Abstract
Risk loci identified through genome-wide association studies have explained about 25% of the phenotypic variations in nonsyndromic orofacial clefts (nsOFCs) on the liability scale. Despite the notable sex differences in the incidences of the different cleft types, investigation of loci for sex-specific effects has been understudied. To explore the sex-specific effects in genetic etiology of nsOFCs, we conducted a genome-wide gene × sex (GxSex) interaction study in a sub-Saharan African orofacial cleft cohort. The sample included 1,019 nonsyndromic orofacial cleft cases (814 cleft lip with or without cleft palate and 205 cleft palate only) and 2,159 controls recruited from 3 sites (Ethiopia, Ghana, and Nigeria). An additive logistic model was used to examine the joint effects of the genotype and GxSex interaction. Furthermore, we examined loci with suggestive significance (P < 1E-5) in the additive model for the effect of the GxSex interaction only. We identified a novel risk locus on chromosome 8p22 with genome-wide significant joint and GxSex interaction effects (rs2720555, p2df = 1.16E-08, pGxSex = 1.49E-09, odds ratio [OR] = 0.44, 95% CI = 0.34 to 0.57). For males, the risk of cleft lip with or without cleft palate at this locus decreases with additional copies of the minor allele (p < 0.0001, OR = 0.60, 95% CI = 0.48 to 0.74), but the effect is reversed for females (p = 0.0004, OR = 1.36, 95% CI = 1.15 to 1.60). We replicated the female-specific effect of this locus in an independent cohort (p = 0.037, OR = 1.30, 95% CI = 1.02 to 1.65), but no significant effect was found for the males (p = 0.29, OR = 0.86, 95% CI = 0.65 to 1.14). This locus is in topologically associating domain with craniofacially expressed and enriched genes during embryonic development. Rare coding mutations of some of these genes were identified in nsOFC cohorts through whole exome sequencing analysis. Our study is additional proof that genome-wide GxSex interaction analysis provides an opportunity for novel findings of loci and genes that contribute to the risk of nsOFCs.
Keywords: craniofacial biology/genetics, gender differences, developmental biology, bioinformatics, craniofacial anomalies, gene expression
Introduction
Orofacial clefts (OFCs) are the most common congenital defect of the head and neck region (Mossey et al. 2009). They are due to incomplete fusion of the facial processes during embryonic development (Smarius et al. 2017). Globally, this developmental anomaly occurs in 1 in 700 live births (Rahimov et al. 2012). There have been reports of different birth incidence rates of this defect, with African populations having an incidence of ~1 in 2,000 live births, Asians 1 in 500, and Europeans 1 in 1,000 (Mossey et al. 2009; Butali et al. 2014). These obvious differences could be a result of the genetic architectures and/or variations among the various populations, the different environmental exposures across geographic regions, or the interactions between genetic risk factors and environmental exposures. Underreporting due to low surveillance could also be a reason for the low incidence in the African population (Butali and Mossey 2009).
Nonsyndromic OFCs (nsOFCs), which include nonsyndromic cleft lip with or without cleft palate (nsCL/P) and nonsyndromic cleft palate only (nsCPO), have multifactorial causes. Genetic factors, environmental factors, and their interactions have been shown to play major roles in the risk of these defects (Beaty et al. 2016). Genome-wide association studies (GWASs) have been carried out in different populations to investigate risk loci associated with nsOFCs (Birnbaum et al. 2009; Grant et al. 2009; Beaty et al. 2010; Mangold et al. 2010; Sun et al. 2015; Leslie et al. 2016; Butali et al. 2019). The first GWAS of nsOFCs in an African population identified 2 novel risk loci for nsCPO on chromosomes 2 and 19 (Butali et al. 2019). Despite the number of GWASs, the genetic contribution to the risk of the disease is yet to be fully understood.
Sex differences can be found in the levels and patterns of gene expression. These differences in the gene expression and their contribution to complex traits have been well established (Khramtsova et al. 2019). A genetic study of schizophrenia identified novel sex-specific up- and downregulated genes that contribute to the risk of this neurodevelopmental disorder (Qin et al. 2016). These sex-dependent genetic effects have also been reported in the traits of model organisms. One such study noted that overexpression of the p53 gene in Drosophila melanogaster significantly increases the life span in males, but a reversed effect is seen in females (Waskar et al. 2009).
Studies have shown that there is a sex predilection to the different cleft types. nsCL/P is twice as common in males than females, while nsCPO is twice as common in females than males (Marazita 2012). A recent investigation of the GxSex interaction in a nsOFC cohort identified a novel risk locus on chromosome 10q21 that increases the risk for nsCL/P in females (Carlson et al. 2018). Therefore, we hypothesize that genome-wide analysis of the GxSex interaction in our African cohort will lead to the discovery of new risk loci associated with nsOFCs. If so, this will explain the sex differences seen in nsOFC subtypes and help explain some missing heritability for nsOFCs in this population. To test this hypothesis, we investigated GxSex interactions using our GWAS case-control genotype data.
Methods
Study Population
The study population is described in our previously published study (Butali et al. 2019). In summary, ethical approvals were obtained at the local institution review boards: University of Iowa (IRB ID No. 201101720), Lagos University Teaching Hospital (ADM/DCST/HREC/VOL.XV/321), Obafemi Awolowo University Teaching Hospital (ERC/2011/12/01), Kwame Nkrumah University of Science and Technology (CHRPE/RC/018/130), and Addis Ababa University (003/10/surg). Patients were among those who took part in the Pan-African Association of Cleft Lip and Palate network for free surgical repair of clefts in Africa.
DNA Extraction and XY Genotyping
Saliva samples were collected from cases and controls with the Oragene saliva tool kit (DNA Genotek). DNA extraction was conducted at the Butali Laboratory. Extracted DNA samples were quantified with Qubit (Thermo Fisher Scientific). Stocks and working aliquots were made for downstream analyses. We confirmed the reported sex with TaqMan XY genotyping, which is a quality control step in the analysis workflow at the Butali Laboratory. Working aliquots (25 µL) with confirmed sex and DNA concentration ≥50 ng/µL were shipped to the Center for Inherited Disease Research for MEGA genotyping (Multi-ethnic Genotyping Array).
Genotyping of Single-Nucleotide Polymorphisms and Data Cleaning
The methods for genotyping, quality control, imputation, and derivation of the principal components (PCs) of ancestry are described in our previously published study (Butali et al. 2019; Appendix). Briefly, >2 million common single-nucleotide polymorphisms (SNPs) and >87,000 rare coding variants in the African population were genotyped with the expanded Illumina MEGA v2 15070954 A2 (genome build 37). Following the genotyping and imputation, we checked for chromosomal anomalies, missingness, batch effects, and relatedness (identity by descent/identity by state) and confirmed the continental ancestry of our cohorts with respect to the HapMap samples with the methods published by Laurie et al. (2010). This was a quality control step and was executed with the R package, SNPRelate, and GENESIS. The final data set analyzed included 3,178 unique samples: 814 cases of nsCL/P, 205 cases of isolated nsCPO, and 2,159 controls (Appendix Table 1A).
Statistical Analysis
We investigated the potential gene-by-sex (GxSex) interactions following a case-control approach that was first introduced by Kraft et al. (2007). An additive genetic logistic model was used to examine these GxSex interactions. In the first step, we examined the joint effect of the genotype (G) and GxSex interaction on risk of nsCL/P.
For these models, coefficient β(G) estimates the additive effects of each additional minor allele; β(SEX), the effect of sex; and β(GxSex), the effect of the interaction term GxSex. The response was coded as 0/1 for control/case response, respectively. β (PC1) through β (PC13) estimate the effects of the principal components (PCs) of ancestry, which control for population stratifications.
We proposed these models as per our hypothesis that GxSex interaction increases the risk of OFC. Following this analysis, SNPs that had suggestive p values (<1.0 × 10–5) were investigated for the effect of GxSex interaction in the second step.
In this step, we examined the effect of GxSex interaction as shown:
Our analyses were based on cleft types: nsCL/P and nsCPO. The choice of these PCs was justified in our previously published GWAS (Butali et al. 2019). The p values were examined for a genome-wide significance threshold of 5.00 × 10−8 and a suggestive threshold of 1.00 × 10–5.
Following the discovery of the genome-wide significant locus, we tested the association between the locus and nsOFCs in male and female clusters separately.
Replication Study
We examined the sex-specific effects of the genome-wide significant locus in an independent cohort. This cohort comprised samples from the same African populations (Nigeria, Ghana, and Ethiopia). A total of 1,858 samples consisting of 428 nsOFC cases and 1,430 controls (Appendix Table 1B) were successfully genotyped with the TaqMan SNP genotyping assay. To evaluate the sex-specific hypothesis proposed, we separated the independent samples into male (n = 899) and female (n = 959) clusters. Then, we tested the association between the SNP and nsOFCs in each cluster. PLINK was used to estimate the odds of nsOFCs in each cluster assuming an additive model.
Identification of GxSex Interaction Candidate Genes via the Topologically Associating Domain
We know that signals resulting from enhancers influence the expression of genes that are within the same topologically associating domain (TAD). We visualized the region in the human reference genome (hg19) by searching for the interaction domain for the index SNP ID (http://3dgenome.fsm.northwestern.edu/view.php). In our search, we used human embryonic stem cells as the query tissue to encompass the unique cells that are involved in the formation of the lip and palate.
SysFACE-Based Craniofacial Gene Expression Analysis
To identify the craniofacially relevant candidate genes among those flanking the genome-wide GxSex interaction significant SNP, we employed the previously used SysFACE database (Systems Tool for Craniofacial Expression-Based Gene Discovery; Liu et al. 2017) to examine the craniofacial expression of genes present in the interval flanking the SNP. The candidate genes in the interval were retrieved flanking 5 Mb upstream and downstream of the SNP with the UCSC browser (human assembly GRCh38/hg38), and the expression and enrichment of their mouse orthologs were evaluated with whole genome transcriptomics-based microarray data sets on mouse craniofacial tissues (maxilla, frontonasal, and palate) at the embryonic and postnatal stages. The gene-level expression profiles were generated with an approach that successfully used transcriptomics data from the FaceBase and NCBI Gene Expression Omnibus repositories as previously described (Liu et al. 2017; PMID: 29805042). Mouse tissue data sets deposited in FaceBase (FB00000106, FB00000348, FB00000349, FB00000350, FB00000351, FB00000352, FB00000353, FB00000107, FB00000254, FB00000264, FB00000468.01, FB00000474.01, FB00000477.01, FB00000905) and NCBI Gene Expression Omnibus (GSE7759, GSE55965, GSE22989, GSE31004, GSE11400) were analyzed, and the expression of the candidate genes was determined. Tissue-enriched expression has been shown to be a good indicator of the potential function in development/homeostasis of the concerned tissue (Anand and Lachke 2017). The data for craniofacial tissue expression (expressed in fluorescence units obtained from microarrays) are presented in a heat map created with in-house python script.
Results
Identification of Genome-wide Significant GxSex Interaction SNPs
Of the 603,101 SNPs examined with the additive model, 93 SNPs had a suggestive significant p value (1.00 × 10−5) or lesser in the joint test and GxSex interaction analysis (Appendix Table 2). Among these SNPs, only rs2720555 was significant in the joint test and GxSex interaction test (p2DF = 1.16E-08, pGxSex = 1.49E-09, OR = 0.44, 95% CI = 0.34 to 0.57; Fig. 1A). The sex-specific allelic distribution of the reference and alternate alleles of this SNP (rs2720555) in cases and controls is presented in Appendix Table 3. This rs2720555 is an upstream transcript variant on chromosome 8p22 (i.e., a variant within the intergenic region of this chromosome; Fig. 1B). Our analysis of the odds of nsOFCs indicated that for an additional minor allele at this locus, the odds of cleft in males is significantly reduced (p < 0.0001, OR = 0.60, 95% CI = 0.48 to 0.74), while the effect is opposite for females (p = 0.0004, OR = 1.36, 95% CI = 1.15 to 1.60; Fig. 1C).
Figure 1.
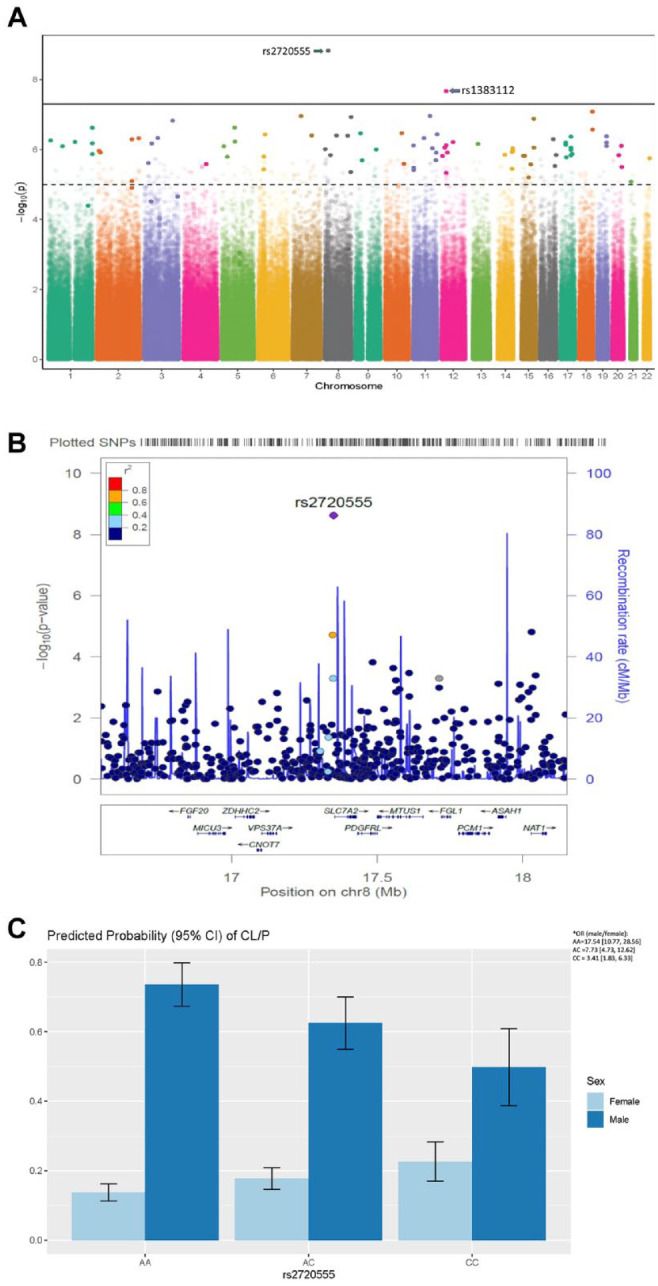
Discovery of novel risk locus through genome-wide GxSex interaction analysis in nsOFCs cohorts. (A) Manhattan plot shows the association between the single-nucleotide polymorphisms (SNPs) in the different chromosomes and the –log10(P). The broken line is the threshold P value (1 × 10–5) below which SNPs are considered to be suggestive, while the bold line is the threshold P value (5 × 10–8) below which SNPs are considered to be genome-wide significant. The SNPs plotted in faded colors are not suggestive or significant (P > 1 × 10–5) in the joint test analysis, while those SNPs in bold are at least suggestive (P < 1 × 10–5) in the joint test analysis. The SNP rs2720555 is the most significant with P values of p2df = 1.16E-08 and pGxSex = 1.49E-09 (odds ratio [OR] = 0.44, 95% CI = 0.34 to 0.57). (B) Locus zoom plot illustrates the genome-wide gene × sex interaction significant SNP (rs2720555) and surrounding SNPs with their associated genes. In addition to the parameters shown on the Manhattan plot, this plot presents the recombination rates at different loci. (C) The 95% CI of the predicted probabilities (which estimate the odds) of nonsyndromic cleft lip with or without cleft palate for an additional alternate allele. For every additional minor allele, the odds of nonsyndromic orofacial clefts in males decrease (P < 0.0001, OR = 0.60, 95% CI = 0.48 to 0.74) while the effect is opposite for females (P = 0.0004, OR = 1.36, 95% CI = 1.15 to 1.60). *The odds of nonsyndromic cleft lip with or without cleft palate for an additional minor allele in males versus females is also reported. As expected, this OR (male/female) decreased from 17.54 for alleles AA to 7.73 when there is a copy of the minor allele (AC); this reduced further to 3.41 for a homozygous minor allele (CC).
Replication Result
A case-control analysis was conducted with PLINK to analyze the sex-specific association between the genome-wide GxSex interaction significant SNPs from our discovery sample and nsOFCs in our replication cohort. We found that the 8p22 locus (SNP rs2720555) significantly increases the risk of nsOFCs in females (p = 0.037, OR = 1.30, 95% CI = 1.02 to 1.65), thus replicating the discovery signal. This association was more significant in the nonsyndromic cleft lip-only (nsCLO) subtype (p = 0.018, OR = 1.62, 95% CI = 1.09 to 2.40). In the male cluster, the 8p22 locus was not significantly associated with nsOFCs (P = 0.29, OR = 0.86, 95% CI = 0.65 to 1.14) or with any cleft subtype (Appendix Table 4).
Genome-wide Significant GxSex Interaction SNP in the TAD with Genes Involved in Craniofacial Development
The TAD defines the interaction between an SNP in a potential enhancer region and a promoter of a nearby gene. All genes within the TAD as the GWAS significant SNP (rs2720555) are potential candidate genes. Therefore, the TAD helps to narrow the search for potential candidate genes. To determine which genes are likely candidates based on their expression in relevant craniofacial structures, we utilized the SysFACE bioinformatics tool.
The GWAS significant SNP (rs2720555) is not within a gene (not an intronic or exonic variant). It is an upstream variant, with the nearest gene located about 3 kb downstream. There are a number of surrounding genes at 5 Mb upstream and downstream of the locus. These surrounding genes are potential candidate genes based on the hypothesis that this locus is within a regulatory region (enhancer) that regulates the expression of surrounding genes within the same TAD (Fig. 2).
Figure 2.
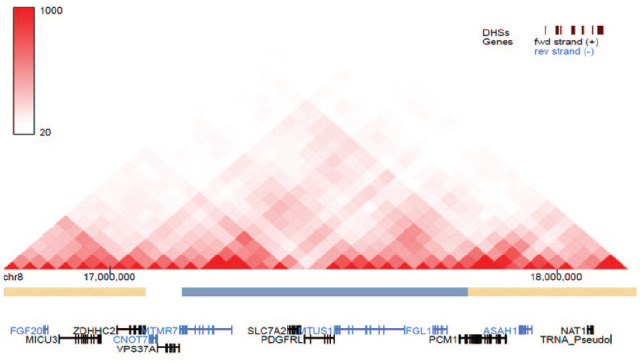
Genes predicted to be in the same topologically associating domain as the genome-wide significant single-nucleotide polymorphism (rs2720555). The single-nucleotide polymorphism is predicted to affect the enhancer-promoter association of the genes listed here. Among these genes are those that are robustly expressed and enriched in different craniofacial tissues during embryonic development.
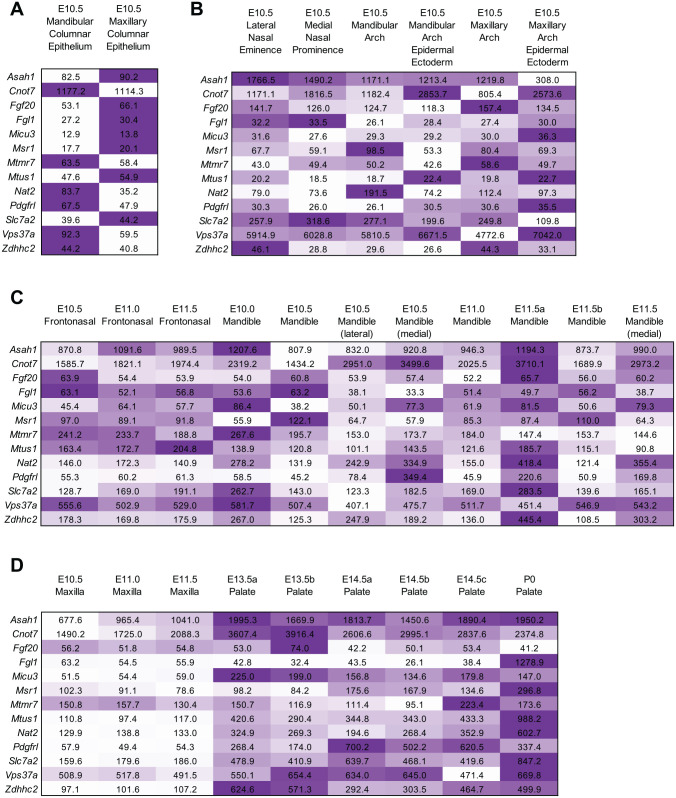
Among the genes that are in the TAD with the genome-wide GxSex Interaction SNP rs2720555 (Fig. 2), the SysFACE bioinformatics resource tool identified those genes that showed high expression in several mouse craniofacial tissues (frontonasal, maxilla, and palate) at developmental stages prior to lip and palate fusion (i.e., prior to embryonic day 12.0 [E12.0]; Fig. 3). This analysis effectively provides the expression of individual candidate genes in maxillary tissue as compared with other tissues. For example, this analysis reveals that Fgf20 exhibits the highest expression in E10.5 maxillary columnar epithelium as compared with other tissues, including mandibular columnar epithelium (Fig. 3A), similar to previously characterized CL/P-associated genes. Furthermore, this analysis indicates that Fgf20 expression is higher in E10.5 maxillary arch and E10.5 maxillary arch epidermal ectoderm as compared with E10.5 mandibular arch and E10.5 mandibular arch epidermal ectoderm (Fig. 3B). Additionally, Fgf20 has the highest expression in E13.5 palate tissue as compared with other tissues at earlier stages (Fig. 3C, D). This analysis notes that some of the other candidates (e.g., Vps37a, Cnot7) are ubiquitously expressed at similar levels in the tissues analyzed. Interestingly, newborn palate exhibits high expression of Fgl1.
Figure 3.
SysFACE analysis of gene expression in mouse craniofacial tissue development. Expression of candidate genes per analysis on (A) FaceBase microarray data sets on platform Affymetrix Mouse Gene 1.0 ST Array, (B) GSE55965 microarray data sets on platform Affymetrix Mouse Gene 1.0 ST Array, and (C, D) GSE7759 microarray data sets on platform Affymetrix Mouse Genome 430 2.0 Array. Heat map is based on expression of individual gene in different tissues (row-wise comparison) per analysis of microarray data sets from different studies. Embryonic (E) and postnatal (P) stages are indicated. Expression of the gene is represented by the intensity of the color. The number indicates the average of the fluorescence signal intensity for the specific gene as determined by microarray meta-analysis.
Whole Exome Sequencing Identified Rare Coding Variants in Potential Candidate Genes
In a bid to identify potentially pathogenic variants in those genes flanking the genome-wide significant GxSex interaction locus that may explain the heritability of nsCL/P, we examined the whole exome sequencing data of nsOFC cohorts available through collaboration. Note that these were from a Brazilian population with nsCL/P and were sequenced at the Baylor-Hopkins Center for Mendelian Genetics. Following this analysis, nonsynonymous coding variants were identified in some of the craniofacially expressed and enriched genes flanking the genome-wide significant locus. These variants are rare, with MAF < 0.01 (Table).
Table.
Variants Identified From Whole Exome Sequencing of Brazilian Cohorts with Nonsyndromic Cleft Lip with or without Cleft Palate.
| Allele | Score | ||||||
|---|---|---|---|---|---|---|---|
| Position a | Reference | Alternate | Gene | HGVS.c | HGVS.p | Sift | CADD |
| 17,400,978 | G | A | SLC7A2 | c.G130A | p.V44I | 0 | 23.9 |
| 17,402,104 | T | C | SLC7A2 | c.T641C | p.L214S | 0 | 29.4 |
| 17,419,606 | A | C | SLC7A2 | c.A1775C | p.K592T | 0.06 | 22.9 |
| 17,613,230 | G | C | MTUS1 | c.C87G | p.Y29X | 0.19 | 34 |
| 17,611,837 | C | T | MTUS1 | c.G1480A | p.V494I | 0 | 19.49 |
| 17,611,796 | G | C | MTUS1 | c.C1521G | p.F507L | 0 | 24.7 |
| 17,611,488 | G | A | MTUS1 | c.C1829T | p.S610L | 0.24 | 21.1 |
| 17,612,145 | T | A | MTUS1 | c.A1172T | p.H391L | 0.03 | 0.058 |
| 17,612,356 | C | T | MTUS1 | c.G961A | p.E321K | 0 | 15.24 |
| 17,542,024 | C | T | MTUS1 | c.G2489A | p.R830Q | 0 | 24.1 |
| 17,613,230 | G | C | MTUS1 | c.C87G | p.Y29X | 0.19 | 34 |
| 17,612,874 | C | T | MTUS1 | c.G443A | p.C148Y | 0.19 | 2.792 |
| 17,513,386 | G | C | MTUS1 | c.C2932G | p.Q978E | 0.07 | 21.9 |
| 17,731,991 | C | G | FGL1 | c.G284C | p.G95A | 0.07 | 23.3 |
| 18,258,279 | A | G | NAT2 | c.A766G | p.K256 | 0.02 | 22.3 |
| 18,257,583 | T | A | NAT2 | c.T70A | p.L24I | 0.02 | 23.0 |
| 18,258,196 | C | T | NAT2 | c.C683T | p.P228L | 0.005901 | 16.43 |
| 17,132,402 | G | A | VPS37A | c.G577A | p.A193T | 0.21 | 23.5 |
| 17,916,923 | C | T | ASAH1 | c.G1016A | p.R339H | 0.4 | 22.9 |
| 17,919,807 | A | G | ASAH1 | c.T677C | p.M226T | 0.04 | 22.8 |
| 17,924,803 | C | G | ASAH1 | c.G356C | p.G119A | 0.45 | 13.95 |
| 17,067,959 | C | G | ZDHHC2 | c.C920G | p.T307S | 0.76 | 16 |
| 17,055,890 | A | T | ZDHHC2 | c.A444T | p.K148N | 0.49 | 17.79 |
These variants are rare and among the top 10% and 1% most deleterious mutations in the human genome (as predicted by the CADD score).
CADD, combined annotation-dependent depletion; HGVS, Human Genome Variation Society; SIFT, sorting intolerant from tolerant. This table is available in color online.
Chromosome 8 for every position.
SIFT score interpretation: green, tolerated; red, deleterious.
Discussion
Based on the available evidence of GxSex interaction in the modification of the risk of nsCL/P (Marazita 2012; Carlson et al. 2018), we investigated how this affects the risk for cleft in our African cohort.
These individuals were part of the first African GWAS reported in 2019 where 2 novel genes, CTNNA2 and SULT2A1, were identified to contribute to the risk of cleft palate only (Butali et al. 2019). The present study identified a novel locus on chromosome 8p22 whose signal was driven strictly by the GxSex interaction. We found that this locus increases the risk of nsOFC in females, but the effect is reversed in males. We replicated the sex-specific effect in an independent female cohort. However, the signal in males could not be replicated. The SNP is in the TAD with a number of genes that show robust expressions in relevant structures during craniofacial morphogenesis in mice. Examination of the available whole exome sequencing data showed rare deleterious mutations in some of these genes in nsCL/P cases.
We characterized the genes in the TAD with this genome-wide significant locus and found several genes that show robust expressions in the craniofacial structures at critical time points during lip and palate development. These expressions were investigated at time points critical for the fusion of these processes in the formation of the lip and palate (Ray and Niswander 2012). The robust expression at these time points and in relevant craniofacial structures suggests that these genes are involved in the development of the lip and palate; thus, they are potential nsOFC candidate genes. Based on this finding, Fgf20 is a top candidate among the lists. Our data suggest that Asah1, Cnot7, and Vps37a play a role in craniofacial development and are potential candidates since they are robustly expressed, albeit ubiquitously, in the craniofacial structures prior to the fusion of the lip during embryogenesis.
FGF20 encodes fibroblast growth factor 20, which is one of the paracrine fibroblast growth factors and belongs to the FGF9 subfamily. These factors bind to fibroblast growth factor receptors: FGF20 binds to FGFR2 and FGFR4 to activate cellular processes important in embryonic development (Weng et al. 2018). Although no cleft phenotype has been associated with FGF20 in human or animal studies, it shows a similar expression pattern within developing craniofacial tissues with the receptor Fgfr2. Fgf20 and Fgfr2 have been detected in the palatal epithelium during embryonic development in mice (Lee et al. 2001). Further studies of Fgfr2 noted that Fgfr2fl/fl mice showed a cleft palate phenotype that was due to the disruption of proliferation in the epithelial cells of the palate (Hosokawa et al. 2009). This is additional evidence of the role of FGF20 in the etiopathogenesis of cleft. However, validation with in situ hybridization on sections of the face is needed to confirm that these genes (FGF20 and other genes reported here) are expressed in the fusing regions of the lip or palate.
Screening for rare variants in genes identified through GWASs helps to discover potentially pathogenic variants. Discovery of these pathogenic variants provides more evidence for the role that these genes play in the risk of nsOFCs. Thus, we screened for these variants in our OFC cohort and found some rare coding variants through whole exome sequencing analysis. Although no pathogenic FGF20 variants were revealed in our cohort, we found deleterious mutations in SLC7A2, MTUS1, NAT2, and ASAH1, which have been shown to be robustly expressed in craniofacial tissues during embryonic development.
The exploration of the sex heterogeneity in the risk of OFCs led to the discovery of a novel risk locus. This heterogeneity may also be due to factors such as candidate genes on the sex chromosomes (TBX22, EFNB1, and DMD) that increase the risk in males more than in females (Marçano et al. 2004; Jugessur et al. 2012; Patel et al. 2013; Skare et al. 2017). This sex difference can be due to the differential timing (between male and female embryos) of the critical stages in craniofacial development. The delay in timing of fusion of the palate in females has been reported to contribute to their increased risk for cleft palate (Burdi and Silvey 1969).
The inability to replicate the male-specific effect of the 8p22 locus is likely due to the small sample size and thus limited power to detect significant association in the independent sample. This may be an indication that this locus does not affect the risk of nsOFCs in males.
In conclusion, our study showed the power of using GxSex interaction in the analysis of complex traits. Despite the selection bias due to low surveillance of this trait in the African population, we carried out the first genome-wide GxSex interaction study and identified a new risk locus associated with OFC in an African population: a historically underrepresented population in genetics study. We have reported an 8p22 locus whose association with nsOFC is driven by GxSex interaction. Craniofacial expression of the genes around this locus during development and identification of potentially pathogenic variants of these genes in a cleft cohort provide strong support for our discovery. More functional studies are required to ascertain the roles played by these genes in development of nsOFCs.
Author Contributions
W. Awotoye, A. Butali, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; C. Comnick, C. Pendleton, E. Zeng, A. Alade, P.A. Mossey, L.J.J. Gowans, M.A. Eshete, W.L. Adeyemo, T. Naicker, C. Adeleke, T. Busch, M. Li, A. Petrin, J. Olotu, M. Hassan, J. Pape, S.E. Miller, P. Donkor, D. Anand, S.A. Lachke, M.L. Marazita, A.A. Adeyemo, J.C. Murray, D. Albokhari, N. Sobreira, contributed to conception, data acquisition, analysis, and interpretation, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, sj-pdf-1-jdr-10.1177_00220345211046614 for Genome-wide Gene-by-Sex Interaction Studies Identify Novel Nonsyndromic Orofacial Clefts Risk Locus by W. Awotoye, C. Comnick, C. Pendleton, E. Zeng, A. Alade, P.A. Mossey, L.J.J. Gowans, M.A. Eshete, W.L. Adeyemo, T. Naicker, C. Adeleke, T. Busch, M. Li, A. Petrin, J. Olotu, M. Hassan, J. Pape, S.E. Miller, P. Donkor, D. Anand, S.A. Lachke, M.L. Marazita, A.A. Adeyemo, J.C. Murray, D. Albokhari, N. Sobreira and A. Butali in Journal of Dental Research
Acknowledgments
We are grateful to the families who voluntarily participated in this study in Ethiopia, Nigeria, and Ghana. We are also grateful to all the administrative and research staff, students, nurses, and resident doctors who assisted with participant recruitment, consent, and data collection. We thank all members of the Butali Laboratory for their helpful comments and suggestions.
Footnotes
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by funds from the National Institutes of Health/National Institute of Dental and Craniofacial Research grants DE024296 to W. Awotoye, DE022378 and DE28300 to A. Butali, and DE024776 to S.A. Lachke.
ORCID iDs: W. Awotoye  https://orcid.org/0000-0002-5223-6846
https://orcid.org/0000-0002-5223-6846
A. Alade  https://orcid.org/0000-0001-9176-0221
https://orcid.org/0000-0001-9176-0221
J. C. Murray  https://orcid.org/0000-0002-8214-6083
https://orcid.org/0000-0002-8214-6083
A supplemental appendix to this article is available online.
References
- Anand D, Lachke SA. 2017. Systems biology of lens development: a paradigm for disease gene discovery in the eye. Exp Eye Res. 156:22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Marazita ML, Leslie EJ. 2016. Genetic factors influencing risk to orofacial clefts: today’s challenges and tomorrow’s opportunities. F1000Res. 5:2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, Liang KY, Wu T, Murray T, Fallin MD, et al. 2010. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet. 42(6):525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, Baluardo C, Ferrian M, de Assis NA, Alblas MA. 2009. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat Genet. 41(4):473–477. [DOI] [PubMed] [Google Scholar]
- Burdi AR, Silvey RG. 1969. Sexual differences in closure of the human palatal shelves. Cleft Palate J. 6:1–7. [PubMed] [Google Scholar]
- Butali A, Adeyemo WL, Mossey PA, Olasoji HO, Onah II, Adebola A, Efunkoya Akintububo A, James O, Adeosun OO, et al. 2014. Prevalence of orofacial clefts in Nigeria. Cleft Palate Craniofac J. 51(3):320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butali A, Mossey PA. 2009. Epidemiology of orofacial clefts in Africa: methodological challenges in ascertainment. Pan Afr Med J. 2:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butali A, Mossey PA, Adeyemo WL, Eshete MA, Gowans LJJ, Busch TD, Jain D, Yu W, Huan L, Laurie CA, et al. 2019. Genomic analyses in African populations identify novel risk loci for cleft palate. Hum Mol Genet. 28(6):1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JC, Nidey NL, Butali A, Buxo CJ, Christensen K, Deleyiannis FW, Hecht JT, Field LL, Moreno-Uribe LM, Orioli IM, et al. 2018. Genome-wide interaction studies identify sex-specific risk alleles for nonsyndromic orofacial clefts. Genet Epidemiol. 42(7):664–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SF, Wang K, Zhang H, Glaberson W, Annaiah K, Kim CE, Bradfield JP, Glessner JT, Thomas KA, Garris M. 2009. A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. J Pediatr. 155(6):909–913. [DOI] [PubMed] [Google Scholar]
- Hosokawa R, Deng X, Takamori K, Xu X, Urata M, Bringas P, Jr, Chai Y. 2009. Epithelial-specific requirement of FGFR2 signaling during tooth and palate development. J Exp Zool B Mol Dev Evol. 312B(4):343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugessur A, Skare Ø, Lie RT, Wilcox AJ, Christensen K, Christiansen L, Nguyen TT, Murray JC, Gjessing HK. 2012. X-linked genes and risk of orofacial clefts: evidence from two population-based studies in Scandinavia. PLoS One. 7(6):e39240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khramtsova EA, Davis LK, Stranger BE. 2019. The role of sex in the genomics of human complex traits. Nat Rev Genet. 20(3):173–190. [DOI] [PubMed] [Google Scholar]
- Kraft P, Yen YC, Stram DO, Morrison J, Gauderman WJ. 2007. Exploiting gene-environment interaction to detect genetic associations. Hum Hered. 63(2):111–119. [DOI] [PubMed] [Google Scholar]
- Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, Bhangale T, Boehm F, Caporaso NE, Cornelis MC, Edenberg HJ, et al. 2010. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol. 34(6):591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Crisera CA, Erfani S, Maldonado TS, Lee JJ, Alkasab SL, Longaker MT. 2001. Immunolocalization of fibroblast growth factor receptors 1 and 2 in mouse palate development. Plast Reconstr Surg. 107(7):1776–1784. [DOI] [PubMed] [Google Scholar]
- Leslie EJ, Liu H, Carlson JC, Shaffer JR, Feingold E, Wehby G, Laurie CA, Jain D, Laurie CC, Doheny KF, et al. 2016. A genome-wide association study of nonsyndromic cleft palate identifies an etiologic missense variant in GRHL3. Am J Hum Genet. 98(4):744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Busch T, Eliason S, Anand D, Bullard S, Gowans LJJ, Nidey N, Petrin A, Augustine-Akpan EA, Saadi I, et al. 2017. Exome sequencing provides additional evidence for the involvement of ARHGAP29 in Mendelian orofacial clefting and extends the phenotypic spectrum to isolated cleft palate. Birth Defects Res. 109(1):27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold E, Ludwig KU, Birnbaum S, Baluardo C, Ferrian M, Herms S, Reutter H, de Assis NA, Chawa TA, Mattheisen M, et al. 2010. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat Genet. 42(1):24–26. [DOI] [PubMed] [Google Scholar]
- Marazita ML. 2012. The evolution of human genetic studies of cleft lip and cleft palate. Annu Rev Genomics Hum Genet. 13:263–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçano AC, Doudney K, Braybrook C, Squires R, Patton MA, Lees MM, Richieri-Costa A, Lidral AC, Murray JC, Moore GE, et al. 2004. TBX22 mutations are a frequent cause of cleft palate. J Med Genet. 41(1):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossey PA, Little J, Munger RG, Dixon MJ, Shaw WC. 2009. Cleft lip and palate. Lancet. 374(9703):1773–1785. [DOI] [PubMed] [Google Scholar]
- Patel PJ, Beaty TH, Ruczinski I, Murray JC, Marazita ML, Munger RG, Hetmanski JB, Wu T, Murray T, Rose M, et al. 2013. X-linked markers in the Duchenne muscular dystrophy gene associated with oral clefts. Eur J Oral Sci. 121(2):63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Liu C, Sodhi M, Lu H. 2016. Meta-analysis of sex differences in gene expression in schizophrenia. BMC Syst Biol. 10 Suppl 1:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimov F, Jugessur A, Murray JC. 2012. Genetics of nonsyndromic orofacial clefts. Cleft Palate Craniofac J. 49(1):73–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray HJ, Niswander L. 2012. Mechanisms of tissue fusion during development. Development. 139(10):1701–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skare Ø, Gjessing HK, Gjerdevik M, Haaland ØA, Romanowska J, Lie RT, Jugessur A. 2017. A new approach to chromosome-wide analysis of X-linked markers identifies new associations in Asian and European case-parent triads of orofacial clefts. PLoS One. 12(9):e0183772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smarius B, Loozen C, Manten W, Bekker M, Pistorius L, Breugem C. 2017. Accurate diagnosis of prenatal cleft lip/palate by understanding the embryology. World J Methodol. 7(3):93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Huang Y, Yin A, Pan Y, Wang Y, Wang C, Du Y, Wang M, Lan F, Hu Z, et al. 2015. Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate. Nat Commun. 6:6414. [DOI] [PubMed] [Google Scholar]
- Waskar M, Landis GN, Shen J, Curtis C, Tozer K, Abdueva D, Skvortsov D, Tavaré S, Tower J. 2009. Drosophila melanogaster p53 has developmental stage-specific and sex-specific effects on adult life span indicative of sexual antagonistic pleiotropy. Aging (Albany NY). 1(11):903–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M, Chen Z, Xiao Q, Li R, Chen Z. 2018. A review of FGF signaling in palate development. Biomed Pharmacother. 103:240–247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jdr-10.1177_00220345211046614 for Genome-wide Gene-by-Sex Interaction Studies Identify Novel Nonsyndromic Orofacial Clefts Risk Locus by W. Awotoye, C. Comnick, C. Pendleton, E. Zeng, A. Alade, P.A. Mossey, L.J.J. Gowans, M.A. Eshete, W.L. Adeyemo, T. Naicker, C. Adeleke, T. Busch, M. Li, A. Petrin, J. Olotu, M. Hassan, J. Pape, S.E. Miller, P. Donkor, D. Anand, S.A. Lachke, M.L. Marazita, A.A. Adeyemo, J.C. Murray, D. Albokhari, N. Sobreira and A. Butali in Journal of Dental Research