Abstract
Quantitative susceptibility mapping (QSM) is an MRI-based, computational method for anatomically localizing and measuring concentrations of specific biomarkers in tissue such as iron. Growing research suggests QSM is a viable method for evaluating the impact of iron overload in neurological disorders and on cognitive performance in aging. Several software toolboxes are currently available to reconstruct QSM maps from 3D GRE MR Images. However, few if any software packages currently exist that offer fully automated pipelines for QSM-based data analyses: from DICOM images to region-of-interest (ROI) based QSM values. Even less QSM-based software exist that offer quality control measures for evaluating the QSM output. Here, we address these gaps in the field by introducing and demonstrating the reliability and external validity of Ironsmith; an open-source, fully automated pipeline for creating and processing QSM maps, extracting QSM values from subcortical and cortical brain regions (89 ROIs) and evaluating the quality of QSM data using SNR measures and assessment of outlier regions on phase images. Ironsmith also features automatic filtering of QSM outlier values and precise CSF-only QSM reference masks that minimize partial volume effects. Testing of Ironsmith revealed excellent intra- and inter-rater reliability. Finally, external validity of Ironsmith was demonstrated via an anatomically selective relationship between motor performance and Ironsmith-derived QSM values in motor cortex. In sum, Ironsmith provides a freely-available, reliable, turn-key pipeline for QSM-based data analyses to support research on the impact of brain iron in aging and neurodegenerative disease.
Keywords: Quantitative susceptibility mapping, Aging, Brain iron, Software, Automated pipeline
1. Introduction
Quantitative susceptibility mapping (QSM) is a computational method for reconstructing maps of local tissue magnetic susceptibility from MRI-based gradient-recalled echo (GRE) magnitude and phase images. QSM capitalizes on the property of certain substances to locally alter magnetic susceptibility in tissue; that is, the degree to which tissue can be magnetized in an external magnetic field. For example, paramagnetic iron increases the magnetic susceptibility of gray matter (GM) in a roughly linear manner to its concentration (Hametner et al., 2018; Langkammer et al., 2012; Sun et al., 2015). In contrast, diamagnetic calcifications reduce the magnetic susceptibility of GM. As such, the strength and polarity (increase or decrease relative to a reference structure) of local tissue magnetic susceptibility can be used to measure local concentrations of specific biomarkers in tissue. For instance, positive magnetic susceptibility relative to cerebrospinal fluid (CSF) is highly correlated with ex vivo determined brain iron concentrations in subcortical structures (Langkammer et al., 2012; Sun et al., 2015) and in cortical regions (Hametner et al., 2018; T. J. Liu et al., 2012).
The ability of QSM to quantify local, in vivo iron concentration in tissue makes it an invaluable tool in the study of iron overload in aging and neurodegenerative disease. Aging is associated with accumulation of non-heme brain iron, which has been linked with oxidative stress, neurodegeneration and cognitive decline (Lauffer, 1992; Wayne Martin et al., 1998; Zecca et al., 2004). Growing research suggests that QSM is a powerful neuroimaging method to identify relationships between aging, increases in brain iron concentration and cognitive health (Bandt et al., 2019; Betts et al., 2016; Darki et al., 2016; Sun et al., 2017; Wang et al., 2017; Zachariou et al., 2020). In addition, QSM is being used to study accumulation of non-heme iron concentrations in neurodegenerative diseases, such as Parkinson’s (He et al., 2015; Mazzucchi et al., 2019; Thomas et al., 2020), Huntington’s (Chen et al., 2019) and Alzheimer’s disease (Acosta-Cabronero et al., 2013; Fan et al., 2020; Ward et al., 2014).
As a result, the previous decade has seen the development of a number of software toolboxes for reconstructing QSM maps from 3D GRE magnitude and phase images (e.g. see list at https://github.com/mathieuboudreau/qsm-tools). The first publicly available software package for QSM reconstruction was MEDI Toolbox (De Rochefort et al., 2010; J. Liu et al., 2012; Liu et al., 2011a, 2011b) which uses a Morphology Enabled Dipole Inversion approach (MEDI) for dipole deconvolution. The MEDI Toolbox remains the predominant method for reconstructing QSM maps in the literature, although other software toolboxes are now publicly available, including COSMOS (Liu et al., 2009), FANSI toolbox (Bilgic et al., 2015; Milovic et al., 2018), QSM Toolbox (Kames et al., 2018), QSMxT (Stewart et al., 2021), SEPIA (Chan and Marques, 2021) and STI suite (Liu et al., 2015). In addition, automated, hardware-based QSM reconstruction solutions are also becoming available. One example is a server-based QSM reconstruction solution developed by the MEDI Toolbox group (Spincemaille et al., 2019) which can automatically reconstruct QSM maps from raw GRE DICOM data as it is collected from an MRI scanner. These are typically not open-source and require custom configured computers/servers to be directly connected to an MRI scanner.
While extremely valuable, the currently available QSM tools do not provide users with automated, standardized pipelines for QSM analyses. Instead, available tools (specifically software toolboxes) are semi-automated and depend on scripts, written (e.g., MEDI Toolbox, FANSI toolbox, QSMxT) or customized by users (e.g., SEPIA; GUI-based), to reconstruct QSM maps from GRE MRI images. In addition, pre-processing of data (e.g., specific naming of input files), applying the toolboxes to groups of participants, filtering of outlier QSM voxels and quality control procedures, such as evaluating the impact of head motion and phase-image artifacts on QSM maps are generally not included.
Further, for most QSM-based studies, reconstructing QSM maps is only the first step in the analyses process. Following reconstruction, QSM values need to be extracted from anatomical regions of interest (ROIs) and the quality of extracted QSM values needs to be assessed. To our knowledge, few QSM toolboxes are currently available for extracting QSM-based brain iron concentrations from anatomical ROIs. Two examples we are aware of are the Johns Hopkins University (JHU) multi-atlas tool (Li et al., 2019) and the QSMxT toolbox (Stewart et al., 2021). Both are valuable, however, neither are fully automated.
The JHU multi-atlas tool is limited to ten subcortical structures and is not fully automated. Further, the JHU multi-atlas tool requires a-priori reconstructed QSM maps and skull-stripped T1 anatomical images as inputs, which must be co-registered. The QSM map inputs must also be scaled so values vary within a predefined range (−0.5 to 0.5 ppm). Finally, the JHU multi-atlas tool is not open source, which prevents users from modifying the toolkit to add more automation and/or functionality (e.g., support for additional anatomical ROIs).
The QSMxT toolkit can extract QSM-based susceptibility values from anatomical ROIs (although a list of supported/tested ROIs is not available on the QSMxT website). Similar to the JHU multi-atlas tool, QSMxT is not fully automated and requires some scripting for batch processing of participants. In addition, partial volume effects related to the segmented ROIs have not been ascertained, quality control measures are generally not included in the QSMxT toolkit and external validity has not been established. However, QSMxT is open-source and is regularly updated. As such, new features and quality control procedures can be added in the future.
In short, few if any software packages currently exist that offer fully automated pipelines for QSM-based data analyses, from raw DICOM images to output of ROI-based QSM values. Even less QSM-based software exist that offer quality control measures and/or procedures for evaluating the QSM outputs. These shortcomings can act as a barrier to research labs who wish to conduct QSM-based studies but do not have the required programming skills to do so. In addition, reliable, valid, fully automated pipelines for QSM analyses are needed to promote direct comparison of results across studies, which should aid scientific replication.
Here, we address these issues by introducing Ironsmith: a comprehensive, open-source, fully automated pipeline for creating and processing QSM maps and extracting QSM-based iron concentrations from subcortical and cortical brain regions (89 ROIs; Appendix A). Ironsmith offers the following unique features in a single pipeline/package. 1) Iron-smith is completely automated. That is, the pipeline does not simply run a series of scripts in a sequence. Instead, every user input is verified for correctness, outputs are evaluated against a list of expected outcomes and even in the event of a failure or software crash, Ironsmith can recover (see section 2.1. Functionality for details). Additionally, Ironsmith can monitor and optimize multiple versions of itself running in parallel which significantly reduces processing time. Importantly, during parallel processing, Ironsmith uses a single input file and provides a single set of output files, irrespective of how many instances of ironsmith are running in parallel. Further, Ironsmith can differentiate if the participants provided as inputs have been analyzed previously or not. As such, a user can simply update a single input file with new/added participants and Ironsmith will update all existing output files with the new/added results only. 2) Ironsmith provides a number of quality control measures and analytic solutions to issues commonly encountered during QSM analyses. Specifically, Ironsmith can automatically identify artifacts on phase images, provides per-ROI SNR measures, can filter out per-ROI outlier QSM values (such as values associated with large veins) and offers a precise, CSF-only reference region for QSM reconstruction to minimize partial volume effects. Lastly 3), Ironsmith provides comprehensive feedback and intelligible error/warning messages. When an error or warning occurs, Ironsmith does not display cryptic messages but instead provides detailed reports of what might have gone wrong and how a user can fix the error/warning.
Ironsmith is designed to process single or groups of participants autonomously with minimal user input and/or supervision, does not depend on user scripts to run analyses and requires little technical/programming knowledge to setup and use. Further, Ironsmith does not rely on custom, in-house algorithms for processing QSM data. Instead, the pipeline uses reliable, third-party software and toolboxes such as MEDI Toolbox (the predominant method for reconstructing QSM maps in the literature; De Rochefort et al., 2010; J. Liu et al., 2012; Liu et al., 2011a, 2011b), to analyze QSM data (complete list of third party software used by Ironsmith can be found in section 2.2. Software requirements and dependencies).
Following the introduction and architecture walkthrough of Iron-smith, we benchmark the pipeline using data from 35, healthy older adults (20 of these used for inter- and intra-rater reliability analyses; see Methods section). Specifically, we (1) test both the intra-rater and inter-rater reliability of Ironsmith-derived QSM values; (2) determine if Ironsmith-derived SNR values are sensitive to sources of noise such as head motion; (3) demonstrate the utility of several Ironsmith features, including identification of outlier regions on phase images, filtering of outlier QSM values and use of a CSF-only, lateral ventricles based reference mask for QSM reconstruction, shown to reduce partial volume effects; and (4) evaluate the external validity of Ironsmith by demonstrating expected associations between QSM values extracted from primary motor cortex and motor performance.
2. Materials and methods
2.1. Functionality
Ironsmith is a fully automated QSM processing pipeline for creating QSM maps and extracting QSM-based iron concentrations from anatomical ROIs, including both subcortical and cortical brain regions (89 GM ROIs included with version 1.2; See Appendix A). Ironsmith is open source, under the GNU general public license (version 3), and was developed entirely using Bash script. The latest version of Ironsmith can be downloaded from Github, together with full documentation and installation instructions. Ironsmith can perform the following tasks:
Automate the process of creating QSM maps from GRE DICOM images using MEDI Toolbox (De Rochefort et al., 2010; J. Liu et al., 2012; Liu et al., 2011a, 2011b).
Register MPR or multi-echo MPR (MEMPR) T1 images to QSM maps and then segment these into 89 ROIs (Appendix A), using Freesurfer (Dale et al., 1999; Desikan et al., 2006).
Extract QSM-based iron concentrations from 89 ROIs, filter outlier voxels from ROIs (default: larger than the 97th percentile of QSM values), and output the results into comma separated variable (CSV) tables.
Calculate SNR (GRE magnitude image based) for each ROI as a measure of quality control for QSM data and output SNR values in CSV tables.
Identify outlier regions on phase images using a median absolute deviation (MAD) based outlier detection process. Calculate the percent overlap between phase-image outlier regions and each of the 89 supported ROIs. Output the results into CSV tables.
Non-linearly warp QSM maps and aligned MPR/MEMPR to MNI152 1mm space. This step allows users to (1) extract QSM values from standard space ROIs not included with Ironsmith and (2) conduct voxelwise analyses (these options are not used/described in the present manuscript, which focuses on extraction of QSM values from native participant space).
Process single or multiple participants in parallel (multiple instances and nohup supported).
2.2. Software requirements and dependencies
Ironsmith requires a Linux distribution with Singularity version 3.5 or higher installed (https://sylabs.io/guides/3.5/admin-guide/installation.html) and Bash Unix shell version 4.2.46(2) or later (GNU coreutils). Ironsmith was tested under native Red Hat Enterprise Linux Workspace release 7.8, Ubuntu 16.04 and on Windows Subsystem for Linux V2 (WSL2), running Ubuntu 18.04. Singularity versions 3.5.2 and 3.5.3 were used for testing. Additionally, Ironsmith requires Matlab (supported versions R2017b to R2019b) and MEDI Toolbox version 01/15/2020 (De Rochefort et al., 2010; J. J. Liu et al., 2012; Liu et al., 2011a, 2011b) if QSM maps need to be reconstructed by Ironsmith from GRE magnitude/phase DICOM images.
In addition to the software requirements described in the previous paragraph, Ironsmith has the following third-party software dependencies: AFNI (Cox, 1996), dcm2niix (Li et al., 2016), Freesurfer (Dale et al., 1999; Desikan et al., 2006) and FSL (Jenkinson et al., 2012). These software packages are all open source and are provided together with Iron-smith in the form of a singularity image and do not need to be separately installed. The singularity image used with Ironsmith was created using Neurodocker. All provided third-party software are subject to their own licenses which are in full effect when using Ironsmith. Please consult the individual licenses of third party software for specific requirements. The singularity image will be updated with future versions of Ironsmith to include up-to-date versions of third-party software as needed.
2.3. Architecture
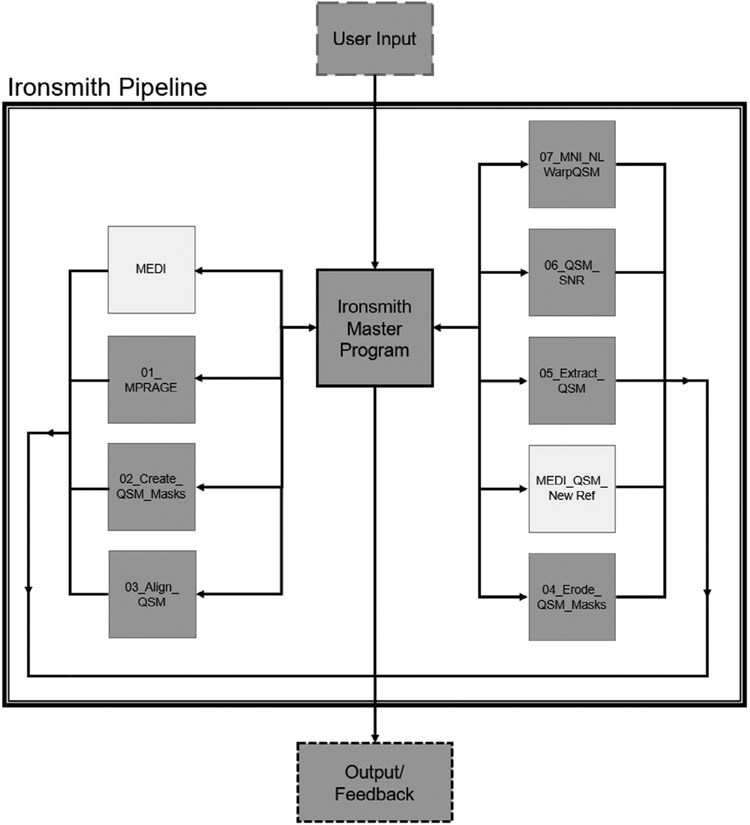
The architecture of Ironsmith is illustrated in Fig. 1. The pipeline is controlled by a master program which oversees nine specialized analyses scripts. The procedure of each of these scripts is described in detail in the sections below. All scripts can output files to disk and provide feedback (including error messages) both on screen and in log files, however only the master program can skip a participant or terminate the pipeline due to errors.
Fig. 1.
Schematic of the Ironsmith pipeline. The central Iron-smith master program accepts all user inputs and controls nine specialized scripts, depicted as orbiting boxes. The specialized scripts process the data, create output files and provide user-feedback. The two specialized scripts depicted with white boxes are optional and used only if QSM maps need to be reconstructed.
2.3.1. Ironsmith input
User input is provided during execution of the Ironsmith master program by running the following shell command: “Ironsmith [MyInput-File.csv] [path to output folder]”. That is, a user needs to provide (1) a CSV formatted text file containing information and MRI file locations for each participant that needs to be processed, and (2) the absolute path to an output directory. The input, a CSV formatted text file, is referred to as “MyInputFile.csv” here for clarity, however, this file can have any name. Additionally, MyInputFile.csv can be created in any text editor or spreadsheet program as long as it is saved as a CSV file and Ironsmith will adjust the file accordingly to comply with Unix line endings. Lastly, the specified output directory will be created, if not present, by Iron-smith and does not need to be created beforehand. The exact format of MyInputFile.csv is described in detail in the full documentation of Ironsmith together with examples, as well as in supplementary material section 2.3.1. (S) Ironsmith input.
Recommended parameters for acquiring 3D multi-echo GRE sequences for QSM reconstruction can be found on the MEDI Toolbox website. In addition to the MEDI recommended parameters, we strongly advise the use of Prescan Normalize - Adaptive Combine (PS-AC), as the coil combine method for GRE sequences collected on Siemens scanners (software version MR VE11A, or newer), as used in the present study. PS-AC can address coil combination issues (see https://cds.ismrm.org/protected/18MProceedings/PDFfiles/4992.html) that can lead to phase artifacts (Bernstein et al., 1994). For Philips MRI scanners, we recommend acquiring GRE images using the SENSE parallel imaging method (KP et al., 1999) which is equivalent to ASPIRE (Eckstein et al., 2018) and can correctly combine phase images from different channels (personal communication, Dr. Xu Li and Hanzhang Lu, Johns Hopkins University). For GE MRI scanners the ASSET (GE version of SENSE) parallel imaging method can be used to acquire GRE images for QSM. For Philips and GE MRI scanners, any software version that supports SENSE/ASSET respectively, should correctly combine phase images from different coils. Ironsmith can detect a number of coil combination related artifacts on phase images and will issue warnings (see section 2.3.10. 06_QSM_SNR). However, we further advise users to visually inspect at least a few of the reconstructed QSM maps for potential artifacts. Alternatively, if correct coil combination cannot be achieved with the default scanner software, then uncombined, raw scanner data can be collected instead and reconstructed using software toolboxes such as ASPIRE (Eckstein et al., 2018; https://github.com/korbinian90/ASPIRE).
2.3.2. Ironsmith master program
Upon execution, the master program first checks the command syntax for errors. If errors are found, the user is prompted to make adjustments and to rerun the Ironsmith command. If the correct syntax is provided, the master program checks whether the software required to process the data is installed and working (e.g., Singularity, Matlab, MEDI Toolbox). Following the software check, all input provided with the shell command is validated. This involves checking whether MyInputFile.csv is formatted correctly, all files and/or directories specified in MyInputFile.csv exist and if files are in the correct MRI modality and/or file type. For instance, if files are indeed in DICOM and/or NIFTI format and/or if the GRE magnitude and phase images provided consist of a sufficient number of volumes/echoes (i.e. a minimum of two) appropriate for QSM reconstruction.
If DICOM files are provided as inputs, Ironsmith will additionally search the DICOM headers for any text (standalone or part of a string) matching the nominal participant label specified in MyInputFile.csv and issue a warning if the label is not found. This participant label check can help verify whether the DICOM files specified in MyInputFile.csv match the participant to be processed. Lastly, the master program ascertains if a participant specified in MyInputFile.csv has already been processed or has partial data in the output folder. Both of these conditions will issue a warning message on screen and a participant will be skipped. If validation is successful, a directory is created under the output folder, corresponding to the nominal text label in MyInputFile.csv of the participant currently being processed. The master program then sequentially executes the scripts described in the sections below in order to process the data. All scripts executed by the master Ironsmith program maintain their own log files which are saved in each participant’s folder within the output directory. Additionally, all feedback provided by Ironsmith, including error/warning messages are logged in dated, log files within the Ironsmith installation directory (under the LogFiles sub-folder). Log files within this folder that are 30 days or older are automatically deleted by Ironsmith to save disk space.
Multiple instances of Ironsmith can be executed in parallel and multiple instances can work on the same MyInputFile.csv and output folder. Under this condition, each instance of Ironsmith will process a different subset of participants from MyInputFile.csv until all participants are processed. Only one set of output files will be created under the output folder and all instances of Ironsmith will work on the same output files. Irrespective of which Ironsmith instance finishes which participants first, the order of participants in output files will be the same as that provided in MyInputFile.csv.
2.3.3. MEDI
This script is optional and is only called by the master program if QSM maps need to be reconstructed for a participant (i.e. if “MEDI_Yes” is indicated in MyInputFile.csv for that participant). The MEDI script creates a Matlab “.m” file, compatible with MEDI Toolbox (De Rochefort et al., 2010; J. J. Liu et al., 2012; Liu et al., 2011a, 2011b) based on information provided in MyInputFile.csv. This Matlab script file is saved under a participant’s folder, inside the output directory. The MEDI script then runs this file in Matlab without launching the graphical interface (-nodisplay, -nosplash, -nodesktop). The MEDI script monitors the Matlab process for errors and evaluates the outputs if successful. If errors occur, the script terminates and yields control back to the master program in order to report error messages to the user. The resulting outputs from the MEDI script are NIFTI files corresponding to a QSM map, a GRE magnitude image and the default MEDI Toolbox QSM reference structure mask (whole-brain cerebrospinal fluid; CSF), segmented internally by MEDI Toolbox. Lastly, the MEDI script, using MEDI Toolbox creates a relative difference field (RDF) image by unwrapping (region growing method; Witoszynskyj et al., 2009) the input phase image and removing the background field (using projections onto dipole fields; De Rochefort et al., 2010; Liu et al., 2011a). This RDF image is saved within a participant’s folder, inside the output directory and is used by a subsequent script (see section 2.3.10. 06_QSM_SNR) to identify potential artifacts on the input phase image.
2.3.4. 01_MPRAGE
This script is responsible for preprocessing anatomical MPR/MEMPR images and for running the Freesurfer segmentation pipeline. It is the first script to be called if QSM maps are already available (i.e. if “MEDI_No” is indicated in MyInputFile.csv for a participant). The script 01_MPRAGE first determines if DICOM or NIFTI images were provided as the MPR/MEMPR input. If DICOMs were provided these are converted to NIFTI using dcm2niix. If the resulting or user provided NIFTI file/s consist/s of more than one volumes/echoes, 01_MPRAGE will assume the data are MEMPR and a root mean square image will be created (RMS) using the mri_concat Freesurfer command with the –rms option. If a single volume/echo is provided, the MPR data will be used as is. Freesurfer is then executed with the recon-all –all option. OpenMP is supported by 01_MPRAGE and will use half of the available CPU cores to significantly speed-up the Freesurfer segmentation process. Once recon-all is complete (between 6–8 h per participant), 01_MPRAGE will check whether all expected output files are present and if the recon-all process finished without errors. Control is then returned to the master Ironsmith program together with information required to provide feedback to the user and in log files (errors or successful completion).
2.3.5. 02_Create_QSM_Masks
This script creates 89 anatomical ROIs, covering both subcortical and cortical brain regions (compete list of ROIs is in Appendix A) from the Freesurfer output of 01_MPRAGE. All files created by this script are saved under a participant’s folder in the output directory. The script first converts the following Freesurfer parcellation files to NIFTI from .mgz volume format, using the Freesurfer mri_convert function and RAS as the –out_orientation option: (a) skull-stripped MPR/MEMPR image, (b) brain mask of skull-stripped MPR/MEMPR image, (c) the main aseg parcellation file, (d) the DKT atlas parcellation file, (e) the white matter (WM) parcellation file, and (f) the a2009s atlas parcellation file. The script then uses these files to create anatomical ROIs using the FSL, fslmaths function with the –bin option and a threshold corresponding to the Freesurfer label number of a particular anatomical structure (https://surfer.nmr.mgh.harvard.edu/fswiki/FsTutorial/AnatomicalROI/FreeSurferColorLUT). The AFNI command 3dmerge is also used by the script to combine anatomical structures into lobes as recommended by Freesurfer (https://surfer.nmr.mgh.harvard.edu/fswiki/CorticalParcellation). A whole-brain WM mask and a mask of the lateral ventricles are also created. These are used by a subsequent script in the pipeline as additional QSM reference structures for reconstruction in MEDI Toolbox. As with previous scripts, 02_Create_QSM_Masks is monitored and control is returned to the master Ironsmith program upon completion for feedback/logging.
It should be noted that zero voxel ROIs are possible during this step and typically indicate issues with the Freesurfer segmentation pipeline for a particular anatomical structure (possibly due to severe head motion artifacts or a significant sized stroke/lesion or other anatomical abnormality). These zero voxel ROIs are communicated to a user both on screen and in log files but will not cause Ironsmith to skip a participant or to exit. Subsequent scripts detect these zero voxel ROIs and can account for them.
2.3.6. 03_AlignQSM
This script registers the Freesurfer derived skull-stripped MPR/MEMPR anatomical image of a participant to its corresponding GRE magnitude image provided in MyInputFile.csv, or created by the MEDI script, using the AFNI function align_epi_anat.py and a local Pearson correlation cost function. Since QSM maps are reconstructed from GRE phase/magnitude images, the previous step effectively registers a participant’s MPR/MEMPR anatomical image to its corresponding QSM map. Ironsmith assumes that GRE magnitude and MPR/MEMPR images have been collected in the same scan session. The pipeline has not been tested for registering MPR/MEMPR images to GRE magnitude images acquired in different scan sessions and might fail without an initial registration performed prior to running Ironsmith. In addition, the resolution of the GRE magnitude image is important for registration. Ironsmith uses the AFNI function align_epi_anat.py and an absolute local Pearson correlation cost function to register an MPR/MEMPR image to a GRE magnitude image. That is, local correlations (both positive and negative) between the MPR/MEMPR and the GRE magnitude image drive the alignment. For this reason, the closer the resolution of a GRE magnitude image to the corresponding MPR/MEMPR image, the higher the local correlations will be for correct alignment which will lead to a more accurate registration.
The transformation matrix resulting from the registration step is then used to register all Freesurfer derived ROIs created by 02_Create_QSM_Masks to the QSM map in native space, using the AFNI function 3dAllineate and a nearest neighbor interpolation method. Lastly, each ROI is resampled to the QSM voxel resolution using the AFNI 3dresample function. All aligned and resampled masks are saved under a participant’s folder in the output directory.
2.3.7. 04_Erode_QSM_Masks
This script erodes the QSM aligned and resampled ROIs (created by previous scripts) by one voxel in order to prevent partial volume effects with surrounding WM. Erosion occurs as follows: for each GM parcellation, a corresponding WM parcellation is also created by Freesurfer. These WM parcellations are processed by all previous scripts and QSM aligned/resampled masks are created from them. These WM masks are first dilated by one voxel and then subtracted from their corresponding GM masks. As such, any potential overlap of a GM ROI with WM is eliminated.
The 04_Erode_QSM_Masks script is also responsible for eroding the lateral ventricle mask by two voxels and the whole-brain WM mask by one voxel in preparation for their use as QSM reference structures by the next script. These erosion parameters were determined from QSM data and analyses presented in our recent work (Zachariou et al., 2021, 2020). The AFNI function 3dmask_tool is used to erode the lateral ventricle and whole-brain WM masks. All eroded masks are saved in the output directory under a participant’s folder and have the suffix “_Erx1”.
2.3.8. MEDI_QSM_New_Ref
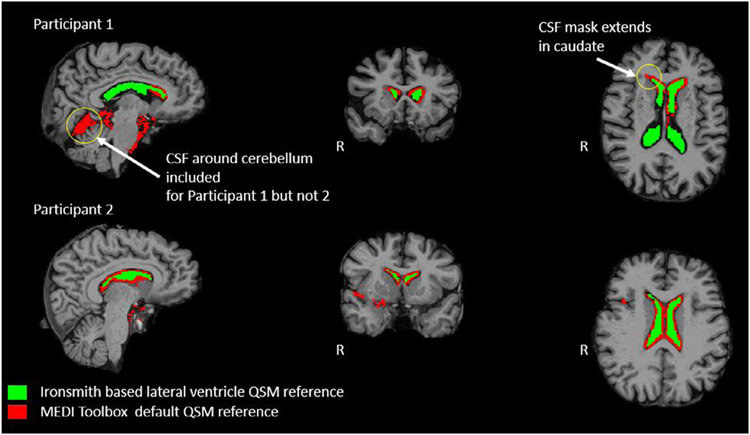
This script is called by the master Ironsmith program if the “MEDI_Yes” flag is provided in MyInputFile.csv for a participant. The script creates a new MEDI Toolbox compatible Matlab “.m” file and reruns MEDI Toolbox to reconstruct two additional QSM maps. One QSM map is reconstructed with the eroded lateral ventricles mask as the reference structure and the other with the eroded whole-brain WM mask as the reference structure. More specifically, MEDI Toolbox calculates average magnetic susceptibility within the provided reference mask and then subtracts this average from each voxel of the entire magnetic susceptibility map to yield the final QSM map. The new Matlab script file is saved in the output directory under a participant’s folder. The two new QSM maps created by this script are used in subsequent benchmark analyses to evaluate the impact of different reference structures on QSM data. Additionally, MEDI Toolbox uses a fairly rough parcellation of whole-brain CSF as the default reference structure for QSM. As such, the default MEDI Toolbox reference mask can vary in size and anatomical location across participants and can sometimes partially overlap with GM and WM regions (Fig. 2). Using Freesurfer-derived eroded lateral ventricle/WM masks as the QSM reference structures will likely improve the quality of QSM data by eliminating this unwanted overlap with parenchyma and by reducing partial volume effects.
Fig. 2.
Comparison between QSM reference masks created by Ironsmith and by MEDI Toolbox. The Fig. depicts T1 anatomical images from two participants used to test Iron-smith. The default MEDI Toolbox QSM reference structure is overlaid in red and the Iron-smith lateral ventricles CSF reference structure is overlaid in green on top of the T1 images.
2.3.9. 05_Extract_QSM
The primary purpose of this script is to extract QSM values from each of the (previously created) GM ROIs, using the AFNI 3dBrickStat function. Only positive QSM values (susceptibility greater than that of the reference structure used; CSF or WM) are considered in averaging. For QSM maps reconstructed using CSF-based reference structures (MEDI Default, CSF-only generated by Ironsmith), excluding negative QSM values from averages can limit QSM signal from myelin, associated with bordering white matter and neuropil (shadow artifacts; see; Kee et al., 2017). These shadow artifacts are induced by anisotropic WM adjacent to GM, which contributes to a dipole incompatible field and manifests as strongly negative QSM values within GM (Kee et al., 2017). It should be noted that the method of excluding negative QSM values to reduce shadow artifacts cannot be used for QSM maps reconstructed using average whole-brain WM as the reference because all GM voxels are positive when this reference is used (see Discussion section for more details).
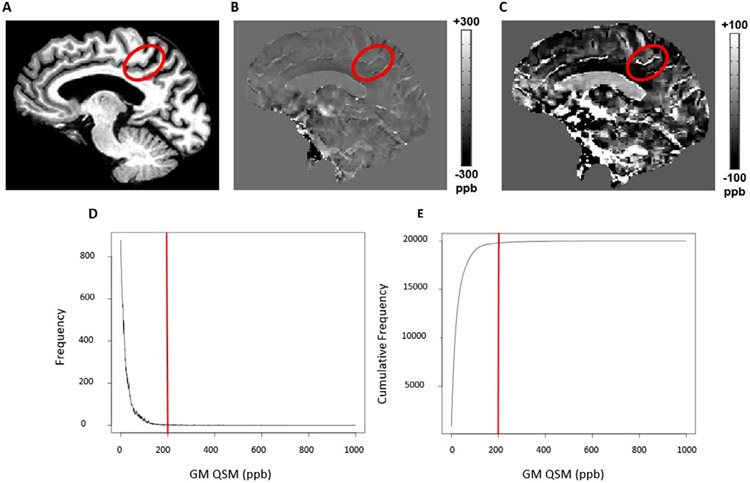
Further, outlier QSM voxels are excluded before averaging. Based on information acquired from pilot testing of Ironsmith, outlier QSM voxels can occur in GM and typically relate to partial overlap between a large vein and a GM ROI (see Fig. 3). Typical QSM values extracted from large veins during pilot testing were fairly uniform and around 200 parts per billion (ppb; Fig.s 3B, 3C; using CSF from the lateral ventricles as the QSM reference structure). In the pilot data, 2.4% of all positive QSM voxels in GM (subcortical and cortical regions) had values greater than or equal to vein-like QSM values (Fig.s 3D, 3E). The 05_Extract_QSM script eliminates these outlier voxels from averages by only including QSM voxels with values less than the 97th percentile of all positive QSM values within an ROI. The percentile cutoff point for outliers can be modified by manually editing the header of the 05_Extract_QSM.sh script file under the Ironsmith installation folder.
Fig. 3.
Ironsmith detection of outlier values associated with veins. A prominent vein is displayed on QSM images of a single representative participant as well as corresponding QSM values. Panel A highlights a sulcus within the left parietal lobe on a T1 anatomical image. Panel B highlights a vein within this sulcus, depicted on a QSM map at a typical contrast threshold of −300 to 300 ppb. Panel C highlights the same vein as in panel B but the contrast threshold of the QSM map is adjusted to vary between −100 to 100 in order to better highlight the vein. Large veins typically have QSM values around 200 ppb. Panel D is a histogram of positive QSM values in GM for the same participant as panels A, B and C. Panel E shows the cumulative frequency distribution of the GM QSM values in D. In both panels D and E the 200 ppb QSM cut-off threshold is highlighted with a solid red line.
Following outlier removal, 05_Extract_QSM calculates average QSM and normalized average QSM values for each GM ROI: average QSM values correspond to the sum of positive QSM values per voxel, divided by the total number of positive voxels. However, average QSM values do not account for differences in head/ROI size between participants (e.g. participants with larger ROIs will tend to have more positive QSM voxels compared to participants with smaller heads/ROIs). To this end, normalized QSM values are additionally computed to correct for variability in head/ROI size across participants. Specifically, normalized QSM values are computed by dividing the sum of all positive QSM voxels within an ROI by the ROI size (all voxels in an ROI). The unit of measure of all extracted QSM values is susceptibility in ppb.
These average QSM values per GM ROI are then written to group CSV formatted text files (in a different column per-ROI) within the output folder, with separate lines/rows in a text file for each participant. Separate CSV files are created for average QSM values and normalized average QSM values and for each QSM map type (corresponding to different reference structures). If a zero voxel ROI is detected, the word “FAIL” is written in the output file at the appropriate column instead of a numerical value.
The secondary purpose of 05_Extract_QSM is to monitor multiple instances of Ironsmith running in parallel and only allow one instance to write to a group output text file at a time. This is achieved via a temporary lifeline file created in the output folder that updates periodically while the script writes QSM data to disk. If another instance of 05_Extract_QSM detects a lifeline file that is not stale (i.e., it is being updated), the script will wait until the lifeline file is removed before accessing a group output file. If the lifeline file is stale (indicating a crash or an issue), 05_Extract_QSM will attempt to identify and correct the damaged output files by reverting these back to the last participant that completed successfully. This error is clearly communicated to the user both on screen and in log files so a corrupted/unfinished participant can be re-processed (see full documentation on how to skip the Freesurfer segmentation step for situations like this). Lastly, 05_Extract_QSM sorts the group output files periodically, so the order of participants in the output files is the same as the one provided in MyInputFile.csv, irrespective of how many instances of Ironsmith are working on the same MyInputFile.csv and output folder.
2.3.10. 06_QSM_SNR
This script has two main functions: (1) it calculates the signal-to-noise ratio (SNR) for each of the (previously created) GM ROIs and (2), identifies median absolute deviation (MAD) based outlier regions on RDF images created by the MEDI script (section 2.3.3), if “MEDI_Yes” has been indicated for a participant in MyInputFile.csv file. A MAD-based outlier detection process is used here because in the presence of outliers, MAD is the most robust measure of dispersion (Leys et al., 2013). The 06_QSM_SNR script then returns the percentage overlap of these outlier regions with the previously created GM ROIs.
For SNR, the GRE magnitude image is used to compute SNR (root mean square of all echoes/volumes of a GRE magnitude image; see section 2.4.2. Imaging protocol) rather than the QSM map because the latter tends to mix noise spatially due to spatial deconvolution. The first step in the process is to calculate the standard deviation (SD) of background voxels in the GRE magnitude image (using an outside-the-head mask) for each participant. The script first creates a binary mask of the entire GRE magnitude image, using the AFNI function 3dAutomask, which consists of all visible structures, including the skull and neck. This GRE head/neck mask is then dilated and contracted several times in order to eliminate potential holes within the mask. Next, the entire mask is dilated by two voxels and then inverted using the AFNI function 3dcalc. After inversion, voxels outside the head/neck (2-voxels away) have a value of one (selected) and voxels inside the head/neck have a value of zero (not selected). Lastly, the bottom half of this inverted mask is discarded along the z-axis. The final outcome is a binary, background image mask (i.e., an outside-the-head binary mask), two voxels away from the skull and away from the phase-encoding direction (top of the head with sagittal slice acquisition, anterior to posterior order). The FSL fslstats function is then used to calculate the SD of the GRE magnitude image within this background mask for each participant.
Once outside-the-head SD is calculated, 06_QSM_SNR extracts average GRE magnitude image intensity from each of the (previously created) GM ROIs, using the AFNI 3dBrickStat function. These per GM ROI averages are then divided by the outside-the-head SD. The noise distribution outside the head, however, is not Gaussian but instead follows the Rayleigh distribution. The SD of a Rayleigh distribution is related to the SD of a Gaussian distribution by a factor of √(2–π/2) (Edelstein et al., 1984), also known as the Rayleigh distribution correction factor. As such, the result of the previous step needs to be multiplied by this Rayleigh distribution correction factor to yield true, per GM ROI SNR values for each participant. These SNR values are stored in the output folder, within a CSV formatted, group SNR file, using the same procedure and error correction method as 05_Extract_QSM.
MAD-based outlier regions on RDF images are identified as follows: first, 06_QSM_SNR calculates the median of an RDF image, constrained within an aligned, Freesurfer-derived whole-brain (WB) mask, eroded by one voxel (created in previous scripts). The median is calculated using the FSL function fslstats. Then, using the same FSL function and WB mask, the median is subtracted from every voxel of the RDF image and the absolute value of the outcome is saved into a new intermediate map/image. The median of this intermediate map/image is then calculated to get the MAD of the RDF image. Positive and negative thresholds for outlier voxels are subsequently calculated and correspond to the median of the RDF image +/− (5 * MAD). These outlier thresholds have been determined by testing the outlier detection procedure on the 35 participants described in section 2.4. Ironsmith testing. Next, a MAD-based outlier mask is created using the AFNI function 3dcalc by identifying all voxels within an RDF image (WB-mask constrained) lower or higher than the positive and negative outlier thresholds calculated in the previous step. Lastly, the percent overlap between a MAD-based outlier mask and each of the previously created anatomical ROIs is calculated and saved in an output file labelled Group_QSM_MAD.csv within the output directory (output directory/Group/ Group_QSM_MAD.csv). Additionally, the 06_QSM_SNR script issues a visual warning, while processing the data (this warning is also saved in the log files for each participant) if the percent overlap between an ROI and the MAD-based outlier mask exceeds 15%.
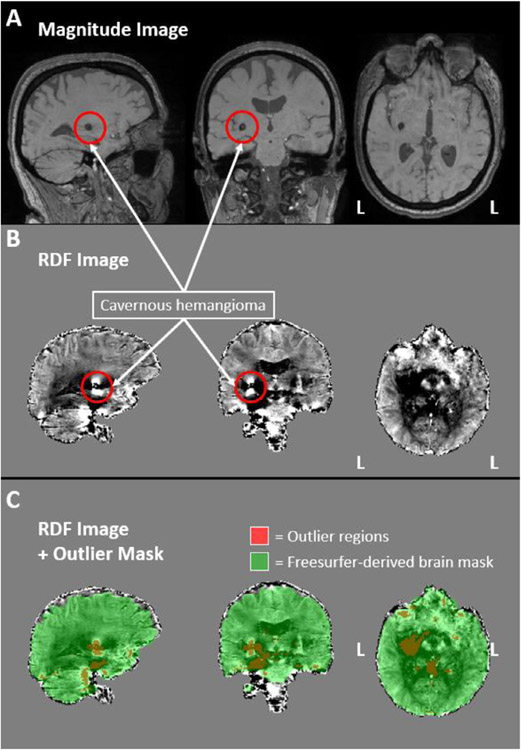
Fig. 4 demonstrates this outlier detection process on a participant (excluded from analyses) with a clinically-confirmed cavernous hemangioma in the right ventral basal ganglia. A hemangioma consists of a growth/mass of blood vessels and is often accompanied by local blood leakage (increased local deoxygenated hemoglobin). Due to the hemorrhage, cavernous hemangiomas are considered regions of strong susceptibility and can cause streaking artifacts on QSM maps (Wei et al., 2015). As can be seen on Fig. 4, the outlier detection process successfully identified the cavernous hemangioma as an outlier region. Importantly, SNR values in ROIs overlapping with the cavernous hemangioma in this participant were not lower compared to the rest of the participant cohort. As such, SNR alone was unable to identify this artifact.
Fig. 4.
Ironsmith-based phase image outlier detection. The magnitude (panel A) and RDF (panel B) images of a participant with a clinically-confirmed cavernous hemangioma are presented, outlined with red circles. Panel C. depicts, in red, the regions identified by Ironsmith as outliers using the MAD-based outlier detection process. The green mask in panel C represents a Freesurfer-derived brain mask used to constrain the outlier detection process.
2.3.11. 07_MNI_NL_WarpQSM
The purpose of this script is to warp the QSM aligned skull-stripped MPR/MEMPR image to MNI space using the MNI ICBM152, 1mm, 6th generation atlas (Grabner et al., 2006) and a non-linear transformation (3dQwarp). The resulting transformation matrices are used to warp all QSM maps created in previous scripts to MNI space. In addition, the transformation matrices are saved in the output folder after warping and example code is made available (within the log file of 07_MNI_NL_WarpQSM) that can align/register additional, user-specified atlas-based ROIs from MNI space to the QSM maps of each participant in native space. Users can use this code to extract QSM-based iron concentrations from template-based ROIs in MNI space that are not included with Ironsmith. The MNI warped QSM maps can be used in voxel-wise analyses outside Ironsmith. Neither voxel-wise analyses, nor are MNI-based ROI analyses used/described further in the present manuscript (which focuses on extraction of ROI values from native participant space).
2.3.12. Ironsmith outputs
Every participant processed through the Ironsmith pipeline has the following outputs, stored within separate sub-folders for each participant under the output directory specified in MyInputFile.csv (example participant shown in Appendix B): (1) A “LogFiles” folder containing text file logs for each of the scripts called by the master Ironsmith program; (2) An “MPR” folder comprised of the Freesurfer recon-all output files. This folder can be used as an optional input to Ironsmith in order to skip the Freesurfer segmentation step of the pipeline (details in the full documentation). Skipping this step can save time if a participant needs to be re-processed; (3) A “QSM” folder comprised of the following: three QSM maps, reconstructed using whole-brain CSF (default of MEDI Toolbox), lateral ventricles CSF or whole-brain WM as the QSM reference structure; all QSM reference structure masks, MNI warped versions of QSM maps; all ROI masks used in the analyses (original, aligned, resampled and eroded versions); all Matlab script files used to reconstruct QSM maps in MEDI Toolbox; all transformation matrices calculated during alignment and warping; all masks created by 06_QSM_SNR.
Additionally, a “Group” folder is created under the output directory consisting of six CSV formatted text files, corresponding to per GM ROI average QSM and normalized average QSM values for each of the three QSM maps created during processing (default MEDI Toolbox, lateral ventricles CSF and whole-brain WM as the QSM reference structures). If per participant QSM maps are provided in MyInputFile.csv then the lateral ventricle and whole-brain WM QSM maps are not created and their group QSM output files will not be present in the Group folder. CSV formatted text files comprised of per GM ROI SNR values and per GM ROI percent overlap with phase-image outlier regions, for each participant can also be found in the Group folder.
2.4. Ironsmith testing
2.4.1. Participants
A subset of 35, healthy older adults from the participant cohort described in Zachariou et al. (2020) were used to test Ironsmith (21 women, age range 66–86 years). The main inclusion criteria for these analyses was enrollment in the University of Kentucky’s Sanders-Brown Center on Aging participant cohort and thus availability of neuropsychological data (Schmitt et al., 2012). Specifically, participants needed to have completed the Comprehensive Trail Making Test (CTMT; Reynolds, 2002) as associations between the Trails-A portion of CTMT (motor performance) and Ironsmith-derived QSM values were explored to assess the external validity of Ironsmith.
All participants provided informed consent under a protocol approved by the Institutional Review Board of the University of Kentucky. All participants were cognitively intact based on clinical consensus diagnosis and scores from the UDS3. Exclusion criteria for our previous MRI study (Zachariou et al., 2020) were significant head injury (defined as loss of consciousness for more than five minutes), heart disease, neurological or psychiatric disorders, claustrophobia, pacemakers, the presence of metal fragments or any metal implants that are incompatible with MRI and diseases affecting the blood (anemia, kidney/heart disease, etc.). Detailed characteristics of the group of participants included in analyses are shown in Table 1.
Table 1.
Group Demographics and Mean Cognitive Measures.
Mean ± standard deviation is shown for participants.
MMSE: Mini-Mental State Exam,.
MoCA: Montreal Cognitive Assessment.
Table 1. Group demographics and mean cognitive scores. The table lists the total number of participants, mean (±sd) for age, male/female distribution and mean (±sd) for Mini-Mental State Exam (MMSE) and Montreal Cognitive Assessment (MoCA) scores.
2.4.2. Imaging protocol
Participants were scanned with a Siemens 3T PRISMA scanner (software version MR VE11C), using a 64-channel head-coil, at the University of Kentucky Magnetic Resonance Imaging and Spectroscopy Center (MRISC). The following sequences were used to test Ironsmith: a 3D multi-echo, T1-weighted anatomical image (MEMPR) and a 3D, multi-echo, gradient-recalled echo (GRE) sequence used for Quantitative Susceptibility Mapping (QSM). Several other sequences were collected during the scanning session related to other scientific questions and are not discussed further here.
The MEMPR sequence had four echoes [repetition time (TR) = 2530 ms, first echo time (TE1) = 1.69 ms, echo time spacing (ΔTE) = 1.86ms, flip angle (FA) = 7°] and covered the entire brain [176 slices, field of view = 256mm, parallel imaging (GRAPPA), acceleration factor = 2, 1mm isotropic voxels, scan duration =5.53 min]. The MEMPR sequence was used to optimize the Freesurfer segmentation step in the Ironsmith pipeline and improve the accuracy of anatomical ROIs (van der Kouwe et al., 2008).
The sequence used for QSM was a flow compensated, multi-echo, 3D spoiled GRE sequence in the sagittal plane with eight echoes (TR/TE1/ΔTE/FA= 24ms/2.98ms/2.53ms/15°). The entire brain was covered [acquisition matrix = 224 × 224 × 144, parallel imaging (GRAPPA), acceleration = 2, 1.2 mm isotropic voxels and scan duration = 6.18 min]. The coil combine mode used was prescan normalize - adaptive combine.
2.4.3. MRI data processing
Raw DICOM images were used as inputs for Ironsmith which handled the entire analyses pipeline as described in section 2.3. (Architecture).
2.5. Statistical analyses
Statistical analyses were performed using SPSS 27 (IBM, Chicago, IL, USA). Analyses included Intraclass Correlation Coefficient (ICC) analyses (using two-way mixed models and absolute agreement as the type of index), mixed effects models (for repeated measures analyses) and linear regression models. ICC analyses focused on the first 20 participants, consistent with reliability analysis protocols performed for the automated biomarker kits of the MarkVCID consortium sites, including our own UKy site (Lu et al., 2021). Mixed effects models and linear regressions used gender and age as covariates. Variance inflation factors (VIF) are provided in linear regression analyses with more than one anatomical ROI included in the model in order to evaluate the degree of collinearity between anatomical regions. All multiple comparisons are reported using the Sidak correction.
All analyses used participant’s normalized, average QSM values within ROIs because they account for ROI size differences between participants and are thus computed as part of the Ironsmith pipeline. Further, analyses on QSM-based iron concentrations in subcortical structures were conducted separately from those in cortical GM, because iron concentration in subcortical brain regions was substantially higher than in cortical brain regions (see section 3.3. Relationship between QSM Values and Gibbs artifacts). Lastly, where applicable, separate analyses were conducted for QSM values reconstructed using each of the following three reference structures: a whole-brain CSF mask generated by MEDI Toolbox (default MEDI Toolbox reference), a lateral ventricles-only CSF mask generated by Ironsmith and a whole-brain WM mask generated by Ironsmith.
3. Results
3.1. Reliability
All Ironsmith scripts as well as the third party pipelines used by Iron-smith are predominantly deterministic. That is, given the same input, Ironsmith should always produce the same output. For example, given the same anatomical dataset, the Freesurfer recon-all pipeline will always produce the exact same segmented ROIs. However, since multiple different pipelines interact throughout the QSM-analyses process and since these are tied together by custom code, the overall deterministic nature of Ironsmith needs to be tested. This section demonstrates/tests the deterministic nature of Ironsmith by running the first 20 participants through the pipeline three times. During this intra-rater reliability analysis, all Ironsmith outputs (e.g. segmented ROIs, QSM maps, QSM reference structure masks, QSM values, etc.) were manually inspected for correctness by the first author (V.Z.). Additionally, to evaluate the turn-key nature of Ironsmith and to explore how the pipeline performs under different operating systems and with different processing options, an inter-rater reliability was conducted using three independent raters (V.Z., C.E.B. and B.T.G.). The different raters run Ironsmith on the same dataset (20 participants) but used different computers, with different operating systems (Ubuntu and Redhat) and slightly different processing options (e.g. raw DICOM inputs vs NIFTI inputs). Average ICCs were then calculated for intra-rater and inter-rater reliability for nine representative subcortical and cortical GM ROIs. As anticipated from the deterministic nature of Ironsmith and included third-party pipelines, all ICC values were equal to 1.
3.2. Relationship between SNR values and participant head motion
The following analyses were conducted to test if SNR values provided by Ironsmith are sensitive to sources of noise that can affect QSM data, such as participant head motion. In 3D GRE images, head motion is closely associated with ringing artifacts. Visually, these closely resemble Gibbs artifacts (Czervionke et al., 1988; Gibbs, 1899). GRE magnitude images and QSM maps from each participant were visually rated by the second author (C.E.B.) on severity of ringing artifacts present on a three-level scale: (1) little-to-no ringing present, (2) moderate ringing present, and (3) significant ringing present (Fig. 5). Average SNR values from subcortical ROIs (caudate, pallidum and putamen) and cortical ROIs (frontal, parietal, occipital and temporal lobar GM) were then used in mixed models analyses to evaluate the relationship between SNR and severity of ringing artifacts. The results from these analyses are summarized in Table 2. As can be seen, lower SNR values were associated with larger ringing artifacts, in both cortical and subcortical brain regions.
Fig. 5.
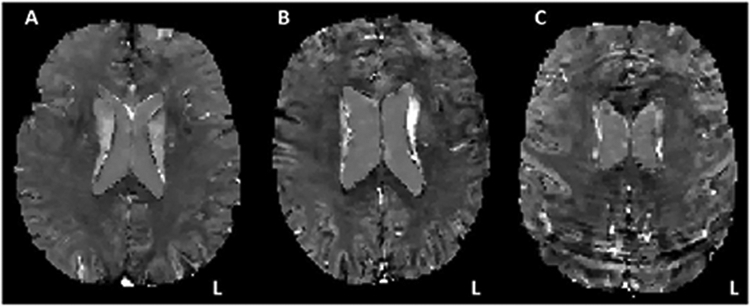
Ringing artifacts on QSM maps. QSM maps are presented from three different study participants containing little-to-no ringing (A), moderate ringing (B) and significant ringing (C).
Table 2.
Mixed models ANOVA results: Relationship between SNR and ringing artifacts.
| Fixed effects: Ringing artifact rating | Subcortical brain regions | Cortical brain regions | ||
|---|---|---|---|---|
| F-stat | p-value | F-stat | p-value | |
| 20.58 | <0.0001** | 22.42 | <0.0001** | |
| Marginal means for SNR | ||||
| Ringing artifact rating | SNR: subcortical brain regions | SNR: Cortical brain regions | ||
| Little-to-no ringing | 66.673 | 66.413 | ||
| Moderate ringing | 48.627 | 48.621 | ||
| Significant ringing | 34.805 | 34.492 | ||
| Pairwise comparisons | ||||
| Subcortical brain regions | Cortical brain regions | |||
| Ringing artifact rating pair | SE | p-value | SE | p-value |
| Little-to-no ringing vs Moderate ringing | 5.058 | 0.003** | 4.842 | 0.002** |
| Moderate ringing vs Significant ringing | 5.914 | 0.071 | 5.662 | 0.048* |
| Significant ringing vs Little-to-no ringing | 5.136 | 0.0001** | 5.136 | 0.0001** |
p < 0.01.
p < 0.05.
3.3. Relationship between QSM values and ringing artifacts
The following analyses were conducted to determine the impact of ringing artifacts on QSM values. Linear mixed effects models were conducted to evaluate the relationship between QSM values and ringing artifacts and how this relationship can change as a function of the reference structure used to reconstruct QSM maps. QSM values extracted from both cortical and subcortical ROIs were used in these analyses. Subcortical QSM comprised average QSM values from the caudate, pallidum and putamen ROIs. Cortical QSM comprised average QSM values from the frontal, parietal, occipital and temporal lobar GM ROIs. Results are summarized in Table 3.
Table 3.
Mixed models ANOVA results: Relationship between subcortical/cortical QSM values and ringing artifacts.
| Fixed effects: Ringing artifact rating | Subcortical brain regions | Cortical brain regions | ||||
|---|---|---|---|---|---|---|
| F-stat | p-value | Average QSM (ppb/mm3) | F-stat | p-value | Average QSM (ppb/mm3) | |
| QSM Reference Structure | ||||||
| Default MEDI Toolbox | 0.931 | 0.402 | 87.25 | 16.89 | <0.0001* | 3.55 |
| Lateral Ventricles | 0.366 | 0.696 | 76.67 | 19.51 | <0.0001* | 2.77 |
| Whole-brain WM | 0.155 | 0.857 | 104.41 | 0.766 | 0.471 | 10.9 |
| Marginal Means (average QSM, ppb/mm3): Cortical brain regions | ||||||
| QSM reference structure | ||||||
| Ringing artifact rating | Default MEDI Toolbox | Lateral ventricles | ||||
| Little-to-no ringing | 2.53 | 2.11 | ||||
| Moderate ringing | 3.32 | 2.40 | ||||
| Significant ringing | 5.9 | 4.63 | ||||
| Pairwise comparisons | ||||||
| QSM reference structure | ||||||
| Default MEDI Toolbox | Lateral ventricles | |||||
| Ringing artifact rating pair | SE | p-value | SE | p-value | ||
| Little-to-no ringing vs Moderate ringing | 0.535 | 0.380 | 0.437 | 0.883 | ||
| Moderate ringing vs Significant ringing | 0.626 | < 0.0001* | 0.511 | <0.0001* | ||
| Significant ringing vs Little-to-no ringing | 0.544 | <0.0001* | 0.443 | <0.0001* | ||
p < 0.01.
In sum, ringing artifacts, likely caused by participant head motion, appear to inflate QSM values in cortical brain regions when using both the default MEDI Toolbox CSF reference and the Ironsmith lateral ventricles CSF reference. QSM values in subcortical brain regions appear less affected by ringing artifacts. No significant correlations were observed between ringing artifact ratings and QSM values for QSM maps reconstructed using whole-brain WM as the QSM reference structure. However, we do not believe this result justifies the use of WM as a reference structure due to a number of problems with its use. Specific issues associated with the use of whole-brain WM as the QSM reference are described in section 3.5. “Possible uses of SNR in QSM data analyses” as well as in the Discussion.
3.4. Relationship between QSM and SNR
The purpose of this analysis was to determine if SNR values generated by Ironsmith are sensitive to the relationship between QSM values and Gibbs artifacts (i.e. motion-related artifacts discussed in the previous section). If so, it may be possible to use the SNR values outputted automatically by Ironsmith as a quality control measure. Linear regressions were conducted between QSM and SNR values from subcortical and cortical brain regions (in separate models) to evaluate their relationship. Results are summarized in Table 4.
Table 4.
Linear regression analyses: QSM and SNR for each QSM reference structure.
| QSM Reference Structure: Default MEDI Toolbox | ||||||
|---|---|---|---|---|---|---|
| Effect | 1 β | R2 | p-value | SE | 95% CI | |
| Subcortical QSM | −0.461 | 0.112 | 0.025* | 0.198 | −0.860 | −0.062 |
| Cortical QSM | −0.076 | 0.504 | < 0.0001** | 0.011 | −0.099 | −0.053 |
| QSM Reference Structure: Lateral Ventricles | ||||||
| Subcortical QSM | −0.393 | 0.088 | 0.048* | 0.193 | −0.782 | −0.004 |
| Cortical QSM | −0.058 | 0.475 | < 0.0001** | 0.009 | −0.077 | −0.039 |
| QSM Reference Structure: Whole-brain WM | ||||||
| Subcortical QSM | −0.368 | 0.079 | 0.062 | 0.192 | −0.755 | 0.019 |
| Cortical QSM | −0.069 | 0.175 | 0.005** | 0.023 | −0.115 | −0.022 |
Standardized coefficients.
p < 0.01.
p < 0.05.
In summary, negative correlations were observed between SNR and QSM values in both subcortical and cortical brain regions, irrespective of QSM reference structure. That is, lower SNR (either due to head motion or due to other sources of MRI noise) tends to be associated with higher QSM values. Cortical brain regions appear to be more susceptible to this effect.
3.5. Possible uses of SNR in QSM data analyses
Previous analyses indicated that Gibbs artifacts, likely driven by participant head motion, artificially inflate QSM values. Fortunately, SNR values outputted automatically by Ironsmith appear to be sensitive to this effect. As such, one way to improve QSM data analyses is to exclude participants with very low SNR values, as outliers.
An alternative approach could be to scale/adjust per-ROI QSM values using corresponding per-ROI SNR. One way to achieve this is to first normalize the per-ROI SNR values to vary between zero and one across participants: SNRsubj, o to 1 = where SNRMin is the lowest SNR in the participant sample and SNRMax is the highest SNR in the participant sample, for a given ROI. That is, participants with high SNR are assigned normalized SNR values closer to one and participants with low SNR are assigned normalized SNR values closer to zero on a per-ROI basis. The second step would be to multiply the normalized, per-ROI SNR values by the corresponding QSM values in each ROI. As a result, for each ROI, QSM values will be lowered for participants with low SNR, whereas QSM values for participants with high SNR will remain relatively unchanged.
To evaluate potential benefits of this proposed SNR-based correction, linear regressions between participant age and QSM values from subcortical and cortical brain regions were conducted with and without applying this correction. Participant age was used in these analyses as age-related increases in non-heme brain iron are well documented (Acosta-Cabronero et al., 2016; Bartzokis et al., 2011; Betts et al., 2016; Darki et al., 2016; Daugherty and Raz, 2015) and, as such, positive correlations between age and QSM values are expected. Participant gender was controlled for in these regressions. Lastly, separate analyses were conducted for each QSM map type (default MEDI Toolbox, lateral ventricle CSF and whole-brain WM as reference structures). Results from these analyses are summarized in Table 5 (gender results are omitted from Table 5 for brevity as gender was not a significant predictor in any of the linear regressions).
Table 5.
Linear regression analyses: participant age and QSM values from subcortical and cortical brain regions, with and without SNR-based correction.
| QSM Reference Structure: Default MEDI TooWox | ||||||
|---|---|---|---|---|---|---|
| Effect | 1 β | r2 | p-value | SE | 95% CI | |
| Subcortical QSM | 0.474 | 0.223 | 0.001** | 0.024 | 0.036 | 0.132 |
| Subcortical QSM (SNR corrected) | 0.346 | 0.119 | 0.019* | 0.057 | 0.024 | 0.255 |
| Cortical QSM | 0.088 | 0.008 | 0.562 | 0.394 | −0.564 | 1.025 |
| Cortical QSM (SNR corrected) | 0.315 | 0.100 | 0.032* | 0.013 | 0.003 | 0.056 |
| QSM Reference Structure: Lateral Ventricles | ||||||
| Subcortical QSM | 0.449 | 0.206 | 0.002** | 0.718 | 0.975 | 3.869 |
| Subcortical QSM (SNR corrected) | 0.368 | 0.135 | 0.012* | 0.202 | 0.123 | 0.937 |
| Cortical QSM | 0.045 | 0.002 | 0.762 | 0.045 | −0.077 | 0.105 |
| Cortical QSM (SNR corrected) | 0.330 | 0.110 | 0.024* | 0.010 | 0.003 | 0.042 |
| QSM Reference Structure: Whole-brain WM | ||||||
| Subcortical QSM | 0.432 | 0.189 | 0.003** | 0.710 | 0.840 | 3.703 |
| Subcortical QSM (SNR corrected) | 0.202 | 0.041 | 0.176 | 0.444 | −0.284 | 1.506 |
| Cortical QSM | 0.014 | 0.0002 | 0.925 | 0.089 | −0.171 | 0.188 |
| Cortical QSM (SNR corrected) | 0.043 | 0.002 | 0.776 | 0.065 | −0.112 | 0.150 |
Standardized coefficients.
p < 0.01.
p < 0.05.
In summary, cortical QSM values appear to benefit from the proposed SNR-based correction: participant age predicted QSM values in cortical brain regions for SNR corrected data only. Subcortical brain regions do not appear to benefit from this correction as participant age predicted QSM-based iron concentration in subcortical brain regions irrespective of whether the data were SNR corrected or not. Further, QSM maps reconstructed using either the default MEDI Toolbox CSF or the lateral ventricles CSF as reference structures appear to be best suited for extracting QSM values from cortical brain regions: participant age did not predict cortical QSM values from QSM maps reconstructed using whole-brain WM as the reference structure. Lastly, all QSM map types performed comparably for QSM data extracted from subcortical brain regions.
To extend and bolster the QSM vs age results presented in the previous analyses, additional regressions were conducted aimed at replicating two specific findings reported in previous literature. These analyses/results can be found in supplementary material section 3.5. (S) Possible uses of SNR in QSM data analyses.
3.6. Overlap of GM ROIs with phase image outlier regions
This analysis evaluates the percent overlap between the Ironsmith derived GM ROIs and outlier regions identified using the Ironsmith phase image quality control (QC) procedure. Across the 35 participants included in the analysis, five bilateral GM ROIs consistently overlapped with phase-image outlier regions: (1) the nucleus accumbens (18% overlap; SE = 9.4), (2) the amygdala (24% overlap; SE = 6.3), (3) the entorhinal cortex (43% overlap; SE = 4.5), (4) the medial orbitofrontal cortex (17% overlap; SE = 2.8) and (5) the parahippocampal gyrus (21% overlap; SE = 3.3). The percent overlap with outlier regions of these five ROIs did not correlate significantly with corresponding SNR: the lowest p-value was for the bilateral medial orbitofrontal cortex ROI (r = 0.26; p = 0.08; SE = 0.15; 95 CI = −0.62 to −0.04). Inspection of these outlier regions revealed no overlap with ringing artifacts, typically caused by participant head motion. Therefore, these phase image outlier regions appear to be distinct from those identified by the SNR QC procedure. Consequently, these ROIs will not be considered in the external validity analyses (section 3.7.) as they could be compromised.
3.7. External validity
This section assesses whether Ironsmith-derived QSM values in specific GM ROIs show expected relationships with specific behavioral measures. That is, are QSM results outputted by Ironsmith useful for research? As this section constitutes a benchmark of the Ironsmith pipeline, only Ironsmith-specific features are tested. Namely, the QSM values used in these analyses are not SNR corrected since Ironsmith does not output SNR corrected QSM values. Further, these analyses include QSM values extracted from QSM maps reconstructed using both the default MEDI Toolbox and Ironsmith-based CSF reference masks. Both the MEDI/Ironsmith CSF-based reference structures are included in order to elucidate which of these two might be better for cortical QSM based analyses.
The primary motor cortex was selected to test the external validity of Ironsmith because this region has a very well-established function—voluntary motor movement—and is known to undergo significant age-related iron accumulation (Betts et al., 2016; Buijs et al., 2016; Hallgren and Sourander, 1958). In addition, previous studies have demonstrated negative associations between iron concentration in the primary motor cortex of Parkinson’s and amyotrophic lateral sclerosis (ALS) patients and motor performance (Acosta-Cabronero et al., 2018; Costagli et al., 2016; Schweitzer et al., 2015). Linear regression analyses were conducted to evaluate whether QSM values extracted from primary motor cortex (precentral gyrus) are associated with worse performance on Trails-A. In addition, since 33 out of the 35 participants included in the analyses were right handed, QSM values extracted from the left precentral gyrus were expected to be more predictive of Trails-A performance compared to those extracted from the right precentral gyrus.
Ironsmith-derived QSM values were also extracted from two control GM ROIs to test the anatomical specificity between Ironsmith-derived QSM values and Trails-A performance. The control regions were selected on the basis of being anatomically proximal to the precentral gyrus but not being primarily involved in voluntary motor function; namely, the superior frontal gyrus (anterior to precentral gyrus) and superior parietal cortex (single ROI corresponding to the superior and inferior parietal lobule parcellation; posterior to precentral gyrus). Separate linear regressions were conducted for left and right hemisphere ROIs to avoid issues of multicollinearity. Age and gender were added as covariates in the regression model. Results are summarized in Table 6.
Table 6.
Linear regression: QSM values from superior frontal, precentral and superior parietal GM ROIs and Trails-A performance.
| QSM Reference Structure: Default MEDI Toolbox | |||||||
|---|---|---|---|---|---|---|---|
| Left hemisphere | |||||||
| Effect | 1 β | r2 | p-value | SE | 95% CI | VIF | |
| L Superior frontal | −0.292 | 0.052 | 0.207 | 1.781 | −5.932 | 1.341 | 2.232 |
| L Precentral | 0.504 | 0.120 | 0.052 | 0.820 | −0.019 | 3.329 | 2.707 |
| L Superior parietal | −0.381 | 0.091 | 0.093 | 1.127 | −4.255 | 0.348 | 2.105 |
| Age | 0.371 | 0.143 | 0.033* | 0.278 | 0.055 | 1.190 | 1.197 |
| Gender | 0.042 | 0.002 | 0.804 | 2.949 | −5.286 | 6.789 | 1.230 |
| Right hemisphere | |||||||
| Effect | 1 β | r2 | p-value | SE | 95% CI | VIF | |
| R Superior frontal | −0.077 | 0.003 | 0.759 | 1.791 | −4.213 | 3.102 | 2.573 |
| R Precentral | 0.206 | 0.026 | 0.377 | 0.948 | −1.085 | 2.786 | 2.171 |
| R Superior parietal | −0.359 | 0.070 | 0.142 | 1.398 | −4.965 | 0.748 | 2.334 |
| Age | 0.392 | 0.151 | 0.028* | 0.284 | 0.076 | 1.238 | 1.189 |
| Gender | −0.123 | 0.018 | 0.468 | 2.948 | −8.187 | 3.854 | 1.164 |
| QSM Reference Structure: Lateral Ventricles | |||||||
| Left hemisphere | |||||||
| Effect | 1 β | r2 | p-value | SE | 95% CI | VIF | |
| L Superior frontal | −0.294 | 0.045 | 0.242 | 2.797 | −9.046 | 2.376 | 2.577 |
| L Precentral | 0.459 | 0.125 | 0.047* | 1.378 | 0.045 | 5.673 | 2.078 |
| L Superior parietal | −0.212 | 0.030 | 0.342 | 1.752 | −5.270 | 1.887 | 2.039 |
| Age | 0.341 | 0.113 | 0.060 | 0.292 | −0.025 | 1.166 | 1.284 |
| Gender | 0.029 | 0.0004 | 0.866 | 2.973 | −5.565 | 6.576 | 1.217 |
| Right hemisphere | |||||||
| Effect | 1 β | r2 | p-value | SE | 95% CI | VIF | |
| R Superior frontal | −0.026 | 0.0003 | 0.924 | 2.835 | −6.061 | 5.519 | 2.915 |
| R Precentral | 0.161 | 0.015 | 0.502 | 1.733 | −2.362 | 4.717 | 2.136 |
| R Superior parietal | −0.249 | 0.034 | 0.315 | 1.957 | −5.995 | 1.998 | 2.268 |
| Age | 0.386 | 0.141 | 0.034* | 0.291 | 0.052 | 1.240 | 1.150 |
| Gender | −0.137 | 0.02 | 0.435 | 3.043 | −8.626 | 3.805 | 1.148 |
Standardized coefficients.
p < 0.05.
In summary, in a model including multiple ROIs, only QSM values extracted from the left precentral gyrus correlated negatively with performance on Trails-A (total time to complete the task). In contrast, iron concentration extracted from five control regions (right precentral gyrus, left and right superior frontal regions and left and right superior parietal regions) did not predict performance on Trails-A. Lastly, results obtained using the default MEDI toolbox and Ironsmith-based CSF reference structures were largely comparable. However, the correlation between QSM values extracted from the left precentral gyrus and Trails-A performance was only marginally significant for QSM maps reconstructed using the default MEDI Toolbox CSF reference structure.
4. Discussion
Ironsmith is a comprehensive, open-source, fully automated pipeline for creating QSM maps and extracting QSM-based iron concentrations from subcortical and cortical brain regions. Ironsmith also provides several useful features such as per-participant CSF-only QSM reference structure masks that minimize partial volume effects, and quality control evaluation of QSM data including (1) per-ROI SNR measures, (2) automatic filtering of outlier QSM values and (3) automatic identification of phase-image outlier regions. Testing of Ironsmith revealed excellent intra- and inter-rater reliability (ICC = 1) and established external validity: in accord with previous studies, QSM values extracted using Iron-smith from both subcortical and cortical brain regions correlated positively with participant age. Further, consistent with previous studies, QSM values extracted from primary motor cortex, but not from control regions, were correlated negatively with motor task performance.
4.1. Ironsmith automates QSM map reconstruction and extraction of ROI-based QSM values
As discussed in the Introduction, at present, few if any software packages exist that offer a single, fully automated pipeline for reconstructing QSM maps and extracting QSM-based data from anatomical ROIs. Importantly, to our knowledge, no QSM-based software exist that offer quality control measures and/or procedures for evaluating the QSM outputs. In response to this gap in QSM research, we developed Iron-smith, which is a comprehensive, open-source, fully automated pipeline for QSM-based data analyses. Ironsmith is designed to process multiple participants in parallel, autonomously with minimal user input and/or supervision and requires minimal programming skills to operate. Iron-smith also offers a series of quality control measures and solutions to issues commonly encountered during QSM analyses. For example, Iron-smith provides per-ROI SNR measures, can automatically identify and report phase-image outlier regions, can automatically filter per-ROI outlier values and offers highly accurate CSF only, QSM reference structure masks that reduce partial volume effects and unwanted overlap with parenchyma (Fig. 2).
4.2. Ironsmith is highly reliable
Testing of Ironsmith revealed excellent intra-rater reliability (ICC = 1). Additional inter-rater reliability analyses yielded equally high ICC values, demonstrating the turn-key nature of the pipeline. These results verify that the Ironsmith pipeline is deterministic and produces replicable results, irrespective of user, which is a significant strength. This high degree of reliability stems from several architectural features of Ironsmith. For example, Ironsmith makes use of well-established and reliable third party software programs (e.g. Freesurfer, AFNI, FSL and MEDI Toolbox). Importantly, the pipelines within Ironsmith that utilize these third party software are all deterministic and will always produce the same output given the same input. Additionally, Ironsmith minimizes human error by performing all preprocessing steps (skull stripping, registration, MEDI Toolbox script generation etc.), and by performing quality controls for each main processing step, automatically without the need for user-intervention. Lastly, the main inputs to Ironsmith are MRI file locations which are evaluated by Ironsmith automatically before it runs. As such, Ironsmith will either run an analysis deterministically if it finds all the appropriate files at the specified file locations, or it will not run and will output specific and detailed error messages.
4.3. Ironsmith computes ROI-based SNR values for assessing the quality of QSM data
ROI-based SNR values provided by Ironsmith closely correspond with visual ratings of ringing artifacts found to be related to participant head motion. As such, these per-ROI SNR values provided by Ironsmith can be used to assess the quality of QSM data. Evaluating the quality of QSM data proved important during testing because the severity of ringing artifacts correlated positively with QSM values in both subcortical and cortical brain regions. Specifically, participant head motion was found to artificially inflate QSM values. Notably, cortical GM QSM values were more affected by participant head motion than subcortical GM QSM values. This latter finding is consistent with previous MRI findings in non-QSM studies, showing that participant head motion more severely affects MR signal near the surface of the brain than near the center of the brain (Baum et al., 2018; Iglesias et al., 2017; Reuter et al., 2015).
In addition to offering quality control assessment, Ironsmith can be used to filter out ROIs or participants with excessive head motion as outliers, specifically those participants with low SNR values. Alternatively, SNR-based corrections may also be applied to the QSM data. One possible SNR-based correction method was explored and showed promising results in the regression analyses between participant age and QSM values extracted from cortical GM regions. This analysis focused on age due to well documented age-related increases in non-heme brain iron (Acosta-Cabronero et al., 2016; Bartzokis et al., 2011; Betts et al., 2016; Darki et al., 2016; Daugherty et al., 2015). Results indicated that SNR-corrected QSM values in cortex were correlated with age, whereas QSM values in cortex not corrected for SNR were not correlated with age. For subcortical brain structures, participant age predicted QSM values irrespective of SNR correction. This latter finding is in line with the asymmetric impact of participant head motion between QSM values extracted from subcortical and cortical brain regions. It should be noted that the SNR-based correction we propose is optional and is not a feature offered with Ironsmith (Ironsmith does not output SNR corrected QSM values). Further, this SNR-based correction is most useful when low SNR participants are present and cannot all be excluded as outliers. If few participants have low SNR values or the range of SNR values is small then the SNR-based correction will not be useful. In this situation, it might be preferable to exclude low SNR participants from analyses as outliers.
4.4. Ironsmith can filter out per-ROI outlier QSM values
As demonstrated in section 2.3. (Architecture), outlier QSM voxels can occur in GM. These outlier voxels have very large QSM values typical of those seen in veins. Indeed, these very large QSM values in GM appear to relate to partial overlap between a GM ROI and large veins (Fig. 3A). Ironsmith can filter-out the majority of these outlier voxels, per ROI, by excluding QSM voxels with values larger than the 97th percentile of all positive QSM values within the ROI. The percentile cutoff point for outliers can be modified by manually editing the header of the 05_Extract_QSM.sh script file under the Ironsmith installation folder. Automatic selection of outlier cutoff points for subcortical/cortical GM may be a feature that can be included in subsequent versions of Ironsmith.
4.5. Ironsmith can identify outlier regions on phase images
As demonstrated in section 4.3., per ROI SNR values can be used to evaluate the quality of QSM data. However, this SNR-based metric may not be entirely representative of the quality of a QSM map as it only depends on GRE magnitude images. For this reason, artifacts that manifest predominately on GRE phase images may not be detected. To address this issue, Ironsmith offers a phase image QC procedure that can identify outlier regions on an RDF image, (an unwrapped phase image with the background field removed). Phase-image specific artifacts, such as heavy streaking, open-ended fringe lines, and missed phase unwraps tend to be more evident on RDF images than raw phase images. These artifacts can then be detected as outlier regions. A MAD-based outlier detection procedure was selected for Ironsmith since in the presence of outliers, MAD is the most robust measure of dispersion (Leys et al., 2013). As indicated in section 3.6. Overlap of GM ROIs with phase image outlier regions, Ironsmith detected outlier regions in our participant cohort. Importantly, these phase image outlier regions did not correlate with corresponding SNR and did not appear to overlap with ringing artifacts. As such, the phase image outlier regions appear to represent sources of noise or artifacts that are distinct from those identified by the SNR QC procedure. Consequently, we advise users to exclude such phase outlier ROIs from analyses.
4.6. Ironsmith demonstrates external validity
Consistent with previous studies, positive correlations were observed between age and Ironsmith-derived QSM values extracted from both subcortical and cortical brain regions (Acosta-Cabronero et al., 2016; Betts et al., 2016; Darki et al., 2016). In addition, negative correlations were observed between QSM values extracted from precentral gyrus (primary motor cortex) and motor task performance. Specifically, Ironsmith-derived QSM values extracted from the left precentral gyrus correlated negatively with performance on Trails-A (total time to complete the task). Conversely, neither Ironsmith-derived QSM values extracted from the right precentral gyrus, nor those extracted from control regions not primarily devoted to voluntary motor movement (superior frontal and superior parietal regions), were found to predict performance on Trails-A.
The negative association between precentral gyrus QSM and motor task performance is consistent with non-heme iron being a potent oxidizer that can contribute to oxidative stress, interfere with neurotransmission and lead to neurodegeneration (Becerril-Ortega et al., 2014; Hare and Double, 2016; Ke and Qian, 2007; Matak et al., 2016; Moos et al., 2007; Zecca et al., 2004). As such, higher iron concentration in primary motor cortex is expected to interfere with motor function (e.g. Acosta-Cabronero et al., 2018; Costagli et al., 2016; Schweitzer et al., 2015). Moreover, our findings show that Ironsmith-derived QSM values demonstrate the expected lateralization effects in that higher iron concentration in left primary motor cortex is expected to be more predictive of Trails-A performance in a sample of predominantly right-handed participants (33 out of 35 participants were right handed), which is what we found. Overall, these findings provide evidence of external validity for the Ironsmith pipeline.
4.7. QSM reference structures offered by ironsmith
Ironsmith offers users the ability to view and evaluate results computed using three different QSM reference structures. Our intention in outputting QSM results from different reference structures is not to encourage users to pick-and-choose the results they ‘like best’. Instead, our intention is just the opposite: to facilitate discussion and further research on potential strengths and weakness between different QSM reference structures and how these could be applied in different QSM-based analyses. Such discussions can aid the adoption of common reference structures across the field of QSM research and thus enhance replication of scientific findings.
Our present findings suggest that CSF-based reference structures (average susceptibility within CSF reference masks; MEDI default or Iron-smith generated) appear to be more appropriate than whole-brain WM reference structures (average susceptibility within the whole-brain WM reference mask, generated by Ironsmith), at least for QSM analyses of cortical brain regions. Specifically, we observed that in linear regression analyses between participant age and QSM, age did not predict susceptibility in cortical GM when whole-brain WM was used as the QSM reference structure, irrespective of SNR-based correction. We suspect shadow artifacts may be contributing to this null finding. As noted in section 2.3.9 (05_Extract_QSM), shadow artifacts are induced by anisotropic WM bordering GM, which contributes to a dipole incompatible field (Kee et al., 2017). When CSF is the reference structure, shadow artifacts manifest as strongly negative QSM values within GM due to the diamagnetic effect of myelin on susceptibility, relative to CSF (e.g., Hametner et al., 2018; Wisnieff et al., 2015). To account for these artifacts, Ironsmith eliminates all negative QSM values from ROI averages under the assumption that these are likely associated with diamagnetic myelin susceptibility. However, relative to whole-brain WM as the reference structure, GM voxels affected by shadow artifacts are positive (i.e. the averaged GM-WM susceptibility in the shadow is slightly less negative than susceptibility in pure WM1). For this reason, Ironsmith cannot remove the shadow artifact from QSM averages when whole-brain WM is the QSM reference and these artifacts may interfere with analyses.
For this reason, as suggested in previous studies (e.g. Straub et al., 2017), we also recommend the use of CSF-based reference structures over WM reference structures, at least until QSM analysis programs evolve viable analytic solutions to deal with shadow artifacts. Further, it should be noted that the use of whole-brain WM as the QSM reference can be problematic for a number of additional reasons. For example, WM health changes with age and with certain diseases both of which can impact magnetic susceptibility. For instance, small vessel disease is associated with white matter hyper-intensities (Pantoni and Garcia, 1995; Wardlaw et al., 2013; Wiseman et al., 2015). As such, by using whole-brain WM as the QSM reference, hyper-intense and non-hyper-intense (abnormal vs normal) WM are implied to have the same magnetic susceptibility which is not the case.
Considering which specific CSF-based reference structures to use, every effort should be made to ensure minimal overlap with parenchema. Toward that end, Ironsmith offers an automated solution to this issue by generating participant specific, eroded lateral ventricles CSF masks that do not overlap with parenchema. These CSF-only reference masks are used by default in the Ironsmith pipeline.
4.8. Limitations of the ironsmith pipeline
Certain limitations of the Ironsmith pipeline should be acknowledged: The SNR values provided by Ironsmith appear to be useful in identifying sources of noise (as described in section 4.3.) but should not be considered a gold-standard SNR measure for parallel imaging MRI (e.g. SENSE, GRAPPA). For the SNR calculations, Ironsmith assumes a Rician noise distribution outside the head which is not the case in parallel image acquisitions. For gold-standard SNR measurements in parallel imaging, at least two approaches have been proposed. These two approaches involve either (1) collection of at least two sets of GRE scans (Goerner and Clarke, 2011) or (2) calculation of the noise amplification factor (g-factor) directly from the reconstruction weights (Breuer et al., 2009). Neither of these approaches are implemented in Ironsmith. The first approach (1) requires users to collect at least two identical QSM scans in direct succession within the same scan session. This may not be practical for many studies in which a full battery of additional, non-QSM sequences must be collected and time-in-scanner is a consideration. The second approach (2) involves a methodologically intensive procedure (Breuer et al., 2009) requiring access to raw scanner data and sensitivity profiles, which may not be easily available/interpretable by users conducting clinical as opposed to methodological studies. In the absence of these gold-standard measures of SNR, our results suggest that Ironsmith-derived SNR values appear to be a useful general metric of multiple sources of noise, including head motion, which may aid in the identification of data sets requiring further inspection.
Additionally, users should be aware that Ironsmith has only been tested with GRE data acquired with a Siemens 3T PRISMA scanner (software version MR VE11C). Although, Ironsmith should function correctly with data acquired with other MRI scanner manufacturers (e.g. Philips, GE), this functionality has not been tested. Lastly, the test-retest reliability of Ironsmith has not been evaluated in the current study. Both of these limitations will be addressed in the near future and the Ironsmith GitHub website will be updated accordingly.
5. Conclusions
Ironsmith is a comprehensive, fully automated, reliable and valid software pipeline for QSM data analyses. Ironsmith is designed to process single or groups of participants autonomously with minimal user input and/or supervision, does not depend on user scripts to run analyses and requires little to no technical/programming knowledge to setup and use. Additionally, Ironsmith provides a number of quality control measures and analytic solutions not typically offered by QSM toolboxes, such as per-ROI SNR measures, automatic identification of outlier regions on phase images, filtering of per-ROI outlier QSM values and a precise, CSF-only reference region for QSM reconstruction. Lastly, Iron-smith is completely open-source and thus free for anyone to use and/or modify. These features allow robust and replicable, user-friendly access to QSM data analyses for researchers who are interested in including QSM in their studies.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (grant numbers NIA R01 AG055449, NIA R01 AG068055, NIA P30 AG072946 and NIGMS S10 OD023573). The content is solely the responsibility of the authors and does not necessarily represent the official views of these granting agencies. The authors thank Shoshana Bardach for help with participant recruitment and Beverly Meacham and Eric Foreman for assisting/conducting the MRI scans. We also thank Drs. Xu Li, Hanzhang Lu, Anders Anderson, Ahmed Bahrani, Pascal Spincemaille and Yi Wang for helpful discussions.
Appendix
Appendix A
L_ = Left hemisphere
R_ = Right hemisphere
LR_ = Bilateral
_GM = Gray matter
LR_Frontal_Lobe_GM
LR_Parietal_Lobe_GM
LR_Occipital_Lobe_GM
LR_Temporal_Lobe_GM
L_CaudalAnteriorCingulate_GM
L_CaudalMiddleFrontal_GM
L_Cuneus_GM
L_DLPFC_GM
L_Entorhinal_GM
L_Frontal_GM
L_Fusiform_GM
L_InferiorParietal_GM
L_AngularGyrus_GM
L_InferiorTemporal_GM
L_Insula_GM
L_IsthmusCingulate_GM
L_LateralOccipital_GM
L_LateralOrbitofrontal_GM
L_Lingual_GM
L_MedialOrbitofrontal_GM
L_MiddleTemporal_GM
L_Occipital_GM_Mask
L_Parietal_GM_Mask
L_Temporal_GM_Mask
L_Parahippocampal_GM
L_Pericalcarine_GM
L_Postcentral_GM
L_PosteriorCingulate_GM
L_Precentral_GM
L_Precuneus_GM
L_RostralMiddleFrontal_GM
L_RostralAnteriorCingulate_GM
L_SuperiorFrontal_GM
L_SuperiorParietal_GM
L_SuperiorTemporal_GM
L_TransverseTemporal_GM
R_CaudalAnteriorCingulate_GM
R_CaudalMiddleFrontal_GM
R_Cuneus_GM
R_DLPFC_GM
R_Entorhinal_GM
R_Frontal_GM_Mask
R_Fusiform_GM
R_InferiorParietal_GM
R_AngularGyrus_GM
R_InferiorTemporal_GM
R_Insula_GM
R_IsthmusCingulate_GM
R_LateralOccipital_GM
R_LateralOrbitofrontal_GM
R_Lingual_GM
R_MedialOrbitofrontal_GM
R_MiddleTemporal_GM
R_Occipital_GM_Mask
R_Parietal_GM_Mask
R_Temporal_GM_Mask
R_Parahippocampal_GM
R_Pericalcarine_GM
R_Postcentral_GM
R_PosteriorCingulate_GM
R_Precentral_GM
R_Precuneus_GM
R_RostralMiddleFrontal_GM
R_RostralAnteriorCingulate_GM
R_SuperiorFrontal_GM
R_SuperiorParietal_GM
R_SuperiorTemporal_GM
R_TransverseTemporal_GM
LR_Accumbens_area
LR_Amygdala
LR_Caudate
LR_Hipp
LR_Pallidum
LR_Putamen
LR_Thalamus_Proper
L_Accumbens_area
L_Amygdala
L_Caudate
L_Hipp
L_Pallidum
L_Putamen
L_Thalamus_Proper
R_Accumbens_area
R_Amygdala
R_Caudate
R_Hipp
R_Pallidum
R_Putamen
R_Thalamus_Proper
Appendix B


Footnotes
Declaration of Competing Interest
The authors declare no competing financial interests.
Credit authorship contribution statement
Valentinos Zachariou: Conceptualization, Methodology, Software, Validation, Formal analysis, Data curation, Writing – original draft, Writing – review & editing, Visualization. Christopher E. Bauer: Validation, Formal analysis, Data curation, Writing – review & editing. David K. Powell: Methodology, Resources. Brian T. Gold: Conceptualization, Methodology, Validation, Formal analysis, Data curation, Writing – review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neuroimage.2021.118835
In Zachariou et al. 2020 we demonstrated that all negative QSM voxels in cortical GM with lateral ventricles CSF as the reference structure become positive when whole-brain WM is used as the QSM reference.
References
- A. Schmitt F, T. Nelson P, Abner E, Scheff S, A. Jicha G, Smith C, Cooper G, Mendiondo M, D. Danner D, J. Van Eldik L, Caban-Holt A, A. Lovell M, J. Kryscio R, 2012. University of Kentucky Sanders-Brown healthy brain aging volunteers: donor characteristics, procedures and neuropathology. Curr. Alzheimer Res 9, 724–733. doi: 10.2174/156720512801322591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta-Cabronero J, Betts MJ, Cardenas-Blanco A, Yang S, Nestor PJ, 2016. In vivo MRI mapping of brain iron deposition across the adult lifespan. J. Neurosci 36, 364–374. doi: 10.1523/JNEUROSCI.1907-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta-Cabronero J, Machts J, Schreiber S, Abdulla S, Kollewe K, Petri S, Spotorno N, Kaufmann J, Heinze H-J, Dengler R, Vielhaber S, Nestor PJ, 2018. Quantitative susceptibility MRI to detect brain iron in amyotrophic lateral sclerosis. Radiology 289, 195–203. doi: 10.1148/radiol.2018180112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta-Cabronero J, Williams GB, Cardenas-Blanco A, Arnold RJ, Lupson V, Nestor PJ, 2013. In vivo quantitative susceptibility mapping (QSM) in Alzheimer’s disease. PLoS ONE 8. doi: 10.1371/journal.pone.0081093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandt SK, de Rochefort L, Chen W, Dimov AV, Spincemaille P, Kopell BH, Gupta A, Wang Y, 2019. Clinical integration of quantitative susceptibility mapping magnetic resonance imaging into neurosurgical practice. World Neurosurg. 122, e10–e19. doi: 10.1016/J.WNEU.2018.08.213. [DOI] [PubMed] [Google Scholar]
- Bartzokis G, Lu PH, Tingus K, Peters DG, Amar CP, Tishler TA, Finn JP, Villablanca P, Altshuler LL, Mintz J, Neely E, Connor JR, 2011. Gender and iron genes may modify associations between brain iron and memory in healthy aging. Neuropsychopharmacology 36, 1375–1384. doi: 10.1038/npp.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum GL, Roalf DR, Cook PA, Ciric R, Rosen AFG, Xia C, Elliott MA, Ruparel K, Verma R, Tunç B, Gur RC, Gur RE, Bassett DS, Satterthwaite TD, 2018. The impact of in-scanner head motion on structural connectivity derived from diffusion MRI. Neuroimage 173, 275–286. doi: 10.1016/j.neuroimage.2018.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerril-Ortega J, Bordji K, Fréret T, Rush T, Buisson A, 2014. Iron overload accelerates neuronal amyloid-β production and cognitive impairment in transgenic mice model of Alzheimer’s disease. Neurobiol. Aging 35, 2288–2301. doi: 10.1016/j.neurobiolaging.2014.04.019. [DOI] [PubMed] [Google Scholar]
- Bernstein MA, Grgic M, Brosnan TJ, Pelc NJ, 1994. Reconstructions of phase contrast, phased array multicoil data. Magn. Reson. Med 32, 330–334. doi: 10.1002/MRM.1910320308. [DOI] [PubMed] [Google Scholar]
- Betts MJ, Acosta-Cabronero J, Cardenas-Blanco A, Nestor PJ, Düzel E, 2016. High-resolution characterisation of the aging brain using simultaneous quantitative susceptibility mapping (QSM) and R2* measurements at 7 T. Neuroimage 138, 43–63. doi: 10.1016/j.neuroimage.2016.05.024. [DOI] [PubMed] [Google Scholar]
- Bilgic B, Chatnuntawech I, Langkammer C, Setsompop K, 2015. Sparse methods for Quantitative Susceptibility Mapping. In: Papadakis M, Goyal VK, Van De Ville D (Eds.), Wavelets and Sparsity XVI. SPIE; doi: 10.1117/12.2188535. [DOI] [Google Scholar]
- Breuer FA, Kannengiesser SAR, Blaimer M, Seiberlich N, Jakob PM, Griswold MA, 2009. General formulation for quantitative G-factor calculation in GRAPPA reconstructions. Magn. Reson. Med 62, 739–746. doi: 10.1002/MRM.22066. [DOI] [PubMed] [Google Scholar]
- Buijs M, Doan NT, Van Rooden S, Versluis MJ, Van Lew B, Milles J, Van Der Grond J, Van Buchem MA, 2016. In vivo assessment of iron content of the cerebral cortex in healthy aging using 7-Tesla T2* -weighted phase imaging. 10.1016/j.neurobiolaging.2016.09.005 [DOI] [PubMed] [Google Scholar]
- Chan KS, Marques JP, 2021. SEPIA—Susceptibility mapping pipeline tool for phase images. Neuroimage 227, 117611. doi: 10.1016/j.neuroimage.2020.117611. [DOI] [PubMed] [Google Scholar]
- Chen L, Hua J, Ross CA, Cai S, van Zijl PCM, Li X, 2019. Altered brain iron content and deposition rate in Huntington’s disease as indicated by quantitative susceptibility MRI. J. Neurosci. Res 97, 467–479. doi: 10.1002/jnr.24358 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costagli M, Donatelli G, Biagi L, Caldarazzo Ienco E, Siciliano G, Tosetti M, Cosottini M, 2016. Magnetic susceptibility in the deep layers of the primary motor cortex in amyotrophic lateral sclerosis. NeuroImage Clin. 12, 965–969. doi: 10.1016/j.nicl.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW, 1996. AFNI: Software for Analysis and Visualization of Functional Magnetic Resonance Neuroimages. COMPUTERS AND BIOMEDICAL RESEARCH. [DOI] [PubMed] [Google Scholar]
- Czervionke LF, Czervionke JM, Daniels DL, Hauhgton VM, 1988. Characteristic features of MR truncation artifacts. Am. J. Roentgenol 151, 1219–1228. doi: 10.2214/ajr.151.6.1219. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI, 1999. Cortical surface-based analysis: I. Segmentation and surface reconstruction. Neuroimage 9, 179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Darki F, Nemmi F, Möller A, Sitnikov R, Klingberg T, 2016. Quantitative susceptibility mapping of striatum in children and adults, and its association with working memory performance. Neuroimage 136, 208–214. doi: 10.1016/j.neuroimage.2016.04.065. [DOI] [PubMed] [Google Scholar]
- Daugherty AM, Haacke EM, Raz N, 2015. Striatal iron content predicts its shrinkage and changes in verbal working memory after two years in healthy adults. J. Neurosci 35, 6731–6743. doi: 10.1523/JNEUROSCI.4717-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty AM, Raz N, 2015. Appraising the role of iron in brain aging and cognition: promises and limitations of MRI methods. Neuropsychol. Rev doi: 10.1007/s11065-015-9292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rochefort L, Liu T, Kressler B, Liu J, Spincemaille P, Lebon V, Wu J, Wang Y, 2010. Quantitative susceptibility map reconstruction from MR phase data using bayesian regularization: validation and application to brain imaging. Magn. Reson. Med 63, 194–206. doi: 10.1002/mrm.22187. [DOI] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ, 2006. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31, 968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Eckstein K, Dymerska B, Bachrata B, Bogner W, Poljanc K, Trattnig S, Robinson SD, 2018. Computationally efficient combination of multi-channel phase data from multi-echo acquisitions (ASPIRE). Magn. Reson. Med 79, 2996–3006. doi: 10.1002/mrm.26963. [DOI] [PubMed] [Google Scholar]
- Edelstein WA, Bottomley PA, Pfeifer LM, 1984. A signal-to-noise calibration procedure for NMR imaging systems. Med. Phys 11, 180–185. doi: 10.1118/1.595484. [DOI] [PubMed] [Google Scholar]
- Fan X, Liu X, Yan L, Mok VCT, Li K, 2020. Assessment of brain iron accumulation in Alzheimer’s disease with quantitative susceptibility mapping. Alzheimer’s Dement 16, e038799. doi: 10.1002/alz.038799. [DOI] [Google Scholar]
- Gibbs JW, 1899. Fourier’s series [3]. Nature doi: 10.1038/059606a0. [DOI] [Google Scholar]
- Goerner FL, Clarke GD, 2011. Measuring signal-to-noise ratio in partially parallel imaging MRI. Med. Phys 38, 5049–5057. doi: 10.1118/1.3618730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabner G, Janke AL, Budge MM, Smith D, Pruessner J, Collins DL, 2006. Symmetric atlasing and model based segmentation: an application to the hippocampus in older adults. In: Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). Springer Verlag, pp. 58–66. doi: 10.1007/11866763_8. [DOI] [PubMed] [Google Scholar]
- Hallgren B, Sourander P, 1958. The effect of age on the non-haemin iron in the human brain. J. Neurochem 3, 41–51. doi: 10.1111/j.1471-4159.1958.tb12607.x. [DOI] [PubMed] [Google Scholar]
- Hametner S, Endmayr V, Deistung A, Palmrich P, Prihoda M, Haimburger E, Menard C, Feng X, Haider T, Leisser M, Köck U, Kaider A, Höftberger R, Robinson S, Reichenbach JR, Lassmann H, Traxler H, Trattnig S, Grabner G, 2018. The influence of brain iron and myelin on magnetic susceptibility and effective transverse relaxation - a biochemical and histological validation study. Neuroimage 179, 117–133. doi: 10.1016/j.neuroimage.2018.06.007. [DOI] [PubMed] [Google Scholar]
- Hare DJ, Double KL, 2016. Iron and dopamine: a toxic couple. Brain 139, 1026–1035. doi: 10.1093/brain/aww022. [DOI] [PubMed] [Google Scholar]
- He N, Ling H, Ding B, Huang J, Zhang Y, Zhang Z, Liu C, Chen K, Yan F, 2015. Region-specific disturbed iron distribution in early idiopathic Parkinson’s disease measured by quantitative susceptibility mapping. Hum. Brain Mapp 36, 4407–4420. doi: 10.1002/hbm.22928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias JE, Lerma-Usabiaga G, Garcia-Peraza-Herrera LC, Martinez S, Paz-Alonso PM, 2017. Retrospective head motion estimation in structural brain MRI with 3D CNNs. In: Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). Springer Verlag, pp. 314–322. doi: 10.1007/978-3-319-66185-8_36. [DOI] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM, 2012. FSL. Neuroimage 62, 782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Kames C, Wiggermann V, Rauscher A, 2018. Rapid two-step dipole inversion for susceptibility mapping with sparsity priors. Neuroimage 167, 276–283. doi: 10.1016/J.NEUROIMAGE.2017.11.018. [DOI] [PubMed] [Google Scholar]
- Ke Y, Qian ZM, 2007. Brain iron metabolism: neurobiology and neurochemistry. Prog. Neurobiol doi: 10.1016/j.pneurobio.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Kee Y, Liu Z, Zhou L, Dimov A, Cho J, De Rochefort L, Seo JK, Wang Y, 2017. Quantitative susceptibility mapping (qsm) algorithms: mathematical rationale and computational implementations. IEEE Trans. Biomed. Eng 64, 2531–2545. doi: 10.1109/TBME.2017.2749298. [DOI] [PubMed] [Google Scholar]
- KP P, M W, MB S, P B, 1999. SENSE: sensitivity encoding for fast MRI. Magn. Reson. Med 42, 952–962. doi: . [DOI] [PubMed] [Google Scholar]
- Langkammer C, Schweser F, Krebs N, Deistung A, Goessler W, Scheurer E, Sommer K, Reishofer G, Yen K, Fazekas F, Ropele S, Reichenbach JR, 2012. Quantitative susceptibility mapping (QSM) as a means to measure brain iron? A post mortem validation study. Neuroimage 62, 1593–1599. doi: 10.1016/j.neuroimage.2012.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffer RB (1992). Iron and Human Disease (1st ed.). CRC Press. 10.1201/9781351073899. [DOI] [Google Scholar]
- Leys C, Ley C, Klein O, Bernard P, Licata L, 2013. Detecting outliers: do not use standard deviation around the mean, use absolute deviation around the median. J. Exp. Soc. Psychol 49, 764–766. doi: 10.1016/J.JESP.2013.03.013. [DOI] [Google Scholar]
- Li X, Chen L, Kutten K, Ceritoglu C, Li Y, Kang N, Hsu JT, Qiao Y, Wei H, Liu C, Miller MI, Mori S, Yousem DM, van Zijl PCM, Faria AV, 2019. Multi-atlas tool for automated segmentation of brain gray matter nuclei and quantification of their magnetic susceptibility. Neuroimage 191, 337–349. doi: 10.1016/j.neuroimage.2019.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Morgan PS, Ashburner J, Smith J, Rorden C, 2016. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J. Neurosci. Methods 264, 47–56. doi: 10.1016/j.jneumeth.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Liu C, Li W, Tong KA, Yeom KW, Kuzminski S, 2015. Susceptibility-weighted imaging and quantitative susceptibility mapping in the brain. J. Magn. Reson. Imaging 42, 23–41. doi: 10.1002/jmri.24768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Liu T, de Rochefort L, Ledoux J, Khalidov I, Chen W, Tsiouris AJ, Wisnieff C, Spincemaille P, Prince MR, Wang Y, 2012. Morphology enabled dipole inversion for quantitative susceptibility mapping using structural consistency between the magnitude image and the susceptibility map. Neuroimage 59, 2560–2568. doi: 10.1016/j.neuroimage.2011.08.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Khalidov I, de Rochefort L, Spincemaille P, Liu J, Tsiouris AJ, Wang Y, 2011a. A novel background field removal method for MRI using projection onto dipole fields (PDF). NMR Biomed. 24, 1129–1136. doi: 10.1002/nbm.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Liu J, de Rochefort L, Spincemaille P, Khalidov I, Ledoux JR, Wang Y, 2011b. Morphology enabled dipole inversion (MEDI) from a single-angle acquisition: comparison with COSMOS in human brain imaging. Magn. Reson. Med 66, 777–783. doi: 10.1002/mrm.22816. [DOI] [PubMed] [Google Scholar]
- Liu T, Spincemaille P, de Rochefort L, Kressler B, Wang Y, 2009. Calculation of susceptibility through multiple orientation sampling (COSMOS): a method for conditioning the inverse problem from measured magnetic field map to susceptibility source image in MRI. Magn. Reson. Med 61, 196–204. doi: 10.1002/mrm.21828. [DOI] [PubMed] [Google Scholar]
- Lu H, Kashani AH, Arfanakis K, Caprihan A, DeCarli C, Gold BT, Li Y, Maillard P, Satizabal CL, Stables L, Wang DJJ, Corriveau RA, Singh H, Smith EE, Fischl B, Kouwe A, Schwab K, Helmer KG, Greenberg SM, 2021. MarkVCID cerebral small vessel consortium: II. Neuroimaging protocols. Alzheimer’s Dement doi: 10.1002/alz.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matak P, Matak A, Moustafa S, Aryal DK, Benner EJ, Wetsel W, Andrews NC, 2016. Disrupted iron homeostasis causes dopaminergic neurodegeneration in mice. Proc. Natl. Acad. Sci. U. S. A 113, 3428–3435. doi: 10.1073/pnas.1519473113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzucchi S, Frosini D, Costagli M, Del Prete E, Donatelli G, Cecchi P, Migaleddu G, Bonuccelli U, Ceravolo R, Cosottini M, 2019. Quantitative susceptibility mapping in atypical Parkinsonisms. NeuroImage Clin. 24, 101999. doi: 10.1016/j.nicl.2019.101999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milovic C, Bilgic B, Zhao B, Acosta-Cabronero J, Tejos C, 2018. Fast nonlinear susceptibility inversion with variational regularization. Magn. Reson. Med 80, 814–821. doi: 10.1002/mrm.27073. [DOI] [PubMed] [Google Scholar]
- Moos T, Nielsen TR, Skjørringe T, Morgan EH, 2007. Iron trafficking inside the brain. J. Neurochem 103, 1730–1740. doi: 10.1111/j.1471-4159.2007.04976.x. [DOI] [PubMed] [Google Scholar]
- Pantoni L, Garcia JH, 1995. The significance of cerebral white matter abnormalities 100 years after Binswanger’s report. A review. Stroke 26, 1293–1301. doi: 10.1161/01.STR.26.7.1293. [DOI] [PubMed] [Google Scholar]
- Reuter M, Tisdall MD, Qureshi A, Buckner RL, van der Kouwe AJW, Fischl B, 2015. Head motion during MRI acquisition reduces gray matter volume and thickness estimates. Neuroimage 107, 107–115. doi: 10.1016/j.neuroimage.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds CR, 2002. Comprehensive trail making test (CTMT). [Google Scholar]
- Schweitzer AD, Liu T, Gupta A, Zheng K, Seedial S, Shtilbans A, Shahbazi M, Lange D, Wang Y, Tsiouris AJ, 2015. Quantitative susceptibility mapping of the motor cortex in amyotrophic lateral sclerosis and primary lateral sclerosis. Am. J. Roentgenol 204, 1086–1092. doi: 10.2214/AJR.14.13459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spincemaille P, Liu Z, Zhang S, Kovanlikaya I, Ippoliti M, Makowski M, Watts R, Rochefort L.de, Venkatraman V, Desmond P, Santin MD, Lehéricy S, Kopell BH, Péran P, Wang Y, 2019. Clinical integration of automated processing for brain quantitative susceptibility mapping: multi-site reproducibility and single-site robustness. J. Neuroimaging 29, 689–698. doi: 10.1111/JON.12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart AW, Robinson SD, O’Brien K, Jin J, Widhalm G, Hangel G, Walls A, Goodwin J, Eckstein K, Tourell M, Morgan C, Narayanan A, Barth M, Bollmann S, 2021. QSMxT: Robust masking and artefact reduction for quantitative susceptibility mapping. bioRxiv 2021.05.05.442850 10.1101/2021.05.05.442850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub S, Schneider TM, Emmerich J, Freitag MT, Ziener CH, Schlemmer HP, Ladd ME, Laun FB, 2017. Suitable reference tissues for quantitative susceptibility mapping of the brain. Magn. Reson. Med 78, 204–214. doi: 10.1002/MRM.26369. [DOI] [PubMed] [Google Scholar]
- Sun H, Walsh AJ, Lebel RM, Blevins G, Catz I, Lu J-Q, Johnson ES, Emery DJ, Warren KG, Wilman AH, 2015. Validation of quantitative susceptibility mapping with Perls’ iron staining for subcortical gray matter. Neuroimage 105, 486–492. doi: 10.1016/j.neuroimage.2014.11.010. [DOI] [PubMed] [Google Scholar]
- Sun Y, Ge X, Han X, Cao W, Wang Y, Ding W, Cao M, Zhang Y, Xu Q, Zhou Y, Xu J, 2017. Characterizing brain iron deposition in patients with subcortical vascular mild cognitive impairment using quantitative susceptibility mapping: a potential biomarker. Front. Aging Neurosci 9, 81. doi: 10.3389/fnagi.2017.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GEC, Leyland LA, Schrag AE, Lees AJ, Acosta-Cabronero J, Weil RS, 2020. Brain iron deposition is linked with cognitive severity in Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry 91, 418–425. doi: 10.1136/jnnp-2019-322042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kouwe AJW, Benner T, Salat DH, Fischl B, 2008. Brain morphometry with multiecho MPRAGE. Neuroimage 40, 559–569. doi: 10.1016/j.neuroimage.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Spincemaille P, Liu Z, Dimov A, Deh K, Li J, Zhang Y, Yao Y, Gillen KM, Wilman AH, Gupta A, Tsiouris AJ, Kovanlikaya I, Chiang GCY, Weinsaft JW, Tanenbaum L, Chen W, Zhu W, Chang S, Lou M, Kopell BH, Kaplitt MG, Devos D, Hirai T, Huang X, Korogi Y, Shtilbans A, Jahng GH, Pelletier D, Gauthier SA, Pitt D, Bush AI, Brittenham GM, Prince MR, 2017. Clinical quantitative susceptibility mapping (QSM): biometal imaging and its emerging roles in patient care. J. Magn. Reson. Imaging doi: 10.1002/jmri.25693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RJ, Zucca FA, Duyn JH, Crichton RR, Zecca L, 2014. The role of iron in brain ageing and neurodegenerative disorders. Lancet. Neurol 13, 1045–1060. doi: 10.1016/S1474-4422(14)70117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardlaw JM, Smith C, Dichgans M, 2013. Mechanisms of sporadic cerebral small vessel disease: insights from neuroimaging. Lancet. Neurol 12, 483–497. doi: 10.1016/S1474-4422(13)70060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne Martin WR, Ye FQ, Allen PS, 1998. Increasing striatal iron content associated with normal aging. Mov. Disord 13, 281–286. doi: 10.1002/mds.870130214. [DOI] [PubMed] [Google Scholar]
- Wei H, Dibb R, Zhou Y, Sun Y, Xu J, Wang N, Liu C, 2015. Streaking artifact reduction for quantitative susceptibility mapping of sources with large dynamic range. NMR Biomed 28, 1294–1303. doi: 10.1002/NBM.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman SJ, Doubal FN, Chappell FM, Valdés-Hernndez MC, Wang X, Rumley A, Lowe GDO, Dennis MS, Wardlaw JM, 2015. Plasma biomarkers of inflammation, endothelial function and hemostasis in cerebral small vessel disease. Cerebrovasc. Dis 40, 157–164. doi: 10.1159/000438494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisnieff C, Ramanan S, Olesik J, Gauthier S, Wang Y, Pitt D, 2015. Quantitative susceptibility mapping (QSM) of white matter multiple sclerosis lesions: interpreting positive susceptibility and the presence of iron. Magn. Reson. Med 74, 564–570. doi: 10.1002/mrm.25420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witoszynskyj S, Rauscher A, Reichenbach JR, Barth M, 2009. Phase unwrapping of MR images using ΦUN – A fast and robust region growing algorithm. Med. Image Anal 13, 257–268. doi: 10.1016/J.MEDIA.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Zachariou V, Bauer CE, Seago ER, Panayiotou G, Hall ED, Butterfield DA, Gold BT, 2021. Healthy dietary intake moderates the effects of age on brain iron concentration and working memory performance. Neurobiol. Aging 106, 183–196. doi: 10.1016/J.NEUROBIOLAGING.2021.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariou V, Bauer CE, Seago ER, Raslau FD, Powell DK, Gold BT, 2020. Cortical iron disrupts functional connectivity networks supporting working memory performance in older adults. Neuroimage 223, 117309. doi: 10.1016/j.neuroimage.2020.117309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca L, Stroppolo A, Gatti A, Tampellini D, Toscani M, Gallorini M, Giaveri G, Arosio P, Santambrogio P, Fariello RG, Karatekin E, Kleinman MH, Turro N, Hornykiewicz O, Zucca FA, 2004. The role of iron and copper molecules in the neuronal vulnerability of locus coeruleus and substantia nigra during aging. Proc. Natl. Acad. Sci. U. S. A 101, 9843–9848. doi: 10.1073/pnas.0403495101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.