Abstract
Objective.
Handcrafted radiomics features or deep learning model-generated automated features are commonly used to develop computer-aided diagnosis schemes of medical images. The objective of this study is to test the hypothesis that handcrafted and automated features contain complementary classification information and fusion of these two types of features can improve CAD performance.
Approach.
We retrospectively assembled a dataset involving 1,535 lesions (740 malignant and 795 benign). Regions of interest (ROI) surrounding suspicious lesions are extracted and two types of features are computed from each ROI. The first one includes 40 radiomic features and the second one includes automated features computed from a VGG16 network using a transfer learning method. A single channel ROI image is converted to three channel pseudo-ROI images by stacking the original image, a bilateral filtered image, and a histogram equalized image. Two VGG16 models using pseudo-ROIs and 3 stacked original ROIs without pre-processing are used to extract automated features. Five linear support vector machines (SVM) are built using the optimally selected feature vectors from the handcrafted features, two sets of VGG16 model-generated automated features, and the fusion of handcrafted and each set of automated features, respectively.
Main Results.
Using a 10-fold cross-validation, the fusion SVM using pseudo-ROIs yields the highest lesion classification performance with area under ROC curve (AUC=0.756±0.042), which is significantly higher than those yielded by other SVMs trained using handcrafted or automated features only (p<0.05).
Significance.
This study demonstrates that both handcrafted and automated futures contain useful information to classify breast lesions. Fusion of these two types of features can further increase CAD performance.
Keywords: Computer-aided diagnosis (CAD), deep transfer learning, convolutional neural network, handcrafted features, feature level fusion, mammography, classification of breast lesions
1. INTRODUCTION
Breast cancer has the highest incident rate and second highest mortality rate among cancers in women [1]. Routine mammographic screening is considered a widely used cost-effective approach to detect breast cancer in its earliest stages, which can help significantly improve cancer treatment efficacy and reduce patients’ mortality rate as demonstrated in many clinical studies [2, 3]. While mammography is the only accepted population-based breast cancer screening tool currently in clinical practice, mammograms are often difficult for radiologists to interpret due to the great heterogeneity of breast lesions and overlapped dense fibro-glandular tissues, which results in a high false positive recall rate. Among the suspicious breast lesions detected in mammograms and recommended for biopsy by radiologists, less than 30% of lesions are actually confirmed as malignant [4]. The high rate of benign biopsies is not only an economic burden, but also results in long-term psychosocial consequences to many women who participate in mammography screening [5]. Thus, improving the accuracy of classifying mammography-detected suspicious lesions to reduce the false-positive recall rate is a pressing clinical challenge.
One method to help improve breast lesion detection and classification, and the accuracy of radiologists is through the assistance of computer-aided detection and/or diagnosis (CAD) schemes. Typically, computer-aided detection schemes are developed and applied to detect and highlight locations of suspicious lesions depicting on mammograms, which may end up overlooked by radiologists, thus help increase lesion detection sensitivity (or reduce false negative rate) [6]. In addition, many other researchers have focused substantial efforts on development and clinical translation of computer-aided diagnosis schemes that aim to classify the suspicious lesions as malignant or benign. While the commercialized computer-aided detection schemes have been routinely used in clinical practice, no computer-aided diagnosis schemes have been used in clinical practice to date to help radiologists more accurately classify between malignant and benign lesions to reduce unnecessary benign biopsies. In this article, we only develop and discuss computer-aided diagnosis (CAD) schemes. All CAD schemes include machine learning classifiers trained using a set of optimal image features extracted using one of two approaches. The first approach, often referred to as traditional CAD, involves extraction of a set of handcrafted radiomics image features to train a machine learning classifier. However, previous research indicates that extraction and selection of a set of optimal handcrafted features varies drastically between studies and is a time intensive, error-prone, and non-trivial task which often leads to increased false positive rates [7–9].
In order to overcome challenges or limitations of the first approach, many researchers investigated a second approach that uses a deep learning model to automatically learn and extract features directly from the image itself, which significantly decreases or eliminates user intervention. The deep learning models applied to the medical images are primarily the deep convolution neural networks (CNN) due to their immense success in many tasks involving computer vision. CNNs differ from traditional artificial neural networks (ANNs) in that they use filters and convolutional operations to transfer all information from neurons in one layer to the neurons in the next hidden layers, which are called convolutional layers. The selected filters and convolutional layers are what make the CNN a powerful tool as it enables it to detect, learn, and recognize different image features or patterns. Specifically, a CNN transforms the input image with a designated number of filters of a set size to produce an output feature map. This feature map will then go through an activation function and act as the input to the next layer. Without activation functions these networks would only be capable of linear feature mapping which would make it nearly impossible to learn features of complex non-linear distributions [10, 11]. Subsampling layers often follow these convolutional layers. The purpose of these layers is to down sample the feature maps to decrease the number of parameters that the network needs to learn and also increases model robustness [12]. This is often done through a max pooling operation, which will take the maximum value from each patch of a designated size. Stacking multiple blocks of convolutional layers followed by a pooling layer is what makes this a deep network. The repetitive layer structure allows for the extraction of increasingly meaningful information while preserving spatial information. After an image goes through multiple blocks of convolution and pooling, it is fed to one or multiple fully connected layers, which capture the relationships within the final feature maps and output a class label.
While deep CNNs have become an immensely powerful tool for many different image classification tasks, there are several limitations that hinder their applications to medical imaging tasks. Firstly, many of these deep learning algorithms are thought of as a black box. This is a key weakness and hurdle when trying to translate these technologies to the clinic. Visualization techniques have been proposed which give insight into the type of features extracted from each convolutional layer [13–15]; the goal of these techniques is to provide some level of explanation for their decision-making process. Second, training of these deep neural networks requires a very large dataset, which is often not available in medical imaging. Transfer learning has emerged as a solution to this problem. Transfer learning involves the transfer of knowledge from one task to another by using the model learned on one task for a separate task [16]. The theory is that if the original model is trained on a very large and diverse dataset, then the model will have a good sense of computer vision, therefore the features learned can be applied to other tasks [17]. Thus, several well established CNNs such as AlexNet, GoogLe-Net, ResNet, VGG16, and others have been pre-trained on the ImageNet dataset and successfully used in a wide variety of computer vision tasks including detection, segmentation, and classification of medical images [18–20]. These pre-trained CNNs can be used as a feature extractor in which the top fully connected layers can be removed, and the output feature map can be flattened into a feature vector that can be used to train another separate machine learning classifier for different medical imaging application tasks [21].
Using these pre-trained CNNs leads to a third problem; there are many fundamental differences between the natural images in the ImageNet dataset used to train the CNNs and medical images [16]. The ImageNet images are natural color images with three channels (RGB), while mammograms are single channel greyscale images. Since the CNNs are trained on three channel RGB images they require this as an input. Stacking the grayscale mammogram into three channels is the most obvious solution, but this may provide redundant information to the network. While many studies have demonstrated potential or success in using transfer learning with the ImageNet dataset for classification of medical images [22–24], more work must be done to explore the role of image pre-processing and deep transfer learning for breast lesion classification using mammograms.
Thus, although over the last decade great research efforts have been made to develop novel traditional and deep learning CAD schemes for detection and diagnosis of diseases [24–27], the existing CAD schemes have their unique characteristics including different advantages and limitations, which have not yet been fully investigated or compared in previous studies [28, 29]. In our research work, we hypothesize that the traditional handcrafted features and the deep learning model generated automated features contain complementary discriminatory information because some of the handcrafted features can closely mimic image features or markers used by radiologists in lesion diagnosis, while automated features have the potential to extract new clinically relevant features that may be invisible or difficult to detect by human eyes. Thus, optimal fusion of these two types of features has potential to increase CAD performance to classify breast lesions. To test this hypothesis, this work aims to develop and evaluate a new fusion CAD scheme with improved mammogram lesion classification performance by combining both handcrafted and automated image features. The rest of the paper is organized as follows. Section 2 describes the information of the image dataset and experimental design including all steps of the proposed fusion CAD scheme. Section 3 reports study results by comparing lesion classification performance of CAD schemes using several machine learning classifiers trained using different sets of features. Section 4 discusses the impact of this study along with limitations and future work. Section 5 concludes this study.
2. METHOD
2.1. Image Dataset
In our research laboratory, we have assembled a retrospective breast cancer screening image database of full-field digital mammograms (FFDM) under an institutional review board (IRB) approved image collection protocol. Each collected study case contains sequential FFDM images acquired in two to six annual screening sessions from 2008 to 2014. All FFDM images were acquired using the Hologic Selenia digital mammography machine (Hologic Inc., Bedford, MA, USA) with a fixed pixel size or spatial resolution of 70μm. Since in developing CAD schemes of mammograms, the high resolution FFDM images are used to detect microcalcifications and subsampled low-resolution images are used to detect soft tissue mass lesions, the original FFDM images are also subsampled using a pixel value averaging method with a 5×5 pixel frame to make image size of 818×666 pixels. Then, the subsampled FFDM image has a pixel size or spatial resolution of 0.35mm. The 12-bit gray level remains the same. From this image database, we have selected and assembled number of subsets of images for performing different CAD tasks including predicting cancer risk, detecting and classifying suspicious breast lesions as reported in our previous research papers[8, 30–33].
In this study, we collected 1,535 craniocaudal (CC) FFDM images from our existing image database. Each image contains a suspiciously soft tissue mass-based lesion that was previously detected by a radiologist during the original image reading and diagnosis in screening environment. All lesions were recommended for biopsy. Based on the pathology examination results of the biopsied lesion samples, 740 lesions were confirmed as malignant, while 795 lesions were confirmed as benign. In each image, the lesion center was marked by the radiologist and recorded in our database. Using the recorded lesion center as a center reference, a square patch or region of interest (ROI) with a size of 64×64 pixels is extracted from each image. Figure 1 shows two FFDM images and corresponding two extracted ROIs in which one depicts a malignant lesion and one depicts a benign lesion.
Figure 1:

A and B display two craniocaudal mammogram images including a malignant and benign lesion, respectively. Red boxes represent the 64×64 patch extracted around the suspicious lesion.
2.2. Image Preprocessing
In order to use deep transfer learning method, we first expand or rescale the original ROI with 64×64 pixels to 224×224 pixels using bilinear interpolation method. In order to generate three channel images suitable for deep transfer learning, we combine the original greyscale image (Io) with two preprocessed variations [34]. Since mammograms are low dose X-ray images, these images may have poor contrast and the brightness may vary greatly between patients [35]. Firstly, a histogram equalization technique is applied to the original greyscale image to normalize and enhance the contrast of the mammogram (IHE). Second, to denoise the mammogram images, we apply a bilateral low-pass filter to the original greyscale image (I0) and generate a new filtered image (IBF). This filter is selected because of its ability to reduce image noise, while effectively preserving edge and other textural information [36].
| (1) |
where and are two Gaussian functions with two different kernel sizes determined by two sigma values, σs and σr, respectively. Wp is a normalization factor:
| (2) |
Thus, in using the bilateral low-pass filter, the first Gaussian low-pass filter in the spatial domain ensures that only the pixels in the area around the central pixel are considered and blurred. The second Gaussian low-pass filter considers the difference in intensity between pixels, which decreases the influence of pixel blurring with the increase of intensity difference and allows for edge preservation as edge locations have large intensity variations. As a result, applying this bilateral filter to mammograms will ensure that only pixels with small intensity variations (i.e., relatively homogeneous breast tissue or internal tumor areas) are blurred to reduce image noise, while tumor edge and other textural information of tumor and surrounding fibro-glandular tissues are preserved. Based on a previous study, the diameter of the pixel neighborhood (Gaussian filter kernel) used in this study is set to 9 and both sigma values (σs and σr) are set to 75 [34]. Therefore, three images, Io, IHE, IBF are stacked to form a pseudo color image (Figure 2), which are fed to the CNN for automated feature extraction.
Figure 2:

Intermediate images in the creation of the pseudo-ROIs. Pseudo-ROI is created by stacking the three greyscale images.
2.3. Deep Transfer Learning Feature Selection
Although several deep learning models have been applied as feature extractors for transfer learning in the medical imaging field, we use a VGG16 network whose weights have been pre-trained on the ImageNet dataset [37] to test hypothesis of this study as this network has performed well in many previous studies [38–41]. The VGG16 network is comprised of 13 convolutional layers followed by two full connected layers and a SoftMax layer [42]. All convolutional layers use a 3×3 kernel and all max-pooling layers have a stride of 2 (Table 1). VGG16 takes a 224×224 3-channel RGB image as an input. For the purpose of this study, the top fully connected layers are removed and a 7×7×512 feature map is extracted after the final max pooling layer. This feature map is then flattened into a 25,088-dimensional feature vector, which can be used to train a machine learning classifier.
Table 1:
VGG-16 Architecture. For this study, block 6 is removed and features are extracted after the final max pooling layer.
| Block | Layer | Size | Filter Size |
|---|---|---|---|
|
| |||
| 1 | Convolution-1 | 224×224×64 | 3×3 |
| Convolution-2 | 224×224×64 | 3×3 | |
| Max pooling | 112×112×64 | - | |
|
| |||
| 2 | Convolution-1 | 112×112×128 | 3×3 |
| Convolution-2 | 112×112×128 | 3×3 | |
| Max pooling | 56×56×128 | - | |
|
| |||
| 3 | Convolution-1 | 56×56×256 | 3×3 |
| Convolution-2 | 56×56×256 | 3×3 | |
| Convolution-3 | 56×56×256 | 3×3 | |
| Max pooling | 28×28×256 | - | |
|
| |||
| 4 | Convolution-1 | 28×28×512 | 3×3 |
| Convolution-2 | 28×28×512 | 3×3 | |
| Convolution-3 | 28×28×512 | 3×3 | |
| Max pooling | 14×14×512 | - | |
|
| |||
| 5 | Convolution-1 | 14×14×512 | 3×3 |
| Convolution-2 | 14×14×512 | 3×3 | |
| Convolution-3 | 14×14×512 | 3×3 | |
| Max pooling | 7×7×512 | - | |
|
| |||
| 6 | Flatten | 25,088 | |
| Dense | 4,096 | ||
| Dense | 4,096 | ||
| Dense | 1,000 | ||
Due to the extremely high dimensionality of this feature vector, a three-step feature reduction pipeline is used to select the optimal feature set. Since the VGG16 network is pre-trained on the ImageNet dataset that comprises a very heterogenous set of natural images, yet the intensity distributions of mammograms are relatively homogenous, a large percentage of the neurons will not be activated when a ROI of mammogram passes through. This would result in many features being inactive (zero) for most images. Thus, in the first step, all features with a variance of 0.01 or less are eliminated.
The second step in the feature selection pipeline takes advantage of a quick and powerful relief-based algorithm, Relief-F, to rank feature importance based on how well that feature does at differentiating between instances that are nearby [43]. It relies on a nearest neighbor approach to do so. Briefly, given a randomly selected instance Ri, represented by an a-dimensional vector, where a is the total number of features, Relief-F searches for the k nearest neighbors from the same class, near hits Hj, and k nearest neighbors from a different class, near misses Mj [44]. Feature weights, W[A], are then updated according to equation 3:
| (3) |
where n is the totally number of training instances. The diff function (equation 4) is used to quantify the difference between the attribute at two nearby instances.
| (4) |
where I is either a nearby hit, Hj, or a nearby miss, Mj. Larger feature weights reflect features that will be more relevant for distinguishing between classes therefore more desirable. The weight of feature A, W[A], will be increased if randomly selected instance Ri and nearby instance I belong to the different classes and the feature values are different. W[A] will be decreased if randomly selected instance Ri and nearby instance I belong to the same class feature values are different. The update in the feature weight will be proportional to the difference between the feature values as seen in equation 4. While original proposals of relief based algorithms describe a relevance threshold value, τ, such that all features with W[A] > τ, will be selected as a relevant feature, it is often more practical to select a set number of features to be considered relevant [45, 46]. In this study, 10 neighbors were used, and the top 300 features were chosen to undergo further feature selection. A more in depth review of relief based algorithms can be found elsewhere [45]. The final step in the feature selection pipeline uses a sequential forward floating feature selector (SFFS) with 10-fold cross validation to select the final optimal feature set [47, 48].
As mentioned previously, VGG16 pretrained on ImageNet takes a 3-channel image as an input. Two methods can be used to convert one-channel grey level image to a three-channel image. Therefore, in addition to extracting features from the pseudo-color-ROI images described in section 3.2, we also extracted features from a second group of input images, namely, stacked-ROI images. The stacked-ROI images are created by stacking the same original greyscale ROI in three channels. Therefore, after using the VGG16 network as a feature extractor, two independent optimal vectors of automated features are created, namely, a pseudo-ROI feature vector and a stacked-ROI feature vector.
2.4. Handcrafted Feature Extraction and Selection
There exists an abundance of CAD schemes which use a wide variety of handcrafted features. We initially computed 40 commonly used features from two separate feature groups. The first group consists of the first order statistical features that describe the distribution of pixel intensities across the image. These include 6 features namely, the mean, maximum, standard deviation, energy, skewness, and kurtosis of pixel intensity values. While first order statistical features provide information about the intensity distribution of the image, they do not provide any insight into the relative spatial positions of these intensities. The second group consists of textural features which describe the spatial arrangement of the intensity distributions. These textural features include those derived from the gray level co-occurrence matrix (GLCM) and the gray level run-length matrix (GLRLM).
The GLCM describes the number of co-occurrences of two pairs of grey level intensities which are a specific distance apart [49]. From the GLCM, 6 features are computed along four angles, 0°, 45°, 90°, and 135°, and at a distance of one pixel, namely, contrast, dissimilarity, homogeneity, ASM, energy, and correlation. The mean and maximum values of these features are computed resulting in 12 GLCM features. The GLRLM describes the number of consecutive pixels that have a specific pixel intensity [50]. From the GLRLM, 11 features are computed along the same four angles, 0°, 45°, 90°, and 135°, namely: short run emphasis, long run emphasis, gray level nonuniformity, run length nonuniformity, run percentage, low gray level run emphasis, high gray level run emphasis, short run low gray level emphasis, short run high gray level emphasis, long run low gray level emphasis, and long run high gray level emphasis. The mean and maximum of each of these features are computed resulting in 22 GLRLM features. Mathematical descriptions of these features are explained in detail elsewhere [9]. After these 40 features were initially extracted, a variance threshold of 0.01 is applied to remove irrelevant features (Figure 3). As a result, 17 features are selected to create an optimal vector of handcrafted features.
Figure 3:

Heatmaps of correlation coefficients of the handcrafted features before and after applying variance thresholding.
2.5. Classification and Evaluation
Five separate machine learning classifiers are built using 5 optimal feature vectors extracted from handcrafted features, two sets of automated deep transfer learning features computed from pseudo-color-ROIs and stacked-ROIs, and fusion between handcrafted features and each set of automated features to test the hypothesis that fusion of handcrafted and deep transfer learning features can improve the performance of using CAD schemes to classify breast lesions as malignant or benign. Although many different types of machine learning classifiers have been used and tested in CAD field, we choose to use a support vector machine (SVM) as SVMs have many advantages as demonstrated in traditional machine learning or CAD field including its higher generalizability.
In this study, SVM1 is trained using only the handcrafted features, SVM2 is trained using the deep transfer learning features, which includes two SVM2s namely, SVM2-pseudo and SVM2-stacked, and SVM3 is trained using a fused feature set containing both the handcrafted and one set of deep transfer learning features, which also includes two SVM3s namely, SVM3-pseudo and SVM3-stacked. To build SVM3, handcrafted features and one set of deep transfer learning features are first combined through concatenation to create a new fusion feature pool. A SFFS algorithm is then used to select the optimal feature set from the fusion feature pool to train SVM3. All five SVMs are built using a linear kernel and trained using a 10-fold cross validation method. L2 regularization with C=1.0 is used to avoid overfitting.
Each trained SVM model is applied to every image in the testing fold of the dataset and generate a prediction score between 0 and 1 to indicate the likelihood of the image depicting a malignant lesion. Prediction scores of all 1,535 images are used to create a receiver operating characteristic (ROC) curve and the area under the ROC curve (AUC) is computed as an evaluation metric. Next, an operating threshold (T= 0.5) is applied on all SVM generated prediction scores to divide all testing images into two classes of malignant and benign lesions, so that the classification accuracy, inducing sensitivity and specificity, can be computed from a confusion matrix. In addition, the statistically significant differences of performance comparison are also computed and determined based on the criterion of p<0.05. A flowchart of this entire experimental design is shown in Figure 4.
Figure 4:
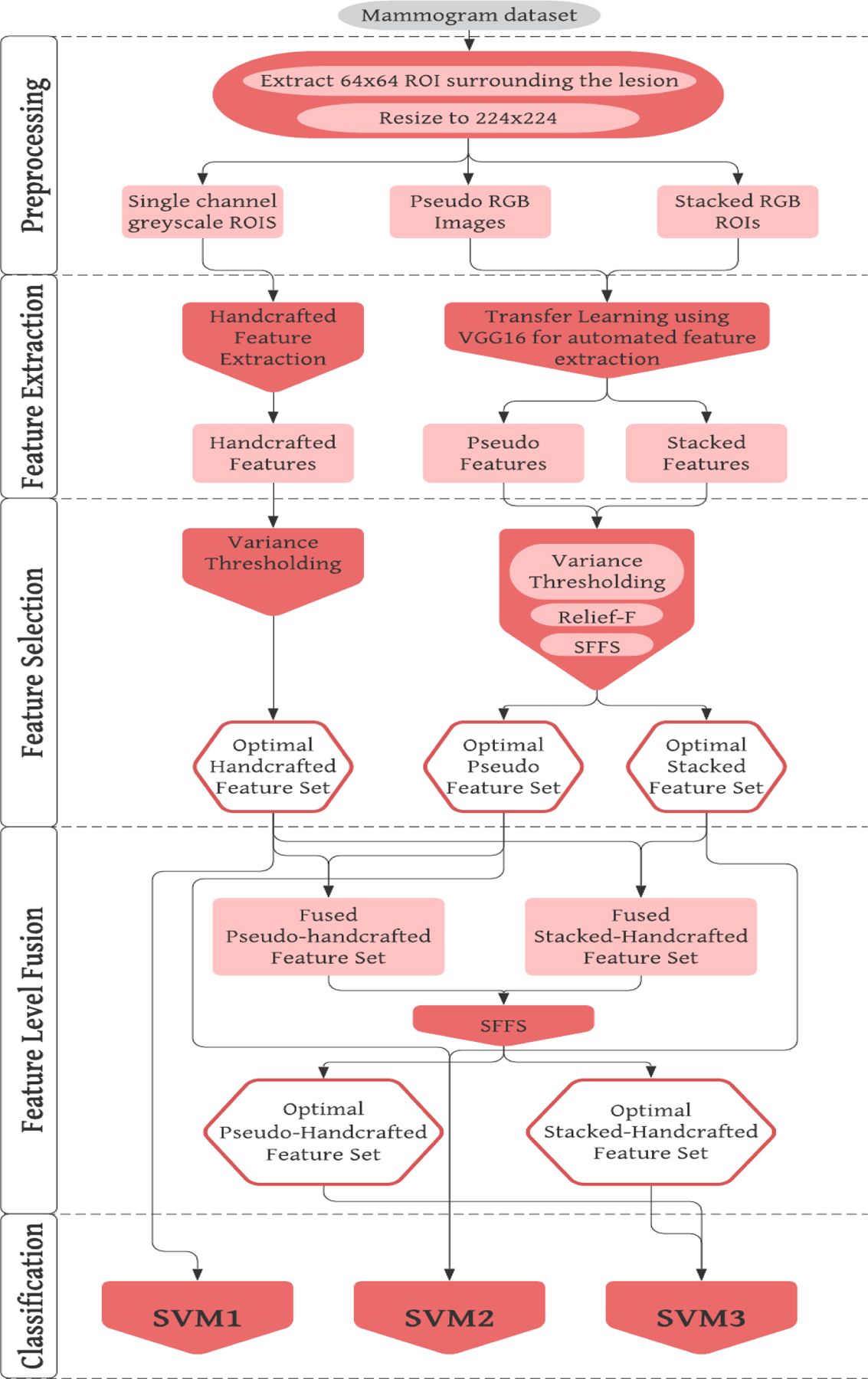
Flowchart of the entire experimental design.
3. RESULTS
Table 2 shows the results of the feature selection pipeline used to reduce the pseudo and stacked ROI feature sets. After variance thresholding about 75% of features were removed from the pseudo-ROI feature set and 70% were removed from the stacked-ROI feature set. This may support the idea that stacking the single channel greyscale image into a 3-channel RGB image provides extra redundant or irrelevant information. In addition, when applying a SFFS algorithm to select optimal features from two fusion pools of features, which include 17 handcrafted features plus 57 or 55 automated features (as shown in Table 2), two optimal fusion feature vectors including 61 and 37 features are generated. These two feature vectors include 9 and 6 handcrafted features, respectively.
Table 2:
Number of the feature selected in each feature selection pipeline.
| Feature Selection Step | Pseudo ROI | Stacked ROI | Handcrafted + Pseudo ROI | Handcrafted + Stacked ROI |
|---|---|---|---|---|
| Initial number of features | 25,088 | 25,088 | 74 | 72 |
| Variance Thresholding | 6,256 | 7,502 | - | - |
| Relief-F | 300 | 300 | - | - |
| SFFS | 57 | 55 | 61 | 37 |
Figure 5 shows 5 ROC curves generated from classification scores of 5 SVMs along with corresponding AUC values. When applying an operation threshold (T = 0.5), each ROI with an SVM-generated classification score ≥ T is classified as a malignant lesion, otherwise, it is classified as a benign lesion. Figure 6 shows 5 confusion matrices generated by 5 SVMs. These confusion matrices represent the sum after 10-fold cross validation. Based on the data shown in Figures 5 and 6, the mean AUC values (along with corresponding standard deviation of 10-fold cross-validation), classification accuracy, sensitivity, and specificity of all five SVMs are summarized in table 3.
Figure 5:

ROC AUC curves for all 5 SVMs.
Figure 6:
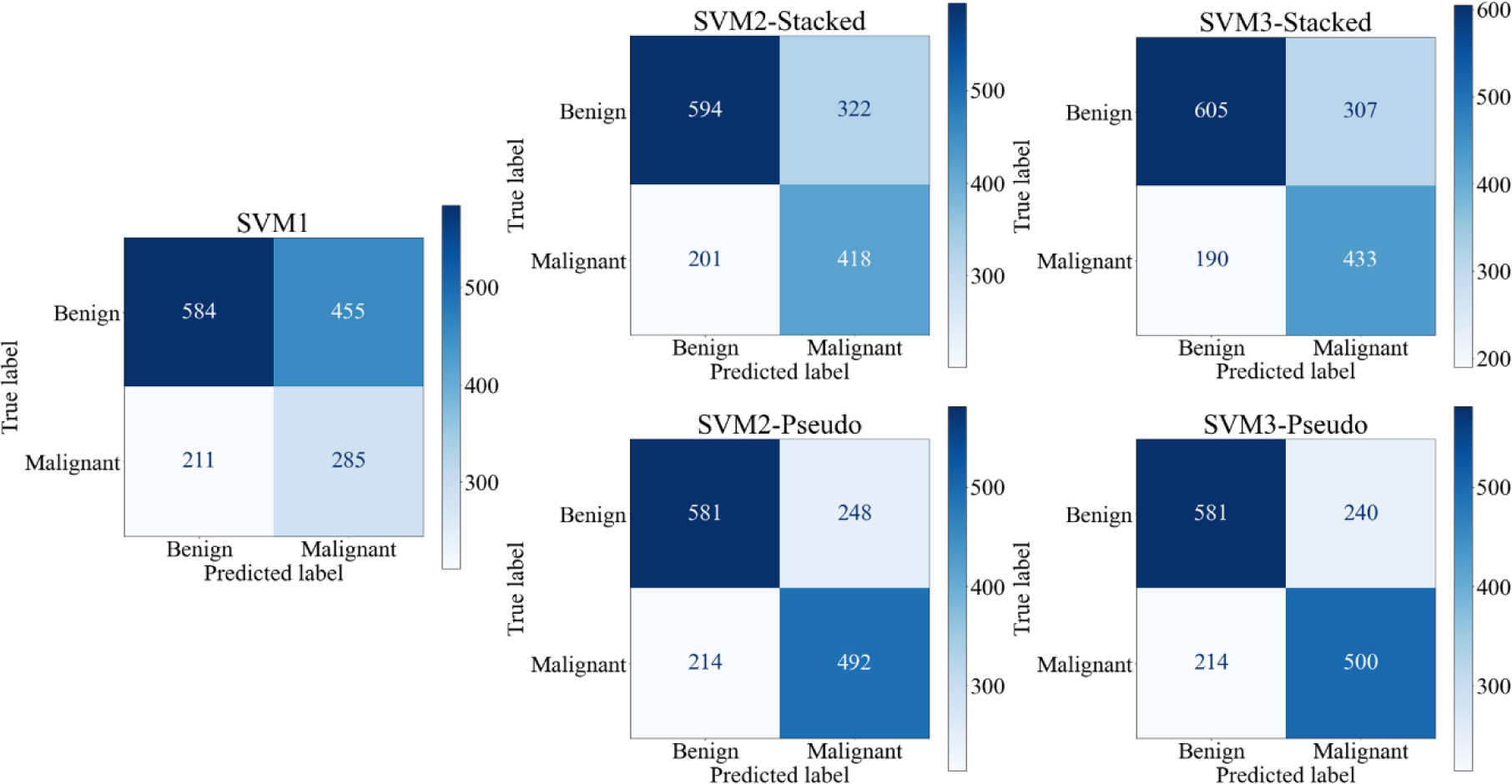
Confusion Matrices of all 5 SVMS. Confusion matrices are the sum of all matrices after 10-fold cross validation
Table 3:
Summary of classification performance indices including mean values and standard deviations of all 5 SVM models after 10-fold cross validation.
| Learning Model | AUC | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|
| SVM1 | 0.596±0.032 | 0.566±0.033 | 0.385±0.055 | 0.735±0.044 |
| SVM2-Stacked | 0.717±0.022 | 0.659±0.030 | 0.565±0.052 | 0.747±0.037 |
| SVM3-Stacked | 0.734±0.017 | 0.676±0.041 | 0.585±0.058 | 0.761±0.032 |
| SVM2-Pseudo | 0.750±0.043 | 0.699±0.036 | 0.665±0.064 | 0.731±0.028 |
| SVM3-Pseudo | 0.756±0.042 | 0.704±0.035 | 0.676±0.061 | 0.731±0.028 |
A paired t-test at an alpha value of 0.05 was used to test for a statistically significant difference in means between classification performance of groups of two SVMs. When analyzing the three SVMs developed using features extracted from the stacked ROIs, we observe that SVM3-stacked, trained using a fused feature vector, performs significantly better than both SVM1, trained using handcrafted features, and SVM2-stacked, trained using automated features, in both AUC and accuracy (Figure 7A and B). When analyzing the three SVMs developed using features extracted from the pseudo-ROIs, we observe that the AUC value yielded by SVM3-psuedo is also significantly higher than AUC values yielded by both SVM1 and SVM2-psuedo (Figure 7C). While the accuracy of SVM3-psuedo is greater than that of SVM2-pseudo, this difference is not significant (p=0.1363) (Figure 7D). Since both feature fusion based SVMs performed better than both SVMs trained using single type of features, the study results validate and support our hypothesis that feature fusion by optimally selecting handcrafted and automated features can create a machine learning classifier with improved classification abilities.
Figure 7:
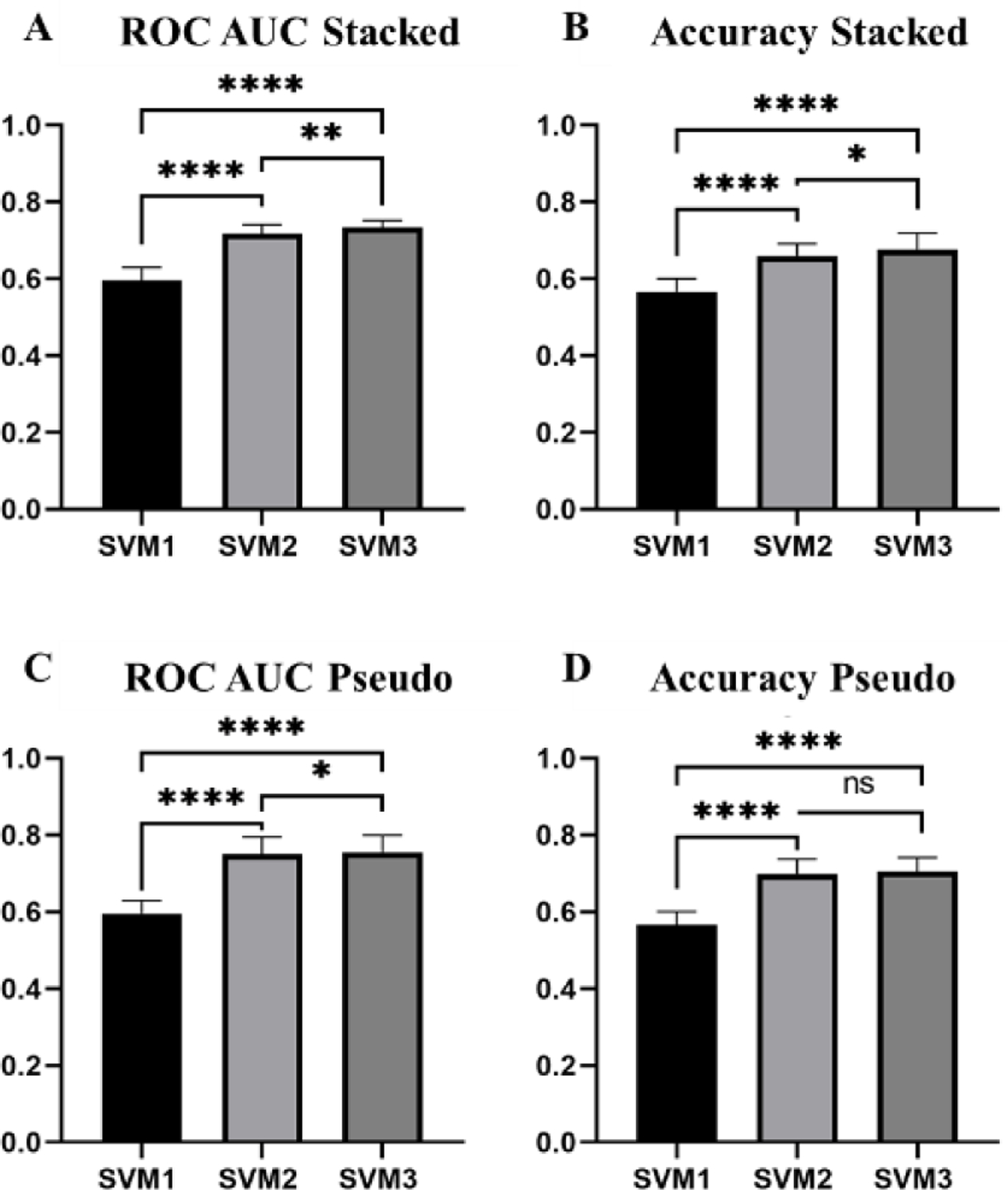
Bar graphs displaying the mean and standard deviation (STD) of all three SVMs for the pseudo-ROIs and stacked ROIs. (**** = p <0.001, **=p<0.01, *=p<0.05)
In addition to develop and evaluate a feature fusion-based SVM classifier for improved performance, we also compared the performance of the SVMs trained using pseudo-ROIs as an input to VGG16 for feature extraction, with the performance of SVM2s trained using stacked-ROIs as an input to VGG16 for feature extraction. While there is only a statistically significant difference between the accuracies of SVM2-stacked and SVM2-psuedo (p=0.0251), SVMs trained using features extracted from pseudo-ROIs achieve a higher AUC and accuracy than SVMs trained using features extracted from stacked-ROIs (Figure 8).
Figure 8:
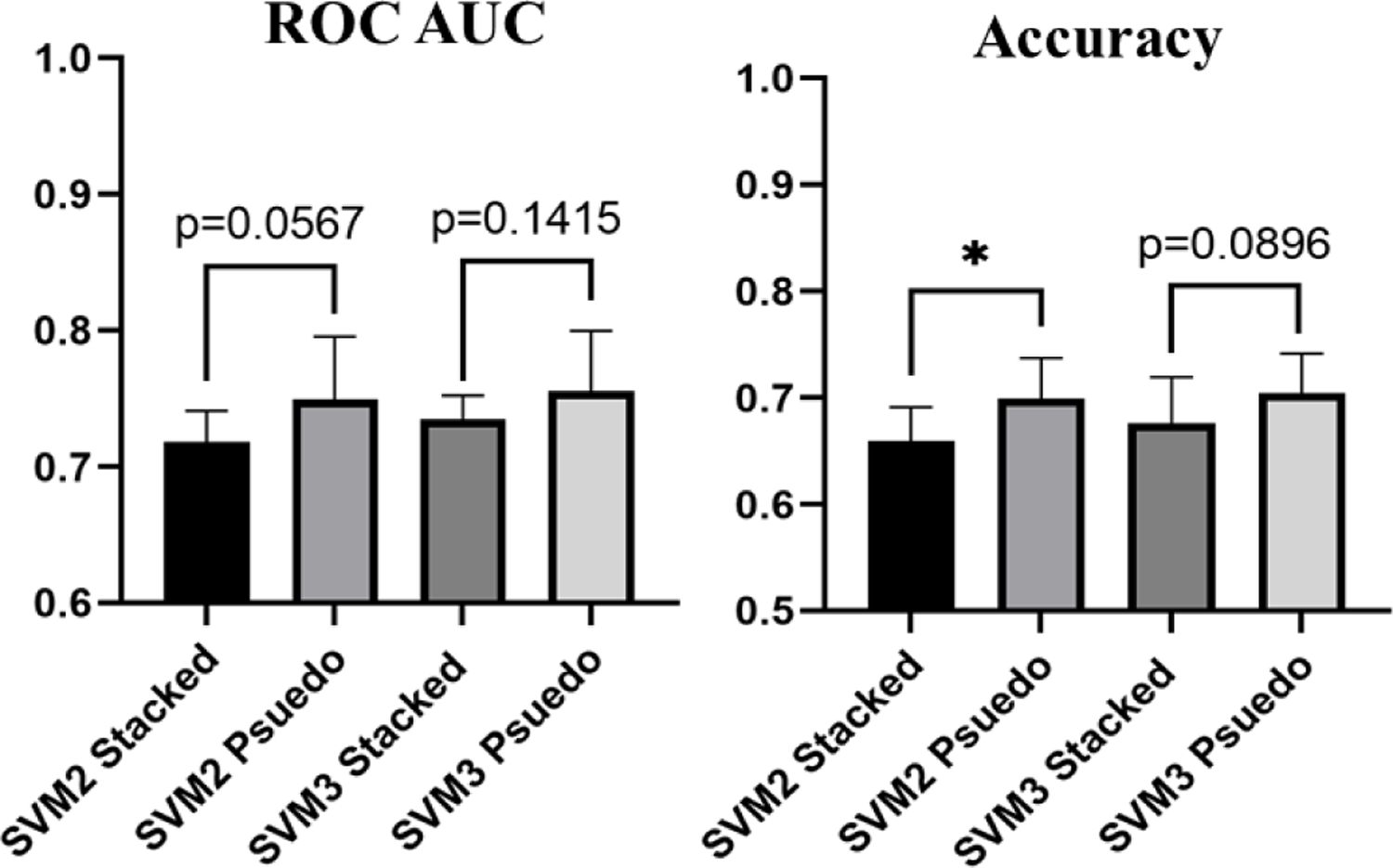
Bar graphs displaying the difference in ROC AUC and Accuracy of SVMs trained using deep transfer learning features extracted based on pseudo-ROIs and stacked-ROIs (*=p<0.05)
In addition, Figure 9 shows 3 blocks or pairs of lesion classification examples. Each pair includes one malignant lesion (marked by Red ROI frame) and one benign lesion (marked by Yellow ROI frame) in the top row. The magnified images of the extracted ROI are shown in the bottom row of the figure. First, the two lesions in the left block are correctly classified by 3 SVMs (SVM1, SVM2-pseudo, and SVM3-pseudo). In this pair of 2 lesions, we can see from the ROIs that the benign mass has a roundish shape and looks relatively uniform when compared to the background, while the malignant lesion appears with spiculated margins and is much brighter than the surrounding tissue. Second, two lesions in the middle block are correctly classified by 2 SVMs (SVM2-pseudo and SVM3-pseudo), but incorrectly classified by SVM1. Since SVM1 is trained on handcrafted features only, this means that using only handcrafted features is not sufficient to make a correct distinction but automated features alone and the combination of the handcrafted and automated features can be more accurate. Third, two lesions in right block may be more subtle and are only correctly classified by one SVM (SVM3-pseudo) and misclassified by SVM1 and SVM2-pseudo. This highlights the fact that handcrafted and automated features do contain complementary information that when used together can better classify suspicious lesions.
Figure 9:
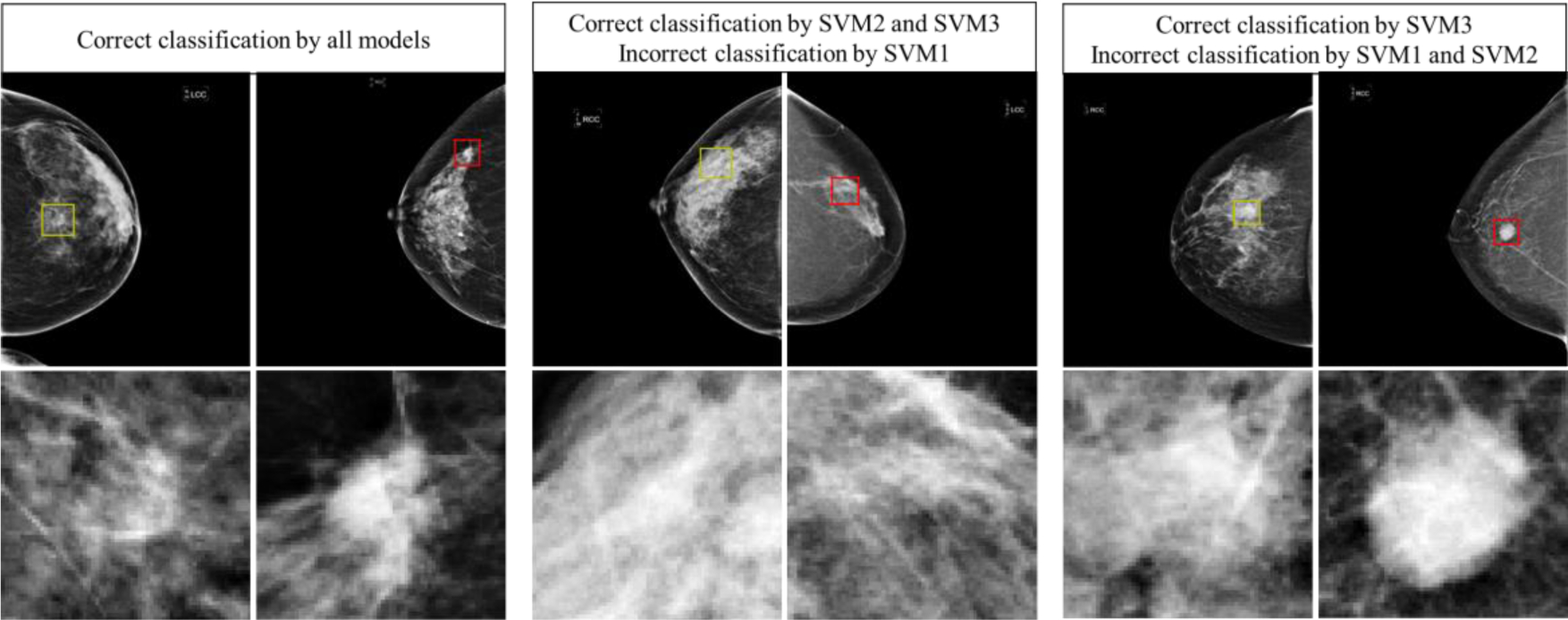
Examples of correct and failed classifications. The top row displays the full CC image while the bottom row shows the ROI that was used for feature extraction. Yellow ROI indicates that the true value is benign while a red ROI indicates that the true value is malignant.
In summary, when using only automated deep learning features with pseudo-ROI input images, the AUC value and classification accuracy of SVM2-pseudo increase 25.8% (from 0.596 to 0.750) and 21.9% (from 0.566 to 0.690), respectively, as compared to SVM1 trained using handcrafted features only. Additionally, when fusion of handcrafted and automated features, AUC value and classification accuracy of SVM3-pseudo are further increased by 0.8% (from 0.750 to 0.756) and 2.1% (from 0.690 to 0.704), respectively, as comparing to SVM2-pseudo.
4. DISCUSSION
This work demonstrates a new CAD scheme for the classification of breast lesions as malignant or benign. This study uses a diverse dataset of 1,535 cases, which is larger than most datasets used in previous CAD studies to classify breast lesions (such as 8 studies summarized in [51], which reported AUC values ranging from 0.70 to 0.87 using datasets involving 38 to 1,076 images). The reported performance of breast lesion classification in this study is not directly comparable to those reported by many previous studies due to the use of different image datasets. However, this new CAD scheme generates the promising performance as comparing to the high rates of false-positive recalls and unnecessary biopsies of benign lesions in current clinical practice. The contribution of this study includes following unique characteristics or research approaches and new interesting observations, which fully support our study hypothesis.
First, many deep learning CAD schemes or studies have been developed and reported in the literature. Previous approaches can be divided into three categories. (1) The studies use deep learning as an end-to-end classifier. For example, using a smaller dataset of 560 FFDM images (280 are malignant and 280 are benign), Qiu et al. developed and tested a deep learning model using a 4-fold cross validation method [52]. The study reported AUC values ranged from 0.696±0.044 to 0.836±0.036 in 4 folds (with average of AUC = 0.790±0.019). (2) The studies that use a deep transfer learning model as feature extractors. For example, Mendel at al. used a pretrained VGG19 model to extract features from 78 biopsy confirmed FFDM cases [53]. Then, by using the extracted features to train a linear SVM classifier using a reduced feature set, the study reported an AUC of 0.76±0.05. This study is somewhat like the SVM2 classifiers trained in our study, which yielded comparable AUC=0.75±0.04 (SVM2-pseudo) while it is tested using a much larger image dataset. (3) The studies fused two classifiers separately trained using handcrafted and automated features at the final decision level. For example, Huynh et al. applied a soft-voting technique to fuse the outputs of two SVMs trained using automated features extracted by an AlexNet model and an SVM trained using traditional CAD features [54]. When applying to 607 ROIs extracted from 219 FFDM cases and using a 5-fold cross-validation method, the study reported an AUC of 0.86±0.01. However, in this study, we investigate a new novel approach that fuse handcrafted and automated features at feature selection level to create an optimal feature set and train a single classifier (i.e., SVM3-pseudo or SVM3-stacked). To the best of our knowledge, such fusion method to develop CAD schemes of medical images has not been reported in the literature.
Second, since using deep transfer learning model as a feature extractor generates a very large feature vector (25,088 from VGG16 model), identifying a small set of optimal features is a difficult but important task. Our experiments indicated that many commonly used feature dimension reduction methods including principal component analysis (PCA) have lower performance when applying to such large feature vector. Thus, in this study, we developed a new feature selection pipeline with 3 steps allows for the successful selection of an optimal set of automated deep learning features from the huge number of initially extracted features. Among these 3 steps, we investigated and identified an optimal and effective approach to use Relief based algorithms, which are unique in that they do not assume independence among features as many other feature selection filter methods do. Relief-F was chosen for this study since we are unaware of what kind of feature interactions exist from the feature map extracted from VGG16. A limitation of Relief-F worth noting is that in an extremely large feature space the identification of a nearest neighbor becomes increasingly random, which leads to a decrease in performance [44, 55]. Iterative RBA such as Iterative Relief [56], Tuned Relief-F(TuRF) [57], VLSRelief-F [55], and more [58], have been developed which improve the performance in very large feature spaces. There is no consensus on what defines an extremely large feature set and when these iterative approaches perform better. We observed no significant difference in the performance of SVM2 or SVM3 when using Relief-F alone and using TuRF wrapped around Relief-F. As a result, the optimal Relief-F algorithm was used to reduce dimensionality of feature space by more than 95% (i.e., reducing the number of features from 6,256 to 300 when using pseudo-ROIs). Combining 3 steps in this feature selection pipeline, the number of features is reduced to 55 or 57 from original 25,088 (as shown in Table 2), which supports building robust SVMs using a large training dataset with 1,382 cases (9 folds of our dataset).
Third, this study supports that using deep transfer learning model generated features has significant advantages over using the traditional CAD handcrafted features since classification performance of SVM2 is significantly higher than SVM1 (i.e., AUC=0.750 for SVM2-pseudo and AUC=0.596 for SVM1). However, our study also demonstrates that the handcrafted features and automated features contain complementary information to classify breast lesions. Thus, using the fusion feature sets including both handcrafted and automated features to train and build SVM3-pseudo and SVM3-stacked enables to further improve lesion classification performance. Since both handcrafted and deep features extracted from mammograms may be able to pick up on details and patterns that cannot be detected with the human eye, classifications made by this fusion-based CAD scheme have the potential to better assist radiologists in reducing the false positive recall rate of mammogram lesion detection by acting as a second reader.
Fourth, we observe that AUC value and classification accuracy are higher when using pseudo-ROIs for feature extraction from deep transfer learning models than simply using 3 stacked ROIs. Few studies have been conducted to investigate how to optimally utilize pseudo-RGB ROIs as inputs for deep CNNs. Overall, these studies show higher lesion classification performance when using ROIs that have been meaningfully pre-processed when the original greyscale image is just stacked in three dimensions [34, 59, 60]. This further solidifies the idea that image preprocessing is a crucial step when utilizing a deep learning network trained on natural images for medical imaging tasks. As many different contrast enhancement techniques exist for processing mammograms [35], future work must be done to better investigate these techniques roles in developing more effective pseudo-RGB images for deep transfer learning.
Despite a large image dataset, promising classification results, and new observations that can help facilitate research effort to further develop and optimize CAD schemes to classify between breast lesions using mammograms, there are also several limitations in this study. First, the dataset used in this study focuses solely on craniocaudal view mammograms. As a result, this is only a region-based CAD scheme. Since in mammography a lesion can often be detected in both craniocaudal (CC) and mediolateral oblique (MLO) views, fusion of classification results from two views has potential to further improve classification performance. Thus, in future studies, we will develop and test a more complete case or lesion-based CAD scheme that fuses the two lesion regions detected on both CC and MLO views. Second, for a proof-of-concept study we only computed 40 handcrafted image features, selected and directly used VGG16 as a deep transfer learning model as a feature extractor, and a standard SVM as a multifeatured-based classifier. Although this an efficient approach to test our hypothesis, the results may not be the best or optimal. More existing radiomics features and/or other features (i.e., local binary patterns) should be explored in handcrafted features, and more advanced deep transfer learning and classification models should be investigated and applied in future studies to improve lesion classification performance. Third, as shown in figure 4, this study used a concatenation method as a feature-level fusion operator. Although the concatenation method is widely used in the CAD field, it may not be the best method. We will further investigate different feature-level and decision-level fusion schemes in future studies. Fourth, we recognize the importance of using balanced dataset of two classes to train machine learning classifier. In this study, our dataset is slightly unbalanced with a ratio of 1.074 (795/740 or 51.8% benign and 48.2% malignant images). Although the imbalanced ratio is relatively small and should not have significant negative impact in training SVM models, we will investigate this issue or impact of using the more balanced image datasets such as adding synthetic FFDM images using a Generative Adversarial Networks (GAN) as demonstrated in a recent study in our lab [61]. Lastly, although we use a standard 10-fold cross-validation method to test CAD performance, its robustness needs to be further validated or tested on multiple different datasets and compared to different traditional or deep learning classifiers in future work.
5. CONCLUSIONS
This study presents a new fusion-based CAD scheme that combines handcrafted features with automated deep transfer learning features aiming to improve the performance of a machine learning classifier in classification of breast lesions as malignant or benign. Although this is a preliminary study with several limitations, to the best of our knowledge, this is the first proof-of-concept study that investigates and demonstrates the feasibility and advantages of optimally fusing handcrafted and two types of deep transfer learning generated automated features extracted from pseudo-ROIs and stacked ROIs to train new machine leaning classifiers to improve accuracy in breast lesion classification. Therefore, this study helps build a solid foundation for us to facilitate future studies and make progress in this CAD field. We currently continue to investigate new approaches to (1) compute both handcrafted features (based on radiomics concept) and automated features (based on improved deep transfer learning models) including using more effective image pre-processing method to produce pseudo images, (2) more effectively post-process the automated features to generate optimal and more robust feature vectors to train machine learning classifiers, and (3) to investigate and apply more effective fusion methods to combine handcrafted and automated features to train machine learning classifiers, which aims to more effectively take or combine advantages of both types of image features.
ACKNOWLEDGMENT
This work is supported in part by Grants of R01CA197150 from the National Cancer Institute, National Institutes of Health, and Graduate Study Scholarship from Stephenson School of Biomedical Engineering, the University of Oklahoma.
REFERENCES
- 1.Siegel RL, Miller KD, and Jemal A, Cancer statistics, 2020. CA Cancer J Clin, 2020. 70(1): p. 7–30. [DOI] [PubMed] [Google Scholar]
- 2.Bleyer A, Baines C, and Miller AB, Impact of screening mammography on breast cancer mortality. Int J Cancer, 2016. 138(8): p. 2003–12. [DOI] [PubMed] [Google Scholar]
- 3.Bleyer A and Welch HG, Effect of Three Decades of Screening Mammography on Breast-Cancer Incidence. New England Journal of Medicine, 2012. 367(21): p. 1998–2005. [DOI] [PubMed] [Google Scholar]
- 4.National Breast Cancer Foundation. Available from: https://www.nationalbreastcancer.org/breast-cancer-biopsy.
- 5.Brodersen J and Siersma VD, Long-term psychosocial consequences of false-positive screening mammography. Ann Fam Med, 2013. 11(2): p. 106–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castellino RA, Computer aided detection (CAD): an overview. Cancer imaging : the official publication of the International Cancer Imaging Society, 2005. 5(1): p. 17–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roth HR, et al. , Improving Computer-Aided Detection Using Convolutional Neural Networks and Random View Aggregation. IEEE Transactions on Medical Imaging, 2016. 35(5): p. 1170–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan M, Pu J, and Zheng B, Reduction of false-positive recalls using a computerized mammographic image feature analysis scheme. Physics in medicine and biology, 2014. 59(15): p. 4357–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang T, et al. , Correlation between CT based radiomics features and gene expression data in non-small cell lung cancer. J Xray Sci Technol, 2019. 27(5): p. 773–803. [DOI] [PubMed] [Google Scholar]
- 10.Zheng Q, et al. , Rethinking the Role of Activation Functions in Deep Convolutional Neural Networks for Image Classification. Engineering Letters, 2020. 28(1). [Google Scholar]
- 11.Rawat W and Wang Z, Deep Convolutional Neural Networks for Image Classification: A Comprehensive Review. Neural Computation, 2017. 29(9): p. 2352–2449. [DOI] [PubMed] [Google Scholar]
- 12.Singh SP, et al. , 3D deep learning on medical images: a review. Sensors, 2020. 20(18): p. 5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeiler MD and Fergus R. Visualizing and understanding convolutional networks. in European conference on computer vision. 2014. Springer. [Google Scholar]
- 14.Adebayo J, et al. , Sanity checks for saliency maps. arXiv preprint arXiv:1810.03292, 2018. [Google Scholar]
- 15.Gilpin LH, et al. Explaining explanations: An overview of interpretability of machine learning. in 2018 IEEE 5th International Conference on data science and advanced analytics (DSAA). 2018. IEEE. [Google Scholar]
- 16.Raghu M, et al. , Transfusion: Understanding transfer learning for medical imaging. arXiv preprint arXiv:1902.07208, 2019. [Google Scholar]
- 17.Alzubaidi L, et al. , Towards a Better Understanding of Transfer Learning for Medical Imaging: A Case Study. Applied Sciences, 2020. 10(13): p. 4523. [Google Scholar]
- 18.Deng J, et al. Imagenet: A large-scale hierarchical image database. in 2009 IEEE conference on computer vision and pattern recognition. 2009. Ieee. [Google Scholar]
- 19.Dhillon A and Verma GK, Convolutional neural network: a review of models, methodologies and applications to object detection. Progress in Artificial Intelligence, 2020. 9(2): p. 85–112. [Google Scholar]
- 20.Zhao B, et al. , A survey on deep learning-based fine-grained object classification and semantic segmentation. International Journal of Automation and Computing, 2017. 14(2): p. 119–135. [Google Scholar]
- 21.Arevalo J, et al. , Representation learning for mammography mass lesion classification with convolutional neural networks. Comput Methods Programs Biomed, 2016. 127: p. 248–57. [DOI] [PubMed] [Google Scholar]
- 22.Paul R, et al. , Deep Feature Transfer Learning in Combination with Traditional Features Predicts Survival Among Patients with Lung Adenocarcinoma. Tomography (Ann Arbor, Mich.), 2016. 2(4): p. 388–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul R, et al. Combining deep neural network and traditional image features to improve survival prediction accuracy for lung cancer patients from diagnostic CT. in 2016 IEEE International Conference on Systems, Man, and Cybernetics (SMC). 2016. IEEE. [Google Scholar]
- 24.Cai L, Gao J, and Zhao D, A review of the application of deep learning in medical image classification and segmentation. Annals of Translational Medicine, 2020. 8(11): p. 713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, et al. , Recent advances and clinical applications of deep learning in medical image analysis. arXiv preprint arXiv:2105.13381, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katzen J and Dodelzon K, A review of computer aided detection in mammography. Clin Imaging, 2018. 52: p. 305–309. [DOI] [PubMed] [Google Scholar]
- 27.Zou L, et al. , A Technical Review of Convolutional Neural Network-Based Mammographic Breast Cancer Diagnosis. Computational and Mathematical Methods in Medicine, 2019. 2019: p. 6509357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan SA and Yong S-P, A comparison of deep learning and hand crafted features in medical image modality classification. 2016 3rd International Conference on Computer and Information Sciences (ICCOINS), 2016: p. 633–638. [Google Scholar]
- 29.Lin W, et al. , Comparison of handcrafted features and convolutional neural networks for liver MR image adequacy assessment. Scientific Reports, 2020. 10(1): p. 20336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heidari M, et al. , Development and Assessment of a New Global Mammographic Image Feature Analysis Scheme to Predict Likelihood of Malignant Cases. IEEE transactions on medical imaging, 2020. 39(4): p. 1235–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan M, et al. , Developing a new case based computer-aided detection scheme and an adaptive cueing method to improve performance in detecting mammographic lesions. Phys Med Biol, 2017. 62(2): p. 358–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan M, et al. , A new approach to develop computer-aided detection schemes of digital mammograms. Phys Med Biol, 2015. 60(11): p. 4413–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirniaharikandehei S, et al. , Applying a new computer-aided detection scheme generated imaging marker to predict short-term breast cancer risk. Phys Med Biol, 2018. 63(10): p. 105005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heidari M, et al. , Improving the performance of CNN to predict the likelihood of COVID-19 using chest X-ray images with preprocessing algorithms. International Journal of Medical Informatics, 2020. 144: p. 104284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu S, et al. , Feature and Contrast Enhancement of Mammographic Image Based on Multiscale Analysis and Morphology. Computational and Mathematical Methods in Medicine, 2013. 2013: p. 716948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paris S, et al. , A gentle introduction to bilateral filtering and its applications, in ACM SIGGRAPH 2007 courses. 2007, Association for Computing Machinery: San Diego, California. p. 1–es. [Google Scholar]
- 37.Simonyan K and Zisserman A, Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556, 2014. [Google Scholar]
- 38.Shen L, et al. , Deep Learning to Improve Breast Cancer Detection on Screening Mammography. Scientific Reports, 2019. 9(1): p. 12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montaha S, et al. , BreastNet18: A High Accuracy Fine-Tuned VGG16 Model Evaluated Using Ablation Study for Diagnosing Breast Cancer from Enhanced Mammography Images. Biology, 2021. 10(12): p. 1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gardezi SJS, et al. Mammogram classification using deep learning features. in 2017 IEEE International Conference on Signal and Image Processing Applications (ICSIPA). 2017. IEEE. [Google Scholar]
- 41.Saber A, et al. , A Novel Deep-Learning Model for Automatic Detection and Classification of Breast Cancer Using the Transfer-Learning Technique. IEEE Access, 2021. 9: p. 71194–71209. [Google Scholar]
- 42.Tammina S, Transfer learning using vgg-16 with deep convolutional neural network for classifying images. International Journal of Scientific and Research Publications (IJSRP), 2019. 9(10): p. 143–150. [Google Scholar]
- 43.Kononenko I and Robnik-Šikonja M, Non-Myopic Feature Quality Evaluation with (R)ReliefF, in Computational Methods of Feature Selection, Liu H and Motoda H, Editors. 2008, Taylor & Francis Group: Boca Raton, Florida. [Google Scholar]
- 44.Robnik-Šikonja M and Kononenko I, Theoretical and Empirical Analysis of ReliefF and RReliefF. Machine Learning, 2003. 53(1): p. 23–69. [Google Scholar]
- 45.Urbanowicz RJ, et al. , Relief-based feature selection: Introduction and review. Journal of Biomedical Informatics, 2018. 85: p. 189–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kira K and Rendell LA. The feature selection problem: Traditional methods and a new algorithm. in Aaai. 1992. [Google Scholar]
- 47.Zongker D and Jain A. Algorithms for feature selection: An evaluation. in Proceedings of 13th International Conference on Pattern Recognition. 1996. [Google Scholar]
- 48.Tan M, Pu J, and Zheng B, Optimization of breast mass classification using sequential forward floating selection (SFFS) and a support vector machine (SVM) model. International journal of computer assisted radiology and surgery, 2014. 9(6): p. 1005–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haralick RM, Shanmugam K, and Dinstein I, Textural features for image classification, IEEE Transaction on Systems, Man and Cybernitics, Vol. SMC, 1973(3 (6)): p. 610. [Google Scholar]
- 50.Tang X, Texture information in run-length matrices. IEEE Trans Image Process, 1998. 7(11): p. 1602–9. [DOI] [PubMed] [Google Scholar]
- 51.Yu X, et al. , Mammographic image classification with deep fusion learning. Scientific Reports, 2020. 10(1): p. 14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qiu Y, et al. , A new approach to develop computer-aided diagnosis scheme of breast mass classification using deep learning technology. Journal of X-ray science and technology, 2017. 25(5): p. 751–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mendel K, et al. , Transfer Learning From Convolutional Neural Networks for Computer-Aided Diagnosis: A Comparison of Digital Breast Tomosynthesis and Full-Field Digital Mammography. Academic radiology, 2019. 26(6): p. 735–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huynh BQ, Li H, and Giger ML, Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. Journal of medical imaging (Bellingham, Wash.), 2016. 3(3): p. 034501–034501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eppstein MJ and Haake P. Very large scale ReliefF for genome-wide association analysis. in 2008 IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology. 2008. [Google Scholar]
- 56.Draper B, Kaito C, and Bins J. Iterative Relief. in 2003 Conference on Computer Vision and Pattern Recognition Workshop. 2003. [Google Scholar]
- 57.Moore JH and White BC. Tuning ReliefF for Genome-Wide Genetic Analysis. 2007. Berlin, Heidelberg: Springer Berlin Heidelberg. [Google Scholar]
- 58.Sun Y, Iterative RELIEF for feature weighting: algorithms, theories, and applications. IEEE Trans Pattern Anal Mach Intell, 2007. 29(6): p. 1035–51. [DOI] [PubMed] [Google Scholar]
- 59.Yunzhi W, et al. A hybrid deep learning approach to predict malignancy of breast lesions using mammograms. in Proc.SPIE. 2018. [Google Scholar]
- 60.Sun W, Zheng B, and Qian W, Automatic feature learning using multichannel ROI based on deep structured algorithms for computerized lung cancer diagnosis. Computers in Biology and Medicine, 2017. 89: p. 530–539. [DOI] [PubMed] [Google Scholar]


