Extended Data Fig. 4:
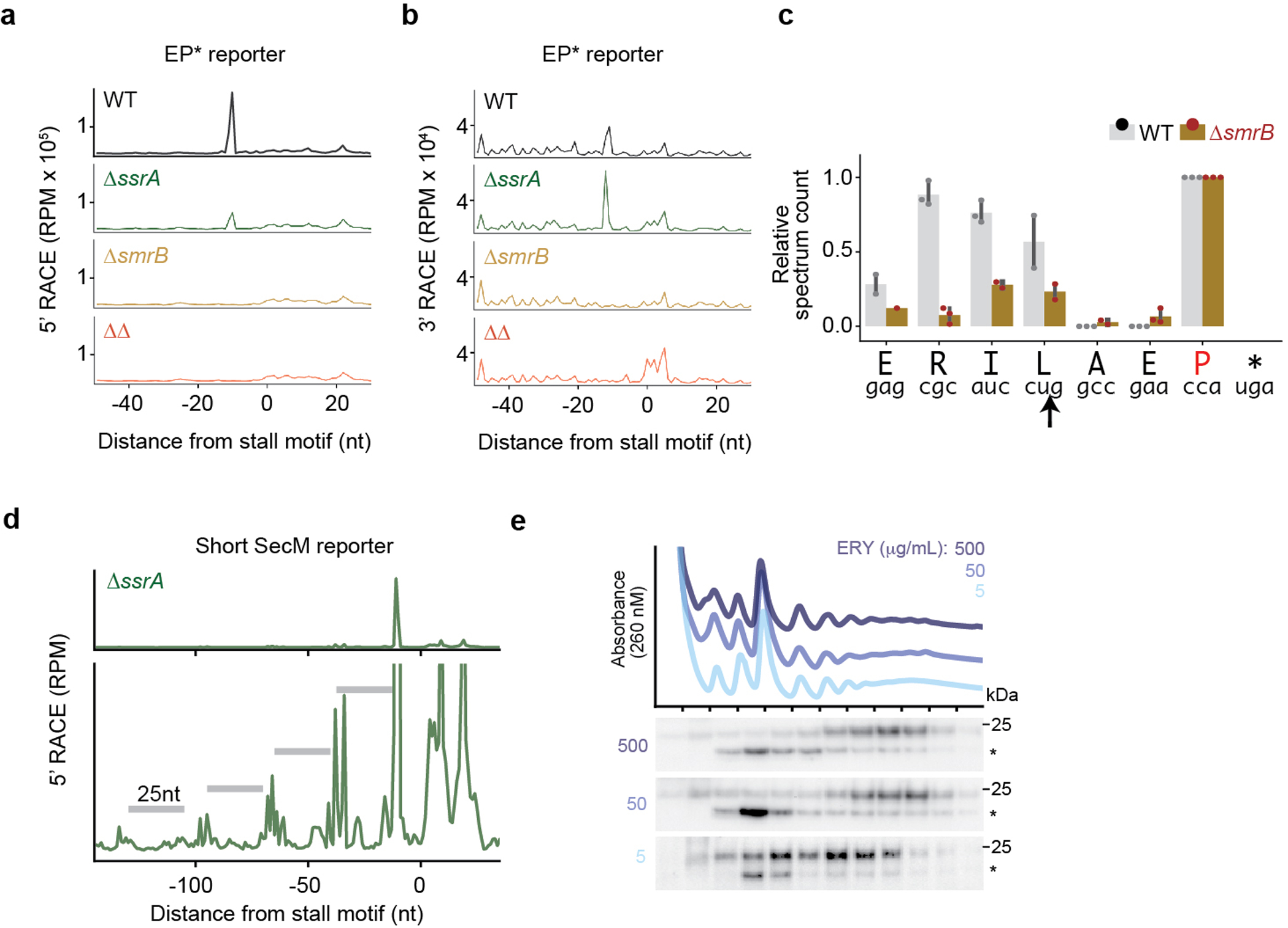
SmrB cleavage, tmRNA tagging, and ribosome collisions. a, The results of 5’-RACE showing the 5’-ends of downstream fragments in reads per million on the EP* reporter. The first nt in the A site codon in the stall motif is designated as zero. b, The results of 3’-RACE showing the 3’-ends of upstream fragments. c, tmRNA tagging sites on the EP* reporter in the wild-type and ΔsmrB strains, corresponding to the residue immediately preceding the tmRNA tag in peptide sequences detected by targeted LC-MS-MS. The relative spectrum count is normalized by the count at the EP* stall site (red) where tmRNA tagging was expected to occur in both the wild-type and ΔsmrB strains. The spectrum count corresponds to the mean and the standard deviation of three replicates. The arrow indicates the SmrB cleavage site demonstrated by 5’-RACE. d, 5’-RACE data on the Short SecM reporter reveal the SmrB cleavage sites as in Fig. 2b, zoomed in to show smaller peaks upstream. e, The distribution of FLAG-SmrB in cells treated with 5, 50, or 500 μg/mL erythromycin (ERY) was determined by fractionation over a sucrose gradient and detected with an anti-FLAG antibody.
