Abstract
Serine integrases are emerging as one of the most powerful biological tools for synthetic biology. They have been widely used across genome engineering and genetic circuit design. However, developing serine integrase-based tools for directly/precisely manipulating synthetic biobricks is still missing. Here, we report SYMBIOSIS, a versatile method that can robustly manipulate DNA parts in vivo and in vitro. First, we propose a ‘keys match locks’ model to demonstrate that three orthogonal serine integrases are able to irreversibly and stably switch on seven synthetic biobricks with high accuracy in vivo. Then, we demonstrate that purified integrases can facilitate the assembly of ‘donor’ and ‘acceptor’ plasmids in vitro to construct composite plasmids. Finally, we use SYMBIOSIS to assemble different chromoprotein genes and create novel colored Escherichia coli. We anticipate that our SYMBIOSIS strategy will accelerate synthetic biobrick manipulation, genetic circuit design and multiple plasmid assembly for synthetic biology with broad potential applications.
INTRODUCTION
DNA is essential for cellular processes and behaviors as the instructions for biological processes are encoded in DNA molecules. Changing DNA sequences to reprogram cells can endow them with new, specialized properties. Over the last two decades, synthetic biology, taking the core mission to (re)design and construct new biological parts, devices and systems, has been rapidly developed to require even more controllable and efficient tools for DNA manipulation and modification (1–4). To meet this demand, synthetic biologists have paid much more attention to DNA-modifying enzymes, which can catalyze DNA sequence variations (e.g. deletion, insertion and mutation) for precise and predictable programming of living organisms (5).
Recombinases are a class of enzymes that catalyze DNA–DNA site-specific recombination events within single or among different DNA strands (6). Serine integrase, a subfamily of recombinase, can recognize and catalyze recombination between two specific DNA sites (∼50 bp for each) called attP (attachment site by phage) and attB (attachment site by bacteria) (7–11). Depending on the attP/attB position and orientation, the DNA fragments can be deleted, integrated, recombined or inverted (7). The features of site specificity, orthogonality, irreversibility, high affinity and high efficiency make serine integrase a powerful genetic tool in synthetic biology (7). For example, serine integrases have been widely used for linear DNA fragment assembly (12), genome engineering (13) and genetic circuit design (14–20). In particular, a series of complicated genetic circuits were constructed recently using serine integrase technologies such as Boolean logic gates (14–19), memory circuits (14–20), state machine (16), binary counter (17), message storage (16,19) and gene expression cascades (20). However, the ‘input’ and ‘output’ signals within one genetic circuit often only have a one-to-one corresponding relationship, which means that a finite number of orthogonal serine integrases can only switch on an equal number of genetic circuits; therefore, the output signals are limited to the number of serine integrases present (16). Moreover, the common problems of inducible recombinase expression leakage and cell toxicity in vivo still remain unsolved (17,18,21).
In synthetic biology, the general strategy for biobrick construction and assembly often uses one of the following methods: restriction enzyme digestion and ligation (22), TOPO cloning (23,24), golden gate assembly (25), gateway cloning (26) and Gibson assembly (27). However, these approaches typically require the preparation of multiple linear DNA fragments in advance by polymerase chain reaction (PCR). Therefore, a method for one-step assembly of circular biobricks (generally considered as plasmids) is highly desirable. It is worth noting that a tyrosine recombinase (another subfamily of recombinase)-based in vitro DNA assembly method ‘ACEMBL’ has been established by using Cre-LoxP recombination (28). Cre recombinase, one of the tyrosine recombinases, cleaves single DNA strands forming covalent 3′-phosphotyrosine bonds with the DNA backbone and rejoins the strands via a Holliday junction-like intermediate state (6). However, if one plasmid contains more than one homologous LoxP site, it can form many unpredictable variants before the next round of assembly (28,29). The exact DNA sequences of all possible variants are subsequently subjected to post-verification and selection by restriction enzyme digestion or DNA sequencing, which limits precise, predictable and efficient manipulation of plasmid assembly. Moreover, unlike serine integrases, the tyrosine recombinase Cre-mediated Cre-LoxP reaction is reversible, often leading to unpredictable variation and reversible degradation (6). In contrast, serine integrases make double-strand breaks in DNA forming covalent 5′-phosphoserine bonds with the DNA backbone before religation (6). Notably, serine integrase-driven DNA recombination is highly directional, orthogonal and irreversible (when no presence of an accessory recombination directionality factor), which makes the system a reliable choice for catalyzing in vitro DNA–DNA recombination (7).
In this study, we introduce SYMBIOSIS (synthetic manipulable biobricks via orthogonal serine integrase systems), a concept describing the closely interdependent relationship between serine integrases and related genetic designs, which need to coexist and work synergistically for desired purposes. Specifically, on the basis of SYMBIOSIS we aim to provide a strategy that can precisely switch on scalable genetic circuits in Escherichia coli and expand the toolkit to establish a method for in vitro multiple plasmid assembly. The foundational principle is that we can manipulate DNA parts through serine integrase-driven DNA recombination. To demonstrate the feasibility of SYMBIOSIS in vivo, we initially constructed three plasmids with constitutive promoters and each of them harbored an orthogonal serine integrase TP901-1, Bxb1 or Phic31, respectively. We defined each above plasmid as an ‘input’ signal once it was transformed into E. coli cells. Meanwhile, seven different chromoproteins were chosen as ‘output’ signals. By observing the changes in the color of E. coli cells, we can determine the ‘on/off’ states of synthetic biobricks. Next, we demonstrated that purified serine integrases were able to assemble ‘donor’ and ‘acceptor’ plasmids in vitro, generating expression vectors for protein synthesis in vivo. More interestingly, we applied our SYMBIOSIS in vitro to construct various plasmids harboring different combinations of chromoprotein genes. When transformed, these genes created some unique colored E. coli cells that, to our knowledge, have never been reported so far.
MATERIALS AND METHODS
Strains, plasmids, primers and reagents
The details of E. coli strains and plasmids used in this study are listed in Supplementary Tables S1, S3 and S5. All plasmid sequences are listed in the Supplementary Data (Excel file). General plasmids were checked for correctness with Sanger sequencing (GENEWIZ) unless otherwise noted. All assembled plasmids (pFB66–pFB72, pFB96–pFB130) were checked for correctness by agarose gel electrophoresis after FastDigest XbaI (Thermo Scientific) digestion. Oligonucleotides were synthesized by GENEWIZ and are listed in Supplementary Table S6. Q5 High-Fidelity DNA Polymerase (New England Biolabs), FastPure Gel DNA Extraction Mini Kit (Vazyme) and NovoRec plus One step PCR Cloning Kit (Novoprotein) were used for molecular cloning. DreamTaq Green PCR Master Mix (Thermo Scientific) was used for colony PCR. Lysogeny broth (LB) liquid medium contains 10 g tryptone (Oxoid), 5 g yeast extract (Oxoid) and 10 g sodium chloride (Titan/Greagent) in 1 l ddH2O. LB agar plate was prepared by adding 15 g agar (Sangon Biotech) per liter LB liquid medium. Antibiotic stocks (1000×) are 100 mg/ml ampicillin, 50 mg/ml kanamycin, 50 mg/ml streptomycin and 34 mg/ml chloramphenicol.
Genetic parts
All genetic parts [promoter, ribosome binding site (RBS), terminator, integrase recognition site, coding sequence (CDS) and 16S rRNA fragment] used in this study are listed in Supplementary Table S2. Short genetic parts (promoter, RBS, terminator and integrase recognition site) were synthesized within the oligonucleotides (GENEWIZ) and inserted into PCR fragments during molecular cloning. 16S rRNA fragments were amplified from E. coli Mach1-T1 genome (GenBank: CP002185.1). The CDS of TP901-1, Phic31, mKate2, CyOFP1 and sfGFP (superfolder green fluorescent protein) were synthesized by GENEWIZ. The CDS of mCherry, core-sz17 and sz18-σ-T3 were gifts from Dr Chao Zhong Lab, ShanghaiTech University. The other CDS were collected from iGEM Registry Distribution Kit (Spring 2016 and Spring 2017 distribution).
Construction of general plasmids
All general plasmids (except assembled plasmids) were constructed by Gibson assembly. In brief, all linear PCR products (Q5 High-Fidelity DNA Polymerase, New England Biolabs) were extracted by gel extraction and then ligated using NovoRec plus One step PCR Cloning Kit (Novoprotein). After ligation, the reaction mixture was added to 50 μl of competent E. coli Mach1-T1 cells, which were incubated overnight on LB agar plate. Then, DreamTaq Green PCR Master Mix (Thermo Scientific) was used for colony PCR. The possible PCR products were sent to GENEWEZ for Sanger sequencing. Especially, the plasmids with repeated sequences were constructed with three to four fragments in one Gibson assembly reaction. For example, pFB26 was constructed with four PCR fragments: fragment 1 [attB(Phic31) homolog-B0034-asPink-B0015-pSB1C3-J23100-attP(TP901-1) homolog] derived from pFB137, fragment 2 [J23100 homolog-attP(TP901-1)-B0015-attB(TP901-1)-attP(Bxb1) homolog], fragment 3 [attB(TP901-1) homolog-attP(Bxb1)-B0015-attB(Bxb1)-attP(Phic31) homolog] and fragment 4 [attB(Bxb1) homolog-attP(Phic31)-B0015-attB(Phic31)-B0034 homolog].
Western blot assay
The E. coli Mach1-T1 strains containing compatible single, double or triple plasmids (pFB10, pFB13, pFB15) were cultivated in 5 ml LB liquid medium at 30°C and 250 rpm for 16 h. Cell pellets were collected in 1.5 ml centrifuge tubes at 5000 × g and 4°C for 10 min. The supernatant was discarded and the pellets were resuspended with 1 ml 1× phosphate-buffered saline (pH 7.4) and lysed by sonication (Q125 sonicator, Qsonica, 10 s on/off, 50% of amplitude, input energy ∼600 J). The lysate was then centrifuged at 12 000 × g and 4°C for 10 min. The total (cell lysate) and soluble (supernatant) fractions were separated by SDS-PAGE (Omni-Easy One-Step PAGE Gel Fast Preparation Kit, EpiZyme), followed by wet transferring to PVDF membrane (Bio-Rad) with 1× transfer buffer [25 mM Tris–HCl, 192 mM glycine and 20% (v/v) methanol in 1 l ddH2O, pH 8.3]. Then, the PVDF membrane was blocked (Protein Free Rapid Blocking Buffer, EpiZyme) for 1 h at room temperature. After washing thrice with TBST for each 5 min, 1:10 000 (TBST buffer based) diluted His-Tag Mouse Monoclonal Antibody (Proteintech) solution was added to the membrane and incubated for 1 h at room temperature. After washing thrice with TBST for each 5 min, 1:10 000 (TBST buffer based) diluted HRP-Goat Anti-Mouse IgG (H + L) Antibody (Proteintech) solution was added to the membrane and incubated for another 1 h at room temperature. After the last washing with TBST thrice for each 5 min, the membrane was visualized using Omni ECL reagent (EpiZyme) under UVP ChemStudio (analytikjena).
Assay of in vivo SYMBIOSIS for ‘keys match locks’ manipulation
Escherichia coli Mach1-T1 strains containing seven different ‘key’ plasmid combinations (pFB10, pFB13, pFB15, pFB10 + pFB13, pFB10 + pFB15, pFB13 + pFB15 and pFB10 + pFB13 + pFB15) worked as ‘keys’, respectively. Each of the strains was transformed with the ‘lock’ plasmids pFB20–pFB26 for evaluating the ‘keys match locks’ (KmL) model. After ‘lock’ transformation, all 49 ‘unlocking’ state strains were incubated overnight on LB agar plates with appropriate antibiotics at 30°C for at least 24 h growth and in vivo recombination. The plates were photographed for recombination efficiency characterization and the matured colonies were then picked up for overnight incubation (each colony in 5 ml LB at 30°C and 250 rpm for 18 h). On the second day, E. coli Mach1-T1 pellets were collected from 3 ml culture in 1.5 ml centrifuge tubes and then photographed. The other 2 ml culture was used for plasmid extraction. After the KmL process, 7 ‘unlocked’ strains would be chromoprotein visualized, and the other 42 strains would remain locked and colorless. For electrotransformation, Gene Pulser Xcell System (Bio-Rad) and Gene Pulser/Micropulser Electroporation Cuvettes, 0.1 cm gap (Bio-Rad) were used in this study.
Visualization and imaging of E. coli containing chromoproteins
Escherichia coli Mach1-T1 strains containing chromoprotein unexpressed/expressed plasmids (pFB20–pFB26, pFB27–pFB33) were visualized and imaged under both bright field and LED transilluminator (SLB-01W UltraSlim, MaestroGen) (see Supplementary Figures S7 and S8).
Recombination efficiency characterization of the KmL process
After the KmL process, matured colonies on the overnight LB agar plates were visualized and characterized with or without appropriate colors. When the ‘unlocked’ state (Supplementary Figure S8) occurred, colored colonies were counted as positive and few colorless colonies were counted as negative. The recombination efficiency was calculated as the positive percentage of total colonies. Each value was calculated with six biological replicates by counting at least 100 colonies on LB agar plates.
Escherichia coli total RNA extraction
Three milliliters of each overnight cultured (30°C, 250 rpm, 18 h) E. coli Mach1-T1 pellet was collected in 1.5 ml centrifuge tube and then added 1 ml Trizol (Sangon Biotech) at room temperature for 10 min. Phenol–chloroform–isoamyl alcohol (0.4 ml, 25:24:1, Aladdin) was then added and shaken vigorously for 15 s. Afterward, the samples were centrifuged at 4°C and 12 000 × g for 10 min. The resulting supernatant (∼600 μl) was transferred to a new tube, mixed with equal volume of isopropanol and kept at room temperature for 20 min. After centrifugation at 4°C and 12 000 × g for 10 min, the supernatant was discarded and the pellet was washed with 1 ml ethanol. Then, the sample was centrifuged again for 3 min at 4°C and 12 000 × g. The supernatant ethanol was discarded and total RNA (white flocculent precipitate) was collected at the bottom of the tube. After drying for 10 min at room temperature, the precipitate was resuspended in 100 μl nuclease-free water (Invitrogen). The concentration of total RNA was measured by the NanoDrop 2000/2000c Spectrophotometer (Thermo Scientific).
RT-qPCR
One microgram of E. coli total RNA was used for reverse transcription to cDNA [Hifair III 1st Strand cDNA Synthesis Kit (gDNA digester plus), Yeasen]. One microliter of cDNA product was used for qPCR (ChamQ SYBR Color qPCR Master Mix, Vazyme) quantification. Bio-Rad CFX96 Touch Real-Time PCR Detection System was used for the experiments and data analysis. For standard curve calculation, pFB141/pFB145 were used as sfGFP/16S rRNA quantification templates. Two plasmids were diluted in the range of 109 to 104 copies for calculation.
Expression and purification of serine integrases
All three CDS of serine integrases TP901-1/Bxb1/Phic31 were constructed into pET22b vector with a C-terminal 6×His tag and named as pFB142/pFB143/pFB144. Three serine integrases used in this study were expressed and purified by the following same protocol. The E. coli strain Rosetta-gami(DE3) pLysS was transformed with the relevant overexpression plasmid. Starter cultures [LB liquid medium containing ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml)] were inoculated from single transformant colonies and grown overnight at 37°C for 16 h. Five-milliliter aliquot of the starter culture was used to inoculate 1 l LB medium containing ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml) in a 2 l conical flask. The cultures were incubated at 37°C with shaking (250 rpm) until OD600 reached 0.6–0.8. The cultures were then cooled down on ice rapidly to 20°C, and isopropyl-β-d-thiogalactopyranoside (IPTG) was added for induction at a final concentration of 0.5 mM (note that expression of TP901-1 and Phic31 at 30°C with 1 mM IPTG led to almost insoluble protein fractions). The cultures were then grown for 16 h at 20°C and 220 rpm. Afterward, cells were harvested by centrifugation (Avanti JXN-26 High-Speed Centrifuge, Beckman Coulter) at 5000 × g and 4°C for 10 min. The collected cell pellet (∼4 g) was resuspended by 25 ml 1× PBS and transferred into 50 ml centrifuge tubes. After a second centrifugation at 5000 × g and 4°C for 10 min, the pellet was resuspended in 45 ml buffer A, containing 20 mM sodium phosphate (pH 7.4), 1 M sodium chloride, 1 mM dithiothreitol (DTT) and 50 mM imidazole. The suspension was cooled on ice and then lysed thrice at 1500 bar by ultrahigh-pressure homogenization (JNBIO). The lysate was then centrifuged at 4°C and 20 000 × g for 30 min. The supernatant was collected and passed through a 0.22 μm filter. The filtered solution was then purified by Ni2+ affinity chromatography using a 1 ml HisTrap FF column (GE Healthcare). The column was equilibrated with 25 ml buffer A [20 mM sodium phosphate (pH 7.4), 1 M sodium chloride, 1 mM DTT and 50 mM imidazole], at a constant flow rate of 1 ml/min. After column equilibration, the filtered protein solution was loaded, followed by washing with buffer A (∼25 ml). Then, bounded proteins were eluted with buffer B (buffer A, but with 500 mM imidazole) and collected into 1.5 ml centrifugal tubes with 0.5 ml elution. Each 0.5 ml elution was analyzed by SDS-PAGE. Eluted protein samples were combined in one tube and desalinated using an ultrafiltration tube (Amicon Ultra 3 kDa molecular weight cutoff, Merck/Millipore) with buffer C [50 mM Tris–HCl (pH 7.5), 2 mM DTT and 2 M sodium chloride]. The desalinated and concentrated protein solution was then mixed with 40% glycerol (v/v in water), with a volume ratio of 1:1. The concentration of the final protein solution was measured at 280 nm by the NanoDrop 2000/2000c Spectrophotometer (Thermo Scientific). Purified proteins were flash frozen by liquid nitrogen and stored at −80°C until further use.
Optimization of in vitro recombination reactions and product analysis
Each of the three purified serine integrases was diluted in buffer D [25 mM Tris–HCl (pH 7.5), 1 mM DTT, 1 M sodium chloride and 20% (v/v) glycerol] to the final concentrations of 10, 1 and 0.1 μM. In a typical reaction, 3 μl of diluted serine integrase was added to a 27 μl reaction containing 400 ng of substrate plasmid (pFB16), 50 mM Tris–HCl (pH 7.5), 0.1 mM EDTA and 100 μg/ml bovine serum albumin. Then, the recombination reaction was incubated at various temperatures (16/30/37°C) and time intervals (10/30/60/90/120/150 min) for optimization. The reactions were terminated by heating at 80°C for 20 min, and then the samples were separated by agarose gel electrophoresis and visualized under UVP ChemStudio (analytikjena). Darker bands (except the plasmid bands) on agarose gel indicated a higher recombination efficiency.
In vitro SYMBIOSIS for multiple plasmid assembly
For each in vitro SYMBIOSIS reaction, only one of the purified serine integrases was used for assembly between one ‘donor’ plasmid and one ‘acceptor series’ plasmid. The ‘acceptor series’ plasmids refer to three groups: ‘empty’ acceptor (pFB62), ‘one donor assembled’ acceptors (pFB66–pFB68, pFB90–pFB95) and ‘double donor assembled’ acceptors (pFB69–pFB71, pFB96–pFB110). Three microliters of diluted serine integrase (10 μM) was added to a 27 μl solution containing ‘donor’ plasmid and ‘acceptor series’ plasmid [each plasmid was added as ‘base pair/10’ ng; for example, 221.8 ng of the plasmid pFB62 (2218 bp) was added per reaction], 50 mM Tris–HCl (pH 7.5), 0.1 mM EDTA and 100 μg/ml bovine serum albumin. Then, the recombination reaction was incubated at 30°C for 120 min, followed by termination with heating at 80°C for 20 min. Afterward, 10 μl of the reaction mixture was added to 100 μl E. coli Mach1-T1 competent cells for chemical transformation. To screen the correctly assembled plasmids, LB agar plates were supplied with multiple antibiotics matched to the multiple antibiotic-resistant genes on the correctly assembled plasmids. Only the colonies harboring assembled plasmids could grow on the plates, while the colonies containing unreacted ‘donor’ or ‘acceptor series’ plasmids did not.
Imaging of colored E. coli pellets
Three milliliters of each overnight cultivated (37°C, 250 rpm, 18 h) E. coli Mach1-T1 cells was collected in 1.5 ml centrifuge tubes and then visualized under bright field and imager (ChemiDoc MP Imaging System, Bio-Rad). For imaging the fluorescence of mCherry, sfGFP and CyOFP1, three different programs were used, respectively, which are Cy5 (light: red Epi illumination, filter: 695/55 filter), Cy2 (light: blue Epi illumination, filter: 530/28 filter) and Cy3 (light: green Epi illumination, filter: 605/50 filter). All the samples were photographed in one photo without post-splicing.
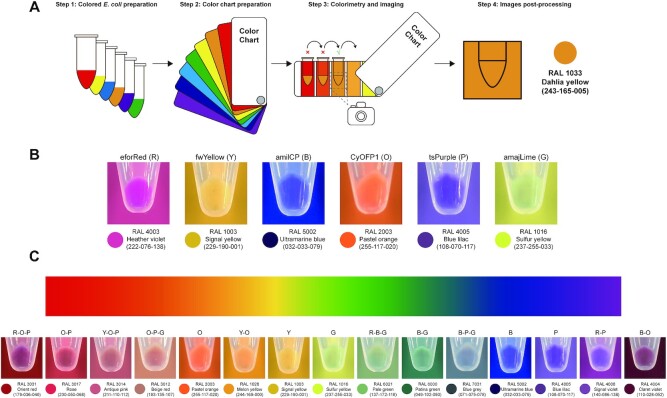
Colorimetry and imaging of composite colored E. coli
A RAL K7 classic color chart was used for colorimetry with composite colored E. coli Mach1-T1 pellets. Three milliliters of each overnight cultivated (37°C, 250 rpm, 18 h) composite colored E. coli Mach1-T1 cells was collected in 1.5 ml centrifuge tubes and then analyzed by colorimetry and imaging under bright field. The tubes were continuously compared to different color charts unless the most matched situation occurred. Then, the photos (the tube was placed in the middle of photo and the matched color chart acted as background) were taken in a mini photo studio (Supplementary Figure S19) using a mobile phone without any image parameter (e.g. brightness, contrast, etc.) adjustments.
RESULTS
Establishing the SYMBIOSIS framework
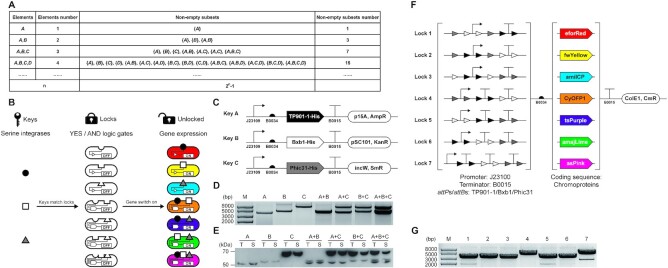
The idea for designing SYMBIOSIS was inspired by the mathematical concept of ‘set’. The elements that constitute sets can be any object. In principle, a finite number of elements can be freely combined and organized as various different nonempty subsets. If there is one element A, it can only form one nonempty subset {A}. When another element B is added, two new nonempty subsets {B} and {A, B} can be formed. Similarly, three elements A, B and C can generate seven nonempty subsets {A}, {B}, {C}, {A, B}, {A, C}, {B, C} and {A, B, C}. By parity of reasoning, the number of n elements can be composed into 2n − 1 different nonempty subsets (30) (Figure 1A).
Figure 1.
Design and validation of the SYMBIOSIS framework. (A) Schematic of the mathematical ‘set’ concept. Nonempty subsets are organized with elements. The number of nonempty subsets increases exponentially with the element number (n) as 2n − 1. (B) Design of the KmL model. Three orthogonal serine integrases (TP901-1: black circle; Bxb1: white square; and Phic31: gray triangle) work as ‘keys’ to match E. coli strains (ovals with gaps) containing seven different ‘lock’ genetic circuits. A correct ‘key–lock’ match can unlock the closed circuit and switch on chromoprotein expression. (C) Architectures of three ‘key’ plasmids. TP901-1, Bxb1 and Phic31 are constructed in three compatible plasmids, respectively, with a weak constitutive promoter J23109 (pFB1–pFB3, see Supplementary Table S4). (D) Compatibility of single, double or triple ‘key’ plasmids in one E. coli strain. Extracted plasmids are digested by XbaI and then separated by agarose gel electrophoresis. (E) Western blot analysis of individual expression and co-expression of TP901-1 (56.5 kDa), Bxb1 (57.2 kDa) and Phic31 (67.9 kDa) labeled with the anti-His antibody. T, total protein; S, soluble protein. (F) Schematic of seven ‘lock’ plasmids. ‘Lock’ biobricks (left brackets) are assembled with a strong promoter J23100, a strong terminator B0015 and attP–attB sites (TP901-1 attP/attB: black triangles; Bxb1 attP/attB: white triangles; and Phic31 attP/attB: gray triangles). Seven different chromoproteins (eforRed, fwYellow, amilCP, CyOFP1, tsPurple, amajLime and asPink, right brackets) work as output signals. The seven ‘lock’ plasmids are constructed using the same vector that is compatible to three ‘key’ plasmids. (G) Plasmid sizes of the seven ‘locks’ (pFB20–pFB26, see Supplementary Table S5) after XbaI digestion. Expected bands of the seven digested plasmids are observed at 3371, 3401, 3356, 3521, 3506, 3509 and 3647 bp, respectively.
Based on the principle of set, we were motivated to establish the SYMBIOSIS framework by matching biological inputs (elements) with visible outputs (subsets) in living E. coli cells. To this end, we first chose three orthogonal serine integrases TP901-1 (8,9), Bxb1 (10) and Phic31 (11), which were defined as input elements A, B and C, respectively. Then, seven chromoproteins were used as output signals in response to three serine integrase inputs. We called this input–output relation as a KmL model (Figure 1B). In the KmL model, only when integrase(s) match the correct logic gates, the ‘locked’ genetic circuits can be switched on, leading to the expression of corresponding chromoproteins in E. coli.
We began our investigation by trying two expression systems, which are l-arabinose inducible AraC-pBAD expression system (31) and anhydrotetracycline inducible TetR expression system (32), to express serine integrases as inputs (‘keys’) together with different ‘lock’ circuits. Unfortunately, our initial experiments were not successful due to the leaky expression of integrases that caused unexpected chromoprotein expression (data not shown). We suspected that a basal leakage of integrase was enough to unlock the genetic circuit, resulting in the activation of gene expression. To tackle this problem and achieve the goal of a strict switch on/off using integrases, we decided to construct three compatible and constitutive expression plasmids for three serine integrases (Figure 1C). In this context, we defined these plasmids as ‘keys’ once they were transformed into E. coli cells. We initially hypothesized that a stronger constitutively active promoter may lead to more integrase expression and higher catalytic efficiency. However, we found that the strongest promoter (J23100) resulted in an unexpected larger plasmid size (>5000 bp) than the correct plasmid (4007 bp) harboring the TP901-1 gene (pFB7, Supplementary Table S5) after ∼1 day of cultivation (Supplementary Figure S1). The exact reason was unclear, but we suspected that overexpression of TP901-1 with the strongest promoter might cause cellular toxicity and therefore plasmid mutation. We then constructed a series of plasmids varying in their promoter strength to express TP901-1 integrase (see pFB7–pFB11 in Supplementary Table S5). The results indicated that, like the strongest promoter (J23100), both the second (J23106) and third (J23105) strong promoters also led to an increased plasmid size on days 4 and 5, respectively (Supplementary Figure S1). In contrast, no plasmid mutations were observed with two low-strength promoters (J23114 and J23109) even after 5 days of serial dilution/cultivation (Supplementary Figure S1). While the above problem did not happen to Bxb1 and Phic31, we finally chose the lowest strength promoter (J23109) for all three plasmids’ construction to avoid potential plasmid mutations (pFB10, pFB13 and pFB15) (Figure 1C). After transformation, each plasmid could stably exist in E. coli, and expressed integrase was active to catalyze the DNA recombination (Figure 1D and Supplementary Figure S2). Notably, two or three compatible plasmids were able to coexist in E. coli for at least 6 days (Supplementary Figure S3). In addition, these plasmids exhibited no obvious cell growth inhibition on E. coli as shown in Supplementary Figure S4. All three integrases were fused with a C-terminal 6×His tag and their expression was analyzed by western blotting. As can be seen in Figure 1E, three integrases could be expressed individually or together in E. coli, demonstrating the compatibility of three plasmids and their ability for constitutive soluble protein expression.
With three integrase ‘keys’ in hand, we next constructed seven ‘lock’ circuits, in which two DNA parts were alterable (Figure 1F). The first part consists of a promoter, terminator(s) and attP/attB sites in different orders. This part determines the matching rule between ‘keys’ and ‘locks’. Only when proper integrases are present, the terminators between relevant attP/attB sites will be deleted, leading to the downstream gene expression (Supplementary Figure S5). The second part is inserted with one of the seven protein coding sequences working as ‘output’ signals. For easy observation and discrimination, we chose seven chromoproteins as our outputs: eforRed (33), fwYellow (34), amilCP (33), CyOFP1 (35), tsPurple (34), amajLime (36) and asPink (34). After plasmid construction, the correct sizes of seven ‘lock’ plasmids were first confirmed by a restriction digestion test (Figure 1G). Since repeated genetic elements (e.g. promoter and terminator) in one plasmid might cause plasmid instability (37), we then carried out a 6-day serial dilution/cultivation to test the stability of each ‘lock’ plasmid (pFB20–pFB26). The results showed that seven ‘lock’ plasmids were stable with expected DNA fragment sizes across at least 6-day cultivation (Supplementary Figure S6). In addition, seven chromoproteins could be expressed with distinct colors in E. coli using normal constitutive expression plasmids (pFB131–pFB137, Supplementary Figure S7). Therefore, they can be used as indicators for different output signals in our following studies.
Evaluating the orthogonality of three ‘keys’ and seven ‘locks’
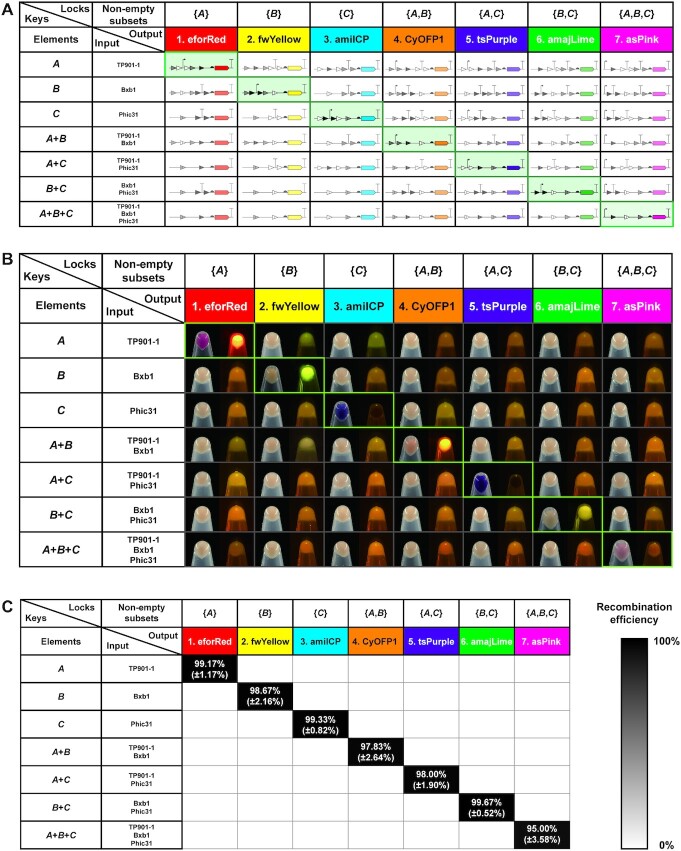
Having successfully constructed the plasmids of three ‘keys’ and seven ‘locks’, we next set out to explore whether ‘keys’ can manipulate ‘locks’ orthogonally. To this end, a 7 × 7 orthogonal table was designed, showing a total of 49 possible combinations with ‘keys’ and ‘locks’ (Figure 2A). In the table, only seven proper matches along the diagonal line (from top left to bottom right) will result in the deletion of terminators between promoter and RBS and activate the subsequent chromoprotein expression. However, the rest of the mismatches will make incorrect organization of promoter and terminator in front of the RBS and thus lead to the failure expression of chromoproteins. To test the orthogonality, we first transformed seven different combinations of ‘key’ plasmids into E. coli to create seven distinct E. coli strains. Then, seven ‘lock’ plasmids were individually transformed into above seven E. coli strains, resulting in a total of 49 E. coli strains with all possible combinations of ‘keys’ and ‘locks’ (for details, see Figure 2B). For comparison, we cultivated all E. coli strains in liquid LB and collected cell pellets in 1.5 ml tubes. Then, the pellets were photographed under bright field and LED transilluminator. The results indicated that our KmL model worked precisely as designed and only correct KmL strains on the diagonal line were able to express chromoproteins (Figure 2B, from top left to bottom right; for the plasmid combinations of seven correct KmL strains, see Supplementary Figure S8). Clearly, colored E. coli pellets were visible under bright field and/or LED transilluminator, whereas other mismatched strains were observed as a wild-type E. coli color. Thus, our results demonstrated that the KmL system is highly orthogonal to one another. In addition, we were curious to know the recombination efficiency of our KmL system. To do this, seven KmL strains were cultivated overnight on LB agar plates and then the colored colonies were counted to calculate the recombination efficiency (Supplementary Figure S8). The data suggested that the efficiency of each KmL was remarkably high, ranging from 95% to nearly 100% (Figure 2C). This is in line with previous reports that orthogonal serine integrases can precisely bind their own recognition sites and efficiently catalyze the DNA recombination (7,16). In addition, we designed another ‘binary to decimal converter’ genetic circuit using three ‘lock’ biobricks that can respond to serine integrase inputs (‘keys’) separately or together, leading to eight different outputs (states) in one E. coli strain. The results also indicated that ‘keys’ could manipulate ‘locks’ with high orthogonality and efficiency (Supplementary Figure S9), suggesting that the KmL model can be potentially applied to other circuit designs.
Figure 2.
Orthogonality and performance of the KmL model. (A) Schematic diagram of the orthogonality matrix. Elements in the first column are input ‘keys’ (A: TP901-1; B: Bxb1; and C: Phic31). Nonempty subsets in the first line represent seven different ‘lock’ plasmids containing chromoproteins as outputs. The 49 organizations of DNA parts shown in the table indicate the ‘key’-switched ‘lock’ plasmids. Only seven proper matches on the diagonal line (from top left to bottom right) can activate the expression of chromoproteins. The remaining mismatches make incorrect organization of DNA parts (i.e. promoter and terminator) leading to no protein expression. (B) Escherichia coli cells are collected and photographed under bright field (left) and LED transilluminator (right). Each E. coli sample is collected in a 1.5 ml centrifuge tube from 3 ml overnight culture. Only cell pellets on the diagonal line (from top left to bottom right) with proper key–lock matches show chromoprotein colors. All samples are repeated at least three times with similar results. (C) Recombination efficiency of the seven proper key–lock matches. Each value (mean ± SD) is calculated with six biological replicates by counting at least 100 colonies on LB agar plates.
Enhancing the SYMBIOSIS system to amplify output signals
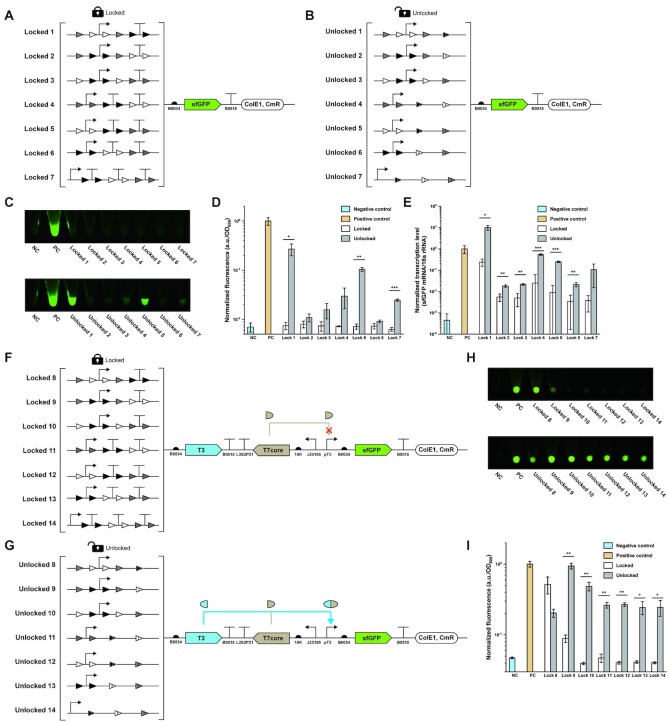
While we have demonstrated the success of using our KmL model for SYMBIOSIS in vivo, we observed that the colors of the seven ‘unlocked’ E. coli pellets were lower to different levels as compared to their positive controls (pFB131–pFB137, Supplementary Figure S10). We hypothesized that the residual attL/attR sites between the promoter and RBS in the 5′-UTR region might form mRNA secondary structures, which serve as steric blocks for ribosome binding and hinder the downstream protein translation (38,39). To test this hypothesis and find out the reason for low protein expression, we next sought to investigate the transcription and translation levels of designed genetic circuits. First, we replaced all chromoprotein coding sequences with the same reporter gene (i.e. sfGFP) for easy quantification and comparison (Figure 3A and B). After cultivation, the fluorescence of each sample was measured. We found that no sfGFP fluorescence was detected from the group of ‘locked’ genetic circuits (Figure 3C, top panel, and D), suggesting no sfGFP expression leakage. When each ‘locked’ circuit was switched on, fluorescent sfGFP was clearly expressed. However, their expression levels varied significantly and were lower than that of the positive control (Figure 3C, bottom panel, and D). This is in agreement with the expression results of chromoproteins (Supplementary Figure S10), indicating that DNA sequence of the 5′-UTR region indeed affected the transcription and/or translation level. To quantify the transcription level, we carried out RT-qPCR experiments to calculate the ratio of sfGFP mRNA/16S rRNA in E. coli (Figure 3E and Supplementary Figures S11–S13). Overall, the amount of sfGFP mRNA from the ‘unlocked’ circuits was much higher than that from the corresponding ‘locked’ ones, which turned out that the terminator(s) located between promoter and RBS significantly blocked the transcription process. Meanwhile, we also noticed that the levels of transcribed sfGFP mRNA from the ‘locked’ circuits were higher than that of the negative control. However, the basal mRNA transcription leakage did not lead to detectable protein expression (Figure 3C, top panel, and D). This is likely because the translation process (e.g. the initiation stage) is completely blocked by the upstream 5′-UTR that contains both terminator(s) and multiple integrase recognition sites. In addition, we found that even if the transcription level of ‘unlocked’ circuit 1 was higher than the positive control (Figure 3E), its sfGFP translation level was still lower (Figure 3D). This result further suggested that residual recognition sites (attL/attR) could reduce the translation efficiency. Furthermore, we predicted the mRNA secondary structures of seven genetic circuits under ‘locked’ and ‘unlocked’ states. The models showed that stem–loop structures near the RBS sites (Supplementary Figures S14 and S15) might limit ribosomes to bind RBS sites (40,41). Taken together, our results demonstrated that diverse residual attL/attR sites (Figure 3B) located in 5′-UTR played different influential roles on both transcription and translation levels (Figure 3D and E).
Figure 3.
Quantitative analysis and enhancement of the SYMBIOSIS system. (A, B) Schematics of ‘locked’ (pFB34–pFB40) and ‘unlocked’ (pFB41–pFB47) plasmids for quantitative analysis (for details of all plasmids, see Supplementary Table S5). (C) sfGFP fluorescence of E. coli pellets. NC, negative control (pSB1C3); PC, positive control (pFB141). Each E. coli sample is collected in a 1.5 ml centrifuge tube from 3 ml overnight culture. All samples are repeated by three biological replicates with similar results. (D) Normalized sfGFP fluorescence values (a.u./OD600) corresponding to panel (C). Fluorescence values are normalized to the one of positive control. Each value (mean ± SD) is calculated with three biological replicates and the error bar represents the SD. Student’s t-tests are used for statistical analysis, and P < 0.05 indicates statistical significance (*P < 0.05, **P < 0.01 and ***P < 0.001). (E) Normalized transcription levels (sfGFP mRNA/16S rRNA) of the sfGFP gene. Transcription levels are normalized to the one of positive control. Each value (mean ± SD) is calculated with three biological replicates and the error bar represents the SD. Student’s t-tests are used for statistical analysis, and P < 0.05 indicates statistical significance (*P < 0.05, **P < 0.01 and ***P < 0.001). (F, G) Schematics of improved ‘locked’ (pFB48–pFB54) and ‘unlocked’ (pFB55–pFB61) plasmids with signal amplification circuits (for details of all plasmids, see Supplementary Table S5). (H) sfGFP fluorescence of E. coli pellets. The experiment is performed in the same way as described in panel (C). (I) Normalized sfGFP fluorescence values (a.u./OD600) corresponding to panel (H). Fluorescence values are normalized to the one of positive control. Each value (mean ± SD) is calculated with three biological replicates and the error bar represents the SD. Student’s t-tests are used for statistical analysis, and P < 0.05 indicates statistical significance (*P < 0.05, **P < 0.01 and ***P < 0.001).
In order to reduce the influence of residual attL/attR sites on the downstream protein translation, we next aimed to use ‘genetic amplifier’ as a potential solution to enhance the output expression level (15). Such strategy can be achieved by using the so-called transcriptional amplifier like the strong bacteriophage T7 RNA polymerase for cascaded signal amplification. However, direct overexpression of T7 RNA polymerase may lead to severe cell toxicity (42). We therefore chose a reported ‘resource allocator’ as our genetic amplifier (43–45). The resource allocator consists of two parts: a nonactive ‘core’ fragment of T7 RNA polymerase and a T3 σ fragment that contains the DNA binding domain. Once the two fragments are expressed, they will bind together to form an active RNA polymerase to transcribe the pT3 promoter-mediated downstream gene sequence. For this, we modified our original ‘lock’ plasmids (Figure 3A) by inserting both T3 σ and T7 core fragments between the ‘locked’ circuits and the sfGFP sequence (pFB48–pFB54, Figure 3F). In this case, the T7 core fragment was expressed from a constitutive promoter J23105. When the circuits were unlocked, T3 σ fragments would be expressed and bind to the T7 core fragments to transcribe the reporter sfGFP gene (Figure 3G). By using this ‘genetic amplifier’ strategy, the sfGFP expression levels were substantially improved. The ‘unlocked’ circuit 9 enabled the expression level as high as the positive control (Figure 3H, bottom panel, and I). However, it was unexpected that ‘locked’ circuit 8 also gave rise to notable sfGFP expression, which is even higher than that of the ‘unlocked’ circuit 8 (Figure 3H and I). We ascribed this observation to two reasons. First, the leaky transcription (see the transcription level of ‘lock’ 1 in Figure 3E) and translation of T3 σ fragments could form sufficient T3–T7 RNA polymerase to activate sfGFP synthesis. Second, overexpression of T3 σ fragments with the ‘unlocked’ circuit 8 might take up too much cellular resources, leading to a low sfGFP expression. Nonetheless, our results demonstrated that (i) the inhibiting effects of residual attL/attR sites if any on protein translation vary based on different DNA sequences (Figure 3D); (ii) the final output signal in most cases can be amplified to a higher level by using a ‘genetic amplifier’ (Figure 3I); and (iii) our SYMBIOSIS system is highly adaptable and can be rationally modified and improved for other synthetic biology applications.
Expanding SYMBIOSIS to assemble multiple plasmids in vitro
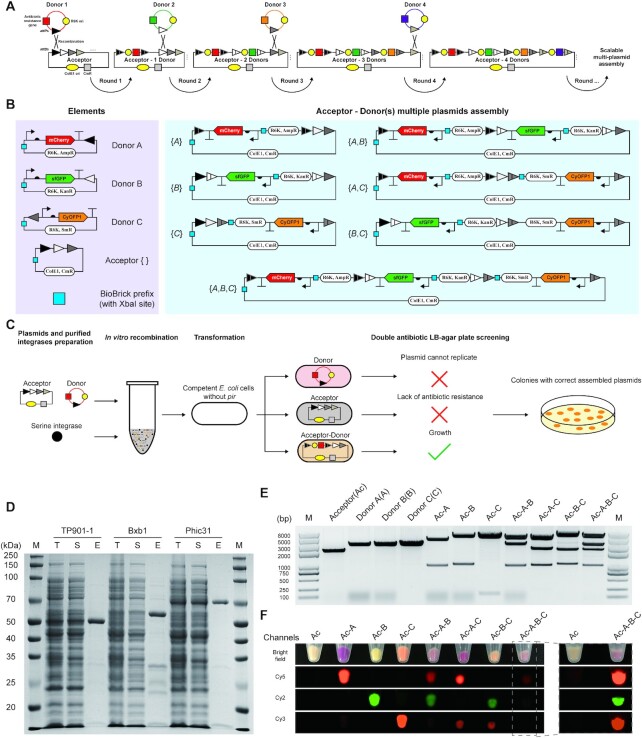
To expand the concept of SYMBIOSIS for synthetic biology applications, we next aimed to establish in vitro SYMBIOSIS for assembling multiple plasmids (Figure 4A), which can be transformed into cells for in vivo protein expression. To do this, two groups of plasmids named ‘acceptor’ and ‘donor’ are initially designed. ‘Acceptor’ plasmids have various attB sites. ‘Donor’ plasmids consist of three parts: (i) the origin of replication (ori) of R6K, which prevents the plasmids from correct replication in E. coli strains without the gene pir (46); (ii) an antibiotic resistance gene that is unique to the specific ‘donor’ plasmids; and (iii) an attP site, which can be recognized by serine integrase and mediates attP–attB site-specific recombination with an ‘acceptor’ plasmid. Furthermore, the ‘donor’ plasmids can carry target biobricks, for example gene coding sequences. From this, the ‘donor’ plasmid can be integrated into the ‘acceptor’ backbone after the first-round recombination, forming a new combined ‘acceptor–donor’ plasmid. This newly constructed plasmid can serve as an ‘acceptor’ in the second-round integration to accept another donor DNA sequence. After several rounds, different composite ‘acceptor–donor(s)’ plasmids can be assembled for the subsequent transformation and gene expression.
Figure 4.
SYMBIOSIS enables in vitro multiple plasmid assembly. (A) Principle and design of scalable ‘acceptor–donor(s)’ assembly. In the first round, the ‘donor 1’ plasmid is integrated into the ‘acceptor’ backbone by one serine integrase, forming a combined ‘acceptor–1 donor’, which can serve as a new ‘acceptor’ in the next round of assembly. The ‘acceptor’ plasmid has different attB sites, which are paired to their cognate attP sites in the ‘donor’ plasmids. ‘Donor’ plasmids use the same R6K ori and each one has its own unique antibiotic resistance gene, allowing for the subsequent plasmid screening. In addition, each ‘donor’ plasmid carries a target biobrick (e.g. gene coding sequence) for desired application. (B) A paradigm of in vitro SYMBIOSIS for multiple plasmid assembly. Left, three ‘donor’ plasmids (pFB63–pFB65) harboring three fluorescent proteins, respectively, and one ‘acceptor’ (pFB62). Right, seven composite plasmids after assembly (pFB66–pFB72). The details of these plasmids are shown in Supplementary Table S5. (C) Schematic workflow of in vitro SYMBIOSIS. (D) SDS-PAGE analysis of three purified serine integrases (TP901-1: 56.5 kDa; Bxb1: 57.2 kDa; and Phic31: 67.9 kDa). T, cell lysate; S, supernatant fraction; E, purified enzyme. (E) DNA fragment sizes of the ‘acceptor’, ‘donors’ and assembled plasmids after XbaI digestion. All expected bands are observed on the agarose gel (from left to right, pFB62: 2218 bp; pFB63: 3284 bp; pFB64: 3243 bp; pFB65: 3276 bp; pFB66: 4516 + 986 bp; pFB67: 4410 + 1051 bp; pFB68: 5341 + 153 bp; pFB69: 4410 + 3349 + 986 bp; pFB70: 5341 + 2451 + 986 bp; pFB71: 5341 + 2345 + 1051 bp; pFB72: 5341 + 3349 + 2345 + 986 bp). (F) Orthogonal fluorescence detection of E. coli with assembled plasmids. Four channels (bright field, Cy5, Cy2 and Cy3) are used to detect the fluorescence of all cell pellet samples. For E. coli containing the triple assembled plasmid (pFB72, Ac-A-B-C), the pellet sample is also independently detected with the negative control of ‘acceptor’ (pFB62, Ac) to show clear fluorescent signals. All samples are repeated by three biological replicates with similar results.
To demonstrate the capability of in vitro SYMBIOSIS, we first constructed three ‘donor’ plasmids with different attP sites and one ‘acceptor’ plasmid containing three sequential attB sites (Figure 4B, left). In each plasmid, a ‘biobrick prefix’ part with one XbaI restriction site was inserted in an appropriate position to check the correctness of assembled plasmids. In addition, each ‘donor’ plasmid contained its own fluorescent protein gene, which can be integrated into the ‘acceptors’ to verify protein expression (Figure 4B, right). As shown in the schematic workflow (Figure 4C), ‘donor’ and ‘acceptor’ plasmids are mixed with purified serine integrases (TP901-1, Bxb1 and Phic31) for plasmid recombination in vitro. Then, the reaction mixture is transformed into E. coli (pir–) competent cells. By double antibiotic resistance screening, only the cells harboring the correctly assembled ‘acceptor–donor’ plasmids can grow on the LB agar plates (Figure 4C).
The expression of three serine integrases was carried out in E. coli Rosetta-gami(DE3) pLysS. However, the initially expressed enzymes of TP901-1 and Phic31 were predominantly insoluble (Supplementary Figure S16). Then, we switched to a low concentration of inducer and a low cultivation temperature (for details, see the ‘Materials and Methods’ section). By doing this, all serine integrases were expressed in soluble forms and could be purified with high purity (Figure 4D). Next, the conditions for in vitro plasmid recombination were investigated. The optimal reaction contained 1 μM of integrase and was performed at 30°C for 120 min (Supplementary Figure S17). In total, seven assembled plasmids were generated (Figure 4B, right) and their correctness was verified by XbaI digestion, each showing the correct digested DNA fragments on the gel (Figure 4E). Afterward, they were each transformed into E. coli cells, respectively, for protein expression. The results showed that all fluorescent protein genes on the relevant plasmids could be successfully expressed in single, in pairs or in trios (Figure 4F). Furthermore, in vitro recombination efficiency was calculated and the value reached 100% from each group of the assembled plasmids (Supplementary Figure S18). These results demonstrated that our SYMBIOSIS system was able to efficiently catalyze multiple plasmid assembly in vitro, providing a promising and reliable approach for plasmid construction.
SYMBIOSIS creates colored E. coli
Following the demonstration of in vitro SYMBIOSIS, we next sought to further extend this proof of concept to create colored E. coli strains by recombination of different chromoprotein genes. While E. coli cells were able to show a featured color with a single chromoprotein (Supplementary Figure S7), E. coli cells with composite colors have not been shown before. Therefore, we chose six different chromoproteins eforRed, fwYellow, amilCP, CyOFP1, tsPurple and amajLime, which represent six basic colors of red, yellow, blue, orange, purple and green, respectively. The goal was to use them to make various double or triple combinations with the hope to generate novel composite colors. To achieve this goal, the selected chromoprotein genes were individually constructed in three ‘donor’ vectors (pFB65 and pFB73–pFB89, Supplementary Table S5). Then, we ran in vitro SYMBIOSIS to generate 15 double and 20 triple combinations, leading to a total of 35 different expression plasmids.
To standardize the colorimetry process, we built up a step-by-step procedure for E. coli imaging and colorimetry (Figure 5A). First, colored E. coli pellets were collected in transparent 1.5 ml Eppendorf tubes. Then, the color of cell pellets was compared to a color chart. When the best standard color was matched, the image was taken in a photo studio (Supplementary Figure S19) and then its RGB parameters were captured by Photoshop on a computer (note that no image parameters such as brightness and contrast were adjusted with Photoshop). This standardization procedure was performed for all color conversions from real cell colors to digital RGB colors. To begin, we compared the colors of E. coli pellets, each containing one of the six basic chromoproteins, with the color chart. We observed that each color matched well with the standard color and could be converted to an RGB color (Figure 5B). Next, we used the six chromoproteins to make different combinations via in vitro SYMBIOSIS, yielding 35 assembled plasmids. Their correctness was verified by XbaI digestion and the sizes of digested DNA fragments were confirmed on the gel (Supplementary Figures S20 and S21). All plasmids were transformed into E. coli for chromoprotein co-expression, generating a variety of colored cells (Supplementary Figures S22 and S23). Most interestingly, some colors have never been observed before in E. coli, for example orient red (pFB118), patina green (pFB107), blue gray (pFB129) and claret violet (pFB105). In particular, we were able to choose representative colors from the 6 basic colors and our 35 composite colors to span the color gradient (Figure 5C). Taken together, these results demonstrated the power of using in vitro SYMBIOSIS to construct large, complex plasmids for defined applications like the creation of novel colored E. coli cells in this work. Moreover, we envision that these colored E. coli strains might be used in different fields such as living pigments for kid’s painting education and living materials for environmental response.
Figure 5.
Creation of colored E. coli by SYMBIOSIS. (A) Workflow for the preparation of colored E. coli cells and colorimetry. Each E. coli sample is collected in a 1.5 ml centrifuge tube from 3 ml overnight culture and matched with the color chart. When the cell color matches the standard color, the image is taken in a mini photo studio (Supplementary Figure S19) and further processed to convert the real cell color to a digital RGB color. (B) Six basic colors of red (R), yellow (Y), blue (B), orange (O), purple (P) and green (G) are generated with six chromoproteins (pFB90–pFB95, Supplementary Table S5), respectively. They are used for double or triple combinations to create colored E. coli cells. (C) Representative colored E. coli cells spanning the color gradient. All samples are repeated by three biological replicates with similar results.
DISCUSSION
We demonstrate a serine integrase-based tool platform, called SYMBIOSIS, which can be used to directly and precisely manipulate DNA parts in vivo and in vitro. In one scenario, we propose a KmL model framework, in which three ‘keys’ (i.e. three orthogonal serine integrases TP901-1, Bxb1 and Phic31) and their combinations can successfully unlock seven genetic circuits with high efficiency in E. coli cells. If the output signal (the protein expression level) is low due to the inhibitory effect of residual attL/attR sites, it can be amplified and enhanced by inserting a ‘genetic amplifier’ to the ‘lock’ plasmids. In another scenario, we apply SYMBIOSIS in vitro to assemble multiple plasmids, which gives rise to a novel method that catalyzes the assembly of circular plasmids rather than linear DNA fragments. Using this strategy, we construct 35 plasmids that carry different combinations of six chromoprotein genes. These plasmids are able to mediate in vivo protein co-expression, creating various colored E. coli cells with some novel and never reported colors such as patina green, blue gray and claret violet.
Overall, our work highlights the potential applications of serine integrases, which will be a promising addition to the current genetic toolkit of synthetic biology. Although we only use three serine integrases to verify the SYMBIOSIS system in this study, such a key concept may be applied to many other orthogonal phage integrases that have been increasingly discovered (30). This will allow researchers to design and manipulate more complex genetic circuits and study their performance in vivo. Using serine integrases, we also establish a new strategy for in vitro multigene assembly based on ‘donor’ and ‘acceptor’ plasmids as direct inputs, but not linear PCR fragments. As compared to a previous ‘ACEMBL’ method (28,29), which enables plasmid fusion via Cre-LoxP recombination but unavoidably generates plasmid variants due to only one kind of LoxP sites available, our SYMBIOSIS technology utilizes orthogonal serine integrases and each of them has a specific recognition site, making the plasmid architecture and the position of inserted genes more predictable. Moreover, we showcase the success of using in vitro SYMBIOSIS to assemble two or three chromoprotein genes on one plasmid for in vivo heterologous co-expression. This endows E. coli cells with featured new colors. As such, other new functionalities might be generated for E. coli to perform desirable applications, for example construction of complex metabolic pathways when co-expression of all enzymes using multiple plasmids in one cell remains difficult or not possible.
Looking forward, we anticipate that SYMBIOSIS together with currently versatile genetic tools will further facilitate DNA manipulation for synthetic biology applications. Especially, our SYMBIOSIS, driven by serine integrases, can address two requirements. First, fewer integrases are able to mediate more genetic circuits in vivo by designing a logic relationship (e.g. the KmL model). Second, multicomponent DNA assembly in vitro enables tunable construction of one plasmid carrying multiple genes for executing in vivo biological functions. Building on these achievements, future developments of SYMBIOSIS could seek to discover and study more serine integrases, which will expand the repertoire of serine integrase-based tools and thus help advance the efforts to design, build and test for fundamental and applied research in synthetic biology.
Supplementary Material
Contributor Information
Fang Ba, School of Physical Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Yushi Liu, School of Physical Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Wan-Qiu Liu, School of Physical Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Xintong Tian, School of Physical Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Jian Li, School of Physical Science and Technology, ShanghaiTech University, Shanghai 201210, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31971348 and 32171427]; Natural Science Foundation of Shanghai [19ZR1477200]; ShanghaiTech University [to J.L.]. Funding for open access charge: ShanghaiTech University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Gardner T.S., Cantor C.R., Collins J.J.. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000; 403:339–342. [DOI] [PubMed] [Google Scholar]
- 2. Elowitz M.B., Leibler S.. A synthetic oscillatory network of transcriptional regulators. Nature. 2000; 403:335–338. [DOI] [PubMed] [Google Scholar]
- 3. Brophy J.A.N., Voigt C.A.. Principles of genetic circuit design. Nat. Methods. 2014; 11:508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meng F., Ellis T.. The second decade of synthetic biology: 2010–2020. Nat. Commun. 2020; 11:5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sheth R.U., Wang H.H.. DNA-based memory devices for recording cellular events. Nat. Rev. Genet. 2018; 19:718–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grindley N.D.F., Whiteson K.L., Rice P.A.. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006; 75:567–605. [DOI] [PubMed] [Google Scholar]
- 7. Merrick C.A., Zhao J., Rosser S.J.. Serine integrases: advancing synthetic biology. ACS Synth. Biol. 2018; 7:299–310. [DOI] [PubMed] [Google Scholar]
- 8. Breüner A., Brøndsted L., Hammer K.. Resolvase-like recombination performed by the TP901-1 integrase. Microbiology. 2001; 147:2051–2063. [DOI] [PubMed] [Google Scholar]
- 9. Stoll S.M., Ginsburg D.S., Calos M.P.. Phage TP901-1 site-specific integrase functions in human cells. J. Bacteriol. 2002; 184:3657–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim A.I., Ghosh P., Aaron M.A., Bibb L.A., Jain S., Hatfull G.F.. Mycobacteriophage Bxb1 integrates into the Mycobacterium smegmatis groEL1 gene. Mol. Microbiol. 2003; 50:463–473. [DOI] [PubMed] [Google Scholar]
- 11. Smith M.C., Till R., Brady K., Soultanas P., Thorpe H., Smith M.C.. Synapsis and DNA cleavage in phiC31 integrase-mediated site-specific recombination. Nucleic Acids Res. 2004; 32:2607–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Colloms S.D., Merrick C.A., Olorunniji F.J., Stark W.M., Smith M.C., Osbourn A., Keasling J.D., Rosser S.J.. Rapid metabolic pathway assembly and modification using serine integrase site-specific recombination. Nucleic Acids Res. 2014; 42:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brown W.R., Lee N.C., Xu Z., Smith M.C.. Serine recombinases as tools for genome engineering. Methods. 2011; 53:372–379. [DOI] [PubMed] [Google Scholar]
- 14. Siuti P., Yazbek J., Lu T.K.. Synthetic circuits integrating logic and memory in living cells. Nat. Biotechnol. 2013; 31:448–452. [DOI] [PubMed] [Google Scholar]
- 15. Bonnet J., Yin P., Ortiz M.E., Subsoontorn P., Endy D.. Amplifying genetic logic gates. Science. 2013; 340:599–603. [DOI] [PubMed] [Google Scholar]
- 16. Roquet N., Soleimany A.P., Ferris A.C., Aaronson S., Lu T.K.. Synthetic recombinase-based state machines in living cells. Science. 2016; 353:aad8559. [DOI] [PubMed] [Google Scholar]
- 17. Zhao J., Pokhilko A., Ebenhöh O., Rosser S.J., Colloms S.D.. A single-input binary counting module based on serine integrase site-specific recombination. Nucleic Acids Res. 2019; 47:4896–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guiziou S., Mayonove P., Bonnet J.. Hierarchical composition of reliable recombinase logic devices. Nat. Commun. 2019; 10:456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zúñiga A., Guiziou S., Mayonove P., Meriem Z.B., Camacho M., Moreau V., Ciandrini L., Hersen P., Bonnet J.. Rational programming of history-dependent logic in cellular populations. Nat. Commun. 2020; 11:4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim T., Weinberg B., Wong W., Lu T.K.. Scalable recombinase-based gene expression cascades. Nat. Commun. 2021; 12:2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fischer K.B., Collins H.K., Callaway E.M.. Sources of off-target expression from recombinase-dependent AAV vectors and mitigation with cross-over insensitive ATG-out vectors. Proc. Natl Acad. Sci. U.S.A. 2019; 116:27001–27010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Di Blasi R., Zouein A., Ellis T., Ceroni F.. Genetic toolkits to design and build mammalian synthetic systems. Trends Biotechnol. 2021; 39:1004–1018. [DOI] [PubMed] [Google Scholar]
- 23. Shuman S. Recombination mediated by vaccinia virus DNA topoisomerase I in Escherichia coli is sequence specific. Proc. Natl Acad. Sci. U.S.A. 1991; 88:10104–10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shuman S. Novel approach to molecular cloning and polynucleotide synthesis using vaccinia DNA topoisomerase. J. Biol. Chem. 1994; 269:32678–32684. [PubMed] [Google Scholar]
- 25. Engler C., Gruetzner R., Kandzia R., Marillonnet S.. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One. 2009; 4:e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hartley J.L. Use of the gateway system for protein expression in multiple hosts. Curr. Protoc. Protein Sci. 2003; Chapter 5:Unit 5.17. [DOI] [PubMed] [Google Scholar]
- 27. Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A. 3rd, Smith H.O.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–345. [DOI] [PubMed] [Google Scholar]
- 28. Bieniossek C., Nie Y., Frey D., Olieric N., Schaffitzel C., Collinson I., Romier C., Berger P., Richmond T.J., Steinmetz M.O.et al.. Automated unrestricted multigene recombineering for multiprotein complex production. Nat. Methods. 2009; 6:447–450. [DOI] [PubMed] [Google Scholar]
- 29. Nie Y., Chaillet M., Becke C., Haffke M., Pelosse M., Fitzgerald D., Collinson I., Schaffitzel C., Berger I.. ACEMBL tool-kits for high-throughput multigene delivery and expression in prokaryotic and eukaryotic hosts. Adv. Exp. Med. Biol. 2016; 896:27–42. [DOI] [PubMed] [Google Scholar]
- 30. Yang L., Nielsen A.A., Fernandez-Rodriguez J., McClune C.J., Laub M.T., Lu T.K., Voigt C.A.. Permanent genetic memory with >1-byte capacity. Nat. Methods. 2014; 11:1261–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guzman L.M., Belin D., Carson M.J., Beckwith J.. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995; 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lutz R., Bujard H.. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and Arac/I1–I2 regulatory elements. Nucleic Acids Res. 1997; 25:1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alieva N.O., Konzen K.A., Field S.F., Meleshkevitch E.A., Hunt M.E., Beltran-Ramirez V., Miller D.J., Wiedenmann J., Salih A., Matz M.V.. Diversity and evolution of coral fluorescent proteins. PLoS One. 2008; 3:e2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liljeruhm J., Funk S.K., Tietscher S., Edlund A.D., Jamal S., Wistrand-Yuen P., Dyrhage K., Gynnå A., Ivermark K., Lövgren J.et al.. Engineering a palette of eukaryotic chromoproteins for bacterial synthetic biology. J. Biol. Eng. 2018; 12:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chu J., Oh Y., Sens A., Ataie N., Dana H., Macklin J.J., Laviv T., Welf E.S., Dean K.M., Zhang F.et al.. A bright cyan-excitable orange fluorescent protein facilitates dual-emission microscopy and enhances bioluminescence imaging in vivo. Nat. Biotechnol. 2016; 34:760–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Matz M.V., Fradkov A.F., Labas Y.A., Savitsky A.P., Zaraisky A.G., Markelov M.L., Lukyanov S.A.. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 1999; 17:969–973. [DOI] [PubMed] [Google Scholar]
- 37. Wan X., Pinto F., Yu L., Wang B.. Synthetic protein-binding DNA sponge as a tool to tune gene expression and mitigate protein toxicity. Nat. Commun. 2020; 11:5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Leppek K., Das R., Barna M.. Functional 5' UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 2018; 19:158–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Seo S.W., Yang J.S., Kim I., Yang J., Min B.E., Kim S., Jung G.Y.. Predictive design of mRNA translation initiation region to control prokaryotic translation efficiency. Metab. Eng. 2013; 15:67–74. [DOI] [PubMed] [Google Scholar]
- 40. Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003; 31:3429–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gruber A.R., Lorenz R., Bernhart S.H., Neuböck R., Hofacker I.L.. The Vienna RNA websuite. Nucleic Acids Res. 2008; 36:W70–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tan S.I., Ng I.S.. New insight into plasmid-driven T7 RNA polymerase in Escherichia coli and use as a genetic amplifier for a biosensor. ACS Synth. Biol. 2020; 9:613–622. [DOI] [PubMed] [Google Scholar]
- 43. Segall-Shapiro T.H., Meyer A.J., Ellington A.D., Sontag E.D., Voigt C.A.. A ‘resource allocator’ for transcription based on a highly fragmented T7 RNA polymerase. Mol. Syst. Biol. 2014; 10:742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fernandez-Rodriguez J., Moser F., Song M., Voigt C.A.. Engineering RGB color vision into Escherichia coli. Nat. Chem. Biol. 2017; 13:706–708. [DOI] [PubMed] [Google Scholar]
- 45. Moser F., Tham E., González L.M., Lu T.K., Voigt C.A.. Light-controlled, high-resolution patterning of living engineered bacteria onto textiles, ceramics, and plastic. Adv. Funct. Mater. 2019; 29:1901788. [Google Scholar]
- 46. Rakowski S.A., Filutowicz M.. Plasmid R6K replication control. Plasmid. 2013; 69:231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.