Abstract
Considering human brain disorders, Major Depressive Disorder (MDD) is seen as a lethal disease in which a person goes to the extent of suicidal behavior. Physical detection of MDD patients is less precise but machine learning can aid in improved classification of disease. The present research included three RNA-seq data classes to classify DEGs and then train key gene data using a random forest machine learning method. The three classes in the sample are 29 CON (sudden death healthy control), 21 MDD-S (a Major Depressive Disorder Suicide) being included in the second group, and 9 MDD (non-suicides MDD) which are included in the third group. With PCA analysis, 99 key genes were obtained. 47.1% data variability is given by these 99 genes. The model training of 99 genes indicated improved classification. The RF classification model has an accuracy of 61.11% over test data and 97.56% over train data. It was also noticed that the RF method offered greater accuracy than the KNN method. 99 genes were annotated using DAVID and ClueGo packages. Some of the important pathways and function observed in the study were glutamatergic synapse, GABA receptor activation, long-term synaptic depression, and morphine addiction. Out Of 99 genes, four genes, namely DLGAP1, GNG2, GRIA1, and GRIA4, were found to be predominantly involved in the glutamatergic synapse pathway. Another substantial link was observed in the GABA receptor activation involving the following two genes, GABBR2 and GNG2. Also, the genes found responsible for long-term synaptic depression were GRIA1, MAPT, and PTEN. There was another finding of morphine addiction which comprises three genes, namely GABBR2, GNG2, and PDE4D. For massive datasets, this approach will act as the gold standard. The cases of CON, MDD, and MDD-S are physically distinct. There was dysregulation in the expression level of 12 genes. The 12 genes act as a possible biomarker for Major Depressive Disorder and open up a new path for depressed subjects to explore further.
Keywords: RNA-Seq data, Machine learning, Random forest, k-nearest neighbor (KNN), Classification, Feature selection
Introduction
Major Depressive Disorder, also known as clinical depression, is a chronic mental disorder that has a significant and adverse effect on human being's quality of life both socially and economically (Murray et al. 2012). It hampers the human body's physical functions, such as appetite, sleep, emotions, which troubles one to perform day-to-day activities. People suffering from this chronic disorder often experience disturbances in cognitive and executive brain functions. The symptoms associated with this chronic disorder are recurring as well as life-threatening (Fekadu et al. 2017). As per the Diagnostic and Statistical Manual of Mental Disorders, 5th edition (DSM- V), the indication of major depressive disorder is the occurrence of depressed mood known as dysphoria and sudden loss of interest in the activities that were pleasurable at some point of the time in the past (Diagnostic and statistical manual of mental disorders (DSM-5®) American Psychiatric Pub. 2013). The most common Major depressive disorder symptoms consist of sad feelings, loss of interest, low concentration, irregular sleeping patterns, insomnia, feeling guilty, ultimately resulting in suicidal thoughts or even suicide (Zarate et al. 2013). Based on the severity of the symptoms, three different parts of MDD can be categorized such as mild, moderate, and severe (Kessler et al. 2003). Often, the symptoms of MDD get confused with other medical conditions such as brain tumors, vitamin deficiency, and thyroid. But, symptoms lasting for two weeks or more should require a medical diagnosis immediately. Some of the effective treatment procedures comprise psychotherapy, medical countermeasures, and electroconvulsive therapy (Zarate et al. 2006).
RNA sequencing uses Next-Generation Sequencing (NGS) capabilities and is a powerful technique to analyze organisms' transcriptome profiling. The recent advancement in NGS technologies has enabled transcriptome sequencing (RNA-Seq) to become one of the most significant experimental approaches in generating a comprehensive catalog of protein-coding genes, non-coding RNAs transcriptional active genome sites. The promising growth is seen in mRNA sequencing by NGS. It has enabled biotechnologists to measure the expression levels of tens to thousands of transcripts simultaneously (Jabeen et al. 2018; Zararsiz et al. 2014). The expression values are utilized to develop expression-based classification algorithms. The diagnosis, disease classification, monitoring at a molecular level, and providing potential markers of the disease becomes much more straightforward and cost-effective (Zararsiz et al. 2014).
Machine-learning (ML) algorithms (Sundararaj 2016, 2019; Sundararaj and Selvi 2021; Kumari et al. 2019; Tarai et al. 2019) proved to be very useful in classification and the prediction of CON, MDD, and MDD-S outputs when applied to a resampling strategy. A predictive utility in detecting structural trends from high-dimensional data obtained from a limited sample size, ML algorithms help in evaluating next-generation sequencing data. This study focuses primarily on the Machine-Learning methods to classify human brain disorder, prediction studies amongst Control, MDD, and MDD-S, which utilizes features derived from transcriptomic (RNA-Seq) data.
Previous studies have examined disease vs. healthy classification tasks using different methods, such as random forests, KNN(Akter et al. 2019). For MDD patients, the neurotransmitter released and receptors in the synapse are lower in depressed people compared to healthy people (Niciu et al. 2014).
The paper is organized as follows: In Sect. 2, the proposed method is described. The detailed explanation of results is presented in Sect. 3. Section 4 concludes the paper.
Methods
RNA sequencing and quantification of expression levels
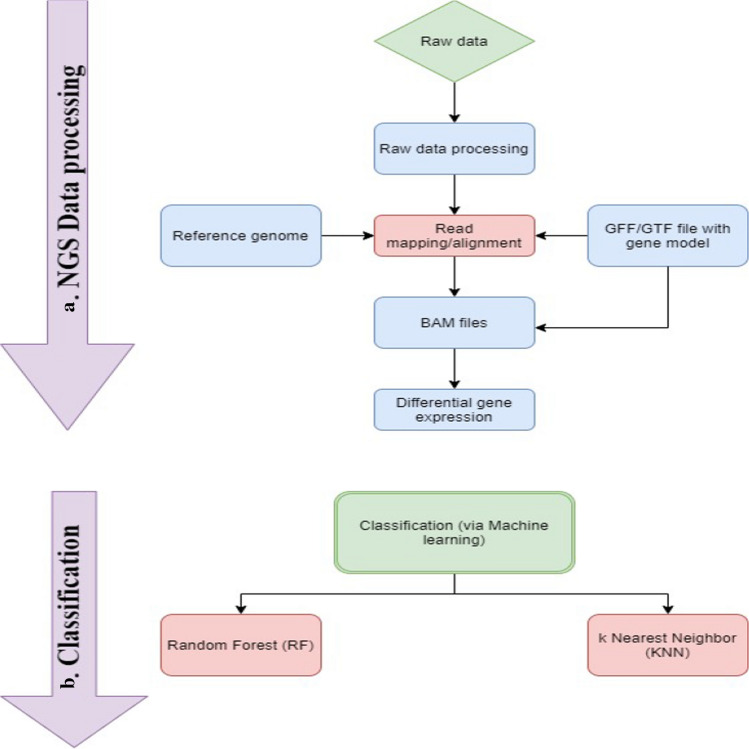
This study contains data from fifty-nine samples of RNA-Seq, which included Control, MDD, and MDD-S. The data source of these samples is available at National Center for Biotechnology Information (NCBI). SRA files from the NCBI was converted to FASTQ file format using the FASTQ-dump program. The raw read quality was checked using FastQC. As part of the analysis, high quality processed reads were mapped to the reference human genome (Human hg19) using HISAT2 (version 2.1.0) for the 59 samples. GFF files were downloaded from the Ensembl for the study to perform a reference-based transcriptomics study (http://www.ensem bl.org/info/data/ftp/index.html). As part of the alignment to the reference in the HISAT2 program, the parameters were included in spliced alignment options and reported algorithms fitted specifically for Cufflinks. Later, the "Cuffdiff" tool was used for the normalization of the RNA-Seq expression data. Figure 1a represents the systematic steps for the RNA-Seq analysis method.
Fig. 1.
Steps included in the pipeline of RNA-Seq data processing and training. a. The NGS data (RNA-Seq dataset) is pre-processed, and transcripts are quantified, subject to differential expression analysis, analysis of co-expression, the study of gene–gene association, or classification of patients with the disease. b. The outcome of step ais subjected to a classification process to classify items by machine learning algorithms according to their attributes
The Fragments Per Kilobase of transcript per Million mapped reads (FPKM) expression values were calculated for the assembled transcript using Cuffdiff v2.2.1. With the FPKM values in Cuffdiff, differential expression genes between 21 MDD-S (subjects with major depressive disorder and suicide), 9 MDD (subjects with MDD and no suicide), against 29 CON (sudden death healthy control) samples were examined. A whole transcriptome gene-level analysis was carried out. The results mentioned throughout the paper are from the analysis of a full dataset of 59 samples. This differentially regulated gene from different compassion was used for PCA.
Analysis of associated genes with suicide and depression by machine learning
The features (i.e. statistical variables or characteristics) for the classification of human brain diseases into CON, MDD, and MDD-S were the expression levels of the gene as FPKM (Normalized count) (i.e., people having the normal, suicide & depressive). Considering that all the classification algorithms used are accurate and appropriate, the quality of this particular human brain disorder categorization can be taken to measure the degree to which the gene expression level differences explain CON, MDD, and MDD-S differences.
Therefore, it has been considered that the genes involved either directly or highly correlated in determining the suicide and depression of the human were the genes with a more informative expression level for the brain disorder classification (CON, MDD, and MDD-S). Identification and selection to obtain a list of genes from gene expression data were carried out to obtain a potential and promising gene list prior to ML algorithm data training. With the most informative genetic feature selected, the ML algorithm was used to classify human brain disease further. The Random forest (RF) and k nearest neighbor (KNN) strategies were used to perform the classifications with ML techniques and feature sets. The methods carried in each of these steps are described below.
Identification and detection of batch effects
Batch effects, which can result from certain causes, such as laboratory conditions, day of processing and technical discrepancies, are common with RNA-Seq data. These batch effects do not get removed by the data normalization technique, thus affects the subsets of genes in different ways (Papiez et al. 2019). Detection and removing these batch effects must improve the analysis accuracy and provide correct conclusions (Leek et al. 2010). Various published high-throughput studies, batch effects were identified and reported, but few methods were available to detect them in high-dimensional expression datasets (Reese 2013). Principal Component Analysis (PCA) is a method commonly used in all available methods (Reese et al. 2013) to reduce data by linear dataset iterations and detect data variance (Yang et al. 2008). The PCA method was used in the current study to reduce the dimensionality of the RNA-Seq expression data. Those components were figured out, which correlated and contributed to data variability, with biological or technical variables. For more stable and accurate PCA results for such a high number of genes (> 70,000), it is advised to perform a dimensionality reduction. Thus, only 624 most varying genes were used from the PCA study based on their level of within-tissue expression. Afterwards, the linear regression analysis was used to adjust the RNA-Seq data for the first two principal components (PC). The correlation cut-off taken was 0.9, and the P-value of 0.0 was considered from the two-dimension after PCA. Finally, 99 genes were taken as the key feature for the 59 samples in the sample classification using ML.
Feature selection
Feature selection is a very vital step in analyzing the sizeable multi-featured data sets effectively and accurately. Over the last few decades, feature selection, also known as subset selection, attribute selection, or variable selection, has been extensively researched and studied by machine learning and statistics communities. An evaluation function was used to assign scores to subsets of a feature in the most common selection model, and then the search algorithm comes into the picture to search for the subset having a high score (Navot et al. 2006).
In this study, the filter approach has been chosen because of its computational speed, scalability, and loose coupling from the specific ML techniques used for the classification. The primary focus of this study was on the objective of identifying the fewer number of genes with the highest classification quality. With an unbiased Random Forest (RF) algorithm based on conditional analysis and deductions, the features' selection was made by using their significance measure (Strobl et al. 2007). This non-linear and nonparametric method is useful in the problems comprising complex interactions whose effects are non-linear and works for data set, which is smaller than a number of the predictors.
Machine-learning algorithms for classification
Two ML algorithms, KNN, and Random forest, were used to classify human brain disorder for Control, MDD, and MDD-S (normal, non-suicide, and suicide condition) using the gene expression levels treated as predictor variables from Next Generation Sequencing (NGS). Figure 1b summarizes the machine‐learning pipeline for MDD diagnosis and prediction using depressive data. Python package, namely NumPy and Scikit-learn, was used for implementing the ML algorithms(Pedregosa et al. 2011). Python has provided an interface to the ML algorithm and a wide variety of classification and regression techniques for the present analysis. Under the same conditions, Python let’s compare various ML algorithms' outcomes and automatically determines the most suitable hyperparameters for ML approaches(Raschka and Mirjalili 2019). The results achieved are therefore extremely accurate, and impartial.
A very well-known ML technique known as Random Forest(RF) has been used in various domains. RF is considered the state-of-the-art ML algorithms (Rodriguez-Galiano et al. 2012) as it yielded great results in most cases. In early 2000 Leo Breiman proposed building a predictor ensemble with some set of decision trees that keep growing in the randomly selected subspaces of data. Despite being so popular and of utmost practical use, very little is known or researched about the mathematical forces that drive the algorithms used. Also, there has been very little finding on the random forests (Biau 2012). The random forest consists of a tree-like hierarchical predictor. Each tree entirely depends on a random vector's values, sampled independently and with the same distribution set for all the trees (Breiman 2001). Growing an ensemble of trees has helped the random forest algorithms vote for the most popular class and made the classifications more accurate. The generation of random vectors plays a vital role in growing these ensembles and governing each tree (Breiman 2001).
Just a single global forecast is taken from the whole 'forest' as a regression average or as a consensus vote in the case of tree classification. The main benefits of the approach are-
Simple and easy interpretation of the results with the use of few predictors in training the model for classification.
Applicable to various problems where there is a very high-order interaction effect or non-linear relationship amongst the variables.
As part of this study, the "random-forest" Python package was used for the analysis (Piles et al. 2019).
Since 1970, the kNN algorithms have been used extensively in numerous applications like pattern recognition and statistical estimation. K-nearest neighbor (kNN) is an algorithm for storing all cases and classifying new instances based on their similarity measure. kNN algorithms are also known by the below names as they are used interchangeably, (i) case-based reasoning, (ii) example-based reasoning, (iii) lazy learning, (iv) k-nearest neighbor, (v) memory-based reasoning and (vi) instance-based learning (Sayad 2010).
KNN is a nonparametric classification method that is majorly divided into two types –
Structure less NN techniques: In this technique, the whole data is classified into training and test sample data. The distance between the training point and sample point is evaluated, and two points with the least distance are called nearest neighbor.
Structure-based NN techniques: This technique is based on the structures of the data like orthogonal structure tree (OST), ball tree, k-d tree, axis tree, nearest future line, and central line (Bhatia 2010). In this study, the "KNN" Python package was used for the analysis.
Prediction model when carried out on the 76,486 transcripts. The PCA was carried out on a total of 76,486 transcripts, which gave 1197 important transcripts by cut off 0.9 in dim1 and p-value 0.0. The model trained gave an accuracy of 85.36% by RF and 68.29% by KNN. The other approach using DGE resulted in better accuracy for the model prediction. The other approach was carried by first identifying the DGE and then applying the PCA on the DGE. The functionally dysregulated gene, when taken for PCA provide better accuracy with the same cut-off in predicting model. Higher accuracy was seen by training the model with PCA on DGE.
Classification performance measure
Performance measurements are available for classification problems: the mean accuracy model has been tested using a confusion matrix. Also, the confusion matrix's look provides a better and simple way to evaluate a classifier's performance. Here, the general idea is to count the occurrences of class A being classified as class B. A row in the confusion matrix represents an actual class, whereas a column represents a predicted class. A classification model's actual performance is evaluated using a confusion matrix (i.e., N * N matrix) where N signifies a number of target classes. This matrix makes a comparison of actual target values and values predicted by the machine learning model. The classification model is based on the notion of fault concept. However, suppose the application of classification models for a particular case leads to the prediction of a class that is quite different from the actual class examples,in that case, there is certainly a classification error. This approach is based on the accuracy taken as a measure for evaluating the classification model's quality. This accuracy can be measured by obtaining the number of correctly classified examples and the total number of cases.
| 1 |
A confusion matrix is a measurement technique or methodology that helps to evaluate the performance of the classification algorithms. There are four important parameters represented in a confusion matrix namely True Positive (TP), True Negative (TN), False Positive (FP), and False Negative (FN). TP signifies that both predicted and expected results are positive, TN signifies that both predicted and expected results are negative. On the other hand, FP, known as Type-I error, signifies predicted results to be positive, but the expected result was found negative and FN, known as Type-II error, signifies predicted results to be negative, but the expected result was found positive (Karthik and Sudha 2020).
TP and TN based on that accuracy are calculated. The accuracy formula of a confusion matrix is below.
| 2 |
Functional annotation of the most informative genes
After PCA, 99 important genes were significant out of 624 dysregulated genes in different comparisons. These 99 genes were taken for functional and pathway annotation using DAVID (The Database for Annotation, Visualization, and Integrated Discovery) a bioinformatics tool (https://david.ncifcrf.gov/, version 6.8). Gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, and significantly enriched biological functions (padj < 0.05) in the list of genes were selected using ClueGO V2.5.6 plug-in Cytoscape v3.8.0 (Bindea et al. 2009; Shannon et al. 2003) as a contributor to the entire classification. For network analysis and functional enrichment analysis, the ClueGO application in Cytoscape was used. The networks were organized into different groups, such as co-expression, physical interaction, genetic interaction, protein domains shared, co-localization, pathway, etc. Pearson correlation was used to create a network as the strength of the association between each pair of genes. The ClueGo algorithm can predict genes or gene products that are closely similar to the original gene list using publicly accessible datasets. A hypergeometric test was conducted using the Benjamini–Hochberg procedure for a functional enrichment analysis with a cut-off of q-values of 0.10.
Results
A total of 59 RNA-Seq sample data was taken from NCBI related to a human brain disorder. The 59 samples taken are Control, MDD & MDD-S, and RNA-Seq analysis was performed. RNA-Seq, gene-level analysis using "Cuffdiff" examines differential expression between 21 MDD-S, 9 MDD, and 29 CON. For Analysis, RNA-Seq raw reads were processed and mapped to the Ensembl GRCh38 human reference genome. Mapping to the reference genome was carried using HISAT2, resulting in BAM files for each of the 59 samples. An average of 30 million reads were obtained, and ~ 90–94% reads were mapped to the genome successfully for the samples. FPKM values were calculated using cufflinks for each sample, and the "Cuffdiff" program was used to retrieve the DGE between control, MDD-S, and MDD. A total of 624 dysregulated genes were found from different sample groups.
Classification of human brain disorder based on transcriptomic data
Multiple approaches have to be tested to select an important gene that would classify the samples with higher accuracy. These genes would play an important role later when one has performed the classification study in the new sample from the patients. For 59 samples, the FPKM gene expression matrix was generated. 76,486 transcripts were observed in the matrix. Two approaches were implemented to find an important list of genes, which defines the different conditions.
In the first approach to eliminate the non-variable data from the matrix with 76,486 transcripts, PCA was performed, and the PC1 &PC2 was studied. It was observed that 1197 transcripts showed higher variability in the data with a cut-off of 0.9 (correlation) in dim1 and a p-value of 0. The PC1 percentage contribution is 32.35%. The 1197 transcript matrix was taken for model training and prediction. The RF and KNN model training algorithm was incorporated to train the data and test the accuracy. With the RF algorithm,38.88% accuracy was obtained for the test data set, and 85.36% accuracy was obtained for the train data set. The KNN machine learning algorithm gave test data accuracy of 27.77% and train data accuracy of 68.29%. With these results, it is observed that the RF algorithm performs better in training the data model and predicting.
In the second approach, we have considered 624 DGE transcripts. So the matrix of 76,486 transcripts was filtered, and a matrix of 624 transcripts was considered in the current second approach. These 624 transcripts are selected because these transcripts are functionally and biologically relevant between the control, MDD-S, and MDD. So training the model with a functionally differential element would be more associative to the sample condition. With PCA's help on the input matrix of 59 samples and 624 transcripts, 99 most significant transcripts were obtained by the cut off value of 0.9 correlation and p-value of 0.0. The training was carried out with RF and KNN algorithms, and the best model was selected. With the RF algorithm 61.11%, test data accuracy was obtained, and the train data accuracy 97.56%. With the KNN algorithm's help, the test data accuracy of 61.11%, and train data accuracy of 76.60% was achieved shown in Table 1. Thus, in the second method RF algorithm was able to give better accuracy. Table 2 shows the confusion matrix.
Table1.
Comparison of classifier accuracy based on different approaches
| ML classifier | 1st Approach with 1197 transcripts | 2nd Approach with 624 DGE transcripts | ||
|---|---|---|---|---|
| Train accuracy | Test accuracy | Train accuracy | Test accuracy | |
| Random forest | 85.36% | 38.88% | 97.56% | 61.11% |
| K- Nearest neighbor | 68.29% | 27.77% | 76.60% | 61.11% |
Table 2.
Confusion matrix for the classification problem of recognizing CON, MDD, and MDD-S
| Actual class | Predicted class | ||
|---|---|---|---|
| Control | MDD | MDD-S | |
| Control | 8 | 0 | 3 |
| MDD | 2 | 0 | 1 |
| MDD-S | 1 | 0 | 3 |
The confusion matrix diagonal elements represent the correct classified examples. Except for the diagonal elements, all other elements indicate incorrectly classified examples to some of the other classes. From Table 2, eight input is correctly classified after training as control. Three times MDD-S shows correct classification. We can observe that three control inputs are wrongly classified. Two inputs are improperly classified as class MDD, and one input is wrongly classified as class MDD-S.
The difference was observed in the model's training with the two methods using 1197 and 99 transcript. In both, the method RF algorithm performed the best and gave the best model fit with 99 most relevant transcripts. Hence, this model would be able to classify the unknown sample if the model is provided with an expression input of the 99 transcripts.
Identification of genes associated with suicide and depression
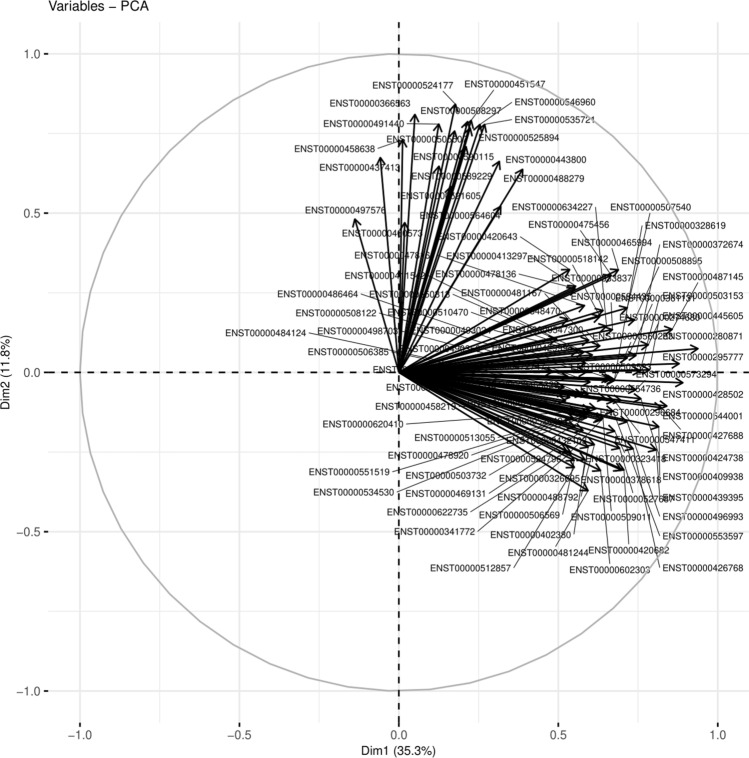
RNA-Seq data analysis was performed, PCA of Differential gene expression data from CON, MDD, and MDD-S gave 99 most important dysregulated genes. The association between control, suicide, and depression sample expression of the human brain was removed after adjusting RNA-Seq data by PC1 (Fig. 2), which explained 35.3% of the variation, while PC2 explained 11.8%. A total of 80 genes are considered from PC1, with the cut-off of p-value 0.0and correlation > 0.5. From PC2 19 genes with cut-off of p-value 0 and correlation > 0.5. So a total of 99 genes were obtained.
Fig. 2.
The figure represents the PCA plot from the differentially expressed gene. The R library used for generating the PCA was FactoMineR. The Ensembl transcript ID for the DEGs contributing to PC1 & PC2 is shown as a data label. The PC1 & PC2 gives data variability of a total 47.1%
Functional analysis of the most informative genes of suicide and depression classification
In this study, we categorized those genes functionally for whom the expression level allowed the best classification among normal and depressive conditions. Based on the contribution to the classification, the DGE genes are selected for PCA. PCA on 624 dysregulated genes resulted in 99 genes that were finally submitted to the DAVID tool for functional enrichment analysis, respectively. Figure 3 lists the most important networks found by CuleGo. Glutamatergic synapse, GABA receptor activation, long-term synaptic depression, Neuro-transmitter receptors, postsynaptic signal transmission, and morphine addiction functions were associated with the most represented network.
Fig. 3.
ClueGo network analysis. The network was constructed using 99 genes as an input to the cytoscape tool. The triangle shows Reactome pathways, the hexagon shows the KEGG pathway, while the circle represents a biological process
For control, MDD-S, and MDD, the following KEGG pathways, were found considerably enriched in genes selected in human brain disorder as the best predictive set for suicide, depression. In this entire course of work, the affected pathway and functional character were studied as well. The study results found the GRIA4 gene to be up-regulated and DLGAP1, GNG2, GRIA1 genes as down-regulated. These genes' major involvement is in the glutamatergic synapse pathway, which holds importance for synaptic signaling. In the dopaminergic synapse pathway, a MAPK10 gene was found as down-regulated. It has been observed that the cAMP signaling pathway and Morphine addiction were altered due to dysregulation of NDUFS4, GABBR2, and PDE4D genes.
Discussion
MDD was studied and diagnosed for the first time in the year 1980. Later during World War II (Comstock and Helsing 1977; Helgason 1964; Lin 1953), mental disorders surveys were conducted. Afterward, the Diagnostic and Statistical Manual of Mental Disorders, Third Edition (DSM-III) criteria, first instrument, and diagnostic Interview Schedule (DIS) were developed and used in the Epidemiologic Catchment Area (ECA) to study subjects with mental disorders(Association 2013; Robins et al. 1981). Researches done earlier have provided in-depth information about MDD and the factors which attribute to it. However, much of the area was unexplored about MDD disease. To distinct MDD, various physiological changes were taken into consideration as well as studied thoroughly(Tremblay et al. 2002). The most common symptoms of MDD are feeling of depressed mood, generalized loss of interest, reduced appetite, and disturbed sleep as well as a spike in suicidal thoughts, cognitive impairment, and loss of memory (Fakhoury 2015; Gamez et al. 2007),constipation, sexual desire, disinterest towards work, crying, suicidal thoughts, and less active speaking and action(Lopez et al. 2006). The early detection techniques of MDD were later advanced, and significant improvement was made in the pharmacological aspect of treating MDD. Along with the previous findings, the unexplored biological mechanism was taken up in research. MDD patients when given the antidepressants in prior research, showed that antidepressants help to block the reuptake of norepinephrine and serotonin. Hence, it proves that norepinephrine and serotonin availability will be more at the synapse (Tremblay et al. 2002).
The RNA-seq data analysis of 59 samples from three individuals provides evidence that the combination of large transcriptomic data with ML allows for the development of robust disease classifiers. Such classifiers help to make an easy detection instrument where hematological expertise is not sufficiently available and/or costly. We propose to re-evaluate ML-based classifiers' application due to the increased utilization of whole-transcriptome sequencing in depression patients' management (Kessler et al. 2016). RNA-seq analysis and ML together can prove to be useful for other diseases as well when we analyze the whole blood or PBMC-derived gene expression profiles or for multiple conditions occurring in parallel.
The current study aimed to understand and address some of the bottlenecks that come in the way of clinical deployment of transcriptomic-based ML tools to diagnose depression in patients. In this study, RNA-Seq data and different prediction algorithms were considered. Even with relatively few training samples, we observed that accurate prediction is possible for the case study. However, depending on the use-case, large training sets can be preferred to achieve high accuracy and yield acceptable positive predictive value.
Even with the existing technologies, our results prove that it is possible to achieve good performance in a near-automated fashion. The cost of running an ML-plus-genomics approach is minimal as the RNA-Seq data is openly available from the previous studies in the public domain. This research indicates that the expense of diagnosis is smaller than the expense of simultaneous use of morphology, immunophenotyping, and cytochemistry in primary MDD diagnostics(Warnat-Herresthal et al. 2020). Therefore, such transcriptome-based ML can be utilized at an earlier point in the disease course when patients with non-specific symptoms report to their primary care physician. Here, ML-based diagnostics will undoubtedly assist in speeding the transfer of the patient. Hence, ML-based diagnostics might assist a faster transfer of the patient to specialized hematology centers for complete diagnostics and therapeutic management.
The field of machine learning involves efforts being made in developing various computational techniques that learn from training data(Schmidt et al. 2019). The training using two approaches RF and KNN shows good accuracy in disease classification (Gaudillo et al. 2019). In this study, both RF and kNN were used to distinguish MDD patients from health communities using multivariate analysis methods. We found that RF achieved the best classification performance. Both kNN and RF display a diagnostic potential for diagnosing MDD, but compared to kNN, RF may be more efficient.
Few studies have highlighted as well as shown the importance of feature selection methods(Saeys et al. 2007) to select the informative genes prior to the classification. Feature selection methods improve classification accuracy by removing redundant and irrelevant features. In the second step of analyses, the predictor variable importance was measured by making use of an unbiased RF algorithm based on conditional inference (Pedregosa et al. 2011). The biggest advantage of this screening method over univariate screening methods is the consideration of interactions between predictor variables.
To remove the non-variable data from the matrix of 76,486 transcripts, PCA was performed and analyzed. The observation indicates that 1197 transcripts displayed greater heterogeneity in the results, with dim1 having a p-value cut-off of 0.0 and a correlation of 0.9. The percentage share from dim1 is 32.35%. For the data (1197 transcript matrix) training, the RF and KNN model training algorithms were used. The RF algorithm shows 38.88%, 85.36% accuracy for test and trained data set, respectively. However, for the test and trained data collection, the KNN algorithm shows 27.77 percent and 68.29 percent accuracy. These observations thus suggest that in the training data model and prediction, the RF algorithm performs better.
This method was explained to produce unbiased variable importance measures, even when predictor variables have different measurement scales or a different number of categories(Strobl et al. 2008). Due to the high correlation between predictors, "Conditional permutation importance" was used as the variable importance measure. After ranking gene expression variables based on this criterion, the two ML algorithms (RF, KNN) were executed. These two ML algorithms' performance was evaluated to classify the extreme human brain on healthy and depressive disorder using gene datasets having the most informative genes. These algorithms were chosen because of their proven excellent performance for classification in various studies(Kuhn and Johnson 2013).
We have considered 624 DEG transcripts in the second model because between the control, MDD-S, and MDD these transcripts are functionally and biologically important. With PCA's aid on the input matrix of 59 samples and 624 transcripts, 99 most significant transcripts were obtained by conducting PCA analysis with a cut-off value of 0.9 correlation and a p-value of 0.0. Again, for data set training, the RF and KNN algorithms were used. The RF algorithm shows 61.11% test data accuracy and 97.56% trained data accuracy. However, KNN algorithms showed 61.11 percent, 76.60 percent test data, and trained data accuracy, respectively. This gives the highest consistency for RF algorithms to train the data and execute the classification.
The classification-based algorithm was used for classifying different samples. The data variable considered is gene expression from transcriptomics. The two approaches are tested with two different methods such as KNN and RF. PCA analysis carried over the differentially expressed gene resulted in 99 genes that were finally used to classify the samples.
Our findings portray the negative effect of non-informative genes on the classification and the need for feature selection. The RNA-Seq data for all 1197 genes from healthy and depressive disorder alone was not effective as it did not allow ML algorithms to identify extreme human brain CON, MDD, MDD-S (mean test data accuracy = 38.88 percent, train data accuracy = 85.36 percent with RF and KNN means test data accuracy = 27.77 percent, train data accuracy = 68.29 percent). However, the performance was significantly improved when the classification was limited to the 99 most informative genes. With 99 genes, the mean test data accuracy of 61.11%, train data accuracy of 97.56% with RF algorithm, and using KNN algorithm test data accuracy of 61.11%, train data accuracy 76.60% was observed. With RF and KNN algorithms for the first method (1197 genes), the performance was slightly reduced when more or less non-informative predictors were included due to inbuilt feature selection. On the other hand, the RF algorithm for the second method (99 genes) gave better accuracy.
A significant difference was observed in the model's training with two methods, namely 1197 and 99 transcripts. The RF algorithm performed best and produced the best model fit with 99 most relevant transcripts on comparing both methods. Hence, this model would classify the unknown sample if the model is provided with the expression input of the 99 transcripts.
Conclusion
Our findings have shown a very promising conclusion that the use of RNA-Seq expression data has the predictive ability to successfully classify human brain disorders into CON, MDD, and MDD-S groups. We used effective ML algorithms to make accurate predictions on complex traits such as suicide and depression based on the transcriptomic information. Among all the ML algorithms tested so far, the RF algorithm has performed better in analyzing different samples of transcriptome data and genes. Hence, we state that the human brain's gene expression data resulted in better classification of human brain disorders.
Data availability
Authors declares no data available.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Akter S, Xu D, Nagel SC, Bromfield JJ, Pelch K, Wilshire GB, Joshi T. Machine learning classifiers for endometriosis using Transcriptomics and Methylomics data. Front Genet. 2019;10:766. doi: 10.3389/fgene.2019.00766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association (2013) Diagnostic and statistical manual of mental disorders (DSM-5®) American Psychiatric Pub.
- Association, A. P. (2013). Diagnostic and statistical manual of mental disorders (DSM-5®): American Psychiatric Pub. [DOI] [PubMed]
- Bhatia, N. (2010). Survey of nearest neighbor techniques. arXiv preprint http://arxiv.org/pdf/1007.0085.
- Biau G. Analysis of a random forests model. J Mach Learn Res. 2012;13(1):1063–1095. [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- Comstock GW, Helsing KJ. Symptoms of depression in two communities. Psychol Med. 1977;6(4):551–563. doi: 10.1017/S0033291700018171. [DOI] [PubMed] [Google Scholar]
- Fakhoury M. New insights into the neurobiological mechanisms of major depressive disorders. Gen Hosp Psychiatry. 2015;37(2):172–177. doi: 10.1016/j.genhosppsych.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Fekadu N, Shibeshi W, Engidawork E. Major depressive disorder: pathophysiology and clinical management. J Depress Anxiety. 2017;6(1):255–257. doi: 10.4172/2167-1044.1000255. [DOI] [Google Scholar]
- Gamez W, Watson D, Doebbeling BN. Abnormal personality and the mood and anxiety disorders: implications for structural models of anxiety and depression. J Anxiety Disord. 2007;21(4):526–539. doi: 10.1016/j.janxdis.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Gaudillo J, Rodriguez JJR, Nazareno A, Baltazar LR, Vilela J, Bulalacao R, Albia J. Machine learning approach to single nucleotide polymorphism-based asthma prediction. PloS one. 2019;14(12):e0225574. doi: 10.1371/journal.pone.0225574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason, T. (1964). Epidemiology of mental disorders in iceland. A psychiatric and demographic investigation of 5395 icelanders. Acta Psychiatrica Scandinavica, 40, SUPPL 173: 171+-171+. [PubMed]
- Jabeen A, Ahmad N, Raza K. Machine learning-based state-of-the-art methods for the classification of rna-seq data In Classification in BioApps. Cham: Springer; 2018. [Google Scholar]
- Karthik S, Sudha M. Predicting bipolar disorder and schizophrenia based on non-overlapping genetic phenotypes using deep neural network. Evol Intell. 2020;14:1–16. [Google Scholar]
- Kessler RC, Barker PR, Colpe LJ, Epstein JF, Gfroerer JC, Hiripi E, Zaslavsky AM. Screening for serious mental illness in the general population. Arch Gen Psychiatry. 2003;60(2):184–189. doi: 10.1001/archpsyc.60.2.184. [DOI] [PubMed] [Google Scholar]
- Kessler RC, van Loo HM, Wardenaar KJ, Bossarte RM, Brenner LA, Cai T, Nierenberg AA. Testing a machine-learning algorithm to predict the persistence and severity of major depressive disorder from baseline self-reports. Mol Psychiatry. 2016;21(10):1366–1371. doi: 10.1038/mp.2015.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M, Johnson K. Applied predictive modeling. New York: Springer; 2013. [Google Scholar]
- Kumari E, Shang Y, Cheng Z, Zhang T. U1 snRNA over-expression affects neural oscillations and short-term memory deficits in mice. Cogn Neurodyn. 2019;13(4):313–323. doi: 10.1007/s11571-019-09528-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, Irizarry RA. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11(10):733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TY. A study of incidence of mental disorders in Chinese and other cultures. Psychiatry. 1953;16:315–335. doi: 10.1080/00332747.1953.11022936. [DOI] [PubMed] [Google Scholar]
- Lopez AD, Mathers CD, Ezzati M, Jamison DT, Murray CJ. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet. 2006;367(9524):1747–1757. doi: 10.1016/S0140-6736(06)68770-9. [DOI] [PubMed] [Google Scholar]
- Murray CJ, Vos T, Lozano R, Naghavi M, Flaxman AD, Michaud C, Aboyans V. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: a systematic analysis for the global burden of disease study 2010. The Lancet. 2012;380(9859):2197–2223. doi: 10.1016/S0140-6736(12)61689-4. [DOI] [PubMed] [Google Scholar]
- Navot, A., Shpigelman, L., Tishby, N., &Vaadia, E. (2006). Nearest neighbor based feature selection for regression and its application to neural activity. In Advances in neural information processing systems (pp. 996–1002
- Niciu MJ, Ionescu DF, Richards EM, Zarate CA. Glutamate and its receptors in the pathophysiology and treatment of major depressive disorder. J Neural Transm. 2014;121(8):907–924. doi: 10.1007/s00702-013-1130-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papiez A, Marczyk M, Polanska J, Polanski A. BatchI: Batch effect Identification in high-throughput screening data using a dynamic programming algorithm. Bioinformatics. 2019;35(11):1885–1892. doi: 10.1093/bioinformatics/bty900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Vanderplas J. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- Piles M, Fernandez-Lozano C, Velasco-Galilea M, González-Rodríguez O, Sánchez JP, Torrallardona D, Quintanilla R. Machine learning applied to transcriptomic data to identify genes associated with feed efficiency in pigs. Genet Sel Evol. 2019;51(1):10. doi: 10.1186/s12711-019-0453-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschka S, &Mirjalili V (2019). Python machine learning: Machine learning and deep learning with Python, scikit-learn, and TensorFlow 2. Packt Publishing Ltd.
- Reese SE, Archer KJ, Therneau TM, Atkinson EJ, Vachon CM, De Andrade M, Eckel-Passow JE. A new statistic for identifying batch effects in high-throughput genomic data that uses guided principal component analysis. Bioinformatics. 2013;29(22):2877–2883. doi: 10.1093/bioinformatics/btt480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese, S. (2013). Detecting and Correcting Batch Effects in High-Throughput Genomic Experiments.
- Robins LN, Helzer JE, Croughan J, Ratcliff KS. National Institute of Mental Health diagnostic interview schedule: Its history, characteristics, and validity. Arch Gen Psychiatry. 1981;38(4):381–389. doi: 10.1001/archpsyc.1981.01780290015001. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Galiano VF, Ghimire B, Rogan J, Chica-Olmo M, Rigol-Sanchez JP. An assessment of the effectiveness of a random forest classifier for land-cover classification. ISPRS J Photogramm Remote Sens. 2012;67:93–104. doi: 10.1016/j.isprsjprs.2011.11.002. [DOI] [Google Scholar]
- Saeys Y, Inza I, Larrañaga P. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23(19):2507–2517. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]
- Sayad S. K nearest neighbors. Toronto: University of Toronto; 2010. [Google Scholar]
- Schmidt J, Marques MR, Botti S, Marques MA. Recent advances and applications of machine learning in solid-state materials science. NPJ Comput Mater. 2019;5(1):1–36. doi: 10.1038/s41524-019-0221-0. [DOI] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl C, Boulesteix AL, Zeileis A, Hothorn T. Bias in random forest variable importance measures: Illustrations, sources and a solution. BMC Bioinformatics. 2007;8(1):25. doi: 10.1186/1471-2105-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl C, Boulesteix AL, Kneib T, Augustin T, Zeileis A. Conditional variable importance for random forests. BMC Bioinformatics. 2008;9(1):307. doi: 10.1186/1471-2105-9-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararaj V. An efficient threshold prediction scheme for wavelet based ECG signal noise reduction using variable step size firefly algorithm. Int J Intell Eng Syst. 2016;9(3):117–126. [Google Scholar]
- Sundararaj V. Optimised denoising scheme via opposition-based self-adaptive learning PSO algorithm for wavelet-based ECG signal noise reduction. Int J Biomed Eng Technol. 2019;31(4):325–345. doi: 10.1504/IJBET.2019.103242. [DOI] [Google Scholar]
- Sundararaj V, Selvi M. Opposition grasshopper optimizer based multimedia data distribution using user evaluation strategy. Multim Tools Appl. 2021;19:1–17. [Google Scholar]
- Tarai S, Mukherjee R, Gupta S, Rizvanov AA, Palotás A, Pammi VC, Bit A. Influence of pharmacological and epigenetic factors to suppress neurotrophic factors and enhance neural plasticity in stress and mood disorders. Cogn Neurodyn. 2019;13:1–19. doi: 10.1007/s11571-019-09522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay LK, Naranjo CA, Cardenas L, Herrmann N, Busto UE. Probing brain reward system function in major depressive disorder: altered response to dextroamphetamine. Arch Gen Psychiatry. 2002;59(5):409–416. doi: 10.1001/archpsyc.59.5.409. [DOI] [PubMed] [Google Scholar]
- Warnat-Herresthal S, Perrakis K, Taschler B, Becker M, Baßler K, Beyer M, Ulas T. Scalable prediction of acute myeloid leukemia using high-dimensional machine learning and blood transcriptomics. Iscience. 2020;23(1):100780. doi: 10.1016/j.isci.2019.100780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Harrington CA, Vartanian K, Coldren CD, Hall R, Churchill GA. Randomization in laboratory procedure is key to obtaining reproducible microarray results. PloS one. 2008;3(11):e3724. doi: 10.1371/journal.pone.0003724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zararsiz G, Goksuluk D, Korkmaz S, Eldem V, Duru IP, Unver T, & Ozturk A (2014). Classification of RNA-Seq data via bagging support vector machines. bioRxiv, 007526.
- Zarate CA, Singh JB, Carlson PJ, Brutsche NE, Ameli R, Luckenbaugh DA, Manji HK. A randomized trial of an N-methyl-D-aspartate antagonist in treatment-resistant major depression. Arch Gen Psychiatry. 2006;63(8):856–864. doi: 10.1001/archpsyc.63.8.856. [DOI] [PubMed] [Google Scholar]
- Zarate CA, Jr, Mathews D, Ibrahim L, Chaves JF, Marquardt C, Ukoh I, Luckenbaugh DA. A randomized trial of a low-trapping nonselective N-methyl-D-aspartate channel blocker in major depression. Biol Psychiat. 2013;74(4):257–264. doi: 10.1016/j.biopsych.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Authors declares no data available.