Abstract
Background
The phase 3 CheckMate 214 trial demonstrated higher response rates and improved overall survival with nivolumab plus ipilimumab versus sunitinib in first-line therapy for advanced clear-cell renal cell carcinoma (RCC). An unmet need exists to identify patients with RCC who are most likely to benefit from treatment with nivolumab plus ipilimumab.
Methods
In exploratory analyses, pretreatment levels of programmed death ligand 1 were assessed by immunohistochemistry. Genomic and transcriptomic biomarkers (including tumor mutational burden and gene expression signatures) were also investigated.
Results
Biomarkers previously associated with benefit from immune checkpoint inhibitor-containing regimens in RCC were not predictive for survival in patients with RCC treated with nivolumab plus ipilimumab. Analysis of gene expression identified an association between an inflammatory response and progression-free survival with nivolumab plus ipilimumab.
Conclusions
The exploratory analyses reveal relationships between molecular biomarkers and provide supportive data on how the inflammation status of the tumor microenvironment may be important for identifying predictive biomarkers of response and survival with combination immunotherapy in patients with RCC. Further validation may help to provide biomarker-driven precision treatment for patients with RCC.
Keywords: kidney neoplasms, tumor biomarkers, immunotherapy, gene expression profiling, inflammation
Background
Nivolumab combined with ipilimumab or the tyrosine kinase inhibitor (TKI) cabozantinib are among several immune checkpoint inhibitor (ICI) regimens approved as first-line treatment for patients with advanced renal cell carcinoma (RCC).1–6 After a 48-month minimum follow-up analysis of CheckMate 214, superior survival benefit was observed with nivolumab plus ipilimumab versus sunitinib, the only double-ICI combination therapy currently approved for patients with RCC.1 7 Given these promising data, there still remains an unmet need to understand the underlying biology of RCC and identify patients who are most likely to respond to treatment.8
Programmed death ligand 1 (PD-L1) expression has shown promise as a biomarker for response to anti-programmed death-1 (PD-1) therapy in a number of cancer types.9 However, PD-L1 expression alone has been a suboptimal biomarker for patients with RCC and its predictive value is, to date, unclear. A number of studies have indicated that PD-L1 expression is negatively prognostic, but not predictive of response to treatment with ICIs alone or in combination with vascular endothelial growth factor (VEGF) inhibitors in patients with RCC.10–12 In CheckMate 214, exploratory analysis showed that longer overall survival (OS) with nivolumab plus ipilimumab versus sunitinib was independent of PD-L1 expression on tumor cells (TCs).1
A number of DNA-based biomarkers have been associated with response to ICIs. High tumor mutational burden (TMB) and the number of small insertions and deletions (tumor indel burden; TIB) are associated with increased numbers of tumor neoantigens presented on major histocompatibility complexes, facilitating immune recognition of the tumor and the development of an antitumor response.13 In addition, human leukocyte antigen (HLA) zygosity may affect the presentation of immunogenic antigens, potentially impacting response to ICIs.14 In RCC, TMB, TIB, and HLA, zygosity have demonstrated limited value in predicting clinical benefit with ICI monotherapy or ICIs in combination with VEGF inhibitors.12 15 16
Mutational inactivation of individual genes, including VHL, PBRM1, BAP1, and SETD2, is frequent in clear-cell RCC.15–18 In particular, results are conflicting regarding the association of PBRM1 mutation with clinical benefit from TKIs. PBRM1 mutation was associated with clinical benefit from ICI monotherapy following prior treatment with VEGF inhibitors,16 19 but not in previously untreated patients, whether given as monotherapy or in combination with VEGF inhibitors.12 15
Gene expression profiling (GEP) can be used to classify disease types and characterize biological processes such as inflammation in the tumor microenvironment (TME).20 21 An angiogenesis (Angio) gene expression signature was found to be predictive for benefit with sunitinib in the IMmotion150 and JAVELIN Renal 101 trials.12 15 In patients with sarcomatoid RCC, low Angio signature scores, poor response to sunitinib, and improved responses to ICIs have been observed in some studies.22 23
Immune-cell (IC) infiltration has been reported to be associated with prognosis in patients with RCC.24 The GEP-based microenvironment cell population (MCP)-counter method has been used to identify neutrophils and endothelial cells as having potential roles in immune suppression that may result in poorer clinical outcomes with nivolumab.25 Improved clinical efficacy with ICIs alone or in combination with VEGF inhibitors has been associated with high scores from a T-effector function (Teff) gene expression signature.12 15 22 High Teff and low Angio gene expression signature scores were associated with improved response rates with nivolumab,25 26 and a combined high score from both Teff and Myeloid signatures was associated with improved progression-free survival (PFS) with atezolizumab plus VEGF inhibitor, but not with avelumab plus VEGF inhibitor.12 15
In summary, there exists extensive literature evaluating PD-L1 expression, genomic mutations, gene expression, and IC infiltration for associations with clinical benefit from ICI and other therapies in multiple tumor types, including RCC. However, associations between these factors and response to nivolumab plus ipilimumab have not yet been fully investigated in patients with RCC. This comprehensive exploratory post-hoc analysis of patients with RCC from the CheckMate 214 phase 3 trial with a 42-month follow-up period represents a thorough investigation of PD-L1, as well as genomic and transcriptomic biomarkers for response to nivolumab plus ipilimumab.
Methods
Trial design and clinical assessments
The CheckMate 214 (NCT02231749) trial design and clinical assessments have been reported previously.1 See online supplemental methods for details.
jitc-2021-004316supp001.pdf (604.2KB, pdf)
Exploratory analysis of patient samples
Histological analysis and biomarker assessments were performed on archival formalin-fixed, paraffin-embedded (FFPE) primary and metastatic tumor samples and baseline blood samples from patients subsequently treated in CheckMate 214 (online supplemental table 1).
Histology and immunohistochemistry
The presence of sarcomatoid features was determined by reviewing local pathology reports accompanying preassessment tumor samples (n=843) and by independent central review (n=1009) of available hematoxylin and eosin–stained slides, as previously described.23
PD-L1 expression was assessed manually by a single pathologist at Covance (Princeton, New Jersey) using the Dako PD-L1 immunohistochemistry (IHC) 28–8 pharmDx assay (Agilent, Santa Clara, California). Tissue sections chosen for analysis were representative of the majority of the tumor. If present, sarcomatoid or rhabdoid areas were selected for analysis. Tissue processing and PD-L1 staining were performed following protocols optimized and approved for FFPE non-squamous non-small cell lung cancer samples.27 PD-L1 expression was quantified as the percentage of PD-L1–expressing TCs (% TC) as described previously for RCC,1 28 and patients were categorized as having tumors with PD-L1 expression <1% and ≥1% TCs. PD-L1 was also quantified on TCs and ICs using the combined positive score (CPS), defined as the number of PD-L1–expressing TCs, lymphocytes, and macrophages, divided by the total number of TCs, and multiplied by 100.29 Patients were categorized as having tumors with CPS <1 or ≥1.
Assessment of genomic status
Germline and somatic genomic statuses were assessed using whole exome sequencing (WES), see online supplemental methods.
TIB, defined as the total number of indels that are commonly called by two variant callers, Strelka30 and TNscope,31 and TMB, defined as the total number of somatic missense mutations,32 were assessed. HLA genotyping was performed on HLA loci A, B, and C, as previously described.14 Patients were classified as heterozygous if all three loci were heterozygous and homozygous if one or more loci were homozygous.
Mutation status
WES data were used to categorize patients as wild type (WT) or mutant (MUT) for all protein-coding genes, based on somatic variant calls of single-nucleotide variants and indels. See online supplemental methods for more details.
Assessment of transcriptomic signatures
Whole transcriptome analysis was performed using RNA-sequencing (RNA-seq) (see online supplemental methods). In the 213 RNA-seq–evaluable samples from treated patients, scores were derived for three IMmotion150 signatures Angio, Teff, and Myeloid,15 the JAVELIN Renal 101 signature (JAVELIN),12 and tumor inflammation signature (TIS).21 Signature scores were calculated as the median value of Z-scored expression for the constituent transcripts.
Estimation of cell type abundance in the TME
The abundance of eight tumor-infiltrating IC populations (CD3 +T cells, CD8 +T cells, cytotoxic lymphocytes, natural killer cells, B lymphocytes, monocytic lineage cells, myeloid dendritic cells, and neutrophils) and two stromal cell populations (endothelial cells and fibroblasts) was assessed within the TME using MCP-counter deconvolution. See online supplemental methods for details.
Gene set enrichment analysis
A Cox proportional-hazards model was used to estimate the association of each gene’s expression with PFS and OS as a continuous variable in RNA-seq–evaluable patients within each treatment arm.
Reverse signed Wald test Z scores from the Cox proportional-hazards models were ranked and then evaluated with the ‘GSEA’ algorithm (Bioconductor V.3.8)33 on the HALLMARK gene sets from the Molecular Signatures Database.33 34
Statistical analyses
All analyses were conducted using SAS (SAS Institute) and R V.3.6.1 unless otherwise stated. Patient characteristics and clinical outcomes were compared between the biomarker-evaluable population and the intent-to-treat population using frequency statistics and descriptive statistics. Survival analyses were conducted using the R survival (V.2.44–1.1) and survminer (https://github.com/kassambara/survminer) (V.0.4.6) packages. HRs and their 95% CIs for associations with PFS or OS in patient subgroups were assessed by Cox proportional-hazards models and were illustrated with forest plots and Kaplan-Meier plots. Descriptive p values were calculated from Wald tests unless otherwise stated and adjusted for multiple hypothesis testing using the Benjamini-Hochberg Procedure35; all comparisons were exploratory, and p values should be interpreted with caution. All comparisons for continuous variables use the two-sided Mann-Whitney test for two groups.
Results
In this exploratory analysis of 1082 treated patients from CheckMate 214, 992 patients were evaluable for PD-L1 expression by IHC, 481 patients were evaluable by WES, and 213 patients were evaluable by RNA-seq (online supplemental tables 1 and 2). Baseline characteristics were similar between each category of biomarker-evaluable patients and the entire treated population (online supplemental table 2). PFS and OS were similar across biomarker-evaluable and treated populations with nivolumab plus ipilimumab and sunitinib (online supplemental figures 1 and 2). Sarcomatoid RCC was identified in 143 patients (13%): 5 with favorable risk and 138 with intermediate/poor risk.23 Of note, the success rate of RNA analysis was low. Factors influencing the availability of RNA-seq data included the age of the tissue (the median sample age at which RNA-seq data passed quality control was 104 days, while 335 samples that failed had a median age of 219 days from sample procurement), the availability of tumor tissue for biomarker analyses allowed by patient informed consent, the amount of tissue available, and tissue quality.
OS was improved with nivolumab plus ipilimumab compared with sunitinib, regardless of PD-L1 expression. The association between PD-L1 expression on TCs and survival was previously reported.36 In this study of patients from CheckMate 214 with a minimum follow-up of 42 months, we expanded this evaluation to include association of combined PD-L1 expression on TCs and ICs by CPS (see Methods) with PFS and OS. In the nivolumab plus ipilimumab arm, 113 of 498 (22.7%) patients were PD-L1–positive by % TC (≥1% TC); in the sunitinib arm, 125 of 494 (25.3%) patients were PD-L1–positive by % TC. When assessed by CPS, 298 of 487 (61.2%) and 298 of 493 (60.4%) patients were PD-L1–positive (CPS ≥1) in the nivolumab plus ipilimumab arm and sunitinib arm, respectively.
PD-L1 assessed by CPS did not provide a greater predictive power than PD-L1 on TCs for PFS with nivolumab plus ipilimumab. For patients with PD-L1 expression on ≥1% TCs, the median PFS was longer in the nivolumab plus ipilimumab arm than in the sunitinib arm (HR, 0.43 (95% CI, 0.31 to 0.60); p<0.0001). A similar observation was made when comparing PFS with nivolumab plus ipilimumab versus sunitinib in patients evaluated by PD-L1 CPS (HR, 0.68 (95% CI, 0.56 to 0.82); p<0.0001). For patients with PD-L1 expression on <1% TCs or PD-L1 CPS <1, PFS was similar in the nivolumab plus ipilimumab and sunitinib arms (p>0.05). Median PFS with nivolumab plus ipilimumab was longer in patients with PD-L1 expression on ≥1% compared with <1% TCs (HR, 0.73 (95% CI, 0.55 to 0.95); p=0.02), while median PFS was similar in CPS ≥1 and in CPS <1 (HR, 1.01 (95% CI, 0.81 to 1.26)) (figure 1A, B). The significant associations observed between PD-L1 expression, assessed using either TC scoring or CPS, and PFS were maintained after adjustment for multiple hypothesis testing (online supplemental table 3).
Figure 1.
PFS and OS by PD-L1 expression assessed by % TCs or CPS. (A) PFS with nivolumab plus ipilimumab vs sunitinib in patients with PD-L1 expression on <1% and ≥1% TCs. (B) PFS with nivolumab plus ipilimumab vs sunitinib in patients with PD-L1 CPS <1 and ≥1. (C) OS with nivolumab plus ipilimumab vs sunitinib in patients with PD-L1 expression on <1% and ≥1% TCs. (D) OS with nivolumab plus ipilimumab vs sunitinib in patients with PD-L1 CPS <1 and ≥1. CPS, combined positive score; IPI, ipilimumab; NIVO, nivolumab; NR, not reached; OS, overall survival; PD-L1, programmed death ligand 1; PFS, progression-free survival; SUN, sunitinib; TC, tumor cell.
PD-L1 assessed by CPS did not provide a greater predictive power than PD-L1 on TCs for OS with nivolumab plus ipilimumab. For patients with PD-L1 expression on ≥1% TCs, OS was longer in the nivolumab plus ipilimumab arm (median not reached) compared with OS in the sunitinib arm (HR, 0.57 (95% CI, 0.40 to 0.82); p<0.01). A similar observation was made for OS when PD-L1 was evaluated by CPS (HR, 0.72 (95% CI, 0.57 to 0.89); p<0.01). For patients with PD-L1 expression on <1% TC, a trend towards longer median OS was observed in the nivolumab plus ipilimumab arm compared with the sunitinib arm (HR, 0.79 (95% CI, 0.64 to 0.97); p=0.02). The same observation was made for OS when PD-L1 CPS <1 (HR, 0.71 (95% CI, 0.52 to 0.96); p=0.03). Significant associations between PD-L1 expression and OS were maintained after adjustment for multiple hypothesis testing (online supplemental table 3). Median OS with nivolumab plus ipilimumab was similar between patients with PD-L1 expression on <1% and ≥1% TCs (HR, 0.99 (95% CI, 0.72 to 1.36)), while median OS appeared shorter in CPS ≥1 than in CPS <1 (HR, 1.40 (95% CI, 1.06 to 1.87)) (figure 1C, D).
The analyses described above were also conducted using % TC cut-offs of ≥5% and ≥10%, as well as CPS cut-offs of ≥5 and ≥10, with no substantive difference in findings (data not shown).
Genomic features did not associate with survival in patients treated with nivolumab plus ipilimumab
To assess the association between mutation burden and survival, WES-evaluable samples from 262 and 219 patients in the nivolumab plus ipilimumab and the sunitinib arms, respectively, were categorized around the median values for TMB (50 missense mutations) and TIB (six indels). For the nivolumab plus ipilimumab and the sunitinib arms, 137 and 108 patients were categorized as having high TMB (≥median) while 125 and 111 had low TMB (<median), respectively. No association was identified between high versus low TMB levels and PFS or OS in either treatment arm (PFS: nivolumab plus ipilimumab HR, 1.05 (95% CI, 0.78 to 1.40), p=0.76; sunitinib HR, 0.88 (95% CI, 0.64 to 1.19), p=0.40; OS: nivolumab plus ipilimumab HR, 1.36 (95% CI, 0.94 to 1.98); p=0.10 vs sunitinib HR, 1.01 (95% CI, 0.70 to 1.45); p=0.96) (figure 2A, B). For the nivolumab plus ipilimumab and the sunitinib arms, 146 and 116 patients were categorized as having high TIB (≥median) while 116 and 103 had low TIB (<median), respectively. TIB was not associated with either PFS or OS with nivolumab plus ipilimumab (PFS: HR, 1.14 (95% CI, 0.85 to 1.53); p=0.38; OS: HR, 1.35 (95% CI, 0.93 to 1.97); p=0.12). However, TIB was positively associated with both PFS and OS with sunitinib (PFS: HR, 0.63 (95% CI, 0.46 to 1.87); p<0.01; OS: HR, 0.70 (95% CI, 0.49 to 1.01); p=0.05) (figure 2A, B). The association between TIB and PFS with sunitinib was maintained after adjustment for multiple hypothesis testing (adjusted p=0.01) and in multivariate analyses accounting for age, sex, International Metastatic RCC Database Consortium criteria, and presence of sarcomatoid features (p=0.04); however, associations between TIB and OS were not maintained in multivariate analyses or after adjustment for multiple hypothesis testing (online supplemental table 4). Patients were also categorized on the basis of HLA zygosity. No differences were observed for PFS or OS with nivolumab plus ipilimumab in patients with homozygous (n=50) or heterozygous (n=212) HLA status (PFS: HR, 1.37 (95% CI, 0.94 to 2.00); p=0.10; OS: HR, 1.47 (95% CI, 0.88 to 2.47); p=0.14) (figure 2A, B).
Figure 2.
Association of genomic biomarkers with survival. (A) Forest plot for PFS comparing high (≥median) vs low TMB, high (≥median) vs low TIB, and heterozygous vs homozygous HLA in the nivolumab plus ipilimumab and the sunitinib arms. (B) Forest plot for OS comparing high (≥median) vs low TMB, high (≥median) vs low TIB, and heterozygous vs homozygous HLA in the nivolumab plus ipilimumab and the sunitinib arms. (C) Representation of mutations found across both treatment arms in seven selected genes. (D) Forest plot of PFS by mutation status. (E) Forest plot of OS by mutation status. Forest plots show HRs and 95% CIs for given comparisons, and p values compare patient subgroups within each treatment arm. Het, heterozygous; HLA, human leukocyte antigen; Homo, homozygous; IPI, ipilimumab; MUT, mutant; NIVO, nivolumab; OS, overall survival; PFS, progression-free survival; SUN, sunitinib; TIB, tumor indel burden; TMB, tumor mutational burden; WES, whole-exome sequencing; WT, wild type.
Associations between mutation status and survival in patients treated with nivolumab plus ipilimumab
The association of individual gene mutation status with OS was also assessed in WES-evaluable patients (n=481). In total, 382 genes that were mutated in ≥10 patients (prevalence >2%) were chosen for the association analysis. Online supplementary figure 3 shows the association between mutation status of those genes and OS in the nivolumab plus ipilimumab arm, based on Cox proportional-hazards models. Mutation of either PTEN or PCDH15 was associated with unfavorable OS with nivolumab plus ipilimumab (p<0.05). This association should be interpreted with caution, given that only 11 patients with PTEN mutations and 4 patients with PCDH15 mutations were identified in WES-evaluable patients who received nivolumab plus ipilimumab (figure 2C), and no associations with mutation status were maintained after adjustment for multiple hypothesis testing (online supplemental table 5). A subset of five genes were selected for further analysis based on the findings of previous publications and known prevalence in RCC (PBRM1, VHL, BAP1, SETD2, and MTOR).12 15–17 The frequency of mutations in these five selected genes was >5% across all treated patients (figure 2C). Although an association was suggested for prolonged PFS, but not OS, with nivolumab plus ipilimumab in patients with PBRM1WT RCC (p=0.04), the mutation status of none of these genes was significantly associated with PFS or OS in the nivolumab plus ipilimumab arm or the sunitinib arm after adjustment for multiple hypothesis testing (figure 2D, E; online supplemental table 5).
Gene expression signatures are related to tumor characteristics in pretreatment biopsies
In total, 109 patients in the nivolumab plus ipilimumab arm and 104 patients in the sunitinib arm were RNA-seq–evaluable. RNA-seq data were analyzed to establish scores for published gene expression signatures, and MCP-counter was used to predict the abundance of cell populations in each sample.
Clustering of all 213 RNA-seq samples based on their signature score revealed overlap in sample classification by three gene expression signatures (JAVELIN Renal 101 Immuno signature (JAVELIN), TIS, and IMmotion150 Teff) despite minimal overlap in gene content (online supplemental figure 4). In addition, lower Angio signature scores were associated with sarcomatoid RCC samples (p=0.02), while no significant relationship was observed between presence of sarcomatoid features and Myeloid (p=0.05) or Teff (p=0.56) signature scores (online supplemental figure 5).
Previously evaluated gene expression signatures were not associated with survival in patients treated with nivolumab plus ipilimumab
In our RNA-seq–evaluable population, we evaluated associations between published gene expression signatures and survival in each treatment arm. Patients were categorized as having high (≥median) or low (<median) gene expression signature scores using the median score across both treatment arms. Higher Angio scores associated with longer PFS and a numerically higher (although not significant) OS in patients receiving sunitinib (median PFS in Angiohigh, 11.14 months (95% CI, 8.38 to 20.96) vs median PFS in Angiolow, 7.0 months (95% CI, 5.06 to 14.52); p=0.02; median OS in Angiohigh, 39.72 months (95% CI, 29.04 to NR) vs median OS in Angiolow, 33.61 months (95% CI, 19.25 to 52.34); p=0.16) (figure 3A–D). However, the association between Angio score and PFS with sunitinib was not maintained after adjustment for multiple hypothesis testing (online supplemental table 6). Angio score did not predict PFS or OS in patients treated with nivolumab plus ipilimumab (median PFS in Angiohigh, 9.89 months (95% CI, 5.62 to 18.43) vs median PFS in Angiolow, 9.69 months (95% CI, 6.41 to 20.76); p=0.35; median OS in Angiohigh, NR (95% CI, 30.16 to NR) vs median OS in Angiolow, 49.15 months (95% CI, 25.33 to NR); p=0.71) (figure 3A–F). Early separation of OS curves for nivolumab plus ipilimumab vs sunitinib was observed (figure 3E, F), but median OS could be unstable as data to establish 95% CIs have not been reached.
Figure 3.
Association of gene expression signatures and molecular subtypes with survival. (A) Association of gene expression signatures with PFS comparing high (≥median) vs low gene expression signature scores in the nivolumab plus ipilimumab and the sunitinib arms. P values compare patient subgroups within each treatment arm. (B) Association of gene expression signatures with OS comparing high vs low gene expression signature scores in the nivolumab plus ipilimumab and the sunitinib arms. P values compare patient subgroups within each treatment arm. PFS according to Angio signature score with (C) nivolumab plus ipilimumab and (D) sunitinib. OS according to Angio signature score with (E) nivolumab plus ipilimumab and (F) sunitinib. Association of Teffhigh/Myeloidlow and Teffhigh/Myeloidhigh with PFS with (G) nivolumab plus ipilimumab and (H) sunitinib. Association of Teffhigh/Myeloidlow and Teffhigh/Myeloidhigh with OS with (I) nivolumab plus ipilimumab and (J) sunitinib. Angio, angiogenesis; IPI, ipilimumab; MUT, mutant; NIVO, nivolumab; NR, not reached; OS, overall survival; PFS, progression-free survival; SUN, sunitinib; Teff, T-effector; TIS, tumor inflammation signature; WT, wild type.
No association between JAVELIN, TIS, Teff, or Myeloid gene expression signature scores with PFS and OS was observed for patients treated with nivolumab plus ipilimumab or sunitinib in the RNA-seq–evaluable population (figure 3A, B). Similarly, no association with PFS or OS was seen in composite analysis of Myeloidhigh versus Myeloidlow patients in the Teffhigh group, or in Teffhigh versus Tefflow patients in the Angiolow group. HRs improved for the association of Myeloidhigh versus Myeloidlow in Teffhigh with PFS and OS compared with HRs observed in the analysis of Myeloid or Teff signatures alone, and a trend for prolonged PFS and OS was observed in patients with high versus low Myeloid scores in Teffhigh patients treated with nivolumab plus ipilimumab but not with sunitinib (figure 3G–J).
Cellular composition of the TME and clinical benefit with nivolumab plus ipilimumab
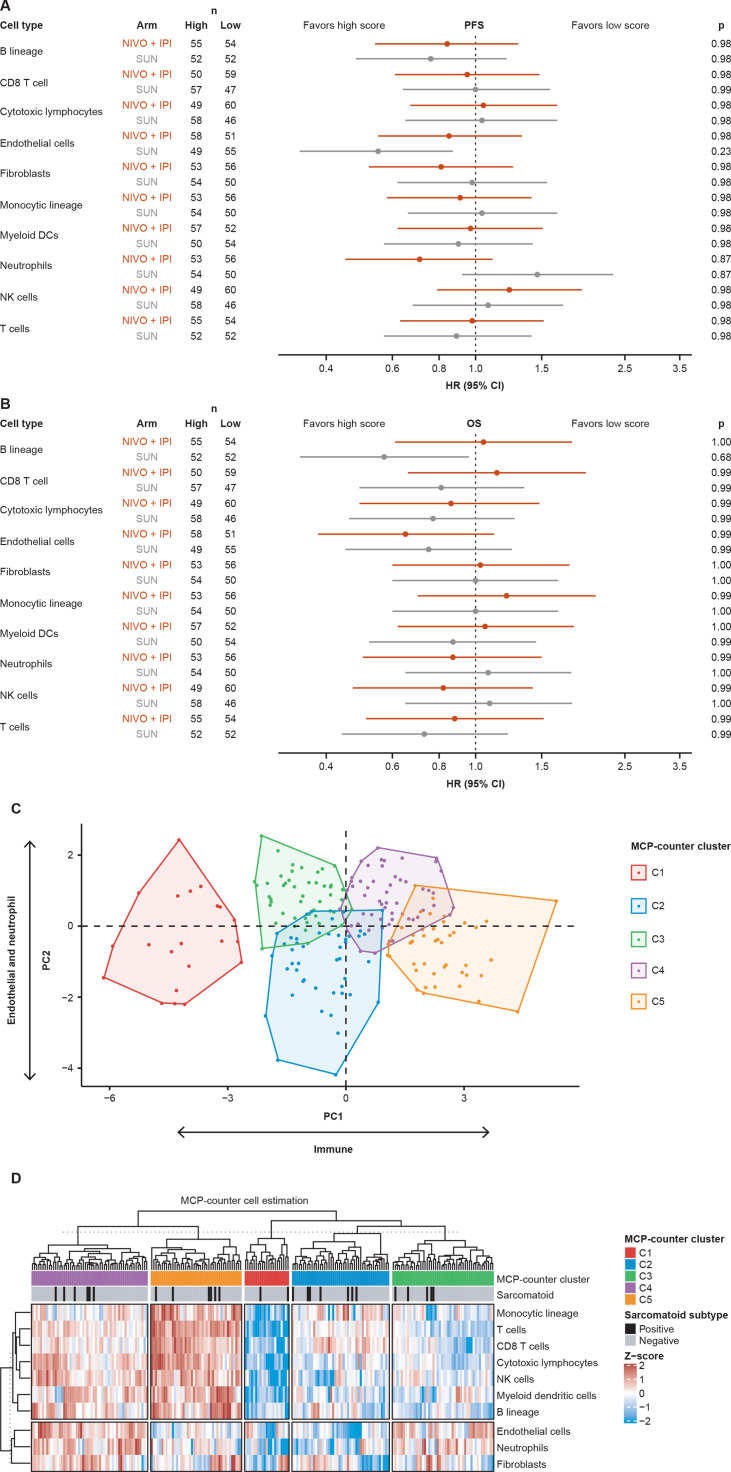
MCP-counter analysis37 was performed on RNA-seq–evaluable samples to quantify the abundance of 10 different cell types in the TME. No associations were observed between these scores and survival, although among patients treated with sunitinib, potential trends were noted for prolonged PFS in patients with high endothelial cell abundance scores and for prolonged OS in patients with high B lineage cell abundance scores (figure 4A, B).
Figure 4.
Analyses of cell populations in the TME estimated in RNA-seq data. (A) HRs for the association of high (≥median) vs low cell abundance scores with PFS. P values compare patient subgroups within each treatment arm and were corrected for multiple hypothesis testing using Benjamini-Hochberg procedure.35 (B) HRs for the association of cell abundance scores with OS. P values compare patient subgroups within each treatment arm. (C) RNA-seq–evaluable samples grouped into five clusters (C1–C5) by unsupervised clustering. (D) Heatmap showing abundance scores for the 10 cell types in each evaluable sample. Sample annotation tracks show the MCP-counter cluster assignment and sarcomatoid status. DC, dendritic cell; IPI, ipilimumab; MCP, microenvironment cell population; NIVO, nivolumab; NK, natural killer; OS, overall survival; PC, principal component; PFS, progression-free survival; SUN, sunitinib; TME, tumor microenvironment.
Unsupervised clustering based on the 10 cell abundance scores identified five distinct clusters of tumor samples (C1–C5), reflecting different compositions of ICs (eg, T cells, natural killer cells, and myeloid dendritic cells) and stromal/immune suppressing components (eg, neutrophils, endothelial cells, and fibroblasts) in the TME of each patient (cluster 1: immune low, cluster 2: stromal/immune suppression low, cluster 3: stromal/immune suppression high and immune low, cluster 4: stromal/immune suppression high and immune high, cluster 5: stromal/immune suppression low and immune high (figure 4C, D)). Only cluster 1 was completely separated from the rest of the clusters by principal component 1 (PC1) and PC2. Differences in PFS and OS with nivolumab plus ipilimumab versus sunitinib did not reach statistical significance when the 213 RNA-seq–evaluable patients were grouped into these five clusters of patients (online supplemental figure 6).
Biological processes associated with survival in patients treated with nivolumab plus ipilimumab
To further investigate biological processes that may drive prolonged survival with nivolumab plus ipilimumab, we used a Cox proportional-hazards model to evaluate the association between gene expression levels and PFS or OS in each treatment arm. Gene set enrichment analysis was then performed on these results to identify HALLMARK gene sets that were significantly enriched in the association analysis (false discovery rate <0.05). For several gene sets, higher expression of transcripts was associated with longer PFS in patients treated with nivolumab plus ipilimumab but with shorter PFS in patients treated with sunitinib (figure 5). These gene sets included Apoptosis, the Reactive Oxygen Species Pathway, IL-6/JAK/STAT3 Signaling, Allograft Rejection, and Inflammatory Response. Interestingly, expression of transcripts from the HALLMARK Adipogenesis and fatty acid metabolism gene sets were positively associated with OS in both nivolumab plus ipilimumab and sunitinib arms, suggesting a potential prognostic value of these gene sets in RCC. In contrast, expression of transcripts from gene sets for E2F targets and MYC targets were negatively associated with OS in both treatment arms.
Figure 5.
HALLMARK gene sets with significant enrichment (FDR <0.05) in the transcripts associated with PFS or OS by a Cox proportional-hazards analysis. Gene sets enriched in transcripts associated with longer PFS in the (A) nivolumab plus ipilimumab or (B) sunitinib arms. Bold typeset indicates gene sets with opposing association in the two arms (ie, transcripts associated with longer PFS in patients treated with nivolumab plus ipilimumab but shorter PFS in those treated with sunitinib). Gene sets enriched in transcripts associated with longer OS in the (C) nivolumab plus ipilimumab and (D) sunitinib arms. FDR, false discovery rate; IPI, ipilimumab; NES, normalized enrichment score; NIVO, nivolumab; OS, overall survival; PFS, progression-free survival; SUN, sunitinib.
Discussion
In CheckMate 214, nivolumab plus ipilimumab demonstrated superior clinical outcomes versus sunitinib in patients with advanced RCC.1 36 An unmet need exists for biomarkers with the potential to identify patients with advanced RCC who are most likely to respond to available treatments.8 The present study used pretreatment tumor samples in exploratory analysis of associations of histological, genomic, and transcriptomic biomarkers with survival outcomes.
There is currently no evidence for the value of PD-L1 alone as a biomarker of response to treatment in RCC. Compared with sunitinib, improved OS in patients with RCC was observed with nivolumab plus ipilimumab regardless of PD-L1 expression level or scoring method used.
Genomic analyses performed in this study showed that, in contrast to previous studies in other tumor types such as non-small cell lung cancer and melanoma,38–40 genomic features such as TMB and TIB did not associate with response to ICIs in CheckMate 214. These findings are consistent with biomarker analyses carried out as part of the JAVELIN Renal 101 trial12 and in CheckMate 010 and CheckMate 02516 in patients with advanced RCC and may be due to the relatively low TMB and TIB levels, as well as their narrow distribution, in RCC samples compared with tumor types where the predictive value of TMB has been reported.41 Other factors may play a more significant role in determining response to ICIs in RCC, or act synergistically with TMB and TIB.
Among the 382 genes alterations assessed in this study for an association between mutation status and response to nivolumab plus ipilimumab, none were found to be associated with survival after adjustment for multiple hypothesis testing. Mutation frequencies for genes that are commonly mutated in RCC were consistent with other reports.12 15–17 Similarly, our results are consistent with other studies that reported no association between VHL, PBRM1, BAP1, and MTOR mutation status and outcomes in patients with RCC treated with first-line PD-L1 inhibitor monotherapy or in combination with TKIs.12 15 Our findings that are suggestive of prolonged PFS with nivolumab plus ipilimumab in patients with PBRM1WT RCC are in contrast to other reports of improved clinical benefit with PD-1 or PD-L1 inhibitor monotherapy or in combination with anti-cytotoxic T lymphocyte antigen-4 inhibitors in patients with PBRM1MUT RCC.16 19 Differences in the reported associations between the mutation status of PBRM1 and treatment efficacy may be influenced by processes within the TME such as T-cell infiltration or immunosuppression.
RNA-seq data from CheckMate 214 were analyzed to evaluate the association between previously published gene expression signatures and clinical benefit with nivolumab plus ipilimumab versus sunitinib. Higher levels of the Angio signature were associated with improved response to sunitinib in agreement with a previous report,15 but are not predictive of PFS and OS with nivolumab plus ipilimumab. In our analysis, Angio scores were lower in sarcomatoid-positive than in sarcomatoid-negative tumors, suggesting that Angio is not the main driver of sarcomatoid RCC, or that Angio may be driven by other factors beyond the Angio signature. A recent report indicates that MYC-regulated transcriptional programs may drive the aggressive nature and poor prognosis associated with sarcomatoid RCC.42
Gene expression signatures of tumor inflammation previously reported to be associated with response to anti-PD-L1 therapy15 26 did not independently predict PFS or OS with nivolumab plus ipilimumab when assessed in pretreatment samples from CheckMate 214. The use of combined gene expression signatures to categorize patients with RCC has previously been shown to be predictive of response to anti-PD-L1 therapy alone and in combination with VEGF inhibitors.12 15 25 Our data also suggest that combining multiple gene expression signatures may improve the ability of gene expression signatures to predict response to treatment with nivolumab plus ipilimumab, and combination signatures such as Angiolow/Teffhigh and Teffhigh/Myeloidhigh may provide further information about biological factors that are important for response. Further studies are needed to optimize the most appropriate gene expression signatures for predicting response to this combination treatment in patients with RCC. Combinations of nivolumab plus ipilimumab have previously demonstrated synergistic antitumor activity.43 44 Evidence suggests that nivolumab plus ipilimumab combination therapy may enhance T-cell infiltration and bypass a need for pre-existing Teff cells in the TME prior to therapy compared with nivolumab monotherapy.44 This may explain the differential association between gene expression signatures representing T-cell inflammation and survival in patients treated with a single ICI (alone or in combination with VEGF inhibitors) and in patients treated with nivolumab plus ipilimumab in this study.12 15 This is further supported by previous transcriptomic analyses from CheckMate 009 that identified ipilimumab-mediated transcripts as baseline predictors of response to nivolumab monotherapy in RCC, suggesting mechanistically how ipilimumab may act in synergy with nivolumab and lead to improved efficacy over ICI monotherapy, regardless of baseline inflammation status.26
The potential immune suppression caused by neutrophils, endothelial cells, and fibroblasts, and their negative impact on clinical outcomes in patients treated with nivolumab monotherapy, were recently reported.25 In the present study, no significant associations between individual cell types and survival were identified with nivolumab plus ipilimumab. Patients with higher levels of ICs and/or lower levels of stromal/immune-suppressive cells could be hypothesized to show improved response to nivolumab plus ipilimumab. Although no significant association with PFS or OS was observed for any of the five patient subgroups derived by unsupervised clustering and characterized by levels of IC and stromal/immune suppression components, these results are suggestive, and further investigation in larger sample numbers may demonstrate the enrichment of clinical benefit with nivolumab plus ipilimumab in some patient subgroups. As previously reported, however, it is possible that combination therapy with nivolumab plus ipilimumab may bypass the effect of immune-suppressive cell types in the TME.45
In the 42-month follow-up of CheckMate 214, PFS curves for the two treatment arms notably diverged at 24 months, with over 30% of patients receiving nivolumab plus ipilimumab achieving long-term PFS.36 In the current study, analysis of gene expression in pretreatment samples suggested that higher baseline expression of particular biological processes may be prognostic for survival in patients with RCC. For example, expression of transcripts corresponding to MYC and E2F targets were negatively associated with OS in both treatment arms, supporting the association of E2F-driven and MYC-driven gene expression with poor prognosis in patients with RCC.46–48 While the association of the HALLMARK fatty acid metabolism gene set with response to sunitinib is in accordance with previously reported data,49 the current study shows that enrichment of the same gene set is also associated with prolonged survival with nivolumab plus ipilimumab, supporting the hypothesis that expression of genes involved in fatty acid metabolism is a prognostic biomarker for RCC.50 This is in contrast to the negative correlation between the HALLMARK fatty acid metabolism gene set and response to nivolumab monotherapy in CheckMate 009, where the majority of patients had received prior therapy with VEGF inhibitors.26 The observed difference may be due to the first-line setting of Checkmate 214 or to expansion of the pool of responsive patients resulting from combination therapy. Other biological processes may be specifically predictive for response and survival with nivolumab plus ipilimumab. For example, the association of the HALLMARK Inflammatory Response gene set with PFS in patients treated with nivolumab plus ipilimumab may help to dissect the role of baseline IC infiltration in durable responses to ICI therapy.51 Expression of transcripts related to IL-6/JAK/STAT3 signaling, which has been shown to be involved in the induction of PD-1 and/or PD-L1 expression,52 53 was also associated with long PFS in patients treated with nivolumab plus ipilimumab. Further investigation and clinical validation of the gene sets associated with longer PFS in patients treated with nivolumab plus ipilimumab may provide additional clues towards identification of prognostic or predictive biomarkers of response to this therapy.
The FFPE samples evaluated in the present study may have originated from primary or metastatic sites; therefore, caution should be used in interpreting the results due to the potential for tumor heterogeneity at the pathological and molecular level. Furthermore, this retrospective study may have been limited by the availability of quality samples for DNA and RNA analysis. While our transcriptomic analyses showed potential associations between gene expression signatures and outcomes to treatment, no distinct profile identifying patients more likely to respond to nivolumab plus ipilimumab was identified. Further studies are warranted to evaluate transcriptomic biomarkers of response to nivolumab plus ipilimumab in patients with RCC.
The results reported here offer insights into the biology of RCC, reveal relationships between molecular biomarkers, and provide supportive data for identifying predictive biomarkers that have either positive or negative associations with response and survival in patients with RCC treated with immunotherapy. Further validation of these exploratory analyses will be conducted in ongoing studies to identify biomarkers predictive of response to ICIs in patients with RCC, with the ultimate aim to provide biomarker-driven precision treatment for patients with RCC.
Acknowledgments
The authors would like to thank the patients and families who made this study possible and the clinical study teams who participated, and Dako, an Agilent Technologies, Inc company, for collaborative development of the PD-L1 IHC 28-8 pharmDx assay. Professional medical writing and editorial assistance were provided by Katerina Pipili, PhD, and Jay Rathi, MA, of Spark Medica, funded by Bristol Myers Squibb.
Footnotes
Twitter: @DrChoueiri
RJM and TKC contributed equally.
Contributors: RJM, TC, DM, TP, MBH, PR-M, and MW-R contributed to the design of the study; RJM, TC, TP, MBH, and PR-M contributed to the acquisition of the data set; RJM, DM, TP, JY, CH, RA, SP-C, MBH, PR-M, and MW-R contributed to the analysis of the data and RJM, TC, DM, TP, YV, SG, JY, CH, SP-C, SSS, MBH, PR-M, and MW-R contributed to their interpretation. All authors reviewed and revised the work and approved the final draft for submission. RJM accepts full responsibility for the finished work and the conduct of the study, had access to the data, and controlled the decision to publish.
Funding: The study was supported by Bristol Myers Squibb. Patients treated at Memorial Sloan Kettering Cancer Center were supported in part by Memorial Sloan Kettering Cancer Center Support Grant/Core Grant (P30 CA008748).
Competing interests: RJM reports consulting fees from Aveo, Calithera, Eisai, Eli Lilly, EMD Serono, Genentech, Merck, Novartis AG, Pfizer, and Roche, and contracted research to employer MSKCC for Bristol Myers Squibb, Eisai, Exelixis, Genentech, Merck, Pfizer, and Roche. TC reports personal and institutional research undertaken for Alexion, Analysis Group, AstraZeneca, Aveo, Bayer, Bristol Myers Squibb/ER Squibb & Sons LLC, Calithera, Cerulean, Corvus, Eisai, Exelixis, F. Hoffmann-La Roche, Foundation Medicine, Genentech, GlaxoSmithKline, Ipsen, Lilly, Merck, Novartis, Peloton, Pfizer, Prometheus Labs, Roche, Roche Products Limited, Sanofi/Aventis, Takeda, and Tracon; consulting/honoraria or advisory roles from Alexion, Analysis Group, AstraZeneca, Aveo, Bayer, Bristol Myers Squibb/ER Squibb & Sons LLC, Cerulean, Corvus, Eisai, EMD Serono, Exelixis, Foundation Medicine Inc, Genentech, GlaxoSmithKline, Heron Therapeutics, Infinity Pharma, Ipsen, Jansen Oncology, IQVIA, Lilly, Merck, NCCN, NiKang, Novartis, Peloton, Pfizer, Pionyr, Prometheus Labs, Roche, Sanofi/Aventis, Surface Oncology, Tempest, and Up-to-Date; travel, accommodations, expenses, and medical writing in relation to consulting, advisory roles, or honoraria; participation in CME-related events by OncLive, PVI, MJH Life Sciences, and in the NCI GU steering committee; owning Pionyr and Tempest stock; and patents filed, royalties, and other intellectual properties related to biomarkers of immune checkpoint inhibitors and ctDNA. TC is also supported in part by the Dana-Farber/Harvard Cancer Center Kidney SPORE and Program, the Kohlberg Chair at Harvard Medical School and the Trust Family, Michael Brigham, and Loker Pinard Funds for Kidney Cancer Research at DFCI. DM reports honoraria from Alkermes, Bristol Myers Squibb, Calithera Biosciences, Eisai, Eli Lilly, EMD Serono, Iovance, Merck, Pfizer, and Werewolf Therapeutics; and research support from Alkermes Inc, Bristol Myers Squibb, Exelixis, Genentech, Merck, Pfizer, and X4 Pharma. TP reports honoraria for advisory/research boards from Astellas, AstraZeneca, Bristol Myers Squibb, Eisai, Exelixis, Incyte, Ipsen, Johnson & Johnson, Merck, Merck Serono, MSD, Novartis, Pfizer, Roche, and Seattle Genetics; institutional grants and funding from Astellas, AstraZeneca, Bristol Myers Squibb, Eisai, Exelixis, Ipsen, Johnson & Johnson, Merck, Merck Serono, MSD, Novartis, Pfizer, Roche, and Seattle Genetics; and fees for travel and accommodation expenses from AstraZeneca, Ipsen, MSD, Roche, and Pfizer. YV reports no conflicts of interest. SG is employed by, and owns stock in, Bristol Myers Squibb. JY is employed by, and owns stock in, Bristol Myers Squibb. CH is employed by Bristol Myers Squibb. RA is employed by, and owns stock in, Bristol Myers Squibb. SP-C is employed by, and a shareholder of, Bristol Myers Squibb. SSS is employed by, and owns stock in, Bristol Myers Squibb. MBM is currently employed by BeiGene and owns stock in Bristol Myers Squibb. MW-R is currently employed by Agios Pharmaceuticals and owns stock in Agios Pharmaceuticals and Bristol Myers Squibb. MBM, MW-R, and PR-M were employees of Bristol Myers Squibb at the time this work was conducted.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. Data in support of the findings of this study, including anonymized raw FASTQ files of WES and RNA-seq data from 87, and 39 patients, respectively, who granted informed consent to share such data, are made available at the European Genome-Phenome Archive (EGA) under accession number EGAS00001005501 and EGAD00001007938. More information on Bristol Myers Squibb’s data sharing policy can be found here: https://www.bms.com/researchers-and-partners/clinical-trials-and-research/disclosure-commitment.html.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This trial was approved by the institutional review board or ethics committee at each site and was conducted according to Good Clinical Practice guidelines, defined by the International Conference on Harmonisation. All the patients provided written informed consent that was based on the Declaration of Helsinki principles. Participants gave informed consent to participate in the study before taking part.
References
- 1.Motzer RJ, Tannir NM, McDermott DF, et al. Nivolumab plus ipilimumab versus sunitinib in advanced renal-cell carcinoma. N Engl J Med 2018;378:1277–90. 10.1056/NEJMoa1712126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rini BI, Plimack ER, Stus V, et al. Pembrolizumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med 2019;380:1116–27. 10.1056/NEJMoa1816714 [DOI] [PubMed] [Google Scholar]
- 3.Mori K, Mostafaei H, Miura N, et al. Systemic therapy for metastatic renal cell carcinoma in the first-line setting: a systematic review and network meta-analysis. Cancer Immunol Immunother 2021;70:265–73. 10.1007/s00262-020-02684-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McFarlane JJ, Kochenderfer MD, Olsen MR, et al. Safety and efficacy of nivolumab in patients with advanced clear cell renal cell carcinoma: results from the phase IIIb/IV CheckMate 374 study. Clin Genitourin Cancer 2020;18:469–76. 10.1016/j.clgc.2020.06.002 [DOI] [PubMed] [Google Scholar]
- 5.Choueiri TK, Motzer RJ. Systemic therapy for metastatic renal-cell carcinoma. N Engl J Med 2017;376:354–66. 10.1056/NEJMra1601333 [DOI] [PubMed] [Google Scholar]
- 6.Choueiri TK, Powles T, Burotto M, et al. Nivolumab plus cabozantinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med 2021;384:829–41. 10.1056/NEJMoa2026982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albiges L, Tannir NM, Burotto M, et al. Nivolumab plus ipilimumab versus sunitinib for first-line treatment of advanced renal cell carcinoma: extended 4-year follow-up of the phase III CheckMate 214 trial. ESMO Open 2020;5:e001079. 10.1136/esmoopen-2020-001079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choueiri TK, Atkins MB, Bakouny Z, et al. Summary from the first Kidney Cancer Research Summit, September 12-13, 2019: a focus on translational research. J Natl Cancer Inst 2021;113:234–43. 10.1093/jnci/djaa064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther 2015;14:847–56. 10.1158/1535-7163.MCT-14-0983 [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez-Vida A, Hutson TE, Bellmunt J, et al. New treatment options for metastatic renal cell carcinoma. ESMO Open 2017;2:e000185. 10.1136/esmoopen-2017-000185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motzer RJ, Escudier B, McDermott DF, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med 2015;373:1803–13. 10.1056/NEJMoa1510665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Motzer RJ, Robbins PB, Powles T, et al. Avelumab plus axitinib versus sunitinib in advanced renal cell carcinoma: biomarker analysis of the phase 3 JAVELIN renal 101 trial. Nat Med 2020;26:1733–41. 10.1038/s41591-020-1044-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turajlic S, Litchfield K, Xu H, et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol 2017;18:1009–21. 10.1016/S1470-2045(17)30516-8 [DOI] [PubMed] [Google Scholar]
- 14.Chowell D, Morris LGT, Grigg CM, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 2018;359:582–7. 10.1126/science.aao4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDermott DF, Huseni MA, Atkins MB, et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat Med 2018;24:749–57. 10.1038/s41591-018-0053-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun DA, Hou Y, Bakouny Z, et al. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nat Med 2020;26:909–18. 10.1038/s41591-020-0839-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerlinger M, Horswell S, Larkin J, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 2014;46:225–33. 10.1038/ng.2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Velasco G, Wankowicz SA, Madison R, et al. Targeted genomic landscape of metastases compared to primary tumours in clear cell metastatic renal cell carcinoma. Br J Cancer 2018;118:1238–42. 10.1038/s41416-018-0064-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miao D, Margolis CA, Gao W, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 2018;359:801–6. 10.1126/science.aan5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danaher P, Warren S, Dennis L, et al. Gene expression markers of tumor infiltrating leukocytes. J Immunother Cancer 2017;5:18. 10.1186/s40425-017-0215-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danaher P, Warren S, Lu R, et al. Pan-cancer adaptive immune resistance as defined by the tumor inflammation signature (TIS): results from the Cancer Genome Atlas (TCGA). J Immunother Cancer 2018;6:63. 10.1186/s40425-018-0367-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rini BI, Huseni M, Atkins MB. Molecular correlates differentiate response to atezolizumab (atezo) + bevacizumab (bev) vs sunitinib (sun): results from a phase III study (IMmotion151) in untreated metastatic renal cell carcinoma (mRCC). Ann Oncol 2018;29 (suppl 8):Abstract LBA31. [Google Scholar]
- 23.Tannir NM, Signoretti S, Choueiri TK, et al. Efficacy and safety of nivolumab plus ipilimumab versus sunitinib in first-line treatment of patients with advanced sarcomatoid renal cell carcinoma. Clin Cancer Res 2021;27:78–86. 10.1158/1078-0432.CCR-20-2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giraldo NA, Becht E, Pagès F, et al. Orchestration and prognostic significance of immune checkpoints in the microenvironment of primary and metastatic renal cell cancer. Clin Cancer Res 2015;21:3031–40. 10.1158/1078-0432.CCR-14-2926 [DOI] [PubMed] [Google Scholar]
- 25.Meylan M, Beuselinck B, Dalban C, et al. 700O kidney ccRCC immune classification (KIC) enhances the predictive value of T effector (Teff) and angiogenesis (Angio) signatures in response to nivolumab (N). Ann Oncol 2020;31:S553. 10.1016/j.annonc.2020.08.772 [DOI] [Google Scholar]
- 26.Ross-Macdonald P, Walsh AM, Chasalow SD, et al. Molecular correlates of response to nivolumab at baseline and on treatment in patients with RCC. J Immunother Cancer 2021;9:e001506. 10.1136/jitc-2020-001506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agilent Technologies . PD-L1 IHC 28-8 pharmDx interpretation manual – non-squamous non-small cell lung cancer (US version), 2015. Available: https://www.agilent.com/cs/library/usermanuals/public/29111_pd-l1-ihc-28-8-interpretation-manual.pdf
- 28.Choueiri TK, Fishman MN, Escudier B, et al. Immunomodulatory activity of nivolumab in metastatic renal cell carcinoma. Clin Cancer Res 2016;22:5461–71. 10.1158/1078-0432.CCR-15-2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei M, Siemers NO, Pandya D, et al. Analyses of PD-L1 and inflammatory gene expression association with efficacy of nivolumab ± ipilimumab in gastric cancer/gastroesophageal junction cancer. Clin Cancer Res 2021;27:3926–35. 10.1158/1078-0432.CCR-20-2790 [DOI] [PubMed] [Google Scholar]
- 30.Kim S, Scheffler K, Halpern AL, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods 2018;15:591–4. 10.1038/s41592-018-0051-x [DOI] [PubMed] [Google Scholar]
- 31.Freed D, Pan R, Aldana R. TNscope: accurate detection of somatic mutations with haplotype-based variant candidate detection and machine learning filtering. bioRxiv 2018. [Google Scholar]
- 32.Chang H, Sasson A, Srinivasan S, et al. Bioinformatic methods and bridging of assay results for reliable tumor mutational burden assessment in non-small-cell lung cancer. Mol Diagn Ther 2019;23:507–20. 10.1007/s40291-019-00408-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liberzon A, Birger C, Thorvaldsdóttir H, et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–25. 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 1995;57:289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 36.Motzer RJ, Escudier B, McDermott DF, et al. Survival outcomes and independent response assessment with nivolumab plus ipilimumab versus sunitinib in patients with advanced renal cell carcinoma: 42-month follow-up of a randomized phase 3 clinical trial. J Immunother Cancer 2020;8:e000891. 10.1136/jitc-2020-000891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Becht E, Giraldo NA, Lacroix L, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol 2016;17:218. 10.1186/s13059-016-1070-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ready N, Hellmann MD, Awad MM, et al. First-line nivolumab plus ipilimumab in advanced non-small-cell lung cancer (CheckMate 568): outcomes by programmed death ligand 1 and tumor mutational burden as biomarkers. J Clin Oncol 2019;37:992–1000. 10.1200/JCO.18.01042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362:eaar3593. 10.1126/science.aar3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chae YK, Viveiros P, Lopes G, et al. Clinical and immunological implications of frameshift mutations in lung cancer. J Thorac Oncol 2019;14:1807–17. 10.1016/j.jtho.2019.06.016 [DOI] [PubMed] [Google Scholar]
- 41.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bakouny Z, Braun DA, Shukla SA, et al. Integrative molecular characterization of sarcomatoid and rhabdoid renal cell carcinoma. Nat Commun 2021;12:808. 10.1038/s41467-021-21068-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med 2015;373:23–34. 10.1056/NEJMoa1504030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selby MJ, Engelhardt JJ, Johnston RJ, et al. Preclinical development of ipilimumab and nivolumab combination immunotherapy: mouse tumor models, in vitro functional studies, and cynomolgus macaque toxicology. PLoS One 2016;11:e0161779. 10.1371/journal.pone.0161779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selby M, Engelhardt J, Lu L-S, et al. Antitumor activity of concurrent blockade of immune checkpoint molecules CTLA-4 and PD-1 in preclinical models. J Clin Oncol 2013;31:Abstract 3061:3061. 10.1200/jco.2013.31.15_suppl.3061 [DOI] [Google Scholar]
- 46.Zhang C, Cui Y, Wang G, et al. Comprehensive analysis of the expression and prognosis for E2Fs in human clear cell renal cell carcinoma. J Healthc Eng 2021;2021:5790416. 10.1155/2021/5790416 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Bellut J, Bertz S, Nolte E, et al. Differential prognostic value of MYC immunohistochemistry in subtypes of papillary renal cell carcinoma. Sci Rep 2017;7:16424. 10.1038/s41598-017-16144-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bailey ST, Smith AM, Kardos J, et al. MYC activation cooperates with Vhl and Ink4a/Arf loss to induce clear cell renal cell carcinoma. Nat Commun 2017;8:15770. 10.1038/ncomms15770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Motzer RJ, Banchereau R, Hamidi H, et al. Molecular subsets in renal cancer determine outcome to checkpoint and angiogenesis blockade. Cancer Cell 2020;38:803–17. 10.1016/j.ccell.2020.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao Z, Liu Y, Liu Q, et al. The mRNA expression signature and prognostic analysis of multiple fatty acid metabolic enzymes in clear cell renal cell carcinoma. J Cancer 2019;10:6599–607. 10.7150/jca.33024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Darvin P, Toor SM, Sasidharan Nair V, et al. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp Mol Med 2018;50:165.. 10.1038/s12276-018-0191-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang N, Zeng Y, Du W, et al. The EGFR pathway is involved in the regulation of PD-L1 expression via the IL-6/JAK/STAT3 signaling pathway in EGFR-mutated non-small cell lung cancer. Int J Oncol 2016;49:1360–8. 10.3892/ijo.2016.3632 [DOI] [PubMed] [Google Scholar]
- 53.Johnson DE, O'Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol 2018;15:234–48. 10.1038/nrclinonc.2018.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-004316supp001.pdf (604.2KB, pdf)
Data Availability Statement
Data are available in a public, open access repository. Data in support of the findings of this study, including anonymized raw FASTQ files of WES and RNA-seq data from 87, and 39 patients, respectively, who granted informed consent to share such data, are made available at the European Genome-Phenome Archive (EGA) under accession number EGAS00001005501 and EGAD00001007938. More information on Bristol Myers Squibb’s data sharing policy can be found here: https://www.bms.com/researchers-and-partners/clinical-trials-and-research/disclosure-commitment.html.