Abstract
Coronavirus disease (COVID-19), a coronavirus-induced illness attributed to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission, is thought to have first emerged on November 17, 2019. According to World Health Organization (WHO). COVID-19 has been linked to 379,223,560 documented occurrences and 5,693,245 fatalities globally as of 1st Feb 2022. Influenza A virus that has also been discovered diarrhea and gastrointestinal discomfort was found in the infected person, highlighting the need of monitoring them for gastro intestinal tract (GIT) symptoms regardless of whether the sickness is respiration related. The majority of the microbiome in the intestines is Firmicutes and Bacteroidetes, while Bacteroidetes, Proteobacteria, and Firmicutes are found in the lungs. Although most people overcome SARS-CoV-2 infections, many people continue to have symptoms months after the original sickness, called Long-COVID or Post COVID. The term “post-COVID-19 symptoms” refers to those that occur with or after COVID-19 and last for more than 12 weeks (long-COVID-19). The possible understanding of biological components such as inflammatory, immunological, metabolic activity biomarkers in peripheral blood is needed to evaluate the study. Therefore, this article aims to review the informative data that supports the idea underlying the disruption mechanisms of the microbiome of the gastrointestinal tract in the acute COVID-19 or post-COVID-mediated elevation of severity biomarkers.
Keywords: Microbiota, SARS-CoV-2, Bacteroides species, Inflammatory cytokines, Lectin RegIII beta
1. Introduction
COVID-19, a coronavirus-induced illness attributed to SARS-CoV-2 transmission, is thought to have first emerged on November 17, 2019 [1]. According to W.H.O., COVID-19 has been linked to 379,223,560 documented occurrences and 5,693,245 fatalities globally as of 1st Feb 2022. Among them, India has recorded over 32,768,880 documented occurrences of COVID-19, the second-highest amount of any nation behind the United States. However, even though India has recorded 438,560 COVID-19-related deaths, the overall number of fatalities is fewer than other settings have documented. It has been claimed that there are several predictors of death among hospitalized patients, including studies published in South Africa, China, Iran, Brazil, and India. There have also been multicountry studies conducted and exploring that growing age, male gender [2], and numerous chronic illnesses (such as asthma, COPD, diabetes, TB, obesity, CKD, hypertension, or CVD) are related to an increased risk of mortality [[3], [4], [5], [6], [7], [8]]. These findings are in line with those from high-income nations and replicated in other countries [9]. According to several studies, coinfections or super-infections with bacteria have contributed to fatality in past viral pandemics, including the 1918 and 2009 influenza pandemics [10,11]. Recent research on Influenza A virus that has also been discovered diarrhea and gastrointestinal discomfort was found in the infected person, highlighting the need of monitoring them for GIT symptoms regardless of whether the sickness is respiration related [12]. The majority of the microbiome in the intestines is Firmicutes and Bacteroidetes, while Bacteroidetes, Proteobacteria, and Firmicutes are found in the lungs [13]. The COVID-19 infection was linked with various symptoms, all of which were related to elevated inflammation on a daily pathological basis characterized as the “cytokine storm” [14]. According to research, an increased state of inflammation has been related to a range of unfavorable systemic consequences, notably oxidative stress [15,16], iron homeostasis dysregulation called ferroptosis [17,18], hypercoagulability, and thrombus formation [19]. Several additional clinical features have been described such as cough, fever, tiredness, and dermatological along with upper gastrointestinal (GI) symptoms [20,21]. On average, 17.6% of COVID-19 infected individuals suffer from GI symptoms, which are more frequent in severe cases of infection [22], such as diarrhea and inflammatory bowel disease (IBD) with colitis [23]. SARS-CoV-2 transmission has pathophysiology that is characterized by elevated inflammatory cytokine synthesis, which culminates in lung damage, organ failure, and death [24,25]. Notably, most people recover from SARS-CoV-2 pathogen-induced infectious disease; many people appear to have a slew of lingering symptoms months after the acute illness has resolved, which is called Long-COVID or Post COVID. The term “post-COVID-19 symptoms” refers to those that occur with or after COVID-19 and last for more than 12 weeks (long-COVID-19), without an alternative diagnosis [[26], [27], [28]]. While the molecular cause, or causes, of Long-COVID remains unknown, the primary hypotheses have focused on virus-induced autoimmune or cellular malfunction. Nevertheless, with current evidence that SARS-CoV-2 genetic material and infections are found in certain patients' weeks to a month after elimination of respiratory illness, an alternate hypothesis that a latent chronic illness is perhaps the cause of long-term COVID manifestations has gotten scant attention. Around 10% of individuals in COVID-19 have long-term concerns, which are being investigated more closely [29]. It was recently discovered that more than 73,000 COVID-19 sufferers had been subjected to a higher dimensional space assessment of their Long-COVID was shown to have a wide range of symptoms, including myocarditis, arrhythmia, Nausea or vomiting, mental confusion, kidney failure, persistent tiredness, joint pain, difficulty in breathing, pulmonary fibrosis, insomnia, throat pain, red eyes, sweating, headache, fevers, cough, hair loss, metabolic syndrome, diabetes, ageusia, depressed mood, anxiety, weight loss, metabolic defect, and gastrointestinal disturbances, among others (Table 1 ) [30,31]. COVID-19 and long-term COVID have been linked with elevated interferon-inducible protein and other cytokines associated with hyperinflammation in the early stage, followed by immunocompromising and low-grade inflammation. Many pharmaceutical companies have produced COVID-19 vaccine that are both safe and effective. Some COVID-19 recipients, however, have experienced nausea, vomiting, and anorexia unrelated to the coagulation and neurological system, according to reports [32]. Some newer medicines are also reported less or ineffective to manage severe cases of COVID19 and deaths; for example, newer medications including chloroquine, remdesivir, ivermectin, lopinavir/ritonavir, cepharanthine, baricitinib, and tenofovir disoproxil fumarate. Due to COVID-19 has only a limited number of available treatment options, it is vital to consider host cytokine pathways and the microbiotas of COVID-19, to better understand SARS-CoV-2 responses and aid in developing innovative therapies for the virus. Throughout the early to late stages of COVID-19, a substantial change in the gut microbiota was seen, with an increase in bacterial diversity and a reduction in the relative abundance of beneficial commensals such as Bacteroides. All together the evidence reinforces the view that a person's gut microbiota affects the individual's vulnerability and the responsiveness to SARS-CoV-2 infection. Therefore, the possible understanding of biological component such as biomarkers of inflammatory, immunological, metabolic activity in peripheral blood is needed to evaluate study. Thus, the goal of this article is to review the informative data that supports the idea underlying the disruption mechanisms of the microbiome of the gastrointestinal tract in the acute COVID-19 or post COVID-mediated elevation of severity biomarkers.
Table 1.
Exhibits COVID-19's long-term health consequences with prevalence data from study.
| Health Consequences | Prevalence in % |
|---|---|
| Fatigue | 58 |
| Joint pain | 19 |
| Ageusia | 23 |
| Headache | 44 |
| Cough | 19 |
| Dyspnea | 24 |
| Depression | 12 |
| Weight loss | 12 |
| Red eyes | 6 |
| Arrythmia | 0.4 |
| Renal failure | 1 |
| Myocarditis | 1 |
| Throat pain | 3 |
| Diabetes mellitus | 4 |
| Digestive disorders | 12 |
| Anxiety | 13 |
| Nausea or vomit | 16 |
| Sweat | 17 |
| Pulmonary fibrosis | 5 |
| Intermittent fever | 11 |
| Dyspnea | 24 |
| Sleep disorder | 11 |
| Mental health related | 7 |
| Hair loss | 25 |
| Metabolic defect | 1 |
2. Selection of literature review
The literature study was performed during the last weeks of August 2021, with a primary focus on PubMed and secondary sources such as Scopus and Google Scholar. The papers were chosen based on their relevance to the worldwide pandemic caused by acute SARSCoV-2 or long-term COVID-19 and how it changes gut microbiota and severity biomarkers mediated consequences. “Long COVID-19″, “gut microbiota,” “COVID-19 biomarkers,” “Bacteroide Spp. in acute COVID-19 or Post COVID-19 manifestations”, and “Mechanism of SARS CoV-2 assault” were some of the keywords utilized in this study. With the works published in late 2019 to 2021 (when the epidemic sifts into pandemic) in mind, many scientific publications were uncovered during this first search. Further, the language filter was applied, choosing only English-language papers.
3. Overview of microbiome or microflora mediated immune-regulatory responses
The majority of bacteria in the intestinal colon (more than 1014 organisms per gram of wet weight) are anaerobes, with Bacteroides species accounting for around 25% of the total [33,34]. For example, it stimulates the release of a pro-inflammatory molecule such as TNF-α and IFN-γ regulatory CD4+ T cells, B-cell expressions, and generation of both T-cell dependent and T-cell independent antibodies [[35], [36], [37]]. As outlined above, Commensals in the gut flora include Bacteroides species, typically present in the stomach, the most commonly detected anaerobic pathogens. They can cause life-threatening conditions such as septicemia, brain and lung infections, as well as intraabdominal abscesses in the intestinal and other organs [38]. Most natural food is composed of biopolymers, such as polysaccharides. Bacteroides and other intestinal bacteria meet a large portion of the host's daily energy requirements by digesting glucose in the gut, which produces an excess of volatile fatty acids that are reabsorbed and utilized by the host as an energy source [39]. The polysaccharide utilization loci (PUL) are a collection of physically related genes clustered around pairs of glycan-binding proteins. Bacteroides species have established an ecological benefit over other bacteria due to their ability to regulate the degradation of a wide variety of glycans in the intestine [40]. According to one research, all Bacteroides species exhibit endo-mucinous and sialidase activities on their cell surfaces, which are believed to be the first phases of O-glycan degradation. CAZymes that trim capping sugars are always likely to turn up, demonstrating the breakdown process by microbiome [41]. Several metabolic modifications have been identified to assist Bacteroides in surviving the environmental changes of the human gastrointestinal tract [40]. It is thought that these creatures have a gene that causes the quantity of oxygen in their stomach to decrease, which allows them and other anaerobic bacteria to grow quicker than other organisms [42,43]. Bacteroides organisms may cause significant disease due to their capacity to escape from the gut following gastrointestinal tract rupture or intestinal surgery. This includes abscess development in various body locations (e.g., the abdominal region, lungs, pelvic liver brain) and induces bacteremia. However, Bacteroides possess an outer membrane macrolide efflux protein and have β-lactam derivatives tetracycline resistance genes [44,45], which allows the commensal bacterium to survive against antimicrobial agents.
The intestinal mucosa produces a large amount of the bactericidal lectin RegIII beta in an inflamed gut. The expression of lectin RegIII beta is substantially increased in response to bacterial colonization of the intestinal and illnesses with pathogenicity, both of which contribute to the progression of inflammation [46]. Because it binds peptidoglycan and lipids, lectin RegIII beta can kill different Gram- (-ve) and Gram- (+ve) bacteria, including commensal and enteropathogenic bacteria in the gastrointestinal tract. One research found that lectin RegIII beta did not protect against gut infection but rather slowed the spread of the illness [47]. If this is the case, the pathogen may receive its assistance [48]. It is also believed that excessive lectin RegIII beta expression alters the composition of intestinal microbiota and contributes to infectious diarrhea by decreasing the number of Bacteroidetes species [49]. Furthermore, gut environmental variables such as microbiota-derived compounds affect gut bacterial viability, well-known and well-established.
According to the microbiota's presence at the locations where viruses enter their host, they can influence the outcome of infection. For instance, antibiotic therapy that eliminates mice's natural flora enhances the animals' vulnerability to the influenza A virus. The method by which the microbiota protects against the virus appears to be indirect—the microbiota activates the inflammasome [50], which is necessary for influenza defense [51]. Afterward, to prepare for influenza-specific T-cell responses, inflammasome activation triggers dendritic cell migration from the lung to the outflow lymph node. Interestingly, applying lipopolysaccharide (LPS) intranasal or intrarectal restores the immune function to influenza in antibiotic-treated rats [50].
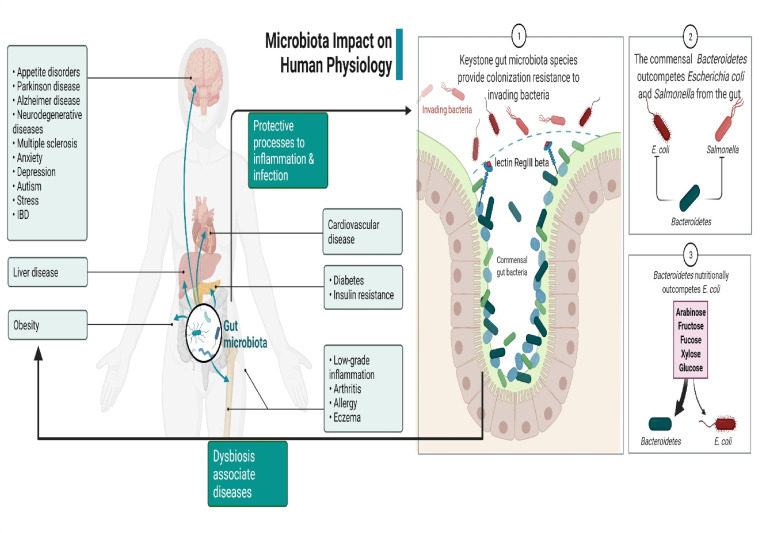
Multiple investigations have been done to evaluate the association between both the gut microbiota and viruses using germ-free rodents (sterile mice born and grown in an antibiotic-free setting) and antimicrobial-treated animals [52]; influences of gut microorganisms on viral infections might be positive, negative, or nonexistent [53]. Either the virion itself may be modified to play these functions, or the host's immune response can be modulated [54]. An increasing amount of attention is being paid to the role that Bacteroides spp. (particularly PSA) play in the immune system's ability to deal with viral infections. In the immunomodulatory investigation, mice were pretreated with PSA and subsequently infected with HSV1 and treated with Acyclovir, the recommended antiviral drug. PSA-treated mice had a higher survival rate than controls (pre-treatment with PBS), and there was also a decrease in brainstem inflammation. The major anti-inflammatory chemical generated by CD4+ and CD8+ T cells was IL-10. It appears that PSA binding to B cells induces these T lymphocytes. There is a possibility that Bacteroides spp. PSA might provide significant anti-inflammatory protection against viral infections [55] (Fig. 1 ).
Fig. 1.
Demonstrate microbiota impact on human physiology and pathophysiology.
4. Plausible signaling pathways of SARS CoV-2 attacks mediated change in microbiota abundance and long-term COVID-19 infection
Low-grade inflammation, defined as a persistent elevation in pro-inflammatory biologicals IL-1/-6, and TNF-alpha, is at the core of all significantly high-risk groups for progressive, long-term COVID-19 pathogenesis. As part of the metabolic syndrome, high blood pressure, overweight, and increased blood sugar increase the risk of diabetes and CVD. They are also thought to be contributing factors to low-grade inflammation. It is believed that SARS-CoV-2 causes hypercytokinemia in metabolic syndrome individuals due to the infiltration of pro-inflammatory lymphocytes and macrophages into the target organs (respiratory tract, central nervous system, kidney, and gastrointestinal tract). According to several research, the inherent and acquired immune systems of SARS CoV-2 infected patients have undergone significant alterations. Patients with acute COVID-19 and long-term COVID suffer from lymphocytopenia and an increase in neutrophils, which seem to be directly related to the severity of the illness and death [56,57]. As a result of severe COVID-19 or long-term COVID, the number of circulating T-lymphocytes (CD4+/CD8+), B, and NK cells, along with monocyte and eosinophil counts, has been shown to drop in patients. At a later stage, elevated levels of pro-inflammatory mediators (IL-1/2/6/8/17, TNF-alpha, GM-CSF, G-CSF, IP-10, CCL3, and MCP-1) were elevated detected in the preponderance of people with acute or long-term COVID [58]. But even though there is a shortage of direct evidence to support pro-inflammatory mediators' involvement in COVID pathology, serum cytokine and chemotherapy levels correlate, as do neutrophil-lymphocyte ratios and the severity of the disease and its associated risks. These things imply that the long-term COVID pathology is inflammatory [14]. It was also shown that individuals with COVID-19 had an excessive concentration of procalcitonin and hs-CRP in their blood, two key inflammatory indicators linked with increased mortality and organ damage risks [59].
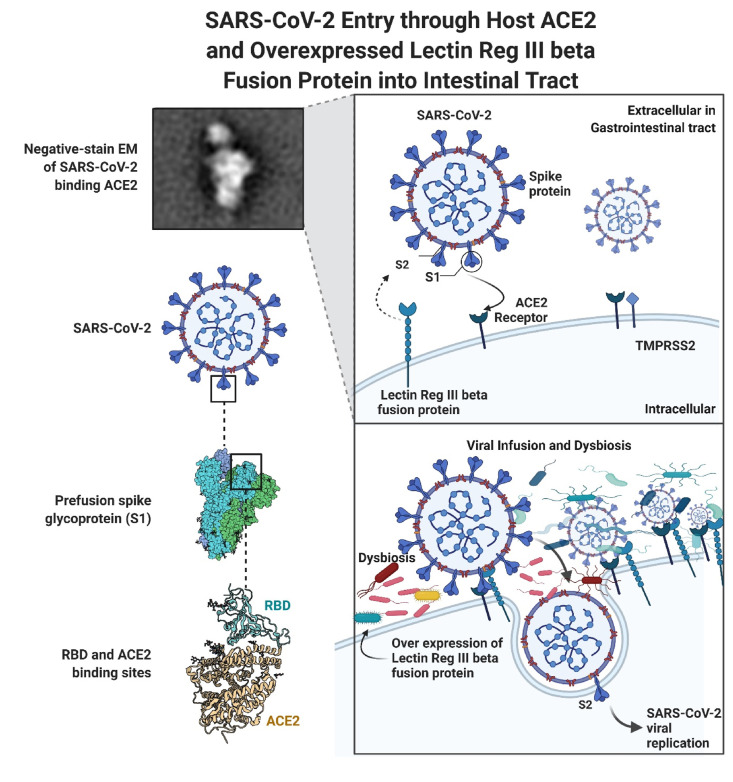
SARS-CoV-2 attacks susceptible sites through the spike (S) glycoprotein, structured as a homotrimer composed of S1 and S2 subunits in each monomer. The pathogenic mechanism involves attaching to cells, inducing S rearrangements in the viral envelope, and fusing the viral particles to the host cellular membranes. In the S1 subunit, the NTD and RBD interact with ACE2 through a region known as the RBM [60,61]. The contradictory relationship between poor ACE2 expression in the respiratory system and infection in other organs, believed to result in extrapulmonary problems, indicates additional receptors such as basigin (CD147), CD209, neuropilin-1, heparan sulfate, and CD209L/CLEC4M, may have a role in viral invasion and proliferation [[62], [63], [64], [65], [66], [67]]. According to new research published in Nature, transmembrane lectins may promote the SARS-CoV-2 infection. As a result, SARS-CoV-2 binds to the intestinal mucosal lectin RegIII beta in an inflamed gastrointestinal tract. Moreover, lectin RegIII beta expression is significantly elevated in response to pathogen binding in the gut, resulting in inflammation. Additionally, the overexpression of lectin RegIII beta seems to have a detrimental effect on the microbiome's composition, resulting in decreased Bacteroidetes spp. Numbers in the gut lumen may play a role(s) in infectious diarrhea. Despite the unexpectedly modest level of expression of ACE2 in the presence of interferon, This observation most likely pertains toward the respiratory to multi-organ damage [68]. ACE2 receptors for SARS CoV-2 are also explored in the intestine, kidneys, heart, and liver. During a recent autopsy examination of 27 people who died due to COVID-19 disease, it was shown that SARS-CoV-2 was shown to be prevalent not just in the lungs but also in the brain and liver, kidneys, and gastrointestinal tract, indicating that multisystem infections may occur. Furthermore, microbiota alterations mediated by lectin RegIII beta affect intestinal metabolism. Vitamin B6 metabolism is the most impacted by the changed metabolic pathways, and lectin RegIII beta expression significantly lowers luminal vitamin B6 levels [49]. Furthermore, Singh et al. [17] speculation of high-level availability of intermediate product called “homocysteine,” in COVID-19 infection, which is recycled by re-methylation (Vit.B12) and trans-sulphuration (Vit.B6) pathway in the human body may be interrupted due to metabolic aggravation through SARS CoV-2. Certain vitamins have been shown to affect the mucosal defenses of the gut. Vitamin D stimulates the synthesis of antimicrobial activity genes such as cathelicidin and -defensin 2 [69], aiding the body's natural defense against pathogen infection during COVID-19 infection. Additionally, vitamin D3 regulates mucosal defence activities; hence, an excess or deficiency of vitamin D3 alters intestine inflammatory responses.
A study comparing the quantitative serum proteome and metabolomic differences between 14 individuals classified as high- or low-risk based on serum lactate dehydrogenase (LDH) expression. These findings demonstrate that the COVID-19 patients with increased circulatory LDH exhibited specific blood coagulation and immunological reactions, such as the complement cascade mediated platelet degranulation, acute inflammatory responses, and several metabolic responses, including several metabolic responses protein ubiquitination, fat metabolism, and pyruvate fermentation. Strong attention was given to activating hypoxic responses in subjects with elevated LDH levels in 40% community [70]. Researchers found that participants with IL-6 levels of 80 pg/ml had a 22-fold higher risk of respiratory failure than those with lower concentrations of this cytokine [71]. Numerous indicators of inflammation and thrombosis (58%) in the population have been identified as predictors of death in severely sick COVID-19 patients. Lymphopenia in 70% of samples (<1000/mmc) was identified as a distinguishing characteristic and considered to be a possible prognostic indicator [72]. D-dimer and IL-6 levels rose as the illness progressed [73] and higher mortality [74]. When IL-6 or D-dimer concentrations were raised, the mortality risk rose 10%. This provides more insight into COVID-19 etiology regarding endothelial-vascular damage and systemic inflammation [75]. Furthermore, A higher risk of mortality from the COVID-19 illness can also be predicted by several biochemical markers, including procalcitonin <0·25 ng/mL, hyperferritinemia, LDH >250 U/L, ESR, and CRP >10 mg/L, are abnormally elevated depending upon age, severity and comorbid conditions such as chronic liver disease, coronary heart disease, diabetes(>400U/L), according to published research [17,72,[76], [77], [78]]. Notably, another meta-analysis study explored the 6 abnormal laboratory markers measured in long-term COVID-19. In 34% of patients, a chest X-ray/CT was abnormal. D-dimer (20%), NT-proBNP (11%), CRP (8%), serum ferritin (8%), procalcitonin (4%), and IL-6 (3%) were all observed to be increased [31].
Many mechanisms, including thrombosis, cause insufficient tissue perfusion, leading to elevated LDH in COVID-19 infection [79]. Furthermore, SARS CoV-2 infected patients' blood LDH was a good predictor of death [80,81]. Based on 75 clinical features (including blood marker concentrations), Yan and co-workers [82] developed an algorithm to predict mortality for 485 individuals with SARS CoV-2 infection. It was revealed that three indicators (blood-borne markers) could accurately predict a patient's death in more than 10 days, with an accuracy rate of more than 90% with elevated lymphocytes, LDH, and CRP. Simply measuring the blood level of LDH might predict COVID-19 mortality.
Although the LDH has been used as a biomarker of cardiac damage since the 1960s, it is important to note that aberrant levels can occur from multiple organ injury, infections, malignancies, MI, sepsis, or cardio-pulmonary defect mediated reduced oxygenation, and an increase in glycolysis [83]. However, according to the study's findings, elevated blood LDH levels were closely connected to a rise in the prevalence of organ failures such as acute kidney injury (AKI), ARDS, and septic shock, among other conditions, through reversible conversion of pyruvate to lactate, which is the final step of aerobic glycolysis. Notably, the progression of damage to the organs and death are associated with raised inflammatory cytokines such as IL-1beta and lactate elevation [84]. Furthermore, it is also common to measure CRP and lactic acid in the blood to detect inflammation. It has been observed that CRP and lactic acid levels are good markers of COVID-19 disease severity and that a higher CRP represents strongly the size of lung abscesses identified by CT [59,85,86]. It has been proven that people suffering from COVID-19 have an extremely high amount of the inflammatory cytokine IL-6, which increases the production of CRP and lactic acid, too [87].
Additionally, studies have shown that sepsis and circulatory shock are associated with an increase in lactic acid [88]; as known, serum lactate levels >4 mmol/L are associated with an increased risk of death in patients [84]. Albumin becomes less predictive of death when a severe inflammatory condition results in elevated lactic acid levels [89]. Moreover, both septic shock and hemorrhagic shock were associated with lactate deposition, and pyruvate levels rose during septic shock. Thus, cells must use glycolysis to produce ATP when OXPHOS is reduced due to hypoxia [90], showing that the raised lactate level was not due to anaerobic metabolism [91]. In other words, carbohydrate metabolic reprogramming of immune cells encourages illness by providing energy for the synthesis and secretion of pro-inflammatory molecules and increasing the amount of serum lactate in the circulation. As a result of lactate accumulation in the mice's bodies, they gained more weight and had a larger body fat percentage while having worse glucose tolerance and insulin sensitivity (Fig. 2 ).
Fig. 2.
Depicts SARS-CoV-2 Entry through Host ACE2 and Overexpressed Lectin Reg III beta Fusion Protein into Intestinal Tract.
5. Dysbiosis in long-term COVID-19 and severity biomarkers focusing towards LDH/lactate
Dysbiosis is produced by enteric pathogen infection, and inflammation profoundly affects the metabolomic profile. Understanding the changes in the gut environment produced by infection and inflammation will enable the development of novel treatment methods to tackle infectious pathogens. Recent research has employed NGS to elucidate the microbial diversity and habitat and the host's response to infection. The presence of gut-associated microflora concerning the lungs and an increase in the pathogenic bacterial community has been linked to worse outcomes in these immune-compromised individuals [[92], [93], [94]], implying that the microbiome may play a role in determining the effects of SARS CoV-2 infection.
COVID-19, H1N1 influenza patients, and healthy controls were compared for their gut microbiome. In COVID-19 patients, opportunistic fungal and bacterial pathogens (Aspergillus, Candida, Actinomyces, Streptococcus, Veillonella, Rothia, and Clostridium) were found to have displaced beneficial microbes/comments such as Bifidobacterium romboutsia, Proteobacteria, Collinsella, Actinobacteria Blautia, and Bacteroides [[95], [96], [97], [98], [99]]. Additional analysis indicated that SARS-CoV-2 assault altered the microbiota ecological environment of the COVID-19 patient population. Furthermore, in contrast to both healthy people and seasonal flu patients, COVID-19 patients have a notable uptick in levels of Lactobacillus, Bifidobacterium, Clostridium, Streptococcus, and a noticeable decrease in Coprococcus, Roseburia, Parabacteroides, Faecalibacterium, Bacteroidetes, and ratios of several other groups of bacteria [100]. There was an association between bacterial species and faecal viral load in SARS-CoV-2 patients during the COVID-19, hospitalization, and discharge. One research discovered a negative relationship between SARS-CoV-2 burden in the feces and the presence of Bacteroides microflora such as B. massiliensis, B. ovatus, B. thetaiotaomicron, and B. dorei in the gastrointestinal tract [[101], [102], [103]]. Notably, this 4 spp. of Bacteroides were linked with a decreased expression of ACE2 in rodents' large intestines [104]. Severe SARS-CoV-2 morbidity and death rates are greatest in older individuals and those with chronic illnesses linked with inflammatory response, such as high blood pressure, adiposity, diabetes, and coronary heart disease [[105], [106], [107]]. In addition, they have a smaller density of Bacteroides species versus nutritionally balanced persons, which is also remarkable [108,109].
COVID-19 participants were shown to have a high pro-inflammatory component IL-18, but not those with seasonal flu. COVID-19 patients with SARS-CoV-2 RNA had higher fecal IL-18 levels than COVID-19 patients who tested negative [100]. These findings suggest that changes in the intestinal microbiota's ecology may be directly related to the production of inflammatory cytokines in the intestinal lumen caused by SARS-CoV-2 and, perhaps, to the initiation of a cytokine storm. Apart from supplying the energy required for immunological systems to produce inflammatory molecules, aerobic glycolysis can result in a significant amount of lactate accumulations, complicating the therapy of COVID-19 patients.
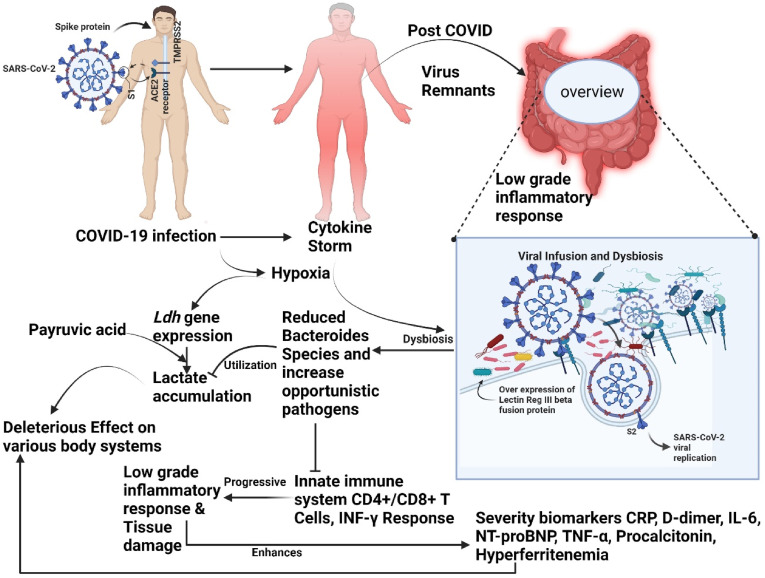
Intestine hypoxia-inducible factor 2alpha (HIF-2a) has been shown to positively regulate gut lactate via regulating the expression of the intestinal Ldha gene in the previous studies. An intestinal HIF-2a deficiency in rodents led to reduced lactate concentration that induces suppression of Bacteroides spp. growth instead of Ruminococcus torques population, Moreover, HIF signaling suppresses apoptosis of neutrophils, increasing their lifespan in inflamed tissue [110]. Although in long-term SARS CoV-2, persistent low-grade hypoxia stimulates the expression of Ldha gene-mediated LDH and maintains lactate levels. However, the pathogenic attachment with lectin RegIII beta expression and low-grade inflammation damage the Bacteroides spp. the population that leads to decline the growth of microbiome. Recent research published in cell metabolism revealed that B. vulgatus displayed a delayed proliferation rate and lesser maximal growth percentage in the cecal sample from Hif2a mutant mice.
Furthermore, from the same study, lactate boosted the expansion of B. vulgatus dose-dependently but did not affect the growth of R. torques at the same dosage. Consistent with this finding, the lactate levels in the total cecal content, ileum, of Hif2a knockout mice were lower [111]. However, hypoxia responses also occur when lactate is accumulated by glycolysis. LDH can maintain cellular homeostasis by balancing lactate secretion via pyruvate fermentation and a series of metabolic control [112]. Although Bacteroides spp. do not use lactate as a carbon source; they can degrade a wide variety of macromolecules (mostly polysaccharides), accelerating their replication. The primary data suggest that lactate may accelerate B. vulgatus growth by increasing polysaccharide consumption. However, pathogenic stimulation of bactericidal lectin RegIII beta lowers the Bacteroides spp.
On the other hand, abundance raises the amount of lactate in the host cell. If the inflamed gut accumulates carbohydrates and metabolites, this can also lead to the depletion of commensal bacteria, especially those of the Bacteroidetes phylum [113]. Considered collectively, our observation indicates that Bacteroides spp. of microbiota may have a therapeutic potential in fighting against SARS-CoV-2 attack by inhibiting host entrance via ACE2 and optimizing the immunological response of the hosts (Fig. 3 ).
Fig. 3.
Exhibits concept toward gut microbiota disruption in COVID-19 or Post-COVID illness association with severity biomarkers.
6. Gut microbiota diversity with CRP and other severity biomarkers in long-term COVID-19
Although high CRP levels indicate bacterial infection, they were determined to be inadequate to prove that viruses cause CRP levels to rise. SARS CoV-2 and post-COVID infection can result in various abnormalities, including changes in CRP and platelet counts resulting from inflammatory reactions and inhibition of bone marrow progenitors. CRP levels, in particular, are indicative of the acute phase reaction severity. Patients with pneumonia typically have significantly elevated CRP values, and elevated CRP levels have been demonstrated to be a powerful predictor of illness in general practice. CRP may be utilized as an inflammatory biomarker, allowing for the tracking of infection progression or treatment impact, and therefore several research on various sources of infection and CRP have been done to date. In the ongoing study, elevated CRP levels were related to a lengthier hospitalization. High CRP levels may be utilized as a predictor of intensive care unit admission or the need for mechanical breathing. Additionally, studies show a distinction between high- and low-risk individuals, which has significant clinical and therapeutic implications for patient management decision-making.
According to research, the gut microbiota is also linked to other unhealthy conditions such as IBD, autoimmune arthritis, atherosclerosis, adiposity, diabetes, inflammatory skin illnesses such as Atopic dermatitis and psoriasis. In individuals with inflammatory bowel disease (IBD) and obesity, for example, there is less bacterial diversity and lesser Bacteroides (Activate CD4 + T cells and reduces inflammation) [114,115]) and higher Firmicutes [116]. Firmicutes generate butyrate while Bacteroidetes produce acetate and propionate, according to MacFarlane & colleagues [117]. In the gastrointestinal tract, butyrate and other short-chain fatty acids (SCFAs) are considered to have a direct anti-inflammatory impact [116]. Through the generation of SCFAs acetate, propionate, and butyrate, several studies have demonstrated gastrointestinal regulatory T cells in limiting local inflammation. These SCFAs also inhibit neutrophil infiltration into tissues by activating the G protein-coupled receptor GPR43 on neutrophils, thus mitigating the inflammatory environment of the gut [118,119].
Additionally, the gut microbiota generates microbial signals necessary for the proper establishment and activation of the host's immune system. Clostridial species, and Bacteroides fragilis, can elicit Treg responses [[120], [121], [122], [123]] and T-cell-dependent and -independent immunoglobulin A reactions at the local level [[120], [121], [122]]. Additional antimicrobial peptides and secretory IgA antibodies help maintain the mucosal barrier [123,124]. However, obese persons have a greater Bacteroidetes/Firmicutes ratio, resulting in a higher butyrate production and a lower generation of propionate and acetate. Furthermore, obese subjects are more prone to develop COVID-19 infection due to higher insulin resistance, lower glucose tolerance, high adiposity, and low-grade inflammation [125].
Moreover, The Firmicutes/Bactroidetes ratio was 0.68 in mild, 0.65 moderates, and 0.58 in severe COVID-19, indicating that reduced Bacteroide species are responsible for declining beneficial functions in severe COVID-19 and long-term COVID patients [126]. Research revealed that serious COVID-19 participants were more prone to be male and had higher CRP serum concentrations than mild-to-moderate COVID-19 patients using Shannon's index diversity for CRP.
Furthermore, utilizing Shannon's index diversity, researchers found a negative connection between the severity of COVID-19 disease and relative microbiome abundance at the genus level [126]. Furthermore, dysbiosis due to declined Bacteroides spp. the population was positively correlated with D-dimer, neutrophils, IL-6, CRP, and LDH in acute and long-term COVID infection [127].
7. Approaches to rectify the COVID-19 and long COVID mediated consequences targeting microbiota
Recently, the scientific, health care, and public sectors have been paying more and more attention to probiotics and prebiotics. Furthermore, the public's perspective of microbial species has shifted from disease-causing entities to a more logical view that incorporates knowledge of microbes' positive functions in modern health. With these advancements, the public acceptability of prebiotics and probiotics started to climb. The sector grew at an anticipated 7%/year, and prebiotics grew at 12.7% within the upcoming eight years [[128], [129], [130]]. Probiotics and prebiotics have varied, heterogeneous, and frequently strain- and compound-specific mechanisms of action. Even though numerous instances have been presented, further research is needed, especially in correspondence descriptions of claimed health consequences and long-term influence [[131], [132], [133]]. Probiotic effector compounds are capable of direct interaction inside and activate a multitude of receptors on the intestinal epithelium, enteroendocrine, immunological systems, along with vagal afferent fibers. These signalings have an effect on the integrity of the intestinal barrier and inflammation (through TLR), as well as systemic consequences via host immunological, nervous systems, and hormonal messengers. It is also worth noting that certain probiotics can break down host compounds, such as bile salts and inhaled foreign chemicals, using enzymes. Most of these compounds are strain-specific and thus produce strain-specific effects [[133], [134], [135]].
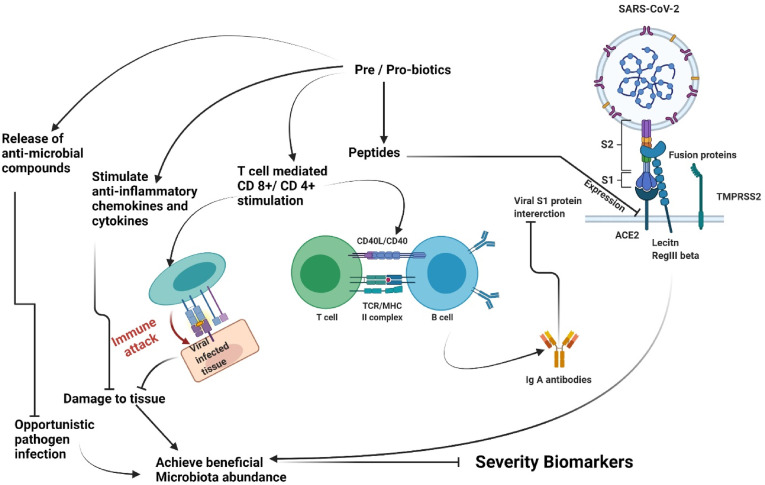
Probiotics have been hypothesized to have antiviral action via inhibiting viral entrance into host cells, secreting antiviral metabolites, and stimulating innate immunity in host cells [136]. Hosts and microbiomes interact via biochemical signaling pathways that are either incorporated in the cell structure or produced as metabolic products by probiotics [134]. It has been suggested that some strains of probiotics might connect to epithelial cells and compete with viruses for binding to cell receptors, limiting viral penetration into the cells [137]. A probiotic byproduct can modify the microflora populations via cross-feeding associations, alterations in the gut environment (— for instance, decreasing the pH), competition for micronutrients and co-receptors, and growth inhibition through the generation of strain-specific antibacterial chemicals such as bacteriocins. These microbiota-specific activities add to probiotics' potential to elicit therapeutic outcomes in conditions characterized by abnormal pathogenic expansion, including genital and oropharyngeal dysbioses [131]. Improving antiviral immune responses in the host, including expression of type 1 interferons and manipulation of inflammatory cytokines, maybe a plausible pathway for probiotics' preventive or curative benefits towards viral infections [[138], [139], [140], [141]]. The innate immune response is critical for viral infection protection. Type 1 IFNs, particularly IFN- -β and IFN- -α, represent key elements of the innate immunological mechanism to viral infection. Type 1 interferons exert their antiviral activity by binding to particular receptors and activating the JAK/STAT signaling pathways, therefore inducing the expression of hundreds of IFN-stimulated genes (ISGs) [142,143]. ISGs modify protein function to control all aspects of viral replication, including destroying viral genetic material and inhibiting translation [144,145]. As a supplement towards treatment, these antiviral probiotics may be an effective way to guard against viral infections.
Prebiotic effects are often mediated by particular microbiota groups consuming the substrate, boosting their development and metabolic activity. Additionally, providing nourishment to one or more groups of bacteria within the microbiome can indirectly impact other microbial species, promoting development via cross-feeding connections and limiting growth via pathogenic elimination. The modifications in microflora composition and metabolite levels caused by prebiotic administration affect host epithelial, nervous, endocrine, and immune signaling, thereby reconciling health benefits such as better immune response, bowel function, satiety, or appetite regulation, bone health, glucose, and lipid metabolism [146]. Byproducts of prebiotic biotransformation in microbes include the propionate, butyrate, acetate, and SCFAs, most of which have been demonstrated to communicate with these host systems and stimulate a range of prebiotic effects (Fig. 4 ).
Fig. 4.
Possible role of pre/pro-biotics in Re-establishment of beneficial gut microbiota abundance in COVID-19 illness.
The disruption caused by influenza and COVID-19 has imposed enormous healthcare, social, and economic costs during the previous decade. When a novel mutant virus appears and an effective vaccine is developed and tested, there may be a window of opportunity for interventions with low-cost and high preventative medications with comprehensive immunoregulatory, anti-inflammatory, and antiviral characteristics. For COVID-19 infection prevention and treatment approaches, prebiotics and probiotics have been proposed as possible ingredients [[147], [148], [149]]. In addition to their ability to alter immunological responses, probiotics are also renowned for their ability to reduce inflammation [150], were found to be effective in preventing illnesses in the past [151], and lowering the likelihood of ventilator-associated pneumonia [149]. One study demonstrated that adding a multi-strain probiotic formulation to conventional treatment decreased morbidity and death in a small cohort of hospitalized COVID-19 patients [152]. As of now, there is no clinical evidence to support probiotics and prebiotics as therapy or prophylactic for severe viral infections. However, probiotics and prebiotics might be investigated in the future in order to determine their efficiency and safety as additional therapies.
8. Conclusion
Numerous studies on nutrition and treatments targeting the microbiome have extended the disciplines of probiotics and prebiotics and many other related interventions. Although a gut will likely continue to be the epicenter of these therapeutics, clinically established applications in the respiratory system, cardiometabolic system, immunological system, oral cavity, skin, neurological system, urogenital tract, and weight-management fields will likely expand. These findings, along with increased involvement in large interventional and population-based research, will provide novel strategies for increasing dietary relevance and therapeutic efficacy and targeting and customizing these treatment approaches to individuals' biology and microbiome. Further data from prospective trials are required to comprehend the pathophysiology of COVID-19 illness and identify the long-term COVID-19 symptom. Symptoms, indicators, and biomarkers of COVID-19 should be known to clinicians to evaluate, diagnose, and stop long-term COVID-19 incidence, eliminate the danger of chronic consequences, and aid patients in regaining their pre-COVID-19 health as quickly as possible. A better understanding of these consequences is required to design innovative, customized cross-sectoral therapies in long-term COVID-19 centers.
Funding
The authors would like to thank the Deanship of Scientific Research at Umm Al-Qura University for supporting this work by Grant Code-224UQU4310387DSR03.
Data and material availability
No datasets were generated or analyzed during the current study.
Ethical approval
Not Applicable.
Consent to participate
Not Applicable.
Consent to publish
Not Applicable.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.The L. Emerging understandings of 2019-nCoV. Lancet. 2020;395:311. doi: 10.1016/S0140-6736(20)30186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh Y., Gupta G., Mishra A., Chellappan D.K., Dua K. Gender and age differences reveal risk patterns in COVID-19 outbreak. Alternative Ther. Health Med. 2020;26:54–55. [PubMed] [Google Scholar]
- 3.Singh Y., Gupta G., Satija S., Negi P., Chellappan D.K., Dua K. RAAS blockers in hypertension posing a higher risk toward the COVID-19. Dermatol. Ther. 2020;33 doi: 10.1111/dth.13501. e13501-e13501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh Y., Ali H., Alharbi K.S., Almalki W.H., Kazmi I., Al-Abbasi F.A., Anand K., Dureja H., Singh S.K., Thangavelu L., Chellappan D.K., Dua K., Gupta G. Drug Dev Res.; 2021. Calcium Sensing Receptor Hyperactivation through Viral Envelop Protein E of SARS CoV2: A Novel Target for Cardio-Renal Damage in COVID-19 Infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh Y., Gupta G., Anand K., Kumar Jha N., Thangavelu L., Kumar Chellappan D., Dua K. Molecular exploration of combinational therapy of orlistat with metformin prevents the COVID-19 consequences in obese diabetic patients. Eur. Rev. Med. Pharmacol. Sci. 2021;25:580–582. doi: 10.26355/eurrev_202101_24614. [DOI] [PubMed] [Google Scholar]
- 6.Crisan-Dabija R., Grigorescu C., Pavel C.A., Artene B., Popa I.V., Cernomaz A., Burlacu A. Tuberculosis and COVID-19: lessons from the past viral outbreaks and possible future outcomes. Cancer Res. J. 2020;2020:1401053. doi: 10.1155/2020/1401053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouazza B., Hadj-Said D., Pescatore K.A., Chahed R. Are patients with asthma and chronic obstructive pulmonary disease preferred targets of COVID-19? Tuberc. Respir. Dis. 2021;84:22–34. doi: 10.4046/trd.2020.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phua J., Weng L., Ling L., Egi M., Lim C.-M., Divatia J.V., Shrestha B.R., Arabi Y.M., Ng J., Gomersall C.D., Nishimura M., Koh Y., Du B. Intensive care management of coronavirus disease 2019 (COVID-19): challenges and recommendations. Lancet Respir. Med. 2020;8:506–517. doi: 10.1016/S2213-2600(20)30161-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.R. Laxminarayan, C.M. B, V.T. G, K.V. Arjun Kumar, B. Wahl, J.A. Lewnard, SARS-CoV-2 infection and mortality during the first epidemic wave in Madurai, south India: a prospective, active surveillance study, Lancet Infect. Dis.. [DOI] [PMC free article] [PubMed]
- 10.Morens D.M., Fauci A.S. The 1918 influenza pandemic: insights for the 21st century. J. Infect. Dis. 2007;195:1018–1028. doi: 10.1086/511989. [DOI] [PubMed] [Google Scholar]
- 11.Shieh W.J., Blau D.M., Denison A.M., Deleon-Carnes M., Adem P., Bhatnagar J., Sumner J., Liu L., Patel M., Batten B., Greer P., Jones T., Smith C., Bartlett J., Montague J., White E., Rollin D., Gao R., Seales C., Jost H., Metcalfe M., Goldsmith C.S., Humphrey C., Schmitz A., Drew C., Paddock C., Uyeki T.M., Zaki S.R. pandemic influenza A (H1N1): pathology and pathogenesis of 100 fatal cases in the United States. Am. J. Pathol. 2009;177:166–175. doi: 10.2353/ajpath.2010.100115. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar D., Gupta G., Jhamb U. IDDF2019-ABS-0014 Gastrointestinal symptoms among hospitalized children admitted with h1n1 infection: a report from a tertiary hospital in north India. Gut. 2019;68 A61-A61. [Google Scholar]
- 13.Zhang D., Li S., Wang N., Tan H.Y., Zhang Z., Feng Y. The cross-talk between gut microbiota and lungs in common lung diseases. Front. Microbiol. 2020;11:301. doi: 10.3389/fmicb.2020.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar Gothwal S., Singh V.B., Shrivastava M., Banseria R., Goyal K., Singh S., Ali H., Saad Alharbi K., Singh Y., Gupta G. Complete blood-count-based inflammatory score (CBCS) of COVID-19 patients at tertiary care center. Alternative Ther. Health Med. 2021;27:18–24. [PubMed] [Google Scholar]
- 15.G. Gupta, R. S., Y. Singh, L. Thangavelu, S.K. Singh, H. Dureja, D.K. Chellappan, K. Dua, Emerging Cases of Mucormycosis under COVID-19 Pandemic in India: Misuse of Antibiotics, Drug Dev Res., (n/a). [DOI] [PMC free article] [PubMed]
- 16.Moore J.B., June C.H. Cytokine release syndrome in severe COVID-19. Science (New York, N.Y.) 2020;368:473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 17.Singh Y., Gupta G., Kazmi I., Al-Abbasi F.A., Negi P., Chellappan D.K., Dua K. SARS CoV-2 aggravates cellular metabolism mediated complications in COVID-19 infection. Dermatol. Ther. 2020;33 doi: 10.1111/dth.13871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kernan K.F., Carcillo J.A. Hyperferritinemia and inflammation. Int. Immunol. 2017;29:401–409. doi: 10.1093/intimm/dxx031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh Y., Fuloria N.K., Fuloria S., Subramaniyan V., Almalki W.H., Gupta G., Shaikh M.A.J., Singh M., Al-Abbasi F.A., Kazmi I. Drug Dev Res.; 2021. Disruption of the Biological Activity of Protease-Activated Receptors2/4 in Adults rather than Children in SARS CoV-2 Virus-Mediated Mortality in COVID-19 Infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta G., Singh Y., Chellappan D., Dua K. Emerging dermatological symptoms in coronavirus pandemic. J. Cosmet. Dermatol. 2020;19:2447–2448. doi: 10.1111/jocd.13466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheung K.S., Hung I.F.N., Chan P.P.Y., Lung K.C., Tso E., Liu R., Ng Y.Y., Chu M.Y., Chung T.W.H., Tam A.R., Yip C.C.Y., Leung K.-H., Fung A.Y.-F., Zhang R.R., Lin Y., Cheng H.M., Zhang A.J.X., To K.K.W., Chan K.-H., Yuen K.-Y., Leung W.K. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159:81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Din A.U., Mazhar M., Waseem M., Ahmad W., Bibi A., Hassan A., Ali N., Gang W., Qian G., Ullah R., Shah T., Ullah M., Khan I., Nisar M.F., Wu J. SARS-CoV-2 microbiome dysbiosis linked disorders and possible probiotics role. Biomed. Pharmacother. 2021;133:110947. doi: 10.1016/j.biopha.2020.110947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banseria R., Shrivastava M., Meena H., Kumar Gothwal S., Meratwal G., Singh V.B., Singh Y., Kazmi I., Al-Abbasi F.A., Almalki Z.S., Khan M.F. Altern Ther Health Med; 2021. Changes in Hematological Parameters by Quantifying HRCT Chest Results in Patients with COVID-19 in Tertiary Care Hospital. [PubMed] [Google Scholar]
- 25.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubin R. As their numbers grow, COVID-19 "long haulers" stump experts. JAMA. 2020;324:1381–1383. doi: 10.1001/jama.2020.17709. [DOI] [PubMed] [Google Scholar]
- 27.Greenhalgh T., Knight M., A'Court C., Buxton M., Husain L. Management of post-acute covid-19 in primary care. BMJ. 2020;370:m3026. doi: 10.1136/bmj.m3026. [DOI] [PubMed] [Google Scholar]
- 28.Del Rio C., Malani P.N. COVID-19-New insights on a rapidly changing epidemic. JAMA. 2020;323:1339–1340. doi: 10.1001/jama.2020.3072. [DOI] [PubMed] [Google Scholar]
- 29.Greenhalgh T., Knight M., A'Court C., Buxton M., Husain L. Management of post-acute covid-19 in primary care. BMJ. 2020;370:m3026. doi: 10.1136/bmj.m3026. [DOI] [PubMed] [Google Scholar]
- 30.Al-Aly Z., Xie Y., Bowe B. High-dimensional characterization of post-acute sequelae of COVID-19. Nature. 2021;594:259–264. doi: 10.1038/s41586-021-03553-9. [DOI] [PubMed] [Google Scholar]
- 31.Lopez-Leon S., Wegman-Ostrosky T., Perelman C., Sepulveda R., Rebolledo P.A., Cuapio A., Villapol S. More than 50 long-term effects of COVID-19: a systematic review and meta-analysis. Sci. Rep. 2021;11:16144. doi: 10.1038/s41598-021-95565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.India T.o. Time of India; India: Sep 10, 2021. Covishield Vaccine Side-Effects: 4 New Side-Effects Discovered with the Astrazeneca COVID Shot. [Google Scholar]
- 33.Salyers A.A. Bacteroides of the human lower intestinal tract. Annu. Rev. Microbiol. 1984;38:293–313. doi: 10.1146/annurev.mi.38.100184.001453. [DOI] [PubMed] [Google Scholar]
- 34.Bäckhed F., Ley R.E., Sonnenburg J.L., Peterson D.A., Gordon J.I. Host-bacterial mutualism in the human intestine. Science (New York, N.Y.) 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 35.Schirmer M., Smeekens S.P., Vlamakis H., Jaeger M., Oosting M., Franzosa E.A., Ter Horst R., Jansen T., Jacobs L., Bonder M.J., Kurilshikov A., Fu J., Joosten L.A.B., Zhernakova A., Huttenhower C., Wijmenga C., Netea M.G., Xavier R.J. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell. 2016;167:1125–1136. doi: 10.1016/j.cell.2016.10.020. e1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belkaid Y., Hand T.W. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schirmer M., Kumar V., Netea M.G., Xavier R.J. The causes and consequences of variation in human cytokine production in health. Curr. Opin. Immunol. 2018;54:50–58. doi: 10.1016/j.coi.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 38.Sánchez E., Laparra J.M., Sanz Y. Discerning the role of Bacteroides fragilis in celiac disease pathogenesis. Appl. Environ. Microbiol. 2012;78:6507–6515. doi: 10.1128/AEM.00563-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hooper L.V., Midtvedt T., Gordon J.I. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 40.Wexler H.M. Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007;20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crouch L.I., Liberato M.V., Urbanowicz P.A., Baslé A., Lamb C.A., Stewart C.J., Cooke K., Doona M., Needham S., Brady R.R., Berrington J.E., Madunic K., Wuhrer M., Chater P., Pearson J.P., Glowacki R., Martens E.C., Zhang F., Linhardt R.J., Spencer D.I.R., Bolam D.N. Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown. Nat. Commun. 2020;11:4017. doi: 10.1038/s41467-020-17847-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eckburg P.B., Bik E.M., Bernstein C.N., Purdom E., Dethlefsen L., Sargent M., Gill S.R., Nelson K.E., Relman D.A. Diversity of the human intestinal microbial flora. Science (New York, N.Y.) 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brook I., Frazier E.H. Aerobic and anaerobic microbiology in intra-abdominal infections associated with diverticulitis. J. Med. Microbiol. 2000;49:827–830. doi: 10.1099/0022-1317-49-9-827. [DOI] [PubMed] [Google Scholar]
- 44.Sóki J., Gal M., Brazier J.S., Rotimi V.O., Urbán E., Nagy E., Duerden B.I. Molecular investigation of genetic elements contributing to metronidazole resistance in Bacteroides strains. J. Antimicrob. Chemother. 2006;57:212–220. doi: 10.1093/jac/dki443. [DOI] [PubMed] [Google Scholar]
- 45.Zafar H., Saier M.H., Jr. Comparative genomics of transport proteins in seven Bacteroides species. PLoS One. 2018;13 doi: 10.1371/journal.pone.0208151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stelter C., Käppeli R., König C., Krah A., Hardt W.D., Stecher B., Bumann D. Salmonella-induced mucosal lectin RegIIIβ kills competing gut microbiota. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miki T., Okada N., Hardt W.-D. Inflammatory bactericidal lectin RegIIIβ: friend or foe for the host? Gut Microb. 2018;9:179–187. doi: 10.1080/19490976.2017.1387344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diard M., Hardt W.D. Evolution of bacterial virulence. FEMS Microbiol. Rev. 2017;41:679–697. doi: 10.1093/femsre/fux023. [DOI] [PubMed] [Google Scholar]
- 49.Miki T., Goto R., Fujimoto M., Okada N., Hardt W.D. The bactericidal lectin RegIIIβ prolongs gut colonization and enteropathy in the streptomycin mouse model for Salmonella diarrhea. Cell Host Microbe. 2017;21:195–207. doi: 10.1016/j.chom.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 50.Ichinohe T., Pang I.K., Kumamoto Y., Peaper D.R., Ho J.H., Murray T.S., Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. U. S. A. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ichinohe T., Lee H.K., Ogura Y., Flavell R., Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J. Exp. Med. 2009;206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kennedy E.A., King K.Y., Baldridge M.T. Mouse microbiota models: comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front. Physiol. 2018;9:1534. doi: 10.3389/fphys.2018.01534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pfeiffer J.K., Sonnenburg J.L. The intestinal microbiota and viral susceptibility. Front. Microbiol. 2011;2:92. doi: 10.3389/fmicb.2011.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCormick A.L., Mocarski E.S. In: Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Arvin A., Roizman B., Mocarski E., Campadelli-Fiume G., Yamanishi K., Moore P.S., Whitley R., editors. Cambridge University Press; Cambridge: 2007. CMV modulation of the host response to infection; pp. 324–338. [PubMed] [Google Scholar]
- 55.Zafar H., Saier M.H. Gut Bacteroides species in health and disease. Gut Microb. 2021;13:1848158. doi: 10.1080/19490976.2020.1848158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J., Li Q., Yin Y., Zhang Y., Cao Y., Lin X., Huang L., Hoffmann D., Lu M., Qiu Y. Excessive neutrophils and neutrophil extracellular traps in COVID-19. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.02063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reusch N., De Domenico E., Bonaguro L., Schulte-Schrepping J., Baßler K., Schultze J.L., Aschenbrenner A.C. Neutrophils in COVID-19. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.652470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Catanzaro M., Fagiani F., Racchi M., Corsini E., Govoni S., Lanni C. Immune response in COVID-19: addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Transduct. Targeted Therapy. 2020;5:84. doi: 10.1038/s41392-020-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan C., Huang Y., Shi F., Tan K., Ma Q., Chen Y., Jiang X., Li X. C-reactive protein correlates with computed tomographic findings and predicts severe COVID-19 early. J. Med. Virol. 2020;92:856–862. doi: 10.1002/jmv.25871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh Y., Fuloria N.K., Fuloria S., Subramaniyan V., Meenakshi D.U., Chakravarthi S., Kumari U., Joshi N., Gupta G. N-terminal domain of SARS CoV-2 spike protein mutation associated reduction in effectivity of neutralizing antibody with vaccinated individuals. J. Med. Virol. 2021;93:5726–5728. doi: 10.1002/jmv.27181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh Y., Gupta G., Satija S., Pabreja K., Chellappan D.K., Dua K. COVID-19 transmission through host cell directed network of GPCR. Drug Dev. Res. 2020;81:647–649. doi: 10.1002/ddr.21674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gustine J.N., Jones D. Immunopathology of hyperinflammation in COVID-19. Am. J. Pathol. 2021;191:4–17. doi: 10.1016/j.ajpath.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amraei R., Yin W., Napoleon M.A., Suder E.L., Berrigan J., Zhao Q., Olejnik J., Chandler K.B., Xia C., Feldman J., Hauser B.M., Caradonna T.M., Schmidt A.G., Gummuluru S., Muhlberger E., Chitalia V., Costello C.E., Rahimi N. 2021. CD209L/L-SIGN and CD209/DC-SIGN Act as Receptors for SARS-CoV-2, bioRxiv : the Preprint Server for Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soh W.T., Liu Y., Nakayama E.E., Ono C., Torii S., Nakagami H., Matsuura Y., Shioda T., Arase H. 2020. The N-Terminal Domain of Spike Glycoprotein Mediates SARS-CoV-2 Infection by Associating with L-SIGN and DC-SIGN, bioRxiv : the Preprint Server for Biology. 2020.2011.2005.369264. [Google Scholar]
- 65.Wang K., Chen W., Zhang Z., Deng Y., Lian J.Q., Du P., Wei D., Zhang Y., Sun X.X., Gong L., Yang X., He L., Zhang L., Yang Z., Geng J.J., Chen R., Zhang H., Wang B., Zhu Y.M., Nan G., Jiang J.L., Li L., Wu J., Lin P., Huang W., Xie L., Zheng Z.H., Zhang K., Miao J.L., Cui H.Y., Huang M., Zhang J., Fu L., Yang X.M., Zhao Z., Sun S., Gu H., Wang Z., Wang C.F., Lu Y., Liu Y.Y., Wang Q.Y., Bian H., Zhu P., Chen Z.N. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct Target Ther. 2020;5:283. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., Smura T., Levanov L., Szirovicza L., Tobi A., Kallio-Kokko H., Österlund P., Joensuu M., Meunier F.A., Butcher S.J., Winkler M.S., Mollenhauer B., Helenius A., Gokce O., Teesalu T., Hepojoki J., Vapalahti O., Stadelmann C., Balistreri G., Simons M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science (New York, N.Y.) 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N., Thacker B.E., Glass C.A., Yang Z., Torres J.L., Golden G.J., Bartels P.L., Porell R.N., Garretson A.F., Laubach L., Feldman J., Yin X., Pu Y., Hauser B.M., Caradonna T.M., Kellman B.P., Martino C., Gordts P.L.S.M., Chanda S.K., Schmidt A.G., Godula K., Leibel S.L., Jose J., Corbett K.D., Ward A.B., Carlin A.F., Esko J.D. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183:1043–1057. doi: 10.1016/j.cell.2020.09.033. e1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lempp F.A., Soriaga L., Montiel-Ruiz M., Benigni F., Noack J., Park Y.-J., Bianchi S., Walls A.C., Bowen J.E., Zhou J., Kaiser H., Joshi A., Agostini M., Meury M., Dellota E., Jaconi S., Cameroni E., Martinez-Picado J., Vergara-Alert J., Izquierdo-Useros N., Virgin H.W., Lanzavecchia A., Veesler D., Purcell L., Telenti A., Corti D. Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies. Nature. 2021 doi: 10.1038/s41586-021-03925-1. [DOI] [PubMed] [Google Scholar]
- 69.Wang T.T., Nestel F.P., Bourdeau V., Nagai Y., Wang Q., Liao J., Tavera-Mendoza L., Lin R., Hanrahan J.W., Mader S., White J.H. Cutting edge: 1,25-dihydroxyvitamin D3 is a direct inducer of antimicrobial peptide gene expression. J. Immunol. 2004;173:2909–2912. doi: 10.4049/jimmunol.173.5.2909. [DOI] [PubMed] [Google Scholar]
- 70.Yan H., Liang X., Du J., He Z., Wang Y., Lyu M., Yue L., Zhang F., Xue Z., Xu L., Ruan G., Li J., Zhu H., Xu J., Chen S., Zhang C., Lv D., Lin Z., Shen B., Zhu Y., Qian B., Chen H., Guo T. medRxiv; 2021. Proteomic and Metabolomic Investigation of COVID-19 Patients with Elevated Serum Lactate Dehydrogenase. 2021.2001.2010.21249333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.AbdelMassih A.F., Kamel A., Mishriky F., Ismail H.-A., El Qadi L., Malak L., El-Husseiny M., Ashraf M., Hafez N., AlShehry N., El-Husseiny N., AbdelRaouf N., Shebl N., Hafez N., Youssef N., Afdal P., Hozaien R., Menshawey R., Saeed R., Fouda R. Is it infection or rather vascular inflammation? Game-changer insights and recommendations from patterns of multi-organ involvement and affected subgroups in COVID-19. Cardiovasc. Endocrinol. Metabol. 2020;9:110–120. doi: 10.1097/XCE.0000000000000211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Magro B., Zuccaro V., Novelli L., Zileri L., Celsa C., Raimondi F., Gori M., Cammà G., Battaglia S., Genova V.G., Paris L., Tacelli M., Mancarella F.A., Enea M., Attanasio M., Senni M., Di Marco F., Lorini L.F., Fagiuoli S., Bruno R., Cammà C., Gasbarrini A. Predicting in-hospital mortality from Coronavirus Disease 2019: a simple validated app for clinical use. PLoS One. 2021;16 doi: 10.1371/journal.pone.0245281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Terpos E., Ntanasis-Stathopoulos I., Elalamy I., Kastritis E., Sergentanis T.N., Politou M., Psaltopoulou T., Gerotziafas G., Dimopoulos M.A. Hematological findings and complications of COVID-19. Am. J. Hematol. 2020;95:834–847. doi: 10.1002/ajh.25829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tang N., Li D., Wang X., Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemostasis. 2020;18:844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cummings M.J., Baldwin M.R., Abrams D., Jacobson S.D., Meyer B.J., Balough E.M., Aaron J.G., Claassen J., Rabbani L.E., Hastie J., Hochman B.R., Salazar-Schicchi J., Yip N.H., Brodie D., O'Donnell M.R. Epidemiology, clinical course, and outcomes of critically ill adults with COVID-19 in New York City: a prospective cohort study. Lancet. 2020;395:1763–1770. doi: 10.1016/S0140-6736(20)31189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang H., Cai S., Li Y., Li Y., Fan Y., Li L., Lei C., Tang X., Hu F., Li F., Deng X. Prognostic factors for COVID-19 pneumonia progression to severe symptoms based on earlier clinical features: a retrospective analysis. Front. Med. 2020;7 doi: 10.3389/fmed.2020.557453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schalekamp S., Huisman M., Dijk R.A.v., Boomsma M.F., Jorge P.J.F., Boer W.S.d., Herder G.J.M., Bonarius M., Groot O.A., Jong E., Schreuder A., Schaefer-Prokop C.M. Model-based prediction of critical illness in hospitalized patients with COVID-19. Radiology. 2021;298:E46–E54. doi: 10.1148/radiol.2020202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang I., Pranata R., Lim M.A., Oehadian A., Alisjahbana B. C-reactive protein, procalcitonin, D-dimer, and ferritin in severe coronavirus disease-2019: a meta-analysis. Ther. Adv. Respir. Dis. 2020;14 doi: 10.1177/1753466620937175. 1753466620937175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lim M.A., Pranata R., Huang I., Yonas E., Soeroto A.Y., Supriyadi R. Multiorgan failure with emphasis on acute kidney injury and severity of COVID-19: systematic review and meta-analysis. Can. J. Kidney Health Dis. 2020;7 doi: 10.1177/2054358120938573. 2054358120938573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yan L., Zhang H.-T., Goncalves J., Xiao Y., Wang M., Guo Y., Sun C., Tang X., Jing L., Zhang M., Huang X., Xiao Y., Cao H., Chen Y., Ren T., Wang F., Xiao Y., Huang S., Tan X., Huang N., Jiao B., Cheng C., Zhang Y., Luo A., Mombaerts L., Jin J., Cao Z., Li S., Xu H., Yuan Y. An interpretable mortality prediction model for COVID-19 patients. Nat. Mach. Intell. 2020;2(2):283–288. [Google Scholar]
- 83.Szarpak L., Ruetzler K., Safiejko K., Hampel M., Pruc M., Kanczuga-Koda L., Filipiak K.J., Jaguszewski M.J. Lactate dehydrogenase level as a COVID-19 severity marker. Am. J. Emerg. Med. 2021;45:638–639. doi: 10.1016/j.ajem.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lu J., Wei Z., Jiang H., Cheng L., Chen Q., Chen M., Yan J., Sun Z. Lactate dehydrogenase is associated with 28-day mortality in patients with sepsis: a retrospective observational study. J. Surg. Res. 2018;228:314–321. doi: 10.1016/j.jss.2018.03.035. [DOI] [PubMed] [Google Scholar]
- 85.Wang L. C-reactive protein levels in the early stage of COVID-19. Med. Maladies Infect. 2020;50:332–334. doi: 10.1016/j.medmal.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peng Y.D., Meng K., Guan H.Q., Leng L., Zhu R.R., Wang B.Y., He M.A., Cheng L.X., Huang K., Zeng Q.T. [Clinical characteristics and outcomes of 112 cardiovascular disease patients infected by 2019-nCoV] Zhonghua Xinxueguanbing Zazhi. 2020;48:450–455. doi: 10.3760/cma.j.cn112148-20200220-00105. [DOI] [PubMed] [Google Scholar]
- 87.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang X., Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lichtenauer M., Wernly B., Ohnewein B., Franz M., Kabisch B., Muessig J., Masyuk M., Lauten A., Schulze P.C., Hoppe U.C., Kelm M., Jung C. The lactate/albumin ratio: a valuable tool for risk stratification in septic patients admitted to ICU. Int. J. Mol. Sci. 2017;18:1893. doi: 10.3390/ijms18091893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Booth A.L., Abels E., McCaffrey P. Development of a prognostic model for mortality in COVID-19 infection using machine learning. Mod. Pathol. 2021;34:522–531. doi: 10.1038/s41379-020-00700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Corcoran S.E., O'Neill L.A. HIF1α and metabolic reprogramming in inflammation. J. Clin. Invest. 2016;126:3699–3707. doi: 10.1172/JCI84431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Douglas J.J., Walley K.R. Metabolic changes in cardiomyocytes during sepsis. Crit. Care. 2013;17:186. doi: 10.1186/cc13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kitsios G.D., Yang H., Yang L., Qin S., Fitch A., Wang X.H., Fair K., Evankovich J., Bain W., Shah F., Li K., Methé B., Benos P.V., Morris A., McVerry B.J. Respiratory tract dysbiosis is associated with worse outcomes in mechanically ventilated patients. Am. J. Respir. Crit. Care Med. 2020;202:1666–1677. doi: 10.1164/rccm.201912-2441OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dickson R.P., Singer B.H., Newstead M.W., Falkowski N.R., Erb-Downward J.R., Standiford T.J., Huffnagle G.B. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat. Microbiol. 2016;1:16113. doi: 10.1038/nmicrobiol.2016.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dickson R.P., Schultz M.J., van der Poll T., Schouten L.R., Falkowski N.R., Luth J.E., Sjoding M.W., Brown C.A., Chanderraj R., Huffnagle G.B., Bos L.D.J. Lung microbiota predict clinical outcomes in critically ill patients. Am. J. Respir. Crit. Care Med. 2020;201:555–563. doi: 10.1164/rccm.201907-1487OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen X., Liao B., Cheng L., Peng X., Xu X., Li Y., Hu T., Li J., Zhou X., Ren B. The microbial coinfection in COVID-19. Appl. Microbiol. Biotechnol. 2020;104:7777–7785. doi: 10.1007/s00253-020-10814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zuo T., Zhang F., Lui G.C.Y., Yeoh Y.K., Li A.Y.L., Zhan H., Wan Y., Chung A.C.K., Cheung C.P., Chen N., Lai C.K.C., Chen Z., Tso E.Y.K., Fung K.S.C., Chan V., Ling L., Joynt G., Hui D.S.C., Chan F.K.L., Chan P.K.S., Ng S.C. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159:944–955. doi: 10.1053/j.gastro.2020.05.048. e948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ferreira C., Viana S.D., Reis F. Gut microbiota dysbiosis-immune hyperresponse-inflammation triad in coronavirus disease 2019 (COVID-19): impact of pharmacological and nutraceutical approaches. Microorganisms. 2020;8:1514. doi: 10.3390/microorganisms8101514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gu S., Chen Y., Wu Z., Chen Y., Gao H., Lv L., Guo F., Zhang X., Luo R., Huang C., Lu H., Zheng B., Zhang J., Yan R., Zhang H., Jiang H., Xu Q., Guo J., Gong Y., Tang L., Li L. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin. Infect. Dis. 2020;71:2669–2678. doi: 10.1093/cid/ciaa709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zuo T., Zhan H., Zhang F., Liu Q., Tso E.Y.K., Lui G.C.Y., Chen N., Li A., Lu W., Chan F.K.L., Chan P.K.S., Ng S.C. Alterations in fecal fungal microbiome of patients with COVID-19 during time of hospitalization until discharge. Gastroenterology. 2020;159:1302–1310. doi: 10.1053/j.gastro.2020.06.048. e1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tao W., Zhang G., Wang X., Guo M., Zeng W., Xu Z., Cao D., Pan A., Wang Y., Zhang K., Ma X., Chen Z., Jin T., Liu L., Weng J., Zhu S. Analysis of the intestinal microbiota in COVID-19 patients and its correlation with the inflammatory factor IL-18. Med. Microecol. 2020;5:100023. doi: 10.1016/j.medmic.2020.100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Forrester J.D., Spain D.A. Clostridium ramosum bacteremia: case report and literature review. Surg. Infect. 2014;15:343–346. doi: 10.1089/sur.2012.240. [DOI] [PubMed] [Google Scholar]
- 102.Yang T., Chakraborty S., Saha P., Mell B., Cheng X., Yeo J.Y., Mei X., Zhou G., Mandal J., Golonka R., Yeoh B.S., Putluri V., Piyarathna D.W.B., Putluri N., McCarthy C.G., Wenceslau C.F., Sreekumar A., Gewirtz A.T., Vijay-Kumar M., Joe B. Gnotobiotic rats reveal that gut microbiota regulates colonic mRNA of Ace2, the receptor for SARS-CoV-2 infectivity. Hypertension. 2020;76:e1–e3. doi: 10.1161/HYPERTENSIONAHA.120.15360. Dallas, Tex. : 1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Elsayed S., Zhang K. Human infection caused by Clostridium hathewayi. Emerg. Infect. Dis. 2004;10:1950–1952. doi: 10.3201/eid1011.040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Geva-Zatorsky N., Sefik E., Kua L., Pasman L., Tan T.G., Ortiz-Lopez A., Yanortsang T.B., Yang L., Jupp R., Mathis D., Benoist C., Kasper D.L. Mining the human gut microbiota for immunomodulatory organisms. Cell. 2017;168:928–943. doi: 10.1016/j.cell.2017.01.022. e911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hill M.A., Mantzoros C., Sowers J.R. Commentary: COVID-19 in patients with diabetes. Metabolism. 2020;107 doi: 10.1016/j.metabol.2020.154217. 154217-154217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fang L., Karakiulakis G., Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir. Med. 2020;8:e21. doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cai Q., Chen F., Wang T., Luo F., Liu X., Wu Q., He Q., Wang Z., Liu Y., Liu L., Chen J., Xu L. Obesity and COVID-19 severity in a designated hospital in shenzhen, China. Diabetes Care. 2020;43:1392–1398. doi: 10.2337/dc20-0576. [DOI] [PubMed] [Google Scholar]
- 108.Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 109.Emoto T., Yamashita T., Sasaki N., Hirota Y., Hayashi T., So A., Kasahara K., Yodoi K., Matsumoto T., Mizoguchi T., Ogawa W., Hirata K.-i. J Atheroscler Thromb.; advpub: 2016. Analysis of Gut Microbiota in Coronary Artery Disease Patients: a Possible Link between Gut Microbiota and Coronary Artery Disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Walmsley S.R., Print C., Farahi N., Peyssonnaux C., Johnson R.S., Cramer T., Sobolewski A., Condliffe A.M., Cowburn A.S., Johnson N., Chilvers E.R. Hypoxia-induced neutrophil survival is mediated by HIF-1α–dependent NF-κB activity. J. Exp. Med. 2005;201:105–115. doi: 10.1084/jem.20040624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu Q., Liang X., Wang K., Lin J., Wang X., Wang P., Zhang Y., Nie Q., Liu H., Zhang Z., Liu J., Pang Y., Jiang C. Cell Metab; 2021. Intestinal Hypoxia-Inducible Factor 2α Regulates Lactate Levels to Shape the Gut Microbiome and Alter Thermogenesis. [DOI] [PubMed] [Google Scholar]
- 112.Lee D.C., Sohn H.A., Park Z.Y., Oh S., Kang Y.K., Lee K.M., Kang M., Jang Y.J., Yang S.J., Hong Y.K., Noh H., Kim J.A., Kim D.J., Bae K.H., Kim D.M., Chung S.J., Yoo H.S., Yu D.Y., Park K.C., Yeom Y.I. A lactate-induced response to hypoxia. Cell. 2015;161:595–609. doi: 10.1016/j.cell.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 113.Zeng M.Y., Inohara N., Nuñez G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017;10:18–26. doi: 10.1038/mi.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lucke K., Miehlke S., Jacobs E., Schuppler M. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J. Med. Microbiol. 2006;55:617–624. doi: 10.1099/jmm.0.46198-0. [DOI] [PubMed] [Google Scholar]
- 115.Prindiville T.P., Sheikh R.A., Cohen S.H., Tang Y.J., Cantrell M.C., Silva J., Jr. Bacteroides fragilis enterotoxin gene sequences in patients with inflammatory bowel disease. Emerg. Infect. Dis. 2000;6:171–174. doi: 10.3201/eid0602.000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lucas López R., Grande Burgos M.J., Gálvez A., Pérez Pulido R. The human gastrointestinal tract and oral microbiota in inflammatory bowel disease: a state of the science review. Apmis. 2017;125:3–10. doi: 10.1111/apm.12609. [DOI] [PubMed] [Google Scholar]
- 117.Fei N., Zhao L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 2013;7:880–884. doi: 10.1038/ismej.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brentani R.R. Biological implications of complementary hydropathy of amino acids. J. Theor. Biol. 1988;135:495–499. doi: 10.1016/s0022-5193(88)80272-8. [DOI] [PubMed] [Google Scholar]
- 119.Smith P.M., Howitt M.R., Panikov N., Michaud M., Gallini C.A., Bohlooly Y.M., Glickman J.N., Garrett W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science (New York, N.Y.) 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hirota K., Turner J.E., Villa M., Duarte J.H., Demengeot J., Steinmetz O.M., Stockinger B. Plasticity of Th17 cells in Peyer's patches is responsible for the induction of T cell-dependent IgA responses. Nat. Immunol. 2013;14:372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Macpherson A.J., Gatto D., Sainsbury E., Harriman G.R., Hengartner H., Zinkernagel R.M. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science (New York, N.Y.) 2000;288:2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 122.Macpherson A.J., Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science (New York, N.Y.) 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 123.Peterson D.A., McNulty N.P., Guruge J.L., Gordon J.I. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 124.Hapfelmeier S., Lawson M.A., Slack E., Kirundi J.K., Stoel M., Heikenwalder M., Cahenzli J., Velykoredko Y., Balmer M.L., Endt K., Geuking M.B., Curtiss R., 3rd, McCoy K.D., Macpherson A.J. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science (New York, N.Y.) 2010;328:1705–1709. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Popkin B.M., Du S., Green W.D., Beck M.A., Algaith T., Herbst C.H., Alsukait R.F., Alluhidan M., Alazemi N., Shekar M. Individuals with obesity and COVID-19: a global perspective on the epidemiology and biological relationships. Obes. Rev. 2020;21:e13128. doi: 10.1111/obr.13128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Moreira-Rosário A., Marques C., Pinheiro H., Araújo J.R., Ribeiro P., Rocha R., Mota I., Pestana D., Ribeiro R., Pereira A., de Sousa M.J., Pereira-Leal J., de Sousa J., Morais J., Teixeira D., Rocha J.C., Silvestre M., Príncipe N., Gatta N., Amado J., Santos L., Maltez F., Boquinhas A., de Sousa G., Germano N., Sarmento G., Granja C., Póvoa P., Faria A., Calhau C. 2021. Gut Microbiota Diversity and C-Reactive Protein Are Predictors of Disease Severity in COVID-19 Patients, bioRxiv : the Preprint Server for Biology. 2021.2004.2020.440658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhou Y., Shi X., Fu W., Xiang F., He X., Yang B., Wang X., Ma W.L. Gut microbiota dysbiosis correlates with abnormal immune response in moderate COVID-19 patients with fever. J. Inflamm. Res. 2021;14:2619–2631. doi: 10.2147/JIR.S311518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jackson S.A., Schoeni J.L., Vegge C., Pane M., Stahl B., Bradley M., Goldman V.S., Burguière P., Atwater J.B., Sanders M.E. Improving end-user trust in the quality of commercial probiotic products. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.00739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chin-Lee B., Curry W.J., Fetterman J., Graybill M.A., Karpa K. Patient experience and use of probiotics in community-based health care settings. Patient Prefer. Adherence. 2014;8:1513–1520. doi: 10.2147/PPA.S72276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mano M.C.R., Neri-Numa I.A., da Silva J.B., Paulino B.N., Pessoa M.G., Pastore G.M. Oligosaccharide biotechnology: an approach of prebiotic revolution on the industry. Appl. Microbiol. Biotechnol. 2018;102:17–37. doi: 10.1007/s00253-017-8564-2. [DOI] [PubMed] [Google Scholar]
- 131.Kleerebezem M., Binda S., Bron P.A., Gross G., Hill C., van Hylckama Vlieg J.E.T., Lebeer S., Satokari R., Ouwehand A.C. Understanding mode of action can drive the translational pipeline towards more reliable health benefits for probiotics. Curr. Opin. Biotechnol. 2019;56:55–60. doi: 10.1016/j.copbio.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 132.Plaza-Diaz J., Ruiz-Ojeda F.J., Gil-Campos M., Gil A. Mechanisms of action of probiotics. Adv. Nutr. 2019;10:S49–S66. doi: 10.1093/advances/nmy063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Monteagudo-Mera A., Rastall R.A., Gibson G.R., Charalampopoulos D., Chatzifragkou A. Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Appl. Microbiol. Biotechnol. 2019;103:6463–6472. doi: 10.1007/s00253-019-09978-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lebeer S., Bron P.A., Marco M.L., Van Pijkeren J.P., O'Connell Motherway M., Hill C., Pot B., Roos S., Klaenhammer T. Identification of probiotic effector molecules: present state and future perspectives. Curr. Opin. Biotechnol. 2018;49:217–223. doi: 10.1016/j.copbio.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 135.Plaza-Diaz J., Ruiz-Ojeda F.J., Gil-Campos M., Gil A. Mechanisms of action of probiotics. Adv. Nutr. 2019;10:S49–S66. doi: 10.1093/advances/nmy063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Biliavska L., Pankivska Y., Povnitsa O., Zagorodnya S. Antiviral activity of exopolysaccharides produced by lactic acid bacteria of the genera pediococcus, leuconostoc and Lactobacillus against human adenovirus type 5. Medicina (Kaunas) 2019;55:519. doi: 10.3390/medicina55090519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mousavi E., Makvandi M., Teimoori A., Ataei A., Ghafari S., Samarbaf-Zadeh A. Antiviral effects of Lactobacillus crispatus against HSV-2 in mammalian cell lines. J. Chin. Med. Assoc. 2018;81:262–267. doi: 10.1016/j.jcma.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 138.Villena J., Vizoso-Pinto M.G., Kitazawa H. Intestinal innate antiviral immunity and immunobiotics: beneficial effects against rotavirus infection. Front. Immunol. 2016;7:563. doi: 10.3389/fimmu.2016.00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Arena M.P., Capozzi V., Russo P., Drider D., Spano G., Fiocco D. Immunobiosis and probiosis: antimicrobial activity of lactic acid bacteria with a focus on their antiviral and antifungal properties. Appl. Microbiol. Biotechnol. 2018;102:9949–9958. doi: 10.1007/s00253-018-9403-9. [DOI] [PubMed] [Google Scholar]
- 140.Zelaya H., Tada A., Vizoso-Pinto M.G., Salva S., Kanmani P., Agüero G., Alvarez S., Kitazawa H., Villena J. Nasal priming with immunobiotic Lactobacillus rhamnosus modulates inflammation–coagulation interactions and reduces influenza virus-associated pulmonary damage. Inflamm. Res. 2015;64:589–602. doi: 10.1007/s00011-015-0837-6. [DOI] [PubMed] [Google Scholar]
- 141.Eguchi K., Fujitani N., Nakagawa H., Miyazaki T. Prevention of respiratory syncytial virus infection with probiotic lactic acid bacterium Lactobacillus gasseri SBT2055. Sci. Rep. 2019;9:4812. doi: 10.1038/s41598-019-39602-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lenschow D.J., Giannakopoulos N.V., Gunn L.J., Johnston C., O'Guin A.K., Schmidt R.E., Levine B., Virgin H.W.t. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J. Virol. 2005;79:13974–13983. doi: 10.1128/JVI.79.22.13974-13983.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pichlmair A., Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 144.Schoggins J.W., Rice C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sadler A.J., Williams B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gibson G.R., Hutkins R., Sanders M.E., Prescott S.L., Reimer R.A., Salminen S.J., Scott K., Stanton C., Swanson K.S., Cani P.D., Verbeke K., Reid G. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017;14:491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 147.Bottari B., Castellone V., Neviani E. Probiotics and covid-19. Int. J. Food Sci. Nutr. 2021;72:293–299. doi: 10.1080/09637486.2020.1807475. [DOI] [PubMed] [Google Scholar]
- 148.Villena J., Kitazawa H. The modulation of mucosal antiviral immunity by immunobiotics: could they offer any benefit in the SARS-CoV-2 pandemic? Front. Physiol. 2020;11 doi: 10.3389/fphys.2020.00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Baud D., Dimopoulou Agri V., Gibson G.R., Reid G., Giannoni E. Using probiotics to flatten the curve of coronavirus disease COVID-2019 pandemic. Front. Public Health. 2020;8:186. doi: 10.3389/fpubh.2020.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Maldonado Galdeano C., Cazorla S.I., Lemme Dumit J.M., Vélez E., Perdigón G. Beneficial effects of probiotic consumption on the immune system. Ann. Nutr. Metab. 2019;74:115–124. doi: 10.1159/000496426. [DOI] [PubMed] [Google Scholar]
- 151.Hao Q., Dong B.R., Wu T. Cochrane Database Syst Rev.; 2015. Probiotics for Preventing Acute Upper Respiratory Tract Infections. [DOI] [PubMed] [Google Scholar]
- 152.d'Ettorre G., Ceccarelli G., Marazzato M., Campagna G., Pinacchio C., Alessandri F., Ruberto F., Rossi G., Celani L., Scagnolari C., Mastropietro C., Trinchieri V., Recchia G.E., Mauro V., Antonelli G., Pugliese F., Mastroianni C.M. Challenges in the management of SARS-CoV2 infection: the role of oral bacteriotherapy as complementary therapeutic strategy to avoid the progression of COVID-19. Front. Med. 2020;7:389. doi: 10.3389/fmed.2020.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]