Abstract
Rad4/XPC recognizes diverse DNA lesions to initiate nucleotide excision repair (NER). However, NER propensities among lesions vary widely and repair-resistant lesions are persistent and thus highly mutagenic. Rad4 recognizes repair-proficient lesions by unwinding (‘opening’) the damaged DNA site. Such ‘opening’ is also observed on a normal DNA sequence containing consecutive C/G’s (CCC/GGG) when tethered to Rad4 to prevent protein diffusion. However, it was unknown if such tethering-facilitated DNA ‘opening’ could occur on any DNA or if certain structures/sequences would resist being ‘opened’. Here, we report that DNA containing alternating C/G’s (CGC/GCG) failed to be opened even when tethered; instead, Rad4 bound in a 180°-reversed manner, capping the DNA end. Fluorescence lifetime studies of DNA conformations in solution showed that CCC/GGG exhibits local pre-melting that is absent in CGC/GCG. In MD simulations, CGC/GCG failed to engage Rad4 to promote ‘opening’ contrary to CCC/GGG. Altogether, our study illustrates how local sequences can impact DNA recognition by Rad4/XPC and how certain DNA sites resist being ‘opened’ even with Rad4 held at that site indefinitely. The contrast between CCC/GGG and CGC/GCG sequences in Rad4-DNA recognition may help decipher a lesion’s mutagenicity in various genomic sequence contexts to explain lesion-determined mutational hot and cold spots.
Keywords: Protein-DNA interaction, DNA damage recognition, sequence impact, nucleotide excision repair, xeroderma pigmentosum, XPC, Rad4, x-ray crystallography, time-resolved fluorescence, fluorescence lifetime, Förster resonance energy transfer, molecular dynamics simulation
1. Introduction
Nucleotide excision repair (NER) is a versatile DNA repair pathway that can remove an extraordinarily broad range of structurally diverse lesions, including UV-induced intra-strand crosslinks and bulky base adducts generated by numerous environmental carcinogens (reviewed in [1–5]). Such DNA lesions, if left unrepaired, can block important cellular functions such as replication and transcription, eventually leading to mutagenesis and various diseases [6]. Dysfunctional NER caused by inherited mutations in the key NER proteins underlies various cancer and neurological syndromes in humans, such as xeroderma pigmentosum and Cockayne syndromes [7]. Importantly, while NER repairs diverse DNA lesions, the efficiency of NER among the lesions varies widely depending on the lesion’s chemical structure, stereochemistry, base modification site, conformation as well as base sequence context in the DNA [5, 8–15]. Certain lesions can even evade NER altogether, and by persisting in cells longer, become highly mutagenic [16]. How NER repairs diverse lesions and what determines their repair efficiencies remain of great interest in the field.
One of the keys to understanding the versatility and variable efficiencies of NER lies in its critical step of lesion recognition. This can occur via two sub-pathways: global genome NER (GGNER) and transcription coupled NER (TC-NER) [17]. TC-NER is initiated by an RNA polymerase stalled at a lesion during transcription and repairs lesions on actively transcribed strands. In GGNER, the lesions are sensed globally by specialized damage sensors such as the UV-damaged DNA binding protein (UV-DDB) E3 ubiquitin ligase complex and the xeroderma pigmentosum C (XPC)-RAD23B-CETN2 complex. UV-DDB is important in sensing UV lesions such as cyclobutane pyrimidine dimers in chromatin, which are then handed over to XPC [18–20]. For many helix-distorting and destabilizing bulky DNA adducts, XPC functions as the primary sensor. The lesion-bound XPC complex in turn recruits the 10-subunit transcription factor IIH complex (TFIIH), which scans the damaged DNA strand for lesion verification using its XPD helicase subunit with the help of XPA and replication protein A (RPA). The damaged nucleotides are eventually cut out by the XPF-ERCC1 and XPG endonucleases as a part of a 24–32 nucleotidelong single-stranded DNA, and the resulting gap in the DNA is finally restored by DNA repair synthesis and ligation.
The mechanism by which XPC recognizes diverse NER lesions has been under extensive investigation. The crystal structures of the yeast XPC-RAD23B ortholog, Rad4-Rad23 (hereafter Rad4), bound to DNA lesions showed that the binding caused two nucleotide pairs harboring the lesion to be flipped out of the DNA duplex and a β−hairpin motif from the β−hairpin domain 3 (BHD3) was inserted into the DNA duplex to fill the gap [21, 22]. Notably, in this ‘open’ structure, Rad4 did not form direct contacts with the damage-containing nucleotides themselves (such as UV-induced 6–4 photoproducts) but exclusively interacted with the nucleotides on the complementary strand. The structures therefore suggested that the protein must recognize the lesions in an indirect manner that relies on features of helix destabilization or distortion induced by a lesion rather than the lesion structure itself [23–26]; this in turn would allow the protein to bind to and recognize many different types of structurally distinct lesions.
Interestingly, we also previously observed that the same ‘open’ structure could be formed when Rad4 was covalently tethered to a non-specific, undamaged DNA sequence containing a stretch of consecutive C/G base pairs (‘CCC/GGG’) [27], and that similar ‘open’-like structures could be formed on the same CCC/GGG DNA even when Rad4 lacked the tips of β−hairpin2 or β−hairpin3 [28]. These ‘open’ or ‘open’-like structures with Rad4 (or the mutants) tethered to non-specific DNA were observed in solution as well as in crystal structures [28]. These studies indicated that the ‘open’ conformation is the thermodynamically more stable conformation whether the DNA is damaged or not; this pointed to a ‘kinetic gating’ mechanism for Rad4/XPC, whereby lesion recognition selectivity arises from the kinetic competition between DNA ‘opening’ and the residence time of Rad4/XPC per site. In this model, a sufficiently long residence time of the protein on one DNA location (e.g., enabled by covalent tethering) can result in the ‘opening’ of the given DNA site that is otherwise non-specific, including even undamaged DNA [27, 29, 30]. However, it remains unknown whether such ‘opening’ would happen for any DNA lesion or sequence, or if there are certain DNA lesions/sequences that resist being ‘opened’ even under a guaranteed long residence time, either because the barrier for opening is simply too high or because the ‘open’ state is not thermodynamically stable. This question has important implications for understanding a longstanding puzzle in NER – why do the recognition and repair efficiencies vary widely even among structurally similar lesions? And how can certain lesions even evade recognition and repair altogether? The repair propensity of a lesion has a profound impact on its mutagenicity since repair resistant ones persist longer in cells and therefore are more likely to survive to replication and cause mutations [16].
Here we present crystallographic and fluorescence lifetime (FLT)-based conformational studies showing that certain DNA sequences can resist ‘opening’ despite a long residence time. A mutant Rad4, which we previously showed could open the CCC/GGG sequence, failed to open an alternating CG/GC-repeat (‘CGC/GCG’) sequence under identical tethering conditions. Notably, the FLT analyses also revealed local pre-melting of the ‘openable’ CCC/GGG DNA but not the ‘opening-resistant’ CGC/GCG sequence. Finally, MD simulations on these DNA duplexes showed that CCC/GGG was more likely than CGC/GCG to access conformations that are closer to the ‘open’ conformation even in the absence of the protein due to the run of guanines. CCC/GGG was also more likely to be distorted towards the ‘open’ structure in binding to intact (‘WT’) Rad4. In contrast, CGC/GCG failed to engage Rad4 in a productive manner and was resistant to Rad4induced initial untwisting/bending.
Altogether, our study indicates that while stalling a protein and prolonging its residence time may facilitate the ‘opening’ of some sites, certain other sites may resist ‘opening’ altogether. Our study showcases the first example of a DNA sequence/structure that fails to be ‘opened’ by Rad4 even under tethered conditions, indicating that for these sites the ‘open’ conformation is thermodynamically disfavored. Importantly, our study demonstrates how local sequence differences can lead to two opposing outcomes in DNA ‘opening’ by Rad4/XPC. In light of recent progress in high-resolution mapping of NER lesions in cellular DNA, our work helps lay the foundation for interpreting such data to decipher the mutagenicity of different lesions at different positions in the genome ‒ mutational hot and cold spots associated with specific lesions [31].
2. Materials and Methods
2.1. Preparation of Rad4–Rad23 complexes.
The intact (‘WT’) and the Δβ−hairpin3 Rad4-Rad23 complex constructs are as published previously [21, 27–30]. Rad4 in both constructs spanned residues 101–632 and contained all four domains involved in DNA binding (Figure 1A). The WT complex has been shown to exhibit the same DNA-binding characteristics as the fulllength Rad4-Rad23 complex [21]. The Δβ−hairpin3 mutant complex (construct name: <137>) lacked the tip of the long β−hairpin in the BHD3 domain of Rad4 (residues 599–605 in the context of the WT construct). For crosslinking experiments, the WT (construct name: <SC32>) and Δβ−hairpin3 mutant (construct name: <SC41b>) also harbored V131C/C132S mutations in Rad4 to introduce site-specific disulfide crosslinking with DNA as done before [27, 28].
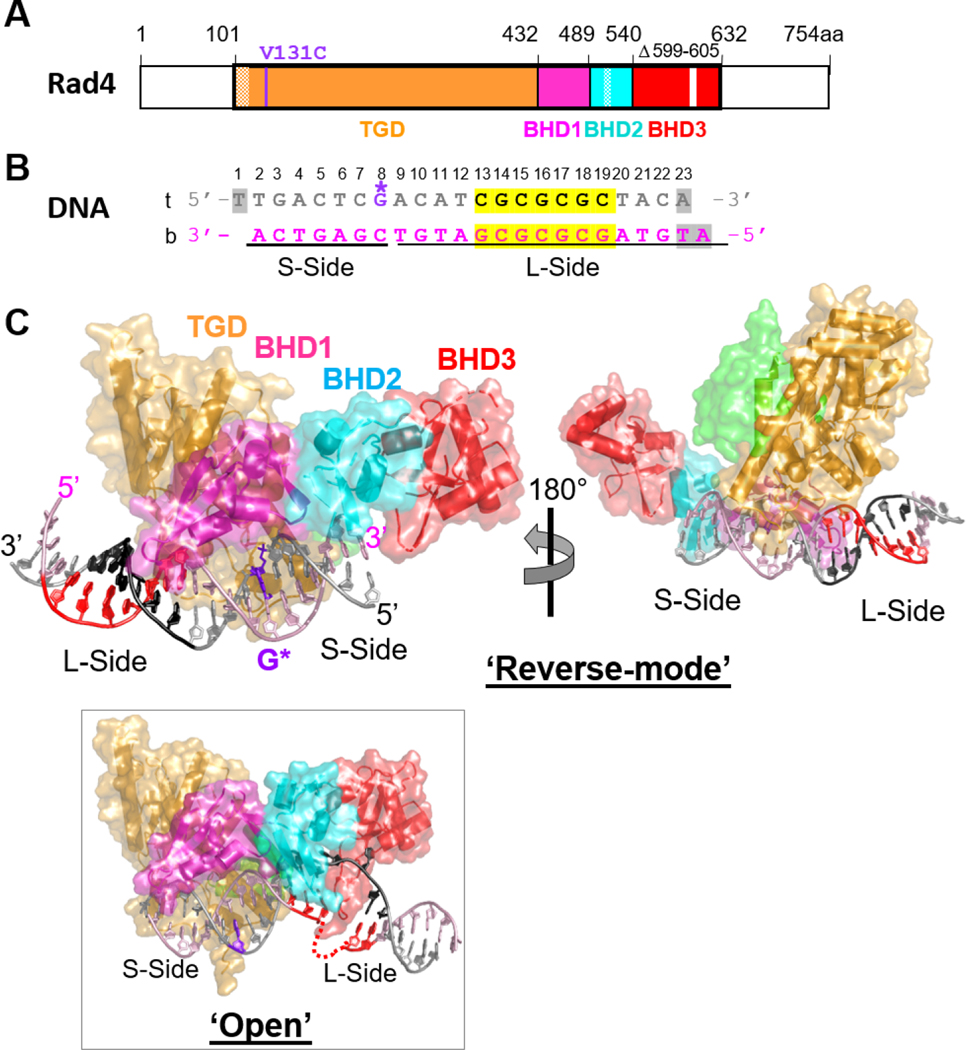
Fig.1. Crystal structure of the Δβ-hairpin3 mutant Rad4–Rad23 complex tethered to DNA containing alternating CG/GC repeat shows ‘reverse mode’ binding.
(A) The crystallized Rad4 construct spans residues 101–632. The transglutaminase domain (TGD) is indicated in orange, β-hairpin domain 1 (BHD1) magenta, BHD2 cyan and BHD3 red. The deleted region in the BHD3 β-hairpin (residues 599–605) is indicated in white. The disordered regions in crystals (residues 101–128, 518–525) are checkered. The V131C point mutation introduced for disulfide crosslinking is in purple. (B) The 23-bp CGC/GCG DNA construct for crystallization. Top strand (‘t’) is in silver and the bottom (‘b’) in pink. The CG/CG repeats are highlighted in yellow and colored black in ‘t’ and red in ‘b’. The disulfide-modified nucleotide, G* in dG8 is shown in purple. The DNA residues with missing electron densities are shaded in gray. The bottom strand was the damage-containing strand in the ‘open’ structures of lesion-bound Rad4 (PDB ID: 2QSG, 6CFI). (C) The ‘reverse- mode’ structure of the Δβ-hairpin3 mutant bound to CGC/GCG DNA duplex (PDB ID: 6UG1). The color scheme is the same as in (A) and (B). Rad23’s Rad4-binding domain (R4BD) is shown in light green. The right panel shows the structure rotated by 180 ° along a vertical axis. (Inset) The ‘open’ structure previously determined with CCC/GGG DNA tethered to the WT Rad4 complex (PDB ID: 4YIR).
All Rad4–Rad23 complexes in the study were co-expressed and purified from baculovirus-infected insect cells using previously described methods [21]. Briefly, the Hi5 insect cells coexpressing Rad4 and Rad23 were harvested two days after infection. After lysis, the proteins were purified using immobilized metal affinity chromatography (Ni-NTA agarose, MCLAB) and then anion-exchange chromatography (Source Q, GE healthcare). For crystallization purposes, the complex was subjected to overnight thrombin digestion at 4 °C, followed by cation exchange (Source S, GE healthcare) and size-exclusion (Superdex200, GE healthcare) chromatography. The pure sample (> 90% by SDS PAGE) was concentrated by ultracentrifugation (Amicon Ultra-15, Millipore) to ~15 mg/ml (185 μM) and stored in 5 mM bis-tris propane–HCl (BTP-HCl), 800 mM NaCl and 5 mM dithiothreitol (DTT), pH 6.8. For competitive EMSA and fluorescence studies, the protein complex was purified without thrombin digestion, thus retaining the UBL domain of Rad23 and a histidine-tag on Rad4 as also previously done [21, 27, 28, 32].
2.2. Synthesis of oligonucleotides and preparation of duplex DNA.
Different oligonucleotides containing disulfide-modified guanine (indicated as G* on the top strand) were prepared by incorporating the 2-F-dI-CE phosphoramidite (Glen Research) at the desired position during solid-phase synthesis. The conversion and deprotection of 2-F-dI were performed according to the guidelines provided by Glen Research (https://www.glenresearch.com/media/productattach/import/tbn/TB_2-F-dI.pdf). Briefly, 2-F-dI-containing oligonucleotides were treated with cystamine (prepared freshly from cystamine hydrochloride and sodium hydroxide) to tether the disulfide group, then deprotected with 1,8diazabicycloundec-7-ene. All synthetic oligonucleotides were purified with HPLC and checked by MALDI-MS. The HPLC-purified bottom strands were purchased from IDT. Oligonucleotides modified with cytosine analogs, tC° and tCnitro were purchased from Biosynthesis, purified by HPLC and verified by MALDI-MS. DNA duplexes were prepared by annealing the top and bottom strands in ratios 1:1.1 for crystallization and 1:1 for fluorescence lifetime (FLT) studies.
2.3. Disulfide crosslinking of the Rad4-Rad23 complexes with double-stranded DNA.
Sitespecific disulfide crosslinking between DNA and the WT or Δβ−hairpin3 Rad4-Rad23 complexes was done as previously described [27, 28]. First, DTT from the protein complex was removed by extensive buffer exchange using a desalting column (Zeba Spin Desalting Column, 40,000 Da molecular weight cut-off, Thermo Scientific) pre-equilibrated in 5 mM BTP-HCl and 800 mM sodium chloride (NaCl), pH 6.8. The protein complex was thereafter incubated with equimolar DNA containing disulfide-modified base (G*) in crosslinking buffer (5 mM BTP-HCl, 100 mM NaCl and 10% glycerol, pH 6.8) at 4°C overnight. To quench the reaction, the samples were treated with 0.1 mM S-methylmethanethiosulfonate (Sigma) at room temperature for 10 min. Subsequently the samples were loaded to 15% SDS-PAGE gel using a loading buffer lacking 2mercaptoethanol and were run at 180 V for 50 min. The crosslinking yield was ~50–60%. The complex was then purified by anion-exchange chromatography (Mono Q, GE healthcare) over a 0–2 M NaCl gradient in 5 mM BTP-HCl and 10% glycerol, pH 6.8. The buffers were degassed by nitrogen purging. Purified crosslinked complexes eluted at 400–480 mM NaCl and were further concentrated by ultrafiltration (Amicon, Millipore) to ~3.5 mg/ml (30 μ M). As previously described [27], the V131C/C132S mutations in Rad4 for crosslinking are at a site separate from the putative ‘open’ site involving BHD2 and BHD3. The C131-dG*8 crosslink is also designed to match the original distance between V131 and DNA (dG8). The V131C/C132S mutations were necessary and sufficient for efficient crosslinking of the purified Rad4–Rad23 complex with the DNA. The presence of seven other cysteines (C276, C354, C355, C463, C466, C509 and C572) whose side chains were exposed on the surface of Rad4 did not affect the crosslinking, further validating the specificity of the DNA binding and crosslinking. In fact, mutating these other cysteine residues to serines decreased the solubility of the protein and did not increase the crosslinking yield with DNA.
2.4. Crystallization of Δβ−hairpin3 mutant crosslinked with the CGC/GCG DNA duplex.
All crystallization trials were set up using the hanging-drop vapor diffusion method at 4 °C in which 1.5 μl of protein-DNA complex was mixed with 1.5 μ l of crystallization buffer (50 mM BTP-HCl, 200 mM NaCl and 10–15% isopropanol pH 6.8) and sealed over 1 ml of crystallization buffer. Among the CGC/GCG DNA sequences of various lengths that we tried (Table S1), the 22-bp DNA (CH9d) did not crystallize; the 24-bp (CH9a) and 25-bp (CH9b) DNA formed showers of needle-like microcrystals, but they did not diffract even after several rounds of optimization. The best crystals were obtained with the 23-bp DNA, first as showers of small plate-like crystals (20 μm) which appeared within a few days. Subsequently these crystals were harvested and used for micro-streak-seeding, which yielded larger crystals that grew to a maximum size of ~70–80 μm in 10–12 days. The crystals were harvested in a harvest buffer (50 mM BTP-HCl, 200 mM NaCl, 3% isopropanol, pH6.8) and submerged for a few seconds in a cryoprotectant buffer (50 mM BTPHCl, 200 mM NaCl, 3% isopropanol and 20–25% MPD, pH 6.8) before being flash-frozen in liquid nitrogen. Diffraction data were collected in LS-CAT, 21-ID-F beamline at 103 K and were processed with the HKL2000 [33]. The data collection statistics are summarized in Table S2.
2.5. Structure determination and refinement.
The structure of the mutant Rad4–Rad23–DNA complex was determined by molecular replacement method using the previous structure of WT Rad4 crosslinked with CCC/GGG (PDB ID: 4YIR) using MOLREP (CCP4) [34]. Several rounds of model building were performed using WinCoot [35], followed by refinement with Phenix [36]. First, the model building and refinement proceeded with the ‘open’ conformation as the model. However, the refinement statistics did not improve beyond R/Rfree: 33%/34% with this model. Furthermore, the electron density of the DNA after the TGD-BHD1 domains appeared completely absent, indicating that the DNA may be bound in a different orientation. The ‘reverse mode’ DNA model was constructed by 180° rotation of the DNA duplex with respect to the crosslinked nucleotide G* (Figure 1B). This yielded improved model geometry and lower R/Rfree: 22%/27%, further confirming the validity of the model. The refinement statistics for the current model is summarized in Table S2. The final structure (PDB ID: 6UG1) contains residues 129–301, 305504, 506–513, 528–598, 606–632 of Rad4 and 255–311 of Rad23. The DNA nucleotides with missing densities are W1 and W23 in the top strand and Y1 and Y2 in the bottom strand. All figures were made using PyMOL Molecular Graphics System, version 2.1.1 (Schrodinger, LLC).
The distance between the crosslinked residues (Cα of V131 in Rad4 and C2 of dG*8 in the top strand of the CGC/GCG DNA) is 9.0 Å, only ~0.1 Å shorter than the distance between equivalent atoms in the non-tethered, lesion-bound structure (9.1 Å between Cα of C131 in Rad4 and C2 of A8 in the top strand of the DNA; PDB code 2QSH), further confirming that the crosslinking did not induce structural distortions.
2.6. DNA conformational distributions obtained from fluorescence lifetime (FLT) measurements of Förster resonance energy transfer (FRET).
DNA duplexes labeled with cytosine analog FRET pair, tC° and tCnitro, or labeled with tC° alone were prepared as described in Methods. The 5 μM samples (DNA and DNA-protein complex) were prepared in phosphatebuffered saline (PBS: 10 mM Na2HPO4, 2 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl pH 7.4) with 1 mM DTT (DTT was omitted from the buffer for disulfide-tethered protein-DNA complexes). Sample volume for each FLT measurement was 45 μl. Fluorescence decay curves for the FRET donor tC° (in the absence and presence of the FRET acceptor tCnitro, which in itself is nonfluorescent) were measured with a DeltaFlex fluorescence lifetime instrument (HORIBA) equipped with a Ti-sapphire laser as an excitation source (Mai Tai HP, Spectra-Physics) as previously done [28]. The beam for tC° excitation was produced by frequency doubling of the fundamental beam (730 nm) and pulse-picking at 4 MHz, which was then passed through a monochromator set at 365 nm (band pass 10 nm). The fluorescence signal emitted at 470 nm (band pass 10 nm) was collected by a Picosecond Photon Detection module (PPD-850, Horiba) using time-correlated single-photon counting (TCSPC) electronics. Fluorescence decay curves were recorded on a 100 ns timescale, resolved into 4,096 channels, to a total of 10,000 counts in the peak channel. All details describing the analyses of the decay traces are in SI Methods.
2.7. Fluorescence lifetime analyses using maximum entropy method (MEM) and Gaussian fitting.
The fluorescence decay traces were analyzed by the maximum entropy method (MEM) using MemExp software [37, 38], as done previously [32]. The MEM analysis yielded a distribution of donor lifetimes with each lifetime component (τD,i for donor-only samples and τDA,i for donor-acceptor labeled samples) having a corresponding amplitude αi. The reproducibility of the distributions obtained from the MEM analyses are illustrated from three independent lifetime measurements on each sample. The lifetime distributions from the MEM analyses were subsequently fitted as a sum of Gaussians. The donor-only samples exhibited a single peak, and the characteristic lifetime of the donor-only sample (τD), was obtained from the peak position of the Gaussian-fitted distribution. The average FRET efficiency for each sample was computed as , were corresponding to each lifetime component (τDA,i). Each Gaussian component in the τDA distribution was used to calculate the average lifetime and FRET efficiency representing that component, and the area under the Gaussian curve was taken as a measure of the fractional population of that component. The results are summarized in Table S3–S5. Errors are indicated with standard deviations (s.d.) from three independent sets of measurements.
2.8. Competition electrophoretic mobility shift assays (EMSA).
Full details are given in the SI Methods.
2.9. MD simulations and structural analyses.
Full details are given in the SI Methods.
3. Results
3.1. Matched DNA with alternating CG/GC repeats fails to be ‘opened’ by tethered Rad4.
In our previous study, we showed that WT Rad4 forms an ‘open’ conformation with a 24-bp matched DNA containing consecutive runs of C/G’s (‘CCC/GGG’) when site-specifically tethered with a covalent disulfide linkage between residue 131 of Rad4 transglutaminase domain (TGD) and nucleotide residue 8 of the DNA top strand [27]. The crosslink was designed to be distant from the putative ‘open’ site involving BHD2 and BHD3 and to have minimal perturbation to the original protein-DNA contacts involving TGD (see Methods). We also recently solved the structure of a mutant Rad4 lacking the β−hairpin3 tip (residues 599–605; Δβ−hairpin3) tethered in the same manner to the CCC/GGG DNA as the WT, which showed a similar, albeit more dynamic (‘open-like’) conformation [28]. These results showed that the Rad4-bound, thermodynamically stable state was structurally identical for both specific and non-specific substrates, and suggested that the mechanism by which a freely-diffusing Rad4/XPC discriminated between damaged and undamaged sites was the ease with which it could form the ‘open’ conformation at a given site before it diffused away – the so-called ‘kinetic gating’ mechanism [27–30].
Under covalently tethered conditions, the ‘opening’ of even matched DNA such as CCC/GGG, which normally is a non-specific substrate for both the intact and the mutant Rad4 [21, 27, 29, 32], was possible because it increased the protein’s residence time on a single register of DNA by limiting its diffusion. However, it was unknown if such ‘opening’ would indeed happen for any sequence or structure given a long residence time (i.e., tethering) or if there were some DNA sequences/structures that were intrinsically resistant to being ‘opened’ even with a guaranteed long residence time.
To address these questions, we set out to determine the structures of Rad4 tethered to DNA sequences other than the ‘openable’ CCC/GGG sequence. Since the Δβ−hairpin3 mutant was still able to form an ‘open-like’ structure with the CCC/GGG sequence like the WT under tethered conditions, we chose to focus on this mutant protein to make direct structural comparisons. After extensive trials with varying DNA sequences and lengths, we obtained crystals with DNA duplexes containing alternating CG/GC over seven nucleotides (hereafter ‘CGC/GCG’). Like CCC/GGG, the CGC/GCG-containing duplexes also behave as non-specific substrates for Rad4 under non-tethered conditions in the competitive gel-shift assays (Figure S1) [22, 27–29, 32, 39].
Crystals with a 23-bp CGC/GCG construct diffracted the best, up to 2.9 Å, among the DNA containing the same CG/GC repeats (Table S1). The crystals belonged to the P1 space group, different from that formed with the CCC/GGG DNA (P41212). Furthermore, the resulting structure was strikingly different from the ‘open’ conformation previously determined with the CCC/GGG DNA or with any of the other lesion-bound specific structures: while the TGD and BHD1 domains were still bound to the double-stranded portion of the DNA as in the ‘open’ conformation, the orientation of the protein with respect to the DNA was reversed by 180°. In such a 180°-reversed mode of binding (hereafter referred to as ‘reverse mode’), the C-terminal BHD2/3 domains faced the short end of the DNA duplex with respect to the tethering site (hereafter ‘S-side’) instead of binding to and ‘opening’ the CGC/GCG site on the long end of the DNA (or ‘L-side’) (Figure 1C). The positioning of BHD2/3 on the ‘S-side’ also indicated that the BHDs would be bent towards and cap the duplex end of the DNA and that an extension of straight B-DNA would be incompatible with this binding conformation. On the other hand, TGD-BHD1 was bound to the ‘L-side’ of the DNA in which the CG/GC repeat sequence maintained its ‘closed’ duplex form.
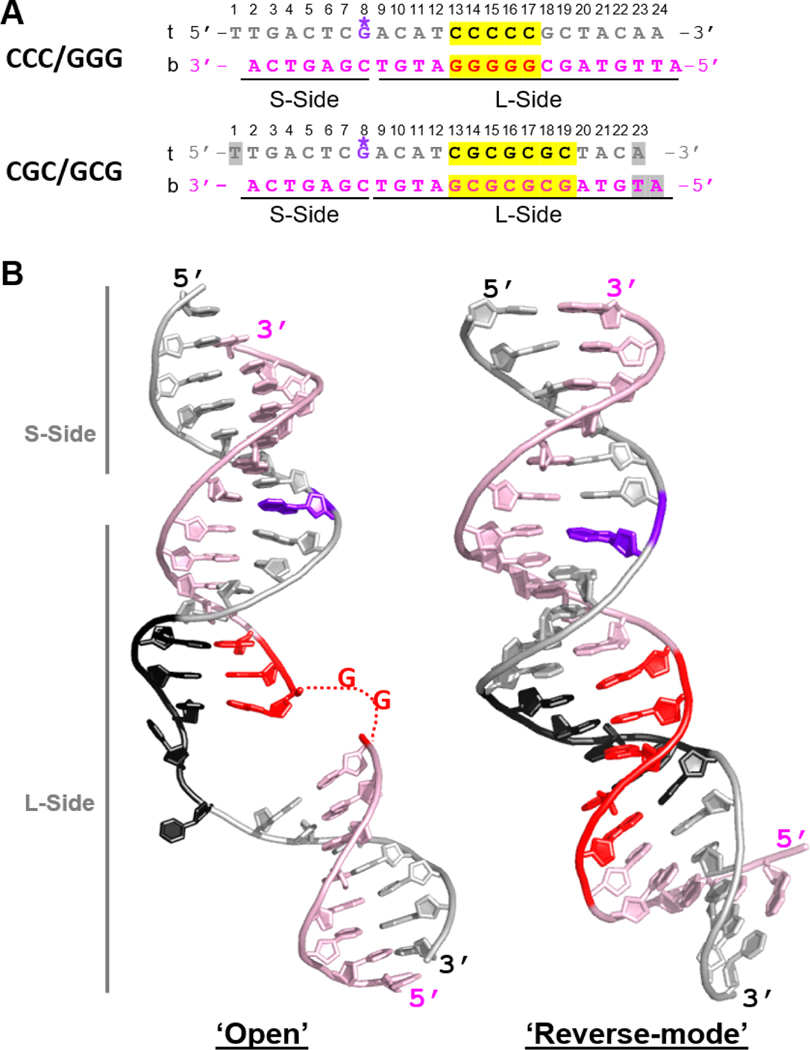
The DNA was also extended beyond TGD-BHD1 and made contacts with the BHD3 of Rad4 in a neighboring unit cell (Figure S2); this interaction seemed more optimal for a 23-bp substrate than other DNA lengths. The structural parts common to the ‘open’/’open-like’ and the ‘reverse mode’ structures superpose within ~0.96 Å RMSD (Figure S3). However, the DNA in the ‘reverse mode’ structure maintained all base pairings without any nucleotide flipping or local unwinding seen in the ‘open’ structure (Figure 2).
Fig.2. Comparison of DNA conformations in ‘open’ versus ‘reverse mode’ structures.
(A) The sequences of the CCC/GGG DNA that forms ‘open’ or ‘open-like’ structures when tethered to Rad4 and of the CGC/GCG DNA forming a ‘reverse mode’ structure. The top (‘t’) and bottom strands (‘b’) are in silver and light pink, respectively; consecutive C/G’s and alternating CG/GC repeats are highlighted in yellow and colored black in ‘t’ and red in ‘b’. The DNA residues with missing electron densities are shaded in gray. The crosslinkable G* is in purple. The short (S-side) and long sides (L-side) of the DNA duplexes are designated with respect to the G* tethering site. (B) (left) The ‘open’ DNA conformation in the WT Rad4CCC/GGG structure (PDB ID: 4YIR); (right) The ‘reverse mode’ DNA shown in the Δβ-hairpin3-CGC/GCG DNA structure (PDB ID: 6UG1).
What causes Rad4 to bind to the CGC/GCG DNA in such a different binding mode from that with CCC/GGG in a complex whose constituents are otherwise the same? We argue that the reason is unlikely to be because of the DNA length difference between the two DNA constructs (23-bp for CGC/GCG vs 24-bp for CCC/GGG) since both lengths could in principle accommodate the ‘open’ and ‘reverse-mode’ structures (see SI Discussion). Thus, a more likely explanation for the different structures stems from the difference in the DNA sequence. We posit that the CGC/GCG sequence is less ‘openable’ by Rad4 than CCC/GGG and that the ‘reverse mode’ structure observed with CGC/GCG is the preferred mode of Rad4 binding to a 23- or 24-mer that resists ‘opening’. Such DNA would thus maintain a duplexed B-DNA conformation while providing accessible DNA ends that help stabilize the ‘reverse mode’ over the other binding mode. What the binding mode would be for a longer ‘opening-resistant’ DNA lacking accessible DNA ends remains to be determined. To further examine the issue of less ‘openable’ DNA, we next turned to fluorescence lifetime (FLT) studies of DNA conformational distributions in solution.
3.2. Matched DNA containing alternating CG/GC is resistant to ‘opening’ by tethered Rad4 in solution.
Although the crystallographic studies showed the 3D structures of the Rad4–DNA complexes in detail, such structures remain as snapshots captured in crystals, and whether the CG/GC repeats are indeed resistant to ‘opening’ by Rad4 in solution remained to be examined. To examine accessible DNA conformations in solution, we turned to fluorescence lifetime-based FRET (FLT-FRET) studies of DNA, without and with Rad4. We have previously used the FLTFRET approach using tCo and tCnitro (cytosine analogs) FRET pair incorporated in DNA to characterize the conformational distributions of DNA with WT and mutant Rad4 in solution [28, 32, 40]. Using this approach, we showed that the matched CCC/GGG DNA indeed adopts ‘open’ conformations when tethered to Rad4, similar to a specific, model lesion substrate (CCC/CCC mismatched DNA) which Rad4 recognizes and ‘opens’ without tethering [28]. Here, we applied this approach to a 24-bp CG/GC-repeat-containing DNA duplex tethered to Rad4. The tC° (FRET donor) and tCnitro (FRET acceptor) probes were introduced to the DNA on either side of the putative ‘open’ site as before [28, 29, 32, 40] and a crosslinkable guanine nucleotide (G*) was introduced to enable tethering of Rad4 as done for crystallization (Figures 3A & S4–S5). The fluorescence lifetime distributions of the DNA and DNA-protein samples are shown in Figure 3 and the results of the conformational analyses as well as DNA sequences used are in Table S3. In the absence of the acceptor (thus no FRET), the donor-only DNA (denoted as CGC/GCGF_D) showed a single lifetime peak corresponding to the intrinsic lifetime of the donor fluorescence (τD= 4.8 ns) (Figure 3B dotted grey; Table S3B); the τD of the DNA also showed no change in the presence of bound Rad4 (Figure 3B dotted blue).
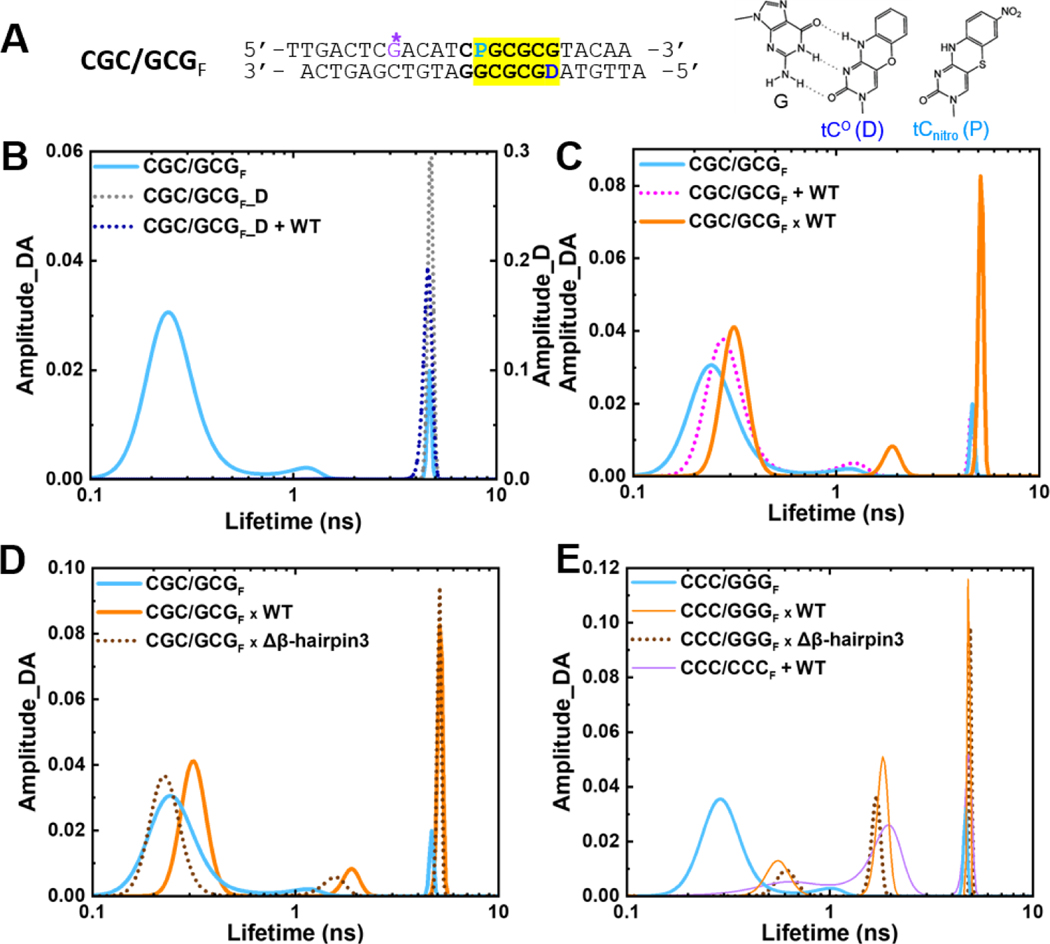
Fig. 3. Conformational distribution of DNA and DNA-protein complexes in solution revealed by fluorescence lifetime measurements.
(A) The donor/acceptor-labeled, CG/GC-repeat DNA construct used for FLT studies (“CGC/GCGF”). The segment containing CG/GC’s is highlighted in yellow. ‘D’ indicates Tc° (FRET donor) and ‘P’ is tCnitro (FRET acceptor). G* is disulfide-modified guanine for tethering. (right) Chemical structures of tC° and tCnitro. As cytosine analogs, they form Watson-Crick type base-pairing with guanine (G), as shown on the left for tC°. (B) FLT distributions of the donor/acceptorlabeled CGC/GCGF (cyan) and the donor-only DNA (CGC/GCGF_D) in the absence (dotted grey) and presence of WT Rad4 (dotted blue). (C) FLT distributions of CGC/GCGF when by itself (cyan), noncovalently bound to (“+”; dotted magenta) or site-specifically tethered with WT Rad4 (“x”; orange). (D) FLT distributions of CGC/GCGF when by itself (cyan), tethered to WT Rad4 (orange) or to Δβ-hairpin3 (dotted brown). (E) FLT distributions of CCC/GGGF when by itself (cyan), tethered to WT Rad4 (orange) or to Δβ-hairpin3 (dotted brown) as well as the mismatch DNA CCC/CCCF + WT (purple). Data in (E) are adopted from our previous study [28]. All amplitudes indicate the normalized, fractional amplitudes of the donor/acceptor (Amplitude_DA) or the donor-only construct (Amplitude_D). Reproducibility of FLT distributions for each sample is shown in Figure S6. DNA sequences containing fluorescent probes and full reports of the lifetimes, fractional amplitudes, FRET efficiencies of each peak as well as the sample’s average FRET efficiencies are in Table S3.
The donor/acceptor-labeled DNA (denoted as CGC/GCGF) showed a major lifetime peak (τDA) at 0.23 ns corresponding to a FRET efficiency (E) of 0.96, with a fractional amplitude (thus fractional population) of 86% (Figure 3B & S6A, cyan; Table S3C), with two minor peaks at 1.1 ns (E = 0.83) and 4.3 ns (E = 0.12) occupying 8% and 6% fractional amplitudes, respectively. The FRET efficiency corresponding to the major peak (0.96) closely matches the FRET computed for an ideal B-DNA conformation with the given probe positions (0.93) [32, 41]. Furthermore, the FLT profile of CGC/GCGF matched well with that of the same DNA with normal G instead of G* (Figure S6B, Table S3D). Altogether, these results confirm that the matched CGC/GCGF adopts a predominantly B-DNA conformation, as observed for several other matched DNA sequences (including CCC/GGG) examined previously with or without G* [28, 32].
Next, we examined the CGC/GCGF DNA in the presence of noncovalently interacting, equimolar WT Rad4. The profile of the matched DNA did not change when Rad4 was added (Figures 3C & S6C, cyan vs. dotted magenta), retaining the majority B-DNA conformation at ~86% fractional population (Table S3C). This result agrees with those previously seen for other matched DNA sequences [28, 32].
Following these, we examined the lifetime profile of the DNA covalently tethered to Rad4 (Figures 3C–D & S6D–E). With both WT and Δβ−hairpin3, the same three major lifetime components were observed as with free DNA with the overall FLT profiles showing: (i) the fractional population in the short lifetime peak at ~0.2–0.3 ns dropped to 65–68% compared with 86% in free DNA, (ii) the medium lifetime peak shifted to 1.6–1.7 ns but retained ~8% fractional population, and (iii) the fractional population in the long lifetime component at 4.8 ns increased to 23–26% compared with 6% in free DNA (Table S3C). These results indicate that tethering did alter the distribution of conformations accessed by DNA, as will be discussed further. However, these changes are not consistent with Rad4-induced ‘open’ conformation, as explained below.
The most notable result from FLT-FRET studies on CGC/GCG DNA tethered to WT Rad4 or Δβ−hairpin3 mutant is that the predominant (short lifetime) peak was unshifted from a position that reflects a FRET E characteristic of B-DNA (E ~0.94–0.95 compared to 0.96 for free DNA). These results are in stark contrast with those previously observed with the ‘open’ complexes whether formed naturally or with tethering [28, 32]. In the ‘open’ complexes, as shown for matched CCC/GGG tethered to WT or Δβ−hairpin3 and for mismatched CCC/CCC bound to WT (Figure 3E), the ~0.3 ns component, characteristic of B-DNA, almost completely disappeared. Instead, the shortest lifetime component was observed at ~0.6–1.0 ns (E ~0.88–0.80) and with a much reduced fractional population of 25–35%; the medium lifetime population, which shifted from ~1.1 ns to ~1.7–1.9 ns, also significantly increased in fractional population, from 8% to 42–47%, and the long lifetime component, at ~4.4–4.8 ns, also increased in fractional population, from 8% to 23–28% (Table S3E). Overall, the FLT-profile of the tethered CCC/GGG shows no B-DNA-like population and overlaps well with Rad4-bound ‘open’ specific complex formed with CCC/CCC mismatched DNA [28], while the tethered CGC/GCG resembles the free DNA more than these ‘open’ DNA conformations.
We now return to the observation that tethering increased the fractional population in the long lifetime component at the expense of the B-DNA like conformation in each of the constructs. This long lifetime component, observed at ~4.0‒4.8 ns, overlaps with the donor-only lifetime of ~4.8 ns. indicating that either we have unpaired donor strands in the double-labeled constructs or that some DNA conformations exhibit an apparent ‘zero-FRET’ component. Notably, a FRET value close to zero is also the computed value for the ‘open’ DNA conformation observed in the crystal structures, given the placement of the tC° and tCnitro probes in these constructs [28, 32]. In previous studies, we ruled out unpaired donor strands as contributing significantly to this ‘zeroFRET’ component, since the component persisted even in the presence of 5-fold excess acceptor strands [32]. Furthermore, our observation that the fractional population in this component increases when tethering is used [28], suggests that this component corresponds to a real DNA conformation in our samples and whose population increases in tethered complexes.
To further probe the origin of this increase, we treated the tethered CGC/GCGF x WT complex with 5 mM dithiothreitol (DTT) which abolishes the disulfide tethering and analyzed the FLT-FRET profiles at different time points after DTT addition (Figure S7A, Table S4A). The long lifetime peak gradually decreased from ~28% to ~6%, as the entire profile also completely returned to that of free DNA after 1 hour. The results were similar for CGC/GCGF x Δβ-hairpin3 complex (Figure S7B, Table S4B). These results confirm that the increase in this longest lifetime population that is concomitant with the decrease in the shortest lifetime (characteristic of B-DNA form) population, is due to the tethering. Therefore, the possibility remains that up to ~20–30% of Rad4-tethered CGC/GCGF DNA are present as significantly distorted conformations in solution. This result is not entirely unexpected as the conformations in solution are much more dynamic and heterogeneous than those obtained from crystal structures, as noted before [28]. Nevertheless, the FLT-FRET studies strongly corroborate the crystallographic structures and show that, under the same tethering condition, the conformations accessed by CGC/GCG DNA are predominantly BDNA-like and distinct from those accessed by CCC/GGG DNA [28], indicating that CG/GC repeat DNA is not prone to be ‘opened’ by Rad4.
3.3. Temperature dependence of fluorescence lifetime distribution shows local pre-melting for CCC/GGG DNA.
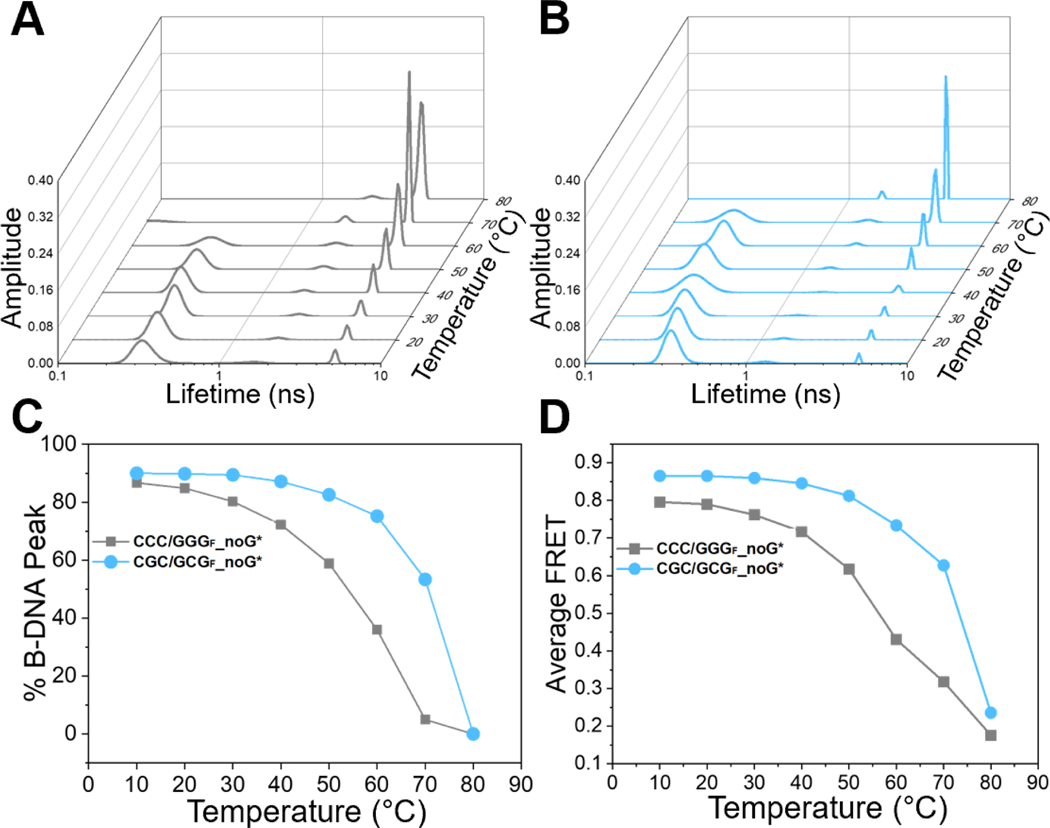
To examine whether the different behaviors exhibited by the two DNA constructs were related to their inherent thermal stabilities, we next performed DNA melting studies in the absence of Rad4. The melting temperatures (Tm) of the two DNA sequences (both 24-bp) were not significantly different from each other (76.5 ± 1.0 for CCC/GGG vs. 74.3 ± 1.8 °C for CGC/GCG) when measured by the UV absorbance changes at 260 nm (Figure S8). This result is not too surprising, since UV absorbance changes report primarily base-pair disruption, and the transition temperature, which reflects global thermal stability against strand separation, is expected to be similar for two sequences of the same length and overall G/C content. However, local stabilities of the DNA may differ, which can in turn affect ‘opening’ by Rad4. Indeed, the UV absorbance melting profile of the ‘openable’ CCC/GGG construct shows a minor ‘premelting’ transition at ~50 °C prior to the relatively broad, dominant ~77 °C transition that reports on the overall separation of the strands, while the ‘open’-resistant CGC/GCG construct shows no such pre-melting transition and a much sharper strand separation transition compared with CCC/GGG (compare Figures S8B and S8C).
To further investigate the local thermal stabilities of the two matched DNA sequences, we took advantage of the fact that tC° and tCnitro FRET probes sense local DNA conformations and measured the FLT distributions and the average FRET of the tC°/tCnitro-labeled DNA constructs in the temperature range from 10 °C to 80 °C (Figure 4, Table S5). Previous studies have shown that the FRET probes themselves do not significantly alter the thermodynamic stability of DNA duplexes [29]. For the CCC/GGG sequence, the low lifetime peak corresponding to the B-DNA conformation (~84 % at 20 °C) started to decrease almost immediately as the temperature was raised above ~20 °C and was reduced to 50 % at ~55 °C (Figure 4A & 4C, grey), well below the Tm ≈ 77 °C measured by UV absorbance (Figure S8). This trend also corroborates with the premelting transition already detected in the UV absorbance melting profile. In contrast, the fractional B-DNA population in the CGC/GCG sequence decreased to 50% at a much higher temperature of ~70 °C (Figure 4B & 4C, cyan), close to the Tm ≈ 74 °C measured by UV absorbance, indicating no pre-melting tendency. These results are mirrored in the average FRET versus temperature profiles (Figure 4D) and show that the sequence containing consecutive C/G’s undergoes local pre-melting at that site before the global melting of the DNA duplex, indicating that it is intrinsically more deformable than the site with alternating CG/GC’s, and hence more readily ‘opened’ by Rad4. These measurements also underscore the high sensitivity of our FLT-FRET approach in capturing local DNA conformations and their changes.
Fig.4. Temperature dependence of FLT distributions shows local pre-melting for CCC/GGG but not for CGC/GCG.
(A-B) FLT distributions with increasing temperatures (10 °C to 80 °C) for donor/acceptor-labeled DNA constructs containing either the CCC/GGG (grey, left) or CGC/GCG (cyan, right) sequence motifs. (C) Fractional population of B-DNA conformation (taken from τ1, the short lifetime component) versus temperature and (D) Average FRET versus temperature, plotted for each DNA. The average FRET was calculated as , where the donor-only lifetimes (τD) for the DNAs were taken as 4.8 ns. The uncertainties reported are standard deviations (s.d.) from 2 independent sets of measurements, and they were usually less than 5%. DNA sequences and plotted values are in Table S5.
3.4. MD simulations of unbound DNA duplexes reveal that the CCC/GGG duplex exhibits inherent structural distortions that foster the ‘open’ conformation.
To further explore the structural factors that may influence the ‘openability’ by Rad4/XPC in detail, we next turned to molecular dynamics simulations and examined the CCC/GGG and CGC/GCG DNA sequences without and with bound Rad4.
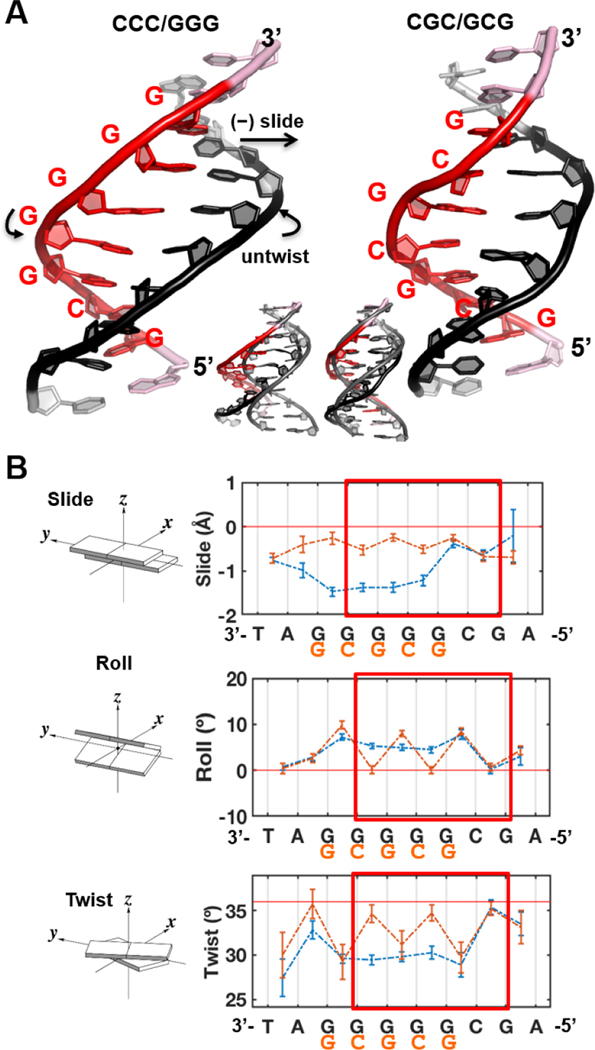
First, we investigated the impact of local sequence identity on the DNA conformations by performing 2 μs MD simulations for 13-mer DNA duplexes containing CCC/GGG and CGC/GCG sequences at their centers. Stable ensembles were achieved in the 0 – 2 μs range of MDs following equilibration (Figure S9). Best representative structures of the unbound CCC/GGG and CGC/GCG 13-mer DNA duplexes are shown in Figure 5A. Our MD simulations revealed the striking impact of the run of guanines in CCC/GGG on the local DNA conformation. The structural ensembles of the CCC/GGG and CGC/GCG duplexes differ prominently in base pair slide, roll, twist, and also in helix bending direction (Figures 5B and S10). We first measured the 6 base pair step parameters: shift, slide, rise, tilt, roll and twist for the structures along the trajectories excluding the more dynamic end base pairs. Shift, rise and tilt did not show much deviation from an ideal B-DNA structure for either CCC/GGG or CGC/GCG sequences (Figure S10A). However, slide, roll, and twist did deviate from those of B-DNA and were quite different in the two sequences (Figure 5B). The CCC/GGG duplex had significant slide per GG step for the run of guanines with an average of ~−1.5 Å, while the CGC/GCG duplex did not deviate much from the ideal B-DNA (0 Å slide) with an average slide of ~−0.2 Å for the CG step and ~−0.4 Å for the GC step. Correlated with its large slide, the CCC/GGG duplex exhibited consistent untwisting over the run of guanines indicated by an average twist angle per GG step of ~30°, which is ~6° lower than the ideal B-DNA value of 36° per step. In contrast, the CGC/GCG duplex showed significant untwisting only at its CG steps with an average twist angle of ~31º, while the average twist angle for the GC steps was ~35°. Correlated with the twist angle, CCC/GGG had a constant average roll angle per GG step of ~8°, while CGC/GCG exhibited roll only at the CG steps with an average value of ~9°.
Fig.5. Intrinsic structural differences of the CCC/GGG and CGC/GCG sequences.
(A) Best representative structures. Overlay with a B-DNA model is shown in the inset. (B) The DNA sequences had prominent impacts on the helix parameters, slide, roll and twist. See Figure S10 for the other parameters, shift, rise and tilt. Illustrations of the base pair step parameters are adapted from 3DNA [109]. The standard deviations of the block averaged means [110, 111] for the parameter values are shown. The twist angle is 36º per step for ideal B-DNA. Note that the sequence labels are from 3’ to 5’. The regions boxed red are centered around the putative ‘open’ site and the end base pairs of the 6-mer regions were used to calculate the (un)twist angle upon the DNA’s initial binding with Rad4.
The base pair step parameters, twist, roll and slide, manifest the pair-wise sequence effects that have been well studied and explicated by Wilma Olson and colleagues [42, 43]. Thus, the CG step and GC step alternate in the CGC/GCG sequence with less slide, lower twist, and greater roll at the CG than the GC step (Figure 5B). However, in the absence of such GC and CG alternations, steric hindrance between guanine amino groups in the run of guanines have the dominant impact (in slide, untwist and roll) on the structure in the CCC/GGG sequence [42, 44].
We followed the dynamic bending of both duplexes by characterizing the DNA bend directions. While the bend angles themselves over the central 10-bp DNA were 13 ± 1° for both sequences (Figure S10B), the bend direction angles were −45 ± 8° for CCC/GGG and −19 ± 10° for CGC/GCG (Figure S10C). This pseudo-dihedral angle adopts more negative values when bending is towards the minor groove around the potential open site. In the ‘open’ conformation crystal structure the bend direction dihedral is −66°, while in the ‘reverse mode’ crystal structure of the Δβ−hairpin3 in complex with the CGC/GCG duplex it is −26°. Therefore, the directions of bending in the free, unbound CCC/GGG and CGC/GCG duplexes tracked with those of the respective DNA bound to Rad4 in the crystal structures.
Lastly, we computed the van der Waals (vdW) energy for base stacking over the 6-mer region centered around the potential ‘open’ site (red box in Figure 5B). The CCC/GGG and CGC/GCG duplexes had a 1.3 kcal/mol difference over the central 6-mer region: −80.3 ± 0.3 kcal/mol for CCC/GGG vs. −81.6 ± 0.2 kcal/mol for CGC/GCG (Figure S10D).
In sum, MD simulations showed that CCC/GGG had higher intrinsic slide, roll, and untwist compared to ideal B-DNA and more bending toward the ‘open’ conformation, accompanied by weaker van der Waals stacking energy compared with CGC/GCG. Such inherent distortions in free CCC/GGG DNA may lead to a higher propensity for the DNA to be ‘opened’ by Rad4 while CGC/GCG DNA could potentially resist such ‘opening’, which we further examined as described below.
3.5. MD simulations of initial binding between Rad4 and the two different DNA sequences.
Next, we asked how the DNA binding with Rad4 is directly impacted by the differences in the CCC/GGG and CGC/GCG sequences. For this purpose, we performed MD simulations on the initial binding process between the WT Rad4 with the CGC/GCG sequence (SI Methods) for comparison with our previously presented results for the CCC/GGG sequence [28]. For the CCC/GGG case, we also present new trajectory-derived data and analyses to pinpoint the sequence-dependent differences. Prior MD simulation studies with different NER lesions have identified several features that are common to lesions repaired efficiently by NER: upon initial binding with Rad4, these lesion-containing duplexes all exhibit significant untwisting, ready engagement of the BHD2 β−hairpin (β−hairpin2) with the DNA minor groove, and capture of a partner base into a groove at the BHD2/BHD3 interface [14, 22]. By contrast, NER-resistant lesions resisted such structural changes. In the current simulations, a stable BHD2 conformation in the minor groove was achieved at ~1 μs for the CCC/GGG sequence, and for both simulations the conformations of the complexes were stable afterwards (Figure S11). Hence, we took the 1 – 2 μs trajectories as the initial binding states for further characterization. The best representative structures of each ensemble are shown in Figure 6A and Movies S1–S2.
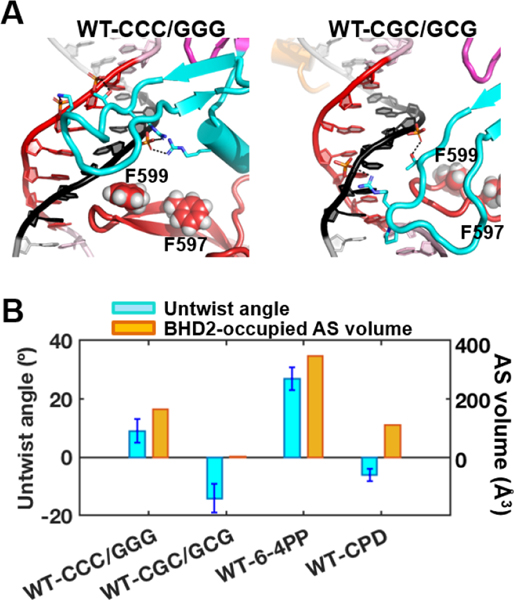
Fig.6. Initial binding of Rad4 to the CCC/GGG and CGC/GCG sequence-containing duplexes.
(A) Best representative structures upon initial binding of Rad4. The structures are shown in cartoon representation color-coded as in Figure 1. Heavy atoms of the BHD2 amino acid side chains and the potential open site’s DNA backbone phosphate groups that form hydrogen bonds are shown in sticks. Hydrogen bonds are shown as black dashed lines. Side chains of two Phe (F599 and F597) in the β−hairpin3 are shown in spheres. The structure for the CCC/GGG case is adapted from Figure 1A in [28]. (B) Potential ‘open’ site untwisting (6-mer) and BHD2 binding into the minor groove were quantified using the untwist angles and the BHD2-occupied AS volumes in the minor groove for the Rad4 binding. The values for the CCC/GGG sequence are from [28], and for the 6–4PP and CPD, they are from [22]. The CCC/GGG sequence shows significant BHD2 binding and modest untwisting, resembling the well-recognized and repaired 6–4PP. In contrast, the CGC/GCG sequence shows no BHD2 binding and slight over-twisting, reminiscent of the poorly recognized/repaired CPD. The standard deviations of block averaged means [110, 111] for the untwist angles are shown. Full details of the block averaging method are given in SI Methods.
First, we computed the untwist angle of the duplex DNA around the potential ‘open’ site (see SI Methods, Figure S12A) [14, 22]. The twist angles between the two base pairs that are 3bp away from either side of the putative ‘open’ site (end base pair of the 6-mer sequence, boxed red in Figure 5B) were computed along the trajectories; the change in the twist angle from that of the initial twist angle was used as a measure of untwist around the ‘opening’ site: Untwist = Twist initial –Twist. The untwist angle thus reflects the extent of untwisting upon achieving the initial binding state. Positive values indicate untwisting and negative values indicate over-twisting. Upon achieving the initial binding state, the CCC/GGG duplex showed modest untwist of 9 ± 4° [28], whereas the CGC/GCG duplex showed over-twisting of −14 ± 5°. The positive value with CCC/GGG thus tracks directionally with our previous result with the NER-proficient 6–4 photoproduct that showed 27 ± 4° of untwisting upon initial Rad4-binding; the value with CGC/GCG, on the other hand, is reminiscent of the NER-resistant CPD that showed slight overtwisting of −6 ± 2° (Figure 6B).
Next, the helix bend angle and the bend directions were analyzed for the same sequence region as in the unbound DNA simulations.
Accompanying the untwisting upon Rad4 initial binding, the CCC/GGG DNA showed a larger bend angle (23 ± 5°) than the CGC/GCG case (12 ± 2°); the latter showed essentially no change in the bend angle upon Rad4 initial binding, while the CCC/GGG bend angle increased notably upon Rad4 initial binding, being 13 ± 1° in unbound DNA (Figures S10B and S12B). Another feature of the CCC/GGG DNA is that its bend becomes further directed towards the ‘open’ structure direction (−66°), with a bend direction angle of −53 ± 7° [28], compared to −45 ± 8° for the unbound DNA. The bend direction of the CGC/GCG DNA (−17 ± 12°) is similar to that of the unbound DNA (−19 ± 10º) and oscillates greatly as it fails to be bound by the β−hairpin2, described below (Figures S10C and S12C).
In accordance with these structural differences in the two DNA duplexes, the CCC/GGG duplex exhibited weakened stacking for the 6-mer sequence, with van der Waals stacking interactions of −77.3 ± 1.3 kcal/mol, 3 kcal/mol higher than its unbound state (−80.3 ± 0.3 kcal/mol, Figures S10D and S12D). However, the CGC/GCG duplex had van der Waals stacking interactions of −81.5 ± 0.4 kcal/mol, unchanged from its unbound state (−81.6 ± 0.2 kcal/mol, Figures S10D and S12D).
Above all, the most conspicuous difference was in the way BHD2 engaged with the DNA. The aforementioned differences in the DNA bending and stacking energies could primarily arise from such differences in BHD2 binding. With CCC/GGG, the β-hairpin2 engaged with the DNA’s minor groove and the interaction was sustained (Movie S1). In contrast, the β-hairpin2 not only failed to engage with but also was expelled from the minor groove of CGC/GCG (Movie S2). These differences were further quantified by the BHD2-occupied alpha space (AS) volume [45]. The computed AS volume reflects the curvature and surface area of the DNA minor groove occupied by BHD2. The AS volume was 165 (Å3) with the CCC/GGG sequence [28] and 0 (Å3) with the CGC/GCG sequence. For comparison, the AS volumes with Rad4 were 349 (Å3) for 64PP and 110 (Å3) for CPD, respectively (Figure 6B) [22]. The representative structures also showed that β−hairpin2 formed four hydrogen bonds with the DNA backbones of CCC/GGG but only two with those of CGC/GCG (Figure 6A). We remark that the free CCC/GGG DNA had already exhibited significant untwisting and slide in a direction that was conducive to accommodating the incoming β-hairpin2 in the minor groove whereas CGC/GCG lacked such intrinsic structural features. Thus, again, such results indicate that the inherent structural properties of CCC/GGG likely promote the ‘opening’ by Rad4 shown in the crystal structures. For CGC/GCG DNA, the exclusion of BHD2 from the minor groove led BHD2 to reside partially in the major groove (Figure 6A), which could further hinder the insertion of the β−hairpin3 into the DNA major groove to form an ‘open’ structure (Figure S12E). These MD simulations are thus consistent with the crystal structures and FLT-FRET-based solution conformational studies and point to a significant resistance to ‘opening’ by the CGC/GCG sequence.
4. Discussion
4.1. Importance of sequence context on lesion recognition and NER.
What determines the repair efficiency of a DNA lesion has been a central question in DNA repair, especially for the NER process which handles an extraordinarily diverse array of lesions. NER-resistant lesions tend to be highly mutagenic, thus highlighting the importance of the variability in NER recognition/repair in understanding the mutagenicity of the lesions [16]. Previous studies have implicated various factors in the lesion recognition step of NER, including DNA conformations, lesion topology, stereochemistry, nature of the adducted base and sequence context [9, 10, 13, 46–55]. For many lesions, the more destabilizing and distorting a lesion is, the better it is recognized and repaired. For instance, the 6–4PP UV-lesion is more destabilizing, distorting and dynamic than CPD when present in a duplex DNA [56–63]. Accordingly, 6–4PP is much more efficiently recognized and repaired by XPC and NER than CPD [15, 48, 64, 65]. Similarly, the recognition and repair rates of thymine-thymine-linked CPD improve dramatically if the CPD is placed against mismatched dT’s instead of matched dA partners [15, 21]. We also previously showed that different mismatch sequences have varying Rad4-binding specificities, which also correlated with the extent of the distortions induced by the mismatches. For instance, the more distorted and conformationally heterogeneous CCC/CCC mismatch had a greater Rad4-recognition specificity than a TAT/TAT mismatch that was more B-DNA like and less heterogeneous. CCC/CCC also exhibited greater destabilization in thermal melting than TAT/TAT [32, 66].
In this study, we delved into a fundamental aspect of what determines such ‘openability’. Our previous observation that even a matched DNA (thus no lesion-induced thermal destabilization) could be ‘opened’ when locally tethered to Rad4 led us to conclude that the ‘open’ conformation was the thermodynamically most stable state for Rad4-bound DNA, whether damaged or not [22, 27, 28]. However, the present study revealed that the propensity and trajectory for ‘opening’ is sequence-dependent (Figure 7). When tethered to the Δβ-hairpin3, CCC/GGG DNA was able to form an ‘open-like’ conformation, while CGC/GCG DNA could not be opened; instead, it accommodated the protein bound in 180°-reversed manner, capping one end of the DNA duplex. As previously noted, this structure is the first of Rad4/XPC bound to DNA that shows a structure that is different from the ‘open’/’open-like’ conformation. Whether the protein would have adopted yet another binding mode if the duplex end was not accessible – for example with a longer DNA duplex – remains unknown. However, the important result here is that certain DNA sequences are more prone to being ‘opened’ than others. Additionally, this study further underscores that the ‘opening’ shown with CCC/GGG was not because the tethering limited the binding orientation of the protein on DNA, as the ‘reverse mode’ would have been allowed just the same if the DNA had failed to ‘open’.
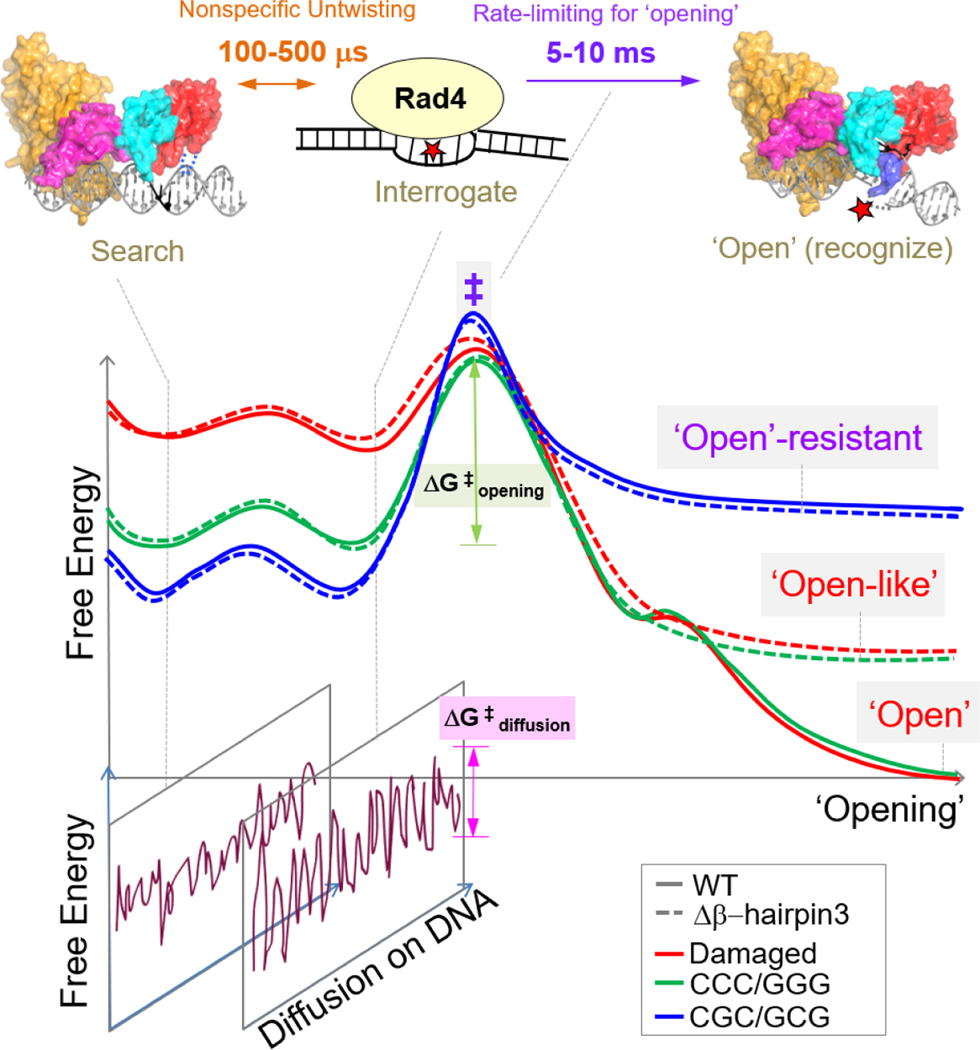
Fig.7.
Proposed DNA ‘opening’ trajectory and ‘kinetic gating’ mechanism of Rad4/XPC. The top panel illustrates distinct binding modes for Rad4/XPC as it searches for, interrogates, and recognizes a damaged site, and the time scales for fluctuations between these modes, based on prior studies [27, 29, 30]. The middle panel shows a schematic free-energy profile along the ‘opening’ trajectory. The faster 100- to 500-μs nonspecific untwisting step entails a smaller energetic barrier than the slower 5-to 10-ms rate-limiting step (‡) of the ‘opening’ process. The rate-limiting step involves sufficiently unwound and bent DNA but with the nucleotides not yet fully or stably flipped out into the BHD2/BHD3 groove [29]. The free energy barrier (ΔG‡ opening) for ‘opening’ damaged DNA (red) is naturally lower than that for undamaged DNA (green) as DNA damage usually destabilizes the B-DNA structure. For Rad4 mutants that are lacking either β-hairpin2 or β-hairpin3, the protein can still overcome ΔG‡ opening to form ‘open-like’ structures [27, 28]. The bottom panel illustrates that, for each step along the ‘opening’ trajectory, there is also a kinetically competing process of diffusion of Rad4/XPC along the DNA, characterized by ΔG‡ diffusion. For undamaged DNA, the high ΔG‡ opening compared with ΔG‡ diffusion favors the protein diffusing away before ‘opening’ a given site, while for damaged DNA this competition favors ‘opening’. When the diffusion of the protein is blocked (e.g., by tethering), DNA containing consecutive C/G’s (CCC/GGG) could be ‘opened’ indicating that the ΔG‡ opening is thermally surmountable and that the ‘open(-like)’ structure was thermodynamically the most stable structure for such DNA. However, this study showed that this is not the case for DNA containing alternating CG/GC repeats: for this DNA, the ‘open(-like)’ structure is no longer the most stable structure, and the DNA retains predominantly B-DNA like conformation even under tethered conditions. We note here that while the free energies of the transition state ensemble (‡) are shown to be approximately the same for all DNA constructs, we cannot rule out that this free energy gets successively larger as we go from damaged to ‘openable’ CCC/GGG to ‘open-resistant’ CGC/GCG.
Further insights into the fundamental factors determining the ‘opening’ propensities of different DNA sequences come from DNA melting studies. The melting temperatures of 24-bp DNA containing the CCC/GGG or CGC/GCG sequences used in our studies were comparable to each other (~77 vs. ~74 °C) when measured with UV absorbance changes (Figures S8); these melting temperatures report on the propensity for strand separation, thus global melting, which is a property of the entire DNA sequence. With FLT-FRET, we probed the temperature-induced changes in local DNA conformations in the vicinity of the CCC/GGG and the CGC/GCG sites and found that CCC/GGG exhibited local ‘softening’ and more readily lost its B-DNA conformation compared with CGC/GCG. Thus, the local sequence that could be opened by Rad4 was intrinsically more deformable. It is worth noting here that global thermal stability of a DNA duplex typically measured with UV absorbance does not always correlate with NER [16, 67, 68]. Indeed, the local thermal stability differences, as showcased here, may provide a better correlation with recognition and repair in NER, as they recapitulate the site-specific, local deformability or ‘openability’ of the DNA.
Critical insights into the structural mechanisms underpinning these observations are provided by MD simulations. In free DNA, CCC/GGG had higher intrinsic slide, roll and untwist and weaker base stacking energy than CGC/GCG or the ideal B-DNA while being more bent towards the ‘open’ structure. Upon the initial binding with WT Rad4, BHD2-induced untwisting led to the approach of β−hairpin3 to the major groove side for further opening of the CCC/GGG sequence, illustrating how Rad4 could take advantage of and amplify the intrinsically higher distortion/distortability and weaker van der Waals stacking energy in the CCC/GGG sequence. In contrast, CGC/GCG failed in untwisting and engaging with the BHD2 hairpin in the minor groove in the presence of Rad4, congruent with both crystal and solution studies showing predominantly ‘closed’ DNA even with tethered Rad4. In general, the CCC/GGG sequence recapitulated the features previously shown with NER-proficient lesions (such as the untwisting and the engagement of BHD2) while CGC/GCG recapitulated those of NER-impaired/resistant lesions such as CPD [14, 22]. However, unlike the bona fide NER lesions such as 6–4PP or a dibenzo[a,l]pyrene-derived dG lesion [14, 22], we did not observe partner base flipping for CCC/GGG, which indicates that the kinetics of DNA opening for the more stable, matched DNA must be slower than those of the 6–4PP lesion even under tethered conditions, consistent with the underlying assumptions of the ‘kinetic gating’ mechanism.
Nevertheless, this work provides key evidence that the sequence context can have a dramatic effect on the function of a DNA repair protein and compels us to take the precise sequence context into consideration when considering the repair propensity and mutagenicity of a lesion in genomic DNA. We posit that the sequences containing runs of G’s may provide a better platform for NER than those with alternating CG/GC repeats, especially when the lesion itself does not provide enough DNA destabilization/distortion (e.g., CPD versus 6–4PP). The number of consecutive G’s required to show impact needs to be determined, but previous MD studies have suggested that the sequence impact on B-DNA conformations and fluctuations likely stretch beyond nearest neighbors to include at least 4–5 base pairs [69, 70]. The sequence impact may also be present even with shorter repeats depending on what the flanking nucleotides are. Recent sequencing technologies that track the formation and excision of NER lesions in their precise genomic DNA contexts provide rich opportunities for further examination (e.g., CPD [71–74], 64PP [72–74], cisplatin-crosslinks [75, 76], and benzo[a]pyrene adducts [77]). Notably, in a recent study by Jiang et al., the NER excision ‘super hotspots’ for CPD (but not for 6–4PP) showed an enrichment in strings of C’s flanking the damaged sites in normal human fibroblasts mostly under GG-NER conditions [74]. It would be interesting to compare these sequence contexts of the repaired CPDs with those of unrepaired or slowly repaired CPDs in the genomic DNA.
4.2. Implications of the ‘openability’ of C/G-rich sequences with consecutive C/G’s.
Several recent studies have implicated XPC/Rad4 complexes in roles outside of the NER repair function, such as in base excision repair (reviewed in [78]; [79–81]) and transcription [81–87]. In light of our study, the sequence impacts we observed may also influence Rad4/XPC-DNA binding in its non-NER functions, especially in a situation where prolonged residence time is allowed (e.g., by interaction with a binding partner and/or by posttranslational modifications). Interestingly, we note that many of the gene regulatory DNA sequences reported to associate with Rad4/XPC possess a GGG-containing consensus sequence. In the study by Reed and colleagues, Rad4-Rad23 was shown to associate with STRE (Stress Response Element) promoter in the absence of UV light to regulate the transcription of several DNA damage repair signaling genes in yeast [88]. The STRE elements are present in the upstream region of many genes, induced under various stress conditions such as osmotic pressure, oxidative stress and heat, and they contain a run of G’s (AGGGG) [89, 90]. Also, previously, Tjian and colleagues reported that XPC serves as a stem cell coactivator required for OCT4/SOX2 transcriptional activation. Intriguingly, the consensus sequences of XPC/RAD23B colocalization includes KLF4 (nCCnCnCCCn) and SP1 (CCCCnCCCCC) [82, 84] that are also enriched with strings of G’s. Le May and colleagues recently reported that XPC colocalizes with RNA polymerase II in the absence of damage and functions as a co-activator for recruiting the ATAC transcription coactivator complex to promoters by interacting with E2F1 [87]. Interestingly, the E2F1 consensus sequence contains runs of G’s (NNGGCGGGAA, http://homer.ucsd.edu/homer/motif/HomerMotifDB/homerResults/E2F1.html).
4.3. Implications of DNA-end binding.
The ‘reverse mode’ structure solved with CGC/GCG featured Rad4 bound to a DNA duplex end. The duplex-end binding by Rad4/XPC has been suggested from previous biochemical and proteomics studies and was shown by atomic force microscopy [27, 49, 91–94], but this is the first time that a 3D structure was determined for such a binding mode. While DNA ends (as in double-strand break) may not be a lesion that Rad4/XPC is involved in repairing through NER in cells, we argue that the propensity to bind to a DNA end can shed light into the mechanism of Rad4/XPC. For instance, a duplex end can present itself as the DNA duplex is further opened (e.g, as a fork structure) during lesion verification in NER [95] or during transcription (see above). The end-bound structure also shows the range of motions accessible to Rad4 while bound to DNA, which may be important in its ability to scan along the DNA while interrogating for a damaged site [29, 30, 96]. In fact, DNA-end binding is common among DNA lesion-binding proteins, e.g., XPA [97, 98], UvrA [99, 100], MutS [101–104], UVDDB [94, 105], AGT [106], AlkA [107].
Conclusion:
Here we examined the structural features of Rad4-DNA using a combination of chemical tethering, x-ray crystallography, fluorescence lifetime-based DNA conformational analyses, as well as MD simulations. By examining DNA ‘opening’ under a condition where the kinetic residence time of the protein on a given DNA site is not a limiting factor, we unveiled how the DNA sequence contexts alone can critically influence the DNA ‘opening’ and ‘openability’ by Rad4/XPC. These findings may be important in explaining the variability in NER efficiencies for diverse lesions in the genomic DNA as well as in understanding mutational hot and cold spot sequences induced by specific environmental carcinogens [31, 108] and the roles of XPC/Rad4 beyond NER such as transcription and base excision repair.
Supplementary Material
Acknowledgments
We thank the staff of Advanced Photon Source LS-CAT beamline for the help with data collection. We also thank the members of the Min, Broyde and Ansari groups for their support.
Funding
National Science Foundation (NSF) [MCB-1412692 to J.-H.M., MCB-060037 to S.B., MCB1715649 to A.A.]; National Institutes of Health (NIH) [R21-ES028384 to J.-H.M, R01-ES025987 to S.B., HG006827 to C.H.]; Baylor University. Funding for open access charge: NSF [MCB1412692 to J.-H.M.]; NIH [R21-ES028384 to J.-H.M, R01-ES025987 to S.B.]. C.H. is a Howard Hughes Medical Institute Investigator.
Footnotes
Accession Numbers
Atomic coordinates and structure factors for the reported crystal structure of the Δβ-hairpin3-CGC/GCG complex have been deposited with the Protein Data Bank under accession number 6UG1.
Conflict of interest
The authors declare no conflict of interest.
References
- [1].Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T, DNA repair and mutagenesis, 2nd ed., ASM Press, Washington, D.C., 2006. [Google Scholar]
- [2].Ganesan A, Hanawalt P, Photobiological Origins of the Field of Genomic Maintenance, Photochem Photobiol, 92 (2016) 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mao P, Wyrick JJ, Roberts SA, Smerdon MJ, UV-Induced DNA Damage and Mutagenesis in Chromatin, Photochem Photobiol, 93 (2017) 216–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Scharer OD, Nucleotide excision repair in eukaryotes, Cold Spring Harb. Perspect. Biol, 5 (2013) a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gillet LC, Scharer OD, Molecular mechanisms of mammalian global genome nucleotide excision repair, Chem Rev, 106 (2006) 253–276. [DOI] [PubMed] [Google Scholar]
- [6].Tiwari V, Wilson DM 3rd, DNA Damage and Associated DNA Repair Defects in Disease and Premature Aging, Am. J. Hum. Genet, 105 (2019) 237–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kraemer KH, Patronas NJ, Schiffmann R, Brooks BP, Tamura D, DiGiovanna JJ, Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: a complex genotype-phenotype relationship, Neuroscience, 145 (2007) 1388–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Buterin T, Hess MT, Luneva N, Geacintov NE, Amin S, Kroth H, Seidel A, Naegeli H, Unrepaired fjord region polycyclic aromatic hydrocarbon-DNA adducts in ras codon 61 mutational hot spots, Cancer Res, 60 (2000) 1849–1856. [PubMed] [Google Scholar]
- [9].Cai Y, Patel DJ, Geacintov NE, Broyde S, Differential nucleotide excision repair susceptibility of bulky DNA adducts in different sequence contexts: hierarchies of recognition signals, J. Mol. Biol, 385 (2009) 30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kropachev K, Kolbanovskii M, Cai Y, Rodriguez F, Kolbanovskii A, Liu Y, Zhang L, Amin S, Patel D, Broyde S, Geacintov NE, The sequence dependence of human nucleotide excision repair efficiencies of benzo[a]pyrene-derived DNA lesions: insights into the structural factors that favor dual incisions, J. Mol. Biol, 386 (2009) 1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu Y, Reeves D, Kropachev K, Cai Y, Ding S, Kolbanovskiy M, Kolbanovskiy A, Bolton JL, Broyde S, Van Houten B, Geacintov NE, Probing for DNA damage with beta-hairpins: similarities in incision efficiencies of bulky DNA adducts by prokaryotic and human nucleotide excision repair systems in vitro, DNA Repair (Amst), 10 (2011) 684–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kropachev K, Kolbanovskiy M, Liu Z, Cai Y, Zhang L, Schwaid AG, Kolbanovskiy A, Ding S, Amin S, Broyde S, Geacintov NE, Adenine-DNA adducts derived from the highly tumorigenic Dibenzo[a,l]pyrene are resistant to nucleotide excision repair while guanine adducts are not, Chem. Res. Toxicol, 26 (2013) 783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cai Y, Patel DJ, Geacintov NE, Broyde S, Dynamics of a benzo[a]pyrene-derived guanine DNA lesion in TGT and CGC sequence contexts: enhanced mobility in TGT explains conformational heterogeneity, flexible bending, and greater susceptibility to nucleotide excision repair, J. Mol. Biol, 374 (2007) 292–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mu H, Zhang Y, Geacintov NE, Broyde S, Lesion Sensing during Initial Binding by Yeast XPC/Rad4: Toward Predicting Resistance to Nucleotide Excision Repair, Chem. Res. Toxicol, 31 (2018) 1260–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sugasawa K, Okamoto T, Shimizu Y, Masutani C, Iwai S, Hanaoka F, A multistep damage recognition mechanism for global genomic nucleotide excision repair, Genes Dev, 15 (2001) 507–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Geacintov NE, Broyde S, Repair-Resistant DNA Lesions, Chem. Res. Toxicol, 30 (2017) 1517–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lans H, Hoeijmakers JHJ, Vermeulen W, Marteijn JA, The DNA damage response to transcription stress, Nat. Rev. Mol. Cell Biol, 20 (2019) 766–784. [DOI] [PubMed] [Google Scholar]
- [18].Matsumoto S, Fischer ES, Yasuda T, Dohmae N, Iwai S, Mori T, Nishi R, Yoshino K, Sakai W, Hanaoka F, Thoma NH, Sugasawa K, Functional regulation of the DNA damage-recognition factor DDB2 by ubiquitination and interaction with xeroderma pigmentosum group C protein, Nucleic Acids Res, 43 (2015) 1700–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Matsumoto S, Cavadini S, Bunker RD, Grand RS, Potenza A, Rabl J, Yamamoto J, Schenk AD, Schubeler D, Iwai S, Sugasawa K, Kurumizaka H, Thoma NH, DNA damage detection in nucleosomes involves DNA register shifting, Nature, 571 (2019) 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yeh JI, Levine AS, Du S, Chinte U, Ghodke H, Wang H, Shi H, Hsieh CL, Conway JF, Van Houten B, Rapic-Otrin V, Damaged DNA induced UV-damaged DNA-binding protein (UV-DDB) dimerization and its roles in chromatinized DNA repair, Proc. Natl. Acad. Sci. U. S. A, 109 (2012) E27372746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Min JH, Pavletich NP, Recognition of DNA damage by the Rad4 nucleotide excision repair protein, Nature, 449 (2007) 570–575. [DOI] [PubMed] [Google Scholar]
- [22].Paul D, Mu H, Zhao H, Ouerfelli O, Jeffrey PD, Broyde S, Min JH, Structure and mechanism of pyrimidine-pyrimidone (6–4) photoproduct recognition by the Rad4/XPC nucleotide excision repair complex, Nucleic Acids Res, 47 (2019) 6015–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gunz D, Hess MT, Naegeli H, Recognition of DNA adducts by human nucleotide excision repair. Evidence for a thermodynamic probing mechanism, J. Biol. Chem, 271 (1996) 25089–25098. [DOI] [PubMed] [Google Scholar]
- [24].Geacintov NE, Broyde S, Buterin T, Naegeli H, Wu M, Yan S, Patel DJ, Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery, Biopolymers, 65 (2002) 202–210. [DOI] [PubMed] [Google Scholar]
- [25].Buterin T, Meyer C, Giese B, Naegeli H, DNA quality control by conformational readout on the undamaged strand of the double helix, Chem Biol, 12 (2005) 913–922. [DOI] [PubMed] [Google Scholar]
- [26].Maillard O, Camenisch U, Clement FC, Blagoev KB, Naegeli H, DNA repair triggered by sensors of helical dynamics, Trends Biochem Sci, 32 (2007) 494–499. [DOI] [PubMed] [Google Scholar]
- [27].Chen X, Velmurugu Y, Zheng G, Park B, Shim Y, Kim Y, Liu L, Van Houten B, He C, Ansari A, Min JH, Kinetic gating mechanism of DNA damage recognition by Rad4/XPC, Nat. Comm, 6 (2015) 5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Paul D, Mu H, Tavakoli A, Dai Q, Chen X, Chakraborty S, He C, Ansari A, Broyde S, Min JH, Tethering-facilitated DNA ‘opening’ and complementary roles of beta-hairpin motifs in the Rad4/XPC DNA damage sensor protein, Nucleic Acids Res, 48 (2020) 12348–12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Velmurugu Y, Chen X, Slogoff Sevilla P, Min JH, Ansari A, Twist-open mechanism of DNA damage recognition by the Rad4/XPC nucleotide excision repair complex, Proc. Natl. Acad. Sci. U. S. A, 113 (2016) E2296–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kong M, Liu L, Chen X, Driscoll KI, Mao P, Bohm S, Kad NM, Watkins SC, Bernstein KA, Wyrick JJ, Min JH, Van Houten B, Single-Molecule Imaging Reveals that Rad4 Employs a Dynamic DNA Damage Recognition Process, Mol. Cell, 64 (2016) 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kucab JE, Zou X, Morganella S, Joel M, Nanda AS, Nagy E, Gomez C, Degasperi A, Harris R, Jackson SP, Arlt VM, Phillips DH, Nik-Zainal S, A Compendium of Mutational Signatures of Environmental Agents, Cell, 177 (2019) 821–836 e816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chakraborty S, Steinbach PJ, Paul D, Mu H, Broyde S, Min JH, Ansari A, Enhanced spontaneous DNA twisting/bending fluctuations unveiled by fluorescence lifetime distributions promote mismatch recognition by the Rad4 nucleotide excision repair complex, Nucleic Acids Res, 46 (2018) 1240–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Otwinowski Z, Minor W, [20] Processing of X-ray diffraction data collected in oscillation mode, in: Carter Charles W. Jr. (Ed.) Methods Enzymol, Academic Press, 1997, pp. 307–326. [DOI] [PubMed] [Google Scholar]
- [34].Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS, Overview of the CCP4 suite and current developments, Acta Crystallogr. D Biol. Crystallogr., 67 (2011) 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot, Acta Crystallogr. D Biol. Crystallogr., 66 (2010) 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, Moriarty NW, Oeffner RD, Poon BK, Prisant MG, Read RJ, Richardson JS, Richardson DC, Sammito MD, Sobolev OV, Stockwell DH, Terwilliger TC, Urzhumtsev AG, Videau LL, Williams CJ, Adams PD, Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix, Acta Crystallogr D Struct Biol, 75 (2019) 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Steinbach PJ, Filtering artifacts from lifetime distributions when maximizing entropy using a bootstrapped model, Analytical Biochemistry, 427 (2012) 102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Steinbach PJ, Ionescu R, Matthews CR, Analysis of Kinetics Using a Hybrid Maximum-Entropy/Nonlinear-Least-Squares Method: Application to Protein Folding, Biophysical Journal, 82 (2002) 2244–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tavakoli A, Paul D, Mu H, Kuchlyan J, Baral S, Ansari A, Broyde S, Min J-H, Light-induced modulation of DNA recognition by the Rad4/XPC damage sensor protein, RSC Chemical Biology, In press (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Tavakoli A, Paul D, Mu H, Kuchlyan J, Baral S, Ansari A, Broyde S, Min JH, Light-induced modulation of DNA recognition by the Rad4/XPC damage sensor protein, RSC Chem Biol, 2 (2021) 523536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Preus S, Kilsa K, Miannay FA, Albinsson B, Wilhelmsson LM, FRETmatrix: a general methodology for the simulation and analysis of FRET in nucleic acids, Nucleic Acids Res, 41 (2013) e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gorin AA, Zhurkin VB, Olson WK, B-DNA twisting correlates with base-pair morphology, J. Mol. Biol, 247 (1995) 34–48. [DOI] [PubMed] [Google Scholar]
- [43].Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB, DNA sequence-dependent deformability deduced from protein-DNA crystal complexes, Proc. Natl. Acad. Sci. U. S. A, 95 (1998) 11163–11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Dlakic M, Harrington RE, Bending and torsional flexibility of G/C-rich sequences as determined by cyclization assays, J. Biol. Chem, 270 (1995) 29945–29952. [DOI] [PubMed] [Google Scholar]
- [45].Rooklin D, Wang C, Katigbak J, Arora PS, Zhang Y, AlphaSpace: Fragment-Centric Topographical Mapping To Target Protein-Protein Interaction Interfaces, J. Chem. Inf. Model, 55 (2015) 1585–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cai Y, Kropachev K, Xu R, Tang Y, Kolbanovskii M, Kolbanovskii A, Amin S, Patel DJ, Broyde S, Geacintov NE, Distant neighbor base sequence context effects in human nucleotide excision repair of a benzo[a]pyrene-derived DNA lesion, J. Mol. Biol, 399 (2010) 397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Mu H, Kropachev K, Wang L, Zhang L, Kolbanovskiy A, Kolbanovskiy M, Geacintov NE, Broyde S, Nucleotide excision repair of 2-acetylaminofluorene- and 2-aminofluorene-(C8)-guanine adducts: molecular dynamics simulations elucidate how lesion structure and base sequence context impact repair efficiencies, Nucleic Acids Res, 40 (2012) 9675–9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Batty D, Rapic’-Otrin V, Levine AS, Wood RD, Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites, J. Mol. Biol, 300 (2000) 275–290. [DOI] [PubMed] [Google Scholar]
- [49].Hey T, Lipps G, Sugasawa K, Iwai S, Hanaoka F, Krauss G, The XPC-HR23B complex displays high affinity and specificity for damaged DNA in a true-equilibrium fluorescence assay, Biochemistry, 41 (2002) 6583–6587. [DOI] [PubMed] [Google Scholar]
- [50].Kusumoto R, Masutani C, Sugasawa K, Iwai S, Araki M, Uchida A, Mizukoshi T, Hanaoka F, Diversity of the damage recognition step in the global genomic nucleotide excision repair in vitro, Mutat. Res, 485 (2001) 219–227. [DOI] [PubMed] [Google Scholar]
- [51].Reardon JT, Mu D, Sancar A, Overproduction, purification, and characterization of the XPC subunit of the human DNA repair excision nuclease, J. Biol. Chem, 271 (1996) 19451–19456. [DOI] [PubMed] [Google Scholar]
- [52].Bunick CG, Miller MR, Fuller BE, Fanning E, Chazin WJ, Biochemical and structural domain analysis of xeroderma pigmentosum complementation group C protein, Biochemistry, 45 (2006) 1496514979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Trego KS, Turchi JJ, Pre-steady-state binding of damaged DNA by XPC-hHR23B reveals a kinetic mechanism for damage discrimination, Biochemistry, 45 (2006) 1961–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ding S, Kropachev K, Cai Y, Kolbanovskiy M, Durandina SA, Liu Z, Shafirovich V, Broyde S, Geacintov NE, Structural, energetic and dynamic properties of guanine(C8)-thymine(N3) cross-links in DNA provide insights on susceptibility to nucleotide excision repair, Nucleic Acids Res, 40 (2012) 2506–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Cai Y, Patel DJ, Broyde S, Geacintov NE, Base sequence context effects on nucleotide excision repair, J Nucleic Acids, 2010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kim JK, Choi BS, The solution structure of DNA duplex-decamer containing the (6–4) photoproduct of thymidylyl(3’-->5’)thymidine by NMR and relaxation matrix refinement, Eur. J. Biochem, 228 (1995) 849–854. [DOI] [PubMed] [Google Scholar]
- [57].Kim JK, Patel D, Choi BS, Contrasting structural impacts induced by cis-syn cyclobutane dimer and (6–4) adduct in DNA duplex decamers: implication in mutagenesis and repair activity, Photochem Photobiol, 62 (1995) 44–50. [DOI] [PubMed] [Google Scholar]
- [58].Dehez F, Gattuso H, Bignon E, Morell C, Dumont E, Monari A, Conformational polymorphism or structural invariance in DNA photoinduced lesions: implications for repair rates, Nucleic Acids Res, 45 (2017) 3654–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kemmink J, Boelens R, Koning TM, Kaptein R, van der Marel GA, van Boom JH, Conformational changes in the oligonucleotide duplex d(GCGTTGCG) x d(CGCAACGC) induced by formation of a cis-syn thymine dimer. A two-dimensional NMR study, Eur. J. Biochem, 162 (1987) 37–43. [DOI] [PubMed] [Google Scholar]
- [60].McAteer K, Jing Y, Kao J, Taylor JS, Kennedy MA, Solution-state structure of a DNA dodecamer duplex containing a Cis-syn thymine cyclobutane dimer, the major UV photoproduct of DNA, J. Mol. Biol, 282 (1998) 1013–1032. [DOI] [PubMed] [Google Scholar]
- [61].Wang CI, Taylor JS, Site-specific effect of thymine dimer formation on dAn.dTn tract bending and its biological implications, Proc. Natl. Acad. Sci. U. S. A, 88 (1991) 9072–9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Park H, Zhang K, Ren Y, Nadji S, Sinha N, Taylor JS, Kang C, Crystal structure of a DNA decamer containing a cis-syn thymine dimer, Proc. Natl. Acad. Sci. U. S. A, 99 (2002) 15965–15970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Jing Y, Kao JF, Taylor JS, Thermodynamic and base-pairing studies of matched and mismatched DNA dodecamer duplexes containing cis-syn, (6–4) and Dewar photoproducts of TT, Nucleic Acids Res, 26 (1998) 3845–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Young AR, Chadwick CA, Harrison GI, Hawk JL, Nikaido O, Potten CS, The in situ repair kinetics of epidermal thymine dimers and 6–4 photoproducts in human skin types I and II, J. Invest. Dermatol, 106 (1996) 1307–1313. [DOI] [PubMed] [Google Scholar]
- [65].Canturk F, Karaman M, Selby CP, Kemp MG, Kulaksiz-Erkmen G, Hu J, Li W, Lindsey-Boltz LA, Sancar A, Nucleotide excision repair by dual incisions in plants, Proc. Natl. Acad. Sci. U. S. A, 113 (2016) 4706–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Chakraborty S, Visualizing Spontaneous DNA Dynamics and Its Role in Mismatch Recognition by Damage Repair Protein Rad4, Doctoral Thesis, (2018). [Google Scholar]
- [67].Cai Y, Geacintov NE, Broyde S, Nucleotide Excision Repair Efficiencies of Bulky Carcinogen-DNA Adducts Are Governed by a Balance between Stabilizing and Destabilizing Interactions, Biochemistry, 51 (2012) 1486–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hilton B, Gopal S, Xu L, Mazumder S, Musich PR, Cho BP, Zou Y, Dissociation Dynamics of XPCRAD23B from Damaged DNA Is a Determining Factor of NER Efficiency, PLoS One, 11 (2016) e0157784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Lavery R, Zakrzewska K, Beveridge D, Bishop TC, Case DA, Cheatham T 3rd, Dixit S, Jayaram B, Lankas F, Laughton C, Maddocks JH, Michon A, Osman R, Orozco M, Perez A, Singh T, Spackova N, Sponer J, A systematic molecular dynamics study of nearest-neighbor effects on base pair and base pair step conformations and fluctuations in B-DNA, Nucleic Acids Res, 38 (2010) 299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Pasi M, Maddocks JH, Beveridge D, Bishop TC, Case DA, Cheatham T 3rd, Dans PD, Jayaram B, Lankas F, Laughton C, Mitchell J, Osman R, Orozco M, Perez A, Petkeviciute D, Spackova N, Sponer J, Zakrzewska K, Lavery R, muABC: a systematic microsecond molecular dynamics study of tetranucleotide sequence effects in B-DNA, Nucleic Acids Res, 42 (2014) 12272–12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Mao P, Smerdon MJ, Roberts SA, Wyrick JJ, Chromosomal landscape of UV damage formation and repair at single-nucleotide resolution, Proc. Natl. Acad. Sci. U. S. A, 113 (2016) 9057–9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Hu J, Adar S, Selby CP, Lieb JD, Sancar A, Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution, Genes Dev, 29 (2015) 948–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Li W, Adebali O, Yang Y, Selby CP, Sancar A, Single-nucleotide resolution dynamic repair maps of UV damage in Saccharomyces cerevisiae genome, Proc. Natl. Acad. Sci. U. S. A, 115 (2018) E3408–E3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Jiang Y, Li W, Lindsey-Boltz LA, Yang Y, Li Y, Sancar A, Super-hotspots and -coldspots in the repair of UV-induced DNA damage in the human genome, J. Biol. Chem, (2021) 100581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Hu J, Lieb JD, Sancar A, Adar S, Cisplatin DNA damage and repair maps of the human genome at single-nucleotide resolution, Proc. Natl. Acad. Sci. U. S. A, 113 (2016) 11507–11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Yimit A, Adebali O, Sancar A, Jiang Y, Differential damage and repair of DNA-adducts induced by anti-cancer drug cisplatin across mouse organs, Nat. Comm, 10 (2019) 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Li W, Hu J, Adebali O, Adar S, Yang Y, Chiou YY, Sancar A, Human genome-wide repair map of DNA damage caused by the cigarette smoke carcinogen benzo[a]pyrene, Proc. Natl. Acad. Sci. U. S. A, 114 (2017) 6752–6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Kumar N, Raja S, Van Houten B, The involvement of nucleotide excision repair proteins in the removal of oxidative DNA damage, Nucleic Acids Res, 48 (2020) 11227–11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Shimizu Y, Iwai S, Hanaoka F, Sugasawa K, Xeroderma pigmentosum group C protein interacts physically and functionally with thymine DNA glycosylase, EMBO J, 22 (2003) 164–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Shimizu Y, Uchimura Y, Dohmae N, Saitoh H, Hanaoka F, Sugasawa K, Stimulation of DNA Glycosylase Activities by XPC Protein Complex: Roles of Protein-Protein Interactions, J Nucleic Acids, 2010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Ho JJ, Cattoglio C, McSwiggen DT, Tjian R, Fong YW, Regulation of DNA demethylation by the XPC DNA repair complex in somatic and pluripotent stem cells, Genes Dev, 31 (2017) 830–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Cattoglio C, Zhang ET, Grubisic I, Chiba K, Fong YW, Tjian R, Functional and mechanistic studies of XPC DNA-repair complex as transcriptional coactivator in embryonic stem cells, Proc. Natl. Acad. Sci. U. S. A, 112 (2015) E2317–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Zhang ET, He Y, Grob P, Fong YW, Nogales E, Tjian R, Architecture of the human XPC DNA repair and stem cell coactivator complex, Proc. Natl. Acad. Sci. U. S. A, 112 (2015) 14817–14822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Fong YW, Inouye C, Yamaguchi T, Cattoglio C, Grubisic I, Tjian R, A DNA repair complex functions as an oct4/sox2 coactivator in embryonic stem cells, Cell, 147 (2011) 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Le May N, Mota-Fernandes D, Velez-Cruz R, Iltis I, Biard D, Egly JM, NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack, Mol. Cell, 38 (2010) 54–66. [DOI] [PubMed] [Google Scholar]
- [86].Compe E, Egly JM, Nucleotide Excision Repair and Transcriptional Regulation: TFIIH and Beyond, Annu Rev Biochem, 85 (2016) 265–290. [DOI] [PubMed] [Google Scholar]
- [87].Bidon B, Iltis I, Semer M, Nagy Z, Larnicol A, Cribier A, Benkirane M, Coin F, Egly JM, Le May N, XPC is an RNA polymerase II cofactor recruiting ATAC to promoters by interacting with E2F1, Nat. Comm, 9 (2018) 2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Zhou Z, Humphryes N, van Eijk P, Waters R, Yu S, Kraehenbuehl R, Hartsuiker E, Reed SH, UV induced ubiquitination of the yeast Rad4-Rad23 complex promotes survival by regulating cellular dNTP pools, Nucleic Acids Res, 43 (2015) 7360–7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Yoshikawa K, Furusawa C, Hirasawa T, Shimizu H, Genome-wide analysis of the effects of location and number of stress response elements on gene expression in Saccharomyces cerevisiae, J. Biosci. Bioeng, 106 (2008) 507–510. [DOI] [PubMed] [Google Scholar]
- [90].Estruch F, Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast, FEMS Microbiol Rev, 24 (2000) 469–486. [DOI] [PubMed] [Google Scholar]
- [91].Lee YC, Cai Y, Mu H, Broyde S, Amin S, Chen X, Min JH, Geacintov NE, The relationships between XPC binding to conformationally diverse DNA adducts and their excision by the human NER system: is there a correlation?, DNA Repair (Amst), 19 (2014) 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Janicijevic A, Sugasawa K, Shimizu Y, Hanaoka F, Wijgers N, Djurica M, Hoeijmakers JH, Wyman C, DNA bending by the human damage recognition complex XPC-HR23B, DNA Repair (Amst), 2 (2003) 325–336. [DOI] [PubMed] [Google Scholar]
- [93].Roche Y, Zhang D, Segers-Nolten GM, Vermeulen W, Wyman C, Sugasawa K, Hoeijmakers J, Otto C, Fluorescence correlation spectroscopy of the binding of nucleotide excision repair protein XPChHr23B with DNA substrates, J Fluoresc, 18 (2008) 987–995. [DOI] [PubMed] [Google Scholar]
- [94].Berthelot V, Mouta-Cardoso G, Hegarat N, Guillonneau F, Francois JC, Giovannangeli C, Praseuth D, Rusconi F, The human DNA ends proteome uncovers an unexpected entanglement of functional pathways, Nucleic Acids Res, 44 (2016) 4721–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Kokic G, Chernev A, Tegunov D, Dienemann C, Urlaub H, Cramer P, Structural basis of TFIIH activation for nucleotide excision repair, Nat. Comm, 10 (2019) 2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Cheon NY, Kim HS, Yeo JE, Scharer OD, Lee JY, Single-molecule visualization reveals the damage search mechanism for the human NER protein XPC-RAD23B, Nucleic Acids Res, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Koch SC, Kuper J, Gasteiger KL, Simon N, Strasser R, Eisen D, Geiger S, Schneider S, Kisker C, Carell T, Structural insights into the recognition of cisplatin and AAF-dG lesion by Rad14 (XPA), Proc. Natl. Acad. Sci. U. S. A, 112 (2015) 8272–8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Beckwitt EC, Jang S, Carnaval Detweiler I, Kuper J, Sauer F, Simon N, Bretzler J, Watkins SC, Carell T, Kisker C, Van Houten B, Single molecule analysis reveals monomeric XPA bends DNA and undergoes episodic linear diffusion during damage search, Nat. Comm, 11 (2020) 1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Wagner K, Moolenaar G, van Noort J, Goosen N, Single-molecule analysis reveals two separate DNA-binding domains in the Escherichia coli UvrA dimer, Nucleic Acids Res, 37 (2009) 1962–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Wagner K, Moolenaar GF, Goosen N, Role of the two ATPase domains of Escherichia coli UvrA in binding non-bulky DNA lesions and interaction with UvrB, DNA Repair (Amst), 9 (2010) 1176–1186. [DOI] [PubMed] [Google Scholar]
- [101].Tessmer I, Yang Y, Zhai J, Du C, Hsieh P, Hingorani MM, Erie DA, Mechanism of MutS searching for DNA mismatches and signaling repair, J. Biol. Chem, 283 (2008) 36646–36654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Yang Y, Sass LE, Du C, Hsieh P, Erie DA, Determination of protein-DNA binding constants and specificities from statistical analyses of single molecules: MutS-DNA interactions, Nucleic Acids Res, 33 (2005) 4322–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Wang H, Yang Y, Schofield MJ, Du C, Fridman Y, Lee SD, Larson ED, Drummond JT, Alani E, Hsieh P, Erie DA, DNA bending and unbending by MutS govern mismatch recognition and specificity, Proc. Natl. Acad. Sci. U. S. A, 100 (2003) 14822–14827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Beckwitt EC, Kong M, Van Houten B, Studying protein-DNA interactions using atomic force microscopy, Semin Cell Dev Biol, 73 (2018) 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Ghodke H, Wang H, Hsieh CL, Woldemeskel S, Watkins SC, Rapic-Otrin V, Van Houten B, Single-molecule analysis reveals human UV-damaged DNA-binding protein (UV-DDB) dimerizes on DNA via multiple kinetic intermediates, Proc. Natl. Acad. Sci. U. S. A, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Tessmer I, Melikishvili M, Fried MG, Cooperative cluster formation, DNA bending and baseflipping by O6-alkylguanine-DNA alkyltransferase, Nucleic Acids Res, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Zhao B, O’Brien PJ, Kinetic Mechanism for the Excision of Hypoxanthine by Escherichia coli AlkA and Evidence for Binding to DNA Ends, Biochemistry, 50 (2011) 4350–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, Ye K, Alipaz J, Bauer MJ, Beare D, Butler A, Carter RJ, Chen L, Cox AJ, Edkins S, Kokko-Gonzales PI, Gormley NA, Grocock RJ, Haudenschild CD, Hims MM, James T, Jia M, Kingsbury Z, Leroy C, Marshall J, Menzies A, Mudie LJ, Ning Z, Royce T, SchulzTrieglaff OB, Spiridou A, Stebbings LA, Szajkowski L, Teague J, Williamson D, Chin L, Ross MT, Campbell PJ, Bentley DR, Futreal PA, Stratton MR, A comprehensive catalogue of somatic mutations from a human cancer genome, Nature, 463 (2010) 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Lu XJ, Olson WK, 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures, Nucleic Acids Res, 31 (2003) 5108–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Flyvbjerg H, Petersen HG, Error estimates on averages of correlated data, The Journal of Chemical Physics, 91 (1989) 461–466. [Google Scholar]
- [111].Yang W, Bitetti-Putzer R, Karplus M, Free energy simulations: use of reverse cumulative averaging to determine the equilibrated region and the time required for convergence, J. Chem. Phys, 120 (2004) 2618–2628. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.