Abstract
During pregnancy, the immune system is modified to allow developmental tolerance of the semi-allogeneic fetus and placenta to term. Pregnant women suffering from stress, anxiety, and depression show dysfunctions of their immune system that may be responsible for fetal and/or newborn disorders, provided that placental gene regulation is compromised. The present study explored the effects of maternal chronic self-perceived stress, anxiety, and depression during pregnancy on the expression of immune-related genes and pathways in term placenta. Pregnancies were clinically monitored with the Beck Anxiety Inventory (BAI) and Edinburgh Postnatal Depression Scale (EPDS). A cutoff threshold for BAI/EPDS of 10 divided patients into two groups: Index group (>10, n = 11) and a Control group (<10, n = 11), whose placentae were sampled at delivery. The placental samples were subjected to RNA-Sequencing, demonstrating that stress, anxiety, and depression during pregnancy induced a major downregulation of placental transcripts related to immune processes such as T-cell regulation, interleukin and cytokine signaling, or innate immune responses. Expression differences of main immune-related genes, such as CD46, CD15, CD8α & β ILR7α, and CCR4 among others, were found in the Index group (P < 0.05). Moreover, the key immune-like pathway involved in humoral and cellular immunity named “Primary immunodeficiency” was significantly downregulated in the Index group compared with Controls. Our results show that mechanisms ruling immune system functions are compromised at the maternal-fetal interface following self-perceived depressive symptoms and anxiety during pregnancy. These findings may help unveil mechanisms ruling the impact of maternal psychiatric symptoms and lead to new prevention/intervention strategies in complicated pregnancies.
Keywords: antenatal stress, immune system, term-placentae, RNA-Seq, human
Mechanisms ruling immune system functions are compromised at the maternal-fetal interface following self-perceived depressive symptoms and anxiety during pregnancy.
Graphical Abstract
Graphical Abstract.
Introduction
During pregnancy, placental permeability and immune transfer, including the transfer of maternal antibodies, depend on many factors, including maternal physical and mental condition [1]. For instance, depression during pregnancy (prenatal depression) has been identified as an important clinical risk factor for the transmission of abnormal health and behavior to the offspring [2].
Past and recent studies demonstrated that psychiatric patients exhibit dysregulated immune responses, suggesting that the immune system is strongly associated with psychosomatic illnesses [3]. Evidence of how immunological factors are affected during major depression and stress has been repeatedly presented in the current literature. Whereas patients with major depressive disorder (MDD) reveal increase in both circulating pro-inflammatory cytokines and their respective receptors [4, 5], treatments that reduce or remit depressive symptomology are correlated with normalization of immune signaling levels [6]. For instance, an overrepresentation of IL-6, IL-8, and type I IFN-induced signaling pathways and an increased expression of a wide range of innate immune genes and proteins such as IL-1β, IL-6, TNFα, TLR3, and TLR4 have been found in post-mortem brain samples from depressed suicide victims [7–9]. Furthermore, meta-analyses indicate that the cytokines IL-1β, IL-6, and TNF are consistent biomarkers of inflammation in patients with psychiatric symptoms, with changes in gene expression correlating with depression and even in response to antidepressant treatment [9–11]. These data provide strong evidence implicating the immune system in depression, which may affect the placenta functions and the fetus.
Exposure of the human fetus to high concentrations of biomarkers for inflammation leads to subsequent effects on the HPA axis, affecting behavior, and cognitive function in the offspring [12–15]. These specific prenatal effects suggest a direct biological effect of the in utero environment during depression on the development of the fetal brain which, in turn, could program an abnormal behavior and biological stress responses in the offspring; underlying psychopathology later in life [14].
Women with chronic self-perceived stress, anxiety, and depression during pregnancy had an increased expression of stress-mediating genes in their term placentas [16]. Considering the relation between these psychiatric disorders and immunity [17], it is pertinent to wonder if these women may have a particularly compromised regulation of their immune system in the placenta. Considering the relevance of the placental interface, stress-related maternal dysfunctions could have long-lasting effects on placental gene expression, lead to dysregulation of the fetal immune system, and contribute to some of the neuropsychological components of the newborn health [17]. Studies integrating these complex networks are required to elucidate the immunological and clinical consequences of the maternal programming of the newborn immune system. Comparison of healthy and complicated pregnancies would provide important opportunities to identify key mechanisms influencing neonatal immunity laying a basis for new or improved health interventions targeting the pregnant women or the child.
The present study therefore tested the hypothesis that transcripts and pathways associated with immune responses would have a differential expression in the term placenta of women suffering from chronic self-perceived anxiety and depression during pregnancy, compared with a control population.
Material and methods
Ethical considerations
Patient integrity was ensured by ethical approvals guaranteeing full pre-consent information and full anonymity. Data were handled at the group level only as all data were coded and treated anonymously. This is an established procedure in Swedish clinical investigations. The study was approved by the Regional Ethical Review Board in Linköping (Dnr 2011/499-31, 2013/355-32) and the Swedish Ethical Review Authority (EPM), Uppsala (Dnr 2020-05429).
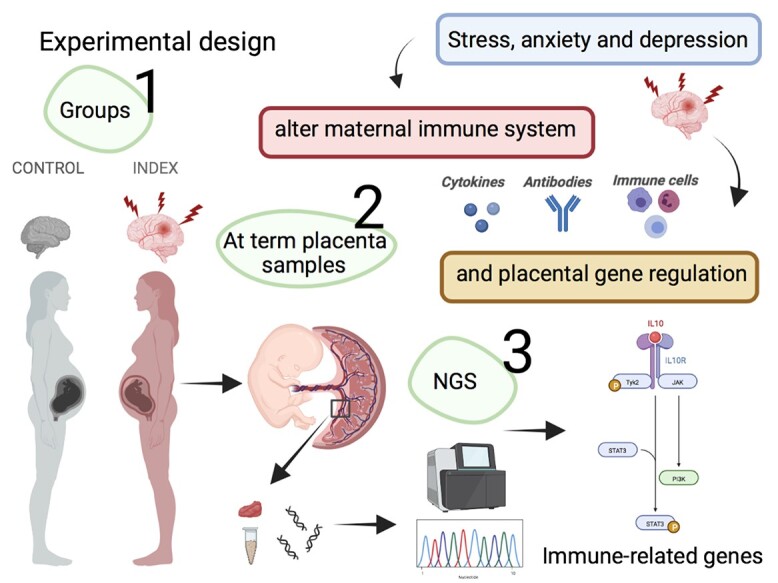
Experimental design
The primary experimental layout of this study has been published [16, 18]. In brief, data were collected from a total of 390 pregnant women attending an antenatal care clinic in southeastern Sweden. The women filled out anxiety and depression inventories during pregnancy, which are considered markers of stress. Self-perceived anxiety symptoms were assessed among women at the same time during pregnancy with the Beck Anxiety Inventory (BAI) [19] and depressive symptoms were assessed with the Edinburgh Postnatal Depression Scale (EPDS) [20]. Both inventories are well known, have been validated in Swedish [21, 22] and are frequently used in research settings as well as in clinical settings [23]. A cutoff score of 10 was used as a measure of symptoms of depression and anxiety during pregnancy for both the EPDS and the BAI. By choosing this threshold, the sensitivity for detecting major depression was close to 100% and the specificity was 82% [24]. Women who had pregnancy complications, including preeclampsia and/or preterm delivery, were excluded. A total of 11 women scoring >10 on both EPDS/BAI—indicating symptoms of depression and anxiety—were included in the Index group. A total of 11 controls who scored <10 on both EPDS/BAI were referred as the Control group. After birth, placentas were immediately collected and frozen until transcriptome analyses. Demographic data of patients included in the present study is shown in Table 1.
Table 1.
Demographic data of the patients included in this study
| Index n (%) | Control n (%) | P-value | |
|---|---|---|---|
| University/college degree | 1.000* | ||
| No | 4 (36.4) | 5 (45.5) | |
| Yes | 7 (63.6) | 6 (54.5) | |
| Born in Sweden | 0.234* | ||
| No | 0 (0.0) | 3 (27.3) | |
| Yes | 11 (100.0) | 8 (72.7) | |
| Married/cohabiting | NA | ||
| No | 0 (0.0) | 0 (0.0) | |
| Yes | 11 (100.0) | 11 (100.0) | |
| Age | 0.586* | ||
| 15–34 | 10 (90.9) | 8 (72.7) | |
| 35- | 1 (9.1) | 3 (27.3) | |
| Employed | 1.000* | ||
| No | 0 (0.0) | 1 (9.1) | |
| Yes | 11 (100.0) | 10 (90.9) | |
| Offspring’s sex | 0.4* | ||
| Female | 5 (45.5) | 8 (72.7) | |
| Male | 6 (54.5) | 3 (27.3) | |
| Mean/SD | Mean/SD | P-value | |
| Gestational length, weeks | 39.91/1.22 | 39.54/1.04 | 0.519** |
| Birthweight, grams | 3650.91/379.37 | 3700.45/647.45 | 0.797** |
*Fisher’s exacttest.
**Mann–Whitney U-test.
Collection and preparation of placenta samples
At-term placentas were placed on ice and samples distant about 5 cm from the confluence of the umbilical cord dissected. This villous parenchyma, approximately 2.5 cm thick, was then perforated to obtain 1 g samples of the fetal (including the chorionic plate), middle, and maternal (including the thin basal plate) regions, snap-frozen, and stored until further analyses.
RNA extraction
Isolation of total RNA was performed from pools of four different placental segments within the same patience using Trizol reagent (Invitrogen, Carlsbad, CA, USA) following manufacturer’s instructions. Briefly, placental samples were placed in tubes containing 1 mL of Trizol reagent and mechanically disrupted using a TissueLyser II (Qiagen, Hilden, Germany), and were then centrifuged at 12 000 × g at 4°C for 10 min. The supernatant was then incubated with bromochloropropane (100 μL/mL homogenized) for 5 min at room temperature (RT). Samples were then centrifuged at 12 000 × g at 4°C for 15 min. The aqueous phases obtained after centrifugation were mixed with isopropanol and RNA precipitation solution (1.2 M NaCl and 0.8 M Na2C6H6O7) and incubated for 10 min at RT. Samples were then centrifuged at 12 000 × g at 4°C for 10 min. After discarding the supernatant, 1 mL of 75% ethanol was added to the pellet fraction and centrifuged at 7500 × g at 4°C for 5 min. The obtained RNA pellets were air dried for 25 min and mixed with 30 μL of RNase-free water. The total RNA obtained was quantified using a NanoDrop ND -1000 (Thermo Fisher Scientific, Fremont, CA, USA) and the quality was determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), giving 8–10 values for the RNA integrity number (RIN).
Transcriptome analyses
The analyses of altered transcripts were performed using the RNA-Seq technique. The TruSeq stranded mRNA library prep kit (Illumina, San Diego, CA, USA) was used for library preparation, following manufacturer’s instructions. Library sequencing was performed using the NextSeq 500/550 High Output kit v2.5 (150 cycles), with sequencing setup of 2 × 75 bp paired-end, with at least 25 million reads per sample. Quantification and quality of the libraries are done using Qubit DNA Assay kit (Invitrogen) and DNA 1000 kit (Agilent) with an Agilent 2100 Bioanalyzer. Trimmomatics (version 0.36) [25] was used to trim adapter sequence, filter the quality (minimum quality of 15 in 4-bases window) and retain only reads longer than 36 bp. Cleaned reads were aligned with STAR [26] on human genome (GRCh38), using GRCh38.84 annotation to extract gene counts per sample. The raw datasets were deposited at Sequence Read Archive (SRA) with the accession number: PRJNA764856 (https://www.ncbi.nlm.nih.gov/sra/PRJNA764856).
RNA-Seq data analysis
The RNA-Seq data were examined using Partek Genomics Suite 7.0 (Partek). Data were first normalized using Robust Multichip Average RMA method [27]. Differently expressed genes (DEGs) between Index and Control groups were established performing a one way-analysis of variance (ANOVA) setting parameters as a fold change (FC) >1 or < −1 with P-value <0.05. To obtain biological meaning of the significantly modified transcripts, an enrichment analysis was performed. Analysis of altered Gene Ontology (GO) terms and pathways were assessed according to DAVID (database for annotation, visualization, and integrated discover), and KEGG (Kyoto Encyclopedia of Genes and Genomes) databases.
RNA-Seq validation
RNA-Seq results were validated by real time quantitative polymerase chain reaction (RT-qPCR) of nine selected DEGs. The RNA samples used for RT-qPCR assay were the same samples used for RNA-Seq analysis. Primers were commercially synthetized and tested (Bio Rad, Hercules, CA, USA). Total RNA was reverse transcribed into cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). qPCR was performed in 10-μL reactions with 5 μL of PowerUp™ SYBR™ Green Master Mix (Applied Biosystems™, CA, USA), 50 nM for each set of primers, 2 μL of synthetized cDNA and water to a final volume of 10 μL. All reactions were carried out using the Real-Time PCR Detection System (CFX96™; Bio-Rad Laboratories, Inc; CA, USA). The thermal cycling profile was 50°C for 2 min, 95°C for 10 min, 40 cycles at 95°C for 15 s, and 60°C for 1 min. Each sample for each gene was run in duplicate. The gene relative expression levels were quantified using the 2−ΔΔct [28] method and two reference genes (GAPDH and ACTB) were used for cDNA normalization.
Immunohistochemistry
Placental samples were fixed in 4% paraformaldehyde for 4 h and embedded in paraffin wax. Tissue blocks were cut into 5 μm thick sections and dewaxed with Histoclear (HS-200, National Diagnostics, USA) followed by rehydration through gradients of ethanol (100%, 95%, 90%, 70% and PBS). For antigen retrieval, Tris-EDTA (pH -9.0) buffer was used. Samples were blocked in Hydrogen Peroxide Block (ab64261, Abcam, Cambridge, UK) and Protein block (ab64261, Abcam, Cambridge, UK) and incubated in a humid chamber at 4°C overnight with the primary monoclonal antibodies, namely CD8B (rabbit monoclonal, ABIN6924225, Antibodies-online GmbH, Aachen, Germany at 1:100 dilution) and IL7R (rabbit monoclonal, ab259806, Abcam, Cambridge, UK, dilution 1:100). For protein detection, rabbit specific HRP/DAB (ABC) Detection IHC Kit (ab64261, Abcam, Cambridge, UK) was used, including incubation with biotinylated goat anti-rabbit secondary antibody at RT for 10 min. A washing step (PBS-0.01% Tween20) of 2 × 10 min was performed after each procedure. Primary antibodies were incubated with CD8B & IL7R Blocking peptides (MBS3231207 and MBS8244306, respectively, mybiosources, San Diego, CA, USA) as negative controls. All slides were counterstained (ab245880, Abcam, Cambridge, UK) and mounted in Aqueous Mounting Medium (ab128982, Abcam, Cambridge, UK). Images were obtained with a Nikon E800 microscope connected to software NIS elements (Nikon Instruments Inc. Tokyo, Japan). DAB staining intensity was calculated in a minimum of five areas per section with the Image J Fiji software and differences in DAB staining between groups were statistically compared using Mann–Whitneytest.
Results
Chronic self-perceived anxiety and depression during pregnancy modifies the placental transcriptome profile with a strong influence in immune system processes
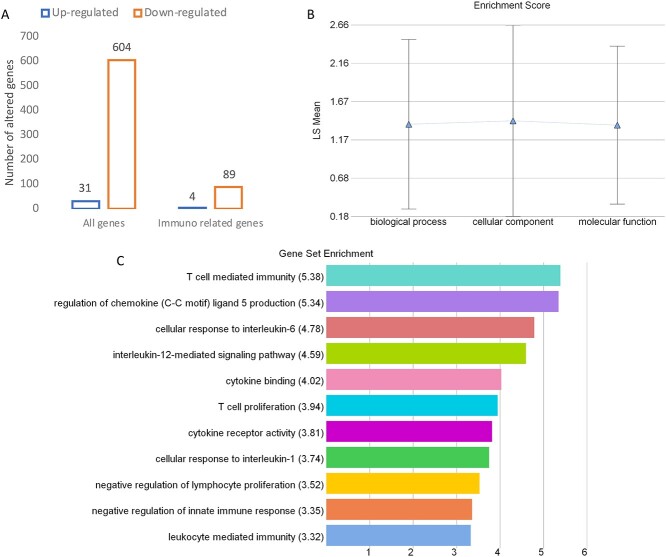
Following statistical analysis of the data yield by RNA-Sequencing, we found that in the Index group, a total of 635 transcripts were differently expressed when compared with the Control group. Overall, most genes appeared clearly downregulated (604 downregulated and only 31 upregulated). From that general panel we found 225 genes with a fold change <−5 and 28 genes with a fold change >5. After an overall enrichment analysis, there was a conspicuous overrepresentation of biological process associated with immune system process. The most significant findings in this gene set were associated with T cell regulation and interleukin and cytokine production (Figure 1).
Figure 1.
Number of total and immune-related genes (A), the functional categories (B), and the biological processes (C) that are affected in placental samples from women displaying self-perceived stress, anxiety, and depression during pregnancy (Index group, n = 11) compared with placental samples from healthy patients (Control group, n = 11). The RNA-Seq data were examined using Partek Genomics Suite 7.0 (Partek). Data were normalized using the RMA method. Differently expressed genes between Index and Control groups were established performing a one way-ANOVA setting parameters as a fold change (FC) >1 or <−1 with P-value <0.05.
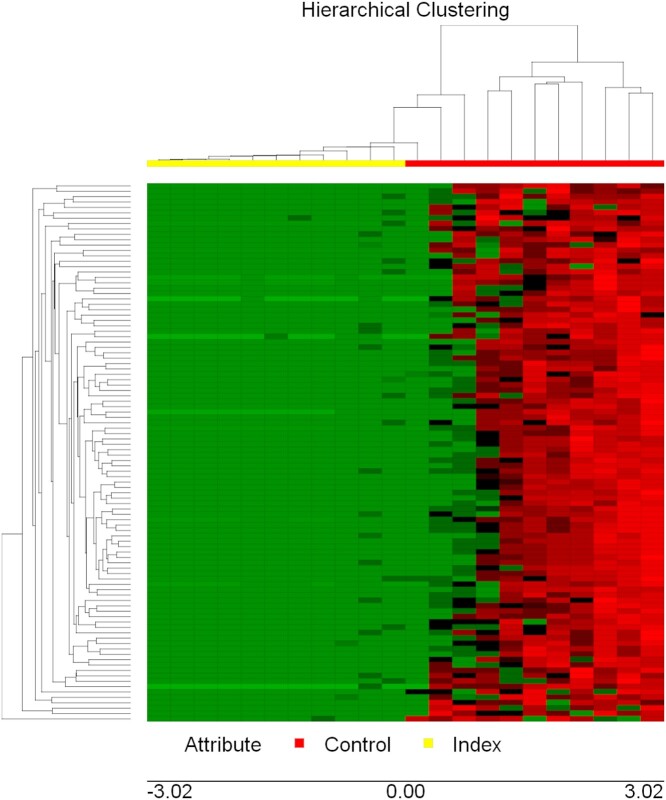
To gain insight into similarities among biological samples, differentially expressed genes were subjected to a hierarchical clustering procedure and presented as heatmaps (Figure 2). The heatmap indicates that the selected differential gene set associates the biological samples correctly into two groups, each representing one of the two clinical conditions (Index vs Control).
Figure 2.
Hierarchical cluster plot of gene expression data of term placenta samples of patients with self-perceived stress, anxiety, and depression (Index group, n = 11) and healthy patients (Control group, n = 11). Both rows and columns are clustered using correlation distance and average linkage. Colors represent mRNA levels (red: higher, green: lower).
From the general gene set, we selected 80 significantly altered genes (Table 2) involved in different biological processes and pathways, with potential roles in immune processes, all of them being downregulated in the Index group (P < 0.05).
Table 2.
Subset of genes associated with immune system processes, differentially expressed in the Index group compared with the Control group (P < 0.05)
| Gene name | Fold change | P-value | Gene name | Fold change | P-value |
|---|---|---|---|---|---|
| ABL1 | −3.23 | 0.02 | IL17Rα | −1.87 | 0.009 |
| ACKR2 | −5.12 | 0.04 | IL7R | −4.82 | 0.02 |
| ADAM10 | −2.12 | 0.03 | IMPDH1 | −11.83 | 0.007 |
| ADGRB1 | −2.61 | 0.02 | IRF7 | −2.34 | 0.02 |
| AP2A2 | −2.54 | 0.007 | JARID2 | −3.96 | 0.01 |
| AP3D1 | −2.74 | 0.04 | KAT8 | −7.67 | 0.03 |
| APEH | −5.37 | 0.006 | KIF22 | −3.24 | 0.004 |
| ARHGAP45 | −4.22 | 0.01 | LAX1 | −3.15 | 0.01 |
| ARHGEF2 | −16.28 | 0.02 | LPCAT1 | −4.13 | 0.02 |
| ATM | −5.66 | 0.04 | MKNK2 | −2.44 | 0.01 |
| BCL11B | −9.73 | 0.04 | MUC17 | −2.01 | 0.04 |
| BSG | −2.16 | 0.04 | MYO10 | −2.02 | 0.01 |
| BTN2A1 | −3.22 | 0.02 | NFE2L1 | −3.28 | 0.04 |
| BTN2A2 | −3.13 | 0.007 | NUB1 | −4.07 | 0.04 |
| BTN2A3 | −4.09 | 0.008 | OPRD1 | −3.78 | 0.01 |
| C4A | −3.66 | 0.02 | PCID2 | −3.58 | 0.02 |
| CACTIN | −3.24 | 0.01 | PIP5K1C | −3.79 | 0.04 |
| CAPZA2 | −2.57 | 0.04 | PMS2P5 | −3.90 | 0.04 |
| CCR4 | −4.33 | 0.03 | PSMD11 | −5.23 | 0.04 |
| CD151 | −3.65 | 0.03 | PSMD13 | −1.96 | 0.02 |
| CD46 | −5.13 | 0.02 | PTPN11 | −6.42 | 0.03 |
| CD8α | −4.23 | 0.005 | RAB31 | −1.64 | 0.01 |
| CD8β | −20.25 | 0.01 | RAB3A | −8.81 | 0.04 |
| CD99L2 | −4.31 | 0.03 | RAB4A | −3.21 | 0.01 |
| CERT1 | −3.54 | 0.03 | RC3H1 | −3.79 | 0.03 |
| CHRNB2 | −2.36 | 0.04 | RPS6 | −1.61 | 0.04 |
| CHRNB4 | −2.84 | 0.03 | SBNO2 | −2.56 | 0.03 |
| CLPB | −5.61 | 0.004 | SCRIB | −13.78 | 0.04 |
| CNN2 | −2.42 | 0.03 | SERPINB9 | −2.84 | 0.002 |
| DAPK3 | −3.57 | 0.01 | SIRPA | −6.27 | 0.03 |
| DSP | −1.97 | 0.04 | SLC16A8 | −24.89 | 0.04 |
| EDN1 | −7.97 | 0.02 | SLC2A5 | −3.86 | 0.02 |
| EEF2 | −1.97 | 0.005 | SOD2 | −5.15 | 0.003 |
| EPPIN | −46.77 | 0.01 | SOX4 | −2.49 | 0.01 |
| FAM3α | −4.76 | 0.02 | SYVN1 | −3.05 | 0.03 |
| FCGR3α | −5.86 | 0.01 | TCF3 | −3.28 | 0.01 |
| FCGR3β | −7.54 | 0.02 | TEK | −4.10 | 0.03 |
| GOLPH3 | −1.77 | 0.02 | TNFRSF13β | −17.41 | 0.03 |
| GRAP2 | −30.82 | 0.01 | TRIM34 | −4.14 | 0.02 |
| ICAM1 | −2.74 | 0.03 | ZBTB7A | −2.64 | 0.0005 |
| IL11Rα | −4.11 | 0.02 | ZDHHC11 | −4.25 | 0.02 |
| IL12Rβ1 | −3.28 | 0.02 |
The offspring’s sex influences placental gene expression
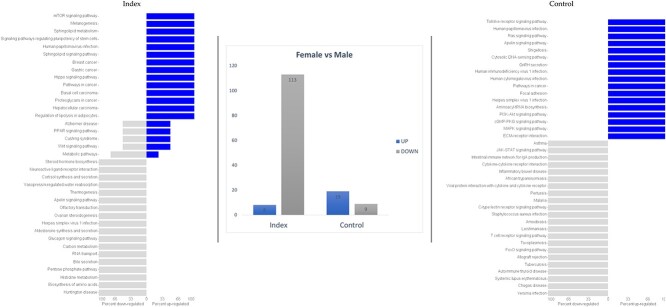
We further evaluated the impact of offspring sex on gene expression among placentae harvested from Index and Control women at term. This analysis demonstrated different responses on gene expression levels between “female” and “male” placentas with a total of 121 and 28 altered genes in the Index and Control groups, respectively (Figure 3). Eight immune-related genes were dysregulated in “female” placentas compared with “male” placentas in response to maternal stress (CLEC5A, VSTM1, IGHV23, IGKC, IGKV2, IGKV2D, IGLC1, and IGLC3).
Figure 3.
Differential gene expression and GO terms in “female” vs “male” placentas from women displaying self-perceived stress, anxiety, and depression during pregnancy (Index group, n = 11) compared with placental samples from healthy patients (Control group, n = 11).
Biological meaning of a selected gene subset
The GO biological process analysis revealed several significantly downregulated biological processes associated with immune functions, such as T cell immunity and proliferation and cytokine and interleukin regulation. The details of these analyses are summarized in Table 3. Moreover, to gain insight into the roles of these immune-related genes, this particular set of immune-related genes was subjected to a functional clustering procedure and presented as a network (Figure 4).
Table 3.
Most significant differently expressed biological processes (P < 0.05) examined with KEGG database in the Index group compared with the Control group
| Biological process | Enrichment score | P-value | Genes altered (%) | Gene list |
|---|---|---|---|---|
| T cell mediated immunity | 5.38 | 0.004 | 16.7 | BTN3A2, BTN3A3, CD46, CD8α |
| Negative regulation of chemokine ligan 5 | 5.34 | 0.004 | 50 | MUL1, SIRPA |
| Cellular response to interleukine-6 | 4.78 | 0.008 | 20 | ABCC2, ICAM1, SBNO2 |
| Interleukine-12 signaling | 4.59 | 0.01 | 10.86 | AIP, BOLA2, CNN2, IL12Rβ1, SOD2 |
| Cytokine binding | 4.02 | 0.01 | 6.56 | A2M, ACKR2, BMPR2, CCR4, IL11Rα, IL12Rβ1, LIFR, TGFBR3L, WFIKKN1 |
| T cell proliferation | 3.94 | 0.01 | 11.11 | ABL1, CD151, RC3H1, RPS6 |
| Cytokine receptor activity | 3.81 | 0.02 | 7.21 | ACKR2, CCR4, IL11Rα, IL12R 1, IL17Rα, IL7R, LIFR |
| Cellular response to interleukine-1 | 3.74 | 0.02 | 7.79 | ABCC2, CACTIN, EDN1, ICAM1, RC3H1, SIRPA |
| Negative regulation of lymphocyte proliferation | 3.52 | 0.02 | 7.41 | ATM, GNRH1, GPNMB, RC3H1, SCRIB, TNFRSF13β |
| Negative regulation of innate immune response | 3.35 | 0.03 | 7.93 | A2M, CACTIN, FAM3α, MUL1, SERPINB9 |
| Leukocyte mediated immunity | 3.32 | 0.03 | 7.05 | BTN3A2, BTN3A3, CD46, CD8α, IRF7, SERPINB9 |
Figure 4.
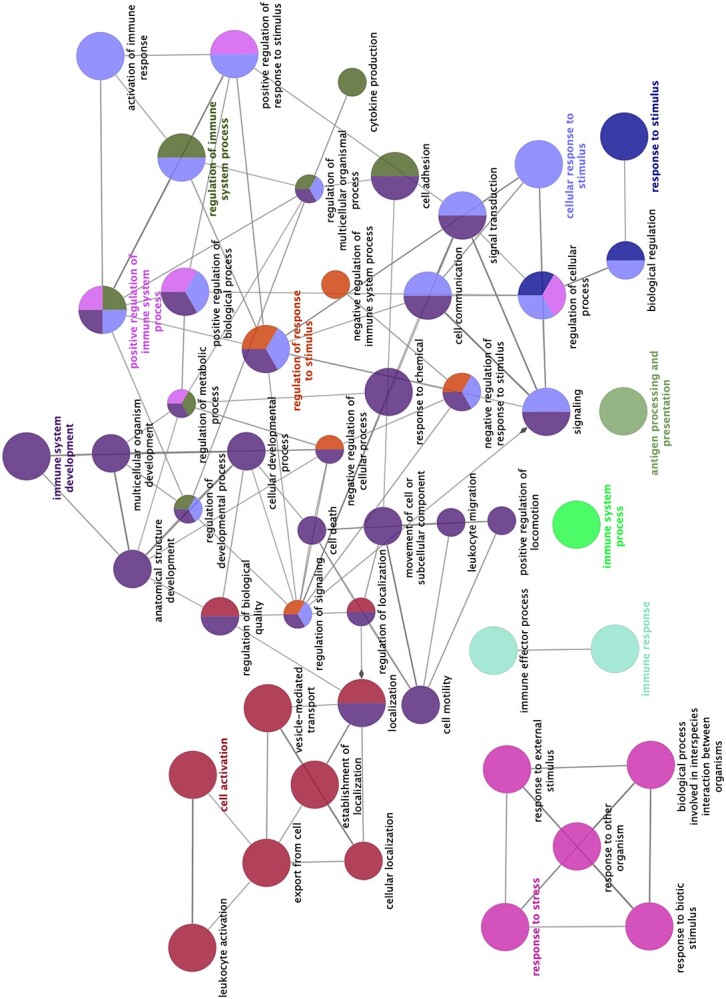
Schematic representation of selected altered transcripts in placentae from women displaying self-perceived stress, anxiety, and depression during pregnancy (Index group, n = 11) compared with placental samples from healthy patients (Control group, n = 11). Analysis of overrepresented biological functions was performed using the Cytoscape v3.0.0 application ClueGo v2.0.3. The following databases were used: GO subgroups biological process represented as circles. Different colors represent different terms which are functionally grouped based on common genes (kappa score). The size of the circles indicates the level of significance, where the largest circles correspond to the highest significance. The following ClueGo parameters were used: GO tree levels, 1–3 (first level = 0); minimum number of genes, 1; GO term fusion; GO term connection restriction (kappa score), 0.4; GO term grouping.
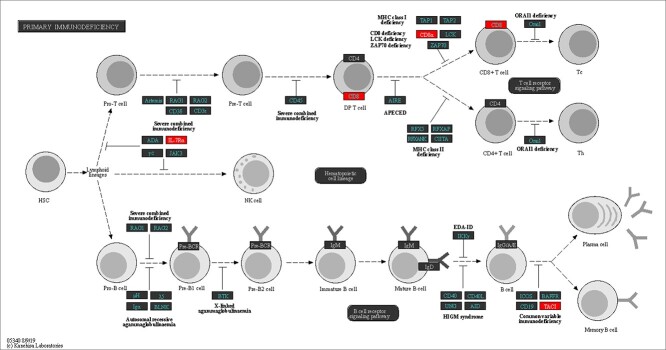
One major immune-related pathway dominated among significantly affected pathways
Following pathway enrichment analysis, we found that one of the key pathways ruling mechanisms for cellular and humoral immune regulation named “Primary immunodeficiency” was significantly altered (P < 0.02) in placentae from the Index group compared with Controls. Several transcripts belonging to this pathway (CD8α, CD8Β, IL7R, and TNFRSF13B) were downregulated in the Index group (Figure 5).
Figure 5.
Schematic representation of the “Primary immunodeficiency” pathway (hsa05340). Downregulated genes (P < 0.05) in placentae from women displaying self-perceived stress, anxiety, and depression during pregnancy (Index group, n = 11) compared with placental samples from healthy patients (Control group, n = 11) are red-marked.
Validation of the RNA-Seq data
We selected nine genes to verify the RNA-Seq results by RT-qPCR. These genes presented similar patterns of expression under both methods (Figure 6), proving that RNA-Seq results were reliable.
Figure 6.
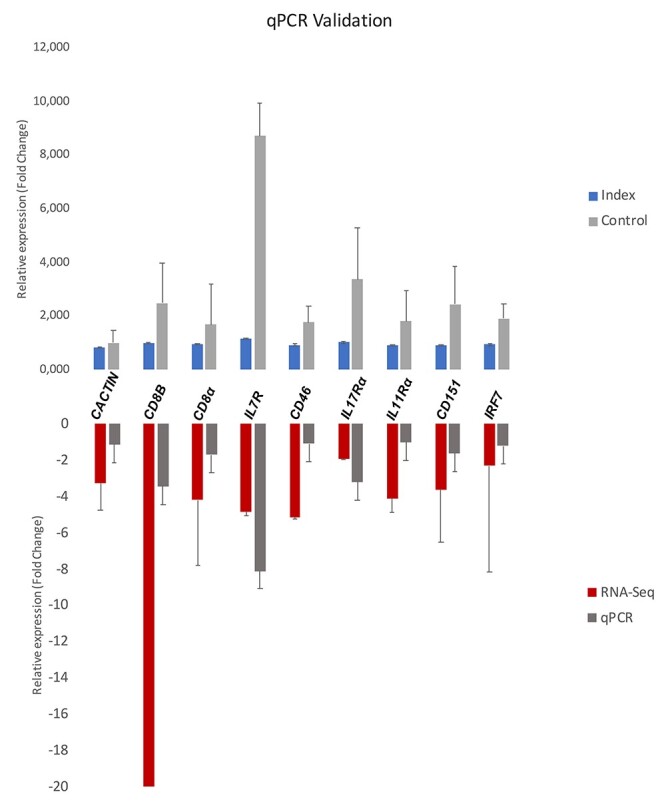
Validation of RNA-Seq results by RT-qPCR of nine differentially expressed genes. The upper graph represents difference of expression of nine immune-related genes between women displaying self-perceived stress, anxiety, and depression during pregnancy (Index group, n = 11) compared with placental samples from healthy patients (Control group, n = 11). Differences in gene expression between groups were statistically compared using Mann–Whitney test. All genes validated were significantly different (P < 0.05) between groups. Values on Y axle represent the relative gene expression (fold change).
Chronic self-perceived anxiety and depression during pregnancy alters the protein expression of two candidate immune-related genes
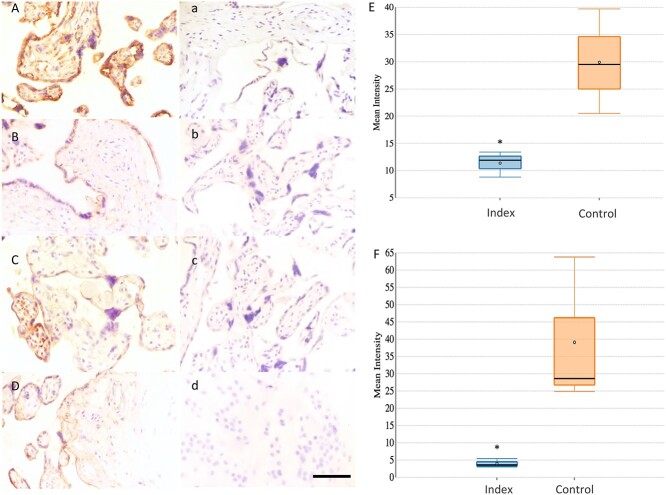
The expression of two candidate proteins (CD8B and IL7R) was assessed by immunohistochemistry to validate the results of gene expression. We confirmed the presence of both proteins within the extra-villous trophoblast of placentas from either studied group (Index and Control groups) (Figure 7Aa–Dd).
Figure 7.
(A–F) Representative immunohistochemical staining for CD8B and IL7R in extra-villous trophoblast of placentas from healthy patients (Control group: A, a- CD8B and C, c- IL7R, n = 4) and from patients that were affected by maternal stress during pregnancy (Index group: B,b- CD8B and D,d- IL7R, n = 4). A-B images show positive staining of CD8B. a-b images are staining controls of CD8B. C-D images show positive staining of IL7R. c-d images are staining controls of IL7R. Size bar 50 μm. E and F show Whisker-box blots depicting the relative quantification of immunolabeling intensity for CD8B and IL7R proteins using Image J software. The box indicates the interquartile range; the horizontal line in the box depicts the median; whiskers indicate the data range. Differences in DAB staining between groups were statistically compared using Mann–Whitney test. Asterisk indicates that the measured differences were statistically significant (P < 0.05).
Protein expression was calculated by measuring DAB staining intensity in a minimum of five areas per section. Following the line of the results obtained for gene expression, the expression of CD8B and IL7R was significantly downregulated (P < 0.05) in placental samples from the Index group compared with the Controls (CD8B: 11.4 ± 0.9 vs 29.9 ± 3.9, respectively and IL7R: 3.9 ± 0.5 vs 39.1 ± 8.7, respectively) (Figure 7E-F). These results suggest the improper protein expression prevailing in placental samples from mothers suffering from stress, anxiety, and depression during pregnancy compared with the Control group.
Discussion
The present study explored the effects of chronic self-perceived stress, anxiety, and depression during pregnancy on the expression of immune-related genes and pathways in term placentae. The results demonstrated the presence of a modified placental transcriptome profile with a strong influence in immune system processes.
Chronic self-perceived anxiety and depression during pregnancy clearly induced a major downregulation of transcripts related to T-cell regulation, interleukins, and cytokine signaling or innate immune response. CD46, CD151,and CD8α&β genes were downregulated in the Index group (FC: −5.14, −3.65, −4.23, and − 20.25, respectively). CD46 encodes the membrane cofactor protein which is a regulatory protein that protects host cells from injury by complement [29]. This protein is commonly known as the “pathogen magnet” [30] and is widely recognized for its roles in linking innate and adaptive immune responses [31, 32] and further related to T-cell biology (reviewed in Kemper et al., 2007). The co-engagement of the T-cell receptor and CD46 by CD4+ T cells leads to the induction of interferon-γ (IFN-γ)-secreting effector T-helper type-1 (Th1) cells (reviewed in Kemper et al., 2007 [33]). Moreover, common variable immunodeficiency was found in CD46-deficient patients, a syndrome characterized by hypogammaglobulinemia [34]. CD46-activated T-cells support B-cell activation, evidenced in a CD46-deficient patient whose T-cells were impaired in promoting immunoglobulin G production by B cells [35]. CD151, a member of tetraspanins, is an important immunomodulator of Natural Killer (NK) cells [36] and a regulator of antioxidants in placentas. Downregulation of this gene has been recently associated with accused oxidative stress and apoptosis in trophoblast cells [37]. CD8α, an adaptive immune gene, has a key role in the “Primary immunodeficiency” pathway, which was downregulated in the Index group.
Primary immunodeficiency disease (PID) is a disorder where components of the immune system are missing or compromised [38]; and likely transferred to the newborn [39]. This pathway involves a group of genes (including CD8α, ILR7α, and TAC1; downregulated in the Index group; FC: −4.23, −4.82, −17.41, respectively) affecting cellular and humoral immunity or nonspecific host defense mechanisms mediated by complement proteins. It would also affect phagocytes and natural killer (NK) cells [40], with responsibility for immune system changes in early life as well as a neuroendocrinological programming of the offspring’s brain, which may trigger future neuropsychiatric and/or autoimmune disorders [41]. Such interpretation harmonizes well with the prior findings of high basal levels of inflammatory cytokines and cortisol in blood, associated with reduced GR activity [42] in the offspring of pregnant women with MDD, which makes them highly vulnerable for developing adult affective behaviors. Such postulation is also in line with prior hypothesis that pregnant women suffering from stress and anxiety show dysfunction of circulating inflammatory cytokines, which may be responsible for fetal and/or newborn disorders [43]. Moreover, it is worth noting that the offspring of pregnant women with MDD have high basal levels of inflammatory cytokines and cortisol in blood, associated with reduced GR activity [42], which makes them highly vulnerable for developing adult affective behaviors.
In the present study, the gene encoding the Chemokine receptor CCR4 was downregulated in the Index group (FC: −4.33). This gene is mainly expressed on näive T-cells [44] and has a key functional role in both cell migration and in cell–cell interaction as well as in the regulation of various immune responses [45]. Lopez et al. (2006) proposed that CCR4 expression increases in the third trimester of healthy pregnancies implicating T-cell recruitment to inflamed areas, while it reduces in neonates from mothers suffering from atopic immune responses alike the anxious/depressive mothers during pregnancy [46]. This suggests a delayed maturation of cellular immune functions [46] whose impact on the placenta and fetus is unknown and needs further researchwork.
The receptor for interleukin 7 (IL7R), which encodes a protein that plays a critical role during lymphocytes development [47, 48], was repressed in the Index group (FC: −4.82). Dysfunction of IL7R has been detected in patients with severe combined immunodeficiency (SCID) leading to chronic inflammatory diseases [49, 50]. Two other interleukin receptors of interest (IL11Rα and IL17Rα) were also decreased in the Index group, which is of interest since it is known that the correct function of these receptors is required for a correct fetoplacental development [51]. In contrast to most interleukin receptors, the expression levels of IL17Rα are functionally significant since high levels of IL17Rα receptor are required for an effective function [52, 53]. Interestingly, pathological dysfunction of IL-17Rα in pregnant mothers affects fetal brain development and subsequently may contribute to the autism spectrum disorder (ASD)-like behavioral phenotypes in offspring [54].
In summary, these findings suggest that symptoms of chronic anxiety and depression during pregnancy have the potential to suppress immune functions that are physiologically regulated by cytokines, chemokines, and interleukins during pregnancy. Conceivably, these alterations are related to modifications in cellular oxidative stress taking place during the pathogenesis of these disorders since a relation between the accumulation of oxygen radicals and immune dysfunction has been reported before [55]. Indeed, an activation of signaling pathways and alteration of the physiology of inflammatory cytokines and chemokines have been associated with high concentrations of reactive oxygen substances (ROS) in several cell populations [56].
In addition, a downregulation of the two main Toll-like receptors (TLRs) gene regulators (Cactin; FC: −3.24 and IRF7; FC: −2.34) was observed in the Index group. It is known that TLRs, which are transmembrane proteins, are known to function as primary sensors of the immune system by recognizing specific microbial motifs and inducing proinflammatory genes that promote innate and adaptive immunity [57]. TLRs regulate gene expression through the activation of transcription factors, such as NF-κB and interferon-regulatory factors. Hence, upregulation of these pathways can lead to inflammatory diseases and is a subject to tight control by negative regulators of innate immune signaling. Cactin acts as a negative regulator of TLRs. Overexpression of Cactin suppresses TLR-induced activation of NF-κB and induction of TLR-responsive genes, whereas knockdown of endogenous Cactin enhances TLR-induction of these responses [58]. The downregulation of Cactin found in the Index group might be explained by the fact that, under stress conditions, TLRs are needed to counteract the effects of the immune system downregulation and create an environment of immune tolerance toward the fetal allograft.
Conclusions
Taken together, our results demonstrate that an immune dysfunction, evidenced as a differential expression of transcripts and pathways associated with immune responses, was present in the term placenta of women displaying symptoms of chronic self-perceived stress, anxiety, and depression during pregnancy. These findings have potential implications for prevention and intervention strategies. However, more studies are needed to investigate the consequences of these maternal disorders on the immunological condition of the offspring.
Acknowledgments
The authors are grateful to Lovisa Holm for punch-sampling of the placentas, Annette Molbaek and Åsa Schippert, from the Genomics Core Facility at LiU for expert assistance with NGS and Jonathan J.M. Landry from EMBL, Genomics Core Facilities, Services & Technology Unit (Germany), for his help with the bioinformatic analyses.
Footnotes
† Grant Support: This research was funded by the Medical Research Council of Southeast Sweden (FORSS projects 114841, 661011, 392061, and 472721), the Swedish doctors union (SLS-157771, SLS-332861), the European Union’s Horizon 2020 research and innovation program under the MSCA (grant agreement No 891663); and the Swedish Research Council FORMAS (Projects 2017-00946 and 2019-00288), Stockholm, Sweden.
Contributor Information
Cristina A Martinez, Department of Biomedical & Clinical Sciences, Obstetrics & Gynaecology, Faculty of Medicine and Health Sciences, Linköping University, Linköping, Sweden.
Ina Marteinsdottir, Department of Medicine and Optometry, Faculty of Health and Life Sciences, Linnaeus University, Kalmar, Sweden.
Ann Josefsson, Department of Biomedical & Clinical Sciences, Obstetrics & Gynaecology, Faculty of Medicine and Health Sciences, Linköping University, Linköping, Sweden.
Gunilla Sydsjö, Department of Biomedical & Clinical Sciences, Obstetrics & Gynaecology, Faculty of Medicine and Health Sciences, Linköping University, Linköping, Sweden.
Elvar Theodorsson, Division of Clinical Chemistry, Department of Biomedical and Clinical Sciences, Faculty of Medicine and Health Sciences, Linköping University, Linköping, Sweden.
Heriberto Rodriguez-Martinez, Department of Biomedical & Clinical Sciences, Obstetrics & Gynaecology, Faculty of Medicine and Health Sciences, Linköping University, Linköping, Sweden.
Data availability
The data underlying this article are available in Sequence Read Archive (SRA) at (https://www.ncbi.nlm.nih.gov/sra/PRJNA764856), and can be accessed with accession number: PRJNA764856.
Author contributions
Conceptualization and design: IM, AJ, GS, ET and HR-M; analysis: CAM; interpretation of the data: IM, CAM, AJ, GS, ET and HR-M; original draft preparation: CAM and HR-M; review and editing: AJ, GS, ET and HR-M; project administration: IM, AJ, GS and ET; funding acquisition: IM. All authors have read and agreed to the published version of the manuscript.
Conflict of interest
The authors have declared that no conflict of interest exists.
References
- 1. Palmeira P, Quinello C, Silveira-Lessa AL, Zago CA, Carneiro-Sampaio M. IgG placental transfer in healthy and pathological pregnancies. Clin Dev Immunol 2012; 2012:985646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Field T. Prenatal depression risk factors, developmental effects and interventions: a review. J Pregnancy Child Heal 2017; 4:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Iwata M, Ota KT, Duman RS. The inflammasome: pathways linking psychological stress, depression, and systemic illnesses. Brain Behav Immun 2013; 31:105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lichtblau N, Schmidt FM, Schumann R, Kirkby KC, Himmerich H. Cytokines as biomarkers in depressive disorder: current standing and prospects. Int Rev Psychiatry 2013; 25:592–603. [DOI] [PubMed] [Google Scholar]
- 5. Anisman H, Hayley S. Inflammatory factors contribute to depression and its comorbid conditions. Sci Signal 2012; 5:pe45. [DOI] [PubMed] [Google Scholar]
- 6. Gazal M, Souza LD, Fucolo BA, Wiener CD, Silva RA, Pinheiro RT, Jansen K, Ghislene G, Oses JP, Kaster MP. The impact of cognitive behavioral therapy on IL-6 levels in unmedicated women experiencing the first episode of depression: a pilot study. Psychiatry Res 2013; 209:742–745. [DOI] [PubMed] [Google Scholar]
- 7. Mostafavi S, Battle A, Zhu X, Potash JB, Weissman MM, Shi J, Beckman K, Haudenschild C, McCormick C, Mei R, Gameroff MJ, Gindes Het al. Type I interferon signaling genes in recurrent major depression: increased expression detected by whole-blood RNA sequencing. Mol Psychiatry 2014; 19:1267–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bufalino C, Hepgul N, Aguglia E, Pariante CM. The role of immune genes in the association between depression and inflammation: a review of recent clinical studies. Brain Behav Immun 2013; 31:31–47. [DOI] [PubMed] [Google Scholar]
- 9. Miller AH. Norman cousins lecture. Mechanisms of cytokine-induced behavioral changes: psychoneuroimmunology at the translational interface. Brain Behav Immun 2009; 23:149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raison CL, Lowry CA, Rook GAW. Inflammation, sanitation, and consternation: loss of contact with coevolved, tolerogenic microorganisms and the pathophysiology and treatment of major depression. Arch Gen Psychiatry 2010; 67:1211–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yirmiya R, Pollak Y, Morag M, Reichenberg A, Barak O, Avitsur R, Shavit Y, Ovadia H, Weidenfeld J, Morag A, Newman ME, Pollmächer T. Illness, cytokines, and depression. Ann N Y Acad Sci 2000; 917:478–487. [DOI] [PubMed] [Google Scholar]
- 12. Seckl JR. Glucocorticoid programming of the fetus; adult phenotypes and molecular mechanisms. Mol Cell Endocrinol 2001; 185:61–71. [DOI] [PubMed] [Google Scholar]
- 13. Seckl JR, Holmes MC. Mechanisms of disease: glucocorticoids, their placental metabolism and fetal ‘programming’ of adult pathophysiology. Nat Clin Pract Endocrinol Metab 2007; 3:479–488. [DOI] [PubMed] [Google Scholar]
- 14. Glover V. Prenatal stress and its effects on the fetus and the child: possible underlying biological mechanisms. Adv Neurobiol 2015; 10:269–283. [DOI] [PubMed] [Google Scholar]
- 15. Segovia SA, Vickers MH, Reynolds CM. The impact of maternal obesity on inflammatory processes and consequences for later offspring health outcomes. J Dev Orig Health Dis 2017; 8:529–540. [DOI] [PubMed] [Google Scholar]
- 16. Martinez CA, Marteinsdottir I, Josefsson A, Sydsjö G, Theodorsson E, Rodriguez-Martinez H. Expression of stress-mediating genes is increased in term placentas of women with chronic self-perceived anxiety and depression. Genes (Basel) 2020; 11:869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goodman JH. Perinatal depression and infant mental health. Arch Psychiatr Nurs 2019; 33:217–224. [DOI] [PubMed] [Google Scholar]
- 18. Marteinsdottir I, Sydsjö G, Faresjö Å, Theodorsson E, Josefsson A. Parity-related variation in cortisol concentrations in hair during pregnancy. BJOG 2021; 128:637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beck AT, Epstein N, Brown G, Steer RA. An inventory for measuring clinical anxiety: psychometric properties. J Consult Clin Psychol 1988; 56:893–897. [DOI] [PubMed] [Google Scholar]
- 20. Cox JL, Holden JM, Sagovsky R. Detection of postnatal depression. Development of the 10-item Edinburgh postnatal depression scale. Br J Psychiatry 1987; 150:782–786. [DOI] [PubMed] [Google Scholar]
- 21. Beck ATSR. Beck Depression Inventory, second ed. Manual, Svensk version (Swedish version); 2005. [Google Scholar]
- 22. Rubertsson C, Borjesson K, Berglund A, Josefsson A, Sydsjo G. The Swedish validation of Edinburgh postnatal depression scale (EPDS) during pregnancy. Nord J Psychiatry 2011; 65:414–418. [DOI] [PubMed] [Google Scholar]
- 23. Josefsson A, Berg G, Nordin C, Sydsjö G. Prevalence of depressive symptoms in late pregnancy and postpartum. Acta Obstet Gynecol Scand 2001; 80:251–255. [DOI] [PubMed] [Google Scholar]
- 24. Harris B, Huckle P, Thomas R, Johns S, Fung H. The use of rating scales to identify post-natal depression. Br J Psychiatry 1989; 154:813–817. [DOI] [PubMed] [Google Scholar]
- 25. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003; 19:185–193. [DOI] [PubMed] [Google Scholar]
- 28. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 29. Liszewski MK, Atkinson JP. Complement regulator CD46: genetic variants and disease associations. Hum Genomics 2015; 9:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cattaneo R. Four viruses, two bacteria, and one receptor: membrane cofactor protein (CD46) as pathogens’ magnet. J Virol 2004; 78:4385–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cardone J, Le Friec G, Kemper C. CD46 in innate and adaptive immunity: an update. Clin Exp Immunol 2011; 164:301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yamamoto H, Fara AF, Dasgupta P, Kemper C. CD46: the ‘multitasker’ of complement proteins. Int J Biochem Cell Biol 2013; 45:2808–2820. [DOI] [PubMed] [Google Scholar]
- 33. Kemper C, Atkinson JP. T-cell regulation:with complements from innate immunity. Nat Rev Immunol 2007; 7:9–18. [DOI] [PubMed] [Google Scholar]
- 34. Fremeaux-Bacchi V, Moulton EA, Kavanagh D, Dragon-Durey M-A, Blouin J, Caudy A, Arzouk N, Cleper R, Francois M, Guest G, Pourrat J, Seligman Ret al. Genetic and functional analyses of membrane cofactor protein (CD46) mutations in atypical hemolytic uremic syndrome. J Am Soc Nephrol 2006; 17:2017–2025. [DOI] [PubMed] [Google Scholar]
- 35. Fuchs A, Atkinson JP, Fremeaux-Bacchi V, Kemper C. CD46-induced human Treg enhance B-cell responses. Eur J Immunol 2009; 39:3097–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Koopman LA, Kopcow HD, Rybalov B, Boyson JE, Orange JS, Schatz F, Masch R, Lockwood CJ, Schachter AD, Park PJ, Strominger JL. Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential. J Exp Med 2003; 198:1201–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Z, Cai B, Cao C, Lv H, Dai Y, Zheng M, Zhao G, Peng Y, Gou W, Wang J, Liu D, Hu Y. Downregulation of CD151 induces oxidative stress and apoptosis in trophoblast cells via inhibiting ERK/Nrf2 signaling pathway in preeclampsia. Free Radic Biol Med 2021; 164:249–257. [DOI] [PubMed] [Google Scholar]
- 38. Bonilla FA, Geha RS. 12. Primary immunodeficiency diseases. J Allergy Clin Immunol 2003; 111:S571–S581. [DOI] [PubMed] [Google Scholar]
- 39. Sheikhbahaei S, Sherkat R, Camacho-Ordonez N, Khoshnevisan R, Kalantari A, Salehi M, Nazemian SS, Nasr-Esfahani MH, Klein C. Pregnancy, child bearing and prevention of giving birth to the affected children in patients with primary immunodeficiency disease; a case-series. BMC Pregnancy Childbirth 2018; 18:299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Candotti F. Gene transfer into hematopoietic stem cells as treatment for primary immunodeficiency diseases. Int J Hematol 2014; 99:383–392. [DOI] [PubMed] [Google Scholar]
- 41. Figueiredo AS, Schumacher A. The T helper type 17/regulatory T cell paradigm in pregnancy. Immunology 2016; 148:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gelman PL, Flores-Ramos M, López-Martínez M, Fuentes CC, Grajeda JPR. Hypothalamic-pituitary-adrenal axis function during perinatal depression. Neurosci Bull 2015; 31:338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lazarides C, Epel ES, Lin J, Blackburn EH, Voelkle MC, Buss C, Simhan HN, Wadhwa PD, Entringer S. Maternal pro-inflammatory state during pregnancy and newborn leukocyte telomere length: a prospective investigation. Brain Behav Immun 2019; 80: 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yoshie O, Matsushima K. CCR4 and its ligands: from bench to bedside. Int Immunol 2015; 27:11–20. [DOI] [PubMed] [Google Scholar]
- 45. Haddeland U, Sletten GB, Brandtzaeg P, Nakstad B. Impaired interleukin (IL)-4-associated generation of CCR4-expressing T cells in neonates with hereditary allergy risk. Clin Exp Immunol 2005; 139:314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. López C, Comabella M, Tintoré M, Sastre-Garriga J, Montalban X. Variations in chemokine receptor and cytokine expression during pregnancy in multiple sclerosis patients. Mult Scler 2006; 12:421–427. [DOI] [PubMed] [Google Scholar]
- 47. Aiello FB, Keller JR, Klarmann KD, Dranoff G, Mazzucchelli R, Durum SK. IL-7 induces myelopoiesis and erythropoiesis. J Immunol 2007; 178:1553–1563. [DOI] [PubMed] [Google Scholar]
- 48. Palmer MJ, Mahajan VS, Trajman LC, Irvine DJ, Lauffenburger DA, Chen J. Interleukin-7 receptor signaling network: an integrated systems perspective. Cell Mol Immunol 2008; 5:79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Inchley CS, Osterholt HCD, Sonerud T, Fjærli HO, Nakstad B. Downregulation of IL7R, CCR7, and TLR4 in the cord blood of children with respiratory syncytial virus disease. J Infect Dis 2013; 208:1431–1435. [DOI] [PubMed] [Google Scholar]
- 50. Liao C-Y, Yu H-W, Cheng C-N, Chen J-S, Lin C-W, Chen P-C, Shieh C-C. A novel pathognic mutation on Interleukin-7 receptor leading to severe combined immunodeficiency identified with newborn screening and whole exome sequencing. J Microbiol Immunol Infect 2020; 53:99–105. [DOI] [PubMed] [Google Scholar]
- 51. Dimitriadis E, Robb L, Liu Y-X, Enders AC, Martin H, Stoikos C, Wallace E, Salamonsen LA. IL-11 and IL-11Ralpha immunolocalisation at primate implantation sites supports a role for IL-11 in placentation and fetal development. Reprod Biol Endocrinol 2003; 1:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shen F, Hu Z, Goswami J, Gaffen SL. Identification of common transcriptional regulatory elements in interleukin-17 target genes. J Biol Chem 2006; 281:24138–24148. [DOI] [PubMed] [Google Scholar]
- 53. Maitra A, Shen F, Hanel W, Mossman K, Tocker J, Swart D, Gaffen SL. Distinct functional motifs within the IL-17 receptor regulate signal transduction and target gene expression. Proc Natl Acad Sci U S A 2007; 104:7506–7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Choi GB, Yim YS, Wong H, Kim S, Kim H, Kim SV, Hoeffer CA, Littman DR, Huh JR. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science 2016; 351:933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pangrazzi L, Balasco L, Bozzi Y. Oxidative stress and immune system dysfunction in autism Spectrum disorders. Int J Mol Sci 2020; 21:3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hsieh H-L, Yang C-M. Role of redox signaling in neuroinflammation and neurodegenerative diseases. Biomed Res Int 2013; 2013:484613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fitzgerald KA, Kagan JC. Toll-like receptors and the control of immunity. Cell 2020; 180:1044–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Atzei P, Gargan S, Curran N, Moynagh PN. Cactin targets the MHC class III protein IkappaB-like (IkappaBL) and inhibits NF-kappaB and interferon-regulatory factor signaling pathways. J Biol Chem 2010; 285:36804–36817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article are available in Sequence Read Archive (SRA) at (https://www.ncbi.nlm.nih.gov/sra/PRJNA764856), and can be accessed with accession number: PRJNA764856.