Abstract
Sterol Δ22-desaturase has been purified from a strain of Candida glabrata with a disruption in the gene encoding sterol 14α-demethylase (cytochrome P-45051; CYP51). The purified cytochrome P-450 exhibited sterol Δ22-desaturase activity in a reconstituted system with NADPH–cytochrome P-450 reductase in dilaurylphosphatidylcholine, with the enzyme kinetic studies revealing a Km for ergosta-5,7-dienol of 12.5 μM and a Vmax of 0.59 nmol of this substrate metabolized/min/nmol of P-450. This enzyme is encoded by CYP61 (ERG5) in Saccharomyces cerevisiae, and homologues have been shown in the Candida albicans and Schizosaccharomyces pombe genome projects. Ketoconazole, itraconazole, and fluconazole formed low-spin complexes with the ferric cytochrome and exhibited type II spectra, which are indicative of an interaction between the azole moiety and the cytochrome heme. The azole antifungal compounds inhibited reconstituted sterol Δ22-desaturase activity by binding to the cytochrome with a one-to-one stoichiometry, with total inhibition of enzyme activity occurring when equimolar amounts of azole and cytochrome P-450 were added. These results reveal the potential for sterol Δ22-desaturase to be an antifungal target and to contribute to the binding of drugs within the fungal cell.
Azole antifungal compounds inhibit cytochrome P-450 sterol 14α-demethylase (Erg11p), a key enzyme in the ergosterol biosynthetic pathway of fungi, resulting in an accumulation of 14α-methylated sterols and a decrease in ergosterol levels, leading to cell growth arrest. Candida glabrata is a pathogenic haploid yeast species which causes fungemia and other systemic infections in humans (12). The widespread use of the azole antifungal compounds due to higher numbers of immunocompromised patients with AIDS, as well as patients undergoing cancer chemotherapy and organ transplantation, has led to the appearance of resistance to these compounds in C. glabrata (20) and fungi in general (9, 10, 13).
Azole antifungal compounds inhibit CYP51 through coordination of the triazole N3 or imidazole N4 of the azole ring with the cytochrome P-450 heme, while hydrophobic N1 substituent groups of the azole interact with the protein in a manner not yet fully understood (17, 22). Disruption of CYP51 in Saccharomyces cerevisiae has revealed the presence of a second cytochrome P-450 species (5), which has been identified as CYP61, sterol Δ22-desaturase (7, 16). These results supported the finding of Hata et al. (2, 3), based on the use of specific inhibitors, that sterol Δ22-desaturase is a cytochrome P-450. The role that this enzyme plays in the overall azole antifungal tolerance in the cell is unknown.
Recently, the genes encoding sterol 14α-demethylase (ERG11) and sterol Δ5,6-desaturase (ERG3) from C. glabrata were cloned and sequenced (1). Deletion of both of these genes resulted in a strain that was aerobically viable and produced 14α-methylfecosterol as its predominant sterol. As in similar strains of S. cerevisiae (21), resistance to azole antifungal compounds was shown. Here we report for the first time the purification and reconstitution of a second cytochrome P-450 from this mutant strain and identify the role of this enzyme as a sterol Δ22-desaturase. Cytochrome P-450 multiplicity is demonstrated, and this enzyme activity is revealed here to be sensitive to azole antifungal compounds. The Schizosaccharomyces pombe and Candida albicans genome projects have revealed genes homologous to CYP61 of S. cerevisiae, and this clan of the cytochrome P-450 superfamily is almost invariably conserved throughout fungi producing ergosterol and potentially in other kingdoms of life (8).
MATERIALS AND METHODS
Strains.
C. glabrata L5DU61 (erg3Δ::LEU2erg11Δ::URA3) was used in this study and was described previously (1). This strain was obtained by disrupting the ERG11 (CYP51) gene of L59D (erg3Δ::LEU2ura3).
Media and growth conditions.
L5DU61 was grown in yeast extract-peptone-dextrose (YEPD) medium containing 10% (wt/vol) glucose, 2% (wt/vol) Bacto Peptone (Difco), and 1% (wt/vol) yeast extract (Difco). Cultures were grown at 37°C, and plate cultures contained YEPD medium, consisting of 2% (wt/vol) Difco Bacto Agar, 2% (wt/vol) glucose, 2% (wt/vol) Bacto Peptone (Difco), and 1% (wt/vol) yeast extract (Difco). In the preparation of biomass for enzyme purification, L5DU61 was grown in 2-liter cultures in 3-liter flasks with shaking (150 rpm). Cells were inoculated at a density of 106/ml and harvested at a density of 5 × 108/ml.
Chemicals.
Unless otherwise specified, chemicals were obtained from Sigma Chemical Co., Poole, Dorset, United Kingdom. Ketoconazole and itraconazole were purchased from Jannssen Pharmaceutica, and fluconazole was from Pfizer.
Preparation of microsomes.
Cells were harvested at a density of 5 × 108/ml and resuspended in buffer A (100 mM potassium phosphate, 20% [vol/vol] glycerol, 1 mM EDTA, and 0.5 mM dithiothreitol; pH 7.2). Following the addition of 20 g of glass beads, cells were homogenized with a disintegrator (Braun GmbH, Mesungen, Germany) operating at 1,500 × g, using four 30-s bursts as well as liquid carbon dioxide cooling. Cell debris, unbroken cells, and glass beads were removed by centrifugation at 1,500 × g for 10 min. All steps after cell breakage were performed at 4°C. Mitochondria were removed by centrifugation at 10,000 × g for 20 min followed by a spin at 100,000 × g for 1 h to produce the microsomal pellet containing cytochrome P-450. The microsomal pellet was resuspended in buffer B (50 mM Tris-HCl and 0.4 M sorbitol; pH 7.2) to a final protein concentration of approximately 10 mg/ml and stored at −80°C until use. Protein concentrations were estimated by using a Sigma bicinchoninic acid kit, and cytochrome P-450 concentrations were determined by reduced carbon monoxide difference spectroscopy according to the method of Omura and Sato (14), using a Philips PU8800 scanning spectrophotometer.
Purification of C. glabrata sterol Δ22-desaturase.
Microsomes were solubilized in 100 mM potassium phosphate buffer with 20% (vol/vol) glycerol, pH 7.2, containing 2% (wt/vol) sodium cholate. After being gently stirred for 1 h, the solution was centrifuged at 100,000 × g for 90 min to pellet membrane material, and the supernatant was diluted with a 20% (vol/vol) glycerol solution to 25 mM potassium phosphate–0.8% (wt/vol) sodium cholate. The supernatant was loaded directly onto an amino-octyl Sepharose column equilibrated with 10 mM potassium phosphate buffer containing 0.8% (wt/vol) sodium cholate, pH 7.2. The column was washed (three times the column volume) with 10 mM potassium phosphate buffer, pH 7.2, containing 0.8% (wt/vol) sodium cholate; a second wash with the same buffer containing 1.2% (wt/vol) sodium cholate and a third wash (twice the column volume) with 100 mM potassium phosphate buffer, pH 7.2, containing 0.5% (wt/vol) sodium cholate were subsequently carried out. Cytochrome P-450 was eluted from the column in this final buffer additionally containing 0.3% (vol/vol) Tween 20. Cytochrome P-450-containing fractions were pooled and dialyzed overnight against 2 liters of 10 mM potassium phosphate buffer, pH 6.8, containing 0.3% (wt/vol) sodium cholate. The sample was then loaded onto a hydroxyapatite column equilibrated with 10 mM potassium phosphate buffer, pH 6.8. The column was washed with 100 ml of 10 mM potassium phosphate buffer, pH 6.8, before the bound hemoproteins were eluted with a step gradient of 10 to 200 mM potassium phosphate buffer, pH 6.8. The elution of hemoproteins was monitored spectrophotometrically at 416 nm. The fractions were eluted with 100 mM potassium phosphate buffer which contained sterol Δ22-desaturase, pooled, and concentrated with an Amicon Centricon 10 microconcentrator. The cytochrome P-450 concentration was assessed by reduced carbon monoxide difference spectroscopy (14), and enzyme purity was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Purified cytochrome P-450 was stored at −80°C until use.
Purification of CPR.
NADPH–cytochrome P-450 reductase (CPR) was purified from C. glabrata microsomes according to the modified procedures described by Venkateswarlu et al. (18), and its activity was determined by the method of Vermillion and Coon (19), using cytochrome c as an electron acceptor. The final preparation showed a single band by SDS-PAGE, a molecular mass of 80,000 Da, and a specific activity of 40 U/mg of protein. One unit of activity was defined as the amount of enzyme resulting in the reduction of 1 μmol of cytochrome c by NADPH in 1 min.
Reconstitution of sterol Δ22-desaturase activity and kinetic analysis.
Ergosta-5,7-dienol, at appropriate concentrations, purified from a polyene-resistant erg5 mutant of S. cerevisiae (15), and 50 μg of dilauroylphosphatidylcholine were mixed and sonicated at low power until a white suspension had formed. Purified C. glabrata sterol Δ22-desaturase (0.2 nmol) and 1 U of CPR were added, and the reaction volume was adjusted to 1 ml by the addition of buffer A. NADPH (final concentration, 1 mM) was added to the mixture to start the reaction. All reaction mixtures were incubated at 37°C for 20 min in a shaking water bath. In control experiments, the involvement of cytochrome P-450 was examined by bubbling carbon monoxide through the enzyme preparation 2 min prior to substrate addition and also by the omission of cytochrome P-450 and NADPH from the reconstituted system. Reactions were stopped by the addition of 3 ml of methanol, and the sterols were extracted following the addition of 2 ml of 60% (wt/vol) potassium hydroxide in water and incubation at 90°C for 2 h. After they had cooled, the saponified mixtures were extracted three times with 5 ml of hexane and dried under nitrogen. Following silylation for 1 h at 60°C with bis(trimethylsilyl)trifluoroacetamide (50 μl, in 50 μl of toluene), sterols were analyzed by gas chromatography-mass spectroscopy (GC-MS) (model VG 12-250 gas chromatograph-mass spectrometer; VG Biotech, Manchester, United Kingdom), using split injections with a split ratio of 20:1. Sterol identification was achieved by reference to relative retention time and mass spectra as reported previously (7). Trimethylsilylated derivatives of ergosta-5,7-dienol and the Δ22-desaturated metabolite (ergosterol) were clearly separated as two clear peaks. The conversion ratio was calculated from the areas of the two peaks, and the activity (in nanomoles of ergosterol formed per minute) was obtained from the amount of substrate added and the conversion ratio. For calculation of enzyme kinetic parameters, linear regression was used in double-reciprocal analyses of activity data.
Inhibition of reconstituted sterol Δ22-desaturase activity.
For inhibition of cytochrome P-450 activity, reconstituted mixtures were prepared as described above and incubated for 20 min at 37°C, but with various amounts of the azole antifungal compounds ketoconazole, itraconazole, and fluconazole, added from 1,000-fold-concentrated stock solutions, present in the reaction mixtures.
RESULTS AND DISCUSSION
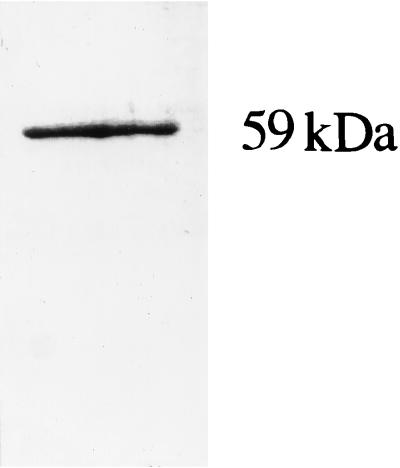
Cytochrome P-450 was still present in the microsomal fraction of C. glabrata L5UD61, a strain containing an erg11 gene disruption, confirming that CYP51 was not the only cytochrome P-450 isoform in microsomes of this pathogenic yeast. Indeed, the specific content of microsomal protein in the cultures indicated that the erg11-disrupted strain contained cytochrome P-450 in amounts equivalent to those of the strain from which it was derived (means ± standard deviations, 0.042 ± 0.011 and 0.051 ± 0.009 nmol of P-450/mg of microsomal protein, respectively). This may reflect a relatively low content of CYP51 under normal conditions of growth or a regulatory effect on the sterol biosynthetic pathway following gene disruption. Cytochrome P-450 was purified from the microsomal fractions of an erg11 erg3 gene-disrupted mutant of C. glabrata by methods similar to those employed for the purification of CYP61 from S. cerevisiae (7). As summarized in Table 1, this method permitted the purification of cytochrome P-450 to a specific content of 13.8 nmol/mg of protein. Upon SDS-PAGE, the cytochrome P-450 form was found to be homogeneous, with a single protein band in the gel (Fig. 1). An apparent monomeric Mr of 59,000 was estimated for this cytochrome. This value corresponds to the Mr of 58,000 for CYP61 purified from S. cerevisiae (7).
TABLE 1.
Purification of cytochrome P-450 from C. glabrata L5UD61a
| Fraction | P-450 concn (nmol) | Protein concn (mg) | Specific P-450 content (nmol/mg of protein) | % Recovery |
|---|---|---|---|---|
| Microsomes | 80.0 | 350.0 | 0.23 | 100 |
| Amino-octyl Sepharose | 37.0 | 4.86 | 7.6 | 46.0 |
| Hydroxyapatite | 12.0 | 0.87 | 13.8 | 15.0 |
Shown are data from a typical experiment for the purification of cytochrome P-450 from C. glabrata L5UD61, containing a gene disruption in cyp51.
FIG. 1.
Purification of C. glabrata sterol Δ22-desaturase. SDS-PAGE analysis of the purified sterol Δ22-desaturase. Protein (2.0 μg) was analyzed on a 10% polyacrylamide–SDS gel and visualized with Coomassie blue. The molecular mass of the cytochrome P-450 was determined by comparison with standard protein molecular mass markers.
The absolute absorption spectrum of purified C. glabrata sterol Δ22-desaturase was similar to that obtained for S. cerevisiae CYP61 and other CYP isozymes. In the oxidized state, the protein exhibited a Soret peak at 417 nm, indicating that it was in the low-spin state. Upon addition of sodium dithionite, the Soret band underwent a slight blue shift. The Soret peak of the reduced carbon monoxide complex was situated at 448 nm, like the microsomal cytochrome P-450 and the purified CYP enzymes from other fungal species.
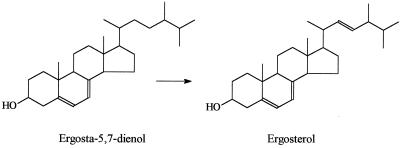
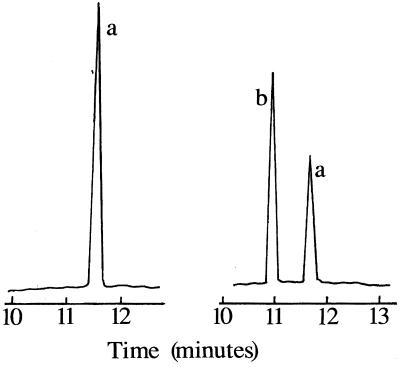
The endogenous enzyme activity of the purified cytochrome P-450 in a reconstituted system with CPR was revealed to be that of a sterol Δ22-desaturase. There was an absolute requirement for cytochrome P-450 and CPR. The purified cytochrome P-450 preparation was capable of metabolizing ergosta-5,7-dienol to ergosterol (Fig. 2) as detected by GC-MS (Fig. 3). The Michaelis constant for ergosta-5,7-dienol, calculated from the Lineweaver-Burk plot, was 12.5 μM, and the maximal enzymatic rate, Vmax, was calculated to be 0.59 nmol of ergosta-5,7-dienol metabolized/min/nmol of P-450. This enzyme was less active at Δ22-desaturation of ergosta-5,7-dienol than was CYP61 from S. cerevisiae (4).
FIG. 2.
The conversion of ergosta-5,7-dienol to ergosterol. In the normal pathway, 22 desaturation may preceed the 25 (28) reduction.
FIG. 3.
Catalytic activities of isolated sterol Δ22-desaturase as monitored by GC. This gas chromatogram, monitoring reconstitution of cytochrome P-450 activity, shows the conversion of ergosta-5,7-dienol (a) to ergosterol (b).
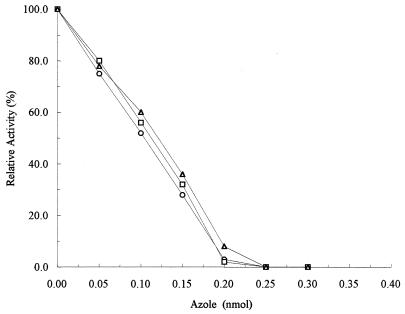
Figure 4 shows the results of experiments in which cytochrome P-450 activity was measured in the presence of increasing amounts of ketoconazole, itraconazole, or fluconazole, respectively. The level of inhibition of enzymatic activity was linearly dependent on the amount of azole present, and total inhibition of enzyme activity occurred at amounts equal to those of the cytochrome P-450 in the reaction mixtures, indicating that the antifungal compounds bound with a one-to-one stoichiometry and with high affinity. The amount of ketoconazole, itraconazole, or fluconazole required for 50% inhibition of enzyme activity was calculated to be approximately 0.1 nmol, indicating that these three azoles have similar potencies. Furthermore, fluconazole was shown to be competitive in its mode of action. A Ki of 0.18 nM was calculated, which is comparable to the value of 0.1 nM obtained for fluconazole inhibition of its target enzyme, C. albicans CYP51, sterol 14α-demethylase (11).
FIG. 4.
Inhibition of sterol Δ22-desaturase-catalyzed activity. Shown is a comparative plot of activity versus azole concentration for purified C. glabrata sterol Δ22-desaturase. Purified cytochrome P-450 was used to examine the inhibitory effects of ketoconazole (○), itraconazole (□), and fluconazole (▵).
Azole antifungal compounds have been identified as potent inhibitors of CYP51 in Candida spp., leading to a decreased availability of ergosterol, accumulation of 14α-methylated sterols, changes in activities of membrane-bound enzymes, and ultimately cell growth arrest (6). However, despite the site-specific mode of action of these compounds, the development of resistance to them in plant and human pathogens has increased and the search for more-efficacious compounds acting at the same target site or new compounds possessing unique modes of action is intense. In this study, the sensitivity of sterol Δ22-desaturase to existing agents was revealed. All of the azole antifungal compounds used bound to the cytochrome P-450 with a high affinity and inhibited enzymatic activity in a one-to-one stoichiometric ratio. Furthermore, comparison to fluconazole inhibitory data for C. albicans CYP51 revealed comparable inhibitory activity (11).
Azole antifungal agents inhibit CYP61 at concentrations equal to those at which CYP51 is inhibited, but following CYP51 inhibition after azole antifungal treatment, 14α-methylated sterols accumulate because the other reactions in the pathway are blocked. Skaggs et al. (16) reported that disruption of CYP61 in S. cerevisiae was not lethal to the cell. However, it has been recently observed that inhibition of sterol Δ22-desaturase in the cereal pathogen Rhynchosporium secalis correlates with cell growth arrest in azole-resistant strains (3a). Data from the fungal genome projects, beginning with S. cerevisiae (8), indicate that this enzyme is most likely encoded by different CYP61 genes. Thus, the overall role that CYP61 may play in future antifungal therapy remains to be clarified in microbial genetic studies coupled with sensitivity measurements. The use of microsomal fractions of Candida spp. in assessing azole binding and displacement to investigate azole antifungal compounds must also be undertaken cautiously since there are multiple cytochromes P-450.
ACKNOWLEDGMENT
This research was supported by The Wellcome Trust.
REFERENCES
- 1.Gerber A, Hitchcock C A, Swartz J E, Pullen F S, Marsden K E, Kwon-Chung K J, Bennett J E. Deletion of the Candida glabrata ERG3 and ERG11 genes: effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob Agents Chemother. 1995;39:2708–2717. doi: 10.1128/aac.39.12.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hata S, Nishino T, Katsuki H, Aoyama Y, Yoshida Y. Two species of cytochrome P450 involved in ergosterol biosynthesis of yeast. Biochem Biophys Res Commun. 1983;116:162–166. doi: 10.1016/0006-291x(83)90395-9. [DOI] [PubMed] [Google Scholar]
- 3.Hata S, Nishino T, Komori M, Katsuki H. Involvement of cytochrome P450 in Δ22-desaturation in ergosterol biosynthesis of yeast. Biochem Biophys Res Commun. 1981;103:272–277. doi: 10.1016/0006-291x(81)91689-2. [DOI] [PubMed] [Google Scholar]
- 3a.Hollomon, D. W. Personal communication.
- 4.Kelly S L, Lamb D C, Baldwin B C, Corran A J, Kelly D E. Characterization of Saccharomyces cerevisiae CYP61, sterol Δ22-desaturase, and inhibition by azole antifungal agents. J Biol Chem. 1997;272:9986–9988. doi: 10.1074/jbc.272.15.9986. [DOI] [PubMed] [Google Scholar]
- 5.Kelly S L, Lamb D C, Baldwin B C, Kelly D E. Benzo(a)pyrene hydroxylase activity in yeast is mediated by P450 other than sterol 14α-demethylase. Biochem Biophys Res Commun. 1993;197:428–432. doi: 10.1006/bbrc.1993.2497. [DOI] [PubMed] [Google Scholar]
- 6.Kelly S L, Lamb D C, Corran A J, Baldwin B C, Kelly D E. Mode of action and resistance to azole antifungals associated with the formation of 14α-methylergosta-8,24(28)-dien-3β,6α-diol. Biochem Biophys Res Commun. 1995;207:910–915. doi: 10.1006/bbrc.1995.1272. [DOI] [PubMed] [Google Scholar]
- 7.Kelly S L, Lamb D C, Corran A J, Baldwin B C, Parks L W, Kelly D E. Purification and reconstitution of activity of Saccharomyces cerevisiae P450 61, a sterol Δ22-desaturase. FEBS Lett. 1995;377:217–220. doi: 10.1016/0014-5793(95)01342-3. [DOI] [PubMed] [Google Scholar]
- 8.Kelly S L, Lamb D C, Kelly D E. Sterol 22-desaturase, cytochrome P45061, possesses activity in xenobiotic metabolism. FEBS Lett. 1997;412:233–235. doi: 10.1016/s0014-5793(97)00785-0. [DOI] [PubMed] [Google Scholar]
- 9.Kerridge D, Fasoli M, Wayman F J. Drug resistance in Candida albicans and Candida glabrata. Ann NY Acad Sci. 1988;544:245–259. doi: 10.1111/j.1749-6632.1988.tb40410.x. [DOI] [PubMed] [Google Scholar]
- 10.Kerridge D, Nicholas R O. Drug resistance in the opportunistic pathogens Candida albicans and Candida glabrata. J Antimicrob Chemother. 1986;18:39–49. doi: 10.1093/jac/18.supplement_b.39. [DOI] [PubMed] [Google Scholar]
- 11.Lamb D C, Kelly D E, Schunck W H, Shyadehi A Z, Akhtar M, Lowe D J, Baldwin B C, Kelly S L. The mutation T315A in Candida albicans sterol 14α-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J Biol Chem. 1997;272:5682–5688. doi: 10.1074/jbc.272.9.5682. [DOI] [PubMed] [Google Scholar]
- 12.Musial C E, Cockerill III F R, Roberts G D. Fungal infections of the immunocompromised host: clinical and laboratory aspects. Clin Microbiol Rev. 1988;1:349–364. doi: 10.1128/cmr.1.4.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholas R O, Burton J A, Kerridge D, Wayman F J. Isolation of mutants of Candida glabrata resistant to miconazole. Crit Rev Microbiol. 1987;15:103–110. doi: 10.3109/10408418709104453. [DOI] [PubMed] [Google Scholar]
- 14.Omura T, Sato R. The carbon monoxide binding pigment of liver microsomes. I. Evidence for its haemoprotein nature. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 15.Parks L W, Bottema C D K, Rodriguez R J, Lewis T A. Yeast sterols—yeast mutants as tools for the study of sterol metabolism. Methods Enzymol. 1985;111:333–346. doi: 10.1016/s0076-6879(85)11020-7. [DOI] [PubMed] [Google Scholar]
- 16.Skaggs B A, Alexander J F, Pierson C A, Schweitzer K S, Chun K T, Koegel C, Barbuch R, Bard M. Cloning and characterization of the Saccharomyces cerevisiae C-22 sterol desaturase gene, encoding a second cytochrome P-450 involved in ergosterol biosynthesis. Gene. 1996;169:105–109. doi: 10.1016/0378-1119(95)00770-9. [DOI] [PubMed] [Google Scholar]
- 17.Vanden Bossche H, Willemsens G, Cools W, Marichal P, Lauwers W. Hypothesis on the molecular basis of the antifungal activity of N-substituted imidazoles and triazoles. Biochem Soc Trans. 1983;11:665–667. doi: 10.1042/bst0110665. [DOI] [PubMed] [Google Scholar]
- 18.Venkateswarlu K, Lamb D C, Kelly D E, Manning N J, Kelly S L. The N-terminal membrane domain of yeast NADPH-cytochrome P450 (CYP) oxidoreductase is not required for catalytic activity in sterol biosynthesis or in reconstitution of CYP activity. J Biol Chem. 1998;273:4492–4496. doi: 10.1074/jbc.273.8.4492. [DOI] [PubMed] [Google Scholar]
- 19.Vermillion J L, Coon M J. Purified liver microsomal NADPH–cytochrome P-450 reductase. Spectral characterization of oxidation-reduction states. J Biol Chem. 1978;253:2694–2704. [PubMed] [Google Scholar]
- 20.Warnock D W, Burke J, Cope N J, Johnson E M, Von Fraunhofer N A, Williams E W. Fluconazole resistance in Candida glabrata. Lancet. 1988;ii:1310. doi: 10.1016/s0140-6736(88)92919-4. [DOI] [PubMed] [Google Scholar]
- 21.Watson P F, Rose M E, King D J, Ellis S W, England H, Kelly S L. Defective sterol C5-6 desaturation and azole resistance: a new hypothesis on the mode of action of azole antifungals. Biochem Biophys Res Commun. 1989;164:1170–1175. doi: 10.1016/0006-291x(89)91792-0. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida Y, Aoyama Y. Interaction of azole antifungal agents with cytochrome P45014DM purified from Saccharomyces cerevisiae microsomes. Biochem Pharmacol. 1987;36:229–235. doi: 10.1016/0006-2952(87)90694-0. [DOI] [PubMed] [Google Scholar]