Abstract
Human apolipoprotein E (ApoE) is a 299-amino acid secreted glycoprotein that binds cholesterol and phospholipids. ApoE exists as three common isoforms (ApoE2, ApoE3, and ApoE4) and heterozygous carriers of the ε4 allele of the gene encoding ApoE (APOE) have a fourfold greater risk of developing Alzheimer’s disease (AD). The enzymes thrombin, cathepsin D, α-chymotrypsin-like serine protease, and high-temperature requirement serine protease A1 are responsible for ApoE proteolytic processing resulting in bioactive C-terminal-truncated fragments that vary depending on ApoE isoforms, brain region, aging, and neural injury. The objectives of the present narrative review were to describe ApoE processing, discussing current hypotheses about the potential role of various ApoE fragments in AD pathophysiology, and reviewing the current development status of different anti-ApoE drugs. The exact mechanism by which APOE gene variants increase/decrease AD risk and the role of ApoE fragments in the deposition are not fully understood, but APOE is known to directly affect tau-mediated neurodegeneration. ApoE fragments co-localize with neurofibrillary tangles and amyloid β (Aβ) plaques, and may cause neurodegeneration. Among anti-ApoE approaches, a fascinating strategy may be to therapeutically overexpress ApoE2 in APOE ε4/ε4 carriers through vector administration or liposomal delivery systems. Another approach involves reducing ApoE4 expression by intracerebroventricular antisense oligonucleotides that significantly decreased Aβ pathology in transgenic mice. Differences in the proteolytic processing of distinct ApoE isoforms and the use of ApoE fragments as mimetic peptides in AD treatment are also under investigation. Treatment with peptides that mimic the structural and biological properties of native ApoE may reduce Aβ deposition, tau hyperphosphorylation, and glial activation in mouse models of Aβ pathology. Alternative strategies involve the use of ApoE4 structure correctors, passive immunization to target a certain form of ApoE, conversion of the ApoE4 aminoacid sequence into that of ApoE3 or ApoE2, and inhibition of the ApoE-Aβ interaction.
Keywords: Alzheimer’s disease, antisense oligonucleotides, apolipoprotein E, dementia, fragments, proteolysis, tau protein, therapeutics
Introduction
The human apolipoprotein E (ApoE) is a 35 kDa glycoprotein encoded in three more frequent isoforms (ApoE2, ApoE3, and ApoE4) and one very rare (ApoE3r) isoform. The four variants (ε2, ε3, ε4, and ε3r) of the human APOE gene coding for the different protein isoforms are determined by four haplotypes,1,2 originated from the allele association of two common single-nucleotide polymorphisms rs429358 (C3,937 → T) and rs7412 (C4,075 → T) at the APOE locus (19q13.32). The APOE2 variant is identified by the haplotype T3,937 − T4,075, the APOE3 variant by the haplotype T3,937 −C4,075, and the APOE4 variant by the haplotype C3,937−C4,075. The fourth haplotype, C3,937 − T4,075, identifies APOE3r.1–3 Each C → T transition of the first base in two CGC codons corresponding at nucleotides 3937 (rs429358) and 4075 (rs7412), encoding arginine (Arg), induces the formation of the TGC codons encoding cysteine (Cys). Therefore, ApoE2 protein isoform (Cys112−Cys158) is encoded by the ε2 haplotype (T3,937−T4,075), the ApoE3 protein isoform (Cys112−Arg158) is encoded by the ε3 haplotype (T3,937−C4,075), and the ApoE4 protein isoform (Arg112− Arg158) is encoded by the ε4 haplotype (C3,937−C4,075). Consequently, the protein isoform ApoE3r (Arg112 − Cys158) is encoded by the ε3r haplotype (C3,937 − T4,075). Notably, both ε3 and ε3r are ∼34 kDa glycoproteins of 299 amino acids (aa), which are indistinguishable from each other by means of the classical protein analysis methods.1,2 In Caucasians, more than 95% of genetic variability in APOE is represented by these four isoforms, 4 and they are probably the most investigated gene variants in human genome.
ApoE is involved in modulating synaptic function,5,6 blood–brain barrier (BBB) integrity,7–9 neuronal receptor recycling, 10 and physiological processes (including cytoskeletal assembly and stability, mitochondrial integrity and function, and dendritic morphology and function) and numerous metabolic pathways such as lipid transport, 11 glucose metabolism,12,13 and insulin signaling.14,15
ApoE was first identified in 1973 as a lipoprotein constituent of very-low-density lipoproteins (VLDL), 16 high-density lipoproteins (HDL), and chylomicrons, as it was established that dietary cholesterol altered APOE distribution in plasma. Thus, ApoE keeps the same structure in both VLDL and HDL and plays a pivotal role for cholesterol and other lipid transport, participating in their redistribution to cells and facilitating their internalization in cells. However, while the role of ApoE in lipid pathophysiology may be associated with normal aging, its function in central nervous system (CNS) pathophysiology needs clarification. Nevertheless, APOE genotype is an established risk factor for Alzheimer’s disease (AD), independent of its role in the pathophysiology of lipid metabolism. 17
HDL and low-density lipoprotein (LDL) are present in individuals with exceptional longevity and their offspring, but their particle sizes are larger compared with controls. 18 ApoE protein is required for healthy cholesterol metabolism and CNS cholesterol transport. In very old individuals, total ApoE levels in plasma seem to correlate with lower total cholesterol and LDL cholesterol levels, which in turn are correlated with allele ε2. 19 The presence of an abnormal cerebrospinal fluid (CSF) lipid profile and lower capacity to deliver neuronal cholesterol has been related to the ε4 allele. 20 Detrimental APOE ε4 allele effects might be managed by nutritional interventions, 21 in particular with a Mediterranean diet including higher intakes of n-3 polyunsaturated fatty acids.22,23 So, in healthy aging and longevity, lipid and cholesterol upkeep is an important factor.
About 25% of total body cholesterol resides in the CNS, playing an important role in synaptic plasticity. 24 Cholesterol metabolism can change with advancing age, and its alteration in the brain may be associated with AD development. 24 In addition, in the hippocampus and cortical areas of AD patients, there is decreased cholesterol compared with age-matched controls. 25 Moreover, modifiable lifestyle factors such as education, alcohol consumption, physical activity, and smoking may attenuate genetic risk for accelerated age-related cognitive decline. The complex interactions between genetics, lifestyle, and cognitive aging may encourage behaviors preserving cognitive health into later life. 26
ApoE is important not only for the pathophysiology of lipid metabolism 27 and CNS, but also for healthy aging and longevity.28–31 Longevity studies focus on lifespan, while healthy aging studies consider healthspan; they are correlated because persons who live long have a tendency to be healthy for much of their lives. 32 Healthy aging can be defined as reaching older age without multimorbidity (i.e. the coexistence of two or more chronic conditions in the same individual) or disabilities, and with intact cognition and/or mobility. 32 The probability of an extreme human lifespan is reduced by the detrimental effects on longevity related to the APOE ε4 allele. 33 In long-lived individuals, the ε2 allele is more frequent than ε4. 34 The elevated frequency of the homozygous ε3/ε3 genotype compared with the heterozygous ε2/ε3 or ε3/ε4 genotypes produces a higher frequency of the ε3 allele in older individuals and their offspring than in controls. Thus, the homozygous ε3/ε3 genotype is the main longevity factor. 35
Since the early 90s, several studies have pointed out that ApoE could have a central role in AD neurodegeneration. Allele ε4, linked to the ApoE4 isoform, represents a key genetic risk factor for noninherited forms of AD (NIAD),36–39 with a semidominant inheritance. 40 Nevertheless, in NIAD, the ApoE2 isoform might exert a protective effect.36,41–43 Although in APOE ε4 carriers the risk for NIAD is definitely increased, the presence of the APOE ε4 allele is not a causal factor for AD pathology.44–46
In AD pathogenesis, ApoE binds and transports amyloid β (Aβ) peptides,47–50 with differential affinity for Aβ according to ApoE isoform. Affinity is highest for lipid-associated ApoE4, intermediate for lipid-associated ApoE3, and lowest for lipid-associated ApoE2.51–54 Accordingly, distinct ApoE isoforms may exert different effects on Aβ aggregation and clearance,51,55–59 and also on Aβ production. 60 ApoE also modulates microglial responses to amyloid plaque pathology61,62 and can affect tauopathy and tau-mediated neurodegeneration.63–65
Thus, distinct ApoE isoforms may either increase the risk for AD48,52 or have a protective role, 66 depending on the different effects of fragments of distinct ApoE isoforms on deposition of Aβ.58,67,68 Brain ApoE is indeed present in smaller fragments (ApoE peptides), which are biologically active. 69 It has been shown that the concentration of specific proteolytic fragments of ApoE is increased in AD brain 70 and that some synthetic peptides of ApoE may be neurotoxic.71,72 The impact of ApoE fragments in AD pathogenesis remains unclear. Differences in the proteolytic processing of distinct ApoE isoforms are also under investigation. In the present narrative review, we have considered biochemical studies about APOE proteolysis, focusing on the enzymes involved in such processes and on the fragments produced by each enzyme. Furthermore, we have focused on recent studies about APOE fragments and their role in AD pathogenesis. In addition, we will review the current development status of different anti-ApoE drugs, and will discuss the feasibility of modulating ApoE processing as a new AD therapeutic approach.
Methods
The present was a narrative review article. We performed separate searches in the US National Library of Medicine (PubMed), Medical Literature Analysis and Retrieval System Online (MEDLINE), EMBASE, Scopus, Ovid, and Google Scholar databases to find original articles of interest. The search strategy used in PubMed and MEDLINE and adapted to the other four electronic sources was based upon searches using the following terms to identify risk exposure (apolipoprotein E OR APOE AND fragments AND enzymes) combined with terms to determine the outcomes of interest [Alzheimer’s disease OR dementia AND (pathogenesis OR development OR treatment OR therapeutics OR drugs OR compounds)]. Identified studies were analyzed for additional references of interest. The last search was performed on October 15, 2021. No language restriction was adopted. Two investigators (F.L.V., P.B.) independently and in duplicate searched for articles, screened titles and abstracts of the retrieved articles, reviewed the full texts, and selected articles for their inclusion. The following inclusion criteria were applied: original studies in cell and animal models and humans. Technical reports, letters to the editor, and systematic and narrative review articles were excluded. Data were cross-checked, any discrepancy was discussed, and disagreements were resolved by a third researcher (D.S.).
The ApoE structure
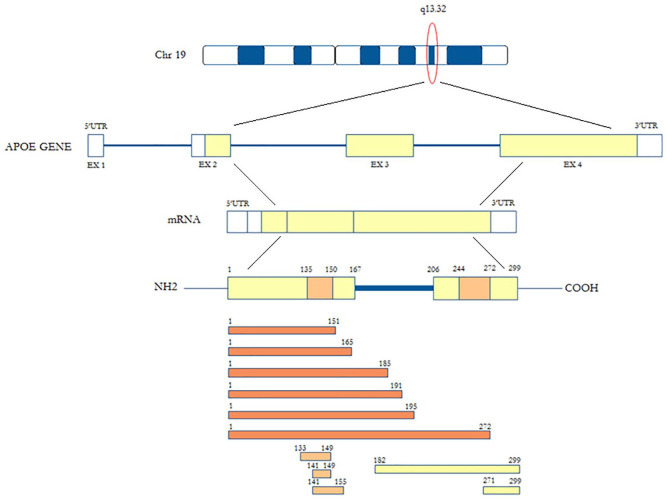
Encoded by the APOE gene at locus 19q13.32, APOE is a 299 aa glycoprotein of 34 kDa, found in several classes of lipoproteins in both humans and other vertebrates. In humans, peripheral and CNS ApoE does not cross the BBB, thus forming two independent ApoE pools with no exchange.73,74 In the periphery, ApoE is produced primarily by hepatocytes, 75 while in the CNS APOE derives mostly from astrocytes, activated microglia, vascular mural cells, and the choroid plexus.76,77 Although it was initially suggested that only astrocytes synthesize ApoE in the brain, 78 subsequent studies have demonstrated that under various physiological and pathological conditions, CNS neurons can express ApoE, even if at lower levels than astrocytes.63,79,80
ApoE is synthetized as a preprotein of 317 aa, with a signal peptide of 18 aa removed in the post-translational phase. ApoE is composed of two structural domains that are linked by an unstructured short hinge region. The NH2-(N)-terminal elongated domain (aa 1-167) forms a 4 α-helix bundle and presents a region rich in basic Arg and lysine (Lys) residues (aa 135-150) that creates the low-density lipoprotein receptor (LDL-R)-binding region. Extracellularly, ApoE binds lipoproteins or lipid complexes and transports them to cells via specific neuronal and glial transmembrane receptors such as LDL-related protein (LRP). 81
The carboxyl-(C)-terminal domain (aa 206-299) is composed of amphipathic α-helices [characteristic of the exchangeable apolipoprotein and contains the lipid-binding region (aa 244-272)]. As previously described, ApoE was identified as a main apolipoprotein/component in HDL particles, VLDL, and chylomicron remnants, being a fundamental ligand in the uptake of these lipoproteins by members of the LDL-R family, except for HDL particles. The structural organization of ApoE in HDL profoundly impacts its ability to regulate cholesterol homeostasis in AD and cardiovascular diseases.
ApoE also serves as the primary cholesterol chaperone in the neuropil and maintains brain cholesterol homeostasis. At residues 141-150, the dual-domain ApoE mimetic peptide Ac-hE18A-NH2 (ligand active site of the protein) co-localizes with a major heparin-binding site. 82 This peptide, the putative receptor-binding region of human ApoE, is covalently linked to a well-characterized class A amphipathic helix, 18A, which has no sequence homology to any other exchangeable apolipoprotein sequences. It demonstrates dramatic ability to reduce plasma cholesterol levels in dyslipidemic mouse and rabbit models. 82
The ApoE3 and ApoE3r structures
The ApoE3 isoform, characterized by the aa combination Cys112 − Arg158, is the major ApoE isoform in humans and is therefore considered the wild-type (or parental) isoform. ApoE3 includes a domain for lipid association between aa 203-266 and a globular domain between aa 1-191 encompassing the LDL-R-binding region.83,84 ApoE3 promotes clearance of triglyceride-rich lipoprotein particles, producing normal plasma lipid levels. Residues 1-167 of the N-terminal domain form an antiparallel 4 α-helix bundle 85 and present the nonpolar aspect of the amphipathic helices. The C-terminal domain encompasses aa 206-299 and has three α-helices presenting a large, exposed hydrophobic surface. Both terminal domains are separated by a hinge region. Through hydrogen bonds and salt bridges the α-helices interact with those in the NH2-terminal helix bundle domain. The LDL-R recognizes a site within the helix bundle domain, while the lipid-binding ability of the protein depends on the C-terminal domain. In helix 4 of the helix bundle domain, there are basic residues (mainly Arg and Lys), corresponding to aa 136-150, that interact with acidic residues present in the ligand-binding domain of the LDL-R family. 86 Arg172 is also necessary for ApoE receptor binding activity. 87
Interestingly, the isoform ApoE3r, characterized by the aa combination Arg112−Cys158, has the same molecular weight and electrical charge of the parental form, ApoE3. ApoE3r has been shown to have normal VLDL receptor binding activity, 1 resulting in an ApoE3-like lipid asset. However, ApoE3r is the rarest isoform across the population worldwide. 1 The appearance of ApoE3r isoform during evolution and its role in lipid metabolism and CNS pathophysiology need to be further elucidated. 2
The ApoE4 structure
The ApoE4 isoform, the second most common allele, differs from ApoE3 in the change of Cys112 to Arg, producing a combination Arg112 − Arg158. The resultant change in structure and stability of both terminal domains results in enhanced lipid binding. In particular, the substitution of a Cys side chain with a positively charged Arg side chain in the N-terminal helix bundle domain is responsible for its destabilization.88,89 At variance with ApoE3, in ApoE4 the presence of Arg at position 112 reconfigures the Arg 61 side chain with formation of a saline bridge with glutamic acid (Glu)255 in the C-terminal region.90,91 Such binding, probably together with other allosteric effects, 92 produces an altered relationship between the N- and C-terminal regions. In particular, the Cys112 to Arg substitution in ApoE4 leads to unfolding of certain helical segments that reduces self-association and is probably related to a reduced ability of ApoE4 to form tetramers, and is expected to enhance the binding of ApoE4 to plasma triglyceride-rich lipoprotein particles and to brain Aβ deposits. 93 A recently suggested mechanism based on lipid binding between the N- and C-terminal domains proposed that separation of the two domains, along with the presence of intrinsically disordered regions, may control protein motions. This mechanism may partly explain why ApoE3 and ApoE4 are functionally different, why lipid may increase the binding of ApoE to its receptor, and why specific residues may be conserved. 94
The ApoE2 structure
The ApoE2 isoform, the third most common allele, differs from ApoE3 by a single aa substitution Arg158 → Cys, producing the aa combination Cys112 − Cys158. Due to this, the salt-bridge between Arg158 and aspartic acid (Asp)154 in the α-helix of ApoE2 (which is present in ApoE3) is absent, while it is present between Asp158 and Arg150. 95 The novel salt-bridge induces a conformational change of Arg150 relative to the other basic residues in the receptor-binding domain, so that it is no longer able to interact with the LDL-R, reducing binding between ApoE2 and the LDL-R, and resulting in reduced clearance of triglyceride-rich lipoproteins associated with type III hyperlipoproteinemia. 17
The role of ApoE in AD pathogenesis
APOE is considered the most important genetic risk factor for sporadic AD. 96 Heterozygous carriers of the APOE ε4 allele have a 4-fold greater risk of developing AD, whereas homozygous APOE ε4 carriers have a 12-fold greater risk, compared with APOE ε3 carriers. Conversely, the relative rare APOE ε2 allele is associated with a 40% lower AD risk and being homozygous for it further reduces the risk. 97 Similarly, homozygous carriers of the rare ‘Christchurch’ mutation of APOE ε3 (R136 S) appear resistant to autosomal-dominant AD. 98 Compared with APOE ε3 homozygotes, cognitively normal APOE ε4 carriers show higher Aβ and tau brain burden while APOEε2 carriers have lower global Aβ burden – however, they do not differ on regional tau burden or tau accumulation over time. 99 Thus, APOE ε4 carriers show earlier, and APOE ε2 carriers later, onset of cognitive impairment compared with APOE ε3 homozygotes. Nevertheless, the effect of APOE genotype on rate of cognitive decline after disease onset remains controversial. 100 Although initial studies linked APOE with Aβ aggregation and clearance, recently it has emerged that the role of APOE in AD pathogenesis involves tau-mediated neurodegeneration, 101 microglia dysfunction,102–104 astrocyte responses,11,105 and BBB disruption.106,107
The ApoE-cutting enzymes
As seen above, under various physiological and pathological conditions, CNS neurons can express ApoE, even if at lower levels than astrocytes.63,79,80 Indeed, various in vivo and in vitro studies suggest that ApoE fragmentation occurs in neurons (not neuroglia) in specific brain regions. ApoE fragments are found only in the neocortex and hippocampus of ApoE-expressing transgenic mice. 63 The proteolytic process usually yields C-terminal-truncated ApoE fragments in an isoform-dependent manner (ApoE4 > ApoE3). 70 Specifically, ApoE4 cleavage produces more neurotoxic fragments than ApoE3, these fragments only being generated by neurons and not astrocytes.63,108 Both murine and human neuronal precursor cells produce increased ApoE levels when challenged with Aβ peptides. 109 In the hippocampus, the formation of intraneuronal phospho-tau-containing filamentous inclusions is stimulated by production of C-terminal-truncated ApoE4 fragments, 63 although the protease(s) responsible for ApoE fragmentation have not been confirmed. It has been reported that Aβ1-42 treatment of Neuro-2a cells transfected with either ApoE3 or ApoE4 cDNA significantly increased C-terminal truncated ApoE3 and ApoE4 levels, suggesting that the peptide activates intracellular protease(s). 70 It has also been demonstrated that intracellular Aβ/amyloid precursor protein (APP) C-terminal fragments raised ApoE levels and C-terminal fragments thereof, 109 leading to the assumption that intracellular Aβ accumulation has a different effect on ApoE than exogenous application of Aβ peptides.
Two classes of proteases have so far been suggested as mediators of ApoE fragmentation, targeting Asp and Ser. Four enzymes appear to play a critical role: cathepsin D (the only aspartic protease identified) and thrombin (a serine protease), α-chymotrypsin-like serine protease, and high-temperature requirement serine protease A1 (HtrA1). Specific ApoE fragments are involved early in neurodegenerative processes impacting deposition of Aβ plaques, tau hyperphosphorylation, and intracellular formation of neurofibrillary tangles (NFTs) (Figure 1).110–112 α-Chymotrypsin-like serine proteases and matrix metalloproteinase (MMP)-9 probably mediate the fragmentation process in the neuropil, while cathepsin D mediates the process in neuronal cytoplasm. 113 Cathepsin D mediated ApoE fragments in the cytoplasm of the neurons and ApoE co-localize to a subset of predominantly senile plaques and some NFTs in the human post-mortem frontal cortex. 114 In studies of serine proteases, ApoE fragments were found in the culture medium but not in cell lysates, which could imply that serine proteases are secreted to mediate fragmentation of ApoE in the extra-neuronal environment.
Figure 1.
Apolipoprotein E (ApoE) fragments generated by cutting of enzymes.
ApoE N-terminal fragments (orange), ApoE central fragments (light orange) and ApoE C-terminal fragments (yellow).
Thrombin
Thrombin, a serine protease belonging to chymotrypsin family, 115 has a 36-residue polypeptide A chain and a 259-residue B chain, linked by a disulfide bridge at the corresponding Cys1 and Cys122 residues.115,116 The A chain may be superfluous to its enzymatic function. 117 This structure presents two basic anion-binding exosites and one Na+-binding site. 118 Anion-binding exosite I (the fibrinogen recognition exosite) is the recognition site for many cofactors and procoagulant substrates (such as fibrinogen and thrombomodulin), and thrombin cleaves protease activated receptor 1 (PAR-1). Exosite II is the heparin-binding site and the principal interaction site of the platelet glycoprotein receptor IbAα. 79
In AD pathophysiology, a role for this enzyme has been suggested, 119 given that AD patients have amyloid deposits, senile plaques, NTFs, and micro-vessels, all containing thrombin. In addition, neuronal cells express prothrombin mRNA,120,121 and it has been observed that PAR-1, PAR-3 and PAR-4 are up-regulated in rat hippocampus. 122 Thrombin cannot process phosphorylated tau protein, resulting in intracellular aggregates of tau protein in hippocampal neurons, but it may induce proteolysis of microtubule-associated tau protein in vitro. 123 Activation of PAR-1/4 and ERK1/2 pathways is implicated in thrombin-induced hyperphosphorylation and aggregation of tau. 124 Hyperphosphorylated tau deposits are neurotoxic and, in the hippocampus, may lead to neuronal apoptosis. 124 In vitro, the thrombin-mediated cleavage of APP promotes Aβ accumulation and amyloid plaque formation.125,126 Although some studies indicated that Aβ neurotoxic effects could be alleviated by thrombin, it is also possible that it may increase Aβ neurotoxicity through intracellular influx of calcium ions and high oxidative stress.127,128 Arg191/alanine (Ala) 192 (major site) and Arg215/Ala216 (minor site) are two thrombin cleavage sites on the ApoE protein. 129 ApoE cleavage by thrombin creates a 22 kDa fragment and a 10–12 kDa fragment. The ApoE N-terminal 22 kDa fragment originating from the ApoE4 isoform shows greater neurotoxicity compared to that originating from ApoE3.71,84
Cathepsin D
The lysosomal enzyme cathepsin D is implicated in unfolded, unused, and damaged protein degradation and is highly expressed in the brain. Through autophagy and endocytosis processes, these damaged proteins are delivered into lysosomes and degraded. 130 Dysregulation of cathepsin D enzymatic activity may lead to accumulation and aggregation of various proteins, and is involved in several proteinopathies. Cathepsins are proteases including several members differentiating for the aa number in the active site: cathepsins B, C, F, H, K, L, O, S, V, W, and X comprise the cysteine cathepsin family; cathepsins A and G, the serine cathepsin family; while D and E are the aspartyl cathepsin family. All human tissues express cathepsins B, L, H, C and D, while A, G, K, S, V, X and W CatD are tissue-specific and based on cell type.131–137 Preprocathepsin D, the synthetized inactive zymogen, is proteolytically cleaved and cathepsin D is activated; preprocathepsin D contains an N-terminal signal peptide, a propeptide, and a catalytic domain.135–137 Cathepsin D is mainly involved in degradation of proteins in lysosomes, but also has key roles in signal transduction pathways, such as activation of enzymatic precursors, prohormones and growth factors, brain-specific antigen processing, neuronal cell homeostasis, and apoptotic processes.138,139 Cathepsin D is a pathology-specific biomarker involved in neurodegenerative disorders such as AD, but is also implicated in cancer, atherosclerosis, and inflammation.140–143 As cathepsin D might have neuroprotective effects by blocking abnormal tau accumulation, cathepsin D deficiency may induce elevation in C-terminally truncated tau variants, thus promoting neurotoxicity. 144
In human ApoE, 13 cathepsin D cutting-sites are found, which preferentially cleave aromatic aa in the C-terminus. 145 Interestingly, there is only one of the 13 cathepsin D cutting-sites at tryptophan (Trp) 210 in the ApoE hinge. This finding confirms the preferential generation of a 22 kDa thrombin fragment with cleavage in the hinge region of ApoE. The predicted mass of the aa 1-210 ApoE peptide is 24 kDa. 146 As such a peptide should present an O-glycosylation site at Thr 194 , the actual molecular mass of the aa 1-210 ApoE peptide should be closer to 25 kDa. 147 Notably, in post-mortem human brain tissue the 24 kDa ApoE fragment co-localizes with Aβ plaques and NFTs. 148
α-Chymotrypsin-like serine protease
α-Chymotrypsin-like serine protease cleaves aromatic and specific hydrophobic aa. ApoE has 67 sites for cleavage by α-chymotrypsin-like serine protease, and in the hinge, region Leu198, Leu203, Trp210, Leu214, and Met218 are identified. Cleavage generates peptides from 23 kDa to 25 kDa (or from 24 to 26 kDa including O-glycosylation) in size. However, the endogenous pattern of ApoE fragmentation may be reflected by the profile of ApoE fragmentation mediated by cathepsin D. 149 The aa 272-299 are included in the C-terminal ApoE4 fragment produced by this enzyme.72,114
High-temperature requirement serine protease A1
The HtrA1 belongs to a four-enzyme subfamily of serine proteases in humans. 150 At variance with HtrA2 (which is a mitochondrial transmembrane protein), HtrA1, HtrA3 and HtrA4 have identical regional composition: an insulin-like growth factor–binding protein domain (IGF-BD), a Kazal motif followed by a trypsin-like catalytic domain (KM), and a PDZ region. 137 Although regulation of HtrA1 activity is still under debate, the IGF-BD and PDZ domains are dispensable for its activity.151,152 Indeed, a feature of an elastase-like serine protease activity is the cleavage of ApoE after valine (Val) 194 by HtrA1, thus generating a 25 kDa N-terminal ApoE fragment encompassing aa 1-195. The PDZ domain can influence HtrA1 protease activity by interacting with a specific substrate, although its activity may be similar to a trypsin-like serine protease.151,153 Notably, HtrA1 can mediate ApoE4 proteolysis of more rapidly than that of ApoE3.154,155
The role of ApoE isoforms in fragment generation
The three ApoE isoforms differ in their physical properties, including lipid-binding capacity, type of lipids bound, domain-domain interactions, and stability. The isoform-dependent secretion of ApoE and its immunomodulatory effects could be attributable to post-translational modifications. 156 ApoE3 and ApoE2 are less sensitive to proteolytic cleavage than ApoE4. 157 The greater instability of ApoE4 is due to its presentation of numerous protease-sensitive sites in the hinge region, including to thrombin, 158 cathepsin D, 114 and HtrA1,155,159 and its vulnerable N- and C-terminal domains. 112 In addition, ApoE 14-20 kDa N-terminal fragments 111 and ApoE 10-15 kDa C-terminal fragments 109 have been also shown to be more frequent in AD patients than in age-matched controls. The 4 α-helix bundle of ApoE4 is partially opened and extended, so exposing the hydrophobic surface of the protein. Therefore, proteolytic enzymes may have greater access to the short hinge region 160 and the hydrophobic region, as compared to other isoforms. These differences may account for the lower AD risk in APOE ε2 carriers.36,161,162 Notably, AD patients carrying the APOE ε2 allele have reduced Aβ deposition in the neocortex 163 and reduced formation of NFTs. 164
Recently, it has been shown that the 25 kDa ApoE fragment may be neuroprotective, and is more abundantly produced in brains of APOE ε3 carriers compared to APOE ε4 carriers. 159 This finding suggests that reduction of ApoE3 fragments with neuroprotective effects may contribute to neurodegeneration in AD, alongside the neurotoxic impact of ApoE4 fragments. Other detailed studies are required to understand the function of ApoE fragments not only in astrocytes and microglia cells, but also in non-neuronal cell types and their exact role in AD development, lipid metabolism, healthy aging and longevity.
ApoE2 contains a Cys158 residue and its substitution with Arg makes this isoform more stable and resistant to thermal and chemical denaturation than ApoE3 and ApoE4. 165 Furthermore, in both these isoforms, the presence of a negative Cys112 residue reduces their propensity for domain interactions compared to ApoE4, which has a positive Arg112 residue. 166 New synapse formation is promoted by increased dendritic outgrowth mediated by ApoE2, thus protecting against AD-related synaptic loss. 110 ApoE protects cells from oxidative injury in vitro, but such activity varies by isoform: ApoE2 is the most effective, ApoE3 is moderately effective, while ApoE4 is the least effective. 167 The immunomodulatory effects of ApoE also vary by isoform, and a new study indicates that isoform status dictates its own the glycosylation state and secretion. 156
The role of ApoE peptides in neurodegeneration and AD
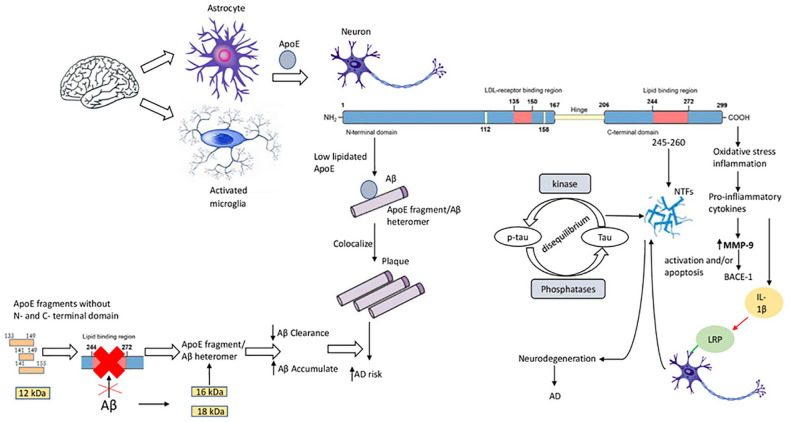
The interaction between ApoE and Aβ is influenced by various factors, including the ApoE isoform and its lipidation, and Aβ levels. 168 In healthy brain, Aβ can associate with ApoE, which is involved in its metabolism and clearance. The brains of both cognitively intact controls and AD patients contain full-length ApoE, high molecular weight (HMW-22–30 kDa) and low molecular weight (LMW-10–20 kDa) fragments in varying quantities, although fragments are more frequent in AD brains than in those of controls. Nonetheless, the proportions of the various fragments are similar in both groups, as well as among the various APOE genotypes. 169 APOE ε4 carriers’ brains have more fragments than non-carriers’ brains.157,170 Each of the post-cleavage ApoE4 domains is associated with a specific pathological process in AD brain: C-terminal domain binds to Aβ and localizes to senile plaques, while the N-terminal region mainly localizes with NFTs. 108 N-terminal truncated ApoE fragments appears to be the major species in Aβ deposits in AD brains. 171 In human brain, the dynamic processes involving cholesterol and Aβ may be impaired by loss of ApoE4 function, 70 given that the accumulation of intraneuronal Aβ peptides is also correlated with the APOE ε4 genotype. 172
Low lipidation of ApoE4 favors its binding to Aβ, which may induce generation of ApoE-fragment/Aβ heteromers, increasing Aβ accumulation and AD risk (Figure 2). 173 It is this ApoE4/Aβ interaction that plays a main role in AD development, while each component in isolation seems less critical. 174 ApoE fragments lacking N-terminus and C-terminus, together with Aβ accumulation, may also favor AD pathology. Such ApoE fragments are without the Aβ transporter-binding domain, and formation of ApoE-fragment/Aβ heteromers decelerates Aβ clearance and favors accumulation. ApoE4 is more efficient than other isoforms in favoring ApoE-fragment/Aβ heteromer formation. 170 Furthermore, ApoE can increase formation of Aβ oligomers.169,175 In AD brain, 3 ApoE fragments have been found – 18 kDa (ApoE18), 16 kDa (ApoE16) and 12 kDa (ApoE12) – with the 18 and 16 kDa forms being hybrid heteromers composed by Aβ1-42 peptides and ApoE middle fragments. One study showed that the formation of Aβ/ApoE16 and Aβ/ApoE12 heteromers (but not those involving ApoE18) seems to correlate with memory deficits in AD. 112 Only the ApoE18 fragment was significantly increased, while ApoE16 and ApoE12 fragments were less elevated. 109 Furthermore, the observation of increased hippocampal Aβ/ApoE18 heteromer suggested that it might serve as a biomarker for AD pathology. 112
Figure 2.
The role of apolipoprotein E (ApoE) peptides in neurodegeneration and Alzheimer’s disease (AD). In the brain, astrocytes and activated microglia synthesize ApoE. In neurons, ApoE is cleaved in the C-terminal domain that binds to amyloid β (Aβ) and localizes to plaques; and in the N-terminal domain that localizes with NFTs. Low ApoE lipidation influences the binding of ApoE4 to Aβ, promoting ApoE-fragment/Aβ heteromer generation. ApoE fragments without N- and C-terminal domains (ApoE 133-149, ApoE 141-149, ApoE 141-155, 12 kDa fragment) do not have the Aβ transporter-binding domain – these fragments (together with Aβ accumulation) favor the formation of ApoE-fragment/Aβ heteromers, decelerating Aβ clearance and favoring Aβ accumulation in AD. The lipidated form of ApoE interacts with NFTs through its amino acid residues 245-260. Imbalance between the activities of tau protein kinases and phosphatases promotes accumulation of tau protein in neurons. C-terminal truncated ApoE fragments induce oxidative stress and inflammation, releasing pro-inflammatory cytokines. Increased MMP-9 leads to β-site amyloid precursor protein–cleaving enzyme-1 (BACE-1) activation and/or apoptosis, and interleukin (IL)-1β interacting with neurons by LDL-related protein receptor induces NFT formation.
ApoE mediates clearance of Aβ-protein from the neuropil by acting as a bridging protein between Aβ, via its C-terminal domain, and LRP, via its N-terminal domain. 176 A study in senile plaques showed that the N-terminal/C-terminal domain interaction is stronger in ApoE4 than in ApoE3. ApoE4, therefore, has a shorter inter-terminal domain distance, but a relatively longer and more exposed hinge, which is more susceptible to proteolysis. Increased hinge proteolysis in ApoE4 leads to more disassociated Aβ-bound C-terminal fragments. These events contribute to the loss of Aβ clearance function in brains with ApoE4, enhancing amyloid deposition, 169 given also that intracellular clearance of Aβ peptides is correlated with the APOE ε4 genotype. 172
ApoE fragmentation presents as a potential AD-related pathological process (Figure 2). C-terminal truncated forms of ApoE induce intracellular NFT-like inclusions in neurons.70,157,177 ApoE fragments associate with or induce the formation of hyperphosphorylated tau and NFTs in an isoform-dependent manner. The lipidated form can also induce NFT formation, both in vitro and in vivo, in murine neocortex and hippocampus70,157 and interacts with NFTs via its aa residues 245-260 70 – these effects occur only in neurons. Cytosolic expression of ApoE4 (272–299) induces NFT-like inclusions in mouse primary-cultured cortical neurons and in human NT2 cells but not in various non-neuronal cells. 70 Pathologic human tau accumulates in neurons due to an imbalance between the activities of tau protein kinases and phosphatases. Full-length ApoE decreases phosphorylation of tau kinases and inhibits tau phosphorylation at Thr 171 and Ser202/Thr205 epitopes.
The ApoE receptor-binding N-terminal domain peptides
The potential neurodegenerative effects of ApoE N-terminal fragments are summarized in Table 1. In the BV2 mouse microglial cell line, the 17 kDa fragment has been reported to promote cell death. 178 It has been reported that intracellular Aβ1-42 accumulation is stimulated by the 19 kDa fragment, which produces reactive oxygen species (ROS) in the SK-N-SH human neuroblastoma cell line.74,179 In this cell line and SW-1783 human astrocytoma cells, the 21 kDa fragment has been also reported to promote MMP9/TIMP1 imbalance, by stimulating IL-1β and reducing IL-10 levels. 180 The 22 kDa thrombin cleavage ApoE4 fragment is also neurotoxic in vitro,103,181 but only at high concentrations – this has not yet been confirmed in vivo. The 22 kDa fragment exhibits receptor-mediated neurotoxicity103,158 and increases intracellular calcium in embryonic chick sympathetic sympathetic ganglia and embryonic rat hippocampal tissue. 181 The 25 kDa fragments 1–195 have been reported to promote neuritogenesis in SK-N-SH/SH-SY5Y human neuroblastoma cells, 159 whereas the 1–272 fragments may induce neurotoxic mitochondrial dysfunction in Neuro-2a mouse neuroblastoma cells. 182
Table 1.
Effect of apolipoprotein E (ApoE) N-terminal peptides in cell and animal models related to Alzheimer’s disease (AD) and differences in ApoE isoforms.
| Organism | Tissue | Fragment | Effect | Model | Note | Reference |
|---|---|---|---|---|---|---|
| Mouse | Neuro-2a neuroblastoma cells | ApoE4 (1-271) 1 (N-terminal) |
In neuronal cells promotes NFT-like inclusion formation | In vitro | ApoE4 showed more inclusions than ApoE3 | Huang et al. 70 |
| Mouse | C57BL/6 J Neuro-2a neuroblastoma cells overexpressing APOE 272-299 | ApoE4 (1-271) 1 (N-terminal) |
Promotes AD-like tau pathology and behavioral disorders | In vivo | Studies conducted only on ApoE4 | Harris et al. 157 |
| Mouse | J20 line of hAPPFAD and overexpressing APOE 272-299 |
ApoE4 (1-271) 1 (N-terminal) |
Decreases Aβ clearance and increases Aβ deposition | In vivo/In vitro | Full-length ApoE3 and ApoE4 expressing mice were able to stimulate Aβ clearance | Bien-Ly et al. 108 |
| Human | Primary neurons | 22 kDa N-terminal | Cytotoxicity | In vitro | Higher effect in ApoE4 than ApoE3 background | Marques et al. 158 |
| Chick | Embryonic sympathetic ganglia |
22 kDa N-terminal | The neurotoxic function receptor- induced | In vitro | ApoE4 fragment was more toxic than ApoE3 | Tolar et al. 72 |
| Rat | Embryonic hippocampal tissue | 22 kDa N-terminal | Determines increased intracellular calcium levels and neurotoxicity | In vitro | Studies conducted only on ApoE4 | Tolar et al. 181 |
| Mouse | BV2-microglia cells | 1-151 2 (17 kDa N-terminal) | Induces increased cell death and trafficking to the nucleus | In vitro | Effects not seen in ApoE3 | Love et al. 178 |
| Human | SK-N-SH human neuroblastoma |
1-165
3
(19 kDa N-terminal) 1-185 3 (21 kDa N-terminal) |
ApoE 1-165 promotes Aβ1-42 intracellular accumulation which induces ROS formation, whereas ApoE 1-185 do not | In vitro | Effects not seen in ApoE3 and ApoE2 | Dafnis et al.
54
Dafnis et al. 179 |
| Human | SK-N-SH neuroblastoma and SW-1783 astrocytoma cells | 1-185
3
(21 kDa N-terminal) |
Induces interleukin-1β and reduces interleukin-10 expression through MMP9/TIMP1 imbalance | In vitro | Only ApoE4 studied | Dafnis et al. 180 |
| Human | SK-N-SH/SH-SY5Y human neuroblastoma cells | 1-195
3
(25 kDa N-terminal) |
Promotes neuritogenesis | In vitro | Only ApoE3 studied | Muñoz et al. 159 |
| Mouse | Neuro-2a neuroblastoma cells |
1-272
2
(N-terminal) |
Exhibits neurotoxicity via mitochondrial dysfunction | In vitro | Only ApoE4 studied | Chang et al. 182 |
1Gene ID: 11816.
2Gene ID: 25728.
3Gene ID: 348.
Aβ, amyloid β; MMP9, matrix metalloproteinase 9; NFT, neurofibrillary tangle; ROS, reactive oxygen species; TIMP1, tissue inhibitor of metalloproteinase 1.
The 22 kDa fragments originated by thrombin cleavage lack the aa 244-272 lipid-binding site and are different from ApoE fragments generated in AD brains. 70 In contrast, this site seems to be essential for fragment-induced neurotoxic effects in vivo all ApoE fragments contain it, 70 and it is implicated in the interplay between ApoE and Aβ peptides.47,183
The ApoE central domain peptides
The potential effects of the ApoE central fragments in neurodegeneration are summarized in Table 2. In BV2-microglia cells, release of tumor necrosis factor-α (TNF-α) and nitric oxide (iNOS) is induced by the 133-149 aa peptide by suppression of microglial activation. 184 This peptide also reduces inflammation response after lipopolysaccharide (LPS) injection in C57BL/6 J blastocysts, 185 suppressing the apoptosis and intracellular calcium increase caused by N-methyl-d-aspartate in primary neuronal-glia cells. 186 In hippocampal slices, this peptide, together with the C-terminal portion of the 141-148 aa peptide, inhibited acetylcholine-induced responses. 187 In Xenopus laevis oocytes, these peptides block the α7 nicotinic acetylcholine receptor (nAChR), disrupting nAChR signaling.188,189 The Arg-rich domain of the C-terminal ApoE 141-148 aa peptide favors the interaction with nAChR and may inhibit α7 nAChRs-mediated responses, 187 as does APOE 133-149 aa. 189 In contrast to ApoE 133-149 and ApoE 141-148, the ApoE 141-149 and ApoE 141-155 residues are associated with deleterious effects such as neurite degeneration 69 and cytotoxicity, 71 respectively. The 141-149 peptide was able to inhibit cell proliferation and showed cytotoxic effects in IL2-dependent T lymphocyte cells, 71 whereas the 141-155 aa peptide may induce neurite degeneration in embryonic chick sympathetic neurons. 69
Table 2.
Effect of apolipoprotein E (ApoE) peptides from central protein domain in cell and animal models related to Alzheimer’s disease (AD) and differences in ApoE isoforms.
| Organism | Tissue | Fragment | Effect | Model | Note | Reference |
|---|---|---|---|---|---|---|
| Chicken | Embryonic sympathetic ganglia | 141-155 1 | Neurite degeneration | In vitro | N/A | Crutcher et al. 69 |
| Human | Interleukin-2-dependent T lymphocytes | 141-149 2 | Cytotoxicity | In vitro | N/A | Clay et al. 71 |
| Mouse | BV2-microglia cells | 133-149 3 | Inhibits the release of TNFα and NO through suppresses microglial activation | In vitro | N/A | Laskowitz et al. 184 |
| Mouse | Primary neuronal-glia cells | 133-149 3 | Suppresses neuronal cell death and calcium influx induced by NMDA | In vitro | N/A | Aono et al. 186 |
| Mouse | C57BL/6 J blastocysts | 133-149 3 | Following LPS administration suppresses inflammatory response | In vivo | N/A | Lynch et al. 185 |
| Rat | Hippocampal slices | 133-149 3 /141-148 4 | Acetylcholine-evoked responses in a dose-dependent manner are inhibited | Ex vivo | N/A | Klein and Yakel 187 |
| Frog | Oocytes | 133-1493/141-148 4 | Block α7 nAChRs disrupting nAChR signaling | Ex vivo | N/A | Gay et al.
188
Gay et al. 189 |
Gene ID: -
Gene ID: 348.
Gene ID: 25728.
Gene ID: 394678.
LPS, lipopolysaccharide; nAChR, nicotinic acetylcholine receptor; NMDA, N-methyl-d-aspartate; NO, nitric oxide; TNFα, tumor necrosis factor α.
The ApoE lipid-binding C-terminal domain peptides
The potential neurodegenerative effects of ApoE peptides originating from the C-terminal protein domain are summarized in Table 3. The C-terminal truncated ApoE fragment neither affects APP expression nor APP processing, but induces formation of NFTs. The C-terminal-derived fragment can stabilize Aβ to form pathogenic Aβ oligomers, but not Aβ fibrils. Furthermore, C-terminal fragments induce oxidative stress and inflammation, stimulating neuronal secretion of the pro-inflammatory cytokines IL-1β and MMP-9. Increased MMP-9 leads to BACE-1 activation and/or apoptosis, and IL-1β promotes NFT formation by interacting with neurons via the LRP receptor. In addition to aberrant activation or repression of tau kinases and phosphatases, the third α-helix of the C-terminal fragment is involved in abnormal tau phosphorylation and formation of NFT-like inclusions. Inclusions similar to NFTs containing the 272-299 aa fragment (also designated as 1-271 aa fragment) 70 are found in neuronal cells, and may cause AD-like tau pathology and behavioral disturbances in Neuro 2-a mouse neuroblastoma cells of transgenic mice C57BL/6 J overexpressing this peptide. 157 In Neuro-2a cells, removal of the first 20 aa, up to the first α-helix, of ApoE4 (272-299) does not modify its ability to induce intracellular NFT-like inclusions. Only a further truncation of its third α-helix decreases the induction of NFT-like inclusions by ∼50%. 70
Table 3.
Effect of apolipoprotein E (ApoE) C-terminal peptides in cell and animal models related to Alzheimer’s disease (AD) and differences in ApoE isoforms.
| Organism | Tissue | Fragment | Effect | Model | Note | Reference |
|---|---|---|---|---|---|---|
| Mouse | Neuro-2a neuroblastoma cells | 182-299
1
(13 kDa C-terminal) |
Inhibits Aβ fibril formation and favors formation and stabilization of Aβ hexameric species | In vitro | ApoE4 generates more 13 kDa fragment than ApoE2 or ApoE3 | Wellnitz et al. 190 |
| Human | Samples of frontal cortex | 224-299
2
(C-terminal) |
Greater accumulation of ApoE C-terminal fragment in the insoluble fraction of tissue homogenate in the severe AD group versus the control group | In vitro | ApoE4 could be a source of the C-terminal fragment accumulation | Wang and Turko 177 |
Gene ID: 11816.
Gene ID: 348.
Aβ, amyloid β.
In transgenic mice hAPPFA, overexpressing 272-299 aa peptide, there was lower Aβ clearance. 108 In Neuro-2a mouse neuroblastoma cells, production and stabilization of hexameric Aβ species may be induced by an ApoE C-terminal 13 kDa fragment that may also inhibit Aβ fibril formation. 190 ApoE fragments escape the secretory and endocytic–lysosome internalization pathways to induce the formation of NFTs and/or to associate with them. In Neuro-2a cells, the neurotoxic ApoE4 fragment is not found in either the Golgi apparatus or endoplasmic reticulum. In contrast, the fragment either interacts with the mitochondria or cytoskeletal elements to form filamentous inclusions containing phosphorylated tau and phosphorylated neurofilament proteins. 182 The mechanisms underlying this ability to evade the recycling pathway remain unknown.
It has been proposed that the LDL receptor–binding domain directs the C-terminal domain in initiating rapid lipid binding, followed by a slower N-terminal domain helix bundle opening, to yield discoidal reconstituted HDL. It appears that the bulky N-terminal domain determines the spatial organization of its C-terminal domain in reconstituted HDL, a finding that has significance for ApoE4, which is more susceptible to proteolytic cleavage in AD brains. 191
The ApoE mimetic peptides
In nature, potentially therapeutic peptides are present as 100 aa short-chain monomers. Such peptides may bind certain membrane receptors and activate specific signaling pathways. 192 Historically, the use of peptides as therapeutic agents has been ignored due to several limitations including dimension, degradation susceptibility, absence of effective delivery methods, fast excretion, poor distribution, low oral bioavailability, reduced cell permeability, and target specificity. 193 In 1953, oxytocin was the first therapeutic peptide synthesized. In 1982, recombinant human insulin was the first such peptide produced through recombinant fermentation. 194 Transformation of peptides into mimetic peptides is one intriguing strategy for using them as therapeutic agents. Mimetic peptides exert effects similar to those of the original molecule, but promise several advantages: increased structural consistency, good cell membrane permeability, and increased stability to proteolytic digestion and target specificity. Peptides have been chemically modified to induce non-natural aa changes, amide bond variations, scaffold rigidity, or hydrophobic residue inclusion.192,193,195
Mimetic peptide studies have been inspired by lipid-binding domains shared from apolipoproteins with a common structure to these mimetic peptides.196,197 Lipoprotein binding activity and clearance have been tested for peptides spanning the ApoE 130-169 region.71,198 Amphipathic α-helices within the C-terminal region are essential to its efflux capacity. 199 The ApoE mimetic peptide (ATI-5361) has been developed on the basis of a 26 aa peptide including residues 238-266.200,201 A peptide composed of a dimer of residues 141-155 with an N-terminal tyrosine residue, designated Y (141-155)2, binds to the LDL receptor. This latter peptide, upon acetylation of its N-terminus, binds to all lipoproteins. 202 In ApoE-/- mice, VLDL and intermediate-density lipoprotein (IDL) clearance is promoted by injection of the acetylated peptide, whereas non-acetylated peptide exerted no such effects.
The ATP-binding cassette transporter A1 (ABCA1) belongs to a large family of such transporters. It is localized on the plasma membrane of several cells where it mediates reverse cholesterol transport and (in particular) cholesterol and phospholipids efflux to extracellular apolipoprotein acceptors, such as ApoA-I, ApoE and small, lipid-poor HDL particles.199,203 This cholesterol efflux function is vital to the prevention of degenerative diseases, since intracellular cholesterol accumulation may have cytotoxic and inflammatory effects, and may induce structural and functional alteration in the plasma membrane.204,205 ABCA1 lipid efflux activity optimization may also be important in protecting against AD.206,207 ApoE lipidation and Aβ clearance could be facilitated by ABCA1 in brain cells208,209 and may protect against cognitive decline, particularly in the presence of ApoE4 phenotype. 209 This observation suggests that therapeutic strategies aimed at targeting ABCA1 with a small peptide might be beneficial to AD patients carrying the APOE ε4 allele. It would be also interesting to study the impact of specific ApoE isoforms in any improvement in glucose utilization and mitochondrial function in the aging brain, and in AD. 210 LPS-induced inflammatory reactions may be modified by polymorphic human APOE alleles, and ApoE mimetic peptides may suppress such responses. Following LPS administration, there is an innate immune response in human subjects with APOE heterozygosity ε3/ε4 compared with homozygosity ε3/ε3. 211 ApoE mimetics such as AchE18A-NH2 may therefore restore or replace ligands in genetically induced hyperlipidemias to enable reduction in atherogenic lipoproteins by heparan sulfate proteoglycan, even in the absence of functional LDL receptors. Therefore, this and similar peptides may be useful in the treatment of dyslipidemias such as familial hyperlipidemia and AD-associated atherosclerosis. 82 In conclusion, the potential role of mimetic peptides in AD seems to deserve further clinical studies.
Anti-ApoE therapeutics in AD
The complete absence of ApoE caused by a rare ablative APOE frameshift mutation may cause abnormal lipoprotein metabolism but normal visual, cognitive, neurological, and retinal function, with normal CSF fluid Aβ and tau protein levels and normal findings on brain magnetic resonance imaging. 212 This observation has encouraged different approaches to interfere with the pathological role of ApoE in AD. 213 We now briefly describe the main approaches being pursued, some of which have reached the clinic.
ApoE2 overexpression
Gene therapy methods may be a strategy to overexpress ApoE2 in APOε4/ε4 carriers through vector administration or liposomal delivery systems. An initial study has shown that single intracerebroventricular injection of an adeno-associated viral (AAV) vector expressing ApoE4 in a mouse model of AD increased brain Aβ deposition and worsened Aβ-mediated synaptotoxicity, whereas increased expression of ApoE2 reduced brain Aβ levels, attenuating peri-plaque synapse loss and neurite dystrophy. 214 These findings were confirmed by a further study in which intracerebral AAV-mediated delivery of ApoE2 markedly reduced Aβ deposition in AD mouse models. 215 Intracisternal administration in nonhuman primates of AAVrh.10hAPOE2-HA, an AAVrh.10 serotype coding for an HA-tagged human APOE2 cDNA sequence, safely mediated wide distribution of ApoE2 in AD relevant regions. 216 An open-label, phase I study (NCT03634007) in 15 AD APOE ε4 homozygotic subjects with evidence of brain amyloid accumulation is presently assessing the safety and tolerability of intracisternal AAVrh.10hAPOE2. The study will assess whether gene therapy can convert CSF ApoE isoform status in APOE ε4 homozygotes from ApoE4 to ApoE2-ApoE4. Recently, a brain-targeted delivery of ApoE2-encoding plasmid DNA (pApoE2) and using glucose transporter-1 (glut-1)-targeted liposomes has also recently suggested. 217 After single tail vein administration of dual-functionalized liposomes, there was a higher transfection of pApoE2 in the C57BL/6 mice brain without signs of toxicity. 217 Using transferrin- and penetratin-modified liposomes, a successful brain delivery of plasmid-encoding ApoE2 was also demonstrated after single intravenous injection in mice. 218
Downregulation of expression of ApoE4
In APP/PS1 transgenic mice homozygous for the APOE ε4 or APOE ε3 allele, intracerebroventricular antisense oligonucleotides (ASOs) reduced ApoE expression and significantly decreased Aβ pathology when given prior to plaque deposition. 219 In the P301 S/ApoE4 mouse model of tauopathy, intracerebroventricular injection of ApoE ASOs reduced ApoE4 protein levels by ~50%, significantly protecting against tau pathology and associated neurodegeneration, decreasing neuroinflammation, and preserving synaptic density. 220
Anti-ApoE immunotherapy
The use of passive immunization targeting certain ApoE isoforms may be an alternative therapeutic strategy. The anti-ApoE4 monoclonal antibody 9D11 bound specifically to brain ApoE4 and not ApoE3. 221 For this monoclonal antibody, a direct intracerebroventricular application prevented ApoE4-driven accumulation of Aβ in hippocampal neurons, while repeated intraperitoneal injections of 9D11 in ApoE4 mice resulted in the formation of ApoE/IgG complexes, associated with reversal of cognitive impairments and ApoE4-driven pathologies, including hyperphosphorylated tau. 221 In APP/PS1 mice, the monoclonal antibody HJ6.3 delivered peripherally to either before or after plaque deposition reduced Aβ levels and fibrillary amyloid pathology, increased microglial activation, and improved spatial-memory performance. 222 Moreover, intraperitoneal HJ6.3 (10 mg/kg/week for 21 weeks) affected Aβ plaques, neuronal network function, and behavior in 7-month-old APP/PS1 mice after plaque onset. 223 In mice producing human ApoE3 and ApoE, central or peripheral administration of 4HAE-4, a monoclonal antibody that targets aggregated ApoE3 and ApoE4, reduced fibrillary amyloid plaque load without reducing cerebral or plasma ApoE levels. 224 Furthermore, this monoclonal antibody increased peri-plaque Aβ phagocytosis, 224 reduced cerebral amyloid angiopathy and amyloid plaque load, and improved cerebrovascular function. 225
ApoE mimetics
As previously discussed, in mouse models of amyloid pathology, treatment with peptides that mimic the structural and biological properties of native ApoE reduces Aβ deposition,226,227 tau hyperphosphorylation, 228 and glial activation.226–228 In male transgenic APP/PS1/APOETR mice, treatment with the ApoE mimetic CN-105 for 40 days beginning at 14–18 weeks old reduced Aβ pathology and rescued memory deficits. 229 Notably, delaying treatment onset to 25–28 weeks produced a less robust effect. At present, in 201 subjects with perioperative neurocognitive disorders, CN-105 is being tested in a double-blind, placebo-controlled study (NCT03802396). 230
ApoE4 structure correctors
The pathological conformation of ApoE4 may result from an interaction between its amino-terminal and carboxy-terminal domains,231,232 and several small organic molecules designed to block this interaction have been tested in vitro.233,234 The ApoE4 structure corrector PH002 was shown to decrease ApoE4 fragmentation, reducing the effects of ApoE4 on Aβ production, tau phosphorylation, and GABAergic neuron degeneration in human iPSC-derived neurons. 235
Conversion of the ApoE4 amino acid sequence into that of ApoE3 or ApoE2
As the three ApoE isoforms only differ structurally by two aa residues, conversion of the ApoE4 sequence into that of ApoE3 or ApoE2 has been considered a logical approach to attenuating its neurotoxic effects. Indeed, conversion of APOE ε4 to APOE ε3 by gene editing significantly altered cellular phenotypes.235,236 As mentioned previously, reductions in ApoE fragmentation, Aβ production, tau phosphorylation, and GABAergic neuron degeneration were observed in iPSC-derived neurons when APOE ε4 was converted to APOE ε3, suggesting that the detrimental effects of ApoE4 could be abolished by gene editing. 235 Similarly, converting APOE ε4 to APOE ε3 attenuates several AD-related phenotypes in glial cells and organoids. This intervention enhances glial cell endocytosis of extracellular Aβ and significantly reduces Aβ deposition in organoids after 6 months of culture. 236 Despite these promising in vitro findings, the in vivo feasibility and clinical translatability of this therapeutic concept appear challenging. 237
Inhibition of the ApoE-Aβ interaction
The synthetic peptide Aβ12-28P, which is homologous to the ApoE-binding site on the full-length Aβ molecule, inhibiting the ApoE-Aβ interaction reduced Aβ deposition25,238,239 and insoluble tau accumulation 25 in AD mouse models, and intraneuronal Aβ accumulation 240 in in vitro primary hippocampal neurons. Moreover, in amyloid mouse models, treatment with Aβ12-28P decreased brain Aβ accumulation, co-deposition of ApoE within Aβ plaques, and neuritic degeneration, with APOE ε2-targeted replacement and APOE ε4-targeted replacement backgrounds. 241
Challenges of ApoE targeting therapeutic approaches
Given the poor clinical results obtained so far in AD with both anti-Aβ and anti-tau drugs, 242 anti-ApoE therapeutics represent a promising alternative approach. The most promising ApoE modulation approaches appear to be increasing ApoE2 expression or increasing ApoE4 clearance by immunotherapy. However, it is not clear whether chronic treatment with anti-ApoE drugs will be limited by the physiological role of this important glycoprotein in cytoskeletal assembly and stability, mitochondrial integrity and function, and dendritic morphology and function. 213 Indeed, clinical failures of unselective anti-Aβ and anti-tau therapies could be explained by their clearing effects of physiological forms of Aβ and tau protein. 243 Ideally, anti-ApoE therapeutics should attack the pathological forms of ApoE without interfering with its physiological functions.
Conclusion
Repeated studies on ApoE and APOE polymorphism have demonstrated their involvement in lipid transport and neurodegeneration. 244 However, such fields of investigation are still quite independent, as ApoE effects in neurodegenerative diseases such as AD cannot be easily explained by ApoE function in lipid metabolism. Meanwhile, the contribution of ApoE fragments is under discussion in various areas of AD pathogenesis and pathophysiology, including neuron interactions, APP processing, tau hyperphosphorylation, oxidative stress, and inflammation.
Recently, a catabolic pathway has been discovered, in which ApoE undergoes a proteolytic process to produce active peptides with neurotoxic or neuroprotective effects. This finding sheds new light on the possible role of ApoE and its peptides, particularly in AD. The influence that ApoE peptides may have in AD pathogenesis is currently under investigation, while their roles in the transport and metabolism of cholesterol and phospholipids are largely unknown. This may be due to ApoE catabolism being restricted to the CNS, as only astrocytes express the enzymes producing ApoE fragmentation. 159
Given the different roles of ApoE isoforms in AD, the relationship between these and ApoE cutting enzymes is also of interest. Production of ApoE peptides depends on the isoform, as distinct effects of ApoE fragments on CNS cells have been suggested. Several studies have investigated the impact of ApoE4-derived peptides, which may promote neurotoxicity, mitochondrial dysfunction, phosphorylation of tau protein, NFT-like inclusions, neurodegeneration, neuronal apoptosis or suppress microglial activation. 245 Mitochondrial dysfunction in AD also varies with APOE genotype, being greater in APOE ε4 than in APOE ε3 carriers. It is tempting to speculate that the impairment of mitochondrial function elicited by the expression of truncated ApoE4 in Neuro-2a cells correlates with the mitochondrial dysfunction observed in AD patients. Consequently, blocking the interaction of ApoE fragments with mitochondria is another potential strategy for inhibiting the detrimental effects of ApoE in AD. 182
Understanding the differences between ApoE isoforms in different cell types will undoubtedly involve use of induced pluripotent stem cells that express physiological levels of endogenous genes, rather than relying on overexpression models. 159 Recent studies have shown the role of ApoE4 in AD neuropathology in vitro. 246 Furthermore, the roles of 12 and 20 kDa ApoE fragments in an ApoE4 background 235 need to be clarified. Although other studies are currently attempting to investigate ApoE fragment levels in an ApoE2 background, 60 little is known about ApoE2 fragmentation. In conclusion, the use of different cell lines with ApoE2, ApoE3r, or ApoE4 backgrounds might explain not only the specific impact of endogenous levels of ApoE fragments from each genotype, but also the involvement of these fragments in lipid metabolism, healthy aging, longevity, neurodegeneration, and AD pathogenesis.
Footnotes
Author contributions: Filomena Lo Vecchio: Conceptualization; Data curation; Formal analysis; Investigation; Methodology; Validation; Visualization; Writing – original draft.
Paola Bisceglia: Data curation; Formal analysis; Investigation; Methodology; Software; Writing – review & editing.
Bruno Pietro Imbimbo: Investigation; Supervision; Validation; Visualization; Writing – original draft; Writing – review & editing.
Madia Lozupone: Methodology; Software; Supervision; Validation; Visualization; Writing – original draft; Writing – review & editing.
Raffaela Rita Latino: Methodology; Software; Supervision; Validation; Visualization; Writing – review & editing.
Emanuela Resta: Software; Validation; Visualization; Writing – review & editing.
Maurizio Leone: Resources; Supervision; Validation; Visualization; Writing – review & editing.
Vincenzo Solfrizzi: Data curation; Methodology; Software; Supervision; Validation; Visualization; Writing – review & editing.
Antonio Greco: Data curation; Funding acquisition; Methodology; Project administration; Resources; Supervision; Validation; Visualization; Writing – review & editing.
Antonio Daniele: Methodology; Supervision; Validation; Visualization; Writing – review & editing.
Mark Watling: Software; Supervision; Validation; Visualization; Writing – review & editing.
Francesco Panza: Conceptualization; Data curation; Investigation; Methodology; Project administration; Resources; Software; Supervision; Validation; Visualization; Writing – original draft; Writing – review & editing.
Davide Seripa: Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Supervision; Validation; Visualization; Writing – original draft; Writing – review & editing.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was fully supported by ‘Ministero della Salute’, I.R.C.C.S. Research Program, Ricerca Corrente 2018–2020, Linea n. 2 ‘Meccanismi genetici, predizione e terapie innovative delle malattie complesse’ and by the ‘5 x 1000’ voluntary contribution to the Fondazione I.R.C.C.S. Ospedale ‘Casa Sollievo della Sofferenza’.
Conflict of interest statement: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
ORCID iD: Francesco Panza  https://orcid.org/0000-0002-7220-0656
https://orcid.org/0000-0002-7220-0656
Contributor Information
Filomena Lo Vecchio, Research Laboratory, Complex Structure of Geriatrics, Department of Medical Sciences, Fondazione IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo, Foggia 71013, Italy.
Paola Bisceglia, Research Laboratory, Complex Structure of Geriatrics, Department of Medical Sciences, Fondazione IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo, Italy.
Bruno Pietro Imbimbo, Department of Research and Development, Chiesi Farmaceutici, Parma, Italy.
Madia Lozupone, Neurodegenerative Disease Unit, Department of Basic Medicine, Neuroscience, and Sense Organs, University of Bari Aldo Moro, Bari, Italy.
Raffaela Rita Latino, Complex Structure of Neurology, Department of Medical Sciences, Fondazione IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo, Italy.
Emanuela Resta, Translational Medicine and Management of Health Systems, University of Foggia, Foggia, Italy.
Maurizio Leone, Complex Structure of Neurology, Department of Medical Sciences, Fondazione IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo, Italy.
Vincenzo Solfrizzi, ‘Cesare Frugoni’ Internal and Geriatric Medicine and Memory Unit, University of Bari ‘Aldo Moro’, Bari, Italy.
Antonio Greco, Department of Neuroscience, Catholic University of the Sacred Heart, Rome, Italy; Neurology Unit, IRCCS Fondazione Policlinico Universitario A. Gemelli, Rome, Italy; Research Laboratory, Complex Structure of Geriatrics, Department of Medical Sciences, Fondazione IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo, Italy.
Mark Watling, CNS & Pain Department, TranScrip Ltd, Reading, UK.
Francesco Panza, Research Laboratory, Complex Structure of Geriatrics, Department of Medical Sciences, Fondazione IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo, Foggia, Italy; Population Health Unit, Healthy Aging Phenotypes Research Unit, ‘Salus in Apulia Study’, National Institute of Gastroenterology ‘Saverio de Bellis’, Research Hospital, Castellana Grotte, Bari 70013, Italy.
Davide Seripa, Research Laboratory, Complex Structure of Geriatrics, Department of Medical Sciences, Fondazione IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo, Italy; Hematology and Stem Cell Transplant Unit, ‘Vito Fazzi’ Hospital, Lecce, Italy.
References
- 1. Seripa D, Matera MG, Daniele A, et al. The missing ApoE allele. Ann Hum Genet 2007; 71: 496–500. [DOI] [PubMed] [Google Scholar]
- 2. Seripa D, D’Onofrio G, Panza F, et al. The genetics of the human APOE polymorphism. Rejuvenation Res 2011; 14: 491–500. [DOI] [PubMed] [Google Scholar]
- 3. Persico AM, D’Agruma L, Zelante L, et al. Enhanced APOE2 transmission rates in families with autistic probands. Psychiatr Genet 2004; 14: 73–82. [DOI] [PubMed] [Google Scholar]
- 4. Nickerson DA, Taylor SL, Fullerton SM, et al. Sequence diversity and large-scale typing of SNPs in the human apolipoprotein E gene. Genome Res 2000; 10: 1532–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arendt T, Schindler C, Brückner MK, et al. Plastic neuronal remodeling is impaired in patients with Alzheimer’s disease carrying apolipoprotein Epsilon 4 allele. J Neurosci 1997; 17: 516–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao J, Fu Y, Yamazaki Y, et al. APOE4 exacerbates synapse loss and neurodegeneration in Alzheimer’s disease patient iPSC-derived cerebral organoids. Nat Commun 2020; 11: 5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bell RD, Winkler EA, Singh I, et al. Apolipoprotein E controls cerebrovascular integrity via cyclophilin A. Nature 2012; 485: 512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Montagne A, Nation DA, Sagare AP, et al. APOE4 leads to blood-brain barrier dysfunction predicting cognitive decline. Nature 2020; 581: 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zlokovic BV. Cerebrovascular effects of apolipoprotein E: implications for Alzheimer disease. JAMA Neurol 2013; 70: 440–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen Y, Durakoglugil MS, Xian X, et al. ApoE4 reduces glutamate receptor function and synaptic plasticity by selectively impairing ApoE receptor recycling. Proc Natl Acad Sci U S A 2010; 107: 12011–12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao J, Davis MD, Martens YA, et al. APOE ε4/ε4 diminishes neurotrophic function of human iPSC-derived astrocytes. Hum Mol Genet 2017; 26: 2690–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Drzezga A, Riemenschneider M, Strassner B, et al. Cerebral glucose metabolism in patients with AD and different APOE genotypes. Neurology 2005; 64: 102–107. [DOI] [PubMed] [Google Scholar]
- 13. Wu T, Dejanovic B, Gandham VD, et al. Complement C3 is activated in human AD brain and is required for neurodegeneration in mouse models of amyloidosis and tauopathy. Cell Rep 2019; 28: 2111–2123.e6. [DOI] [PubMed] [Google Scholar]
- 14. Geijselaers SLC, Aalten P, Ramakers IHGB, et al. Association of cerebrospinal fluid (CSF) insulin with cognitive performance and CSF biomarkers of Alzheimer’s disease. J Alzheimers Dis 2018; 61: 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao N, Liu CC, Van Ingelgom AJ, et al. Apolipoprotein E4 impairs neuronal insulin signaling by trapping insulin receptor in the endosomes. Neuron 2017; 96: 115–129.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shore VG, Shore B. Heterogeneity of human plasma very low density lipoproteins. Separation of species differing in protein components. Biochemistry 1973; 12: 502–507. [DOI] [PubMed] [Google Scholar]
- 17. Mahley RW, Rall SC., Jr. Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet 2000; 1: 507–537. [DOI] [PubMed] [Google Scholar]
- 18. Barzilai N, Atzmon G, Schechter C, et al. Unique lipoprotein phenotype and genotype associated with exceptional longevity. JAMA 2003; 290: 2030–2040. [DOI] [PubMed] [Google Scholar]
- 19. Muenchhoff J, Song F, Poljak A, et al. Plasma apolipoproteins and physical and cognitive health in very old individuals. Neurobiol Aging 2017; 55: 49–60. [DOI] [PubMed] [Google Scholar]
- 20. Liu CC, Kanekiyo T, Xu H, et al. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol 2013; 9: 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ordovas JM, Lopez-Miranda J, Mata P, et al. Gene-diet interaction in determining plasma lipid response to dietary intervention. Atherosclerosis 1995; 118(Suppl.): S11–S27. [PubMed] [Google Scholar]
- 22. Grimm MOW, Michaelson DM, Hartmann T. Omega-3 fatty acids, lipids, and ApoE lipidation in Alzheimer’s disease: a rationale for multi-nutrient dementia prevention. J Lipid Res 2017; 58: 2083–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bos MM, Noordam R, Blauw GJ, et al. The ApoE ε4 isoform: can the risk of diseases be reduced by environmental factors? J Gerontol A Biol Sci Med Sci 2019; 74: 99–107. [DOI] [PubMed] [Google Scholar]
- 24. Yanagisawa K. Cholesterol and pathological processes in Alzheimer’s disease. J Neurosci Res 2002; 70: 361–366. [DOI] [PubMed] [Google Scholar]
- 25. Liu S, Breitbart A, Sun Y, et al. Blocking the apolipoprotein E/amyloid β interaction in triple transgenic mice ameliorates Alzheimer’s disease related amyloid β and tau pathology. J Neurochem 2014; 128: 577–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reas ET, Laughlin GA, Bergstrom J, et al. Effects of APOE on cognitive aging in community-dwelling older adults. Neuropsychology 2019; 33: 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science 1988; 240: 622–630. [DOI] [PubMed] [Google Scholar]
- 28. Siest G, Pillot T, Regis-Bailly A, et al. Apolipoprotein E: an important gene and protein to follow in laboratory medicine. Clin Chem 1995; 41: 1068–1086. [PubMed] [Google Scholar]
- 29. Mahley RW, Huang Y, Rall SC. Pathogenesis of type III hyperlipoproteinemia (dysbetalipoproteinemia): questions, quandaries, and paradoxes. J Lipid Res 1999; 40: 1933–1949. [PubMed] [Google Scholar]
- 30. Seripa D, Franceschi M, Matera MG, et al. Sex differences in the association of apolipoprotein E and angiotensin-converting enzyme gene polymorphisms with healthy aging and longevity: a population-based study from Southern Italy. J Gerontol A Biol Sci Med Sci 2006; 61: 918–923. [DOI] [PubMed] [Google Scholar]
- 31. Seripa D, Panza F, Franceschi M, et al. Non-apolipoprotein E and apolipoprotein E genetics of sporadic Alzheimer’s disease. Ageing Res Rev 2009; 8: 214–236. [DOI] [PubMed] [Google Scholar]
- 32. Brooks-Wilson AR. Genetics of healthy aging and longevity. Hum Genet 2013; 132: 1323–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sebastiani P, Gurinovich A, Nygaard M, et al. APOE alleles and extreme human longevity. J Gerontol A Biol Sci Med Sci 2018; 74: 44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith JD. Apolipoproteins and aging: emerging mechanisms. Ageing Res Rev 2002; 1: 345–365. [DOI] [PubMed] [Google Scholar]
- 35. Feng J, Xiang L, Wan G, et al. Is APOE ε3 a favourable factor for the longevity: an association study in Chinese population. J Genet 2011; 90: 343–347. [DOI] [PubMed] [Google Scholar]
- 36. Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 1993; 261: 921–923. [DOI] [PubMed] [Google Scholar]
- 37. Chartier-Harlin MC, Parfitt M, Legrain S, et al. Apolipoprotein E, epsilon 4 allele as a major risk factor for sporadic early and late-onset forms of Alzheimer’s disease: analysis of the 19q13.2 chromosomal region. Hum Mol Genet 1994; 3: 569–574. [DOI] [PubMed] [Google Scholar]
- 38. Farrer LA, Cupples LA, Haines JL, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer disease meta analysis consortium. JAMA 1997; 278: 1349–1356. [PubMed] [Google Scholar]
- 39. Masullo C, Daniele A, Seripa D, et al. Apolipoprotein E genotype in sporadic early- and late-onset Alzheimer’s disease. Dement Geriatr Cogn Disord 1998; 9: 121–125. [DOI] [PubMed] [Google Scholar]
- 40. Genin E, Hannequin D, Wallon D, et al. APOE and Alzheimer disease: a major gene with semi-dominant inheritance. Mol Psychiatry 2011; 16: 903–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Corder EH, Saunders AM, Risch NJ, et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet 1994; 7: 180–184. [DOI] [PubMed] [Google Scholar]
- 42. Panza F, Solfrizzi V, Torres F, et al. Apolipoprotein E in Southern Italy: protective effect of epsilon 2 allele in early- and late-onset sporadic Alzheimer’s disease. Neurosci Lett 2000; 292: 79–82. [DOI] [PubMed] [Google Scholar]
- 43. Chiang GC, Insel PS, Tosun D, et al. Hippocampal atrophy rates and CSF biomarkers in elderly APOE2 normal subjects. Neurology 2010; 75: 1976–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mayeux R, Saunders AM, Shea S, et al. Utility of the apolipoprotein E genotype in the diagnosis of Alzheimer’s disease. Alzheimer’s disease centers consortium on apolipoprotein E and Alzheimer’s disease. N Engl J Med 1998; 338: 506–511. [DOI] [PubMed] [Google Scholar]
- 45. Richard F, Amouyel P. Genetic susceptibility factors for Alzheimer’s disease. Eur J Pharmacol 2001; 412: 1–12. [DOI] [PubMed] [Google Scholar]
- 46. Zick CD, Mathews CJ, Roberts JS, et al. Genetic testing for Alzheimer’s disease and its impact on insurance purchasing behavior. Health Aff 2005; 24: 483–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Strittmatter WJ, Saunders AM, Schmechel D, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A 1993; 90: 1977–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fagan AM, Holtzman DM. Astrocyte lipoproteins, effects of apoE on neuronal function, and role of apoE in amyloid-beta deposition in vivo. Microsc Res Tech 2000; 50: 297–304. [DOI] [PubMed] [Google Scholar]
- 49. Bentley N, Ladu M, Rajan C, et al. Apolipoprotein E structural requirements for the formation of SDS-stable complexes with beta-amyloid-(1-40): the role of salt bridges. Biochem J 2002; 366: 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Arélin K, Kinoshita A, Whelan CM, et al. LRP and senile plaques in Alzheimer’s disease: colocalization with apolipoprotein E and with activated astrocytes. Brain Res Mol Brain Res 2002; 104: 38–46. [DOI] [PubMed] [Google Scholar]
- 51. Tokuda T, Calero M, Matsubara E, et al. Lipidation of apolipoprotein E influences its isoform-specific interaction with Alzheimer’s amyloid beta peptides. Biochem J 2002; 348: 359–365. [PMC free article] [PubMed] [Google Scholar]
- 52. Deane R, Sagare A, Hamm K, et al. ApoE isoform-specific disruption of amyloid beta peptide clearance from mouse brain. J Clin Invest 2008; 118: 4002–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Petrlova J, Hong HS, Bricarello DA, et al. A differential association of apolipoprotein E isoforms with the amyloid-β oligomer in solution. Proteins 2011; 79: 402–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dafnis I, Stratikos E, Tzinia A, et al. An apolipoprotein E4 fragment can promote intracellular accumulation of amyloid peptide beta 42. J Neurochem 2010; 115: 873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ma J, Yee A, Brewer HB, Jr, et al. The amyloid-associated proteins α1-antichymotrypsin and apolipoprotein E promote the assembly of the Alzheimer β-protein into filaments. Nature 1994; 372: 92–94. [DOI] [PubMed] [Google Scholar]
- 56. Wisniewski T, Castano EM, Golabek A, et al. Acceleration of Alzheimer’s fibril formation by apolipoprotein E in vitro. Am J Pathol 1994; 145: 1030–1035. [PMC free article] [PubMed] [Google Scholar]
- 57. Koistinaho M, Lin S, Wu X, et al. Apolipoprotein E promotes astrocyte colocalization and degradation of deposited amyloid-β peptides. Nat Med 2004; 10: 719–726. [DOI] [PubMed] [Google Scholar]
- 58. Castellano JM, Kim J, Stewart FR, et al. Human ApoE isoforms differentially regulate brain amyloid-β peptide clearance. Sci Transl Med 2011; 3: 89ra57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Huang YA, Zhou B, Wernig M, et al. ApoE2, ApoE3, and ApoE4 differentially stimulate APP transcription and Aβ secretion. Cell 2017; 168: 427–441.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dafnis I, Raftopoulou C, Mountaki C, et al. ApoE isoforms and carboxyl-terminal-truncated apoE4 forms affect neuronal BACE1 levels and Aβ production independently of their cholesterol efflux capacity. Biochem J 2018; 475: 1839–1859. [DOI] [PubMed] [Google Scholar]
- 61. Sala Frigerio C, Wolfs L, Fattorelli N, et al. The major risk factors for Alzheimer’s disease: age, sex, and genes modulate the microglia response to Aβ plaques. Cell Rep 2019; 27: 1293–1306.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ulrich JD, Ulland TK, Mahan TE, et al. ApoE facilitates the microglial response to amyloid plaque pathology. J Exp Med 2018; 215: 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Brecht W, Harris F, Chang S, et al. Neuron-specific apolipoprotein E4 proteolysis is associated with increase tau phosphorylation in brains of transgenic mice. J Neurosci 2004; 24: 2527–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shi Y, Holtzman DM. Interplay between innate immunity and Alzheimer disease: APOE and TREM2 in the spotlight. Nat Rev Immunol 2018; 18: 759–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhao N, Liu C-C, Van Ingelgom AJ, et al. APOE ε2 is associated with increased tau pathology in primary tauopathy. Nat Commun 2018; 9: 4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bruinsma IB, Wilhelmus MMM, Kox M, et al. Apolipoprotein E protects cultured pericytes and astrocytes from D-Abeta(1-40)-mediated cell death. Brain Res 2010; 1315: 169–180. [DOI] [PubMed] [Google Scholar]
- 67. Huynh TV, Davis AA, Ulrich JD, et al. Apolipoprotein E and Alzheimer’s disease: the influence of apolipoprotein E on amyloid-β and other amyloidogenic proteins. J Lipid Res 2017; 58: 824–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Robert J, Button EB, Yuen B, et al. Clearance of beta-amyloid is facilitated by apolipoprotein E and circulating high-density lipoproteins in bioengineered human vessels. Elife 2017; 6: e29595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Crutcher KA, Clay MA, Scott SA, et al. Neurite degeneration elicited by apolipoprotein E peptides. Exp Neurol 1994; 130: 120–126. [DOI] [PubMed] [Google Scholar]
- 70. Huang Y, Liu XQ, Wyss-Coray T, et al. Apolipoprotein e fragments present in Alzheimer’s disease brains induce neurofibrillary tangle-like intracellular inclusions in neurons. Proc Natl Acad Sci U S A 2001; 98: 8838–8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Clay MA, Anantharamaiah GM, Mistry JM, et al. Localization of a domain in apolipoprotein E with both cytostatic and cytotoxic activity. Biochemistry 1995; 34: 11142–11151. [DOI] [PubMed] [Google Scholar]
- 72. Tolar M, Marques MA, Harmony JA, et al. Neurotoxicity of the 22 kDa thrombin-cleavage fragment of apolipoprotein E and related synthetic peptides is receptor-mediated. J Neurosci 1997; 17: 5678–5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Linton MF, Gish R, Hubl ST, et al. Phenotypes of apolipoprotein B and apolipoprotein E after liver transplantation. J Clin Invest 1991; 88: 270–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Li Z, Shue F, Zhao N, et al. APOE2: protective mechanism and therapeutic implications for Alzheimer’s disease. Mol Neurodegener 2020; 15: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Elshourbagy NA, Liao WS, Mahley RW, et al. Apolipoprotein E mRNA is abundant in the brain and adrenals, as well as in the liver, and is present in other peripheral tissues of rats and marmosets. Proc Natl Acad Sci U S A 1985; 82: 203–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Xu Q, Bernardo A, Walker D, et al. Profile and regulation of apolipoprotein E (ApoE) expression in the CNS in mice with targeting of green fluorescent protein gene to the ApoE locus. J Neurosci 2006; 26: 4985–4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Huynh T-V, Wang C, Tran AC, et al. Lack of hepatic ApoE does not influence early Aβ deposition: observations from a new APOE knock-in model. Mol Neurodegener 2019; 14: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Boyles J, Pitas R, Wilson E, et al. Apolipoprotein E associated with astrocytic glia of the central nervous system and with nonmyelinating glia of the peripheral nervous system. J Clin Invest 1985; 76: 1501–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Diedrich JF, Minnigan H, Carp RI, et al. Neuropathological changes in scrapie and Alzheimer’s disease are associated with increased expression of apolipoprotein E and cathepsin D in astrocytes. J Virol 1991; 65: 4759–4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Boschert U, Merlo-Pich E, Higgins G, et al. Apolipoprotein E expression by neurons surviving excitotoxic stress. Neurobiol Dis 1999; 6: 508–514. [DOI] [PubMed] [Google Scholar]
- 81. Mahley RW. Central nervous system lipoproteins: ApoE and regulation of cholesterol metabolism. Arter Thromb Vasc Biol 2016; 36: 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sharifov OF, Nayyar G, Garber DW, et al. Apolipoprotein E mimetics and cholesterol-lowering properties. Am J Cardiovasc Drugs 2011; 11: 371–381. [DOI] [PubMed] [Google Scholar]
- 83. Datta G, Chaddha M, Garber DW, et al. The receptor binding domain of apolipoprotein E, linked to a model class A amphipathic helix, enhances internalization and degradation of LDL by fibroblasts. Biochemistry 2000; 39: 213–220. [DOI] [PubMed] [Google Scholar]
- 84. Datta G, Garber DW, Chung BH, et al. Cationic domain 141-150 of ApoE covalently linked to a class A amphipathic helix enhances atherogenic lipoprotein metabolism in vitro and in vivo. J Lipid Res 2001; 42: 959–966. [PubMed] [Google Scholar]
- 85. Wilson C, Wardell MR, Weisgraber KH, et al. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science 1991; 252: 1817–1822. [DOI] [PubMed] [Google Scholar]
- 86. Weisgraber KH. Apolipoprotein E: structure-function relationships. Adv Protein Chem 1994; 45: 249–302. [DOI] [PubMed] [Google Scholar]
- 87. Morrow JA, Arnold KS, Dong J, et al. Effect of arginine 172 on the binding of apolipoprotein E to the low density lipoprotein receptor. J Biol Chem 2000; 275: 2576–2580. [DOI] [PubMed] [Google Scholar]
- 88. Morrow JA, Segall ML, Lund-Katz S, et al. Differences in stability among the human apolipoprotein E isoforms determined by the amino-terminal domain. Biochemistry 2000; 39: 11657–11666. [DOI] [PubMed] [Google Scholar]
- 89. Morrow JA, Hatters DM, Lu B, et al. Apolipoprotein E4 forms a molten globule. A potential basis for its association with disease. J Biol Chem 2002; 277: 50380–50385. [DOI] [PubMed] [Google Scholar]
- 90. Dong LM, Wilson C, Wardell MR, et al. Human apolipoprotein E. Role of arginine 61 in mediating the lipoprotein preferences of the E3 and E4 isoforms. J Biol Chem 1994; 269: 22358–22365. [PubMed] [Google Scholar]
- 91. Dong LM, Weisgraber KH. Human apolipoprotein E4 domain interaction. Arginine 61 and glutamic acid 255 interact to direct the preference for very low density lipoproteins. J Biol Chem 1996; 271: 19053–19057. [DOI] [PubMed] [Google Scholar]
- 92. Frieden C, Garai K. Concerning the structure of ApoE. Protein Sci 2013; 22: 1820–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chetty PS, Mayne L, Lund-Katz S, et al. Helical structure, stability, and dynamics in human apolipoprotein E3 and E4 by hydrogen exchange and mass spectrometry. Proc Natl Acad Sci U S A 2017; 114: 968–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Frieden C, Wang H, Ho CMW. A mechanism for lipid binding to apoE and the role of intrinsically disordered regions coupled to domain-domain interactions. Proc Natl Acad Sci U S A 2017; 114: 6292–6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Dong LM, Parkin S, Trakhanov SD, et al. Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia. Nat Struct Biol 1996; 3: 718–722. [DOI] [PubMed] [Google Scholar]
- 96. Serrano-Pozo A, Das S, Hyman BT. APOE and Alzheimer’s disease: advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol 2021; 20: 68–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Reiman EM, Arboleda-Velasquez JF, Quiroz YT, et al. Exceptionally low likelihood of Alzheimer’s dementia in APOE2 homozygotes from a 5,000-person neuropathological study. Nat Commun 2020; 11: 667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Arboleda-Velasquez JF, Lopera F, O’Hare M, et al. Resistance to autosomal dominant Alzheimer’s disease in an APOE3 Christchurch homozygote: a case report. Nat Med 2019; 25: 1680–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Salvadó G, Grothe MJ, Groot C, et al. Differential associations of APOE-ε2 and APOE-ε4 alleles with PET-measured amyloid-β and tau deposition in older individuals without dementia. Eur J Nucl Med Mol Imaging 2021; 48: 2212–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Suzuki K, Hirakawa A, Ihara R, et al. Effect of apolipoprotein E ε4 allele on the progression of cognitive decline in the early stage of Alzheimer’s disease. Alzheimers Dement 2020; 6: e12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Therriault J, Benedet AL, Pascoal TA, et al. Association of apolipoprotein E e4 with medial temporal tau independent of amyloid-β. JAMA Neurol 2020; 77: 470–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Krasemann S, Madore C, Cialic R, et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 2017; 47: 566–581.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Shi Y, Manis M, Long J, et al. Microglia drive APOE-dependent neurodegeneration in a tauopathy mouse model. J Exp Med 2019; 216: 2546–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Chen Y, Hong T, Chen F, et al. Interplay between microglia and Alzheimer’s disease-focus on the most relevant risks: APOE genotype, sex and age. Front Aging Neurosci 2021; 13: 631827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Chung WS, Verghese PB, Chakraborty C, et al. Novel allele-dependent role for APOE in controlling the rate of synapse pruning by astrocytes. Proc Natl Acad Sci U S A 2016; 113: 10186–10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Blanchard JW, Bula M, Davila-Velderrain J, et al. Reconstruction of the human blood-brain barrier in vitro reveals a pathogenic mechanism of APOE4 in pericytes. Nat Med 2020; 26: 952–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Riphagen JM, Ramakers IHGM, Freeze WM, et al. Linking APOE-ε4, blood-brain barrier dysfunction, and inflammation to Alzheimer’s pathology. Neurobiol Aging 2020; 85: 96–103. [DOI] [PubMed] [Google Scholar]
- 108. Bien-Ly N, Andrews-Zwilling Y, Xu Q, et al. C-terminal-truncated apolipoprotein (apo) E4 inefficiently clears amyloid-beta (Abeta) and acts in concert with Abeta to elicit neuronal and behavioral deficits in mice. Proc Natl Acad Sci U S A 2011; 108: 4236–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Saul A, Wirths O. Endogenous apolipoprotein E (ApoE) fragmentation is linked to amyloid pathology in transgenic mouse models of Alzheimer’s disease. Mol Neurobiol 2017; 54: 319–327. [DOI] [PubMed] [Google Scholar]
- 110. Dumanis SB, Tesoriero JA, Babus LW, et al. ApoE4 decreases spine density and dendritic complexity in cortical neurons in vivo. J Neurosci 2009; 29: 15317–15322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Hebert LE, Bienias JL, Aggarwal NT, et al. Change in risk of Alzheimer disease over time. Neurology 2010; 75: 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Elliott DA, Tsoi K, Holinkova S, et al. Isoform-specific proteolysis of apolipoprotein-E in the brain. Neurobiol Aging 2011; 32: 257–271. [DOI] [PubMed] [Google Scholar]
- 113. Amponsah AE, Feng B, Guo R, et al. Fragmentation of brain apolipoprotein E (ApoE) and its relevance in Alzheimer’s disease. Rev Neurosci 2020; 31: 589–603. [DOI] [PubMed] [Google Scholar]
- 114. Zhou W, Scott SA, Shelton SB, et al. Cathepsin D-mediated proteolysis of apolipoprotein E: possible role in Alzheimer’s disease. Neuroscience 2006; 143: 689–701. [DOI] [PubMed] [Google Scholar]
- 115. Bode W, Mayr I, Baumann U, et al. The refined 1.9 A crystal structure of human alpha-thrombin: interaction with D-Phe-Pro-Arg chloromethylketone and significance of the Tyr-Pro-Pro-Trp insertion segment. EMBO J 1989; 8: 3467–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Bode W. Structure and interaction modes of thrombin. Blood Cells Mol Dis 2006; 36: 122–130. [DOI] [PubMed] [Google Scholar]
- 117. DiBella EE, Maurer MC, Scheraga HA. Expression and folding of recombinant bovine prethrombin-2 and its activation to thrombin. J Biol Chem 1995; 270: 163–169. [DOI] [PubMed] [Google Scholar]
- 118. Huntinghton JA. Molecular recognition mechanisms of thrombin. J Thromb Haemost 2005; 3: 1861–1872. [DOI] [PubMed] [Google Scholar]
- 119. Akiyama H, Ikeda K, Kondo H, et al. Thrombin accumulation in brains of patients with Alzheimer’s disease. Neurosci Lett 1992; 146: 152–154. [DOI] [PubMed] [Google Scholar]
- 120. Arai T, Miklossy J, Klegeris A, et al. Thrombin and prothrombin are expressed by neurons and glial cells and accumulate in neurofibrillary tangles in Alzheimer disease brain. J Neuropathol Exp Neurol 2006; 65: 19–25. [DOI] [PubMed] [Google Scholar]
- 121. Grammas P, Samany PG, Thirumangalakudi L. Thrombin and inflammatory proteins are elevated in Alzheimer’s disease microvessels: implications for disease pathogenesis. J Alzheimer Dis 2006; 9: 51–58. [DOI] [PubMed] [Google Scholar]
- 122. Pompili E, Nori SL, Geloso MC, et al. Trimethyltin-induced differential expression of par subtypes in reactive astrocytes of the rat hippocampus. Brain Res Mol Brain Res 2004; 122: 93–98. [DOI] [PubMed] [Google Scholar]
- 123. Suo Z, Wu M, Citron BA, et al. Persistent protease-activated receptor 4 signaling mediates thrombin-induced microglial activation. J Biol Chem 2003; 278: 31177–31183. [DOI] [PubMed] [Google Scholar]
- 124. Coughlin SR. Protease-activated receptors start a family. Proc Natl Acad Sci U S A 1994; 91: 9200–9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Davis-Salinas J, Saporito-Irwin SM, Donovan FM, et al. Thrombin receptor activation induces secretion and nonamyloidogenic processing of amyloid beta-protein precursor. J Biol Chem 1994; 269: 22623–22627. [PubMed] [Google Scholar]
- 126. Brewer G. Thrombin causes cell spreading and redistribution of beta-amyloid immunoreactivity in cultured hippocampal neurons. J Neurochem 1996; 67: 119–130. [DOI] [PubMed] [Google Scholar]
- 127. Smith-Swintosky VL, Zimmer S, Fenton JW, et al. Opposing actions of thrombin and protease nexin-1 on amyloid beta-peptide toxicity and on accumulation of peroxides and calcium in hippocampal neurons. J Neurochem 1995; 65: 1415–1418. [DOI] [PubMed] [Google Scholar]
- 128. Pike CJ, Vaughan PJ, Cunningham DD, et al. Thrombin attenuates neuronal cell death and modulates astrocyte reactivity induced by beta-amyloid in vitro. J Neurochem 1996; 66: 1374–1382. [DOI] [PubMed] [Google Scholar]
- 129. Wetterau JR, Aggerbeck LP, Rall SC, Jr, et al. Human apolipoprotein E3 in aqueous solution. I. Evidence for two structural domains. J Biol Chem 1988; 263: 6240–6148. [PubMed] [Google Scholar]
- 130. Vidoni C, Follo C, Savino M, et al. The role of cathepsin d in the pathogenesis of human neurodegenerative disorders. Med Res Rev 2016; 36: 845–870. [DOI] [PubMed] [Google Scholar]
- 131. Wex T, Levy B, Smeekens SP, et al. Genomic structure, chromosomal localization, and expression of human cathepsin W. Biochem Biophys Res Commun 1998; 248: 255–261. [DOI] [PubMed] [Google Scholar]
- 132. Brömme D, Li Z, Barnes M, et al. Human cathepsin v functional expression, tissue distribution, electrostatic surface potential, enzymatic characterization, and chromosomal localization. Biochemistry 1999; 38: 2377–2385. [DOI] [PubMed] [Google Scholar]
- 133. Turk B, Turk D, Turk V. Lysosomal cysteine proteases: more than scavengers. Biochim Biophys Acta 2000; 1477: 98–111. [DOI] [PubMed] [Google Scholar]
- 134. Lecaille F, Kaleta J, Brömme D. Human and parasitic papain-like cysteine proteases: their role in physiology and pathology and recent developments in inhibitor design. Chem Rev 2002; 102: 4459–4488. [DOI] [PubMed] [Google Scholar]
- 135. Hasilik A, Neufeld EF. Biosynthesis of lysosomal enzymes in fibroblasts. Synthesis as precursors of higher molecular weight. J Biol Chem 1980; 255: 4937–4945. [PubMed] [Google Scholar]
- 136. Gieselmann V, Pohlmann R, Hasilik A, et al. Biosynthesis and transport of cathepsin D in cultured human fibroblasts. J Cell Biol 1983; 97: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Erickson AH. Biosynthesis of lysosomal endopeptidases. J Cell Biochem 1989; 40: 31–41. [DOI] [PubMed] [Google Scholar]
- 138. Minarowska A, Minarowski L, Karwowska A, et al. Regulatory role of cathepsin D in apoptosis. Folia Histochem Cytobiol 2007; 45: 159–163. [PubMed] [Google Scholar]
- 139. Benes P, Vetvicka V, Fusek M. Cathepsin D – many functions of one aspartic protease. Crit Rev Oncol Hematol 2008; 68: 12–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Reiser J, Adair B, Reinheckel T. Specialized roles for cysteine cathepsins in health and disease. J Clin Invest 2010; 120: 3421–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Conus S, Simon HU. Cathepsins and their involvement in immune responses. Swiss Med Wkly 2010; 140: w13042. [DOI] [PubMed] [Google Scholar]
- 142. Masson O, Bach AS, Derocq D, et al. Pathophysiological functions of cathepsin D: targeting its catalytic activity versus its protein binding activity? Biochimie 2010; 92: 1635–1643. [DOI] [PubMed] [Google Scholar]
- 143. Tan GJ, Peng ZK, Lu JP, et al. Cathepsins mediate tumor metastasis. World J Biol Chem 2013; 4: 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Khurana V, Elson-Schwab I, Fulga TA, et al. Lysosomal dysfunction promotes cleavage and neurotoxicity of tau in vivo. PLoS Genet 2010; 6: e1001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Minarowska A, Gacko M, Karwowska A, et al. Human cathepsin D. Folia Histochem Cytobiol 2008; 46: 23–38. [DOI] [PubMed] [Google Scholar]
- 146. Marques MA, Owens PA, Crutcher KA. Progress toward identification of protease activity involved in proteolysis of apolipoprotein E in human brain. J Mol Neurosci 2004; 24: 73–80. [DOI] [PubMed] [Google Scholar]
- 147. Zanni EE, Kouvatsi A, Hadzopoulou-Cladaras M, et al. Expression of ApoE gene in Chinese hamster cells with a reversible defect in O-glycosylation. Glycosylation is not required for ApoE secretion. J Biol Chem 1989; 264: 9137–9140. [PubMed] [Google Scholar]
- 148. Green RC, Cupples LA, Go R, et al. Risk of dementia among white and African American relatives of patients with Alzheimer disease. JAMA 2002; 287: 329–336. [DOI] [PubMed] [Google Scholar]
- 149. Marques MA, Crutcher KA. Apolipoprotein E-related neurotoxicity as a therapeutic target for Alzheimer’s disease. J Mol Neurosci 2003; 20: 327–337. [DOI] [PubMed] [Google Scholar]
- 150. Clausen T, Kaiser M, Huber R, et al. Htra proteases: regulated proteolysis in protein quality control. Nat Rev Mol Cell Biol 2011; 12: 386–368. [DOI] [PubMed] [Google Scholar]
- 151. Truebestein L, Tennstaedt A, Mönig T, et al. Substrate-induced remodeling of the active site regulates human Htra1 activity. Nat Struct Mol Biol 2011; 18: 386–388. [DOI] [PubMed] [Google Scholar]
- 152. Eigenbrot C, Ultsch M, Lipari MT, et al. Structural and functional analysis of Htra1 and its subdomains. Structure 2012; 20: 1040–1050. [DOI] [PubMed] [Google Scholar]
- 153. Runyon ST, Zhang Y, Appleton BA, et al. Structural and functional analysis of the PDZ domains of human HtrA1 and HtrA3. Protein Sci 2007; 16: 2454–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Tamboli IY, Heo D, Rebeck G.W. Extracellular proteolysis of apolipoprotein E (ApoE) by secreted serine neuronal protease. PLoS One 2014; 9: e93120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Chu Q, Diedrich JK, Vaughan JM, et al. Htra1 proteolysis of ApoE in vitro is allele selective. J Am Chem Soc 2016; 138: 9473–9478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Hasel P, Liddelow SA. Isoform-dependent APOE secretion modulates neuroinflammation. Nat Rev Neurol 2021; 17: 265–266. [DOI] [PubMed] [Google Scholar]
- 157. Harris FM, Brecht WJ, Xu Q, et al. Carboxyl-terminal-truncated apolipoprotein E4 causes Alzheimer’s disease-like neurodegeneration and behavioral deficits in transgenic mice. Proc Natl Acad Sci U S A 2003; 100: 10966–10971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Marques MA, Tolar M, Harmony JA, et al. A thrombin cleavage fragment of apolipoprotein E exhibits isoform-specific neurotoxicity. Neuroreport 1996; 7: 2529–2532. [DOI] [PubMed] [Google Scholar]
- 159. Muñoz SS, Li H, Ruberu K, et al. The serine protease Htra1 contributes to the formation of an extracellular 25-kDa apolipoprotein E fragment that stimulates neuritogenesis. J Biol Chem 2018; 293: 4071–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Wernette-Hammond ME, Lauer SJ, Corsini A, et al. Glycosylation of human apolipoprotein E. The carbohydrate attachment site is threonine 194. J Biol Chem 1989; 264: 9094–9101. [PubMed] [Google Scholar]
- 161. Wisniewski T, Frangione B. Apolipoprotein E: a pathological chaperone protein in patients with cerebral and systemic amyloid. Neurosci Lett 1992; 135: 235–238. [DOI] [PubMed] [Google Scholar]
- 162. Kim J, Basak JM, Holtzman DM. The role of apolipoprotein E in Alzheimer’s disease. Neuron 2009; 63: 287–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Nagy Z, Esiri MM, Jobst KA, et al. Influence of the apolipoprotein E genotype on amyloid deposition and neurofibrillary tangle formation in Alzheimer’s disease. Neuroscience 1995; 69: 757–761. [DOI] [PubMed] [Google Scholar]
- 164. Morris CM, Benjamin R, Leake A, et al. Effect of apolipoprotein E genotype on Alzheimer’s disease neuropathology in a cohort of elderly Norwegians. Neurosci Lett 1995; 201: 45–47. [DOI] [PubMed] [Google Scholar]
- 165. Zhong N, Weisgraber KH. Understanding the association of apolipoprotein E4 with Alzheimer disease: clues from its structure. J Biol Chem 2009; 284: 6027–6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Xu Q, Brecht WJ, Weisgraber KH, et al. Apolipoprotein E4 domain interaction occurs in living neuronal cells as determined by fluorescence resonance energy transfer. J Biol Chem 2004; 279: 25511–25516. [DOI] [PubMed] [Google Scholar]
- 167. Miyata M, Smith JD. Apolipoprotein e allele-specific antioxidant activity and effects on cytotoxicity by oxidative insults and beta-amyloid peptides. Nat Genet 1996; 14: 55–61. [DOI] [PubMed] [Google Scholar]
- 168. Tai LM, Mehra S, Shete V, et al. Soluble apoE/Aβ complex: mechanism and therapeutic target for APOE4-induced AD risk. Mol Neurodegener 2014; 9: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Jones PB, Adams KW, Rozkalne A, et al. Apolipoprotein E: isoform specific differences in tertiary structure and interaction with amyloid-β in human Alzheimer brain. PLoS One 2011; 6: e1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Mouchard A, Boutonnet MC, Mazzocco C, et al. ApoE-fragment/Aβ heteromers in the brain of patients with Alzheimer’s disease. Sci Rep 2019; 9: 3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Aizawa Y, Fukatsu R, Takamaru Y, et al. Amino-terminus truncated apolipoprotein E is the major species in amyloid deposits in Alzheimer’s disease-affected brains: a possible role for apolipoprotein E in Alzheimer’s disease. Brain Res 1997; 768: 208–214. [DOI] [PubMed] [Google Scholar]
- 172. Christensen DZ, Schneider-Axmann T, Lucassen PJ, et al. Accumulation of intraneuronal Abeta correlates with ApoE4 genotype. Acta Neuropathol 2010; 119: 555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Tai LM, Bilousova T, Jungbauer L, et al. Levels of soluble apolipoprotein E/amyloid-β (Aβ) complex are reduced and oligomeric Aβ increased with APOE4 and Alzheimer disease in a transgenic mouse model and human samples. J Biol Chem 2013; 288: 5914–5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Mormino EC, Betensky RA, Hedden T, et al. Amyloid and ApoE E4 interact to influence short-term decline in preclinical Alzheimer disease. Neurology 2014; 82: 1760–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Hashimoto T, Serrano-Pozo A, Hori Y, et al. Apolipoprotein E, especially apolipoprotein E4, increases the oligomerization of amyloid beta peptide. J Neurosci 2012; 32: 15181–15192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Baranello RJ, Bharani KL, Padmaraju V, et al. Amyloid-beta protein clearance and degradation (ABCD) pathways and their role in Alzheimer’s disease. Curr Alzheimer Res 2015; 12: 32–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Wang M, Turko IV. Mass spectrometry quantification revealed accumulation of C-terminal fragment of apolipoprotein E in the Alzheimer’s frontal cortex. PLoS One 2013; 8: e61498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Love JE, Day RJ, Gause JW, et al. Nuclear uptake of an amino-terminal fragment of apolipoprotein E4 promotes cell death and localizes within microglia of the Alzheimer’s disease brain. Int J Physiol Pathophysiol Pharmacol 2017; 9: 40–57. [PMC free article] [PubMed] [Google Scholar]
- 179. Dafnis I, Argyri L, Sagnou M, et al. The ability of apolipoprotein E fragments to promote intraneuronal accumulation of amyloid beta peptide 42 is both isoform and size-specific. Sci Rep 2016; 6: 30654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Dafnis I, Tzinia AK, Tsilibary EC, et al. An apolipoprotein E4 fragment affects matrix metalloproteinase 9, tissue inhibitor of metalloproteinase 1 and cytokine levels in brain cell lines. Neuroscience 2012; 210: 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Tolar M, Keller JN, Chan S, et al. Truncated apolipoprotein E (ApoE) causes increased intracellular calcium and may mediate ApoE neurotoxicity. J Neurosci 1999; 19: 7100–7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Chang S, ran Ma T, Miranda R, et al. Lipid- and receptor-binding regions of apolipoprotein E4 fragments act in concert to cause mitochondrial dysfunction and neurotoxicity. Proc Natl Acad Sci U S A 2005; 102: 18694–18699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Cho HS, Hyman BT, Greenberg SM, et al. Quantitation of apoE domains in Alzheimer disease brain suggests a role for apoE in Abeta aggregation. J Neuropathol Exp Neurol 2001; 60: 342–349. [DOI] [PubMed] [Google Scholar]
- 184. Laskowitz DT, Thekdi AD, Thekdi SD, et al. Downregulation of microglial activation by apolipoprotein E and ApoE-mimetic peptides. Exp Neurol 2001; 167: 74–85. [DOI] [PubMed] [Google Scholar]
- 185. Lynch JR, Tang W, Wang H, et al. ApoE genotype and an ApoE-mimetic peptide modify the systemic and central nervous system inflammatory response. J Biol Chem 2003; 278: 48529–48533. [DOI] [PubMed] [Google Scholar]
- 186. Aono M, Bennett E, Kim K, et al. Protective effect of apolipoprotein E-mimetic peptides on n-methyl-daspartate excitotoxicity in primary rat neuronal-glial cell cultures. Neuroscience 2003; 116: 437–445. [DOI] [PubMed] [Google Scholar]
- 187. Klein RC, Yakel JL. Inhibition of nicotinic acetylcholine receptors by apolipoprotein E-derived peptides in rat hippocampal slices. Neuroscience 2004; 127: 563–567. [DOI] [PubMed] [Google Scholar]
- 188. Gay EA, Klein RC, Yakel JL. Apolipoprotein E-derived peptides block alpha7 neuronal nicotinic acetylcholine receptors expressed in Xenopus oocytes. J Pharmacol Exp Ther 2006; 316: 835–842. [DOI] [PubMed] [Google Scholar]
- 189. Gay EA, Bienstock RJ, Lamb PW, et al. Structural determinates for apolipoprotein E-derived peptide interaction with the alpha7 nicotinic acetylcholine receptor. Mol Pharmacol 2007; 72: 838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Wellnitz S, Friedlein A, Bonanni C, et al. A 13 kDa carboxy-terminal fragment of ApoE stabilizes abeta hexamers. J Neurochem 2005; 94: 1351–1360. [DOI] [PubMed] [Google Scholar]
- 191. Kothari S, Bala N, Patel AB, et al. The LDL receptor binding domain of apolipoprotein E directs the relative orientation of its C-terminal segment in reconstituted nascent HDL. Biochim Biophys Acta Biomembr 2021; 1863: 183618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Vlieghe P, Lisowski V, Martinez J, et al. Synthetic therapeutic peptides: science and market. Drug Discov Today 2010; 15: 40–56. [DOI] [PubMed] [Google Scholar]
- 193. Vagner J, Qu H, Hruby VJ. Peptidomimetics, a synthetic tool of drug discovery. Curr Opin Chem Biol 2008; 12: 292–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Puttagunta AL, Toth EL. Insulin lispro (Humalog), the first marketed insulin analogue: indications, contraindications and need for further study. CMAJ 1998; 158; 506–511. [PMC free article] [PubMed] [Google Scholar]
- 195. Gentilucci L, Tolomelli A, Squassabia F. Peptides and peptidomimetics in medicine, surgery and biotechnology. Curr Med Chem 2006; 13: 2449–2466. [DOI] [PubMed] [Google Scholar]
- 196. Segrest JP, Jackson RL, Morrisett JD, et al. A molecular theory of lipid-protein interactions in the plasma lipoproteins. FEBS Lett 1974; 38: 247–258. [DOI] [PubMed] [Google Scholar]
- 197. Chung BH, Anatharamaiah GM, Brouillette CG, et al. Studies of synthetic peptide analogs of the amphipathic helix. correlation of structure with function. J Biol Chem 1985; 260: 10256–10262. [PubMed] [Google Scholar]
- 198. Dyer CA, Cistola DP, Parry GC, et al. Structural features of synthetic peptides of apolipoprotein E that bind the LDL receptor. J Lipid Res 1995; 36: 80–88. [PubMed] [Google Scholar]
- 199. Vedhachalam C, Narayanaswami V, Neto N, et al. The C-terminal lipid-binding domain of apolipoprotein E is a highly efficient mediator of ABCA1-dependent cholesterol efflux that promotes the assembly of high-density lipoproteins. Biochemistry 2007; 46: 2583–2593. [DOI] [PubMed] [Google Scholar]
- 200. Hafiane A, Bielicki JK, Johansson JO, et al. Apolipoprotein e derived HDL mimetic peptide ati-5261 promotes nascent HDL formation and reverse cholesterol transport in vitro. Biochim Biophys Acta 2014; 1842: 1498–512. [DOI] [PubMed] [Google Scholar]
- 201. Bielicki J, Zhang H, Cortez Y, et al. A new HDL mimetic peptide that stimulates cellular cholesterol efflux with high efficiency greatly reduces atherosclerosis in mice. J Lipid Res 2010; 51: 1496–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Nikoulin IR, Curtiss LK. An apolipoprotein E synthetic peptide targets to lipoproteins in plasma and mediates both cellular lipoprotein interactions in vitro and acute clearance of cholesterol-rich lipoproteins in vivo. J Clin Invest 1998; 101: 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Heinecke JW. Small HDL promotes cholesterol efflux by the ABCA1 pathway in macrophages: implications for therapies targeted to HDL. Circ Res 2015; 116: 1101–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Westerterp M, Murphy AJ, Wang M, et al. Deficiency of ATP-binding cassette transporters A1 and G1 in macrophages increases inflammation and accelerates atherosclerosis in mice. Circ Res 2013; 112: 1456–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Tabas I, Garcia-Cardena G, Owens GK. Recent insights into the cellular biology of atherosclerosis. J Cell Biol 2015; 209: 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Boehm-Cagan A, Michaelson D. Reversal of ApoE4-driven brain pathology and behavioral deficits by bexarotene. J Neurosci 2014; 34: 7293–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Nordestgaard LT, Tybjaerg-Hansen A, Nordestgaard BG, et al. Loss-of-function mutation in ABCA1 and risk of Alzheimer’s disease and cerebrovascular disease. Alzheimers Dement 2015; 11: 1430–1438. [DOI] [PubMed] [Google Scholar]
- 208. Wahrle SE, Jiang H, Parsadanian M, et al. Abca1 is required for normal central nervous system ApoE levels and for lipidation of astrocyte-secreted ApoE. J Biol Chem 2004; 279: 40987–40993. [DOI] [PubMed] [Google Scholar]
- 209. Fitz NF, Cronican AA, Saleem M, et al. Abca1 deficiency affects Alzheimer’s disease-like phenotype in human ApoE4 but not in ApoE3-targeted replacement mice. J Neurosci 2012; 32: 13125–13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Bielicki J. Abca1 agonist peptides for the treatment of disease. Curr Opin Lipidol 2016; 27: 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Gale SC, Gao L, Mikacenic C, et al. ApoEpsilon4 is associated with enhanced in vivo innate immune responses in human subjects. J Allergy Clin Immunol 2014; 134: 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Mak AC, Pullinger CR, Tang LF, et al. Effects of the absence of apolipoprotein E on lipoproteins, neurocognitive function, and retinal function. JAMA Neurol 2014; 71: 1228–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Yamazaki Y, Zhao N, Caulfield TR, et al. Apolipoprotein E and Alzheimer disease: pathobiology and targeting strategies. Nat Rev Neurol 2019; 15: 501–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Hudry E, Dashkoff J, Roe AD, et al. Gene transfer of human Apoe isoforms results in differential modulation of amyloid deposition and neurotoxicity in mouse brain. Sci Transl Med 2013; 5: 212ra161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Zhao L, Gottesdiener AJ, Parmar M, et al. Intracerebral adeno-associated virus gene delivery of apolipoprotein E2 markedly reduces brain amyloid pathology in Alzheimer’s disease mouse models. Neurobiol Aging 2016; 44: 159–172. [DOI] [PubMed] [Google Scholar]
- 216. Rosenberg JB, Kaplitt MG, De BP, et al. AAVrh.10-mediated APOE2 central nervous system gene therapy for APOE4-associated Alzheimer’s disease. Hum Gene Ther Clin Dev 2018; 29: 24–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Arora S, Layek B, Singh J. Design and validation of liposomal ApoE2 gene delivery system to evade blood-brain barrier for effective treatment of Alzheimer’s disease. Mol Pharm 2021; 18: 714–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Dos Santos Rodrigues B, Kanekiyo T, Singh J. ApoE-2 brain-targeted gene therapy through transferrin and penetratin tagged liposomal nanoparticles. Pharm Res 2019; 36: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Huynh TV, Liao F, Francis CM, et al. Age-dependent effects of apoE reduction using antisense oligonucleotides in a model of β-amyloidosis. Neuron 2017; 96: 1013–1023.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Litvinchuk A, Huynh TV, Shi Y, et al. Apolipoprotein E4 reduction with antisense oligonucleotides decreases neurodegeneration in a tauopathy model. Ann Neurol 2021; 89: 952–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Luz I, Liraz O, Michaelson DM. An anti-apoE4 specific monoclonal antibody counteracts the pathological effects of apoE4 in vivo. Curr Alzheimer Res 2016; 13: 918–929. [DOI] [PubMed] [Google Scholar]
- 222. Kim J, Eltorai AE, Jiang H, et al. Anti-apoE immunotherapy inhibits amyloid accumulation in a transgenic mouse model of Aβ amyloidosis. J Exp Med 2012; 209: 2149–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Liao F, Hori Y, Hudry E, et al. Anti-ApoE antibody given after plaque onset decreases Aβ accumulation and improves brain function in a mouse model of Aβ amyloidosis. J Neurosci 2014; 34: 7281–7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Liao F, Li A, Xiong M, et al. Targeting of nonlipidated, aggregated apoE with antibodies inhibits amyloid accumulation. J Clin Invest 2018; 128: 2144–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Xiong M, Jiang H, Serrano JR, et al. APOE immunotherapy reduces cerebral amyloid angiopathy and amyloid plaques while improving cerebrovascular function. Sci Transl Med 2021; 13: eabd7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Vitek MP, Christensen DJ, Wilcock D, et al. APOE-mimetic peptides reduce behavioral deficits, plaques and tangles in Alzheimer’s disease transgenics. Neurodegener Dis 2012; 10: 122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Handattu SP, Monroe CE, Nayyar G, et al. In vivo and in vitro effects of an apolipoprotein E mimetic peptide on amyloid-β pathology. J Alzheimers Dis 2013; 36: 335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Ghosal K, Stathopoulos A, Thomas D, et al. The apolipoprotein-E-mimetic COG112 protects amyloid precursor protein intracellular domain-overexpressing animals from Alzheimer’s disease-like pathological features. Neurodegener Dis 2013; 12: 51–58. [DOI] [PubMed] [Google Scholar]
- 229. Krishnamurthy K, Cantillana V, Wang H, et al. ApoE mimetic improves pathology and memory in a model of Alzheimer’s disease. Brain Res 2020; 1733: 146685. [DOI] [PubMed] [Google Scholar]
- 230. VanDusen KW, Eleswarpu S, Moretti EW, et al. The MARBLE study protocol: modulating ApoE signaling to reduce brain inflammation, delirium, and postoperative cognitive dysfunction. J Alzheimers Dis 2020; 75: 1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Mahley RW, Huang Y. Small-molecule structure correctors target abnormal protein structure and function: structure corrector rescue of apolipoprotein E4-associated neuropathology. J Med Chem 2012; 55: 8997–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Chen HK, Ji ZS, Dodson SE, et al. Apolipoprotein E4 domain interaction mediates detrimental effects on mitochondria and is a potential therapeutic target for Alzheimer disease. J Biol Chem 2011; 286: 5215–5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Chen HK, Liu Z, Meyer-Franke A, et al. Small molecule structure correctors abolish detrimental effects of apolipoprotein E4 in cultured neurons. J Biol Chem 2012; 287: 5253–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Brodbeck J, McGuire J, Liu Z, et al. Structure-dependent impairment of intracellular apolipoprotein E4 trafficking and its detrimental effects are rescued by small-molecule structure correctors. J Biol Chem 2011; 286: 17217–17226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Wang C, Najm R, Xu Q, et al. Gain of toxic apolipoprotein E4 effects in human ipsc-derived neurons is ameliorated by a small-molecule structure corrector. Nat Med 2018; 24: 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Lin YT, Seo J, Gao F, et al. ApoE4 causes widespread molecular and cellular alterations associated with Alzheimer’s disease phenotypes in human iPSC-derived brain cell types. Neuron 2018; 98: 1141–1154.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Tagalakis AD, Dickson JG, Owen JS, et al. Correction of the neuropathogenic human apolipoprotein E4 (APOE4) gene to APOE3 in vitro using synthetic RNA/DNA oligonucleotides (chimeraplasts). J Mol Neurosci 2005; 25: 95–103. [DOI] [PubMed] [Google Scholar]
- 238. Sadowski M, Pankiewicz J, Scholtzova H, et al. A synthetic peptide blocking the apolipoprotein E/beta-amyloid binding mitigates beta-amyloid toxicity and fibril formation in vitro and reduces beta-amyloid plaques in transgenic mice. Am J Pathol 2004; 165: 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Sadowski MJ, Pankiewicz J, Scholtzova H, et al. Blocking the apolipoprotein E/amyloid-beta interaction as a potential therapeutic approach for Alzheimer’s disease. Proc Natl Acad Sci U S A 2006; 103: 18787–18792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Kuszczyk MA, Sanchez S, Pankiewicz J, et al. Blocking the interaction between apolipoprotein E and Aβ reduces intraneuronal accumulation of Aβ and inhibits synaptic degeneration. Am J Pathol 2013; 182: 1750–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Pankiewicz JE, Guridi M, Kim J, et al. Blocking the apoE/Aβ interaction ameliorates Aβ-related pathology in APOE ε2 and ε4 targeted replacement Alzheimer model mice. Acta Neuropathol Commun 2014; 2: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Imbimbo BP, Lozupone M, Watling M, et al. Discontinued disease-modifying therapies for Alzheimer’s disease: status and future perspectives. Expert Opin Investig Drugs 2020; 29: 919–933. [DOI] [PubMed] [Google Scholar]
- 243. Kent SA, Spires-Jones TL, Durrant CS. The physiological roles of tau and Aβ: implications for Alzheimer’s disease pathology and therapeutics. Acta Neuropathol 2020; 140: 417–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Huang Y, Mahley RW. Apolipoprotein E: structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiol Dis 2014; 72: 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Rohn TT, Catlin LW, Coonse KG, et al. Identification of an amino-terminal fragment of apolipoprotein E4 that localizes to neurofibrillary tangles of the Alzheimer’s disease brain. Brain Res 2012; 1475: 106–115. [DOI] [PubMed] [Google Scholar]
- 246. Wang C, Xiong M, Gratuze M, et al. Selective removal of astrocytic APOE4 strongly protects against tau-mediated neurodegeneration and decreases synaptic phagocytosis by microglia. Neuron 2021; 109: 1657–1674.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]