
HiCAR is a sensitive and accurate method to identify significant chromatin interactions anchored on open chromatin regions.
We applied MAPS (Juric et al., 2019) to identify the statistically significant cRE-anchored interactions identified by HiCAR. First, we evaluated the sensitivity of HiCAR interactions called by MAPS for detecting “known” chromatin interactions defined by well-established methods in matched cell types. We processed the in situ Hi-C data using two widely-used methods, HiCCUPS (Durand et al., 2016) and FitHiC2 (Kaul et al., 2020). We also processed the relevant published HiChIP and PLAC-seq datasets using MAPS (including H1 hESC H3K4me3 PLAC-seq, H9 hESC CTCF HiChIP, and GM1878 SMC1A HiChIP datasets) (Dekker et al., 2017; Lyu et al., 2018; Mumbach et al., 2016). Due to the lower sequencing depth of some public datasets, we called chromatin interactions at 10kb rather than 5 kb resolution and identified 142,325 and 97,840 significant HiCAR interactions (MAPS FDR <0.01) in H1 hESC and GM12878 (Table S2), respectively.
By visual examination, we found that HiCAR interactions exhibit a similar pattern with chromatin loops and interactions identified by in situ Hi-C, PLAC-seq, and HiChIP in the same cell types (Fig 3A, hESC; Fig S3A, GM12878). HiCAR interactions highly overlap with the loops identified by HiCCUPS and FitHiC2 using in situ Hi-C data (Fig S3B). We further selected a set of “testable” Hi-C loops and HiChIP/PLAC-seq interactions that have at least one anchor overlapping the open chromatin peaks defined by HiCAR 1D peaks. In H1 hESC, HiCAR identified 92%, 81% and 69% of the “testable” loops/interactions called by in situ Hi-C, H3K4me3 PLAC-seq, and CTCF HiChIP in hESCs, respectively (Fig S3C). Similarly, in GM12878 cells, HiCAR identified 78% and 89% of the “testable” loops/interactions called by in situ Hi-C and SMC1A HiChIP, respectively (Fig S3D). These results demonstrated that HiCAR is highly sensitive in detecting “known” chromatin loops/interactions identified by existing methods such as in situ Hi-C, HiChIP, and PLAC-seq.
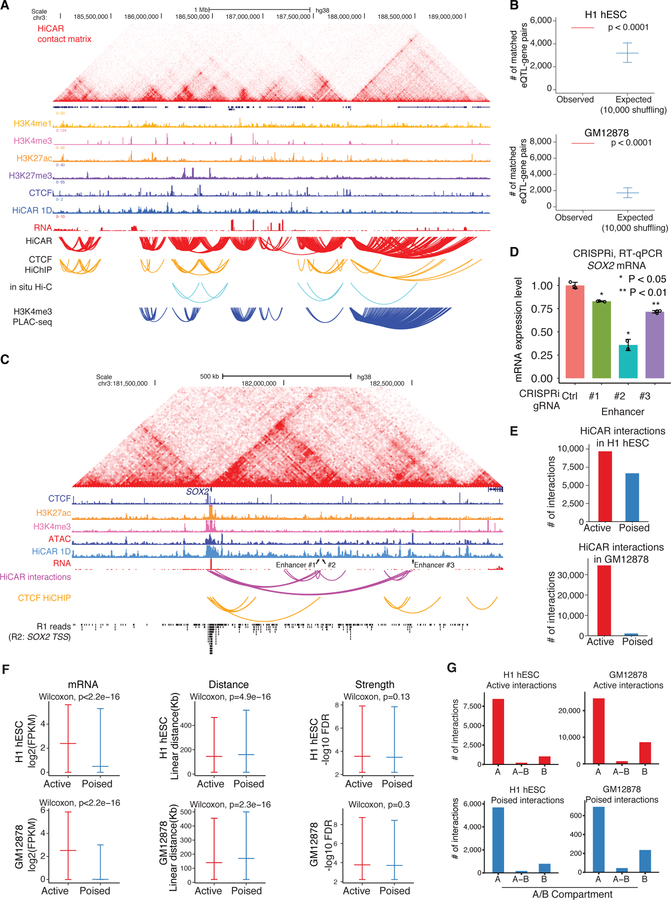
Figure 3. HiCAR is a robust and sensitive method to identify open chromatin anchored cRE interactions.
(A) HiCAR chromatin contact matrices are shown as heatmaps along with ChIP-seq tracks of H3K4me1, H3K4me3, H3K27ac, H3K27me3, CTCF; RNA-seq; and 1D open chromatin track derived from HiCAR R2 reads in H1 hESC. The arch tracks represent the chromatin loops and interactions called from HiCAR, CTCF HiChIP, H3K4me3 PLAC-seq, and in situ Hi-C as indicated. (B) The number of eQTL-gene pairs overlapping with observed HiCAR interactions (red), and (blue) randomly sampled pairwise DNA regions (10,000 times shuffling, with controlled linear genomic distance matching with HiCAR interactions distance) in H1 hESC (top) and GM12878 (bottom). One-sided empirical p-value <0.0001. (C) HiCAR contact matrix and indicated ChIP-seq, RNA-seq, and HiCAR 1D tracks surrounding SOX2 locus in H1 hESC. Arch tracks: H1 hESC HiCAR interactions (purple) and H9 hESC CTCF HiChIP interactions (yellow). The bottom track shows the aggregated R1 reads whose R2 reads overlap with the 2kb SOX2 transcription start site (TSS). The arrowheads point to enhancer #1 (chr3: 182,139,814–182,140,635), #2 (chr3: 182,143,023–182,143,349), and #3 (chr3: 182,500,129–182,500,831) for CRISPRi experiment shown in (D). (D) The sgRNAs were designed to recruit dCas9-KRAB to the candidate enhancers of SOX2 in H1 hESC. The non-targeting sgRNA was used as control. After CRISPRi, for each condition, three biological replicates were collected and SOX2 mRNA was analyzed by RT-qPCR. P values are calculated by a two-tailed Student’s t test. (E) The total number of active (red) and “poised” (blue) HiCAR interactions identified in H1 hESC (top) and in GM12878 (bottom). (F) We took the active (red) versus “poised” (blue) interactions identified from H1 hESC (top panels) and GM12878 (bottom panels) to compared: (left) the mRNA level (log2 transformed FPKM) of genes with promoters overlapping with anchors; (middle) the linear genomic distance between pairwise anchors; and (right). the interaction “strength” quantified by −log10 FDR (output from MAPS). The P values are calculated from the Wilcoxon rank-sum test. (G) We counted the number of active (top) versus “poised” (bottom) HiCAR interactions with their pairwise anchors located within the A, B, or across A-B compartments in H1 hESC (left) and GM12878 (right). See also Figure S3, Figure S4, Table S2, Table S3, and Table S4.
Next, we assessed the accuracy of HiCAR for identifying functional cRE-anchored interactions. Based on the loop exclusion model, chromatin loops prefer convergent CTCF motif orientations at loop anchors (Rao et al., 2014). Therefore, we examined CTCF motif orientation located on the anchors of HiCAR interactions in H1 hESC and GM12878 cells. In hESCs, we found that 63% of HiCAR interactions harbor convergent CTCF motifs on their anchors, consistent with the ratio observed using publicly available PLAC-seq (60%) (Fig S3E, blue bar). Similarly, in GM12878, 76% of HiCAR interactions harbor the convergent CTCF motifs on their anchors, consistent with the ratio (75%) based on SMC1A HiChIP interactions (Fig S3F). Interestingly, in both H1 hESC and GM12878, a higher percentage of in situ Hi-C loops are anchored on convergent CTCF motifs compared with loops identified by HiCAR and PLAC-seq or HiChIP (Fig S3E, S3F). We reasoned that this difference is because HiCCUPS uses a local background model for loop calling and therefore only identifies the most significant loop summits among a cluster of loops/interactions. Together, these results indicate that HiCAR can identify cRE interactions with high accuracy and sensitivity.
HiCAR identifies functional cRE interactions controlling gene expression.
HiCAR interactions are significantly enriched for expression quantitative trait loci (eQTL) and their associated genes in human pluripotent stem cells (hPSC) and GM12878 cells (DeBoever et al., 2017; The GTEx Consortium, 2015) (Fig 3B, empirical p value < 0.0001). This finding supports the idea that the chromatin interactions identified by HiCAR are involved in gene expression control. To test this hypothesis, we selected three putative enhancers interacting with SOX2 TSS and tested their function using CRISPR interference (CRISPRi). As shown in Fig 3C, enhancers #1, #2, and #3 are located 428kb, 431kb, and 788kb from the SOX2 TSS, respectively. Enhancer #1 and #2 also anchor on the CTCF-loop identified from CTCF HiChIP data using H9 hESC (Fig 3C). Upon silencing each individual enhancer, SOX2 expression was significantly reduced compared to the hESC expressing dCas9-KRAB and non-targeting control sgRNA (Fig 3D). These results indicate that the cRE interactions identified by HiCAR directly regulate gene expression.
The transcriptionally “poised” cREs exhibit extensive spatial interaction activity.
In H1 hESCs, we found that HiCAR can indeed effectively enriched the PETs associated with distinct epigenetic features, including chromatin accessibility, CTCF binding, and modification of H3K4me1, H3K4me3, H3K27ac and H3K27me3 (Fig S4A, S4B). Since we are particularly interested in the cRE-interactions overlapping with active mark H3K27ac and repressive/poised mark H3K27me3, we focused on these interactions in the following analysis. Using MAPS, we identified 9,692 interactions (5kb resolution) with at least one anchor overlapping with H3K27ac ChIP-seq peaks in H1 hESC (Fig 3E, red bar). We also identified 6,662 interactions (5kb resolution) overlapping with the H3K27me3 peaks (Fig 3E, blue bar; Fig S4C, “poised” interactions on GATA6 locus). Hereafter, we define these two types of interactions as “active” versus “poised” interactions, respectively. The interactions overlapping with both H3K27ac and H3K27me3 peaks were excluded from our analysis. Applying the same criteria, we identified 34,545 “active” and 1,116 “poised” interactions in GM12878 cells at 5 kb resolution (Fig 3E). Interestingly, a substantially greater proportion of “poised” interactions were observed in hESCs (19%, 6,662 out of 34,399 interactions) compared to GM12878 cells (2%, 1,116 out of 48,516 interactions) (Fig 3E, Fig S5A). The over-representation of “poised” interactions in H1 hESCs suggests that this type of interaction is particularly important for pluripotency. This observation is consistent with prior findings on H3K27me3-marked chromatin interactions in mouse embryonic stem cells and embryos (Cruz-Molina et al., 2017; Ghavi-Helm et al., 2014; Lonfat et al., 2014; Montavon et al., 2011; Ngan et al., 2020).
Through integration of HiCAR and additional ChIP-seq data we were able to identify both active and “poised” cREs from one single assay. Therefore, these interactions can be directly compared to each other without considering biases such as ChIP pulldown efficiency or batch effect. In both H1 and GM12878s, we found that genes with promoters located on the anchors of active interactions indeed show significantly higher expression levels compared to those with “poised” interactions (Fig 3F, left, Wilcoxon rank-sum, p < 2.2e-16). However, their interaction “strength” (quantified by −log10 FDR calculated by MAPS) was indistinguishable (Fig 3F, right, Wilcoxon rank-sum p = 0.13 and 0.3 in H1 hESC and GM12878, respectively). Interestingly, the linear genomic distance of active interactions was significantly shorter than that of “poised” interactions (Fig 3F, middle, median distance 135 kb versus 175 kb in H1, Wilcoxon rank-sum, p = 4.9e-16; 135 kb versus 155 kb in GM12878; Wilcoxon rank-sum, p = 2.3e-16). Additionally, it is well established that the mammalian genome can be compartmentalized into transcriptionally active A and repressive B compartments (Lieberman-Aiden et al., 2009). Interestingly, when we examined the compartment distribution of active and “poised” interactions in H1 hESC and GM12878s we found that both types of interaction are significantly enriched in compartment A and depleted in B (Fig 3G, Fig S5B, Table S4). Taken together, these results show that HiCAR can capture chromatin interactions anchored on accessible cREs that are active and “poised’, and that the “poised” cREs are associated with significant spatial interactions comparable with active cREs.
“Poised” cRE interactions are associated with developmentally silenced genes and Polycomb Repressive Complex proteins.
To explore the potential biological function of active and “poised” cRE interactions captured by HiCAR, we performed gene ontology (GO) analysis of genes with promoters located on active versus “poised” interactions anchors (Table S5). We found that GO terms related to essential cellular functions, such as nucleosome organization, DNA replication, and protein transport, are significantly enriched on the active interaction anchors in both H1 hESC and GM12878s (Fig S5C). Interestingly, GO terms related to cell lineage specific functions, such as stem cell proliferation and maintenance in H1 hESC and lymphocyte/B cell proliferation/differentiation in GM12878, were only enriched in the relevant cell type (Fig S5C). In H1 hESC, the genes on the anchors of “poised” interactions are enriched for GO terms related to development of brain, limb, cardiac, muscle, and leukocyte/lymphocyte. Interestingly, GO terms related to leukocyte/lymphocyte differentiation were enriched on the anchors of “poised” cRE interactions in H1 hESCs, but on anchors of active cRE interactions in GM12878 cells (Fig S5C).
We also carried out transcription factor (TF) motif enrichment analysis on the open chromatin sequences of active or “poised” interaction anchors. We found that the motifs of CTCF and BORIS (also known as CTCFL) were significantly enriched on the active interactions in both cell types (Fig S5D). By contrast, the motifs of lineage specific TFs, such as PU.1, IRF8, RUNX, IRF3/4 in GM12878 cells and OCT4, SOX2, NANOG in hESC, were specifically enriched in the relevant cells expressing these lineage specific TFs. Furthermore, the “poised” cRE interactions in hESC were: (1) significantly enriched for PRC2 complex protein binding (Fig S5E), and (2) associated with significantly broader H3K27me3 peaks (Fig S5F, Wilcoxon, P < 2.2e-16). These results are consistent with studies showing that Polycomb-group proteins can mediate long-range chromatin interactions anchored on H3K27me3 regions in mouse embryonic stem cells (Cai et al.; Cruz-Molina et al., 2017; Denholtz et al., 2013; Joshi et al., 2015; Kraft et al., 2020; Ngan et al., 2020; Schoenfelder et al., 2015). The enrichment of lineage specific genes and TF motifs on active and “poised” cRE interactions suggests that both interactions are involved in cell type specific gene regulation.
Gene promoters exhibit silencer-like function to repress the distal genes expression via promoter-promoter interactions.
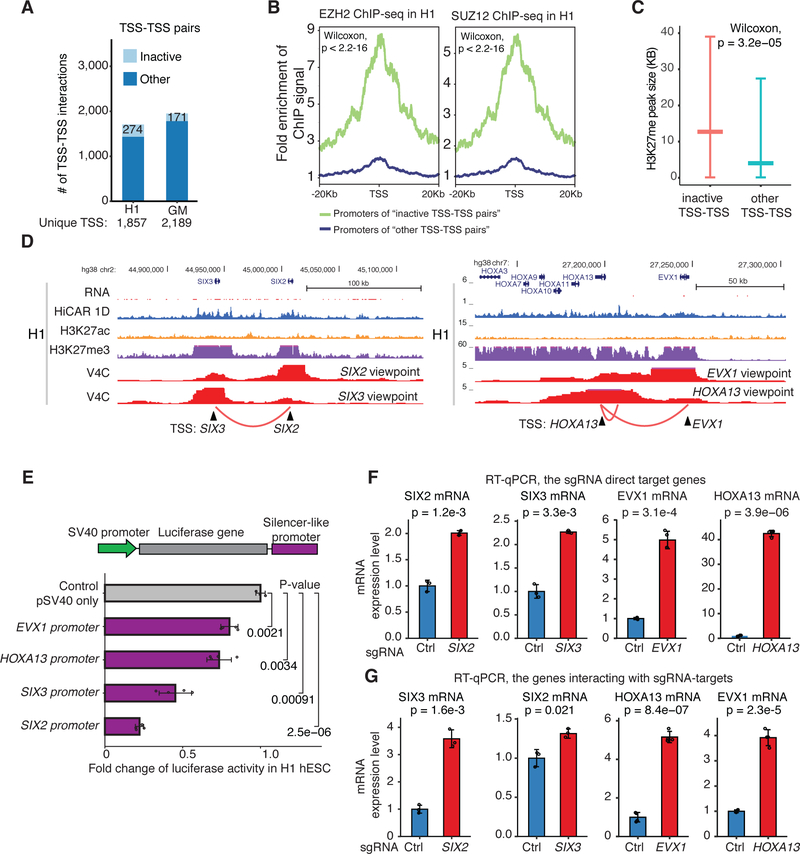
In the mammalian genome, many promoters can regulate the expression of other genes by acting as distal enhancers via long-range promoter-promoter interactions (Dao et al., 2017; Diao et al., 2017; Engreitz et al., 2016; Li et al., 2012; Ren et al., 2021). Using HiCAR, we also identified 1,706 and 1,950 promoter-promoter interactions (5kb resolution) in H1 hESC and GM12878 cells, respectively (Fig 4A). These interactions involve 1,875 and 2,189 unique TSS in H1 and GM12878, respectively. Interestingly, we found 274 and 171 interactions connecting pairwise inactive gene promoters (RNA-seq RPKM < 1 for both genes) in H1 hESCs (277 unique TSS) and GM12878 cells (171 unique TSS), respectively (Fig 4A). Consistent with previous findings (Joshi et al., 2015; Schoenfelder et al., 2015), in hESCs the promoters of “poised” TSS-TSS interactions are significantly enriched with binding of PRC2 proteins (Fig 4B, EZH2 and SUZ12), and overlapping with broader H3K27me3 peaks (Fig 4C).
Figure 4. Promoters act as silencer-like elements of distal genes via promoter-promoter interactions.
(A) 1,706 and 1,950 TSS-TSS interactions are identified in H1 hESC and GM12878, respectively. These interactions include 274 and 171 interactions between the inactive gene promoters (light blue). The rest of TSS-TSS interactions are defined as “other” (dark blue). (B) We took the TSS from the “inactive” (green line) and “other” TSS-TSS pairs (blue line). from ach TSS. EZH2 (left) and SUZ12 (right) ChIP-seq signal centered +/− 20kb of the selected TSS was calculated. Fold change: ChIP-seq reads of every 100 bp bin was calculated by comparing to those of genome background 20kb away from TSS. Wilcoxon test P-value < 2.2e-16. (C) The size (kilobase, kb) of H3K27me peaks in H1 hESC overlapping with the “inactive” and “other” TSS defined in (A). Wilcoxon test P-value = 3.2e-05. (D) Genome browser screenshot illustrating the inactive TSS-TSS interactions between SIX3 and SIX2 promoters (left), and EVX1 and HOXA13 promoters (right) in H1 hESC cells. The genome browser tracks include virtual 4C (V4C), HiCAR interactions, ChIP-seq of H3K27ac and H3K27me3, HiCAR 1D open chromatin profile, and RNA-seq. (E) The genomic sequence corresponding to the promoters of EVX1, HOXA13, SIX3 and SIX2 was cloned downstream of the luciferase gene in pGL3-Promoter reporter construct. H1 hESC cells were transiently transfected, and whole cell extract was subjected to luciferase assay. The data was collected from three biological replicates. P-values: two-tailed Student’s t-test. (F, G) For CRISPRa experiments, H1 hESC were infected by lentiviral co-expressing VP64-dCas9-VP64 and sgRNAs targeting the promoters of SIX2, SIX3, EVX1, and HOXA13. The non-targeting sgRNA was used as negative control. H1 hESC infected by lentiviral were selected by Puromycin for 3 days. 10-days after infection, total RNA was extracted and subjected to RT-qPCR analysis to assess the mRNA levels of indicated genes. (F) mRNA changes of sgRNA direct target genes; (F) mRNA changes of the genes that are not directly targeted by sgRNA, but interacting with the promoters targeted by the sgRNAs. The data was collected from three biological replicates. P-values: two-tailed Student’s t-test. See also Figure S5, Table S4, and Table S4.
Intrigued by the fact that gene promoters can act as enhancers of distal genes, we asked whether gene promoters can also exert silencer-like function. In hESC, the promoters of SIX3-SIX2 and HOXA13-EVX1 form significant interactions (Fig 4D). First, we performed classical plasmid-based luciferase reporter assays. The genomic fragments corresponding to the promoters of SIX2, SIX3, HOXA13, and EVX1 genes were cloned downstream of the luciferase reporter gene of pGL3-Promoter Vector (GenBank Accession Number U47298). We transiently transfected the plasmids into H1 hESCs and found that all four tested promoter sequences significantly repress reporter expression (Fig 4E). We also performed RT-qPCR analysis to test whether these cloned promoters initiate antisense transcripts from the plasmids and found that the antisense transcripts were expressed at very low levels (RT-qpCR CT value > 35). Given that these promoters act as distal elements of a linked promoter (SV40) and reduce SV40 transcriptional activity, by definition (Brand et al., 1985; Ogbourne and Antalis, 1998; Segert et al., 2021), these promoter sequences can be termed silencer-like elements in hESC.
To determine the function of the fours “poised” promoters in the native chromatin environment, we carried out CRISPR activation (CRISPRa) experiment by co-expressing transcriptional activator VP64-dCas9-VP64 (Kwon et al., 2020) and promoter-targeting sgRNAs in hESC. We designed sgRNAs to target the promoters of SIX2, SIX3, EVX1 and HOXA13, and used non-targeting sgRNAs as negative control. As expected, CRISPRa induced the expression of the direct target genes of the sgRNAs (Fig 4F). Interestingly, the expression of the genes that are not directly targeted by sgRNAs but interact with the sgRNA-target TSS, were also significantly upregulated (Fig 4G). To ensure that the observed phenotype is caused specifically by TSS-TSS interactions, we also examined the expression of genes neighboring SIX2, SIX3, EVX1 and HOXA13, namely HOXA4 and HIBADH. As expected, CRISPRa of HOXA13 or EVX1 TSS does not activate these nearby genes (Fig S5G). Interestingly, CRISPRa of SIX3 and SIX2 TSS induced upregulation of their respective neighboring genes CAMKMT and SRBD1 (Fig S5H, bottom). Notably, the CAMKMT promoter is 584kb away from the SIX3 promoter, while the distance between promoters of SIX2 and SRBD1 is 601kb. Intrigued by these results, we further examined the H1 hESC HiCAR contact matrix on SIX2 and SIX3 loci. We found that the promoters of SIX3 and SIX2 interact with promoters of CAMKMT and SRBD1 at higher frequency than background (Fig S5H, the black circles on HiCAR heatmap). These results suggest that the “poised” promoters of SIX2 and SIX3 also exhibit silencer-like function to repress the expression of distal genes (CAMKMT and SRBD1), likely through TSS-TSS interactions. Taken together, these results demonstrate that the “poised” gene promoters indeed play a silencer-like role to repress the transcription of distal genes in the native chromatin via promoter-promoter interactions.
HiCAR is capable of analyzing chromatin accessibility and looping using low-input primary human cells.
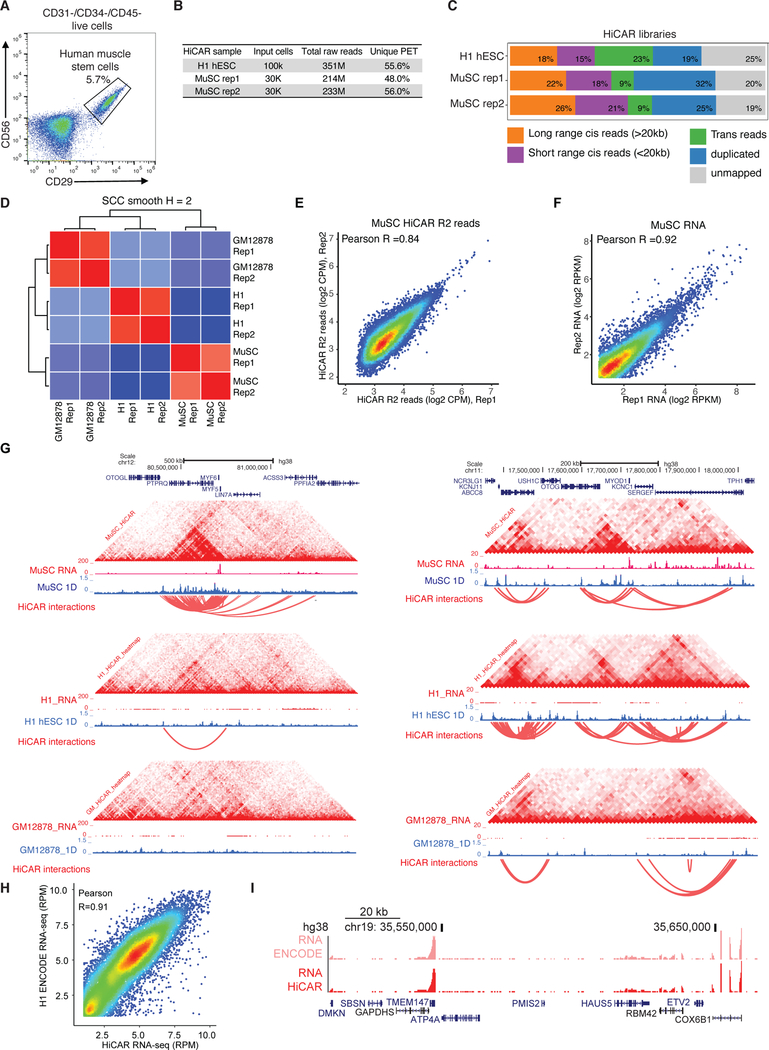
To demonstrate the application of HiCAR on low-input primary cells and clinical samples, we applied HiCAR to 30,000 primary human muscle stem cells (MuSC) purified from skeletal muscle biopsies (Fig 5A). As shown in Table S2, the MuSC HiCAR libraries are highly complex (Fig 5B, 48% and 56% unique PETs), and contain a large proportion of long-range cis-PETs (Fig 5C, 22% and 26%). Notably, the chromatin contact matrices and the open chromatin R2 reads of the MuSC HiCAR libraries are highly reproducible between two biological replicates (Fig 5D HiCRep SCC = 0.89, 5E PCC = 0.84). From a total of 233 million uniquely mapped PETs of MuSC, we called 46,244 1D peaks (MACS2 FDR < 0.01) using HiCAR R2 reads. Further analysis using HOMER showed that these open chromatin sequences are significant enriched with motifs of CTCF (p value = 1e-7158), MYF5 (p value = 1e-1252), and MYOD1 (p value = 1e-956). Using MAPS, we identified 25,693 significant cRE interactions in MuSC at 10kb resolution (Table S2). In addition to HiCAR analysis, we also collected the “leftover” RNAs from the HiCAR procedure to make RNA-seq libraries using SMART-seq3 (Hagemann-Jensen et al., 2020). At MYF5 and MYOD1 loci, two master regulators of MuSC, we observed highly cell-type specific chromatin interaction patterns (HiCAR interactions and contact matrix), chromatin accessibility (HiCAR 1D peaks), and transcriptome (RNA-seq, Fig 5F) compared to those observed in H1 and GM12878 (Fig 5G). These results clearly demonstrated that HiCAR is versatile and broadly applicable for simultaneous analysis of chromatin accessibility and looping using the same batch of low input cells from primary tissue or clinical samples.
Fig 5. HiCAR is compatible with SMART-seq for multi-omics analysis of the same low-input primary cells.
(A) The CD31−/CD34−/CD45−/CD56+/CD29+ human muscle stem cells (MuSCs) were isolated from human skeletal muscle and purified by FACS sorting. About 30,000 MuSCs were used for HiCAR analysis. The “leftover” RNAs were also collected from the same batch of MuSC cells for SMART-seq3. (B, C, D) Quality control analysis of HiCAR libraries of human MuSC, H1 hESC, and GM12878. Human MuSC HiCAR R2 reads (E) and the RNA-seq reads (F) are highly reproducible between the two biological replicates. (G) The genome browser screenshots showing HiCAR contact matrix, RNA-seq, and HiCAR 1D open chromatin tracks of human MuSC, H1 hESC, and GM12878 cells at MYF5 (left) and MYOD1 (right) loci. (H, I) The “leftover” polyA mRNAs were collected from the H1 hESC used for HiCAR experiment, and subjected to RNA-seq library construction following SMART-seq2 protocol. (H) Scatter plots showing the correlation of the reads counts of HiCAR/SMART-seq2 RNA-seq data versus ENCODE bulk H1 hESC RNA-seq data. (I) The representative genome browser view of (H). See also Table S2.
Discussion:
We demonstrate here that HiCAR is a robust, sensitive, and cost-effective method for detecting accessible chromatin and cRE interaction using low-input cells and less sequencing reads. Importantly, HiCAR identifies chromatin contacts with an efficiency comparable to that of in situ Hi-C over all genomic distances. The technical advantages of HiCAR are multifold. First, HiCAR captures cRE-anchored interactions in a more comprehensive way than existing methods (ChIA-PET, HiChIP, PLAC-seq) because it does not rely on antibodies to enrich cREs associated with only one specific protein or histone modification. Second, compared to Trac-looping (Lai et al., 2018), Ocean-C (Li et al., 2018), and HiCoP (Zhang et al., 2020), HiCAR requires 100 – 1000 fold fewer input cells and yields highly complex libraries. Third, HiCAR is cost-effective and requires remarkably less sequencing depth (9–19%) than Hi-C for high-resolution loop calling. Because of the 4C-seq-like library cloning strategy, HiCAR does not require the expensive Biotin-conjugated dNTP to capture the in situ ligated DNAs, which further reduces costs. Fourth, using 30,000 primary cells we demonstrated that HiCAR can be used to analyze small samples from primary tissues.
In this study, we also demonstrate that HiCAR can be combined with SMART-seq (Hagemann-Jensen et al., 2020; Picelli et al., 2014) protocols for transcriptome, chromatin accessibility, and genome organization analysis using the same low-input cells. In hESC, we collected the “leftover” poly-A mRNA from the same batch of cells used for HiCAR experiments, and used these RNAs to make RNA-seq libraries following SMART-seq2 (Picelli et al., 2014) protocol. We found that our H1 hESC RNA-seq data correlate very well with the H1 hESC bulk RNA-seq data generated by ENCODE, both at genome-wide scale (Fig 5H, Pearson correlation R = 0.91) and by visualization on genome browser (Fig 5I). In our analysis of primary human MuSCs (Fig 5A), the RNA-seq data generated using the MuSC “leftover” RNA are also highly reproducible (Fig 5F, Pearson Correlation R = 0.92). Notably, our MuSC RNA-seq results show strong RNA signals on the marker genes, MYF6, MYF5, and MYOD1 (Fig 5G, MuSC RNA tracks). These results confirm the feasibility of combining the HiCAR procedure with SMART-seq protocol to obtain multiple types of data using the same batch of low-input primary cells. Such a multi-omics “co-assay” would be particularly useful for analyzing complex tissue samples collected from animal models or human biopsies. Given the intrinsic cellular heterogeneity of complex tissues or biopsies, the cells collected from the same tissue and subjected to separated assays may represent very different cell populations. Therefore, the option of combining HiCAR and SMART-seq protocols not only enables the best use of precious low-input cells collected from primary tissue or clinical samples, but also provides an attractive alternative approach for simultaneous analysis of chromatin accessibility, genome architecture, and gene expression.
Another important conclusion of this work is that “poised” gene promoters can function as silencer-like elements to repress the expression of distal genes via long-range promoter-promoter looping. Together with the recent findings that many promoters can act as distal enhancers of other genes (Dao et al., 2017; Diao et al., 2017; Engreitz et al., 2016; Schmitt et al., 2016), our results reveals the complexity, or from another point of view, the underlying simplicity of gene regulation principles, that is, a single DNA sequence can encode different types of regulatory functions, including being a promoter for immediate genes, or an enhancer or silencer for neighboring- or distal-genes via linear chromatin proximity or long-range chromatin interactions.
In conclusion, we present HiCAR as a robust, sensitive, and cost-effective assay for comprehensive analysis of chromatin organization associated with accessible chromatin sites. Our results also uncover the unexpected role of “poised” gene promoters in exerting silencer-like function to repress distal gene expression via promoter-promoter interactions.
Limitations:
We note that the open chromatin peaks identified by HiCAR R2 reads are slightly different from peaks called by regular ATAC-seq. More than 70% of HiCAR 1D peaks overlap with the high confidence ATAC-seq peaks (with smaller MACS2 p-value). However, the less confident ATAC-seq peaks (with larger MACS2 p-value) are often missed from the 1D peaks called from HiCAR R2 reads. These data suggest HiCAR is less sensitive to capture weak open chromatin peaks compared to ATAC-seq. Interestingly, we found that 42% (H1 hESC) and 26% (GM12878) of HiCAR unique 1D peaks can be explained by binding of genome architecture proteins CTCF/Cohesin. Compared to regular ATAC-seq, the capture of R2 reads in the HiCAR library requires the in situ ligation between Tn5 adapter and the spatially proximal DNA sequences. Therefore, we speculate that the HiCAR protocol favors open chromatin sequences that are proximal to the anchor sequences of chromatin looping. This may likely cause some bias in HiCAR libraries to enrich the open chromatin regions bound by genome architecture protein CTCF and Cohesin.
We also acknowledge that the “leftover” RNAs collected from HiCAR procedure and used for SMART-seq undergo harsh treatment such as crosslinking and reverse-crosslinking, SDS treatment, high-temperature, and several steps of physical separation. All these steps are harmful for RNA integrity and could potentially introduce bias to the transcriptome analysis. Since low-input RNA-seq can be performed with a very small number of cells, in the analysis of abundant homogeneous cells (such as H1 hESC and GM12878), carrying out a regular RNA-seq analysis is a better choice rather than collecting the “leftover” RNAs for transcriptome analysis. Since the SMART-seq protocol is not an integral part of the HiCAR protocol, it is beyond the scope of this study to further discuss the details of RNA-seq library preparation.
STAR Methods text:
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yarui Diao (yarui.diao@duke.edu).
Materials availability
The DNA constructs generated in this study are available upon request. The other reagents used in this study are commercially available and detailed in the key resource table.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CD31-Alexa Fluor 488 | BioLegend | 303110 |
| Anti-CD34-FITC | BioLegend | 343503 |
| Anti-CD29-APC | BioLegend | 303008 |
| Anti-NCAM-Biotin | BioLegend | 318319 |
| Bacterial and virus strains | ||
| NA | ||
| Biological samples | ||
| Lower extremity human skeletal muscle samples (collected from a female, 67-year-old individual) | Duke University Medical Center | NA |
| Chemicals, peptides, and recombinant proteins | ||
| Matrigel | Corning | 354230 |
| mTeSR™ Plus | STEMCELL Technologies | 05825 |
| Accutase | BioLegend | 423201 |
| Chitin column | Bio-Rad | 7372522 |
| Amicon Ultracel 30K | Millipore | UFC903024 |
| Dialysis membrane tube | Spectra/Por | D1614-11 |
| NEBuffer 3.1 | NEB | B7203S |
| CviQI | NEB | R0639L |
| T4 DNA ligase (400U/μl) | NEB | M0202S |
| 20mg/ml BSA | NEB | B9000S |
| Proteinase K | Thermo Fisher | AM2546 |
| Phenol:Chloroform:Isoamyl Alcohol (25:24:1, v/v,) | Spectrum | 136112-00-0 |
| T4 DNA polymerase | NEB | M0203L |
| NlaIII | NEB | R0125L |
| SPRI beads | Beckman | B23319 |
| DNA clean & concentrator kit | Zymo | D4013 |
| Gel extraction using DNA recovery kit | Zymo | D4002 |
| Roche Complete Protease Inhibitor | Sigma | 5056489001 |
| PmeI | NEB | R0560L |
| RNaseOUT | Invitrogen | 10777-019 |
| Q5® High-Fidelity DNA Polymerase | NEB | m0491L |
| FuGENE® HD Transfection Reagent | Promega | E2311 |
| Anza™ 13 Esp3I | Invitrogen | IVGN0136 |
| Critical commercial assays | ||
| iTaq™ Universal SYBR® Green Supermix | Bio-Rad | 1725122 |
| Dual-Luciferase® Reporter Assay System | Promega | E1910 |
| Deposited data | ||
| HiCAR H1 | This study | GEO: GSE162819 |
| HiCAR GM12878 | This study | GEO: GSE162819 |
| HiCAR human muscle stem cells | This study | Synapse:syn26841404 |
| For published genomics datasets, see Table S3 | Table S3 | NA |
| Experimental models: Cell lines | ||
| H1 hESC | WiCell | WA01 |
| Human lymphoblastoid cell lines | Coriell Institute | GM12878 |
| Experimental models: Organisms/strains | ||
| NA | ||
| Oligonucleotides | ||
| Custom primers for HiCAR, see Table S1 | Table S1 | NA |
| sgRNA Oligos, see Table S1 | Table S1 | NA |
| Recombinant DNA | ||
| pTXB1-Tn5 | Addgene | 60240 |
| Lentiguide-puro | Addgene | 52963 |
| Lenti-dCas9-KRAB-blast | Addgene | 89567 |
| Lenti dCAS-VP64_Blast | Addgene | 61425 |
| pGL3-Promoter | Promega | E1761 |
| pRL-SV40 | Promega | E2231 |
| Software and algorithms | ||
| BWA | (Li and Durbin 2010) | http://bio-bwa.sourceforge.net/bwa.shtml |
| Pairtools | Python package | https://github.com/mirnylab/pairtools |
| HiGlass | (Kerpedjiev et al. 2018) | https://github.com/higlass |
| EnrichedHeatmap | (Gu et al. 2018) | https://github.com/jokergoo/EnrichedHeatmap |
| Cooltools | Python package | https://github.com/open2c/cooltools |
| HiCRep | (Yang et al. 2017) | https://github.com/qunhualilab/hicrep |
| Pairsqc | (Lee et al. 2021) | https://github.com/4dn-dcic/pairsqc |
| MACS2 | (Zhang et al. 2008) | https://github.com/macs3-project/MACS |
| Chip-R | (Newell et al. 2021) | https://github.com/rhysnewell/ChIP-R/ |
| Gimme | (van Heeringen and Veenstra 2011) | https://github.com/vanheeringen-lab/gimmemotifs |
| MAPS | (Juric et al. 2019) | https://github.com/ijuric/MAPS |
| Juicer:HiCCUPS | (Durand et al. 2016) | https://github.com/aidenlab/juicer |
| FitHiC2 | (Kaul, Bhattacharyya, and Ay 2020) | https://github.com/ay-lab/fithic |
| HiCAR nextflow pipeline | (Ou, Ewels, and Bot,. 2022) | DOI: 10.5281/zenodo.5889172 |
| Clusterprofile | (Yu et al. 2012) | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| Other | ||
| Detailed protocol for the preparation of HiCAR library | In this study | See Supplemental information, Method S1 |
Data and code availability
Raw and processed H1 hESC and GM12878 HiCAR data have been deposited at Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) and are publicly available under the accession numbers GSE162819. The processed human MuSC HiCAR data are available at synapse (https://www.synapse.org/#!Synapse:syn26841404). All the public genomic data used in this study has been listed in Table S3.
All original code used for HiCAR analysis is publicly available at Nextflow (https://nf-co.re/hicar) and has been deposited at Zenodo (Ou et al., 2022). The DOI of the released code is listed in the key resources table.
Any additional information regarding data and code required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines and culture conditions
H1 hESCs (WiCell, WA01) were cultured in Matrigel (Corning, 354230) coated plates with Stabilized feeder-free maintenance medium mTeSR™ Plus (STEMCELL Technologies, #05825). mTeSR™ Plus was changed every other day. GM12878 cells were cultured in suspension using RPMI 1640 with 15% FBS in T-75 flasks (200,000–800,000 cells/ml). Cells were harvested at the end of day2. For crosslinking, cells were washed once by PBS, then treated by Accutase (BioLegend, #423201) for 10mins at 37°C. After removing the Accutase, cells were resuspended by DMEM. Formaldehyde was added to the final concentration of 1%, incubated at room temperature for 10mins. Glycine was added to the final concentration of 0.2M, incubated at room temperature for 10mins to quench formaldehyde. Fixed cells were pelleted by centrifugation for 5 min at 4°C and washed with ice-cold PBS once
Human skeletal muscle samples
Lower extremity human skeletal muscle samples were obtained with informed consent from an patient (female, 67-year-old) undergoing a surgical procedure in accordance with a research protocol approved by the Duke University Institutional Review Board (IRB#Pro00065709).
Method Details
Tn5 Purification
Briefly, Rosetta DE3 cells transformed with Tn5 expression plasmid pTXB1-Tn5 (Addgene #60240) were cultured in 500ml LB and incubated at 16°C overnight for protein induction. The bacteria were collected by centrifuge and resuspend by pre-cooled HEGX (40mM Hepes-KOH pH 7.2, 1.6M NaCl, 2 mM EDTA, 20% Glycerol, 0.4% Triton-X100, Roche Complete Protease Inhibitor), sonicated to release the protein. PEI (10% PEI, 4.44% HCl, 800mM NaCl, 20mM Hepes, 0.3mM EDTA, 0.2% Triton X-100, pH 7.2) were then added to the lysate in dropwise to precipitate the E. coli DNA. The lysate was then centrifuged and the supernatant was loaded to Chitin column (Bio-Rad, #7372522). The column was rotated at 4°C for 2–3h then washed by HEGX buffer. 15ml HEGX buffer containing 100mM DTT was added to elute the protein. The column was incubated for another 24 hr at 4°C. The elution fraction was collected and concentrated to about 1ml by Amicon Ultracel 30K (Millipore, #UFC903024), then dialyzed twice by 1L dialysis buffer (100 HEPES-KOH pH 7.2, 0.2 M NaCl, 0.2 mM EDTA, 2 mM DTT, 0.2% Triton X-100, 20% glycerol) for 24h using dialysis membrane tube (Spectra, D1614–11). Then the protein was added 80% glycerol to a final concentration of 50%.
Tn5 transposase assembly
To assemble Tn5, 50μl of 200μM ME-rev and 50μl of 200μM BfaI-truseqR1-pmeI-nextera7 (Table S1) were annealed by the following program: 95°C 5min, cool to 14°C with a slow ramp 1°C /min. The annealed adaptor was mixed with Tn5 Transposase in 1: 1.5 molar ratio, the mixture was mixed by pipette and incubated at room temperature for 30mins.
Detailed HiCAR protocol
Step1. Nuclei preparation and tagmentation:
100,000 crosslinked cells were treated by 400μl NPB (PBS containing 5% BSA, 1mM DTT, 0.2% IGEPAL, Roche Complete Protease Inhibitor, 12.5μl RNaseOUT) at 4°C for 15min to isolate the nuclei. After centrifugation, the supernatant containing cytoplasm RNA was saved for future RNA-seq analysis. The isolated nuclei were resuspended in 350μl 2X TB buffer (66mM Tris-AC pH 7.8, 132mM K-AC, 20mM Mg-AC, 32% DMF), 335μl water and 15μl assembled Tn5 transposome. The oligos used for Tn adaptors are listed in Table S1). Next, nuclei are rotated at 37°C for 1.5h. 350μl of 40mM EDTA was added to stop the reaction. After washing the nuclei once by 0.075% BSA, the nuclei were treated by 32.5μl water, 5μl 10X NEBuffer 3.1 (NEB, # B7203S), 12.5μl 2% SDS at 62ºC for 10 minutes. After centrifugation at 850g for 5min, the supernatant containing nuclei RNA was collected for future RNA-seq library construction. The nuclei were resuspended in 100μl H2O, 14μl 10X NEBuffer3.1, 25μl 10% Triton X-100, and incubated at 37°C for 15min to quench SDS.
Step 2. CviQI digestion and in situ ligation
The nuclei were washed by 1ml 1.1X NEBuffer 3.1, then treated by 90μl 1.1X NEBuffer 3.1 containing 100U CviQI (NEB, #R0639L) and 1μl of 20μM TruseqR1 oligo (Table S1) at room temperature for 2h. After digestion, we added 48μl 10X T4 ligation buffer, 6μl T4 DNA ligase (400U/μl, NEB, #M0202S), 2.4μl 20mg/ml BSA (NEB, #B9000S), 40μl 10% Triton X-100, 283.6 μl H2O), into the reaction and rotated the nuclei at room temperature for 4h.
Step 3. Reverse crosslink and DNA purification
After centrifugation at 2000g for 5min, the supernatant was discarded. The nuclei were resuspended in 200μl of 2XRCB (100 mM Tris-HCl pH 8.0, 100 mM NaCl, 0.4% SDS), incubated the nuclei at 68°C for at least 1.5h to reverse crosslink. The DNA was purified by ethanol precipitation followed by 80% ethanol wash. The DNA was dissolved by 21μl 10mM Tris-HCl (pH8.0).
Step 4. NlaIII digestion and circularization
The purified DNA was incubated with 4μl 10mM dNTP, 5μl 10X CutSmart buffer 1.5μl T4 DNA polymerase (NEB, # M0203L) and 20.5μl H2O at room temperature for 30min to repair the Tn5 transposition gap. Next, the reaction was incubated at 75°C for 20min to inactivate T4 DNA polymerase. After that, 1μl NlaIII (NEB, # R0125L) were added into the sample followed by incubation at 37°C for 1h. The digested DNA was purified by 0.9X (45μl) volume SPRI beads (Beckman, # B23319), and dissolved in 80μl 10mM Tris-HCl (pH8.0) buffer. Next, the DNA was diluted to 1ng/μl and circulated in T4 Ligation Buffer by T4 DNA ligase (400U/μl, NEB, #M0202S). The sample is mixed and incubated at room temperature for at least 2h. The DNA was purified by DNA clean & concentrator kit (Zymo, #D4013) and eluted in 16μl water.
Step 5. PmeI digestion and PCR
14μl purified DNA was mixed with 1.7μl 10X CutSmart buffer, 1.3μl PmeI and incubated at 37°C for 1h to digest DNA. Then 16μl 5X Q5 buffer, 1.6μl 10mM dNTP, 1.6μl primer1 (Table S1) (10μM Nextera-pcr-i7–10-L), 1.6μl primer2 (Table S1) (10μM NEB primer i501), 0.8μl Q5 polymerase (NEB, #m0491L) and 58.4μl water was added into the sample. The PCR library amplification was performed using the following program (step 1: 72 °C 5 min, 98 °C 30 s; step 2: 98 °C 10 s, 59 °C 30 s, 72 °C 45s, repeating step 2 for an additional 11 cycles; step 3: 72°C 5 min and 4°C forever). After PCR, the DNA product between 300–750bp was purified by gel extraction using DNA recovery kit (Zymo, #D4002) for deep sequencing.
(Optional) Step 6. SMART-seq based RNA-seq libraries constructed using RNAs collected from HiCAR procedure.
The cytoplasmic and nuclei RNA fraction was combined, added with 1X volume of 2XRCB (100 mM TrisHCl pH 8.0, 100 mM NaCl, 0.4% SDS, RNase OUT), and incubated at 68°C for at least 1.5h for reverse crosslinking. Next, the RNA was purified by Phenol:Chloroform:Isoamyl Alcohol (25:24:1, v/v, Spectrum, #136112–00-0) extraction and ethanol precipitation. The sample was dissolved in 21μl 10mM Tris-HCl (pH8.0). Then the sample was treated by 0.5μl DNaseI at 37°C for 30min to remove DNA in solution. The RNA was purified by 2X volume of SPRI beads, dissolved by 20μl 10mM Tris-HCl (pH8.0). Then 2.3μl RNA was taken out to make an RNAseq library using Smart-seq2 protocol.
Purification of primary human skeletal muscle stem cell
Lower extremity human skeletal muscle samples were obtained with informed consent from an patient (female, 67-year-old) undergoing a surgical procedure in accordance with a research protocol approved by the Duke University Institutional Review Board (IRB#Pro00065709). Muscle samples were placed in DMEM with pen/strep on ice and brought to the laboratory for immediate processing and cell dissociation. Muscle was cleaned of tendon and fat and minced into pieces less than 1mm3 using sterile scissors and razor blades. Minced muscle was transferred to a 50mL conical tube for enzymatic dissociation with 0.05% Pronase (Roche) in DMEM with pen/strep for 1 hour at 37°C with slow continuous mixing. After triturating the sample for 10–15 times with a cannula, dissociated cells were resuspended in DMEM plus 10% FBS with pen/strep and filtered through 100mm vacuum filter. Cells were resuspended in Ham’s F-10 media with 10% horse serum and stained for FACS with the following antibodies: anti-CD31-Alexa Fluor 488 (clone WM59, BioLegend, #303110), anti-CD34-FITC (clone 581, BioLegend, #343503), anti-CD45-Alexa Fluor 488 (clone HI30, Invitrogen, #MHCD4520), anti-CD29-APC (clone TS2/16, BioLegend, #303008), anti-NCAM-Biotin (clone HCD56, BioLegend, #318319). Cells were washed, resuspended in Ham’s F-10 with 10% horse serum, and PE/Cy7 streptavidin (BioLegend, #318319) and PI was added. Cells were washed and resuspended in Ham’s F-10 with 10% horse serum for FACS. Satellite cells were isolated by FACS using a Sony SH800S Cell Sorter. CD31−/CD34−/CD45−/PI− cells were then gated on CD56 and CD29,and the CD56+/CD29+ satellite cells were sorted and fixed for HiCAR as described above.
Luciferase reporter assay
The Promoter fragments of SIX2, SIX3, EVX1, and HOXA13 were PCR amplified using the primers listed in Table S1 and cloned into the pGL3-promoter vector (Promega, Cat#: E1751) downstream of SV40 promoter and reporter gene. The purified reporter constructs were co-transfected to H1 hESC with Renilla plasmid pRL-SV40 using FuGENE® HD Transfection Reagent following manufacture’s instruction and described in our previous study (Diao et al., 2016). Briefly, 50,000 dissociated H1 hESC cells were seeded per well in a 48-well plate in mTeSR1 medium supplemented with Rock inhibitor Y-27632 for 12 hours. Next, 480μg of luciferase reporter constructs was mixed with 20ng of control Renilla plasmid pRL-SV40 and 1.5μl of prewarmed FuGENE® HD Transfection Reagent for transient transfection for cells per well. Added directly to medium, and mixed immediately. 48 hours after transfection, the cells were collected and subjected to luciferase reporter assay using the Dual-Luciferase® Reporter Assay System (Promega, Cat# E1910) on the SpectraMax® M5 Microplate Reader following manufacturer’s instructions.
CRISPRa and CRISPRi perturbation
The sgRNA sequences were designed by CHOPCHOP (Labun et al., 2019) and the primers used for sgRNA-expressing constructs cloning are listed in the Table S1. The sgRNA sequences were cloned into the Lentiviral vector expressing both sgRNA and dCas9-KRAB (Addgene #67620) or VP64-dCas9-VP64 (Addgene# #59791). Lentivirus was packed, purified, and used for H1 hESC infection following the protocol described in our previous study (Diao et al., 2017). After 10 days of Puromycin (2mg/ml) selection, the cells were collected and total RNA was extracted for RT-qPCR analysis. The sequences of qPCR primers were listed in Table S1.
QUANTIFICATION AND STATISTICAL ANALYSIS
HiCAR data processing
HiCAR datasets were processed following the distiller pipeline (https://github.com/mirnylab/distiller-nf). Briefly, reads were aligned to hg38 reference genome using bwa mem (Li and Durbin, 2010) with flags −SP. Alignments were parsed, and paired end tags (PET) were generated using the pairtools (https://github.com/mirnylab/pairtools). PET with low mapping quality (MAPQ < 10) was filtered out. PET with the same coordinate on the genome or mapped to the same digestion fragment were removed. Uniquely mapped PETs were flipped as side 1 with the lower genomic coordinate and aggregated into contact matrices in the cooler format using the cooler tools(Abdennur and Mirny, 2020) at delimited resolution (5kb, 10kb, 50kb, 100kb, 250kb, 500Kb, 1Mb, 25MB, 50MB,100MB). The dense matrix data were extracted from cooler files and visualized using HiGlass (Kerpedjiev et al., 2018). The R1 and R2 reads signals around TSS or peaks were calculated with EnrichedHeatmap (Gu et al., 2018) before PET flipping.
Hi-C matrix correlation SCC (stratum-adjusted correlation coefficient)
The similarity between different Hi-C datasets were measured by HiCRep(Yang et al., 2017). The stratum adjusted correlation coefficient (SCC) is calculated on a per chromosome basis using HiCRep on 100 kb resolution data with a max distance of 5 Mb. The SCC was calculated as a weighted average of stratum-specific Pearson’s correlation coefficients.
Compartments A and B, directionality and Insulation score
Compartmentalization, directionality index and insulation score was assessed using cooltools (https://github.com/mirnylab/cooltools). Briefly, eigenvector decomposition was performed on cis contact maps at 100-kb resolution. The first three eigenvectors and eigenvalues were calculated, and the eigenvector associated with the largest absolute eigenvalue was chosen. An identically binned track of GC content was used to orient the eigenvectors. The insulation score and directionality Index were computed by Cooltools using ‘find_insulating_boundaries’ and ‘directionality’ functions, respectively.
Contact probability decaying curve
The curves of contact probability as a function of genomic separation were generated by pairsqc following the 4DN pipeline (https://github.com/4dn-dcic/pairsqc) (Lee et al., 2021). Briefly, the genome is binned at log10 scale at an interval of 0.1. For each bin, contact probability is computed as number of reads/number of possible reads/bin size.
HiCAR 1D open chromatin peak processing
Unique mapped HiCAR DNA library R2 reads were extracted and processed to be compatible as MACS2 (Zhang et al., 2008) input BED files. MACS2 was used to identify open chromatin regions following the ENCODE pipeline (https://github.com/ENCODE-DCC/atac-seq-pipeline) with the following parameters: “−q 0.01 --shift 150 --extsize −75--nomodel −B --SPMR --keep-dup all ”. The reproducible peaks from multiple experimental replicates are identified using Chip-R (Newell et al., 2021).
CTCF motif orientation analysis
CTCF ChIP-seq peak list of H1 and GM12878 was downloaded from ENCODE (accession No. ENCFF821AQO and ENCFF485CGE, respectively) and searched for CTCF sequence motifs using gimme (van Heeringen and Veenstra, 2011) and CTCF motif (MA0139.1) from the JASPAR database (Fornes et al., 2020). We then selected a subset of interactions with both ends containing either a single CTCF motif or multiple CTCF motifs in the same direction. The frequency of all possible directionality of CTCF motif pairs, convergent, tandem and divergent are evaluated.
Chromatin interaction calling
For HiCAR, PLAC-seq and HiChIP datasets, we used the MAPS(Juric et al., 2019) to call the significant chromatin interactions. First, paired-end tags were extracted from cooler datasets at 5KB or 10Kb resolution using the “cooler dump” function with parameters: “−t pixels −H --join”. The interaction anchor bins were defined by the ATAC peaks or corresponding ChIP-seq peaks called using MACS2(Zhang et al., 2008). MAPS applied a positive Poisson regression-based approach to normalize systematic biases from restriction enzyme cut sites, GC content, sequence mappability, and 1D signal enrichment. We grouped interactions that were located within 15 kb of each other at both ends into clusters and classified all other interactions as singletons. We retained only interactions with 6 or more and normalized contact frequency (raw read counts/expected read counts) >= 2 and the significant interactions were defined by FDR < 0.01 for clusters and FDR < 0.0001 for singletons. For in situ Hi-C dataset, the .hic file is downloaded from 4DN data portal (accession No. 4DNES2M5JIGV) and significant chromatin interactions at 10Kb resolution is identified by HiCCUPS (Durand et al., 2016) with the following parameters: “−r 10000 −k KR −f .1,.1 −p 4,2 −i 7,5 −t 0.02,1.5,1.75,2 −d 20000,20000” and FitHiC2 (Kaul et al., 2020) using default parameters.
Comparison between eQTL-TSS association and HiCAR interaction
To test the enrichment for HiCAR identified interactions in significant eQTL-TSS association, we first obtain the eQTL-TSS associations in H1 hESC and GM12878 from the previous study(DeBoever et al., 2017). To assess the significance of the enrichment, we generated a null distribution by creating a simulated interaction datasets by resampling the same number of interactions at random from distance-matched interactions (with 10,000 repeats). The empirical P-value was computed by comparing the observed overlapping number with the null distribution.
Gene Ontology enrichment analysis
We used Clusterprofile (Yu et al., 2012) to examine whether particular gene sets were enriched in certain gene lists. GO categories with “BH” adjusted p value < 0.05 were considered as significant.
Key resources table
Supplementary Material
Figure S1. HiCAR identifies high-confident chromatin accessibility and genome architecture in both H1 hESC and GM12878 cells. Related to Figure 1.
(A) We compared the open chromatin peaks called by MAPS2 using HiCAR R2 reads and regular ATAC-seq data in H1 hESC (left) and GM12878 (right) cells. Boxplot showing the distribution of the MACS2 P-value of the overlapping peaks shared by HiCAR and ATAC-seq (red box), and the peaks unique to ATAC-seq (blue box). Wilcoxon rank-sum test was used for statistical analysis to compute P value. (B) The HiCAR unique 1D peaks overlap with DHS sites (DNase) and ChIP-seq peaks of CTCF, RAD21, H3K27ac, H3K4me3, H3K4me1 in H1 hESC (left) and GM12878 cells (right). (C) We count the number of HiCAR R2 reads (blue), R1 reads (red), and in situ Hi-C (black) reads within +/− 3kb window centered at GM12878 ATAC-seq peaks. The HiCAR R1, R2 and Hi-C reads are normalized against sequence depth (reads per million, RPM). The average reads count per ATAC-seq peak was plotted as signal coverage surrounding the +/− 3kb window of ATAC-seq peaks in y-axis. (D) A representative genome browser view showing the reads signals of the public GM12878 ATAC-seq data (top, light blue) and GM12878 HiCAR R2 reads (bottom, dark blue). (E) Venn diagram showing open chromatin peaks called by MAPS2 using GM12878 HiCAR R2 reads (HiCAR 1D peaks, pink) and GM12878 ATAC-seq (blue). (F) The sequence depth normalized contact matrices of HiCAR (top right, above the diagonal) and in situ Hi-C (bottom left, below the diagonal) data from GM12878 at successive zoom-in views. The GM12878 in situ Hi-C data was obtained from the 4DN data portal. The color represents sequence depth normalized reads signal (counts per million mapped reads). The Compartment Score, Directionality index, computed from in situ Hi-C and HiCAR data were plotted underneath the contact matrices as indicated. (G) HiCRep was employed to compute the similarity of chromatin contact matrices generated by HiCAR and in situ Hi-C from H1 hESC and GM12878. SCC values are computed using HiCrep. (H-J) Scatter plots and Pearson correlation coefficient (PCC) comparing the compartment scores (H), directionality index (I), and Insulation score (J) calculated from GM12878 HiCAR versus those from GM12878 in situ Hi-C. PCC: (K) Plot showing chromatin contact frequency (y-axis) as a function of linear genomic distance (x-axis) measured by HiCAR (red curve) and in situ Hi-C (blue curve) in GM12878 cells.
Figure S2. The virtual 4C analysis of HiCAR data. Related to Figure 1.
(A) Schematic illustration comparing HiCAR (top) and 4C-seq (bottom) experimental protocols. After the Tn5 tagmentation in HiCAR, both HiCAR and 4C-seq require two rounds of restriction enzyme digestion followed by ligation and circularization steps, to produce the circularized DNA template for PCR library amplification. 4C-seq requires a pair of primers recognizing the specific “bait” sequence, while HiCAR utilizes a pair of primers annealing to the Tn5 adaptor sequence. (B) The genome browser screenshot showing the virtual 4C (V4C) contact profile extracted from HiCAR data centered on two open chromatin peaks in H1 hESC. The H1 hESC HiCAR interaction matrices are shown as heatmaps along with HiCAR interaction arch tracks. The dashed lines represent the randomly sampled PET with R2 reads (blue) overlapping with the “viewpoint” open chromatin peaks (2kb). The red dots denote the R1 reads from the sampled PETs. (C) The v4C contact profile derived from HiCAR (red tracks) and in situ Hi-C (blue tracks) data of H1 hESC centered on SIX3 (left) and HAND1 (right) locus, respectively.
Figure S3. HiCAR is a robust and sensitive method to identify open chromatin anchored cRE interactions in H1 hESC and GM12878 cells. Related to Figure 3.
(A) GM12878 HiCAR contact matrices are shown on the top as heatmap along with genome browser tracks of H3K4me3, H3K27ac, H3K27me3, CTCF ChIP-Seq, RNA-seq and HiCAR R2 reads (1D open chromatin profile) from GM12878. The arch tracks represent the chromatin loops and interactions identified from HiCAR, in situ Hi-C and SMC1a HiChIP data from GM12878. (B) Venn diagram of HiCAR MAPS interactions compared to in situ Hi-C loops called by FitHiC2 (left box) and HiCCUPS (right box) in both H1 hESC and GM12878 cells. (C, D) We examined the orientation of the CTCF motifs located on the pairwise anchors of each chromatin loop and interactions. The length of the color bar indicates the percentage of convergent (blue), tandem (pink) and divergent (grey) CTCF motif pairs among the tested HiCCUPS loops and MAPS interactions in (C) hESC and (D) GM12878 cells. (E, F) The Hi-C, HiChIP, and PLAC-seq loops and interactions with at least one anchor overlapping with HiCAR 1D open chromatin peaks are defined as the “testable” loops/interactions. The percentage of the “testable” loops/interactions that overlap with HiCAR interaction were calculated to estimate the sensitivity of HiCAR interaction calling in (E) hESC and (F) GM12878 cells.
Figure S4: HiCAR can effectively enrich open chromatin sequences associated with distinct epigenetic marks. Related to Figure 3.
(A) Using sequence depth normalized H1 hESC HiCAR (top) and in situ Hi-C (bottom) contact matrix, the paired-end reads counts from the indicated sub chromatin contact matrix (10kb bin) were aggregated. The sub chromatin contact matrices are entered on the indicated ATAC-seq and ChIP-seq peaks of H1 hESC, and extend +/− 250kb window. Color key: normalized reads counts (RPM). (B) Boxplot showing that the coverage of H1 hESC HiCAR library R2 reads across the human genome at indicated marks at 1kb fixed bins. The number of HiCAR R2 reads overlapping with 1kb ChIP-seq peaks of CTCF, H3K4me1, H3K4me3, H3K27ac, ATAC-seq and HiCAR 1D peaks were calculated and shown as boxplot. The “poised” peaks were defined as H3K27me3 ChIP-seq peaks overlapping with HiCAR 1D peaks. All the 1kb bins not overlapping with any of the above mentioned peaks are selected as the background. The Wilcox test were performed between background bins and bins overlapped with ChIP-seq, ATAC-seq or HiCAR 1D peaks. (C) Genome browser screenshot showing HiCAR contact matric, 1D peaks, RNA signal, and ChIP-seq signals of H3K27ac and H3K27me3 on GATA6 locus in H1 hESC. Right: The zoomed view including virtual 4C (V4C) tracks centered on 2kb GATA6 TSS.
Figure S5. HiCAR captures open chromatin anchored “active” versus “poised” interactions in H1 hESC and GM12878 cells. Related to Figure 4.
(A) Pie chart showing the percentage of active (orange) and “poised” interactions (purple) called in H1 hESC (left) and GM12878 (right) HiCAR data at 5kb resolution. (B) Based on the genome-wide distribution of A/B compartment in H1 hESC and GM12878 cells, we calculated the “expected” number of active or “poised” interactions that are located within A or B compartment in H1 hESC and GM12878. We also counted the actual number of active (left) and “poised” (right) interactions detected in compartment A or B, and compared to that of “expected” numbers in H1 hESC and GM12878. Y-axis: log2 transformed fold change of observed versus expected active (red) and “poised” (blue) interactions in H1 hESC and GM12878 cells. (C) We selected the genes with promoters overlapping with the anchor sequences of active or “poised” interactions detected in H1 hESC and GM12878 cells. The resulting gene lists were subjected to Gene Ontology analysis. Color key: P value from clusterProfile hypergeometric test. (D) We selected the HiCAR 1D peaks overlapping with anchor sequences of active or “poised” interactions detected in H1 hESC and GM12878 cells. Transcription-factor (TF) motif enrichment analysis was performed using HOMER on the open chromatin sequences (HiCAR 1D peaks) overlapping with interaction anchors. Color key: P value output from HOMER. (E, F) We selected the “poised” interactions from H1 hESC, and defined the rest of interactions as “other” interactions. (E) We expanded +/− 25kb from the center of the “poised” and “other” anchors, and calculated the ChIP-seq signal enrichment of EZH2 (left) and SUZ12 (right) surrounding these anchor regions. The ChIP-seq reads within +/− 25kb of anchors (100bp bin) was calculated, and compared to the average ChIP-seq signal outside 25kb away from anchors. The orange and grey curves represent ChIP-seq signal enrichment on the “poised” and “other” anchor, respectively. Wilcoxon test P value < 2.2e-16. (F) The size (kilobase, kb) of H3K27me peaks overlapping with “poised” versus “other” anchors of HiCAR interactions in H1 hESC were shown as box plot. Wilcoxon test P value < 2.2e-16. (G, H) CRISPRa experiments were carried to induce activation of promoter sequences of SIX2, SIX3, EVX1, and HOXA13. The non-targeting sgRNA was used as negative control. H1 hESC were infected by lentivirus co-expressing VP64-dCas9-VP64 and sgRNA, selected by Puromycin for 3 days, and collected for total RNA extraction 10-days post infection. The mRNA level of the indicated genes were quantified by RT-qPCR analysis. The data was collected from three biological replicates. P-values: two-tailed Student’s t-test. The H1 hESC HiCAR contact matrix is shown (H, top panel).
Methods S1. Detailed HiCAR protocol (for 30,000 to 100,000 input cells), related to STAR Methods.
Table S1: Oligo and DNA sequences used in this study. Related to Figure 1.
Table S2: Summary of all the HiCAR data and full list of MAPS interactions from H1 hESCs, GM12878 cells, and primary human muscle stem cells. Related to Figure 2, Figure 3, and Figure 5.
Table S3: The list of public genomic datasets used in this study. Related to Figure 1, Figure 2, and Figure 3.
Table S4: Active and “poised” HiCAR interactions in A and B compartments in H1 hESC and GM12878 cells. Related to Figure 3 and Figure 4.
Table S5: In H1 hESC and GM12878 cells, the genes whose promoters are located on the anchors of active and “poised” cRE-interactions were identified, and subjected to Gene Ontology (GO) enrichment analysis. Related to Figure 4.
Highlights:
HiCAR captures chromatin accessibility and looping from the same input cells.
HiCAR is cost effective and can be applied to low-input clinical samples.
HiCAR is compatible with SMART-seq for multi-omics analysis of DNA and RNA.
The “poised” promoters exhibit silencer-like function via promoter-promoter looping.
Acknowledgements:
We thank Drs. Brigid Hogan (Duke University) and David Gorkin (Emory University) for feedback on the manuscript. This work is supported by the new lab startup fund from Regeneration Next Initiative (current Duke Regeneration Center) (to Y.D.), Duke Whitehead Scholarship (to Y.D.), NIH 4D Nucleome Consortium U01HL156064 (to Y.D.), the NHGRI Genomic Innovator Awards R35HG011328 (to Y.D.) and UL1TR002553 (to K.W.S.). Y. X. is supported by Duke Regeneration Next Initiative postdoctoral fellowship.
Footnotes
Declaration of interests:
The authors declare no competing financial interests. Duke University and the authors (Y.D., X.W., and Y.X.) hold the HiCAR patent under U.S. Provisional Patent Application No. 63/108,565.
Inclusion and diversity statement:
We worked to ensure gender balance, ethnic or other types of diversity in the recruitment of human subjects. We worked to ensure diversity in experimental samples through the selection of the cell lines. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list. The authors of this paper include contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Additional Resources
Detailed Protocol:
A detailed step-by-step HiCAR protocol is provided in Method S1.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Abdennur N, and Mirny LA (2020). Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics 36, 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Mendelson Cohen N, Szabo Q, Fritsch L, Papadopoulos GL, Lubling Y, Xu X, Lv X, Hugnot J-P, Tanay A, et al. (2017). Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell 171, 557–572.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, and Crawford GE (2008). High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Breeden L, Abraham J, Sternglanz R, and Nasmyth K (1985). Characterization of a “silencer” in yeast: A DNA sequence with properties opposite to those of a transcriptional enhancer. Cell 41, 41–48. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Zhang Y, Loh YP, Tng JQ, Lim MC, Cao Z, Raju A, Lieberman-Aiden E, Li S, Manikandan L, et al. H3K27me3-rich genomic regions can function as silencers to repress gene expression via chromatin interactions. [DOI] [PMC free article] [PubMed]
- Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, et al. (2017). An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Molina S, Respuela P, Tebartz C, Kolovos P, Nikolic M, Fueyo R, van Ijcken WFJ, Grosveld F, Frommolt P, Bazzi H, et al. (2017). PRC2 Facilitates the Regulatory Topology Required for Poised Enhancer Function during Pluripotent Stem Cell Differentiation. Cell Stem Cell 20, 689–705.e9. [DOI] [PubMed] [Google Scholar]
- Dao LTM, Galindo-Albarrán AO, Castro-Mondragon JA, Andrieu-Soler C, Medina-Rivera A, Souaid C, Charbonnier G, Griffon A, Vanhille L, Stephen T, et al. (2017). Genome-wide characterization of mammalian promoters with distal enhancer functions. Nat. Genet. 49, 1073–1081. [DOI] [PubMed] [Google Scholar]
- Davies JOJ, Telenius JM, McGowan SJ, Roberts NA, Taylor S, Higgs DR, and Hughes JR (2016). Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat. Methods 13, 74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, Hilton JA, Jain K, Baymuradov UK, Narayanan AK, et al. (2018). The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 46, D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBoever C, Li H, Jakubosky D, Benaglio P, Reyna J, Olson KM, Huang H, Biggs W, Sandoval E, D’Antonio M, et al. (2017). Large-Scale Profiling Reveals the Influence of Genetic Variation on Gene Expression in Human Induced Pluripotent Stem Cells. Cell Stem Cell 20, 533–546.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Belmont AS, Guttman M, Leshyk VO, Lis JT, Lomvardas S, Mirny LA, O’Shea CC, Park PJ, Ren B, et al. (2017). The 4D nucleome project. Nature 549, 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denholtz M, Bonora G, Chronis C, Splinter E, de Laat W, Ernst J, Pellegrini M, and Plath K (2013). Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell 13, 602–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao Y, Li B, Meng Z, Jung I, Lee AY, Dixon J, Maliskova L, Guan K-L, Shen Y, and Ren B (2016). A new class of temporarily phenotypic enhancers identified by CRISPR/Cas9-mediated genetic screening. Genome Res. 26, 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao Y, Fang R, Li B, Meng Z, Yu J, Qiu Y, Lin KC, Huang H, Liu T, Marina RJ, et al. (2017). A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat. Methods 14, 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden NH, Broome LR, Dudbridge F, Johnson N, Orr N, Schoenfelder S, Nagano T, Andrews S, Wingett S, Kozarewa I, et al. (2014). Unbiased analysis of potential targets of breast cancer susceptibility loci by Capture Hi-C. Genome Res. 24, 1854–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES, and Aiden EL (2016). Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst 3, 95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, and Lander ES (2016). Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R, Yu M, Li G, Chee S, Liu T, Schmitt AD, and Ren B (2016). Mapping of long-range chromatin interactions by proximity ligation-assisted ChIP-seq. Cell Res. 26, 1345–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M, Baranasic D, et al. (2020). JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire-Pritchett P, Schoenfelder S, Várnai C, Wingett SW, Cairns J, Collier AJ, García-Vílchez R, Furlan-Magaril M, Osborne CS, Fraser P, et al. (2017). Global reorganisation of cis-regulatory units upon lineage commitment of human embryonic stem cells. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. (2009). An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 462, 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi-Helm Y, Klein FA, Pakozdi T, Ciglar L, Noordermeer D, Huber W, and Furlong EEM (2014). Enhancer loops appear stable during development and are associated with paused polymerase. Nature 512, 96–100. [DOI] [PubMed] [Google Scholar]
- Greenwald WW, Li H, Benaglio P, Jakubosky D, Matsui H, Schmitt A, Selvaraj S, D’Antonio M, D’Antonio-Chronowska A, Smith EN, et al. (2019). Subtle changes in chromatin loop contact propensity are associated with differential gene regulation and expression. Nat. Commun. 10, 1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M, and Ishaque N (2018). EnrichedHeatmap: an R/Bioconductor package for comprehensive visualization of genomic signal associations. BMC Genomics 19, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann-Jensen M, Ziegenhain C, Chen P, Ramsköld D, Hendriks G-J, Larsson AJM, Faridani OR, and Sandberg R (2020). Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat. Biotechnol. 38, 708–714. [DOI] [PubMed] [Google Scholar]
- van Heeringen SJ, and Veenstra GJ (2011). GimmeMotifs: a de novo motif prediction pipeline for ChIP-sequencing experiments. Bioinformatics 27, 270–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JR, Roberts N, McGowan S, Hay D, Giannoulatou E, Lynch M, De Gobbi M, Taylor S, Gibbons R, and Higgs DR (2014). Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat. Genet. 46, 205–212. [DOI] [PubMed] [Google Scholar]
- Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, Cairns J, Wingett SW, Várnai C, Thiecke MJ, et al. (2016). Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell 167, 1369–1384.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerkovic I, and Cavalli G (2021). Understanding 3D genome organization by multidisciplinary methods. Nat. Rev. Mol. Cell Biol. [DOI] [PubMed] [Google Scholar]
- Joshi O, Wang S-Y, Kuznetsova T, Atlasi Y, Peng T, Fabre PJ, Habibi E, Shaik J, Saeed S, Handoko L, et al. (2015). Dynamic Reorganization of Extremely Long-Range Promoter-Promoter Interactions between Two States of Pluripotency. Cell Stem Cell 17, 748–757. [DOI] [PubMed] [Google Scholar]
- Juric I, Yu M, Abnousi A, Raviram R, Fang R, Zhao Y, Zhang Y, Qiu Y, Yang Y, Li Y, et al. (2019). MAPS: Model-based analysis of long-range chromatin interactions from PLAC-seq and HiChIP experiments. PLoS Comput. Biol. 15, e1006982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul A, Bhattacharyya S, and Ay F (2020). Identifying statistically significant chromatin contacts from Hi-C data with FitHiC2. Nat. Protoc. 15, 991–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerpedjiev P, Abdennur N, Lekschas F, McCallum C, Dinkla K, Strobelt H, Luber JM, Ouellette SB, Azhir A, Kumar N, et al. (2018). HiGlass: web-based visual exploration and analysis of genome interaction maps. Genome Biol. 19, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft K, Yost KE, Murphy S, Magg A, Long Y, Corces RM, Granja JM, Mundlos S, Cech TR, Boettiger A, et al. (2020). Polycomb-mediated Genome Architecture Enables Long-range Spreading of H3K27 methylation. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krietenstein N, Abraham S, Venev SV, Abdennur N, Gibcus J, Hsieh T-HS, Parsi KM, Yang L, Maehr R, Mirny LA, et al. (2020). Ultrastructural Details of Mammalian Chromosome Architecture. Mol. Cell 78, 554–565.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon JB, Vankara A, Ettyreddy AR, Bohning JD, and Gersbach CA (2020). Myogenic Progenitor Cell Lineage Specification by CRISPR/Cas9-Based Transcriptional Activators. Stem Cell Reports 14, 755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, and Valen E (2019). CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47, W171–W174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai B, Tang Q, Jin W, Hu G, Wangsa D, Cui K, Stanton BZ, Ren G, Ding Y, Zhao M, et al. (2018). Trac-looping measures genome structure and chromatin accessibility. Nat. Methods 15, 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Vitzthum C, Alver BH, and Park PJ (2021). Pairs and Pairix: a file format and a tool for efficient storage and retrieval for Hi-C read pairs. [DOI] [PMC free article] [PubMed]
- Li H, and Durbin R (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. (2012). Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Jia L, Cao Y, Chen Q, and Li C (2018). OCEAN-C: mapping hubs of open chromatin interactions across the genome reveals gene regulatory networks. Genome Biol. 19, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonfat N, Montavon T, Darbellay F, Gitto S, and Duboule D (2014). Convergent evolution of complex regulatory landscapes and pleiotropy at Hox loci. Science 346, 1004–1006. [DOI] [PubMed] [Google Scholar]
- Lu L, Liu X, Huang W-K, Giusti-Rodríguez P, Cui J, Zhang S, Xu W, Wen Z, Ma S, Rosen JD, et al. (2020). Robust Hi-C Maps of Enhancer-Promoter Interactions Reveal the Function of Non-coding Genome in Neural Development and Diseases. Mol. Cell 79, 521–534.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu X, Rowley MJ, and Corces VG (2018). Architectural Proteins and Pluripotency Factors Cooperate to Orchestrate the Transcriptional Response of hESCs to Temperature Stress. Mol. Cell 71, 940–955.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, Wingett SW, Andrews S, Grey W, Ewels PA, et al. (2015). Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 47, 598–606. [DOI] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, and Duboule D (2011). A regulatory archipelago controls Hox genes transcription in digits. Cell 147, 1132–1145. [DOI] [PubMed] [Google Scholar]
- Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, and Chang HY (2016). HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell R, Pienaar R, Balderson B, Piper M, Essebier A, and Bodén M (2021). ChIP-R: Assembling reproducible sets of ChIP-seq and ATAC-seq peaks from multiple replicates. Genomics 113, 1855–1866. [DOI] [PubMed] [Google Scholar]
- Ngan CY, Wong CH, Tjong H, Wang W, Goldfeder RL, Choi C, He H, Gong L, Lin J, Urban B, et al. (2020). Chromatin interaction analyses elucidate the roles of PRC2-bound silencers in mouse development. Nat. Genet. 52, 264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, and Duboule D (2011). The dynamic architecture of Hox gene clusters. Science 334, 222–225. [DOI] [PubMed] [Google Scholar]
- Ogbourne S, and Antalis TM (1998). Transcriptional control and the role of silencers in transcriptional regulation in eukaryotes. Biochem. J 331 ( Pt 1), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J, Ewels P, and Bot N-C (2022). jianhong/nf-core-hicar: nf-core/hicar PreRelease v1.0.1.
- Oudelaar AM, Beagrie RA, Gosden M, de Ornellas S, Georgiades E, Kerry J, Hidalgo D, Carrelha J, Shivalingam A, El-Sagheer AH, et al. (2020). Dynamics of the 4D genome during in vivo lineage specification and differentiation. Nat. Commun. 11, 2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, and Sandberg R (2014). Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181. [DOI] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Wang M, Li B, Jamieson K, Zheng L, Jones IR, Li B, Takagi MA, Lee J, Maliskova L, et al. (2021). Parallel characterization of cis-regulatory elements for multiple genes using CRISPRpath. Sci Adv 7, eabi4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznikoff WS (2003). Tn5 as a model for understanding DNA transposition. Mol. Microbiol. 47, 1199–1206. [DOI] [PubMed] [Google Scholar]
- Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, and Corces VG (2018). Organizational principles of 3D genome architecture. Nat. Rev. Genet. 19, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AD, Hu M, Jung I, Xu Z, Qiu Y, Tan CL, Li Y, Lin S, Lin Y, Barr CL, et al. (2016). A Compendium of Chromatin Contact Maps Reveals Spatially Active Regions in the Human Genome. Cell Rep. 17, 2042–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Sugar R, Dimond A, Javierre B-M, Armstrong H, Mifsud B, Dimitrova E, Matheson L, Tavares-Cadete F, Furlan-Magaril M, et al. (2015). Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat. Genet. 47, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segert JA, Gisselbrecht SS, and Bulyk ML (2021). Transcriptional Silencers: Driving Gene Expression with the Brakes On. Trends Genet. 37, 514–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JM, Giresi PG, Davis IJ, and Lieb JD (2012). Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat. Protoc. 7, 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, and de Laat W (2006). Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 38, 1348–1354. [DOI] [PubMed] [Google Scholar]
- Song M, Yang X, Ren X, Maliskova L, Li B, Jones IR, Wang C, Jacob F, Wu K, Traglia M, et al. (2019). Mapping cis-regulatory chromatin contacts in neural cells links neuropsychiatric disorder risk variants to target genes. Nat. Genet. 51, 1252–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium (2015). The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilarrasa-Blasi R, Soler-Vila P, Verdaguer-Dot N, Russiñol N, Di Stefano M, Chapaprieta V, Clot G, Farabella I, Cuscó P, Kulis M, et al. (2021). Dynamics of genome architecture and chromatin function during human B cell differentiation and neoplastic transformation. Nat. Commun. 12, 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Werken HJG, Landan G, Holwerda SJB, Hoichman M, Klous P, Chachik R, Splinter E, Valdes-Quezada C, Oz Y, Bouwman BAM, et al. (2012). Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat. Methods 9, 969–972. [DOI] [PubMed] [Google Scholar]
- Yang T, Zhang F, Yardımcı GG, Song F, Hardison RC, Noble WS, Yue F, and Li Q (2017). HiCRep: assessing the reproducibility of Hi-C data using a stratum-adjusted correlation coefficient. Genome Res. 27, 1939–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, and Ren B (2017). The Three-Dimensional Organization of Mammalian Genomes. Annu. Rev. Cell Dev. Biol. 33, 265–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, and He QY (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li Z, Bian S, Zhao H, Feng D, Chen Y, Hou Y, Liu Q, and Hao B (2020). HiCoP, a simple and robust method for detecting interactions of regulatory regions. Epigenetics Chromatin 13, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, et al. (2006). Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 38, 1341–1347. [DOI] [PubMed] [Google Scholar]
- Zheng H, and Xie W (2019). The role of 3D genome organization in development and cell differentiation. Nat. Rev. Mol. Cell Biol. 20, 535–550. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. HiCAR identifies high-confident chromatin accessibility and genome architecture in both H1 hESC and GM12878 cells. Related to Figure 1.
(A) We compared the open chromatin peaks called by MAPS2 using HiCAR R2 reads and regular ATAC-seq data in H1 hESC (left) and GM12878 (right) cells. Boxplot showing the distribution of the MACS2 P-value of the overlapping peaks shared by HiCAR and ATAC-seq (red box), and the peaks unique to ATAC-seq (blue box). Wilcoxon rank-sum test was used for statistical analysis to compute P value. (B) The HiCAR unique 1D peaks overlap with DHS sites (DNase) and ChIP-seq peaks of CTCF, RAD21, H3K27ac, H3K4me3, H3K4me1 in H1 hESC (left) and GM12878 cells (right). (C) We count the number of HiCAR R2 reads (blue), R1 reads (red), and in situ Hi-C (black) reads within +/− 3kb window centered at GM12878 ATAC-seq peaks. The HiCAR R1, R2 and Hi-C reads are normalized against sequence depth (reads per million, RPM). The average reads count per ATAC-seq peak was plotted as signal coverage surrounding the +/− 3kb window of ATAC-seq peaks in y-axis. (D) A representative genome browser view showing the reads signals of the public GM12878 ATAC-seq data (top, light blue) and GM12878 HiCAR R2 reads (bottom, dark blue). (E) Venn diagram showing open chromatin peaks called by MAPS2 using GM12878 HiCAR R2 reads (HiCAR 1D peaks, pink) and GM12878 ATAC-seq (blue). (F) The sequence depth normalized contact matrices of HiCAR (top right, above the diagonal) and in situ Hi-C (bottom left, below the diagonal) data from GM12878 at successive zoom-in views. The GM12878 in situ Hi-C data was obtained from the 4DN data portal. The color represents sequence depth normalized reads signal (counts per million mapped reads). The Compartment Score, Directionality index, computed from in situ Hi-C and HiCAR data were plotted underneath the contact matrices as indicated. (G) HiCRep was employed to compute the similarity of chromatin contact matrices generated by HiCAR and in situ Hi-C from H1 hESC and GM12878. SCC values are computed using HiCrep. (H-J) Scatter plots and Pearson correlation coefficient (PCC) comparing the compartment scores (H), directionality index (I), and Insulation score (J) calculated from GM12878 HiCAR versus those from GM12878 in situ Hi-C. PCC: (K) Plot showing chromatin contact frequency (y-axis) as a function of linear genomic distance (x-axis) measured by HiCAR (red curve) and in situ Hi-C (blue curve) in GM12878 cells.
Figure S2. The virtual 4C analysis of HiCAR data. Related to Figure 1.
(A) Schematic illustration comparing HiCAR (top) and 4C-seq (bottom) experimental protocols. After the Tn5 tagmentation in HiCAR, both HiCAR and 4C-seq require two rounds of restriction enzyme digestion followed by ligation and circularization steps, to produce the circularized DNA template for PCR library amplification. 4C-seq requires a pair of primers recognizing the specific “bait” sequence, while HiCAR utilizes a pair of primers annealing to the Tn5 adaptor sequence. (B) The genome browser screenshot showing the virtual 4C (V4C) contact profile extracted from HiCAR data centered on two open chromatin peaks in H1 hESC. The H1 hESC HiCAR interaction matrices are shown as heatmaps along with HiCAR interaction arch tracks. The dashed lines represent the randomly sampled PET with R2 reads (blue) overlapping with the “viewpoint” open chromatin peaks (2kb). The red dots denote the R1 reads from the sampled PETs. (C) The v4C contact profile derived from HiCAR (red tracks) and in situ Hi-C (blue tracks) data of H1 hESC centered on SIX3 (left) and HAND1 (right) locus, respectively.
Figure S3. HiCAR is a robust and sensitive method to identify open chromatin anchored cRE interactions in H1 hESC and GM12878 cells. Related to Figure 3.
(A) GM12878 HiCAR contact matrices are shown on the top as heatmap along with genome browser tracks of H3K4me3, H3K27ac, H3K27me3, CTCF ChIP-Seq, RNA-seq and HiCAR R2 reads (1D open chromatin profile) from GM12878. The arch tracks represent the chromatin loops and interactions identified from HiCAR, in situ Hi-C and SMC1a HiChIP data from GM12878. (B) Venn diagram of HiCAR MAPS interactions compared to in situ Hi-C loops called by FitHiC2 (left box) and HiCCUPS (right box) in both H1 hESC and GM12878 cells. (C, D) We examined the orientation of the CTCF motifs located on the pairwise anchors of each chromatin loop and interactions. The length of the color bar indicates the percentage of convergent (blue), tandem (pink) and divergent (grey) CTCF motif pairs among the tested HiCCUPS loops and MAPS interactions in (C) hESC and (D) GM12878 cells. (E, F) The Hi-C, HiChIP, and PLAC-seq loops and interactions with at least one anchor overlapping with HiCAR 1D open chromatin peaks are defined as the “testable” loops/interactions. The percentage of the “testable” loops/interactions that overlap with HiCAR interaction were calculated to estimate the sensitivity of HiCAR interaction calling in (E) hESC and (F) GM12878 cells.
Figure S4: HiCAR can effectively enrich open chromatin sequences associated with distinct epigenetic marks. Related to Figure 3.
(A) Using sequence depth normalized H1 hESC HiCAR (top) and in situ Hi-C (bottom) contact matrix, the paired-end reads counts from the indicated sub chromatin contact matrix (10kb bin) were aggregated. The sub chromatin contact matrices are entered on the indicated ATAC-seq and ChIP-seq peaks of H1 hESC, and extend +/− 250kb window. Color key: normalized reads counts (RPM). (B) Boxplot showing that the coverage of H1 hESC HiCAR library R2 reads across the human genome at indicated marks at 1kb fixed bins. The number of HiCAR R2 reads overlapping with 1kb ChIP-seq peaks of CTCF, H3K4me1, H3K4me3, H3K27ac, ATAC-seq and HiCAR 1D peaks were calculated and shown as boxplot. The “poised” peaks were defined as H3K27me3 ChIP-seq peaks overlapping with HiCAR 1D peaks. All the 1kb bins not overlapping with any of the above mentioned peaks are selected as the background. The Wilcox test were performed between background bins and bins overlapped with ChIP-seq, ATAC-seq or HiCAR 1D peaks. (C) Genome browser screenshot showing HiCAR contact matric, 1D peaks, RNA signal, and ChIP-seq signals of H3K27ac and H3K27me3 on GATA6 locus in H1 hESC. Right: The zoomed view including virtual 4C (V4C) tracks centered on 2kb GATA6 TSS.
Figure S5. HiCAR captures open chromatin anchored “active” versus “poised” interactions in H1 hESC and GM12878 cells. Related to Figure 4.
(A) Pie chart showing the percentage of active (orange) and “poised” interactions (purple) called in H1 hESC (left) and GM12878 (right) HiCAR data at 5kb resolution. (B) Based on the genome-wide distribution of A/B compartment in H1 hESC and GM12878 cells, we calculated the “expected” number of active or “poised” interactions that are located within A or B compartment in H1 hESC and GM12878. We also counted the actual number of active (left) and “poised” (right) interactions detected in compartment A or B, and compared to that of “expected” numbers in H1 hESC and GM12878. Y-axis: log2 transformed fold change of observed versus expected active (red) and “poised” (blue) interactions in H1 hESC and GM12878 cells. (C) We selected the genes with promoters overlapping with the anchor sequences of active or “poised” interactions detected in H1 hESC and GM12878 cells. The resulting gene lists were subjected to Gene Ontology analysis. Color key: P value from clusterProfile hypergeometric test. (D) We selected the HiCAR 1D peaks overlapping with anchor sequences of active or “poised” interactions detected in H1 hESC and GM12878 cells. Transcription-factor (TF) motif enrichment analysis was performed using HOMER on the open chromatin sequences (HiCAR 1D peaks) overlapping with interaction anchors. Color key: P value output from HOMER. (E, F) We selected the “poised” interactions from H1 hESC, and defined the rest of interactions as “other” interactions. (E) We expanded +/− 25kb from the center of the “poised” and “other” anchors, and calculated the ChIP-seq signal enrichment of EZH2 (left) and SUZ12 (right) surrounding these anchor regions. The ChIP-seq reads within +/− 25kb of anchors (100bp bin) was calculated, and compared to the average ChIP-seq signal outside 25kb away from anchors. The orange and grey curves represent ChIP-seq signal enrichment on the “poised” and “other” anchor, respectively. Wilcoxon test P value < 2.2e-16. (F) The size (kilobase, kb) of H3K27me peaks overlapping with “poised” versus “other” anchors of HiCAR interactions in H1 hESC were shown as box plot. Wilcoxon test P value < 2.2e-16. (G, H) CRISPRa experiments were carried to induce activation of promoter sequences of SIX2, SIX3, EVX1, and HOXA13. The non-targeting sgRNA was used as negative control. H1 hESC were infected by lentivirus co-expressing VP64-dCas9-VP64 and sgRNA, selected by Puromycin for 3 days, and collected for total RNA extraction 10-days post infection. The mRNA level of the indicated genes were quantified by RT-qPCR analysis. The data was collected from three biological replicates. P-values: two-tailed Student’s t-test. The H1 hESC HiCAR contact matrix is shown (H, top panel).
Methods S1. Detailed HiCAR protocol (for 30,000 to 100,000 input cells), related to STAR Methods.
Table S1: Oligo and DNA sequences used in this study. Related to Figure 1.
Table S2: Summary of all the HiCAR data and full list of MAPS interactions from H1 hESCs, GM12878 cells, and primary human muscle stem cells. Related to Figure 2, Figure 3, and Figure 5.
Table S3: The list of public genomic datasets used in this study. Related to Figure 1, Figure 2, and Figure 3.
Table S4: Active and “poised” HiCAR interactions in A and B compartments in H1 hESC and GM12878 cells. Related to Figure 3 and Figure 4.
Table S5: In H1 hESC and GM12878 cells, the genes whose promoters are located on the anchors of active and “poised” cRE-interactions were identified, and subjected to Gene Ontology (GO) enrichment analysis. Related to Figure 4.
Data Availability Statement
Raw and processed H1 hESC and GM12878 HiCAR data have been deposited at Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) and are publicly available under the accession numbers GSE162819. The processed human MuSC HiCAR data are available at synapse (https://www.synapse.org/#!Synapse:syn26841404). All the public genomic data used in this study has been listed in Table S3.
All original code used for HiCAR analysis is publicly available at Nextflow (https://nf-co.re/hicar) and has been deposited at Zenodo (Ou et al., 2022). The DOI of the released code is listed in the key resources table.
Any additional information regarding data and code required to reanalyze the data reported in this paper is available from the lead contact upon request.