CRISPR editing of T cells shows safety and promise in improving the effectiveness of gene therapy in a small trial of three cancer patients.
In a recent address to the annual meeting for the American Association for the Advancement of Science, Bill Gates touted the myriad of advances that gene therapies could bring to the global health-care market. In particular, he hailed CRISPR as one of the catalysts bringing modern science to a new era of discovery and personalized medical solutions.1
As if on cue, Carl June's team at the Abramson Cancer Center of the University of Pennsylvania, led by Edward Stadtmauer, have published promising results from their first-in-human, Phase I clinical trial of “CRISPR-engineered T cells in patients with refractory cancer.”2 The report was featured on the cover of Science under the headline “Human CRISPR.”
To assess the safety and feasibility of CRISPR-modified cancer immunotherapy, the Penn team delivered transgenic T-cell receptor (TCR) specific for the NY-ESO-1 tumor antigen (found in myeloma, melanoma, and sarcoma) to autologous T cells derived from three seriously ill cancer patients (Fig. 1). To increase efficacy further and decrease the toxicity of engineered T cells, this was performed in tandem with delivery of CRISPR ribonucleoprotein complexes containing guide RNAs (gRNA) targeting two TCR chains—TCRα (TRAC) and TCRβ (TRBC)—as well as programmed death protein-1 (PDCD1) genes.2 This parallel strategy was devised to decrease the chance of self-reactive T cells by eliminating mis-pairing of NY-ESO-1 TCR with endogenous TCRα/β chains. Moreover, disruption of PDCD1 was exercised to reduce T-cell exhaustion over time.
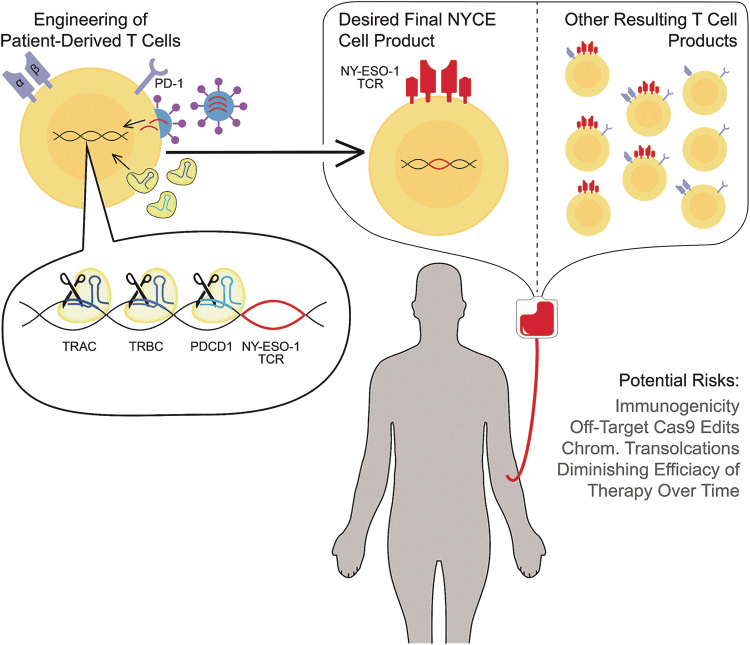
FIG. 1.
Multiplex CRISPR-Cas9 and lentiviral editing of T cells. Schematic of genetic engineering of patient-derived T cells depicting lentiviral transduction of the NY-ESO-1 TCR transgene and delivery of Cas9/gRNA ribonucleoprotein complexes targeting genes for endogenous TCR chains—TCRα and TCRβ—and PD-1 (TRAC, TRBC, and PDCD1, respectively). The final desired T cell product has a synthetic NY-ESO-1 TCR and no TCAR, TCBR, or PD-1, but other editing products include T cells with mixed edits. Important safety considerations for clinical application include chromosomal translocations and off-target Cas9 edits, as well as immune response to the delivered products. gRNA, guide RNA; TCR, T-cell receptor; PD-1, programmed death protein-1; Chrom., Chromosomal. (Image adapted from Stadtmauer et al.2)
For medical applications, reducing the chance of toxicity of infused cell products is the top priority. Our existing CRISPR toolset will most likely cause some degree of off-target indels or translocations in the genome of final cellular products, requiring decisions to be made about the trade-offs of those edits. Stringent measures need to be in place to detect them and perform long-term follow-up of patients to include examination of those edits.
Moreover, due to the risk of preexisting immune response against Cas proteins, additional precautions need to be in place before or during infusion. These include screening for preexisting humoral and cellular immune response in patients, concurrent transient immunosuppression, eliminating the remaining traces of CRISPR ribonucleoproteins before transfusion, as well as developing less immunogenic Cas9 variants.3 Along these lines, the Penn team evaluated NY-ESO-1–transduced CRISPR 3X edited cells (NYCE) cells under several rigorous parameters before infusion. These criteria included evaluation of transduction efficiency of the NYE-ESO-1 TCR transgene, knockout frequency by multiplexed CRISPR-Cas9, on- and off-target editing, and residual presence of Cas9.
Although off-target mutations for gRNAs were detected in certain genes (such as CLIC2), as the genes were silent in T cells, the edits were considered safe. In addition, chromosomal translocations were present in manufactured cells, but the frequency of these translocations diminished as manufacturing time increased, suggesting these translocations did not cause a growth advantage over an extended expansion period. The final T cell product showed minimal levels of Cas9 protein (<0.75 fg per cell), and the subjects did not exhibit humoral immunity to Cas9 after infusion.
Following treatment, NYCE cells did not elicit any serious adverse effects in patients. They showed up to 9 months of persistent engraftment (average decay half-life of 84 days), a significant improvement over TCR-only trials (1 week). This varied patient to patient, however, with editing and transduction efficiency correlating to steady-state levels of engraftment. Successful targeting of cancer cells was validated through bone marrow or tumor biopsies, indicating trafficking of NYCE cells to these locations.
As the study continued, the frequency of chromosomal translocations decreased to the limit of detection, save for the rearrangement of TRBC1:TRBC2, which created a 9.3 kb deletion. To characterize their therapy further, Stadtmauer et al. further evaluated their most successful patient (determined by engraftment rates and tumor regression) 4 months after infusion and found that 40% of this patient's peripheral blood-circulating T cells displayed at least one mutation out of the four target edits.2
The most successful outcome for this study was safe infusion of engineered T cells and stable integration 3–9 months after adoptive cell delivery. Although two patients received additional treatment for their cancer and one patient later succumbed to progressive disease, all patients safely tolerated the infusion of NYCE cells.
Despite the positive headlines, there is still much work to be done to improve this therapy for successful cancer treatment. As this clinical trial was originally approved in 2016, the therapy can benefit from recent CRISPR-Cas advances. As the most successful patient saw the highest level of gene-edited cells initially and in the long term, we can tentatively infer that higher levels of mutated cells can result in improvement of disease (in this case, the patient showed a sustained 50% decrease in a cancerous mass).
It is critical that future trials rely upon the use of improved Cas9 variants with higher editing fidelity and efficiency or improved delivery to ensure a more favorable outlook.4,5 Moreover, although the frequency of chromosomal translocations diminished over time in NYCE cells, it is still necessary to eliminate these instances wherever possible.
In all, while the Penn group's approach did not ultimately result in abatement of the refractory cancer in these patients, it did display the safe application of multiplexed CRISPR-Cas9 systems parallel to transgene transduction. That's a promising advance in the pursuit of effective cancer immunotherapies.
References
- 1. Gates B. My message to America's top scientists. GatesNotes, February 14, 2020. [Google Scholar]
- 2. Stadtmauer EA, Fraietta JA, Davis MM, et al. . CRISPR-engineered T cells in patients with refractory cancer. Science 2020;376:eaba7365. DOI: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ferdosi SR, Ewaisha R, Moghadam F, et al. . Multifunctional CRISPR-Cas9 with engineered immunosilenced human T cell epitopes. Nat Commun 2019;10:1842. DOI: 10.1038/s41467-019-09693-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee JK, Jeong E, Lee J, et al. . Directed evolution of CRISPR-Cas9 to increase its specificity. Nat Commun 2018;9:3048. DOI: 10.1038/s41467-018-05477-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang CH, Lee KC, Doudna JA. Applications of CRISPR-Cas enzymes in cancer therapeutics and detection. Trends Cancer 2018;4:499–512. DOI: 10.1016/j.trecan.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]