Abstract
Infection (either community acquired or nosocomial) is a major cause of morbidity and mortality in critical care medicine. Sepsis is present in up to 30% of all ICU patients. A large fraction of sepsis cases is driven by severe community acquired pneumonia (sCAP), which incidence has dramatically increased during COVID-19 pandemics. A frequent complication of ICU patients is ventilator associated pneumonia (VAP), which affects 10–25% of all ventilated patients, and bloodstream infections (BSIs), affecting about 10% of patients. Management of these severe infections poses several challenges, including early diagnosis, severity stratification, prognosis assessment or treatment guidance. Digital PCR (dPCR) is a next-generation PCR method that offers a number of technical advantages to face these challenges: it is less affected than real time PCR by the presence of PCR inhibitors leading to higher sensitivity. In addition, dPCR offers high reproducibility, and provides absolute quantification without the need for a standard curve. In this article we reviewed the existing evidence on the applications of dPCR to the management of infection in critical care medicine. We included thirty-two articles involving critically ill patients. Twenty-three articles focused on the amplification of microbial genes: (1) four articles approached bacterial identification in blood or plasma; (2) one article used dPCR for fungal identification in blood; (3) another article focused on bacterial and fungal identification in other clinical samples; (4) three articles used dPCR for viral identification; (5) twelve articles quantified microbial burden by dPCR to assess severity, prognosis and treatment guidance; (6) two articles used dPCR to determine microbial ecology in ICU patients. The remaining nine articles used dPCR to profile host responses to infection, two of them for severity stratification in sepsis, four focused to improve diagnosis of this disease, one for detecting sCAP, one for detecting VAP, and finally one aimed to predict progression of COVID-19. This review evidences the potential of dPCR as a useful tool that could contribute to improve the detection and clinical management of infection in critical care medicine.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13054-022-03948-8.
Keywords: Critically ill patients, Digital PCR, Infection diagnosis, Prognosis and treatment guidance, Host response
Introduction
Infection in critical care medicine
Infectious pathology represents a leading cause of admission to the intensive care units (ICU). Sepsis (defined by the presence of a dysregulated host response to infection inducing organ dysfunction) is present in up to 30% of all ICU patients, as recently reported by Sakr et al. in a large study with 10,000 patients from 730 ICUs [1]. One of the leading causes of sepsis is severe community acquired pneumonia (sCAP) of bacterial or viral origin [2]. Current Coronavirus disease 2019 (COVID-19) pandemics has largely boosted the cases of sCAP all over the world.
In turn, infection is one of the most frequent complications in patients who are critically ill. Compromise of body’s physical barriers by invasive devices, surgical aggression or traumatic injury, disruption of the mucosa, pressure sores, ventilator-induced lung injury, immune suppression, poor nutritional state, the use of broad-spectrum antibiotics which alter the commensal microbiota, combined with the increased exposition to opportunistic (often multi-drug resistant, MDR) pathogens [3], all represent predisposing factors favouring ICU acquired infections [4]. In fact, approximately 19.2% of ICU patients develop infections compared to approximately 5.2% of infections developed by patients staying in all other hospital wards [3, 5]. Ventilator-associated pneumonia (VAP) affects 10–25% of all ventilated patients after at least 48 h on mechanical ventilation [6]. Other frequent nosocomial infections affecting critically ill patients are catheter-associated urinary tract infection, bloodstream infection (BSIs), skin and wound infections, sinusitis, and gastrointestinal infection (often with Clostridium difficile) [4]. Clinical management of these infectious diseases or complications of the critically ill patient faces several challenges, including early diagnosis with microorganism identification, severity stratification, prognosis assessment and treatment guidance. Digital polymerase chain reaction (dPCR) is a next-generation PCR method that represents an opportunity to address these challenges.
dPCR: technical principles and applications
dPCR has emerged as a promising technology that might fill in the current gaps of other standard or emerging diagnostic technologies employed in microbiology (Table S1 – additional file 1). dPCR is based on the division of the PCR mastermix (all components including DNA or RNA targets) into thousands of partitions. PCR amplification of target genes occurs in each individual partition, acting as an individual microreactor [7]. These partitions can be created using a number of different mechanisms, such as emulsified microdroplets suspended in oil (droplet digital PCR, ddPCR), manufactured microwells, or microfluidic valving [8]. The distribution of target sequences in the partitions is detected by fluorescence at endpoint. Quantification of target genes is estimated based on Poisson’s distribution, by calculating the ratio of positive partitions (presence of fluorescence) over the total number of partitions [9]. This technology has several advantages: i) it is less affected by PCR inhibitors than other standard or real-time PCR (qPCR) methods, as target sequences are concentrated in the microreactors; ii) it also offers a high reproducibility of the results; iii) it provides an absolute quantification of the target sequence without the need for standard curves; and iv) it has an improved analytical sensitivity ideal for detecting microbial genes, for species identification or for genes conferring antimicrobial resistance or higher pathogenicity. dPCR also presents some limitations: i) it is unable to distinguish between viable and non-viable microorganisms (an inconvenient which affects all PCR-based methods); ii) it might have different sensitivity for different types of microorganisms; iii) it needs specialized training; and iv) it has a high cost, particularly to acquire the devices. This represents a major drawback for applications in low or middle income countries, for example during the COVID-19 pandemics [7, 9]. In spite its limitations, the previously mentioned dPCR properties make it an ideal tool for clinical applications in the field of microbiology and infectious diseases [7]. In this article we reviewed the existing evidence on the use of dPCR to improve the clinical management of infection in critical care medicine.
Methods
We searched PudMed combining the following MESH terms: “digital PCR”, “digital droplet PCR”, “droplet digital PCR” and “droplet PCR” with “ICU”, “critical AND infection”, “critically ill AND infection”, “severe infection”, “critical care”, “pneumoniae”, “ventilator-associated pneumoniae”, “ventilator”, “ventilation”, “sepsis”, “septic shock”, “bloodstream infections”, “skin and soft tissue infections”, “necrotizing fasciitis”, “peritonitis”, “invasive pulmonary aspergillosis”. We found a total of 487 PubMed articles. Only articles in English were considered. Using PMID we excluded duplicated articles appearing in more than one search, obtaining a total of 198 articles. We screened these articles for relevance and excluded 166 for the reasons mentioned in Fig. 1. Finally, thirty-two articles were included in this review (Table 1). These articles were further divided in two groups, one focused on the pathogens—diagnosis of infection, prognosis and treatment guidance (n = 23)—and the other focused on the host response to infection (n = 9).
Fig. 1.
PRISMA diagram
Table 1.
Articles included in the review depicting the applications of dPCR for infection diagnosis and management in critical care medicine
| Study | Year | Usage | Entity | Objective | References |
|---|---|---|---|---|---|
| Hu et al. | 2021 | Bacterial identification in blood or plasma | Sepsis | Identification of DNA from bacterial pathogens and antimicrobial resistance genes in blood from patients BSI | [10] |
| Yamamoto et al. | 2018 | Bacterial identification in blood or plasma | Sepsis | Identification of Mycobacterium tuberculosis through the detection of circulating cell-free DNA | [14] |
| Shin et al. | 2021 | Bacterial identification in blood or plasma | BSI | Diagnosis of Gram-Negative pathogens and antimicrobial resistance genes in plasma from patients with BSIs | [15] |
| Zheng et al. | 2021 | Bacterial identification in blood or plasma | Sepsis | Identification of DNA from Acinetobacter baumannii and Klebsiella pneumoniae in blood from patients BSI | [16] |
| Chen et al. | 2021 | Fungal identification in blood | BSI | Diagnosis of Candida spp in blood from patients with BSI | [18] |
| Zhou et al. | 2021 | Bacterial and fungal identification in other clinical samples | Pleural or peritoneal infections | Identification of pathogens from pleural or peritoneal infections | [19] |
| Simms et al. | 2021 | Viral identification | COVID-19 | Confirmation of detection of SARS-CoV-2 in renal allograft and lung tissue (initially detected by immunohistochemistry) | [20] |
| Alteri et al. | 2020 | Viral identification | COVID-19 | Quantification of SARS-CoV-2 viral load in plasma of patients with negative qPCR results | [21] |
| Jiang et al. | 2020 | Viral identification | COVID-19 | Quantification of SARS-CoV-2 in plasma and hospital environment | [22] |
| Ziegler et al. | 2019 | Quantification of microbial burden to assess severity, prognosis and treatment guidance | Sepsis | Quantification of DNA load overtime in patients with Staphylococcus aureus BSIs | [29] |
| Ziegler et al. | 2019 | Quantification of microbial burden to assess severity, prognosis and treatment guidance | Sepsis | Quantification of DNA from bacterial pathogens load (16S rDNA) overtime in patients BSI to access patients’ progression | [30] |
| Bialasiewicz et al. | 2019 | Quantification of microbial burden to assess severity, prognosis and treatment guidance | Sepsis | Quantification of DNA in blood from Capnocytophaga canimorsus to access patients progression | [31] |
| Dickson et al. | 2020 | Quantification of microbial burden to assess severity, prognosis and treatment guidance | VAP | Quantification of bacteria DNA burden in the lung and association with disease progression and outcomes | [32] |
| Goh et al. | 2020 | Quantification of microbial burden to assess severity, prognosis and treatment guidance |
Sepsis sCAP |
Quantification of EBV (to detect reactivation) in patients with sepsis to monitor disease progression | [33] |
| Veyer et al. | 2021 | Quantification of microbial burden to assess severity, prognosis and treatment guidance | COVID-19 | Quantification of SARS-CoV-2 viral load in plasma and correlation with disease severity | [34] |
| Chen et al. | 2021 | Quantification of microbial burden to assess severity, prognosis and treatment guidance | COVID-19 | Quantification of SARS-CoV-2 viral load in plasma and correlation with disease severity | [35] |
| Bermejo-Martin et al. | 2020 | Quantification of microbial burden to assess severity, prognosis and treatment guidance | COVID-19 | Quantification of SARS-CoV-2 viral load in plasma and correlation with disease severity | [36] |
| Ram-Mohan et al. | 2021 | Quantification of microbial burden to assess severity, prognosis and treatment guidance | COVID-19 | Quantification of SARS-CoV-2 viral load in plasma and correlation with disease severity | [37] |
| Tedim et al. | 2021 | Quantification of microbial burden to assess severity, prognosis and treatment guidance | COVID-19 | Quantification of SARS-CoV-2 viral load in plasma, comparison with qPCR | [38] |
| Martin-Vicente et al. | 2022 | Quantification of microbial burden to assess severity, prognosis and treatment guidance | COVID-19 | Quantification of SARS-CoV-2 viral load in plasma | [39] |
| Bruneau et al. | 2021 |
Quantification of microbial burden to assess severity, prognosis and treatment guidance Host Response |
COVID-19 | Quantification of SARS-CoV-2 viral load and host biomarkers in plasma to predict disease severity | [40] |
| Chanderraj et al. | 2022 | Microbial ecology studies | Sepsis | Quantification of bacterial density in rectal swabs and risk of extraintestinal infection | [41] |
| Brooks et al. | 2018 | Microbial ecology studies | Microbiological burden and microbiome | Quantification of total microbiological burden in hospital neonates ICU and correction with microbiome establishment | [42] |
| Tamayo et al. | 2014 | Host Response | Sepsis | Quantification of the expression of the constant region of the mu heavy chain of IgM in blood to differentiate sepsis from SIRS | [46] |
| Almansa et al. | 2019 | Host Response | Sepsis | Gene expression ratio between MMP8 or LCN2 with HLA-DRA to differentiate surgical patients with sepsis from those with no sepsis | [47] |
| Almansa et al. | 2018 | Host Response | Sepsis | Ratio between HLA-DRA expression and procalcitonin to differentiate surgical patients with sepsis from those with no sepsis | [48] |
| Link et al. | 2020 | Host Response | Sepsis | Quantification of miRNA in blood for the early diagnosis of sepsis | [49] |
| Martin-Fernandez et al. | 2020 | Host Response | Sepsis | Quantification of emergency granulopoiesis gene expression to stratify severity in patients with infection, sepsis and septic shock | [50] |
| Menéndez et al. | 2019 | Host Response | sCAP | Gene expression levels of the immunological synapse genes to identify patients with sCAP | [52] |
| Almansa et al. | 2018 | Host Response | VAP | Gene expression levels of the immunological synapse genes to identify VAP | [53] |
| Busani et al. | 2020 | Host Response | Sepsis | Quantification of mtDNA in patients with Septic shock cause by MDR pathogens predicts disease severity | [54] |
| Sabbatinelli et al. | 2021 | Host Response | COVID-19 | Quantification of miRNA associated with inflammation and aging (Inflammaging) to predict COVID-19 disease progression | [56] |
DNA, deoxyribonucleic acid; BSIs, bloodstream infections; spp, Species; COVID-19, Coronavirus disease 2019; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; qPCR, real-time polymerase chain reaction; VAP, ventilator-associated pneumoniae; sCAP, severe community acquired pneumoniae; EBV, Epstein Barr Virus; rDNA, ribosomal deoxyribonucleic acid; ICU, intensive care unit; IgM, immunoglobulin M; SIRS, Systemic inflammatory response syndrome; MMP8, matrix metalloproteinase-8; LCN2, Lipocalin 2; HLA-DRA, major histocompatibility complex class II; miRNA, micro ribonucleic acid; mtDNA, mitochondrial deoxyribonucleic acid; MDR: multidrug-resistant
Evidence on the use of dPCR for the diagnosis and management of infection in critical care medicine (Fig. 2)
Fig. 2.
Summary of the existing evidence on the applications of dPCR in the field of infection in Critical Care Medicine
Applications of dPCR targeting microbial genes
The gold standard for the detection of bacterial and fungal pathogens still relies on culture based methods that present a long turnaround time, and often yield low positivity rates [10]. For viral pathogens the reference method is frequently qPCR [11–13]. As previously stated, dPCR presents several advantages making it an ideal technique for the detection and quantification of microbial genes (Additional file 1: Table S1).
Bacterial identification in blood or plasma
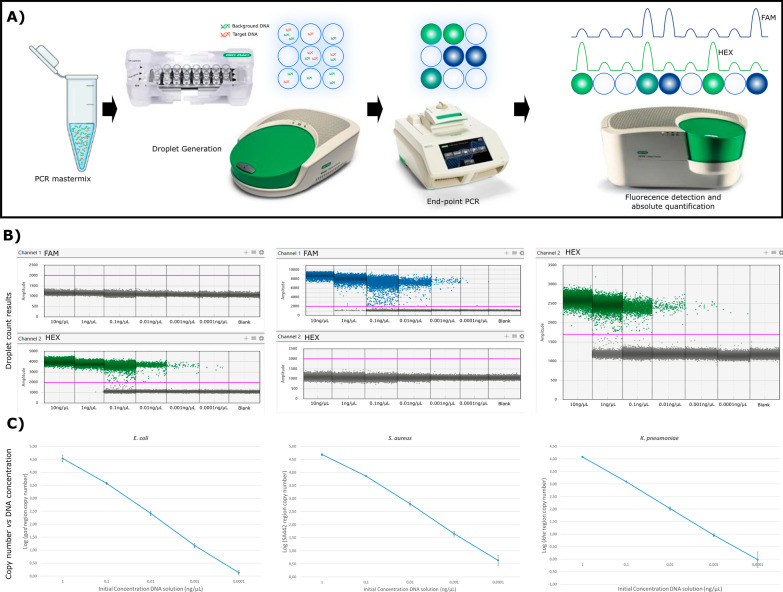
dPCR has been used in critical care medicine for the detection of different bacterial pathogens in septic patients or patients with a suspected BSI [10, 14–16]. Yamamoto et al. successfully diagnosed a septic patient with a Mycobacterium tuberculosis (MTB) disseminated infection by detecting MTB complex-specific sequences in total cell-free DNA (cfDNA) in plasma. dPCR was employed after sputum, urine, and blood samples all tested negative by COBAS TaqMan MTB and MAI tests (Roche Diagnostics) and TSPOT.TB test (Oxford Immunotec) and mycobacterial culture [14]. These results indicate that dPCR is more sensitive than the other molecular and culture methods, and that dPCR could serve as a less invasive diagnostic tool for MTB infections [14]. In two other studies [15, 16] dPCR was used to detect major BSI Gram-negative pathogens in cfDNA isolated from plasma of critically ill patients. Shin et al. [15] developed a dPCR assay able to detect four major Gram-Negative pathogens and four common antimicrobial resistance (AMR) genes. Zheng et al. [16] work focused on only two of the most common MDR Gram-Negative pathogens. These assays report a time from sample collection to result of three to four hours and a detection limit of one Colony-forming Unit/ml of bacteria in the blood [15, 16]. The study by Shin et al. indicated that dPCR was also more sensible than qPCR [15]. Hu et al. [10] compared the detection of pathogens and AMR genes by dPCR with metagenomic Next-Generation Sequencing (mNGS) and with blood culture using samples from a cohort of septic patients with suspicion of BSIs. dPCR showed a great potential to identify the pathogens most commonly associated with BSIs as well as AMR genes, as it was faster and more sensible than mNGS and blood culture [10]. In these previous studies [10, 15, 16], clinical validation revealed that dPCR method was superior to blood culture in terms of specificity, sensitivity, and turnaround time, representing a promising method for the early and accurate diagnosis of BSIs [10, 15, 16]. Our group has established a ddPCR assay capable of detecting and quantifying the housekeeping genes of important nosocomial bacterial species in ICUs, such as Klebsiella pneumoniae, Escherichia coli and Staphylococcus aureus [17]. These assays are compatible with duplexing, show low replication variability and very low limit of detection (less than 1 pg of DNA) (Fig. 3). Ongoing work is focused on applying the developed ddPCR assays directly to blood and designing a panel that allows testing as many samples at the same time as possible.
Fig. 3.
dPCR detection assay for E. coli, K. pneumoniae and S. aureus. A ddPCR workflow; B ddPCR results displayed as droplets of different fluorescence amplitude; C copy number of E. coli, S. aureus and K. pneumoniae in different DNA of different initial DNA concentration
The main limitation of dPCR compared with blood culture or mNGS, but not with qPCR, is that it can only detect the pathogens included in the dPCR panels. Nevertheless, the results obtained support that early identification of MDR pathogens by dPCR can improve treatment outcomes [10, 15, 16].
Fungal identification in blood
dPCR has been also tested for the detection of candidemia [18], showing again higher specificity and sensitivity than blood culture and qPCR (both methods yielding negative results in these patients). Detection of Candida spp in blood by dPCR allowed the implementation of the appropriated antifungal treatment, which translated into patients’ improvement, supporting the use of dPCR for the early diagnosis of candidemia [18].
Bacterial and fungal identification in other clinical samples
dPCR has also been applied to the detection of pathogens in clinical samples other than blood. For instance, Zhou et al. [19] tested dPCR to detect bacteria and fungi in pleural and peritoneal fluids of critically ill patients, comparing the obtained results with culture methods. Compared to the gold standard cultures of pleural and peritoneal fluids, dPCR showed a sensitivity of 96% and 93%, specificity of 87% and 60%, a positive predictive value of 92% and 87% and a negative predictive value of 93% and 75%, respectively. These results support dPCR as a rapid and sensitive alternative for the detection of pleural and peritoneal fungal infections [19].
Viral identification
dPCR has been also tested for viral diagnosis. Simms et al. [20] used commercial Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) dPCR kits to detect this virus in a patient diagnosed with sepsis ten days after a renal transplant. SARS-CoV-2 was detected on the renal allograft and native lung tissue post-mortem by dPCR but not by qPCR [20] indicating a higher sensitivity of the former method. Two other studies confirmed the higher sensitivity of dPCR compared with qPCR to detect SARS-CoV-2 [21, 22] as they identified this virus in symptomatic cases and asymptomatic carriers with low viral copy number (less than 2,000 copies/mL) [22], being these cases negative by qPCR. In the study by Alteri et al., SARS-CoV-2 infection diagnosed by dPCR was confirmed by detection of SARS-CoV-2 IgG at a later stage of the illness [21].
Quantification of microbial burden to assess severity, prognosis and treatment guidance
The possibility to quantify microbial genes by dPCR makes it an ideal method to stratify severity, assess prognosis and guide treatment. Time to positivity of blood culture has been suggested as a surrogate marker of the bacterial load present in the blood and of poor clinical outcome in BSI [23–25]. Some studies have also associated persistent BSI, detected in follow-up blood cultures, as a marker for endovascular complications and mortality risk [26–28].
Two studies published by Ziegler et al. [29, 30] showed that dPCR could be used to quantify bacterial DNA, either using species-specific genes [29, 30] or 16S ribosomal DNA (rDNA) [30] in blood of critically ill patients. Results showed that a high initial 16S rDNA load was associated with both sepsis at admission and mortality, which indicates a potential clinical value for the quantification of bacterial DNA in blood at patient admission [30]. The monitoring of patients’ bacterial load during the course of BSI showed that non-survivors had significantly higher DNA loads than survivors. The authors also found that high DNA load tended to be associated to prolonged C-reactive protein elevation and lymphopenia. The benefit of monitoring clinical progression determining DNA load was further demonstrated by Bialasiewicz et al. [31]. In this study, the authors assessed the effectiveness of antimicrobial therapy by quantifying bacterial DNA load at day zero and five, showing a great decrease in DNA load over time [31].
Another study by Dickson et al. [32] used dPCR to quantify the bacterial DNA burden in the lung at ICU admission, by assessing the concentration of 16S rDNA in mini-bronchoalveolar lavage. This study showed that patients with the highest lung bacterial DNA burden at the baseline were less likely to be extubated and alive at 7, 14, 21, and 28 days than patients with low bacterial DNA burden [32].
dPCR has also demonstrated its value to assess severity and prognosis in severe viral infections. Goh et al. [33] quantified, by dPCR, the viral load of Epstein-Barr Virus (EBV) in the plasma of patients with sCAP-derived sepsis. The hypothesis was that EBV reactivation and viral load levels might be a useful biomarker of patient immunosuppression and prognosis. The authors found that EBV reactivation was common in septic patients, that EBV viral load increased over time, was associated to longer ICU stays and increased organ failure. These findings are consistent with the concept that viral reactivation is a consequence of immune compromise in sepsis [33]. dPCR has also demonstrated its potential for the detection and quantification of SARS-CoV-2 in plasma of COVID-19 patients [34–40]. The percentage of patients with SARS-CoV-2 RNAaemia varies amongst studies (74% to 92%) [34, 35, 38], but available evidence supports that SARS-CoV-2 RNAaemia correlates with disease severity (RNAaemia prevalence ranges from 2% in outpatients, 27–53% in mild-to-moderate patients and 78–88% in critically ill patients) [34, 36, 40]. Viral RNA load in plasma correlates with key signatures of dysregulated host responses, suggesting a major role of uncontrolled viral replication in the pathogenesis of critical illness caused by SARS-CoV-2 [35, 36, 39]. It is also a predictor of severe outcome [35, 37, 39]. dPCR has been compared with qPCR for the detection of SARS-CoV-2 RNAaemia, with one study showing that dPCR is more sensitive than qPCR [37] and the other showing that both could indifferently be used [38]. This disagreement is probably related with the qPCR method used for the detection of SARS-CoV-2 RNAemia.
Microbial ecology studies
Chanderraj et al. [41] used dPCR to quantify bacterial DNA in rectal swabs from hospitalized patients and compared bacterial density and bacterial community composition (using 16S rDNA gene and amplicon sequencing), with clinical exposures (e.g., antibiotics and comorbidities), and subsequent risk of culture-confirmed extra-intestinal infection. This study showed that bacterial DNA density was associated to clinical comorbidities and age, and exposure to piperacillin-tazobactam. It was also predictive of infection during hospitalization [41]. In the same line, Brooks et al. [42] quantified bacterial DNA load by dPCR (through 16S rDNA), to evaluate if the bacterial density in an ICU environment influenced the establishment of the microbiome in hospitalized premature infants. The authors showed that bacterial DNA load and diversity varied between surfaces. Room-specific microbiome signatures were detected, suggesting that the microbes seeding ICU surfaces are sourced from reservoirs within the room, possible shaping hospitalized infants gut microbiome [42].
Applications of dPCR targeting host response
Results coming from high-throughput technologies such as microarrays or next-generation sequencing, which are able to analyse the entire human transcriptome, have revealed the existence of specific host response signatures potentially useful to improve diagnosis [43], severity stratification and prognosis assessment [44, 45] of severe infection. dPCR is making real the promise of translating these signatures into the clinical practice. A work from our group was pioneer in exploring the potential use of dPCR to diagnose sepsis, evidencing that the gene expression ratio between the constant region of the mu heavy chain of IgM and CD20 yielded an area under the receiver operating curve (AUROC) of 0.72 to differentiate sepsis from systemic inflammatory response syndrome (SIRS) [46]. In turn, Almansa et al. evidenced that the transcriptomic ratios between matrix metalloproteinase-8 (MMP8) or Lipocalin 2 (LCN2), which are two genes coding for the proteins contained in the neutrophil granules, with the major histocompatibility complex class II, DR alpha molecule (HLA-DRA), yielded AUROCs > 0.89 to differentiate between sepsis and SIRS [47]. The combination of gene expression profiling by dPCR with standard biomarkers commonly used in the clinical practice is also an exciting avenue of research, already explored in another work from Almansa et al., which evidenced that the combination of procalcitonin and HLA-DRA expression levels outperformed the former biomarker to detect sepsis. In this work, procalcitonin yielded an AUROC of 0.80 and the ratio Procalcitonin/HLA-DRA of 0.85 [48]. In a small study, Link et al. explored the potential of microRNA (miRNA) expression quantification by dPCR to diagnose sepsis. These authors found that miR-26b-5p yielded an AUROC of 0.80 to differentiate critically ill patients with sepsis from those with no sepsis [49]. dPCR has been also employed to evaluate the magnitude of the biological processes occurring during sepsis that are difficult to quantify with the currently available methods. For example, in a cohort of patients with infection, sepsis or septic shock, Martin-Fernandez et al. evidenced a progressive increase in the expression levels of emergency granulopoiesis related genes with severity [50]. Another signature of sepsis and sCAP is the depressed expression of those genes involved in the immunological synapse between antigen-presenting cells and T cells [51]. Menéndez et al. demonstrated that dPCR is an useful method to evidence the depressed expression of three of these genes [HLA-DRA, CD40 Ligand (CD40LG) and CD28] in patients with CAP presenting with organ failure [52], while Almansa et al. evidenced that profiling the expression levels of immunological synapse genes was also useful to identify VAP, yielding AUROCs of 0.82 for CD40LG, 0.79 for inducible T cell costimulator (ICOS), 0.78 for CD28 and 0.74 for CD3E [53]. Regarding prognosis, Busani et al. used dPCR to evidence the potential role of mitochondrial DNA as predictor of mortality in patients with septic shock due to MDR bacteria [54]. In turn, Almansa et al. showed that gene expression levels of HLA-DRA quantified by dPCR were an independent predictor of mortality in sepsis [47]. Cajander et al. had already proposed to use expression levels of this gene to identify those sepsis patients that could benefit from immunostimulatory drugs [55]. More recently, using dPCR, Bruneau et al.revealed that expression levels of a circulating ubiquitous RNA (RNase P) correlated with disease severity, invasive mechanical ventilation status and survival in patients with COVID-19 [40]. Also in severe COVID-19, Sabbatinelli et al. found that low levels in plasma of the inflamm-aging associated miRNA miR-146a were associated with no response to tocilizumab [56]. Metabolomics is a relatively new “-omic” that has demonstrated its use in the management of septic patients (e.g. lactate). The combination of this “-omics” approach with the transcriptomics markers measured by dPCR might be useful to determine patient severity, predict the need for mechanical ventilation and mortality [57].
Conclusions
This review evidences the potential of dPCR as a useful tool that could contribute to improve the diagnosis and clinical management of infection in critical care medicine. Although most of the published works consist of pilot/ exploratory studies, they show the potential of dPCR, supporting the development of further, larger studies aimed to validate the use of this technology in this field.
Supplementary Information
Additional file 1: Table S1. Comparison of techniques to detect microorganisms that can be employed to diagnose the most common infections affecting critically ill patients. Table containing description of emerging and current techniques to detect microorganism that can be applied to the most common critically ill patients’ infections, including the most important advantages and disadvantages.
Abbreviations
- AMR
Antimicrobial resistance genes
- AUROC
Area under the receiver operating curve
- BSIs
Bloodstream infections
- CD40LG
CD40 Ligand
- cfDNA
Cell-free deoxyribonucleic acid
- COVID-19
Coronavirus disease 2019
- ddPCR
Droplet digital polymerase chain reaction
- DNA
Deoxyribonucleic acid
- dPCR
Digital polymerase chain reaction
- EBV
Epstein-Barr Virus
- HLA-DRA
Major histocompatibility complex class II, DR alpha molecule
- ICOS
Inducible T Cell Costimulator
- ICU
Intensive care unit
- IgG
Immunoglobulin G
- IgM
Immunoglobulin M
- LCN2
Lipocalin 2
- MDR
Multidrug-resistance
- miRNA
Micro ribonucleic acid
- MMP8
Matrix metalloproteinase-8
- mNGS
Metagenomic Next-Generation Sequencing ()
- MTB
Mycobacterium tuberculosis
- mtDNA
Mitochondrial DNA
- PCR
Polymerase chain reaction
- PCT
Procalcitonin
- qPCR
Real-time polymerase chain reaction
- rDNA
Ribosomal deoxyribonucleic acid
- RNA
Ribonucleic acid
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- sCAP
Severe community acquired pneumonia
- SIRS
Systemic inflammatory response syndrome
- VAP
Ventilator-associated pneumonia
Authors' contributions
JFBM designed the review. IM and APT looked for bibliography and wrote the manuscript. APT, IM, AF developed the ddPCR works; MDG and JME collected the strains used for ddPCR pilot study. All authors read and approved the final manuscript.
Funding
This work was funded by Instituto de Salud Carlos III (ISCIII), (Project Code PI19/00590), co-funded by European Regional Development Fund (ERDF) · European Social Fund (ESF), "A way to make Europe"/"Investing in your future") (JFBM), and also by a Research Grant 2020 from ESCMID (APT). APT, IM and AF salaries were funded by the Sara Borrell Research Grant (CD18/00123), Río Hortega (CM18/00157) and PFIS (FI20/00278) Research Grants, respectively, from ISCIII and co-funded by ERDF/ESF, "A way to make Europe"/"Investing in your future").
Availability of data and materials
The dataset supporting the conclusions of this article is included within the article (and its additional file).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sakr Y, Jaschinski U, Wittebole X, Szakmany T, Lipman J, Ñamendys-Silva SA, et al. Sepsis in intensive care unit patients: worldwide data from the intensive care over nations audit. Open forum Infect Dis. 2018;5. [DOI] [PMC free article] [PubMed]
- 2.Aliberti S, Dela Cruz CS, Amati F, Sotgiu G, Restrepo MI. Community-acquired pneumonia. Lancet (London, England) 2021;398:906–919. doi: 10.1016/S0140-6736(21)00630-9. [DOI] [PubMed] [Google Scholar]
- 3.Strich JJR, Palmore TTN. Preventing Transmission of Multidrug-Resistant Pathogens in the Intensive Care Unit. Infect Dis Clin North Am. 2017;31:535–550. doi: 10.1016/j.idc.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin SJ, Yost RJ. Infectious diseases in the critically ill patients. J Pharm Pract. 2011;24:35–43. doi: 10.1177/0897190010388906. [DOI] [PubMed] [Google Scholar]
- 5.Suetens C, Latour K, Kärki T, Ricchizzi E, Kinross P, Moro ML, et al. Prevalence of healthcare-associated infections, estimated incidence and composite antimicrobial resistance index in acute care hospitals and long-term care facilities: results from two European point prevalence surveys, 2016 to 2017. Euro Surveill. 2018;23. [DOI] [PMC free article] [PubMed]
- 6.Cillóniz C, Torres A, Niederman MS. Management of pneumonia in critically ill patients. BMJ. 2021;375:e065871. doi: 10.1136/bmj-2021-065871. [DOI] [PubMed] [Google Scholar]
- 7.Salipante SJ, Jerome KR. Digital PCR-an emerging technology with broad applications in microbiology. Clin Chem. 2020;66:117–123. doi: 10.1373/clinchem.2019.304048. [DOI] [PubMed] [Google Scholar]
- 8.Sreejith KR, Ooi CH, Jin J, Dao DV, Nguyen NT. Digital polymerase chain reaction technology—recent advances and future perspectives. Lab Chip. 2018;18:3717–3732. doi: 10.1039/c8lc00990b. [DOI] [PubMed] [Google Scholar]
- 9.Quan PL, Sauzade M, Brouzes E. dPCR: a technology review. Sensors (Basel). 2018;18. [DOI] [PMC free article] [PubMed]
- 10.Hu B, Tao Y, Shao Z, Zheng Y, Zhang R, Yang X, et al. A comparison of blood pathogen detection among droplet digital PCR, metagenomic next-generation sequencing, and blood culture in critically ill patients with suspected bloodstream infections. Front Microbiol. 2021;12:641202. doi: 10.3389/fmicb.2021.641202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchan BW, Ledeboer NA. Emerging technologies for the clinical microbiology laboratory. Clin Microbiol Rev. 2014;27:783–822. doi: 10.1128/CMR.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walter JM, Wunderink RG. Severe respiratory viral infections: new evidence and changing paradigms. Infect Dis Clin North Am. 2017;31:455–474. doi: 10.1016/j.idc.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung J, Seo E, Yoo RN, Sung H, Lee J. Clinical significance of viral-bacterial codetection among young children with respiratory tract infections: findings of RSV, influenza, adenoviral infections. Medicine (Baltimore) 2020;99:e18504. doi: 10.1097/MD.0000000000018504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto M, Ushio R, Watanabe H, Tachibana T, Tanaka M, Yokose T, et al. Detection of Mycobacterium tuberculosis-derived DNA in circulating cell-free DNA from a patient with disseminated infection using digital PCR. Int J Infect Dis. 2018;66:80–82. doi: 10.1016/j.ijid.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 15.Shin J, Shin S, Jung SH, Park C, Cho SY, Lee DG, et al. Duplex dPCR system for rapid identification of gram-negative pathogens in the blood of patients with bloodstream infection: a culture-independent approach. J Microbiol Biotechnol. 2021;31:1481–1489. doi: 10.4014/jmb.2103.03044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Y, Jin J, Shao Z, Liu J, Zhang R, Sun R, et al. Development and clinical validation of a droplet digital PCR assay for detecting Acinetobacter baumannii and Klebsiella pneumoniae in patients with suspected bloodstream infections. Microbiologyopen. 2021;10. [DOI] [PMC free article] [PubMed]
- 17.Zaragoza R, Ramírez P, López-Pueyo MJ. Nosocomial infections in intensive care units. Enferm Infecc Microbiol Clin. 2014;32:320–327. doi: 10.1016/j.eimc.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Chen B, Xie Y, Zhang N, Li W, Liu C, Li D, et al. Evaluation of droplet digital PCR assay for the diagnosis of candidemia in blood samples. Front Microbiol. 2021;12:700008. doi: 10.3389/fmicb.2021.700008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou F, Sun S, Sun X, Chen Y, Yang X. Rapid and sensitive identification of pleural and peritoneal infections by droplet digital PCR. Folia Microbiol (Praha) 2021;66:213–219. doi: 10.1007/s12223-020-00834-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simms EL, Chung H, Oberding L, Muruve DA, McDonald B, Bromley A, et al. Post-mortem molecular investigations of SARS-CoV-2 in an unexpected death of a recent kidney transplant recipient. Am J Transplant. 2021;21:2590–2595. doi: 10.1111/ajt.16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alteri C, Cento V, Antonello M, Colagrossi L, Merli M, Ughi N, et al. Detection and quantification of SARS-CoV-2 by droplet digital PCR in real-time PCR negative nasopharyngeal swabs from suspected COVID-19 patients. PLoS ONE. 2020;15:e0236311. doi: 10.1371/journal.pone.0236311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Y, Wang H, Hao S, Chen Y, He J, Liu Y, et al. Digital PCR is a sensitive new technique for SARS-CoV-2 detection in clinical applications. Clin Chim Acta. 2020;511:346–351. doi: 10.1016/j.cca.2020.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blot F, Schmidt E, Nitenberg G, Tancrède C, Leclercq B, Laplanche A, et al. Earlier positivity of central-venous- versus peripheral-blood cultures is highly predictive of catheter-related sepsis. J Clin Microbiol. 1998;36:105–109. doi: 10.1128/jcm.36.1.105-109.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khatib R, Riederer K, Saeed S, Johnson LB, Fakih MG, Sharma M, et al. Time to positivity in Staphylococcus aureus bacteremia: possible correlation with the source and outcome of infection. Clin Infect Dis. 2005;41:594–598. doi: 10.1086/432472. [DOI] [PubMed] [Google Scholar]
- 25.Kim J, Gregson DB, Ross T, Laupland KB. Time to blood culture positivity in Staphylococcus aureus bacteremia: association with 30-day mortality. J Infect. 2010;61:197–204. doi: 10.1016/j.jinf.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Fernández-Cruz A, Marín M, Kestler M, Alcalá L, Rodriguez-Créixems M, Bouza E. The value of combining blood culture and SeptiFast data for predicting complicated bloodstream infections caused by Gram-positive bacteria or Candida species. J Clin Microbiol. 2013;51:1130–1136. doi: 10.1128/JCM.02882-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi SH, Chung JW. Time to positivity of follow-up blood cultures in patients with persistent Staphylococcus aureus bacteremia. Eur J Clin Microbiol Infect Dis. 2012;31:2963–2967. doi: 10.1007/s10096-012-1648-0. [DOI] [PubMed] [Google Scholar]
- 28.Hsu MS, Huang YT, Hsu HS, Liao CH. Sequential time to positivity of blood cultures can be a predictor of prognosis of patients with persistent Staphylococcus aureus bacteraemia. Clin Microbiol Infect. 2014;20:892–898. doi: 10.1111/1469-0691.12608. [DOI] [PubMed] [Google Scholar]
- 29.Ziegler I, Cajander S, Rasmussen G, Ennefors T, Mölling P, Strålin K. High nuc DNA load in whole blood is associated with sepsis, mortality and immune dysregulation in Staphylococcus aureus bacteraemia. Infect Dis (London, England) 2019;51:216–226. doi: 10.1080/23744235.2018.1562205. [DOI] [PubMed] [Google Scholar]
- 30.Ziegler I, Lindström S, Källgren M, Strålin K, Mölling P. 16S rDNA droplet digital PCR for monitoring bacterial DNAemia in bloodstream infections. PLoS One. 2019;14. [DOI] [PMC free article] [PubMed]
- 31.Bialasiewicz S, Duarte TPS, Nguyen SH, Sukumaran V, Stewart A, Appleton S, et al. Rapid diagnosis of Capnocytophaga canimorsus septic shock in an immunocompetent individual using real-time Nanopore sequencing: a case report. BMC Infect Dis. 2019;19:660. doi: 10.1186/s12879-019-4173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dickson RP, Schultz MJ, Van Der Poll T, Schouten LR, Falkowski NR, Luth JE, et al. Lung microbiota predict clinical outcomes in critically Ill patients. Am J Respir Crit Care Med. 2020;201:555–563. doi: 10.1164/rccm.201907-1487OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goh C, Burnham KL, Ansari MA, de Cesare M, Golubchik T, Hutton P, et al. Epstein-Barr virus reactivation in sepsis due to community-acquired pneumonia is associated with increased morbidity and an immunosuppressed host transcriptomic endotype. Sci Rep. 2020;10. [DOI] [PMC free article] [PubMed]
- 34.Veyer D, Kernéis S, Poulet G, Wack M, Robillard N, Taly V, et al. Highly sensitive quantification of plasma SARS-CoV-2 RNA shelds light on its potential clinical value. Clin Infect Dis. 2020; [DOI] [PMC free article] [PubMed]
- 35.Chen L, Wang G, Long X, Hou H, Wei J, Cao Y, et al. Dynamics of blood viral load is strongly associated with clinical outcomes in coronavirus disease 2019 (COVID-19) patients: a prospective cohort study. J Mol Diagn. 2021;23:10–18. doi: 10.1016/j.jmoldx.2020.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bermejo-Martin JF, González-Rivera M, Almansa R, Micheloud D, Tedim AP, Domínguez-Gil M, et al. Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID-19. Crit Care. 2020;24:691. doi: 10.1186/s13054-020-03398-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ram-Mohan N, Kim D, Zudock EJ, Hashemi MM, Tjandra KC, Rogers AJ, et al. SARS-CoV-2 RNAemia predicts clinical deterioration and extrapulmonary complications from COVID-19. Clin Infect Dis. 2022;74:218–226. doi: 10.1093/cid/ciab394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tedim AP, Almansa R, Domínguez-Gil M, González-Rivera M, Micheloud D, Ryan P, et al. Comparison of real-time and droplet digital PCR to detect and quantify SARS-CoV-2 RNA in plasma. Eur J Clin Invest. 2021;51:e13501. doi: 10.1111/eci.13501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin-Vicente M, Almansa R, Martínez I, Tedim AP, Bustamante E, Tamayo L, et al. Low anti-SARS-CoV-2 S antibody levels predict increased mortality and dissemination of viral components in the blood of critical COVID-19 patients. J Intern Med. 2022;291:232–240. doi: 10.1111/joim.13386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bruneau T, Wack M, Poulet G, Robillard N, Philippe A, Puig P-L, et al. Circulating ubiquitous RNA, a highly predictive and prognostic biomarker in hospitalized COVID-19 patients. Clin Infect Dis. 2021; [DOI] [PMC free article] [PubMed]
- 41.Chanderraj R, Brown CA, Hinkle K, Falkowski N, Woods RJ, Dickson RP. The bacterial density of clinical rectal swabs is highly variable, correlates with sequencing contamination, and predicts patient risk of extraintestinal infection. Microbiome. 2022;10. [DOI] [PMC free article] [PubMed]
- 42.Brooks B, Olm MR, Firek BA, Baker R, Geller-McGrath D, Reimer SR, et al. The developing premature infant gut microbiome is a major factor shaping the microbiome of neonatal intensive care unit rooms. Microbiome. 2018;6. [DOI] [PMC free article] [PubMed]
- 43.Sweeney TE, Shidham A, Wong HR, Khatri P. A comprehensive time-course-based multicohort analysis of sepsis and sterile inflammation reveals a robust diagnostic gene set. Sci Transl Med. 2015;7. [DOI] [PMC free article] [PubMed]
- 44.Scicluna BP, van Vught LA, Zwinderman AH, Wiewel MA, Davenport EE, Burnham KL, et al. Classification of patients with sepsis according to blood genomic endotype: a prospective cohort study. Lancet Respir Med. 2017;5:816–826. doi: 10.1016/S2213-2600(17)30294-1. [DOI] [PubMed] [Google Scholar]
- 45.Sweeney TE, Perumal TM, Henao R, Nichols M, Howrylak JA, Choi AM, et al. A community approach to mortality prediction in sepsis via gene expression analysis. Nat Commun. 2018;9. [DOI] [PMC free article] [PubMed]
- 46.Tamayo E, Almansa R, Carrasco E, Ávila-Alonso A, Rodríguez-Fernández A, Wain J, et al. Quantification of IgM molecular response by droplet digital PCR as a potential tool for the early diagnosis of sepsis. Crit Care. 2014;18. [DOI] [PMC free article] [PubMed]
- 47.Almansa R, Ortega A, Ávila-Alonso A, Heredia-Rodríguez M, Martín S, Benavides D, et al. Quantification of immune dysregulation by next-generation polymerase chain reaction to improve sepsis diagnosis in surgical patients. Ann Surg. 2019;269:545–553. doi: 10.1097/SLA.0000000000002406. [DOI] [PubMed] [Google Scholar]
- 48.Almansa R, Martín S, Martin-Fernandez M, Heredia-Rodríguez M, Gómez-Sánchez E, Aragón M, et al. Combined quantification of procalcitonin and HLA-DR improves sepsis detection in surgical patients. Sci Rep. 2018;8. [DOI] [PMC free article] [PubMed]
- 49.Link F, Krohn K, Burgdorff AM, Christel A, Schumann J. Sepsis diagnostics: intensive care scoring systems superior to MicroRNA biomarker testing. Diagnostics (Basel, Switzerland). 2020;10. [DOI] [PMC free article] [PubMed]
- 50.Martin-Fernandez M, Vaquero-Roncero LM, Almansa R, Gómez-Sánchez E, Martín S, Tamayo E, et al. Endothelial dysfunction is an early indicator of sepsis and neutrophil degranulation of septic shock in surgical patients. BJS open BJS Open. 2020;4:524–534. doi: 10.1002/bjs5.50265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davenport EE, Burnham KL, Radhakrishnan J, Humburg P, Hutton P, Mills TC, et al. Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study. Lancet Respir Med Lancet Respir Med. 2016;4:259–271. doi: 10.1016/S2213-2600(16)00046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Menéndez R, Méndez R, Almansa R, Ortega A, Alonso R, Suescun M, et al. Simultaneous depression of immunological synapse and endothelial injury is associated with organ dysfunction in community-acquired pneumonia. J Clin Med. 2019;8. [DOI] [PMC free article] [PubMed]
- 53.Almansa R, Nogales L, Martín-Fernández M, Batlle M, Villareal E, Rico L, et al. Transcriptomic depression of immunological synapse as a signature of ventilator-associated pneumonia. Ann Transl Med. 2018;6:415–415. doi: 10.21037/atm.2018.05.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Busani S, De Biasi S, Nasi M, Paolini A, Venturelli S, Tosi M, et al. Increased plasma levels of mitochondrial DNA and normal inflammasome gene expression in monocytes characterize patients with septic shock due to multidrug resistant bacteria. Front Immunol. 2020;11. [DOI] [PMC free article] [PubMed]
- 55.Cajander S, Tina E, Bäckman A, Magnuson A, Strålin K, Söderquist B, et al. Quantitative real-time polymerase chain reaction measurement of HLA-DRA gene expression in whole blood is highly reproducible and shows changes that reflect dynamic shifts in monocyte surface HLA-DR expression during the course of sepsis. PLoS One. 2016;11. [DOI] [PMC free article] [PubMed]
- 56.Sabbatinelli J, Giuliani A, Matacchione G, Latini S, Laprovitera N, Pomponio G, et al. Decreased serum levels of the inflammaging marker miR-146a are associated with clinical non-response to tocilizumab in COVID-19 patients. Mech Ageing Dev. 2021;193. [DOI] [PMC free article] [PubMed]
- 57.Evangelatos N, Bauer P, Reumann M, Satyamoorthy K, Lehrach H, Brand A. Metabolomics in sepsis and its impact on public health. Public Health Genomics. 2017;20:274–285. doi: 10.1159/000486362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Comparison of techniques to detect microorganisms that can be employed to diagnose the most common infections affecting critically ill patients. Table containing description of emerging and current techniques to detect microorganism that can be applied to the most common critically ill patients’ infections, including the most important advantages and disadvantages.
Data Availability Statement
The dataset supporting the conclusions of this article is included within the article (and its additional file).