Abstract
Mutant KRAS is a key driver in colorectal cancer (CRC) and promotes Myc translation and Myc-dependent stress adaptation and proliferation. Here, we report that the combination of two FDA-approved drugs Bortezomib and Everolimus (RAD001) (BR) is highly efficacious against mutant KRAS CRC cells. Mechanistically, the combination, not single agent, rapidly depletes Myc protein, not mRNA, and leads to GCN2- and p-eIF2α-dependent cell death through the activation of extrinsic and intrinsic apoptotic pathways. Cell death is selectively induced in mutant KRAS CRC cells with elevated basal Myc and p-eIF2α and is characterized by CHOP induction and transcriptional signatures in proteotoxicity, oxidative stress, metabolic inhibition, and immune activation. BR-induced p-GCN2/p-eIF2α elevation and cell death are strongly attenuated by MYC knockdown and enhanced by MYC overexpression. The BR combination is efficacious against mutant KRAS patient derived organoids (PDO) and xenografts (PDX) by inducing p-eIF2α/CHOP and cell death. Interestingly, an elevated four-gene (DDIT3, GADD45B, CRYBA4 and HSPA1L) stress signature is linked to shortened overall survival in CRC patients. These data support that Myc-dependent stress adaptation drives the progression of mutant KRAS CRC and serves as a therapeutic vulnerability, which can be targeted using dual translational inhibitors.
Supplementary Information
The online version contains supplementary material available at 10.1186/s43556-022-00070-7.
Keywords: Mutant KRAS, Myc, eIF2α, Colorectal cancer, Everolimus, Bortezomib
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide, with an estimated 1.2 million new cases and over 600,000 death annually [1]. Genetic alternations in oncogenes and tumor suppressors cooperate with epigenetic alterations to drive colorectal carcinogenesis [2]. KRAS is the most frequently mutated oncogene in human cancer. A comprehensive up to date analysis revealed that ~ 19% of cancer patients harbor RAS mutations and over 85% are in KRAS, equivalent to ~ 3.4 million new cases per year worldwide [3]. Mutational activation of KRAS is an early event in CRC development and occurs in about 40–50% of cases, with hot spots in codons 12, 13, 61, and 146 [2, 4]. Mutant KRAS is correlated with poor prognosis in CRC and resistance to Epidermal Growth Factor Receptor (EGFR) antibodies, and immune checkpoint blocker such as anti-PD1 [4]. Unfortunately, attempts to directly target mutant KRAS have limited success so far in the clinic. While selective KRAS G12C and G13C inhibitors demonstrated some promise in lung cancer patients recently [5, 6], these mutations however are present in less than 5% of CRCs. Therefore, targeting KRAS mutated CRCs remains a challenge.
Deregulated mRNA translation and protein synthesis is a common node of oncogenesis, as specialized proteins are required to sustain cancer hallmarks such as increased proliferation, altered metabolism, metastasis, and resistance to cell death or immune attack [7–10]. Mutant KRAS or BRAF increases proliferation and translation of many targets including oncogenic ones such as Myc, Bcl-xL and MMPs. This is thought to be mediated largely through the dissociation of Eukaryotic Translation Initiation Factor 4E (eIF4E) from its inhibitor 4EBPs upon their phosphorylation by MAPK/ERK and PI3K/mTOR/AKT signaling [11]. Elevated protein synthesis and demand on quality control in cancer cells lead to increased phosphorylation of Eukaryotic Translation Initiation Factor 2 alpha (eIF2α) (S51, p-eIF2α), the core regulator of the “integrated stress response” (ISR) [12]. There are four known eIF2α kinases in mammals which are activated by distinct and overlapping stresses. Among them, general control nonderepressible 2 (GCN2) is the most ancient and activated by amino acid starvation. A modest increase in p-eIF2α inhibits cap-dependent translation, while facilitating the translation of stress-related transcription factors such as Activating Transcription Factor 4 (ATF4) and C/EBP Homologous Protein (CHOP, encoded by DDIT3) to promote adaptation and survival through widespread changes in transcription, translation, metabolism, and myriad effectors [12, 13]. Failure to adapt leads to prolonged p-eIF2α elevation and CHOP induction, and subsequent cell death through apoptotic mediators such as death receptor 5 (DR5), p53 Upregulated Modulator of Apoptosis (PUMA), and Phorbol-12-myristate-13-acetate-induced protein 1 (also called NOXA) [12, 13].
Myc is a master regulator of oncogenic growth and metabolism [14–17], in part through elevated p-eIF2α and p-eIF4E/4EBP1 that promotes stress adaptation and survival of cancer cells [18–20]. This Myc-translation axis was therefore suggested as an exploitable vulnerability particularly in metabolically active tumors with mutated or amplified KRAS [16, 21]. However, targeting a single Myc target [14, 15], metabolic pathway, or a single step in mRNA translation [7, 8], yielded little clinical efficacy in most solid tumors. For example, inhibitors against mRNA Cap binding, eIF4A helicase [7, 8], or p-eIF2α inducing agents such as proteasome (Bortezomib and Ixazomib) [12, 22] and HSP90 inhibitors [23, 24], lack potency or selectivity. On the other hand, many kinase inhibitors target translation indirectly by blocking the phosphorylation of eIF4E or 4EBP [7, 8]. These include allosteric mTOR inhibitors Everolimus (i.e., RAD001) and Temsirolimus, and EGFR antibodies or inhibitors. However, mutant KRAS and BRAF represents a major resistance mechanism to single agent in CRC preclinical models or patients [25–28] due to complex feedback activation of survival pathways [4, 29]. Dual inhibition of ERK/MAPK and PI3K/mTOR signaling generally produces unacceptable normal tissue toxicity [7, 8].
We reasoned that Myc-driven stress adaptation is a critical survival mechanism in mutant KRAS CRCs, and Myc translation might be an useful therapeutic target [30]. In this study, we focused on FDA-approved agents and discovered that the combination of Bortezomib and Everolimus synergistically and selectively kills mutant KRAS CRC cells at concentrations where single agents had little or no toxicity. The efficacy of this combination is validated using patient derived organoids (PDOs) and xenografts (PDXs). Mechanistically, we showed that mutant KRAS-dependent vulnerability is mediated through elevated Myc, and its ablation leads to GCN2/p-eIF2α -dependent cell death with profound trancriptional signatures of proteotoxcity, oxidative stress and metabolic suppression. Our study provides a potential way to improve the treatment of mutant KRAS CRCs by targeting the deregulated Myc-ISR axis.
Results
Bortezomib and Everolimus synergistically kill mutant KRAS CRC cells
To identify potential combination targeting mutant KRAS in CRC, we first performed a targeted drug screen using 11 translational inhibitors and HCT116 cells. These included pathway agents indirectly targeting p-4EBP1 or p-eIF4E, and ones targeting the cap binding complex (eIF4A, eIF4F assembly), or the 43S preinitiation complex (eIF2α). As expected, these agents showed massive differences in IC50 as single agent (over 20,000-fold, from low nM to sub mM). Cap analog 4Ei-1 was the least potent (at 500 µM or higher), while the p-eIF2α inducer bortezomib and eIF4A inhibitor were among the most potent (10–20 nM, Table S1). Direct or indirect kinase inhibitors had low to modest toxicity with IC50s ranging between 20–100 µM. The combination of Bortezomib (B) and Everolimus (R) (BR, hereafter) showed strong synergy in suppressing cell growth (Fig. 1a). Both are FDA-approved drugs and chosen for further study.
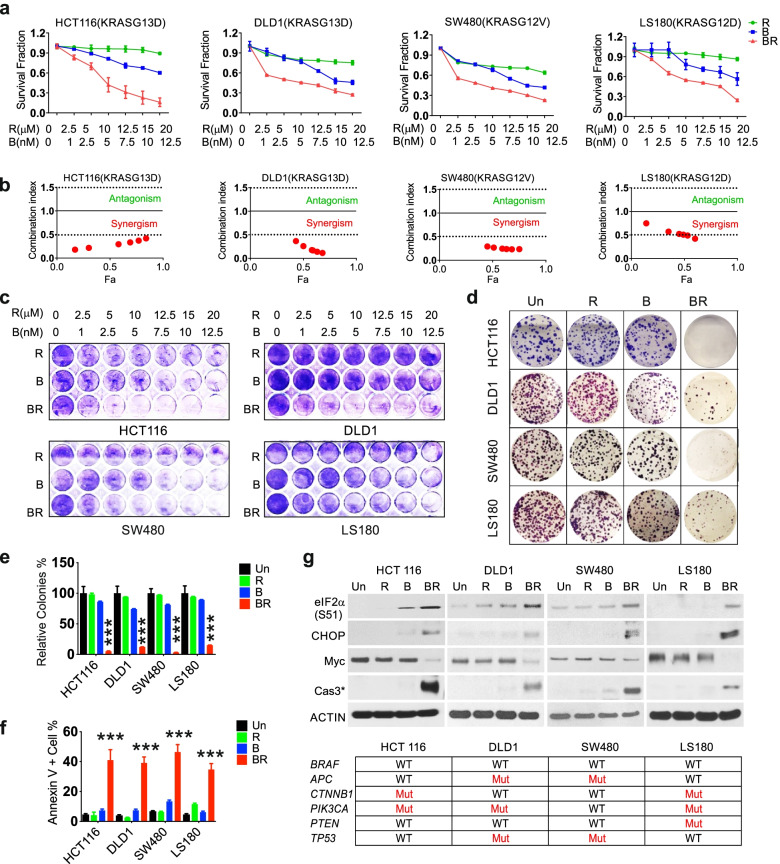
Fig. 1.
Bortezomib and Everolimus show synergistic antitumor effects in mutant KRAS CRC cells. Mutant KRAS HCT116, DLD1, SW480 and LS180 cells were treated with vehicle (untreated, Un), Bortezomib (B), Everolimus (R), or their combination (BR, 5 nM and 10 µM or specified ranges). a Cell growth at 48 h was assessed using MTS assay. b Calculated combination index (CI). The CI presents synergism (CI < 1), additive effect (CI = 1), and antagonism (CI > 1). Red area presents synergism, and green area presents antagonism. Fa represents fraction affected. c Attached cells were visualized by crystal violet at 48 h. (D) Colony formation assay. Cells were treated for 24 h and replated in drug free medium for 14 days before crystal violet staining. e The relative number of colonies as in (d) was normalized to untreated group (100%). f Apoptosis was quantitated by Annexin V + cells using flow cytometry. g Indicated proteins at 24 h was detected using western blotting. ACTIN is used as loading control. The status of major drivers is shown below. e, f, values are mean + s.d. (n = 3). ***p < 0.001 (Student’s t-test, two tailed). B, R vs. BR
We validated the synergy of BR using three additional KRAS mutant CRC cells (DLD1, SW480 and LS180) and viability assay with combination index calculated (CI < 0.5) (Fig. 1a-b). The BR combination (B 5 nM and R 10 µM) strongly suppressed cell growth at 48 h (Fig. 1c), and long-term clonogenicity on day 14 with 24 h exposure. In contrast, either agent alone at the same doses showed little or no toxicity (Fig. 1d-e). The combination potently induced apoptosis as evidenced by increased Annexin V positive cells at 48 h (Fig. 1f and Fig. S1a) and cleaved caspase-3 at 24 h in all four lines (Fig. 1g). Compared to either agent, BR induced much higher levels of phosphorylation of eIF2α (S51) and CHOP, and a strong reduction in Myc protein in all four lines (Fig. 1g). These KRAS mutated lines are all WT for BRAF but otherwise vary in mutational status of APC, CTNNB1 (β-catenin), PIK3CA, PTEN, and TP53 (Fig. 1g and Table S2). We further confirmed induction of prolonged ISR with elevated ATF4, GRP78/BIP and spliced XBP1s in HCT 116 cells by BR, not by single agent (Fig. S1b). BR and single agent reduced 4E-BP1 phosphorylation (S65/70) similarly, but had limited effect on 4E-BP1 (T37/46) or S6 phosphorylation (S235/236) (Fig. S1b). These results demonstrated that BR potently induces Myc ablation and stress-associated cell death in mutant KRAS cells.
The BR combination induces p-eIF2α-dependent killing of mutant KRAS CRC cells
To further explore the mechanisms underlying BR-induced cell killing, we conducted RNA-Seq analysis on HCT116 cells. Differentially expressed genes (DEGs) included 1231 upregulated and 1285 downregulated by two-fold and more at 24 h (p ≤ 0.005) (Fig. 2a). Gene ontology (GO) analysis indicated that upregulated genes are highly enriched in pathways for proteostress and unfolded protein response (UPR), including multiple HSP family members and classical ISR markers CHOP and Growth Arrest and DNA Damage-Inducible Protein 45B and 34 (GADD45B and GADD 34) (Fig. 2a-b). Other highly upregulated pathways included extrinsic apoptosis, oxidative stress, and, surprisingly, immunity (Fig. 2b). The top 10 enriched pathways in downregulated genes were related to metabolism and cell cycle (Fig. S2a). Gene set enrichment analysis (GSEA) confirmed positive enrichment in misfolded proteins and leukocyte activation among others (Fig. S2b and data deposit).
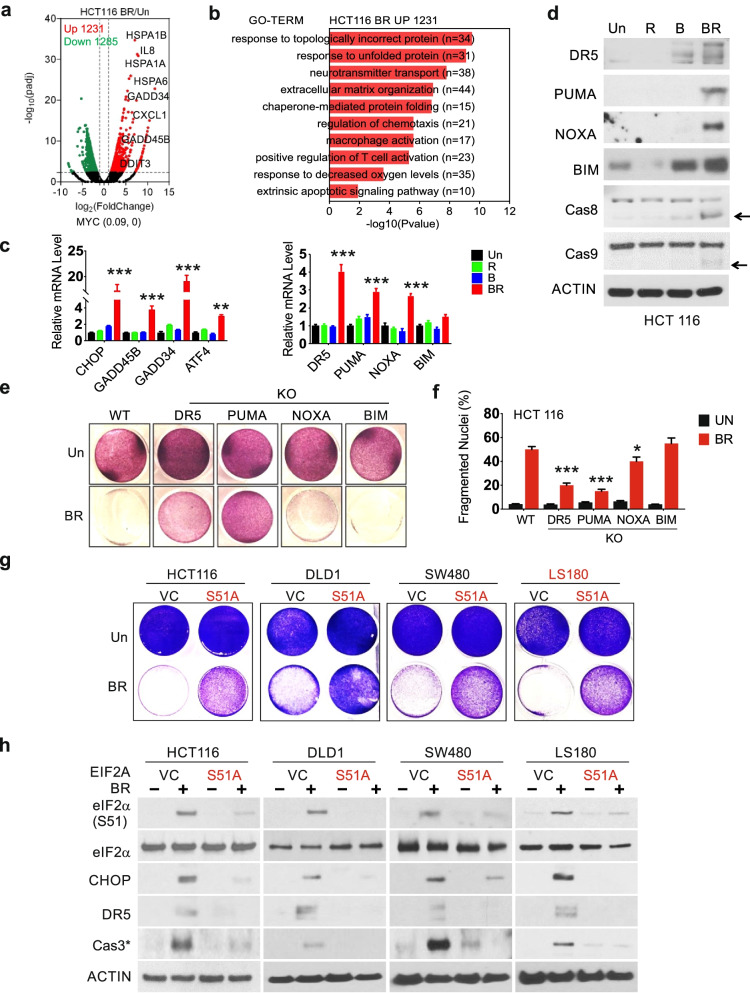
Fig. 2.
The BR combination induces p-eIF2α-dependent apoptosis of mutant KRAS CRC cells. The indicated cells were treated with vehicle (Un, Untreated), Bortezomib (B), Everolimus (R), or the combination (BR, 5 nM and 10 µM). a Differential genes induced by BR in HCT116 cells at 24 h visualized by volcano plot. Upregulated (red) or downregulated (green) genes (fold change ≥ 2, p < 0.005). Selected upregulated genes are shown. b Top 10 enriched non-overlapping pathways in upregulated genes (1231) identified by GO. c qRT-PCR analysis of the indicated markers at 24 h. d Western blotting of the indicated proteins at 24 h. Arrows indicate cleaved and active caspase. Actin was used as the loading control. e Attached cells at 48 h were visualized by crystal violet staining. f Apoptosis at 48 h was analyzed by nuclear fragmentation assay. g Cells were transfected with vector control (VC) or EIF2AS51A expression construct for 24 h, replated for 24 h, and treated by the BR combination for 48 h. Attached cells were visualized by crystal violet staining. h Western blotting of indicated proteins at 24 h. c and f, values are mean + s.d. (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t-test, two tailed). B, R vs. BR, or WT vs. KO
qRT-PCR analysis confirmed the induction of the proximal ISR regulators CHOP(DDIT3), GADD45B, GADD34, and ATF4, as well as apoptotic effectors DR5, PUMA, NOXA, and BIM by the combination, but not single agents in HCT 116 cells (Fig. 2c). The induction of apoptotic proteins was also confirmed by western blotting with cleaved casapase-8 and -9 (Fig. 2d). Using isogenic HCT 116 cells deficient in one of these four apoptotic effectors [31, 32], we found that BR-induced cell killing is abrogated by DR5 knockout (KO) or PUMA KO, but minimally affected by NOXA KO or BIM KO (Fig. 2e-f).
Prolonged p-eIF2α elevation and CHOP induction leads to cell death [12]. p-eIF2α can be inhibited by the overexpression of a phosphorylation defective and dominant negative mutant allele (serine 51 to alanine, herein referred to as S51A) [10, 28]. The expression of eIF2AS51A significantly blocked BR-induced cell loss, with marked reduction in p-eIF2α, CHOP, DR5, and cleaved caspase-3 (Fig. 2g-h), and expression of ISR targets CHOP, GADD45B, GADD34, ATF4, and apoptotic effectors (DR5, PUMA, NOXA and BIM) (Fig. S2c). Inhibition of apoptosis was confirmed by nuclear fragmentation assay and flow cytometry (Fig. S2d-f). These data demonstrate that sustained p-eIF2α leads to the killing of mutant KRAS CRC cells through the activation of intrinsic and extrinsic apoptotic pathways upon BR treatment.
The BR combination triggers Myc-dependent activation of GCN2
The strong metabolic suppression by BR observed at 24 h (Fig. S2a) prompted us to examine the role of GCN2, a classical metabolic stress sensor and eIF2α kinase [13]. Elevation of p-eIF2α was observed at 2–4 h in HCT 116, DLD1, SW480, and LS180 cells (Fig. 3a), in parallel with elevated p-GCN2 in 3 of 4 lines. Despite decreased Myc protein (Fig. 1g), little or no change in MYC mRNA was detected at 24 h in any line (Fig. S3a). Interestingly, loss of Myc and 4EBP1 (S65/70, not T37/46) was rapid and near complete at one hour, when p-GCN2, p-eIF2α and ATF4 just began to rise, continuing to 4 and 24 h in HCT 116 cells (Fig. 3b, Fig. S3b). We detected no change in total eIF4E or p-eIF4E (S209) and decreased p-PERK, the UPR sensor (Fig. 3b). The levels of CHOP and death effectors such as DR5 and cleaved caspase-3 began to rise only at or after 12 h (Fig. S3b). Consistent with Myc loss, significant enrichment of Myc down-regulated targets was evident at 24 h (Fig. 3c).
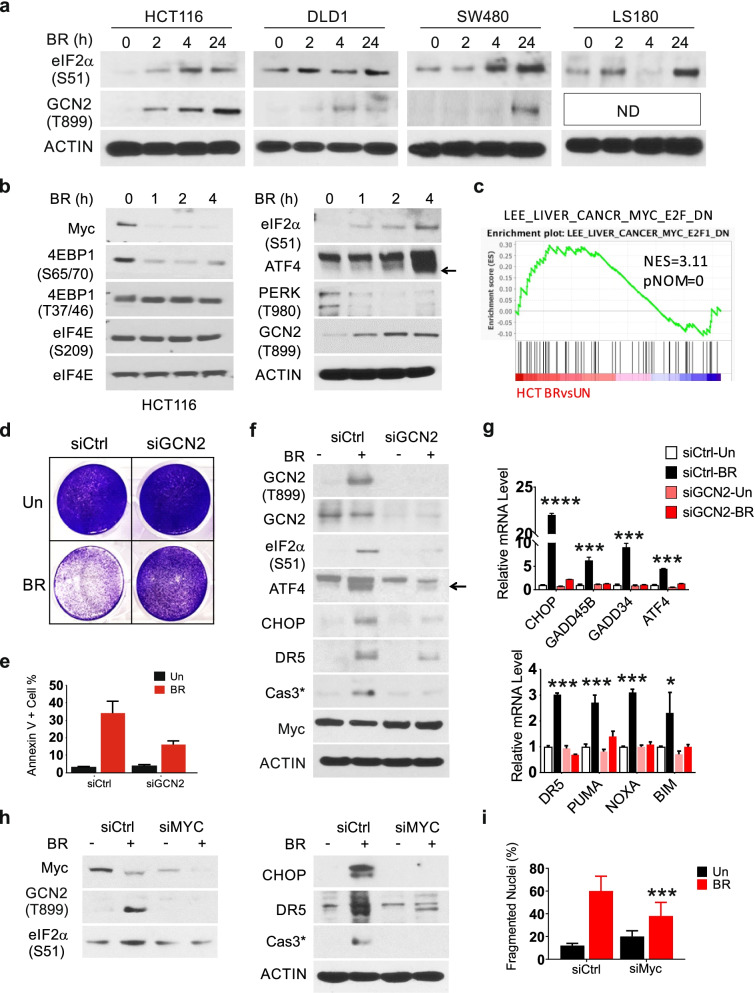
Fig. 3.
The BR combination triggers Myc- and GCN2-dependent apoptosis. Indicated cells were treated with vehicle (Un), or the Bortezomib (B), and Everolimus (R) combination (BR, 5 nM and 10 µM). a Western blotting of indicated proteins at 0, 2, 4 and 24 h. ND, not detected. b Western blotting of indicated proteins at 0, 1, 2 and 4 h. (c). GSEA of differential genes in the indicated pair (C2 dataset). The indicated gene set is shown with NES (normalized enrichment score) and corresponding p-value. d Cells were transfected with either scramble or GCN2 siRNA for 24 h, replated for 24 h and treated with BR for 48 h. Attached cells were visualized by crystal violet staining. e Quantification of Annexin V + cells. f Western blotting of indicated proteins, and (g) qRT-PCR analysis of indicated genes at 24 h. The arrow indicates the specific lower band. h Cells were transfected with either scrambled (Ctrl) or MYC siRNA and treated with BR as in d, analyzed by western blotting at 24 h and i apoptosis at 48 h. g, i, values are mean + s.d. (n = 3). *p < 0.05, ***p < 0.001, ****p < 0.0001 (Student’s t-test, two tailed). Control (Ctrl) vs. siRNA
We further examined the role of GCN2 and Myc in BR-induced cell death. GCN2 siRNA markedly reduced BR-induced growth inhibition and apoptosis (Fig. 3d-e, Fig, S3c), as well as the levels of p-eIF2α, ATF4, CHOP, caspase-3 cleavage, or ISR targets, with a minor effect on Myc loss (Fig. 3f-g). MYC siRNA strongly reduced BR-induced cell death, p-GCN2/p-eIF2α/CHOP and caspase-3 cleavage (Fig. 3h-i). These results suggest that acute Myc loss in mutant KRAS CRC cells impairs stress adaptation and leads to GCN2/p-eIF2α-dependent metabolic crisis and cell death.
The BR combination promotes mutant KRAS-Myc selective proteotoxicity
We further determined if elevated Myc in mutant KRAS CRC cells [30] is the target of BR. Compared to mutant KRAS CRC cells (n = 4), WT KRAS CRC cells (n = 4) showed higher IC50, reduced apoptosis, and lower induction of stress (p-eIF2α, CHOP, GADD45B), DR5, and cleaved caspase-3 (Fig. 4a-b, Fig. S4a-b). To minimize the influence of genetic background, we took advantage of isogenic mutant and WT KRAS CRC cell pairs previously established [33]. We confirmed higher BR sensitivity in mutant KRAS isogenic cells (HCT116, DLD1 and SW48) by growth suppression and apoptosis induction (Fig. 4c-d). qRT-PCR indicated induction of some ISR regulators and apoptotic effector genes (8) by BR in WT KRAS isogenic cells, albeit at much reduced levels compared to those in mutant KRAS counterparts (Fig. 4e). Myc protein, not MYC mRNA, was effectively ablated by BR within all lines (Fig. 4e-f). Isogenic WT KRAS cells showed lower basal Myc and p-eIF2α, and lacked the induction in CHOP, apoptotic targets, or cleaved caspase-3 at 24 h (Fig. 4f).
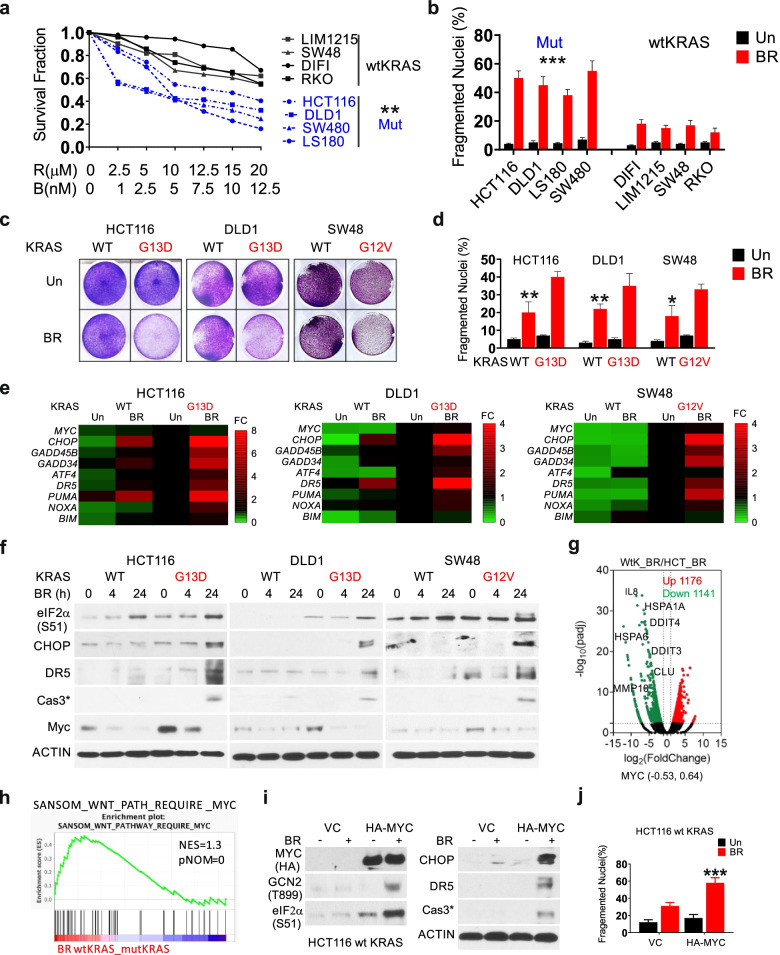
Fig. 4.
The BR combination induces mutant KRAS-selective stress hyperactivation and cell death. CRC cells with either WT or mutant KRAS were treated with vehicle (Un), or the Bortezomib (B) and Everolimus (R) combination (BR, 5 nM and 10 µM, or as specified). a Dose response of 8 cell lines at 48 h was assessed by MTS assay. Dotted (blue) or solid lines represented cells with MUT or wtKRAS. b Apoptosis at 48 h was analyzed by nuclear fragmentation assay. c Attached cells at 48 h were visualized by crystal violet staining. d Apoptosis at 48 h was analyzed by nuclear fragmentation assay. e qRT-PCR analysis of indicated genes at 24 h visualized by heatmap. The expression was normalized to untreated isogenic mutant KRAS cells (1). f Western blotting of indicated proteins at 0, 4 and 24 h. g DEGs (FC ≥ 2, p < 0.005) in BR-treated WT vs. mut KRAS HCT 116 cells at 24 h visualized by volcano plot. Selected down-regulated genes (green) in WT KRAS cells are shown. h GSEA of differential genes in the indicate pair (C2 dataset). The indicated gene set is shown with NES and corresponding p-value. i Cells were transfected with either empty vector (VC) or HA-MYC plasmid for 24 h, replated for 24 h, and treated with BR. Western blotting of indicated proteins at 24 h, and j apoptosis at 48 h analyzed by nuclear fragmentation assay. a, b, d, j, values are mean + s.d. (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (Student’s t-test, two tailed). WT vs. mut KRAS BR group or cell line, or VC vs. HA-MYC
We then used RNA-seq to compare global transcriptional response of isogenic WT and mutant KRAS HCT 116 cells to BR treatment. Consistent with the lack of acute stress or death, WT KRAS cells displayed a drastically reduced global transcriptomic response (Fig. S4c). BR-treated WT KRAS cells showed profound decrease of pathways in proteostress, intrinsic and extrinsic apoptosis, and HIF1α and immunity (-Log10 (pvalue) from 15–5) (Fig. 4g, Fig. S4d), and increase of pathways in development, cell cycle, metabolism, and chromosome segregation (Fig. S4e). Interestingly, top BR differential genes appeared to be regulated at opposite directions in this isogenic pair (Fig. S4f-g). Despite little or no change in MYC mRNA, GSEA showed negative enrichment of Wnt-Myc in BR-treated mutant KRAS cells (Fig. 4g-h). Overexpression of MYC in WT KRAS cells increased p-GCN2/p-eIF2α/CHOP, and DR5 and cleaved caspase-3 at 24 h and cell death at 48 h upon BR treatment, with (Fig. 4i-j). Together with results from MYC siRNA, these data demonstrate a fundamental role of Myc in regulating metabolic and transcriptomic response in mutant KRAS CRC cells.
The BR combination is effective against mutant KRAS CRC PDOs
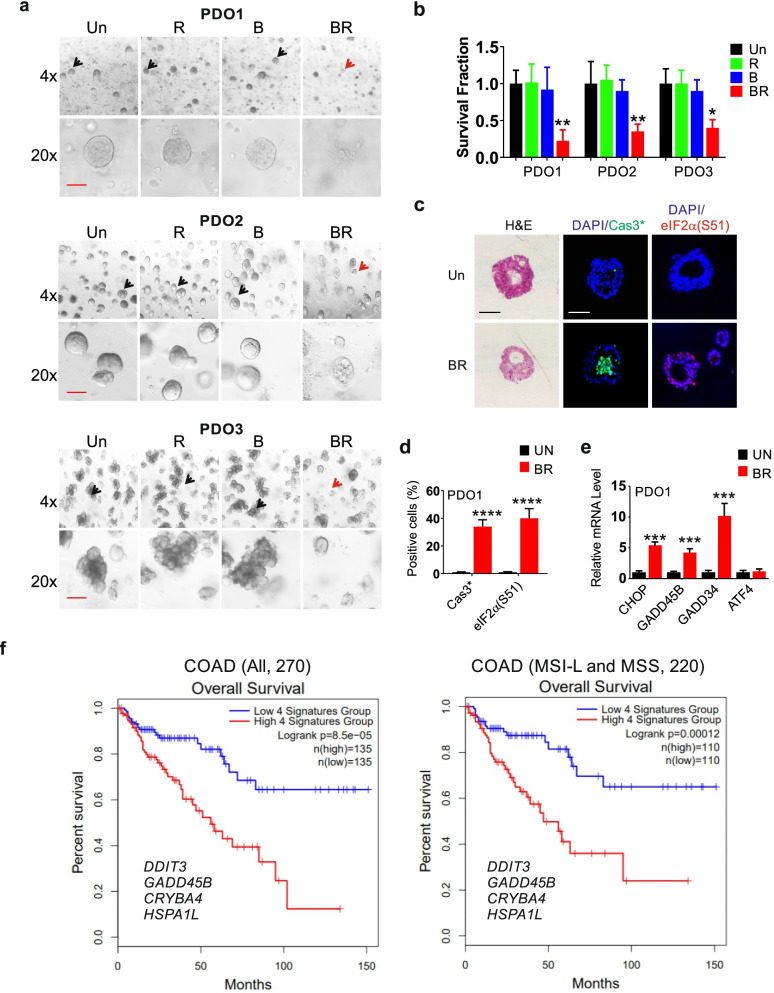
Patient derived organoids (PDOs) retain the architecture, genotype, and phenotype of the patient’s primary tumor and provide a rapid in vitro model for drug testing [34]. We then tested the efficacy of BR using mutant KRAS CRC PDOs. These PDOs had two distinct morphologies as cysts or clusters [35], and did not differ significantly in growth (Fig. S5a). PDOs were significantly more sensitive to the BR combination, compared to single agent (Fig. 5a-b). The BR treatment induced marked apoptosis (cleaved-caspase-3) in the center of PDO, along with highly elevated p-eIF2α and expression of ISR effectors at 24 h (Fig. 5c-e, Fig. S5b).
Fig. 5.
The BR combination induces ISR hyperactivation and killing of PDOs. CRC PDOs were treated with vehicle (Un), Bortezomib (B), Everolimus (R), or their combination (BR, 7.5 nM and 15 µM). a Representative images of PDOs at 48 h. Live and dead organoids are indicated by black and red arrows, respectively. Scale bar = 100 µM. b The growth of PDOs treated as in (A) was quantified by 3D cell viability assay. c Representative images of H&E, and cleaved caspase 3 and p-eIF2α IF at 24 h in Paraffin-embedded PDO1 sections. Scale bar = 50 µM. d Quantitation of cleaved-caspase 3 and p-eIF2α IF in c. A minimum of 30 organoids were analyzed for each condition. e qRT-PCR analysis at 24 h with values normalized to un (1). b, d, e, values are mean + s.d. (n = 3, 8-well pool). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (Student’s t-test, two tailed). Un vs. BR. f Correlation of four stress-gene signature and OS in the COAD (ALL) or the MSL-L (n = 54) and MSS (n = 182) subgroups. Log-rank test
Our data support that mutant KRAS is linked to elevated Myc and ISR in CRC. Mutant KRAS was not significantly correlated with microsatellite instability (MSI) status, and correlated with shortened median overall survival (OS) in CRC patients (59.93 vs. 78.73 months) (n = 1965, cBioportal) (Fig. S5c-d). However, ISR effectors are numerous and regulated by oncogenic drivers other than mutant KRAS or Myc [10, 28]. We therefore examined potential prognostic values of ISR targets using a web-based database GEPIA2 (Gene Expression Profiling Interactive Analysis) [36]. Higher expression of CHOP(DDIT3), GADD45B, CRYBA4, and HSPA1L was associated with shortened overall survival (OS) in TCGA (The Cancer Genome Atlas) COAD cohort (n = 270) (Fig. S5e). CHOP(DDIT3), GADD45B, CRYBA4 were induced by BR in mutant KRAS CRC cells (Figs. 2 and 4). Remarkably, the 4-gene signature predicts OS better than any single gene (Log-rank p = 7.1e-05, HR (Hazard ratio) = 2.7, p(HR) = 0.001384) in this cohort (n = 270), as well as in the MSI-L (n = 52) and microsatellite stable (MSS) (n = 184) subsets (Log-rank p = 0.00012, HR = 2.9, p(HR) = 0.00023) (Fig. 5f, Fig.S5e).
The BR combination shows potent efficacy in mutant KRAS MSS CRC PDXs
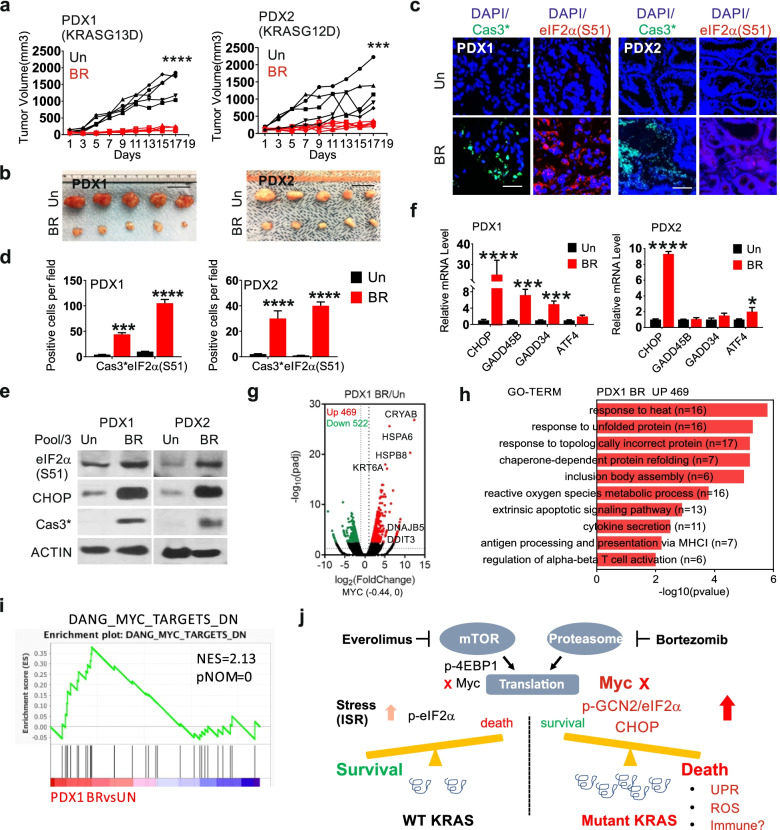
The above data suggest that Myc-driven stress adaptation might be a selective target in aggressive CRCs. Patient derived xenograft (PDX) models preserved tumor histology and heterogeneity [37], and were used to test the efficacy of the BR combination. We selected two PDXs (mutant KRAS and MSS). PDX1 (KRASG13D) and PDO1 were originated from the same tumor. PDX2 (KRASG12D) is therapy resistant and from a metastatic lesion of deceased patient after multiple lines of chemo- and targeted therapies (FOLFIRI, FOLFOX, 5-FU, bevacizumab, onalespib, indimitecan, selumetinib and MK-2206). PDX1 and PDX2 were highly responsive to the BR treatment (Fig. 6a-b, Fig. S6a), showing massive loss of proliferation (Ki67 index) and reduction in cellularity upon BR treatment (Fig. S6b-c). The BR treatment was well tolerated and induced a slight and transient weight loss compared to the control group, which recovered by day 9 (Fig. S6d).
Fig. 6.
The BR combination kills mutant KRAS MSS CRC PDXs. NSG were randomized into control and treatment groups when average PDX volume reached around 100 mm3. Mice were treated with vehicle (Un), or the combination of Everolimus (oral gavage, 10 mg/kg) and Bortezomib (i.p., 0.5 mg/kg) (BR) every other day starting day 1. Tumors were harvested for analysis. a Individual tumor volume was calculated and plotted (n = 5) with b representative images on day 17. Scale Bar = 2 cm. c Representative cleaved caspase 3 and p-eIF2α IF with DAPI counterstain (blue) on day 4. Scale Bar = 100 µM. d Quantification of cleaved-caspase 3 and p-eIF2α IF as (c) in 3 randomly chosen 400X fields. e Western blotting of indicated proteins on day 4. N = 3 (pooled). f qRT-PCR of indicated makers on day 4. The values were normalized to Un (1). N = 3 (pooled mRNA). g BR DEGs in PDX1 on day 4 visualized by volcano plot. Upregulated (red) or down regulated (green) genes (fold ≥ 2, p < 0.005). Selected upregulated genes are shown. h Top 10 enriched non-overlapping pathways identified by GO in upregulated genes (469). i. GSEA of differential genes in PDX1 (C2 dataset). The indicated gene set is shown with NES and corresponding p-value. a, d, f, values are mean + s.d. (n = 3 or as specified). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (Student’s t-test, two tailed). Vehicle (Un) vs. BR. j Working model. Targeting Myc-driven stress in KRAS mutated CRCs. Compared to WT KRAS CRCs, mutant KRAS CRCs show elevated basal Myc and metabolic stress. Acute ablation of Myc protein by the BR combination impairs their adaptation, leading to sustained ISR (p-GCN2/p-eIF2α/CHOP) and cell death associated with UPR, reactive oxygen species (ROS) production, and immune activation
To monitor drug-induced acute response in vivo, we first analyzed several ISR and cell death markers in PDXs on day 4, which is 24 h after the second BR treatment. BR group showed significant increases in p-eIF2α, CHOP and cleaved caspase-3 by staining and western blotting (Fig. 6c-e). qRT-PCR confirmed the induction of ISR effectors (Fig. 6f, Fig. S6e), and a similar pattern between PDO1 and PDX1. We used RNA-Seq to assess BR-induced global transcriptomic changes in PDX1, and identified 469 upregulated and 522 downregulated genes (filtered for human reads,twofold or more, p ≤ 0.005). Upregulated genes were highly enriched in proteo- and oxidative stress, apoptosis, and immunity (Fig. 6g-h), while downregulated genes were highly enriched in metabolism such as oxidative phosphorylation and nucleotide and ATP biosynthesis (Fig. S6f). Upregulated genes shared by HCT 116 and PDX1 were highly enriched in proteostress and extrinsic apoptotic pathway (Fig. S6g-h). Like cell line studies, BR treatments significantly enriched Myc down-regulated targets (Fig. 6i), with little effect on MYC mRNA (Fig. 6g). These data collectively support that the BR combination targets Myc-dependent stress adaptation in mutant KRAS CRCs to promote ISR-dependent metabolic crisis and cell death (Fig. 6j).
Discussion
Targeting mutant KRAS is a major clinical challenge and the “holy grail” in cancer therapy. Mutant KRAS is linked to poor prognosis in CRC, and promotes resistance to EGFR antibodies [4] and anti-PD-1 [38]. Resistance is in part mediated through feedback activition of adaptive responses to avoid drug or immune-mediated cell killing [4, 39]. Here, we report a highly effective combination of two FDA-approved drugs, Bortezomib and Everolimus, against mutant KRAS CRC cells, PDOs, and PDXs. Mechanistically, the BR combination, but not single agents, ablates high Myc levels in mutant KRAS CRC, leading to unresolvable proteostress and cell death with a transcrptomic signature charaterized by proteotoxicity, oxidative stress, metabolic suppression, and immune activation (Fig. 6j). The vulerability to this drug combination is seletive to the mutant KRAS-Myc axis as demonstrated using isogenic and nonisogenic CRC cells. Our study therefore provides a potentially new therapeutic strategy to target Myc in mutant KRAS CRCs using FDA-approved drugs.
KRAS is a membrane-bound GTPase that cycles between GTP-bound active and GDP-bound inactive forms. Most oncognic mutations affect this on–off switch and lock the protein in the active form to drive cell proliferation, apoptosis resistance, and metastasis [4]. KRAS mutations are biochemcially distinct and appear to influence CRC patient outcomes [5], making it challening to develope allele spcific RAS inhibitors. Despite some encouraging data on G12C and G13C inhibitors in lung cancer [5, 6], G12D and G12V mutations are the most prevalent KRAS mutations in CRC [3] associated with worse overall survival [40]. Indirectly targeting mutant KRAS also has not had much succes [4, 29]. Our data suggest elevated Myc and protesostress as a druggable vunerability across mutant KRAS CRC cell lines, PDXs and PDOs using BR combination. Elevated proteostress is also reported in cancers with amplified KRAS [21] or MSI-high [41]. KRAS mutations are prevalent in pancreatic cancer (90%), lung cancer (20–30%), and endometrial cancer (18%) [3]. It is tempting to sepculate that this combination might be effective in other epithelial cancers with mutant KRAS.
Myc is a master regulator of oncogenic growth through extensive transcriptional and translational networks, and cooperates with a variety of cofactors [16, 42, 43]. Myc promotes stress adaptation and survival of cancer cells by increased autophagy [44], preservation of bioenergetics [45] through transient inhibition of RNA pol II-mediated transcription [46] or protein synthesis through GCN2/p-eIF2α-dependent negative feedback [28]. Myc translation is highly regulated and enhanced by mutant KRAS [30, 47]. However, this Myc-translation feed-forward loop is notoriously difficult to break [14, 15]. Our data support that Myc ablation is likely required to disrupt these protective mechanisms in mutant KRAS cells ‘addicted” to Myc (Fig. 6j). Consistent with this model, BR, but not single agents, rapidly ablates Myc protein, not mRNA, leading to sustained induction of p-GCN/p-eIF2α/CHOP. MYC siRNA decreased sensitivity while MYC over-expression increased sensitivity through the p-GCN/p-eIF2α/CHOP axis in mutant and WT KRAS isogenic cells, respectively. The combination is therefore necessary to push mutant KRAS CRC cells out of Myc-dependent adaptive “Goldilocks Zone” [8] and into metabolic crisis and cell death. However, the role of other eIF2α kinases in cell death cannot be ruled out due to their well-documented crosstalk and shared downstream targets [7, 12, 13, 48].
CRCs are heterogeous and can be classified into several molecular groups based on gene expression [49]. The majority of CRC are MSI-L and MSS, associated with a high Wnt/Myc signature, and do not respond to immune checkpoint inhibitors such as anti-PD-1 [50, 51]. Overwhelming evidence supports the cooperation of mutant KRAS and Myc in metabolic reprogramming and therapeutic resistance through the tumor microenvironment [18–20, 52]. Here, we found that the 4-gene ISR signature (DDIT3, GADD45B, CRYBA4, and HSPA1L) is strongly associated with poor prognosis in CRC patients, but also with sensitivity to BR in vitro and in vivo. Our model (Fig. 6j) helps explain this paradox. As mutant KRAS cells become dependent on elevated Myc and ISR (p-GCN2/p-eIF2α) for metabolic adaptation and immune suppression, acute Myc ablation breaks this state by inducing even higher and sustained ISR and cell death (Fig. 6j). Mutant KRAS-Myc strongly increases the range of transcriptional response and level of p-eIF2α to BR treatment, but it remains likely that additional BR targets are involved given the large number of Myc cofactors [16, 42, 43]. Elevated p-eIF2α is a suggested marker of immunogenic cell death (ICD) [53, 54]. The use of syngeneic models can help better understand drug-induced synthetic lethality for diffucult targets [55] through the tumor microenvironment (TME). The challenge however remains to develop mechanism-based clinical biomarkers distinct from activated oncogenes. Our data support that PDO and PDX might be useful in this regard, as those from the same patient showed similar stress-related pathway gene expression changes.
In summary, our study demonstrates a critical role of Myc-mediated stress adaptation in the surival of mutant KRAS CRC. The potent efficacy of the combination of FDA-approved mTOR and proteasome inhibitors is mediated through Myc ablation and induction of p-GCN2/p-eIF2α-dependent cell killing. With multiple FDA-approved agents in each class, the prevelance of KRAS mutations, and mechanistic biomarkers, it would be interesting to design clinical trials to evaluate potential benefit of BR or similar combinations in CRC patients.
Material and methods
Approval and protocols
All methods were performed in accordance with the relevant guidelines and regulations. The protocols for the use of recombinant DNA and animals included IBC201700136 and IACUC# 17071072. The protocol of establishing PDO/PDX includes REN11110076/IRB0411047.
Cell culture, treatment, and transfection
The human colorectal cancer cell lines, including HCT116, DLD1, SW480, LS180, LIM1215, SW48, DIFI and RKO were obtained from the American Type Culture Collection (Manassas, VA, USA) ATCC. Isogenic KRAS pairs HCT116 (WT, G13D), DLD1 (WT, G13D) and SW48 (WT/G12V) cell lines were obtained from Bert Vogelstein at Johns Hopkins University [33]. PUMA KO [31], DR5 KO, NOXA KO, BIM KO [32, 56] HCT 116 cells were generated in the lab. Information on major drivers or isogenic cell lines are found in Supplementary Materials (Table S1). Cell lines were regularly monitored for absence of Mycoplasma, approximately every 6 months. Any cell line is used for less than 2 months (10 or fewer passages) in culture upon thawing from LN tank. All cell lines were cultured in McCoy’s 5A modified medium (Invitrogen, Carlsbad, CA, Cat#16,600–082) supplemented with 10% defined fetal bovine serum (Hyclone, Logan, UT, Cat #SH3007103), 100 units/mL penicillin, and 100 µg/mL streptomycin (Invitrogen, Carlsbad, CA, Cat#15,140,148) unless noted otherwise. More details on drug treatment and transfection are found in the supplemental material.
Cell viability was performed using the MTT assay kit (Promega, Madison, WI, #G3580) as described [30]. In brief, cells were seeded in 96-well plates at a density of 1 × 104 cells/well and treated with different agents for 48 h before the analysis. Combination index(CI) and fraction affected(Fa) values were calculated using Compusin software (https://www.combosyn.com).
Crystal violet staining. Following various treatment, attached cells or clones were stained and with crystal violet (Sigma, St. Louis, MO, Cat# C0775) (3.7% Paraformaldehyde, 0.05% crystal violet in distilled water and filtered at 0.45 um before use) (2). For colony formation assays, equal numbers of cells were subjected to various treatments and plated into 12-well plates at different dilutions. Colonies were visualized by crystal violet staining 14 days after plating. Each assay was conducted in triplicate and repeated three times.
Apoptosis assays
Cell death and apoptosis was analyzed by nuclear staining with cells harvested from 12-well plates and Hoechst 33,258 (Invitrogen, Cat# 40,045), and Annexin V/propidium iodide (PI) followed by flow cytometry as described [57]. Experiments were repeated on two or more occasions (different days) with similar results. Adherent and floating cells were harvested, stained with Hoechst 33,258 (Invitrogen), and analyzed for apoptosis by nuclear staining and counting cells with condensed and fragmented nuclei. At least 300 cells were analyzed for each treatment. Annexin V/propidium iodide (PI) staining was performed. Flow cytometry plots and quantitation were based on the analysis of 20,000 cells for each condition. Results from one representative experiment are shown with fraction (%) of indicated population.
Western blotting and quantitative Reverse transcription PCR (qRT-PCR)
Western blotting was performed as previously described [26, 30]. Details on antibodies are found in the Supplementary Materials (Table S4). Total RNA was isolated from cells or tissues using Mini RNA Isolation II Kit (Zymo Research, Orange, CA, Cat# R1054) according to the manufacturer’s protocol. One μg of total RNA was used to generate cDNA using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA, Cat# 18,064–014). Details on primers are found in the Supplementary Materials (Table S5). For organoids, pooling of 8 or more wells for each condition is necessary to prepare enough lysates or total RNA. For tumors, lysates were pooled from 3 randomly chosen tumors in each group. cDNA was synthesized from RNA pooled from 3 randomly chosen tumors in each group.
Patient derived CRC organoids (PDOs)
PDOs were established using surgically resected de-identified CRC tissues from the Biospecimen Core (PBC) at University of Pittsburgh with tissue collection under informed consent and usage approved by the Institutional Review Board at the University of Pittsburgh. CRC organoids were cultured in Matrigel (Corning) incubated with advanced DMEM/F12 (Invitrogen) medium with supplements including 50% (v/v) L-WRN-conditioned medium containing Wnt3a, R-spondin, and Noggin prepared as described [30, 58]. More details on medium, passage and treatment are found in supplemental materials.
Patient derived CRC xenografts (PDX)
Animal experiments were approved by the University of Pittsburgh Institutional Animal Care and Use Committee. All methods were performed in accordance with the relevant guidelines and regulations. Patient-derived xenograft (PDX) tumors were established and propagated in 5–6-week-old female NOD.Cg-Prkdcscid Il2rgtm1Wjl/Saju (NSG) mice (Jackson Laboratory, Bar Harbor, ME) as described (4, 10) using samples collected with IRB approval and obtained from the NCI.
Tumor-bearing mice were randomized into untreated and treated groups. Mice were treated with BR Injection every other day. B was given by intraperitoneal (IP) injection at 0.5 mg/kg, R was given by oral gavage (OG) at 10 mg/kg. Tumor growth was monitored by calipers, and tumor volumes were calculated according to the formula 1/2 × length × width2. Ethical endpoint was defined as a time point when a tumor reached 2 cm or more in any dimension. Tumor tissues were analyzed for histology, staining, protein, and mRNA expression. Selected tumors were pooled to prepare protein or RNA which was then used for western blotting, RT-PCR and RNA-seq. More Details on PDX establishment and treatment and analysis are found in supplemental materials.
RNA sequencing (RNA-Seq)
Total RNA was prepared from cells and tissues using TRIzol RNA Isolation Reagents (Sigma, St. Louis, MO, Cat# 15,596,026) according to manufacturer’s instructions. Library construction, RNA sequencing (RNA-seq), and data analysis were performed by Novogene using the IIIumina HiSeq platform. Sample quality was assessed by HTSeq v0.6.1 to the count the read numbers mapped of each gene. FPKM (fragments per kilobase of transcript per million mapped reads) of each gene was calculated based on the length of a gene and read counts mapped to this gene. For PDX samples, Flow B was used to filter and map human reads with Fragments Per Kilobase Per Million (FPKM) calculated based on all mapped reads. For samples without biological replicates, readcount was adjusted by TMM, then differential expression significant analysis was performed by using the edgeR package, while the significant criterion are both p value < 0.005 and |log2(Fold Change)|> 1 (i.e., twofold).
Bioinformatics
More details on differential expression analysis and visualization such as Volcano plots, Gene Ontology (GO), Gene Set Enrichment Analysis (GSEA) and Venn diagram are found in supplemental materials.
Data deposit
Analyzed RNA-seq data on differential gene expression, including raw readcount and normalized abundance of all called genes in paired samples (DEG, no cutoff), and differentially expressed genes (DEG_all, including both ups and downs, with indicated cutoff), additional gene list (s) and analyses(GSEA) are deposited at DRYAD. This dataset has been assigned a unique identifier or DOI (https://doi.org/10.5061/dryad.sf7m0cg6h) for free public access upon the publication of manuscript.
Statistical analysis
Statistical analyses were carried out using GraphPad Prism software (VIII, GraphPad Software, Inc., La Jolla, CA). Comparisons between two groups were made by two-tailed, unpaired t test. Differences were considered significant if the probability of the difference occurring by chance was less than 5 in 100 (p < 0.05). The means ± one standard deviation (s.d.) were displayed in the figures. Sample size was determined using a combination of published work and power calculations.
Supplementary Information
Additional file 1: Supplementary Table S1. IC50s of translation-targeted agents in HCT 116 cells. Supplementary Table S2. Cell line information. Supplementary Table S3. chemicals, siRNA, and other key reagents. Supplementary Table S4. Antibodies used in the study. Supplementary Table S5. qRT-PCR primers used (Human). Supplementary Figure S1. The BR combination induces apoptosis in mutant KRAS CRC cells. Supplementary Figure S2. The BR combination induces metabolic suppression and prolonged ISR. Supplementary Figure S3. BR induces ISR hyperactivation without affecting MYC mRNA. Supplementary Figure S4. BR induces mutant KRAS-selective ISR hyperactivation. Supplementary Figure S5. BR induces ISR-associated killing of CRC PDOs. Supplementary Figure S6. BR induces ISR hyperactivation and metabolic crisis in mutant KRAS CRC PDXs.
Acknowledgements
Not applicable.
Code availability
Not applicable.
Abbreviations
- CRC
Colorectal cancer
- EGFR
Epidermal Growth Factor Receptor
- ISR
The Integrated Stress Response
- eIF2α
Eukaryotic Translation Initiation Factor 2 alpha
- eIF4E
Eukaryotic Translation Initiation Factor 4E
- GCN2
General Control Nonderepressible 2
- ATF4
Activating Transcription Factor 4
- CHOP
C/EBP Homologous Protein
- PERK
Protein Kinase RNA-Like ER Kinase
- UPR
Unfolded Protein Response
- ROS
Reactive Oxygen Species
- GADD45B
Growth Arrest and DNA Damage-Inducible Protein 45 Beta
- GADD34
Growth Arrest And DNA Damage-Inducible Protein 34
- DR5
Death receptor 5
- PUMA
P53 Upregulated Modulator of Apoptosis (encoded by BBC3)
- PMAIP1, or NOXA
Phorbol-12-myristate-13-acetate-induced protein 1
- BIM
Bcl-2-like protein 11
- MSI
Microsatellite instability
- MSS
Microsatellite stable (MSS)
- TCGA
The Cancer Genome Atlas
- HR
Hazard ratio
- PDO
Patient derived organoids
- PDX
Patient derived xenografts
- IP injection
Intraperitoneal injection
- OG
Oral gavage
- RNA-Seq
RNA sequencing
- DEGs
Differentially expressed genes
- GO
Gene Ontology
- GSEA
Gene Set Enrichment Analysis
- NES
Normalized enrichment score
Authors’ contributions
Conception and design: HR and JY. Development of methodology: HR, BJL and LS. Data acquisition and analysis: HR, YP, LC and JY. Administrative and technical support (i.e., animal, key material, and facilities): RS, XL, LZ and JY. Review and critical comments: BJL, CK, LZ and JY. Writing: HR and JY. Study supervision: JY. All authors read and approved the final manuscript.
Funding
This work is supported in part by NIH grant R01CA215481 and UPMC HCC funds (JY), R01LM012011 (XL), 1K99LM013089-01A (LC), the Early Detection Research Network U01-CA152753 (RES), and R01CA217141, R01CA236271, R01CA247231 and R01CA248112 (LZ). This project used the UPMC Hillman Cancer Center shared glassware, animal, and cell and tissue imaging facilities that were supported, in part, by National Cancer Institute award P30CA047904.
Availability of data and materials
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq data and associated analyses have been deposited at DRYAD with a unique identifier or DOI (10.5061/dryad.sf7m0cg6h).
Declarations
Ethics approval and consent to participate
Studies on recombinant DNA were approved by Institutional Biosafety Committee (IBC). Animal experiments were approved by the University of Pittsburgh Institutional Animal Care and Use Committee (IACUC). The use of human tissues (de-identified) in PDO or PDX studies was approved by Institutional Review Board (IRB). Informed consent was obtained from all individual participants included in the study. All samples used in this study were deidentified.
Consent for publication
All authors have reviewed the data and agreed on submission. Additional informed consent was obtained from all individual participants for whom identifying information is included for any subsequent study for publication.
Competing interests
The authors declare no conflicts of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prior IA, Hood FE, Hartley JL. The Frequency of Ras Mutations in Cancer. Cancer Res. 2020;80(14):2969–2974. doi: 10.1158/0008-5472.CAN-19-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knickelbein K, Zhang L. Mutant KRAS as a critical determinant of the therapeutic response of colorectal cancer. Genes Dis. 2015;2(1):4–12. doi: 10.1016/j.gendis.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mullard A. Cracking KRAS. Nat Rev Drug Discov. 2019;18(12):887–891. doi: 10.1038/d41573-019-00195-5. [DOI] [PubMed] [Google Scholar]
- 6.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25(3):272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 7.Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Targeting the translation machinery in cancer. Nat Rev Drug Discov. 2015;14(4):261–278. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- 8.Pelletier J, Graff J, Ruggero D, Sonenberg N. Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res. 2015;75(2):250–263. doi: 10.1158/0008-5472.CAN-14-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt S, Denk S, Wiegering A. Targeting Protein Synthesis in Colorectal Cancer. Cancers (Basel) 2020;12(5):1298. doi: 10.3390/cancers12051298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martineau Y, Azar R, Bousquet C, Pyronnet S. Anti-oncogenic potential of the eIF4E-binding proteins. Oncogene. 2013;32(6):671–677. doi: 10.1038/onc.2012.116. [DOI] [PubMed] [Google Scholar]
- 12.Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011;13(3):184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castilho BA, Shanmugam R, Silva RC, Ramesh R, Himme BM, Sattlegger E. Keeping the eIF2 alpha kinase Gcn2 in check. Biochim Biophys Acta. 2014;1843(9):1948–1968. doi: 10.1016/j.bbamcr.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 14.Lin CJ, Nasr Z, Premsrirut PK, Porco JA, Jr, Hippo Y, Lowe SW, et al. Targeting synthetic lethal interactions between Myc and the eIF4F complex impedes tumorigenesis. Cell Rep. 2012;1(4):325–333. doi: 10.1016/j.celrep.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pourdehnad M, Truitt ML, Siddiqi IN, Ducker GS, Shokat KM, Ruggero D. Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proc Natl Acad Sci U S A. 2013;110(29):11988–11993. doi: 10.1073/pnas.1310230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dang CV. A Time for MYC: Metabolism and Therapy. Cold Spring Harb Symp Quant Biol. 2016;81:79–83. doi: 10.1101/sqb.2016.81.031153. [DOI] [PubMed] [Google Scholar]
- 17.Baluapuri A, Wolf E, Eilers M. Target gene-independent functions of MYC oncoproteins. Nat Rev Mol Cell Biol. 2020;21(5):255–267. doi: 10.1038/s41580-020-0215-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robichaud N, Sonenberg N. Translational control and the cancer cell response to stress. Curr Opin Cell Biol. 2017;45:102–109. doi: 10.1016/j.ceb.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Cubillos-Ruiz JR, Bettigole SE, Glimcher LH. Tumorigenic and Immunosuppressive Effects of Endoplasmic Reticulum Stress in Cancer. Cell. 2017;168(4):692–706. doi: 10.1016/j.cell.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tameire F, Verginadis II, Koumenis C. Cell intrinsic and extrinsic activators of the unfolded protein response in cancer: Mechanisms and targets for therapy. Semin Cancer Biol. 2015;33:3–15. doi: 10.1016/j.semcancer.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerr EM, Gaude E, Turrell FK, Frezza C, Martins CP. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature. 2016;531(7592):110–113. doi: 10.1038/nature16967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hetz C, Chevet E, Harding HP. Targeting the unfolded protein response in disease. Nat Rev Drug Discov. 2013;12(9):703–719. doi: 10.1038/nrd3976. [DOI] [PubMed] [Google Scholar]
- 23.He K, Zheng X, Zhang L, Yu J. Hsp90 inhibitors promote p53-dependent apoptosis through PUMA and Bax. Mol Cancer Ther. 2013;12(11):2559–2568. doi: 10.1158/1535-7163.MCT-13-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tong J, Tan S, Nikolovska-Coleska Z, Yu J, Zou F, Zhang L. FBW7-Dependent Mcl-1 Degradation Mediates the Anticancer Effect of Hsp90 Inhibitors. Mol Cancer Ther. 2017;16(9):1979–1988. doi: 10.1158/1535-7163.MCT-17-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chattopadhyay N, Berger AJ, Koenig E, Bannerman B, Garnsey J, Bernard H, et al. KRAS Genotype Correlates with Proteasome Inhibitor Ixazomib Activity in Preclinical In Vivo Models of Colon and Non-Small Cell Lung Cancer: Potential Role of Tumor Metabolism. PLoS One. 2015;10(12):e0144825. doi: 10.1371/journal.pone.0144825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He K, Zheng X, Li M, Zhang L, Yu J. mTOR inhibitors induce apoptosis in colon cancer cells via CHOP-dependent DR5 induction on 4E-BP1 dephosphorylation. Oncogene. 2016;35(2):148–57. doi: 10.1038/onc.2015.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He K, Chen D, Ruan H, Li X, Tong J, Xu X, et al. BRAFV600E-dependent Mcl-1 stabilization leads to everolimus resistance in colon cancer cells. Oncotarget. 2016;7(30):47699–47710. doi: 10.18632/oncotarget.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt S, Gay D, Uthe FW, Denk S, Paauwe M, Matthes N, et al. A MYC-GCN2-eIF2alpha negative feedback loop limits protein synthesis to prevent MYC-dependent apoptosis in colorectal cancer. Nat Cell Biol. 2019;21(11):1413–1424. doi: 10.1038/s41556-019-0408-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Yu J. Role of apoptosis in colon cancer biology, therapy, and prevention. Curr Colorectal Cancer Rep 2013;9(4). DOI: 10.1007/s11888-013-0188-z. [DOI] [PMC free article] [PubMed]
- 30.Ruan H, Li X, Xu X, Leibowitz BJ, Tong J, Chen L, et al. eIF4E S209 phosphorylation licenses myc- and stress-driven oncogenesis. Elife. 2020;9:e60151. doi: 10.7554/eLife.60151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu J, Wang Z, Kinzler KW, Vogelstein B, Zhang L. PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proc Natl Acad Sci U S A. 2003;100(4):1931–1936. doi: 10.1073/pnas.2627984100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan X, Tong J, Wang YJ, Fletcher R, Schoen RE, Yu J, et al. BET inhibitors potentiate chemotherapy and killing of SPOP-mutant colon cancer cells via induction of DR5. Cancer Res. 2019;79(6):1191–1203. doi: 10.1158/0008-5472.CAN-18-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325(5947):1555–1559. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer. 2018;18(7):407–418. doi: 10.1038/s41568-018-0007-6. [DOI] [PubMed] [Google Scholar]
- 35.Song X, Shen L, Tong J, Kuang C, Zeng S, Schoen RE, et al. Mcl-1 inhibition overcomes intrinsic and acquired regorafenib resistance in colorectal cancer. Theranostics. 2020;10(18):8098–8110. doi: 10.7150/thno.45363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morral C, Stanisavljevic J, Hernando-Momblona X, Mereu E, Alvarez-Varela A, Cortina C, et al. Zonation of Ribosomal DNA Transcription Defines a Stem Cell Hierarchy in Colorectal Cancer. Cell Stem Cell. 2020;26(6):845–861.e12. doi: 10.1016/j.stem.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao W, Overman MJ, Boutin AT, Shang X, Zhao D, Dey P, et al. KRAS-IRF2 Axis Drives Immune Suppression and Immune Therapy Resistance in Colorectal Cancer. Cancer Cell. 2019;35(4):559–572.e7. doi: 10.1016/j.ccell.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruan H, Leibowitz BJ, Zhang L, Yu J. Immunogenic cell death in colon cancer prevention and therapy. Mol Carcinog. 2020 doi: 10.1002/mc.23183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imamura Y, Morikawa T, Liao X, Lochhead P, Kuchiba A, Yamauchi M, et al. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorectal cancers. Clin Cancer Res. 2012;18(17):4753–4763. doi: 10.1158/1078-0432.CCR-11-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGrail DJ, Garnett J, Yin J, Dai H, Shih DJH, Lam TNA, et al. Proteome Instability Is a Therapeutic Vulnerability in Mismatch Repair-Deficient Cancer. Cancer Cell. 2020;37(3):371–386.e12. doi: 10.1016/j.ccell.2020.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lorenzin F, Benary U, Baluapuri A, Walz S, Jung LA, von Eyss B, et al. Different promoter affinities account for specificity in MYC-dependent gene regulation. Elife. 2016;5:e15161. doi: 10.7554/eLife.15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baluapuri A, Hofstetter J, DudvarskiStankovic N, Endres T, Bhandare P, Vos SM, et al. MYC Recruits SPT5 to RNA Polymerase II to Promote Processive Transcription Elongation. Mol Cell. 2019;74(4):674–687.e11. doi: 10.1016/j.molcel.2019.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hart LS, Cunningham JT, Datta T, Dey S, Tameire F, Lehman SL, et al. ER stress-mediated autophagy promotes Myc-dependent transformation and tumor growth. J Clin Invest. 2012;122(12):4621–4634. doi: 10.1172/JCI62973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tameire F, Verginadis II, Leli NM, Polte C, Conn CS, Ojha R, et al. ATF4 couples MYC-dependent translational activity to bioenergetic demands during tumour progression. Nat Cell Biol. 2019;21(7):889–899. doi: 10.1038/s41556-019-0347-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dejure FR, Royla N, Herold S, Kalb J, Walz S, Ade CP, et al. The MYC mRNA 3'-UTR couples RNA polymerase II function to glutamine and ribonucleotide levels. EMBO J. 2017;36(13):1854–1868. doi: 10.15252/embj.201796662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grabocka E, Bar-Sagi D. Mutant KRAS Enhances Tumor Cell Fitness by Upregulating Stress Granules. Cell. 2016;167(7):1803–1813.e12. doi: 10.1016/j.cell.2016.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep. 2016;17(10):1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27(4):450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lizardo DY, Kuang C, Hao S, Yu J, Huang Y, Zhang L. Immunotherapy efficacy on mismatch repair-deficient colorectal cancer: From bench to bedside. Biochim Biophys Acta Rev Cancer. 2020;1874(2):188447. doi: 10.1016/j.bbcan.2020.188447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruan H, Leibowitz BJ, Zhang L, Yu J. Immunogenic cell death in colon cancer prevention and therapy. Mol Carcinog. 2020;59(7):783–793. doi: 10.1002/mc.23183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kepp O, Semeraro M, Bravo-San Pedro JM, Bloy N, Buque A, Huang X, et al. eIF2alpha phosphorylation as a biomarker of immunogenic cell death. Semin Cancer Biol. 2015;33:86–92. doi: 10.1016/j.semcancer.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 54.Galluzzi L, Buque A, Kepp O, Zitvogel L, Kroemer G. Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol. 2017;17(2):97–111. doi: 10.1038/nri.2016.107. [DOI] [PubMed] [Google Scholar]
- 55.Setton J, Zinda M, Riaz N, Durocher D, Zimmermann M, Koehler M, et al. Synthetic Lethality in Cancer Therapeutics: The Next Generation. Cancer Discov. 2021 doi: 10.1158/2159-8290.CD-20-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tong J, Zheng X, Tan X, Fletcher R, Nikolovska-Coleska Z, Yu J, et al. Mcl-1 Phosphorylation without Degradation Mediates Sensitivity to HDAC Inhibitors by Liberating BH3-Only Proteins. Cancer Res. 2018;78(16):4704–4715. doi: 10.1158/0008-5472.CAN-18-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu J, Yue W, Wu B, Zhang L. PUMA sensitizes lung cancer cells to chemotherapeutic agents and irradiation. Clin Cancer Res. 2006;12(9):2928–2936. doi: 10.1158/1078-0432.CCR-05-2429. [DOI] [PubMed] [Google Scholar]
- 58.Leibowitz BJ, Yang L, Wei L, Buchanan ME, Rachid M, Parise RA, et al. Targeting p53-dependent stem cell loss for intestinal chemoprotection. Sci Transl Med. 2018;10(427):eaam7610. doi: 10.1126/scitranslmed.aam7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Table S1. IC50s of translation-targeted agents in HCT 116 cells. Supplementary Table S2. Cell line information. Supplementary Table S3. chemicals, siRNA, and other key reagents. Supplementary Table S4. Antibodies used in the study. Supplementary Table S5. qRT-PCR primers used (Human). Supplementary Figure S1. The BR combination induces apoptosis in mutant KRAS CRC cells. Supplementary Figure S2. The BR combination induces metabolic suppression and prolonged ISR. Supplementary Figure S3. BR induces ISR hyperactivation without affecting MYC mRNA. Supplementary Figure S4. BR induces mutant KRAS-selective ISR hyperactivation. Supplementary Figure S5. BR induces ISR-associated killing of CRC PDOs. Supplementary Figure S6. BR induces ISR hyperactivation and metabolic crisis in mutant KRAS CRC PDXs.
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq data and associated analyses have been deposited at DRYAD with a unique identifier or DOI (10.5061/dryad.sf7m0cg6h).