Abstract
Exosomes are membrane bound extracellular vesicles that play an important role in many biological processes. While they have great application value, exosome isolation is still considered a major scientific challenge. In the present study, a novel separation strategy for exosomes is proposed based on the specific interaction between immobilized peptide ligands and phosphatidylserine moieties which are highly abundant on the surface of exosomes. With the new affinity method, intact model exosomes can be recovered with a high yield in a short processing time. The purity of exosome samples enriched from serum by the affinity method is far higher than that isolated by ultrafiltration, and similar to that obtained by density gradient centrifugation and ultracentrifugation. Moreover, the variety of contaminants co-isolated by the affinity method is relatively low due to its specific separation principle. Proteomics analysis of exosomes isolated by the affinity method from the serum of healthy, hepatocellular carcinoma patients, and intrahepatic cholangiocarcinoma patients was performed to prove the applicability of this method. In conclusion, our novel strategy shows characteristics of easy preparation, high specificity, and cost-effectiveness, and provides a promising approach for exosome isolation which should have wide applications.
Keywords: Exosome, Isolation, Phosphatidylserine, Peptide ligand, Hepatic cancer, Proteomics
Graphical abstract
Highlights
-
•
Exosomes can be isolated based on specific affinity between peptide ligands and PS.
-
•
Exosomes isolated by new affinity method have a high purity and simple contaminants.
-
•
Exosomes can be utilized as low invasive diagnostic tools for HCC and CCA diagnosis.
-
•
Exosomes are suitable candidates for discriminating HCC and CCA from healthy control.
-
•
Differential proteins can be analyzed to explore the mechanism of hepatic cancers.
1. Introduction
Exosomes are bilipid membrane bound extracellular vesicles which have typical cup-shaped structure, with sizes ranging from 30 to 200 nm, and they are released by various cell types during normal physiological functions or during acquired abnormalities [1,2]. In general, exosomes are rich in numerous bodily fluids, including serum, urine, and interstitial fluid, and contain an assortment of biomolecules of endosomal origin that vary with the state of the cell and they play important roles in intercellular communication, immune response, and cancer pathogenesis [[1], [2], [3]]. Because of these communication capabilities, exosomes have emerged as drug delivery carriers, disease biomarkers, and potential therapeutic targets [1,4,5]. However, while the application potential of exosomes is increasing, exosome isolation remains a significant impediment to many future applications. Thus, the challenge is to obtain exosomes of sufficiently high purity while maintaining an intact structure and preserving the enclosed biomarkers for downstream analysis.
To date, a range of isolation methods have been reported that utilize different techniques to capture exosomes, including ultracentrifugation (UC), density gradient centrifugation (DGC), ultrafiltration (UF), size-exclusion chromatography, polymerized precipitation, antibodies and affinity agents, and several microfluidic chip-based methods [[6], [7], [8]]. Several of these methods have also been simplified into commercial kits [[9], [10], [11]]. The most widely adopted methods are UC and DGC, which separate exosomes according to their size and density. However, co-isolation of contaminating vesicles with similar properties to exosomes and the requirements for expensive equipment and tedious operating processes limit the applicability of the isolated exosomes in subsequent studies and large-scale clinical sample processing [[10], [11], [12], [13]]. For increased specificity and high purity, exosome isolation using antibodies and other affinity agents can be a good alternative. In immunoaffinity methods, exosomes are captured via the binding of membrane surface receptor molecules, such as CD9 and CD63, to specific monoclonal antibodies immobilized on the surface of a solid matrix. Despite the simplicity of this kind of isolation, it suffers from a high cost and poor reproducibility during the elution of fully functional exosomes [6,14,15]. Additional affinity-based methods utilizing interactions involving membrane groups or molecules have also been explored. For example, exosomes can be easily isolated from human serum by taking advantage of the specific interaction between TiO2 and phosphate groups on the lipid bilayer of exosomes; however, the ubiquitous nature of phosphate groups results in the co-isolation of several contaminants [16].
To develop a method with the combined advantages of high specificity, low cost, and simplicity, small molecules specifically present on the membrane of exosomes could be explored to target. While the lipid composition of exosomes is generally cell-type dependent, exosome membrane content is known to diverge from that of its cell of origin; for example, phosphatidylserine (PS) is known to be exosome specific [17]. In flow cytometry experiments using FITC-annexin V-labeled exosomes, PS moieties were demonstrated to be exposed on the outer surface of exosome membranes. This can be explained by the absence of an organized cytoskeleton in exosomes (even though actin is detected) and the presence of a calcium-dependent phospholipid scramblase, resulting in a higher flip-flop of lipids between the two leaflets of the exosome bilipid membrane as compared with the plasma membrane [18]. This phenomenon facilitates several downstream functions of exosomes; thus, exosomes are internalized by other types of cells because of the externalized PS moieties, and the blocking of PS moieties by annexin V significantly inhibits exosome internalization and their downstream functions [[19], [20], [21]]. The presence of externalized PS on exosomes has been exploited in a number of isolation methods, and annexin V, Tim4, and recombinant nanobodies with a strong affinity for PS have all been employed to capture exosomes [[22], [23], [24], [25]]. For example, by exploiting the known affinity between annexin V and PS, exosomes derived from several cancer cell lines and a normal cell line have been captured with average efficiencies of 90.19 ± 5.70 and 38.43 ± 15.80%, respectively [22], and this observation is consistent with several studies in which authors have revealed that tumor-derived exosomes overexpress PS in their outer membrane leaflet as compared to exosomes from normal cells, therefore, PS could be utilized in cancer diagnostic applications [26,27]. However, protein implementation has also many issues in clinical applications because of the cost of production, endogenous competition, and the restrained diffusion to the targeted sites [28]. A viable alternative to large proteins is the use of smaller peptides to target PS with high affinity and specificity. Burtea et al. screened two phage display-derived PS targeting peptides (LIKKPF and PGDLSR) [29]. LIKKPF has a similar structure to the human transient receptor potential Ca2+ channel 5 which is involved in the regulation of membrane PS asymmetry and in the apoptotic signal. Derivatives of these PS-specific peptides (CLIKKPF, Kd = 1.9 μmol L−1; CPGDLSR, Kd = 7.9 μmol L−1) have been demonstrated to bind to membranes containing externalized PS and apoptotic cells with high affinity and specificity in the presence of Ca2+ binding buffer [30]. Moreover, both of these peptides have been modified to carry a fluorescent tag at the cysteine residue to demonstrate specific labeling of membranes containing externalized PS. While the potential of peptide-PS affinity binding was strongly confirmed by the authors, this interesting principle has not been exploited for exosome isolation thus far.
In the present work, we assessed the possibility and applicability of using the affinity interaction between PS and CLIKKPF (the higher affinity peptide) to capture exosomes. First, CLIKKPF was immobilized on the surface of silica microspheres (SiO2-pep). Next, this novel material was used to capture model exosome samples from Hela cell culture medium, and the efficiency of this method was compared with three classical methods (UF, UC, DGC) using serum samples. To expand the applications of our SiO2-pep affinity method and further demonstrate its value, we used this method to isolate exosomes from the serum of healthy, hepatocellular carcinoma (HCC), and intrahepatic cholangiocarcinoma (CCA) patients. The proteomic profiles of exosomes were then compared to identify accurate, low-invasive biomarkers for the early and differential diagnosis of HCC and CCA, and to further explore the underlying molecular mechanisms of these two hepatic cancers.
2. Experimental
2.1. Synthesis and characterization of SiO2-pep microspheres
SiO2 microspheres (Global Chromatography, Suzhou, China) were calcined at 600 °C for 5 h, then activated in a hydrochloric acid solution (12 wt%) at 102 °C for 12 h. After activation, the microspheres were washed thoroughly with ultrapure water to achieve neutral pH, and then dried at 60 °C. Next, the microspheres were dispersed in anhydrous toluene with 3-methacryloxypropyltrimethoxysilane (KH570) and incubated at 112 °C for 24 h. After completion of this reaction, the microspheres were washed with toluene and diethyl ether and then dried at 60 °C. The microspheres were then reacted with peptide CLIKKPF (PopChem peptide Co., Ltd, Hefei, China; purity >98%) in the presence of azobisisobutyronitrile (AIBN) (both at a concentration of 200 μg mL−1) in ethanol at 60 °C for 24 h. Finally, the SiO2-pep microspheres were rinsed with ether and dried at 60 °C for 6 h. The synthesized SiO2-pep microspheres were then stored at 4 °C until use.
All experimental conditions throughout the reaction process were optimized. During the process of optimizing the reaction conditions, the densities of silanol groups and KH570 were measured by titration with NaOH solution, and the density of peptide ligand CLIKKPF was measured using a BCA kit (the processing steps and the densities formula 1, 2, and 3 are mentioned in the experimental section of supporting information, respectively). Finally, the most suitable conditions were adopted for the final synthesis process as detailed above. The optimized SiO2-pep microspheres were then characterized.
The optimized SiO2-pep microspheres were sputtered with gold for scanning electron microscopy (SEM, S-4800 Hitachi, Japan) imaging. The Fourier transform infrared (FT-IR, Nicolet 6700, Thermo Fisher, USA) spectrum was assessed using KBr pellet. The Brunauer-Emmett-Teller surface area (Nitrogen adsorption apparatus, Micromeritics ASAP 2020; accelerated surface area and porosimetry system, Micromeritics Headquarters, USA) was evaluated by analyzing the adsorption isotherm in the 0.01–0.99 relative pressure (p/p0) range. The Zeta potential of microspheres (1 mg mL−1) dispersed in NaCl (5 m mol L−1) was measured using a size and zeta potential analyzer (omni, Brookhaven, USA). The mass percentages of elements (C, H, N, and S) were measured by elemental analysis (Vario EL Cube, Elementar, Germany). The bonding densities of KH570 groups and peptides in the optimized SiO2-pep microspheres were then calculated from the results of elemental analysis and BET specific surface area (formula 4 and formula 5 shown in supporting information Text S2).
2.2. SiO2-pep-based adsorption of model exosomes and specificity verification
Model exosomes (100 μL; total protein concentration, ∼0.16 mg mL−1) were added to SiO2-pep microspheres (5 mg) in Ca2+ binding buffer (10 mmol L−1 HEPES, pH 7.4; 140 mmol L−1 NaCl; 2.5 mmol L−1 CaCl2), and the mixture was incubated in the dark at 37 °C for 1 h on a thermos-shaker (Thermofisher, USA) to allow sufficient attachment. After simple centrifugation, the supernatant was discarded and the pellet was thoroughly washed thrice with PBS to remove non-specific molecules adsorbed on the microsphere surface. In parallel, three control groups were prepared (SiO2-pep + cell lysis, SiO2-pep + model exosome lysis, and KH570–SiO2 + model exosomes) to verify specific adsorption of exosomes on the SiO2-pep microspheres.
The sample preparation and the three controls were then visualized by SEM, transmission electron microscope (TEM, JEM-2100F JEOL, Japan), and immunofluorescence (IF). For IF analysis, samples were blocked in 5% BSA/TBST for 1 h at room temperature, and then incubated with CD81 rabbit monoclonal antibody (1:200, Beyotime, China) overnight at 4 °C. Subsequently, the microspheres were then stained with FITC-conjugated goat anti-rabbit secondary antibody (CWBIO, China; 1:25) for 1 h at room temperature. Images were acquired with a Leica TCS SP8 confocal microscope and processed using LAS AF 3.0 software. The same imaging parameters were used for the experimental group and the control groups.
2.3. Elution of model exosomes adsorbed on SiO2-pep microspheres and optimization of adsorption and elution conditions
Model exosomes (100 μL; total protein concentration, ∼0.16 mg mL−1) were mixed with various amounts of SiO2-pep microspheres (1 mg, 2 mg, 4 mg, 6 mg, 8 mg, and 10 mg) in Ca2+ binding buffer, the subsequent adsorption and washing procedures were as described in section 2.2. Next, 10% NH3·H2O solution (100 μL) was used to elute exosomes absorbed on the SiO2-pep microspheres at 4 °C for 120 min. NH3 in the elution solution was then removed in a vacuum oven at 25 °C for 10 min. Finally, total protein concentration in the elution solution was measured using a BCA kit. The procedure was repeated three times for each sample.
In addition, optimal adsorption time (20 min, 40 min, 60 min, 80 min, 100 min, and 120 min), concentration of eluent (6%, 8%, 10%, 12%, and 14% NH3·H2O), and elution time (20 min, 40 min, 60 min, 80 min, 100 min, and 120 min) were also evaluated. All other steps in the procedure were as detailed above.
The recovery rate was calculated as follows:
where, A is total protein concentration in the elution solution and B is total protein concentration in the model exosome solution.
Additional characterizations of the eluted exosomes, including size range by nanoparticle tracking analysis (NTA), Western blot (WB) for marker proteins, and TEM, were conducted as described in the supporting information.
2.4. Separation of serum exosomes using the SiO2-pep affinity method and comparison with UF, UC, and DGC methods
Serum samples from healthy individuals were purchased from Yuanye Bio-Technology Co., Ltd (Shanghai, China) and filtered with a 0.22 μm filter membrane. Diluted serum samples (5% in PBS) were then mixed with SiO2-pep microspheres in Ca2+ binding buffer. The exosome isolation procedures for adsorption and washing were as described in section 2.2. Serum exosomes adsorbed on the SiO2-pep microspheres then were eluted using a 12% NH3·H2O solution. Finally, the “SiO2-pep-exosomes” were characterized by NTA and TEM.
Serum exosomes isolated using the four different methods (UF, UC, DGC, and SiO2-pep affinity) were lysed by RIPA (containing 1 mmol L−1 PMSF). Equal amounts of total protein (20 μg) were then analyzed by SDS-PAGE gel stained through Coomassie brilliant blue staining, western blotting for CD9 marker protein (1:1000, Beyotime, China) and GAPDH (1:2000, Beyotime, China), and proteomics analysis, respectively.
Next, Venn diagrams were constructed using the proteins identified in the four groups by mass spectrometry analysis, and these were compared with proteins in the exosome database (ExoCarta database, http://www.exocarta.org). The quantities of nineteen common exosome proteins present in the exosome database and contaminating proteins identified in the exosome samples were normalized using the corresponding protein quantity obtained by UF as a reference. Finally, log2-fold differences in the quantities of the nineteen exosome markers and contaminating proteins were compared and plotted.
2.5. Proteomics analysis of exosomes isolated from healthy, HCC patient, and CCA patient serum samples using the SiO2-pep affinity method
All procedures were approved by the ETHICS committee of Shanghai Eastern Hepatobiliary Surgery Hospital (Shanghai, China). Blood samples from healthy donors, HCC patients, and CCA patients were obtained from consenting donors. The peripheral blood samples were collected in tubes and allowed to sit at room temperature for 1 h. The tubes were then centrifuged at 2000×g for 10 min, and the separated serum was stored at −80 °C until use.
Healthy, HCC patient, and CCA patient serum samples were processed as described in section 2.4 to isolate serum exosomes using the SiO2-pep affinity method. After lysis by RIPA (containing 1 mmol L−1 PMSF), the protein compositions of serum exosomes were analyzed using mass spectrometry. Each group contained nine samples.
2.6. Data analysis for biological information
First, the proteins which exist in the exosome proteins database (ExoCarta database) were screened from all identified proteins. Next, we performed a pair-wise comparison of protein expression between the three experimental groups. All proteins with an expression ratio ≥50% in both groups (or with an expression ratio in one group of 0 and an expression ratio ≥50% in the other group) were retained and proteins with missing value ≤ 50% were filled with the mean of the same group of samples. Normalized data for the potential proteins of interest (credible exosome proteins) were obtained by median normalization and log2 logarithm conversion. A boxplot and density map of the data before and after normalization were then generated. All visual presentations and statistical analysis are based on the normalized data for the credible exosome proteins. Unsupervised principal component analysis (PCA) and a hierarchical clustering dendrogram of sample Euclidean Distance were used to determine whether the quantitative data for credible exosome protein expression could discriminate between the three groups. Using the median normalized quantitative data of credible exosome proteins, Adj. P-values were calculated through moderate t-statistic with GraphPad Prism8. All credible exosome proteins that were differentially expressed between the three groups were selected using an Adj. P-value < 0.05 and a Fold Change <0.5 (for down-regulated proteins) or > 2 (for up-regulated proteins). Volcano Plots between pairs of groups were then generated with GraphPad Prism8. In each pair-wise analysis, differentially-expressed credible exosome proteins were visualized using a heat map, and an unsupervised hierarchical cluster analysis was performed.
Finally, biological information on the differentially-expressed credible exosome proteins was searched from free online databases, including GO enrichment analysis (top10 of each classification according to -log10 P-value) through FunRich software with GO database, KEGG pathway analysis (top15 according to -log10 P-value) at http://kobas.cbi.pku.edu.cn/kobas3/genelist/, and PPI (https://string-db.org/). On the basis of the resulting biological information, a bar chart and bubble chart were drawn with Origin 2019b.
3. Results and discussion
3.1. Preparation and characterization of SiO2-pep microspheres
In the present study, SiO2 microspheres were functionalized by “thiol-ene” click reaction with peptide CLIKKPF to yield “SiO2-pep” microspheres and used for the subsequent purification and enrichment of exosomes [31]. The favorable characteristics of SiO2 microspheres, which include their large specific surface area, good dispersion, and good stability, have resulted in them becoming the most commonly used separation matrix in the separation field. Because we intended to capture exosomes derived from biological samples, we selected a particle size of 20 μm, which had an ideal specific surface area (288.599 m2 g−1) and was easy to collect by centrifugation. The specific binding affinity between peptide CLIKKPF and PS molecules was first verified in a fluorescence plate assay, with phosphatidylglycerol (PG) molecules used as a control (Text S1 and Fig. S1; Kd for PS, 8.94 μmol L−1; Kd for PG, 138 μmol L−1).
Fig. 1A shows a schematic of SiO2-pep microsphere modification. The main synthetic process can be divided into three steps: (i) SiO2 microspheres were calcined to remove residual groups formed during the synthesis of the microspheres (marked as “SiO2”), then activated by hydrochloric acid solution to recover silanol groups on the surface (marked as “SiO2–OH”). (ii) “SiO2–OH” microspheres were modified by KH570 via a silanization reaction (marked as “KH570–SiO2”). (iii) A “thiol-ene” click reaction was performed between “KH570–SiO2” microspheres and the peptide CLIKKPF in the presence of the initiator AIBN to yield immobilized peptide ligand microspheres (SiO2-pep). The whole reaction process is initiated by free radicals, and is easy to operate under mild conditions.
Fig. 1.
Preparation of SiO2-pep microspheres and characterization of “SiO2”, “SiO2–OH”, “KH570–SiO2”, and “SiO2-pep” microspheres. (A) Schematic of SiO2-pep microsphere modification; (B) FT-IR spectra.
To achieve the maximum bonding density of peptide ligand CLIKKPF, all conditions in the whole reaction process were investigated. For example, the optimal “thiol-ene” click reaction conditions, including reaction temperature, concentration of peptide ligand CLIKKPF, and concentration of initiator AIBN were shown in Fig. S2. SiO2-pep microspheres were then synthesized under the most suitable conditions, and subsequently characterized to demonstrate successful immobilization of the peptide ligand CLIKKPF.
According to SEM (Fig. S3), the products were spherical and monodispersed, and the diameter of SiO2 and SiO2-pep microspheres were 20.98 ± 1.56 μm and 21.46 ± 2.11 μm, respectively, which indicates that there were no obvious morphological differences before and after the successive reactions. FT-IR spectral analysis (Fig. 1B) provides evidence of the successful immobilization of the peptide ligand CLIKKPF on the surface of the SiO2 microspheres. The appearance of the νSi-OH band at 977 cm−1 in the “SiO2–OH” spectrum demonstrates that the silanol groups lost during calcination were recovered. After modification by KH570, which have a carbon-carbon double bond and a carbonyl group, a δ-C O band at 1704 cm−1 could be observed in the spectrum. Following conjugation of the peptide ligand, a slight decrease in the intensity of the peak νC-H of the terminal alkenyl at 943.9 cm−1 was observed, which was consistent with the carbon-carbon double bond of “KH570–SiO2” participating in the “thiol-ene” click reaction with the thiol group of the peptide.
To further verify the FT-IR spectrum results, zeta potential measurements were performed (Table S1). In neutral aqueous solution, the absolute value of the zeta potential of “SiO2–OH” increased compared to “SiO2”, reflecting the recovery of surface silanol groups. After modification by KH570, we noticed a further increase, probably due to the hydrolysis of unreacted KH570 methoxy groups into OH groups. A reduction in the absolute value of zeta potential was observed in the final product carrying the peptide ligand (SiO2-pep). This observation can be explained by the fact that the peptide is positively charged at neutral pH.
For a quantitative estimation of the grafting efficiency of KH570 and peptide ligand CLIKKPF, an elemental analysis was performed (Table S2). The bonding densities of KH570 and the peptide were estimated using formula 4 and formula 5 (shown in supporting information Text S2), respectively, yielding 2.0437 and 0.0905 μmol m−2, respectively. The bonding density value of KH570 was similar to that of other silanization reactions, such as the classical stationary phase C18 used in chromatographic separation [32,33]. Although the bonding density of peptide ligand CLIKKPF was much lower than that of KH570 group, probably due to the high molecular weight of the peptide ligand causing steric hindrance, the number of peptide ligand CLIKKPF on the surface of microspheres was proved to be enough for the adsorption of exosomes (shown in the latter sections); we speculate that even higher densities will not show a higher adsorption capacities due to the relatively large size of exosomes occupying a considerable area of the particles surface.
3.2. Verification of SiO2-pep-based specific adsorption of model exosomes
Exosomes obtained from Hela cell culture medium by classic UC were used as model exosomes to verify that the synthesized SiO2-pep microspheres could successfully and specifically capture exosomes, which is the key purpose of this study. The model exosomes were obtained from a simple source through the most classic method at high purity and with few contaminants, so they could be considered as self-made “standard products”. As demonstrated in Fig. S4, the validity of this method for obtaining model exosomes was confirmed by measuring their size range by NTA, detecting exosomal marker proteins CD81 and TSG101 by WB, and observing morphological features by TEM [10,12].
In addition to model exosomes mixed with SiO2-pep microspheres, three negative control groups were designed. The first two negative controls were HeLa cell lysis and exosome lysis solution (neither of which contained exosomes with intact vesicle structures). These control samples were mixed with SiO2-pep microspheres as previously described to demonstrate that the intact exosomes could be specifically adsorbed onto the surface of SiO2-pep microspheres through their PS molecules. The third negative control involved processing the model exosomes with KH570–SiO2 microspheres (without the final grafting of the peptide ligand CLIKKPF). The results were compared with those obtained using model exosomes and SiO2-pep microspheres to elucidate the origin of the interaction between the microspheres and exosomes, i.e. whether the interaction was non-specific or due to affinity binding between peptides on the surface of SiO2 microspheres and PS moieties on the outer leaflet of exosomes.
After incubation and repeated washing of the model exosomes with SiO2-pep and of the three negative controls, the samples were analyzed with SEM, TEM, and IF. As shown in the SEM images (Fig. 2A), the surfaces of SiO2-pep microspheres were rough, indicating that the SiO2-pep microspheres had bound particulates, while the surfaces of microspheres in all three negative control groups were still smooth. To identify the bound particulates, the particulates were also imaged by TEM (Fig. 2B). Typical cup-shaped vesicular structures of ∼100 nm in diameter were easily discerned. Finally, IF experiments were performed using antibodies that specifically recognize transmembrane protein CD81, a commonly used exosomal marker protein. As shown in Fig. 2C, strong fluorescence signals were easily discernible at the surface of SiO2-pep microspheres incubated with exosome solution after staining with fluorescent antibodies targeting CD81; in contrast, only minimal background fluorescence was observed in the control groups. Together, these results reveal that exosomes with intact vesicle structures could be specifically captured by SiO2-pep microspheres through peptide CLIKKPF ligand on their surface.
Fig. 2.
Verification of SiO2-pep-based specific adsorption of model exosomes. (A) SEM image showing the model exosomes on the surface of SiO2-pep microspheres. Compared to the three negative controls, only SiO2-pep microspheres incubated with model exosomes demonstrated a roughness on their surface; (B) TEM image of a section of a SiO2-pep microsphere-bound exosome; (C) IF image showing green spots on SiO2-pep microspheres-exosome samples, reflecting the binding of FITC antibody to CD81, thus the presence of exosomes only in this sample compared to the three negative controls.
3.3. Optimization of SiO2-pep-based exosome isolation and characterization of the eluted exosomes
To maximize the adsorption/recovery efficiency of our method, we optimized several isolation/elution conditions, including the amount of SiO2-pep microspheres, incubation time, concentration of eluent, and elution time.
As demonstrated in Fig. 3A and B, the recovery rate reached a maximum of approximately 70% (model exosomes with ∼0.7 μg total proteins per mg SiO2-pep microspheres) when the amount of SiO2-pep microspheres was 8 mg and the incubation time was 60 min; at extended incubation times, the recovery rate stayed constant. For the elution conditions and recovery of the adsorbed exosomes from the surface of SiO2-pep microspheres, the optimum elution solvent was chosen as NH3·H2O for two reasons: first, PS molecules have two negative and one positive charges in neutral solution (the pH of binding buffer and PBS are both 7.4), but only two negative charges in the alkaline solution (-NH3+ is converted to –NH2), and this change in the charge of PS molecules/amino groups may have an effect on the bind between the peptide ligands and PS molecules; and second, NH3 is easily removed by evaporation. As shown in Fig. 3C and D, a maximum recovery rate of approximately 72% was obtained in a 12% NH3·H2O with an elution time of 80 min; further increase in the elution time did not result in any further increase in the recovery rate. These optimized four factors were chosen for the following experiments.
Fig. 3.
Optimization of the adsorption and elution conditions for exosome isolation. Isolation efficiency of model exosomes as a function of (A) the amount of SiO2-pep microspheres, (B) adsorption time, (C) concentration of the elution buffer NH3·H2O, and (D) elution time. When the amount of SiO2-pep microspheres, adsorption time, concentration of eluent NH3·H2O, and elution time were optimized at 8 mg, 60 min, 12%, and 80 min, respectively, the recovery of model exosomes peaked at 72%.
Characterization of the eluted exosomes was performed by checking their size, marker proteins, and microscopic structure. The size range measured by NTA (Fig. 4A) revealed that the X10, X50, and X90 of eluted exosomes were 78.40 nm, 127.20 nm, and 216.10 nm, respectively, and the size range was centered at 135.00 nm, which conforms with the defined size range of exosomes [1,2,9]. In addition, the X90 of eluted exosomes was less than that of the original model exosomes (directly obtained from HeLa cell culture medium by UC; Fig. S4), which reveals that contaminants of larger particle size co-isolated with model exosomes were further removed during the capture process in the SiO2-pep microspheres method. Finally, the exosomal marker proteins CD81 and TSG101 were both detected by WB (Fig. 4B), and a typical exosome membrane structure was observed in TEM image (Fig. 4C), confirming that intact exosomes can be isolated using SiO2-pep microspheres without altering their vesicular structure.
Fig. 4.
Characterization of the eluted exosomes. (A) NTA of eluted exosomes: X10, 78.40 nm; X50, 127.20 nm; and, X90, 216.10 nm; (B) WB of exosomal marker proteins CD81 and TSG101; (C) TEM image of eluted exosomes; the eluted exosomes are indicated by red arrows and have a typical membrane structure without noticeable destruction.
Taking together, using optimal method conditions (amount of SiO2-pep microspheres, 8 mg; adsorption time, 60 min; concentration of NH3·H2O, 12%; and elution time, 80 min), we were able to recover a high yield (72%) of model exosomes while maintaining their intact structure in about 2 h using a simple process.
3.4. Separation of serum exosomes using the SiO2-pep affinity method and comparison with the UF, UC, and DGC methods
To further evaluate the reliability of exosome isolation using SiO2-pep microspheres, this method was applied to real samples. The serum samples are the most common samples in the clinical practice, and due to their complex composition and the presence of many interferences, it is difficult to isolate exosomes with high purity and use them for subsequent clinical analysis. Moreover, the limited volume of serum samples and high throughput analysis renders classical isolation methods, such as UC and other similarly time-consuming methods, unsuitable for serum exosome separation; hence, a simple and efficient method is required. The results of our pilot experiments investigating adsorption between SiO2-pep microspheres and model exosomes demonstrate that this method can capture exosomes with simple steps in a short time, and it is worthwhile testing its efficacy against UF, UC, and DGC.
After demonstrating the ability of SiO2-pep microspheres to specifically isolate exosomes from serum samples through affinity binding towards peptides on their surface rather than non-specific adsorption (Text S3, Fig. S5), as conducted with the model exosomes, and after the serum exosomes isolated by SiO2-pep affinity method were characterized by NTA and TEM (Fig. S6), we evaluated the purity and contaminants of exosome samples isolated from serum by SiO2-pep affinity method, and by three other methods through WB of exosome marker protein CD9 and SDS-PAGE gel, and the protein compositions of exosomes using mass spectrometry analysis.
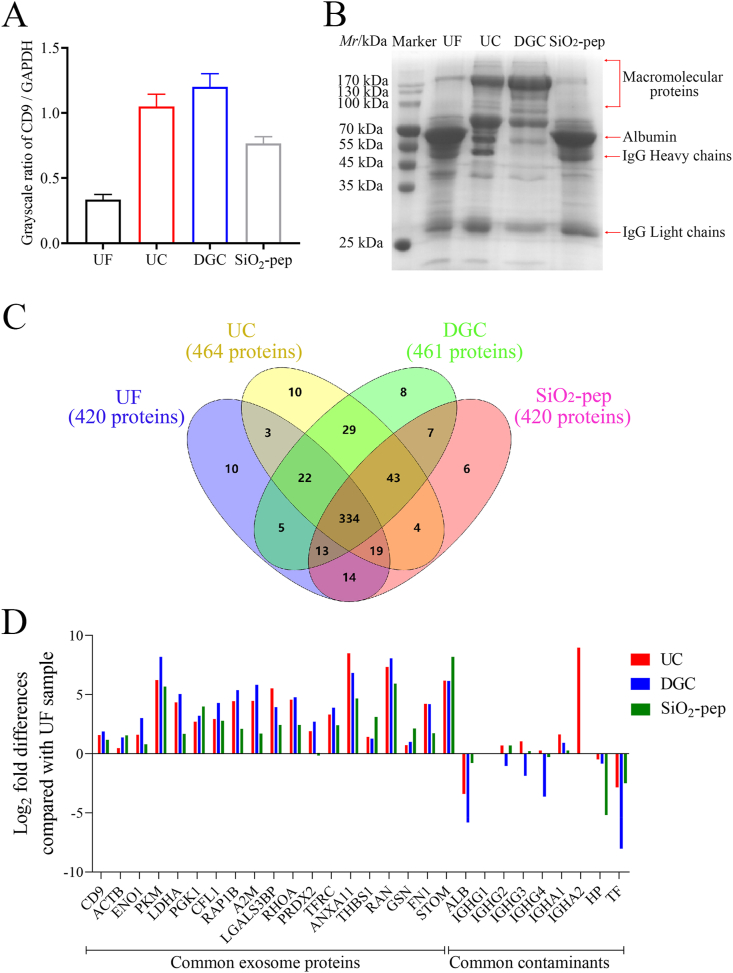
First, we assessed the purity of the isolated exosomes by WB; CD9 was chosen as a marker because, according to a previous research, several standard exosome markers, including CD63, CD81, TSG101, flotillins, and ALIX, are not well represented in exosomes isolated from human plasma, while CD9 is easily detected [34]. The WB results (Fig. S7) reveal that exosome samples isolated by all four methods possessed the exosome marker protein CD9. The grayscale images of band CD9 and GAPDH (which is commonly used as an internal reference for exosomes) were then quantified using the software ImageJ. The grayscale ratio of CD9 to GAPDH (Fig. 5A) reflects the purity of exosomes to a reasonable extent [35,36]. From this relative quantification, we observed that among the exosome samples isolated by the four methods, the purity of the sample isolated by DGC was the highest and this was only slightly higher than that separated by UC. The purity of exosome samples captured by the SiO2-pep affinity method was approximately 64% of that isolated by DGC. Last comes UF with only 28% grayscale ratio compared to DGC. The standard deviations of the grayscale ratio values of exosome samples isolated by UF, UC, DGC, and the SiO2-pep affinity method were 0.04, 0.09, 0.10, and 0.05, respectively, which indicates that all four methods demonstrate good reproducibility.
Fig. 5.
Separation of serum exosomes using the SiO2-pep affinity method and comparison with those isolated by UF, UC, and DGC methods. (A) Grayscale ratio of bands CD9 and GAPDH in WB images of different exosome samples quantified by ImageJ software, which reflects the purity of exosomes to a reasonable extent; (B) SDS-PAGE gel image illustrating the main contaminating protein compositions of different exosome samples; (C) Venn diagram of identified proteins in different exosome samples using mass spectrometry analysis; (D) Log2-fold differences in the quantities of common exosomal proteins and contaminating proteins in different exosome samples.
In previous studies, it was revealed that, irrespective of the content of exosomes isolated from complex body fluids and tissues by the same method, the protein bands visible to the naked eye in SDS-PAGE gel images were almost identical in location and grey degree, and were similar to bands in the SDS-PAGE gel image of proteins in the original body fluids and tissue lysates (samples which did not undergo exosomes isolation). This can be explained by the low content of exosomes in body fluids and tissues, and their low distinct individual protein content [[37], [38], [39], [40]]. Thus, the protein bands visible to the naked eye in SDS-PAGE gel images do not generally represent exosome protein bands, and most represent contaminating proteins from bodily fluids and tissues. As a consequence, SDS-PAGE images can be used as a rough estimation of what contaminants are co-isolated from the serum with the exosomes. The main contaminating proteins in exosome samples isolated by the four methods are shown in the SDS-PAGE gel image (Fig. 5B). The exosome samples isolated by the four different methods demonstrated different contamination trends due to their different isolation principles. Interestingly, the exosome sample isolated by the SiO2-pep affinity method had fewer macromolecular contaminating proteins than the other methods, either because they could not penetrate the 100 kDa ultrafiltration membrane in the UF method, e.g., C3 (187 kDa), C4 (192.6 kDa), and immunoglobulins (150–200 kDa), or because the macromolecular contaminating proteins were contained in a complex of similar size and density to exosomes leading to their co-separation in UC and DGC, such as APOB (515.3 kDa) from very low-density lipoproteins (VLDL, 30–80 nm; 0.930–1.006 g cm−3) and the chylomicrons (CM; 75–1200 nm; < 0.930 g cm−3) which demonstrate a particle size overlap with exosomes, or A2M (163.2 kDa) from high-density lipoproteins (HDL, 5–12 nm, 1.063–1.210 g cm−3) which demonstrates a particle density overlap with exosomes [11]. In addition, the exosome samples showed prominent albumin (ALB, ∼66 kDa) and IgG (∼25 kDa for the light chains and ∼50 kDa for the heavy chains) contamination probably because of interactions with the surfaces of exosomes, with the exception of DGC since the non-physiological experimental conditions used (the sucrose density gradient medium) and high centrifugal forces (also used during UC) interfere with these interactions [40]. The high observed contamination with ALB in UF may be explained by ALB binding to other molecules causing an increase in their molecular size and thus increasing co-isolation. During the SiO2-pep affinity method, ALB may complex with PS molecules or other substances containing PS, thus explaining its high content [41].
Finally, we performed nano-LC-MS/MS and label-free quantitative analysis to determine the protein composition of serum exosome samples isolated by the four methods. As shown in Figs. 5C and 420 proteins, 464 proteins, 461 proteins, and 420 proteins were identified in serum exosome samples isolated by UF, UC, DGC, and the SiO2-pep affinity method, respectively. Of these, 55.17%, 58.80%, 56.58%, and 55.33% of identified proteins in each group (respectively) were exosomal-proteins in the exosome database (ExoCarta database). Moreover, more than 60% (334 proteins) of the total identified proteins (527 proteins) were identified in all four groups, 227 of which have an exosomal origin. Thus, each method isolated exosomes with similar and/or different contaminants.
To further evaluate the protein composition of the serum exosome samples, we compared the quantities of common exosomal proteins and common contaminating proteins identified in these samples [16]. We listed nineteen typical exosome markers belonging to the top 100 proteins that are commonly identified in exosomes (ExoCarta database), and nine common contaminants in serum — highly abundant proteins that represent approximately 85% of the total protein mass, including ALB, IgG (IGHG1 – IGHG4), IgA (IGHA1 and IGHA2), haptoglobin (HP), and transferrin (TF) [42]. After normalization using the corresponding protein quantity obtained by UF as a reference, the log2-fold differences in the quantities of the nineteen exosome markers and nine common contaminants in samples from the other three methods were evaluated. In the resulting histogram (Fig. 5D), the positive values indicate that the content of a certain protein in the exosome sample isolated by one of the other three methods was higher than that of the corresponding protein in the UF-enriched exosome sample (and the content increased with an increase in the absolute value). Conversely, the negative values indicate that the content of a certain protein in the exosome sample isolated by one of the other three methods was lower than that of the corresponding protein in the UF-enriched exosome sample (and the content decreased with an increase in the absolute value). The log2 quantity values of most of the exosome markers were positive in the UC-, DGC-, and SiO2-pep affinity method-enriched samples. The majority of the log2 quantity values were larger in the DGC-enriched sample, followed by the UC-enriched sample. Interestingly, the log2 quantity values obtained using the SiO2-pep affinity method were very similar; this trend correlates with the detection of marker CD9 from the WB results (Fig. 5A). Moreover, several log2 quantity values obtained with the SiO2-pep affinity method were even larger compared with the other methods. In contrast, the log2 quantity values of most of the common contaminants were negative in the UC-, DGC-, and SiO2-pep affinity method-enriched samples. As demonstrated in the SDS-PAGE image (Fig. 5B), ALB contamination was more prominent in SiO2-pep affinity method-enriched exosome samples compared with samples isolated by UC and DGC, but remained markedly below that of UF. As discussed above, this can be simply explained by considering the principles of the SiO2-pep affinity method. Strikingly, the SiO2-pep affinity method had the lowest content of other contaminants (e.g., HP, which forms higher order oligomers and binds to hemoglobin in the blood to form complexes) [43].
To sum up, each enrichment method for exosome samples from serum inevitably co-separates contaminants, but the type and content is method dependent due to the different separation principles. Combining the results of the WB and protein composition analysis of serum exosome samples, the purity of SiO2-pep affinity method-enriched exosome samples was far higher than that of the UF-enriched samples, and approached that of the DGC- and UC-enriched samples, and the DGC and UC are the most commonly used methods for exosome isolation with a high purity but at comparatively low yield compared with other methods in the literature. In the present study, the final protein yields from exosome samples obtained using the UF, UC, DGC, and SiO2-pep affinity methods (after 1 h, 15 h, 32 h, and 2 h, respectively) were 22.10 μg, 0.16 μg, 0.05 μg, and 0.72 μg protein per microliter of serum, respectively. In the other hand, the applications of the DGC and UC methods are limited by the complicated processing and the need for expensive instrumentation, especially in case of large-scale samples, and the harsh conditions may be detrimental to the structural integrity of exosomes. Fortunately for SiO2-pep affinity method, because of its specific separation principle, contaminants in the serum exosomes generally belong to the same category, i.e. the variety of contaminants co-separated is decreased compared with the other three methods. Therefore, despite a small sacrifice in purity, the SiO2-pep affinity method offers a simplified operating process, a relatively high yield, a narrower range of contaminants, and the possibility of high throughput, at a relatively low-cost. These undeniable advantages make the SiO2-pep affinity method more suitable for processing large numbers of clinical samples. Table 1 details a direct comparison of the different methods for the isolation of exosomes.
Table 1.
Comparison of different methods for the isolation of exosomes.
| Methods | UF | UC | DGC | SiO2-pep affinity |
|---|---|---|---|---|
| Processing time (h) | 1 | 15 | 32 | 2 |
| Yield (μg protein in exosome samples per microliter of serum) | 22.10 | 0.16 | 0.05 | 0.72 |
| Purity (grayscale ratio of CD9/GAPDH compared to DGC, %) | 28 | 87 | 100 | 64 |
| Reproducibility (SD of grayscale ratio of CD9/GAPDH) | 0.04 | 0.09 | 0.10 | 0.05 |
| Main contaminants | Macromolecular contaminants, VLDL, CM, HDL, ALB, IgG | Macromolecular contaminants, VLDL, CM, IgG | Macromolecular contaminants, HDL | ALB, IgG, Complexes containing PS |
| Advantages | Simple, fast, does not rely on equipment [6] | High purity, High sample capacity [6] | High purity [6] | Simple, fast, cheap, high yield, relatively high purity, single contamination, can preserve vesicular structure |
| Disadvantages | Low purity, loss of exosomes during the process [6] | Cumbersome process, time consuming, requires expensive equipment, may destroy vesicular structure, low yield, not suitable for high-throughput analysis [6] | Cumbersome process, time consuming, requires expensive equipment, may destroy vesicular structure, low yield, not suitable for high-throughput analysis [6] | Purity needs further improvement, low sample capacity |
3.5. Proteomics of exosomes isolated from healthy, HCC patient, and CCA patient serum samples using the SiO2-pep affinity method and its application in cancer diagnosis and typing
HCC is a very common cancer with an incidence increasing every year, and CCA has a high mortality rate due to its difficulty of diagnosis [44,45]. Detection of cancer at its onset and identification of type is critical for subsequent treatment and for improving survival rates. Current diagnostic methods such as radiation tests and biopsies can be harmful and are comparatively long. Therefore, the discovery of new biomarkers for cancer diagnoses which can be used in low invasive methods is an imperative. Exosomes are widespread in body fluids, and these provide valuable information for disease diagnosis because they carry biomolecules such as proteins and nucleic acids which reflect the state of the organism. Thus, exosomes represent a good target for biomarker discovery. In the present study, the proteomes of SiO2-pep affinity method-enriched exosomes from healthy individuals, HCC patients, and CCA patients were analyzed to expand and verify the application value of this new method.
For the simple and rapid differentiation of healthy individuals (Con), HCC patients, and CCA patients, the proteomes of exosomes isolated from the serum of these three groups using the SiO2-pep affinity method (nine samples per group) were analyzed by nano-LC-MS/MS, and a quantitative comparison was conducted using a label-free quantification method (intensity-based absolute quantification). A total of 695 proteins were detected in the three groups, of which 438 proteins were in the exosome database (ExoCarta database). These proteins were further screened into 370 credible proteins, and these were compared to identify biomarkers and to provide biological information. After median normalization and log2 logarithmic conversion of the quantitative expression data from the 370 credible proteins, only a few outlier data points were observed, and the median of the data tended to the center in the boxplots (Figs. S8A and S8B). The density maps (Figs. S8C and S8D) demonstrated that the data before normalization were strongly skewed, while the normalized data tended to display normal distributions. Both visual presentations demonstrated the necessity of data normalization, which facilitates subsequent statistical analyses. PCA analysis (Fig. 6A) and a hierarchical clustering dendrogram of sample Euclidean Distance (Fig. 6B) revealed that the Con, HCC, and CCA groups were well separated, indicating a significant difference between the three groups.
Fig. 6.
Discrimination between Con, HCC, and CCA groups, and biological information concerning differentially-expressed proteins in pair-wise comparisons. (A) PCA and (B) Hierarchical clustering dendrogram of the Con, HCC, and CCA groups demonstrated that the three groups can be well separated by the quantitative expression normalization data of identified credible exosome proteins; (C) GO enrichment analysis of differentially-expressed proteins in each pair-wise comparison (top10 of each classification according to -log10 P-value). Bubble diagrams of KEGG pathways (top15 according to -log10 P-value in each comparison group) analyzed using differentially-expressed proteins in the (D) HCC vs. Con groups, (E) CCA vs. Con groups, and (F) HCC vs. CCA pair-wise comparisons (the size of each bubble represents the number of differentially-expressed proteins involved in this pathway, while the color represents the P-value).
To screen for significantly up- or down-regulated proteins in each group, a moderate t-statistic analysis was performed. In total, 60 up-regulated and 23 down-regulated proteins were identified in the HCC group (compared to the Con group), and 54 up-regulated and 17 down-regulated proteins were identified in the CCA group (compared to the Con group). In a pair-wise comparison of the two cancer groups (HCC vs. CCA), 15 up-regulated and 15 down-regulated proteins were identified in the HCC group (moderate t-statistic, Adj. P-Value < 0.05, FC > 2 or <0.5; Figs. S9A, S9B, S9C; Table S3). Next, each pairwise comparison was displayed in a heat map and evaluated by unsupervised hierarchical cluster analysis using normalized quantitative expression data for the differential proteins (Fig. S10). As expected, the groups could be well separated, thus demonstrating the potential of using exosomal protein profiling for biomarker screening of HCC patients and CCA patients to improve cancer diagnosis and typing.
Finally, we analyzed the functions of the differentially-expressed proteins and their involvement in biological processes and pathways. According to the GO enrichment analysis, KEGG pathway analysis, and protein-protein interaction results on differentially-expressed proteins in each pair-wise comparison (Figs. 6C, 6D, 6E, 6F and S11), the functions, processes, and pathways enriched by these proteins mainly include natural immune response, blood coagulation, inflammatory response, and receptor binding, such as neutrophil degranulation, cornification, and complement and coagulation cascades, which are related to tumor promotion, suppression, and metastasis. By comparing the biological information on the differentially-expressed proteins in serum exosomes isolated by our SiO2-pep affinity method, we can further deepen our understanding of the mechanisms of the occurrence and progression of these two cancers, and identify reliable biomarkers and potentially new drug actionable targets [46,47].
4. Conclusion
In the present study, we explored a novel separation strategy for exosomes based on specific interactions between peptide ligand immobilized silica microspheres (SiO2-pep) and PS moieties on the surface of exosomes. Model exosomes isolated from Hela cell medium by ultracentrifugation were used as adsorption samples to investigate the specificity of adsorption onto the surface of SiO2-pep microspheres, and to optimize the isolation conditions. We found that exosomes with an intact structure could be isolated and recovered in a short processing time (about 2 h). Next, our novel method was used to extract exosomes from serum, and these were compared with those isolated using three well-established methods. Evaluation of exosome purity through WB, examination of contaminating proteins through SDS-PAGE, and analysis of exosomal protein composition revealed that the purity of SiO2-pep affinity-enriched exosomes was comparable to that of exosomes isolated by DGC and UC, and markedly higher than that of exosomes isolated by UF. Moreover, the yield of SiO2-pep affinity-enriched exosomes was higher and the range of contaminants was decreased (compared with DGC and UC). Finally, our novel SiO2-pep affinity method for the isolation of exosomes was used to search for potential biomarkers and to explore the mechanisms of HCC and CCA by analyzing the proteomes of serum exosomes from Con, HCC, and CCA groups by nano-LC-MS/MS.
Our method provides a novel approach to isolating exosomes by targeting high density PS molecules on the surface of exosomes with immobilized small peptide molecules; despite a small sacrifice in purity compared to the UC and DGC methods, the SiO2-pep affinity method potentially has a wide application because of its advantages. These include its simple operation process, high yield, and low-cost. The resulting exosomes can be used in deeper functional experiments, as drug delivery carriers, or be lysed directly for proteomics and metabolomic analyses.
It is inevitable to introduce contaminants into the exosome samples during the process of isolation independently of the used method. However, by controlling the composition of the contaminants, the experimental effects of contaminants can be greatly reduced, and measures may be developed for further purification. During the SiO2-pep affinity isolation of serum exosomes, the main contaminants introduced were complexes containing PS (in addition to common contaminants such as ALB and IgG). This problem needs further exploration, for instance, we can first dissociate these complexes before the capture process. Another way to reach higher purity of exosome samples is the combination of several methods with different principles to have an orthogonal capture. Finally, our study provides a new research direction, which does not rely on the physical properties of exosomes or on expensive high molecular weight marker proteins. The targeting of specific molecules on the exosome surface using small molecules with high affinity such as peptides or oligonucleotides warrants further investigation in the future.
Funding
This work was supported by the National Natural Science Foundation of China (81874307, 21874088); Shanghai Science and Technology Commission Scientific Research Project (18142200700, 19142203100, 20142200400); the Open Project Program of Engineering Research Center of Cell & Therapeutic Antibody, Ministry of Education, Shanghai Jiao Tong University (19X110020009-005); Startup Fund for Youngman Research at SJTU (19X100040029).
CRediT authorship contribution statement
Kaige Yang: Conceptualization, Methodology. Mengqi Jia: Validation, Writing – original draft, preparation. Soumia Cheddah: Writing – review & editing. Zhouyi Zhang: Formal analysis. Weiwei Wang: Investigation. Xinyan Li: Methodology, Investigation. Yan Wang: Conceptualization, Writing – review & editing. Chao Yan: Conceptualization, Investigation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
Not applicable.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.12.017.
Contributor Information
Xinyan Li, Email: lixy@sjtu.edu.cn.
Yan Wang, Email: wangyan11@sjtu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478) doi: 10.1126/science.aau6977. 640-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hessvik N.P., Llorente A. Current knowledge on exosome biogenesis and release. Cellular Mol. Life Sci. 2018;75(2):193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greening D.W., Gopal S.K., Xu R., Simpson R.J., Chen W.S. Exosomes and their roles in immune regulation and cancer. Seminars Cell Develop. Biol. 2015;40:72–81. doi: 10.1016/j.semcdb.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Dai W.H., Su L., Lu H.T., Dong H.F., Zhang X.J. Exosomes-mediated synthetic Dicer substrates delivery for intracellular Dicer imaging detection. Biosens. Bioelectron. 2020:151. doi: 10.1016/j.bios.2019.111907. [DOI] [PubMed] [Google Scholar]
- 5.Chang Y.H., Wu K.C., Harn H.J., Lin S.Z., Ding D.C. Exosomes and stem cells in degenerative disease diagnosis and therapy. Cell Transplantat. 2018;27(3):349–363. doi: 10.1177/0963689717723636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu L.L., Zhu J., Liu J.X., Jiang F., Ni W.K., Qu L.S., Ni R.Z., Lu C.H., Xiao M.B. A comparison of traditional and novel methods for the separation of exosomes from human samples. Biomed Res. Int. 2018 doi: 10.1155/2018/3634563. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diaz G., Bridges C., Lucas M., Cheng Y., Schorey J.S., Dobos K.M., Kruh-Garcia N.A. Protein digestion, ultrafiltration, and size exclusion chromatography to optimize the isolation of exosomes from human blood plasma and serum. Jove-J. Visualiz. Exp. 2018;(134) doi: 10.3791/57467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He L.Q., Zhu D., Wang J.P., Wu X.Y. A highly efficient method for isolating urinary exosomes. Int. J. Molecular Med. 2019;43(1):83–90. doi: 10.3892/ijmm.2018.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel G.K., Khan M.A., Zubair H., Srivastava S.K., Khushman M., Singh S., Singh A.P. Comparative analysis of exosome isolation methods using culture supernatant for optimum yield, purity and downstream applications. Scientific Rep. 2019;9 doi: 10.1038/s41598-019-41800-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helwa I., Cai J.W., Drewry M.D., Zimmerman A., Dinkins M.B., Khaled M.L., Seremwe M., Dismuke W.M., Bieberich E., Stamer W.D., Hamrick M.W., Liu Y.T. A comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents. Plos One. 2017;12(1) doi: 10.1371/journal.pone.0170628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian Y., Gong M.F., Hu Y.Y., Liu H.S., Zhang W.Q., Zhang M.M., Hu X.X., Aubert D., Zhu S.B., Wu L., Yan X.M. Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J. Extracellular Vesicles. 2020;9(1) doi: 10.1080/20013078.2019.1697028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livshts M.A., Khomyakova E., Evtushenko E.G., Lazarev V.N., Kulemin N.A., Semina S.E., Generozov E.V., Govorun V.M. Isolation of exosomes by differential centrifugation: theoretical analysis of a commonly used protocol. Scientific Rep. 2015;5 doi: 10.1038/srep17319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li K., Wong D.K., Hong K.Y., Raffai R.L. Cushioned–density gradient ultracentrifugation (C-DGUC): a refined and high performance method for the isolation, characterization, and use of exosomes. Extracellular RNA. 2018:69–83. doi: 10.1007/978-1-4939-7652-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Theodoraki M.N., Yerneni S.S., Hoffmann T.K., Gooding W.E., Whiteside T.L. Clinical significance of PD-L1(+) exosomes in plasma of head and neck cancer patients. Clinic. Cancer Res. 2018;24(4):896–905. doi: 10.1158/1078-0432.CCR-17-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang L.G., Kong M.Q., Zhou S., Sheng Y.F., Wang P., Yu T., Inci F., Kuo W.P., Li L.J., Demirci U., Wang S.Q. An integrated double-filtration microfluidic device for isolation, enrichment and quantification of urinary extracellular vesicles for detection of bladder cancer. Scientific Rep. 2017;7 doi: 10.1038/srep46224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao F.Y., Jiao F.L., Xia C.S., Zhao Y., Ying W.T., Xie Y.P., Guan X.Y., Tao M., Zhang Y.J., Qin W.J., Qian X.H. A novel strategy for facile serum exosome isolation based on specific interactions between phospholipid bilayers and TiO2. Chem. Sci. 2019;10(6):1579–1588. doi: 10.1039/c8sc04197k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skotland T., Sandvig K., Llorente A. Lipids in exosomes: current knowledge and the way forward. Prog. Lipid Res. 2017;66:30–41. doi: 10.1016/j.plipres.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Record M., Carayon K., Poirot M., Silvente-Poirot S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochimica Et Biophysica. Acta-Molecul. Cell Biol. Lipids. 2014;1841(1):108–120. doi: 10.1016/j.bbalip.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Keller S., Konig A.K., Marme F., Runz S., Wolterink S., Koensgen D., Mustea A., Sehouli J., Altevogt P. Systemic presence and tumor-growth promoting effect of ovarian carcinoma released exosomes. Cancer Letters. 2009;278(1):73–81. doi: 10.1016/j.canlet.2008.12.028. [DOI] [PubMed] [Google Scholar]
- 20.Morelli A.E., Larregina A.T., Shufesky W.J., Sullivan M.L.G., Stolz D.B., Papworth G.D., Zahorchak A.F., Logar A.J., Wang Z.L., Watkins S.C., Falo L.D., Thomson A.W. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood. 2004;104(10):3257–3266. doi: 10.1182/blood-2004-03-0824. [DOI] [PubMed] [Google Scholar]
- 21.Zakharova L., Svetlova M., Fomina A.F. T cell exosomes induce cholesterol accumulation in human monocytes via phosphatidylserine receptor. J. Cellular Physiol. 2007;212(1):174–181. doi: 10.1002/jcp.21013. [DOI] [PubMed] [Google Scholar]
- 22.Kang Y.T., Purcell E., Palacios-Rolston C., Lo T.W., Ramnath N., Jolly S., Nagrath S. Isolation and profiling of circulating tumor-associated exosomes using extracellular vesicular lipid-protein binding affinity based microfluidic device. Small. 2019;15(47) doi: 10.1002/smll.201903600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu H., Liao C., Zuo P., Liu Z., Ye B.C. Magnetic-based microfluidic device for on-chip isolation and detection of tumor-derived exosomes. Anal Chem. 2018;90(22):13451–13458. doi: 10.1021/acs.analchem.8b03272. [DOI] [PubMed] [Google Scholar]
- 24.Nakai W., Yoshida T., Diez D., Miyatake Y., Nishibu T., Imawaka N., Naruse K., Sadamura Y., Hanayama R. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Scientific Rep. 2016;6 doi: 10.1038/srep33935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kooijmans S.A.A., Gitz-Francois J.J.J.M., Schiffelers R.M., Vader P. Recombinant phosphatidylserine-binding nanobodies for targeting of extracellular vesicles to tumor cells: a plug-and-play approach. Nanoscale. 2018;10(5):2413–2426. doi: 10.1039/c7nr06966a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma R., Huang X., Brekken R.A., Schroit A.J. Detection of phosphatidylserine-positive exosomes for the diagnosis of early-stage malignancies. Br J Cancer. 2017;117(4):545–552. doi: 10.1038/bjc.2017.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haraszti R.A., Didiot M.C., Sapp E., Leszyk J., Shaffer S.A., Rockwell H.E., Gao F., Narain N.R., DiFiglia M., Kiebish M.A., Aronin N., Khvorova A. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J Extracell Vesicles. 2016;5:32570. doi: 10.3402/jev.v5.32570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shih C.L., Chong K.Y., Hsu S.C., Chien H.J., Ma C.T., Chang J.W.C., Yu C.J., Chiou C.C. Development of a magnetic bead-based method for the collection of circulating extracellular vesicles. New Biotechnol. 2016;33(1):116–122. doi: 10.1016/j.nbt.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Burtea C., Laurent S., Lancelot E., Ballet S., Murariu O., Rousseaux O., Port M., Elst L.V., Corot C., Muller R.N. Peptidic targeting of phosphatidylserine for the MRI detection of apoptosis in atherosclerotic plaques. Molecular Pharmaceutic. 2009;6(6):1903–1919. doi: 10.1021/mp900106m. [DOI] [PubMed] [Google Scholar]
- 30.Kapty J., Banman S., Goping I.S., Mercer J.R. Evaluation of phosphatidylserine-binding peptides targeting apoptotic cells. J. Biomolecular Screen. 2012;17(10):1293–1301. doi: 10.1177/1087057112453313. [DOI] [PubMed] [Google Scholar]
- 31.Hoyle C.E., Bowman C.N. Thiol-ene click chemistry. Angew Chem Int Ed Engl. 2010;49(9):1540–1573. doi: 10.1002/anie.200903924. [DOI] [PubMed] [Google Scholar]
- 32.Miyabe K., Guiochon G. Influence of the modification conditions of alkyl bonded ligands on the characteristics of reversed-phase liquid chromatography. J. Chromatography A. 2000;903(1–2):1–12. doi: 10.1016/s0021-9673(00)00891-8. [DOI] [PubMed] [Google Scholar]
- 33.Kailasam K., Muller K. Physico-chemical characterization of MCM-41 silica spheres made by the pseudomorphic route and grafted with octadecyl chains. J. Chromatography A. 2008;1191(1–2):125–135. doi: 10.1016/j.chroma.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 34.Hoshino A., Kim H.S., Bojmar L., Gyan K.E., Cioffi M., Hernandez J., Zambirinis C.P., Rodrigues G., Molina H., Heissel S., Mark M.T., Steiner L., Benito-Martin A., Lucotti S., Di Giannatale A., Offer K., Nakajima M., Williams C., Nogues L., Vatter F.A.P., Hashimoto A., Davies A.E., Freitas D., Kenific C.M., Ararso Y., Buehring W., Lauritzen P., Ogitani Y., Sugiura K., Takahashi N., Aleckovic M., Bailey K.A., Jolissant J.S., Wang H.J., Harris A., Schaeffer L.M., Garcia-Santos G., Posner Z., Balachandran V.P., Khakoo Y., Raju G.P., Scherz A., Sagi I., Scherz-Shouval R., Yarden Y., Oren M., Malladi M., Petriccione M., De Braganca K.C., Donzelli M., Fischer C., Vitolano S., Wright G.P., Ganshaw L., Marrano M., Ahmed A., DeStefano J., Danzer E., Roehrl M.H.A., Lacayo N.J., Vincent T.C., Weiser M.R., Brady M.S., Meyers P.A., Wexler L.H., Ambati S.R., Chou A.J., Slotkin E.K., Modak S., Roberts S.S., Basu E.M., Diolaiti D., Krantz B.A., Cardoso F., Simpson A.L., Berger M., Rudin C.M., Simeone D.M., Jain M., Ghajar C.M., Batra S.K., Stanger B., Bui J., Brown K.A., Rajasekhar V.K., Healey J.H., de Sousa M., Kramer K., Sheth S., Baisch J., Pascual V., Heaton T.E., La Quaglia M.P., Pisapia D.J., Schwartz R., Zhang H.Y., Liu Y., Shukla A., Blavier L., DeClerck Y.A., LaBarge M., Bissell M.J., Caffrey T.C., Grandgenett P.M., Hollingsworth M.A., Bromberg J., Costa-Silva B., Peinado H., Kang Y.B., Garcia B.A., O'Reilly E.M., Kelsen D., Trippett T.M., Jones D.R., Matei I.R., Jarnagin W.R., Lyden D. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. 2020;182(4):1044–+. doi: 10.1016/j.cell.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Y.Y., Luo Y.B., Fang Z.H., Teng W.J., Yu Y.C., Tian J.H., Guo P., Xu R.Z., Wu J.C., Li Y. Mechanism of sanguinarine in inhibiting macrophages to promote metastasis and proliferation of lung cancer via modulating the exosomes in A549 cells. Oncotargets Therap. 2020;13:8989–9003. doi: 10.2147/OTT.S261054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang C., Guo W.B., Zhang W.S., Bian J., Yang J.K., Zhou Q.Z., Chen M.K., Peng W., Qi T., Wang C.Y., Liu C.D. Comprehensive proteomics analysis of exosomes derived from human seminal plasma. Andrology. 2017;5(5):1007–1015. doi: 10.1111/andr.12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L., Chen R.J., Kemper S., Cong M., You H., Brigstock D.R. Therapeutic effects of serum extracellular vesicles in liver fibrosis. J. Extracellular Vesicles. 2018;7(1) doi: 10.1080/20013078.2018.1461505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santucci L., Bruschi M., Del Zotto G., Antonini F., Ghiggeri G.M., Panfoli I., Candiano G. Biological surface properties in extracellular vesicles and their effect on cargo proteins. Scientific Rep. 2019;9 doi: 10.1038/s41598-019-47598-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burkova E.E., Dmitrenok P.S., Bulgakov D.V., Vlassov V.V., Ryabchikova E.I., Nevinsky G.A. Exosomes from human placenta purified by affinity chromatography on sepharose bearing immobilized antibodies against CD81 tetraspanin contain many peptides and small proteins. Iubmb Life. 2018;70(11):1144–1155. doi: 10.1002/iub.1928. [DOI] [PubMed] [Google Scholar]
- 40.Burkova E.E., Grigor'eva A.E., Bulgakov D.V., Dmitrenok P.S., Vlassov V.V., Ryabchikova E.I., Sedykh S.E., Nevinsky G.A. Extra purified exosomes from human placenta contain an unpredictable small number of different major proteins. Int. J. Molecular Sci. 2019;20(10) doi: 10.3390/ijms20102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Vusse G.J. Albumin as fatty acid transporter. Drug Metabolism Pharmacokinetic. 2009;24(4):300–307. doi: 10.2133/dmpk.24.300. [DOI] [PubMed] [Google Scholar]
- 42.Zolotarjova N., Martosella J., Nicol G., Bailey J., Boyes B.E., Barrett W.C. Differences among techniques for high-abundant protein depletion. Proteomics. 2005;5(13):3304–3313. doi: 10.1002/pmic.200402021. [DOI] [PubMed] [Google Scholar]
- 43.Andersen C.B.F., Stodkilde K., Saederup K.L., Kuhlee A., Raunser S., Graversen J.H., Moestrup S.K. Haptoglobin. Antioxid Redox Signal. 2017;26(14):814–831. doi: 10.1089/ars.2016.6793. [DOI] [PubMed] [Google Scholar]
- 44.Massarweh N.N., El-Serag H.B. Epidemiology of hepatocellular carcinoma and intrahepatic cholangiocarcinoma. Cancer Control. 2017;24(3) doi: 10.1177/1073274817729245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banales J.M., Inarrairaegui M., Arbelaiz A., Milkiewicz P., Muntane J., Munoz-Bellvis L., La Casta A., Gonzalez L.M., Arretxe E., Alonso C., Martinez-Arranz I., Lapitz A., Santos-Laso A., Avila M.A., Martinez-Chantar M.L., Bujanda L., Marin J.J.G., Sangro B., Macias R.I.R. Serum metabolites as diagnostic biomarkers for cholangiocarcinoma, hepatocellular carcinoma, and primary sclerosing cholangitis. Hepatology. 2019;70(2):547–562. doi: 10.1002/hep.30319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang C.Y., Peng L., Zhang Y.Q., Liu Z.Y., Li W.L., Chen S.L., Li G.C. The identification of key genes and pathways in hepatocellular carcinoma by bioinformatics analysis of high-throughput data. Medical Oncol. 2017;34(6) doi: 10.1007/s12032-017-0963-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arbelaiz A., Azkargorta M., Krawczyk M., Santos-Laso A., Lapitz A., Perugorria M.J., Erice O., Gonzalez E., Jimenez-Aguero R., Lacasta A., Ibarra C., Sanchez-Campos A., Jimeno J.P., Lammert F., Milkiewicz P., Marzioni M., Macias R.I.R., Marin J.J.G., Patel T., Gores G.J., Martinez I., Elortza F., Falcon-Perez J.M., Bujanda L., Banales J.M. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology. 2017;66(4):1125–1143. doi: 10.1002/hep.29291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.