Abstract
The histone acetyltransferase p300 (also known as KAT3B) is a general transcriptional coactivator that introduces the H3K27ac mark on enhancers triggering their activation and gene transcription. Genome-wide screenings demonstrated that a large fraction of long non-coding RNAs (lncRNAs) plays a role in cellular processes and organ development although the underlying molecular mechanisms remain largely unclear (1,2). We found 122 lncRNAs that interacts directly with p300. In depth analysis of one of these, lncSmad7, is required to maintain ESC self-renewal and it interacts to the C-terminal domain of p300. lncSmad7 also contains predicted RNA-DNA Hoogsteen forming base pairing. Combined Chromatin Isolation by RNA precipitation followed by sequencing (ChIRP-seq) together with CRISPR/Cas9 mutagenesis of the target sites demonstrate that lncSmad7 binds and recruits p300 to enhancers in trans, to trigger enhancer acetylation and transcriptional activation of its target genes. Thus, these results unveil a new mechanism by which p300 is recruited to the genome.
INTRODUCTION
The transcriptional coactivator p300 (also known as KAT3B) identified as adenovirus E1A-associated 300-kD protein (3) and its paralog, the cyclic-AMP response element binding protein (CBP, also known as KAT3A) isolated as a coactivator of the transcription factor CREB (4) are key histone acetyltransferases (HAT) required for the acetylation of chromatin at enhancers and promoters (5). They are large multidomain proteins playing important roles in cell development and in signal transduction pathways. Both p300 and CBP interact with, and acetylate, hundreds of proteins including chromatin modifiers and transcription factors (6–8) whose dimerization has been shown to activate p300 by trans-autoacetylation (9).
CBP and p300 are ubiquitously expressed while the enhancers of developmental genes are differentially regulated in different cell types. Thus, CBP and p300 activities must be finely regulated by recruitment to their target enhancers and by activation of their enzymatic activity. Interestingly, CBP bound to enhancers has been shown to interact with enhancer RNAs (eRNAs) via its HAT domain to activate its catalytic activity (10).
Long non-coding RNAs (lncRNAs) are a class of non-translated polyadenylated transcripts longer than 200 nucleotides that have been implicated in a plethora of diverse cellular processes (11–15). Although initially thought to act mainly on the site of their transcription in in cis, many lncRNAs have been shown to bind to different sites on the genome (16,17), interact with transcription factors or chromatin modifiers to regulate transcription (17–25) and lncRNAs exhibit cell type- and developmental stage-specific expression patterns (26,27). For these reasons, lncRNAs represent ideal candidates as general regulators of enhancers of developmental genes.
To understand the contribution of lncRNAs to the function of p300 we screened the p300-RNA interactome in embryonic stem cells (ESCs) by UV photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP). We found that p300 interacts directly with many different lncRNAs. We further demonstrate that one of them lncSmad7 recruits p300 to the genome by the formation of lncRNA-DNA triple-helix in trans in proximity of enhancer regions. By coupling chromatin isolation by RNA precipitation followed by sequencing (ChIRP-seq) with transcriptomic data we identified a number of lncSmad7 direct target genes. Our results provide evidences that lncRNAs are involved in p300 targeting to the genome, hence providing a novel molecular mechanism for its binding specificity.
MATERIALS AND METHODS
Cell culture
Mouse embryonic stem cells (ESCs) (E14, genotype: male) were cultured on gelatin-coated plates (0.,1% gelatin, Sigma-Aldrich) in high-glucose DMEM (Invitrogen) supplemented with 15% heat-inactivated FBS (Millipore), 0.1 mM NEAA (Invitrogen), 1 mM Sodium pyruvate (Invitrogen), 0.1 mM 2-mercaptoethanol, 1500 U ml−1 LIF (GeneSpin), 25 U ml−1 penicillin and 25 μg ml−1 streptomycin (28)
lncSmad7–/– generation
lncSmad7 KO clone was generated by using a CRISPR-Cas9-based approach for inserting polyA signal. A donor plasmid containing a 5′HA with bGH polyA signal into exon3, the NeoR cassette and a 3′HA was built by cloning PCR fragments of lncSmad7 genomic region from ESCs, into a modified version of PGKneolox2DTA plasmid (Addgene #13443). Briefly, a 1142 bp 3′HA (from +1460 to +2601 of lncSmad7 gene) was amplified with primers Fwd3′HA and Rev3′HA and cloned NheI/SalI into the PGKneolox2DTA vector. The 5′HA was generated by cloning into a fragment of 1078 bp (+195/+1272) containing at its 3′end the HindIII and NdeI restriction sites (primers Fwd5′HA-part1/Rev5′HA-part1), a 400 bp fragment engineered to contain a 5′ HindIII site, the bGH polyA signal, a 193 bp region lncSmad7 gene (from +1267 to +1459) and a 3′ NdeI site, by using an assembling vector (TOPO-TA invitrogen). The latter fragment was obtained by overlap extension PCR between a PCR product corresponding to the 193 bp homology region (Primers Fwd5′HA-part2/Rev5′HA-part2) and a bGH polyA sequence amplified with a reverse primer containing a 5′end complementary the homology region (primers FwdPolyA/RevPolyA). The complete 5′HA fragment was then cloned MluI/NdeI into the 3′HA-PGKneolox2DTA vector. The plasmid containing sgRNA targeting to the third exon of lncSmad7 was designed using the CRISPR Design Tool (http://crispr.mit.edu/). Oligonucleotides corresponding to the two strands the sgRNA were annealed and cloned into the BbsI-digested gRNA backbone (BB) previously cloned into TOPO™TA vector (Invitrogen) (Addgene #42335). These plasmids, together with the Cas9-containing plasmid (Addgene #41815), were co-transfected into ESCs using Lipofectamine Transfection 2000 Reagent (Thermo Fisher), according to manufacturer's protocol. ESCs were selected with G418 (Sigma) after 48 h and seeded as a single cell and following expansion. Clones with the desired insertion were confirmed by PCR, followed by Sanger sequencing.
Mutant TTS generation
sgRNAs targeting the TTS were designed using the GPP sgRNA Design tool (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design). Mutant TTS clones of Id1 (#1.1, #1.2) and Mllt11 (#2.1, #2.2) were generated by using a CRISPR/Cas9-mediated homologous recombination with donor PCR fragments containing a deletion of the Id1 and Mllt11 TTS: 18 and 25 bp, respectively. The deleted sequences were analyzed by HOMER motif analysis (http://homer.ucsd.edu) and by inspection of the ENCODE transcription factor binding (44). These analysis did not identify motifs for DNA binding factors. Fragments of the genomic region from ESCs adjacent to target TTS were amplified by PCR using overlapping oligos and joined by extended PCR. These plasmids, together with the Cas9-containing plasmid (Addgene #41815), were co-transfected into ESCs using Lipofectamine Transfection 2000 Reagent (Thermo Fisher), according to manufacturer's protocol. Id1 and Mllt11 mutant TTS clones were identified by PCR and confirmed by sequencing.
DNA constructs and shRNA
For lncSmad7 expression, full-length lnSmad7 (1–2687) and lncSmad7 fragments (part 1: 1–831; part 2: 832–1734; part 3: 1735–2687) were cloned into pCCLsin.PPT.hPGK vector. Mutant plasmids of lncSmad7 in triplex forming regions (TFS#1 and TFS#2) were generated by QuikChange II XL Site-Directed Mutagenesis kit from lncSmad7 part 3 (1735–2687). Smad7 expression vector was kindly provided by Xin-Hua Feng's laboratory (29). p300 domains (N-terminal, HAT, C-terminal) were cloned into pcDNA3.1 vector in frame with N-term Flag tag. Short hairpin RNA (shRNA) constructs were designed using the TRC hairpin design tool (http://www.broadinstitute.org/rnai/public/seq/search), choosing the hairpin sequences provided in Supplementary Table S4. Oligonucleotides were cloned into pLKO.1 vector (Addgene #10878). Transient transfections of the constructs were performed using Lipofectamine 2000 Transfection Reagent (Invitrogen) according to the manufacturer's protocol.
Protein extraction and western Blot
For total extracts, the cells were resuspended in F-buffer (10 mM Tris–HCl pH 7.0, 50 mM NaCl, 30 mM Na-pyrophosphate, 50 mM NaF, 1% Triton X-100, protease inhibitors). Nuclear extracts were performed as described. Briefly, ESCs were lysed in Isotonic buffer (20 mM HEPES pH 7.5, 100 mM NaCl, 250 mM Sucrose, 5 mM MgCl2, 5 mM ZnCl2) supplemented with 1% NP40. The isolated nuclei pellets were resuspended in F-buffer and sonicated for 3 pulses (3 cycles, 30 s on/off). Extracts were quantified using BCA assay (Pierce) and were run on SDS-PAGE gels in Biorad Mini-PROTEAN chambers, according to the manufacturer's protocol. Gels were transferred to nitrocellulose membranes, blocked in 5% milk in TBST for 1 h at RT rocking platform and incubated with specific primary antibodies overnight 4°C, followed by 5 times washes with TBST and probed with secondary antibody for 1 h at RT and later developed by using ECL reagent (GE Healthcare Amersham).
PAR-CLIP
ESCs, ∼200 × 106, were acutely treated for 16 h with 100 μM 4-thioridine (4-SU) (Sigma) and were cross-linked using 150 mJ/cm2 of 365 nm UV light in a UVP crosslinker (Analytic Jena) as described (30,31). PAR-CLIP experiments were carried out from nuclear extracts by using Isotonic buffer. The isolated nuclei pellets were lysed on ice with NP40 Lysis buffer (150 mM KCl, 50 mM HEPES pH 7.5, 2 mM EDTA, 1 mM NaF, 0.5% NP40, 0.5 mM DTT, protease inhibitors). The cleared lysates were subjected to SUPERase-In digestion using RNase A/T1 cocktail enzyme mix (Ambion) for 3 min at 37°C to improve IP efficiency. The cross-linked p300-RNA complexes are immunoprecipitated with p300 antibody (Millipore) bound to BSA blocked protein G-Dynabeads (Life technologies). Beads were washed 3 times with IP wash buffer (300 mM KCl, 50 mM HEPES, pH 7.5, 0,05% NP40, 0.5 mM DTT, SUPERase-In, protease inhibitors) and resuspended in dephosphorylation buffer by adding Recombinant Shrimp Alkaline Phosphatase (rSAP, NEB) for 15 min at 37°C to dephosphorylate the RNA. The beads were washed with Phosphatase wash buffer (50 mM Tris–HCl, pH 7.5, 20 mM EGTA, 0.5% NP40, SUPERase-In, protease inhibitors) and T4-PNK buffer (50 mM NaCl, 50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 5 mM DTT, SUPERase-In, protease inhibitors) and were incubated with non-radioactive ATP. The protein–RNA complexes were separated by Novex Bis–Tris 4–12% polyacrylamide gel (Invitrogen) for 55 min at 220 V. p300-RNA bands that correspond to the expected size of p300 revealed in Input were excised from the nitrocellulose membrane, eluted by incubating with RNA elution buffer (50 mM NaCl, 50 mM HEPES pH7.0, 1 mM EDTA, 2 mM CaCl2, 1% SDS, SUPERase-In) and digested with Proteinase K (4 mg/ml, NEB) and 3.5 M urea for 30 min at 55°C. RNA was purified using acidic phenol/chloroform (ThermoFisher) followed by ethanol precipitation.
PAR-CLIP library preparation
PAR-CLIP libraries from two independent experiments were prepared using NEB next small RNA library set for Illumina sequencing (NEB), according to manufacturers instructions. To remove potential PCR duplicates we used Unique Molecular Identifier (UMI) during library preparation. The libraries were checked by Bioanalyzer (Agilent) for quality.
PAR-CLIP analysis
After de-multiplexing, sequencing reads were processed using UMI-tools v1.0.0 (32) in order to remove adaptor sequences, keep the small-RNA insert and extract the UMI, which was removed from the read sequence and appended to the read name. A minimum UMI length of 6 nucleotides and a maximum of three mismatches for the complete search pattern was allowed (command: umi_tools extract –extract-method = regex –bc-pattern = ‘.+(?P < discard_1 > AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC(?P < discard_2 > .{7})){s< = }(?P < umi_1 > .{6})(?P < discard_3 > .+)’). Reads not containing the UMI sequence (i.e. too long small-RNA insert) were excluded. These processed reads were aligned to mouse reference genome (mm10/GRCm38) using Bowtie v1.2.3 (33) (options: -S -a -v 2 -m 10 –best –strata -q). Aligned reads were next de-duplicated using the UMI-tools dedup utility (default parameters)], exploiting the UMI sequence present in read name. The resulting unique alignments were used to find protein-RNA interaction sites with PARalyzer v1.5 (34), requiring a minimum read length of 13 nt, a minimum count of five reads for generating groups, clusters and kernel density estimation (KDE); a minimum cluster size of 8 nucleotides; a minimum conversion count of 3 T > C conversion for at least one conversion site; allowing at most one non conversion mismatch; filtering reads overlapping repetitive elements. The remaining parameters were set with default values. Intersection of PAR-CLIP clusters with gene loci and genomic regions (derived from our custom gene annotation) was made in the order: exons > introns > promoters (1 kb upstream of TSS) > intergenic using in-house Perl and R scripts. Genome-wide PAR-CLIP signals were computed as read per millions (RPM) using the deepTool (35) bamCoverage utility (options: –binSize 1 –normalizeUsing CPM) and summarized over genomic regions using the deepTools multiBigwigSummary utility. All replicates, controls and pooled datasets were processed identically. boxplots and scatterplots were made using custom R scripts.
RNA fluorescent in situ hybridization (RNA-FISH)
RNA-FISH analysis was performed with multiple 5′-biotinylated probes (Eurofins MWG Operon) (Supplementary Table S4). 4 × 104 WT and lncSmad7 KO ESCs were plated on 0.1% gelatin-coated glass slide chambers. Cells were fixed in 4% paraformaldehyde and permeabilized with 0.5% TRITON X-100. Cells were then saturated in saturation buffer (1% BSA, 1 μg yeast tRNA) for 2 h at room temperature. Cells were hybridized at 37°C for 3 h with 30 ng biotinylated probes in hybridization buffer (2× SSC, 25% formamide, 100 mg/ml dextran) in a humidified chamber protected from light. After incubation, RNA probes were revealed using Streptavidin Alexa Fluor 488 conjugate (Life Technologies). 0.5 mg/ml DAPI was used to visualize cell nuclei and the images were acquired using Leica TCS SP5 confocal microscope. ESCs treated with RNase A (50 μg/ml) for 30 min at 37°C prior to the hybridization step, were used as control samples.
RNA antisense purification (RAP)
RAP was performed according to the procedure described (18) using 40 × 106 ESCs. Briefly, ESCs were cross-linked using 0.8 J/cm2 of 254 nm UV light in a UVP crosslinker (Analytic Jena). Nuclear extracts were prepared as described in Native RIP protocol. lncSmad7 pull-down was performed using antisense biotinylated ssDNA probes (90 mer) that tile across the length of the target lncSmad7 (Supplementary Table S4) or lacZ as control in Hybridization Buffer (10 mM Tris–HCl pH 7.5, 5 mM EDTA, 500 mM LiCl, 0.2% SDS, 0.1% sodium deoxycholate, 4 M urea, SUPER-In, protease inhibitors) at 67°C for 2 h. The capture was carried out by Streptavidin-coupled Dynabeads (Invitrogen) followed by 5 times washes. Beads were resuspended in Benzonase elution buffer and the lncSmad7 associated nuclear proteins were precipitated with 10% of tricholoroacetic acid (TCA) and cold acetone. Samples were analyzed by Western Blot with p300 antibody (Millipore).
RNA in vitro transcription
Templates for in vitro transcription reactions were PCR amplified using AccuPrime Taq DNA Polymerase (Invitrogen) with a T7 Promoter Primer and confirmed to be the expected size by agarose gel electrophoresis. In vitro transcription reactions were carried out using a T7-FlashScribe Transcripion Kit (CellScript) with biotin–16-UTP (Roche) for 1 h at 37°C. DNA templates were then removed by digestion with DNase I for 20 min at 37°C. RNA probes were purified using acidic phenol/chloroform (ThermoFisher) and ethanol precipitated. Purified RNA probes were quantified by Qubit (ThermoFisher) and checked by Bioanalyser (Agilent).
RNA pull-down assay
Biotinylated lncSmad7 and lacZ fragments were refolded in vitro in 3.3× Folding Buffer (333 mM HEPES pH 7.9; 333 mM NaCl) at 37°C for 10 min before addition of 100 mM MgCl2 and additional incubation at 37°C for 20 (36). The lncSmad7 was divided in three biotinylated fragments: part 1 (1–831), part 2 (832–1734) and part 3 (1735–2687). The RNAs were incubated at 60°C for 10 min and slowly cooled to 4°C. For the pull-down assay, nuclear extracts from 6 × 106 ESCs were incubated with 2 μg of biotinylated RNA and 40 μl of streptavidin-coupled Dynabeads (Invitrogen) in RNA pull-down buffer (150 mM KCl, 25 mM Tris–HCl pH 7.4, 0.5 mM DTT, 0.5% NP40, 50 U ml−1 SUPERase-In, protease inhibitors) for 2 h at 4°C. Beads were washed with RNA pull-down buffer for 3 times. RNA-associated proteins were eluted and detected by western Blot.
RNA-seq analysis
Total RNA from WT, lncSmad7 and lncSmad7 KO ESCs transfected with either lncSmad7 (condition R) or Smad7 was isolated using TRIzol reagent (Invitrogen), according to the manufacturer's protocol.
Quantity and quality of the starting RNA were checked by Qubit and Bioanalyzer (Agilent). ∼2 μg of total RNA were subjected to poly(A) selection, and libraries were prepared using the TruSeq RNA Sample Prep Kit (Illumina) following the manufacturer's instructions. Sequencing was performed on the Illumina NextSeq 500 platform. Sequencing reads were aligned to mouse reference genome (UCSC mm10/GRCm38) using STAR v2.7.7a0 (37) (with parameters –outFilterMismatchNmax 999 –outFilterMismatchNoverLmax 0.04) and providing a list of known splice sites extracted from GENCODE M25 comprehensive annotation. Gene expression levels were quantified with featureCounts v1.6.3 (38) (options: -t exon -g gene_name) using GENCODE M25 basic gene annotation. Multi-mapped reads were excluded from quantification. Gene expression counts were next analyzed using the edgeR package (39). Normalization factors were calculated using the trimmed-mean of M-values (TMM) method (implemented in the calcNormFactors function) and RPKM were obtained using normalized library sizes and gene length. After filtering lowly expressed genes (below 1 CPM in four or more samples), differential expression analysis was carried out by fitting a GLM to all groups and performing LF test for the interesting pairwise contrasts. Genes were considered as significantly differentially expressed when having log FC > 0.5 and FDR < 0.05 in each relevant comparison. The putative direct targets of lncSmad7 were identified using the KO condition as reference and selecting genes significantly upregulated (i.e. log FC > 0.5 and FDR < 0.05) in the WT-KO contrast, significantly upregulated in R-KO and excluding genes that were significantly upregulated in KO_Smad7-KO. An average expression greater than 1 RPKM in WT condition was also required. RPKM values were scaled as Z-scores across samples before computing distances. Gene expression heatmaps were generated using the ComplexHeatmap R package (40).
Chromatin immunoprecipitation assay (ChIP)
Approximately 20 × 106 ESCs were cross-linked by addition of formaldehyde to 1% for 10 min at RT, quenched with 0.125 M glycine for 5 min at RT. Nuclear extracts were prepared as described in Native RIP protocol and were sonicated using the Bioruptor Twin (Diagenode) (20 cycles, 30s on/off) at high-power setting in SDS ChIP Buffer (20 mM Tris–HCl pH 8.0, 10 mM EDTA, 0.5% SDS and protease inhibitors). Cell lysate was diluted with ChIP dilution buffer (20 mM Tris–HCl pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton) before the immunoprecipitation with 2 μg of Histone H3, acetyl-K27 antibody (Abcam) or 2 μg of p300 antibody (Millipore) overnight at 4°C on a rotator. Protein–DNA complexes were immobilized on protein G-Dynabeads (Life technologies) and washed 6 times with RIPA buffer (500 mM LiCl, 50 mM HEPES–KOH pH 7.6, 1 mM EDTA, 1% NP-40, 0.7% Na-deoxycholate). The de-crosslinking was performed at 65°C overnight. De-crosslinked DNA was purified using QiaQuick PCR Purification Kit (Qiagen), according to the manufacturer's instruction. For sequencing of H3K27ac ChIP, ∼10 ng of purified ChIP DNA from two replicates of WT and two replicates of lncSmad7 KO ESCs were end-repaired, dA-tailed, and adaptor-ligated using the NEBNext ChIP-seq Library Prep Master Mix Set (NEB), according to the manufacturer's instructions. The H3K27ac ChIP-seq data and DNA from p300 ChIP were analyzed by ChIP-qPCR using SYBR GreenER kit (Invitrogen), on target genomic regions (Supplementary Table S4). qPCR reactions were performed on a Rotor-Gene Q 2plex HRM Platform (Qiagen, 9001560). The data are expressed as a percentage of the DNA Input.
ChIP-seq analysis
Following quality controls (performed with FastQC v0.11.2), sequencing reads were processed with Trim Galore! v0.5.0 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore) to perform quality and adapter trimming (parameters: –stringency 3 –q 20). Trimmed reads were next analyzed with the ENCODE Transcription Factors and Histone Modifications ChIP-seq pipeline 2 (v1.6.1, available from https://github.com/ENCODE-DCC/chip-seq-pipeline2), using default software and parameter settings for the ‘Histone Modifications’ processing mode. Briefly, reads were aligned to the mouse reference genome (UCSC mm10) using Bowtie2 (33). Duplicated, multi-mapping and poor-quality alignments were discarded, and peak calling was performed using MACS2 (41), using the input DNA as control library. Signal tracks were generated as fold enrichment over control for both individual and pooled replicates using MACS2. Both WT and lncSmad7 KO (KO) sample groups were processed identically.
Differentially H3K27-acetylated regions between KO and WT samples were identified using DiffBind (v3.0.3) (42), starting from a merged set of peaks obtained from overlapped peaks between biological replicates in each group (overlap idr conservative thresholded peaks from ENCODE pipeline) and the processed alignments for both IP and control samples. The following parameters were used for normalization and statistical analysis: normalize = DBA_NORM_LIB, library = DBA_LIBSIZE_PEAKREADS, background = BKGR_FALS, AnalysisMethod = DBA_EDGER. Peaks with FDR ≤ 0.05 and log FC < 0 were considered as significantly de-acetylated regions in KO samples as compared to WT.
Chromatin isolation by RNA purification (ChIRP)
ChIRP experiments were performed using the described protocol (43). A total of 48 antisense oligonucleotide biotinylated probes against lncSmad7 were designed with Stellaris Probe Designer and were split for the ChIRP experiments in two subsets (Odd and Even). LacZ-specific probes were used as negative controls in ChIRP-qPCR. Briefly, 5 × 107 lncSmad7-overexpressing ESCs and lncSmad7 KO cells were cross-linked with 1% formaldehyde for 10 min at RT and processed to obtain nuclear extracts as described in Native RIP protocol. The isolated nuclei pellets were lysed with 1% SDS Lysis buffer (50 mM Tris pH 7.0, 10 mM EDTA, 1% SDS, 1 mM DTT, protease inhibitors). The lysate was sonicated using the Bioruptor Twin (Diagenode) (20 cycles, 30 s on/off) and incubated with biotinylated probes in hybridization buffer (500 mM NaCl, 1% SDS, 50 mM Tris–HCl, pH 7.0, 1 mM EDTA, 15% formamide, protease inhibitors cocktail, PMSF and RNase inhibitor) for 4 h at 37°C with shaking. After hybridization and washes with ChIRP wash buffer (2× SSC, 0.5% SDS, 1 mM DTT, protease inhibitors), the complexes were recovered by incubation with MyOne Streptavidin C1 Dynabeads (Invitrogen) in a total volume of 1 ml and associated DNA fragments from 900 ul of bead samples were eluted with a cocktail of RNase A (Sigma-Aldrich) and RNase H (Invitrogen) at 37°C in DNA elution buffer (50 mM NaHCO3, 1% SDS, 200 mM NaCl) followed by reverse crosslinked at 65°C overnight, acidic phenol/chloroform (ThermoFisher) extraction and ethanol precipitated. Eluted DNA was subject to qPCR and high-throughput sequencing. In parallel, RNA was isolated in a buffer with 95% formamide and 10 mM EDTA pH 8.0 for 10 min at 90°C followed by TRIzol extraction (Invitrogen), from 100 ul of bead samples. Eluted RNA was subject to RT-qPCR analysis as reported for the detection of enriched transcript (Supplementary Table S4). ChIRP-qPCR from mutant TTS clones of Id1 and Mllt11 compared to the WT were carryout using SYBR GreenER kit (Invitrogen), on target genomic regions (Supplementary Table S4). qPCR reactions were performed on a Rotor-Gene Q 2plex HRM Platform (Qiagen, 9001560). The data are expressed as a percentage of the DNA input.
ChIRP-seq library preparation
ChIRP libraries from two biological replicates were prepared from ∼20 ng of purified ChIRP DNA using NEBNext® Ultra™ II DNA Library Prep Kit for Illumina (NEB), according to manufacturer's instructions and were checked by Bioanalyzer (Agilent) for quality.
ChIRP-seq analysis
Following adaptor/base-quality trimming, reads shorter than 60 bp were discarded. Processing of even and odd ChIRP-seq samples was performed with the ENCODE pipeline 2 (v1.6.1,https://github.com/ENCODE-DCC/chip-seq-pipeline2) as described for ChIP-seq data, using the default software and parameters settings for ‘Transcription Factors’ mode (i.e. narrow peak signals) and the UCSC mm10 reference genome. To determine a consensus set of lncSmad7 genomic binding sites conserved in both the ‘even’ and ‘odd’ libraries, the following strategy was adopted: first, peak calling was performed in all the replicated lncSmad7 WT and KO samples using MACSv2 (41), with parameters: –nomodel –shift 0 –extsize 200 -q 0.001, using deduplicated, quality-filtered alignments and the respective input RNA as control. Next, a consensus set of binding regions was determined by taking overlapping peak calls across replicates and excluding peaks called in at least one KO sample (bedtools v2.29.2 intersect utility). Finally, a peak quality filter was applied, retaining peaks that were covered by more than 20 reads in all WT samples as the final set of lncSmad7 binding sites. The annotation and genomic distribution of lncSmad7 peaks was analyzed using the Genomic Association Test (GAT) software testing for association with histone modifications (retrieved from ENCODE for mESC cell line E14) and the annotated list of Candidate cis-Regulatory Elements (cCRE) for mESC cell line E14 available from the SCREEN database (44). Associations with Q-value <0.05 were considered as statistically significant. Heatmaps for signal visualization were generated using deeptools v3.3.1 (33).
Triplex forming predictions
Triplexator version 1.3.2 (45) was used (with parameters -l 10 -L -1 -e 20 -E -1 -fm 0) to find predicted TPX involving the RNA of lncSmad7 and the DNA of each ChIRP-seq peak. Randomized sequences where obtained starting from ChIRP peaks with bedtool shuffle, excluding true ChIRP peaks and exons as reported in GENCODE M25 basic gene annotation.
Triplexator, triplexes and LongTarget comparison
The comparison of Triplexator with other triplex prediction softwares was performed. The algorithm Triplexes (46) produced identical results. Indeed, the authors of triplexes designed the algorithm using the same strategy of Triplexator. A software developed with independent and different strategies was then considered, namely LongTarget (47). Out of 9313 ChIRP peaks 81% of them show a triplexator TTS with score greater or equal than 9, the same percentage of peaks is predicted to have a triplex with mean stability ≥3.25 by LongTarget. The two software are in great agreement (P-value < 2e–16, Fisher's Exact Test). The parameters used were: -ni 10 -ds 8 -lg 10 for longTarget and -l 10 -e 20 -L 1 -E -1 -fm 0 for Triplexator.
Gene–regulatory region association and identification of lncSmad7 targets
To identify putative regulatory regions controlled by lncSmad7, the merged set of H3K27ac marked regions (see ChIP-seq analysis) with a ChIRP-seq peaks. The closest ChIRP-seq peak was then linked to each H3K27ac defined region, and the association to their target genes was next performed considering both proximal and distal loci. In particular, for each gene, the transcription start sites of all transcripts (TSSs) plus distal promoter interacting regions (PIRs) reported in (48) were retrieved, and a regulatory region (H3K27ac plus ChIRP-seq peak) was associated to all genes having a TTS or PIR within 10 kb. The lncSmad7-target genes were then retrieved from the list of lncSmad7 transcriptionally regulated genes (see RNA-seq analysis), considering those genes having a regulatory region with reduced levels of H3K27ac in KO (FDR ≤ 0.05 and log FC < 0, see ‘ChIP-seq analysis’) and a TPX forming score of at least 8 for their respective ChIRP-seq peak (see ‘Triplex forming predictions’).
In vivo dimethyl sulfate (DMS) probing and library preparation
Targeted DMS-MaPseq analysis of lncSmad7 was carried out as previously described (35) with slight modification. ESCs were resuspended in 1 ml of RNA probing buffer (RPB) (50 mM HEPES pH 7.9; 140 mM NaCl; 3 mM KCl) and pre-equilibrated at 37°C for 5 min. After diluting DMS 1:6 in 100% ethanol, it was added to a final concentration of 100 mM. After gentle vortexing, probing was allowed to proceed at 37°C for 2 min with moderate shaking. Reactions were quenched by addition of DTT to a final concentration of 0.7 M. Cells were collected by centrifugation and total RNA extracted with TRIzol reagent (Invitrogen). After phase separation, the aqueous phase was added to 1 ml of 100% ethanol and the RNA purified on RNA Clean & Concentrator-5 columns (Zymo Research, #R1016). For denatured control samples, RNA was directly extracted from ESCs pellets with TRIzol and treated with DMS after denaturation at 95°C for 1 min, in a buffer containing 55% formamide. Samples were then purified on RNA Clean & Concentrator-5 columns. Reverse transcription (RT) of total DMS-probed RNA was performed using the TGIRT-III enzyme (InGex, #TGIRT50). 3 μg of total RNA, 1 μl 10 mM dNTPs and 0.5 μl of a 50 μM pool of the specific reverse primers (Supplementary Table S4) were added, and the mixture was heated to 70°C for 5 min and immediately placed on ice for 1 min. 2 μl of 5× RT buffer (250 mM Tris–HCl pH 8.3; 375 mM KCl; 15 mM MgCl2, DTT to 5 mM, 10 U SUPERase-In and 100 U TGIRT-III) were added. Reverse transcription was then allowed to proceed at 25°C for 5 min, followed by 2 h at 57°C. RNA was degraded by adding 1 μl 5 M NaOH and heating to 95°C for 3 min and cDNA was subsequently purified on RNA Clean & Concentrator-5 columns. The resulting cDNAs were then specifically amplified via PCR. The sequence of lncSmad7 was divided into seven (partially overlapping) tiling fragments, for which specific forward and reverse primers were used. Primer pairs were split into two subsets (odd and even), designed in such a way that the amplicons obtained from the odd set would cover the pairing regions of the primers from the even set, and vice versa. Odd and even fragments (#1, #3, #5, #7 and #2, #4, #6, respectively) were separately combined into equimolar pools, and fragmented by sonication to yield fragments in the range of 100–300 bp. 10 ng of sonicated DNA was subjected to library preparation using NEBNext ChIP-Seq Library Prep Master Mix Set for Illumina (New England Biolabs, #E6240L) according to manufacturer's instructions, but omitting size selection.
DMS-MaPseq data analysis
Analysis of DMS-MaPseq data has been conducted using RNA Framework v2.6.9 (49) (https://github.com/dincarnato/RNAFramework). Reads pre-processing and mapping has been performed using the rf-map module (parameters: -ctn -cmn 0 -cqo -cq5 20 -b2 -mp ‘–very-sensitive-local’). Reads were trimmed of terminal Ns and low-quality bases (Phred < 20). Reads with internal Ns were discarded. Mapping was performed using the ‘very-sensitive-local’ preset of Bowtie2. Mutations were then counted using the rf-count module (parameters: -m -na -md 3 -ni). A mask file containing the sequences of primer pairing regions was passed along (through the -mf parameter). Generated RC files, from both the even and odd sets, containing per base mutation counts and coverage, were then combined in a single RC file using the rf-rctools module (mode: merge). Data normalization was independently performed for the in vivo and denatured control samples using the rf-norm module (parameters: -sm 4 -nm 2 -n 1000), by setting the minimum base coverage to 1000×. The final reactivity was then calculated as in (49), by dividing the winsorized in vivo reactivities over the winsorized denatured reactivities. Structure modeling was then performed using the rf-fold module, as previously described (36).
In vitro binding RNA–DNA triplex assay
The in vitro triplex pull-down assay of lncSmad7 was performed as described lncRNAs (50). 1 pmol of biotin-labeled lncSmad7 RNA region (1735–2687 nt) were incubated with 100 fmoles of PCR fragments (Supplementary Table S4) in hybridization buffer (20 mM KCl, 10 mM Tris–HCl pH 7.5, 10 mM MgCl2, 0.05% Tween 20 and SUPERase-In) for 1 h at room temperature. RNA-DNA complexes were then incubated with streptavidin-coated Dynabeads (Life technologies) for 2 h at 37°C, immobilized and washed 3 times with Wash Buffer (15 mM KCl, 10 mM Tris pH 7.5, 5 mM MgCl2). Beads were resuspended in wash buffer with RNaseA and incubated 30 min at 37°C. RNA-associated DNA was analyzed by qPCR using SYBR GreenER kit (Invitrogen) on a Rotor-Gene Q 2plex HRM Platform (Qiagen, 9001560) by using target regions (Supplementary Table S4).
EMSA
0.2 pmol of biotin-labeled dsDNA (annealed oligos) of the TTS#1 or TTS#2 region were incubated with 80, 40 molar excess of respective lncSmad7 fragments showing the TFS#1 and TFS#2 in Triplex Forming Buffer (25 mM NaCl, 10 mM Tris–HCl pH 7.5, 10 mM MgCl2) for 2 h at 25°C. Complexes were run on 10% TBE gels containing 10 mM MgCl2, 60 min, 150 V and developed using Chemiluminescent Nucleic Acid Detection Module Kit (89880).
RT-qPCR analysis
Total RNA was extracted using TRIzol reagent (Invitrogen) and quantified by Nanodrop (Thermo Scientific). Real-Time PCR (RT-qPCR) was performed as previously described (51) using the SensiFAST SYBR NO-ROX One-Step (BIOLINE) following the manufacturer's instructions. Briefly, RT-qPCR reactions were performed on a Rotor-Gene Q 2plex HRM Platform (Qiagen, 9001560) and relative gene expression levels were determined using calculated concentration values, normalized to β-actin and Gapdh as reference genes. Primers used were reported in Supplementary Table S4.
Statistic and reproducibility
Sample size n refers to the number of independent experiments or biological replicates, shown as dots, as indicated in the figure legends. GraphPad PRISM 8 software was used for statistical analysis. All error bars represent the standard deviation (SD). Statistical tests include Student's t-test and ANOVA test. P-values are reported in the plots (*P < 0.05; **P < 0.005; ***P < 0.0005, ****P < 0.0001). All experiments were performed independently at least two times, as indicated.
Antibodies
Antibodies were purchased from Abcam (anti-histone H3 acetyl K27 ab4729), from Millipore (anti-p300 05-257 and anti-vinculin SAB4200080); from Thermo Fisher Scientific (anti-Smad7 42-0400); normal Mouse IgG 12-371, anti-Flag M2 F3165; anti- β-actin A5441).
RESULTS
p300 binds lncRNAs in ESCs
To identify direct interaction of p300 with non-coding RNAs we performed p300 PAR-CLIP with 4-thiouridine (4-SU) incorporation (10,30,31) sequencing p300 PAR-CLIP libraries from two biological replicates (Supplementary Figure S1A–C). After removing potential PCR duplicates generated during library preparation by using a Unique Molecular Identifier (UMI) (Supplementary Figure S1D), p300-interacting regions were identified by peak calling using the PARalyzer method (34) and reads bearing T-to-C transitions. The p300 PAR-CLIP biological replicates showed a strong signal correlation (Pearson's coefficient = 0.98 for common regions between replicates) (Supplementary Figure S1E). Our analysis identified 122 lncRNAs with signal significantly higher in exons, which corresponds of 10,1% of all p300 associated transcripts (Figure 1A, Supplementary Figure S1F and Supplementary Table S1). The interacting domain revealed by PAR-CLIP showed a median size of 19 bp (Supplementary Figure S1G).
Figure 1.
p300 C-terminal domain binds lncRNAs in ESCs. (A) PAR-CLIP pie chart showing the categories of p300-associated transcripts in the nucleus of ESCs. (B) lncSmad7 RAP-enriched p300 in ESCs. Western blot showing the levels of endogenous p300 pulled down with streptavidin beads by using lncSmad7 antisense biotinylated oligonucleotides. A set of antisense biotinylated probes against lacZ serves as negative control. Bars represent the mean and SD of n = 2 independent experiments. P values calculated against control condition for each experiment by using t-test (*P < 0.05; **P < 0.005; ***P < 0.0005, ****P < 0.0001). (C) Pull-down assays reporting the lncSmad7 association with p300. lncSmad7 and lacZ RNA fragments were biotinylated by in vitro transcription, refolded, and incubated with ES nuclear lysates. Top: schematic representation of lncSmad7 fragments used in the RNA-pull down assay. Bottom: Western blot of p300 in nuclear samples pulled-down by lncSmad7 biotinylated fragments. lacZ transcript serves as an internal control. (D) IGV screenshot of lncSmad7 transcript from two pooled p300 PAR-CLIP biological replicates in ESCs. The box highlights the interaction site indicating the p300 binding sequence. p300 and IgG PAR-CLIP are scaled to the same level. The histone modifications mark the lncSmad7 promoter and gene body. H3K4me3 and H3K36me3 are from ENCODE ChIP dataset. The lncSmad7 sequence interacting with p300 is reported in blue from PARalyzer analysis. (E) Schematic representation of p300 structure and the Flag-tagged constructs used in the RNA-protein interaction assay. (F) Flag-tagged RIP of p300 domains in ESCs followed by RT-qPCR. lncSmad7 association with p300 N-terminal (1–1047 aa), p300 HAT (1048–1663 aa), p300 C-terminal (1664–2414 aa). Gapdh used as negative control. The analysis is normalized to ESCs transfected with empty Flag overexpression vectors (Flag-mock). Bars represent the mean and SD of n = 4 independent experiments. P values calculated against Flag-mock condition for each experiment by using ANOVA test (*P < 0.05; **P < 0.005; ***P < 0.0005, ****P < 0.0001). Primers used are reported in Supplementary Table S4.
The 3′ end of lncSmad7 interacts with the C-terminal domain of p300
Among the p300 interacting lncRNAs, we focused our attention on lncSmad7, a lncRNA previously shown to be induced by TGF-β pathway in mammary epithelial cell lines and included in a functional screening by knockdown to identify lincRNAs acting in the circuitry of pluripotency and differentiation (14,52,53). We further validated the interaction of lncSmad7 with p300 by RNA antisense purification coupled to Western-blot (RAP-WB) (18) in vivo (Figure 1B and Supplementary Figure S1H). The pulldown by streptavidin pulldown using in vitro biotinylated fragments incubated with ESC nuclear extracts showed that lncSmad7 interacts with its last exon and the PAR-CLIP analysis identified the p300 direct interacting sequence within the 3′ end of lncSmad7 (Figure 1C, D).
To map p300 domains responsible for the interaction with lncSmad7, we cloned p300 into three Flag-tagged segments, encompassing the three main domains (N-terminal, HAT, and C-terminal), that we expressed in ESCs. This analysis showed that lncSmad7 interacts with the C-terminal domain of p300 (Figure 1E, F and Supplementary Figure S2A). Taken together th above results demonstrate that lncSmad7 3′ end interacts with p300 on its C-terminal domain.
lncSmad7 is a chromatin associated lncRNA involved in ESC pluripotency
RNA-fluorescence in situ-hybridization (RNA-FISH) revealed that lncSmad7 is a predominantly nuclear lncRNA (Figure 2A). In agreement with the RNA-FISH, biochemical subcellular fractionation confirmed the nuclear localization of lncSmad7, further revealing that it is mainly enriched in the chromatin fraction (Figure 2B).
Figure 2.

Chromatin-enriched lncSmad7 is involved in ESC pluripotency. (A) RNA fluorescent in situ hybridization (RNA-FISH) of lncSmad7 in ESCs. lncSmad7 KO ESCs serves as negative control. Representative confocal images are depicted as a composite image of green channel (lncSmad7) and blue channel (DAPI) as well as merged channels. DAPI serves as a nuclear counterstain. Scale bar: 10 μm. (B) Expression analysis of lncSmad7 in subcellular fractionation of ESCs by RT-qPCR. Percentage ratio of lncSmad7 in chromatin (green), nuclear (blue) and cytoplasmic (gray) over the whole total lncSmad7 expression levels represents the distribution of lncSmad7 in ESCs. Internal controls of subcellular fractionation are U1 for nucleus and β-actin for cytoplasm. Bars represent the mean and SD of n = 3 independent experiments. Primers used are reported in Supplementary Table S4. (C) Alkaline phosphatase (AP) staining of WT, lncSmad7 KO cells and rescued ESC colonies. Shown rescued condition refers to lncSmad7 KO cells transiently transfected with the full-length lnSmad7 cloned into pCCLsin.PPT.hPGK vector. Top: Representative images of clonal assay performed in ESCs. Bottom: quantification of AP positive and AP negative cells. Bars represent the mean and SD of n = 3 independent experiments. P values calculated against WT condition for each experiment by using ANOVA test (*P < 0.05; **P < 0.005; ***P < 0.0005, ****P < 0.0001). (D) Immunostaining for the pluripotency marker Oct4 in WT, lncSmad7 KO ESCs and rescued ESCs. Shown rescued condition refers to lncSmad7 KO cells transiently transfected with the full-length lnSmad7 cloned into pCCLsin.PPT.hPGK vector. Representative images of two independent experiments as shown.
To further analyze lncSmad7 role in ESCs we generated a knockout by inserting a poly(A) signal within the third exon of lncSmad7, to lead to premature termination of transcription and to avoid the perturbation of putative regulatory elements within the region (Supplementary Figure S2B, C).
Upon knockout of lncSmad7, ESCs adopted an epithelial-like morphology, with loss of cell-cell contacts, reduction of AP-positive staining and loss of the core pluripotency factor Oct4 (Figure 2C, D). To verify whether this phenotype is due to the lack of lncSmad7 we performed a control rescuing experiment by the transient transfection of a plasmid expressing the full-length lncSmad7 confirming it plays a regulatory role in ESCs.
lncSmad7 recruits p300 to the upstream Smad7 enhancer
As most lncRNAs regulate the transcription of their neighbouring genes in cis, we first asked whether lncSmad7 would affect the expression of its nearby genes.
Analysis of differentially expressed genes within 1 Mb of the lncSmad7 locus in WT versus knockout cells revealed that Smad7 was the only gene of this region to be negatively affected by the loss of lncSmad7 (Figure 3A and Supplementary Figure S3A). Importantly, Smad7 expression was fully rescued by the ectopic lncSmad7 expression from a plasmid, demonstrating that lncSmad7 exerts its activating function in its mature form (Figure 3A). Short hairpin RNA-mediated silencing of lncSmad7 provided orthogonal validation that ectopic lncSmad7 RNA regulates Smad7 expression (Supplementary Figure S3B).
Figure 3.
lncSmad7 regulates Smad7 expression by forming RNA–DNA triplexes. (A) Heatmap representing the expression levels of protein-coding genes within lncSmad7 locus (1 Mb) in WT, lncSmad7 KO and rescued ESCs from the RNA-seq data. Shown rescued condition refers to lncSmad7 KO ESCs transiently transfected with the full-length lnSmad7 cloned into pCCLsin.PPT.hPGK vector. Expression values from two biological replicates (#1 and #2) are represented as CPM. Scale of red indicates the expression levels. (B) IGV profiles of lncSmad7 and Smad7 transcripts associated with histone modification mapped reads in ESCs. lncSmad7 and Smad7 RNA-seq profiles in lncSmad7 KO cells and in transfected ESCs with lncSmad7 (KO + lncSmad7) compared to WT ESCs from two biological replicates. The H3K27ac ChIP-seq profiles represent the pooled of two independent experiments from WT and lncSmad7 KO conditions, respectively. The polyA insertion site in the third exon of lncSmad7 is highlighted in green. The TTS are indicated in red bars. All the conditions are scaled to the same level. The Smad7 enhancer region is highlighted by red box. RNA-seq and ChIP-seq of H3K27ac are from the present work and are scaled to the same group levels. ChIP-seq of H3K4 methylations, DNase-seq and p300 are from ENCODE. TTS, Triplex Target DNA sites. (C) Electrophoretic Mobility Shift Assays (EMSA) showing the mobility of lncSmad7 TFS#1 (left) and TFS#2 (right) with biotin-labeled dsDNA probes according to Triplexator prediction. For each TFS are shown: incubation of increasing amounts (80- and 40- fold molar excess) of lncSmad7 TFS#1 and TFS#2 with biot-dsDNA probes containing triplex target sites (TTS), respectively. Reactions with biot- dsDNA probes and a 40-molar excess of TFS treated with 0.5 U RNase H (H) or with 0.5 ng RNase A (A). TFS, Triplex Forming Site. (D) Quantification by qPCR of lncSmad7-associated DNA via Hoogsteen base pairing. Schematic view of transcribed biotinylated last 1kb of lncSmad7 (1735–2687 nt) harboring Triplex Forming Site (TFS), TFS#1 and TFS#2, used to capture the indicated DNA double-stranded region near the Smad7 enhancer. Biotinylated RNA used as negative control highlighted in gray. The dashed arrow lines represent the DNA regions: the DNA containing the TTS not treated and generated in the presence of deaza-7-dATP and deaza-7-dGTP and an intronic region as negative control. Bars represent the mean and SD of n = 4 independent experiments. P values calculated against NT condition for each experiment by using ANOVA test (*P < 0.05; **P < 0.005; ***P < 0.0005, ****P < 0.0001). Primers used are reported in Supplementary Table S4. (E) Schematic representation of WT and TFS mutants (mutTFS#1 and mutTFS#2) of lncSmad7. Black boxes represent exons, gray box indicates the part3 of lncSmad7 (1735–2687 nt) showing the validated TFS#1 and TFS#2 with the WT sequences in black, mutated nucleotides in blue. (F) qPCR analysis of p300 ChIP experiments in WT, lncSmad7 KO and rescued ESCs. lncSmad7 KO ESCs are transiently transfected with WT lncSmad7 fragment (1735–2687 nt, part3) and TFS mutants (mut TFS#1 and mut TFS#2) of part3 (1735–2687 nt). The data are expressed as a percentage of the DNA inputs. Bars represent the mean and SD of n = 4 independent experiments. P values calculated against WT condition for each experiment by using ANOVA test (*P < 0.05; **P < 0.005; ***P < 0.0005, ****P < 0.0001). Primers used are reported in Supplementary Table S4. (G) RT-qPCR analysis showing the Smad7 levels after ectopic expression of mut TFS#1 and mut TFS#2 of lncSmad7 (1735–2687 nt, part3) in lncSmad7 KO ESCs. The analysis is normalized to β-actin as reference gene and on the WT condition. Bars represent the mean and SD of n = 4 independent experiments. P values calculated against control condition for each experiment by using ANOVA test (*P < 0.05; **P < 0.005; ***P < 0.0005, ****P < 0.0001). Primers used are reported in Supplementary Table S4.
Analysis of the epigenetic context of Smad7 region, revealed the presence of an enhancer residing within the seventh intron of lncSmad7, characterized by presence of H3K4me1 and H3K27ac marks, absence of H3K4me3, and coordinated binding of several pluripotent transcription factors such as Oct4, Sox2, Nanog, Stat3, as well as p300 (54–57) (Supplementary Figure S3C). The knockout of lncSmad7 resulted into a loss of p300 binding and H3K27ac levels on this enhancer that were rescued by the ectopic expression of lncSmad7 (Figure 3B, Supplementary Figure S3D). Taken together the above results demonstrate that mature lncSmad7 transcript is required to activate the acetylation of the enhancer and the transcription of Smad7 gene.
lncSmad7 binds the DNA next to Smad7 enhancer by forming RNA–DNA triplex
To get insights into the molecular mechanism underlying the lncSmad7-dependent activation of Smad7 transcription, we first mapped, by deletions, the minimal lncSmad7 region able to rescue Smad7 expression within the 3′-most 952 nt on the lncSmad7 (Supplementary Figure S3E). Next, we analyzed lncSmad7 in vivo RNA structure by targeted RNA antisense purification combined with dimethyl sulfate (DMS) mutational profiling (36,58). Surprisingly, we found that the last exon of lncSmad7, that includes the p300 binding site, is mostly unfolded in vivo (Supplementary Figure S3F).
Next, we hypothesized that the lack of structure might be instrumental to mediate the interaction of lncSmad7 with the genome. To this end, we inspected lncSmad7 sequence by Triplexator (45), a tool able to identify putative triplex-forming sequences (TFSs) on the lncRNA. We identified four such segments (TFS#1–4), of which TFS#1 and TFS#2 predicted to form Hoogsteen base-pairing with two putative Triplex Target Sites (TTSs) present on the genome next to the Smad7 enhancer. We verified the RNA-DNA triplex formation by electrophoretic mobility shift assays (EMSA) with TFS#1 or TFS#2 RNA oligonucleotides and the putative TTS biotinylated DNAs. For both oligonucleotides we observed the appearance of a shifted band corresponding to the RNA-DNA triplex, that was resistant to treatment by RNase H (59), but readily disappeared after incubation with RNase A (Figure 3C) indicating that the interaction is via Hoogsteen base-pairing.
To independently validate the triplex formation, we further performed the in vitro hybridization between the biotinylated 3′-most 952 nt-long lncSmad7 fragment harbouring the TFSs and the DNA fragment comprising the identified TTSs followed by capture with streptavidin beads. Accordingly, qPCR analysis confirmed that lncSmad7 binds to the target DNA with high efficiency (Figure 3D). Notably, triplex formation was affected by the presence of 7-deaza-purine nucleotides, which impairs the ability to form Hoogsteen base-pairing, and no interactions was observed with an adjacent intronic DNA sequence. To further verify that lncSmad7 was able to bind and recruit p300 in vivo on this genome tract via triplex formation, we transfected KO ESCs with either the wild type lncSmad7 or with the TFS mutants (Figure 3E). While, the wild type lncSmad7 could rescue the p300 binding and Smad7 expression, mutations of the TFSs significantly impaired lncSmad7 activity (Figure 3F, G and Supplementary Figure S3G). These results demonstrate that lncSmad7 recruits p300 to the Smad7 enhancer via the formation of RNA-DNA triplex.
lncSmad7 controls the expression of key stemness regulators, independently of Smad7
The above results provide direct evidence for a role of lncSmad7 in regulating Smad7 expression. The observation that a plasmid expressing lncSmad7 could rescue the phenotype indicates that, differently from lncRNA Khps1 which has been previously demonstrated to acting only in cis (50) lncSmad7 acts in trans and suggest that it might bind and regulate the transcription also in other genomic loci. In agreement with this hypothesis lncSmad7 knockout resulted in the reduction of acetylation in several genomic loci (Supplementary Figure S4A). To verify the hypothesis that lncSmad7 regulates the transcription of other genes besides Smad7, we first sought to uncouple the contribution to gene expression of lncSmad7 from those regulated by Smad7 (60). To this end, we compared the RNA expression profiles of WT ESCs with lncSmad7 KO, and the lncSmad7 KO ectopically expressing either lncSmad7 or Smad7 (Supplementary Table S2). Knockout of lncSmad7 affected the expression of about 2500 genes. Interestingly, lncSmad7 transient expression in KO cells was able to rescue the expression of almost 50% of them (1188), of which 760 were dependent on Smad7 ectopic expression (Figure 4A and Supplementary Figure S4B, C). Importantly, lncSmad7 rescued 428 genes independently from Smad7 (Supplementary Figure S4B, D). In agreement, the downregulated genes included, among others, nuclear factors involved in ESC pluripotency such as: Id1, a gene involved in the prevention of premature differentiation (61,62); the Yamanaka factor c-Myc that in ESCs contributes to the stemness as an independent module and controls the expression of the Polycomb PRC2 complex (63,64); and Srsf3, a key regulator of pluripotency and oocyte integrity (65,66) (Supplementary Figure S4B). Thus, the above data, together with the phenotypic characterization (Figure 2C, D), indicate that lncSmad7 is a regulator of gene expression which contributes to ESC pluripotency.
Figure 4.
Trans-acting lncSmad7 binds chromatin to acetylate enhancer regions. (A) Venn diagram showing the number of differentially expressed genes that are down-regulated in lncSmad7 KO ESCs and rescued by lncSmad7 expression and/or Smad7 expression. The putative targets of lncSmad7 were identified using the lncSmad7 KO condition as reference and selecting genes significantly upregulated (log FC > 0.5 and FDR < 0.05) in the WT contrast, significantly upregulated in KO + lncSmad7-KO and excluding genes that were significantly upregulated in KO + Smad7-KO. (B) Barplot comparing the number of ChIRP-seq peaks predicted to be bound by lncSmad7 (at different score cutoff) with the expected number according to analogous random regions. (C) Annotation of lncSmad7 binding site across genomic regions using the Genomic Association Test (GAT) software (32). Left panel: lncSmad7 ChIRP-seq peak enrichment and depletion in histone modifications marked regions. Right panel: lncSmad7 ChIRP-seq peaks enrichment and depletion in Candidate cis-Regulatory Elements (cCRE) for ESC cell line E14 available from the SCREEN database. (D) Heatmap of ±5 kb genomic windows centered on lncSmad7 ChIRP-seq peaks from WT and lncSmad7 KO ESCs showing promoters/enhancers (from encode cCREs) near (1 kb) lncSmad7 ChIRP peaks with H3K27ac and H3K4me3 encode histone data. Random lncSmad7 ChIRP-seq peaks as control. (E) Barplot showing Odds Ratios of Fisher's Exact Test for over-representation of direct regulatory evidences occurring in genes that are down-modulated upon lncSMAD7 deletion, with respect to genes that are not transcriptionally affected. ChIRP = presence of a ChIRP-seq peak nearby the respective gene promoter or associated enhancer, marked by H3K7ac. ChIRP + reduced H3K27ac in KO = ChIRP-seq peak nearby a promoter or enhancer coupled with significantly reduced levels of H3K27ac signals in KO with respect to WT samples (P-value = 2.12e–14). (F) Heatmap showing the expression (Z-score, logCMP) of some representative lncSmad7 target genes in WT, lncSmad7 KO and rescued conditions (KO + lncSmad7 and KO + Smad7) from two independent experiments in ESCs. For each lncSmad7 target gene, gene expression levels, H3K27 acetylation value and RNA–DNA triplex forming score of regulatory regions are shown.
lncSmad7 is a trans-acting lncRNA regulating enhancer acetylation and transcriptional activation
To identify lncSmad7 direct targets we performed chromatin isolation by RNA purification followed by sequencing (ChIRP-seq) (43) in ESCs using two independent replicates each with even and odd probes (Supplementary Figure S4E, F). This analysis identified 9313 lncSmad7 binding sites on the genome. ChIRP-seq peaks showed significant overlap with triplex forming regions predicted by two independent programs (Figure 4B and Supplementary Figure S4G). The lncSmad7 binding sites were enriched for chromatin marks of active enhancers enriched at acetylated regions (Figure 4C, D).
By crossing ChIRP-seq at TTS together with the reduction H3K27ac of downregulated genes in lncSmad7 KO we identified 60 bona fide lncSmad7 direct targets genes in trans (Figure 4E, F, Supplementary Table S3 and Supplementary Figure S5A).
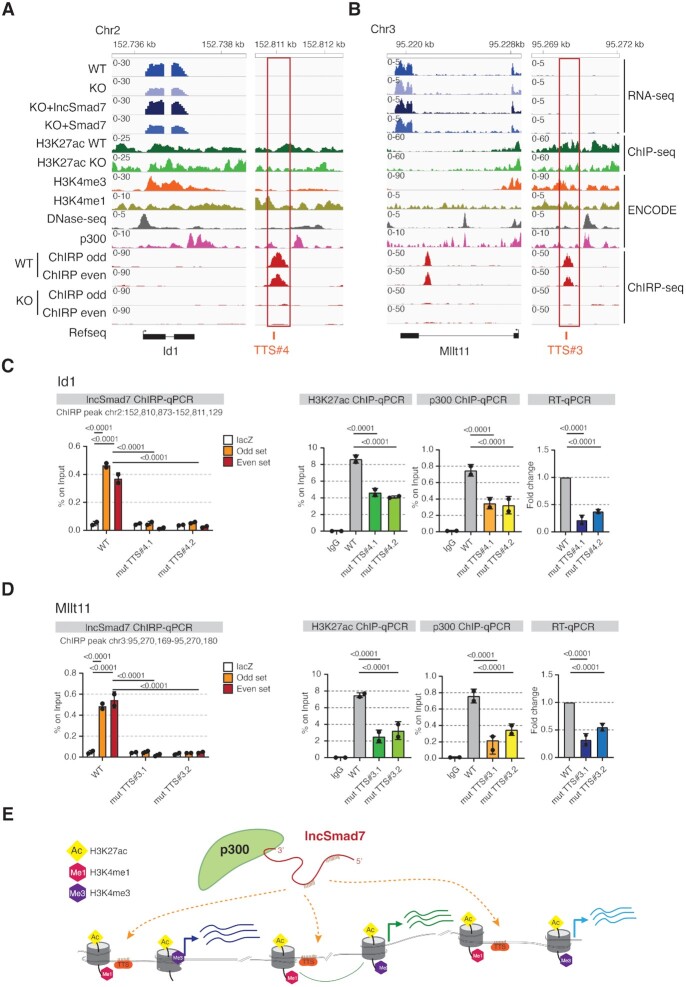
To unambiguously prove the binding of lncSmad7 in trans we performed, by CRISPR/Cas9, two independent mutants at the enhancers of Id1 and of Mllt11 genes corresponding to TTS ChIRP-seq peaks (Figure 5A, B and Supplementary Figure S5B). The TTS mutants impair lncSmad7 binding in both sites, affecting a significant reduction of lncSmad7 binding, p300 occupancy, H3K27ac, and expression of the corresponding genes (Figure 5C, D and Supplementary Figure S5C, D).
Figure 5.
lncSmad7 regulate acetylation levels by forming RNA-DNA triplexes. (A, B) Genomic view of two selected lncSmad7 target transcripts, Id1 and Mllt11, with the associated enhancer regions in ESCs, respectively. The enhancer regions are highlighted by red boxes. The TTS regions at the level of lncSmad7 ChIRP peaks are indicated in red bars. RNA-seq and ChIP-seq of H3K27ac are represented as pooled of two independent experiments and are scaled to the same level. RNA-seq, H3K27ac ChIP-seq and ChIRP-seq are from the present work, ChIP-seq of H3K4 methylations, p300 and DNase-seq are from ENCODE. (C, D) qPCR analysis of Id1 and Mllt11 mutant TTS clones (#4.1, #4.2 and #3.1, 3.2#, respectively) compared to the WT on the indicated Id1 and Mllt11 enhancer regions regulated by lncSmad7. lncSmad7 ChIRP-qPCR of lncSmad7 binding on Id1 and Mllt11 selected peaks from mutant TTS clones respect to the WT. Data are representative of ChIRP odd and ChIRP even compared to lacZ and Input samples. Bars represent the mean and SD of n = 2 independent experiments. P values calculated against control condition for each experiment by using t-test (*P < 0.05; **P < 0.005; ***P < 0.0005, ****P < 0.0001). qPCR analysis of H3K27ac and p300 ChIP experiments from Id1 and Mllt11 mutant TTS clones compared to the WT. The ChIP-qPCR data are expressed as a percentage of the DNA inputs. RT-qPCR analysis showing the Id1 and Mllt11 expression levels in mutant TTS clones compared to the WT ESCs. The analysis is normalized to β-actin and on the WT condition. Bars represent the mean and SD of n = 2 independent experiments. P values calculated against WT condition for each experiment by using t-test (*P < 0.05; **P < 0.005; ***P < 0.0005, ****P < 0.0001). Primers used are reported in Supplementary Table S4. (E) Model of p300 recruitment by lncSmad7 on genomi loci. The enhancers and promoters are marked by histone modifications. The RNA–DNA triplexes derived from the indicate regions in the lncSmad7 transcripts (brown bars) and the TTS regions near the enhancers (orange) of target genes.
DISCUSSION
The acetyltransferase p300 is a transcriptional coactivator that, being expressed ubiquitously, introduces the H4K27ac virtually in all enhancers on the genome. However, H3K27ac enrichment at enhancers shows a high degree of cell-type specificity which is mainly due to its precise recruitment to its target genes and by the activation of its catalytic activity once bound to enhancers via diverse molecular interactions.
In this study, we focused on lncSmad7 a lncRNA whose function and chromatin localization show it is a general regulator of gene expression in trans by interacting with p300 C-terminal domain. Our experiments strongly suggest that lncSmad7 recruits p300 to the genome at enhancers/promoters via Hoogsteen base pairing to acetylate H3K27 (Figure 5E). Our results, demonstrating that lncSmad7 can recruit p300 to enhancers complement the data showing that, once CBP is bound at enhancers, its acetyltransferase activity is activated by interaction with eRNAs on the HAT domain (10). Recently it has been shown that p300 activation is induced by trans-autoacetylation of an autoinhibitory loop (9,10).
We also observed that other lncRNAs interact directly to p300 expanding the repertoire of p300 interactors for a more accurate regulation of gene expression implying that cell- and developmental-specific lincRNAs are good candidates for the regulation of gene expression to contribute to cell specification.
Indeed, a large number of tissue-specific lncRNAs have now been identified and many of them are involved in cell differentiation exerting their functions through various mechanisms mainly based on interactions with different biomolecules (67).
The ability of lncRNAs to form triple helix suggest that the interaction of an RNA with its cognate DNA target sequences is a general mechanism for RNA-mediated target site recognition on the genome and several examples have shown lncRNAs binding via triple helix on specific DNA sites (43,46,50,68–75).
Altogether, our findings expand the repertoire of p300-interacting RNAs, and provide new insights into the mechanistic aspects underlying the regulation of enhancer activity. Our results highlight the role of lncRNAs as important players in gene regulation.
DATA AVAILABILITY
RIP-seq, PAR-CLIP data, RNA-seq, ChIP-seq and ChIRP-seq data of this study have been deposited in the Gene Expression Omnibus (GEO) database under the accession code: GSE154738. Datasets used for comparative analysis were obtained from Gene Expression Omnibus by downloading the following datasets: GSE11431, ENCODE.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: M.M. designed and performed most experiments, analyzed and validated the data and wrote the draft. A.L., E.M., A.L.T. and I.M. performed computational analysis L.M.S. and D.I. performed the lncRNA structure and revised the draft, G.M., S.R., I.P., M.S., F.C. and F.A. performed part of the experiments. S.O. designed experiments, provided supervision, funding acquisition, wrote and revised drafts.
Contributor Information
Mara Maldotti, Dipartimento di Scienze della Vita e Biologia dei Sistemi and MBC, Università di Torino, Via Nizza 52, 10126 Torino, Italy; Italian Institute for Genomic Medicine (IIGM), Sp142 Km 3.95, 10060 Candiolo (Torino), Italy.
Andrea Lauria, Dipartimento di Scienze della Vita e Biologia dei Sistemi and MBC, Università di Torino, Via Nizza 52, 10126 Torino, Italy; Italian Institute for Genomic Medicine (IIGM), Sp142 Km 3.95, 10060 Candiolo (Torino), Italy.
Francesca Anselmi, Dipartimento di Scienze della Vita e Biologia dei Sistemi and MBC, Università di Torino, Via Nizza 52, 10126 Torino, Italy; Italian Institute for Genomic Medicine (IIGM), Sp142 Km 3.95, 10060 Candiolo (Torino), Italy.
Ivan Molineris, Dipartimento di Scienze della Vita e Biologia dei Sistemi and MBC, Università di Torino, Via Nizza 52, 10126 Torino, Italy; Italian Institute for Genomic Medicine (IIGM), Sp142 Km 3.95, 10060 Candiolo (Torino), Italy.
Annalaura Tamburrini, Dipartimento di Scienze della Vita e Biologia dei Sistemi and MBC, Università di Torino, Via Nizza 52, 10126 Torino, Italy; Italian Institute for Genomic Medicine (IIGM), Sp142 Km 3.95, 10060 Candiolo (Torino), Italy.
Guohua Meng, Dipartimento di Scienze della Vita e Biologia dei Sistemi and MBC, Università di Torino, Via Nizza 52, 10126 Torino, Italy; Italian Institute for Genomic Medicine (IIGM), Sp142 Km 3.95, 10060 Candiolo (Torino), Italy.
Isabelle Laurence Polignano, Dipartimento di Scienze della Vita e Biologia dei Sistemi and MBC, Università di Torino, Via Nizza 52, 10126 Torino, Italy; Italian Institute for Genomic Medicine (IIGM), Sp142 Km 3.95, 10060 Candiolo (Torino), Italy.
Mirko Giuseppe Scrivano, Dipartimento di Scienze della Vita e Biologia dei Sistemi and MBC, Università di Torino, Via Nizza 52, 10126 Torino, Italy.
Fabiola Campestre, Dipartimento di Scienze della Vita e Biologia dei Sistemi and MBC, Università di Torino, Via Nizza 52, 10126 Torino, Italy.
Lisa Marie Simon, Dipartimento di Scienze della Vita e Biologia dei Sistemi and MBC, Università di Torino, Via Nizza 52, 10126 Torino, Italy.
Stefania Rapelli, Dipartimento di Scienze della Vita e Biologia dei Sistemi and MBC, Università di Torino, Via Nizza 52, 10126 Torino, Italy; Italian Institute for Genomic Medicine (IIGM), Sp142 Km 3.95, 10060 Candiolo (Torino), Italy.
Edoardo Morandi, Dipartimento di Scienze della Vita e Biologia dei Sistemi and MBC, Università di Torino, Via Nizza 52, 10126 Torino, Italy.
Danny Incarnato, Department of Molecular Genetics, Groningen Biomolecular Sciences and Biotechnology Institute (GBB), University of Groningen, Nijenborgh 7, 9747 AG Groningen, The Netherlands.
Salvatore Oliviero, Dipartimento di Scienze della Vita e Biologia dei Sistemi and MBC, Università di Torino, Via Nizza 52, 10126 Torino, Italy; Italian Institute for Genomic Medicine (IIGM), Sp142 Km 3.95, 10060 Candiolo (Torino), Italy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Associazione Italiana Ricerca sul Cancro (AIRC) [IG 2017 Id. 20240 to S.O.]; PRIN 2017; IIGM institutional funding. Funding for open access charge: AIRC.
Conflict of interest statement. None declared.
REFERENCES
- 1. Joung J., Engreitz J.M., Konermann S., Abudayyeh O.O., Verdine V.K., Aguet F., Gootenberg J.S., Sanjana N.E., Wright J.B., Fulco C.P.et al.. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. 2017; 548:343–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sarropoulos I., Marin R., Cardoso-Moreira M., Kaessmann H.. Developmental dynamics of lncRNAs across mammalian organs and species. Nature. 2019; 571:510–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eckner R., Ewen M.E., Newsome D., Gerdes M., DeCaprio J.A., Lawrence J.B., Livingston D.M.. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Gene Dev. 1994; 8:869–884. [DOI] [PubMed] [Google Scholar]
- 4. Chrivia J.C., Kwok R.P.S., Lamb N., Hagiwara M., Montminy M.R., Goodman R.H.. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993; 365:855–859. [DOI] [PubMed] [Google Scholar]
- 5. Sheikh B.N., Akhtar A.. The many lives of KATs — detectors, integrators and modulators of the cellular environment. Nat. Rev. Genet. 2019; 20:7–23. [DOI] [PubMed] [Google Scholar]
- 6. Weinert B.T., Narita T., Satpathy S., Srinivasan B., Hansen B.K., Schölz C., Hamilton W.B., Zucconi B.E., Wang W.W., Liu W.R.et al.. Time-Resolved analysis reveals rapid dynamics and broad scope of the CBP/p300 acetylome. Cell. 2018; 174:231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang F., Marshall C.B., Ikura M.. Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis: structural and functional versatility in target recognition. Cell. Mol. Life Sci. 2013; 70:3989–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dyson H.J., Wright P.E.. Role of intrinsic protein disorder in the function and interactions of the transcriptional coactivators CREB-binding protein (CBP) and p300*. J. Biol. Chem. 2016; 291:6714–6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ortega E., Rengachari S., Ibrahim Z., Hoghoughi N., Gaucher J., Holehouse A.S., Khochbin S., Panne D.. Transcription factor dimerization activates the p300 acetyltransferase. Nature. 2018; 562:538–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bose D.A., Donahue G., Reinberg D., Shiekhattar R., Bonasio R., Berger S.L.. RNA binding to CBP stimulates histone acetylation and transcription. Cell. 2017; 168:135–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Long Y., Wang X., Youmans D.T., Cech T.R.. How do lncRNAs regulate transcription?. Sci. Adv. 2017; 3:eaao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Engreitz J.M., Haines J.E., Perez E.M., Munson G., Chen J., Kane M., McDonel P.E., Guttman M., Lander E.S.. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016; 539:452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kopp F., Mendell J.T.. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018; 172:393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G., Young G., Lucas A.B., Ach R., Bruhn L.et al.. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011; 477:295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ulitsky I., Bartel D.P.. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013; 154:26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. West J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I., Tolstorukov M.Y., Kingston R.E.. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014; 55:791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.-C., Hung T., Argani P., Rinn J.L.et al.. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010; 464:1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McHugh C.A., Russell P., Guttman M.. Methods for comprehensive experimental identification of RNA-protein interactions. Genome Biol. 2014; 15:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Holmes Z.E., Hamilton D.J., Hwang T., Parsonnet N.V., Rinn J.L., Wuttke D.S., Batey R.T.. The Sox2 transcription factor binds RNA. Nat. Commun. 2020; 11:1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang Y.W., Flynn R.A., Chen Y., Qu K., Wan B., Wang K.C., Lei M., Chang H.Y.. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. Elife. 2014; 3:e02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A.et al.. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011; 472:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hendrickson D.G., Kelley D.R., Tenen D., Bernstein B., Rinn J.L.. Widespread RNA binding by chromatin-associated proteins. Genome Biol. 2016; 17:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gomez J.A., Wapinski O.L., Yang Y.W., Bureau J.-F., Gopinath S., Monack D.M., Chang H.Y., Brahic M., Kirkegaard K.. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the Interferon-γ locus. Cell. 2013; 152:743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Long Y., Hwang T., Gooding A.R., Goodrich K.J., Rinn J.L., Cech T.R.. RNA is essential for PRC2 chromatin occupancy and function in human pluripotent stem cells. Nat. Genet. 2020; 52:931–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grote P., Wittler L., Hendrix D., Koch F., Währisch S., Beisaw A., Macura K., Bläss G., Kellis M., Werber M.et al.. The tissue-specific lncRNA Fendrr Is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 2013; 24:206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L.. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Gene Dev. 2011; 25:1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Flynn R.A., Chang H.Y.. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014; 14:752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neri F., Rapelli S., Krepelova A., Incarnato D., Parlato C., Basile G., Maldotti M., Anselmi F., Oliviero S.. Intragenic DNA methylation prevents spurious transcription initiation. Nature. 2017; 543:72–77. [DOI] [PubMed] [Google Scholar]
- 29. Yu Y., Gu S., Li W., Sun C., Chen F., Xiao M., Wang L., Xu D., Li Y., Ding C.et al.. Smad7 enables STAT3 activation and promotes pluripotency independent of TGF-β signaling. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:10113–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garzia A., Meyer C., Morozov P., Sajek M., Tuschl T.. Optimization of PAR-CLIP for transcriptome-wide identification of binding sites of RNA-binding proteins. Methods. 2017; 118:24–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano Jr., M., Jungkamp A.-C., Munschauer M.et al.. Transcriptome-wide identification of RNA-Binding protein and MicroRNA target sites by PAR-CLIP. Cell. 2010; 141:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith T., Heger A., Sudbery I.. UMI-tools: modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res. 2017; 27:491–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Corcoran D.L., Georgiev S., Mukherjee N., Gottwein E., Skalsky R.L., Keene J.D., Ohler U.. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011; 12:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramírez F., Dündar F., Diehl S., Grüning B.A., Manke T.. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014; 42:W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Simon L.M., Morandi E., Luganini A., Gribaudo G., Martinez-Sobrido L., Turner D.H., Oliviero S., Incarnato D.. In vivo analysis of influenza a mRNA secondary structures identifies critical regulatory motifs. Nucleic Acids Res. 2019; 47:7003–7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 39. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gu Z., Eils R., Schlesner M.. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016; 32:2847–2849. [DOI] [PubMed] [Google Scholar]
- 41. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu D.-Y., Bittencourt D., Stallcup M.R., Siegmund K.D.. Identifying differential transcription factor binding in ChIP-Seq. Front. Genet. 2015; 6:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chu C., Qu K., Zhong F.L., Artandi S.E., Chang H.Y.. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-Chromatin interactions. Mol. Cell. 2011; 44:667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Encode Project Consortium Moore J.E., Purcaro M.J., Pratt H.E., Epstein C.B., Shoresh N., Adrian J., Kawli T., Davis C.A., Dobin A.et al.. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020; 583:699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Buske F.A., Bauer D.C., Mattick J.S., Bailey T.L.. Triplexator: detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res. 2012; 22:1372–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kuo C.-C., Hänzelmann S., Cetin N.S., Frank S., Zajzon B., Derks J.-P., Akhade V.S., Ahuja G., Kanduri C., Grummt I.et al.. Detection of RNA–DNA binding sites in long noncoding RNAs. Nucleic Acids Res. 2019; 47:gkz037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. He S., Zhang H., Liu H., Zhu H.. LongTarget: a tool to predict lncRNA DNA-binding motifs and binding sites via Hoogsteen base-pairing analysis. Bioinformatics. 2015; 31:178–186. [DOI] [PubMed] [Google Scholar]
- 48. Sauvageau M., Goff L.A., Lodato S., Bonev B., Groff A.F., Gerhardinger C., Sanchez-Gomez D.B., Hacisuleyman E., Li E., Spence M.et al.. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013; 2:e01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Incarnato D., Morandi E., Simon L.M., Oliviero S.. RNA framework: an all-in-one toolkit for the analysis of RNA structures and post-transcriptional modifications. Nucleic Acids Res. 2018; 46:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Postepska-Igielska A., Giwojna A., Gasri-Plotnitsky L., Schmitt N., Dold A., Ginsberg D., Grummt I.. LncRNA khps1 regulates expression of the Proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol. Cell. 2015; 60:626–636. [DOI] [PubMed] [Google Scholar]
- 51. Fagnocchi L., Cherubini A., Hatsuda H., Fasciani A., Mazzoleni S., Poli V., Berno V., Rossi R.L., Reinbold R., Endele M.et al.. A Myc-driven self-reinforcing regulatory network maintains mouse embryonic stem cell identity. Nat. Commun. 2016; 7:11903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Arase M., Horiguchi K., Ehata S., Morikawa M., Tsutsumi S., Aburatani H., Miyazono K., Koinuma D.. Transforming growth factor-β-induced lncRNA-Smad7 inhibits apoptosis of mouse breast cancer JygMC(A) cells. Cancer Sci. 2014; 105:974–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mintz P.J., Cardó-Vila M., Ozawa M.G., Hajitou A., Rangel R., Guzman-Rojas L., Christianson D.R., Arap M.A., Giordano R.J., Souza G.R.et al.. An unrecognized extracellular function for an intracellular adapter protein released from the cytoplasm into the tumor microenvironment. Proc. Nat. Acad. Sci. U.S.A. 2009; 106:2182–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J.et al.. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008; 133:1106–1117. [DOI] [PubMed] [Google Scholar]
- 55. Song C.-X., Szulwach K.E., Dai Q., Fu Y., Mao S.-Q., Lin L., Street C., Li Y., Poidevin M., Wu H.et al.. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013; 153:678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chronis C., Fiziev P., Papp B., Butz S., Bonora G., Sabri S., Ernst J., Plath K.. Cooperative binding of transcription factors orchestrates reprogramming. Cell. 2017; 168:442–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen C., Morris Q., Mitchell J.A.. Enhancer identification in mouse embryonic stem cells using integrative modeling of chromatin and genomic features. BMC Genomics. 2012; 13:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zubradt M., Gupta P., Persad S., Lambowitz A.M., Weissman J.S., Rouskin S.. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat. Methods. 2017; 14:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nakamura H., Oda Y., Iwai S., Inoue H., Ohtsuka E., Kanaya S., Kimura S., Katsuda C., Katayanagi K., Morikawa K.. How does RNase H recognize a DNA.RNA hybrid?. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:11535–11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Meng G., Lauria A., Maldotti M., Anselmi F., Polignano I.L., Rapelli S., Donna D., Oliviero S.. Genome-Wide analysis of Smad7-mediated transcription in mouse embryonic stem cells. Int. J. Mol. Sci. 2021; 22:13598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Malaguti M., Migueles R.P., Blin G., Lin C.-Y., Lowell S.. Id1 stabilizes epiblast identity by sensing delays in nodal activation and adjusting the timing of differentiation. Dev. Cell. 2019; 50:462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Aloia L., Gutierrez A., Caballero J.M., Croce L.D.. Direct interaction between Id1 and Zrf1 controls neural differentiation of embryonic stem cells. EMBO Rep. 2015; 16:63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kim J., Woo A.J., Chu J., Snow J.W., Fujiwara Y., Kim C.G., Cantor A.B., Orkin S.H.. A myc network accounts for similarities between embryonic stem and cancer cell transcription programs. 2010; 143:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Neri F., Zippo A., Krepelova A., Cherubini A., Rocchigiani M., Oliviero S.. Myc regulates the transcription of the PRC2 gene to control the expression of developmental genes in embryonic stem cells. Mol. Cell. Biol. 2012; 32:840–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Do D.V., Strauss B., Cukuroglu E., Macaulay I., Wee K.B., Hu T.X., Igor R.D.L.M., Lee C., Harrison A., Butler R.et al.. SRSF3 maintains transcriptome integrity in oocytes by regulation of alternative splicing and transposable elements. Cell Discov. 2018; 4:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ratnadiwakara M., Archer S.K., Dent C.I., Mozos I.R.D.L., Beilharz T.H., Knaupp A.S., Nefzger C.M., Polo J.M., Anko M.-L.. SRSF3 promotes pluripotency through Nanog mRNA export and coordination of the pluripotency gene expression program. Elife. 2018; 7:e37419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Azad F.M., Polignano I.L., Proserpio V., Oliviero S.. Long noncoding RNAs in human stemness and differentiation. Trends Cell Biol. 2021; 31:542–555. [DOI] [PubMed] [Google Scholar]
- 68. Sentürk Cetin N., Kuo C.-C., Ribarska T., Li R., Costa I.G., Grummt I.. Isolation and genome-wide characterization of cellular DNA:RNA triplex structures. Nucleic Acids Res. 2019; 47:2306–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kalwa M., Hänzelmann S., Otto S., Kuo C.-C., Franzen J., Joussen S., Fernandez-Rebollo E., Rath B., Koch C., Hofmann A.et al.. The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic Acids Res. 2016; 44:10631–10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. West J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I., Tolstorukov M.Y., Kingston R.E.. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014; 55:791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Blank-Giwojna A., Postepska-Igielska A., Grummt I.. lncRNA KHPS1 activates a poised enhancer by triplex-dependent recruitment of epigenomic regulators. Cell Rep. 2019; 26:2904–2915. [DOI] [PubMed] [Google Scholar]
- 72. Goñi J.R., Cruz X., Orozco M.. Triplex-forming oligonucleotide target sequences in the human genome. Nucleic Acids Res. 2004; 32:354–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mondal T., Subhash S., Vaid R., Enroth S., Uday S., Reinius B., Mitra S., Mohammed A., James A.R., Hoberg E.et al.. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA–DNA triplex structures. Nat. Commun. 2015; 6:7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Alfeghaly C., Behm-Ansmant I., Maenner S.. Study of genome-wide occupancy of long non-coding RNAs using chromatin isolation by RNA purification (ChIRP). Methods Mol. Biol. 2021; 2300:107–117. [DOI] [PubMed] [Google Scholar]
- 75. Alfeghaly C., Sanchez A., Rouget R., Thuillier Q., Igel-Bourguignon V., Marchand V., Branlant C., Motorin Y., Behm-Ansmant I., Maenner S.. Implication of repeat insertion domains in the trans-activity of the long non-coding RNA ANRIL. Nucleic Acids Res. 2021; 49:4954–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RIP-seq, PAR-CLIP data, RNA-seq, ChIP-seq and ChIRP-seq data of this study have been deposited in the Gene Expression Omnibus (GEO) database under the accession code: GSE154738. Datasets used for comparative analysis were obtained from Gene Expression Omnibus by downloading the following datasets: GSE11431, ENCODE.