Abstract
When Mycobacterium bovis BCG and Staphylococcus aureus were plated on agar containing increasing concentrations of fluoroquinolone, colony numbers exhibited a sharp drop, followed by a plateau and a second sharp drop. The plateau region correlated with the presence of first-step resistant mutants. Mutants were not recovered at concentrations above those required for the second sharp drop, thereby defining a mutant prevention concentration (MPC). A C-8-methoxy group lowered the MPC for an N-1-cyclopropyl fluoroquinolone.
Fluoroquinolones are potent antibacterial agents that have DNA gyrase and DNA topoisomerase IV as their intracellular targets (for a review, see reference 4). Since these targets are essential enzymes that are conserved among bacterial species, fluoroquinolones are effective against a broad range of bacteria. However, many pathogens, such as Staphylococcus aureus, Pseudomonas aeruginosa, and Neisseria gonorrhoeae, readily acquire resistance through the stepwise accumulation of mutations in genes encoding the two topoisomerases (for a review, see references 4 and 9). Resistance can severely limit the clinical usefulness of fluoroquinolones, and many new derivatives have been synthesized in an attempt to find more effective compounds (for examples, see references 8, 12, and 13). We recently noticed that several new compounds differ in their abilities to restrict the selection of resistant mutants (3, 15). Since this restriction was not readily predicted by the standard bacteriostatic assay for potency (15), it was not obvious how fluoroquinolone structure influences the development of resistant bacterial populations. To help define the relationships between fluoroquinolone structure and resistance, we examined the effect of fluoroquinolone concentration on the recovery of resistant mutants. As described below, we observed a complex relationship that depended on the bacterial species being tested and on the structure of the fluoroquinolone.
Two bacterial species were examined for colony formation on fluoroquinolone-containing agar. One organism, Mycobacterium bovis BCG, has DNA gyrase as its primary fluoroquinolone target (3), while the other, S. aureus, has DNA topoisomerase IV as its primary target (1, 5, 6, 10). M. bovis BCG was cultured with Middlebrook 7H9 liquid medium enriched with 10% albumin-dextrose complex and 0.05% Tween 80; Middlebrook 7H10 agar plates were used for single-colony isolation (7). S. aureus was cultured in CY liquid medium and grown into colonies on GL agar plates (11). Both organisms were grown to stationary phase in liquid medium, concentrated by centrifugation (5,000 × g for 25 min), and then resuspended in fresh medium. Aliquots containing up to 1011 cells were applied to agar plates containing various fluoroquinolone concentrations; plates were incubated at 37°C for 30 to 42 days for M. bovis BCG and 2 days for S. aureus. All colonies regrew on agar containing the selecting fluoroquinolone. Fluoroquinolones were obtained from Sigma Biochemical Corp. (norfloxacin), Miles Laboratories (ciprofloxacin), and Parke-Davis Pharmaceutical Co. (PD161148 and PD160793). Fluoroquinolone structures are shown in Fig. 1.
FIG. 1.
Fluoroquinolone structures. The arrow shown in PD161148 indicates the C-8 position.
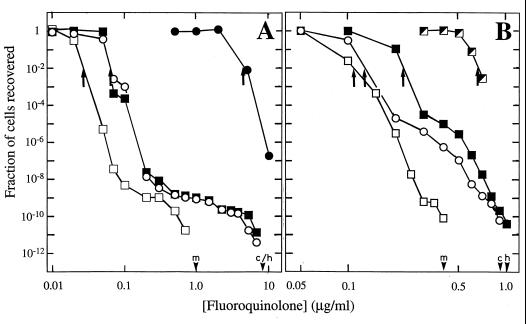
As the fluoroquinolone concentration in agar plates increased, two sharp declines were observed in the fractions of CFU recovered when wild-type cells were applied to plates (Fig. 2). The first decline, which occurred around the MIC for 99% of input cells (MIC99; Fig. 2), we attributed to the inhibition of wild-type cell growth. Above the MIC, a concentration range existed in which the recovery of CFU declined more gradually. The concentration range spanned by this part of the response, the plateau region, varied from one fluoroquinolone to another. For example, the region occurred over a much narrower concentration range for a C-8-methoxy compound (PD161148) than for its C-8-H derivative (PD160793) or for ciprofloxacin (Fig. 2). Since the C-8-methoxy compound is more active than its C-8-H derivative or ciprofloxacin against first-step mutants (3, 14, 15), we attributed the plateau region of the curve to the presence of resistant mutants. As shown in Table 1, a nucleotide sequence analysis of DNA from colonies recovered from ciprofloxacin-containing agar revealed alterations in the quinolone resistance-determining region of GyrA (M. bovis BCG) and ParC (GrlA; S. aureus). In the case of S. aureus, some colonies that were recovered at low concentrations of ciprofloxacin lacked an expected mutation in the quinolone resistance-determining region of ParC. Genes associated with resistance in these colonies have not yet been identified.
FIG. 2.
Effect of fluoroquinolone concentration on selection of resistant mutants. M. bovis BCG isolate KD1295 (A) and S. aureus MT5 (B) were plated on agar containing the indicated concentrations of PD161148, a C-8-methoxy compound (open squares), PD160793, a C-8-H derivative (filled squares), or ciprofloxacin (open circles). Panel A also shows the responses of the M. bovis BCG first-step mutant CX1 (3) to ciprofloxacin (filled circles); in panel B, half-filled squares show the responses of a first-step parC (Cipr) mutant of S. aureus (strain KD1806) to treatment with the indicated concentrations of PD160793. After incubation to allow growth, colonies were counted, and the fraction of the input number was determined. In the experiments shown, up to 1011 cells were applied to agar plates. The MIC99 for each compound is indicated by arrows. Small arrowheads on the abscissa indicate the MPC1010 for the C-8-methoxy compound (m), C-8-H compound (h), and ciprofloxacin (c).
TABLE 1.
Resistance alleles selected by plating on ciprofloxacin-containing agar
| Bacterial species | Ciprofloxacin concn (μg/ml) | Amino acid change(s)a |
|---|---|---|
| M. bovis BCG | 0.5 | A90V |
| 2.25 | D94G, D94N | |
| 3.75 | D94N, D94N | |
| 5.25 | D94N, D94N | |
| S. aureus | 0.2 | NIb, NI |
| 0.3 | NI, S80Y | |
| 0.4 | NI, S80Y | |
| 0.6 | S80Y, S80Y |
Mutations in GyrA (M. bovis BCG) or ParC (S. aureus) contain a letter representing the wild-type amino acid followed by the codon number and a letter representing the amino acid in the mutant. Abbreviations: A, alanine; D, aspartic acid; N, asparagine; G, glycine; Y, tyrosine; V, valine; NI, not identified.
The second sharp drop in mutant recovery (Fig. 2) occurred once the fluoroquinolone concentration was sufficient to block the growth of first-step mutants. For example, the MIC for first-step mutants of M. bovis BCG and S. aureus occurred at about the same fluoroquinolone concentration as the second drop in mutant recovery when wild-type cells were challenged (Fig. 2). The plateau region seen with M. bovis BCG and S. aureus differed in two ways. First, the plateau extended over a greater range of fluoroquinolone concentration for M. bovis BCG. This observation is consistent with the greater difference in quinolone sensitivity between wild-type cells and first-step mutants for M. bovis BCG (13- to 90-fold [3]) than for S. aureus (about 2-fold [14]). Second, the number of mutants recovered in the plateau region was 4 orders of magnitude higher for S. aureus (Fig. 2), indicating that the S. aureus population contained many more first-step mutants. Differences in growth rates and in the target compositions of the two species may contribute to these findings: S. aureus contains both topoisomerase IV and gyrase, while M. bovis BCG probably contains only the latter (an examination of the genomic nucleotide sequence of M. tuberculosis, a closely related organism, failed to identify genes likely to encode topoisomerase IV [2]).
The addition of a C-8-methoxy group to an N-1-cyclopropyl fluoroquinolone lowered the concentration at which the second drop occurred and shortened the plateau region. The effect of adding the methoxy group was about a 10-fold decrease in the case of M. bovis BCG and about a 2.5-fold decrease for S. aureus (Fig. 2). These data suggest that the methoxy group lowers the concentration required to prevent mutants from being recovered, a parameter we call the mutant prevention concentration (MPC). The MPC is estimated by determining the minimal antibiotic concentration that results in recovery of no mutants when large numbers of cells are applied to antibiotic-containing agar plates (the use of large numbers of cells, on the order of 1010 for M. bovis BCG, ensures that the restrictive antibiotic concentration blocks the growth of first-step mutants). The MPC depends on the number of cells applied; consequently, a subscript is added to indicate the number of cells tested (e.g., MPC1010 for 1010 cells applied to plates). This qualification allows data from different organisms, antibiotics, and laboratories to be compared. As with MIC determination, fluoroquinolone concentrations in agar plates can be standardized by having successive concentrations differ by twofold. When smaller increments are used, the MPC can be expressed as a range between the highest concentration at which mutants are recovered and the lowest at which they are not. For example, we performed five independent experiments with S. aureus and obtained the following values for the MPC1010 of ciprofloxacin, expressed as the range defined above: 0.7 to 0.8, 0.8 to 0.9, 0.6 to 0.7, 0.8 to 0.9, and 0.8 to 0.9 μg/ml. When only the upper number in each range is considered, the standard deviation was about 10%. Fluoroquinolones that display superior bacteriostatic activity (low MIC) are often quite effective at preventing the selection of resistant mutants (low MPC). However, the relationship is not proportional (Table 2).
TABLE 2.
Relationship between MIC and MPC in S. aureus
In conclusion, plots comparing antibiotic concentration to the fraction of input cells recovered as resistant colonies discriminate among compounds and simplify identification of those compounds least likely to allow bacterial populations to become resistant. Such a measurement requires no information on the nature of the resistance alleles. The assay can be simplified by determining the minimum concentration that allows no mutants to be recovered when large numbers of cells are applied to agar plates. This concentration, the MPC, should be useful for establishing therapeutic antibiotic regimens, particularly for the long-term treatment of immunodeficient patients.
Acknowledgments
We thank M. Gennaro, S. Kayman, B. Kreiswirth, and I. Smith for critical comments on the manuscript. We also thank D. Hooper for providing wild-type S. aureus strain MT5.
This work was supported by NIH grant AI35257.
Footnotes
Publication 65 from the Public Health Research Institute TB Center.
REFERENCES
- 1.Blanche F, Cameron B, Bernard F-X, Maton L, Manse B, Ferrero L, Ratet N, Lecoq C, Goniot A, Bisch D, Crouzet J. Differential behaviors of Staphylococcus aureus and Escherichia coli type II DNA topoisomerases. Antimicrob Agents Chemother. 1996;40:2714–2720. doi: 10.1128/aac.40.12.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S, Eiglmeier K, Gas S, Barry C E, Tekaia F, Babcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 3.Dong Y, Xu C, Zhao X, Domagala J, Drlica K. Fluoroquinolone action against mycobacteria: effects of C-8 substituents on growth, survival, and resistance. Antimicrob Agents Chemother. 1998;42:2978–2984. doi: 10.1128/aac.42.11.2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrero L, Cameron B, Crouzet J. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39:1554–1558. doi: 10.1128/aac.39.7.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrero L, Cameron B, Manse B, Lagneaux D, Crouzet J, Famechon A, Blanche F. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase. IV. A primary target of fluoroquinolones. Mol Microbiol. 1994;13:641–653. doi: 10.1111/j.1365-2958.1994.tb00458.x. [DOI] [PubMed] [Google Scholar]
- 7.Jacobs W R, Kalpana G V, Cirillo J D, Pascopella L, Snapper S B, Udani R A, Jones W, Barletta R G, Bloom B R. Genetic systems in mycobacteria. Methods Enzymol. 1991;204:537–555. doi: 10.1016/0076-6879(91)04027-l. [DOI] [PubMed] [Google Scholar]
- 8.Klopman G, Fercu D, Renau T E, Jacobs M R. N-1-tert-butyl-substituted quinolones: in vitro anti-Mycobacterium avium activities and structure-activity relationship studies. Antimicrob Agents Chemother. 1996;40:2637–2643. doi: 10.1128/aac.40.11.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamura S. Mechanisms of quinolone resistance. J Infect Chemother. 1997;3:128–138. [Google Scholar]
- 10.Ng E Y, Trucksis M, Hooper D C. Quinolone resistance mutations in topoisomerase IV: relationship to the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:1881–1888. doi: 10.1128/aac.40.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Novick R P, Brodsky R. Studies on plasmid replication. I. Plasmid incompatibility and establishment in Staphylococcus aureus. J Mol Biol. 1972;68:285–302. doi: 10.1016/0022-2836(72)90214-8. [DOI] [PubMed] [Google Scholar]
- 12.Renau T E, Gage J W, Dever J A, Roland G E, Joannides E T, Shapiro M A, Sanchez J P, Gracheck S J, Domagala J M, Jacobs M R, Reynolds R C. Structure-activity relationships of quinolone agents against mycobacteria: effect of structural modifications at the 8 position. Antimicrob Agents Chemother. 1996;40:2363–2368. doi: 10.1128/aac.40.10.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Renau T E, Sanchez J P, Gage J W, Dever J A, Shapiro M A, Gracheck S J, Domagala J M. Structure-activity relationships of the quinolone antibacterials against mycobacteria: effect of structural changes at N1 and C7. J Med Chem. 1996;39:729–735. doi: 10.1021/jm9507082. [DOI] [PubMed] [Google Scholar]
- 14.Zhao X, Wang J-Y, Xu C, Dong Y, Zhou J, Domagala J, Drlica K. Killing of Staphylococcus aureus by C-8-methoxy fluoroquinolones. Antimicrob Agents Chemother. 1998;42:956–958. doi: 10.1128/aac.42.4.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao X, Xu C, Domagala J, Drlica K. DNA topoisomerase targets of the fluoroquinolones: a strategy for avoiding bacterial resistance. Proc Natl Acad Sci USA. 1997;94:13991–13996. doi: 10.1073/pnas.94.25.13991. [DOI] [PMC free article] [PubMed] [Google Scholar]