Abstract
Nucleosomes, the structural building blocks of chromatin, possess 2-fold pseudo symmetry which can be broken through differential modification or removal of one copy of a pair of sister histones. The resultant asymmetric nucleosomes and hexasomes have been implicated in gene regulation, yet the use of these noncanonical substrates in chromatin biochemistry is limited, owing to the lack of efficient methods for their preparation. Here, we report a strategy that allows the orientation of these asymmetric species to be tightly controlled relative to the underlying DNA sequence. Our approach is based on the use of truncated DNA templates to assemble oriented hexasomes followed by DNA ligation and, in the case of asymmetric nucleosomes, addition of the missing heterotypic histones. We show that this approach is compatible with multiple nucleosome positioning sequences, allowing the generation of desymmetrized mononucleosomes and oligonucleosomes with varied DNA overhangs and heterotypic histone H2A/H2B dimer compositions. Using this technology, we examine the functional consequences of asymmetry on BRG1/BRM associated factor (BAF) complex-mediated chromatin remodeling. Our results indicate that cancer-associated histone mutations can reprogram the inherent activity of BAF chromatin remodeling to induce aberrant chromatin structure.
Graphical Abstract
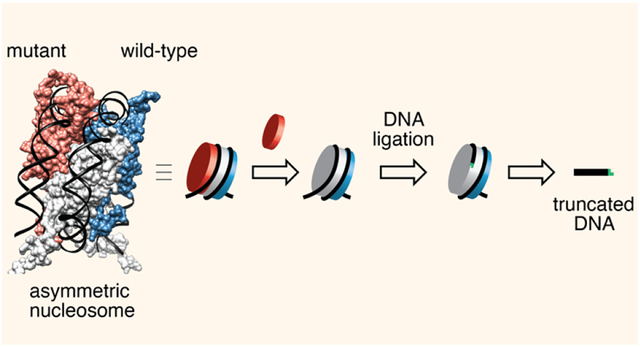
INTRODUCTION
Eukaryotic genomes are packaged into a nucleoprotein complex termed chromatin. The repeating unit of this supramolecular polymer, the nucleosome, has two-fold pseudo symmetry which arises from the C2-symmetrical assembly of two copies of histones H2A, H2B, H3, and H4 into the histone octamer1. Desymmetrization of nucleosome structure is known to occur when only one copy of a histone protein undergoes post-translational modification (PTM) or histone variant exchange2,3,4. This is thought to provide an additional layer of combinatorial diversity to the histone landscape, contributing to the control of gene expression4. Nucleosome symmetry can also be broken by somatic mutation of histone proteins, which have been shown to occur in a wide variety of human cancers5,6,7,8, or through the formation of subnucleosomal structures termed hexasomes - which lack one H2A/H2B dimer and are thus constitutionally asymmetric (Fig. 1A)2,9,10,11. The mechanisms by which chromatin-regulating enzymes interpret hexasomes and asymmetric nucleosomes to elicit downstream biological effects remain largely undefined.
Figure 1. Nucleosome desymmetrization.
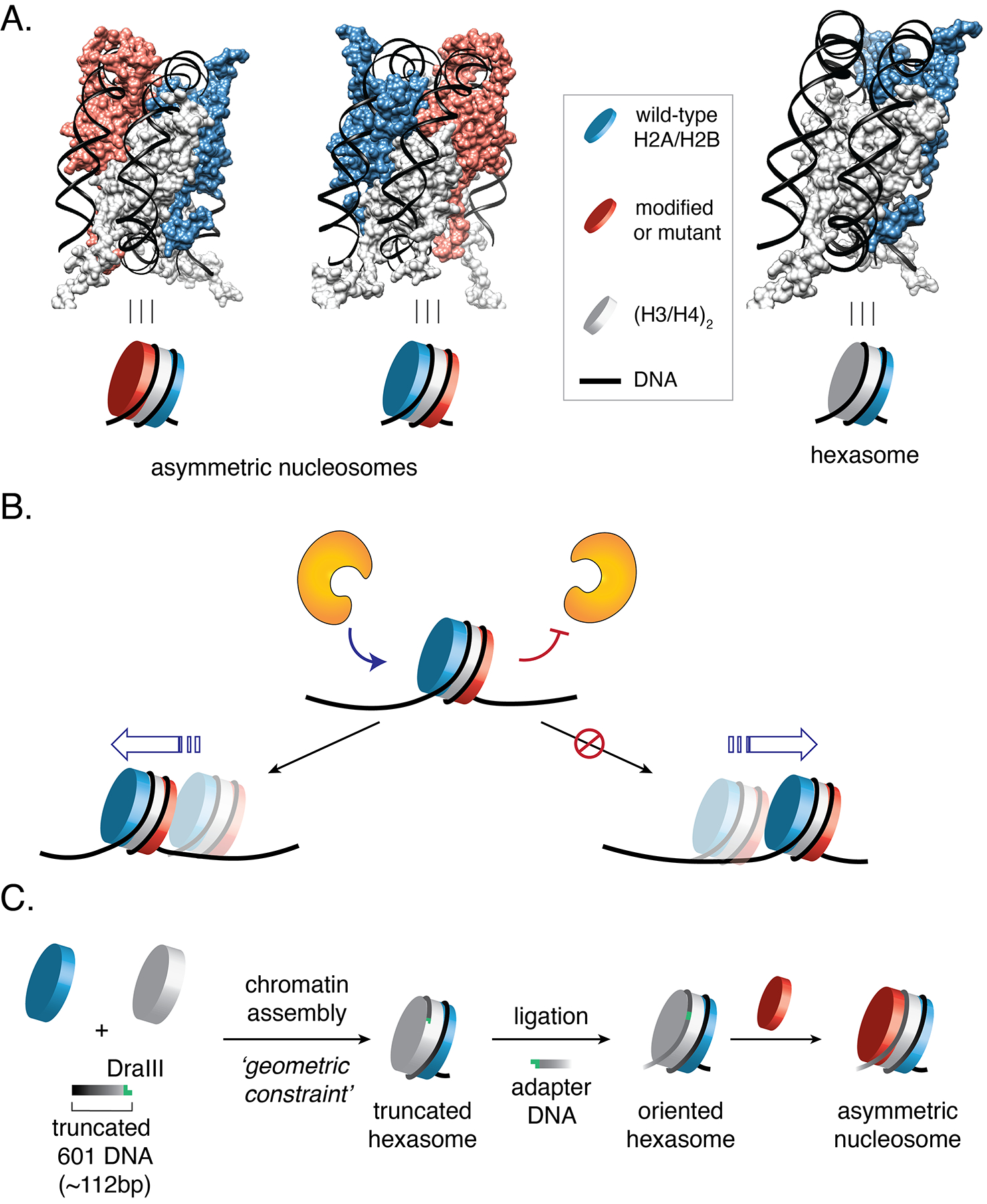
(A) Structural models of asymmetric nucleosomes featuring distinct symmetry-related (‘sister’) H2A/H2B dimers and a hexasome lacking one H2A/H2B dimer. Models derived from a previous nucleosome structure, PDB: 1kx5. (B) Asymmetric nucleosomes and hexasomes can influence the outcome of molecular transactions that operate on chromatin. Shown is a hypothetical scenario where desymmetrization alters the directionality of nucleosome remodeling. (C) Schematic of the DNA-template editing strategy developed in this study for synthesis of oriented hexasomes and asymmetric nucleosomes.
While nucleosome desymmetrization is an inherent feature of chromatin structure in both normal and disease states, biochemical studies employing these substrates are hampered by the lack of practical methods for their preparation. In a typical nucleosome reconstitution, the (H3/H4)2 tetramer is thought to first assemble onto DNA to generate a tetrasome species, followed by stepwise incorporation of two H2A/H2B dimers to yield hexasome intermediates and the final nucleosomes (Fig. S1A)12. This assembly pathway makes access to asymmetric nucleosomes nontrivial, with current approaches requiring elaborate protein chemistry manipulations and multistep purification workflows4,13,14. Further complicating matters is the orientation of an asymmetric histone octamer or hexamer with respect to the underlying DNA sequence. Controlling this directionality is highly desirable when studying many aspects of chromatin biochemistry, for example the impact of asymmetry on remodeling processes (Fig. 1B)15,16. Currently, the only route to oriented, asymmetric nucleosome substrates hinges on an intrinsic property of the commonly used Widom 601 nucleosome positioning sequence which, as a consequence of bias in the sequence content (i.e. high frequency in periodic TA and GC steps on the proximal side (Fig. S1B)), favors the generation of oriented hexasomes17,18,19. Bowman and co-workers exploited this feature to develop a two-step protocol for the synthesis of oriented asymmetric nucleosomes carrying chemically distinct H2A/H2B dimer histones (Fig. S1C)18. This approach requires empirical optimization of the dimer-tetramer ratios in the first step (see below) and either electrophoretic18 or affinity purification8 to separate the desired hexasomes or asymmetric nucleosomes from contaminating symmetric nucleosomes which are also generated in the first step of the process. Moreover, this strategy is not compatible with DNA sequences that lack the aforementioned directing features of Widom-601.
Motivated by the need for a more flexible route to oriented hexasomes and asymmetric nucleosomes, we report here an approach based on the use of truncated DNA templates that set both the composition and orientation of the nucleoprotein complex (Figs. 1C, S1D). We show that this template editing strategy can be used to access a variety of asymmetric species including those deposited on non-Widom 601 DNA. The utility of our method is showcased through the synthesis of a novel FRET-based designer nucleosome system, which enabled investigation into the impact of asymmetry on the activity of the mammalian SWI/SNF (BAF) chromatin remodeling complex.
RESULTS AND DISCUSSION
Development of a DNA Templated Editing Strategy To Synthesize Hexasomes and Asymmetric Nucleosomes.
We began our studies by considering the unique structural properties of hexasomes (Fig. 2). Previous work has shown that ~40 bp of DNA becomes unwrapped from the histone core as a consequence of the missing H2A/H2B dimer in the complex11,20. We reasoned that this ‘unleashed’ DNA does not influence the generation of hexasomes and that it would therefore be possible to selectively generate these subnucleosomal complexes using appropriately truncated DNA. To test this idea, a series of truncated 601 DNA sequences were generated in which increasing numbers of nucleotides (X = 0, 15, 18, 25, 35) were systematically removed from the template (Figs. 3A, S2A). Note, since H2A/H2B dimers preferentially interact with the TA-rich side of the 601 sequence18, we initially truncated from the side distal to this region (Fig. S1D). As expected15,18, native gel electrophoresis indicated the formation of both hexasome 1a and nucleosome 2a when full length 601 DNA was used in the reconstitution (Fig. 3A, X = 0). Lowering the dimer to tetramer ratio was found to improve the hexasome/nucleosome ratio, albeit at the expense of generating a slow mobility-shift species which we presume to be an undesired tetrasome (i.e. H3/H4 tetramer + DNA). The use of truncated 601 DNA sequences in the assembly also gave rise to two major species, which we characterized as nucleosomes 2b-e and hexasomes 1b-e (Fig. 3A and S2B). Excitingly, and consistent with our design, the hexasome selectivity increases as we shorten the 601 DNA, with virtually no nucleosome products observed when we removed 35 nts from the end of the Widom sequence (i.e., hexasome 1e) (Figs. 3A, S2C and S2D).
Figure 2. Structural properties of hexasomes.
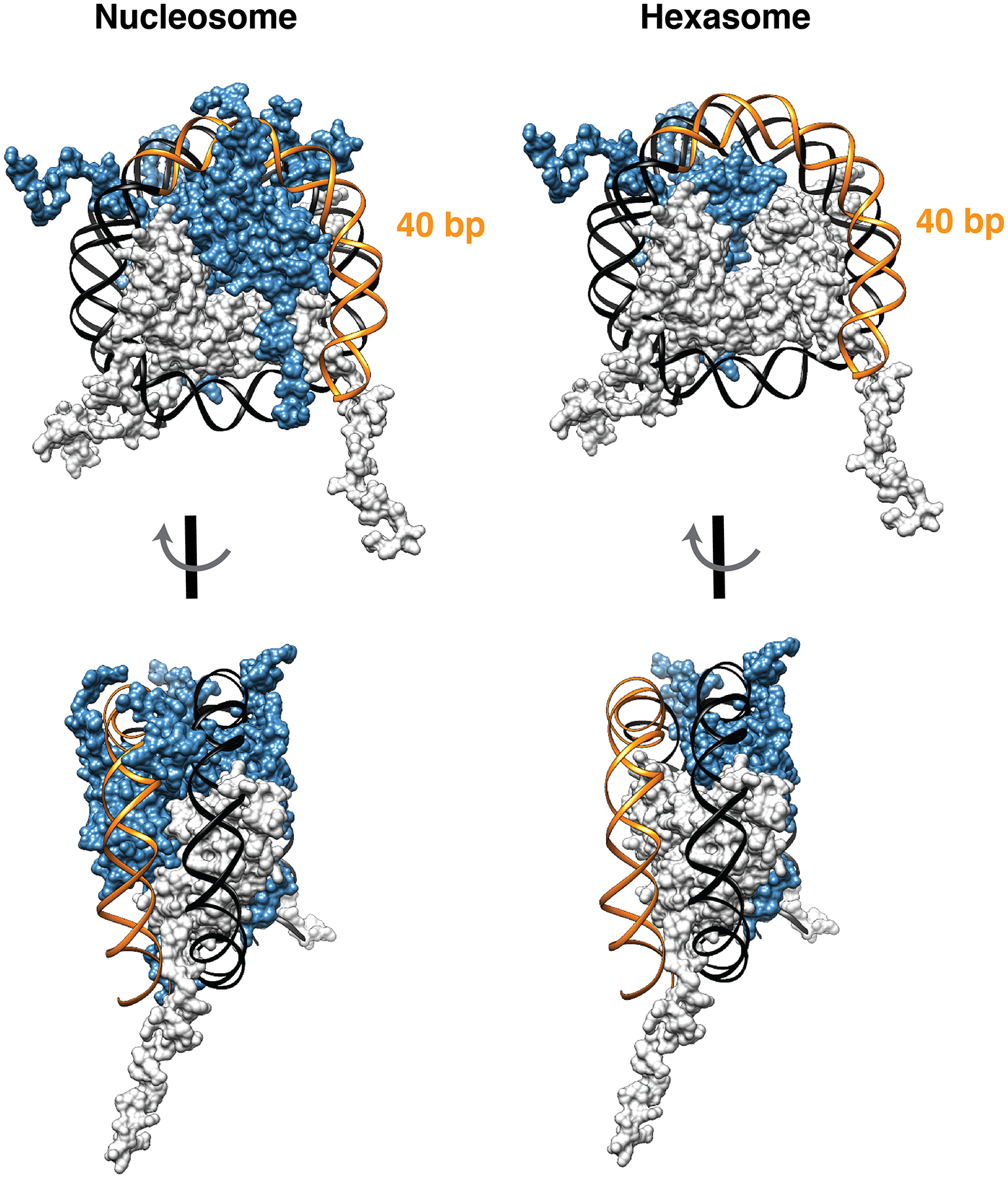
The hexasome structures (right panels) were generated by removing a dimer from the nucleosome structures (left panels) (PDB: 1kx5). The 40-base pair DNA segment that does not interact with the histone core in the context of the hexasome is highlighted in orange.
Figure 3. Synthesis of hexasomes and asymmetric nucleosomes via the DNA templated editing strategy.
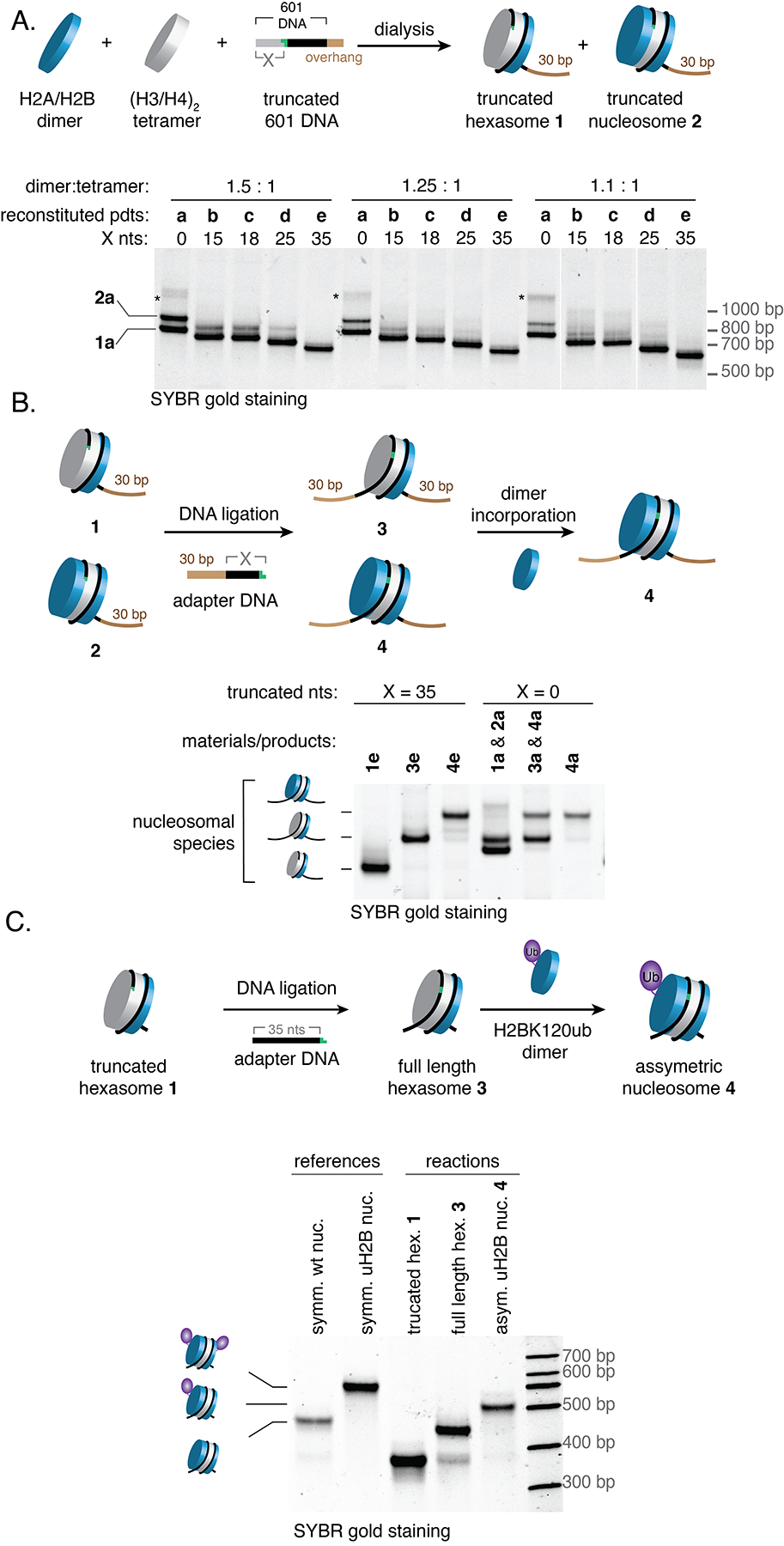
(A) Truncated 601 DNAs enable reconstitution of oriented hexasomes. Truncated 601 DNAs (X = 0, 15, 18, 25, 35) were combined with histone dimers and tetramers at the indicated ratios. The resulting truncated hexasome 1 and truncated nucleosome (2) products were analyzed by native gel electrophoresis (7% acrylamide) with SYBR gold staining. * = putative tetrasomes. (B) Truncated hexasomes/nucleosomes mixtures 1, 2 were converted to full-length hexasomes/nucleosomes 3, 4 in a two-step process involving ligation of a DNA adapter followed by the incorporation of a second H2A/H2B dimer. Products (X = 0, 35) were analyzed by native gel electrophoresis (7% acrylamide) with SYBR gold staining. The crude reaction mixtures involving 1a (X = 0) and 1e (X = 35) are shown. (C) Incorporation of an H2A/uH2B dimer to a hexasome to generate an asymmetric uH2B nucleosome. Note that the hexasome used in the example does not have a 30 bp overhang. Analyses were performed by native gel (5.5% acrylamide) with SYBR gold staining. The mobility shift of the asymmetric uH2B nucleosome is in-between that of symmetric wildtype and symmetric uH2B nucleosomes (references) as revealed by DNA staining. The exchange of the H2A/uH2B dimer with the dimer in the hexasome to generate the symmetrical uH2B nucleosome did not occur. uH2B = H2BK120ub; symm. = symmetric; nuc. = nucleosome; hex. = hexasome; asym. = asymmetric.
We then turned to the conversion of reconstituted truncated products into hexasomal species containing the full-length Widom 601 DNA. For this we employed a T4-mediated DNA ligation step, taking advantage of a DraIII-generated sticky end in the double stranded template DNA to which we could attach an adapter containing the missing 601 sequence and a 30 bp overlap. Thus, the ligated products will have 30 bp flanking DNA either side of the 601 sequence (i.e. 30N30). As expected, the mixture containing full length hexasome 1a and nucleosome 2a underwent ligation to yield two corresponding 30N30 products as indicated by gel electrophoresis (Fig. 3B, X = 0). The same ligation reaction was then applied to truncated hexasomes, 1d (X=25) and 1e (X=35), leading to the efficient generation of identical mobility shift products corresponding to 30N30 hexasomes, 3d and 3e (Fig. 3B, S2E). Gratifyingly, hexasome 3e could then be assembled into nucleosome 4e, by addition of a second H2A/H2B dimer equivalent to the ligated products (Fig. 3B). We then extended our method to the generation of oriented asymmetric nucleosomes by incorporating a dimer carrying H2B ubiquitylated on lysine 120 (abbreviated to H2BK120ub, or uH2B) in the second step. Importantly, native gel analysis indicated the generation of the desired heterotypic species with virtually no exchange between the second and first histone dimers (Fig. 3C).
Synthesis of Oriented Hexasomes, Asymmetric Nucleosomes and Dinucleosomes.
Next, we conducted experiments to explore the scope of the approach. Oriented hexasomes and asymmetric nucleosomes carrying different flanking DNAs on the distal side were easily accessed by varying the length of the adapter DNAs used in the ligation step, including an overhang containing an entire Widom 601 sequence (Fig. 4A). Likewise, different flanking DNAs on the proximal side were prepared by varying the truncated 601 sequence (Fig. S3A). The approach also permitted facile generation of asymmetric nucleosomes containing fluorescent probes on the DNA and histones with specific orientations (Fig. S3B). These designer chromatins are attractive substrates for nucleosome mobility assays (see below).
Figure 4. DNA template editing strategy enables access to a variety of oriented hexasomes, nucleosomes and dinucleosomes.
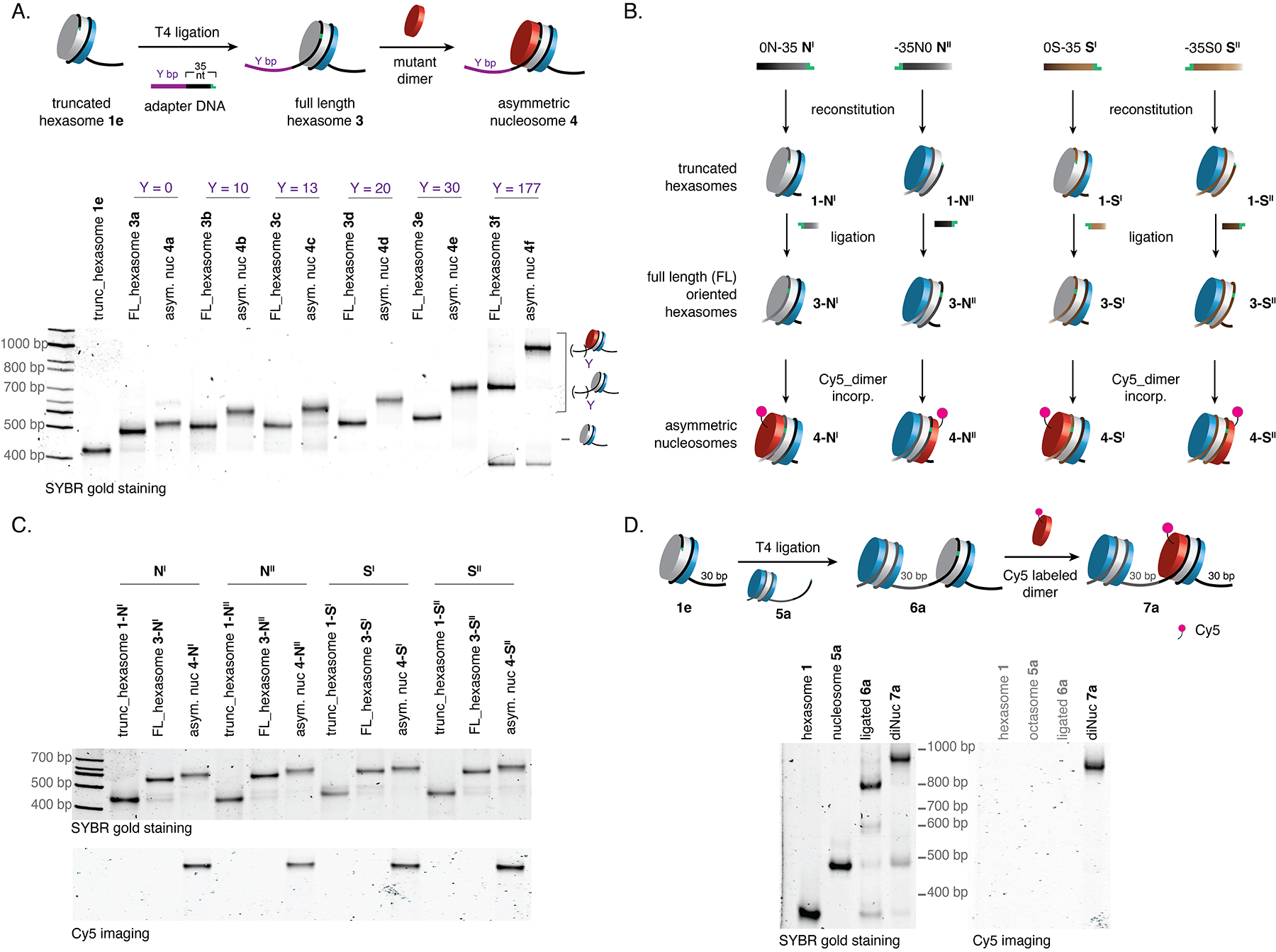
(A) Synthesis of oriented hexasomes and asymmetric nucleosomes carrying different flanking overhangs. The truncated hexasome (1e, X = 35) was ligated with adapter DNAs with variable length, followed by the incorporation of the second H2B/H2B dimer containing an H2A triple mutant (H2AE61A,D90A,E92A; red). Analyses of crude reaction mixtures were performed by native gel electrophoresis (5.5% acrylamide) with SYBR gold staining. (B) Schematic showing the stepwise synthesis of different configurations of oriented hexasomes and asymmetric nucleosomes from truncated Widom 601 sequences (NI and NII) and 5S DNA sequences (SI and SII) with a Cy5 labeled H2A/H2B dimer used in the final step. (C) Native gel analyses (5.5% acrylamide) showing the generation of different oriented hexasomes and asymmetric nucleosomes from the truncated 601 sequences and 5S sequences. Top gel: staining of total DNA with SYBR gold dye to assess the formation of truncated hexasomes, ligated hexasomes and nucleosomes. Bottom gel: imaging of Cy5 signal to assess the incorporation of labeled dimers to generate asymmetric nucleosomes. (D) Synthesis of dinucleosomes carrying a hexasome unit 6a or an asymmetric nucleosome unit 7a. The truncated hexasome (1e, X = 35) was ligated with nucleosome 5a to generate 6a, which was then transformed into asymmetric dinucleosome 7a by the incorporation of a Cy5 labeled dimer. Analyses of crude reaction mixtures were performed by native gel electrophoresis (5.5% acrylamide) with SYBR gold staining and Cy5 imaging. trunc. = truncated; FL = full length; incorp. = incorporation; diNuc = dinucleosome.
Given the efficiency of the hexasome assembly, we asked whether our strategy could be applied to Widom 601-sequences truncated from the TA-rich side (the proximal side), or to other nucleosome positioning sequences such as 5S DNA21. To test this, we prepared two truncated 601 DNA templates, NI and NII, corresponding to truncation from the TA-poor side and TA-rich side, respectively, of a template with no flanking overhangs (Fig. 4B and S4A). Similarly, we generated two truncated 5S DNA sequences, SI and SII (Figs. 4B and S4A). Each of these templates was successfully used in the generation of oriented hexasomes and asymmetric nucleosomes following our general protocol (Figs. 4B,C). Use of H2A/uH2B dimers in the second step indicated a negligible level of dimer exchange in the process (Fig. S4B).
Many chromatin factors simultaneously engage multiple nucleosomes as part of their function22,23. Thus, we wondered whether our technology could be extended beyond a mononucleosome context. To this end, we asked whether oriented nucleosome-hexasome species 6 could be generated by T4-mediated ligation of truncated hexasome 1 and preformed nucleosome, 5 (Fig. 4D). This reaction was found to proceed smoothly and, due to its modularity, allowed the linker length between the two units to be easily varied (Fig. S5A). Importantly, we found that the hexasome unit in 6 could assimilate a heterotypic histone dimer (in this case, Cy5 labeled) to furnish the corresponding dinucleosomes, 7, in which the orientation of (and the linker length between) the asymmetric and symmetric building blocks are defined (Figs. 4D, S5A,B). To our knowledge, this is the first time designer chromatins of this type have been prepared, an advance that will allow investigations into the role of asymmetry to be extended to processes that require multinucleosomal substrates.
Acidic Patch Mutations Reprogram the Inherent Remodeling Activity of cBAF.
Having established the robustness of our method, we then turned our attention to applying the approach to a specific biochemical question, namely how asymmetric nucleosomes carrying cancer-associated histone mutations are interpreted by the BRG1/BRM associated factor (BAF) family of chromatin remodeling complex. BAF complexes are the human analogs of the yeast SWI/SNF (SWItch/Sucrose Non-Fermentable) ATP-dependent chromatin remodeling family that regulate chromatin accessibility to facilitate transcription24. These megadalton-sized molecular machines can be classified into three sub-families: canonical BAF (cBAF), polybromo-associated BAF (PBAF) and non-canonical BAF (ncBAF)25. BAF complexes, which are mutated in over 20% of human cancers26, are known to respond to various histone features including cancer-associated mutations27. However, the impact of asymmetry on this interplay has not been investigated. In particular, we were interested in a symmetry-related pair of nucleosomal epitopes defined by acidic residues on each H2A/H2B dimer. These so-called ‘acidic patches’ are known to engage in a bilateral interaction with cBAF28,29,30. While mutation of both acidic patches has been shown to abolish the remodeling activity of all three human BAF complexes27, it is unclear how mutation of just one of the epitopes affects BAF activity. This is especially relevant for cancer-associated histone mutations which lead to nucleosome desymmetrization due to their stochastic incorporation into chromatin along with the more abundant wild-type versions5,6.
To probe this question, we prepared a series of nucleosomes carrying mutations centered on a key epitope of the acidic patch, the arginine anchor (defined by residues H2AE61,D90,E92), that forms a docking site for nucleosome engagement31,32,15. Four different nucleosomes were generated, each positioned on DNA template such that there is no overhang on one side and a 60 bp overhang on the other (i.e. 0N60): 1) asymmetric syn-Nucmut, which is an asymmetric nucleosome containing a mutant arginine anchor proximal to the longer overhang; 2) asymmetric anti-Nucmut, an asymmetric nucleosome containing a mutant arginine anchor distal to the long overhang; 3) symmetric-Nucmut with the mutations on both sides; 4) symmetric-Nucwt carrying two wild type copies of the H2A/H2B dimer (Fig. S6A). We strategically installed a Cy5 dye on one histone dimer and a Cy3 dye on one side of the DNA for monitoring of the remodeling reactions using fluorescence-based assays.
By following the Cy5 signal in electrophoretic mobility shift assays (EMSAs), we observed that cBAF remodeled the end positioned symmetric-Nucwt to generate multiple slower mobility shift nucleosomal species, indicating that the purified cBAF was active and functional on these templates (Fig. 5A, top left panel). Importantly, at equilibrium, the major components of the reaction mixture corresponded to end positioned nucleosomes, consistent with previous findings that SWI/SNF prefers to move nucleosomes from centered to end positions33,34,35,36. As expected27, mutations of both acidic patches abolished cBAF remodeling activity (Fig. 5A, top right panel). Remarkably, cBAF operated differently on the two oriented asymmetric syn- and anti-nucleosomes (Fig. 5A, bottom panels). The former asymmetric nucleosome yielded similar remodeled species as those observed for Nucwt (Fig. 5A, bottom left vs top left). However, different from Nucwt, the predominate form generated corresponded to a centered nucleosome species, suggesting that the acidic patch mutant can alter the behavior of cBAF remodeling (Fig. 5A, top left vs bottom left, Fig. S6B). Importantly, a similar result was observed using an asymmetric nucleosome carrying the cancer-associated mutation, H2AE92K (Fig. S6C). By contrast, the asymmetric anti-Nucmut substrate did not generate slower-migrating species when subjected to cBAF remodeling (Fig. 5A, bottom right). Rather, we observed a decrease in the Cy5 signal (Fig. 5A, S6D), suggesting a loss of the labeled mutant dimer. Consistent with this interpretation, we observed the generation of a faster migrating species by native gel electrophoresis (Fig. S6E). These results suggest that cBAF requires at least one native acidic patch for its function. In agreement with this idea, we found that cBAF can remodel hexasomes carrying one wild-type acidic patch, whereas the corresponding acidic patch mutant was a poor substrate for the enzyme (Fig. S6F).
Figure 5. Acidic patch mutations reprogram the inherent remodeling activity of cBAF.
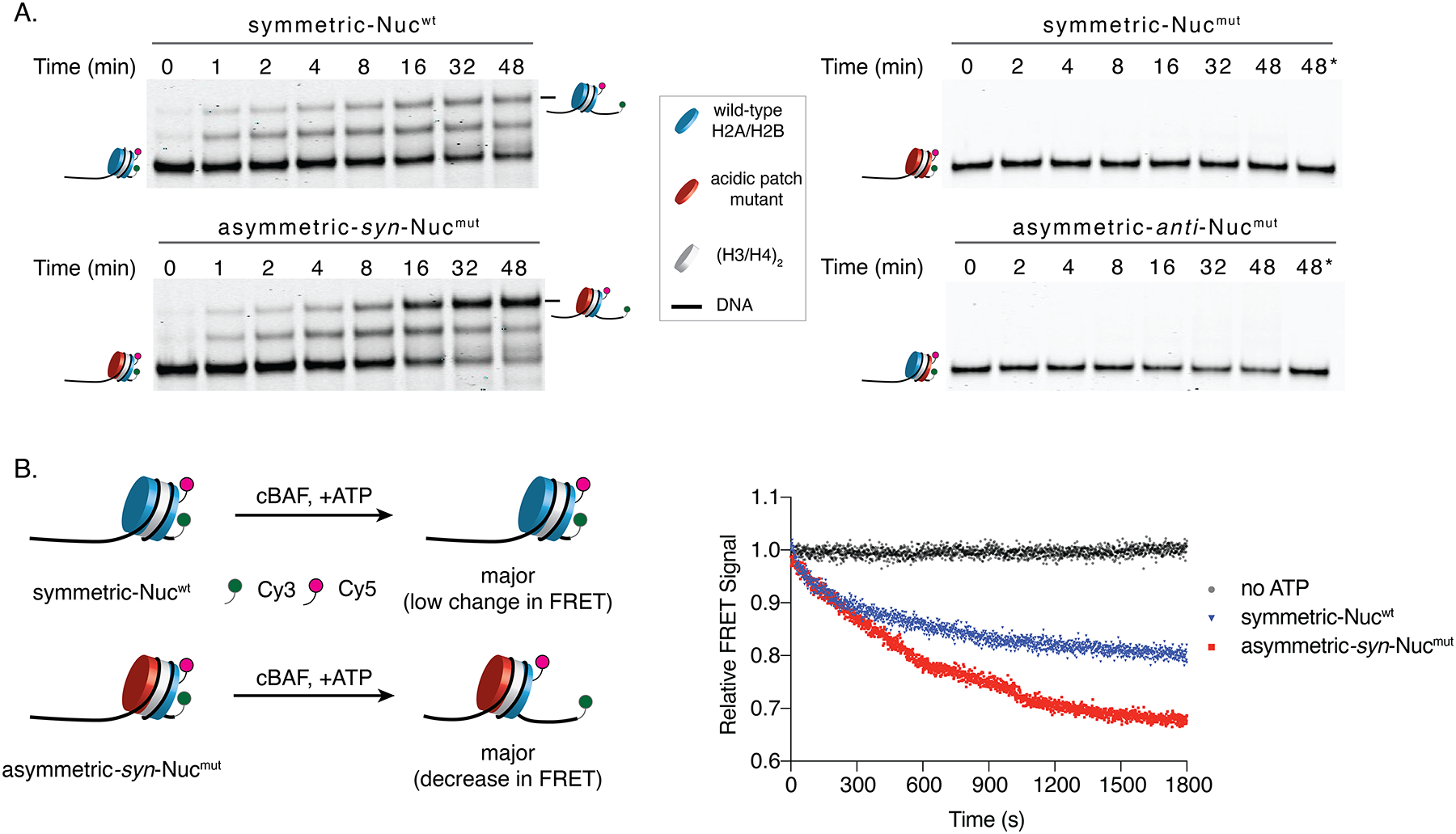
(A) Remodeling activity of cBAF on indicated symmetric and asymmetric nucleosomes as readout by EMSA. cBAF was incubated with an end-positioned nucleosome containing a 60 bp overhang (0N60) in the presence of ATP. Aliquots of the reaction mixtures were evaluated at indicated time points by monitoring the Cy5 signal on native gel (representative of 3 independent experiments). Acidic patch mutant: H2AE61AD90AE92A. (*) Reaction conducted for indicated time without ATP. (B) Left: Schematic of the FRET-based remodeling assays used symmetric (Nucwt) and asymmetric (syn-Nucmut) mononucleosomes containing the Cy3-Cy5 pair. Right: Remodeling activity of cBAF on Nucwt and syn-Nucmut as assessed by the ensemble FRET assay. cBAF was incubated with indicated substrates in the presence and absence of ATP. An average of normalized FRET signals (n=3 independent experiments) as a function of time is shown.
The EMSA data suggests that cBAF preferentially translocates asymmetric nucleosomes in the direction that generates longer flanking DNAs proximal to the native acidic patch. Notably, this trend is opposite to what is observed in the ISWI family of chromatin remodelers15,16. To further demonstrate the effect of the acidic patch mutations on the directionality of cBAF-mediated nucleosome sliding, we measured the ensemble fluorescence resonance energy transfer (FRET) signal between the Cy3-labeled DNAs and the Cy5-labeled histone dimers as the function of chromatin remodeling (Figs. 5B, S6G). A significantly larger time- and ATP-dependent decrease in FRET was observed when monitoring cBAF remodeling of asymmetric syn-Nucmut as compared to the wild-type substrate. This is consistent with a higher tendency to generate center position species from the end position in the case of asymmetric syn-Nucmut. Together, these results indicate that mutations in the acidic patch can reprogram the inherent remodeling activity of cBAF. Since BAF complexes play critical roles in defining the architecture of chromatin at regulatory sites (e.g. promoters, enhancers), malfunction in BAF remodeling as the consequence of asymmetric acidic patch mutations at these sites would be expected to lead to altered transcriptional programs. By the same token, histone PTMs and variants, many of which can activate or inhibit BAF complex-mediated remodeling27, could also lead to differential remodeling effects depending on whether they are incorporated symmetrically or asymmetrically into nucleosomes.
CONCLUSIONS
In conclusion, we have developed a general strategy for the preparation of hexasomes and asymmetric nucleosomes with defined orientations relative to the underlying DNA sequence. A key feature of the method is use of a truncated nucleosome positioning sequence that favors the generation of hexasome intermediates, which can then be further elaborated into a variety of asymmetric species, including oriented dinucleosomes. This modular template editing approach was used to examine the impact of nucleosome desymmetrization on the activity of the cBAF chromatin remodeling complex, revealing that asymmetric incorporation of cancer-associated histone mutations can alter the directional bias of remodeling. More generally, we imagine that the procedures described herein will help illuminate the functional impact of asymmetry in the regulation of a wide range of epigenetic processes.
Supplementary Material
ACKNOWLEDGMENT
This work was funded by NIH-NCI grants P01 CA196539 and R01 CA240768. H.T.D. was supported by a Jane Coffins Child Memorial Fund Postdoctoral Fellowship. N.M. is supported by NIH K99/R00 K99CA237855 and C.K. by National Institutes of Health Grant 1DP2CA195762-01 and the Mark Foundation for Cancer Research Emerging Leader Award. We thank R. E. Thompson for providing Pfu-Sso7d polymerase. We thank J. D. Bagert for providing uH2B dimer. We thank members of the Muir laboratory and Kadoch laboratory for helpful discussions.
Footnotes
Supporting Information
Experimental details and supplementary figures.
C.K. is the Scientific Founder, fiduciary Board of Directors member, Scientific Advisory Board member, shareholder, and consultant for Foghorn Therapeutics, Inc. (Cambridge, MA), serves on Scientific Advisory Boards of Nereid Therapeutics, Nested Therapeutics, and is a consultant for Cell Signaling Technologies. The other authors declare no competing financial interests.
REFERENCES
- (1).Luger K; Mäder AW; Richmond RK; Sargent DF; Richmond TJ Crystal Structure of the Nucleosome Core Particle at 2.8 Å Resolution. Nature 1997, 389 (6648), 251–260. 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- (2).Rhee HS; Bataille AR; Zhang L; Pugh BF Subnucleosomal Structures and Nucleosome Asymmetry across a Genome. Cell 2014, 159 (6), 1377–1388. 10.1016/j.cell.2014.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Li S; Shogren-Knaak MA Cross-Talk between Histone H3 Tails Produces Cooperative Nucleosome Acetylation. Proc. Natl. Acad. Sci 2008, 105 (47), 18243–18248. 10.1073/pnas.0804530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Voigt P; LeRoy G; Drury WJ; Zee BM; Son J; Beck DB; Young NL; Garcia BA; Reinberg D Asymmetrically Modified Nucleosomes. Cell 2012, 151 (1), 181–193. 10.1016/j.cell.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Lewis PW; Müller MM; Koletsky MS; Cordero F; Lin S; Banaszynski LA; Garcia BA; Muir TW; Becher OJ; Allis CD Inhibition of PRC2 Activity by a Gain-of-Function H3 Mutation Found in Pediatric Glioblastoma. Science 2013, 340 (6134), 857–861. 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Lu C; Jain SU; Hoelper D; Bechet D; Molden RC; Ran L; Murphy D; Venneti S; Hameed M; Pawel BR; Wunder JS; Dickson BC; Lundgren SM; Jani KS; De Jay N; Papillon-Cavanagh S; Andrulis IL; Sawyer SL; Grynspan D; Turcotte RE; Nadaf J; Fahiminiyah S; Muir TW; Majewski J; Thompson CB; Chi P; Garcia BA; Allis CD; Jabado N; Lewis PW Histone H3K36 Mutations Promote Sarcomagenesis through Altered Histone Methylation Landscape. Science 2016, 352 (6287), 844–849. 10.1126/science.aac7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Nacev BA; Feng L; Bagert JD; Lemiesz AE; Gao J; Soshnev AA; Kundra R; Schultz N; Muir TW; Allis CD The Expanding Landscape of ‘Oncohistone’ Mutations in Human Cancers. Nature 2019, 567 (7749), 473–478. 10.1038/s41586-019-1038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Bagert JD; Mitchener MM; Patriotis AL; Dul BE; Wojcik F; Nacev BA; Feng L; Allis CD; Muir TW Oncohistone Mutations Enhance Chromatin Remodeling and Alter Cell Fates. Nat. Chem. Biol 2021, 17 (4), 403–411. 10.1038/s41589-021-00738-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Kireeva ML; Walter W; Tchernajenko V; Bondarenko V; Kashlev M; Studitsky VM Nucleosome Remodeling Induced by RNA Polymerase II: Loss of the H2A/H2B Dimer during Transcription. Mol. Cell 2002, 9 (3), 541–552. 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- (10).Kuryan BG; Kim J; Tran NNH; Lombardo SR; Venkatesh S; Workman JL; Carey M Histone Density Is Maintained during Transcription Mediated by the Chromatin Remodeler RSC and Histone Chaperone NAP1 in Vitro. Proc. Natl. Acad. Sci 2012, 109 (6), 1931–1936. 10.1073/pnas.1109994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Arimura Y; Tachiwana H; Oda T; Sato M; Kurumizaka H Structural Analysis of the Hexasome, Lacking One Histone H2A/H2B Dimer from the Conventional Nucleosome. Biochemistry 2012, 51 (15), 3302–3309. 10.1021/bi300129b. [DOI] [PubMed] [Google Scholar]
- (12).Hansen JC; van Holde KE; Lohr D The Mechanism of Nucleosome Assembly onto Oligomers of the Sea Urchin 5 S DNA Positioning Sequence. J. Biol. Chem 1991, 266 (7), 4276–4282. [PubMed] [Google Scholar]
- (13).Lechner CC; Agashe ND; Fierz B Traceless Synthesis of Asymmetrically Modified Bivalent Nucleosomes. Angew. Chem. Int. Ed 2016, 55 (8), 2903–2906. 10.1002/anie.201510996. [DOI] [PubMed] [Google Scholar]
- (14).Mitchener MM; Muir TW Janus Bioparticles: Asymmetric Nucleosomes and Their Preparation Using Chemical Biology Approaches. Acc. Chem. Res 2021, 54 (16), 3215–3227. 10.1021/acs.accounts.1c00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Dao HT; Dul BE; Dann GP; Liszczak GP; Muir TW A Basic Motif Anchoring ISWI to Nucleosome Acidic Patch Regulates Nucleosome Spacing. Nat. Chem. Biol 2020, 16 (2), 134–142. 10.1038/s41589-019-0413-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Levendosky RF; Bowman GD Asymmetry between the Two Acidic Patches Dictates the Direction of Nucleosome Sliding by the ISWI Chromatin Remodeler. eLife 2019, 8, e45472. 10.7554/eLife.45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Chua EYD; Vasudevan D; Davey GE; Wu B; Davey CA The Mechanics behind DNA Sequence-Dependent Properties of the Nucleosome. Nucleic Acids Res 2012, 40 (13), 6338–6352. 10.1093/nar/gks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Levendosky RF; Sabantsev A; Deindl S; Bowman GD The Chd1 Chromatin Remodeler Shifts Hexasomes Unidirectionally. eLife 2016, 5, e21356. 10.7554/eLife.21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Brehove M; Shatoff E; Donovan BT; Jipa CM; Bundschuh R; Poirier MG DNA Sequence Influences Hexasome Orientation to Regulate DNA Accessibility. Nucleic Acids Res 2019, 47 (11), 5617–5633. 10.1093/nar/gkz283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Rychkov GN; Ilatovskiy AV; Nazarov IB; Shvetsov AV; Lebedev DV; Konev AY; Isaev-Ivanov VV; Onufriev AV Partially Assembled Nucleosome Structures at Atomic Detail. Biophys. J 2017, 112 (3), 460–472. 10.1016/j.bpj.2016.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Thåström A; Lowary PT; Widlund HR; Cao H; Kubista M; Widom J Sequence Motifs and Free Energies of Selected Natural and Non-Natural Nucleosome Positioning DNA Sequences. J. Mol. Biol 1999, 288 (2), 213–229. 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- (22).Poepsel S; Kasinath V; Nogales E Cryo-EM Structures of PRC2 Simultaneously Engaged with Two Functionally Distinct Nucleosomes. Nat. Struct. Mol. Biol 2018, 25 (2), 154–162. 10.1038/s41594-018-0023-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Dechassa ML; Sabri A; Pondugula S; Kassabov SR; Chatterjee N; Kladde MP; Bartholomew B SWI/SNF Has Intrinsic Nucleosome Disassembly Activity That Is Dependent on Adjacent Nucleosomes. Mol. Cell 2010, 38 (4), 590–602. 10.1016/j.molcel.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Wang W; Xue Y; Zhou S; Kuo A; Cairns BR; Crabtree GR Diversity and Specialization of Mammalian SWI/SNF Complexes. Genes Dev 1996, 10 (17), 2117–2130. 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- (25).Mashtalir N; D’Avino AR; Michel BC; Luo J; Pan J; Otto JE; Zullow HJ; McKenzie ZM; Kubiak RL; St. Pierre R; Valencia AM; Poynter SJ; Cassel SH; Ranish JA; Kadoch C Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell 2018, 175 (5), 1272–1288.e20. 10.1016/j.cell.2018.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kadoch C; Hargreaves DC; Hodges C; Elias L; Ho L; Ranish J; Crabtree GR Proteomic and Bioinformatic Analysis of Mammalian SWI/SNF Complexes Identifies Extensive Roles in Human Malignancy. Nat. Genet 2013, 45 (6), 592–601. 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Mashtalir N; Dao HT; Sankar A; Liu H; Corin AJ; Bagert JD; Ge EJ; D’Avino AR; Filipovski M; Michel BC; Dann GP; Muir TW; Kadoch C Chromatin Landscape Signals Differentially Dictate the Activities of MSWI/SNF Family Complexes. Science 2021, 373 (6552), 306–315. 10.1126/science.abf8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Valencia AM; Collings CK; Dao HT; Pierre RS; Cheng Y-C; Huang J; Sun Z-Y; Seo H-S; Mashtalir N; Comstock DE; Bolonduro O; Vangos NE; Yeoh ZC; Dornon MK; Hermawan C; Barrett L; Dhe-Paganon S; Woolf CJ; Muir TW; Kadoch C Recurrent SMARCB1 Mutations Reveal a Nucleosome Acidic Patch Interaction Site That Potentiates MSWI/SNF Complex Chromatin Remodeling. Cell 2019, 179 (6), 1342–1356.e23. 10.1016/j.cell.2019.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Mashtalir N; Suzuki H; Farrell DP; Sankar A; Luo J; Filipovski M; D’Avino AR; St. Pierre R; Valencia AM; Onikubo T; Roeder RG; Han Y; He Y; Ranish JA; DiMaio F; Walz T; Kadoch C A Structural Model of the Endogenous Human BAF Complex Informs Disease Mechanisms. Cell 2020, 183 (3), 802. 10.1016/j.cell.2020.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).He S; Wu Z; Tian Y; Yu Z; Yu J; Wang X; Li J; Liu B; Xu Y Structure of Nucleosome-Bound Human BAF Complex. Science 2020, 367 (6480), 875–881. 10.1126/science.aaz9761. [DOI] [PubMed] [Google Scholar]
- (31).McGinty RK; Tan S Nucleosome Structure and Function. Chem. Rev 2015, 115 (6), 2255–2273. 10.1021/cr500373h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).McGinty RK; Tan S Principles of Nucleosome Recognition by Chromatin Factors and Enzymes. Curr. Opin. Struct. Biol 2021, 71, 16–26. 10.1016/j.sbi.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Saha A; Wittmeyer J; Cairns BR Chromatin Remodeling by RSC Involves ATP-Dependent DNA Translocation. Genes Dev 2002, 16 (16), 2120–2134. 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Kassabov SR; Zhang B; Persinger J; Bartholomew B SWI/SNF Unwraps, Slides, and Rewraps the Nucleosome. Mol. Cell 2003, 11 (2), 391–403. 10.1016/S1097-2765(03)00039-X. [DOI] [PubMed] [Google Scholar]
- (35).Fan H-Y; He X; Kingston RE; Narlikar GJ Distinct Strategies to Make Nucleosomal DNA Accessible. Mol. Cell 2003, 11 (5), 1311–1322. 10.1016/S1097-2765(03)00192-8. [DOI] [PubMed] [Google Scholar]
- (36).Flaus A; Owen-Hughes T Dynamic Properties of Nucleosomes during Thermal and ATP-Driven Mobilization. Mol. Cell. Biol 2003, 23 (21), 7767–7779. 10.1128/MCB.23.21.7767-7779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


