Abstract
The complete nucleotide sequence of the plasmid-mediated MIR-1 β-lactamase gene confirms its relationship to chromosomally located ampC genes of Enterobacter cloacae. blaMIR-1 is not part of a typical gene cassette but does lie near an element that could be involved in its capture on a plasmid.
MIR-1, one of the first plasmid-mediated class C (group 1) β-lactamases to be characterized, was distinguished from class A extended-spectrum β-lactamases that also confer resistance to oxyimino-β-lactams by the resistance it provided to α-methoxy β-lactams and its insensitivity to such class A inhibitors as clavulanic acid and tazobactam (10). In confirmation of this assignment, a sequence of 150 bp of blaMIR-1 proved 90% identical to sequence at the C terminus of the class C gene of Enterobacter cloacae P99 (10). Subsequently, ACT-1, another plasmid-mediated β-lactamase, was discovered, fully sequenced, and found also to be more closely related to genes for AmpC β-lactamases of E. cloacae than to those of other gram-negative organisms (3). The sequence of blaMIR-1 and neighboring DNA has now been completed.
The blaMIR-1 gene was cloned from plasmid pMG230 with KpnI as a 4.1-kb insert into vector plasmid pMLC28 to produce plasmid pMG231 and subcloned in the same vector with AccI and PstI on a 1.4-kb fragment as plasmid pMG232 (10). Cycle sequencing with Taq polymerase was initiated with M13/pUC sequencing primers from the termini of the insert in pMG232 and continued by primer walking until both DNA strands had been sequenced. Additional nucleotides upstream and downstream from the coding region were added by sequencing the larger insert in pMG231.
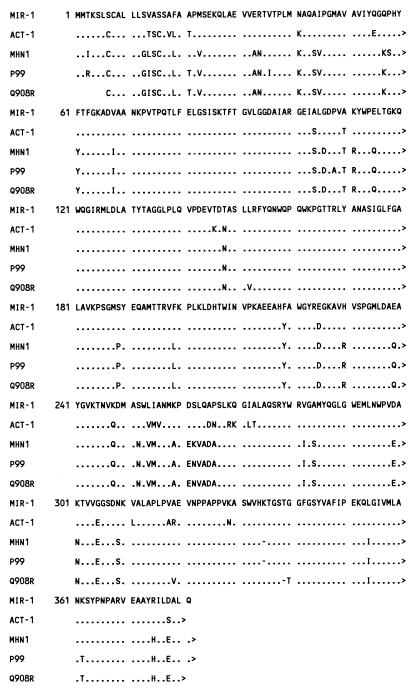
The nucleotide sequence of the blaMIR-1 region is shown in Fig. 1. A BLAST search (1) indicated that DNA from nucleotides 1 to 823 is 98% identical to sequence from Pseudomonas pseudoalcaligenes that resembles an insertion sequence transposase (4) and is also related to ORF341, a component of unidentified function associated with several integrons (12, 13). Nucleotides 824 to 927 are 90% or more identical to DNA upstream from chromosomally located E. cloacae ampC genes (5) and also plasmid-mediated ACT-1 (3), while nucleotides 928 to 2073 encode MIR-1 β-lactamase. Nucleotides 1872 to 2021 are identical to the fragment of blaMIR-1 reported previously (10). A comparison of the amino acid sequences of MIR-1, ACT-1, and three E. cloacae chromosomally mediated AmpC β-lactamases (5) is shown in Fig. 2. The amino acid sequence of MIR-1 most closely resembled that of ACT-1, differing in only 31 of 381 residues (91% identity), and has closer identity to AmpC genes of E. cloacae (85 to 86%) than to those of Citrobacter freundii (74%) or Escherichia coli (72%) (6, 11).
FIG. 1.
Nucleotide sequence of blaMIR-1 gene and neighboring DNA.
FIG. 2.
Comparison of amino acid sequences of MIR-1, ACT-1 (3), and E. cloacae MHN1, P99, and Q908R AmpC (5) β-lactamases. The dots indicate identical amino acid residues, and the hyphens indicate no amino acid residue at that position. The active site serine is at position 86.
How a normally chromosomally located β-lactamase gene was acquired by pMG230 is not yet known, but attempts to demonstrate transposition (8) of blaMIR-1 have not been successful. The sequence shown in Fig. 1 lacks the 59-base element associated with gene cassettes that have become integron associated (7). Whether the nearby DNA encoding a potential transposase reflects the presence of an insertion sequence involved in gene capture will require further study.
The AmpC gene of E. cloacae is usually expressed at a low but inducible level. Some plasmid-mediated ampC genes are inducible (2), but MIR-1 in E. coli is not (10). E. coli, however, lacks the ampR locus necessary for induction (6). Accordingly, pMG230 was transferred to E. coli SNO3/pNU311. SNO3 is ampC8 and so cannot express E. coli chromosomal β-lactamase, while pNU311 carries the ampR (but not ampC) gene of Citrobacter freundii (9). MIR-1 was not inducible with cefoxitin or imipenem in this strain either, suggesting that the few nucleotide differences between the promoter regions of blaMIR-1 and E. cloacae chromosomal AmpC genes are responsible for the escape from induction control.
Nucleotide sequence accession number.
The sequence of blaMIR-1 has been submitted to GenBank under accession no. M37839.
Acknowledgments
This work was supported in part by a Merit Review award from the VA/DoD Collaborative Research Program on Mechanisms of Emerging Pathogens.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;17:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnaud G, Arlet G, Verdet C, Gaillot O, Lagrange P H, Philippon A. Salmonella enteritidis: AmpC plasmid-mediated inducible β-lactamase (DHA-1) with an ampR gene from Morganella morganii. Antimicrob Agents Chemother. 1998;42:2352–2358. doi: 10.1128/aac.42.9.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford P A, Urban C, Mariano N, Projan S J, Rahal J J, Bush K. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob Agents Chemother. 1997;41:563–569. doi: 10.1128/aac.41.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis, J. K., C. C. Somerville, and J. C. Spain. 3 October 1997, posting date. Cloning and molecular characterization of two genes encoding hydroxylaminobenzene mutase activity from Pseudomonas pseudoalcaligenes JS45. [Online.] http://www.ncbi.nlm.nih.gov/htbin-post/entrez. [11 May 1999, last date accessed.]
- 5.Galleni M, Lindberg F, Normark S, Cole S, Honore N, Joris B, Frere J-M. Sequence and comparative analysis of three Enterobacter cloacae ampC β-lactamase genes and their products. Biochem J. 1988;250:753–760. doi: 10.1042/bj2500753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grundström T, Jaurin B. Overlap between ampC and frd operons on the Escherichia coli chromosome. Proc Natl Acad Sci USA. 1982;79:1111–1115. doi: 10.1073/pnas.79.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall R M, Brookes D E, Stokes H W. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol Microbiol. 1991;5:1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 8.Jacoby G A, Sutton L. Properties of plasmids responsible for extended-spectrum β-lactamase production. Antimicrob Agents Chemother. 1991;35:164–169. doi: 10.1128/aac.35.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindberg F, Westman L, Normark S. Regulatory components in Citrobacter freundii ampC β-lactamase induction. Proc Natl Acad Sci USA. 1985;82:4620–4624. doi: 10.1073/pnas.82.14.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papanicolaou G A, Medeiros A A, Jacoby G A. Novel plasmid-mediated β-lactamase (MIR-1) conferring resistance to oxyimino- and α-methoxy β-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1990;34:2200–2209. doi: 10.1128/aac.34.11.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sawai T, Yamaguchi A, Tsukamoto K. Amino acid sequence, active-site residue, and effect of suicide inhibitors on cephalosporinase of Citrobacter freundii GN346. Rev Infect Dis. 1988;10:721–725. doi: 10.1093/clinids/10.4.721. [DOI] [PubMed] [Google Scholar]
- 12.Stokes H W, Tomaras C, Parsons Y, Hall R M. The partial 3′-conserved segment duplications in the integrons In6 from pSa and In7 from pDGO100 have a common origin. Plasmid. 1993;30:39–50. doi: 10.1006/plas.1993.1032. [DOI] [PubMed] [Google Scholar]
- 13.Valentine C R, Heinrich M J, Chissoe S L, Roe B R. DNA sequence of direct repeats of the sulI gene of plasmid pSa. Plasmid. 1994;32:222–227. doi: 10.1006/plas.1994.1059. [DOI] [PubMed] [Google Scholar]