Abstract
Cyclodextrin-based controlled delivery materials have previously been developed for controlled release of different therapeutic drugs. In this study a supramolecular hydrogel made from cyclodextrin based macromonomers were subjected to molecular imprinting to investigate the impact on release kinetics and drug loading, when compared with non-imprinted, or alternately-imprinted hydrogels. Mild synthesis conditions were used to molecularly imprint three antibiotics, novobiocin, rifampicin, and vancomycin, and to test two different hydrogel chemistries. The release profile and drug loading of the molecularly imprinted hydrogels were characterized using ultraviolet spectroscopy over a period of 35 days and compared to non-imprinted, and alternately-imprinted hydrogels. While only modest differences were observed in the release rate the antibiotics tested, a substantial difference was observed in the total drug-loading amount possible for hydrogels releasing drugs which had been templated by those drugs. Hydrogels releasing drugs which had been templated by other drugs did not show improved release or loading. Analysis by FTIR did not show substantial incorporation of drug into the polymer. Lastly, bioactivity assays confirmed long-term stability and release of incorporated antibiotics.
Keywords: Drug Delivery, Molecular Imprinting, Infection, Affinity, Cyclodextrin
Introduction
Of all the risks and adverse outcomes that are associated with surgery and medical implantation, post-surgical infections pose the greatest threat to surgeons and their patients. Even in the most routine operations, the risk of infection persists. [1, 2] Current surgical techniques of combating infection include pre-soaking of grafts, gauze and other equipment with antibiotic, as well as irrigation with antibiotic-containing solutions. [3, 4] However, the efficacy of these methods is limited in that they have a very small therapeutic window because the antibiotic is washed out of the system rather quickly. Researchers have responded to this problem through the introduction of specially designed biomaterials and device coatings that deliver locally administered drugs through various polymers. [5]
Recently, promising results have been demonstrated with the development of an affinity-based cyclodextrin delivery platform which releases antibiotics at a therapeutically desired rate.[5–9] This affinity-based system utilizes the unique properties of β-cyclodextrin (CD), a cyclic oligomer with a relatively hydrophobic interior and a relatively hydrophilic exterior, to create a guest-host complex through various secondary molecular interactions including hydrogen bonding, van der Waals bonding, and hydrophobic interactions. This guest-host complex, delays the release of the guest antibiotic beyond that capable by diffusion alone. Previous experiments conducted by Thatiparti et al. have shown a release of antibiotics beyond 45 days. [6] In vivo studies using animal models have also shown the drug delivery hydrogel to effectively prevent Staphylococcus aureus mesh infections. [8]
This study aims to further enhance the loading and delivery of these supramolecular hydrogels through the use of molecular imprinting. Molecular imprinting is a technique used in hydrogel synthesis to create networks with specially designed template-shaped cavities that have an increased affinity for the specified target molecule. Along with secondary molecular interactions between cyclodextrin and its guest molecule, molecular imprinting allows an additional mechanism to control drug release. Molecularly imprinted materials are frequently used in analytical chemistry and pharmacology. [10] Some of the earliest reports of imprinted materials that provided a sustained therapeutic release mechanism were made by the Norell et al. [11] Here, Theophylline, a drug used in the treatment of asthma, was molecularly imprinted into a polymeric systems using a non-covalent technique using methacrylic acid (MAA) as the functional monomer and ethylene glycol dimethacrylate (EGDMA) as the cross-linker. Since then other groups have achieved similar success imprinting different drugs using various techniques into polymeric systems; however, only showing modest improvements in drug delivery. [12–14]
Interestingly, while Park et al. proposed the use of molecular imprinting of supramolecular hydrogels for drug delivery of small molecules in 2007, [15] there has been virtually no progress in that direction since then. Nevertheless, the field of molecular imprinting has continued to grow, but growth has primarily been in two areas: molecular detection/sensing, and separation. [16, 17] The separations field faces the same challenge as the drug delivery field did, in generating high affinities toward small molecules using only linear polymeric systems of low-affinity components, so investigations have continued using novel matrix components including binary and ternary deep eutectic solvents, [18] ionic liquids, [19] and zwitterionic polymers. [20] The growth that has occurred in biomedical materials has been through templating proteins. Due to the larger size, and higher molecular complexity of proteins vs. small molecule drugs, stronger affinities are possible, and significant advances have been made. These have included generating templates for receptor mimics, [21] fluorescent sensors, [22] pseudoimmunoassays, [23] ligands for targeted drug delivery, [24] protein therapeutics, [25] and others. [26]
A technique to attain higher affinities to small molecule templates has been to use a matrix component with some inherently higher affinity potential, like cyclodextrins. Previous studies by Asanuma et al. involving the imprinting of cyclodextrin concluded that the imprinting promoted the binding activity toward the template compared with non-imprinted polymer. [27] It was noted that the molecular imprinted cyclodextrin also provided enantiomer selectivity for the host molecule. Similar studies also showed a preferential formation of ordered cyclodextrin have shown similar success utilizing cyclodextrin-based molecular imprinted polymers. [28–33] One limit of such systems is that they utilize linear polymers of cyclodextrin with limited chain mobility and imprinting capacity. It is theorized that a supramolecular hydrogel, such as we study here, would have infinite molecular mobility and provide a more thorough imprinting.
Previous modeling studies operated on an anticipated 1:1 binding of CD with the antibiotics in this study.[5–9] If this is the case, then it is predicted that molecular imprinting will not substantially improve affinity, but may alter other properties, such as total drug loading. However considering these molecules are larger than the cavities within CD (in the case of vancomycin, it is much more so), it is possible they form higher order (e.g. 2:1; 3:1, 4:1) complexes, in which case affinity, and therefore release rate could be substantially altered (Figure 1). In this study we molecularly imprinted three different antibiotics: vancomycin, rifampicin, and novobiocin, into cyclodextrin-based hydrogels, and examined the impact on: drug loading; release rate; and antimicrobial bioactivity.
Figure 1.
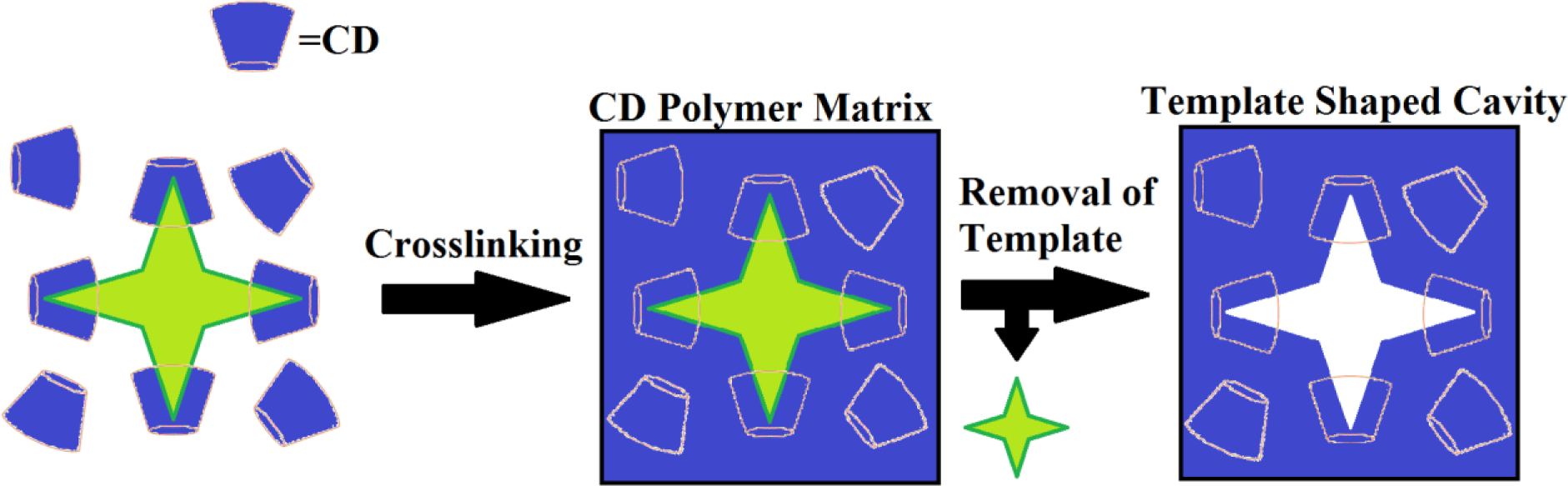
Basic molecular imprinting scheme for higher order complexes, demonstrating how templating could result in geometrically higher affinities, than a linear polymer could.
Methods and Materials
Materials
β-cyclodextrin macromonomer (CD lightly crosslinked with epichlorohydrin with final MW =2–15 kDa) was purchased from CTD, Inc. (High Springs, FL), and was dried under vacuum at 100 °C for 24 h and stored in a desiccator before use. Ethylene glycol diglycidyl ether (EGDE) and 1,6- diisocyanatohexane (HDI) (Aldrich, St. Louis, MO) were used as received. Rifampicin (rifampin) (RM) was purchased from Fisher Scientific. Novobiocin (NB) sodium salt and vancomycin (VM) hydrochloride was purchased from MP Biomedicals, Inc. (Solon, OH). N,N-Dimethylformamide (DMF) was obtained from Applied Biosystems (Foster City, CA, USA) and was dried over calcium hydride and distilled in high vacuum. All other reagents were purchased from Fisher Scientific and used as received.
Supramolecular Hydrogel Synthesis of Novobiocin/Rifampicin Molecularly Imprinted CD-HDI Networks
Molecularly imprinted β-Cyclodextrin hydrogel disks were prepared with lightly epichlorohydrin-crosslinked macromonomer having molecular weights between 2,000–15,000. Before synthesis could begin, the β-CD macromonomer was dried in a vacuum oven at 90°C for 2 hours. 1 g of β-CD was then dissolved in 3mL of dimethylformamide (DMF) along with various concentrations of either novobiocin (NB) or rifampicin (RM) at 3, 6, 9, and 12 weight % based on CD. Once the imprinted supramolecular hydrogel formed around the drugs, the structure was locked in by adding 288 μL of hexamethylene diisocyanate (HDI) crosslinker (1:0.32 molar ratio). The mixture was cured in a 55mm dish and the hydrogels at room temperature for five days. Non-molecularly imprinted β-CD hydrogel disks were made using the same procedure without adding drug (No MI). Depending on the amount of drug used in templating, the final hydrogels were named: No MI, NB 3% MI, NB 6% MI, NB 9% MI; NB 12% MI, RM 3% MI, RM 6% MI, RM 9% MI or RM 12% MI. Once cured the hydrogels were then punched into 8mm disks and thoroughly washed of drug and leftover reactants using DMF/Water for a period of 3 weeks. UV spectroscopy was done periodically on aliquots of the wash solution in order to confirm the complete removal of drug. Once it was confirmed that the drug was completely removed, the disks were then dried for 24hr. The result was an insoluble network hydrogel with a chemistry described in Figure 2. Final hydrogels were named according to the scheme in Table 1.
Figure 2.
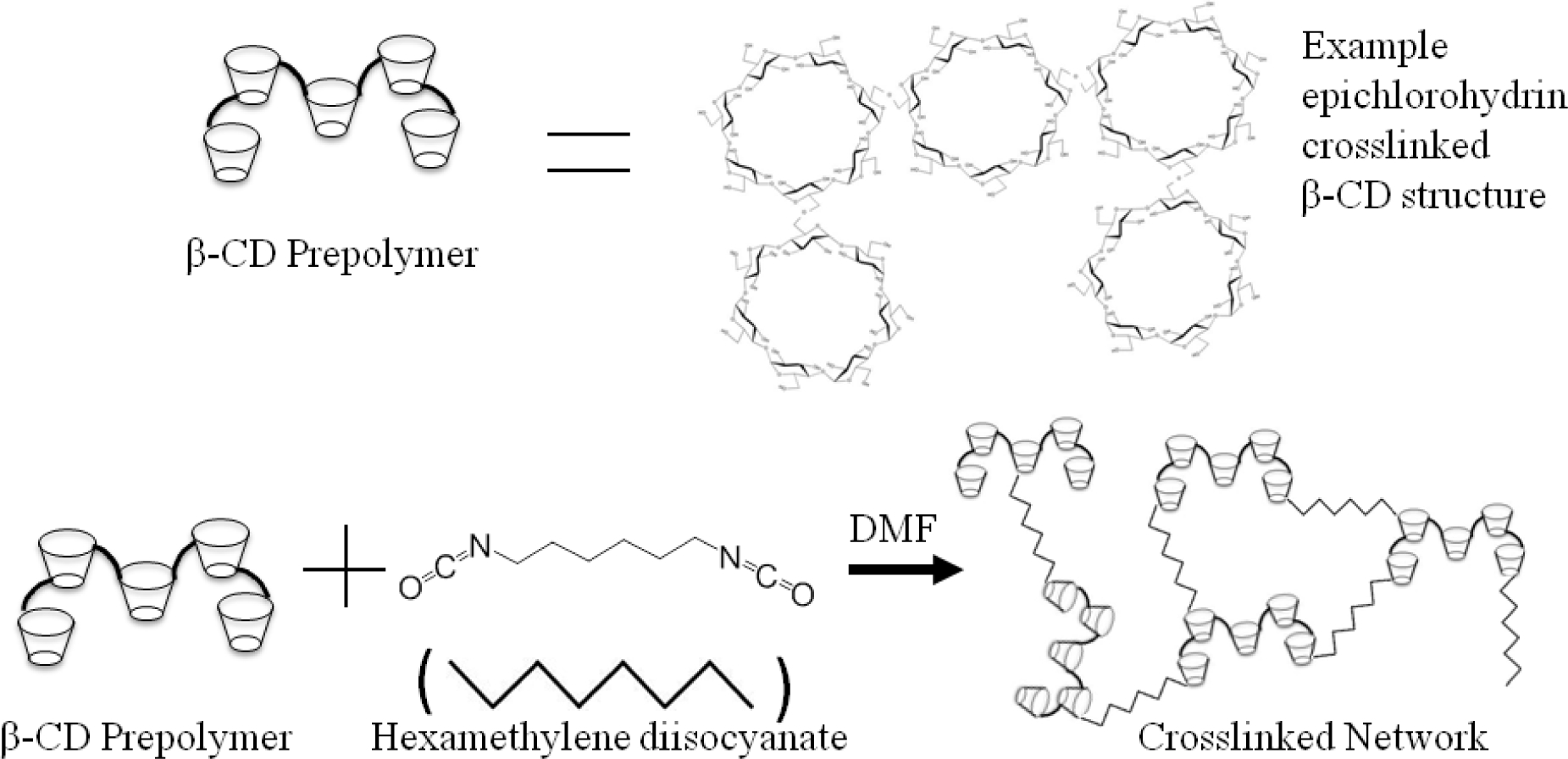
Chemical Synthesis of CD-HDI Networks
Table 1:
Hydrogel Naming Scheme for Imprinted Hydrogels
| Amount of Imprinted Drug | None | 3% | 6% | 9% | 12% | Hydrogel |
|---|---|---|---|---|---|---|
|
| ||||||
| Novobiocin (NB) | No MI* | NB3%MI | NB6%MI | NB9%MI | NB12%MI | CD-HDI |
| Rifampicin (RM) | No MI* | RM3%MI | RM6%MI | RM9%MI | NB12%MI | CD-HDI |
| Vancomycin (VM) | No MI** | VM3%MI | VM6%MI | VM9%MI | VM12%MI | CD-EGDE |
|
| ||||||
| Total Hydrogels | 2 | 3 | 3 | 3 | 3 | Two types |
The No MI controls for NB and RM templated hydrogels were made with CD-HDI
the No MI control for VM templated hydrogels was made with CD-EGDE.
Synthesis of Vancomycin Molecularly Imprinted CD Supramolecular Hydrogel Networks
Due to the poor solubility of vancomycin in DMF, and reactivity of HDI in water, it was not feasible to create a CD-HDI network for the vancomycin-templated materials. Instead, a CD hydrogel with different crosslinker (ethylene glycol diglycidyl ether, EGDE) chemistry was used. Similarly to the CD-HDI hydrogels, molecularly imprinted β-Cyclodextrin-EGDE hydrogels were prepared with lightly epichlorohydrin-crosslinked macromonomer, and dried in a vacuum oven at 90°C for 2 hours. 1 g of β-CD was then dissolved in 3mL of 0.2M potassium hydroxide along with various amounts of specified vancomycin (3, 6, 9, 12 weight % based on CD). 800 μL of ethylene glycol diglycidyl ether (EGDE) crosslinker was then added. The mixture was then added to a 55mm dish and the hydrogels were then cured at room temperature for five days. Non-molecularly imprinted β-CD disks were made using the same procedure without adding drug. Once cured the hydrogels were then punched into 8mm disks and thoroughly washed of drug and leftover reactants using DI water for a period lasting over three weeks. UV spectroscopy was done periodically on aliquots of the wash solution in order to confirm the complete removal of drug. Once it was confirmed that the drug was completely removed, the disks were then dried for 24hr. The result was an insoluble supramolecular network hydrogel with a chemistry described in Figure 3. Final hydrogels were named according to the scheme in Table 1.
Figure 3.
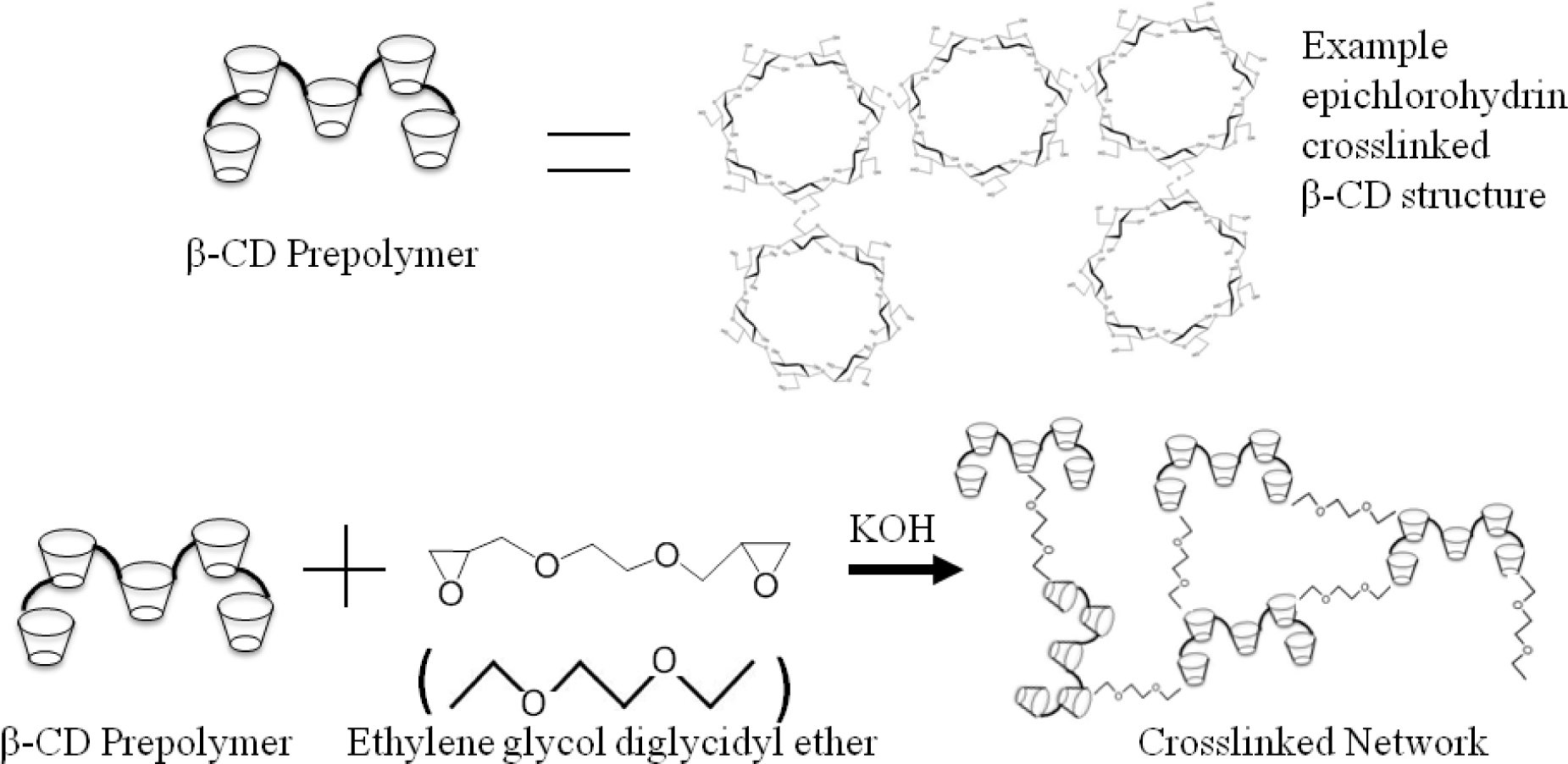
Chemical Synthesis of CD-EGDE Networks
Drug Loading
The 8mm punched disks of the different CD hydrogels were pre-weighed, then hydrodynamically loaded with their respective template drugs (e.g. RM loading into RM9%MI), or one of the alternate drugs not used in that templating (e.g. VM loading into RM9%MI). This was done by placing the dried disks into separate 20cc sample vials containing the different drugs (rifampicin, novobiocin, or vancomycin) at the same concentration (5% w/v). The disks were loaded in these vials for 72 hr at room temperature, and pH 7.4. The disks were then removed from the loading solution and air dried.
Drug Release Studies
Release of drug from the hydrogel disks was determined by UV/VIS spectroscopy. Periodic aliquots of solution were analyzed over a period of 35 days in order to capture and characterize the release profile. First, the λmax, the wavelength of maximum absorbance, was determined for novobiocin, rifampicin, and vancomycin as 306nm, 473nm, 282nm respectively. These results were consistent with what was reported in previous literature. [6, 7] Once the λmax was determined, a standard curve with increasing known amounts of drug (0–50μg/ml) was generated. Using linear regression, a linear equation was generated to determine the amount of drug in an unknown sample and calibration curves were made.
For the release studies three 8mm diameter disks for each condition were weighed, and then placed in 20cc sample vials. 10 mL of phosphate buffer saline (1X PBS) was then added separately to each vial and the vials were placed in an incubator at 37°C with mild agitation. 500μL aliquots were then taken periodically, replaced with fresh PBS, and the drug concentration was determined in the sample using spectroscopy. Once it was determined that no more drug was being released (~35 days), statistical analysis was performed to determine the amount of drug released from the hydrogels as a function of time. Similarly the percent loading was determined by comparing the total amount of drug released to the initial weight of the dry drug loaded disks. This relationship can be described mathematically through a simple equation, with all samples performed in triplicate:
Fourier-Transform Infrared (FTIR) Spectroscopic Analysis
Infrared (IR) spectroscopy was carried out using an Excalibur FTS 3000 Fourier-Transform Infrared (FTIR) Spectrophotometer (BioRad, Hercules, CA, USA). The samples were prepared by grinding dry samples before and after drug loading, and after extensive drug release and non-polar solvent washing (~2 mg) with potassium bromide (KBr) powder (~100 mg) and then compressing the mixtures to form scanning samples. Scans were run from 4000 to 600cm−1.
The drug-templated and non-templated hydrogels were characterized through FTIR which is shown in Figure 4(a–d). The following important peaks can be seen: for CD, 3600–3000 (ν(O-H), 2928cm−1 (ν(C-H); while for the crosslinks, 3600–3000 (ν(O-H), 2928cm−1 (ν(C-H), 1701 (ν(C=O) and 1541cm-1(δ(N-H). The existence of 1701 (ν(C=O) and 1541cm-1(δ(N-H) peaks demonstrates crosslinking by LTI, and formation of the urethane bond. The indication of the formation of rifampin:CD complex (CD-LTI(1:1)-RM) is shown in Figure 2b, where it can be clearly seen that the FTIR spectrum for the complex is almost identical to that of CD alone (Figure 2c). On the other hand, aromatic region peaks of RM at around 700–800cm−1 have disappeared, which suggests low background RM. [34–36]
Figure 4.
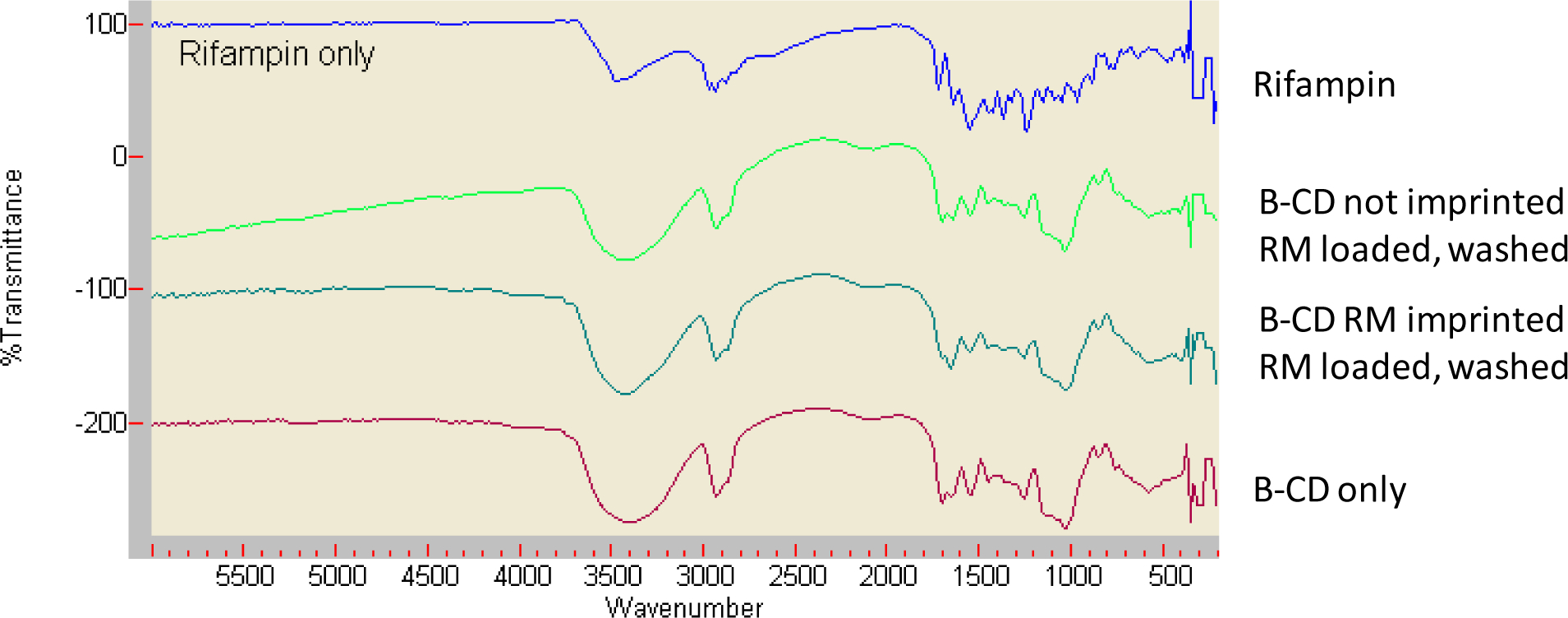
FTIR spectra of (a) Pure rifampicin (RM), (b) CD polymer made without imprinting, then loaded with RM, then subjected to extensive non-polar solvent washing and drug release, (c) CD polymer made from RM imprinting, loaded with RM, and subjected to extensive non-polar solvent washing and drug release, (d) Pure CD polymer. The existence of 1701 and 1541cm−1(δ(N-H) peaks, indicates the CD polymer urethane bond. Unique RM peaks (e.g. aromatic region peaks of rifampin at around 700–800cm−1) demonstrate low background presence of RM.
Kirby Bauer Disk Diffusion Susceptibility Test
To ensure that the bactericidal activity of the molecularly imprinted hydrogels was not compromised a Kirby-Bauer disk diffusion susceptibility test was performed. The purpose of the Kirby-Bauer assay is to determine the sensitivity or resistance of pathogenic bacteria to various antimicrobials and antimicrobial releasing compounds. The disk diffusion method of Kirby and Bauer is considered a viable alternative to broth dilution methods.
In this particular study, trypticase agar plates were made and were seeded with, Staphylococcus aureus as it is one of the most common causes of medical device failure and patient infections. (S. aureus (Seattle 1945 strain) kindly provided by Dr. Ed Greenfield, Case Western Reserve University). Once the bacteria was seeded, three molecularly imprinted disks, (NB9%MI, RM9%MI, and VM9%MI loaded with their original templating antibiotics) were placed on the plate and were allowed to incubate for 24 hours. After incubation, the zones of inhibition were measured using calipers and recorded. The disks were then placed on a fresh new agar plate seeded with bacteria. This procedure was repeated daily for a period of 52 days and was run in triplicate.
Statistical Analysis
All data were processed using Microsoft Excel 2003 software and the results were produced as mean/standard deviation of at least three experiments. Statistical analyses were performed using one-way ANOVA with Minitab (Minitab Inc. State College PA, USA). A p-value smaller than 0.05 was considered statistically significant.
Results:
Supramolecular Hydrogel Synthesis
Both imprinted and non-imprinted supramolecular hydrogels were successfully synthesized. The No-MI hydrogels of each type served as the non-imprinted controls for their hydrogel type in this study. Varying concentrations of well-mixed, imprinted drug (3, 6, 9, 12 weight % based on CD) were made in order to evaluate the impact of the amount of molecular imprinting. Higher concentrations of imprinting was thought to correlate with more template shaped cavities, with a limit established at ~10%, to avoid formation of larger sized voids where drug was too close to other drug. Previous experiments have utilized heating in order to initiate crosslinking/molecular stabilization. [6, 11] However, in this case both MI hydrogels and No MI controls were allowed to cure at room temperature in order to avoid denaturing, and to preserve the chemical structure of the antibiotics (and therefore the templates). It is important to note that in spite of extensive washing in various solvents to remove drug, a small amount of drug remained in the samples, as evidenced in the rifampicin hydrogels by a slight color change in both the templated and the later drug-loaded hydrogels. However this amount is similar to the amount of RM remaining in non-templated polymers as determined by colorimetric spectroscopy (data not shown) and FTIR analysis (Figure 4). Past work by our lab has shown that that any drug remaining in the device was unavailable for further delivery even after extensive washing with organic solvents, so total drug loading was calculated from the maximum released amount, rather than maximum loaded amount. [37, 38]
Fourier-Transform Infrared (FTIR) Results
The drug-templated and non-templated hydrogels were characterized through FTIR which is shown in Figure 4(a–d). The following important peaks can be seen: for CD, 3600–3000 (ν(O-H), 2928cm−1 (ν(C-H); while for the crosslinks, 3600–3000 (ν(O-H), 2928cm−1 (ν(C-H), 1701 (ν(C=O) and 1541cm-1(δ(N-H). The existence of 1701 (ν(C=O) and 1541cm-1(δ(N-H) peaks demonstrates crosslinking by LTI, and formation of the urethane bond. Fully delivered and washed samples were almost identical to that of CD alone, regardless as to whether the polymer was made through imprinting or not imprinting. Specifically, aromatic region peaks of RM at around 700–800cm−1 have disappeared, which suggests low background RM. [34–36]
Drug Release and Drug Loading Studies
Each of the three drugs was loaded into hydrogels templated with that drug as well as their corresponding No MI control. The moderately hydrophobic, small-molecule drug novobiocin, has been previously shown to show a sustained delivery over 30+ days from non-imprinted hydrogels. [27] Release rates of novobiocin from imprinted hydrogels was shown to follow a similar profile, with the only difference being a reduction of the initial burst, but reaching 100% delivery at approximately the same time point (~30 days) (Figure 5). The reduction of initial burst scaled with increasing amount of imprint molecule, where within 120 hrs, 80±1.2% of novobiocin was released from the NB12%MI hydrogels, but 90±1.7% from the No MI hydrogels. In addition to the slight changes in release rate, there were more significant changes in total loading, where after 72 hours loading, No MI hydrogels loaded with 10±0.2% vancomycin, while NB12%MI hydrogels loaded with over twice the normal maximum, at 20±0.6% drug.
Figure 5. Cumulative Release and Total Loading of Novobiocin from Novobiocin- Templated Hydrogels.
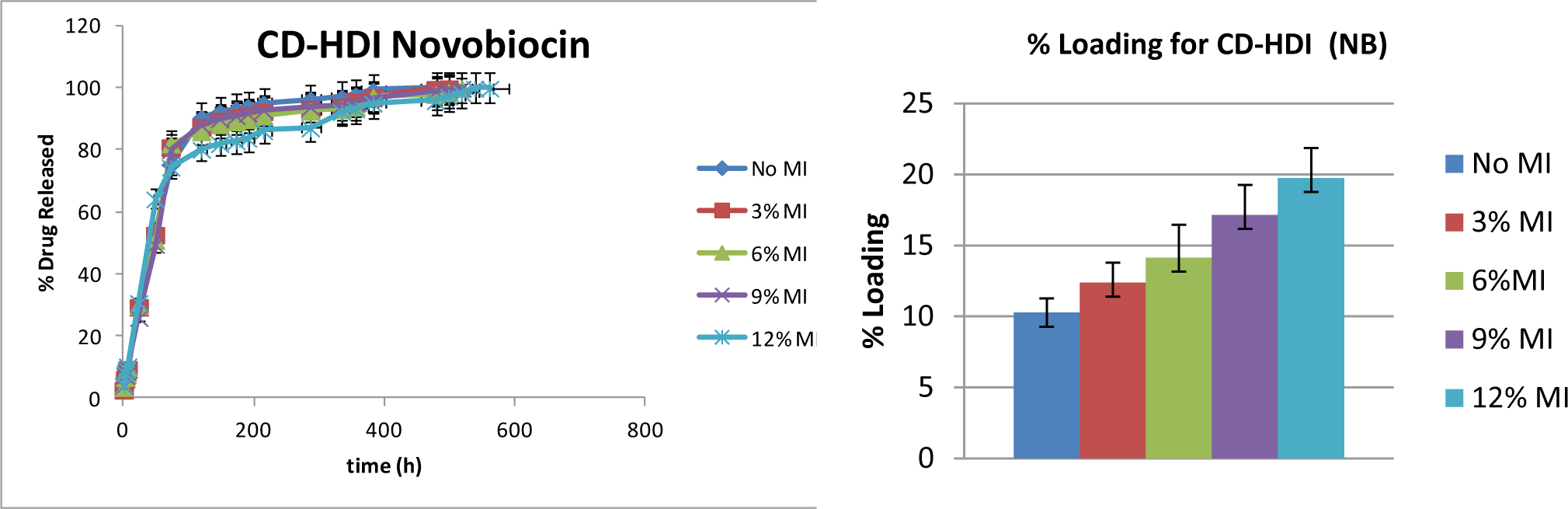
The panel on the left shows overall release as a function of time for the various novobiocin-loaded hydrogels. The panel on the right shows percent loading for each MI hydrogel, where higher MI % corresponds to higher loading (p< 0.05). Samples are in triplicate. Error bars show ± standard deviation.
The change to the highly hydrophilic, large molecule vancomycin was believed to have a higher potential for higher order (e.g. 2:1, 3:1 or 4:1) complex formation with cyclodextrin due to the large amount of the molecule not residing within one CD complex; and therefore show the highest change in affinity due to molecular imprinting. Previous work using similar hydrogels show a sustained delivery over 10+ days from non-imprinted hydrogels. [27] Release rates of vancomycin from imprinted hydrogels was shown to follow a similar profile, with the only difference being a reduction of the initial burst, but reaching 100% delivery at approximately the same time point (~10 days) (Figure 6). The reduction of initial burst was noticeable earlier than that of novobiocin, and again scaled with increasing amount of imprint molecule, where within the first 24 hrs, 56±0.7% of vancomycin was released from the VM12%MI hydrogels, but 75±1.3% from the No MI hydrogels. In addition to these changes in release rate, there were more significant changes in total loading, where after 72 hours loading, No MI hydrogels loaded with 17±0.3% vancomycin, while VM12%MI hydrogels loaded with 22±0.5% drug.
Figure 6. Cumulative Release and Total Loading of Vancomycin from Vancomycin-Templated Hydrogels.
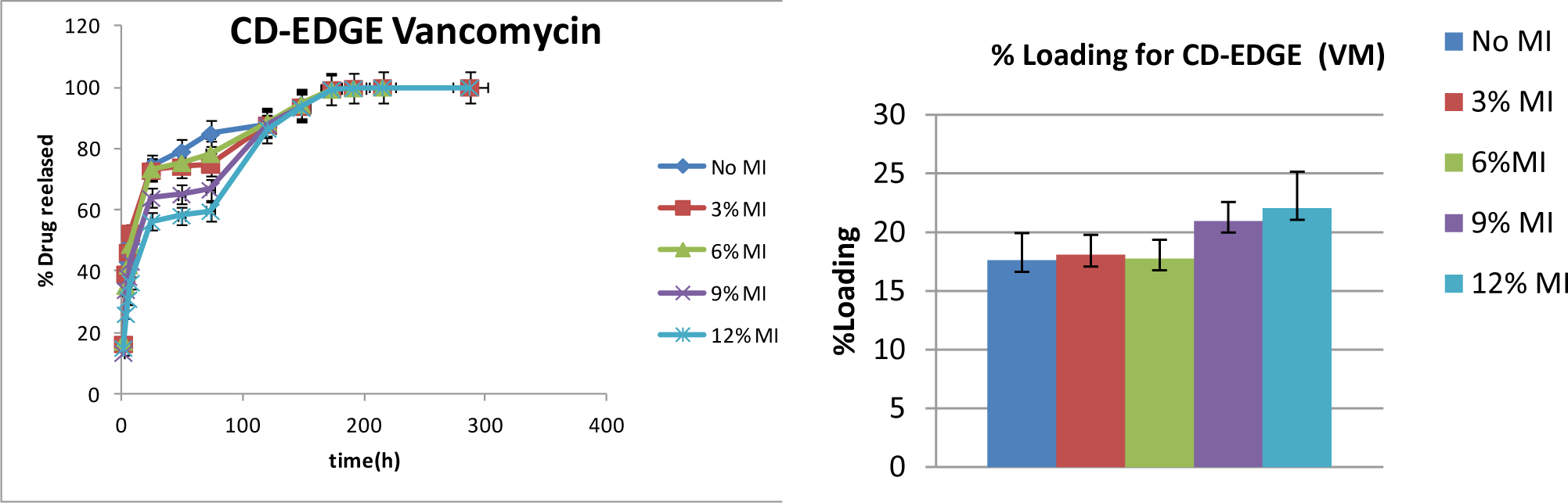
The panel on the left shows overall release as a function of time for the various vancomycin-loaded hydrogels. The panel on the right shows percent loading for each MI hydrogel, where higher MI % corresponds to higher loading (p< 0.05). Samples are in triplicate. Error bars show ± standard deviation.
Lastly, in the change to the more hydrophobic drug, rifampicin, it was unclear how much the release rate would be dependent on affinity, and how much on partitioning due to its lower solubility. Therefore it was not clear that release rate would be impacted by molecular imprinting, unless through formation of possible higher order complexes. Previous work using similar hydrogels show a sustained rifampicin delivery over 30+ days from non-imprinted hydrogels. [27] Release rates of rifampicin from imprinted hydrogels was shown to follow a similar profile, with the only difference being small reduction of the initial burst, but reaching 100% delivery at approximately the same time point as non-imprinted hydrogels (Figure 7). While the reduction of initial burst was noticeable within the first 24 hrs, it was found to be statistically insignificant (49±0.6% of rifampicin was released from the RM12%MI hydrogels, but 50±0.4% from the No MI hydrogels). While changes in release rate of rifampicin were less significant; as with the other drugs, there were still significant changes in total loading, where after 72 hours loading, No MI hydrogels loaded with 7±0.06% rifampicin, while RM12%MI hydrogels loaded with 10±0.07% drug.
Figure 7. Cumulative Release and Total Loading of Rifampicin from Rifampicin-Templated Hydrogels.
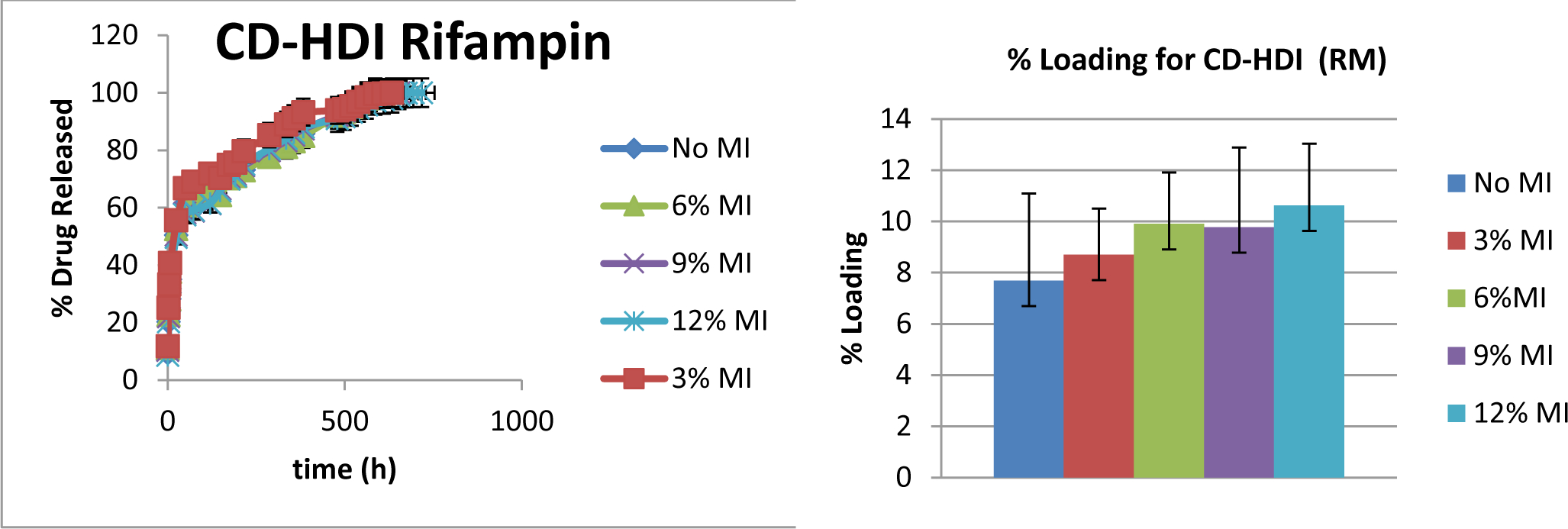
The panel on the left shows overall release as a function of time for the various rifampicin-loaded hydrogels. The panel on the right shows percent loading for each MI hydrogel, where higher MI % corresponds to higher loading (p< 0.05). Samples are in triplicate. Error bars show ± standard deviation.
Release Studies from Hydrogels Templated with Alternate Drugs
In addition to release from hydrogels templated with that same drug, we evaluated release from hydrogels templated with an alternate drug. A second set of release studies was run in which each set of molecularly imprinted hydrogels was loaded with the other two drugs used in this study. This was to determine if the molecular imprinting was specific to that chemistry (in which case hydrogels templated with alternate drugs should show properties similar to non-templated hydrogels), or if hydrogels simply incorporated non-specific voids (in which case release/loading properties could be the same regardless of the templating drug). For simplicity, in this particular study only the 9% MI hydrogels were tested. The resulting release profiles for the various molecularly imprinted disks can be seen in Figure 8, while changes in total drug-loading shown in Figure 9.
Figure 8. Release of Drugs from Alternately Templated Hydrogels.
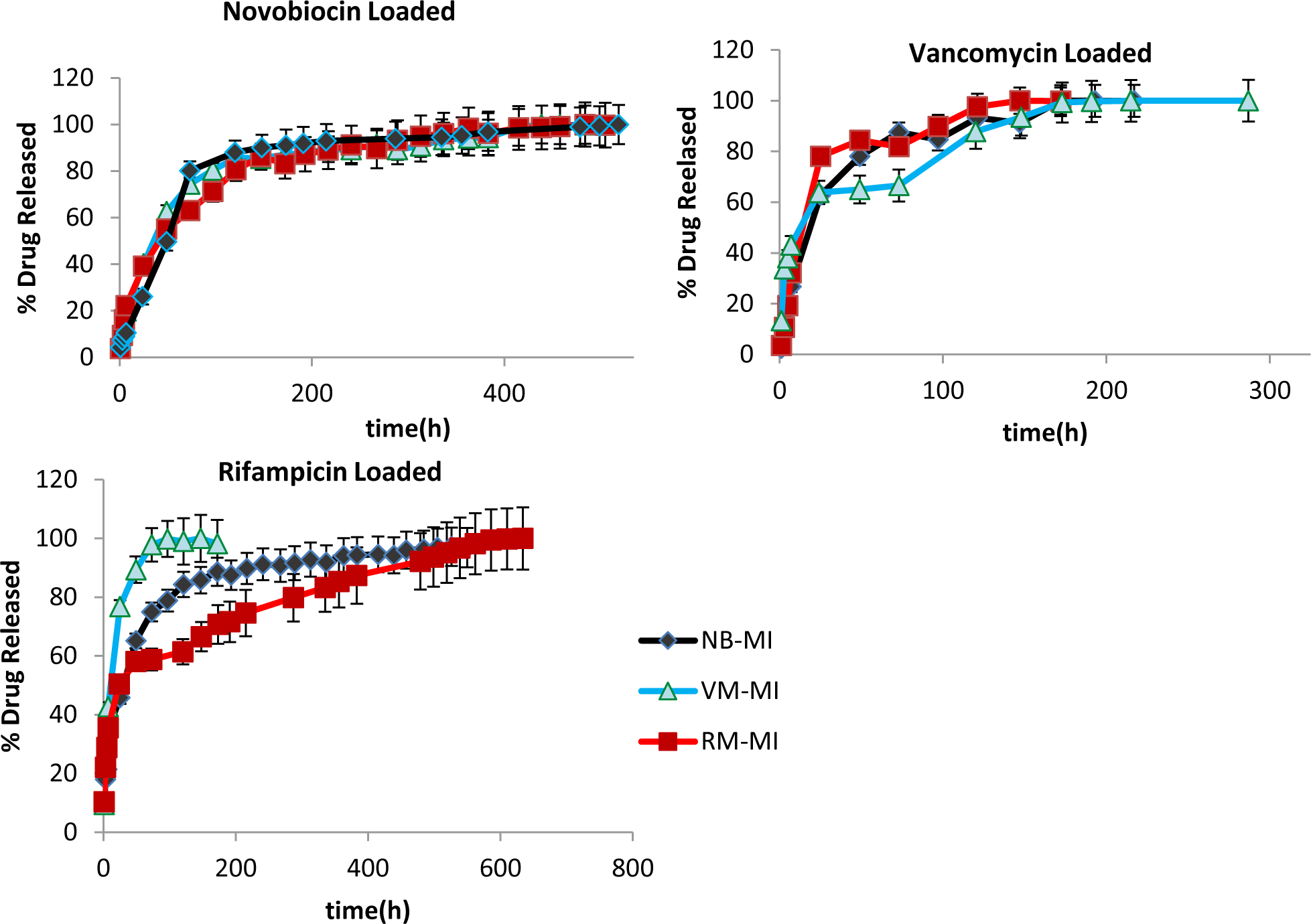
Shown above are the overall release profiles over time for the various drug loaded hydrogels. Novobiocin (top left) showed the least amount of change, where release was statistically comparable, regardless of templating molecule. Vancomycin (top right) showed some change, where release from vancomycin-templated hydrogels was statistically different, particularly in early time points. However release from rifampicin was significantly different depending on whether the hydrogel was templated with the large, hydrophilic VM; the small, moderately hydrophobic NB; or with RM itself (p<0.05). All the hydrogels used were initially imprinted with 9 wt% drug (9% MI). Samples are in triplicate. Error bars show ± standard deviation.
Figure 9. Total Drug-Loading in Alternately Templated Hydrogels.
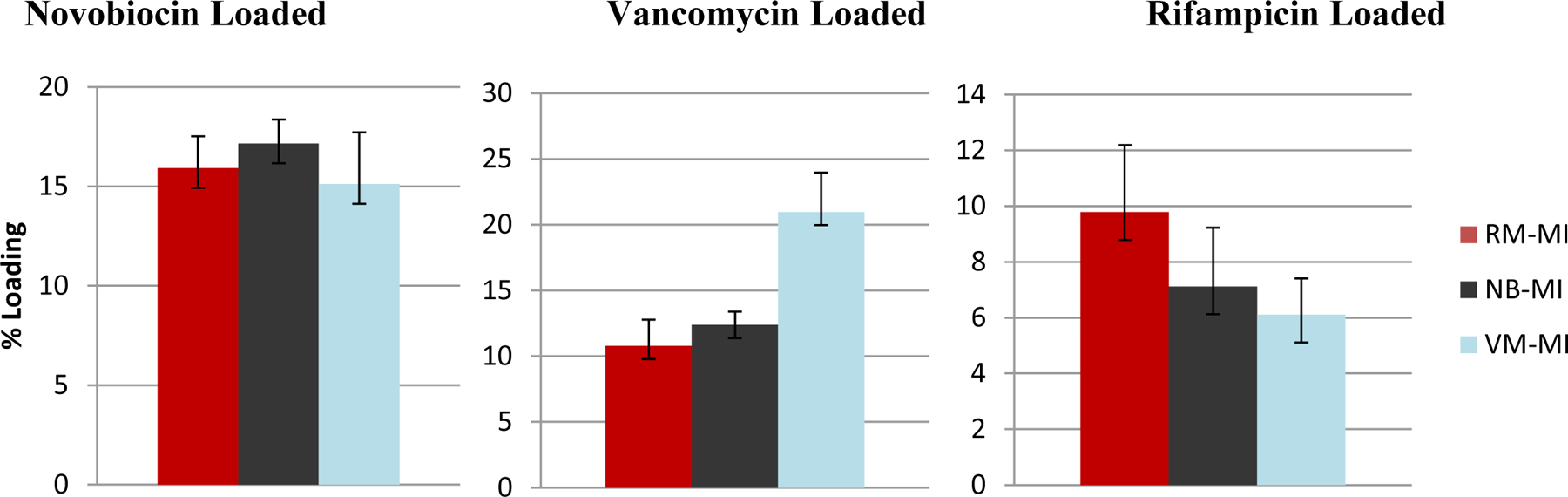
Shown above are the total drug loading amounts over 72hrs for the various drug-loaded hydrogels. All three drug: novobiocin (top left), vancomycin (top right) and rifampicin (bottom) showed total drug-loading that was statistically significantly different depending on whether the hydrogel was templated with VM; NB; or RM (p<0.05) All the hydrogels used were initially imprinted with 9 wt% drug (9% MI). Samples are in triplicate. Error bars show ± standard deviation.
In the case of the novobiocin-loaded hydrogels, there was no significant change in the overall release profiles, regardless of the templated drug used in making the hydrogel (NB, VM or RM templated). In the case of the large, hydrophilic drug, vancomycin, changes in release rates were seen both in early release, as well as time at completion: (within 7 hrs, 32±0.3% of vancomycin from the RM-templated hydrogels was released while 19±0.1% of vancomycin was released from the VM-templated hydrogels), and release of vancomycin was essentially complete within 121hrs for RM-templated hydrogels, but not complete until about 173 hrs for VM-templated hydrogels.
However, the greatest change was seen in the case of the more hydrophobic rifampicin loaded hydrogels, a much more significant change can be seen both initially and ultimately, depending on the templating drug. Within 24 hrs, 76±0.8% of rifampicin from the VM-templated hydrogels was released while 50±0.4% of rifampicin was released from the RM-templated hydrogels; and release of rifampicin was essentially complete within 73hrs for VM-templated hydrogels, ~500 hrs for NB-templated hydrogels, but not complete until >600 hrs for RM-templated hydrogels.
In addition to the release studies, the total drug-loading was calculated for each set of imprinted hydrogels as well and can be seen in Figure 8.
While the impact varied, in all three cases, the template drug had the highest loading capacity in its respective template imprinted hydrogel. Novobiocin showed the least change, with 17% loading in NB-templated hydrogels; 15.9% in RM-templated hydrogels, and 15.1% in VM-templated hydrogels. Rifampin showed a greater change with 10% loading in RM-templated hydrogels, 7.1% loading in NB-templated hydrogels, and 6.1% in VM-templated hydrogels. Vancomycin showed the greatest change with 21% loading in VM-templated hydrogels, 12% loading in NB-templated hydrogels, and 10% loading in RM-templated hydrogels.
Bioactivity Assay
A final test was performed to ensure that drug stability, and therefore antimicrobial activity was maintained over the course of the study, and not impacted by molecularly imprinted hydrogels. A Kirby-Bauer disk diffusion susceptibility test was performed. The results of this study can be seen in Figure 10 below. Previous studies from similar, non-templated hydrogels showed bioactivity out to 45 days. [27] In all cases the antimicrobial activity of the loaded drug was preserved, and lasted longer than previously reported. In this study the samples were followed out to 52 days. As predicted the high water solubility of vancomycin led to a more rapid release of molecule, and faster drop in antimicrobial activity of the drug-loaded disk, being fully depleted by day 24. By day 52, the rifampicin and novobiocin loaded imprinted hydrogel (using a 9% MI), still showed cleared zones of 4.3mm and 1.8mm respectively.
Figure 10. Kirby-Bauer Disk Diffusion Susceptibility Test of Templated Hydrogels.
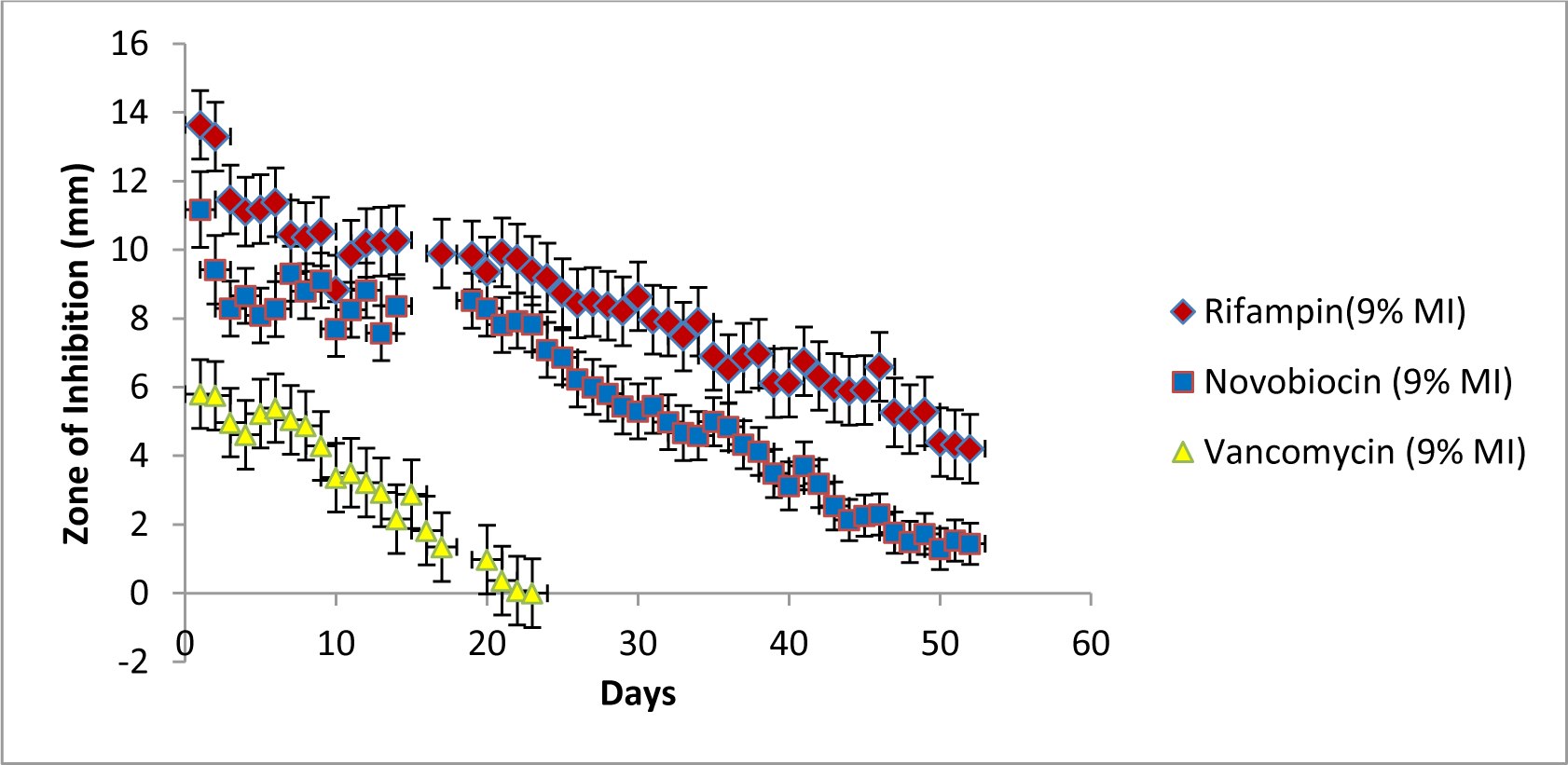
Shown above are the total drug loading amounts over 72hrs for the various drug-loaded hydrogels. All three drug: novobiocin (top left), vancomycin (top right) and rifampicin (bottom) showed total drug-loading that was statistically significantly different depending on whether the hydrogel was templated with VM; NB; or RM (p<0.05) All the hydrogels used were initially imprinted with 9 wt% drug (9% MI).
Discussion
In this work, molecularly imprinted and non- imprinted CD-based supramolecular hydrogels were successfully synthesized. Crosslinking to lock in molecular structure was performed at room temperature in order to avoid denaturing the antibiotic and therefore the template. Past work delivering these, and similar drugs had assumed a 1:1 CD:drug complex. If a higher order complex could form (e.g. 2:1, 3:1, 4:1) this could lead to a geometrically increasing affinity. Ultimately, higher affinities and higher order complexes were not readily apparent as there were not substantial changes in drug delivery rate from hydrogels templated with differing amounts of the same drug. This could be due to a combination of factors. Higher order complexes are dependent on two variables, distance between affinity groups, and strength of affinity groups. For the large drug, vancomycin, it is possible that chemical group with the highest affinity was already complexed with the first available CD, and that all remaining chemical groups only had very low affinities to other CDs. On the other hand the smaller drugs, novobiocin and rifampin may have not been large enough to span between two CD pockets. The only major change seen in delivery rates was when rifampicin was loaded into hydrogels templated with drugs other than rifampicin, where the biggest change was seen when the small, hydrophobic rifampicin was loaded into hydrogels templated with the large, hydrophilic drug vancomycin.
Nevertheless, in spite of seeing only modest changes in most release rates, substantial changes in total drug-loading were detected in all conditions. This was both dependent on the amount of templating molecule (right panels in Figures 4–6), as well as in the chemical nature of the templating molecule (Figure 8). Further, these results were consistent across hydrogel chemistries from CD hydrogels crosslinked with the diisocyanate, HDI, to a hydrogel crosslinked with the diglycidyl ether, EGDE. So while a drug might not be able to span from CD pocket to CD pocket, other parts of the CD molecule are able to participate in forming the shape/chemistry of the drug template.
In regards to templating, it was noticed (e.g. by residual color change in the rifampicin templated hydrogels) that some drug was either statistically unable to come out of the devices in the timecourse or solvent conditions of the experiment; or possibly were chemically bound to the hydrogel during the crosslinking reactions of the templating process. Along with the color change, it was noted that the imprinted hydrogels appeared to be slightly softer and more fragile, than non-imprinted hydrogels, as they were being punched into 8mm disks (data not shown). This suggests that some of the hydrogel crosslinking was disrupted, perhaps due to the crosslinkers attacking the secondary hydroxyl groups of the antibiotic as well as the many primary and secondary hydroxyls of the CD compromising the hydrogel network. While cyclodextrin polymers formed with the same crosslinkers (in the absence of antibiotics) have been well-characterized, [6, 7] while we have shown negligible incorporation of RM in the molecular structure of the polymer (Figure 4), future work could involve further characterization (e.g. atomic analysis, NMR) of the materials to see if there were other differences between templated and non-templated hydrogels to confirm presence of these new chemistries. However the latter will be challenging due to the predicted low number of drug molecules (<0.001%) involved.
Overall these preliminary results show that the higher versatility of molecular imprinting using supramolecular hydrogels has displayed a positive effect in improving an already favorable drug delivery system. The results showed a preferential loading for the template drug as evidenced by the increase in % loading without negatively altering the therapeutic release window. The Kirby-Bauer assay further showed that overall bioactivity was not compromised. Future work will also focus on using imprinting in order to try to deliver larger molecules, such as proteins, which may form higher order complexes, and that would not normally have been delivered with the non-imprinted CD hydrogel system.
Acknowledgements
The authors would like to thank Jeffrey Halpern PhD, and Andrew Fu PhD for their assistance in interpreting the release profiles, as well as Catherine Gormley for assistance in writing and editing. The authors would also like to thank the Support of Undergraduate Research and Creative Endeavors (SoURCE) at Case Western Reserve University, an NIH Research Facilities Construction Grant (C06 RR12463-01), as well as and from T32DK083251-09 for support (NAR).
References
- [1].Engelsman AF, van der Mei HC, Ploeg RJ, Busscher HJ, Biomaterials 2007, 28, 2314. [DOI] [PubMed] [Google Scholar]
- [2].Garibaldi RA, Cushing D, Lerer T, The American Journal of Medicine 1991, 91(3B), 158S. [DOI] [PubMed] [Google Scholar]
- [3].Vertullo CJ, Quick M, Jones A, Grayson JE, Arthroscopy : the journal of arthroscopic & related surgery : official publication of the Arthroscopy Association of North America and the International Arthroscopy Association 2012, 28, 337. [DOI] [PubMed] [Google Scholar]
- [4].Falagas ME, Vergidis PI, Clinical Microbiology and Infection 2005, 11, 862. [DOI] [PubMed] [Google Scholar]
- [5].Wang NX, von Recum HA, Macromolecular bioscience 2011, 11, 321. [DOI] [PubMed] [Google Scholar]
- [6].Thatiparti TR, Shoffstall AJ, von Recum HA, Biomaterials 2010, 31, 2335. [DOI] [PubMed] [Google Scholar]
- [7].Thatiparti TR, von Recum HA, Macromolecular bioscience 2010, 10, 82. [DOI] [PubMed] [Google Scholar]
- [8].Harth KC, Rosen MJ, Thatiparti TR, Jacobs MR, Halaweish I, Bajaksouzian S, Furlan J, von Recum HA, The Journal of surgical research 2010, 163, 337. [DOI] [PubMed] [Google Scholar]
- [9].Fu AS, Thatiparti TR, Saidel GM, von Recum HA, Annals of biomedical engineering 2011, 39, 2466. [DOI] [PubMed] [Google Scholar]
- [10].Cunliffe D, Kirby A, Alexander C, Advanced drug delivery reviews 2005, 57, 1836. [DOI] [PubMed] [Google Scholar]
- [11].Norell MC, Andersson HS, 1998. [DOI] [PubMed]
- [12].Demirel M, Sevin SB, Say R, Yazan Y, Journal of Pharmaceutical Sciences 2009, 32, 147. [Google Scholar]
- [13].Alvarez-Lorenzo C, Hiratani H, Gomez-Amoza JL, Martinez-Pacheco R, Souto C, Concheiro A, Journal of Pharmaceutical Sciences 2002, 91, 2182. [DOI] [PubMed] [Google Scholar]
- [14].Hiratani H, Alvarez-Lorenzo C, Journal of Controlled Release 2002, 83, 223. [DOI] [PubMed] [Google Scholar]
- [15].Chaterji S, Kwon IK, Park K, Progress in polymer science 2007, 32, 1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yoshikawa M, Tharpa K, Dima SO, Chemical reviews 2016, 116, 11500. [DOI] [PubMed] [Google Scholar]
- [17].Zaidi SA, Biomaterials science 2017, 5, 388. [DOI] [PubMed] [Google Scholar]
- [18].Fu N, Liu X, Li L, Tang B, Row KH, Journal of separation science 2017, 40, 2286. [DOI] [PubMed] [Google Scholar]
- [19].Lu X, Yang Y, Zeng Y, Li L, Wu X, Biosensors & bioelectronics 2018, 99, 47. [DOI] [PubMed] [Google Scholar]
- [20].Saeki T, Sunayama H, Kitayama Y, Takeuchi T, Langmuir : the ACS journal of surfaces and colloids 2018. [DOI] [PubMed] [Google Scholar]
- [21].Pan J, Chen W, Ma Y, Pan G, Chemical Society reviews 2018, 47, 5574. [DOI] [PubMed] [Google Scholar]
- [22].Yang Q, Li J, Wang X, Peng H, Xiong H, Chen L, Biosensors & bioelectronics 2018, 112, 54. [DOI] [PubMed] [Google Scholar]
- [23].Chen C, Luo J, Li C, Ma M, Yu W, Shen J, Wang Z, Journal of agricultural and food chemistry 2018, 66, 2561. [DOI] [PubMed] [Google Scholar]
- [24].Liu S, Bi Q, Long Y, Li Z, Bhattacharyya S, Li C, Nanoscale 2017, 9, 5394. [DOI] [PubMed] [Google Scholar]
- [25].Paul PK, Treetong A, Suedee R, Acta pharmaceutica 2017, 67, 149. [DOI] [PubMed] [Google Scholar]
- [26].Clegg JR, Zhong JX, Irani AS, Gu J, Spencer DS, Peppas NA, Journal of biomedical materials research. Part A 2017, 105, 1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Asanuma H, Akiyama T, Kajiya K, Hishiya T, Komiyama M, Analytica Chimica Acta 2000, 435, 25. [Google Scholar]
- [28].Alexander C, Andersson HS, Andersson LI, Ansell RJ, Kirsch N, Nicholls IA, O’Mahony J, Whitcombe MJ, Journal of molecular recognition : JMR 2006, 19, 106. [DOI] [PubMed] [Google Scholar]
- [29].Asanuma H, Kakazu M, Shibata M, Hishiya T, Chemical Communications 1997, 0, 1971. [Google Scholar]
- [30].Chmurski K, Temeriusz A, Bilewicz R, Analytical Chemistry 2003, 75, 5687. [DOI] [PubMed] [Google Scholar]
- [31].Hishiya T, Akiyama T, Asanuma H, Komiyama M, Journal of Inclusion Phenomena 2002, 44, 365. [Google Scholar]
- [32].Hishiya T, Asanuma H, Komiyama M, Journal of the American Chemical Society 2002, 124, 570. [DOI] [PubMed] [Google Scholar]
- [33].Sreenivasan K, Journal of Applied Polymer Science 1998, 70, 15. [Google Scholar]
- [34].Bilensoy E, Rouf MA, Vural I, Sen M, Hincal AA, AAPS PharmSciTech 2006, 7, E38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].He D, Deng P, Yang L, Tan Q, Liu J, Yang M, Zhang J, Colloids and surfaces. B, Biointerfaces 2013, 103, 580. [DOI] [PubMed] [Google Scholar]
- [36].Narra K, Dhanalekshmi U, Rangaraj G, Raja D, Senthil Kumar C, Neelakanta Reddy P, Baran Mandal A, Iranian journal of pharmaceutical research : IJPR 2012, 11, 715. [PMC free article] [PubMed] [Google Scholar]
- [37].Cyphert EL, Learn GD, Hurley SK, Lu CY, von Recum HA, Advanced healthcare materials 2018, e1800812. [DOI] [PubMed] [Google Scholar]
- [38].Cyphert EL, Zuckerman ST, Korley JN, von Recum HA, Acta biomaterialia 2017, 57, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]


