Abstract
Coronavirus disease 2019 (COVID-19) caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is considered one of the worst pandemic outbreaks worldwide. This ongoing pandemic urgently requires rapid, accurate, and specific testing devices to detect the virus. We report a simple electrochemical biosensor based on a highly specific synthetic peptide to detect SARS-CoV-2 Spike protein. Unlike other reported electrochemical biosensors involving nanomaterials or complex approaches, our electrochemical platform uses screen-printed gold electrodes functionalized with the thiolated peptide, whose interaction with the Spike protein is directly followed by Electrochemical Impedance Spectroscopy. The electrochemical platform was Spike protein concentration-dependent, with high sensitivity and reproducibility and a limit of detection of 18.2 ng/mL when tested in Spike protein commercial solutions and 0.01 copies/mL in lysed SARS-CoV-2 particles. The label-free biosensor successfully detected the Spike protein in samples from infected patients straightforwardly in only 15 min. The simplicity of the proposed format combined with an on-demand designed peptide opens the path for detecting other pathogen-related antigens.
Keywords: SARS-CoV-2, Synthetic peptide, Impedimetric biosensor, Infectious disease
Graphical abstract
1. Introduction
The rapid expansion of the coronavirus 2019 (COVID-19), a disease caused by the Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2), was declared a pandemic in 2020 and continues today to threaten public health systems worldwide [[1], [2], [3], [4]]. The current diagnostic recommended by the World Health Organization (WHO) is based on the detection of the SARS-CoV-2 viral RNA by reverse transcriptase real-time polymerase chain reaction (RT-PCR) [5]. Although the RT-PCR is the gold standard method, it is time-consuming and requires specialized testing facilities with highly qualified personnel, which decreases the high number of daily samples essential to slow its spread. Besides, immunoassays based on the detection of viral structural proteins and serological methods that detect antibodies have some limitations related to low sensitivity, reduced efficiency and cross-reactivity. Therefore, developing accurate, specific and sensitive devices for the simple and rapid detection of the SARS-CoV-2 and early diagnosis of COVID 19 has a pivotal role in controlling the viral outbreak [[5], [6], [7]].
Previous reports have found that Electrochemical Impedance Spectroscopy (EIS)-based biosensors rapidly detect bioreceptors associated with SARS-CoV-2 infection, with high sensitivity, specificity and reliability [[8], [9], [10], [11], [12], [13], [14], [15], [16], [17]]. In such biosensors, bioreceptors are anchored on the working electrode surface to directly bind to target molecules specifically, generating changes in the interfacial properties in a concentration-dependent manner [[18], [19], [20]]. The SARS-CoV-2 has also been detected electrochemically by immunosensors [14,[21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32]] and genosensors [[33], [34], [35], [36]]. Furthermore, outstanding features of biosensors have been exploited in the detection of other viruses [37,38], pathogens [39], other infections [18], cancer biomarkers [[40], [41], [42]] and other diseases-related biomarkers [43] demonstrating to be promising analytical tools that can help to solve current diagnosis limitations [44].
Here, we report the first peptide-based biosensor for simple monitoring free Spike protein and SARS-CoV-2 viral particles in COVID-19 positive patients by EIS. Peptides are specific sequences of amino acids with similar selectivity, specificity and chemical nature as proteins but smaller sizes, higher stability against denaturation, cost-effectiveness, accessibility, easiness of modification, and more extensive chemical versatility [45,46]. Therefore, peptides are ideal candidates to substitute proteins as bioreceptor in (bio)recognition and biosensing events. The biosensor uses a synthetic thiolated peptide bioreceptor chemisorbed at the working electrode of a screen-printed gold electrode (SPAuE) that directly interacted with the Spike protein and whose interaction was detected and quantified by EIS. The resultant device showed high sensitivity and reproducibility, low limit of detection (LOD) and a linear dynamic range of clinical relevance. Besides, the peptide-based biosensor was highly specific for SARS-CoV-2 and detected the Spike protein from COVID-19 positive patients in clinical samples. This is the first time the peptide has been incorporated in a transducer platform for SARS-CoV-2 sensing purposes of remarkable analytical performance. Performance, simplicity, and low cost not only underline the great potential of our biosensor for detecting the virus at the clinical level but for the on-demand design of peptide-based sensing platforms for detecting other viruses, pathogens, and disease biomarkers.
2. Materials and methods
2.1. Reagents
A targeting synthetic peptide with 23 amino acids length, {MPA}(PEG4)IEEQAKTFLDKFNHEAEDLFYQS sequence, purity ≥95%, MW: 3137.42 g/mol and a thiol-linker was synthesized on-demand by GenScript U.S.A. Inc (New Jersey, U.S.A.). SARS-CoV-2 (2019-nCoV) Spike recombinant protein (protein S, purity 90% by SDS-Page) (MW: 76.5 kDa) was acquired by SinoBiological (Pennsylvania, USA); recombinant human beta-1,4-galactosyltransferase-5 glycoprotein (β-1,4-GALT-5) (cod Ab160437) was purchased from Abcam (Cambridge, UK); receptor binding domain Spike protein (RBD) was acquired by Active Motif (California, USA); potassium ferricyanide (III) (K3[Fe(CN)6]), potassium hexacyanoferrate (II) trihydrate (K4[Fe(CN)6].3H2O), tris(2-carboxyethyl) phosphine hydrochloride (TCEP) and 6-mercapto-1-hexanol (MCH) were purchased from Merck Millipore (Darmstadt, Germany). Dipotassium hydrogen phosphate (K2HPO4), disodium hydrogen phosphate (Na2HPO4) and potassium nitrate (KNO3) were acquired from PanReac AppliChem (Darmstadt, Germany). Potassium dihydrogen phosphate (KH2PO4), potassium chloride (KCl) and sodium chloride (NaCl) were obtained from J.T.Baker® (Xalostoc, Mexico). Sulphuric acid (H2SO4) was purchased from Honeywell FlukaTM (Seelve, Germany). RIPA (Radioimmunoprecipitation) lysis buffer system composed of protease inhibitor cocktail in dimethyl sulfoxide (DMSO), 100 mM Sodium orthovanadate in water, 200 mM phenylmethanesulfonyl fluoride (PMSF) in DMSO and 1X lysis buffer pH 7.4 were acquired from ChemCruz (Dallas, U.S.A.). Sterile and nuclease-free water was acquired from VWR Life Science (Ohio, USA) used to dilute each sample. All reagents and commercial samples were used without purification and the solutions were prepared using deionized water of 18.3 mΩ cm.
2.2. Devices and equipment
All electrochemical measurements were performed with a three-electrode cell configuration SPAuE (ref. 220BT, from Metrohm) in a potentiostat galvanostat VersaSTAT 3 with a single channel with an impedance mode. The spectra were analyzed using VersaStudio 2.61.3 software. The chips consist of a 4 mm gold working electrode, a gold counter electrode, and a silver pseudo-reference electrode, respectively printed on the same strip.
2.3. Assembly of the peptide-based biosensor
SPAuEs were pre-treated according to the previously reported protocol to clean and activate the surface, i.e., cyclic voltammetry (CV) in 50 μl of 0.1 M H2SO4, scanning at a potential between 1.6 and −0.2 V (vs. Ag pseudo-reference electrode) at a scan rate of 0.1 V/s until the voltammogram becomes stable (approximately 10 consecutive cycles). The effective area and roughness factor were estimated by considering that the gold surface's reductive peak has a charge of 400 μC/cm2 [47] and the gold electrode has a geometry area of 12.56 mm2.
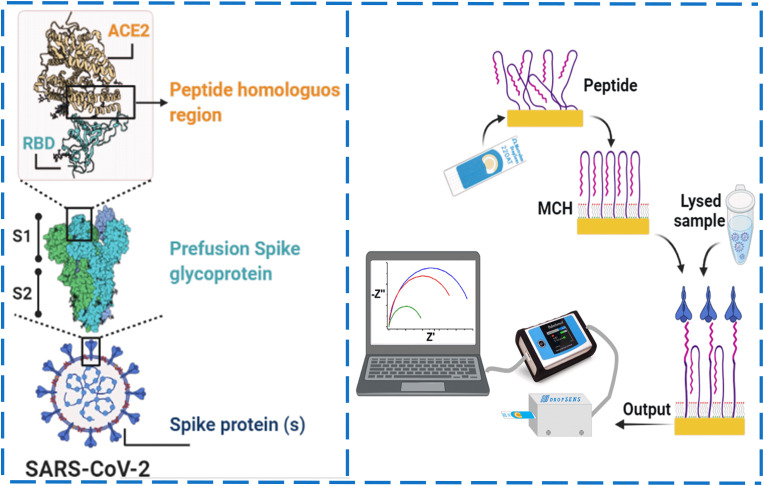
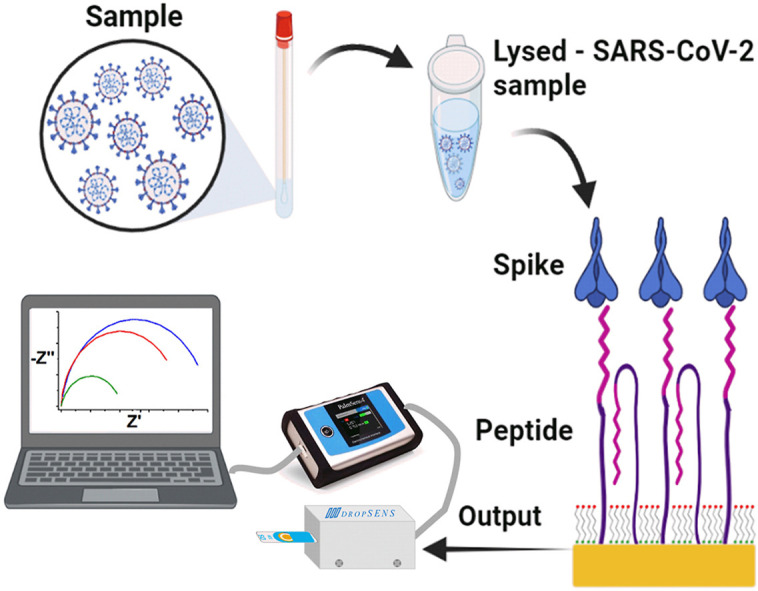
1.59 mM of synthetic peptide stock solution was prepared by dissolving 1 mg of the peptide in 200 μL of nuclease-free water. TCEP was added at a final concentration of 0.5 mM to reduce thiol groups from the peptide that may be oxidized by incubation at 37 °C and 1100 rpm for 30 min. The pre-treated SPAuE was incubated in 6 μL of the TCEP-pretreated peptide solution for 1 h at room temperature, followed by 6 μL 0.01 mM MCH for 10 min to orient the peptide reactive places and block nonspecific sites. The modified SPAuE was then incubated in a Spike protein solution at 37 °C and 50 rpm for 15 min in a wet chamber in a Thermo Scientific MAXQ 4450 shaker for the peptide-protein interaction. The peptide biosensor assembly steps and detection mechanism are illustrated in Scheme 1 .
Scheme 1.
Schematic representation of the peptide-based impedimetric biosensor. Structure with different regions of the Spike protein, including the RBD region base for designing the synthetic peptide used as biorecognition molecule in the biosensor (left) and step-by-step of the biosensor assembly, i.e., formation of a peptide-based mixed self-assembled monolayer, interaction with a lysed sample and readout (right).
2.4. Electrochemical measurements
The electrochemical performance of the as-functionalized platform was evaluated by CV in the 5 mM [Fe(CN)6]3-/4-/KNO3 redox pair in a potential window between +0.4 V and −0.2 V, at a scanning rate of 0.05 V/s for 5 consecutive cycles. This measurement was made initially with the bare electrode and later after each step of the functionalization process.
Electrochemical impedance spectroscopy (EIS) was carried out in the 5 mM [Fe(CN)6]3-/4-/KNO3 redox pair by applying a potential of +0.19 V in frequencies ranging from 50 kHz to 0.05 Hz at 0.010 V of amplitude. The impedance data were fitted with the EIS Spectrum Analyzer software using the Levenberg-Marquardt algorithm. The impedance measurements were performed after each step of the biosensor development, i.e., functionalization of the SPAuE surface by chemisorption of the peptide, blocking the electrodes with MCH and incubation with Spike protein. Calibration curves were built with increasing concentrations of Spike protein and SARS-CoV-2 viral particles and the limit of detection (LOD) was calculated as LOD = 3σDblank/slope.
2.5. Selectivity and specificity studies and quantification of spike protein in human samples
The selectivity of the biosensor was assessed by comparing the response against supernatants of the target (SARS-CoV-2 viral particles inactivated by UV-light) and supernatants of other coronaviruses such as Middle East Respiratory Syndrome (MERS) and SARS-CoV at equivalent concentrations. We also evaluated the specificity of the peptide-based biosensor against region binding domain (RBD) protein solutions and glycoproteins such as beta-1,4-galactosyltransferase-5 (β-1,4-GALT-5).
All viral supernatant samples were prepared at a concentration of 104 copies/mL dissolved in a lysis buffer containing 10 μl of PMSF solution, 10 μl of sodium orthovanadate solution and 20 μl of cocktail solution of inhibitor protease per ml of 1X RIPA lysis buffer. The samples were vortexed for 1 min and then sonicated for 1.5 min to promote the disruption of the nuclear membrane and favor the shedding of Spike protein molecules on the surface of the viral particles. The lysed solutions were dropped on the modified electrodes and incubated at 37 °C and 50 rpm for 15 min in a wet chamber. The biosensor response was interrogated by EIS, as mentioned above. Paired t-test and a 1-way ANOVA with a 99% level of statistical significance (ANOVA) were performed using Excel software to evaluate the statistical significance between the samples.
Nasopharyngeal swab clinical sample solutions in universal transport medium (UTM) from three COVID-19 negative and nine positive patients were donated by the Tropical Diseases Study and Control Program (PECET - by its Spanish acronym) laboratory after testing by RT-PCR in a biosafety lab (BSL)-2 facility following established biosafety and ethical standards and inactivation by UV-light. The RT-PCR protocol is briefly described in the next subsection. Before analyzing the samples with the peptide-based biosensor, they were lysed according to the protocol mentioned above. It facilitated the viral protein release and diluted 100-fold its concentration and interferent species from the samples to fit well in the calibration curve.
2.6. Quantification of viral RNA from clinical samples by RT-PCR
Samples were obtained by nasopharyngeal swabs and immediately deposited in a UTM, followed by RNA extraction from 200 to 300 μl of samples on the same day. The extraction was done by automated extraction using the king Fisher Flex robot and the MagMAXTM Viral/Pathogen II (MVP II) Nucleic Acid Isolation kit (Thermo Fisher) or using a manual extraction with the Quick RNA Viral Kit (Zymo Research). The amplification reaction was done using the Berlin protocol, with modifications as detailed in the Supporting Information section (SI). The Human Research Bioethics Committee from the University of Antioquia governed by Resolution 008430, October 4, 1993, Ministry of Health from Colombia endorsed the project 'Nanobiosensors for rapid detection of SARS-CoV-2′ in an approval certificate number 20-109-897.
3. Results and discussion
3.1. Development of the detection platform
We selected a thiolated synthetic peptide as a bioreceptor to develop the biosensor reported herein, inspired by the high interaction between the RBD region of the SARS-CoV-2 and the angiotensin-converting enzyme (ACE)-2 host receptor protein [48,49]. Recent crystallographic studies revealed that the amino acid residues, mainly through polar residues present in the α1 helix of the peptidase domain of the ACE-2 protein, are the key to the interfacial interaction with the RBD protein of SARS-CoV-2 [4,50]. It has been reported that multiple variants of the virus are related to variations in the hydrogen bonds and amino acid residues involved in the RBD region-ACE-2 protein interaction. However, we propose a peptide-based biosensor concept that can be readily re-designed on-demand based on new changes of the amino acid content of the virus by conserving a similar length chain, terminal functional group, and optimized functionalization conditions before assembling in a device.
Therefore, a synthetic peptide was designed considering the previously reported peptide sequence [4] but changing the N-terminus of the peptide for a thiol group to anchor the peptide to the SPE surface. The amino acids of length and composition are homologous to the sequence of the α1 helix of the peptidase domain of the ACE-2 protein that expects to interact specifically with the RBD while minimizing unspecific interactions with other proteins. The peptide-modified electrode detected the Spike protein expressed at the outermost surface of viral nanoparticles in a spiked buffered solution and lysates from SARS-CoV-2 viral particles, as shown below. The structure of this synthetic peptide consists of 23 amino acids that specifically recognize the Spike protein, a segment of polyethylene glycol (PEG) as a spacer and a C-terminal modification of mercaptopropionic acid (MPA) to link to the transducer platform. The synthetic peptide is highly specific for a side branch of the RBD region of the SARS-CoV-2 Spike protein [51]. We assembled the impedimetric biosensor by chemisorbing the thiolated peptide on an SPAuE surface previously activated by CV in acidic conditions. The activated electrodes showed an effective area and roughness factor of 231.62 ± 11.42 mm2 and 18.44 ± 0.91, respectively. An efficient, reproducible, and well-controlled self-assembled monolayer of the peptide was formed at the SPAuE surface, followed by treatment with MCH characterized by CV and EIS. Incubation with MCH allowed to block nonspecific binding sites on the SPAuE surface and better orient the synthetic peptide perpendicular to the surface to point to the RBD region of the Spike protein [[52], [53], [54]].
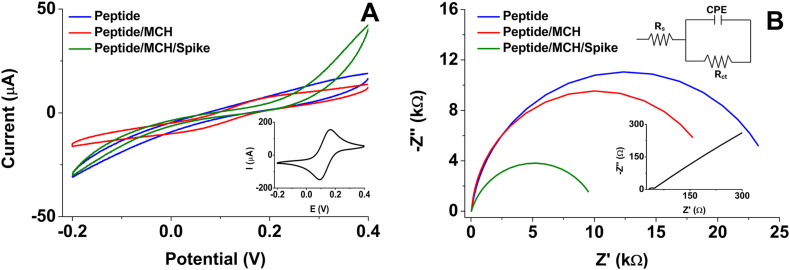
Fig. 1 shows comparative CV and EIS plots from the step-by-step electrode modification process. For example, Fig. 1A shows the current intensity decreased after chemisorbing the peptide at the SPAuE due to the formation of a peptide layer that limits the electrons transfer at the electrode-solution interface as compared with the bare electrode. Yet, the incorporation of MCH favored the interaction with the Spike protein noticeably. Fig. 1B shows differences in electron transfer of the transducer platform after each modification step represented by the Nyquist plot (-Z″ vs. Z′). Fitting electrochemical parameters from EIS by a Randles equivalent circuit (Inset Fig. 1B) was used to estimate the charge-transfer resistance (Rct), the electrolyte resistance (Rs), the constant phase element (CPE), which depends on a pre-exponential factor (P) and an exponent (n) [[55], [56], [57], [58]], as summarized in Table 1 .
Fig. 1.
A) Cyclic voltammograms at a scan rate of 50 mV/s and B) Nyquist plot at 50 kHz to 0.05 Hz frequency range and 10 mV amplitude (inset is the Randles equivalent circuit) for each modification step recorded in 5 mM [Fe(CN)6]3-/4-/KNO3 redox probe. The bare electrodes are in the insets of both figures.
Table 1.
Electrochemical characterization of the modified electrodes. Data from EIS experiments. Charge-transfer resistance (Rct), electrolytic solution resistance (Rs), constant phase element (CPE) with pre-exponential factor (P) and exponent (n) and Chi-squared function (χ2).
| Electrode | Rct (kΩ) | Rs (Ω) | CPE |
Χ2 x 10−3 | |
|---|---|---|---|---|---|
| P x 10−5 | n | ||||
| SPAuE | 1.3 ± 0.2 | 35.1 ± 1.6 | 3.4 | 0.867 | 1.6 |
| Peptide | 25.0 ± 0.8 | 28.0 ± 2.8 | 2.0 | 0.845 | 0.3 |
| Peptide/MCH | 21.9 ± 0.1 | 35.7 ± 0.9 | 1.0 | 0.925 | 1.0 |
| Peptide/MCH/Spike protein 0.1 μg/mL | 10.9 ± 0.6 | 34.2 ± 1.5 | 1.1 | 0.866 | 0.7 |
The EIS from the bare electrode in inset Fig. 1B shows that the semicircle diameter in the Nyquist plot is very similar to the charge transfer resistance, Rct = 1.3 ± 0.2 kΩ and that the Rs = 35.1 ± 1.6 Ω is very low. The bare electrode also shows Warburg diffusion at high frequencies, lost after modification with the peptide (Inset Fig. 1B). In contrast, Rct increased three orders of magnitude (up to 25.0 ± 0.8 kΩ) after immobilizing the peptide at the electrode surface due to an increased interface thickness that insulated the conductive support and blocked the electron transfer from the electrochemical probe and the surface. This change also indicates the successful chemisorption of the thiolated peptide at the electrode surface. Treatment of the SPAuE with MCH induced a decrease in the insulation and Rct value (Fig. 1B) related to the reorganization of the amino acid chain of the peptide that went away from the transducer generating more free spaces for easier access of the electrochemical probe. We further evaluated the interaction of the peptide-modified electrode with synthetic Spike protein. When the peptide-electrode was incubated with a protein solution at 37 °C for 15 min, the protein binding event induced a decrease of the Rct to a value of 10.9 ± 0.6 kΩ due to re-orientation of the synthetic peptide end segment during the Spike protein recognition event [51]. P values in Table 1 showed a decrease in the capacitive behavior of the system, and in the meantime, the n values for modified electrodes were less than one, consistent with a double layer pseudo-interfacial capacitance at the electrode/electrolyte interface [59].
Although the use of the theoretical model of the Randles circuit for the adjustment of experimental data is widely extended; it is crucial to recognize that this model does not consider various factors involved in the biosensing process of the Spike protein, such as binding affinity, leaching, denaturation, transport phenomena, surface defects, among others; that may affect the results. Nevertheless, the very small values of Chi-squared function (χ2) for the best-fitted equivalent circuit, lower than 1.6 × 10−3, demonstrated that the curve showed herein and the experimental data from the mathematical fitting agreed (See S.I Fig. S1 A-C). Table 1 summarizes all the fitted electrochemical parameters. These results suggest that the anchorage of the peptide and the interaction with the Spike protein decreased the capacitive current on modified-electrodes and decreased the charge transfer resistance, which indicates that the peptide-Spike protein recognition event was successful.
3.2. Optimization of the analytical conditions
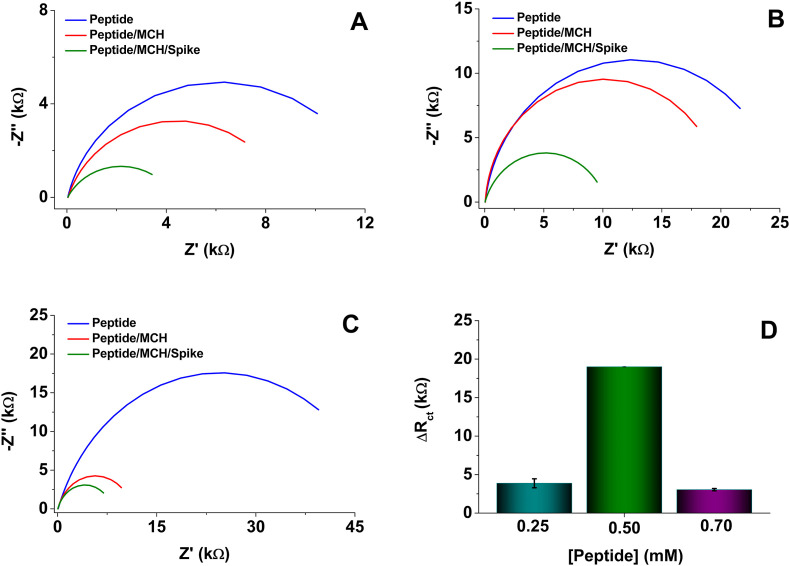
Once the functionality of the biosensor platform was established, the peptide concentration was optimized as a function of the increase in the difference of the charge transfer resistance (ΔRct) (Fig. 2 ). The behavior of the functionalized electrode was evaluated with different concentrations of the peptide after blocking with MCH and incubation with 1.0 μg/mL of a Spike protein solution. The ΔRct for 0.25 and 0.7 mM concentrations were lower (Fig. 2A –C) than for 0.50 mM of peptide solution (Fig. 2B). These differences may be associated with the organization degree of the peptide at the modified-electrode surface. Accordingly, too low concentrations of peptides seem to generate poor self-assembly of the monolayers, thus affecting the Spike protein recognition event. In contrast, too high concentration of peptide molecules could generate overlapping of peptide multilayers that limit the interaction of the specific segment of the peptide and the Spike protein. Fig. 2D shows the comparative results where chemisorption of the bioreceptor at 0.50 mM produced the highest values of ΔRct. At this concentration, a suitable amount of peptide should be immobilized on the electrode surface and be accessible for interaction with the Spike protein in solution. Furthermore, a high peptide density can improve surface blocking and decrease nonspecific adsorption of proteins; therefore, such concentration was selected for the following experiments.
Fig. 2.
Nyquist plots recorded using the redox probe 5 mM [Fe(CN)6]3-/4-/KNO3. Frequency from 50 kHz to 0.05 Hz and amplitude 10 mV. Peptide concentration: A) 0.25, B) 0.50 and C) 0.70 mM. D) The difference in the charge transfer resistance for the different concentrations of the synthetic peptide, with 0.01 mM MCH and interaction with 1.0 μg/mL Spike protein.
The peptide incubation time was evaluated in a range from 30 to 120 min and overnight, finding a higher ΔRct value for 60 min incubation time (See Fig. S2A in SI), in correlation with the formation of a well-organized monolayer at the electrode surface. The concentration and blocking time with MCH were also systematically optimized after modifying the electrode-anchored peptide and comparing its interaction with the Spike protein. The difference in charge transfer resistance was greater for concentrations of 0.01 mM MCH after 10 min of blocking (See Fig. S2 B–C in SI). Concentrations higher than 0.01 mM MCH may generate over reorganization of the monolayer and displacement of the peptide molecules that may move away from the transducer, affecting the Spike protein recognition event.
Finally, although the changes in ΔRct studied at different incubation times at 37 °C showed a maximum value at 30 min, 15 min produced comparable results and better than the longer time at 25 °C (See Fig. S2D in SI). Therefore, the incubation temperature selected for the subsequent tests was 37 °C.
At lower temperatures, the RBD protein has a closed conformation that disables binding to the peptide due to its burying receptor-binding residues [59]. In summary, the optimized conditions are the incubation of the SPAuE in 0.5 mM of peptide for 60 min, blocking with 0.01 mM MCH for 10 min and interaction with Spike protein solutions for 15 min at 37 °C, indicating the easiness of the biosensor assembly of the format proposed and the possibility of a reliable interrogation of the Spike protein concentration in only 15 min once the electrodes are sensitized.
3.3. Direct detection of spike protein using the impedimetric biosensor
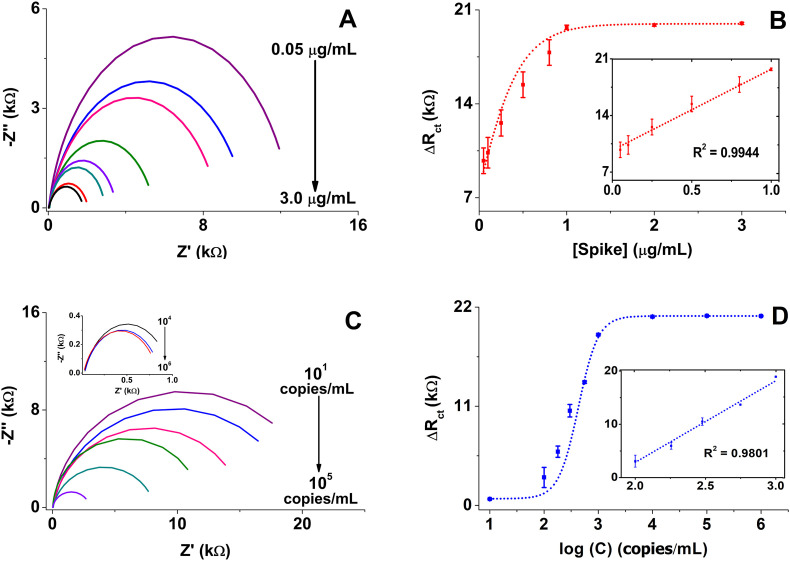
Having the analytical parameters optimized, we evaluated the analytical performance of the peptide-based biosensor by analyzing different dilutions of the Spike protein ranging from 0.05 to 3.0 μg/mL as detailed in the Materials and Methods. Remarkably, charge transference resistance (Rct) decreased (Fig. 3 A) and the difference in charge transfer resistance (ΔRct) increased (Fig. 3B) with increasing concentrations of Spike protein solution due to the reorganization of the peptide structure during the Spike protein biorecognition event, as mentioned above.
Fig. 3.
A) and C) Nyquist plots obtained in 5 mM [Fe(CN)6]3-/4-/KNO3 as redox probe, with 50 kHz to 0.05 Hz frequency range and 10 mV amplitude at 0.05, 0.1, 0.25, 0.5, 0.8, 1.0, 2.0 and 3.0 μg/mL Spike protein and 101, 102, 1.8 × 102, 3.0 × 102, 5.6 × 102, 103 copies/mL and Inset: 104, 105, 106 copies/mL lysate SARS-CoV-2 viral particles, respectively; B) and D) The resultant calibration curves and inset is the linear regression between the concentration of Spike protein and lysate SARS-CoV-2 viral particles, respectively. Bars are mean values from 3 independent biosensors.
The data in Fig. 3B show a trend line, with the inset showing the linear dependence described by the equation ΔRct = 10017 [Spike] + 9702 and a correlation coefficient R2 = 0.9944. The peptide-based biosensor showed a high dependence on the concentration of Spike protein with a linear response of 0.05–1.0 μg/mL, a sensitivity of 10.2 kΩ mL/μg and a LOD of 18.2 ng/mL, whose LOD are higher than 1 fg/mL [60] and 2.9 ng/mL and 14.6 μA mL/ng [61], but comparable with 66 pg/mL and 14.9%/nM [62]. Remarkably, the signal of our peptide-based biosensor was Spike protein concentration-dependent after only 15 min sample incubation.
In an attempt to approach a real scenario, we construct a calibration curve estimating the ΔRct for viral particles of SARS-CoV-2 inactivated by UV-light (Fig. 3C and D). The data showed a linear correlation between 102 to 103 copies/mL with LOD of 0.01 copies/mL described by the equation ΔRct = 15308.5 log C – 27759.1 and a correlation coefficient R2 = 0.9801 (Inset Fig. 3D). Considering the Spike protein levels [63,64] and viral particles on human samples [14]; our results indicate that the peptide-based biosensor can detect Spike protein in solution and viral particles at clinically relevant concentrations as will be shown below.
3.4. Specificity, storage stability, reproducibility, and repeatability studies
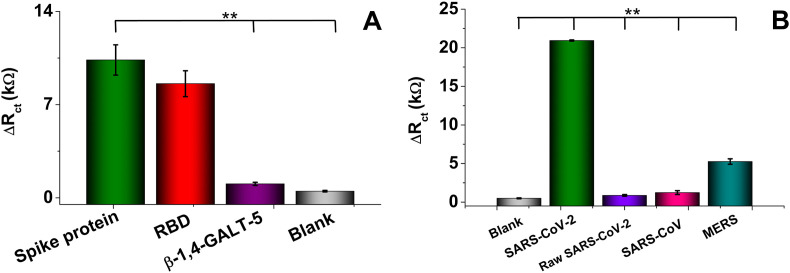
The specificity of the peptide-based biosensor is essential to reduce or avoid a false positive assignment. We considered herein 0.1 μg/mL of solutions that contain the Spike protein and compared it with those with RBD or β-1,4-GALT-5 glycosylated protein (Fig. 4 A). The Spike protein is a transmembrane protein containing two subunits, S1 and S2. S1 mainly contains an RBD responsible for recognizing the ACE-2 host cell surface receptor, and the S2 region contains essential elements needed for membrane fusion once in the host cell [65]. The platform's resultant ΔRct against the Spike protein and RBD, measured by EIS, exhibited a similar response with no statistically significant differences (p values are greater than 0.01). It means the RBD-peptide interaction of free RBD and RBD in the S1 region of the Spike protein are comparable. In contrast, the response was differential concerning β-1,4-GALT-5 and negative control with a very low β-1,4-GALT-5-peptide interaction, which could be explained by the presence of sialic acid in the β-1,4-GALT-5 [66] known as a receptor that binds to the S1 region of the Spike protein [65]. Subsequently, we estimated the specificity of the synthetic peptide-based biosensor against lysates from MERS and SARS-CoV, showing a high response when interrogated with 104 copies/μL of lysed SARS-CoV-2 viral particles respect raw SARS-CoV-2, lysed MERS and SARS-CoV, with statistically significant differences (p < 0.01) (Fig. 4B). In contrast, raw SARS-CoV-2, SARS-CoV, and negative control showed a similar response with no statistically significant differences (p > 0.01), indicating the need for lysis to facilitate the peptide-Spike protein interaction. Yet, overall, the results demonstrate the high specificity of the peptide-sensitized transducer platform only for the Spike protein (in lysed solutions).
Fig. 4.
Performance of peptide-based biosensor in the presence of analogous proteins and viruses. A) Response of buffer spiked with 0.1 μg/mL Spike protein, RBD, and β-1,4-GALT-5; and B) Specificity assessment in the supernatant of 104 copies/mL viral particles (raw and lysate) SARS-CoV-2 and lysate SARS-CoV and MERS samples. Statistical significance was obtained using one-way ANOVA with adjusted p-values of p < 0.01 (∗∗).
Long-term stability is also a significant feature in developing biosensors, especially when thinking about a translational approach. Next, the stability of the resulting peptide-based biosensor was analyzed; the modified-electrodes were stored at 4 °C in a humid chamber and eventually tested their response with 0.1 μg/mL Spike protein solutions for 30 days and analyzed based on the 3σ criteria (See Fig. S3A in SI). The stability was estimated by potting a control chart considering the first day of the study as the initial value. The upper and lower control limits were set at three times this value's standard deviation (3σ). The finding confirms that the response of the peptide-based biosensors was in between the control limits and retained 73.6% of its initial response when tested up to 20 days of storage, which reveals the relative long-term stability of the biosensors related to the mixed self-assembled monolayer at the modified-electrodes surface.
Finally, we checked the reproducibility and repeatability of the developed biosensor by interrogating a 0.1 μg/mL Spike solution. Five peptide-based biosensors were prepared independently and their impedimetric responses were measured, three times each, under identical experimental conditions (See Fig. S3B in SI). The intra- and inter-electrode relative standard deviation (RSD) was estimated to be 4.1 (n = 5) and 2.2% (n = 3), which indicates the high repeatability and reproducibility of the biosensors developed, respectively.
3.5. Performance of peptide-based biosensor in clinical samples
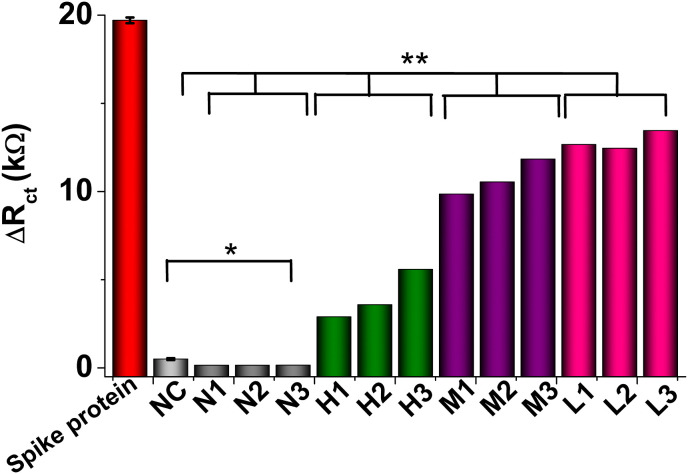
The biosensor was challenged with nasopharyngeal swab samples from nine COVID-19 positive patient samples and three negative samples. The clinical diagnostic feasibility of the peptide-based biosensor was evaluated by analyzing the lysed diluted samples with different CT (cycle threshold) values, tested by RT-PCR, as reported in Tables 2 and SI. It is important to remark that RT-PCR is the gold standard technique for diagnosing COVID-19 based on detecting viral RNA from SARS-CoV-2 and used herein to estimate the samples' viral loading, i.e., lower the CT higher the viral loading. Rather than RNA detection, our peptide-based device was designed to detect the Spike protein from viral swab particles (Table 2 in SI). Therefore, although results from RT-PCR can't be directly compared with ours, they are clear evidence of a positive or negative sample. Fig. 5 shows that the biosensor can differentiate between positive and negative nasopharyngeal swab samples concerning the controls and differentiate the viral loading, i.e., high, middle, or low. It was evident that the biosensor is responding to the presence of Spike protein in COVID-19 positive samples with statistically significant differences (p < 0.01) concerning the negative samples and the negative control and no statistically significant differences between the negative samples and the negative control (p > 0.01). These results agree with those from RT-PCR, thus demonstrating the biosensor's great potential for detecting the Spike protein from SARS-CoV-2 and its correlation with a rapid diagnosis of COVID-19.
Fig. 5.
Analysis of nasopharyngeal swabs samples. Signal change measured with the peptide-based biosensor using positive and negative for COVID-19 samples. The bars represent the mean charge transfer resistance change (Rct) extracted from the Nyquist plot for a group of samples with similar CT measured by RT-PCR. ∗∗ Indicates positive samples with statistically significant differences (p < 0.01) concerning the negative samples and the negative control and ∗ no statistically significant differences between the negative samples and the negative control (p > 0.01).
4. Conclusion
We developed the first peptide-based impedimetric biosensor that detected and quantified Spike protein and viral particles of SARS-CoV-2 in human samples, straightforwardly in only 15 min. The sensing platform uses a synthetic peptide inspired by the interaction of the RBD region of the SARS-CoV-2 and the ACE-2 host receptor protein minimizing unspecific interactions with other proteins. The high performance of the label-free peptide-based biosensor detected a concentration of Spike protein and viral particles of SARS-CoV-2 of clinical relevance. The resultant device recognized Spike proteins in clinical swab samples and discriminated patients diagnosed with COVID-19 and healthy individuals. Overall, the peptide-based biosensor can sensitively and selectively detect the SARS-CoV-2 Spike protein and viral particles from clinical samples. Since the method can be performed with portable voltammetric analyzers, it can provide significant potential as point-of-care systems in the COVID-19 testing and be employed by mobile healthcare teams.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Dayana Soto reports financial support was provided by the University of Antioquia - Colombia Ministry of Science Technology and Innovation. Jahir Orozco reports financial support was provided by the University of Antioquia - Colombia Ministry of Science Technology and Innovation.
Acknowledgments
JO and DS thank the financial support provided by MINCIENCIAS, the University of Antioquia and the Max Planck Society through the Cooperation agreement 566-1, 2014; and EPM and Ruta N for hosting the Max Planck Tandem Groups. The authors acknowledge MINCIENCIAS for partial support from the project Nanobiosensors to detect SARS-CoV-2 rapidly (Cod. 1115101576765). The authors also thank the Tropical Diseases Study and Control Program (PECET) laboratory, the Immunovirology and the Gastrohepatology Groups from the University of Antioquia for donating the clinical samples and for letting us use the BSL-2 facilities, respectively.
Biographies
Dayana Soto is a chemist, M.Sc. in Chemical Sciences and currently a Ph.D. student in Chemical Sciences at the University of Antioquia (UdeA, Colombia). Since 2017, she has been part of the electrochemical biosensing research line from the Max Planck Tandem Group in Nanobioengineering at the UdeA. Her areas of interest include nano(bio)sensors, electrochemistry, nanomaterials, enzymatic catalysis, surface chemistry, characterization techniques and development of analytical detection tools of biomedical interest.
Jahir Orozco is a chemist from the University of Antioquia (Colombia), with a Ph.D. in chemistry from the University of Barcelona and the Institute for Microelectronics of Barcelona (2008). He completed three postdoctoral stays for seven years at the Banyuls sur Mer Oceanological Observatory (France), the Department of Nanoengineering at UCSD (USA) and the Catalan Institute for Nanoscience and Nanotechnology (Spain). He is currently the group leader of the Max Planck Tandem Group in Nanobioengineering (UdeA) and an editorial board member of the Molecules journal. His areas of interest include nano(bio)technology, nano(micro)carriers, nanomotors, nano (micro)devices, nanomaterials, (bio)sensors, electrochemistry, and the development of analytical tools for environmental and biomedical applications.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aca.2022.339739.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Idili A., Parolo C., Alvarez-Diduk R., Merkoçi A. Rapid and efficient detection of the SARS-CoV-2 Spike protein using an electrochemical aptamer-based sensor. ACS Sens. 2021;6:3093–3101. doi: 10.1021/acssensors.1c01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Chailapakul O., Chaiyo S. Paper-based electrochemical biosensor for diagnosing COVID-19: detection of SARS-CoV-2 antibodies and antigen, Biosens. Bioelectron. 2021;176 doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yousefi H., Mahmud A., Chang D., Das J., Gomis S., Chen J.B., Wang H., Been T., Yip L., Coomes E., Li Z., Mubareka S., Mcgeer A., Christie N., Gray-Owen S., Cochrane A., Rini J.M., Sargent E.H., Kelley S.O. Detection of SARS-CoV-2 viral particles using direct, reagent-free electrochemical sensing. Cite This J. Am. Chem. Soc. 2021;143:1722–1727. doi: 10.1021/jacs.0c10810. [DOI] [PubMed] [Google Scholar]
- 4.Zhang G., Pomplun S., Loftis A.R., Tan X., Loas A., Pentelute B.L. The first-in-class peptide binder to the SARS-CoV-2 Spike protein. bioRxiv. 2020:1–20. [Google Scholar]
- 5.Zhang M., Li X., Pan J., Zhang Y., Zhang L., Wang C., Yan X., Liu X., Lu G. Ultrasensitive detection of SARS-CoV-2 spike protein in untreated saliva using SERS-based biosensor. Biosens. Bioelectron. 2021;190 doi: 10.1016/j.bios.2021.113421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mojsoska B., Larsen S., Olsen D.A., Madsen J.S., Brandslund I., Alatraktchi F.A. Rapid SARS-CoV-2 detection using electrochemical immunosensor. Sensors. 2021;21:1–11. doi: 10.3390/s21020390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yousefi H., Mahmud A., Chang D., Das J., Gomis S., Chen J.B., Wang H., Been T., Yip L., Coomes E., Li Z., Mubareka S., Mcgeer A., Christie N., Gray-Owen S., Cochrane A., Rini J.M., Sargent E.H., Kelley S.O. Detection of SARS-CoV-2 viral particles using direct, reagent-free electrochemical sensing. J. Am. Chem. Soc. 2021;143:1722–1727. doi: 10.1021/jacs.0c10810. [DOI] [PubMed] [Google Scholar]
- 8.Aydın E.B., Aydın M., Sezgintürk M.K. New impedimetric sandwich immunosensor for ultrasensitive and highly specific detection of spike receptor binding domain protein of SARS-CoV-2. ACS Biomater. Sci. Eng. 2021;7:3874–3885. doi: 10.1021/acsbiomaterials.1c00580. [DOI] [PubMed] [Google Scholar]
- 9.Witt S., Rogien A., Werner D., Siegenthaler J., Lesiyon R., Kurien N., Rechenberg R., Baule N., Hardy A., Becker M. Boron doped diamond thin films for the electrochemical detection of SARS-CoV-2 S1 protein. Diam. Relat. Mater. 2021;118 doi: 10.1016/j.diamond.2021.108542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abrego-Martinez J.C., Jafari M., Chergui S., Pavel C., Che D., Siaj M. Aptamer-based electrochemical biosensor for rapid detection of SARS-CoV-2: nanoscale electrode-aptamer-SARS-CoV-2 imaging by photo-induced force microscopy. Biosens. Bioelectron. 2022;195 doi: 10.1016/j.bios.2021.113595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z., Pandey R., Li J., Gu J., White D., Stacey H.D., Ang J.C., Steinberg C.-J., Capretta A., Filipe C.D.M., Mossman K., Balion C., Miller M.S., Salena B.J., Yamamura D., Soleymani L., Brennan J.D., Li Y. High affinity dimeric aptamers enable rapid electrochemical detection of Wild-Type and B.1.1.7 SARS-CoV-2 in unprocessed saliva. Angew. Chem. Int. Ed. 2021;60:24266–24274. doi: 10.1002/anie.202110819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X., Qin Z., Fu H., Li T., Peng R., Li Z., Rini J.M., Liu X. Enhancing the performance of paper-based electrochemical impedance spectroscopy nanobiosensors: an experimental approach. Biosens. Bioelectron. 2021;177 doi: 10.1016/j.bios.2020.112672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soares J.C., Soares A.C., Rodrigues V.C., Oiticica P.R.A., Raymundo-Pereira P.A., Bott-Neto J.L., Buscaglia L.A., de Castro L.D.C., Ribas L.C., Scabini L., Brazaca L.C., Correa D.S., Mattoso L.H.C., de Oliveira M.C.F., de Carvalho A.C.P.L.F., Carrilho E., Bruno O.M., Melendez M.E., Oliveira O.N. Detection of a SARS-CoV-2 sequence with genosensors using data analysis based on information visualization and machine learning techniques. Mater. Chem. Front. 2021;5:5658–5670. [Google Scholar]
- 14.Rashed M.Z., Kopechek J.A., Priddy M.C., Hamorsky K.T., Palmer K.E., Mittal N., Valdez J., Flynn J., Williams S.J. Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaccariotto G.C., Silva M.K.L., Rocha G.S., Cesarino I. A novel Method for the detection of SARS-CoV-2 based on graphene-impedimetric immunosensor. Materials. 2021;14:4230. doi: 10.3390/ma14154230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avelino K.Y.P.S., dos Santos G.S., Frías I.A.M., Silva-Junior A.G., Pereira M.C., Pitta M.G.R., de Araújo B.C., Errachid A., Oliveira M.D.L., Andrade C.A.S. Nanostructured sensor platform based on organic polymer conjugated to metallic nanoparticle for the impedimetric detection of SARS-CoV-2 at various stages of viral infection. J. Pharm. Biomed. Anal. 2021;206 doi: 10.1016/j.jpba.2021.114392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ali M.A., Hu C., Jahan S., Yuan B., Saleh M.S., Ju E., Gao S.-J., Panat R. Sensing of COVID-19 antibodies in seconds via aerosol Jet nanoprinted reduced-graphene-oxide-coated 3D electrodes. Adv. Mater. 2021;33 doi: 10.1002/adma.202006647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Echeverri D., Garg M., Varón Silva D., Orozco J. Phosphoglycan-sensitized platform for specific detection of anti-glycan IgG and IgM antibodies in serum. Talanta. 2020;217 doi: 10.1016/j.talanta.2020.121117. [DOI] [PubMed] [Google Scholar]
- 19.Kiew L.V., Chang C.Y., Huang S.Y., Wang P.W., Heh C.H., Te Liu C., Cheng C.H., Lu Y.X., Chen Y.C., Huang Y.X., Chang S.Y., Tsai H.Y., Kung Y.A., Huang P.N., Hsu M.H., Leo B.F., Foo Y.Y., Su C.H., Hsu K.C., Huang P.H., Ng C.J., Kamarulzaman A., Yuan C.J., Bin Shieh D., Shih S.R., Chung L.Y., Chang C.C. Development of flexible electrochemical impedance spectroscopy-based biosensing platform for rapid screening of SARS-CoV-2 inhibitors. Biosens. Bioelectron. 2021;183 doi: 10.1016/j.bios.2021.113213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torres M.D.T., de Araujo W.R., de Lima L.F., Ferreira A.L., de la Fuente-Nunez C. Low-cost biosensor for rapid detection of SARS-CoV-2 at the point of care. Matter. 2021;4:2403–2416. doi: 10.1016/j.matt.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ehsan M.A., Khan S.A., Rehman A. Screen-printed graphene/carbon electrodes on Paper substrates as impedance sensors for detection of coronavirus in nasopharyngeal fluid samples. Diagnostics. 2021;11:1030. doi: 10.3390/diagnostics11061030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Idili A., Parolo C., Alvarez-Diduk R., Merkoçi A. Rapid and efficient detection of the SARS-CoV-2 Spike protein using an electrochemical aptamer-based sensor. ACS Sens. 2021;6:3093–3101. doi: 10.1021/acssensors.1c01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muñoz J., Pumera M. 3D-Printed COVID-19 immunosensors with electronic readout. Chem. Eng. J. 2021;425 doi: 10.1016/j.cej.2021.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Il Kim S. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 25.Karakuş E., Erdemir E., Demirbilek N., Liv L. Colorimetric and electrochemical detection of SARS-CoV-2 spike antigen with a gold nanoparticle-based biosensor. Anal. Chim. Acta. 2021;1182 doi: 10.1016/j.aca.2021.338939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahmati Z., Roushani M., Hosseini H., Choobin H. Electrochemical immunosensor with Cu2O nanocube coating for detection of SARS-CoV-2 spike protein. Microchim. Acta. 2021;188:1–9. doi: 10.1007/s00604-021-04762-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fabiani L., Saroglia M., Galatá G., De Santis R., Fillo S., Luca V., Faggioni G., D'Amore N., Regalbuto E., Salvatori P., Terova G., Moscone D., Lista F., Arduini F. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: a reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yousefi H., Mahmud A., Chang D., Das J., Gomis S., Chen J.B., Wang H., Been T., Yip L., Coomes E., Li Z., Mubareka S., Mcgeer A., Christie N., Gray-Owen S., Cochrane A., Rini J.M., Sargent E.H., Kelley S.O. Detection of SARS-CoV-2 viral particles using direct, reagent-free electrochemical sensing. J. Am. Chem. Soc. 2021;143:1722–1727. doi: 10.1021/jacs.0c10810. [DOI] [PubMed] [Google Scholar]
- 29.Shao W., Shurin M.R., Wheeler S.E., He X., Star A. Rapid detection of SARS-CoV-2 antigens using high-purity semiconducting single-walled carbon nanotube-based field-effect transistors. ACS Appl. Mater. Interfaces. 2021;13:10321–10327. doi: 10.1021/acsami.0c22589. [DOI] [PubMed] [Google Scholar]
- 30.Singh N.K., Ray P., Carlin A.F., Magallanes C., Morgan S.C., Laurent L.C., Aronoff-Spencer E.S., Hall D.A. Hitting the diagnostic sweet spot: point-of-care SARS-CoV-2 salivary antigen testing with an off-the-shelf glucometer. Biosens. Bioelectron. 2021;180 doi: 10.1016/j.bios.2021.113111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vezza V.J., Butterworth A., Lasserre P., Blair E.O., MacDonald A., Hannah S., Rinaldi C., Hoskisson P.A., Ward A.C., Longmuir A., Setford S., Farmer E.C.W., Murphy M.E., Corrigan D.K. An electrochemical SARS-CoV-2 biosensor inspired by glucose test strip manufacturing processes. Chem. Commun. 2021;57:3704–3707. doi: 10.1039/d1cc00936b. [DOI] [PubMed] [Google Scholar]
- 32.Vadlamani B.S., Uppal T., Verma S.C., Misra M. Functionalized TiO2 nanotube-based electrochemical biosensor for rapid detection of SARS-CoV-2. Sensors. 2020;20:5871. doi: 10.3390/s20205871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao H., Liu F., Xie W., Zhou T.C., OuYang J., Jin L., Li H., Zhao C.Y., Zhang L., Wei J., Zhang Y.P., Li C.P. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sensor. Actuator. B Chem. 2021;327 doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farzin L., Sadjadi S., Sheini A. A nanoscale genosensor for early detection of COVID-19 by voltammetric determination of RNA-dependent RNA polymerase (RdRP) sequence of SARS-CoV-2 virus. Mikrochim. Acta. 2021;188:1–12. doi: 10.1007/s00604-021-04773-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alafeef M., Dighe K., Moitra P., Pan D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano. 2020;14:17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng Y., Pan Y., Sun Z., Li J., Yi Y., Yang J., Li G. An electrochemical biosensor for sensitive analysis of the SARS-CoV-2 RNA. Biosens. Bioelectron. 2021;186 doi: 10.1016/j.bios.2021.113309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cajigas S., Alzate D., Orozco J. Gold nanoparticle/DNA-based nanobioconjugate for electrochemical detection of Zika virus. Microchim. Acta. 2020;187:1–10. doi: 10.1007/s00604-020-04568-1. [DOI] [PubMed] [Google Scholar]
- 38.Alzate D., Cajigas S., Robledo S., Muskus C., Orozco J. Genosensors for differential detection of Zika virus. Talanta. 2020;210 doi: 10.1016/j.talanta.2019.120648. [DOI] [PubMed] [Google Scholar]
- 39.Orozco J., Medlin L.K. Electrochemical performance of a DNA-based sensor device for detecting toxic algae. Sensor. Actuator. B Chem. 2011;153:71–77. [Google Scholar]
- 40.Hasan J., Pyke A., Nair N., Yarlagadda T., Will G., Spann K., Yarlagadda P.K.D.V. Antiviral nanostructured surfaces reduce the viability of SARS-CoV-2. ACS Biomater. Sci. Eng. 2020;6:4858–4861. doi: 10.1021/acsbiomaterials.0c01091. [DOI] [PubMed] [Google Scholar]
- 41.Kaya S.I., Ozcelikay G., Mollarasouli F., Bakirhan N.K., Ozkan S.A. Recent achievements and challenges on nanomaterial based electrochemical biosensors for the detection of colon and Lung cancer biomarkers. Sensor. Actuator. B Chem. 2022;351 [Google Scholar]
- 42.Torrente-Rodríguez R.M., Martín C.M.-S., Gamella M., Pedrero M., Martínez-Bosch N., Navarro P., de Frutos P.G., Pingarrón J.M., Campuzano S. Electrochemical immnosensing of ST2: a checkpoint target in Cancer diseases. Biosensors. 2021;11:1–13. doi: 10.3390/bios11060202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghorbani F., Abbaszadeh H., Mehdizadeh A., Ebrahimi-Warkiani M., Rashidi M.-R., Yousefi M. Biosensors and nanobiosensors for rapid detection of autoimmune diseases: a review. Microchim. Acta. 2019;186:1–11. doi: 10.1007/s00604-019-3844-4. 2019 18612. [DOI] [PubMed] [Google Scholar]
- 44.Kumari V., Rastogi S., Sharma V. Nanotechnol. Life Sci.; 2019. Emerging Trends in Nanobiosensor; pp. 419–447. [Google Scholar]
- 45.Liu Q., Wang J., Boyd B.J. Peptide-based biosensors. Talanta. 2015;136:114–127. doi: 10.1016/j.talanta.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 46.Karimzadeh A., Hasanzadeh M., Shadjou N., de la Guardia M. Peptide based biosensors. TrAC Trends Anal. Chem. 2018;107:1–20. [Google Scholar]
- 47.Orozco J., Jiménez-Jorquera C., Fernández-Sánchez C. Gold nanoparticle-modified ultramicroelectrode arrays for biosensing: a comparative assessment. Bioelectrochemistry. 2009;75:176–181. doi: 10.1016/j.bioelechem.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 48.Wan J., Xing S., Ding L., Wang Y., Gu C., Wu Y., Rong B., Li C., Wang S., Chen K., He C., Zhu D., Yuan S., Qiu C., Zhao C., Nie L., Gao Z., Jiao J., Zhang X., Wang X., Ying T., Wang H., Xie Y., Lu Y., Xu J., Lan F. Human-IgG-neutralizing monoclonal antibodies block the SARS-CoV-2 infection. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang J., Petitjean S., Derclaye S., Koehler M., Zhang Q., Dumitru A., Soumillion P., Alsteens D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020;11:1–10. doi: 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang G., Pomplun S., Loftis A.R., Loas A., Pentelute B.L. The first-in-class peptide binder to the SARS-CoV-2 Spike protein. bioRxiv. 2020;1–19 [Google Scholar]
- 52.Ferreira D.C., Batistuti M.R., Bachour B., Mulato M. Aptasensor based on screen-printed electrode for breast cancer detection in undiluted human serum. Bioelectrochemistry. 2021;137 doi: 10.1016/j.bioelechem.2020.107586. [DOI] [PubMed] [Google Scholar]
- 53.Hashem A., Hossain M.A.M., Marlinda A.R., Al Mamun M., Simarani K., Johan M.R. Nanomaterials based electrochemical nucleic acid biosensors for environmental monitoring: a review. Appl. Surf. Sci. Adv. 2021;4 [Google Scholar]
- 54.Xia N., Wang X., Yu J., Wu Y., Cheng S., Xing Y., Liu L. Design of electrochemical biosensors with peptide probes as the receptors of targets and the inducers of gold nanoparticles assembly on electrode surface. Sensor. Actuator. B Chem. 2017;239:834–840. [Google Scholar]
- 55.Dominguez-Benetton X., Sevda S., Vanbroekhoven K., Pant D. The accurate use of impedance analysis for the study of microbial electrochemical systems. Chem. Soc. Rev. 2012;41:7228–7246. doi: 10.1039/c2cs35026b. [DOI] [PubMed] [Google Scholar]
- 56.Kumar B., Bhalla V., Singh Bhadoriya R.P., Suri C.R., Varshney G.C. Label-free electrochemical detection of malaria-infected red blood cells. RSC Adv. 2016;6:75862–75869. [Google Scholar]
- 57.Singal S., Srivastava A.K., Dhakate S., Biradar A.M., Rajesh R. Electroactive graphene-multi-walled carbon nanotube hybrid supported impedimetric immunosensor for the detection of human cardiac troponin-I. RSC Adv. 2015;5:74994–75003. [Google Scholar]
- 58.Soto D., Alzate M., Gallego J., Orozco J. Electroanalysis of an Iron@Graphene-carbon nanotube hybrid material. Electroanalysis. 2018;30 [Google Scholar]
- 59.Rath S.L., Kumar K. Investigation of the effect of temperature on the structure of SARS-CoV-2 Spike protein by molecular dynamics simulations. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.583523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mavrikou S., Moschopoulou G., Tsekouras V., Kintzios S. Development of a portable, ultra-rapid and ultra-sensitive cell-based biosensor for the direct detection of the SARS-CoV-2 S1 spike protein antigen. Sensors. 2020;20:3121. doi: 10.3390/s20113121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beduk T., Beduk D., Ilton De Oliveira Filho J., Zihnioglu F., Cicek C., Sertoz R., Arda B., Goksel T., Turhan K., Salama K.N., Timur S. Rapid point-of-care COVID-19 diagnosis with a gold-nanoarchitecture-assisted laser-scribed graphene biosensor. Anal. Chem. 2021;93:8585–8594. doi: 10.1021/acs.analchem.1c01444. [DOI] [PubMed] [Google Scholar]
- 62.Abrego-Martinez J.C., Jafari M., Chergui S., Pavel C., Che D., Siaj M. Aptamer-based electrochemical biosensor for rapid detection of SARS-CoV-2: nanoscale electrode-aptamer-SARS-CoV-2 imaging by photo-induced force microscopy. Biosens. Bioelectron. 2022;195 doi: 10.1016/j.bios.2021.113595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song G., He W., Callaghan S., Anzanello F., Huang D., Ricketts J., Torres J.L., Beutler N., Peng L., Vargas S., Cassell J., Parren M., Yang L., Ignacio C., Smith D.M., Voss J.E., Nemazee D., Ward A.B., Rogers T., Burton D.R., Andrabi R. Cross-reactive serum and memory B-cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. Nat. Commun. 2021;12:1–10. doi: 10.1038/s41467-021-23074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C.B.E.M., Bosch B.-J., Drosten C., Koopmans M.P.G., Haagmans B.L. Severe Acute respiratory Syndrome coronavirus 2−Specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 2020;26:1478. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen S., Tan T.H.P., Tan Y.-J. Expression, glycosylation, and modification of the spike (S) glycoprotein of SARS CoV. Methods Mol. Biol. 2007;379:127–135. doi: 10.1007/978-1-59745-393-6_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shishkanova T.V., Broncová G., Němečková Z., Vrkoslav V., Král V., Matějka P. Molecular frameworks of polymerized 3-aminobenzoic acid for chemical modification and electrochemical recognition. J. Electroanal. Chem. 2019;832:321–328. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.