Abstract
The dormant population of ovarian primordial follicles is determined at birth and serves as the reservoir for future female fertility. Yet our understanding of the molecular, biochemical, and cellular processes underpinning primordial follicle activation remains limited. The survival of primordial follicles relies on the correct complement and morphology of granulosa cells, which provide signaling factors essential for oocyte and follicular survival. To investigate the contribution of granulosa cells in the primordial-to-primary follicle transition, gene expression profiles of granulosa cells undergoing early differentiation were assessed in a murine model. Ovaries from C57Bl/6 mice were enzymatically dissociated at time-points spanning the initial wave of primordial follicle activation. Post-natal day (PND) 1 ovaries yielded primordial granulosa cells, and PND4 ovaries yielded a mixed population of primordial and primary granulosa cells. The comparative transcriptome of granulosa cells at these time-points was generated via Illumina NextSeq 500 system, which identified 131 significantly differentially expressed transcripts. The differential expression of eight of the transcripts was confirmed by RT-qPCR. Following biological network mapping via Ingenuity Pathway Analysis, the functional expression of the protein products of three of the differentially expressed genes, namely FRZB, POD1, and ZFX, was investigated with in-situ immunolocalization in PND4 mouse ovaries was investigated. Finally, evidence was provided that Wnt pathway antagonist, secreted frizzled-related protein 3 (FRZB), interacts with a suppressor of primordial follicle activation WNT3A and may be involved in promoting primordial follicle activation. This study highlights the dynamic changes in gene expression of granulosa cells during primordial follicle activation and provides evidence for a renewed focus into the Wnt signaling pathway’s role in primordial follicle activation.
Keywords: folliculogenesis, granulosa cells, RNAseq, infertility, premature ovarian insufficiency
Comparing transcriptomes of two populations of granulosa cells: those from primordial follicles and a mixed group enriched for granulosa cells from primary follicles. Frzb expression increased in primary granulosa cells relative to primordial, and FRZB interacts with primordial follicle activation suppressor WNT3A.
Introduction
Primordial follicle activation is integral to the fertility of sexually reproducing females as it is necessary for the follicle development and is the committing step to ovulation and subsequent fertilization. The number of primordial follicles within the ovaries defines the age of menopause in women, and therefore the end of a woman’s fertility. Premature ovarian insufficiency (POI) is the early cessation or absence of ovarian function due to a reduction in the pool of primordial follicles in women before the age of 40 [1]. POI occurs in 1–3% of women, and a common cause of POI is a rapid acceleration in the rate of activation of primordial follicles [2–4]. The process of primordial follicle activation is complex, involving massive parallel molecular, cellular, and biochemical events, with limited characterization [5, 6]. Granulosa cells communicate between neighboring granulosa cells and intracellularly with the oocyte to coordinate primordial follicle activation [7]; however, the extent of their role in this process is yet to be determined. Importantly, as primordial follicles activate, there must be a sufficient number of granulosa cells supporting the oocyte, and they must all transition correctly to the activated, cuboidal morphological form for successful development [8, 9]. Improving our understanding of how the rate of primordial follicle activation is controlled is imperative to improving fertility outcomes for women at risk of premature infertility and POI.
The research in human primordial follicle activation has been hampered by small samples sizes and limitations in tissue (reviewed in [10]). The use of non-human models overcomes a number of the limitations associated with the study of human samples and is instrumental to our current understanding of primordial follicle activation. [10]. Much of the research underpinning our understanding of primordial follicle activation has focused on protein interactions and single gene knockouts in mice (reviewed in [6]). Briefly, it has been established that phosphoinositide 3-kinase (PI3K) stimulates primordial follicle activation in oocytes, and this is assisted by mammalian target of rapamycin complex 1 (mTORC1) and Forkhead box L2 (FOXL2) in the granulosa cells [11]. Consequently, well-known factors, which maintain the quiescence of primordial follicles include anti-Mullerian hormone (AMH), and phosphatase and tensin homolog deleted on chromosome 10 (PTEN) [12]. Animal studies provide important groundwork with advantages such as functional studies via conditional knockouts, drug testing, and robust characterization through omics technologies. In this study, granulosa cells isolated from mouse neonatal ovaries were utilized both before and during the initial wave of primordial follicle activation to obtain large quantities of granulosa cells originating from primordial and primary follicles, respectively. These two distinct populations of cells were subjected to RNA-seq to compare the gene expression profiles of primordial and activating granulosa cells and identify novel factors contributing to follicle activation. This study presents the first detailed transcriptome comparison of granulosa cells from mouse primordial and activating follicles, we were able to annotate the expression occurring within granulosa cells and observed a small but significant shift in the transcriptome between the two populations of these cells. From this comprehensive dataset, we identified the Wnt pathway inhibitor, secreted frizzle-related protein 3 (FRZB) was significantly increased in expression in the population of granulosa cells isolated during the initial wave of primordial follicle activation, compared to granulosa cells isolated before activation and representing primordial follicles. Additionally, we determined an interaction between FZRB and WNT3A these granulosa cells. Our findings represent the commencement of a renewed investigation into the role of Wnt signaling factors in primordial follicle activation.
Methods
Ethics
C57Bl/6 mice were supplied by the University of Newcastle Animal Services Unit under ethics approval number A-2018-803. All experiments involving the use of animals were conducted in accordance with the Institutes’ Animal Care and Ethics Committee guidelines.
Tissue and cell collection
Mice were sacrificed at post-natal days one (PND1) and four (PND4). These time points were selected to coincide with immediately prior to the initial wave of primordial follicle activation when the ovary is comprised of only primordial follicles (PND1), and during the initial wave of primordial follicle activation when there is an enriched population of follicles undergoing activation (PND4) [13, 14]. See Supplementary Material S1 for follicle counts of PND1 and PND4 ovaries for proportions of primordial and primary follicles. Follicles that develop during the initial wave of primordial follicle activation do contribute to post-pubertal fertility [15] and thus are used to investigate primordial follicle activation. This approach maximizes the quantity and homogeneity of the follicle types of interest retrieved for analysis that would otherwise be confounded by later-stage, gonadotropin-dependent follicles. Other biological processes do occur in the postnatal mouse ovary (reviewed in [16]); however, the magnitude of primordial follicle activation taking place in the PND4 ovary is unparalleled by any other point in ovary development and thus makes PND1 and PND4 ovaries an effective way to explore primordial follicle activation.
Mice were housed under a controlled lighting regime (12 h light: 12 h dark) at 21–22°C and supplied with food and water ad libitum. Neonatal mice were euthanized by asphyxiation with carbon dioxide, followed by decapitation. Ovaries were collected immediately after euthanasia and either snap-frozen, fixed in 10% neutral buffered formalin for ≤10 h, or placed into 37°C Leibovitz L-15 media supplemented with 1% fetal bovine serum (FBS) for immediate granulosa cell collection. For a detailed description of the methodology and rationale behind time point selection and granulosa cell collection, see [17].
Briefly, the outer tissue and bursa were removed from the ovary, ovaries then transferred into Dulbecco Modified Eagle/F-12 medium (DMEM/F12), containing 0.02% collagenase, and 0.02% DNase I. Replicates contained between 4 and 8 ovaries, though some replicates contained pooled samples of up to 15 ovaries, as specified where appropriate. After incubation for 45–60 minutes at 37°C in 5% CO2 with gentle pipetting every 15 minutes, cells were retrieved by centrifugation (1500 g, 3 min), supernatant removed, and incubated with 0.1% trypsin/EDTA at 37°C in 5% CO2 for 15–20 minutes. Cells were recovered by centrifugation (1500 g, 3 min), and resuspended in ovary culture media (containing DMEM/F12 with 0.1% bovine serum albumen, 4% penicillin–streptomycin, 5% fetal bovine serum, 0.5% ITS-G and 3% L-glutamine). An aliquot of cells was removed for cell quantification, and the remaining cells incubated in ovary culture media with 0.5% ascorbic acid 18–24 hours at 37°C for either collection or fixing. When collecting granulosa cells, cells were incubated with 0.1% trypsin/EDTA for five minutes, collected (800 g, 15 min) and washed 3 × 5 minutes at 1200 g in Hanks′ Balanced Salt Solution, counted, and stored at −80°C until use. Cell populations were enriched for the cell target of interest, granulosa cells, rather than the other major cell type in neonatal ovaries, the oocytes, and stromal cells, as determined by oocyte and stromal counts using immunofluorescent targeting of DDX4 and Vimentin, respectively. Counts of 300 cells across four biological replicates indicate an average abundance of 0.4% oocytes and 0.8% stromal cells present in isolated granulosa cell samples. Granulosa cell viability prior to freezing and/or downstream experiments was determined via Trypan blue (Invitrogen, Carlsbad, USA; cat no. T102872) and accepted at ≥80% viability.
RNA extraction and RT-qPCR
Granulosa cell RNA was extracted using the RNeasy Plus Micro Kit (Qiagen, Hilden, Germany; cat no. 74034) as per the manufacturer’s instructions. Post-extraction, cDNA was synthesized by reverse transcription with the SuperScript IV VILO system (Invitrogen, Carlsbad, USA; cat no. 11766050). Quantitative real-time PCR (RT-qPCR) was performed in triplicate on cDNA with a reaction equivalent to 20 ng of total RNA. Predesigned and validated gene-specific TaqMan Gene Expression Assays (Life Technologies, Carlsbad, USA) were used for RT-qPCR. Each TaqMan gene expression assay contained gene specific, exon spanning forward and reverse primers for each of the genes of interest (Avpr1a (Mm00444092_m1), Cdkn2b (Mm00483241_m1), Ddr2 (Mm00445615_m1), Fam171a1 (Mm01332727_m1), Frzb (Mm00441378_m1), Ifitm1 (Mm00850040_g1), Rpl19 (Mm02601633_g1), Tcf21 (Mm00448961_m1), Zfx (Mm03053842_s1)) and fluorogenic minor groove binder probes consisting of a target-specific oligonucleotide labelled with a fluorescent dye FAM (6-Carboxyfluorescein) or VIC (2′-chloro-7′phenyl-1,4-dichloro-6-carboxy-fluorescein), and a non-fluorescent quencher. Data were normalized to the expression of the transcript encoding 60S ribosomal protein L19 (Rpl19). Triplicate expression values of each gene was set relative to the reference gene via the ΔΔCT method [18] and is presented as the mean ± SEM with statistical analysis determined by unpaired Student t-test.
RNA sequencing and mapping transcripts
Eight, libraries were sequenced on the Illumina NextSeq 2 × 75bp high output via Auckland Genomics at the University of Auckland. The RNA samples consisted of four replicates each of two groups of mouse granulosa cells (isolated from PND1 and PND4), each replicate contained pooled samples of cells isolated from 4–15 ovaries. The bioinformatics team at Auckland Genomics Facility performed the following analyses on the RNA sequencing output. The overall quality of the data was visualized using the program FastqQC [19]. TrimGalore [20] was used to check and remove adapters from the sequences. Sequences with a Phred score less than 30 were trimmed. Reads with a length shorter than 25 base pairs were discarded. Hisat2 [21, 22] was used to map the cleaned reads to the mouse transcriptome (grcm38). Output sam files from hisat2 were then sorted by genomic position and converted to bam format using samtools sort [23]. To generate expression estimates for each sample, the mapped reads were then assembled into transcripts using StringTie. Differential expression analysis was performed using the R software package Ballgown [24]. Low abundance transcripts with a variance less than one across the samples were removed. To view the similarity between the samples, the Pearson correlation and distance between the samples were calculated. Differential gene expression between the two treatments was calculated using FPKM (fragments per kilobase million) values. The data were then sorted by their adjusted p-value (false discovery rate) and absolute log2 fold change values.
In silico analysis of gene expression
The analysis of genes expressed within PND1 and PND4 neonatal mouse granulosa cells was undertaken in silico using a number of techniques. Briefly, transcript abundance data were assessed via volcano plots to visualize trends associated with differentially accumulating genes in the primordial granulosa cells compared to activating granulosa cells (PND1 versus PND4). The threshold for significant differentially expressed transcripts was defined as having a two-fold change of FPKM in either direction (log2 fold change ±1), and a significance level of P ≤ 0.05. Consistency of gene expression among biological replicates for transcripts above the determined threshold was visualized via a heat map of expression (FPKM) for each gene of each replicate normalized to the PND1 average FPKM for the respective gene. Differential expression that did not meet the threshold of statistical significance, but relevant to demonstrate the cell type of interest being captured are presented in Table 2.
Table 2.
Average number of fragments per kilobase million (FPKM) for notable genes detected via RNAseq from granulosa cells isolated at PND1 and PND4.
| Gene symbol | Gene name | Relevance | Average reads PND1 (FPKM) | Average reads PND4 (FPKM) |
|---|---|---|---|---|
| Inha | Inhibin alpha | Marker of granulosa cells | 44.4 | 58.9 |
| Gata4 | GATA binding protein 4 | Marker of granulosa cells | 71 | 64.4 |
| Amh | Anti-Mullerian hormone | Marker of mature granulosa cells | 0.06 | 3.4 |
| Akt1 | AKT serine/threonine kinase 1 | Promotes primordial follicle activation | 130.1 | 138.8 |
| Bmp4 | Bone morphogenetic protein 4 | Promotes primordial follicle activation | 12.6 | 13.4 |
| Lhx8 | LIM Homeobox 8 | Suppresses primordial follicle activation | 2 | 0.9 |
Ingenuity pathway analysis (IPA) was used to explore biological networks for differentially expressed genes with significance P ≤ 0.05. IPA queries a large database of experimental observations between molecules and uses this information to construct biological networks that represent cause-effect relationships between mammalian genes, proteins, and their functions with probability calculations [25]. The granulosa cell dataset of significantly differentially expressed genes (Supplementary Material S2) was interrogated for enrichment of functional pathways using bioinformatic enrichment tools available via the Database for Annotation, Visualization and Integrated Discovery (DAVID; v6.8) [26, 27] then, separate gene lists according to the clusters identified in the heatmap were assessed with the same tools. DAVID Gene Ontology (GO) annotation tools were utilized for exploration of biological process [28] in differentially expressed genes, GO identifiers identified within the dataset were also subject to enrichment analysis to determine their representation [29], for full list of DAVID GO terms and identifiers returned for this dataset, see Supplementary Material S3.
Immunolocalization
For immunofluorescence of mouse ovaries, tissues were washed with and embedded in Optimal Cutting Temperature (OCT) compound (ProSciTech, IA018), then snap-frozen in dry ice. Blocks were serially sectioned (5 μm thick) with a cryostat (Leica Biosystems) onto Superfrost Plus slides (ThermoFisher; cat no. 4951PLUS4) three ovaries per section for biological triplicate. Before use, slides were dried to room temperature for 5 min and rehydrated in PBS. Heat mediated antigen retrieval was performed by warming slides for 30 min at 65°C in 10 mM sodium citrate buffer (pH 6). To prevent non-specific antibody binding, sections were blocked in phosphate-buffered saline solution with 5% donkey serum and 1% bovine serum albumen for 1.5 h at room temperature. Sections were probed with antibodies in Table 1 (antibody of interest, colocalized with a granulosa cell marker, GATA4 or FOXL2) and incubated overnight at 4°C. After washing, sections were incubated with secondary antibodies Alexa Fluor 488 donkey anti-rabbit IgG (ThermoFisher; cat no. A21206), Alexa Fluor 555 donkey anti-goat IgG (Abcam, cat no. A21432) at a dilution of 1:200 for 1 h at room temperature. Slides were counterstained with 4′-6-Diamidino-2-phenylindole (DAPI) and mounted in anti-fade reagent Mowiol (13% w/v Mowiol4–88, 33% w/v glycerol, 66 mM Tris (pH 8.5), 2.5% w/v 1, 4 diazobcyclo-[2.2.2] octane). Images were taken on an Olympus Fluoview 1000-IX81 confocal microscope (Olympus, Center Valley, USA) under fluorescent optics.
Table 1.
Antibody details.
| Antibody target | Species raised in | Dilution | Company | Catalogue number |
|---|---|---|---|---|
| FOXL2 | Goat | 1:100 | Abcam | ab5096 |
| FRZB | Goat | 1:50 | ThermoFisher | PA5–47793 |
| GATA4 | Rabbit | 1:100 | Abcam | ab84593 |
| POD1 | Rabbit | 1:100 | Bios USA | BS-8688R |
| WNT3A | Rabbit | 1:100 | Abcam | ab28472 |
| ZFX | Rabbit | 1:200 | ThermoFisher | PA5–34376 |
For immunocytochemistry of fixed granulosa cells, wells were permeabilized in phospho-buffered saline (PBS) with 0.01% Triton-X then blocked for 2 h at room temperature in PBS containing 10% donkey serum and 3% bovine serum albumen. Cells were probed overnight at 4°C with anti-FRZB antibody (ThermoFisher; cat no. PA5–47793), and anti-WNT3A antibody (Abcam, Cambridge, UK, cat no. ab28472), each at a dilution of 1:200. After washing, sections were incubated with secondary antibodies: Alexa Fluor 488 donkey anti-rabbit IgG (ThermoFisher; cat no. A21206), or Alexa Fluor 555 donkey anti-goat IgG (Abcam, cat no. A21432) at a dilution of 1:200 for 1 h at room temperature. Cells were counterstained with 4′-6-Diamidino-2-phenylindole (DAPI) and a layer of PBS was added to prevent drying out. Images were taken on an EVOS FL (AMF4300; Thermofisher).
Duolink proximity ligation assay
Protein interactions were detected via Duolink proximity ligation assay kit (Merck, DUO92105) using an anti-rabbit plus probe and an anti-goat minus probe as according to the manufacturer’s instructions. Briefly, cells isolated by dissociation were fixed for immunocytochemistry as above, then blocked with the kit blocking solution for 1 h at room temperature. Antibodies were applied at a ratio of 1:100 with kit antibody diluent solution and incubated overnight at 4°C. Following this, Duolink probes were added to cells at a ratio of 1:5 with kit diluent solution and incubated for 1 h at 37°C in a humidified chamber and excess was washed in kit wash buffer. Cells were incubated in a humidified chamber heated to 37°C in kit solutions for ligation of probes (30 min), and amplification of the signal (100 min), then cells were counterstained with 4′-6-Diamidino-2-phenylindole (DAPI). The plus and minus probes were bound to the anti-WNT3A antibody, and antibody anti-FRZB, respectively. Where the distance between the two bound probes is <40 nm a red signal is generated, which indicates an interaction of proteins of interest [30].
Results
Subtle changes in granulosa cell gene expression during primordial follicle activation
To investigate granulosa cell transcriptional changes associated with primordial follicle activation, RNA-seq analysis was performed on large quantities of granulosa cells isolated from primordial (PND1) ovaries or activating and primary follicles (PND4). A comparison between the PND1 and PND4 date sets did not reveal any transcripts with a false discovery rate ≤ 0.1. A total of 11 784 mapped genes were identified from RNA transcripts during bioinformatics processing, including genes characteristic of granulosa cells and primordial follicle activation (Table 2). There was a total of 131 transcripts differentially expressed (log2 fold change ±1; P ≤ 0.05) between PND1 and PND4 granulosa cells (Figure 1). By comparing the transcripts in the PND1 granulosa cells with those found in the PND4 granulosa cells, 82 RNA transcripts were identified as significantly more abundant whilst 49 transcripts were identified as significantly more abundant in PND4 granulosa cells compared to PND1 (Figure 1); for a complete list of differentially expressed genes, see Supplementary Material S2. This represents a very small (1.1%) change in transcripts from the total number identified between these two fundamental developmental stages.
Figure 1.
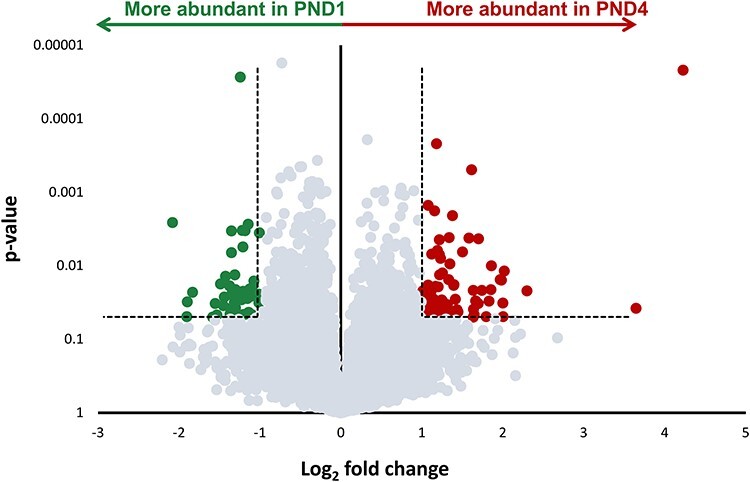
Differential gene expression profiling of neonatal mouse granulosa cells. The volcano plot indicates the differential expression between RNA transcripts from postnatal day 1 (PND1), and postnatal day 4 (PND4) granulosa cells (n = 4 replicates from each group). Mapped genes expressed in higher abundance in PND1 granulosa cell RNA transcripts compared to PND4 granulosa cells are colored green. Genes expressed in higher abundance in PND4 granulosa cells compared to PND1 are colored red. Dashed line indicates the threshold for differential expression; genes with high statistical significance (P ≤ 0.05), and log2 fold change of greater than ±1. Points that fall below these thresholds are colored grey.
A heat map was generated to illustrate the expression (normalized to the average FPKM of the PND1 samples) of each gene within the threshold of significantly differentially expressed genes across each of the sample replicates (Figure 2A). The top biological process occurring in differentially expressed genes were identified using gene ontology (GO) tools (Figure 2B). These processes are reflective of the dynamic events occurring before and during a mass wave of primordial follicle activation, namely transcription (GO:006351), cell differentiation/proliferation (GO:0030154/GO:0008284), phosphorylation (GO:0006468), and genes relating to early development (GO:0007275). Two clusters of differentially abundant genes are apparent from the heatmap; Cluster 1 comprising of transcripts that are more abundant in the PND4 granuolsa cells compared to the PND1, and Cluster 2 of transcripts with a decreased abundance in PND4 relative to PND1 granulosa cells. Within Cluster 1, among the top GO annotation of biological processes in terms of the percentage of genes represented within the dataset (Figure 2C) are those associated with development and growth; multicellular organism development (GO:0007275), positive regulation of cell proliferation (GO:0008284), spreading (GO:1900026), and migration (GO:0010634). These processes signify activation occurring, which is governed by the upregulation of signaling pathways encompassing both the activation and repression of genes and proteins, while granulosa cells differentiate from their squamous to cuboidal structure [6, 11], as is reflective of ovarian events at this time. Cluster 2 (Figure 2D) contains notable biological processes: meiotic cell cycle (GO:0051321), negative regulation of cell migration (GO:0030336), oogenesis (GO:0048477), and positive regulation of translation (GO:0045727). These biological processes are indicative of primordial follicles, known to be regulated by genes associated with development of the ovary [16] that establish the ovarian reserve after germ cell nest breakdown, and are the exclusive follicle type in the ovaries at PND1 [13, 31].
Figure 2.
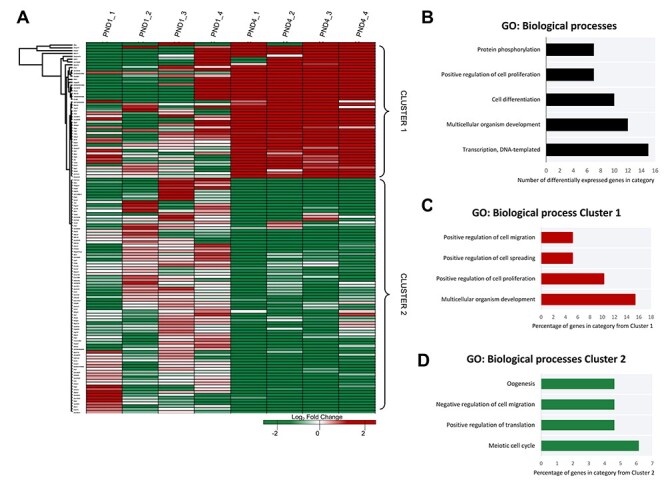
Gene expression clustering of neonatal mouse granulosa cells. (A) A heatmap differential expression within the defined threshold (P ≤ 0.05, log2 fold change of ± ≥ 1, n = 131) depicts the consistency of expression of mapped transcripts identified as having significant between biological replicates. Different genes are represented in different rows, and different replicates in different columns. Expression values (fragments per kilobase million) are represented as a color scale from green for lower expressions to red for higher expressions and normalized to the average FPKM for the PND1 group. Differentially expressed genes were annotated via gene ontology (GO) analysis in DAVID (v6.8). (B) GO information of the biological process categories with highest count of genes from the differentially expressed genes. Clusters of differential expression were identified from the heat map and annotated via gene ontology (GO) analysis in DAVID (v6.8). (C) GO information of four biological process categories from Cluster 1 are presented as the percentage of genes from within that cluster belonging to a given category, and similarly (D) the GO information of biological process categories from Cluster 2 for four notable categories with the greatest percentage. For full list of DAVID GO terms and identifiers returned for this dataset, see Supplementary Material S3.
Validation of differentially expressed genes
To verify the differential gene expression between primordial granulosa cells and granulosa cells undergoing primordial follicle activation in light of the false discovery rate, eight candidate genes were selected for validation of RNA-seq data using RT-qPCR. Genes above the threshold of significance for differential expression were chosen, while also selecting genes with varying degrees of differential expression within the threshold. Four genes decreased abundance in PND1 (primordial) granulosa cells compared to PND4 (activating) granulosa cells (frizzled related protein [Frzb], transcription factor 21 [Tcf21], discoidin domain receptor 2 [Ddr2], and zinc finger protein x-linked [Zfx]), and conversely, four genes exhibiting increased abundance in PND1 granulosa cells relative to PND4 (Interferon-induced transmembrane protein 1 [Ifitm1], Family with sequence similarity 171 member A1 [Fam171a1], Cyclin-dependent kinase inhibitor 2B [Cdkn2b], arginine vasopressin receptor 1A [Avpr1a]), see Table 3. Experiments were performed on at least six pooled biological replicates (n = 4–15 animals per sample). The transcript encoding the 60S ribosomal protein L19 (Rpl19) was used as an endogenous control in all RT-qPCR analyses and confirmed the differential expression of all eight candidate genes while following a parallel trend to the RNA-seq data (Figure 3). Six out of the eight genes validated remained significant (P ≤ 0.05) following Bonferroni correction for multiple comparisons. Collectively, these results validate the accuracy our RNA-seq dataset. Importantly this outcome strengthens our justification for the use of this valuable dataset to explore the differential temporal gene expression between mouse neonatal granulosa cells, representative of primordial and activating subtypes.
Table 3.
Targets selected for validation of RNAseq.
| Gene symbol | Absolute fold change | Abundance in PND4 relative to PND1 | P-value | Gene name |
|---|---|---|---|---|
| Frzb | 3.56 | Increased | 0.023 | Frizzle-related protein |
| Tcf21 | 2.7 | Increased | 0.014 | Transcription factor 21 |
| Ddr2 | 2.59 | Increased | 0.019 | Discoidin domain receptor 2 |
| Zfx | 2.5 | Increased | 0.027 | Zinc finger protein x-linked |
| Ifitm1 | 4.05 | Decreased | 0.012 | Interferon-induced transmembrane protein1 |
| Fam171a1 | 3.95 | Decreased | 0.016 | Family with sequence similarity 171 member A1 |
| Cdkn2b | 3.91 | Decreased | 0.016 | Cyclin-dependent kinase inhibitor 2B |
| Avpr1a | 2.99 | Decreased | 0.004 | Arginine vasopressin receptor 1A |
Figure 3.
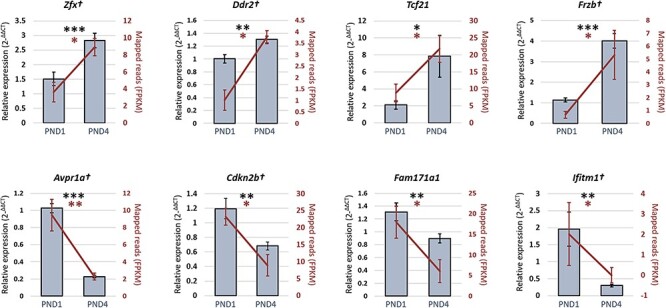
The RT-qPCR validation of differentially expressed genes within neonatal mouse granulosa cells. In verifying the RNA-seq data (represented on the secondary y-axis by the fragment per kilobase of exon per million, FPKM, for each mapped read), eight genes that displayed significantly different levels of expression in PND1 granulosa cells compare to PND4 granulosa cells (see Table 3) were selected for orthogonal validation using RT-qPCR. Validation experiments were performed in triplicate using ≥ six distinct pools of biological samples (n = 4–15 animals per sample), statistical significance was investigated via a student t-test. The 60S ribosomal protein L19 gene was employed as an endogenous control to normalize the expression levels of target genes. Six out of 8 genes remained significant after Bonferroni correction for multiple comparison and are indicated by †. Data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Validating protein expression in the mouse ovary
Following gene expression validation, the functional protein expression of notable genes from the RNA-seq dataset was investigated. Through utilizing the Ingenuity Pathway Analysis (IPA) network analysis feature, a number of genes from the dataset were identified to encode proteins that interact with molecules demonstrated to be involved in primordial follicle activation. A proportion of the IPA-generated network map is presented in Figure 4 and identifies six molecules from the dataset interacting through binding or regulation and includes two of those validated in via RT-qPCR (FRZB, and ZFX). Primordial follicle activation component TGFBR from the dataset was also linked within the network. To explore a potential relationship with primordial follicle activation, the two molecules from the pathway analysis network that had been validated at the gene expression level were selected for in situ immunolocalization in neonatal mouse ovaries (Figure 5), granulosa cells staged according to morphology, described elsewhere [32]. Podocyte-expressed 1 (POD1) did not feature in the IPA-generated network, but was also selected for further investigation, as the expression of its gene (Tcf21) increased 3.7-fold (P < 0.05) in PND4 granulosa cells compared to PND1 granulosa cells as determined by RT-qPCR. The relationship of POD1 to primordial follicle activation that made it of interest is depicted in addition to the network map in Figure 4. ZFX protein was localized to the oocyte cytoplasm of both primordial and primary follicles and was also observed in the extracellular space proximal to the granulosa cells of follicles (Figure 5A). There was some expression of POD1 in all granulosa cells, but POD1 protein was more intensely localized in the nuclei of some granulosa cells (Figure 5B). Oocytes were also observed to have punctate expression of POD1. FRZB was expressed in the oocyte cytoplasm (Figure 5C) and exhibited differential localization in granulosa cells.
Figure 4.
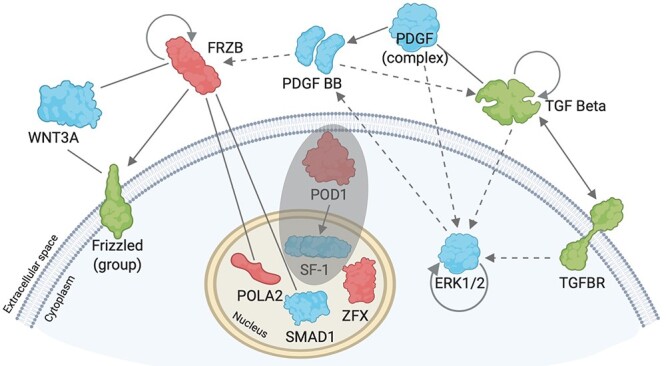
Schematic network diagram of biological network. Pathway analysis (IPA) network explorer was used to generate a biological network of molecules (or groups of molecules) from the dataset, based on their connectivity with other molecules both within the dataset and connected with the literature. A segment of the IPA-generated image was re-created using BioRender.com. Molecules detected within the dataset are color coded according to their differential expression (red = increased differential expression in PND1 granulosa cells compared to PND4 granulosa cells, green = decreased differential expression in PND1 granulosa cells compared to PND4 granulosa cells). Blue molecules are those generated by the IPA network explorer. Line between molecules indicates binding, solid arrow indicates direct regulation, dashed arrow indicates indirect regulation, and curved arrow within molecules indicates self-regulation. Shaded circle indicates molecules added to the diagram, which were not generated by the IPA network explorer.
Figure 5.
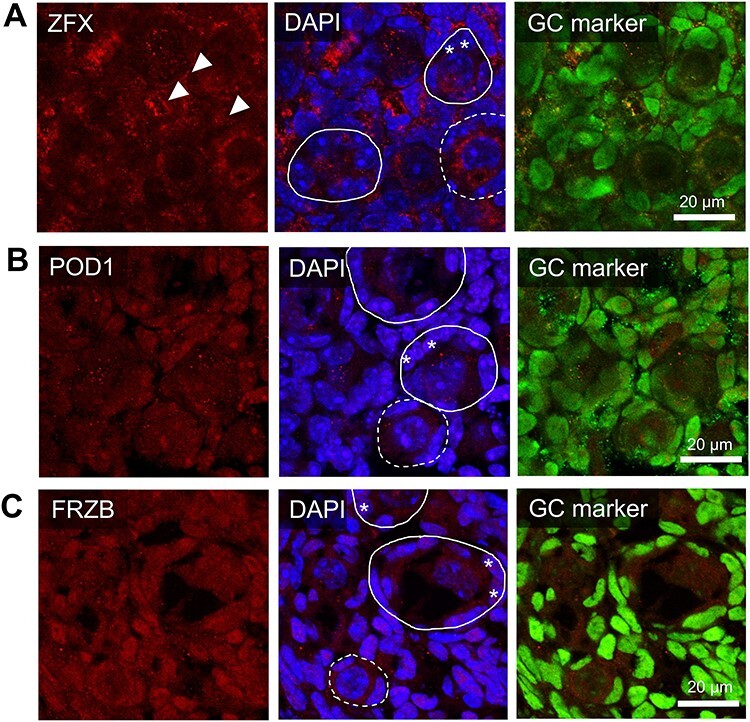
Expression and localization of proteins of interest within the PND4 ovary. The in situ immunofluorescent expression of three proteins of interest (A-C) were explored in the neonatal mouse ovaries and were co-localized with a nuclear marker (DAPI, blue), a granulosa cell marker (GATA4 or FOXL2, green), and either (A) ZFX (B) POD1 or (C) FRZB in red. Representative images from PND4 selected as they include populations of primordial, activating and primary follicles. Representative images are indicative of n = 4–6 biological replicates of both PND1 and PND4 performed in triplicate. Images taken at 60× magnification, scale bars represent 20 μm with dotted circles outlining primordial follicles, solid lines outlining activating or primary follicles. Arrows indicate extracellular staining regions; asterisks indicate activating granulosa cells.
The expression and localization of FRZB was further investigated in neonatal mouse ovaries and granulosa cells to identify possible links to the process of primordial follicle activation considering it was placed among the top known genes in differential expression due to its 3.5-fold increase (P < 0.001) in gene expression in PND4 granulosa cells compared to PND1, and its protein localization to granulosa cells (Figure 5C). Additionally, FRZB was of interest due to its known interaction with 35% of the molecules within the IPA-generated network (Figure 4), and its canonical role as a Wnt-pathway inhibitor.
The expression of FRZB was investigated in granulosa cells and to validate its role in the Wnt pathway via WNT3A, a known suppressor of primordial follicle activation [33] was also investigated. WNT3A protein localization has not previously been reported in neonatal ovaries, only its gene expression [34]. Immunocytochemistry was performed on isolated PND4 granulosa cells to determine if there is an association between FRZB and WNT3A. As in the whole ovary, FRZB was detected in both the nuclear and cytoplasmic regions of the isolated PND4 granulosa cells. FRZB co-localized with WNT3A in the granulosa cytoplasm (Figure 6A), the expression of WNT3A was intense and punctate adjacent to the nuclear region of the cell. Duolink proximity ligation assay demonstrated that FRZB and WNT3A are expressed in close proximity and likely interacting in this peri-nuclear region (Figure 6B). Taken together, findings establish Wnt pathway inhibitor Frzb was observed to display differential expression in granulosa cells from primordial and activating follicles. Additionally, FRZB protein is localized to activating granulosa cells and interacts with WNT3A in these cells.
Figure 6.
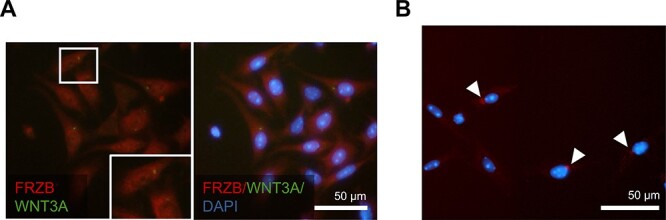
FRZB interacts with WNT3A in granulosa cells. (A) Immunocytochemistry of granulosa cells isolated from a post-natal day (PND) 4 mouse ovary. Frzb (red) co-localized with Wnt pathway activator WNT3A. Insets show zoomed image of boxed area. (B) Duolink proximity ligation assay of granulosa cells isolated from PND4 mouse ovary, red dot indicates proteins are interacting; representative images are indicative of n = 3 biological replicates performed in triplicate. Images taken at 40× magnification. Scale bar represents 50 μm.
Discussion
Primordial follicle activation involves the layering of many signaling networks, and a complete picture of this process is essential to our understanding of female fertility [10]. In granulosa cells, these signaling pathways initiate and control the complex remodeling of cells from a quiescent, flattened shape toward cuboidal structures, and involves the dispatching and receiving of signals and small molecules from the oocyte [6, 7]. Studies of global gene expression profiles increase our understanding of primordial follicle activation by identifying departures from, and extensions to known signaling pathways when they are combined with downstream computational or functional analyses. This project utilized the initial wave of primordial follicle activation in the neonatal mouse ovary to successfully study the granulosa cells during the primordial to primary transition. This study is the first to describe the transcriptome of populations of mouse granulosa cells representing primordial and activating follicles, and we have identified a number of genes differentially expressed in these cells, which reflect novel interactions potentially contributing to primordial follicle activation and early follicle development in the mouse. Our data highlight the transcription factor, ZFX, as important for ovary development and primordial follicle activation, and introduces FRZB as a new potential regulator in primordial follicle activation.
Overall, the gene expression changes observed were subtle, which may be a consequence of masking from the proportion of granulosa cells from primordial follicles in the activating (PND4) sample. However, our results resemble findings from other transcriptome studies of primordial and primary granulosa cells in humans, mice primates, pigs and sheep, which also detect ≤1000 genes when significance thresholds are introduced [35–40]. The use of proteomic analyses to correlate with transcriptomic data would complement this research, as rodent studies on the proteome of early ovary development [41, 42] have yet to substantially contribute to the understanding of primordial follicle activation. One limitation of the dataset was the false discovery rate above commonly accepted value of 0.1 [43]. False discovery rate predicts the type I error among features deemed significant, so the risk of rejecting a true null is possible but is mitigated by downstream multiple testing correction and biological validation of genes. Thus, in the case of the current dataset, we interpret our data with caution and validated changes biologically, with RT-qPCR experiments mirroring expression changes of the RNA-seq. The advantage of this study over previous studies characterizing gene expression characterization in young stage follicles is the targeted focus on the granulosa cell, and the use of relatively large populations of cells to more accurately reflect the heterogeneity within these cells. Where granulosa cell transcriptome studies have been performed on human samples, they have been small in sample size and/or utlilize microarray assays [44–47], both of which have limitations in identifying unique genes and accounting for sample heterogeneity. Furthermore, this study presents a granulosa cell-focused investigation of primordial follicle activation in mice. Previous mouse model studies using bulk RNA-seq to capture primordial follicle activation or early ovarian time points analyze a compendium of cell types and do not focus specifically on the granulosa cell to granulosa cell transition [48, 49]. Single cell sequencing has, so far, been underutilized in the study of primordial follicle activation and lacking depth in the analysis of granulosa cells during this important physiological event [39].
In the PND4 mouse ovary, POD1 protein was expressed in the nuclei of granulosa cells, consistent with previous reports of POD1 expression increasing postnatally in the mouse [50]. This study provides further evidence, demonstrating that the expression of Tcf21 also increases by a factor of 3.7 in granulosa cells from PND4 ovaries. This group also identified POD1 as a negative regulator of steroidogenic factor-1 (SF-1), an important regulator of granulosa cell regulated ovary development [51], suggesting a possible role in the differentiation of granulosa cells. Further studies focusing on whether there is a link between POD1-induced repression of SF-1 and activation of primordial follicles would be valuable, as the SF-1 gene Nr5a1 was also detected in the granulosa cells of our RNAseq experiment (for entire dataset, see GEO accession No. GSE162927).
ZFX is a transcription factor previously speculated to be involved in POI as it occurs in the POI-critical region of the X chromosome [52], and early research in mice showed a reduced ovarian reserve when Zfx was deleted, but the mechanism by which this occurs is not known [53]. The extracellular expression of ZFX in the ovaries was notable considering that in other somatic cells, ZFX is associated with promoting cell proliferation through cell cycle control [54, 55], but was not in the nucleus of activating granulosa cells, which are known to proliferate. Our results demonstrate that Zfx expression is upregulated in granulosa cells from PND4 mice during primordial follicle activation. Our protein localization studies indicate that ZFX is predominately expressed in oocyte and extracellular space suggesting the subcellular reorganization of the transcription factor. The bidirectional communication between oocytes and granulosa cells is a critical feature of folliculogenesis and follicular survival [7, 56]. These findings provide a renewed advocacy for investigating the role of ZFX to gain insight into primordial follicle activation and POI in the context of ZFX and oocyte-granulosa cell–cell communications.
An important finding in this study was the 3.5-fold increase in mRNA expression of Frzb in PND4 granulosa cells indicative of activating granulosa cells. Importantly, FRZB directly and indirectly has association with or influences cellular function of molecules known to be involved in primordial follicle activation including Wnt ligands like WNT3A, in addition to ERK1/2, and the TGFb superfamily [5, 33, 57], providing further evidence to investigate its role in primordial follicle activation. This study also provides compelling evidence for an interaction between FRZB and WTN3A in granulosa cells. This is consistent with previous findings identifying two sites on the FRZB protein on which WNT3A may bind [58]. WNT3a has been implicated in the maintenance of primordial follicle quiescence through FOXO3 [33]. When Wntless (gene required for secretion of all WNT ligands) was deleted in granulosa cells with an Sf1-cre mouse line, the transition from squamous to cuboidal granulosa cell morphology, a hallmark of primordial follicle activation, was impaired and coupled with the lack of translocation of FOXO3 [59]. This study provides novel evidence linking FRZB to the potential Wnt signaling cascade involved in primordial follicle activation through its ability to bind to WNT3A in the granulosa cells. A recent human granulosa cell transcriptome study observed a downregulation of Wnt family signaling in granulosa cells from primordial follicles compared to primary follicle granulosa cells [38], indicating that WNT3A, and by extension FRZB, may have a conserved role in the human ovary. Taken together with our findings, this emphasizes the need for further studies to determine the role of FRZB in primordial follicle activation, including a detailed investigation into a potential FRZB-WNT3A-FOXO3a relationship.
One limitation of the method of granulosa cell isolation to model processes occurring during primordial follicle activation is evident in the functions related to development (multicellular organism development), and genes observed in PND1 samples (such as Dazl). This is likely to be due to the remnants of naive pre-granulosa cells adjacent to germ cell nests, the structural precursors of primordial follicles. Germ cell nests begin to break down in the hours post-birth in mice, and contain pre-granulosa cells surrounding clusters of multiple germ cells, eventually giving rise to the single-oocyte encapsulated primordial follicle [31], thus ovaries from a PND1 (~24 h post birth) mice may contain residual germ cell nests.
This study has utilized a method for the isolation of mouse granulosa cells for the study of primordial follicle activation by taking advantage of the initial wave of primordial follicle activation in neonatal ovaries. The transcriptome of primordial and activating granulosa cells presented in this study has identified a range of factors previously associated with primordial follicle activation, which signify that future research could be well-spent redoubling efforts into extricating the roles of these factors, such as SF-1, ZFX, and WNT3A, and their up- and down-stream effectors. Co-immunoprecipitation assays for example, would be valuable to validate the binding between WNT3A and FRZB, in addition to rigorous assessment of the proteins via quantifying localization or via proteomics and phosphoproteomics. The findings in this study are enhanced by the validation of transcriptome data via RT-qPCR, confirming the dramatic changes in gene expression of granulosa cells during primordial follicle activation. Additionally, novel evidence is presented for Wnt antagonist Frzb as a potential regulator among the genetic network controlling primordial follicle activation through its protein interaction with murine primordial follicle suppressor WNT3A in granulosa cells. Understanding the factors, which dictate the maintenance of primordial follicle quiescence or their activation into primary follicles will inform our knowledge of early follicle development and may lead to future diagnostic methods and treatment regimens for women with infertility and premature ovarian insufficiency.
Author contributions
EAF collected the samples, performed all laboratory experiments (excluding RNA-seq, library prep and mapping), and in silico data analysis. EAF contributed to study design, wrote the manuscript, analyzed the data and created the figures. ERF contributed to sample preparation and collection, and prepared samples for immunolocalization experiments. SDR, EAM, and ELB contributed to interpretation of the data, the preparation, and critical feedback of the manuscript. EAM funded the RNA-seq and bioinformatics. JMS conceived and designed the study. JMS funded the remainder of the study, contributed to data analysis and manuscript preparation and had the final decision on the manuscript.
Supplementary Material
Acknowledgements
The authors would like to acknowledge V. Fan, N. Poonawala-Lohani, and L. Williams from Auckland Genomics, University of Auckland for their assistance in preparing transcriptomic data for analysis. Thank you to David Skerrett-Byrne for assistance with in silico analysis features.
Footnotes
† Grant Support: This project has been funded by the Australian National Health and Medical Research Council (G1600095) and the Hunter Medical Research Institute Bob and Terry Kennedy Children’s Research Project Grant in Pregnancy & Reproduction (G1501433 and G1801335).
Contributor Information
Emmalee A Ford, Priority Research Centre for Reproductive Science, Schools of Biomedical Science & Pharmacy and Environmental & Life Sciences, University of Newcastle, Callaghan, New South Wales, Australia; Hunter Medical Research Institute, New Lambton Heights, New South Wales, Australia.
Emily R Frost, Priority Research Centre for Reproductive Science, Schools of Biomedical Science & Pharmacy and Environmental & Life Sciences, University of Newcastle, Callaghan, New South Wales, Australia; Hunter Medical Research Institute, New Lambton Heights, New South Wales, Australia; Stem Cell Biology and Developmental Genetics Laboratory, The Francis Crick Institute, London, UK.
Emma L Beckett, Hunter Medical Research Institute, New Lambton Heights, New South Wales, Australia; School of Environmental & Life Sciences, Faculty of Science, University of Newcastle, Callaghan, New South Wales, Australia.
Shaun D Roman, Priority Research Centre for Reproductive Science, Schools of Biomedical Science & Pharmacy and Environmental & Life Sciences, University of Newcastle, Callaghan, New South Wales, Australia; Hunter Medical Research Institute, New Lambton Heights, New South Wales, Australia; Priority Research Centre for Drug Development, University of Newcastle, Callaghan, New South Wales, Australia.
Eileen A McLaughlin, Priority Research Centre for Reproductive Science, Schools of Biomedical Science & Pharmacy and Environmental & Life Sciences, University of Newcastle, Callaghan, New South Wales, Australia; School of Science, Western Sydney University, Penrith, Australia; School of Biological Sciences, Faculty of Science, University of Auckland, Auckland, New Zealand.
Jessie M Sutherland, Priority Research Centre for Reproductive Science, Schools of Biomedical Science & Pharmacy and Environmental & Life Sciences, University of Newcastle, Callaghan, New South Wales, Australia; Hunter Medical Research Institute, New Lambton Heights, New South Wales, Australia.
Conflict of interest
The authors declare that there is no conflict of interest.
References
- 1. Shelling AN. Premature ovarian failure. Reproduction 2010; 140:633–641. [DOI] [PubMed] [Google Scholar]
- 2. Golezar S, Ramezani Tehrani F, Khazaei S, Ebadi A, Keshavarz Z. The global prevalence of primary ovarian insufficiency and early menopause: a meta-analysis. Climacteric 2019; 22:403–411. [DOI] [PubMed] [Google Scholar]
- 3. Nelson LM. Primary ovarian insufficiency. N Engl J Med 2009; 360:606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haller-Kikkatalo K, Uibo R, Kurg A, Salumets A. The prevalence and phenotypic characteristics of spontaneous premature ovarian failure: a general population registry-based study. Hum Reprod 2015; 30:1229–1238. [DOI] [PubMed] [Google Scholar]
- 5. Adhikari D, Liu K. Molecular mechanisms underlying the activation of mammalian primordial follicles. Endocr Rev 2009; 30:438–464. [DOI] [PubMed] [Google Scholar]
- 6. Zhang H, Liu K. Cellular and molecular regulation of the activation of mammalian primordial follicles: somatic cells initiate follicle activation in adulthood. Hum Reprod Update 2015; 21:779–786. [DOI] [PubMed] [Google Scholar]
- 7. Eppig JJ. Reproduction: oocytes call, granulosa cells connect. Curr Biol 2018; 28:R354–r356. [DOI] [PubMed] [Google Scholar]
- 8. Gougeon A, Chainy GB. Morphometric studies of small follicles in ovaries of women at different ages. J Reprod Fertil 1987; 81:433–442. [DOI] [PubMed] [Google Scholar]
- 9. Matsuda F, Inoue N, Manabe N, Ohkura S. Follicular growth and atresia in mammalian ovaries: regulation by survival and death of granulosa cells. J Reprod Dev 2012; 58:44–50. [DOI] [PubMed] [Google Scholar]
- 10. Ford E, Beckett EL, Roman S, McLaughlin EA, Sutherland J. Advances in human primordial follicle activation and premature ovarian insufficiency. Reproduction 2020; 159:R15–R29. [DOI] [PubMed] [Google Scholar]
- 11. Chen Y, Yang W, Shi X, Zhang C, Song G, Huang D. The factors and pathways regulating the activation of mammalian primordial follicles in vivo. Front Cell Dev Biol 2020; 8:1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reddy P, Zheng W, Liu K. Mechanisms maintaining the dormancy and survival of mammalian primordial follicles. Trends Endocrinol Metab 2010; 21:96–103. [DOI] [PubMed] [Google Scholar]
- 13. Kerr JB, Duckett R, Myers M, Britt KL, Mladenovska T, Findlay JK. Quantification of healthy follicles in the neonatal and adult mouse ovary: evidence for maintenance of primordial follicle supply. Reproduction 2006; 132:95–109. [DOI] [PubMed] [Google Scholar]
- 14. Kerr JB, Myers M, Anderson RA. The dynamics of the primordial follicle reserve. Reproduction 2013; 146:R205. [DOI] [PubMed] [Google Scholar]
- 15. McGee EA, Hsueh AJ. Initial and cyclic recruitment of ovarian follicles. Endocr Rev 2000; 21:200–214. [DOI] [PubMed] [Google Scholar]
- 16. Edson MA, Nagaraja AK, Matzuk MM. The mammalian ovary from genesis to revelation. Endocr Rev 2009; 30:624–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frost ER, Ford EA, Taylor G, Boeing S, Beckett EL, Roman SD, Lovell-Badge R, McLaughlin EA, Sutherland JM. Two alternative methods for the retrieval of somatic cell populations from the mouse ovary. Mol Hum Reprod 2021; 27:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C T method. Nat Protoc 2008; 3:1101. [DOI] [PubMed] [Google Scholar]
- 19. Andrews S. FastQC: a Quality Control Tool for High Throughput Sequence Data. Version 0.11. 2. Babraham Institute, Cambridge, UK: http://www.bioinformatics.babraham.ac.uk/projects/fastqc2014. [Google Scholar]
- 20. Lindgreen S. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res Notes 2012; 5:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015; 12:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 2016; 11:1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fu J, Frazee AC, Collado-Torres L, Jaffe AE, Leek JT. Ballgown: flexible, isoform-level differential expression analysis. R Package Version 2018; 2. https://rdrr.io/bioc/ballgown/. [Google Scholar]
- 25. Krämer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics (Oxford, England) 2014; 30:523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang Da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44. [DOI] [PubMed] [Google Scholar]
- 28. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DPet al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 2019; 47:D419–d426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius K-J, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson L-G. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 2006; 3:995–1000. [DOI] [PubMed] [Google Scholar]
- 31. Pepling ME. From primordial germ cell to primordial follicle: mammalian female germ cell development. Genesis 2006; 44:622–632. [DOI] [PubMed] [Google Scholar]
- 32. Sutherland JM, Frost ER, Ford EA, Peters AE, Regan NL, Seldon AN, Mihalas BP, Russell DL, Dunning KR, McLaughlin EA. Janus kinase JAK1 maintains the ovarian reserve of primordial follicles in the mouse ovary. Mol Hum Reprod 2018; 24:533–542. [DOI] [PubMed] [Google Scholar]
- 33. Li L, Ji SY, Yang JL, Li XX, Zhang J, Zhang Y, Hu ZY, Liu YX. Wnt/beta-catenin signaling regulates follicular development by modulating the expression of Foxo3a signaling components. Mol Cell Endocrinol 2014; 382:915–925. [DOI] [PubMed] [Google Scholar]
- 34. Harwood BN, Cross SK, Radford EE, Haac BE, De Vries WN. Members of the WNT signaling pathways are widely expressed in mouse ovaries, oocytes, and cleavage stage embryos. Dev Dyn 2008; 237:1099–1111. [DOI] [PubMed] [Google Scholar]
- 35. Arraztoa JA, Zhou J, Marcu D, Cheng C, Bonner R, Chen M, Xiang C, Brownstein M, Maisey K, Imarai M, Bondy C. Identification of genes expressed in primate primordial oocytes. Hum Reprod 2005; 20:476–483. [DOI] [PubMed] [Google Scholar]
- 36. Bonnet A, Bevilacqua C, Benne F, Bodin L, Cotinot C, Liaubet L, Sancristobal M, Sarry J, Terenina E, Martin P, Tosser-Klopp G, Mandon-Pepin B. Transcriptome profiling of sheep granulosa cells and oocytes during early follicular development obtained by laser capture microdissection. BMC Genomics 2011; 12:417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bonnet A, Le Cao KA, Sancristobal M, Benne F, Robert-Granie C, Law-So G, Fabre S, Besse P, De Billy E, Quesnel H. In vivo gene expression in granulosa cells during pig terminal follicular development. Reproduction 2008; 136. [DOI] [PubMed] [Google Scholar]
- 38. Ernst EH, Franks S, Hardy K, Villesen P, Lykke-Hartmann K. Granulosa cells from human primordial and primary follicles show differential global gene expression profiles. Hum Reprod 2018; 33:666–679. [DOI] [PubMed] [Google Scholar]
- 39. Niu W, Spradling AC. Two distinct pathways of pregranulosa cell differentiation support follicle formation in the mouse ovary. Proc Natl Acad Sci 2020; 117:20015–20026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang J-J, Ge W, Zhai Q-Y, Liu J-C, Sun X-W, Liu W-X, Li L, Lei C-Z, Dyce PW, De Felici M. Single-cell transcriptome landscape of ovarian cells during primordial follicle assembly in mice. PLoS Biol 2020; 18:e3001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang N, Zhang P, Guo X, Xie J, Huo R, Wang F, Chen L, Shen J, Zhou Z, Shi Q. Comparative proteome profile of immature rat ovary during primordial follicle assembly and development. Proteomics 2009; 9:3425–3434. [DOI] [PubMed] [Google Scholar]
- 42. Xu M, Che L, Yang Z, Zhang P, Shi J, Li J, Lin Y, Fang Z, Che L, Feng B. Proteomic analysis of fetal ovaries reveals that primordial follicle formation and transition are differentially regulated. Biomed Res Int 2017; 2017:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Korthauer K, Kimes PK, Duvallet C, Reyes A, Subramanian A, Teng M, Shukla C, Alm EJ, Hicks SC. A practical guide to methods controlling false discoveries in computational biology. Genome Biol 2019; 20:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hasegawa A, Kumamoto K, Mochida N, Komori S, Koyama K. Gene expression profile during ovarian folliculogenesis. J Reprod Immunol 2009; 83:40–44. [DOI] [PubMed] [Google Scholar]
- 45. Kezele PR, Ague JM, Nilsson E, Skinner MK. Alterations in the ovarian transcriptome during primordial follicle assembly and development. Biol Reprod 2005; 72:241–255. [DOI] [PubMed] [Google Scholar]
- 46. Kristensen SG, Ebbesen P, Andersen CY. Transcriptional profiling of five isolated size-matched stages of human preantral follicles. Mol Cell Endocrinol 2015; 401:189–201. [DOI] [PubMed] [Google Scholar]
- 47. Yoon SJ, Kim KH, Chung HM, Choi DH, Lee WS, Cha KY, Lee KA. Gene expression profiling of early follicular development in primordial, primary, and secondary follicles. Fertil Steril 2006; 85:193–203. [DOI] [PubMed] [Google Scholar]
- 48. Bian X, Xie Q, Zhou Y, Wu H, Cui J, Jia L, Suo L. Transcriptional changes of mouse ovary during follicle initial or cyclic recruitment mediated by extra hormone treatment. Life Sci 2020; 264:118654. [DOI] [PubMed] [Google Scholar]
- 49. Terren C, Munaut C. Molecular basis associated with the control of primordial follicle activation during transplantation of cryopreserved ovarian tissue. Reprod Sci 2020; 28:1257–1266. [DOI] [PubMed] [Google Scholar]
- 50. Tamura M, Kanno Y, Chuma S, Saito T, Nakatsuji N. Pod-1/Capsulin shows a sex-and stage-dependent expression pattern in the mouse gonad development and represses expression of Ad4BP/SF-1. Mech Dev 2001; 102:135–144. [DOI] [PubMed] [Google Scholar]
- 51. Pelusi C, Ikeda Y, Zubair M, Parker KL. Impaired follicle development and infertility in female mice lacking steroidogenic factor 1 in ovarian granulosa cells. Biol Reprod 2008; 79:1074–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Persani L, Rossetti R, Cacciatore C, Bonomi M. Primary ovarian insufficiency: X chromosome defects and autoimmunity. J Autoimmun 2009; 33:35–41. [DOI] [PubMed] [Google Scholar]
- 53. Luoh S-W, Bain PA, Polakiewicz RD, Goodheart ML, Gardner H, Jaenisch R, Page DC. Zfx mutation results in small animal size and reduced germ cell number in male and female mice. Development 1997; 124:2275–2284. [DOI] [PubMed] [Google Scholar]
- 54. Wu S, Lao X-Y, Sun T-T, Ren L-L, Kong X, Wang J-L, Wang Y-C, Du W, Yu Y-N, Weng Y-R. Knockdown of ZFX inhibits gastric cancer cell growth in vitro and in vivo via downregulating the ERK-MAPK pathway. Cancer Lett 2013; 337:293–300. [DOI] [PubMed] [Google Scholar]
- 55. Harel S, Tu EY, Weisberg S, Esquilin M, Chambers SM, Liu B, Carson CT, Studer L, Reizis B, Tomishima MJ. ZFX controls the self-renewal of human embryonic stem cells. PLoS One 2012; 7:e42302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. El-Hayek S, Yang Q, Abbassi L, FitzHarris G, Clarke HJ. Mammalian oocytes locally remodel follicular architecture to provide the Foundation for Germline-Soma Communication. Curr Biol 2018; 28:1124–1131.e1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ding X, Zhang X, Mu Y, Li Y, Hao J. Effects of BMP4/SMAD signaling pathway on mouse primordial follicle growth and survival via up-regulation of Sohlh2 and c-kit. Mol Reprod Dev 2013; 80:70–78. [DOI] [PubMed] [Google Scholar]
- 58. Bovolenta P, Esteve P, Ruiz JM, Cisneros E, Lopez-Rios J. Beyond Wnt inhibition: new functions of secreted frizzled-related proteins in development and disease. J Cell Sci 2008; 121:737–746. [DOI] [PubMed] [Google Scholar]
- 59. Habara O, Logan CY, Kanai-Azuma M, Nusse R, Takase HM. WNT signaling in pre-granulosa cells is required for ovarian folliculogenesis and female fertility. Development 2021; 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


