Abstract
The blood oxygenation level-dependent (BOLD)-based resting-state functional magnetic resonance imaging (rsfMRI) has been widely used as a non-invasive tool to map brain-wide connectivity architecture. However, the neural basis underpinning the resting-state BOLD signal remains elusive. In this study, we combined simultaneous calcium-based fiber photometry with rsfMRI in awake animals to examine the relationship of the BOLD signal and spiking activity at the resting state. We observed robust couplings between calcium and BOLD signals in the dorsal hippocampus as well as other distributed areas in the default mode network (DMN), suggesting that the calcium measurement can reliably predict the rsfMRI signal. In addition, using the calcium signal recorded as the ground truth, we assessed the impacts of different rsfMRI data preprocessing pipelines on functional connectivity mapping. Overall, our results provide important evidence suggesting that spiking activity measured by the calcium signal plays a key role in the neural mechanism of resting-state BOLD signal.
Keywords: Resting-state fMRI, GCaMP, Preprocessing, Awake, Rat
1. Introduction
Over the past three decades, the effort to study brain activity has benefited significantly from functional neuroimaging approaches. In particular, blood oxygenation level-dependent (BOLD) functional magnetic resonance imaging (fMRI) has been used as a popular non-invasive technique for mapping brain functional activity in humans and animals at both task and resting states. It is generally believed that the BOLD signal is spatiotemporally coupled to neural activity through neurovascular coupling (Hillman, 2014; Kim and Ogawa, 2012). However, the exact neural basis underpinning the BOLD signal during stimulated and resting states is still an area of intensive and extensive research.
Neural activity can be broadly considered as the combination of spiking activity and local field potential (LFP). Spiking activity primarily reflects the suprathreshold neuronal output whereas LFP results mostly from weighted average of synchronized synaptic inputs (Kim and Ogawa, 2012). Uncovering the exact relationship between different kind of neural activity and the BOLD signal is critical as it can not only provide insight into the neural mechanism of hemodynamic changes, but also assist us to better interpret functional patterns observed with fMRI. For instance, using simultaneous electrophysiology and fMRI in monkeys, Logothetis et al. showed that under visual stimulation, BOLD fMRI response better correlated with the corresponding LFP than multi-unit activity (MUA) (Logothetis et al., 2001; Logothetis and Wandell, 2004). This result suggests that the BOLD signal might better reflect synaptic input in neuronal activities than spiking output (but also see reports of BOLD correlations with spiking activity (Heeger and Ress, 2002; Rees et al., 2000), and correlations with both spiking and LFP activities (Mukamel et al., 2005) during task states).
Unlike relatively consistent results for the neural basis of the BOLD signal during tasks, findings regarding the neural underpinning of the BOLD signal at the resting state are more diverse. Different studies reported mixed neural correlates of resting-state BOLD signal including band-limited power of LFP in different frequency ranges, and modulations of spiking activity (He and Liu, 2008; Hiltunen et al., 2014; Logothetis et al., 2012; Lu et al., 2007; Mantini et al., 2007; Pan et al., 2013; Shmuel and Leopold, 2008; Thompson et al., 2014; Wang et al., 2012; Winder et al., 2017). Although in all these studies neural correlates with the resting-state BOLD signal were reported, it is far from being clear how the BOLD signal arises from spontaneous neural activity at rest. Elusive understanding of the neural origin of spontaneous hemodynamics has highlighted a critical gap in the interpretation of rsfMRI data in terms of the underlying neural activity.
To bridge this gap in rsfMRI research, a couple of obstacles need to be overcome. First, simultaneous fMRI and electrophysiology recording—the common method used to probe the coupling between the BOLD and neural activities (Logothetis et al., 2001; Mukamel et al., 2005; Rees et al., 2000; Viswanathan and Freeman, 2007), is technically challenging particularly at high magnetic field (Carmichael et al., 2012) due to large electrical artifacts generated in electrophysiological recordings during fMRI scans. Electromagnetic artifacts in electrophysiological data also make it difficult to obtain neuron-type information, which limits studies of cellular mechanisms underlying rsfMRI signal. Second, rigorous validation of specific rsfMRI data preprocessing pipelines based on the underlying neural activity is lacking, making the quantification of resting-state functional connectivity (RSFC) somewhat arbitrary. This problem is particularly prominent when studying spontaneous neural activity, as non-neural artifacts in the rsfMRI siganl cannot simply be averaged out based on the experimental paradigm (Florin and Baillet, 2015; Jorge et al., 2015).
Calcium-based fiber photometry has recently emerged as a popular technique to measure neural activity (Chen et al., 2019; He et al., 2018; Lake et al., 2020; Liang et al., 2017; Pais-Roldán et al., 2020; Schulz et al., 2012; Schwalm et al., 2017; Tong et al., 2019; Wang et al., 2018). It measures calcium-dependent fluorescence signal through optic fibers that mainly reflects spiking activity (Grienberger and Konnerth, 2012). With minimally invasive optical access of recorded cells, it allows convenient simultaneous neural-fMRI recording that is insensitive to MRI-related interference (Schlegel et al., 2018). In addition, well-established genetic tools allow GCaMP (i.e. a genetically encoded calcium fluorescent indicator) to be expressed in a specific neuron type (e.g. excitatory neurons or inhibitory neurons (Mollinedo-Gajate et al., 2019), or even astrocytes (Otsu et al., 2015; Wang et al., 2018)). This high cell-type specificity makes it possible to dissect the contributions of separate neuron populations to the BOLD signal. In addition, even though the subcellular accumulation of the GCaMP sensor may induce cell death in a small portion of neurons (Gu et al., 2018; Yang et al., 2018), calcium indicators can still be stable and show consistent sensitivity for months in most cells (Chen et al., 2013; Gu et al., 2018; Liang et al., 2017), which makes GCaMP a reliable tool that can be readily used in longitudinal studies.
As an extension of previous work (Schwalm et al., 2017; Wang et al., 2018), in the present study we utilized simultaneous GCaMP and fMRI recordings in awake animals (Liang et al., 2017; Ma et al., 2020) to investigate the potential relationship between spiking activity and BOLD signal at rest. Collecting multi-modal data at the awake state eliminates the confounding effects of anesthesia on both neuronal and vascular activities (Paasonen et al., 2020; Gao et al., 2017). Notably, aforementioned literature studies were predominantly conducted in anesthetized animals (He et al., 2018; Lake et al., 2020; Liang et al., 2017; Schwalm et al., 2017; Tong et al., 2019; Wang et al., 2018). The diversity in neural correlates of the rsfMRI signal reported likely result from differences in the type (Paasonen et al., 2018; Tsurugizawa and Yoshimaru, 2021) and dose (Hamilton et al., 2017; Ma et al., 2016) of anesthetics used, given that anesthesia profoundly changes both neural and vascular activities. Indeed, both short-term and long-term impact of anesthesia on the rsfMRI signal have been repeatedly demonstrated (Paasonen et al., 2018; Pradier et al., 2021; Stenrros et al., 2021). To fill this gap, here we investigate the coupling relationship between GCaMP and rsfMRI signals in the dorsal hippocampus (dHP). dHP was selected considering its dense reciprocal connections with the cortex and its pivotal role in cognitive function (Fanselow and Dong, 2010). Using the measured GCaMP signal as the ground truth, we further evaluate the efficacies of separate rsfMRI data preprocessing steps.
2. Methods and materials
2.1. Animals
Data used in this study were collected from 23 male Long Evans (LE) rats. Rats were obtained from Charles River Laboratory (Wilmington, MA) and housed in home cages for at least 7–10 days before surgery. Food and water were provided ad libitum. Room temperature was kept at 22–24 °C with a 12 h light: 12 h dark cycle. All experiments were approved by the Institutional Animal Care and Use Committee of the Pennsylvania State University.
2.2. Surgery
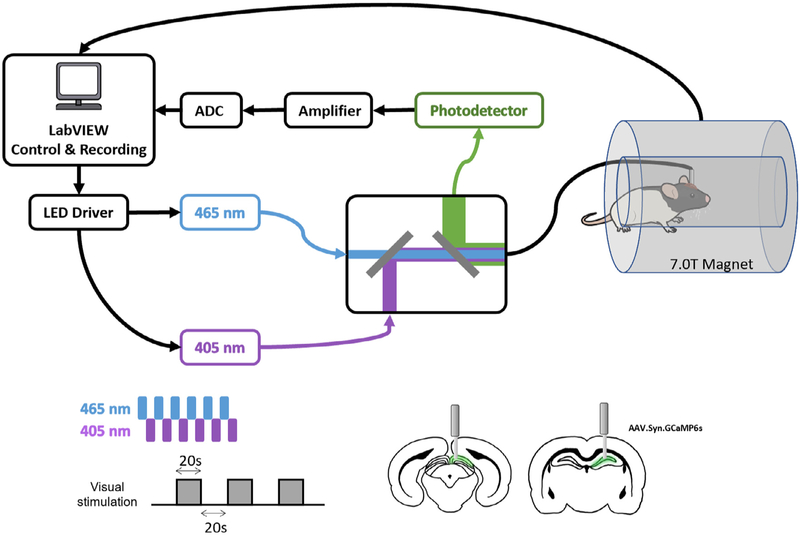
Aseptic stereotaxic surgeries were performed for intracranial virus injection and fiber optic cannula implantation. Before surgery, rats (n = 7 for the superior colliculus [SC] group; n = 16 for the dHP group; 300 g-450 g) were initially anesthetized with isoflurane, followed by intramuscular (IM) injections of ketamine (40 mg/kg) and xylazine (12 mg/kg). Baytril (2.5 mg/kg) and Dexamethasone (0.5 mg/kg) were administered subcutaneously as the antibiotic and anti-inflammatory drugs. Buprenorphine (1.0 mg/kg) was injected subcutaneously as long-term post-surgery analgesia. The rat was first intubated with an endotracheal catheter (Rivera et al., 2005) and ventilated with oxygen (PhysioSuite, Kent Scientific Corporation). During the surgery, heart rate, respiration rate, CO2 level and SpO2 level were monitored (MouseSTAT Jr., Kent Scientific Corporation) and the body temperature was maintained at 37 °C by a warming pad (PhysioSuite, Kent Scientific Corporation). The rat was then head fixed on a stereotaxic platform (David Kopf Instruments, Tujunga, CA). The scalp was infiltrated with bupivacaine (4 mg/kg) as local analgesia. One anterior-posterior incision was made along the midline of the scalp and craniotomies were made for viral injection and fiber optic cannula implantation. AAV5/9.Syn.GCaMP6s.WPRE.SV40 (800–1000nl, at titer ≥ 7 × 1012 vg/mL, Addgene, Watertown, MA) was injected into the dHP (coordinates: AP −3.5, ML +2, DV −2.5, −2.7, and −2.9) or SC (coordinates: AP −6.5, ML + 0.8, DV −2.8, −3.0, −3.3, and −3.5), respectively, through a glass pipette tip fitted with a micropipette syringe (Hamilton Company, Reno, NV) (Fig. 1). After injection, a fiber optic cannula (400 nm core, NA = 0.50, Thorlabs, Newton, NJ) was advanced to the injection site for the dHP group (coordinates: AP −3.5, ML +2, DV −2.5) or the SC group (coordinates: AP −6.5, ML +0.8, DV −2.9). To fix the implanted fiber, dental adhesive (Vetabond, 3M, St. Paul, MN) and dental cement (ParaBond, COLTENE, Cuyahoga Falls, OH) were applied over the skull.
Fig. 1.
Schematic diagram of the experimental setup and paradigm.
2.3. Acclimation
After recovery from surgery, the rat was acclimated in a mock scanning environment for 7 days to minimize motion and stress during rsfMRI (Dopfel and Zhang, 2018; Gao et al., 2017). The rat was briefly anesthetized under isoflurane (2–4%) in a chamber and remained anesthetized with a nose cone. The rat head and body were then restrained in an acclimation restrainer. The rat was allowed to wake up before being placed into a mock MRI scanner where prerecorded sounds of MRI pulse sequences were played. The acclimation duration was gradually increased from 15 min/day to 60 min/day (i.e. 15, 30, 45, 60, 60, 60 and 60 min/day, respectively). This acclimation procedure has been confirmed to provide sufficient habituation in rats during fMRI (King et al., 2005; Zhang et al., 2010; Liang et al., 2011, 2014; Ma et al., 2018b).
2.4. Optical setup
A dual-wavelength GCaMP fiber photometry system (Doric Lenses Inc., Quebec, Canada) was used for calcium signal recording (Kim et al., 2016) (Fig. 1). Ca2+ dependent and Ca2+ independent fluorescence proteins were excited by two alternating low power LEDs at the wavelength 465 nm and an isosbestic wavelength 405 nm, respectively. They were modulated at 400 Hz with 40% duty cycle for each wavelength using LabVIEW (National Instruments, Austin, TX). The two LED excitation sources were combined in a mini-cube (Doric Lenses Inc., Quebec, Canada) via a dichroic mirror and delivered to the implanted fiber optic cannula through a low-autofluorescence mono-fiber optic patchcord cable (400 μm core, NA = 0.48, 7 m long, Doric Lenses Inc., Quebec, Canada). The light power at the tip of the fiber delivered to the animal was maintained at 10 μW and 5 μW for 465 nm and 405 nm, respectively. The emitted fluorescent light from the implanted brain region passed through the same patchcord cable and the mini-cube via another dichroic mirror, then through a single-mode fiber launch (Thorlabs, Newton, NJ) with a microscope objective (40 × 0.65 NA, Olympus, Center Valley, PA), and focused onto a photoreceiver (MiniSM 20,025, SensL Technologies, Somerville, MA). The collected signal from the photoreceiver was subsequently amplified, bandpass filtered (0.3–1 kHz, Dagan Corp., Minneapolis, MN) and recorded using LabVIEW (National Instruments, Austin, TX) with an NI-DAQ board (10 kHz sampling rate, National Instruments, Austin, TX). TTL signals from the two LEDs and the MRI scanner during simultaneous calcium and fMRI recordings were recorded.
2.5. fMRI experiment
All rsfMRI data were simultaneously collected with the GCaMP signal on a 7T magnet (Agilent, CA, USA), which was interfaced to a Bruker console and equipped with a Bruker gradient (Bruker, Germany) at the High Field MRI Facility at the Pennsylvania State University. Structural images were acquired to confirm the location of the fiber optic cannula. For functional data, T2*-weighted images were obtained using the single-shot gradient echo echo-planner-imaging (GE-EPI) sequence with the following parameters: echo time (TE) = 15 ms, repetition time (TR) = 1 s, flip angle = 60°, slice thickness = 1 mm, slice number = 20, in-plane matrix size = 64 × 64, FOV = 3.2 × 3.2 cm2, 600 vol each scan. Three consecutive scans were collected for each rat for each rsfMRI session. For task fMRI sessions, low-power blue light, controlled by the same NI-DAQ board used for GCaMP signal recording, was delivered through the optical fiber to the rat’s left eye (i.e. contralateral to the fiber implantation side in the SC) as the visual stimulus. During simultaneous GCaMP-fMRI data collection, visual stimulation was presented using a 20s-light-ON and 20s-light-OFF paradigm.
2.6. Calcium data preprocessing
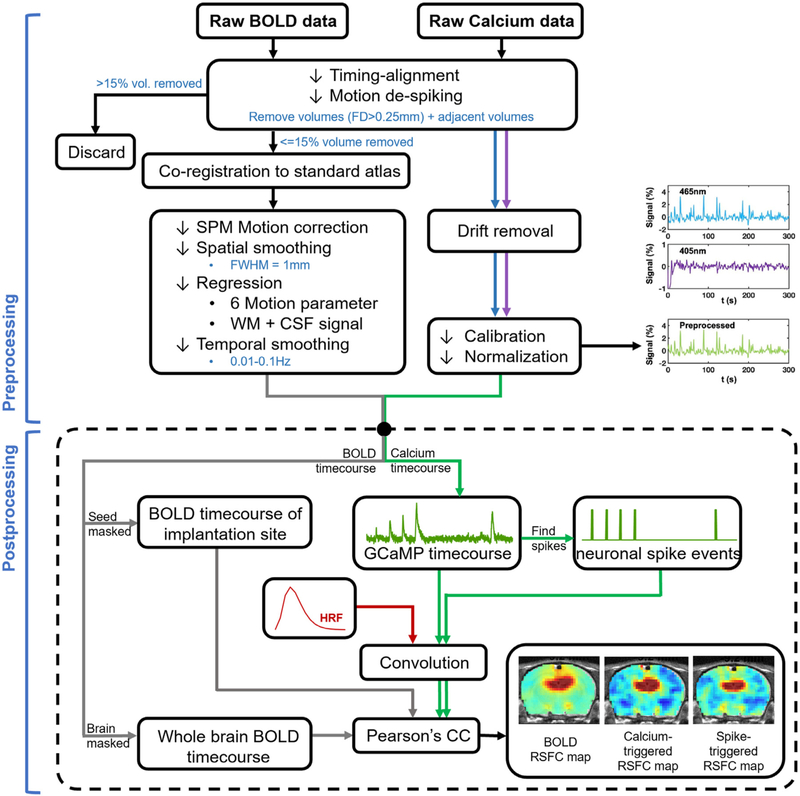
A python-based pipeline was used for calcium signal preprocessing (Martianova et al., 2019). Photometry signals from 465 nm and 405 nm were first detrended to remove signal drifting. Subsequently, the calcium-dependent 465 nm signal was corrected for calcium-independent isosbestic 405 nm signal by linear regression, and then converted to percentage change.
To identify calcium peaks in each scan, a peak detection procedure was performed on preprocessed GCaMP signal based on the peak amplitude and shape of GCaMP signal profiles. The shape of GCaMP profile was taken into account to eliminate large GCaMP signal changes induced by non-neuronal sources such as motion, given that they exhibit highly different signal profiles. First, we used all GCaMP time series to generate a template of GCaMP peaks. For each GCaMP scan, segments surrounding local GCaMP peaks (peak amplitude > one standard deviation of the time series; segment duration = 4 s) were identified. All these segments were pooled together and k-means clustered based on their temporal profiles (k = 6). The averaged GCaMP profile from the cluster with the highest occurrence rate was selected and used as the GCaMP template. Subsequently, this GCaMP template was cross correlated to sliding windows (window length = 4 s, step size = 1 s) in each GCaMP time series, and windows with the correlation coefficient > 0.6 and peak amplitude > one standard deviation were deemed as real GCaMP peaks (Fig. 2). To avoid ambiguity, GCaMP segments that were temporally overlapping were excluded. GCaMP peaks identified by this procedure well agreed with the GCaMP response reported (Figs. 2 and 3), and this procedure was not very sensitive to the parameters we used. Finally, the time points corresponding to local maxima of these GCaMP peaks were defined as the occurrence of neuronal spike events (Fig. 2).
Fig. 2.
Flowchart of fMRI and calcium data processing.
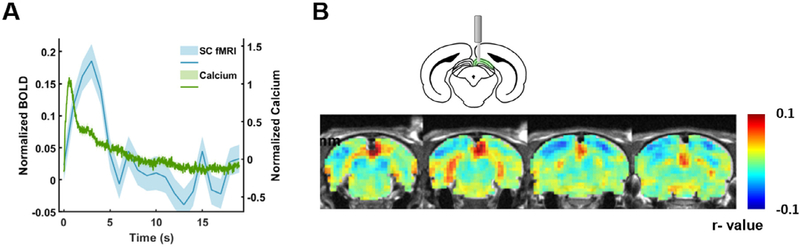
Fig. 3.
Correlation between visual stimulation-evoked GCaMP and BOLD signals in the SC. (A) Averaged calcium (green) and BOLD (blue) signals across all stimulation epochs. Shaded area: SEM. (B) Spatial correlation map between calcium predicted BOLD and measured BOLD signals.
2.7. fMRI data preprocessing
A MATLAB-based pipeline was used for fMRI data preprocessing (Liang et al., 2012, 2013; Liu et al., 2020; Ma et al., 2018b). fMRI data were first temporally synchronized with the preprocessed GCaMP signal based on simultaneously recorded TTLs (Fig. 2). The first 10 vol of each fMRI scan and the corresponding GCaMP timepoints were removed to ensure steady-state MR signal. Framewise displacement (FD) was calculated to estimate the motion level for each EPI vol according to the method descripted in (Power et al., 2012), and adjusted for the size of the rat brain. Volumes with FD > 0.25 mm and their adjacent volumes as well as the corresponding GCaMP timepoints were removed. Scans with more than 15% volumes scrubbed were excluded from further analysis. Subsequently, rsfMRI images were manually co-registered to a predefined template using in-house MATLAB software (Liu et al., 2020). This co-registration only involved rigid-body transformation. Co-registered rsfMRI images were then motion corrected using SPM 12 and spatially smoothed with a gaussian kernel (FWHM = 1 mm, twice of the in-plane voxel size). Signals from six motion parameters, as well as the white matter (WM) and cerebrospinal fluid (CSF) were regressed out. At last, band-pass temporal filtering (0.01–0.1 Hz) was performed. To compare rsfMRI data preprocessing methods based on the GCaMP data, we also employed 6 different preprocessing pipelines as followed: (1) motion regression, (2) motion regression + temporal filtering (TF), (3) motion, WM, CSF regression, (4) motion, WM, CSF regression + TF, (5) motion, WM, CSF, global signal (GS) regression, and (6) motion, WM, CSF, GS regression + TF. For data presented in all other figures, case 4 was used.
2.8. Data analysis
For each scan, the BOLD-driven RSFC map of the dHP was generated by voxelwise correlating the averaged time series extracted from 18 voxels below the implanted fiber with BOLD time series of all voxels in the brain. To obtain GCaMP-related RSFC maps, the GCaMP signal was first down sampled to 1 s (i.e. TR) for each scan. GCaMP-triggered and spike event-triggered RSFC maps were then generated by voxelwise correlating the brain-wide rsfMRI signal with the time series of the corresponding GCaMP signal as well as neuronal spike events convolved with a hemodynamic response function (HRF), respectively. To account for a faster HRF in rats relative to humans, instead of the default HRF in SPM, we used an HRF with a shorter onset time and time to peak reported in a recent study (Tong et al., 2019). The final BOLD-driven, GCaMP-triggered, and spike-triggered RSFC maps were generated by averaging the corresponding maps across scans and animals.
3. Results
3.1. Simultaneous GCaMP fiber photometry and BOLD fMRI in awake rodents
In this study, we examined the coupling between neural spiking activity and the resting-state BOLD signal in awake rats. To accomplish this goal, we established a platform for GCaMP-based fiber photometry using dual wavelength excitation, with simultaneous whole-brain fMRI measurement.
We first confirmed the validity of this platform. AAVs expressing GCaMP protein was stereotaxically injected into the SC and an optical fiber was chronically implanted into the same region. After 4–6 weeks of recovery and protein expression (Supplementary Fig. S1), we observed robust calcium signal increase recorded in the SC evoked by visual stimulation (Fig. 3 and Supplementary Fig. S1). The temporal profile of the GCaMP response is consistent with the GCaMP signal pattern reported in the literature, and is appreciably faster than the BOLD activation simultaneously recorded from the same region. Fig. 3B showed the GCaMP-derived spatial BOLD map, generated by the GCaMP signal convolved with an HRF, and then voxelwise correlated to the BOLD signal across the whole brain. The map obtained clearly demonstrated region-specific correspondence between calcium-derived BOLD and measured BOLD signals at the implanted site. These data validated the platform of simultaneous GCaMP and fMRI signal recordings in awake animals.
3.2. Spatiotemporal coupling between calcium and bold signals at the resting state
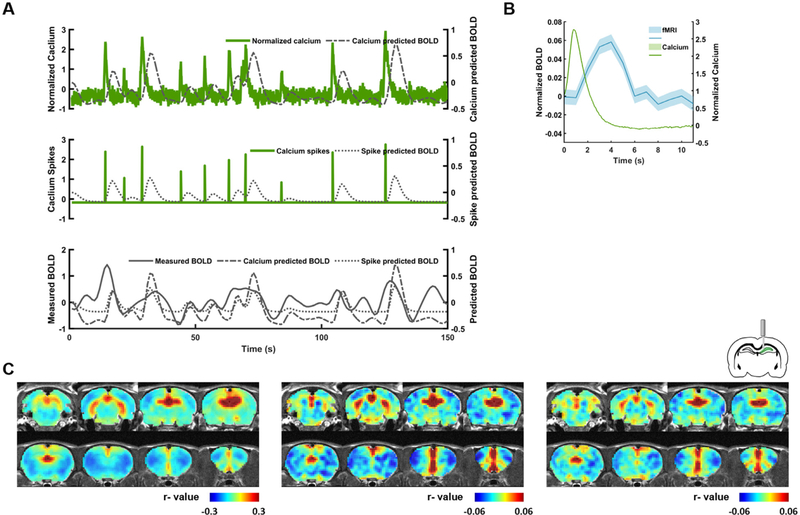
We next explored the relationship between calcium and BOLD signals at the resting state. AAVs expressing GCaMP protein was stereotaxically injected into the dHP. The expression and location of GCaMP protein were verified by histology in each animal (Supplementary Fig. S2). Simultaneously measured calcium and BOLD data from the dHP indicate a clear coupling between these two signals, reflected by the correspondence between the spontaneous calcium- and spike-event-predicted BOLD signals and the measured BOLD signal in the dHP (Fig. 4A). On average, calcium spikes were followed by appreciable BOLD peaks in the dHP as shown in Fig. 4B. To examine the whole-brain spatial patterns of dHP RSFC obtained by BOLD as well as calcium data, we respectively generated the dHP seed maps using the measured BOLD signal as well as BOLD signals derived from the calcium signal/spike events convolved with the HRF (Fig. 4 and Supplementary Fig. S3). We found that the BOLD dHP seed map (Fig. 4C, left) was highly consistent with the calcium signal-triggered (Fig. 4C, middle, spatial correlation coefficient = 0.75) and spike-event-triggered (Fig. 4C, right, spatial correlation coefficient = 0.72) dHP seed maps. These results suggest the calcium signal is strongly coupled with the BOLD signal at rest in awake animals.
Fig. 4.
Correlation between calcium and BOLD signals at the resting state. (A) Representative (top) calcium, (middle) calcium spike, and (bottom) BOLD signals measured in the dHP at the resting state. (B) Averaged calcium (green) and BOLD (blue) signals across all spontaneous calcium peaks identified. Shaded area: SEM. (C) dHP RSFC maps generated using (left) acquired dHP BOLD signal, (middle) calcium-predicted BOLD signal, and (right) spike train-predicted BOLD signal. Statistical results are shown in Fig. S3.
To rule out the possibility that the high spatial similarities (spatial correlations = 0.75/0.72) observed in Fig. 4C might originate from inherent anatomical information or other systematic bias, we performed a simulation analysis. Sham calcium signals with random values drawn from a normal distribution with the same mean and standard deviation as collected calcium signals were generated for each scan. These sham calcium signals were then used to calculate the corresponding RSFC maps in the same manner. The spatial similarities between the BOLD-driven and sham calcium-driven RSFC map (spatial correlation coefficient = 0.031, Supplementary Fig. S4) as well as the sham calcium spike-driven RSFC map (correlation coefficient = 0.039, Supplementary Fig. S4) were much lower compared to the corresponding maps derived from the collected calcium data (correlation coefficient = 0.75/0.72), indicating that the high spatial similarities observed in Fig. 4C indeed resulted from the tight coupling between the calcium and BOLD activity.
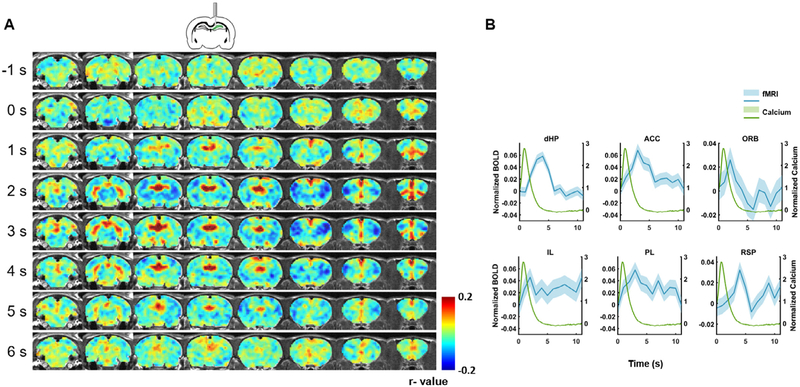
To further examine the spatiotemporal dynamics of spontaneous calcium-triggered BOLD signal, we plotted the spatiotemporal profile of the BOLD response around calcium spikes in the dHP. Fig. 5A showed the averaged whole-brain BOLD amplitude map before and after detected calcium spikes. Multiple brain regions, including the dHP, exhibited appreciable changes in BOLD amplitude that corresponded to calcium peaks. Fig. 5B showed the temporal profiles of the BOLD response in several regions in the default-mode network (DMN). Most DMN regions, including dHP, anterior cingulate cortex (ACC), orbitofrontal cortex (ORB), infralimbic cortex (IL), prelimbic cortex (PL), and retrosplenial area (RSP) showed clear and significant increases in the BOLD signal after calcium peaks in the dHP, again suggesting a strong coupling between the calcium signal in one node and the BOLD signal in distributed nodes within the same functional brain network.
Fig. 5.
Brain-wide BOLD response before and after spontaneous calcium peaks in the dHP. (A) Spatiotemporal patterns of BOLD signal around calcium peaks in the dHP. Each row represents an averaged whole-brain BOLD map in the rostral-caudal direction at a time point before or after spontaneous calcium spikes. Zero second corresponds to the calcium peak time. (B) BOLD profiles in default mode network regions after spontaneous calcium peaks. dHP: dorsal hippocampus. ACC: anterior cingulate cortex. ORB: orbital frontal cortex. IL: infralimbic cortex. PL: prelimbic cortex. RSP: retrosplenial cortex. Shaded regions: SEM.
3.3. Comparison of different fMRI preprocessing pipelines based on BOLD-calcium coupling
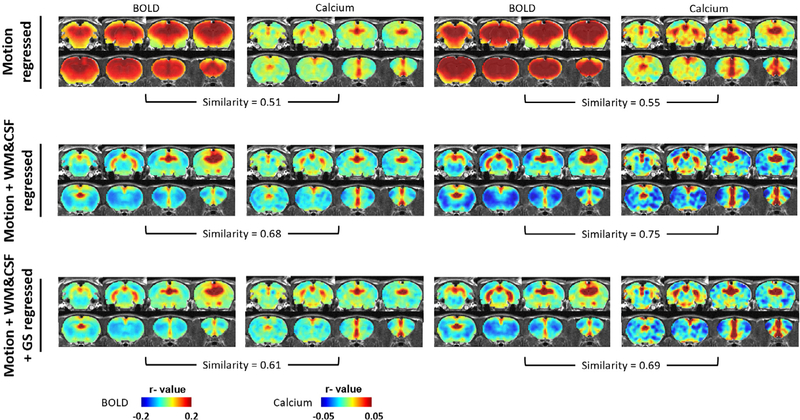
Based on the assumption that the recorded GCaMP signal provides a reliable measurement of neural spiking activity in the dHP, we next explored the efficacy of different nuisance signal regression and temporal filtering steps on rsfMRI data preprocess. Using the calcium signal-derived dHP seed maps as the standard, we compared BOLD-based seed maps obtained under 6 different preprocessing pipelines which all included the steps of motion scrubbing, motion correction and spatial smoothing: (1) motion regression, (2) motion regression + TF, (3) motion, WM&CSF regression, (4) motion, WM&CSF regression + TF, (5) motion, WM&CSF, GS regression, and (6) motion, WM&CSF, GS regression + TF. Fig. 6 showed that calcium signal-based seed maps were generally consistent across different preprocess strategies, corroborating the notion that the calcium signal provided a reliable measurement of neuronal activity in the dHP and was relatively insensitive to various non-neural artifacts. However, the BOLD-based seed maps were highly sensitive to individual preprocessing steps. Without WM/CSF or GS regression, the spatial extent of ‘connected’ brain regions in BOLD-based seed maps lacked spatial specificity compared to the corresponding calcium-based seed maps (Fig. 6, top row). This difference persisted regardless of whether temporal filtering was applied. In contrast, WM/CSF and/or GS regression considerably improved the spatial specificity in BOLD-based seed maps. In all cases, TF seemed to increase the contrast-noise ratio but did not appreciably affect the spatial similarity between the BOLD-based and calcium-based seed maps. Finally, by calculating the spatial correlational coefficient between BOLD-based and calcium-derived seed maps for these 6 preprocessing strategies, Case 4 (motion, WM&CSF regression + TF) yielded the highest spatial similarity, and hence we believe this preprocessing pipeline might provide better performance for rsfMRI data preprocessing.
Fig. 6.
Comparison of BOLD and calcium predicted RSFC maps between different rsfMRI data preprocessing methods.
4. Discussion
fMRI has been widely employed to map brain network dynamics in both humans and animals. Understanding the relationships between neuronal, vascular and BOLD signals is essential for the appropriate interpretation of fMRI results in relation to the corresponding neuronal activity. In the present study, we established a simultaneous dual-wavelength calcium fiber photometry and BOLD fMRI setup in awake rodents. By using this platform, we examined the coupling relationship between calcium activity and BOLD response under both visually evoked and resting states. Our results demonstrate significant correspondence between the calcium and BOLD signals, suggesting that spiking activity plays a key role in the neural mechanism of BOLD signal.
Recently, simultaneous calcium-based fiber photometry with BOLD fMRI has emerged as a popular method to record neural activities inside the MRI scanner. Unlike simultaneous electrophysiology-fMRI recordings, calcium-based fiber photometry does not suffer from the inherent electromagnetic interference from MRI scanning (Schlegel et al., 2018). Despite its prominent advantages, most previous GCaMP-fMRI studies used anesthesia to immobilize animals during the measurement of neural and BOLD activities (He et al., 2018; Lake et al., 2020; Tong et al., 2019). Since numerous events involved in neural and vascular dynamics can be affected by anesthesia, this confounding factor can significantly affect the quantification of neurovascular coupling and interpretation of brain functional imaging signals. To avoid this issue, in the present study we established the simultaneous calcium fiber photometry and fMRI in awake animals (Liang et al., 2017; Ma et al., 2020).
The finding of significant correspondence between calcium and BOLD signals at the resting state is consistent with previous reports in cortical (Lake et al., 2020; Pais-Roldán et al., 2020; Schlegel et al., 2018; Schwalm et al., 2017; Wang et al., 2018) but not subcortical regions (Tong et al., 2019). For instance, cortex-wide BOLD activity was found to be directly correlated with locally recorded slow oscillations of calcium waves (Schwalm et al., 2017). In addition, Schlegel and colleagues showed symmetric and region-specific coupling between spontaneous calcium and BOLD fluctuations in the somatosensory cortex at the resting state (Schlegel et al., 2018). Global fMRI signal fluctuations were also shown to be correlated with cortical population activity in the form of calcium transients, which were directly linked to pupil dynamics (Pais-Roldán et al., 2020). A more recent study demonstrated significant correlation between cortex-wide calcium signal and fMRI under both evoked and resting state (Lake et al., 2020). In addition to neuronal activity, evoked astrocytic Ca2+ signals in the cortex were found to be coupled with positive BOLD signals whereas intrinsic astrocytic Ca2+ signals were linked to negative BOLD signals, demonstrating a brain state-dependent coupling of Ca2+ with BOLD signals (Wang et al., 2018). In contrast to the cortex, weak coupling between resting-state BOLD and calcium signals was observed in the SC and lateral geniculate nucleus (LGN) (Tong et al., 2019). This discrepancy might be due to different physiological conditions under which the calcium and BOLD signals were acquired. In addition, difference in anatomical locations could contribute to this difference. Indeed, primary sensory relay nuclei such as SC and LGN generally exhibit lower baseline firing rates compared to the hippocampus and cortical regions. As a result, lower spontaneous calcium signal in the SC and LGN might lead to weaker coupling between calcium and BOLD signals in these regions.
In addition to strong coupling of resting-state BOLD and calcium signals at the GCaMP recording site (i.e. dHP), GCaMP-triggered spatiotemporal BOLD maps showed prominent coactivation patterns between spatially distant regions in the same functional network of DMN. DMN is a well-known large-scale brain functional network that includes several high-level cognitive areas such as medial prefrontal cortex (mPFC), posterior cingulate cortex (PCC), and hippocampus. A growing number of studies have found compelling evidence that DMN exists in multiple species including chimpanzees (Rilling et al., 2007), macaque monkeys (Vincent et al., 2007), mouse (Stafford et al., 2014) as well as rats (Lu et al., 2012; Ma et al., 2018a). In the present study, we observed a clear correlation between calcium predicted BOLD and acquired BOLD signals not only in the dHP, but also in other distributed DMN regions such as the ACC and ORB. (Fig. 5). These spatiotemporal coactivation patterns triggered by dHP calcium spikes clearly suggest that DMN is indeed a functional network with coordinated activity between separate nodes (Tu et al., 2021b, 2021a).
Using the calcium signal recorded as the ground truth, we also compared the efficacy of different rsfMRI data preprocessing pipelines. The resting-state BOLD signal is often contaminated by non-neuronal sources including both physiological and non-physiological noise, such as head motion, respiration, cardiac cycles and MR system instability (Caballero-Gaudes and Reynolds, 2017; Esteban et al., 2019; Power et al., 2017). These nuisance signals can compound rsfMRI time series and thereby affect the quantification of RSFC. Therefore, it is crucial to implement a robust preprocessing pipeline to minimize the contribution of nuisance noise without affecting the neural component in the BOLD signal. By comparing to the corresponding calcium signal-derived dHP seed maps, here we demonstrate that WM/CSF regression and/or GS regression can improve the spatial specificity in BOLD-based seed maps (Fig. 6). Among the 6 different combinations of processing steps we tested, rsfMRI data preprocessed with the pipeline of motion regression, WM/CSF regression and TF yielded the highest spatial similarity to the calcium-derived seed map, suggesting that this preprocessing pipeline might provide functional connectivity mapping result most correlated to neuronal activity.
4.1. Limitations
First, in this study, we selected AAVs that expressed GCaMP in all neuron types. As a result, we were not able to dissect the contributions of different neural populations to the resting-state BOLD signal. Indeed, both excitatory and inhibitory neurons can contribute to the BOLD signal (Logothetis et al., 2001; Mukamel et al., 2005; Rees et al., 2000; Viswanathan and Freeman, 2007). In addition, increased flux of Na+, K+, and/or Ca2+ caused by increased neural activity may induce signaling to astrocytes and other neuronal cells that can send vasoactive signals to nearby vessels (Kim and Ogawa, 2012). With the recent advancement of genetically encoded calcium indicators, which can be selectively expressed in different cell types including excitatory neurons, inhibitory neurons (Mollinedo-Gajate et al., 2019) and even astrocytes (Otsu et al., 2015), further dissecting more specific neural contributions to the BOLD signal becomes feasible. Moreover, emerging technologies on voltage sensitive indicators may offer an alternative solution, although these techniques still suffer from low SNR at this point (Kulkarni and Miller, 2017). Second, by comparing the GCaMP-based and BOLD-based seed maps, we demonstrated that regression of WM/CSF signal from rsfMRI is important in rsfMRI data preprocessing. However, whether the WM/CSF signal contains neuronally related signal remains controversial (Ding et al., 2018; Logothetis and Wandell, 2004). Evidence of strong correlation between functional cortical volumes and signals in specific, segmented WM tracts in both resting and task states was demonstrated (Ding et al., 2018). Therefore, further investigation on the potential impact of WM regression on rsfMRI preprocessing is still needed. Third, in the present study we used the slow version of GCaMP (i.e. GCaMP6s). Although GCaMP6s had higher signal-to-noise ratio than GCaMP6f, it might be too slow to resolve neuronal spiking, but rather represents integrated neuronal activity. Therefore, the GCaMP signal we obtained could also contain synaptic input activity (Schlegel et al., 2018).
Supplementary Material
Acknowledgments
The present study was partially supported by National Institute of Neurological Disorders and Stroke (R01NS085200), National Institute of Mental Health (RF1MH114224), and National Institute of General Medical Sciences (R01GM141792). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Declaration of Competing Interest
none.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neuroimage.2022.118960.
Data and code availability statement
This manuscript did not use publicly available datasets.
Raw data associated with any figures can be provided upon request.
There is no restrictions on data availability.
References
- Caballero-Gaudes C, Reynolds RC, 2017. Methods for cleaning the BOLD fMRI signal. NeuroImage 154, 128–149. doi: 10.1016/j.neuroimage.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael DW, Vulliemoz S, Rodionov R, Thornton JS, McEvoy AW, Lemieux L, 2012. Simultaneous intracranial EEG-fMRI in humans: Protocol considerations and data quality. NeuroImage 63, 301–309. doi: 10.1016/j.neuroimage.2012.05.056. [DOI] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, Svoboda K, Kim DS, 2013. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Sobczak F, Chen Y, Jiang Y, Qian C, Lu Z, Ayata C, Logothetis NK, Yu X, 2019. Mapping optogenetically-driven single-vessel fMRI with concurrent neuronal calcium recordings in the rat hippocampus. Nat. Commun. 10, 1–12. doi: 10.1038/s41467-019-12850-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Huang Y, Bailey SK, Gao Y, Cutting LE, Rogers BP, Newton AT, Gore JC, 2018. Detection of synchronous brain activity in white matter tracts at rest and under functional loading. Proc. Natl. Acad. Sci. USA 115, 595–600. doi: 10.1073/pnas.1711567115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopfel D, Zhang N, 2018. Mapping stress networks using functional magnetic resonance imaging in awake animals. Neurobiol. Stress 9, 251–263. doi: 10.1016/j.ynstr.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban O, Markiewicz CJ, Blair RW, Moodie CA, Isik AI, Erramuzpe A, Kent JD, Goncalves M, DuPre E, Snyder M, Oya H, Ghosh SS, Wright J, Durnez J, Poldrack RA, Gorgolewski KJ, 2019. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat. Methods 16, 111–116. doi: 10.1038/s41592-018-0235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanselow MS, Dong HW, 2010. Are the dorsal and ventral hippocampus functionally distinct structures? Neuron doi: 10.1016/j.neuron.2009.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin E, Baillet S, 2015. The brain’s resting-state activity is shaped by synchronized cross-frequency coupling of neural oscillations. NeuroImage 111, 26–35. doi: 10.1016/j.neuroimage.2015.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YR, Ma Y, Zhang Q, Winder AT, Liang Z, Antinori L, Drew PJ, Zhang N, 2017. Time to wake up: studying neurovascular coupling and brain-wide circuit functionin the un-anesthetized animal. NeuroImage 153, 382–398. doi: 10.1016/j.neuroimage.2016.11.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienberger C, Konnerth A, 2012. Imaging Calcium in Neurons. Neuron 73, 862–885. doi: 10.1016/j.neuron.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Gu X, Chen W, You J, Koretsky AP, Volkow ND, Pan Y, Du C, 2018. Long-term optical imaging of neurovascular coupling in mouse cortex using GCaMP6f and intrinsic hemodynamic signals. NeuroImage 165, 251–264. doi: 10.1016/j.neuroimage.2017.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton C, Ma Y, Zhang N, 2017. Global reduction of information exchange during anesthetic-induced unconsciousness. Brain Struct. Funct. 222, 3205–3216. doi: 10.1007/s00429-017-1396-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Liu Z, 2008. Multimodal Functional Neuroimaging: integrating Functional MRI and EEG/MEG. IEEE Rev. Biomed. Eng. 1, 23–40. doi: 10.1109/RBME.2008.2008233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Wang M, Chen X, Pohmann R, Polimeni JR, Scheffer K, Rosen BR, Kleinfeld D, Yu X, 2018. Ultra-slow single-vessel BOLD and CBV-based fMRI spatiotemporal dynamics and their correlation with neuronal intracellular calcium signals. Neuron 97, 925–939. doi: 10.1016/j.neuron.2018.01.025, e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeger DJ, Ress D, 2002. What does fMRI tell us about neuronal activity? Nat. Rev. Neurosci doi: 10.1038/nrn730. [DOI] [PubMed] [Google Scholar]
- Hillman EMC, 2014. Coupling mechanism and significance of the BOLD signal: a status report. Annu. Rev. Neurosci. doi: 10.1146/annurev-neuro-071013-014111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltunen T, Kantola J, Elseoud AA, Lepola P, Suominen K, Starck T, Nikkinen J, Remes J, Tervonen O, Palva S, Kiviniemi V, Matias Palva J, 2014. Infra-slow EEG fluctuations are correlated with resting-state network dynamics in fMRI. J. Neurosci 34, 356–362. doi: 10.1523/JNEUROSCI.0276-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorge J, Grouiller F, Ipek Ö, Stoermer R, Michel CM, Figueiredo P, van der Zwaag W, Gruetter R, 2015. Simultaneous EEG-fMRI at ultra-high field: artifact prevention and safety assessment. NeuroImage 105, 132–144. doi: 10.1016/j.neuroimage.2014.10.055. [DOI] [PubMed] [Google Scholar]
- Kim CK, Yang SJ, Pichamoorthy N, Young NP, Kauvar I, Jennings JH, Lerner TN, Berndt A, Lee SY, Ramakrishnan C, Davidson TJ, Inoue M, Bito H, Deisseroth K, 2016. Simultaneous fast measurement of circuit dynamics at multiple sites across the mammalian brain. Nat. Methods 13, 325–328. doi: 10.1038/nmeth.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SG, Ogawa S, 2012. Biophysical and physiological origins of blood oxygenation level-dependent fMRI signals. J. Cereb. Blood Flow Metab. doi: 10.1038/jcbfm.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JA, Garelick TS, Brevard ME, Chen W, Messenger TL, Duong TQ, Ferris CF, 2005. Procedure for minimizing stress for fMRI studies in conscious rats. J. Neurosci. Methods 148, 154–160. doi: 10.1016/j.jneumeth.2005.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni RU, Miller EW, 2017. Voltage Imaging: pitfalls and potential. Biochemistry 56, 5171–5177. doi: 10.1021/acs.biochem.7b00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake EMR, Ge X, Shen X, Herman P, Hyder F, Cardin JA, Higley MJ, Scheinost D, Papademetris X, Crair MC, Constable RT, 2020. Simultaneous cortex-wide fluorescence Ca2+ imaging and whole-brain fMRI. Nat. Methods 17, 1262–1271. doi: 10.1038/s41592-020-00984-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, King J, Zhang N, 2012. Anticorrelated resting-state functional connectivity in awake rat brain. NeuroImage 59, 1190–1199. doi: 10.1016/j.neuroimage.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, King J, Zhang N, 2011. Uncovering intrinsic connectional architecture of functional networks in awake rat brain. J. Neurosci. 31, 3776–3783. doi: 10.1523/JNEUROSCI.4557-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, King J, Zhang N, 2014. Neuroplasticity to a single-episode traumatic stress revealed by resting-state fMRI in awake rats. NeuroImage 103, 485–491. doi: 10.1016/j.neuroimage.2014.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Li T, King J, Zhang N, 2013. Mapping thalamocortical networks in rat brain using resting-state functional connectivity. NeuroImage 83, 237–244. doi: 10.1016/j.neuroimage.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Ma Y, Watson GDR, Zhang N, 2017. Simultaneous GCaMP6-based fiber photometry and fMRI in rats. J. Neurosci. Methods 289, 31–38. doi: 10.1016/j.jneumeth.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Perez PD, Ma Z, Zhiwei Ma, Dopfel D, Cramer S, Tu W, Zhang N, 2020. An open database of resting-state fMRI in awake rats. NeuroImage 220. doi: 10.1016/j.neuroimage.2020.117094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logothetis NK, Eschenko O, Murayama Y, Augath M, Steudel T, Evrard HC, Besserve M, Oeltermann A, 2012. Hippocampal-cortical interaction during periods of subcortical silence. Nature 491, 547–553. doi: 10.1038/nature11618. [DOI] [PubMed] [Google Scholar]
- Logothetis NK, Pauls J, Augath M, Trinath T, Oeltermann A, 2001. Neurophysiological investigation of the basis of the fMRI signal. Nature 412, 150–157. doi: 10.1038/35084005. [DOI] [PubMed] [Google Scholar]
- Logothetis NK, Wandell BA, 2004. Interpreting the BOLD signal. Annu. Rev. Physiol. doi: 10.1146/annurev.physiol.66.082602.092845. [DOI] [PubMed] [Google Scholar]
- Lu H, Zou Q, Gu H, Raichle ME, Stein EA, Yang Y, 2012. Rat brains also have a default mode network. Proc. Natl. Acad. Sci. USA 109, 3979–3984. doi: 10.1073/pnas.1200506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Zuo Y, Gu H, Waltz JA, Zhan W, Scholl CA, Rea W, Yang Y, Stein EA, 2007. Synchronized delta oscillations correlate with the resting-state functional MRI signal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Hamilton C, Zhang N, 2016. Dynamic connectivity patterns in conscious and unconscious brain. Brain Connect. 7. doi: 10.1089/brain.2016.0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Ma Z, Liang Z, Neuberger T, Zhang N, 2020. Global brain signal in awake rats. Brain Struct. Funct. 225, 227–240. doi: 10.1007/s00429-019-01996-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Ma Y, Zhang N, 2018a. Development of brain-wide connectivity architecture in awake rats. NeuroImage 176, 380–389. doi: 10.1016/j.neuroimage.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Perez P, Ma Z, Liu Y, Hamilton C, Liang Z, Zhang N, 2018b. Functional atlas of the awake rat brain: a neuroimaging study of rat brain specialization and integration. NeuroImage 170, 95–112. doi: 10.1016/j.neuroimage.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantini D, Perrucci MG, del Gratta C, Romani GL, Corbetta M, Raichle ME, 2007. Electrophysiological signatures of resting state networks in the human brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianova E, Aronson S, Proulx CD, 2019. Multi-fiber photometry to record neural activity in freely-moving animals. J. Vis. Exp. 2019. doi: 10.3791/60278. [DOI] [PubMed] [Google Scholar]
- Mollinedo-Gajate I, Song C, Knöpfel T, 2019. Genetically encoded fluorescent calcium and voltage indicators. 10.1007/164_2019_299 [DOI] [PubMed] [Google Scholar]
- Mukamel R, Gelbard H, Arieli A, Hasson U, Fried I, Malach R, 2005. Neuroscience: coupling between neuronal firing, field potentials, and fMRI in human auditory cortex. Science 309, 951–954. doi: 10.1126/science.1110913. [DOI] [PubMed] [Google Scholar]
- Otsu Y, Couchman K, Lyons DG, Collot M, Agarwal A, Mallet JM, Pfrieger FW, Bergles DE, Charpak S, 2015. Calcium dynamics in astrocyte processes during neurovascular coupling. Nat. Neurosci. 18, 210–218. doi: 10.1038/nn.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paasonen J, Stenroos P, Salo RA, Kiviniemi V, Gröhn O, 2018. Functional connectivity under six anesthesia protocols and the awake condition in rat brain. NeuroImage 172, 9–20. doi: 10.1016/j.neuroimage.2018.01.014. [DOI] [PubMed] [Google Scholar]
- Paasonen J, Laakso H, Pirttimäki T, Stenroos P, Salo RA, Zhurakovskaya E, Lehto LJ, Tanila H, Garwood M, Michaeli S, Idiyatullin D, Mangia S, Gröhn O, 2020. Multi-band SWIFT enables quiet and artefact-free EEG-fMRI and awake fMRI studies in rat. NeuroImage 206. doi: 10.1016/j.neuroimage.2019.116338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pais-Roldán P, Takahashi K, Sobczak F, Chen Y, Zhao X, Zeng H, Jiang Y, Yu X, 2020. Indexing brain state-dependent pupil dynamics with simultaneous fMRI and optical fiber calcium recording. Proc. Natl. Acad. Sci. USA 117, 6875–6882. doi: 10.1073/pnas.1909937117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan WJ, Thompson GJ, Magnuson ME, Jaeger D, Keilholz S, 2013. Infraslow LFP correlates to resting-state fMRI BOLD signals. NeuroImage 74, 288–297. doi: 10.1016/j.neuroimage.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE, 2012. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage 59, 2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Plitt M, Laumann TO, Martin A, 2017. Sources and implications of whole-brain fMRI signals in humans. NeuroImage 146, 609–625. doi: 10.1016/j.neuroimage.2016.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradier B, Wachsmuth L, Nagelmann N, Segelcke D, Kreitz S, Hess A, Pogatzki-Zahn EM, Faber C, 2021. Combined resting state-fMRI and calcium recordings show stable brain states for task-induced fMRI in mice under combined ISO/MED anesthesia. NeuroImage 245. doi: 10.1016/j.neuroimage.2021.118626. [DOI] [PubMed] [Google Scholar]
- Rees G, Friston K, Koch C, 2000. A direct quantitative relationship between the functional properties of human and macaque V5. Nat. Neurosci. 3, 716–723. doi: 10.1038/76673. [DOI] [PubMed] [Google Scholar]
- Rilling JK, Barks SK, Parr LA, Preuss TM, Faber TL, Pagnoni G, Bremner JD, Votaw JR, 2007. A comparison of resting-state brain activity in humans and chimpanzees. Proc. Natl. Acad. Sci. 104, 17146–17151. doi: 10.1073/pnas.0705132104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera B, Miller S, Brown E, Price R, 2005. A novel method for endotracheal intubation of mice and rats used in imaging studies. Contemp. Topics Lab. Anim. Sci. 44, 52–55. [PubMed] [Google Scholar]
- Schlegel F, Sych Y, Schroeter A, Stobart J, Weber B, Helmchen F, Rudin M, 2018. Fiber-optic implant for simultaneous fluorescence-based calcium recordings and BOLD fMRI in mice. Nat Protoc 13, 840–855. doi: 10.1038/nprot.2018.003. [DOI] [PubMed] [Google Scholar]
- Schulz K, Sydekum E, Krueppel R, Engelbrecht CJ, Schlegel F, Schröter A, Rudin M, Helmchen F, 2012. Simultaneous BOLD fMRI and fiber-optic calcium recording in rat neocortex. Nat. Methods 9, 597–602. doi: 10.1038/nmeth.2013. [DOI] [PubMed] [Google Scholar]
- Schwalm M, Schmid F, Wachsmuth L, Backhaus H, Kronfeld A, Jury FA, Prouvot PH, Fois C, Albers F, van Alst T, Faber C, Stroh A, 2017. Cortex-wide BOLD fMRI activity reflects locally-recorded slow oscillation-associated calcium waves. eLife 6, 1–27. doi: 10.7554/eLife.27602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenroos P, Pirttimaki T, Paasonen J, Paasonen E, Salo RA, Koivisto H, Natunen T, Makinen P, Kuulasmaa T, Hiltunen M, Tanila H, Grohn O, 2021. Isoflurane affects brain functional connectivity in rats 1 month after exposure. Neuroimage 234, 117987. doi: 10.1016/j.neuroimage.2021.117987. [DOI] [PubMed] [Google Scholar]
- Shmuel A, Leopold DA, 2008. Neuronal correlates of spontaneous fluctuations in fMRI signals in monkey visual cortex: implications for functional connectivity at rest. Hum. Brain Mapp. 29, 751–761. doi: 10.1002/hbm.20580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford JM, Jarrett BR, Miranda-Dominguez O, Mills BD, Cain N, Mihalas S, Lahvis GP, Lattal KM, Mitchell SH, David SV, Fryer JD, Nigg JT, Fair DA, 2014. Large-scale topology and the default mode network in the mouse connectome. Proc. Natal. Acad. Sci. 111, 18745–18750. doi: 10.1073/pnas.1404346111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson GJ, Pan WJ, Magnuson ME, Jaeger D, Keilholz SD, 2014. Quasi-periodic patterns (QPP): large-scale dynamics in resting state fMRI that correlate with local infraslow electrical activity. NeuroImage 84, 1018–1031. doi: 10.1016/j.neuroimage.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong C, Jkun Dai, Chen Y, Zhang K, Feng Y, Liang Z, 2019. Differential coupling between subcortical calcium and BOLD signals during evoked and resting state through simultaneous calcium fiber photometry and fMRI. NeuroImage 200, 405–413. doi: 10.1016/j.neuroimage.2019.07.006. [DOI] [PubMed] [Google Scholar]
- Tsurugizawa T, Yoshimaru D, 2021. Impact of anesthesia on static and dynamic functional connectivity in mice. NeuroImage 241. doi: 10.1016/j.neuroimage.2021.118413. [DOI] [PubMed] [Google Scholar]
- Tu W, Ma Z, Ma Y, Dopfel D, Zhang N, 2021a. Suppressing anterior cingulate cortex modulates default mode network and behavior in awake rats. Cerebral Cortex 31, 312–323. doi: 10.1093/cercor/bhaa227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu W, Ma Z, Zhang N, 2021b. Brain network reorganization after targeted attack at a hub region. NeuroImage 237. doi: 10.1016/j.neuroimage.2021.118219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, van Essen DC, Zempel JM, Snyder LH, Corbetta M, Raichle ME, 2007. Intrinsic functional architecture in the anaesthetized monkey brain. Nature 447, 83–86. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- Viswanathan A, Freeman RD, 2007. Neurometabolic coupling in cerebral cortex reflects synaptic more than spiking activity. Nat. Neurosci. 10, 1308–1312. doi: 10.1038/nn1977. [DOI] [PubMed] [Google Scholar]
- Wang L, Saalmann YB, Pinsk MA, Arcaro MJ, Kastner S, 2012. Electrophysiological low-frequency coherence and cross-frequency coupling contribute to BOLD connectivity. Neuron 76, 1010–1020. doi: 10.1016/j.neuron.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, He Y, Sejnowski TJ, Yu X, 2018. Brain-state dependent astrocytic Ca2+ signals are coupled to both positive and negative BOLD-fMRI signals. Proc. Natl. Acad. Sci. USA 115, E1647–E1656. doi: 10.1073/pnas.1711692115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder AT, Echagarruga C, Zhang Q, Drew PJ, 2017. Weak correlations between hemodynamic signals and ongoing neural activity during the resting state. Nat. Neurosci. 20. doi: 10.1038/s41593-017-0007-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Liu N, He Y, Liu Y, Ge L, Zou L, Song S, Xiong W, Liu X, 2018. Improved calcium sensor GCaMP-X overcomes the calcium channel perturbations induced by the calmodulin in GCaMP. Nat. Commun 9. doi: 10.1038/s41467-018-03719-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Rane P, Huang W, Liang Z, Kennedy D, Frazier JA, King J, 2010. Mapping resting-state brain networks in conscious animals. J. Neurosci. Methods 189, 186–196. doi: 10.1016/j.jneumeth.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This manuscript did not use publicly available datasets.
Raw data associated with any figures can be provided upon request.
There is no restrictions on data availability.