Abstract
Aims
Cardiac allograft vasculopathy (CAV) is the major long‐term complication after heart transplantation, leading to mortality and re‐transplantation. As available non‐invasive biomarkers are scarce for CAV screening, we aimed to identify a proteomic signature for CAV.
Methods and results
We measured urinary proteome by capillary electrophoresis coupled with mass spectrometry in 217 heart transplantation recipients (mean age: 55.0 ± 14.4 years; women: 23.5%), including 76 (35.0%) patients with CAV diagnosed by coronary angiography. We randomly and evenly grouped participants into the derivation cohort (n = 108, mean age: 56.4 ± 13.8 years; women: 22.2%; CAV: n = 38) and the validation cohort (n = 109, mean age: 56.4 ± 13.8 years; women: 24.8%, CAV: n = 38), stratified by CAV. Using the decision tree‐based machine learning methods (extreme gradient boost), we constructed a proteomic signature for CAV discrimination in the derivation cohort and verified its performance in the validation cohort. The proteomic signature that consisted of 27 peptides yielded areas under the curve of 0.83 [95% confidence interval (CI): 0.75–0.91, P < 0.001] and 0.71 (95% CI: 0.60–0.81, P = 0.001) for CAV discrimination in the derivation and validation cohort, respectively. With the optimized threshold of 0.484, the sensitivity, specificity, and accuracy for CAV differentiation in the validation cohort were 68.4%, 73.2%, and 71.6%, respectively. With adjustment of potential clinical confounders, the signature was significantly associated with CAV [adjusted odds ratio: 1.31 (95% CI: 1.07–1.64) for per 0.1% increment in the predicted probability, P = 0.012]. Diagnostic accuracy significantly improved by adding the signature to the logistic model that already included multiple clinical risk factors, suggested by the integrated discrimination improvement of 9.1% (95% CI: 2.5–15.3, P = 0.005) and net reclassification improvement of 83.3% (95% CI: 46.7–119.5, P < 0.001). Of the 27 peptides, the majority were the fragments of collagen I (44.4%), collagen III (18.5%), collagen II (3.7%), collagen XI (3.7%), mucin‐1 (3.7%), xylosyltransferase 1 (3.7%), and protocadherin‐12 (3.7%). Pathway analysis performed in Reactome Pathway Database revealed that the multiple pathways involved by the signature were related to the pathogenesis of CAV, such as collagen turnover, platelet aggregation and coagulation, cell adhesion, and motility.
Conclusions
This pilot study identified and validated a urinary proteomic signature that provided a potential approach for the surveillance of CAV. These proteins might provide insights into CAV pathological processes and call for further investigation into personalized treatment targets.
Keywords: Heart transplantation, Cardiac allograft vasculopathy, Biomarkers, Proteomics, Diagnosis
Introduction
Heart transplantation is the ultimate therapeutic option for patients with end‐stage heart failure. The survival time post‐heart transplantation has been remarkably prolonged due to progress in immunosuppression in recent decades. 1 However, within 7 to 13 years after heart transplantation, around 50% of heart transplantation recipients are affected by cardiac allograft vasculopathy (CAV) characterized by the progressive diffuse intimal thickening of the coronary arteries. 2 , 3 Patients with CAV are exposed to a 5.7‐fold higher risk of death or re‐transplantation compared with recipients without CAV. 4 Although various risk factors, such as inflammation, thrombosis, endothelial dysfunction, and fibrosis have been demonstrated to be associated with CAV, the pathogenesis remains incompletely understood. 5 , 6 , 7 , 8 , 9 , 10 , 11 , 12 To date, the routine surveillance of CAV is via invasive diagnostic techniques. Non‐invasive diagnostic biomarkers are scarce in clinical practice. 1 , 13 , 14
With the advent of precision medicine, the bottom‐up proteomic approach has been extensively implemented in cardiovascular research. 15 , 16 , 17 It enables us to identify thousands of proteins from a small volume of biofluids with no prior assumption. Nevertheless, the acquired massive, multi‐dimensional proteomic data also brings new challenges to the biomarker discovery in large‐scale cohorts due to the complex, formidable data analysis. 18 To address this issue, machine learning has been recently introduced to process and analyse proteomic data. 19 , 20 , 21
Embracing these cutting‐edge approaches, we aimed to identify and validate a urinary proteomic signature of CAV in a relatively large heart transplantation cohort and to provide perspectives on the underlying mechanism of CAV.
Methods
Study cohort
This study was conducted in compliance with the Helsinki declaration for research in humans and was approved by the Ethics Committee of the University Hospitals Leuven [approval numbers B322201421186 (S56384) and B322201421045 (S56472)]. Written informed consents were obtained from all participants. The recruitment was started in 2014 at the University Hospital Leuven. All surviving heart transplantation recipients were invited to provide a 5 mL midstream urine sample during their regular follow‐up visit at the hospital. Patients with CAV were included in the study if they had been diagnosed with CAV before or within 3 months of the urinary sample collection. For the non‐CAV group, patients were required to have a negative coronary angiographic result within 3 months of the urine sample collection or two negative results before and after urine sample collection. Of 305 heart transplant recipients, 88 patients were excluded because their coronary angiography results were uncertain (n = 28), or they were deemed ineligible according to the inclusion criteria (n = 60): CAV diagnosed after 3 months of urine sample collection (n = 3), or non‐CAV diagnosed beyond a 3 month interval of urine sample collection and without a repetitive angiography afterwards (n = 57). Thus, a total of 217 patients were included in the current study, 76 of which were CAV cases. To develop and validate the proteomic classifier, participants were divided into a derivation cohort (n = 108) and a validation cohort (n = 109) stratified by random sampling with equal distribution of CAV cases (Figure 1 ).
Figure 1.
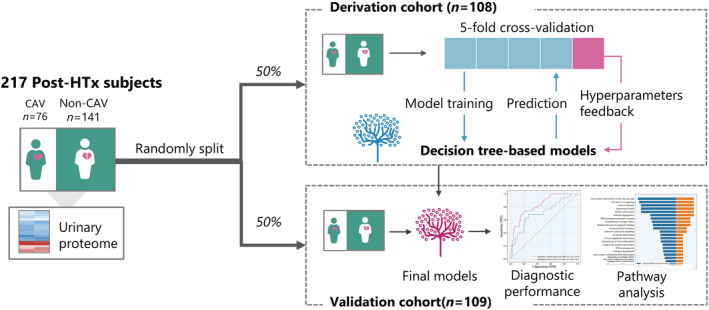
Schematic diagram of the study design. In total, 217 post‐heart transplantation (HTx) patients were included in this study and divided into a derivation and a validation cohort by stratified random sampling. The derivation cohort was further divided into 5‐folds for cross‐validation when training the decision tree‐based machine learning model for the discrimination of cardiac allograft vasculopathy (CAV). The diagnostic performance of the final model was evaluated in the validation cohort and enrichment pathway analysis was applied.
All recipients were prescribed lifelong immunosuppressive agents that consisted of calcineurin inhibitors, antimetabolites, steroids, and mammalian target of rapamycin (mTOR) inhibitors. Most patients received double or triple‐drug regimens for maintenance treatment, and it usually included a calcineurin inhibitor, an antimetabolite with or without methylprednisolone. Few patients were maintained by regimens that combined an mTOR inhibitor (everolimus) with a calcineurin inhibitor or an antimetabolite. For calcineurin inhibitors, cyclosporine was replaced with tacrolimus around 2000, while antimetabolite was changed from azathioprine to mycophenolate mofetil. Patients who did not experience rejection episodes were weaned off steroids during 6 to 12 months.
Diagnosis of cardiac allograft vasculopathy
The surveillance of CAV by coronary angiography was recommended by the International Society for Heart and Lung Transplantation (ISHLT). 13 Coronary angiography was scheduled at the first year after heart transplantation, at three‐year intervals during the first 10 year, and at 5 year intervals thereafter. Additional angiographies were performed as clinically indicated. According to ISHLT criteria, CAV was classified as CAV0, and CAV1 to 3. 13 CAV0 indicated no significant angiographic lesion detected; CAV1 denoted mild angiographic lesion without allograft dysfunction, including <50% stenosis of the left main coronary artery, or <70% stenosis of any primary vessel (the proximal and middle 33% of the left anterior descending artery, the left circumflex, and right coronary artery), or <70% stenosis (including diffuse narrowing) in any branch of three systems (the distal 33% of the left anterior descending artery, the left circumflex, and right coronary artery); CAV2 was defined as moderate angiographic lesions without allograft dysfunction, including <50% stenosis of the left main coronary artery, or ≥70% stenosis of a single primary vessel, or ≥70% isolated stenosis in branches of two systems; CAV3 referred to severe angiographic lesions, including ≥50% stenosis of the left main coronary artery, or ≥70% stenosis of 2 or 3 primary vessels, or ≥70% isolated stenosis of branches in all three systems, or CAV1 or CAV2 with allograft dysfunction. Allograft dysfunction was defined as left ventricular ejection fraction ≤45% or restrictive physiology determined by right atrial pressure >12 mmHg, pulmonary capillary wedge pressure >25 mmHg, or cardiac index <2 L/min/m2. Coronary angiography reports were retrospectively retrieved. Due to the limited cases of CAV2 and CAV3, patients were classified into CAV or non‐CAV groups for analysis.
Urinary proteomics
Urine samples were bio‐banked until assayed. Detailed information on sample preparation, proteome analysis by capillary electrophoresis coupled to mass spectrometry, data processing, and sequencing of the peptides were described in previous publications 22 , 23 and are included in the methods section of the supplements. Due to sparsity and multicollinearity of high dimension data and the risk of overfitting, peptides were excluded when being detectable in less than 90% of all participants or had a high correlation with other peptides (Pearson correlation coefficient <0.7). 24 Under these circumstances, 89 peptides were eventually analysed.
Machine learning model
The extreme gradient boost (XGBoost), a recently developed machine learning algorithm, was applied in this study. XGBoost is an ensemble method based on decision trees under the gradient boosting framework. 25 The algorithm starts from a weak decision tree and boosts by sequentially stacking succeeding decision trees to minimize the error of previous trees by optimizing the weights of misclassified samples. Moreover, this algorithm also regularizes parameters to avoid overfitting. The detailed information on the model construction is described in the supplements. Briefly, the proteomic data of the derivation cohort was used to train the model for the discrimination of patients with and without CAV. The derivation cohort was further randomly shuffled into 5‐folds for cross‐validation to evaluate internal discrimination performance and generalization performance. The final model was verified by an unforeseen validation cohort (Figure 1 ). The model construction was performed with Python, version 3.9 (https://www.python.org) and Scikit‐learn package, version 0.24.1 (https://scikit‐learn.org). 26
Statistical analysis
The statistical analysis was completed in SAS, version 9.4 (SAS Institute Inc., Cary, NC, USA). The means and proportions of clinical characteristics were compared by t test, Wilcoxon rank‐sum test or the Fisher's test where appropriate. Statistical significance was a two‐sided P value of 0.05. The predetermined sample size was calculated assuming that the area under the curve (AUC) of the proteomic classifier was at least 0.70. To achieve a power of 0.90 and a probability of type I error of 0.05 using a two‐sided test with a CAV to non‐CAV ratio of 2, the minimal sample size requested was 96 patients per cohort. 27 , 28
Univariate logistic models were applied to identify potential covariables. Then, statistically significant clinical variables were summarized in a propensity score. In the multivariable‐adjusted logistic regression model, the association between CAV and the urinary proteomic classifier was analysed after the adjustment of the propensity score. The association was presented as odds ratio (OR) every 0.1% increment in the predicted probability. The correlations of the proteomic classifier with high‐sensitivity troponin T (hsTnT), estimated glomerular filtration rate (eGFR) were analysed by the Spearman correlation and multivariable‐adjusted linear regression. The associations of the proteomic signature with the rejection episodes, the use of antiplatelet agents, and diabetes mellitus were determined by logistic regression analysis.
To evaluate the potential utility of the classifier for CAV detection, a predicted probability threshold was used to dichotomize the predicted probabilities into a binary prediction status (1/0). The optimized probability threshold was determined by maximizing Youden's index (sensitivity + specificity − 1) in the validation cohort. The diagnostic performance of the urinary proteomic classifier was evaluated by AUC, sensitivity, specificity, accuracy, positive predictive value, and negative predictive value. The added diagnostic value of the classifier was assessed by the integrated discrimination improvement (IDI) and the net reclassification improvement (NRI). 29 , 30 IDI refers to the difference in discrimination slopes before and after adding the proteomic classifier into a reference model. Specifically, the discrimination slope was the difference between the predicted probability of patients with and without CAV. For NRI, if P(up/CAV) is the percentage of patients with CAV whose predicted probability increased by adding the urinary proteomic classifier and if P(up/non‐CAV) is the percentage of patients without CAV whose predicted probability is increased, the continuous NRI equals 2 × [P(up/CAV) − P(up/non‐CAV)]. The 95% confidence interval (CI) was calculated by 1000 times bootstrap methods.
To interpret the biological function of the parental proteins from peptides included in the proteomic classifier, the enrichment pathway analysis was performed with Online Reactome Pathway Database, version 75 (https://reactome.org). 31 The P value of the annotated pathways was corrected by Bonferroni correction and considered as statistically significant if P < 0.05.
Results
Characteristics of study cohort
The age (±SD) of the 217 recipients averaged 55.0 ± 14.4 years. Of 217 patients, 51 (23.5%) were female, 76 (35.0%) were diagnosed with CAV, 173 (79.7%) underwent transplantation due to ischaemic cardiomyopathy or dilated cardiomyopathy, 192 (88.5%) had hypertension, 47 (21.7%) had diabetes mellitus. For the immunosuppressive regimens, 210 (96.8%) received calcineurin inhibitors, 181 (83.4%) were prescribed with antimetabolites, and 14 (6.5%) received mTOR inhibitors. Among all the patients, 57 (26.3%) were prescribed with antiplatelet agents, 8 (3.7%) with anticoagulants. The interval between heart transplantation and urine sample collection was a median of 7.6 years [interquartile range (IQR): 4.0–13.8]. The characteristics did not differ between the derivation and validation cohorts (Table 1 , P ≥ 0.13), except for the proportion of ischaemic cardiomyopathy (41.7% vs. 28.4%, P = 0.047). Compared with non‐CAV patients, patient with CAV were older, had longer years after heart transplantation, a higher prevalence of severe renal dysfunction and higher usage of antiplatelet agents (P ≤ 0.013, Table S1 ) in both derivation and validation cohorts, whereas the indications for transplantation, the prevalence of hypertension and diabetes mellitus, donor age, same donor–recipient sex, body mass index, immunosuppressive regimen, use of anticoagulants, serum total cholesterol, and left ventricular ejection fraction were similar (P ≥ 0.063, Table S1 ).
Table 1.
Clinical characteristics of participants
| Characteristics | Derivation cohort (n = 108) | Validation cohort (n = 109) | P |
|---|---|---|---|
| Number (%) with characteristic | |||
| Male | 84 (77.8) | 82 (75.2) | 0.75 |
| Same sex as donor | 22 (20.6) | 27 (25.0) | 0.52 |
| Indication for transplantation | |||
| Ischaemic cardiomyopathy | 45 (41.7) | 31 (28.4) | 0.047 |
| Dilated cardiomyopathy | 46 (42.6) | 51 (46.8) | 0.59 |
| Others | 17 (15.7) | 27 (24.8) | 0.13 |
| Smoking history | 71 (66.4) | 64 (58.7) | 0.30 |
| Hypertension | 93 (86.1) | 99 (90.8) | 0.86 |
| Diabetes mellitus | 26 (24.1) | 21 (19.3) | 0.26 |
| Treatments | |||
| Methylprednisolone | 25 (23.2) | 28 (25.7) | 0.41 |
| Calcineurin inhibitors | 105 (97.2) | 105 (96.3) | 0.75 |
| Antimetabolites | 93 (86.1) | 88 (80.7) | 0.36 |
| mTOR inhibitors | 4 (3.7) | 10 (9.2) | 0.17 |
| Statins | 98 (90.7) | 103 (94.5) | 0.31 |
| Antiplatelet agents | 29 (26.9) | 28 (25.7) | 0.88 |
| Anticoagulants | 3 (2.8) | 5 (4.6) | 0.72 |
| Mean ± SD or median (IQR) of characteristic | |||
| Age (years) | 56.4 ± 13.8 | 53.6 ± 15.0 | 0.15 |
| Donor age (years) | 36.6 ± 12.7 | 34.4 ± 12.9 | 0.20 |
| Years after transplantation (years) | 8.2 (4.2, 14.3) | 7.5 (3.9, 13.2) | 0.50 |
| Body mass index (kg/m2) | 25.0 ± 3.9 | 25.8 ± 4.3 | 0.28 |
| Systolic blood pressure (mmHg) | 140 ± 19 | 140 ± 18 | 0.97 |
| Diastolic blood pressure (mmHg) | 84 ± 10 | 85 ± 12 | 0.33 |
| Biochemistry | |||
| Serum total cholesterol (mmol/L) | 4.02 ± 0.73 | 3.98 ± 0.69 | 0.74 |
| HDL‐C (mmol/L) | 1.49 ± 0.43 | 1.46 ± 0.43 | 0.57 |
| LDL‐C (mmol/L) | 1.97 ± 0.60 | 1.90 ± 0.55 | 0.76 |
| Triglycerides (mmol/L) | 1.19 ± 0.71 | 1.32 ± 0.79 | 0.13 |
| Serum creatinine (mg/dL) | 1.33 ± 0.42 | 1.36 ± 0.46 | 0.69 |
| eGFR (mL/min/1.73 m2) | 63.1 ± 26.3 | 61.1 ± 21.5 | 0.95 |
| Left ventricular ejection fraction (%) | 58.9 ± 2.8 | 59.3 ± 2.7 | 0.31 |
Abbreviations: CAV, cardiac allograft vasculopathy; eGFR, estimated glomerular filtration rate; HDL‐C, high‐density lipoprotein cholesterol; IQR, interquartile range; LDL‐C, low‐density lipoprotein cholesterol; mTOR, the mechanistic target of rapamycin; SD, standard deviation.
Continuous values are presented as mean ± SD or median (IQR) and categorical variables as numbers (percentage). History of smoking referred to inhaling tobacco on a daily basis in the past; hypertension was an office blood pressure of ≥140 mmHg systolic or ≥90 mmHg diastolic or use of antihypertensive drugs; diabetes mellitus was a fasting/random blood glucose of ≥126/200 mg/dL or the use of antidiabetic drugs; calcineurin inhibitors included cyclosporine and tacrolimus; antimetabolites consisted of azathioprine and mycophenolate mofetil; mTOR inhibitors included everolimus; eGFR was estimated using the chronic kidney disease epidemiology collaboration creatinine equation.
The diagnostic performance of the proteomic classifier
Among 89 analysed peptides, the XGBoost model eventually selected 27 peptides to establish the classifier for discrimination of patients with CAV from those without CAV. The 27 peptides were listed in Table S2 , and their contribution to the classifier was assessed by the importance score. As shown in Figure 2 , the AUCs for CAV diagnosis were 0.83 (95% CI: 0.75–0.91, P < 0.0001) and 0.71 (95% CI: 0.60–0.81, P = 0.001) in the derivation cohort and the validation cohort, respectively. With the optimized threshold of 0.484, the classifier achieved the maximum of Youden's index of 0.42 in the validation cohort. With this threshold, its sensitivity, specificity, and accuracy were 73.7%, 71.4%, and 72.2% in the derivation cohort, and 68.4%, 73.2%, and 71.6% in the validation cohort (Table 2 ). The negative predictive value was over 80% and the positive predictive value was around 58% in both cohorts.
Figure 2.
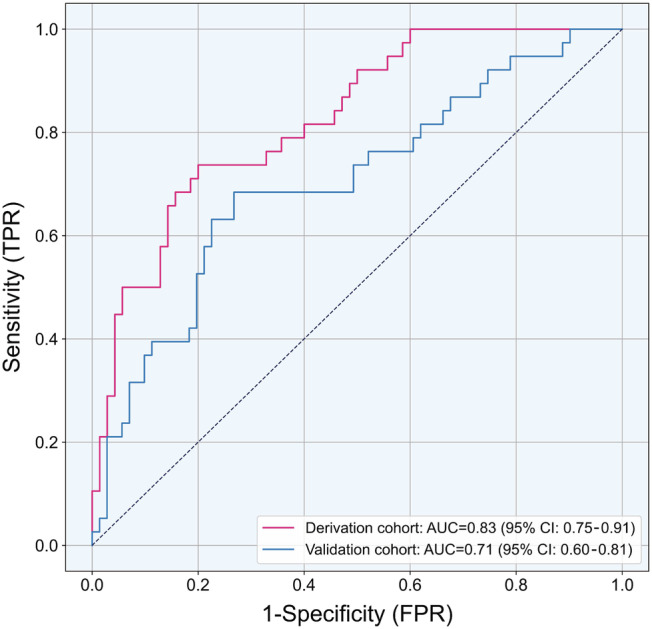
Receiver operating characteristic (ROC) curves of the urinary proteomic classifier for the diagnosis of cardiac allograft vasculopathy. AUC, area under the receiver operating characteristic curve; TPR, true positive rate; FPR, false positive rate.
Table 2.
Diagnostic performance of the urinary proteomic classifier for non‐invasive assessment of cardiac allograft vasculopathy
| Parameters | The derivation cohort | The validation cohort |
|---|---|---|
| Sensitivity, % | 73.7 | 68.4 |
| Specificity, % | 71.4 | 73.2 |
| Positive predictive value, % | 58.3 | 57.8 |
| Negative predictive value, % | 83.3 | 81.3 |
| Accuracy, % | 72.2 | 71.6 |
Abbreviation: AUC, area under the receiver operating characteristic curve.
The optimal threshold of the predicted probability of the urinary proteomic classifier was 0.484, obtained by maximizing Youden's index in the validation cohort.
The subgroup analysis by the prescription of antiplatelet agents showed similar results. For patients without the treatment of antiplatelet agents, the proteomic signature offered an AUC of 0.82 (95% CI: 0.71–0.92, P = 0.0001) and 0.72 (95% CI: 0.59–0.86, P = 0.001) in the derivation cohort and the validation cohort, respectively. The corresponding AUCs were 0.86 (95% CI: 0.72–0.99, P < 0.0001), and 0.71 (95% CI: 0.51–0.90, P = 0.039) for patients treated with antiplatelet agents.
The associations of the proteomic classifier with cardiac allograft vasculopathy and clinical variables
In univariate analysis, every 0.1% increment of the predicted probability of the proteomic classifier was associated with a 1.60 (95% CI: 1.34–1.98, P < 0.0001) and 1.32 (95% CI: 1.13–1.58, P = 0.001) fold‐increase in the odds of having CAV in the derivation and validation cohort, respectively. Age, donor age, years after heart transplantation, history of hypertension, diabetes mellitus, diastolic blood pressure, serum creatinine, and the use of antiplatelet agents and methylprednisolone (P ≤ 0.043) were significantly associated with CAV (Table S3 ). Forcing sex into the model, all the significant variables were combined into a propensity score. With adjustment for the propensity score, every 0.1% increment in the predicted probability significantly increased the odds of CAV [ORs: 1.46 (95% CI: 1.20–1.83, P = 0.0003) in the derivation cohort; 1.31 (1.07–1.64, P = 0.012) in the validation cohort, respectively].
The proteomic signature was positively correlated with the conventional biomarkers of myocardial injury, hsTnT (r = 0.35, P < 0.0001) whereas inversely correlated with eGFR (r = −0.37, P < 0.0001). These correlations were still significant after adjustment for sex, age (P ≤ 0.007). Within 6 months of urine sample collection, 10 patients experienced a rejection episode determined by myocardial biopsy. However, the proteomic signature was not significantly associated with the rejection episode [OR: 0.99 (95% CI: 0.81–1.23), P = 0.97]. The association between the proteomic signature and the use of antiplatelet agents was not significant with adjustment of age [OR: 1.06 (95% CI: 0.95–1.19), P = 0.27]. The same was observed for diabetes mellitus as well [unadjusted OR: 1.10 (95% CI: 0.99–1.22), P = 0.09].
Improvements in diagnostic accuracy for cardiac allograft vasculopathy
The capability of CAV detection increased after adding the proteomic classifier into the reference model in the validation cohort. The reference model consisted of the clinical variables that were significantly associated with CAV, including age, donor age, years after heart transplantation, history of hypertension, diabetes mellitus, diastolic blood pressure, serum creatinine, and the use of methylprednisolone. Sex was forced into the reference model for clinical coherence. Because the use of antiplatelet agents usually started after the diagnosis of CAV, it was not included in the reference model. As shown in Table 3 , the model extended for the proteomic classifier significantly improved the diagnostic accuracy for CAV, as suggested by NRI of 80.5% (95% CI: 41.7–116.9, P < 0.0001) in overall, 36.8% (95% CI: 4.9–67.1, P = 0.018) for recipients with CAV and 43.7% (95% CI: 22.7–63.3, P < 0.0001) for recipients without CAV. Consistent with this, IDI was 9.9% (3.5–16.2, P = 0.002) and the relative IDI was 33.4% (10.7–65.1, P = 0.014), indicating the proteomic signature substantially improved the diagnostic accuracy of CAV.
Table 3.
Improvements in diagnosis accuracy of cardiac allograft vasculopathy upon addition of the urinary proteomic classifier
| Improvements in diagnosis accuracy | ||
|---|---|---|
| % (95% CI) | P | |
| Continuous NRI | ||
| NRICAV | 36.8 (4.9–67.1) | 0.018 |
| NRInon‐CAV | 43.7 (22.7–63.3) | <0.0001 |
| NRI | 80.5 (41.7–116.9) | <0.0001 |
| IDI | ||
| IDICAV | 6.4 (0.7–11.9) | 0.026 |
| IDInon‐CAV | 3.5 (0.5–6.2) | 0.021 |
| IDI | 9.9 (3.5–16.2) | 0.002 |
| Relative IDI | 33.4 (10.7–65.1) | 0.014 |
Abbreviation: CAV, cardiac allograft vasculopathy; CI, confidence interval; IDI, integrated discrimination improvement; NRI, net reclassification improvement.
The reference models included age, sex, donor age, years after heart transplantation, history of hypertension, diabetes mellitus, diastolic blood pressure, serum creatinine, and the use of methylprednisolone. Improvement of the diagnostic accuracy was evaluated in the validation cohort.
IDI refers to the difference in discrimination slopes before and after adding proteomic classifier into a reference model. Specifically, the discrimination slope was the difference between the predicted probability with CAV and without CAV. If P(up/CAV) is the percentage of patients with CAV whose predicted probability increased by adding the urinary proteomic classifier and if P(up/non‐CAV) is the percentage of patients without CAV whose predicted probability is increased, the continuous NRI equals 2 × [P(up/CAV) − P(up/non‐CAV)].
Pathway enrichment analysis
The proteomic classifier included 22 peptides annotating to 7 different proteins and 5 unknown peptides (Table S2 ). The annotated proteins included collagen I (12, 44.4%), collagen II alpha I chain (1, 3.7%), collagen III alpha I chain (5, 18.5%), collagen XI alpha 1 chain (1, 3.7%), mucin‐1 subunit alpha (1, 3.7%), xylosyltransferase 1 (1, 3.7%), and protocadherin‐12 (1, 3.7%). With the Reactome pathway enrichment analysis, the biological processes of these proteins were predominately relevant to collagen biosynthesis and degradation, platelet aggregation and blood coagulation, cell‐extracellular matrix interaction, as well as cell adhesion and motility (Figure 3 and Table S4 ).
Figure 3.
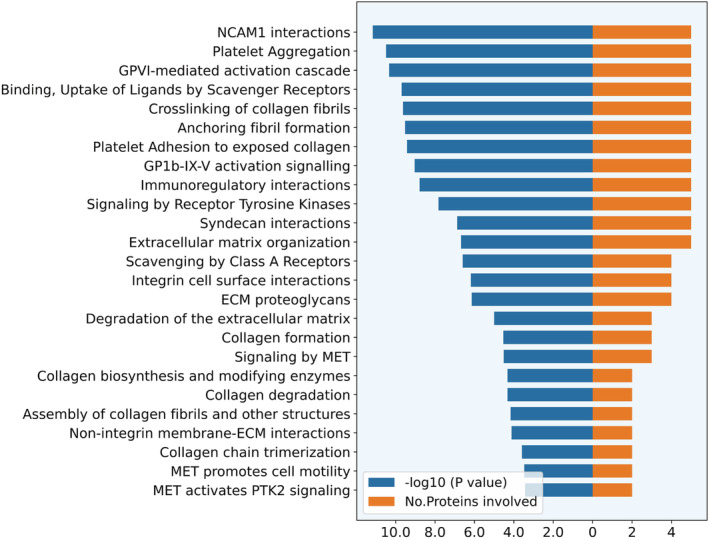
Enrichment pathways highlighted for cardiac allograft vasculopathy. The pathways were ranked by −log10 adjusted P values as shown by the blue bars. The corresponding numbers of overlapped proteins with urinary proteomic classifier are presented as orange bars. ECM, extracellular matrix; GPVI, glycoprotein VI; GP1b‐IX‐V, platelet glycoprotein 1b‐V‐IX complex; IL, interleukin; MET, mesenchymal epithelial transition; NCAM1, neural cell adhesion molecule 1; PTK2, protein tyrosine kinase 2.
Discussion
To the best of our knowledge, this is the first study investigating non‐invasive biomarkers for the discrimination of CAV in a well‐powered cohort, which also provide insights into the underlying mechanisms of CAV by pathway enrichment. In the present study, we constructed a novel urinary proteomic classifier for CAV diagnosis by machine learning and confirmed its performance in a validation cohort. Our main findings could be summarized as follows: (i) The established urinary proteomic classifier provided an AUC of 0.71 and a negative predictive value over 80% for the detection of CAV; (ii) the prediction of the classifier was independently associated with CAV after adjustment for potential clinical confounders; (iii) the urinary classifier significantly improved the diagnostic accuracy for CAV over and beyond the clinical risk factors with an NRI of 80.5% and an IDI of 9.9%; (iv) the enriched pathways involved collagen turnover, platelet aggregation, coagulation, cell‐extracellular matrix interaction, cell adhesion, and motility, providing important perspectives on the pathological mechanism of CAV.
The proteomic signature achieved an AUC of 0.83 for the detection of CAV in the derivation cohort, and it offered an AUC of 0.71 when generalizing to the validation cohort. The decreased performance might be explained by the variances between the two cohorts. In fact, prior to the construction of the proteomic signature, we used random sampling to divide the participants into the derivation and validation cohorts to eliminate the potential influence of the variances. Consequently, the important clinical characteristics, as shown in Table 1 , were basically balanced between the cohorts (P ≥ 0.13), except that the proportion of transplantation indication due to ischaemic cardiomyopathy was higher in the derivation cohort (41.7% vs. 28.4, P = 0.047). However, ischaemic cardiomyopathy as the transplantation indication did not show a significant difference between CAV group and non‐CAV group, as shown in Table S1 . Moreover, it was not significantly associated with CAV [unadjusted OR = 1.13 (0.63–2.02); P = 0.68]. Although the characteristics of the participants were well balanced, the residual variance among cohorts may exist and interfere with the performance of the signature in the validation cohort. Collaboration with other heart transplantation centres to extend the sample size would improve the generalized performance. Nonetheless, the proteomic signature developed with 217 heart transplantation patients had a decent performance, with an AUC of >0.71, suggesting its diagnostic value for CAV. This was further confirmed in the multivariate‐adjusted logistic regression analysis and supported by the improvement of NRI and IDI.
Previous studies reported several biomarkers related to CAV, including vascular endothelial growth factor (VEGF), plasma cytokines, C‐reactive protein, high‐sensitive troponin T, and micro‐RNA 628‐5p. 32 , 33 , 34 , 35 , 36 For instance, a study investigated 55 serum angiogenesis‐related proteins in 33 heart transplantation recipients (17 with CAV and 16 without CAV) using assay kit, and revealed that the levels of VEGF‐A, VEGF‐C, and platelet factor‐4 were higher in the CAV group. 32 In addition, a few studies have attempted to investigate the plasma proteomics of CAV. One study analysed the plasma proteome profile by labelled mass spectrometry in 19 heart transplantation patients (10 CAV and 9 non‐CAV). Using the elastic net regression model, they constructed a classifier with 18 plasma proteins. 37 The classifier consisted of proteins mainly involved in immune response, complement activation. Another targeted proteomics study compared the concentrations of a set of serum proteins between 12 heart recipients with CAV and 10 without CAV by Slow Off‐rate Modified Aptamer assay. 38 There were 14 differentially expressed proteins between CAV and non‐CAV groups, involved in apoptosis, cell injury, platelet aggregation, coagulation, and inflammation. Indeed, despite no overlapping proteins between the plasma and urinary proteomic signatures, some biological pathways were shared such as platelet aggregation, coagulation, and the immune process. The dissimilar proteomics signatures are probably ascribed to the differences in protein constitutions between plasma and urine, the quantified proteomics approaches, statistic methods, the variation between studied cohorts.
Overall, most of the proteins in our proteomic signature were involved in collagen turnover, platelet aggregation, coagulation, and cell adhesion and motility. It appeared that immunological processes and smooth muscle cell (SMC) proliferation that have been considered to associate with the pathogenesis of CAV were not predominant. 39 One possible explanation might be the interference from the lifelong immunosuppressive regimens after heart transplantation that suppress the host immune response against the donor graft, including interference in activated T cells, DNA synthesis in T and B cells, and anti‐inflammation. 1 In general, most recipients receive immunosuppressive regimens including calcineurin inhibitors, antimetabolites inhibitors, steroids, and mTOR inhibitors. Notably, mTOR inhibitors have been demonstrated to be beneficial in the prevention of CAV by impeding SMC proliferation. 40 In our cohort, the prescription of mTOR inhibitors was more prevalent among CAV patients (10.5% vs. 5.7%). On the other hand, the proteomic signature was related to immune response and SMC proliferation to a certain extent. In pathway analysis, it enriched immunoregulatory interactions between a lymphoid and a non‐lymphoid cell. As the major component in the proteomic signature, collagen I is considered to promote the migration of SMCs and proliferation probably mediated by integrin. 41 , 42
Fibrosis is one of the major pathogenesis of CAV. 11 Triggered by various factors, such as inflammation, ischaemia, the fibrotic accumulation of extracellular matrix (ECM) initiates from the intima and gradually invades to the media. Notably, the trivial intimal thickening can already be observed at 6 months after transplantation. 43 In a previous study with 27 heart transplantation patients, the early stage CAV measured by the coronary intima‐media thickness ratio was correlated with the cardiac interstitial fibrosis quantified by the endomyocardial biopsy. 44 Along the same lines, our study found that CAV was associated with urinary peptides of collagen I and III, which are the major fibrotic components of ECM. Although future studies are required to verify this, the finding highlights the possibility of urinary collagen fragments being used as a surrogate for the fibrosis process of CAV.
In addition, our study demonstrated that platelet activation and coagulation were associated with the pathological processes of CAV. An animal experiment showed that the presence of fibrin or antithrombin reactivity in the coronary capillary was more pronounced in CAV. 6 Similarly, a study investigating coronary morphology by serial intravascular ultrasounds in 132 heart transplantation recipients suggested that frequent thrombosis in coronary arteries may lead to the progression of CAV. 12 Recently, emerging evidence illustrated that antiplatelet therapy might reduce the risk of CAV or attenuate its progression. 45 , 46 A recently published randomized controlled trial showed that early aspirin intervention seems to slow down the progression of CAV. 45 Of note, in our cohort, over 80% of patients with moderate to severe CAV received antiplatelet drugs, whereas the proportion dropped to 32% in patients with mild CAV. The prescription of antiplatelet drugs might be attributed to their benefits in ischaemic heart disease. Despite that several studies showed that the exposure of antiplatelet agents was associated with a lower risk of CAV, 45 , 46 its benefit for prevention of CAV requires the accumulating evidence, especially large, randomized clinical trials. Lacking the recommendation from the ISHLT, 13 , 47 antiplatelet agents in our transplantation centre was prescribed when clinically indicated. Whether antiplatelet strategies could attenuate the progression of CAV and prolong survival times after heart transplantation, including its effects in subgroups with different CAV severities, needs to be addressed. In addition to the benefits in attenuating the hypercoagulable state, the novel anticoagulants targeting factor Xa also reduce the activation of protease‐activated receptors that could stimulate fibrosis and inflammation. 48 Future studies assessing the effect of the novel anticoagulants on CAV prevention might be of great interest.
Mucin‐1 is a transmembrane glycoprotein and provides a protective layer for epithelial cells under physiological conditions. 49 A previous study indicated that urinary mucin‐1 is inversely associated with renal function after heart transplantation. 50 In our study, mucin‐1 remained significantly associated with CAV after adjustment of renal function. Another protein in the proteomic signature is xylosyltransferase‐1, which is an essential enzyme for proteoglycan biosynthesis. 51 An experimental study observed the significant upregulation of xylosyltransferase‐1 in cardiac fibroblasts from human dilated cardiomyopathy tissue samples. 51 In addition, deficiency in protocadherin‐12, a member of the cadherin superfamily with a role in cell adhesion, was associated with reduced thickness of elastic properties and mild hypotension in mice. 52 However, little is known about the role of these proteins in CAV pathogenesis and future studies might enhance the understanding of the underlying mechanisms of CAV.
Clinical implications
Given its relatively high negative predictive value (81.3%), a proteomic classifier value of ≤0.484 indicated a low risk for CAV. This non‐invasive approach might provide an alternative when recipients are exposed to a higher risk of contrast‐induced nephropathy. Notably, chronic kidney disease is the common comorbidity after heart transplantation. 53 In our cohort, the prevalence of moderate to severe chronic kidney disease (eGFR <60 mL/min/1.73 m2) was as high as 53.5%. Besides, a recent retrospective study on 17,587 post‐heart transplantation recipients reported that around 11% of the recipients developed solid malignancy between 1 and 5 years after heart transplantation. 54 Apart from chronic immunosuppression and older age, radiation exposure might be another risk factor of malignancy. Of note, the heart transplantation recipients were exposed to a mean radiation dose of 8.4 mSv per year, 3.5‐fold more than the general population who underwent imaging procedures. 55 Cardiac procedures (52%) and computed tomography (25%) were the largest radiation sources. Therefore, to a certain extent, using non‐invasive proteomic biomarkers might reduce radiation exposure for heart recipients and physicians.
The incidence of CAV increases with the time after heart transplantation, achieving 10% at 1 year, 44% at 10 years and 59% at 20 years. 4 The surveillance of CAV by coronary angiography is regularly performed at the first year and at 3 year intervals in the following 9 years. This study investigated the urinary proteomics in patients who underwent heart transplantation after 7.6 years (IQR: 4.0–13.8) ago. Given these conditions, it might be rational to screen CAV with urinary proteomic analysis by every 3 years within 4 to 14 years after heart transplantation. However, future studies are warranted to assess this timeframe. The merits of urinary proteomics analysis include the comfort and convenience for the patients, because it just requires a 5 mL fresh urine sample. The profile of urinary proteins is stable enough to conduct proteomic analysis when it is appropriately stored. 56 The feasibility of this approach has been shown in a multicentre randomized controlled trial PRIORITY. 57 Urine samples will be shipped to a central lab for measurement and the results will be available within 3 days after receiving samples. The cost effectiveness of urinary proteomic markers has already been proven for chronic kidney disease. 58
The proteomic signature was significantly associated with CAV independent of clinical variables, including diabetes mellitus and renal function. Despite the association between the proteomic signature and eGFR, introducing the proteomic signature could further improve the diagnostic accuracy for CAV patients, including those with renal dysfunction, as suggested by the IDI and NRI. As the developed signature was not significantly associated with diabetes, it is applicable for patients with or without diabetes.
Strengths and limitations
One of the major strengths of the study is the relatively large sample size to develop and validate the proteomic classifier. It is crucial to validate whether the diagnostic performance of a biomarker could generalize in an independent cohort. Another strength is that the association of a novel biomarker with CAV was assessed after adjustment for potential confounders, including age and years after transplantation. Furthermore, we introduced the NRI and IDI to evaluate whether the urinary proteomic biomarkers improved the diagnostic accuracy on top of known risk factors. In addition, we incorporated a machine learning model algorithm into high‐throughput proteomics, formulating a pipeline for future proteomic biomarker discovery.
Nonetheless, our study must be interpreted within the context of its limitations. First, the proteomic signature was not correlated with the severity of CAV defined by the ISHLT (P = 0.89). This was attributed to the limited cases of moderate to severe CAV for performing machine learning. Of 76 CAV cases, 53 (69.7) were diagnosed with Grade 1, 15 (19.7%) with Grade 2, and 8 (10.5%) with Grade 3. As a result, we trained a proteomic signature based on the discrimination of all CAV cases. We will continue to collect the moderate and severe CAV cases in the future. Second, although the diagnostic value of the urinary proteomic study has been assessed by this retrospective study, further study is required to determine its prognostic value. Our next study will be focusing on the predictive value of this novel signature. Besides, the pathogenesis of CAV is rather complicated, which involves multiple pathological processes and is hardly fully elucidated with a single biomarker. However, with the proteomic approach, we could comprehensively identify a panel of proteins related to CAV without prior assumptions, which is also a major distinction from conventional protein analysis. 15 , 16 Nonetheless, the mechanisms annotated by proteomics and pathway analysis still require clear, well‐designed experiments to verify. Last, this is a single‐centre study, and replication of the current findings in other heart transplantation cohorts are needed.
Conclusions
In conclusion, this study developed and validated a novel proteomic signature for the diagnosis of CAV. These proteomic biomarkers have the potential to be used as non‐invasive screening tools for CAV after further validation in other cohorts. Moreover, the novel urinary proteins and pathways shed light on the pathophysiological process of CAV, which might be used to guide future treatments.
Conflict of interest
Dr Mischak is cofounder and a shareholder of Mosaiques Diagnostics, GmbH. Dr He and Dr Latosinska are employees of Mosaiques Diagnostics. The other authors declare no conflicts of interest.
Funding
The European Union (HEALTH‐F7‐305507 HOMAGE) and the European Research Council (Advanced Researcher Grant 2011‐294713‐EPLORE and Proof‐of‐Concept Grant 713601‐uPROPHET) and Internal Funds KU Leuven (STG‐18‐00379) currently support the Studies Coordinating Centre in Leuven. APPREMED (URL: www.appremed.org) received a non‐binding grant from OMRON Healthcare Co., Ltd., Kyoto, Japan.
Supporting information
Table S1. Characteristics of participants in the derivation cohort and validation cohort.
Table S2. The list of peptides involving the urinary proteomic classifier.
Table S3. The clinical variables associated with CAV in univariate logistic analysis.
Table S4. The enrichment pathways highlighted by the urinary proteomic classifier.
Acknowledgements
The authors gratefully acknowledge the clerical contribution of Renilde Wolfs.
Wei, D. , Trenson, S. , Van Keer, J. M. , Melgarejo, J. , Cutsforth, E. , Thijs, L. , He, T. , Latosinska, A. , Ciarka, A. , Vanassche, T. , Van Aelst, L. , Janssens, S. , Van Cleemput, J. , Mischak, H. , Staessen, J. A. , Verhamme, P. , and Zhang, Z.‐Y. (2022) The novel proteomic signature for cardiac allograft vasculopathy. ESC Heart Failure, 9: 1216–1227. 10.1002/ehf2.13796.
References
- 1. Stehlik J, Kobashigawa J, Hunt SA, Reichenspurner H, Kirklin JK. Honoring 50 years of clinical heart transplantation in circulation: in‐depth state‐of‐the‐art review. Circulation 2018; 137: 71–87. [DOI] [PubMed] [Google Scholar]
- 2. Khush KK, Potena L, Cherikh WS, Chambers DC, Harhay MO, Hayes D Jr, Hsich E, Sadavarte A, Singh TP, Zuckermann A, Stehlik J, International Society for Heart and Lung Transplantation . The International Thoracic Organ Transplant Registry of the International Society for Heart and Lung Transplantation: 37th adult heart transplantation report‐2020; focus on deceased donor characteristics. J Heart Lung Transplant 2020; 39: 1003–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lund LH, Edwards LB, Kucheryavaya AY, Benden C, Christie JD, Dipchand AI, Dobbels F, Goldfarb SB, Levvey BJ, Meiser B, Yusen RD, Stehlik J. The Registry of the International Society for Heart and Lung Transplantation: thirty‐first official adult heart transplant report—2014; focus theme: retransplantation. J Heart Lung Transplant 2014; 33: 996–1008. [DOI] [PubMed] [Google Scholar]
- 4. Van Keer JM, Van Aelst LNL, Rega F, Droogne W, Voros G, Meyns B, Vanhaecke J, Emonds MP, Janssens S, Naesens M, Van Cleemput J. Long‐term outcome of cardiac allograft vasculopathy: importance of the International Society for Heart and Lung Transplantation angiographic grading scale. J Heart Lung Transplant 2019; 38: 1189–1196. [DOI] [PubMed] [Google Scholar]
- 5. Lee JH, Okada K, Khush K, Kobayashi Y, Sinha S, Luikart H, Valantine H, Yeung AC, Honda Y, Fearon WF. Coronary endothelial dysfunction and the index of microcirculatory resistance as a marker of subsequent development of cardiac allograft vasculopathy. Circulation 2017; 135: 1093–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Labarrere CA, Ortiz MA, Ruzmetov N, Sosa MJ, Campana G, Terry C, Baldridge LA, Antonopoulos R, DiCarlo HL. Microvascular thrombosis and cardiac allograft vasculopathy in rat heart transplantation. J Heart Lung Transplant 2006; 25: 1213–1222. [DOI] [PubMed] [Google Scholar]
- 7. Hollenberg SM, Klein LW, Parrillo JE, Scherer M, Burns D, Tamburro P, Oberoi M, Johnson MR, Costanzo MR. Coronary endothelial dysfunction after heart transplantation predicts allograft vasculopathy and cardiac death. Circulation 2001; 104: 3091–3096. [DOI] [PubMed] [Google Scholar]
- 8. Labarrere CA, Lee JB, Nelson DR, Al‐Hassani M, Miller SJ, Pitts DE. C‐reactive protein, arterial endothelial activation, and development of transplant coronary artery disease: a prospective study. Lancet 2002; 360: 1462–1467. [DOI] [PubMed] [Google Scholar]
- 9. Vassalli G, Gallino A, Weis M, von Scheidt W, Kappenberger L, von Segesser L, Goy JJ, Working Group Microcirculation of the Eurpean Society of Cardiology . Alloimmunity and nonimmunologic risk factors in cardiac allograft vasculopathy. Eur Heart J 2003; 24: 1180–1188. [DOI] [PubMed] [Google Scholar]
- 10. Song G, Zhang J, Jiang D, Wang X, Xu J. The experimental study of cardiac allograft vasculopathy and myocardial fibrosis in rats. Transplant Proc 2008; 40: 2712–2715. [DOI] [PubMed] [Google Scholar]
- 11. Yamani MH, Haji SA, Starling RC, Tuzcu EM, Ratliff NB, Cook DJ, Abdo A, Crowe T, Secic M, McCarthy P, Young JB. Myocardial ischemic‐fibrotic injury after human heart transplantation is associated with increased progression of vasculopathy, decreased cellular rejection and poor long‐term outcome. J Am Coll Cardiol 2002; 39: 970–977. [DOI] [PubMed] [Google Scholar]
- 12. Matsuo Y, Cassar A, Li J, Flammer AJ, Choi BJ, Herrmann J, Gulati R, Lennon RJ, Kang SJ, Maehara A, Kitabata H, Akasaka T, Lerman LO, Kushwaha SS, Lerman A. Repeated episodes of thrombosis as a potential mechanism of plaque progression in cardiac allograft vasculopathy. Eur Heart J 2013; 34: 2905–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mehra MR, Crespo‐Leiro MG, Dipchand A, Ensminger SM, Hiemann NE, Kobashigawa JA, Madsen J, Parameshwar J, Starling RC, Uber PA. International Society for Heart and Lung Transplantation working formulation of a standardized nomenclature for cardiac allograft vasculopathy‐2010. J Heart Lung Transplant 2010; 29: 717–727. [DOI] [PubMed] [Google Scholar]
- 14. Kransdorf EP, Kobashigawa JA. Biomarkers for cardiac allograft vasculopathy: still searching after all these years. Transplantation 2017; 101: 28–29. [DOI] [PubMed] [Google Scholar]
- 15. Leopold JA, Loscalzo J. Emerging role of precision medicine in cardiovascular disease. Circ Res 2018; 122: 1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lam MP, Ping P, Murphy E. Proteomics research in cardiovascular medicine and biomarker discovery. J Am Coll Cardiol 2016; 68: 2819–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gerszten RE, Asnani A, Carr SA. Status and prospects for discovery and verification of new biomarkers of cardiovascular disease by proteomics. Circ Res 2011; 109: 463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Domon B, Aebersold R. Challenges and opportunities in proteomics data analysis. Mol Cell Proteomics 2006; 5: 1921–1926. [DOI] [PubMed] [Google Scholar]
- 19. Agranoff D, Fernandez‐Reyes D, Papadopoulos MC, Rojas SA, Herbster M, Loosemore A, Tarelli E, Sheldon J, Schwenk A, Pollok R, Rayner CFJ, Krishna S. Identification of diagnostic markers for tuberculosis by proteomic fingerprinting of serum. Lancet 2006; 368: 1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sweatt AJ, Hedlin HK, Balasubramanian V, Hsi A, Blum LK, Robinson WH, Haddad F, Hickey PM, Condliffe R, Lawrie A, Nicolls MR, Rabinovitch M, Khatri P, Zamanian RT. Discovery of distinct immune phenotypes using machine learning in pulmonary arterial hypertension. Circ Res 2019; 124: 904–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoogeveen RM, Pereira JPB, Nurmohamed NS, Zampoleri V, Bom MJ, Baragetti A, Boekholdt SM, Knaapen P, Khaw KT, Wareham NJ, Groen AK, Catapano AL, Koenig W, Levin E, Stroes ESG. Improved cardiovascular risk prediction using targeted plasma proteomics in primary prevention. Eur Heart J 2020; 41: 3998–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mischak H, Kolch W, Aivaliotis M, Bouyssié D, Court M, Dihazi H, Dihazi GH, Franke J, Garin J, de Peredo AG, Iphöfer A, Jänsch L, Lacroix C, Makridakis M, Masselon C, Metzger J, Monsarrat B, Mrug M, Norling M, Novak J, Pich A, Pitt A, Bongcam‐Rudloff E, Siwy J, Suzuki H, Thongboonkerd V, Wang LS, Zoidakis J, Zürbig P, Schanstra JP, Vlahou A. Comprehensive human urine standards for comparability and standardization in clinical proteome analysis. Proteomics Clin Appl 2010; 4: 464–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mischak H, Vlahou A, Ioannidis JP. Technical aspects and inter‐laboratory variability in native peptide profiling: the CE‐MS experience. Clin Biochem 2013; 46: 432–443. [DOI] [PubMed] [Google Scholar]
- 24. Altman N, Krzywinski M. The curse(s) of dimensionality. Nat Methods 2018; 15: 399–400. [DOI] [PubMed] [Google Scholar]
- 25. Chen TQ, Guestrin C. XGBoost: a scalable tree boosting system. Kdd'16: Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining. 2016:785–794.
- 26. Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J. Scikit‐learn: machine learning in Python. J Mach Learn Res 2011; 12: 2825–2830. [Google Scholar]
- 27. Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982; 143: 29–36. [DOI] [PubMed] [Google Scholar]
- 28. Hajian‐Tilaki K. Sample size estimation in diagnostic test studies of biomedical informatics. J Biomed Inform 2014; 48: 193–204. [DOI] [PubMed] [Google Scholar]
- 29. Pencina MJ, D'Agostino RB Sr, D'Agostino RB Jr, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med 2008; 27: 157–172 discussion 207–112. [DOI] [PubMed] [Google Scholar]
- 30. Pencina MJ, D'Agostino RB Sr, Demler OV. Novel metrics for evaluating improvement in discrimination: net reclassification and integrated discrimination improvement for normal variables and nested models. Stat Med 2012; 31: 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, Sidiropoulos K, Cook J, Gillespie M, Haw R, Loney F, May B, Milacic M, Rothfels K, Sevilla C, Shamovsky V, Shorser S, Varusai T, Weiser J, Wu G, Stein L, Hermjakob H, D'Eustachio P. The reactome pathway knowledgebase. Nucleic Acids Res 2020; 48: D498–D503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Daly KP, Seifert ME, Chandraker A, Zurakowski D, Nohria A, Givertz MM, Karumanchi SA, Briscoe DM. VEGF‐C, VEGF‐A and related angiogenesis factors as biomarkers of allograft vasculopathy in cardiac transplant recipients. J Heart Lung Transplant 2013; 32: 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Daly KP, Stack M, Eisenga MF, Keane JF, Zurakowski D, Blume ED, Briscoe DM. Vascular endothelial growth factor A is associated with the subsequent development of moderate or severe cardiac allograft vasculopathy in pediatric heart transplant recipients. J Heart Lung Transplant 2017; 36: 434–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Przybylek B, Boethig D, Neumann A, Borchert‐Moerlins B, Daemen K, Keil J, Haverich A, Falk C, Bara C. Novel cytokine score and cardiac allograft vasculopathy. Am J Cardiol 2019; 123: 1114–1119. [DOI] [PubMed] [Google Scholar]
- 35. Hognestad A, Endresen K, Wergeland R, Stokke O, Geiran O, Holm T, Simonsen S, Kjekshus JK, Andreassen AK. Plasma C‐reactive protein as a marker of cardiac allograft vasculopathy in heart transplant recipients. J Am Coll Cardiol 2003; 42: 477–482. [DOI] [PubMed] [Google Scholar]
- 36. Garrido IP, Garcia‐Lara J, Pinar E, Pastor‐Pérez F, Sánchez‐Mas J, Valdés‐Chavarri M, Pascual‐Figal DA. Optical coherence tomography and highly sensitivity troponin T for evaluating cardiac allograft vasculopathy. Am J Cardiol 2012; 110: 655–661. [DOI] [PubMed] [Google Scholar]
- 37. Lin D, Cohen Freue G, Hollander Z, John Mancini GB, Sasaki M, Mui A, Wilson‐McManus J, Ignaszewski A, Imai C, Meredith A, Balshaw R, Ng RT, Keown PA, Robert McMaster W, Carere R, Webb JG, McManus B, Biomarkers in Transplantation Team , Networks of Centres of Excellence, Centres of Excellence for Commercialization and Research‐Prevention of Organ Failure Centre of Excellence . Plasma protein biosignatures for detection of cardiac allograft vasculopathy. J Heart Lung Transplant 2013; 32: 723–733. [DOI] [PubMed] [Google Scholar]
- 38. Almufleh A, Zhang L, Mielniczuk LM, Stadnick E, Davies RA, Du Q, Rayner K, Liu PP, Chih S. Biomarker discovery in cardiac allograft vasculopathy using targeted aptamer proteomics. Clin Transplant 2020; 34: e13765. [DOI] [PubMed] [Google Scholar]
- 39. Rahmani M, Cruz RP, Granville DJ, McManus BM. Allograft vasculopathy versus atherosclerosis. Circ Res 2006; 99: 801–815. [DOI] [PubMed] [Google Scholar]
- 40. Eisen HJ, Tuzcu EM, Dorent R, Kobashigawa J, Mancini D, Valantine‐von Kaeppler HA, Starling RC, Sørensen K, Hummel M, Lind JM, Abeywickrama KH, Bernhardt P. Everolimus for the prevention of allograft rejection and vasculopathy in cardiac‐transplant recipients. N Engl J Med 2003; 349: 847–858. [DOI] [PubMed] [Google Scholar]
- 41. Koyama H, Raines EW, Bornfeldt KE, Roberts JM, Ross R. Fibrillar collagen inhibits arterial smooth muscle proliferation through regulation of Cdk2 inhibitors. Cell 1996; 87: 1069–1078. [DOI] [PubMed] [Google Scholar]
- 42. Hollenbeck ST, Itoh H, Louie O, Faries PL, Liu B, Kent KC. Type I collagen synergistically enhances PDGF‐induced smooth muscle cell proliferation through pp60src‐dependent crosstalk between the alpha2beta1 integrin and PDGFbeta receptor. Biochem Biophys Res Commun 2004; 325: 328–337. [DOI] [PubMed] [Google Scholar]
- 43. Ramzy D, Rao V, Brahm J, Miriuka S, Delgado D, Ross HJ. Cardiac allograft vasculopathy: a review. Can J Surg 2005; 48: 319–327. [PMC free article] [PubMed] [Google Scholar]
- 44. van Heeswijk RB, Bastiaansen JAM, Iglesias JF, Degrauwe S, Rotman S, Barras JL, Regamey J, Lauriers N, Tozzi P, Yerly J, Ginami G, Stuber M, Hullin R. Quantification of myocardial interstitial fibrosis and extracellular volume for the detection of cardiac allograft vasculopathy. Int J Cardiovasc Imaging 2020; 36: 533–542. [DOI] [PubMed] [Google Scholar]
- 45. Kim M, Bergmark BA, Zelniker TA, Mehra MR, Stewart GC, Page DS, Woodcome EL, Smallwood JA, Gabardi S, Givertz MM. Early aspirin use and the development of cardiac allograft vasculopathy. J Heart Lung Transplant 2017; 36: 1344–1349. [DOI] [PubMed] [Google Scholar]
- 46. Bjerre KP, Clemmensen TS, Berg K, Poulsen SH, Hvas AM, Grove EL, Løgstrup BB, Jakobsen L, Thim T, Kristensen SD, Eiskjær H. Platelet aggregation and response to aspirin therapy in cardiac allograft vasculopathy. J Heart Lung Transplant 2020; 39: 371–378. [DOI] [PubMed] [Google Scholar]
- 47. Costanzo MR, Dipchand A, Starling R, Anderson A, Chan M, Desai S, Fedson S, Fisher P, Gonzales‐Stawinski G, Martinelli L, McGiffin D. The International Society of Heart and Lung Transplantation guidelines for the care of heart transplant recipients. J Heart Lung Transplant 2010; 29: 914–956. [DOI] [PubMed] [Google Scholar]
- 48. Borensztajn K, Peppelenbosch MP, Spek CA. Factor Xa: at the crossroads between coagulation and signaling in physiology and disease. Trends Mol Med 2008; 14: 429–440. [DOI] [PubMed] [Google Scholar]
- 49. Nath S, Mukherjee P. MUC1: A multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med 2014; 20: 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huang QF, Zhang ZY, Van Keer J, Trenson S, Nkuipou‐Kenfack E, Yang WY, Thijs L, Vanhaecke J, Van Aelst LN, Van Cleemput J, Janssens S. Urinary peptidomic biomarkers of renal function in heart transplant recipients. Nephrol Dial Transplant 2018; 34: 1336–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Prante C, Milting H, Kassner A, Farr M, Ambrosius M, Schön S, Seidler DG, Banayosy AE, Körfer R, Kuhn J, Kleesiek K, Götting C. Transforming growth factor beta1‐regulated xylosyltransferase I activity in human cardiac fibroblasts and its impact for myocardial remodeling. J Biol Chem 2007; 282: 26441–26449. [DOI] [PubMed] [Google Scholar]
- 52. Philibert C, Bouillot S, Huber P, Faury G. Protocadherin‐12 deficiency leads to modifications in the structure and function of arteries in mice. Pathol Biol (Paris) 2012; 60: 34–40. [DOI] [PubMed] [Google Scholar]
- 53. Kolsrud O, Karason K, Holmberg E, Ricksten SE, Felldin M, Samuelsson O, Dellgren G. Renal function and outcome after heart transplantation. J Thorac Cardiovasc Surg 2018; 155: 1593–1604.e1. [DOI] [PubMed] [Google Scholar]
- 54. Youn JC, Stehlik J, Wilk AR, Cherikh W, Kim IC, Park GH, Lund LH, Eisen HJ, Kim DY, Lee SK, Choi SW, Han S, Ryu KH, Kang SM, Kobashigawa JA. Temporal trends of de novo malignancy development after heart transplantation. J Am Coll Cardiol 2018; 71: 40–49. [DOI] [PubMed] [Google Scholar]
- 55. Noor M, Shekhdar J, Banner NR. Radiation exposure after heart transplantation: trends and significance. J Heart Lung Transplant 2011; 30: 309–314. [DOI] [PubMed] [Google Scholar]
- 56. Fliser D, Novak J, Thongboonkerd V, Argilés À, Jankowski V, Girolami MA, Jankowski J, Mischak H. Advances in urinary proteome analysis and biomarker discovery. J Am Soc Nephrol 2007; 18: 1057–1071. [DOI] [PubMed] [Google Scholar]
- 57. Tofte N, Lindhardt M, Adamova K, Bakker SJL, Beige J, Beulens JWJ, Birkenfeld AL, Currie G, Delles C, Dimos I, Francová L, Frimodt‐Møller M, Girman P, Göke R, Havrdova T, Heerspink HJL, Kooy A, Laverman GD, Mischak H, Navis G, Nijpels G, Noutsou M, Ortiz A, Parvanova A, Persson F, Petrie JR, Ruggenenti PL, Rutters F, Rychlík I, Siwy J, Spasovski G, Speeckaert M, Trillini M, Zürbig P, von der Leyen H, Rossing P, Zimmermann S, Rädisch B, Hävemeier A, Busmann A, Wittkop U, Neuhaus B, Ax‐Smolarski R, Zieglschmid V, Bollweber E, Wölk H, Curovic VR, Tougaard NH, Eickhoff MK, Pilemann‐Lyberg S, Winther SA, Rosenlund SV, Hansen TW, von Scholten BJ, Hansen CS, Zobel EH, Laursen JC, Theilade S, Jelstrup L, Juhl TR, Riis D, Hermann JA, Lundgaard AG, Halkjær MLD, Aabo L, Frost Lerche T, Lajer M, Stefansen RJ, Campbell MA, Durban A, Raad J, Prigge M, Schiemann M, Wilson R, Kean S, Douglas E, Surtees P, Gant C, Yeung SMH, Hagedoorn I, Flynn J, Galloway J, Brooksbank K, Aparicio C, Iliev IP, Nones F, Lo Bue F, Melacini D, Cugini D, Prandini S, Lecchi V, Yakymchuk S, Gherardi G, Villa A, Villa D, Gaspari F, Cannata AN, Ferrari S, Stucchi N, Albrechtová Š, Eldeik E, Amanaki R, Fernandez‐Fernandez B, Sanchez‐Rodriguez J, Vázquez C, Sanz AB, Sanchez‐Niño MD, Ramos AM, Gonzalo MÁ, Schmidt U, Selim G, Gjorgovski T, Stratrova SS, Stojceva‐Taneva O, Schutten‐Westerneng P, Wierbos B, Huvers F, de Bruin AK, Lapauw B, de Man E, Rokegem K, Inion S, Kreutzmann K, Dewettinck I, Boukens‐de Graaf C, Clerc‐de Jong F, Entius J, Nannings M, van Steenderen S, Petry FW, Kilic C. Early detection of diabetic kidney disease by urinary proteomics and subsequent intervention with spironolactone to delay progression (PRIORITY): a prospective observational study and embedded randomised placebo‐controlled trial. Lancet Diabetes Endocrinol 2020; 8: 301–312. [DOI] [PubMed] [Google Scholar]
- 58. Critselis E, Vlahou A, Stel VS, Morton RL. Cost‐effectiveness of screening type 2 diabetes patients for chronic kidney disease progression with the CKD273 urinary peptide classifier as compared to urinary albumin excretion. Nephrol Dial Transplant 2018; 33: 441–449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Characteristics of participants in the derivation cohort and validation cohort.
Table S2. The list of peptides involving the urinary proteomic classifier.
Table S3. The clinical variables associated with CAV in univariate logistic analysis.
Table S4. The enrichment pathways highlighted by the urinary proteomic classifier.


