ABSTRACT
Treatment response to clopidogrel is associated with CYP2C19 activity through the formation of the active H4 metabolite. The aims of this study were to develop a physiologically based pharmacokinetic (PBPK) model of clopidogrel and its metabolites for populations of European ancestry, to predict the pharmacokinetics in the Japanese population by CYP2C19 phenotype, and to investigate the effect of clinical and demographic factors. A PBPK model was developed and verified to describe the two metabolic pathways of clopidogrel (H4 metabolite, acyl glucuronide metabolite) for a population of European ancestry using plasma data from published studies. Subsequently, model predictions in the Japanese population were evaluated. The effects of CYP2C19 activity, fluvoxamine coadministration (CYP2C19 inhibitor), and population‐specific factors (age, sex, BMI, body weight, cancer, hepatic, and renal dysfunction) on the pharmacokinetics of clopidogrel and its metabolites were then characterized. The predicted/observed ratios for clopidogrel and metabolite exposure parameters were acceptable (twofold acceptance criteria). For all CYP2C19 phenotypes, steady‐state AUC0‐τ of the H4 metabolite was lower for the Japanese (e.g., EM, 7.69 [6.26–9.45] ng·h/ml; geometric mean [95% CI]) than European (EM, 24.8 [20.4–30.1] ng·h/ml, p < .001) population. In addition to CYP2C19‐poor metabolizer phenotype, fluvoxamine coadministration, hepatic, and renal dysfunction were found to reduce H4 metabolite but not acyl glucuronide metabolite concentrations. This is the first PBPK model describing the two major metabolic pathways of clopidogrel, which can be applied to populations of European and Japanese ancestry by CYP2C19 phenotype. The differences between the two populations appear to be determined primarily by the effect of varying CYP2C19 liver activity.
Keywords: clopidogrel, CYP2C19, inter‐ethnic differences, metabolites, PBPK modelling
The contribution of intrinsic and extrinsic factors to clopidogrel PK explored and clinical observations were confirmed. Lower H4 metabolite AUC was predicted in Japanese populations compared to European populations.

Abbreviations
- ACS
acute coronary syndrome
- AUC
area under the plasma concentration‐time curve
- CES1
carboxylesterase 1
- Cmax
peak plasma concentration
- EM
extensive metaboliser
- GFR
glomerular filtration rate
- IM
intermediate metaboliser
- IV
intravenous
- PBPK
physiologically‐based pharmacokinetic model
- PCI
percutaneous coronary intervention
- PM
poor metaboliser
- UM
ultrarapid metaboliser
1. INTRODUCTION
Ethnic diversity in drug exposure and response is an important consideration in global drug development. One of the key sources of intrinsic pharmacokinetic variability is genetic polymorphisms of drug metabolizing enzymes and differences in the prevalence of variant alleles associated with altered enzyme function in different populations. 1 Among the many drug metabolizing enzymes, inter‐ethnic differences in the frequency of variant CYP2C19 alleles of functional consequence are well recognized. 2
CYP2C19 is involved in the metabolism of a number of drugs used routinely in clinical practice including antidepressants (e.g., citalopram), and protein pump inhibitors (e.g., omeprazole). 3 , 4 CYP2C19*2 and CYP2C19*3 are the most common alleles with decreased activity, while CYP2C19*17 is associated with increased CYP2C19 activity. 5 The frequencies of the decreased activity CYP2C19 alleles are much lower in populations of European ancestry (14.7% and 0.2%, respectively) than East Asian populations (28.3% and 7.3%, respectively). By contrast, the increased activity allele (CYP2C19*17) is more common in European (21.6%) than East Asian (2.1%) populations. 6 , 7 Consequently, inter‐ethnic differences in the pharmacokinetics of CYP2C19 substrates have been observed. 4 , 8
Clopidogrel is an antiplatelet drug that is widely used to reduce the risk of thrombosis, myocardial infarction, and mortality in patients with acute coronary syndromes (ACS) and those undergoing percutaneous coronary intervention (PCI). 9 Clopidogrel is a prodrug that is rapidly absorbed, with an absorption of at least 50% based on metabolite data in the urine. 10 A large proportion of the absorbed drug is metabolized (about 85%) by carboxylesterase 1 (CES1) to form clopidogrel carboxylic acid, which is then glucuronidated via UGT2B7 to the inactive acyl glucuronide, a potent CYP2C8 inhibitor. 11 The remaining proportion of the drug (15%) is metabolized by CYP2C19, CYP2B6, and CYP1A2 to form 2‐oxo‐clopidogrel, which is further metabolized to the pharmacologically active thiol H4 metabolite (Figure 1). 12
FIGURE 1.
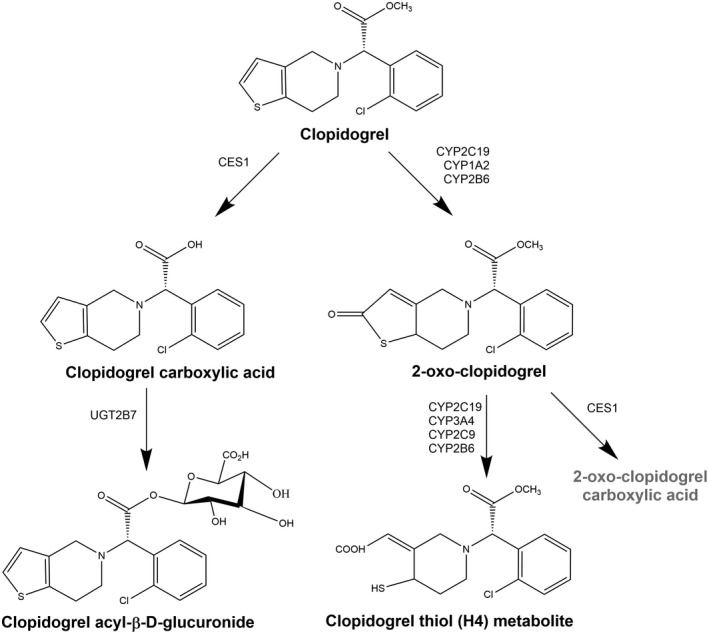
The metabolic pathways of clopidogrel
Treatment response to clopidogrel is strongly associated with CYP2C19 activity through the formation of the H4 metabolite. 9 ACS patients with CYP2C19 decreased activity alleles have significantly lower concentrations of the active H4 metabolite, diminished platelet inhibition, and a higher rate of adverse cardiovascular events indicating lesser efficacy. 8 , 13 In 2010, the US Food and Drug Administration (FDA) added a boxed warning to the Product Information for clopidogrel on reduced antiplatelet effects in individuals with two loss‐of‐function CYP2C19 alleles (*2, *3). 10 , 14 , 15 Since there is a higher prevalence of CYP2C19 decreased activity alleles in populations of East Asian and Pacific Islander ancestry, this places these populations at a higher risk for lack of efficacy and cardiovascular events at the currently recommended doses.
The use of newer antiplatelet drugs such as ticagrelor or prasugrel is recommended for subjects who carry the reduced function CYP2C19 alleles. 15 , 16 In Japan, however, the use of clopidogrel is still recommended for preventing reinfarction following PCI in patients with ACS, 17 even though about 75% of Japanese patients with ACS carry at least one CYP2C19 decreased activity allele. 18 At present, the same dose of oral clopidogrel (ACS, 300 mg loading dose followed by 75 mg once daily; recent myocardial infarction/stroke or established peripheral arterial disease, 75 mg once daily) is recommended in the United States, Europe, and markets in East Asia, including Japan. 19
Physiologically based pharmacokinetic (PBPK) modelling is a key component of model‐informed drug development (MIDD) and can be used to comprehensively investigate factors influencing drug exposure by considering the drug's physiochemical properties and human physiological variables (e.g., body weight, organ size, frequency of drug metabolizing enzymes). 20 PBPK models have been developed to describe the pharmacokinetics of clopidogrel and its metabolites in healthy populations of European ancestry. 21 , 22 , 23 However, it is unclear whether these PBPK models can predict the pharmacokinetics of clopidogrel and metabolites in the Japanese population. Therefore, the aims of this study were (i) to develop and verify a PBPK model including the two major metabolic pathways of clopidogrel in a population of European ancestry, (ii) to apply and evaluate the final PBPK model in a Japanese population, and (iii) to investigate the effect of CYP2C19 activity and various other intrinsic factors on the formation of active and inactive metabolites of clopidogrel.
2. METHODS
2.1. PBPK model development and evaluation
A minimal PBPK model for clopidogrel and its four metabolites (carboxylic acid metabolite, acyl glucuronide metabolite, 2‐oxo‐clopidogrel metabolite, and H4 metabolite) was developed using the Simcyp Simulator v18.2 (Simcyp Division, Certara UK Limited, Sheffield, UK). Figure 2 provides an overview of the model building strategy, which was developed in accordance with the recommended best practices for building, evaluating, and reporting PBPK models for regulatory submissions 24 , 25 , 26 (see Supplementary Methods).
FIGURE 2.
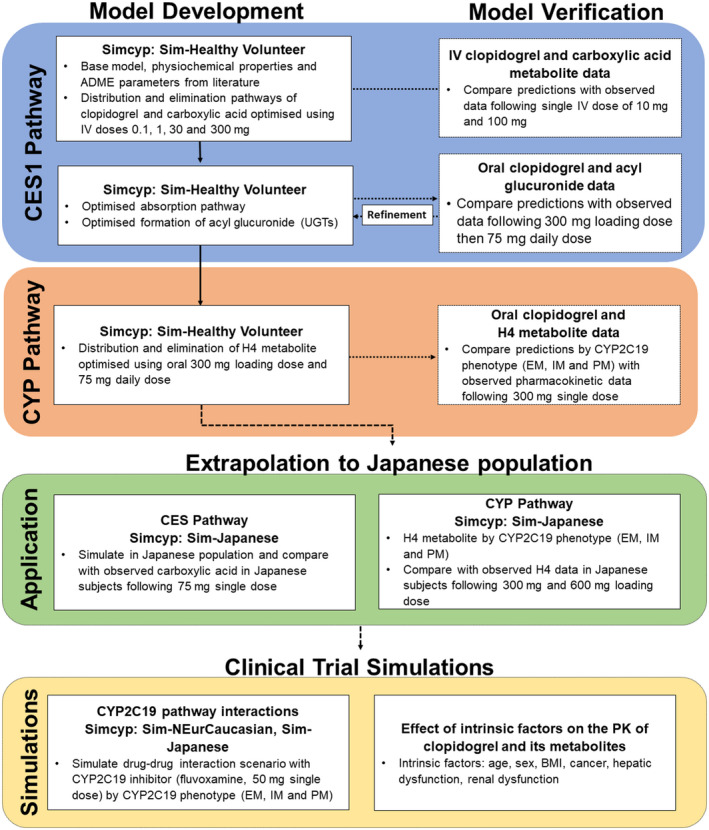
Workflow used for the PBPK model development and verification in populations of European ancestry, model application (extrapolation to a Japanese population) as well as simulations to investigate the effect of CYP2C19 activity, and other intrinsic factors on the pharmacokinetics of clopidogrel and its metabolites
PBPK model performance was evaluated by comparing simulated model‐derived pharmacokinetic estimates (tmax, time to reach peak plasma concentration; Cmax, peak plasma concentration; AUC, area under plasma concentration–time curve) with observed pharmacokinetic parameters. 23 , 27 , 28 , 29 , 30 The predicted/observed ratio of pharmacokinetic parameters was calculated. An acceptance criteria of 0.5‐ to 2.0‐fold was applied indicating that predictions were within a twofold range of the observed values. 24 The predictive performance of the model was also assessed by visual inspection of the overlaid predicted and observed pharmacokinetic profiles (geometric mean, 90% prediction interval).
2.1.1. Clinical data
Mean concentration–time profiles of clopidogrel, 23 , 28 the carboxylic acid metabolite, 19 , 28 the acyl glucuronide metabolite, 23 and the H4 metabolite 29 , 30 in subjects predominately of European ancestry were obtained from the literature to develop and verify the predictive performance of the PBPK model (Table 1) using WebPlotDigitizer v4.1 (Automeris LLC. San Francisco, California, USA). 31
TABLE 1.
Summary of the clinical data in healthy subjects from the literature used to build and verify the PBPK model of clopidogrel and its metabolites (the carboxylic acid metabolite, acyl glucuronide metabolite, and the H4 metabolite)
| Population | n | Dose and dosage form | Concentration‐time data | CYP2C19 phenotype | Data use | Reference |
|---|---|---|---|---|---|---|
|
96% Caucasian 4% Afro‐Caribbean |
18 | Oral: 300 mg loading then 75 mg once daily | H4 metabolite | Not reported | Model development | Farid et al. 29 |
|
94% Caucasian, 6% African |
96 |
IV bolus: 0.1, 1, 30 mg IV infusion: 300 mg a |
Clopidogrel, carboxylic acid metabolite | Not reported | Model development | Cushing et al. 28 |
| 48 |
IV bolus: 10 mg IV infusion: 100 mg b |
Clopidogrel, carboxylic acid metabolite | Not reported | Model verification | ||
| 100% Caucasian | 9 | Oral: 300 mg loading then 75 mg once daily | Clopidogrel, acyl glucuronide metabolite | Not reported | Model development and verification | Tornio et al. 23 |
|
90% Caucasian, 7% African, 2% Asian, 1% mixed |
74 |
Oral: 300 mg (single dose) |
H4 metabolite summary c (Cmax, AUC0‐24) |
EM (*1/*1) n = 56 IM (*1/*2) n = 17 PM (*2/*2) n = 1 |
Model verification | Brandt et al. 27 |
| 100% Japanese | 12 |
Oral: 75 mg (single dose) |
Carboxylic acid metabolites | Not reported | Model application | Clopidogrel (Plavix) Product Information 19 |
| 100% Japanese | 27 |
Oral: 300 mg loading then 75 mg once daily Oral: 600 mg loading then 150 mg once daily |
H4 metabolite |
EM (*1/*1) n = 9 IM (*1/*2, *1/*3) n = 9 PM (*2/*2, *2/*3, *3/*3) n = 9 |
Model application | Kobayashi et al. 30 |
Abbreviations: EM, extensive metabolizer; IM, intermediate metabolizer; PM, poor metabolizer.
Duration of infusion was 8 min.
Duration of infusion was 4 min.
Pharmacokinetic summaries for the H4 metabolite were used to compare observations by CYP2C19 phenotype.
The pharmacokinetics of the H4 metabolite by CYP2C19 phenotype for the extensive metabolizers (EMs; CYP2C19*1/*1), intermediate metabolizers (IMs; CYP2C19*1/*2, CYP2C19*1/*3), and poor metabolizers (PMs; CYP2C19*2/*2, CYP2C19*2/*3, CYP2C19*3/*3) were evaluated in healthy subjects of European 27 and Japanese ancestry. 30 Simulations of the pharmacokinetics of clopidogrel and its metabolites for CYP2C19 UMs (UMs;CYP2C19*1/*17, CYP2C19*17/*17) were also conducted, but results could not be compared against clinical data, as only data from subjects of European ancestry were available. 8
2.1.2. The CES1 metabolic pathway
The CES1‐mediated metabolic pathway was built using clopidogrel and carboxylic acid metabolite concentration–time data following an intravenous (IV) dose of clopidogrel. 28 The data obtained from Cushing et al. 28 were split into a training dataset (IV clopidogrel doses; 0.1, 1, 30, and 300 mg) to develop the model and a verification dataset (IV clopidogrel doses; 10 mg and 100 mg) to evaluate the performance of the model. The initial estimate of CES1‐mediated clearance was based on a previous study, 32 and optimized using IV data. 28 Additional elimination pathways of clopidogrel (P‐glycoprotein) and carboxylic acid (UGTs) were investigated (see Supplementary Methods, Table S1).
2.1.3. The CYP450 pathways
CYP‐mediated metabolism of clopidogrel was based on a previous PBPK model. 21 As the 2‐oxo‐clopidogrel metabolite is also metabolized by CES1 (fm 50%), kinetic information was obtained from Zhu et al. 32 In addition, a sensitivity analysis was performed to confirm 50% metabolism via the CES1 pathway in the population of European ancestry.
In Simcyp (v18.2), the mean CYP2C19 liver abundances for EM and PM individuals in the healthy European population (Sim‐Healthy Volunteer) are 14 pmol/mg protein and 0 pmol/mg protein, respectively. The mean CYP2C19 liver abundance for UM and IM individuals was set to 18 pmol/mg protein and 10 pmol/mg protein, as reported previously 21 (see Table S2).
H4 metabolite pharmacokinetic data 29 were used to optimize the H4 metabolite pharmacokinetic parameters. Results reported by Brandt et al. 27 were used to verify the model predictions in subjects of European ancestry by CYP2C19 phenotype (EM, IM, and PM). Only H4 metabolite data were available to develop this pathway (Table S4).
2.2. Model application: Extrapolation to a Japanese population
The PBPK model was applied to a virtual Japanese population following a single oral dose of either 75 mg 19 or 300/600 mg clopidogrel to assess model performance for the prediction of carboxylic acid or the H4 metabolite, 30 respectively.
The mean CYP2C19 liver abundance for EMs in the Sim‐Japanese population was 4.1 pmol/mg protein (Simcyp v18.2). An approach used by Suzuki et al., 33 Zhou et al., 34 and Higashimori et al. 35 to fit CYP2C19 substrates to Japanese data was to assume the same CYP2C19 liver abundance values for each CYP2C19 phenotype as the European population. This approach was used for the present analysis to improve the pharmacokinetic predictions of the H4 metabolite in the Japanese population (see Table S2). The simulated clopidogrel and metabolite concentrations were compared with observed concentrations in Japanese subjects as published by Kobayashi et al. 30
2.3. Simulations: Co‐administration of clopidogrel and fluvoxamine in populations of European and Japanese ancestry
To investigate the contribution of CYP2C19 to the formation of the metabolites of clopidogrel, a drug–drug interaction (DDI) scenario was implemented with fluvoxamine. 36 Fluvoxamine (Simcyp: SV‐Fluvoxamine) is a strong inhibitor of both CYP2C19 and CYP1A2 and a moderate inhibitor of CYP3A and CYP2D6. The effect of coadministration of fluvoxamine (50 mg daily) with clopidogrel (300 mg loading dose followed by 75 mg daily) on the AUC for the dosing interval on Day 3 (AUC0‐τ) was investigated by CYP2C19 phenotype (EM, IM, and PM) in the general European (Simcyp: Sim‐NEurCaucasian) and Japanese (Simcyp: Sim‐Japanese) populations. Simulations were also performed for CYP2C19 UMs to compare the relative effect on the pharmacokinetics of clopidogrel and its metabolites.
2.4. Simulation: Effect of intrinsic factors on the pharmacokinetics of clopidogrel, acyl glucuronide, and H4 metabolite
The effect of various intrinsic factors on the pharmacokinetics of clopidogrel and its metabolites was investigated using the Sim‐NEurCaucasian population as the comparator group (see Table S5). The population characteristics were 50% female, aged 20–50 years, BMI 18–29 kg/m2, renal function of eGFR > 90 ml/min, normal hepatic function, and CYP2C19 PM frequency of 2.4%. The virtual population libraries in Simcyp (healthy geriatric population, Sim‐Geriatric NEC; obese population, Sim‐Obese; morbidly obese population, Sim‐Morbidly Obese; cancer population, Sim‐Cancer; cirrhosis populations based on Child–Pugh scores for mild [Sim‐CirrhosisCP‐A], moderate [Sim‐CirrhosisCP‐B], and severe [Sim‐CirrhosisCP‐C] disease; renal impairment populations for moderate [Sim‐RenalGFR_30‐60] and severe [Sim‐RenalGFR_less30] disease) were used to determine the steady‐state AUC0‐τ (Day 3) of clopidogrel, the acyl glucuronide metabolite and the H4 metabolite in each population group following the standard dosage regimen (300 mg loading dose, 75 mg daily). The virtual population used to represent mild renal impairment (Stage 2 renal function) was developed using the Sim‐NEurCaucasian population and modifying higher mean creatinine values (male, 114 µmol/L; female, 105 µmol/L to obtain a glomerular filtration rate (GFR) of 60–90 ml/min/1.73 m2). The statistical analyses used to evaluate the effect of intrinsic factors are shown in the Supplementary methods.
2.5. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY, 37 and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/20. 38
3. RESULTS
3.1. CES1 metabolic pathway
The final PBPK model parameter values are shown in Table 2 and a comparison of the present PBPK model with published PBPK models of clopidogrel is shown in Table S3. The mean fraction of clopidogrel metabolized via each pathway (fm%) was 82% for CES1, 3% for CYP1A2, 2% for CYP2B6, 2% for CYP2C19, and 11% for the additional pathway(s). Following a single IV clopidogrel dose of 300 mg, the mean systemic plasma clearance of clopidogrel was 84 L/h, which comprises a hepatic clearance of 61 L/h, metabolic renal clearance (renal CES1) of 13 L/h, and an additional clearance via other routes of 10 L/h.
TABLE 2.
Physiochemical and in vitro ADME parameters used for clopidogrel and its metabolites (carboxylic acid metabolite, acyl glucuronide metabolite, 2‐oxo‐clopidogrel metabolite, H4 metabolite) in the PBPK model
| Parameters | Clopidogrel | Carboxylic acid | Acyl glucuronide | 2‐oxo‐clopidogrel | H4 metabolite | |
|---|---|---|---|---|---|---|
| Physiochemical properties | MW (g/mol) | 321.8 22 , 23 | 307.8 34 | 483.9 22 , 23 | 337.8 21 | 355.8 21 |
| Log P | 2.58 22 , 23 | 3.54 34 | 1.35 22 , 23 | 2.96 21 | 3.60 21 | |
| Compound type | Monoprotic base 22 , 23 | Monoprotic acid 34 | Ampholyte 22 , 23 | Monoprotic acid 21 | Diprotic acid 21 | |
| pKa/pKb | 4.6 21 | 1.81 34 | 2.65, 4.23 22 , 23 | 3.41 21 | 3.2, 5.1 21 | |
| Blood/plasma ratio | 0.72 21 | 1 c | 1 22 , 23 | 1 21 | 0.82 21 | |
| fup | 0.02 21 | 0.1 d | 0.1 22 , 23 | 0.031 21 | 0.018 21 | |
| Absorption | Absorption model | First order | — | — | — | — |
| fa | 0.49 a | — | — | — | — | |
| ka (1/h) | 1.0 a | — | — | — | — | |
| Peff,man (10−4 cm/s) | 7.26 b | — | — | — | — | |
| fugut | 0.02 21 | — | — | — | — | |
| Distribution | Distribution model | Minimal PBPK | Minimal PBPK | Minimal PBPK | Minimal PBPK | Minimal PBPK |
| Vss (L/kg) | 5 22 , 23 | 0.20 d | 0.25 22 , 23 | 0.1 21 | 0.14 g | |
|
SAC: CLin (L/h) CLout (L/h) Vsac (L/kg) |
3.98 a |
26.6 d 4.1 d 0.1 d |
2.0 e 0.00001 e 0.167 e |
— — — |
— — — |
|
| Elimination | Elimination type | Enzyme kinetics | Enzyme kinetics | CL 10 L/h (60% CV) 22 , 23 | Enzyme kinetics | CL 90 L/h g (60% CV) |
| Enzymes |
CES1, HLS9 CLint 50 000 a Tissue scalar (liver/kidney) 1.0/0.1 |
UGT2B7 11 Vmax 50.5, KM 20.9 fumic 0.8 |
CYP2B6 21 Vmax 2.48, Km 1.62 fumic 0.18 |
|||
|
CYP1A2 21 Vmax 2.27, Km 1.58, fumic 0.015 |
UGT2B17 11 Vmax 511, KM 181, fumic 0.8 |
CYP2C19 21 Vmax 9.06, Km 12.1, fumic 0.18 |
||||
|
CYP2B6 21 Vmax 7.66, Km 2.08, fumic 0.015 |
CYP2C9 21 Vmax 0.855, Km 18.1, fumic 0.18 |
|||||
|
CYP2C19 21 Vmax 7.52, Km 1.12, fumic 0.015 |
CYP3A4 21 Vmax 3.63, Km 27.8, fumic 0.18 |
|||||
|
CES1 21 Vmax 210 f , Km 2.4, fumic 1 |
||||||
| Hepatic uptake scalar = 1 | Hepatic uptake scalar = 1 | Hepatic uptake scalar = 5 22 | Hepatic uptake scalar = 2 | Hepatic uptake scalar = 1 | ||
| Additional systemic CL (L/h) | 10 |
References/data sources indicated in superscript.
Abbreviations: CLin, clearance into compartment; CLint, intrinsic clearance (µl/min/mg protein); CLout, clearance out from compartment; CV, coefficient of variation; fa, fraction available from dosage; fugut, fraction unbound in enterocytes; fumic, fraction unbound in microsomes; fup, fraction unbound in plasma; HLS9, human liver S9 fractions; ka, absorption rate constant; Km, Michaelis–Menten constant (µM); Log P, logarithm of the octanol–water partition coefficient (lipophilicity); MW, molecular weight; Peff,man, effective permeability in man; pKa/pKb, negative logarithm of the acid or base dissociation constant; SAC, single adjusting compartment; Vmax, maximum reaction velocity (pmol/min per pmol); Vsac, volume of single adjusting compartment; Vss, volume of distribution at steady state.
Parameter estimated using the Simcyp parameter estimation module based on mean IV carboxylic acid data. 28
Simcyp predicted effective permeability (Peff,man) in humans using the built‐in mechanistic permeability (MechPeff) model.
Assumed blood/plasma ratio of 1.
Parameter estimated using the Simcyp parameter estimation module based on mean IV carboxylic acid data. 28
Parameter estimated using the Simcyp parameter estimation module based on mean oral acyl glucuronide data. 23
Sensitivity analysis to ensure 50% fraction metabolized via CES1 using oral H4 metabolite data. 29
Parameter estimated using the Simcyp parameter estimation module based on mean oral H4 metabolite data. 29
The pharmacokinetic parameter values for the IV clopidogrel PBPK model were within the predefined acceptance criteria (Table 3). The observed concentration versus time profiles were within the 90% prediction interval of the simulated profiles (Figure 3). Similarly, for the carboxylic acid metabolite, Cmax and AUC0‐24 were within the acceptance criteria. In contrast, tmax was overpredicted for the 10 and 100 mg doses (predicted/observed ratio of 2.28 and 2.63).
TABLE 3.
PBPK predictions versus observed pharmacokinetic parameters for clopidogrel and carboxylic acid metabolite following an IV bolus dose of 10 mg and an IV infusion of 100 mg clopidogrel
| Clopidogrel dose | Parameters | Geometric mean (95% CI) | Predicted/observed ratio | |
|---|---|---|---|---|
| PBPK predicted |
Observed 28 (n = 24) |
|||
|
10 mg b (n = 24) |
Clopidogrel | |||
| tmax a (h) | 0.01 (0.01, 0.01) | 0.02 (0.02, 0.08) | 0.58 a | |
| Cmax (ng/ml) | 367 (259, 521) | 504 (318, 690) | 0.73 | |
| AUC0‐24 (ng·h/ml) | 120 (117, 123) | 92 (76, 108) | 1.30 | |
| Carboxylic acid metabolite | ||||
| tmax a (h) | 0.38 (0.06, 7.08) | 0.17 (0.08, 0.33) | 2.28 | |
| Cmax (ng/ml) | 278 (235, 329) | 523 (459, 587) | 0.53 | |
| AUC0‐24 (ng·h/ml) | 1653 (1539, 1774) | 1427 (1267, 1587) | 1.16 | |
|
100 mg c (n = 24) |
Clopidogrel | |||
| tmax a (h) | 0.07 (0.07, 0.07) | 0.08 (0.08, 0.12) | 0.84 | |
| Cmax (ng/ml) | 2696 (2021, 3595) | 3433 (2506, 4360) | 0.79 | |
| AUC0‐24 (ng·h/ml) | 1199 (1172, 1226) | 954 (878, 1030) | 1.26 | |
| Carboxylic acid metabolite | ||||
| tmax a (h) | 0.44 (0.10, 7.14) | 0.17 (0.17, 0.33) | 2.63 | |
| Cmax (ng/ml) | 2776 (2349, 3280) | 4878 (4386, 5370) | 0.57 | |
| AUC0‐24 (ng·h/ml) | 16 763 (15 614, 17 999) | 13 065 (11 831, 14 299) | 1.28 | |
Abbreviations: AUC0‐24, area under the plasma concentration–time curve from time zero to 24 h; Cmax, peak plasma concentration; tmax, time to maximum plasma concentration.
tmax is shown as median (range).
10 mg IV dose was administered as a bolus with assumed duration of 30 s. 28
100 mg IV dose was administered as an infusion with a duration of 4 min. 28
FIGURE 3.
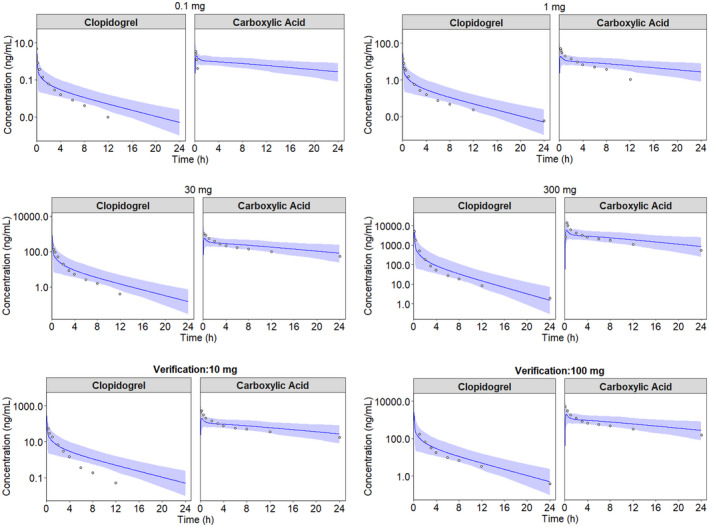
Simulated and observed plasma concentration versus time profiles of clopidogrel and the primary carboxylic acid metabolite in healthy subjects of European ancestry (sim‐Healthy Volunteer) following single IV doses of clopidogrel. Four dose levels were used to develop the model (0.1 mg, 1 mg, 30 mg, and 300 mg), whereas two dose levels were used for model verification (10 mg and 100 mg). The circles represent observed concentrations, 28 the solid blue lines represent the geometric means for the total virtual populations and the shaded areas represent the 90% prediction intervals
A first‐order absorption model was used for the oral PBPK model of clopidogrel, with fa of 0.49 and ka of 1.0 L/h. Except for tmax, the predictions of the clopidogrel and the acyl glucuronide metabolite following a single oral dose of clopidogrel were within the acceptance criteria (Table 4).
TABLE 4.
PBPK predictions versus observed pharmacokinetic parameters for clopidogrel and its acyl glucuronide metabolite and H4 metabolite following a 300‐mg single oral dose of clopidogrel in a healthy population of European ancestry
| CYP2C19 phenotype | Parameter | Geometric mean (CV%) or geometric mean (95% CI) | Predicted/observed ratio | Reference | |
|---|---|---|---|---|---|
| PBPK predicted | Observed | ||||
| No CYP2C19 phenotype data | Clopidogrel |
Tornio et al. (2014) 23 (n = 9) |
|||
| tmax a (h) | 0.45 (0.07, 2.05) | 1.5 (0.92, 2.0) | 0.30 | ||
| Cmax (ng/ml) | 3.4 (72) | 2.9 (97) | 1.17 | ||
| AUC0‐10 (ng·h/ml) | 7.7 (67) | 6.0 (89) | 1.28 | ||
| Acyl Glucuronide Metabolite | |||||
| tmax a (h) | 1.42 (0.68, 5.94) | 1.5 (0.92, 2.0) | 0.95 | ||
| Cmax (ng/ml) | 1592 (62) | 1957 (35) | 0.81 | ||
| AUC0‐10 (ng·h/ml) | 9165 (56) | 7311 (48) | 1.25 | ||
| No CYP2C19 phenotype data | H4 Metabolite |
Farid et al. (2007) (n = 18) |
|||
| Cmax (ng/ml) | 38 (127) | 66 (39.9) | 0.58 | ||
| AUC0‐24 (ng·h/ml) | 69 (123) | 77 (30.7) | 0.90 | ||
| H4 Metabolite |
Brandt et al. (2007) 27 (n = 74) |
||||
| EM (n = 56) | Cmax (ng/ml) | 40 (33, 49) | 58 (56, 61) | 0.69 | |
| AUC0‐24 (ng·h/ml) | 73 (60, 88) | 76 (71, 81) | 0.96 | ||
| IM (n = 17) | Cmax (ng/ml) | 33 (27, 40) | 35 (33, 38) | 0.94 | |
| AUC0‐24 (ng·h/ml) | 57 (47, 69) | 42 (39, 44) | 1.36 | ||
| PM (n = 1) | Cmax (ng/ml) | 23 (19, 29) | 28 (n = 1) | 0.82 | |
| AUC0‐24 (ng·h/ml) | 41 (33, 50) | 27 (n = 1) | 1.52 | ||
Abbreviations: AUC0‐10, area under the plasma concentration–time curve from time zero to 10 h; AUC0‐24, area under the plasma concentration–time curve from time zero to 24 h; Cmax, peak plasma concentration; EM, extensive metabolizer (CYP2C19*1/*1); IM, intermediate metabolizer (CYP2C19*1/*2); PMs, poor metabolizer (CYP2C19*2/*2); tmax, time to maximum plasma concentration.
tmax is shown as median (range).
3.2. CYP pathway
There were good predictions of H4 concentrations for the population of European ancestry (Table 4). This CYP pathway was verified using a second dataset of European ancestry by predicting H4 concentrations for each CYP2C19 phenotype. 27 There was good agreement between the predicted concentrations for the European CYP2C19 EMs, IMs, and PMs and the concentrations observed by Brandt et al. 27 (Table 4). The AUC0‐τ of the H4 metabolite for CYP2C19 UMs was similar to that of the CYP2C19 EMs (Figure S1).
3.3. Predicting pharmacokinetics in Japanese
The observed and predicted tmax and Cmax of the carboxylic acid metabolite were highly variable and were underpredicted for the Japanese population 19 (Table 5), although the predicted AUC0‐48 values were within the acceptance criteria. Clopidogrel and other metabolite concentrations were not reported for this study 19 (Table S6).
TABLE 5.
Predictions of carboxylic acid metabolite pharmacokinetic parameters in Japanese subjects compared to observed values 19 following a 75‐mg single oral dose of clopidogrel
| Parameters | Mean ± SD | Predicted/observed ratio | |
|---|---|---|---|
| PBPK predicted a | Observed (n = 12) | ||
|
tmax a (h) |
0.77 ± 0.53 | 1.9 ± 0.8 | 0.41 |
|
Cmax (ng/ml) |
772 ± 328 | 2290 ± 460 | 0.34 |
|
AUC0‐48 (ng·h/ml) |
8567 ± 4350 | 8460 ± 1360 | 1.01 |
Abbreviations: AUC0‐48, area under the plasma concentration–time curve from time zero to 48 h; Cmax, peak plasma concentration; tmax, time to maximum plasma concentration.
tmax is shown as median (range).
There was good agreement between the observed and predicted concentrations of the H4 metabolite by CYP2C19 phenotype in Japanese subjects following oral doses of either 300 mg or 600 mg clopidogrel (Figure 4). The pharmacokinetic parameters were within a twofold range of the observed parameters (Table 6). The PM/EM AUC0‐24 ratio of the H4 metabolite for the Japanese population was well predicted at both the 300 mg and 600 mg clopidogrel doses (observed: 0.40 vs. 0.37, respectively; predicted:0.38 vs. 0.40, respectively).
FIGURE 4.
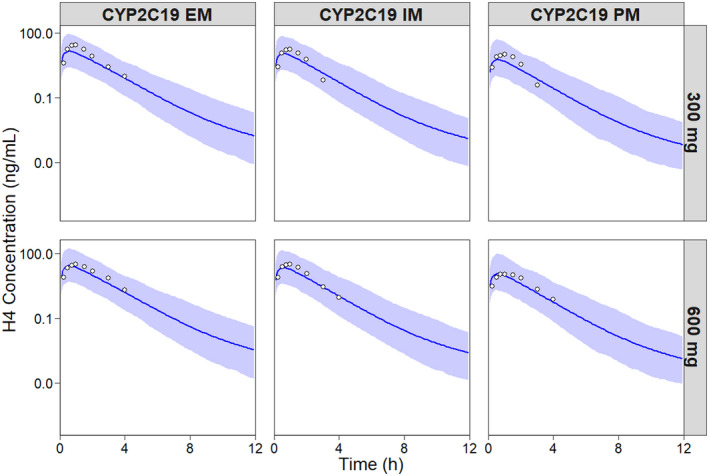
Simulated and observed concentration versus time profiles of the H4 metabolite stratified by CYP2C19 phenotype in the Japanese population following a single oral dose of 300 mg and 600 mg clopidogrel. The circles represent observed concentrations, 30 solid blue lines represent the geometric means for the total virtual population and the shaded areas represent the 90% prediction intervals
TABLE 6.
Predictions of H4 metabolite pharmacokinetic parameters in Japanese subjects compared to observed values following 300 mg or 600 mg single oral doses of clopidogrel
| Dose | CYP2C19 phenotype group | Parameters | Geometric mean (95% CI) | Predicted/observed ratio | |
|---|---|---|---|---|---|
| PBPK predicted | Observed 30 | ||||
| 300 mg | H4 Metabolite | ||||
|
EM (n = 9) |
tmax a (h) | 0.56 (0.32, 1.15) | 0.75 (0.5, 1.5) | 0.75 | |
| Cmax (ng/ml) | 15 (12, 18) | 30 (22, 37) | 0.50 | ||
| AUC0‐24 (ng·h/ml) | 26 (22, 32) | 40 (27, 53) | 0.65 | ||
|
IM (n = 9) |
tmax a (h) | 0.55 (0.30, 1.10) | 1.0 (0.75, 1.5) | 0.55 | |
| Cmax (ng/ml) | 12 (8.89, 13) | 20 (16, 23) | 0.60 | ||
| AUC0‐24 (ng·h/ml) | 20 (16, 24) | 26 (21, 30) | 0.77 | ||
|
PM (n = 9) |
tmax a (h) | 0.56 (0.30, 1.10) | 1.0 (0.50, 1.50) | 0.56 | |
| Cmax (ng/ml) | 6.0 (4.83, 7.51) | 11 (8.13, 15) | 0.55 | ||
| AUC0‐24 (ng·h/ml) | 10 (8.26, 12.8) | 16 (12, 20) | 0.63 | ||
| Ratio | AUC Ratio: PM/EM | 0.38 | 0.40 | 0.95 | |
| 600 mg |
EM (n = 9) |
tmax a (h) | 0.56 (0.30, 1.15) | 1.0 (0.75, 3.0) | 0.56 |
| Cmax (ng/ml) | 28 (23, 34) | 33 (17, 49) | 0.85 | ||
| AUC0‐24 (ng·h/ml) | 50 (41, 61) | 61 (43, 79) | 0.82 | ||
|
IM (n = 9) |
tmax a (h) | 0.56 (0.30, 1.10) | 1.0 (0.75, 2.5) | 0.56 | |
| Cmax (ng/ml) | 22 (18, 28) | 32 (18, 46) | 0.69 | ||
| AUC0‐24 (ng·h/ml) | 39 (31, 48) | 51 (34, 67) | 0.76 | ||
|
PM (n = 9) |
tmax a (h) | 0.57 (0.30, 1.10) | 1.0 (0.50, 2.0) | 0.57 | |
| Cmax (ng/ml) | 12 (9.4, 15) | 12 (8.7, 15) | 1.0 | ||
| AUC0‐24 (ng·h/ml) | 20 (16, 25) | 23 (17, 28) | 0.87 | ||
| Ratio | AUC Ratio: PM/EM | 0.40 | 0.37 | 1.08 | |
Abbreviations: AUC0‐24, area under the plasma concentration–time curve from time zero to 24 h; Cmax, peak plasma concentration; EM, extensive metabolizer (CYP2C19*1/*1); IM, intermediate metabolizer (CYP2C19*1/*2); PMs, poor metabolizer (CYP2C19*2/*2, CYP2C19*2/*3 and CYP2C19*3/*3); tmax, time to maximum plasma concentration.
tmax is shown as median (range).
Despite accounting for CYP2C19 phenotype, the H4 metabolite AUC24 was significantly lower for the Japanese population (300 mg Day 1; CYP2C19 EM, 26 [22–32] ng·h/ml; geometric mean [95% CI]; Table 6) than for the European population of the same CYP2C19 phenotype (73 [60–88] ng·h/ml; Table 4). In a sensitivity analysis for body weight (see Figure S2), the steady‐state AUC0‐τ of the H4 metabolite was independent of body weight and was found to be lower than the European population for any of the CYP2C19 phenotypes.
3.4. Effect of CYP2C19 inhibitor fluvoxamine
Coadministration of fluvoxamine (50 mg daily) with clopidogrel (300 mg loading dose followed by 75 mg daily) reduced the steady‐state AUC0‐τ of the active H4 metabolite but had no effect on the steady‐state AUC0‐τ of clopidogrel and the acyl glucuronide metabolite (Figure 5). The geometric mean H4 AUC0‐τ ratio (clopidogrel + fluvoxamine)/(clopidogrel only) for populations of European ancestry was 0.20 (0.17–0.23, 95% CI), 0.31 (0.26–0.37), and 0.31 (0.26, 0.37) for CYP2C19 EM, IM, and PM, respectively. Similarly, the H4 AUC0‐τ ratio for the effect of fluvoxamine in the Japanese population was 0.11 (0.09–0.13), 0.12 (0.10–0.14), and 0.18 (0.15–0.22) for CYP2C19 EM, IM, and PM, respectively. Therefore, CYP2C19 inhibition is similar for both the Japanese and European populations of the same phenotype.
FIGURE 5.
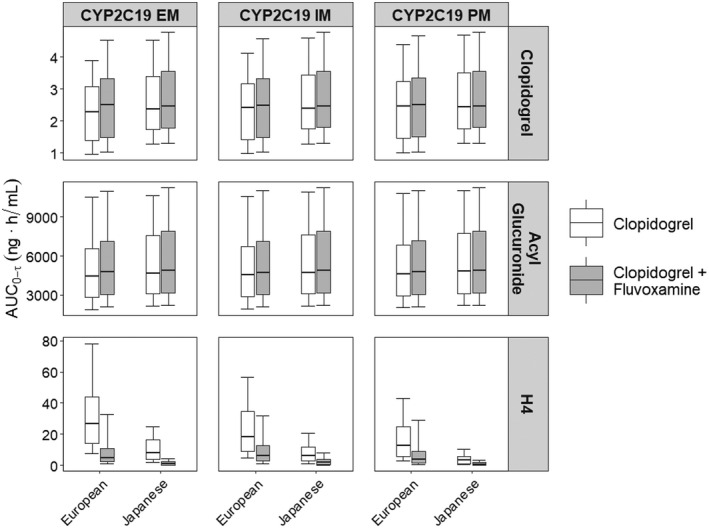
Whisker‐box plots of AUC0‐τ at steady‐state (Day 3, τ = 24 h) of clopidogrel, acyl glucuronide metabolite, and H4 metabolite in populations of European and Japanese ancestry following multiple doses of clopidogrel alone (300 mg loading dose, 75 mg daily) or coadministered with fluvoxamine (50 mg once daily) by CYP2C19 phenotype. The boxes represent the interquartile range with median shown as a solid line; whiskers represent the 10th and 90th percentiles
3.5. Effect of intrinsic factors on clopidogrel, acyl glucuronide, and H4 metabolite
The effects of intrinsic factors on the steady‐state AUC0‐τ of clopidogrel, the acyl glucuronide metabolite, and the H4 metabolite are shown in Figures 6, 7, 8 respectively. For clopidogrel, older age (age 65–92 years) was associated with a significantly higher exposure (p < .001) and there was a trend towards higher acyl glucuronide concentrations; however, this did not reach significance (Figure 6). Obesity (BMI >35 kg/m2) was associated with a lower clopidogrel AUC0‐τ (p < .001) but was not associated with changes in acyl glucuronide (Figure 7) and H4 metabolite concentrations (Figure 8) relative to the Sim‐NEurCaucasian population. For clopidogrel, the acyl glucuronide metabolite, and the H4 metabolite, there were no differences for the AUC0‐τ estimates between the Sim‐NEurCaucasian and the Cancer population, which describes those with altered metabolic and organ function due to neoplastic diseases. Hepatic dysfunction (Child–Pugh B and C) and renal dysfunction (Stage 3 and Stage 4/5) were associated with significantly lower H4 exposures (Figure 8). Lastly, none of the intrinsic factors evaluated influenced the AUC0‐τ of the acyl glucuronide metabolite (Figure 7).
FIGURE 6.
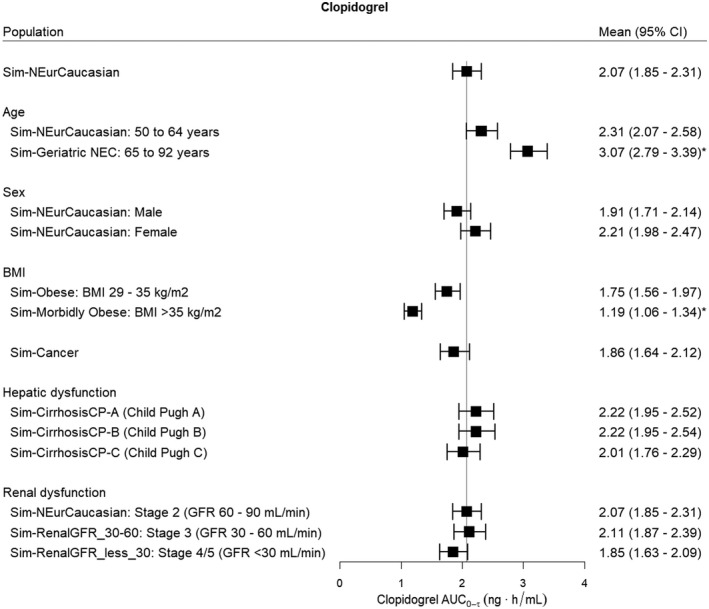
The effect of intrinsic factors on the steady‐state AUC0‐τ (Day 3, τ = 24 h) of clopidogrel (300 mg oral loading dose, 75 mg daily). The reference line indicates the mean clopidogrel AUC0‐τ for the Sim‐NEurCaucasian population, a general population of European ancestry (aged 20–50 years, 50% females. BMI 18–29 kg/m2, normal hepatic function and healthy renal function >90 ml/min, CYP2C19 PM frequency = 2.4%). BMI, body mass index; GFR, glomerular filtration rate. Statistical significance compared to Sim‐NEurCaucasian population at *p < .001
FIGURE 7.
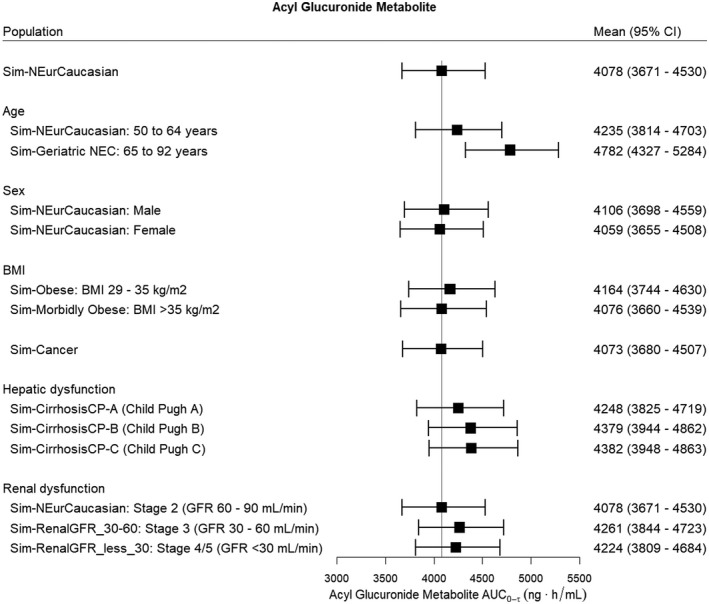
The effect of intrinsic factors on the steady‐state AUC0‐τ (Day 3, τ = 24 h) of the acyl glucuronide metabolite for a clopidogrel oral loading dose of 300 mg then 75 mg daily. The reference line indicates the mean acyl glucuronide AUC0‐τ for the Sim‐NEurCaucasian population, a general population of European ancestry (aged 20–50 years, 50% females, BMI 18–29 kg/m2, normal hepatic function and healthy renal function >90 ml/min, CYP2C19 PM frequency = 2.4%). BMI, body mass index; GFR, glomerular filtration rate. No groups were statistically different from the Sim‐NEurCaucasian population
FIGURE 8.
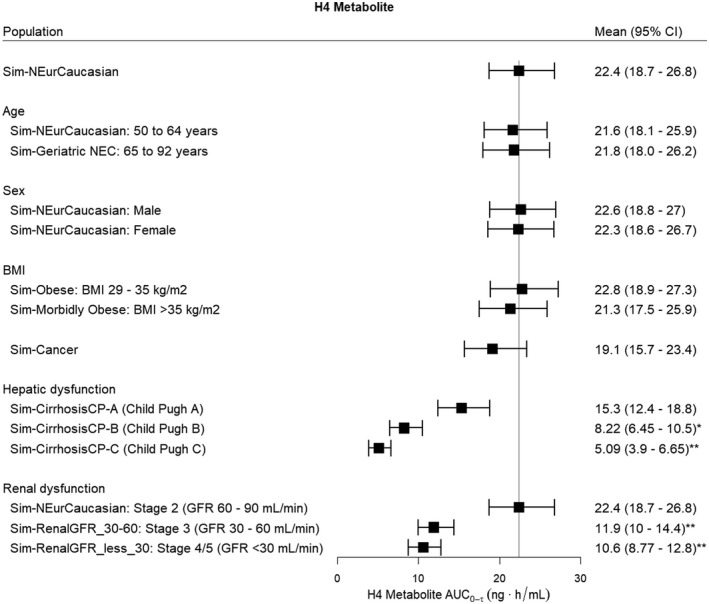
The effect of intrinsic factors on the steady‐state AUC0‐τ (Day 3, τ = 24 h) of the H4 metabolite following clopidogrel (300 mg oral loading dose, 75 mg daily). The reference line indicates the mean H4 metabolite AUC0‐τ for the Sim‐NEurCaucasian population, a general population of European ancestry (aged 20–50 years, 50% females, BMI 18–29 kg/m2, normal hepatic function and healthy renal function >90 ml/min, CYP2C19 PM frequency = 2.4%). BMI, body mass index; GFR, glomerular filtration rate. Statistical significance compared to the Sim‐NEurCaucasian population at *p < .01 and **p < .001
4. DISCUSSION
CYP2C19 genotyping is recommended to predict response to clopidogrel, guide the selection of antiplatelet therapy, and improve patient outcomes following percutaneous coronary intervention (PCI). 39 , 40 This is particularly important in East Asian populations due to the higher frequency of CYP2C19 decreased activity alleles (*2, *3) compared to populations of European ancestry. In this analysis, a PBPK model for clopidogrel and its metabolites was developed that can be used to evaluate the impact of CYP2C19 activity and other intrinsic factors on the pharmacokinetics of clopidogrel and its metabolites in populations of European and Japanese ancestry. The modeling approach outlined here also illustrates how one can address clinical and regulatory questions in global drug development.
4.1. Comparisons with other clopidogrel PBPK models evaluated in populations of European ancestry
The present PBPK model of clopidogrel is an improvement to published PBPK models that only describe one of the major metabolic pathways of clopidogrel. 21 , 22 , 23 The model proposed by Djebli et al. 21 considered CYP2C19 phenotype; however, this model was built using the data from Simon et al., 8 which consisted of European (72.5%), Chinese (20%), and Japanese (7.5%) subjects. 8 Moreover, potential ethnic differences in pharmacokinetics were not considered in the model, and a higher total clearance was obtained for the H4 metabolite (i.e., 500 L/h vs. 90 L/h). The reproducibility of our results is reflected by previous findings based on data from healthy Amish Caucasian individuals (n = 605), 41 which estimated a similar H4 metabolite clearance (93 L/h).
A recently published PBPK model for clopidogrel by Liu et al. 42 considered both major metabolic pathways. However, the CES1‐mediated metabolism of clopidogrel (CES1 CLint 300 µl/min/mg protein 42 ) contributed to 5% of the overall metabolism of clopidogrel, which is not plausible given that CES1 is the major metabolic pathway. Another PBPK‐PD model also investigated the influence of CES1 and CYP2C19, CYP3A4 and CYP2C9 activity on the H4 metabolite concentrations. 43 Nonetheless, these models did not investigate metabolites formed via the CES1‐mediated pathway (carboxylic acid, acyl glucuronide) or explored the effect of other intrinsic factors (age, sex, body weight, and organ impairment). 42 , 43
4.2. Extrapolation and prediction of the pharmacokinetics in a Japanese population
The pharmacokinetics of clopidogrel, the carboxylic acid as well as the acyl glucuronide and H4 metabolites were well predicted for populations of European ancestry. For the Japanese population, the pharmacokinetics of the H4 metabolite and the AUC0‐48 of the carboxylic acid metabolite were within the acceptance criteria, although the tmax and Cmax of the carboxylic acid metabolite were underpredicted. Additionally, there was large inter‐subject variability in the pharmacokinetics of the H4 metabolite for both populations. Since CES1 is the major enzyme involved in the metabolism of clopidogrel to the primary metabolite 2‐oxo‐clopidogrel, and the metabolism of 2‐oxo‐clopidogrel, 32 , 44 CES1 activity may indirectly contribute to the pharmacokinetic variability observed in the H4 metabolite by affecting the concentrations of 2‐oxo‐clopidogrel. CES1 is highly abundant in the liver and there is significant interindividual variability in the expression and activity of CES1, related to genetic polymorphisms. 45 The most clinically relevant CES1 variant is CES1 428G>A (rs71647871), a loss‐of‐function single‐nucleotide polymorphism, which has been associated with higher concentrations of the H4 metabolite and improved antiplatelet effects. 41 , 46 , 47 However, the CES1 428G>A SNP is of low frequency (3.7% Europeans, 0% Asians) 48 and will make very limited contribution to population differences in pharmacokinetics.
4.3. Effect of varying CYP2C19 activity
The predictions of H4 metabolite exposures were within the acceptance criteria for the Japanese population when applying the European CYP2C19 liver abundance values (see Table S2). This approach has been reported by three other recent PBPK analyses of CYP2C19 substrates (lansoprazole, escitalopram, voriconazole, tofacitinib omeprazole 33 , 34 , 35 ), which have also applied the European CYP2C19 liver abundance values to Japanese populations. Other PBPK models of CYP2C19 substrates have used different CYP2C19 liver abundance values (EMs, 4.7 pmol/mg protein, 4 8 pmol/mg protein 49 ) to obtain better pharmacokinetic predictions for the Japanese population. In Simcyp Version 18.2, the mean CYP2C19 liver abundance value for EMs in the Japanese population (4.1 pmol/mg protein) was based on data obtained from a small sample of livers (n = 29) of unspecified CYP2C19 genotype. 50 This highlights the importance of obtaining reliable physiological parameter values for each population and the need for further studies to confirm the CYP2C19 liver abundance for the Japanese population by CYP2C19 genotype in order to improve predictions of the pharmacokinetics of CYP2C19 substrates.
The H4 metabolite concentrations were lower for the CYP2C19 IM and PM phenotype groups compared to the CYP2C19 EM group in both European and Japanese populations. Regardless of CYP2C19 phenotype, the H4 metabolite concentrations were lower in the population of Japanese ancestry than the general population of European ancestry. The pharmacokinetics of H4 was found to be similar across body weight groups (see Figure S2). Nevertheless, an antiplatelet effect of clopidogrel was still observed in Japanese patients with ACS across all CYP2C19 phenotypes, even though the effectiveness of clopidogrel was lower in the IM and PM groups. 18 , 51
There was no effect of CYP2C19 phenotype on the pharmacokinetics of the acyl glucuronide metabolite in both populations of European and Japanese ancestry. This is expected as CYP2C19‐mediated metabolism of clopidogrel is minor (about 5%) compared to CES1‐mediated metabolism of clopidogrel (85–90%). 9 Therefore, there is no increased risk of clopidogrel DDIs with CYP2C8 substrates in patients who are CYP2C19 PMs.
The pharmacokinetics of the H4 metabolite was only evaluated for three CYP2C19 phenotype groups (EM, IM, and PM) for the populations of European 27 and Japanese 30 ancestry. With the identification of the CYP2C19*17 allele, which is associated with increased CYP2C19 activity, 5 the Clinical Pharmacogenetics Implementation Consortium (CPIC) 16 , 52 has updated its guidelines to standardize inferred CYP2C19 phenotype groups based on functional data and an individual's CYP2C19 genotype. The five phenotype groups include the ultrarapid (UR, CYP2C19*17/*17), rapid (RM, CYP2C19*1/*17), normal (NM, CYP2C19*1/*1), intermediate (IM) and poor (PM) metabolizers of CYP2C19. 16 , 52 In the study of Brandt et al., 27 subjects classified as CYP2C19 EM based on the absence of the decreased activity alleles (*2, *3) may have been CYP2C19 EMs, RMs, or UMs. However, in in the present analysis as well as previous reports, 8 the H4 metabolite AUC was similar in the UMs and EMs. Furthermore, Lee et al. 53 reported that the risk of atherothrombotic events was not significantly different between CYP2C19 UMs and CYP2C19 EMs, indicating that the clinical utility of CYP2C19*17 to guide clopidogrel antiplatelet therapy is limited.
4.4. Effect of other intrinsic factors
The simulations of clopidogrel and its metabolites in patient populations with defined demographics and concomitant diseases align with the observations and dosage recommendations in the product information for clopidogrel. 10 , 19 The Product Information reports that the plasma concentrations of the main circulating metabolite (carboxylic acid metabolite) are significantly higher in elderly (≥75 years) compared to young healthy volunteers. 10 This is supported by the current simulations showing 30% higher carboxylic acid concentrations and a trend towards higher acyl glucuronide concentrations (1.2‐fold) in the older patients (≥65 years) compared to the general population. Conversely, our simulations showed no effect of age on H4 metabolite concentrations, which supports the product information recommendation that no dosage adjustment is needed for older patients (≥65 years) as no differences in platelet aggregation and bleeding time were observed. 10 In addition, these findings (clopidogrel; 2.31 [2.07–2.58] ng.h/ml, mean [95% CI]; H4 metabolite, 21.6 [18.1–25.9] ng.h/ml) align with observed steady‐state AUC0‐τ of clopidogrel (3.63 ± 3.23 ng/ml, mean ± SD) and H4 metabolite (19.5 ± 8.69 ng/ml) in older subjects of European ancestry (62.9 ± 8.3 years, n = 50) with the CYP2C19 EM phenotype. 54
Despite controlling for CYP2C19 phenotype and body weight, the H4 metabolite concentrations were lower in the populations of Japanese ancestry compared to those of European ancestry, indicating other population‐specific factors also contribute to the difference observed. In fact, the current analyses also indicate that significantly lower H4 metabolite exposures are observed for populations with moderate (GFR 60–90 ml/min) to severe renal impairment (GFR < 30 ml/min) and in patients with hepatic impairment relative to the general European population. Changes associated with renal impairment are caused by the marked reduction in CYP expression due to the accumulation of uremic toxins, which can modulate CYP activity, particularly CYP2B6, CYP2C9, and CYP3A4. 55 A large systematic analysis of the association between post‐treatment platelet function and renal function reported that chronic kidney disease correlated with poor response to clopidogrel treatment, 56 which supports our findings in the population with severe renal impairment. In addition, the product information describes that lower plasma concentrations of the main circulating metabolite (carboxylic acid metabolite) were observed in patients with severe renal impairment and associated with lower ADP‐induced platelet aggregation (25%) compared to healthy volunteers. 10
In the population with hepatic impairment, altered pharmacokinetics is associated with a decreased functional liver size, reduced functional hepatocytes, altered hepatic blood flow, and a reduction in CYP expression. 57 Yet, the pharmacokinetics of clopidogrel and carboxylic acid in patients with liver cirrhosis (n = 12, Child–Pugh Class A or B) were similar to patients without liver cirrhosis (n = 12), 58 which supports our results. This is in agreement with previous reports showing comparable efficacy of clopidogrel, based on the inhibition of ADP‐induced platelet aggregation in subjects with and without liver cirrhosis. 10 , 58 The effect of liver cirrhosis on the pharmacokinetics of the H4 metabolite is not known and further studies are required to confirm these findings. Currently, clopidogrel should be used with caution in patients with severe renal or hepatic impairment. 10
4.5. Limitations
We acknowledge that there are some limitations to the current investigation, which may need to be carefully considered to allow broader generalization of the findings. First, to optimize the pharmacokinetic parameters, individual data were not available. Instead, mean pharmacokinetic data were obtained from controlled clinical studies of small sample sizes, which may not capture all variability in larger populations and which have not reported concentrations of all four clopidogrel metabolites of interest. 24 In addition, inter‐study variability in each ethnic group may arise from differences in study design and analytical methods used to quantify the concentrations of clopidogrel and its metabolites, leading to potential confounding or contributing lower precision of the parameter estimates. 59
4.6. Potential applications
The US FDA 10 and Japan PMDA 19 product information for clopidogrel both recommend alternative antiplatelet therapy for subjects considered CYP2C19 PMs, while the CPIC guidelines recommend alternative antiplatelet therapy also for CYP2C19 IMs if there are no contraindications for other antiplatelet therapies. 16 In contrast, the Dutch Pharmacogenetics Working Group (DPWG) suggest doubling the dose of clopidogrel (600 mg loading dose, 150 mg daily) for CYP2C19 IMs. 60 As CYP2C19 PMs are not producing the H4 metabolite, doubling the dose of clopidogrel has limited benefits and does not improve platelet aggregation. 61
Clinically significant differences in pharmacokinetics between populations of East Asian and European ancestry are not common and, with the exception of antitumor drugs, characterization of the potential for inter‐ethnic differences takes place during the late stages of drug development. 62 The availability of virtual populations including baseline demographic, genetic, clinical, and ethnicity traits presents an opportunity for the prediction of the pharmacokinetics of a new drug in these ethnic groups early in development. However, the reliability of these predictions will also require an understanding of the distribution of the important physiological parameters identified (e.g., drug metabolizing enzyme [DME] activity) across ethnic groups of interest. As the clopidogrel example indicates, reliable information on the expression and activity of DMEs such as CYP2C19 in specific genotype/phenotype groups across ethnic groups is not currently available. Until further population profiling has been conducted to provide accurate information in specific ethnic groups, uncertainty will need to be factored into model predictions, taking into account historical data where appropriate. While pharmacokinetic variation across ethnic groups may often be explained by other factors than geographical ancestry or genetic factors, model predictions should be confirmed by prospective clinical pharmacokinetic data.
5. CONCLUSION
To our knowledge, this is the first time an integrated PBPK model including the two major metabolic pathways of clopidogrel has been developed and verified for populations of European ancestry that could be used as a tool to support the extrapolation of pharmacokinetics to the Japanese population. Moreover, our analysis suggests that differences between the two populations appear to be determined primarily by the effect of varying CYP2C19 liver activity. These results also highlight the contribution of other intrinsic and extrinsic factors known to cause interindividual variability in systemic exposure. CYP2C19 PM phenotype, coadministration with fluvoxamine as well as hepatic and renal impairment were associated with lower H4 metabolite concentrations. These differences in exposure may have clinical implications, even though clopidogrel or acyl glucuronide concentrations are unlikely to be altered in these conditions. It is important to emphasize that increasing the dose of clopidogrel would not improve therapeutic response for CYP2C19 PMs and alternative antiplatelet therapy is therefore recommended.
CONFLICTS OF INTEREST
All authors declare no conflicts of interest that could have influenced the submitted work.
AUTHOR CONTRIBUTION
JD performed the analysis and wrote the manuscript. JD, RN, ODP, and AG were involved in the study design, data interpretation, and writing of the manuscript. JD, RN, AP, ODP, and AG reviewed and approved the final manuscript.
Supporting information
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge valuable PBPK modelling advice from Dr Gaohua Lu, at the time a member of Systems Modelling and Translational Biology, GlaxoSmithKline R&D Stevenage, United Kingdom.
Duong JK, Nand RA, Patel A, Della Pasqua O, Gross AS. A physiologically based pharmacokinetic model of clopidogrel in populations of European and Japanese ancestry: An evaluation of CYP2C19 activity. Pharmacol Res Perspect. 2022;10:e00946. doi: 10.1002/prp2.946
Funding information
No funding was received for this work.
DATA AVAILABILITY STATEMENT
Data sharing not applicable as no new datasets were generated.
REFERENCES
- 1. Yasuda SU, Zhang L, Huang SM. The role of ethnicity in variability in response to drugs: focus on clinical pharmacology studies. Clin Pharmacol Ther. 2008;84:417‐423. [DOI] [PubMed] [Google Scholar]
- 2. Botton MR, Whirl‐Carrillo M, Del Tredici AL, et al. PharmVar GeneFocus: CYP2C19. Clin Pharmacol Ther. 2020:352‐366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dorji PW, Tshering G, Na‐Bangchang K. CYP2C9, CYP2C19, CYP2D6 and CYP3A5 polymorphisms in South‐East and East Asian populations: a systematic review. J Clin Pharm Ther. 2019;44:508‐524. [DOI] [PubMed] [Google Scholar]
- 4. Feng S, Cleary Y, Parrott N, et al. Evaluating a physiologically based pharmacokinetic model for prediction of omeprazole clearance and assessing ethnic sensitivity in CYP2C19 metabolic pathway. Eur J Clin Pharmacol. 2015;71:617‐624. [DOI] [PubMed] [Google Scholar]
- 5. Sim SC, Risinger C, Dahl ML, et al. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther. 2006;79:103‐113. [DOI] [PubMed] [Google Scholar]
- 6. Whirl‐Carrillo M, McDonagh EM, Hebert JM, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92:414‐417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Genomes Project C , Auton A, Brooks LD, Durbin RM, et al. A global reference for human genetic variation. Nature. 2015;526:68‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Simon T, Bhatt DL, Bergougnan L, et al. Genetic polymorphisms and the impact of a higher clopidogrel dose regimen on active metabolite exposure and antiplatelet response in healthy subjects. Clin Pharmacol Ther. 2011;90:287‐295. [DOI] [PubMed] [Google Scholar]
- 9. Jiang XL, Samant S, Lesko LJ, Schmidt S. Clinical pharmacokinetics and pharmacodynamics of clopidogrel. Clin Pharmacokinet. 2015;54:147‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. US Food and Drug Administration . Prescribing Information. Plavix; 2010. Accessed April 12, 2021. https://www.accessdata.fda.gov/drugsatfda_docs/label/2010/020839s048lbl.pdf [Google Scholar]
- 11. Kahma H, Filppula AM, Neuvonen M, et al. Clopidogrel carboxylic acid glucuronidation is mediated mainly by UGT2B7, UGT2B4, and UGT2B17: implications for pharmacogenetics and drug‐drug interactions. Drug Metab Dispos. 2018;46:141‐150. [DOI] [PubMed] [Google Scholar]
- 12. Sangkuhl K, Klein TE, Altman RB. Clopidogrel pathway. Pharmacogenet Genomics. 2010;20:463‐465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mega JL, Close SL, Wiviott SD, et al. Cytochrome p‐450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360:354‐362. [DOI] [PubMed] [Google Scholar]
- 14. Mega JL, Simon T, Collet JP, et al. Reduced‐function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta‐analysis. JAMA. 2010;304:1821‐1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. US Food and Drug Administration . FDA Drug Safety Communication: Reduced Effectiveness of Plavix (Clopidogrel) in Patients Who are Poor Metabolizers of the Drug. 2010. Accessed November 10, 2020. https://www.fda.gov/drugs/postmarket‐drug‐safety‐information‐patients‐and‐providers/fda‐drug‐safety‐communication‐reduced‐effectiveness‐plavix‐clopidogrel‐patients‐who‐are‐poor
- 16. Scott SA, Sangkuhl K, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013;94:317‐323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Murasaki K. Guidelines for management of anticoagulant and antiplatelet therapy in cardiovascular disease (JCS 2009). Nihon Rinsho. 2011;69(Suppl 9):567‐571. [in Japanese]. [PubMed] [Google Scholar]
- 18. Nagashima Z, Tsukahara K, Morita S, et al. Platelet reactivity in the early and late phases of acute coronary syndromes according to cytochrome P450 2C19 phenotypes. J Cardiol. 2013;62:158‐164. [DOI] [PubMed] [Google Scholar]
- 19. Japan Pharmaceuticals and Medical Devices Agency (PMDA) . Sanofi Plavix (Clopidogrel) Product Information. 2020. Accessed December 7, 2020. https://www.pmda.go.jp/PmdaSearch/iyakuDetail/ResultDataSetPDF/780069_3399008F1025_1_28
- 20. Zhao P, Zhang L, Grillo JA, et al. Applications of physiologically based pharmacokinetic (PBPK) modeling and simulation during regulatory review. Clin Pharmacol Ther. 2011;89:259‐267. [DOI] [PubMed] [Google Scholar]
- 21. Djebli N, Fabre D, Boulenc X, Fabre G, Sultan E, Hurbin F. Physiologically based pharmacokinetic modeling for sequential metabolism: effect of CYP2C19 genetic polymorphism on clopidogrel and clopidogrel active metabolite pharmacokinetics. Drug Metab Dispos. 2015;43:510‐522. [DOI] [PubMed] [Google Scholar]
- 22. Shebley M, Fu W, Badri P, Bow D, Fischer V. Physiologically based pharmacokinetic modeling suggests limited drug‐drug interaction between clopidogrel and dasabuvir. Clin Pharmacol Ther. 2017;102:679‐687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tornio A, Filppula AM, Kailari O, et al. Glucuronidation converts clopidogrel to a strong time‐dependent inhibitor of CYP2C8: a phase II metabolite as a perpetrator of drug‐drug interactions. Clin Pharmacol Ther. 2014;96:498‐507. [DOI] [PubMed] [Google Scholar]
- 24. Shebley M, Sandhu P, Emami Riedmaier A, et al. Physiologically based pharmacokinetic model qualification and reporting procedures for regulatory submissions: a consortium perspective. Clin Pharmacol Ther. 2018;104:88‐110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. European Medicines Agency . Guideline on the Reporting of Physiologically Based Pharmacokinetic (PBPK) Modelling and Simulation. 2018. Accessed October 18, 2020. https://www.ema.europa.eu/en/documents/scientific‐guideline/guideline‐reporting‐physiologically‐based‐pharmacokinetic‐pbpk‐modelling‐simulation_en.pdf
- 26. US Food and Drug Administration . Guidance for Industry: Physiologically Based Pharmacokinetic Analyses—Format and Content. 2018. Accessed October 18, 2020. https://www.fda.gov/media/101469/download
- 27. Brandt JT, Close SL, Iturria SJ, et al. Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. J Thromb Haemost. 2007;5:2429‐2436. [DOI] [PubMed] [Google Scholar]
- 28. Cushing DJ, Souney PF, Cooper WD, et al. Pharmacokinetics and platelet aggregation inhibitory effects of a novel intravenous formulation of clopidogrel in humans. Clin Exp Pharmacol Physiol. 2012;39:3‐8. [DOI] [PubMed] [Google Scholar]
- 29. Farid NA, Payne CD, Small DS, et al. Cytochrome P450 3A inhibition by ketoconazole affects prasugrel and clopidogrel pharmacokinetics and pharmacodynamics differently. Clin Pharmacol Ther. 2007;81:735‐741. [DOI] [PubMed] [Google Scholar]
- 30. Kobayashi M, Kajiwara M, Hasegawa S. A randomized study of the safety, tolerability, pharmacodynamics, and pharmacokinetics of clopidogrel in three different CYP2C19 genotype groups of healthy Japanese subjects. J Atheroscler Thromb. 2015;22:1186‐1196. [DOI] [PubMed] [Google Scholar]
- 31. Rohatgi A. WebPlotDigitizer. Version 4.1. Automeris LLC; 2018. Accessed March 14, 2019. https://automeris.io/WebPlotDigitizer/ [Google Scholar]
- 32. Zhu HJ, Wang X, Gawronski BE, Brinda BJ, Angiolillo DJ, Markowitz JS. Carboxylesterase 1 as a determinant of clopidogrel metabolism and activation. J Pharmacol Exp Ther. 2013;344:665‐672. [DOI] [PubMed] [Google Scholar]
- 33. Suzuki M, Tse S, Hirai M, Kurebayashi Y. Application of physiologically‐based pharmacokinetic modeling for the prediction of tofacitinib exposure in Japanese. Kobe J Med Sci. 2017;62:E150‐E161. [PMC free article] [PubMed] [Google Scholar]
- 34. Zhou L, Sharma P, Yeo KR, et al. Assessing pharmacokinetic differences in Caucasian and East Asian (Japanese, Chinese and Korean) populations driven by CYP2C19 polymorphism using physiologically‐based pharmacokinetic modelling. Eur J Pharm Sci. 2019;139:105061. [DOI] [PubMed] [Google Scholar]
- 35. Higashimori M, Shimada H, Ichikawa K, Zhou D. Physiologically based pharmacokinetic modeling to predict exposures in healthy Japanese subjects with different CYP2C19 phenotypes: Esomeprazole case study. Int J Clin Pharmacol Ther. 2020;58:29‐36. [DOI] [PubMed] [Google Scholar]
- 36. US Food Drug Administration . Drug Development and Drug Interactions: Table of Substrates, Inhibitors and Inducers. 2020. Accessed December 10, 2020. https://www.fda.gov/drugs/drug‐interactions‐labeling/drug‐development‐and‐drug‐interactions‐table‐substrates‐inhibitors‐and‐inducers#table2‐1
- 37. Harding SD, Sharmanm JL, Faccenda E, et al. The IUPHAR/BPS guide to PHARMACOLOGY in 2019: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Res. 2018;46:D1091‐D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Alexander SP, Fabbro D, Kelly E, et al. The concise guide to pharmacology 2021/22: enzymes. Br J Pharmacol. 2021;178(Suppl 1):S313‐S411. [DOI] [PubMed] [Google Scholar]
- 39. Cavallari LH, Beitelshees AL, Blake KV, et al. The IGNITE Pharmacogenetics Working Group: an opportunity for building evidence with pharmacogenetic implementation in a real‐world setting. Clin Transl Sci. 2017;10:143‐146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Klein MD, Williams AK, Lee CR, Stouffer GA. Clinical utility of CYP2C19 genotyping to guide antiplatelet therapy in patients with an acute coronary syndrome or undergoing percutaneous coronary intervention. Arterioscler Thromb Vasc Biol. 2019;39:647‐652. [DOI] [PubMed] [Google Scholar]
- 41. Jiang XL, Samant S, Lewis JP, et al. Development of a physiology‐directed population pharmacokinetic and pharmacodynamic model for characterizing the impact of genetic and demographic factors on clopidogrel response in healthy adults. Eur J Pharm Sci. 2016;82:64‐78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu S, Wang Z, Tian X, Cai W. Predicting the effects of CYP2C19 and carboxylesterases on vicagrel, a novel P2Y12 antagonist, by physiologically based pharmacokinetic/pharmacodynamic modeling approach. Front Pharmacol. 2020. 10.3389/fphar.2020.591854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu RJ, Kong WM, An XF, Zou JJ, Liu L, Liu XD. Physiologically‐based pharmacokinetic‐pharmacodynamics model characterizing CYP2C19 polymorphisms to predict clopidogrel pharmacokinetics and its anti‐platelet aggregation effect following oral administration to coronary artery disease patients with or without diabetes. Front Pharmacol. 2020;11:593982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bouman HJ, Schomig E, van Werkum JW, et al. Paraoxonase‐1 is a major determinant of clopidogrel efficacy. Nat Med. 2011;17:110‐116. [DOI] [PubMed] [Google Scholar]
- 45. Wang D, Zou L, Jin Q, Hou J, Ge G, Yang L. Human carboxylesterases: a comprehensive review. Acta Pharm Sin B. 2018;8:699‐712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lewis JP, Horenstein RB, Ryan K, et al. The functional G143E variant of carboxylesterase 1 is associated with increased clopidogrel active metabolite levels and greater clopidogrel response. Pharmacogenet Genomics. 2013;23:1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tarkiainen EK, Holmberg MT, Tornio A, et al. Carboxylesterase 1 c.428G>A single nucleotide variation increases the antiplatelet effects of clopidogrel by reducing its hydrolysis in humans. Clin Pharmacol Ther. 2015;97:650‐658. [DOI] [PubMed] [Google Scholar]
- 48. Zhu HJ, Patrick KS, Yuan HJ, et al. Two CES1 gene mutations lead to dysfunctional carboxylesterase 1 activity in man: clinical significance and molecular basis. Am J Hum Genet. 2008;82:1241‐1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gong J, Iacono L, Iyer RA, Humphreys WG, Zheng M. Physiologically‐based pharmacokinetic modelling of a CYP2C19 substrate, BMS‐823778, utilizing pharmacogenetic data. Br J Clin Pharmacol. 2018;84:1335‐1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Inoue K, Yamazaki H, Imiya K, Akasaka S, Guengerich FP, Shimada T. Relationship between CYP2C9 and 2C19 genotypes and tolbutamide methyl hydroxylation and S‐mephenytoin 4'‐hydroxylation activities in livers of Japanese and Caucasian populations. Pharmacogenetics. 1997;7:103‐113. [DOI] [PubMed] [Google Scholar]
- 51. Ogawa H, Isshiki T, Kimura T, et al. Effects of CYP2C19 allelic variants on inhibition of platelet aggregation and major adverse cardiovascular events in Japanese patients with acute coronary syndrome: The PRASFIT‐ACS study. J Cardiol. 2016;68:29‐36. [DOI] [PubMed] [Google Scholar]
- 52. Caudle KE, Dunnenberger HM, Freimuth RR, et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet Med. 2017;19:215‐223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee CR, Thomas CD, Beitelshees AL, et al. Impact of the CYP2C19*17 allele on outcomes in patients receiving genotype‐guided antiplatelet therapy after percutaneous coronary intervention. Clin Pharmacol Ther. 2021;109:705‐715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Karazniewicz‐Lada M, Krzyzanska D, Danielak D, et al. Impact of genetic variants of selected cytochrome P450 isoenzymes on pharmacokinetics and pharmacodynamics of clopidogrel in patients co‐treated with atorvastatin or rosuvastatin. Eur J Clin Pharmacol. 2020;76:419‐430. [DOI] [PubMed] [Google Scholar]
- 55. Rowland Yeo K, Aarabi M, Jamei M, Rostami‐Hodjegan A. Modeling and predicting drug pharmacokinetics in patients with renal impairment. Expert Rev Clin Pharmacol. 2011;4:261‐274. [DOI] [PubMed] [Google Scholar]
- 56. Htun P, Fateh‐Moghadam S, Bischofs C, et al. Low responsiveness to clopidogrel increases risk among CKD patients undergoing coronary intervention. J Am Soc Nephrol. 2011;22:627‐633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rodighiero V. Effects of liver disease on pharmacokinetics. An update. Clin Pharmacokinet. 1999;37:399‐431. [DOI] [PubMed] [Google Scholar]
- 58. Slugg PH, Much DR, Smith WB, Vargas R, Nichola P, Necciari J. Cirrhosis does not affect the pharmacokinetics and pharmacodynamics of clopidogrel. J Clin Pharmacol. 2000;40:396‐401. [DOI] [PubMed] [Google Scholar]
- 59. Laporte‐Simitsidis S, Girard P, Mismetti P, Chabaud S, Decousus H, Boissel JP. Inter‐study variability in population pharmacokinetic meta‐analysis: when and how to estimate it? J Pharm Sci. 2000;89:155‐167. [DOI] [PubMed] [Google Scholar]
- 60. Dutch Pharmacogenetics Working Group (DPWG) . Pharmacogenetic Guidelines, Netherlands. Clopidogrel – CYP2C19. 2017. Accessed September 10, 2021. http://kennisbank.knmp.nl
- 61. Collet JP, Hulot JS, Anzaha G, et al. High doses of clopidogrel to overcome genetic resistance: the randomized crossover CLOVIS‐2 (Clopidogrel and Response Variability Investigation Study 2). JACC Cardiovasc Interv. 2011;4:392‐402. [DOI] [PubMed] [Google Scholar]
- 62. Rajman I, Hirano M, Honma W, Zhao S. New paradigm for expediting drug development in Asia. Drug Discov Today. 2020;25:491‐496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
Data sharing not applicable as no new datasets were generated.


