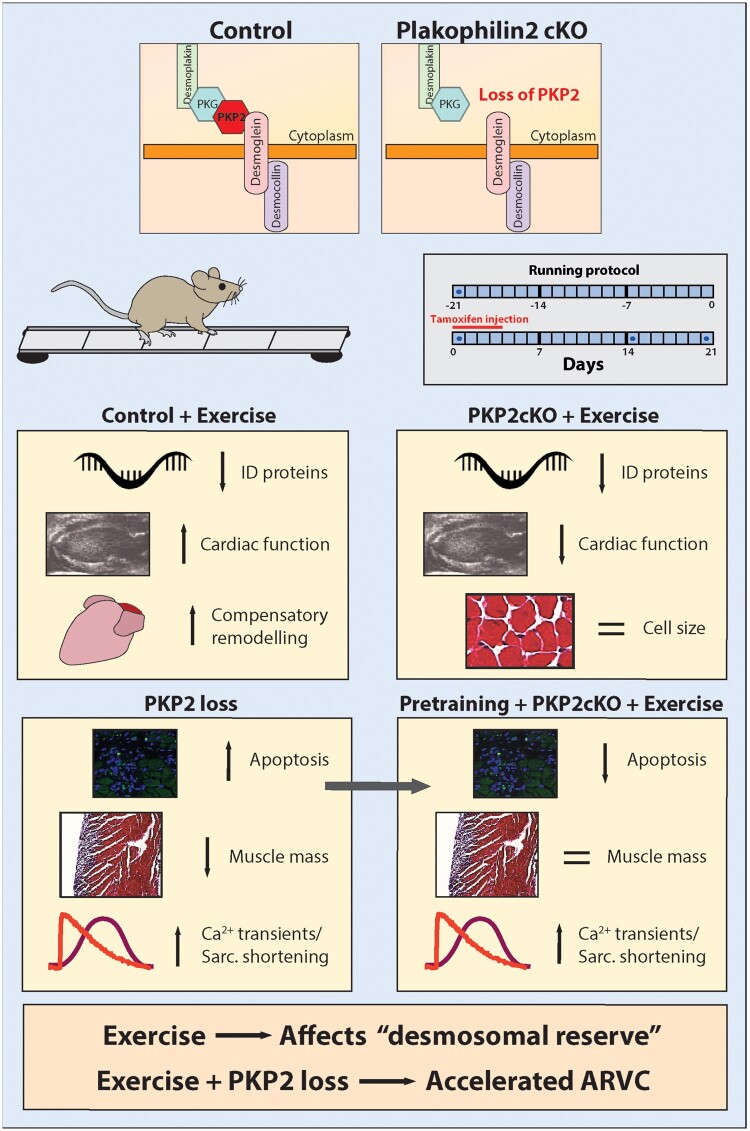
Abstract
Aims
Exercise increases arrhythmia risk and cardiomyopathy progression in arrhythmogenic right ventricular cardiomyopathy (ARVC) patients, but the mechanisms remain unknown. We investigated transcriptomic changes caused by endurance training in mice deficient in plakophilin-2 (PKP2cKO), a desmosomal protein important for intercalated disc formation, commonly mutated in ARVC and controls.
Methods and results
Exercise alone caused transcriptional downregulation of genes coding intercalated disk proteins. The changes converged with those in sedentary and in exercised PKP2cKO mice. PKP2 loss caused cardiac contractile deficit, decreased muscle mass and increased functional/transcriptomic signatures of apoptosis, despite increased fractional shortening and calcium transient amplitude in single myocytes. Exercise accelerated cardiac dysfunction, an effect dampened by pre-training animals prior to PKP2-KO. Consistent with PKP2-dependent muscle mass deficit, cardiac dimensions in human athletes carrying PKP2 mutations were reduced, compared to matched controls.
Conclusions
We speculate that exercise challenges a cardiomyocyte “desmosomal reserve” which, if impaired genetically (e.g., PKP2 loss), accelerates progression of cardiomyopathy.
Keywords: Arrhythmogenic right ventricular cardiomyopathy, ARVC, Plakophilin-2, Desmosomes, Exercise
Graphical Abstract
See the editorial comment for this article ‘Cardiac desmosomal reserve: another piece of the exercise-induced arrhythmogenic cardiomyopathy puzzle?’, by Kristina H. Haugaa and Christine Rootwelt-Norberg, https://doi.org/10.1093/eurheartj/ehab8733.
Translational perspective
Endurance exercise leads to changes in the heart that can evolve from beneficial to deleterious (‘athlete’s heart’). Exercise also accelerates disease progression in patients with arrhythmogenic right ventricular cardiomyopathy, a condition often caused by mutations in the desmosomal protein plakophilin-2 (PKP2). Limitations of experimental models notwithstanding, we speculate that exercise challenges a ‘desmosomal reserve’ which, if insufficient, accelerates cardiomyopathy progression. Moreover, exercise in PKP2-deficient hearts is not followed by a compensatory trophic effect but rather, by increased apoptosis. The latter impedes the heart of an athlete with low desmosomal reserve from matching increased demand, leading to cell damage and reduced muscle mass which, if exercise persists, causes a clinically apparent cardiomyopathy.
Introduction
Plakophilin-2 (PKP2) is classically defined as a protein of the desmosome, an intercellular adhesion structure residing in the cardiac intercalated disc (ID). Recent studies demonstrate that in addition to cell–cell adhesion, PKP2 and its ID partners also translate information initiated at the site of cell–cell contact into intracellular signals that modulate electrical and transcriptional pathways fundamental to homeostasis.1–4 Thus, PKP2 not only participates in intercellular interactions but also regulates the function of the individual cell.
Mutations in the gene coding for PKP2 are associated with most cases of familial arrhythmogenic right ventricular cardiomyopathy (ARVC),5 , 6 a disease characterized by fibrofatty infiltration of right ventricular (RV) predominance, ventricular arrhythmias, and high propensity for sudden death in the young. Furthermore, in the case of ARVC, exercise significantly increases the risk for developing the cardiomyopathy, for its progression to failure, and for the occurrence of arrhythmias and sudden death.7 , 8
Exercise, predominantly aerobic or endurance training, leads to a compensatory adaptation of the heart muscle, necessary to maintain increased demand. High-intensity endurance exercise (heretofore ‘training’) can provoke remodelling that impacts on cardiac structure and function. Particular features of an athlete’s heart, including RV dysfunction and arrhythmias, can resemble those found in ARVC.9 , 10 It is tempting to speculate that the points of convergence of these two phenotypes (athlete’s heart and ARVC) are more than coincidental and that they could be associated with commonalities in the transcriptional reprogramming triggered by each condition.
Our objective was to compare, at the transcriptomic and functional levels, the effect of training vis-a-vis PKP2 deficiency. We used murine hearts to test and validate the hypothesis that there is convergence between exercise-dependent and PKP2-dependent transcriptomes and as such, the co-existence of both variables (exercise and PKP2 deficiency) is catastrophic to the heart. Functionally, we tested the hypothesis that impaired cardiac contractility in trained PKP2-deficient hearts is consequent to impaired sarcomere shortening (SS). After refuting this hypothesis, we validated the alternative, namely, that impaired cardiac contractility was consequent to loss of muscle mass. Our data lead us to propose that, in murine hearts, exercise challenges a ‘desmosomal reserve’ which, if impaired genetically (e.g. PKP2 loss), accelerates cardiomyopathy progression. Accordingly, we found that exercise in PKP2-deficient human hearts was not followed by a compensatory trophic effect. Altogether, our results support the notion that low desmosomal reserve (e.g. a pathogenic PKP2 mutation) impedes the heart of an athlete from matching increased demand, leading to cell damage and reduced muscle mass which, if exercise persists, causes a clinically apparent cardiomyopathy.
Methods
Terminology
‘Control’ (‘Ctrl’) refers to age- and gender-matched, tamofifen (TAM)-injected C57BL/6 PKP2fl/fl/αMHC-CreERT2−/− (Cre−). ‘PKP2cKO’ refers to TAM-injected age- and gender-matched C57BL/6 PKP2fl/fl/αMHC-CreERT2−/+ (Cre+) mice. ‘Sedentary’ refers to animals, control, or PKP2cKO, not subjected to a running protocol. ‘Trained’ refers to animals, control, or PKP2cKO, that underwent a 6-week running (training) programme (Supplementary material online, Figure SI). Animals exercised for only 3 weeks (Supplementary material online, Figure SI) are referred to as ‘3-week trained’ and the protocol as ‘3-week training’. For differential transcriptomes, we indicate the specific subtraction that is made in each of the datasets. These are: ‘Sed.PKP2cKO-Sed.Ctrl’ (to study the effect of loss of PKP2 on the transcriptome of sedentary animals), ‘Trained.PKP2cKO-Trained.Ctrl’ (to define the effect of loss of PKP2 on the transcriptome of trained animals) and ‘Trained.Ctrl-Sed.Ctrl’ (to characterize the effect of training on the transcriptome of control mice).
Exercise protocol in mice
Experiments were performed in adult male mice, 3–4 months of age at the start of training, and control littermates.3 Experiments were approved under NYU-IACUC protocol IA16-01021. Mice underwent two separate treadmill running protocols adapted from11–13: (i) 3 weeks of running before first tamoxifen (TAM) injection and then three additional weeks of running (total 6 weeks of exercise; Supplementary material online, Figure SI; group ‘trained’). (ii) Running starting at time of TAM injection, lasting 21 days (3-week training; Supplementary material online, Figure SI).
RNAseq analysis
We obtained two new differential transcriptomes: (i) Trained.Ctrl-Sed.Ctrl and (ii) Trained.PKP2cKO-Trained.Ctrl. Details in the Supplementary material online.
Weighted-gene network analysis
We explored a correlation between our data and the PKP2 gene network described from the human transcriptome14 using R-based weighted correlation network analysis (WGCNA; R version 3.4.3) and RNAseq data from human left ventricular (LV) tissue available through the Genotype-Tissue Expression v7 (GTEx) consortium (https://www.gtexportal.org/home/datasets, last accessed 14 January 2021).
Echocardiography
Echocardiographic images were acquired and analysed in B-mode and M-mode on a VEVO 2100 machine (VisualSonics Inc.) as previously described.3 Of note, an additional group of four PKP2wt/wt/alphaMHC-CreERT2wt/+, TAM-injected mice were followed for 35 days to rule out possible effects of Cre on phenotype. There were no changes across time in any of the echocardiographic parameters (Supplementary material online, Figure SII).
Histology
Paraffin-embedded four-chamber heart sections were fixed and prepared with Masson Trichrome staining for histology analysis. Measurements of tissue and cell size cross-sectional analyses were performed using custom-made algorithms in ImageJ. Terminal dUTP Nick-End Labeling (TUNEL) staining was performed on paraffin sections (5 µM thick) and images acquired, aligned and quantified using a Keyence BZ-X800 Fluorescent microscope.
Patient population and imaging
The patient population was extracted from the Johns Hopkins ARVC Registry based on the following criteria: carrier of a pathogenic/likely pathogenic PKP2 variant; definite, borderline, or possible diagnosis; class C athlete by the 36th Bethesda Conference of sport15 and available history of training at the time of cardiac magnetic resonance imaging (MRI). These patients were 1:1 matched for age, gender, body surface area, and hours of training/week with a cohort of healthy controls who underwent a cardiac MRI as participants in a research study.16 All measurements were performed by one experienced observer (M.B.).
Dissociation of single myocytes, Ca2+ imaging, and sarcomere shortening
Isolated murine ventricular myocytes were obtained by enzymatic dissociation 21 days post-TAM, as in Ref.4 Myocytes were harvested from both ventricles. Additional considerations and detailed methods for Ca2+ imaging and SS in Supplementary material online.
Statistical analysis
Functional, echocardiographic, and histological parameters are presented as mean and standard deviation. Comparisons were first evaluated by hierarchical analysis17 as specified in figure legends, and statistical significance determined by Student’s t-test, analysis of variance or non-parametric statistics as indicated. Additional details in Supplementary material online, Methods. Data on patients are presented as frequency (percentage), or mean standard deviation. Comparison between groups was made using Student’s t-test or Mann–Whitney U test, as appropriate. All data were analysed using the IBM SPSS statistical package v25.0 or GraphPad Prizm v8.0. SS data analysis was performed using the IonWizard (IonOptix, Westwood, MA, USA) software.
Results
Exercise-induced changes in cardiac function and in the cardiac transcriptome of control mice
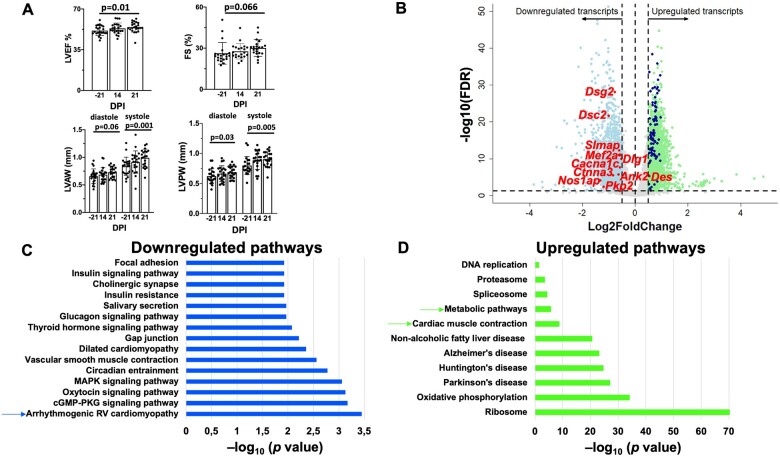
To establish an adequate reference point, we first tested for changes in cardiac function caused by training control animals for 6 weeks (Supplementary material online, Figure SI). As shown in Figure 1A, after 6 weeks of training, we observed an increase in LV ejection fraction (LVEF) and fractional shortening (FS), as well as in thickness of LV walls both in diastole and in systole. Additional echocardiographic parameters are presented in Supplementary material online, Figure SIII. These changes were consistent with cardiac remodelling consequent to endurance training.9 , 18 As a next step, we characterized the associated transcriptome. Mice were euthanized after the last run and hearts immediately processed for RNA extraction. A group of sedentary age-matched animals was used for comparison. The differential transcriptome is presented in Figure 1B. Positive and negative numbers in the x-axis reflect transcripts up-regulated and down-regulated by training, respectively, relative to the same gene in the sedentary group. A principal component analysis (PCA) is presented in Supplementary material online, Figure SIV (complete dataset forming the volcano plot in Supplementary material online, Table S1). A total of 2185 transcripts were significantly down-regulated by training [thresholds of | log2FC | >0.5 and 5% false discovery rate (FDR < 0.05)]. Of particular note is the down-regulation of Pkp2, as well as of desmocollin-2 (Dsc2) and desmoglein-2 (Dsg2) caused by exercise alone, as well as the down-regulation of other genes of the ID (Nos1ap, Ctnna3), scaffolding proteins for ion channels (Dlg1, coding for Sap97; Ank2, coding for ankyrin-B; Slmap, coding for sarcolemma associated protein), transcription regulators relevant to cardiac remodelling (Mef2a), and ion channel-forming proteins (Cacna1c). The above-mentioned transcripts are marked in red in the volcano plot. Quantitative immunofluorescence confirmed changes in protein abundance for selected transcripts (Supplementary material online, Figure SV). Unbiased Kyoto Encyclopedia of Genes and Genomes (KEGG) functional pathway analysis was performed in the 2185 down-regulated transcripts (Figure 1C). Consistent with the observed down-regulation of desmosomal genes, the most significant down-regulated pathway was ‘ARVC’. A similar analysis for the 2260 up-regulated transcripts is presented in Figure 1D. We observed an up-regulation of the ‘cardiac muscle contraction’ pathway, which included genes coding for cytoskeletal proteins, such as Myh7, Cacng6, Fxyd2, Myl2, Tnnc1, Tnni3, and members of the COX, UQCR, and TPM family genes, as well as a significant up-regulation of genes involved in cell respiration and mitochondrial metabolism (Supplementary material online, Table S2). Also of note is the up-regulation of the gene coding for the desmosome-anchored, cardiac intermediate filament, desmin (Des), also confirmed at the protein level (Supplementary material online, Figure SV). Overall, we found that, consistent with the functional data, exercise leads to significant up-regulation of structural and functional proteins likely to facilitate both contractility, and aerobic metabolism, whereas an unexpected down-regulation of ID, scaffolding, and ion channel proteins was noted (Supplementary material online, Table S1).
Figure 1.
Functional and transcriptomic characteristics of murine hearts after exercise. (A) Echocardiographic parameters in 21 control mice undergoing 6 weeks of endurance treadmill training. FS, left ventricular fractional shortening; LVAW, left ventricular anterior wall; LVEF, left ventricular ejection fraction; LVPW, left ventricular posterior wall. Student’s t-test to compare baseline (−21) to effect of PKP2cKO 21 dpi. (B) Transcriptome of hearts from trained mice (6-week treadmill running protocol; n = 3) compared with that of sedentary controls (n = 6). Volcano plot of up-regulated (green) or down-regulated (blue) transcripts. The terms ‘up-regulated’ or ‘down-regulated’ refer to more or less abundance, respectively, of a given transcript in the hearts of mice that followed the training protocol, vs. the sedentary controls. Inclusion criteria: | Log2FC | > 0.5 and false discovery rate < 0.05. Dots in grey: transcripts excluded by criteria. The position of transcripts for desmosomal proteins plakophilin-2 (PKP2), desmocollin-2 (DSC2), and desmoglein-2 (DSG-2) together with additional genes of interest are noted with red and with purple dots. (C and D) KEGG (Kyoto Encyclopedia Genes and Genomes)-based identification of down-regulated and up-regulated pathways, respectively, in the differential transcriptome of trained control murine hearts.
The differential transcriptome of control trained animals (Trained.Ctrl-Sed.Ctrl) converges with that observed after deletion of plakophilin-2 in sedentary animals (Sed.PKP2cKO-Sed.Ctrl)
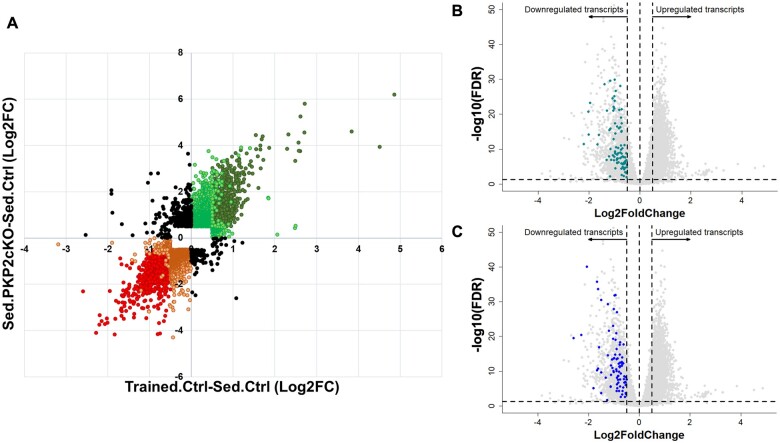
Genetic deletion of PKP2 is known to alter the cardiac transcriptome.3 We therefore compared the training-induced differential transcriptome of control animals (Trained.Ctrl-Sed.Ctrl), where PKP2 transcript abundance was decreased, with that obtained after loss of PKP2 expression in sedentary animals (Sed.PKP2cKO-Sed.Ctrl).3 The complete datasets were plotted on a Cartesian map where the magnitude of the change for a given transcript observed in one data set (exercise; x-axis) was compared against the magnitude observed in the other dataset (PKP2cKO; y-axis). The results are shown in Figure 2A. Notice the large clustering of data points in the 1st (top right) and 3rd (bottom left) quadrants, where the changes were in the same direction (92.8% of all transcripts). Only 7.2% of the total set of transcripts appears in the 2nd and 4th quadrants; namely, they denote transcripts for which the direction of change in one dataset was opposite to that of the other. These results support the notion that these two stressors (training and PKP2-KO) share common molecular pathways.
Figure 2.
Additive effects and preserved directionality of transcriptomic changes induced by exercise and by PKP2 knockdown. (A) Cartesian map where each dot represents the log2Fc value of a given transcript in the ‘trained control’ dataset (X-axis; differential transcriptome of Trained.Ctrl-Sed.Ctrl; same dataset as in Figure 1C and D) against the log2FC recorded for the same transcript in the sedentary PKP2cKO dataset (Sed.PKP2cKO-Sed.Ctrl dataset; Y-axis; dataset previously reported in Ref.3). Notice that, as an overall trend, transcriptomic changes caused by training in controls showed the same direction as those caused by loss of PKP2 in sedentary animals. Red circles in bottom left quadrant show transcripts significantly down-regulated (false discovery rate <0.05), with absolute log2FC values ≥0.5, both in the trained control and in the sedentary PKP2cKO dataset. Similarly, dark green circles in upper right quadrant correspond to transcripts significantly up-regulated (false discovery rate <0.05) with absolute log2FC values >0.5 in both datasets. Orange and light green circles show transcripts with absolute log2FC values >0.5 in one dataset but not in the other, regardless of their significance. Orange and light green data points also represent data where Log2FC’s were >0.5 but the value did not reach statistical significance in one or both of the datasets. Black circles denote data points for which the directionality of the change in one dataset was opposite to that of the other. The latter amounted to only 7.2% of the complete dataset. Red and orange data points correspond to 44.3% and 55.7% of the total data points in the 3rd quadrant, respectively. Green and light green data points correspond to 41.3% and 58.7% of the total data points in the 1st quadrant, respectively. (B and C) Grey dots represent differential transcriptome after exercise (same as in Figure 1A). Turquoise (B) and blue dots (C) mark transcripts identified by Montnach et al. 14 as components of the human PKP2 gene network.
To further explore similarities between training-dependent and PKP2-dependent transcriptomes, we compared transcripts significantly down-regulated by training (Figure 1B–D and Supplementary material online, Table S1) with PKP2-centric human gene network modules identified by Montnach et al. 14 through analysis of the GTEx database. The primary module in Montnach et al. 14 included 108 transcripts; 87 of those (80.5%) were also significantly down-regulated in the training control (Trained.Ctrl-Sed.Ctrl) dataset. These transcripts are graphically represented in the volcano plots of the Trained.Ctrl-Sed.Ctrl differential transcriptome, in Figure 2B. The second module in Montnach et al. 14 included 95 transcripts; 90.5% (86 transcripts) of these were significantly down-regulated by training (Figure 2C). We conclude that, embedded in the transcriptome of trained control animals (Trained.Ctrl-Sed.Ctrl), there is the down-regulation of a subjacent PKP2-related gene network. Accordingly, we posit that a further reduction in PKP2 abundance can switch exercise from beneficial to deleterious to the health of the heart.
Exercise accelerates cardiomyopathy progression in PKP2cKO mice
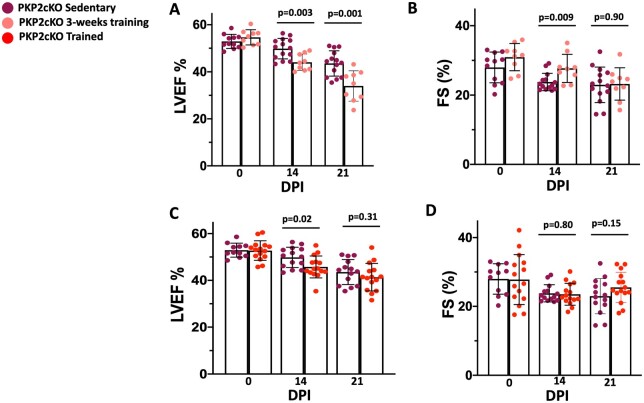
PKP2cKO mice were subjected to a 3-week treadmill running program (3-week training). Animals were injected with TAM on Day 1 of the running protocol. Echocardiographic parameters were measured on Days 0, 14, and 21 post-TAM and compared with data from PKP2cKO sedentary animals. Exercise in PKP2cKO mice caused a rapid decrease in LV function and progression towards heart failure, already manifest at 14 dpi. The LVEF decreased further at 21 dpi, reaching values significantly reduced when compared with those observed in PKP2cKO-sedentary animals (Figure 3A and B).3 Additional echocardiographic parameters are provided in Supplementary material online, Figures SVIA and B and SVIIA–D. Altogether, the data confirm that, while exercise alone can improve cardiac function, a PKP2 deficit drives the phenotype further into the cardiomyopathic state if both stressors (exercise and PKP2-KO) are present.
Figure 3.
Echocardiographic changes in trained PKP2cKO mice compared with sedentary PKP2cKO mice. (A and B) Left ventricular ejection fraction (LVEF) and fractional shortening changes in 3-week trained PKP2-cKO compared with PKP2-cKO sedentary mice. (C and D) Left ventricular ejection fraction and fractional shortening changes between trained (i.e. animals completed 6 weeks of treadmill running) and sedentary PKP2-cKO mice. Purple: sedentary (n = 11 at 0 dpi, 14 at 14 and 21 dpi). Pink: 3-week training (n = 9). Red: trained mice (n = 15). DPI: days post-TAM injection. ‘0’ in C and D reflects the baseline pre-exercise in trained mice (3 weeks before TAM injection). Sedentary data in A and B also displayed in C and D to facilitate comparison. Top bars indicate P-values by unpaired Student’s t-test.
Training prior to TAM injection reduces the impact of loss of plakophilin-2 expression
The improved contractility that results from exercise is a slow and cumulative adaptive process. We examined whether a period of adaptation to exercise prior to loss of PKP2 would have an extended protective effect on the function of a heart that is then made deficient in PKP2. Animals in this group ran for 3 weeks prior to TAM injection and three additional weeks after TAM (‘trained’ group; Supplementary material online, Figure SI). As shown in Figure 3C, echocardiographic measurements showed that LVEF started to significantly decrease around 14 dpi (5 weeks of exercise). Importantly, the deleterious effect observed when 3-week training and loss of PKP2 expression were combined from the outset (Figure 3A and B) was not apparent in the 6-week trained hearts (3 weeks pre-TAM; 3 weeks post-TAM), where echocardiographic parameters at 21 dpi were not different from those recorded from sedentary PKP2cKO animals (Figure 3C and D, Supplementary material online, Figures SVIC and D and SVIIE–H).
Though training prior to TAM injection partly contained the catastrophic drop in function otherwise observed in 3-week trained PKP2cKO animals, the cardiac mechanical function of PKP2cKO animals that completed the 6-week training period still departed from that of the trained control group, where we actually observed improved LVEF and a tendency towards improved FS as a result of training (Figure 1A). We surmised that reduction in contractile function in trained PKP2cKO mice could be due to at least one of two not mutually exclusive factors: reduced contractility of individual myocytes, or reduced myocardial mass. Each variable was analysed separately.
Training increases Ca2+ transient amplitude and sarcomere shortening in plakophilin-2-deficient myocytes
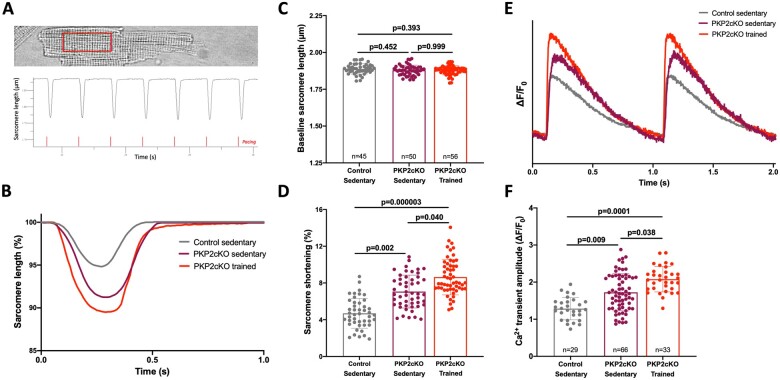
We characterized SS in isolated myocytes from PKP2cKO mice at 21 days dpi. Three groups were studied: sedentary control, sedentary PKP2cKO, and trained PKP2cKO. Cells were paced at 1 Hz. Sarcomere length was measured within a region of interest that included 15–20 z-lines (see Figure 4A) and measurements averaged over 15 cycles. An example of measurements of sarcomere length during seven cycles is shown in Figure 4A and exemplary traces in Figure 4B. Cumulative data are depicted in Figure 4C and D. No difference between groups was observed in resting sarcomere length (Figure 4C). Yet, SS was significantly larger in PKP2-deficient cells, and this increase was even more noticeable in cells from trained animals (Figure 4D).
Figure 4.
Sarcomere length and calcium transient measurements in cardiac myocytes of sedentary and trained PKP2cKO mice. (A) Top: Example of cell region of interest (red square). Bottom: Example of sarcomere length recording, collected in Ionoptix, and analysed with IonWizard. Red lines highlight electrical stimulation; cells were paced for 20–30 s before the start of the recording to achieve steady-state. (B) Exemplary sarcomere shortening traces from sedentary control (grey line), sedentary PKP2cKO (purple line), and trained PKP2cKO myocytes (red line) 21 days post-TAM injection (21 dpi). (C and D) Baseline sarcomere length (C) and sarcomere shortening (D) in sedentary control (grey bar and symbols), sedentary PKP2cKO (purple bar and symbols), and trained PKP2cKO myocytes (red bar and symbols) 21 dpi. Numbers inside bars in C indicate number of cells and apply to both C and D panels. (E) Examples of electrically evoked Ca2+ transients in PKP2cKO myocytes 21 dpi, maintained at room temperature and in standard Tyrode solution. Grey: Myocyte from sedentary control mouse. Purple: Myocyte from sedentary PKP2cKO mouse 21 dpi. Red: Myocyte from trained PKP2cKO mouse 21 dpi. (F) Average Ca2+ transient amplitude in sedentary control (grey bar and symbols), sedentary (purple bar and symbols), and trained (red bar and symbols) PKP2cKO myocytes 21 days dpi. Numbers inside bars indicate number of cells. Data from 4 to 5 mice per group, presented as mean ± SD. Statistical tests: Hierarchical test was used for all parameters (details in Methods section).
We have previously demonstrated that loss of PKP2 leads to increased Ca2+ transient amplitude.4 In Figure 4D–F, we show that this increase is even more pronounced in mice that have completed the 6-week training period. This increase and the consequent increase in free Ca2+ availability for contraction is consistent with the increased SS observed in individual myocytes. Training led to increased SS and Ca2+ transient amplitude also in control cardiomyocytes (Supplementary material online, Figure SVIII). More importantly, the increased SS of isolated cells was not consistent with the decreased FS and LVEF observed at the organ level in trained PKP2cKO hearts. We then examined the alternative hypothesis, namely, that the effects at the organ level associate with reduced muscle mass, caused by PKP2 deficiency.
Muscle content in the ventricular free walls was reduced in PKP2cKO trained hearts
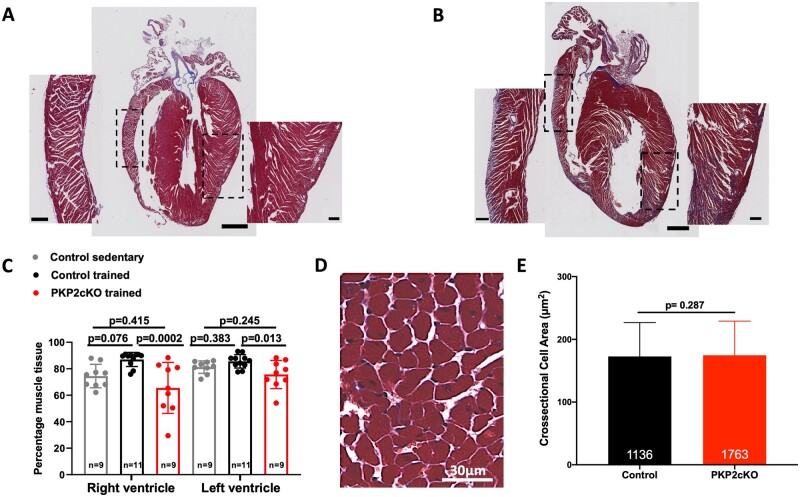
Hearts were paraffin-fixed and divided along the cardiac long axis for visualization of free ventricular walls and septum. Masson trichrome staining allowed us to distinguish muscle vs. non-muscle tissue. An image analysis tool was used for unbiased quantification of the proportion of muscle within the section (see ‘Methods’ and Supplementary material online, Figure SIX). Trained PKP2cKO hearts showed a reduction in muscle content, more prominently in the right ventricle, when compared with trained controls (Figure 5A–C).
Figure 5.
Effect of 6-week training on cardiac histology. (A and B) Examples of Masson Trichrome stained longitudinal axis sections of a control (A) and a PKP2cKO trained heart (B). Scale bars: 1000 µm. Dotted squares indicate area showed at higher magnification on the respective sides. Scale bars: 200 µm. (C) Percentage of healthy muscle tissue detected by Masson Trichrome staining of control sedentary (grey bars and symbols), control trained (black bars and symbols), and PKP2cKO trained (red bars and symbols). Number of hearts indicated inside each bar. One-way analysis of variance. (D) Representative example of left ventricular histology section of PKP2cKO trained heart used for cross-sectional area analysis. (E) Cross-sectional area of left ventricular myocytes in control (black bars) and trained PKP2cKO mice (red bars). Number of cells measured per group is noted inside each bar (from n = 10 control and n = 7 PKP2cKO mice, respectively). Mann–Whitney U test for non-parametric samples. Individual symbols are not shown given the total n values included (over 1000 per bar).
To determine whether reduction in muscle content included changes in the size of individual myocytes, we analysed cross-sectional cell areas from selected regions where the orientation of the cut was transverse to that of the cell axis (example in Figure 5D). Depletion of PKP2 in sedentary myocytes does not influence cardiomyocyte size, as shown in Supplementary material online, Figure SX. Cell cross-sectional area was not different between trained control and PKP2cKO groups (Figure 5E). Overall, our data indicate that trained PKP2cKO hearts are unable to maintain contractile function due to loss of muscle mass, despite the increased contractility of individual myocytes.
Conservation of differential transcriptomes of trained PKP2cKO (Trained.PKP2cKO-Trained.Ctrl), trained controls (Trained.Ctrl-Sed.Ctrl), and sedentary PKP2cKO (Sed.PKP2cKO-Sed.Ctrl) hearts
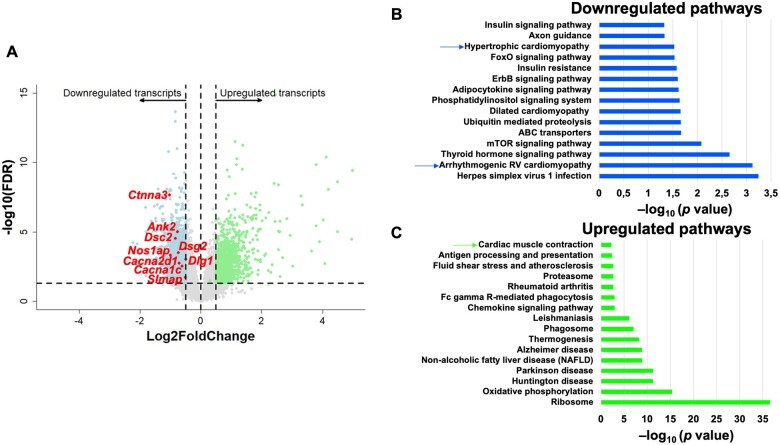
We previously characterized the differential transcriptome of sedentary PKP2cKO vs. sedentary control hearts (Sed.PKP2cKO-Sed.Ctrl).3 Here, we extended our study to obtain the differential transcriptome of trained PKP2cKO hearts compared with exercised controls (Trained.PKP2cKO-Trained.Ctrl). All mice were subjected to the 6-week running regimen (Supplementary material online, Figure SI). Hearts were harvested 21 dpi. A total of 12 602 annotated transcripts from the Ensembl reference database were differentially expressed. The corresponding PCA and the complete dataset are provided in Supplementary material online, Figure SXI and in Supplementary material online, Table S3, respectively. Applying the criteria of FDR <0.05 and absolute log2 fold change >0.5 resulted in 3195 transcripts differentially expressed, with 1445 transcripts down-regulated and 1750 transcripts up-regulated in the trained PKP2cKO group. The volcano plot is presented in Figure 6A; KEGG functional pathway analysis of down-regulated and up-regulated genes is presented in Figure 6B and C, respectively.
Figure 6.
Differential transcriptome of murine trained PKP2cKO hearts. Transcriptome of trained control hearts (Trained.Ctrl-Sed.Ctrl) was compared with that of trained PKP2cKO mice (Trained.PKP2cKO-Trained.Ctrl). (A) Volcano plot of up-regulated (green) or down-regulated (blue) transcripts. Inclusion criteria: Log2FC ± 0.5 and false discovery rate < 0.05. Up-regulated or down-regulated refers to more or less abundance, respectively, of a given transcript in the PKP2cKO hearts. Dots in grey: transcripts not meeting inclusion criteria. Some of the down-regulated transcripts in the ‘hypertrophic cardiomyopathy’ pathway are noted in the plot. (B and C) KEGG (Kyoto Encyclopedia Genes and Genomes)-based identification of down-regulated and up-regulated pathways, respectively, in the differential transcriptome of trained PKP2cKO hearts.
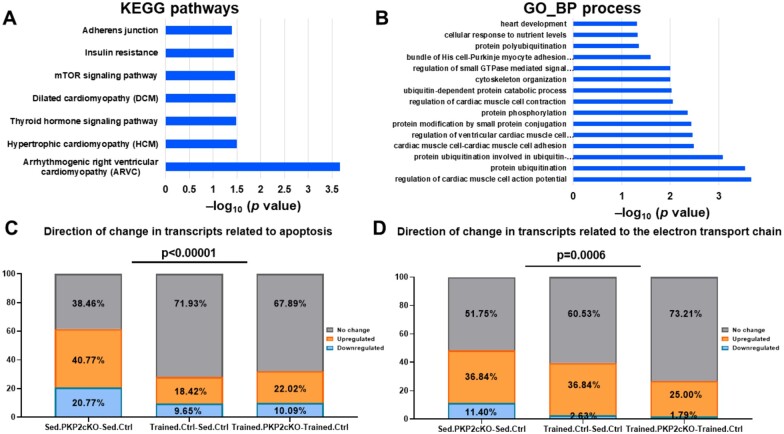
There was extensive conservation of modified transcripts between the three groups studied (Trained.Ctrl-Sed.Ctrl, Sed.PKP2cKO-Sed.Ctrl, and Trained.PKP2cKO-Trained.Ctrl). Indeed, there were 1084 transcripts down-regulated in the Trained.Ctrl-Sed.Ctrl dataset that were also down-regulated in the trained Trained.PKP2cKO-Trained.Ctrl group. Of those, 916 transcripts were also down-regulated in the Sed.PKP2cKO-Sed.Ctrl dataset. In this converging group, there was a dominant representation of the ‘ARVC’ pathway (Figure 7A) and of functional processes associated with the regulation of electrical and mechanical activity (Figure 7B and Supplementary material online, Tables S4 and S5). Analysis of converging transcripts reveals down-regulation of genes coding for proteins of the ID (Dsc2, Dsg2, Nos1ap, Ctnna3), scaffolding proteins (Ank2, Dlg1), and of proteins associated with excitation-contraction coupling (Cacna1c, Cacna2d1, Slmap). Interestingly, a highly significant difference was observed in the expression of genes involved in apoptotic pathways. From a total of 136 genes associated with apoptosis and included in the KEGG pathway, 61.5% showed a significant change in the Sed.PKP2cKO-Sed.Ctrl group, while this fraction was significantly lower (32.1%) in the Trained.PKP2cKO-Trained.Ctrl group (Figure 7C). Concurrent with the observation above, TUNEL staining in sedentary PKP2cKO was more abundant than that observed in PKP2cKO trained and in controls (Supplementary material online, Figure SXII). Similarly, in PKP2cKO animals, training limited the down-regulation of genes involved in the electron transport chain (ETC) compared with sedentary PKP2cKO group (comparing data from Trained.PKP2cKO-Trained.Ctrl against those obtained from Sed.PKP2cKO-Sed.Ctrl; Figure 7D, Supplementary material online, Tables S6 and S7). Of note, a similar analysis for expression of genes involved in the glycolytic pathway yielded no significant difference between groups (Supplementary material online, Figure SXIII).
Figure 7.
Comparative analysis of transcriptomes in three experimental groups studied (trained control, sedentary PKP2cKO, and trained PKP2cKO). (A and B) KEGG (Kyoto Encyclopedia Genes and Genomes) and GO (Biological process-based) pathways obtained from analysis of transcripts that were down-regulated in all three groups. (C) Stack bars representing the direction of change for transcripts under the KEGG term ‘apoptotic pathways’ (136 genes) for the three groups studied (sedentary PKP2cKO, trained control, trained PKP2cKO). Numbers within each block in the bar indicate the per cent of genes that were either down-regulated (blue block), up-regulated (orange block), or showed no significant change (grey block). Exercise significantly reduced the number of apoptosis-related transcripts that were modified consequent to loss of PKP2 deletion (left bar vs. right bar; P-value < 0.00001). In fact, the per cent of transcripts modified by training in the PKP2cKO group was similar to that modified by training in controls (right bar vs. middle bar). The upper bar shows the statistical significance in a comparison between all groups (χ2 test). (D) A comparison for transcripts included under the KEGG term ‘electron transport chain’ showed a similar trend (P = 0.00059).
Cardiac dimensions in human athletes with diagnosis of arrhythmogenic right ventricular cardiomyopathy differ from those in control athletes
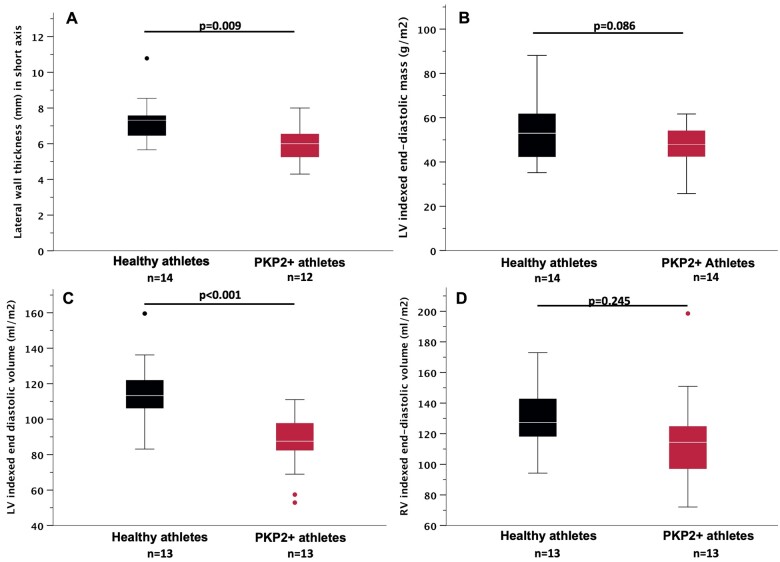
Overall, the data presented would predict, all fundamental limitations notwithstanding, that exercise-induced changes in heart mass that have been reported in human athletes would not be detectable in hearts of athletes with PKP2 mutations. To test for the latter, we identified 14 patients carrying PKP2 mutations, the majority (9/14) reaching a definite diagnosis of ARVC according to the 2010 Task Force Criteria19 and all having a cardiac MRI as part of their diagnostic workup. These patients were 1:1 matched for age, gender, body surface area, and hours of training per week to a cohort of healthy athletes who underwent an MRI as participants in a research study.16 Clinical features and genetic variants are summarized in Supplementary material online, Tables S8 and S9. The data show that ARVC athletes did not develop the same level of LV wall thickness as control athletes (7.3 ± 1.3 mm vs. 6.0 ± 1.0 mm, P = 0.009, Figure 8A). In addition, there was a trend towards a smaller LV indexed end-diastolic mass (Figure 8B) and significantly lower LV indexed end-diastolic volume (EDV; P < 0.001, Figure 8C) in PKP2 mutations carriers. Indexed RV EDV was not different between the groups (Figure 8D). Of note, previous studies20 have shown that LV to RV EDV ratio differs in physiologic and pathologic ventricular remodelling. Representative MRI examples are provided in Supplementary material online, Figure SXIV. While in physiologic conditions the ratio is fairly balanced (LV/RV EDV = 0.9–1.1), this is not observed in the presence of ARVC-causing mutations (LV/RV EDV < 0.9). In our study population, PKP2+ athletes manifested a trend towards maladaptive remodelling [LV/RV EDV 0.82 (interquartile range: 0.58–0.95) vs. 0.91 (interquartile range: 0.86–0.94) in controls, P = 0.06].
Figure 8.
Cardiac magnetic resonance in arrhythmogenic right ventricular cardiomyopathy patients and healthy controls with history of comparable athletic training. (A) Short-axis recording of lateral wall thickness. (B) Left ventricular indexed end-diastolic mass. (C) Left ventricular indexed end-diastolic volume. (D) Right ventricular indexed end-diastolic volume. Black: healthy athletes. Red: PKP2-positive athletes. Student’s t-test. Boxplots: median and interquartile range. Whiskers: 1.5× interquartile range, with outliers indicated as single dots.
Discussion
We have examined the consequences of exercise training (treadmill running) in mice. We found that both in control and in PKP2-deficient animals, training reduced the abundance of ID protein transcripts. Yet, in controls, cardiac function improved whereas in PKP2cKO animals, cardiac function was impaired. We propose that a normal heart expresses more desmosomal proteins than the minimum necessary (i.e. it has a large ‘desmosomal reserve’). As such, it tolerates their reduced abundance (caused by training) without falling into a pathological state (Supplementary material online, Figure SXVA). Yet, if a genetic modification reduces the desmosomal reserve at baseline, further exercise-induced reduction is not tolerated, thus accelerating the ARVC phenotype (see Supplementary material online, Figure SXVB). Thus, it is a main message of this study that exercise reduces desmosomal reserve, and that further impairment of desmosomal reserve by genetic means can affect heart function (Graphical abstract).
Graphical Abstract.
Exercise challenges a cardiomyocyte “desmosomal reserve” which, if impaired genetically (e.g., Plakophilin2 loss), can cause accelerated progression of the cardiomyopathy.
In the second part of our study, we investigated the cellular mechanisms leading to impaired contractility in exercised PKP2-deficient animals. To our surprise, we found that loss of PKP2 actually improves (rather than decreases) SS. On the other hand, loss of PKP2 caused apoptosis and reduced muscle mass, exacerbated by exercise (Supplementary material online, Figure SXVB). Our observations in mice led us to assess myocardial mass parameters in human athletes. We found that exercise-induced remodelling of cardiac dimensions, which normally occurs in athletes, is not present in athletes with a PKP2 deficiency. In other words, consistent with the observations in mice, PKP2 deficiency in humans impairs the necessary exercise-induced trophic response. The latter, we suggest, can set the stage for an acceleration of the ARVC phenotype. Overall, our data concur with extensive evidence showing detrimental effects of exercise in the setting of desmosomal deficiency in humans7 , 8 and in mice21–23 and provide mechanistic insight into its molecular origins.
Training prior to plakophilin-2 loss has a cardioprotective effect
Our data showed that training for 3 weeks previous to KO of PKP2 dampened the progression of the cardiomyopathy, compared with that of animals that exercised without previous training. The latter does not have direct translational value for ARVC patients. Yet, it may relate to the importance of proper, gradual training for non-athletes that engage in an endurance exercise program. Furthermore, training in a normal individual is known to have a positive effect on myocyte respiration. So, we speculate that an animal that has trained before the loss of PKP2 has a more efficient cell respiration at the start. Thus, despite the fact that training would negatively impact (decrease) the desmosomal reserve, the more efficient cell respiration would partly counter the challenge to the desmosomal reserve and the heart would better tolerate PKP2 deficiency before a threshold for apoptosis is reached (Supplementary material online, Figure SXV). Mechanistically, the protective effect may result from strong expression of genes involved in cell respiration/ETC detected in trained controls (Trained.Ctrl-Sed.Ctrl), which persisted in the trained PKP2cKO animals (Trained.PKP2cKO-Trained.Ctrl), vis-a-vis the down-regulation of genes in the same pathway in the PKP2cKO sedentary animals (Sed.PKP2cKO-Sed.Ctrl). Previous studies have suggested that metabolic derangement underlies the cellular pathogenesis of ARVC24 and that mitochondrial dysfunction represents a substrate for the disease.25 Our data show that training provoked an up-regulation of genes involved in oxidative phosphorylation and electron transport and that this change was also apparent in the trained PKP2cKO group. Regarding the optimal exercise strategy in humans with PKP2 deficiency, the issue needs to be considered in the context of current clinical knowledge about harmful exercise intensity.26
The PKP2cKO trained group showed an abundance of transcripts related to mitochondrial function, and a less vulnerable phenotype. We cannot establish cause-effect between these two variables. Yet, given the importance of mitochondrial function on muscle performance, we can speculate that they are related. Yet, other possibilities cannot be excluded; among them, mice may develop better skeletal muscle performance after the first 3 weeks of training, thus enabling training for the last 3 weeks (i.e. after TAM injection) with less cardiovascular effort. Also, a reduced stress on the mice as they are more adapted to the training regime, could help the outcome.
Our data refute the hypothesis that loss of plakophilin-2 impairs sarcomere shortening of isolated cells
We have previously shown that the arrhythmogenic phenotype of ARVC results, at least in part, from molecular changes intrinsic to the cell and not (or not only) from cell separation due to loss of cell–cell adhesion.3 Furthermore, the transcriptome of sedentary PKP2cKO hearts (in Ref.3) shows up-regulation of cytoskeletal components including the actin cytoskeleton (Myh7, Myh6), the intermediate filaments (increased desmin), and the microtubules (Tuba1a-c, Vash1, Ttl). Thus, it was our a priori hypothesis that molecular changes inside the cell would impact negatively on the ability of individual myocytes to shorten. This hypothesis, however, was not supported by our data. In fact, our results showed an increase in SS in PKP2-deficient myocytes, an effect actually amplified in trained animals. Thus, our data refuted the hypothesis as postulated. On the other hand, our data were consistent with the alternative hypothesis, namely, that the cardiomyopathic phenotype is not consequent to an impaired ability of the myocyte to contract (quite the contrary) but rather, to a reduction in the population of contractile myocytes that sum their work to eject the blood during systole.
It should be noted that sarcomere length was measured in unloaded myocytes, given the technical difficulties involved in conducting measurements in tension-exposed cardiomyocytes. We acknowledge this as a limitation of our study, as myocytes in situ are subjected to stretch and increased tension. Future experiments will be necessary to examine the sequitur hypothesis that though the ability of the unloaded myocyte to shorten was in fact heightened by the loss of PKP2, the contractility of loaded myocytes subject to tension may be impaired when compared with controls.
The increased amplitude of Ca2+ transients observed in trained and in sedentary animals4 provides a possible mechanistic explanation for the increased SS observed. Furthermore, it suggests that a dysregulation of intracellular Ca2+ is present and heightened in these animals and can contribute to an increased likelihood of life-threatening arrhythmias. Future experiments will also contemplate measuring Ca2+ fluorescence in Langendorf-perfused hearts using methods as those previously described.27
Limitations of the PKP2cKO murine model
There is, at present, no animal or cellular model that ‘recapitulates’ human ARVC. The latter stands to reason, given fundamental differences between the human heart and that of other species, as well as the impossibility of recreating a disease of the fully differentiated multi-cellular heart by studying immature cardiac myocytes in a dish, even if derived from human blood. There is, however, the possibility of studying endophenotypes of ARVC-relevant genes by knockout methods. Under that premise we have used a cardiomyocyte specific, TAM-activated PKP2 knockout model (dubbed PKP2cKO)3 to learn about the cardiac endophenotype of a gene that, when mutated in humans, causes ARVC. Similarly, to our knowledge, rodent models do not recapitulate an athlete’s heart. Yet, it is possible to subject mice to endurance exercise and analyse transcriptional features that, if amplified, could underlie structural and functional features of the athlete’s heart. Extrapolation to the adaptation of the human heart be it consequent to exercise, to PKP2 deficiency or to their combination, needs to be done with caution.
It should be noted that control animals were Cre-negative and received a TAM injection. This was done to facilitate comparison to the Cre-positive group (PKP2cKO). We have no reason to believe that the outcome of the control animals is affected by the TAM injection, given that these animals present no phenotype and TAM is injected 21 days before any analysis is conducted. Yet, future experiments will be pursued in non-TAM-injected animals to completely discard any possible artefactual effect of TAM exposure.
Cardiac remodelling in plakophilin-2-deficient human athletes differs from that observed in controls
We carried out a preliminary evaluation of the effects of exercise in cardiac dimensions in athletes, compared with those with similar age, sex, and level of training but carrying a pathogenic/likely pathogenic PKP2 mutation. We observed significant differences suggesting an inability in PKP2-deficient patients to properly compensate with a trophic response against increased workload. The interpretability of our data is limited by the relatively low number of cases included, given the low community prevalence of athletes with a desmosomal gene mutation and the challenge of matching imaging data collected at a comparable level of training both in patients and in controls. This limitation notwithstanding, the results were consistent with the notion that humans with PKP2 deficiency do not follow a cardiac remodelling process comparable to age, sex- and training-matched controls.
Summary and conclusions
Our data document an exercise-induced transcriptomic switch that, on the one hand, can limit the abundance of proteins of the ID including desmosomes, and on the other hand, provides increased transcript copies that can facilitate cell energy production. While the former challenges the tolerance of the heart to further decreases in desmosomal abundance (or to stress), the latter may help prevent, if properly timed, the rapid progression of an exercise-induced cardiomyopathy in the PKP2-deficient heart. Our work also documents, for the first time, an increase in SS consequent to PKP2 loss, an effect seemingly paradoxical when compared with the reduced organ contractile function. This leads us to conclude that the PKP2-dependent mechanical dysfunction of the ventricles is not cell-based but rather, consequent to a reduction in the overall muscle mass and/or an impeded remodelling process expected to occur as a result of exercise. Our work is limited to the analysis of an experimental model (exercise and/or PKP2 deficiency in mice), but provides grounds to design (and preliminarily interpret) studies in the human population. Nonetheless, extrapolation of our data to the analysis of patients with an ARVC phenotype needs to be done with caution.
Supplementary material
Supplementary material is available at European Heart Journal online.
Author contributions
Study conceptual design, manuscript preparation, manuscript editing: MC, GML, CvO, MD. Data collection; data analysis; figure preparation: MC, GML, CvO, MZ, MPH, SC, FS, KM, KD, XL, ATR, MB, CJ, NP, BV. Critical revision of manuscript: MD, AH, ALG, BV, HC.
Funding
Work supported by National Institutes of Health grants RO1-HL134328, RO1-HL136179, and RO1-HL145911 (M.D.), a Transatlantic Network of Excellence from the Leducq Foundation (M.D.), a Transformational Project award from the American Heart Association 18TPA34230006 (M.C.), an American Heart Association Postdoctoral Fellowship 20POST35120519 (G.M.M.-L.), the Wilton W. Webster Fellowship in Pediatric Electrophysiology from Heart Rhythm Society (2019–20, MPH; 2020–21, CvO), the Dutch Heart Foundation 2018R008 (CvO), a National Heart Foundation Future Leader Fellowship 102206 (A.L.G.), the Netherlands Heart Foundation grant 2015T058 and UMC Utrecht Fellowship Clinical Research Talent (ATR), and grant 2016T042/Hartstichting (N.K.) and a postdoctoral fellowship Ubprograma Atracció de Talent—Contractes Postdoctorals de la Universitat de València (F.S.). The Johns Hopkins ARVD/C Program (C.J. and H.C.) is supported by the Leonie-Wild Foundation, the Leyla Erkan Family Fund for ARVD Research, the Dr Francis P. Chiramonte Private Foundation, the Dr Satish, Rupal, and Robin Shah ARVD Fund at Johns Hopkins, the Bogle Foundation, the Healing Hearts Foundation, the Campanella family, the Patrick J. Harrison Family, the Peter French Memorial Foundation, and the Wilmerding Endowments and by grant UL1 TR003098.
Conflict of interest: H.C. is a consultant for Medtronic Inc. and St. Jude Medical/Abbott. H.C. receives research support from Boston Scientific Corp. and C.J. receives salary support from this grant. All other authors do not have conflict of interests to disclose.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
Supplementary Material
Contributor Information
Marina Cerrone, The ‘Leon Charney’ Division of Cardiology, New York University Grossmann School of Medicine, 435 East 30th Street, NSB 707, New York, NY 10016, USA.
Grecia M Marrón-Liñares, The ‘Leon Charney’ Division of Cardiology, New York University Grossmann School of Medicine, 435 East 30th Street, NSB 707, New York, NY 10016, USA.
Chantal J M van Opbergen, The ‘Leon Charney’ Division of Cardiology, New York University Grossmann School of Medicine, 435 East 30th Street, NSB 707, New York, NY 10016, USA.
Sarah Costa, Division of Cardiology, University Heart Center Zurich, Rämistrasse 100, Zurich CH-8091, Switzerland.
Mimount Bourfiss, Department of Cardiology, Division of Heart and Lungs, University Medical Center Utrecht and The Netherlands Heart Institute, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands.
Marta Pérez-Hernández, The ‘Leon Charney’ Division of Cardiology, New York University Grossmann School of Medicine, 435 East 30th Street, NSB 707, New York, NY 10016, USA.
Florencia Schlamp, The ‘Leon Charney’ Division of Cardiology, New York University Grossmann School of Medicine, 435 East 30th Street, NSB 707, New York, NY 10016, USA.
Fabian Sanchis-Gomar, Department of Physiology, Faculty of Medicine, University of Valencia and INCLIVA Biomedical Research Institute, C. de Menéndez y Pelayo, 4, 46010 Valencia, Spain.
Kabir Malkani, The ‘Leon Charney’ Division of Cardiology, New York University Grossmann School of Medicine, 435 East 30th Street, NSB 707, New York, NY 10016, USA.
Kamelia Drenkova, The ‘Leon Charney’ Division of Cardiology, New York University Grossmann School of Medicine, 435 East 30th Street, NSB 707, New York, NY 10016, USA.
Mingliang Zhang, The ‘Leon Charney’ Division of Cardiology, New York University Grossmann School of Medicine, 435 East 30th Street, NSB 707, New York, NY 10016, USA.
Xianming Lin, The ‘Leon Charney’ Division of Cardiology, New York University Grossmann School of Medicine, 435 East 30th Street, NSB 707, New York, NY 10016, USA.
Adriana Heguy, Genome Technology Center, Department of Pathology, New York University Grossmann School of Medicine, 550 First Avenue, New York, NY 10016, USA.
Birgitta K Velthuis, Department of Radiology, University Medical Center Utrecht, Heidelberglaan 100, 3584 CX Utrecht, the Netherlands.
Niek H J Prakken, Department of Radiology, University Medical Center Groningen, University of Groningen, Hanzeplein 1, 9713 GZ Groningen, The Netherlands.
Andre LaGerche, Clinical Research Domain, Baker Heart and Diabetes Institute, 75 Commercial Rd, Melbourne VIC 3004, Australia and National Centre for Sports Cardiology, St Vincent's Hospital Melbourne, Building C, 41 Victoria Parade, Fitzroy VIC 3065, Australia.
Hugh Calkins, Division of Cardiology, Johns Hopkins Hospital, 1800 Orleans St, Baltimore, MD 21287, USA.
Cynthia A James, Division of Cardiology, Johns Hopkins Hospital, 1800 Orleans St, Baltimore, MD 21287, USA.
Anneline S J M Te Riele, Department of Cardiology, Division of Heart and Lungs, University Medical Center Utrecht and The Netherlands Heart Institute, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands.
Mario Delmar, The ‘Leon Charney’ Division of Cardiology, New York University Grossmann School of Medicine, 435 East 30th Street, NSB 707, New York, NY 10016, USA.
References
- 1. Chen SN, Gurha P, Lombardi R, Ruggiero A, Willerson JT, Marian AJ. The hippo pathway is activated and is a causal mechanism for adipogenesis in arrhythmogenic cardiomyopathy. Circ Res 2014;114:454–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lombardi R, da Graca Cabreira-Hansen M, Bell A, Fromm RR, Willerson JT, Marian AJ. Nuclear plakoglobin is essential for differentiation of cardiac progenitor cells to adipocytes in arrhythmogenic right ventricular cardiomyopathy. Circ Res 2011;109:1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cerrone M, Montnach J, Lin X et al. Plakophilin-2 is required for transcription of genes that control calcium cycling and cardiac rhythm. Nature Comm 2017;8:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim JC, Perez-Hernandez Duran M, Alvarado FJ et al. Disruption of Ca(2+)i homeostasis and Cx43 hemichannel function in the right ventricle precedes overt arrhythmogenic cardiomyopathy in PKP2-deficient mice. Circulation 2019;140:1015–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Corrado D, Link MS, Calkins H. Arrhythmogenic right ventricular cardiomyopathy. N Engl J Med 2017;376:61–72. [DOI] [PubMed] [Google Scholar]
- 6. Austin KM, Trembley MA, Chandler SF et al. Molecular mechanisms of arrhythmogenic cardiomyopathy. Nat Rev Cardiol 2019;16:519–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. James CA, Bhonsale A, Tichnell C et al. Exercise increases age-related penetrance and arrhythmic risk in arrhythmogenic right ventricular dysplasia/cardiomyopathy-associated desmosomal mutation carriers. J Am Coll Cardiol 2013;62:1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sawant AC, Bhonsale A, Te Riele AS et al. Exercise has a disproportionate role in the pathogenesis of arrhythmogenic right ventricular dysplasia/cardiomyopathy in patients without desmosomal mutations. J Am Heart Assoc 2014;3:e001471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Prior DL, La Gerche A. The athlete’s heart. Heart 2012;98:947–955. [DOI] [PubMed] [Google Scholar]
- 10. Heidbuchel H. The athlete's heart is a proarrhythmic heart, and what that means for clinical decision making. Europace 2018;20:1401–1411. [DOI] [PubMed] [Google Scholar]
- 11. Derbre F, Gomez-Cabrera MC, Nascimento AL et al. Age associated low mitochondrial biogenesis may be explained by lack of response of PGC-1alpha to exercise training. Age (Dordr) 2012;34:669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sanchis-Gomar F, Pareja-Galeano H, Martinez-Bello VE. PPARgamma agonist pioglitazone does not enhance performance in mice. Drug Test Anal 2014;6:922–929. [DOI] [PubMed] [Google Scholar]
- 13. Davies KJ, Packer L, Brooks GA. Biochemical adaptation of mitochondria, muscle, and whole-animal respiration to endurance training. Arch Biochem Biophys 1981;209:539–554. [DOI] [PubMed] [Google Scholar]
- 14. Montnach J, Agullo-Pascual E, Tadros R, Bezzina CR, Delmar M. Bioinformatic analysis of a plakophilin-2-dependent transcription network: implications for the mechanisms of arrhythmogenic right ventricular cardiomyopathy in humans and in boxer dogs. Europace 2018;20:iii125–iii132. [DOI] [PubMed] [Google Scholar]
- 15. Mitchell JH, Haskell W, Snell P, Van Camp SP. Task Force 8: classification of sports. J Am Coll Cardiol 2005;45:1364–1367. [DOI] [PubMed] [Google Scholar]
- 16. Prakken NH, Teske AJ, Cramer MJ et al. Head-to-head comparison between echocardiography and cardiac MRI in the evaluation of the athlete’s heart. Br J Sports Med 2012;46:348–354. [DOI] [PubMed] [Google Scholar]
- 17. Sikkel MB, Francis DP, Howard J et al. Hierarchical statistical techniques are necessary to draw reliable conclusions from analysis of isolated cardiomyocyte studies. Cardiovasc Res 2017;113:1743–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vega RB, Konhilas JP, Kelly DP, Leinwand LA. Molecular mechanisms underlying cardiac adaptation to exercise. Cell Metab 2017;25:1012–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marcus FI, McKenna WJ, Sherrill D, Basso C et al. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the task force criteria. Circulation 2010;121:1533–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luijkx T, Velthuis BK, Prakken NH et al. Impact of revised Task Force Criteria: distinguishing the athlete’s heart from ARVC/D using cardiac magnetic resonance imaging. Eur J Prev Cardiol 2012;19:885–891. [DOI] [PubMed] [Google Scholar]
- 21. Kirchhof P, Fabritz L, Zwiener M et al. Age- and training-dependent development of arrhythmogenic right ventricular cardiomyopathy in heterozygous plakoglobin-deficient mice. Circulation 2006;114:1799–1806. [DOI] [PubMed] [Google Scholar]
- 22. Fabritz L, Hoogendijk MG, Scicluna BP et al. Load-reducing therapy prevents development of arrhythmogenic right ventricular cardiomyopathy in plakoglobin-deficient mice. J Am Coll Cardiol 2011;57:740–750. [DOI] [PubMed] [Google Scholar]
- 23. Chelko SP, Asimaki A, Andersen P et al. Central role for GSK3beta in the pathogenesis of arrhythmogenic cardiomyopathy. JCI Insight 2016;1:e85923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim C, Wong J, Wen J et al. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature 2013;494:105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van Opbergen CJM, den Braven L, Delmar M, van Veen TAB. Mitochondrial dysfunction as substrate for arrhythmogenic cardiomyopathy: a search for new disease mechanisms. Front Physiol 2019;10:1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lie OH, Dejgaard LA, Saberniak J et al. Harmful effects of exercise intensity and exercise duration in patients with arrhythmogenic cardiomyopathy. JACC Clin Electrophysiol 2018;4:744–753. [DOI] [PubMed] [Google Scholar]
- 27. Singh JK, Barsegyan V, Bassi N et al. T-tubule remodeling and increased heterogeneity of calcium release during the progression to heart failure in intact rat ventricle. Physiol Rep 2017;5:e13540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.